Background: Protein-protein interaction between HCN1 and tip-link protocadherin 15CD3 suggests a role for HCN1 in hair-cell mechanotransduction.
Results: HCN1 and HCN2 form alternate ternary protein complexes with hair-cell stereociliary proteins.
Conclusion: Alternate protein-protein interactions reflect separation of binding events for stereociliary proteins implicated in mechanotransduction.
Significance: HCN1 is the only channel component identified as binding to a stereociliary tip-link protein.
Keywords: Cell Biology, Electron Microscopy (EM), Ion Channels, Protein Complexes, Protein-Protein Interactions, HCN Channels, Polarized Epithelial Cells, Protein Folding and Immunoprecipitation, Protocadherin 15
Abstract
A unique coupling between HCN1 and stereociliary tip-link protein protocadherin 15 has been described for a teleost vestibular hair-cell model and mammalian organ of Corti (OC) (Ramakrishnan, N. A., Drescher, M. J., Barretto, R. L., Beisel, K. W., Hatfield, J. S., and Drescher, D. G. (2009) J. Biol. Chem. 284, 3227–3238). We now show that Ca2+-dependent interaction of the organ of Corti HCN1 and protocadherin 15 CD3 is mediated by amino-terminal sequence specific to HCN1 and is not replicated by analogous specific peptides for HCN2 or HCN4 nor by amino-terminal sequence conserved across HCN isoforms utilized in channel formation. Furthermore, the HCN1-specific peptide binds both phosphatidylinositol (3,4,5)-trisphosphate and phosphatidylinositol (4,5)-bisphosphate but not phosphatidylinositol 4-phosphate. Singly isolated cochlear inner and outer hair cells express HCN1 transcript, and HCN1 and HCN2 protein is immunolocalized to hair-cell stereocilia by both z-stack confocal and pre-embedding EM immunogold microscopy, with stereociliary tip-link and subcuticular plate sites. Quantitative PCR indicates HCN1/HCN2/HCN3/HCN4 = 9:9:1:89 in OC of the wild-type mouse, with HCN4 protein primarily attributable to inner sulcus cells. A mutant form of HCN1 mRNA and protein is expressed in the OC of an HCN1 mutant, corresponding to a full-length sequence with the in-frame deletion of pore-S6 domains, predicted by construct. The mutant transcript of HCN1 is ∼9-fold elevated relative to wild-type levels, possibly representing molecular compensation, with unsubstantial changes in HCN2, HCN3, and HCN4. Immunoprecipitation protocols indicate alternate interactions of full-length proteins; HCN1 can interact with protocadherin 15 CD3 and F-actin-binding filamin A forming a complex that does not include HCN2, or HCN1 can interact with HCN2 forming a complex without protocadherin 15 CD3 but including F-actin-binding fascin-2.
Introduction
A role for HCN12 in hair cell function was first suggested by the detection of HCN1 mRNA by in situ hybridization and determination of full-length HCN1 mRNA sequence in a vestibular model hair cell preparation isolated from the trout saccule (1). Evidence of mammalian cochlear hair cell expression of HCN1 transcript came from analysis of rat λ ZAP cDNA libraries of inner and outer cochlear hair cells (2). Protein-protein interaction protocols indicated that the cytoplasmic amino terminus of HCN1 binds the cytoplasmic carboxyl terminus of stereociliary tip-link protein protocadherin 15-protocadherin 15a-like protein in a trout saccular hair cell preparation and protocadherin 15 CD3 in rat organ of Corti preparations (2). The binding for organ of Corti proteins was Ca2+-dependent and specific to protocadherin 15 CD3 and not replicated by protocadherin 15 CD1. HCN1 protein was immunolocalized to stereocilia for the teleost vestibular hair cell model and mammalian cochlear hair cell model with light microscopy (2), consistent with putative stereociliary binding sites for tip-link proteins protocadherin 15a in teleosts (3) and protocadherin 15 CD3 in mammals (4). Given that the stereociliary tip-link proteins are presumed to represent the structural transduction apparatus in hair cell mechanotransduction and have been hypothesized to physically link to the mechanosensory transduction channel(s) (5), the identification of an ion channel component (HCN1) that actually interacts with a tip-link protein is novel.
New studies with direct analysis of pooled samples of singly isolated cochlear inner and outer hair cells, z-stack confocal and immunogold protein immunolocalization, and protein-protein interaction protocols have now yielded results consistent with the specific nature of the protein-protein interaction of HCN1 amino terminus and protocadherin 15 CD3 carboxyl terminus in cochlear hair cells. We describe evidence for the inclusion of HCN2 in a cochlear hair cell HCN ion channel and the molecular mechanisms available for separate interaction of HCN1 and HCN2 with stereociliary F-actin-binding proteins.
EXPERIMENTAL PROCEDURES
Surface Plasmon Resonance (SPR) Analysis of Binding between HCN1 and Protocadherin 15 CD3 Expressed in Rat Organ of Corti
Affinity-purified HCN1-N fusion polypeptide (ligand) was immobilized on a CM5 research grade sensor chip by amine coupling (2). Binding analysis was carried out by injecting the affinity-purified CD3-C fusion peptide (analyte) into the flow cells (ligand and reference cell), and the interaction (response units) between analyte and ligand was recorded. The determination of Ca2+ dependence of binding was based upon analysis of Ca2+ levels in experimental solutions by inductively coupled plasma mass spectroscopy (for details see Ref. 2). In the examination of possible binding by the other HCN isoforms, protocadherin 15 CD3 served as ligand, with the amino-terminal sequence conserved across HCN isoforms as well as HCN2 and HCN4 amino terminus-specific sequences serving as analytes. The expression primers for insertion of rat HCN2-specific amino-terminal sequence, aa 1–134 (NP_446136; supplemental Fig. 1), into the pRSET-A vector were upstream gatGGATCCgaatggatgcgcgcgggggcggc and downstream gagccgcgcggcagccagtgaGAATTCgaa. (In addition, upstream and downstream primers were designed for alternate codon use to express peptide for the native HCN2-specific amino terminus, reducing gc content, see supplemental Fig. 2). Primers for rat HCN4 amino terminus-specific sequence aa 1–213 (NP_067690; supplemental Fig. 1) were upstream gctaGGATCCatggacaagctgccgccgtccat and downstream tgggccagagcggcttcatGAATTCactgc (restriction sites are capitalized).
HCN1 Lipid Binding
Initially, phospholipid binding to the HCN1 amino terminus was examined with membrane strips (Echelon Biosciences, Salt Lake City, UT). The strips were incubated for 1 h in PBS with 1% nonfat dry milk at room temperature. Purified rat HCN1-specific amino-terminal peptide (5 μg/ml), epitope-tagged with Xpress peptide tag, was then added to a fresh PBS + 1% milk containing protease inhibitors (Sigma) and incubated overnight at 4 °C on a rotary shaker. The next day, the strips were washed in PBS buffer three times (10 min each) and incubated in anti-Xpress antibody (1:7,000 dilution for 1 h at RT) to detect the fusion tag of the HCN1 domains. After washing in PBS buffer, the strips were incubated in donkey anti-mouse-HRP secondary antibody (1:10,000 for 1 h) for chemiluminescence detection using the detection kit (GE Healthcare). The binding was further characterized with SPR. The purified HCN1-specific amino terminus was immobilized on a CM5 sensor chip (Biacore, Piscataway, NJ), and a lipid preparation containing 10 μm phosphatidylinositol (4,5)-bisphosphate (PIP2) tri-ammonium salt (Avanti Polar Lipids, Alabaster, AL) or dipalmitoyl phosphatidylinositol (3,4,5)-trisphosphate (PIP3; Echelon Biosciences) was injected as analyte in either the presence of Ca2+ (92 μm) or absence of Ca2+ (1 mm EGTA). Alternatively, an L1 sensor chip (Biacore) was used to capture PIP2 and PIP3 as ligands in binding to the HCN1-specific amino terminus as analyte (10–100 nm) in SPR analysis.
Yeast Two-hybrid Co-transformation of HCN2 Carboxyl Terminus and Fascin-2
The carboxyl terminus of rat HCN2 (GI:50878266, nucleotides 1,371–2,627) was PCR-amplified from the rat organ of Corti cDNA with the upstream primer ggccGAATTCcggcgccagtaccaggagaa and the downstream primer gtatGGATCCtcacaagttggaagagag, cloned into a yeast two-hybrid bait vector pGBKT7, and sequence-verified. Rat fascin-2 sequence (GI:157818956; nucleotides 1,144–1,644) was likewise amplified from a rat organ of Corti cDNA preparation with the upstream primer agtcaGAATTCaacgcaagtttctgccaacac and the downstream primer acgtGGATCCatattcccagagagcttccccc, cloned into prey vector pGADT7, and sequence-verified. Plasmids were mixed (1 μg each), with co-transformation in yeast strain AH109 according to Clontech protocols. Yeast was plated on quadruple dropout media containing X-α-Gal and incubated at 30 °C for 3–5 days (2).
Isolation of Inner and Outer Hair Cells from the Cochlea of the Adult Rat
Cochleas from ACI Black Agouti rats (4–6 weeks of age) were dissected in physiological saline (6) on ice to yield an organ-of-Corti fraction that included the inner hair cells and three rows of outer hair cells attached to the osseous spiral lamina, minus tectorial membrane. The organ of Corti sample was incubated in 1.5 mg/ml trypsin at 37 °C in dissecting saline for 15 min. After trituration with a 100-μl HPLC syringe, the cell suspension was transferred to the well of a microscope slide positioned on an inverted microscope stage that had been cooled to 8 °C. Inner and outer hair cells were identified by visual inspection at ×200 magnification (Fig. 1) and drawn up by gentle negative pressure into a 50-μm tip of a micropipette connected by polyethylene tubing to a screw-driven syringe. The micropipette was then quickly withdrawn from the fluid. The fluid containing each hair cell was expelled into a 1.5-ml microcentrifuge tube and placed on dry ice for future analysis. The separate hair cells collected in this way were pooled to provide adequate signal for studies of hair-cell biochemistry and molecular biology.
FIGURE 1.
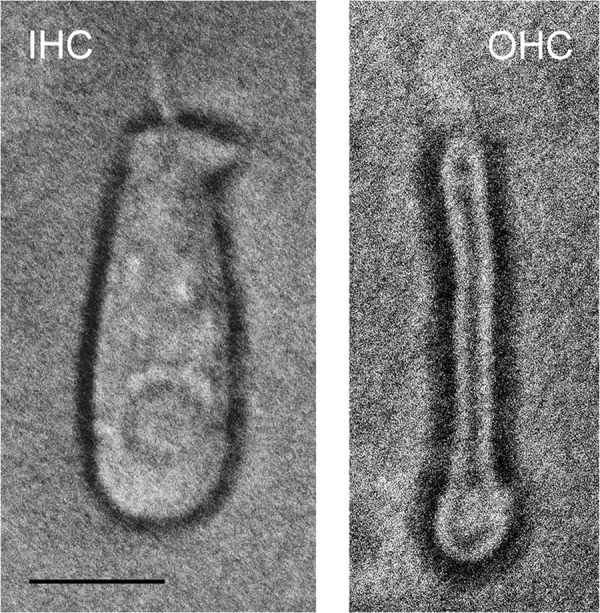
Isolation of inner and outer hair cells from the adult rat organ of Corti as described in the text and microscopically observed (×200 magnification). Left panel, IHC. Right panel, OHC. Scale bar, 10 μm (IHC and OHC).
Amplification of Cochlear Hair Cell Message
25–30 singly isolated outer hair cells and a similar number of inner hair cells were separately pooled and stored at −80 °C. The cells were thawed on ice for 5 min, and the first-strand reaction was carried out as described by the manufacturer (“Make Your Own Mate & Plate Library System,” Clontech). One μl of random hexamer primers, CDSIII-6 (all reagents were from Clontech), was added to 3 μl of RNase-free water containing pooled cells. The sample tube was then incubated at 72 °C for 2 min and transferred to ice, and the remaining reagents (first-strand synthesis buffer, DTT, dNTPs, and Moloney murine leukemia virus reverse transcriptase, or for selected experiments, SMARTScribe Reverse Transcriptase) were added. The mixture was incubated at room temperature for 10 min, followed by incubation at 42 °C for 10 min. One μl of SMART III-modified oligonucleotide primer was added, mixed, and centrifuged briefly. This step was followed by a 1-h incubation at 42 °C and 10 min at 75 °C to terminate the reaction. Then, at room temperature, 1 μl of RNase H (2 units) was added and incubated at 37 °C for 20 min. The entire 10 μl was used for “long distance” PCR (Clontech). For the latter, a 50-μl reaction was set up using the Advantage GC2 polymerase mix, with 5′ and 3′ PCR primers provided by the Clontech library system kit. PCR amplification was carried out as follows: a hold at 95 °C for 30 s, 25 cycles of 95 °C for 15 s alternating with 68 °C for 2 min, concluding with a final extension at 68 °C for 2 min. The double-stranded cDNA was stored at −80 °C.
Further amplification of the cDNA was accomplished by a PCR utilizing gene-specific primers that crossed introns designed with Accelrys software (San Diego) (HCN1 upstream, gtgcagtggtgagaatcttc, and downstream, gctggtaacttgtggaatgac; nested primers for HCN1 upstream, gggaaacagtattcctacgc, and downstream, actgcctccttgaagaatcc; HCN2 upstream, aagttctccctgcggatgttt, and HCN2 downstream, acgatccagggcgccgtggtctcg). Two μl of template were used per 50-μl reaction. PCR conditions were as follows: a hold at 95 °C for 2 min, 50–65 cycles of 95 °C for 20 s, 62 °C for 20–25 s, and 72 °C for 30–45 s, with final extension at 72 °C for 2–5 min. Water was substituted for cDNA in negative controls. All amplification products were sequenced for molecular identification.
Confocal Microscopy and Immunofluorescence
The temporal bones from 5- to 6-week-old B629SF2/J wild-type mice were rapidly harvested, and the cochleas were isolated and perfused with fixative, 4% paraformaldehyde, 0.1% glutaraldehyde at 4 °C, entering the round window and exiting at the helicotrema. The labyrinthine tissues were isolated, washed with PBS three times for 10 min, pretreated in 0.3% Triton X-100 (T-8787, Sigma) in PBS for 5–10 min, incubated in 0.1% sodium borohydride for 30 min, blocked in normal sera (corresponding to the species used to raise the secondary antibodies) for 30 min at room temperature, and incubated with primary antibody at 4 °C overnight. The tissues were washed three times (5 min each) and incubated at room temperature in secondary antibodies for 30 min each (rhodamine-phalloidin 60 min), washed three times in PBS, and mounted as surface preparations. The primary antibodies included affinity-purified goat anti-HCN1 (targeting the carboxyl terminus of mouse HCN1, sc-19706, Santa Cruz Biotechnology, Santa Cruz, CA; 1:50), mouse monoclonal antibody raised to a fusion protein, corresponding to aa 778–910 of rat HCN1 (ab84816, Abcam, Cambridge, MA, 1:100), and rabbit anti-human HCN2 amino-terminal aa 147–161 crossing to mouse (APC-030, Alomone Labs, Jerusalem, Israel, 1:200). Secondary antibodies included donkey anti-goat-568 (A11057, Molecular Probes, 1:2,000), chick anti-rabbit-488 (A21441, Molecular Probes, 1:2,000), and goat anti-mouse-568 (A11031, Molecular Probes, 1:2,000). Rhodamine-phalloidin was added at 1:100 (Molecular Probes). Surface preparations were viewed, and 1-μm z-stack optical slices were captured with a Zeiss meta LSM 510 with 543 nm excitation and 565–615 emission (red) and 488 nm excitation and 500–550 nm emission (green). Fluorescence and 3,3′-diaminobenzidine (DAB) detection of immunoreactivity for filamin A (MAB1678, Chemicon/Millipore, Billerica, MA) in the rat organ of Corti was carried out as described previously (2, 7). Preabsorption negative controls for Alomone HCN1 and HCN2 primary antibodies were carried out with DAB detection on 4–5-μm sections of rat cochlea with a chicken anti-rabbit IgG-B secondary antibody that was mouse/human-absorbed (SC-2986, Santa Cruz Biotechnology) with results compared with those for nonpreabsorbed antibody. Preabsorption of the antibodies with the respective antigenic peptides resulted in a parallel decrease in immunostaining of stereocilia and the cell body of hair cells.
Pre-embedding Immunogold Electron Microscopy
Pre-embedding immunogold was used to maintain the antigenicity in tissues potentially lost in post-embedding procedures during resin polymerization by heat prior to immunocytochemical analysis. The temporal bones from 2-month-old Black Agouti ACI rats of mixed gender were rapidly harvested, and the cochleas were isolated and perfused with fixative, 4% paraformaldehyde, 0.1% glutaraldehyde at 4 °C, entering the round window and exiting at the helicotrema. Following fixation for 1 h, the cochlear tissues were washed three times with PBS, pretreated with 0.3% Triton X-100 for 5–10 min and 0.1% sodium borohydride for 30 min, and blocked with 5% normal serum (corresponding to the species in which the secondary antibody was raised) for 30 min at room temperature. The cochlear tissues were incubated in primary antibody overnight at 4 °C, followed by incubation with secondary antibodies for 30 min at room temperature. The tissues were exposed to 0.5% OsO4 in PBS for 10 min and dehydrated in a graded ethanol series, cleared in propylene oxide, and embedded in Epon. Semi-thin sections were examined with a Zeiss EM 900 electron microscope. Primary antibodies included a rabbit polyclonal raised to rat HCN1 amino terminus, aa 6–24 (APC-056, Alomone; 1:25), and a rabbit polyclonal raised to amino acids 147–161 of human HCN2, identical to rat HCN2 sequence (APC-030, Alomone; 1:200). The secondary antibody was 10 nm gold-conjugated goat anti-rabbit IgG (G3779, Sigma) at 1:50. For negative controls, we substituted equivalent rabbit IgG for rabbit HCN1 and HCN2 primary antibodies, which is appropriate for affinity-purified primary antibodies.
Quantitative PCR: Characterization of HCN Isoform Expression in Organ of Corti of Wild Type and HCN1 Mutant
Organ of Corti fractions from 5- to 6-week-old B6129SF2/J wild-type mice and HCN1 mutant B6;129-HCN1tm2Kndl/J mice (six animals each, The Jackson Laboratory, Bar Harbor, ME) were isolated in ice-cold dissecting saline containing RNase inhibitor and the mRNA converted to cDNA by standard techniques, including a DNase step to remove genomic DNA (6). Full-length HCN1 cDNA amplification was carried out with upstream primer atgcagaggcagttcacctccatgc and downstream primer gctcccgaggacatctgtctgaacaag.
Quantitative RT-PCR was carried out in 25-μl reactions in three sets containing two replicates each. Power SYBR Green PCR MasterMix (Applied Biosciences, Carlsbad, CA) was prepared as instructed in the user protocol. Reactions were carried out in an Opticon DNA Engine (Bio-Rad). Primers were designed with Accelrys software targeting individual HCN isoforms so as not to cross to other HCN isoforms or to other proteins and checked with two-sequence BLAST alignment comparison. Furthermore, we obtained sequence verification that specific primers applied to the organ of Corti cDNA actually yielded the expected individual HCN isoform. Specific primers designed with Accelrys software included the following: HCN1 control upstream agtcacatgctgtgcattg and downstream tccagagactggatcaaagc; HCN1 mutant upstream gggatggctgtcttcagttcctg and downstream tgtcagctggtaacttgtgg; HCN2 upstream agattgtgaacttcaactgcc and downstream tctcggatgatgtaatctccag; HCN3 upstream actacgagcatcgctaccag and downstream gggtcagcatgagcaaacag; HCN4 upstream atgaacaccgctaccaaggc and downstream tgtcacaaagttgggatctgc; GAPDH upstream agtatgtcgtggagtctactg and downstream ggttcacacccatcacaaac.
To estimate copy number, we used primer pairs for each HCN isoform in PCRs containing known amounts of the template. Templates were generated by PCR amplification of a larger portion of each sequence that contained the target sequence and cloned in pGEMT-easy vector. Isolated plasmids were sequence-verified before the reaction and quantified by the Qubit fluorescent system (Invitrogen). A standard curve for each primer pair was generated from control reactions with 115–115,000 copies of plasmids containing the respective HCN inserts. Duplicate reactions were carried out three times under identical conditions, and the Ct values for each set were averaged and plotted on an Excel sheet. The values of the standard curve were calculated by Excel as follows: HCN1 mutant: y = −3.53x + 42.65, R2 = 0.9923; HCN1 control: y = −3.79x + 43.75, R2 = 0.9937; HCN2: y = −3.45x + 41.75, R2 = 0.994; HCN3: y = −3.76x + 43.72, R2 = 0.9965; and HCN4: y = −3.47x + 42.97, R2 = 0.9957.
Western Blot Analysis of HCN1 Protein Fragments in the HCN1 Mutant
Thirty five μg of brain protein from the HCN1 mutant (21-day-old, B6;129-HCN1tm2Kndl/J, The Jackson Laboratory) was loaded into wells of a 1× SDS-polyacrylamide gel, according to protocols described previously (2). The Western blot was probed with a rabbit polyclonal antibody targeting the amino terminus of rat HCN1 that crossed to mouse (18/19 aa identity, APC-056, Alomone, 1:2,000). Alternatively, the Western blot was probed with affinity-purified goat polyclonal antibody targeting sequence near the carboxyl terminus of mouse HCN1 (sc-19706, Santa Cruz Biotechnology; 1:1,000). The secondary antibodies tagged with horseradish peroxidase were as follows: donkey anti-rabbit IgG (sc-2317, Santa Cruz Biotechnology, 1:10,000) and donkey anti-goat IgG (sc-2033, Santa Cruz Biotechnology, 1:6,000). Chemiluminescence was used for detection. For negative controls, rabbit IgG or goat IgG equivalent to the primary antibodies was used to probe Western blots of brain from the HCN1 mutant.
Immunoprecipitation
Rat brain lysate was prepared by sonication of the tissue (Ultrasonicator W375 Cell Disruptor, Qsonica, Newtown, CT) on ice in PBST containing 0.1% Triton X-100/Tween 20 and 1× protease inhibitor mixture (G6521, Promega, Madison, WI) followed by centrifugation (20,900 × g for 5 min) at 4 °C. The supernatant was diluted in PBST + protease inhibitor containing 10% glycerol, brought to a concentration of 1 mg/ml protein, and pretreated with protein A-agarose beads saturated in PBST to remove nonspecific binding proteins from the lysate. After centrifugation at 20,900 × g for 5 min at 4 °C to remove the beads (and nonspecific binding proteins), the supernatant was utilized for immunoprecipitation overnight (4 °C) with mouse anti-human filamin A (clone PM6/317, MAB1678, Chemicon), which crosses to rat sequence, rabbit anti-human HCN2 (targeting aa 147–161, UniProtKB/Swiss Prot Q9UL51, identical to rat HCN2 sequence, APC-030, Alomone), or goat anti-fascin-2 (EB08002, Everest Biotech, UK). Fresh protein A-agarose was added for 30 min at 4 °C, and the beads were pelleted by centrifugation (1,370 × g), washed five times in cold 1× PBST + protease inhibitor containing 10% glycerol, and heat-denatured in SDS gel loading dye in boiling water for 10 min prior to electrophoresis (2). PVDF membrane blots of the gel were blocked with 1% nonfat milk in PBST (and 1% BSA for protocadherin 15 CD3) and incubated with primary antibodies against filamin A (Chemicon), HCN1 (Santa Cruz Biotechnology), protocadherin 15 CD3 (Covance, Princeton, NJ, custom chick antibody raised against rat protocadherin 15 CD3 carboxyl-terminal epitope, SSTGEDSAPESQRSRTHKPSGPSN,GenBankTM accession number XP_001079990), or HCN2 (Alomone). HCN1 and protocadherin 15 CD3 were analyzed in brain lysate immunoprecipitated with HCN2, and HCN2 and HCN1 (and protocadherin 15 CD3) were analyzed in brain lysate immunoprecipitated with fascin-2. HRP-conjugated secondary antibodies (Santa Cruz Biotechnology) were diluted (1:6,000–1:10,000) in blocking solutions consisting of nonfat milk (1%) in PBST (plus 1% BSA for protocadherin 15 CD3) plus 2% serum corresponding to the species in which the secondary antibody was raised. The respective proteins were detected with Western Lightning chemiluminescence (2). Negative controls included the following: (a) brain lysate (no immunoprecipitation) + beads + primary and secondary antibodies for detection; (b) IgG-specific immunoprecipitation of lysate for each of the primary antibodies used for immunoprecipitation + beads + primary and secondary antibodies for detection; (c) IgG + beads (no lysate) + primary and secondary antibodies for detection.
RESULTS
Molecular Requirements for HCN1 Amino Terminus Binding to Protocadherin 15 CD3 Determined with Rat Organ of Corti Proteins
Previously, we obtained evidence for Ca2+-dependent binding of the amino terminus of HCN1 in its entirety to the carboxyl terminus of protocadherin 15 CD3 with KD = 5.26 × 10−8 at 61 μm Ca2+ (2). The amino termini of the HCN isoforms include an isoform-specific sequence followed by an isoform-conserved sequence up to the first transmembrane domain (supplemental Fig. 1). The specific amino termini for the HCN2-HCN4 isoforms have limited identity to HCN1, ranging from 10 to 15%, with amino acid similarity ranging from 15 to 20%. A mechanism for Ca2+ dependence would reside in the HCN1-specific terminus as opposed to amino termini of other HCN isoforms. The HCN1-specific amino terminus incorporates an EF hand-like sequence (aa 11–18), SRD (aa 13–15), and DGC aa 16–18, cited as Ca2+-binding motifs, which are not replicated in specific amino-terminal sequences for HCN3 and HCN4. HCN2 includes SRD at aa 64–67 but does not include the motif DGN. In contrast to the specific amino-terminal sequences, the conserved regions of the amino termini exhibit 96% identity across all HCN isoforms.
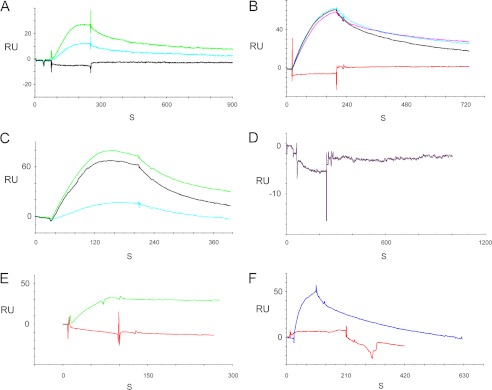
In the present investigation, the HCN1-specific amino-terminal sequence was found to be sufficient for Ca2+-dependent binding to protocadherin 15 CD3 by SPR analysis (Fig. 2, A and B, compared with C; see Table 1), with a KD value for the HCN1-specific amino-terminal sequence of 3.75 × 10−8 m compared with a KD value of 4.05 × 10−8 m for the full HCN1 amino terminus, both at 100 μm Ca2+. The HCN1 conserved amino terminus did not bind (Fig. 2D) nor was there any interaction between the specific nonconserved amino terminus of HCN2 or HCN4 with protocadherin 15 CD3 (Fig. 2, E and F, respectively).
FIGURE 2.
Molecular requirements for HCN amino terminus binding to protocadherin 15 CD3 determined with SPR for rat organ of Corti proteins. A, SPR for HCN1-specific (nonconserved) amino terminus sequence as follows: protocadherin 15 CD3 as ligand, HCN1-specific (nonconserved) amino terminus as analyte at 100 nm; 100 μm Ca2+ (light green), 1 mm EGTA (turquoise); buffer control (100 μm Ca2+, black). B, HCN1-specific amino terminus as analyte at 100 nm; 100 μm Ca2+ (pink, black, and turquoise, three repeats); buffer control (red). C, HCN1 full-length amino terminus as analyte at 100 nm; 100 μm Ca2+ (green), 26.5 μm Ca2+ (black), 1 mm EGTA (turquoise). D, conserved HCN1 amino-terminal sequence as analyte at 200 nm; 100 μm Ca2+. E, HCN2-specific amino terminus as analyte at 100 nm, 100 μm Ca2+ (red); HCN1-specific amino terminus at 100 nm, 100 μm Ca2+ (green). There is no binding of the HCN2-specific amino-terminal sequence to protocadherin 15 CD3. F, HCN4-specific (nonconserved) amino terminus as analyte at 100 nm, 100 μm Ca2+ (red); HCN1-specific (nonconserved) amino terminus as analyte at 100 nm, 100 μm Ca2+ (blue). There is no binding of HCN4 to protocadherin 15 CD3. A–F, three SPR determinations were performed for each condition/construct. (For KD values, see Table 1.) RU, response units.
TABLE 1.
Interaction of HCN1 amino-terminal region with binding partners
Rate constants for association and dissociation (mean ± S.E., n = 3 for each value) were obtained from original surface plasmon resonance plots by the use of BIAevaluation software (Biacore, Piscataway, NJ). Best fits of curves were determined corresponding to a Langmuir binding model (62). Equilibrium binding constants were calculated from the ratio of means of the rate constants. “Specific” HCN1-N (HCN1 amino-terminal sequence) included aa 1–78, and “full” HCN1-N encompassed aa 1–127. Ca2+ concentration was 100 μm for binding between HCN1-specific amino terminus and HCN1 full-length amino terminus and protocadherin 15 CD3. Experiments carried out examining binding between HCN1-specific amino terminus and phospholipids included Ca2+ at 92 μm. Kinetic constants for phospholipid binding were determined with HCN1 as analyte and phospholipid as ligand. PCDH CD3, protocadherin 15 CD3.
| Binding partner | KA | KD | ka | kd |
|---|---|---|---|---|
| m−1 | m | m−1 s−1 | s−1 | |
| PCDH CD3 (specific HCN1-N) | 2.66 × 107 | 3.75 × 10−8 | 2.21 ± 0.70 × 105 | 8.30 ± 2.78 × 10−3 |
| PCDH CD3 (full HCN1-N) | 2.47 × 107 | 4.05 × 10−8 | 6.12 ± 1.74 × 104 | 2.48 ± 0.80 × 10−3 |
| PIP3 (specific HCN1-N) | 3.64 × 108 | 2.75 × 10−9 | 9.92 ± 6.06 × 105 | 2.73 ± 0.61 × 10−3 |
| PIP2 (specific HCN1-N) | 2.96 × 107 | 3.38 × 10−8 | 1.03 ± 0.34 × 105 | 3.49 ± 1.81 × 10−3 |
Binding of the Rat HCN1-specific Amino Terminus to Phospholipids
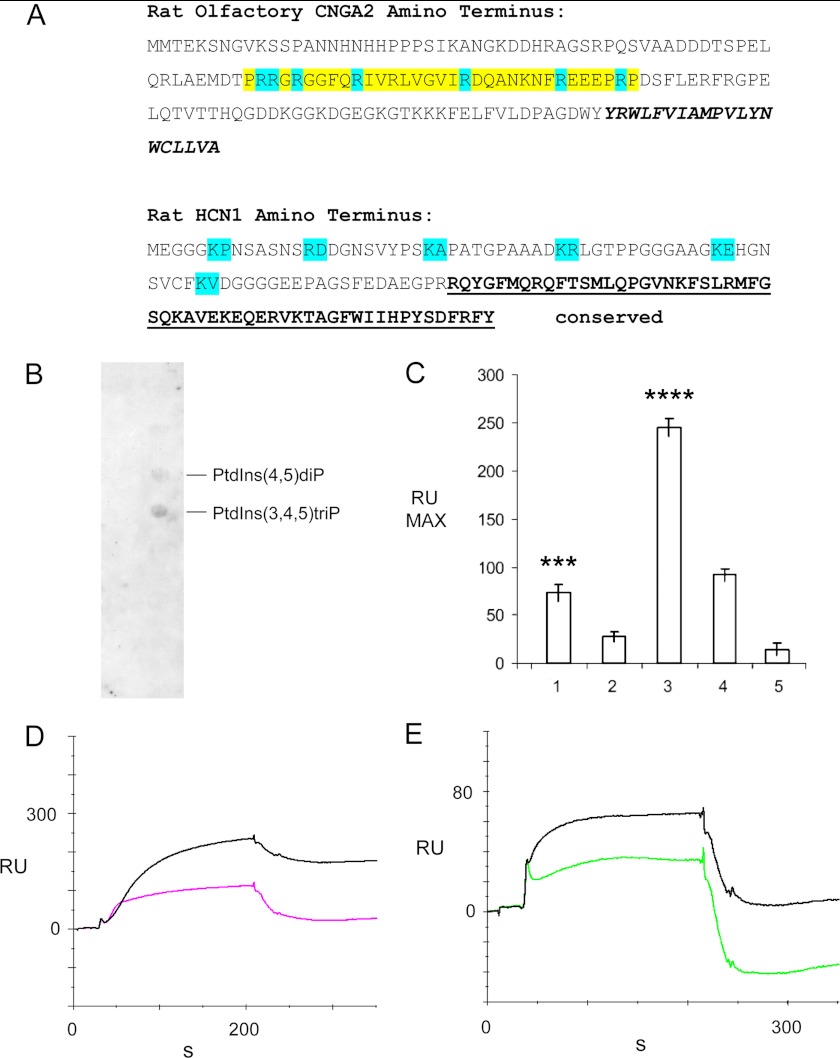
The amino termini of other cyclic nucleotide-gated channels bind phospholipids, thus modulating the current. For example, residues 61–90 of CNGA2 are necessary for PIP3 binding and suppression of CNGA2 currents, putatively mediated by positively charged arginine residues (Fig. 3A). The HCN1-specific amino terminus also includes those positively charged aa residues putatively involved in CNGA2 lipid binding (Fig. 3A). With membrane lipid strip analysis and SPR, binding was observed for the HCN1-specific amino terminus with PIP3 and PIP2, but not with other phospholipids (Fig. 3, B–E). SPR indicated Ca2+ dependence of the binding. The KD value for PIP3, as ligand, binding to the HCN1-specific amino terminus, as analyte, was 2.75 × 10−9 m (Table 1, 92 μm Ca2+). The comparable KD value for PIP2 was 3.38 × 10−8 m (Table 1, 92 μm Ca2+).
FIGURE 3.
Interaction between the HCN1-specific amino terminus with PIP3 and PIP2. A, rat olfactory CNGA2 amino terminus: residues 61–90 (highlighted in yellow) are necessary for PIP3 binding and suppression of CNG channel currents (arginine residues putatively used in CNGA2 binding to PIP3 are highlighted in blue). Bold italicized sequence is first membrane spanning region. Rat HCN1 amino terminus: positively charged aa residues putatively involved in lipid binding are highlighted in blue. HCN1 amino terminus: sequence that is conserved across HCN isoforms is boldface and underlined. B, membrane lipid strip analysis of binding to the specific, nonconserved amino terminus of rat HCN1. Of the 16 lipid components, binding was observed for PIP3 and PIP2. C, SPR for interaction of HCN1 and phospholipids is as follows: bar 1, PIP2 + Ca2+; bar 2, PIP2 + EGTA; bar 3, PIP3 + Ca2+; bar 4, PIP3 + EGTA; bar 5, buffer; means ± S.E. are indicated. ***, PIP2 + Ca2+ versus PIP2 + EGTA, p < 0.01; ****, PIP3 + Ca2+ versus PIP3 + EGTA, p < 0.001 (two-tailed t test for difference of means for small samples, n = 5). The cytoplasmic amino terminus of rat HCN1 was expressed and purified as a histidine-tagged fusion peptide and used as ligand in surface plasmon resonance analysis on a CM5 sensor chip. 10 μm of each phosphatidylinositol served as analyte in either 92 μm Ca2+ or 1 mm EGTA. Three experiments were carried out, each with n = 5, yielding similar results. D, representative sensorgrams showing interaction of rat HCN1-specific amino terminus (ligand) with PIP3 (10 μm) (analyte) in either 92 μm Ca2+ (black) or in 1 mm EGTA (pink). E, representative sensorgram illustrating interaction of PIP2 (10 μm) (analyte) in either 92 μm Ca2+ (black) or 1 mm EGTA (green) with HCN1-specific amino terminus (ligand). D and E, five SPR determinations were performed for each condition/construct per experiment, and multiple experiments were carried out. RU, response units.
Amplification of Cochlear Hair Cell HCN1 and HCN2 cDNA
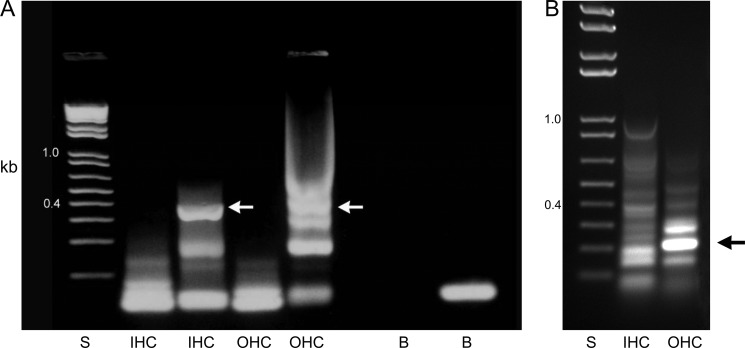
The question of HCN1 transcript expression by cochlear inner and outer hair cells, in addition to protein expression previously examined (2), was further addressed by application of specific primers to the cDNA from pooled samples of individually isolated and morphologically documented inner and outer hair cells from the adult rat cochlea. Primers designed to cross introns elicited amplification of the product of predicted size, 397 bp, from the cDNA of multiple sets of inner and outer hair cells from the rat cochlea (Fig. 4A). The IHC product gave 100% nucleotide identity to rat HCN1. The OHC product was amplified with a nested primer that yielded a PCR product with 100% identity to rat HCN1 nucleotide sequence. Specific primers for HCN2 elicited amplification of HCN2 cDNA of predicted size, 209 bp, from cochlear outer hair cells with 100% nucleotide identity to rat HCN2 sequence (Fig. 4B).
FIGURE 4.
Amplification of HCN1 and HCN2 cDNA from cochlear inner and outer hair cells. A, agarose gel for HCN1 PCR indicating amplification from the cochlear inner and outer hair cell cDNA of predicted 397-bp product crossing intron (white arrows). Nucleotide sequencing of uncloned rat HCN1 amplification product from cochlear IHC indicated 100% identity to rat HCN1. Nested primers applied in PCR to the OHC 397-bp product elicited amplification of HCN1 cDNA with 100% identity to rat HCN1 nucleotide sequence. The adjacent lanes (IHC and OHC) with a second set of primers for HCN1 yielded negative results. B, agarose gel for amplification of HCN2 cDNA from rat cochlear outer hair cells (209 bp, crossing intron, black arrow) with 100% nucleotide identity to rat HCN2. S, 1-kb standards; B, water blanks.
Confocal Immunofluorescence for HCN1 and HCN2 in Organ of Corti of the Adult Mouse
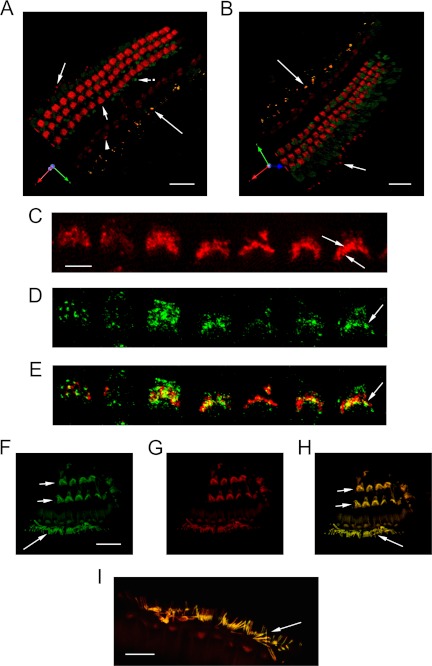
HCN1 protein was immunolocalized to cochlear hair cells within the organ of Corti with both z-stack confocal microscopy and pre-embedding electron microscopy for multiple primary antibodies. In a three-dimensional reconstruction of 43 z-stack 1-μm optical slices of a surface preparation of mouse organ of Corti (Fig. 5, A and B), HCN1 immunofluorescence (red) from the use of the Abcam HCN1 antibody was localized to OHC stereocilia. The optical slices used for reconstruction would extend to subcuticular sites. Immunofluorescence for inner hair cell stereocilia regions was visible but more sparse (Fig. 5, A and B, red). HCN1 protein was also immunolocalized to type I afferents beneath the inner hair cell and in type II afferents beneath the OHC. There was considerably less immunofluorescence for HCN2 (Fig. 5, A and B, green) (Alomone antibody) than for HCN1 in the cochlear outer hair cells for this combination of antibodies. Strong signal for HCN2, however, was found in extensions of Deiters' supporting cells adjacent to outer hair cells at the level of the reticular lamina in the three-dimensional reconstruction (Fig. 5A). HCN2 immunofluorescence co-localized with HCN1 in type I afferents below the IHC (Fig. 5, A and B, overlap in yellow).
FIGURE 5.
z-Stack confocal immunofluorescence analysis of HCN1 and HCN2 protein expression in the organ of Corti of the adult mouse. A and B, three-dimensional reconstruction of 43 1-μm confocal optical slices, indicating immunofluorescence (red) for HCN1 (Abcam 1:50) and HCN2 (green) (Alomone, 1:200) in the organ of Corti of the adult mouse. Immunofluorescence for HCN1 was localized to stereociliary arrays for outer hair cells, which in the reconstruction extended to subcuticular sites (A, short arrow). Immunofluorescence (red) for inner hair cell stereociliary regions was visible but more sparse (A, arrowhead). HCN2 immunoreactivity (green) co-localized with HCN1 in afferents beneath the inner hair cells (A and B, yellow, long arrows). Afferents beneath outer hair cells contained primarily immunofluorescence for HCN1 (A and B, red, medium arrows). The situation that was set up was potentially one of possible competition between HCN1 and HCN2 primary antibodies. Regions of intense immunoreactivity for HCN2 appeared adjacent to the outer hair cells, consistent in position to extensions of Deiters' cells (A, short arrow with dot). Scale bars for A and B, 20 μm. B, second position of three-dimensional reconstruction of z-stack confocal optical slices. The results were consistent with differential distribution of HCN1 and HCN2 in cells of the organ of Corti and differential expression at intracellular sites. C, 1-μm optical section 3 μm in from beginning position of the z-stack, illustrating HCN1 immunofluorescence (red, Santa Cruz Biotechnology) in cochlear outer hair cell stereociliary arrays, with arrows pointing to two individual inner rows of stereocilia in the stereociliary array. Scale bars for C–E, 4 μm. D, 1-μm optical section (paired with C), showing HCN2 immunofluorescence (green, Alomone) in OHC stereocilia (arrow). E, confocally determined overlap (yellow) of HCN1 (C) with HCN2 (D) in stereocilia of OHC (arrow). F, z-stack confocal imaging of HCN2 immunofluorescence (green) in cochlear outer hair cell stereocilia (short arrows) and inner hair cell stereocilia (medium arrow), 0.3-μm optical section (0.6 μm into z-stack) for HCN2 and phalloidin combination. Scale bars for F–H, 20 μm. G, rhodamine-coupled phalloidin detection of F-actin (red) in cochlear hair cell stereocilia. H, confocal overlap of HCN2 with phalloidin (yellow) in cochlear outer hair cell stereocilia (short arrows) and inner hair cell stereocilia (medium arrow). I, magnified view of HCN2 overlap with phalloidin in IHC stereocilia (arrow). Scale bar, 10 μm.
Similar results for immunolocalization of HCN1 to outer hair cell stereocilia were obtained for a second source of primary antibody for HCN1 (Santa Cruz Biotechnology) (Fig. 5C, red, 1-μm optical section). HCN1 immunofluorescence was localized to the inner portion of the stereociliary array in the classic inverted W formation, which would correspond to the tops of the shorter stereocilia. Immunofluorescence for HCN2 (Fig. 5D, green, 1-μm optical section) (Alomone) overlapped that of HCN1 in outer hair cell stereocilia (Fig. 5E, yellow) and in addition appeared to extend beyond immunofluorescence for HCN1 at stereociliary sites.
In the above localization experiments, both HCN1 and HCN2 antibodies were applied together. In contrast, considerably more immunoreactivity was observed for the same HCN2 primary antibody in stereociliary arrays of both outer and inner cochlear hair cells when this antibody was applied separately and only compared with rhodamine-conjugated phalloidin detecting F-actin in secondary antibody incubations (Fig. 5, F–I). The differences in HCN2 immunoreactivity with the same antibody and concentration may reflect competition by HCN1 and HCN2 primary antibodies at the HCN antigenic sites (Fig. 5, A–E) in the first comparison as opposed to lack of competition in the second comparison, with the HCN2 antibody the only primary antibody in the latter localizations of HCN2 and phalloidin (Fig. 5, F–I).
Electron Microscopic Immunolocalization of HCN1 and HCN2 in Cochlear Hair Cells of the Adult Rat
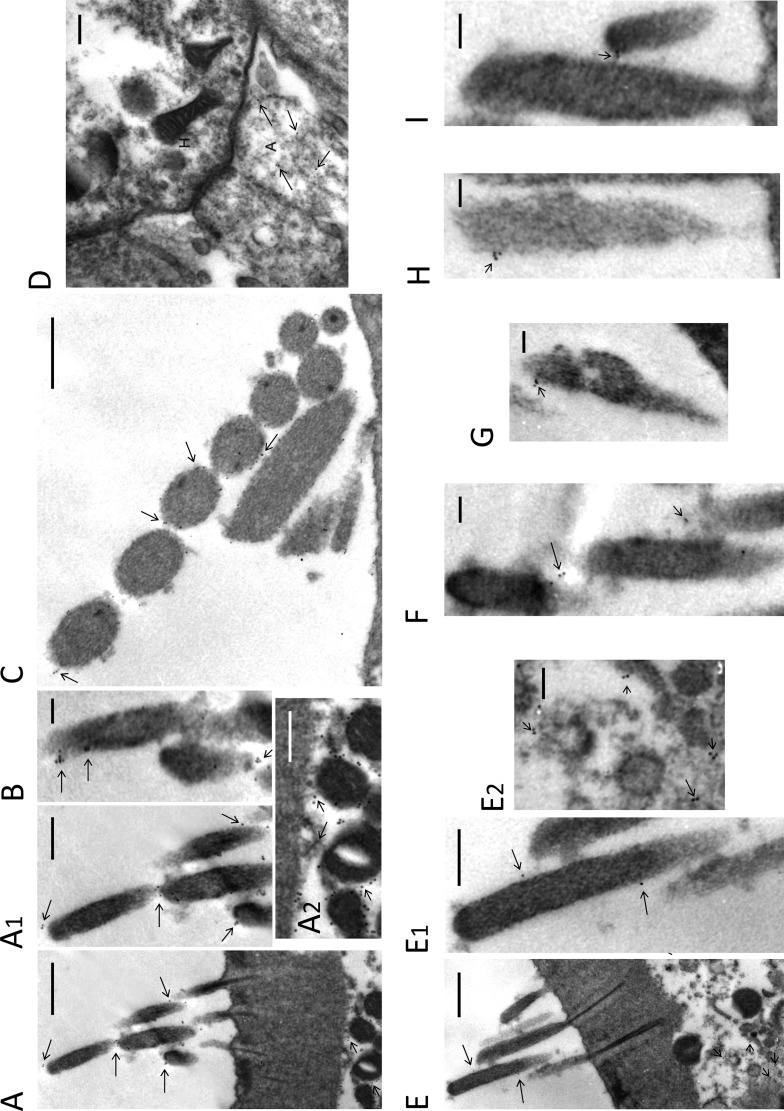
Increased resolution of HCN1 and HCN2 isoform immunolocalization was sought with EM immunogold in a second rodent species. Pre-embedding protocols were utilized to maximize antigenicity potentially lost in post-embedding protocols requiring high temperatures to polymerize embedding media. Immunogold for HCN1 (with the Alomone primary antibody) was localized in OHC to apical (and lateral) sites of stereocilia and concentrated below the cuticular plate (Fig. 6, A, A1, A2, and B) with no gold in IgG negative controls (supplemental Fig. 3C). HCN1 immunogold was also found on IHC stereocilia (Fig. 6C and supplemental Fig. 3A), at sites cross-linking the stereocilia. HCN1 immunogold was localized to type II afferents at the base of the cochlear OHC (Fig. 6D and supplemental Fig. 3B), consistent with results from three-dimensional reconstruction of z-stack optical sections of HCN1 immunofluorescence obtained with the Abcam HCN1 primary antibody for adult mice.
FIGURE 6.
Pre-embedding immunogold detection of HCN1 and HCN2 in hair cells of the organ of Corti in the adult rat. A and B, HCN1 immunogold (Alomone antibody) is found at apical and lateral sites on stereocilia of outer hair cells (arrows) (magnified in A1) not dissimilar in position to that reported for tip-link proteins (4). Also note that HCN1 immunogold in A is concentrated at subcuticular plate sites (short arrows) of outer hair cells (magnified in A2). Scale bars, 500 nm for A; 250 nm for A1 and A2; 100 nm for B. C, HCN1 immunogold (arrows, Alomone primary antibody) is localized in IHC to filaments extending at lateral positions, possibly cross-connecting stereocilia. Scale bar, 300 nm. Magnified view is shown in supplemental Fig. 3A. D, HCN1 immunogold (arrows, Alomone primary antibody) was found in type II afferent endings (A) on OHC (H), as was observed with confocal immunofluorescence with a different HCN1 antibody (Abcam) and different rodent (mouse). Scale bar, 200 nm. Magnified view is shown in supplemental Fig. 3B; HCN2 immunogold was found at lateral positions on taller stereocilia of OHC in the adult rat (E, arrows; magnified in E1) as well as at sites at the top of shorter stereocilia (F–I, arrows). As with HCN1, immunogold for HCN2 was also found at sites beneath the cuticular plate (E, short arrows, magnified in E2), but unlike HCN1, HCN2 was not found in type II afferent dendrites at the base of OHC (not illustrated), consistent with results from confocal microscopy. Scales bars, 500 nm for E; 250 nm for E1; 100 nm for E2, and F–I.
HCN2 immunogold was localized to apical (Fig. 6, E–I) as well as midlateral (Fig. 6, E and E1) positions on OHC stereocilia with some evidence of labeling of adjacent taller stereocilia (Fig. 6, E1 and F) as well as labeling of putative tip-link sites at the top of shorter stereocilia (Fig. 6, G and I). Again, as for HCN1, HCN2 immunogold was present at subcuticular plate sites (Fig. 6, E and E2), but unlike HCN1, HCN2 was not found in type II afferents at the base of cochlear OHC (not illustrated), a result also consistent with findings from confocal microscopy. Taken together, the immunohisto/cytochemical localizations for HCN2 suggest the possibility that HCN2 may have more than one stereociliary site.
Quantitative PCR for HCN Isoform Expression in the Organ of Corti of HCN1 Wild-type and Mutant (HCN1−/−) Mice
A mouse mutant for HCN1, HCN1−/−, created by an in-frame deletion of pore + S6 transmembrane domain (8), was examined for both transcript and protein expression of mutant HCN1. Although there appears to be no question that the slower component of HCN channels (Ih) in the mutant reflects the removal of full-length HCN1, the initial “instantaneous” HCN current, Iinst, remains in the mutant mouse (9). The question that arises for this in-frame deletion is whether a mutated version of HCN1 protein is produced and, in particular, whether the HCN1 amino terminus might be still expressed, be available to bind to protocadherin 15 CD3 (2), and be detected by primary antibodies targeting the HCN1 amino terminus. Furthermore, we wondered whether removal of full-length HCN1 in the HCN1−/− mouse would be accompanied by compensatory quantitative changes in expression in the organ of Corti of a mutant form of HCN1 and the remaining HCN isoforms, HCN2, HCN3, and HCN4.
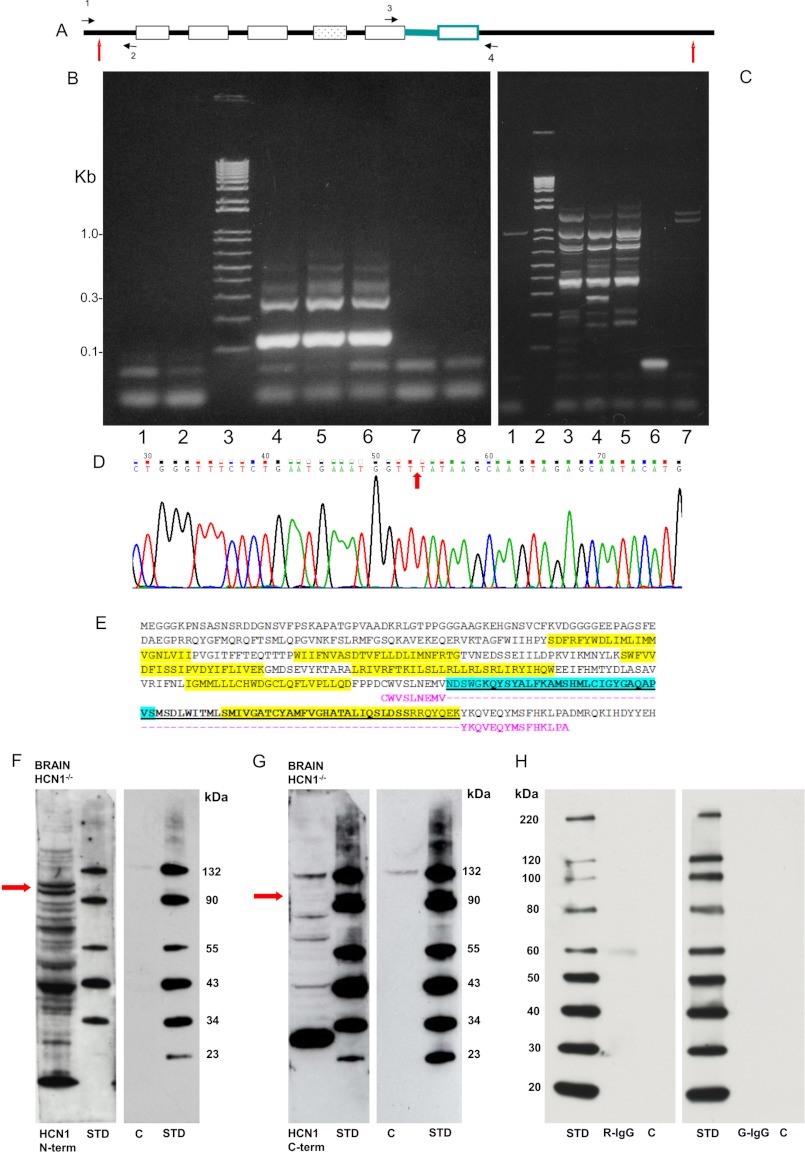
Primers were designed to encompass the deleted pore + transmembrane S6 domain (Fig. 7A, black horizontal arrows 3 and 4), and separately the full amino terminus of HCN1 (Fig. 7A, black horizontal arrows 1 and 2), and applied in PCR to HCN1−/− brain cDNA. The PCR product covering the deleted region clearly corresponded to the predicted size product (120 bp; wild type would be 340 bp, see Fig. 7B) with contiguous mRNA sequence before and after the deletion of the pore and sixth transmembrane region corresponding to exon 4 (Fig. 7D). The amino acid sequence that would be expressed for the in-frame deletion predicted by sequencing of the PCR product for HCN1 in the mutant is indicated in Fig. 7E (highlighted in pink). The PCR product predicted for the wild-type amino-terminal sequence (379 bp) was amplified from HCN1−/− brain cDNA (Fig. 7C), yielding the wild-type HCN1 nucleotide sequence.
FIGURE 7.
Expression of transcript and protein in the HCN1 mutant, HCN1−/− (8). A, diagrammatic representation of HCN1 with primer positions for RT-PCR amplification of the amino-terminal cDNA indicated by black arrows 1 and 2 (upstream primer gggatggctgtcttcagttcctg; downstream primer ccacaagttaccagctgaca, Δ379 bp). Black arrows 3 and 4 designate primers encompassing the pore and S6 transmembrane domains (upstream primer atggaaggcggcggcaaacccaa; downstream primer gcaggcttctggattatccatccg, Δ120 bp). Amino- and carboxyl-terminal positions of HCN1 amino acid sequence targeted by primary antibodies in Western blots (Fig. 8, F and G) are indicated by red vertical arrows, respectively. B, agarose gel separation of cDNA products amplified from brain cDNA of the HCN1−/− mutant mouse with primers situated in front and back of the in-frame deletion of the pore filter + S6 transmembrane domain. Lanes 1 and 2 are water blanks; lane 3, standards; lanes 4–6 replicates with predicted 120-bp products; lanes 7 and 8, −RTs). C, agarose gel separation of cDNA amplified (379 bp) from brain of HCN1−/− for full amino terminus of HCN1; lane 1, −RT; lane 2, standards; lanes 3–5, three separate + RT preparations; lane 6, water blank; and lane 7, −RT. D, arrow shows splice location between exons 3 and 5 (exon 4 is deleted in the knock-out) in contiguous nucleotide sequence of HCN1 in brain cDNA for HCN1−/− mutant. E, HCN1 wild-type amino acid sequence is presented. Transmembrane regions are highlighted in yellow. The underlined sequence, including exon 4 (boldface), is deleted in HCN1−/−. The pore is highlighted in blue; the S6 transmembrane region (underlined) is highlighted in yellow. Contiguous amino acid sequence in HCN1−/− is in pink. F and G, Western blots demonstrating protein fragments of HCN1 remaining in brain (35-μg quantities) of HCN1−/− as determined by the HCN1 amino-terminal targeting antibody (F, Alomone primary antibody raised in rabbit) and HCN1 carboxyl-terminal targeting antibody (G, Santa Cruz Biotechnology primary antibody raised in goat), evidence of full-length protein translation minus the pore + S6. Red arrows show doublet bands, present in both blots, representing longest fragments consistent with predicted mass for HCN1 in HCN1−/− mutant of 93.6 kDa (lower band) and presumably a glycosylated version (10) (upper band). H, IgG negative controls, left panel, Western blots of HCN1−/− brain lysate probed with equivalent amounts of rabbit IgG (R-IgG) as used in F, and right panel, equivalent goat IgG (G-IgG) as used in G. For F–H, C = Western negative control with brain lysate, secondary antibody, but no primary antibody. STD = standards.
To investigate possible HCN1 peptide expression in the brain of the in-frame HCN1−/− mutant, we carried out Western blots with antibodies targeting HCN1 amino- and carboxyl- terminal amino acid sequences. Western blots performed for 35-μg quantities of HCN1−/− brain protein (considerably more than the 15-μg quantities originally analyzed (8)) demonstrated that HCN1 protein fragments remained in the HCN1 mutant whether detected by an amino-terminal targeting antibody (Fig. 7F) or a carboxyl-terminal targeting antibody (Fig. 7G). Western blots of HCN1−/− brain protein in negative controls, probed with the equivalent amounts of corresponding IgGs, were unlabeled (Fig. 7H). These results were consistent with full-length protein translation minus the pore + S6, plus HCN1 peptide fragments. (Fig. 7, F and G, red arrows show doublet bands, present in both blots, representing longest fragments, with the lower band consistent with predicted mass of mutant HCN1 protein, 93.6 kDa, and the upper band, a glycosylated version as reported for wild type; Ref. 10.)
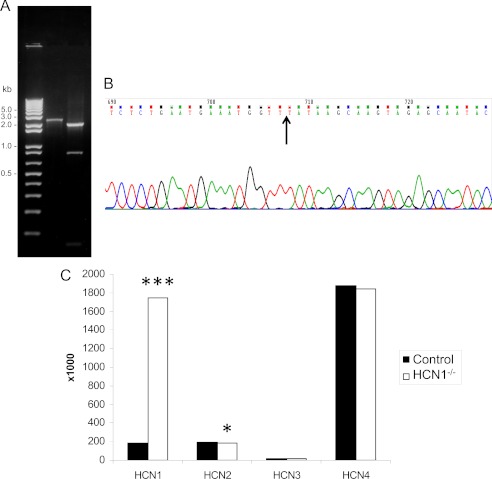
Full-length transcript expression of HCN1 was also examined for an isolated organ of Corti fraction from the cochlea of the HCN1−/− mutant. Agarose gel separation of HCN1 products from RT-PCR indicated reduced size for the HCN1−/− sequence corresponding to the deletion of pore and S6 compared with wild-type expression (Fig. 8A). Nucleotide sequences for HCN1 in the organ of Corti cDNA from the mutant demonstrated again, as for brain, contiguous mRNA sequence before and after the in-frame deletion of the pore filter and sixth transmembrane region (Fig. 8B, arrow shows splice location between exons 3 and 5; exon 4 is deleted in the mutant). Quantitative PCR indicated transcript expression of HCN1/HCN2/HCN3/HCN4 = 9:9:1:89 in organ of Corti of the wild-type mouse. Immunoreactivity for HCN4 protein in the organ of Corti of the wild type is primarily attributable to non-hair cell sources, i.e. the inner sulcus cells (with DAB detection, not illustrated), presumably correlating with the relatively elevated level of transcript for HCN4 in wild-type organ of Corti. Levels of the mutant form of HCN1 in the organ of Corti of the HCN1−/− were elevated by a factor of nine times (900%) the level of HCN1 transcript in wild-type mice, with HCN2, HCN3, and HCN4 levels little changed for the HCN1−/− mutant versus wild type (Fig. 8C).
FIGURE 8.
Quantitative PCR analysis of HCN isoform expression in organ of Corti of adult mouse for wild type and HCN1−/−. A, agarose gel resolution of PCR product for full-length HCN1 cDNA in organ of Corti for control (lane 2, predicted Δ = 2,363 bp) versus mutant (lane 3, predicted Δ = 2,144 bp), demonstrating full-length sequence in mutant with the in-frame deletion. B, nucleotide sequence for organ of Corti HCN1 in the HCN1−/−, demonstrating again, as for brain, contiguous mRNA sequence before and after the in-frame deletion of the pore filter and sixth trans-membrane region. Arrow shows splice location between exons 3 and 5 (exon 4 is deleted in the mutant). C, quantitative PCR for HCN isoforms in morphologically defined mouse cochlear organ of Corti subfraction (6) for HCN1−/− versus control. *** indicates p = 0.0026 for HCN1 in HCN1 mutant versus control ΔCt by unpaired, two-tailed t test. * indicates p = 0.054 for HCN2 in HCN1 mutant versus control. Five sets of experiments each with two replicates per point.
Actin-linked Protein-Protein Interactions for HCN1 and HCN2 by Immunoprecipitation for the Wild Type (Rat)
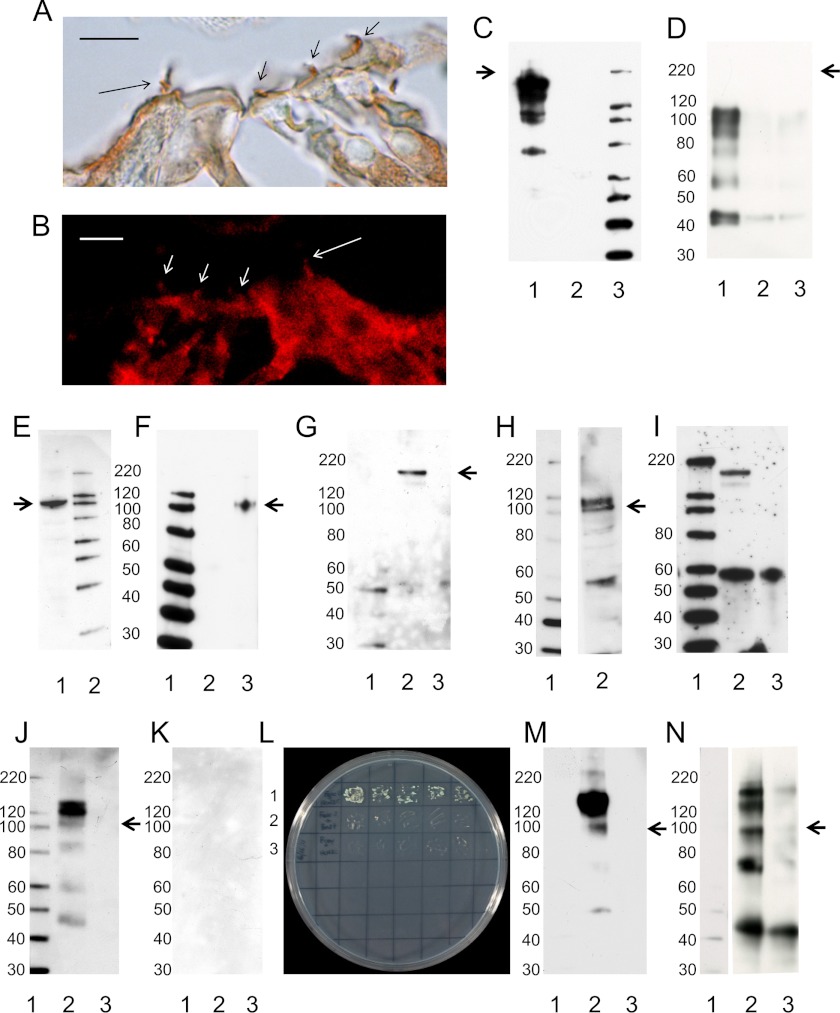
The possibility of a molecular pathway via protein-protein interactions linking the HCN isoform HCN1 to F-actin-binding filamin A was explored. Immunoreactivity to filamin A was localized to stereocilia of cochlear inner and outer hair cells (Fig. 9, A and B). Immunoprecipitation protocols with anti-filamin A primary antibody carried out on rat brain lysates indicated that a ternary complex can be formed between full-length filamin A and HCN1 and protocadherin 15 CD3 (Fig. 9, C, F, and G; supplemental Fig. 4, A and B for additional IgG negative controls), a complex that did not include HCN2 (Fig. 9I). In contrast, anti-HCN2 primary antibody immunoprecipitated HCN1 (Fig. 9J; supplemental Fig. 4C for IgG negative control), consistent with HCN1-HCN2 channel formation via the conserved amino terminus (11), and the complex immunoprecipitated by HCN2 did not contain protocadherin 15 CD3 (Fig. 9K). In addition, evidence was obtained that HCN2 itself interacts, via its carboxyl terminus, with F-actin-binding fascin-2 (Fig. 9, L and M, and supplemental Fig. 4D for IgG negative control) in a complex that includes HCN1 (Fig. 9N) but not protocadherin 15 CD3 (not illustrated).
FIGURE 9.
Immunoprecipitation by primary antibodies to filamin A, HCN2, and fascin-2. Immunoreactivity for filamin A detected by DAB (A) and immunofluorescence (B) (mouse MAB1678 raised to human filamin A crossing to rat; 1:1,000, clone PM6/317, Chemicon) was localized to stereocilia of both inner hair cells (long arrows) and outer hair cells (short arrows) in rat cochlea. Scale bars for A and B, 10 μm. C, lane 1, full-length filamin A (detected with filamin A primary antibody 1:1,000) in immunoprecipitation complex arising from the use of anti-filamin A for immunoprecipitation (1:100) from rat brain lysate. This antibody recognizes unprocessed filamin A (270–280 kDa, arrow, as well as 170-, 150-, and 120-kDa cleavage fragments (Chemicon)). Lane 2, negative control with brain lysate, without anti-filamin A immunoprecipitation + beads + primary and secondary antibodies; lane 3, standards. D, additional negative control for C. Immunoprecipitation with mouse IgG as negative control probed with antifilamin A. Lane 1, mouse IgG immunoprecipitation of brain lysate + beads, probed with filamin A primary + donkey anti-mouse secondary. Lane 2, mouse IgG (no immunoprecipitation) + beads, probed with filamin A primary + donkey anti-mouse secondary. Lane 3, denatured mouse IgG electrophoresis, probed with filamin A primary + donkey anti-mouse secondary. No protein was observed corresponding to molecular mass of filamin A (arrow). E, Western blot of HCN1 (104 kDa, arrow) in rat brain lysate (1:50, sc-19706, Santa Cruz Biotechnology), lane 1; lane 2, standards. F, immunoprecipitation by anti-filamin A of full-length HCN1. Lane 1, standards; lane 2, negative control with brain lysate without anti-filamin A immunoprecipitation + beads + primary and secondary antibodies; lane 3, HCN1 immunoprecipitated with anti-filamin A (arrow, 104 kDa), detected with goat anti-mouse HCN1 (carboxyl terminus) which crosses to rat HCN1 sequence (sc-19706 Santa Cruz Biotechnology). G, complex of filamin A and HCN1 also contained protocadherin 15 CD3. Lane 1, standards; lane 2, full-length protocadherin 15 CD3 (189 kDa, arrow) immunoprecipitated with anti-filamin A detected with custom primary antibody (1:10,000) (arrow). Goat anti-chick IgY-HRP (Santa Cruz Biotechnology) was used as the secondary antibody (1:10,000). Lane 3, negative control without anti-filamin A immunoprecipitation but with brain lysate and protein A beads and primary and secondary antibodies. H, Western blot of HCN2 in rat brain lysate. Lane 1, standards; lane 2, HCN2 detected with a rabbit polyclonal antibody (1:200, Alomone). Predicted molecular masses of 95 and 127 kDa corresponding to unglycosylated (arrow) and glycosylated HCN2, respectively. I, HCN2 is not detected in the complex immunoprecipitated by anti-filamin A. Lane 1, standards; lane 2, HCN2 bands observed in Western (H) are not detected in immunoprecipitated complex. Bands at 170–180 kDa may correspond to protocadherin 15 CD3, given that 5 of 15 aa in the epitope targeted by the HCN2 antibody are identical in protocadherin 15 CD3, and further given that protocadherin 15 CD3 is highly concentrated in the complex immunoprecipitated by anti-filamin A (G). J, anti-HCN2 immunoprecipitates HCN1 from rat brain lysate. Lane 1, standards; lane 2, anti-HCN2 primary antibody (1:33, Alomone) immunoprecipitates HCN1 unglycosylated (104 kDa, arrow) and glycosylated (120 and 130 kDa) forms detected with anti-HCN1 primary antibody (Santa Cruz Biotechnology); lane 3, negative control for immunoprecipitation with all components except anti-HCN2 antibody for immunoprecipitation (brain lysate and beads). K, anti-HCN2 immunoprecipitation of HCN1 complex does not include protocadherin 15 CD3. Lane 1, standards; lane 2, anti-protocadherin 15 CD3 primary antibody (1:7,500), goat anti-chick secondary antibody (1:5,000); lane 3, negative control without anti-HCN2 antibody for immunoprecipitation (brain lysate and beads). No bands were evident for either experimental (lane 2) or negative control (lane 3) with goat anti-chick IgY as the secondary antibody. In a second protocol, a bovine anti-chick secondary antibody (1:5,000) detected an ∼170-kDa protein in both experimental and negative control (not illustrated). L, HCN2 binds to fascin-2 by yeast two-hybrid co-transformation. Row 1, HCN2 carboxyl terminus in bait vector pGBKT7 plus fascin-2 in prey vector pGADT7; row 2, negative control with fascin-2 in prey construct plus empty bait construct; row 3, negative control with HCN2 in bait construct plus empty prey construct. The co-transformation screening was performed once, and then the desired colony was re-plated onto another selection media in triplicate, with negative controls. The same concentrations of yeast were plated in the grids for rows 1-3. M, anti-fascin-2 immunoprecipitation of HCN2. Lane 1, standards; lane 2, 95 kDa (arrow) and 127-kDa bands, corresponding to unglycosylated and glycosylated HCN2, respectively; lane 3 negative control without anti-fascin 2 antibody in immunoprecipitation (brain lysate and beads). N, HCN1 is in fascin-2 immunoprecipitation complex along with HCN2. Lane 1, standards; lane 2, unglycosylated (arrow) and glycosylated forms of HCN1 at 120 kDa. Lane 3, goat IgG immunoprecipitation negative control, with beads and all components except anti-fascin-2 for immunoprecipitation. Three or more immunoprecipitation experiments were carried out for each protein in a given complex.
DISCUSSION
HCN (hyperpolarization-activated, cyclic nucleotide-gated) cation currents (12) include two components as follows: Iinst, a voltage-independent instantaneous current (13–20) requiring low physiological (10 mm) concentrations of [Cl−]i (20), and Ih, a slowly activating voltage-dependent and inwardly rectifying cation current (21), which is not dependent on [Cl−]i (20). Iinst and Ih have been further distinguished on the basis of differential responses to prior hyperpolarization (20) and to extracellular Cs+. [Cs+]o is a potent inhibitor of Ih but not of Iinst (18), except in the circumstance of increases of Iinst with prior hyperpolarization (Ref. 20; however, see Ref. 9). Furthermore, only Ih is sensitive to a mutation of the GYG motif in the selectivity filter of the pore, i.e. this motif and portion of the pore are not required for Iinst (22). Given that Iinst can be regulated separately from Ih (13, 18, 20, 22–24), the possibility has been suggested that Iinst arising from HCN isoform expression could represent a “second configuration of recombinant HCN channels” from that underlying Ih (18) or derive from separate populations of ion channels from those contributing to Ih (23).
Mechanical stimuli can affect HCN channel function. HCN1 would be associated with mechanical stimuli through its interaction with filamin A (25), an actin-binding protein that does not interact with either HCN2 or HCN4. Mechanical strain in actin networks differentially regulates protein-protein interactions of filamin A, specifically to the cytoplasmic tails of β-integrins and FilGAP (26), providing a “direct and specific molecular basis for cellular mechanotransduction” (26). Stretch can be a stimulus for HCN2, reported to accelerate hyperpolarization-induced activation as well as depolarization-induced deactivation of Ih derived from heterologous expression of HCN2 (24). Hypo-osmotic cell swelling induces activation of HCN2, dependent on the integrity of actin (27). The molecular pathways underlying the association of HCN2 with actin are unknown (27). HCN2 underlies mechanically driven action potentials in nociceptors mediating pain sensation (28).
The effect of mechanical stimulation on HCN channels in inner ear hair cells is unknown. A possible role in stereociliary function would relate to activation of HCN channels with hyperpolarization and/or contribution to the initial very rapid cation influx of MET. It is interesting that Iinst is dependent on the frequency of hyperpolarizing pulses (20). Ih in other sensory receptor cells is suggested to have a modulatory role, providing a “tunable element” (24), and for rod and cone photoreceptor cells, providing a mechanism to make the light response more transient (9, 29). HCN1, the predominant HCN isoform expressed in cone photoreceptors, modulates photocurrent input, speeding up the light response (29).
Previously, we found that the cytoplasmic amino terminus of HCN1, the primary full-length HCN isoform expressed in trout saccular hair cells (1), binds the cytoplasmic carboxyl-terminal domain of a tip-link protein, protocadherin 15a-like protein (2). HCN1 was immunolocalized to discrete sites on saccular hair-cell stereocilia, consistent with gradated distribution expected for tip-link sites of protocadherin 15a. HCN1 message was also detected in cDNA libraries of rat cochlear inner and outer hair cells, and HCN1 protein was immunolocalized to cochlear hair-cell stereocilia. As predicted by the trout hair cell model, the amino terminus of rat organ of Corti HCN1 was found by yeast two-hybrid analysis to bind the carboxy intracellular terminus of protocadherin 15, specifically protocadherin 15 CD3, a tip-link protein thought to be associated with hair cell mechanosensory transduction. Binding between HCN1 and protocadherin 15 CD3 was confirmed with pulldown assays and surface plasmon resonance analysis, both predicting dependence on Ca2+. In the presence of calcium chelators, binding between HCN1 and protocadherin 15 CD3 was characterized by a KD = 2.39 × 10−7 m. Ca2+ at 26.5–68.0 μm promoted binding, with a KD = 5.26 × 10−8 m (at 61 μm Ca2+). Binding by deletion mutants of protocadherin 15 CD3 pointed to aa 158–179 (GenBankTM accession number XP_238200), with homology to the comparable region in trout hair-cell protocadherin 15a-like protein, as necessary for binding to HCN1. Amino terminus binding of HCN1 to HCN1, hypothesized to underlie HCN1 channel formation (11), was also found to be Ca2+-dependent, although the binding was skewed toward a lower effective maximum [Ca2+] than for the HCN1 interaction with protocadherin 15 CD3. We hypothesized that competition may therefore exist in vivo between the two binding sites for HCN1, with binding of HCN1 to protocadherin 15 CD3 favored between 26.5 and 68 μm Ca2+ (2), concentrations of calcium associated with the mechanotransduction channel closure during adaptation (30).
Further characterization of HCN1 binding to protocadherin 15 CD3 emphasizes the specificity of the protein-protein interaction. Localization of both transcript and protein for HCN1 to cochlear hair cells has been confirmed. The amino-terminal peptide sequence specific to HCN1 is sufficient for HCN1 interaction with protocadherin 15 CD3. Neither the amino-terminal sequence found in HCN1 that is conserved across HCN isoforms, nor the HCN2- or HCN4-specific amino termini, will substitute, as evidenced by SPR analysis. The first 77-amino-terminal amino acids of HCN1 therefore appear to be key for the interaction and consequent molecular interaction. A phospholipid interaction for the specific amino terminus of HCN1 has also been identified as follows: PIP3 and PIP2 bind with KD values of 2.75 × 10−9 and 3.38 × 10−8 m, respectively. PIP2 is reported to enhance the opening of hyperpolarization-activated cyclic nucleotide-regulated (HCN) channels (apparently for Iinst and Ih), shifting activation gating to more positive voltages (31). Coincidentally, PIP2 is required for both hair cell mechanotransduction and adaptation (32).
Further resolution of immunolocalization of HCN1 protein (2) has been obtained in this study with z-stack confocal immunofluorescence microscopy and three-dimensional reconstruction for commercially available HCN1 antibodies applied to the mouse cochlea. Immunolabeling for each antibody yielded the same result, localization of HCN1 protein to the stereocilia of cochlear hair cells. Pre-embedding electron microscopic (EM) immunogold protocols for rat cochlea with a third commercially available antibody confirmed the results for HCN1 obtained with z-stack confocal microscopy for the mouse and additionally confirmed that HCN1 protein is concentrated in subcuticular plate regions of cochlear outer hair cells, not dissimilar to the localization of protocadherin 15 (33), which binds to HCN1.
Evidence for a second HCN isoform, HCN2, at stereociliary sites was obtained with both confocal and EM immunolocalizations. The immunogold localization sites of HCN1 and HCN2 on stereocilia were noted to be similar to those of tip-link proteins protocadherin 15 CD3 and cadherin 23 (5). The determination that immunogold labeling of stereocilia and subcuticular plate was correlated, for both HCN1 and HCN2, in pre-embedding immunogold EM in this study speaks to the overall specificity of the approach. With three-dimensional reconstruction of z-stack optical slices, we confirmed co-localization of HCN1 and HCN2 in type I afferents (34) and provided new information with both z-stack confocal and immunogold microscopic protocols that HCN1 is also found in type II afferents beneath the outer hair cells.
Evidence from direct analysis of isolated cochlear hair cells indicates that both outer and inner hair cells express the HCN1 transcript, a finding that is consistent with the results from immunolocalization of the HCN1 protein, and it confirms previous results for HCN1 obtained with λ ZAP cDNA libraries (2). Theoretically, HCN2 and HCN4 (and HCN3) could participate in HCN channel formation along with HCN1 in hair cells via protein-protein interactions of their conserved amino-terminal sequence (2, 11). Molecular results in this study place HCN2 (transcript) in cochlear outer hair cells, and immunogold localization points to HCN2 protein in outer hair cell stereocilia, consistent with the possibility that both HCN1 and HCN2 participate in channel formation in the outer hair cell stereocilia. Positive immunolocalization results for HCN isoforms in cochlear inner hair cell stereocilia have been obtained by others, however, with the immunohistochemical specificity questioned by the previous authors due to difficulties in antibody preabsorption (34).
Expression of HCN Isoforms by Quantitative PCR in the Organ of Corti of Wild-type Mice and HCN1 Mutant
An organ of Corti tissue fraction isolated from the mouse cochlea was analyzed by quantitative PCR. This organ of Corti fraction has been morphologically characterized and is free of stria vascularis and lateral wall, spiral limbus (and tectorial membrane), and cell bodies of the spiral ganglion (6). For the wild-type organ of Corti (mouse), HCN1/HCN2/HCN3/HCN4 = 9:9:1:89. According to our immunohistochemical analysis, HCN4 protein within the organ of Corti would be primarily attributable to inner sulcus cells.
Transcript expression was also examined in an HCN1 mutant created by an in-frame deletion of the pore + the sixth transmembrane region (8). We obtained evidence for this mutant of expression of full-length HCN1 mRNA with the in-frame deletion in both brain and organ of Corti. Quantitative PCR applied to the subdissected cochlear organ of Corti indicated an ∼9-fold elevation of mutant HCN1 transcript in HCN1−/− relative to wild-type levels of HCN1 in controls, with unsubstantial changes of HCN2, HCN3, and HCN4 transcript levels.
Western analysis of HCN1 protein in the mutant, albeit for larger amounts of protein than originally analyzed (8), indicated the existence of full-length channel protein (minus the deleted region) as well as HCN1 protein fragments detected by HCN1 primary antibody, presumably the consequence of the fact that the deletion of the pore and S6 was in-frame. This detection was not dependent on the nature of the primary HCN1 antibodies that separately targeted either the amino-terminal (9) or the carboxyl-terminal amino acid sequence, the latter 3′ to the in-frame deletion. The remaining HCN1 fragments (and full-length sequence lacking the deleted region) would be targeted by HCN1 primary antibodies, consistent with observed hair cell stereociliary immunostaining in the HCN1 mutant (35), thus further validating hair cell stereociliary immunoreactivity that was observed for the wild type in the present study and in a previous investigation (2). The amino terminus of HCN1 in the mutant would be available to bind other HCN isoforms (HCN2) in channel formation as well as bind to protocadherin 15 CD3, given the position of the deletion. Abbreviated and truncated versions of HCN isoforms with intact amino termini can form physiologically functional channels in combination with other full-length HCN isoforms (36). In the heart, HCN2 is truncated with only 57% of its full-length sequence expressed, and it apparently forms a physiologically active channel in combination with HCN4 (36).
Electrophysiological investigations of the HCN1 mutant indicate loss of HCN1 in the slow component Ih (9, 37) without diminishment of the initial instantaneous current, Iinst (8, 9). No information on the two current components is available from in vitro heterologous expression of the HCN1 mutant construct. Ion channel compensation has been reported in the HCN1 mutant. There is a parallel reduction in the magnitude of IKL and Ih in bushy and octopus cells of the ventral cochlear nucleus of the HCN1 mutant, and correlative behavior is also observed in the wild type (37). Ih and IKx complementary conductance changes have also been reported in wild-type rod photoreceptors (29). GABAA current in the cortical pyramidal neurons of the HCN1 mutant is increased and hypothesized to replace Ih (38).
Protocadherin 15 Isoforms
A recent publication (39) indicates deficits in the auditory brainstem response and deafness attributable to the deletion of the intracellular cytoplasmic domain of protocadherin 15 CD2 but not in comparable mutants for protocadherin 15 CD3 or protocadherin 15 CD1. The tip-links for all three splices appeared unaltered, presumably due to the fact that the amino-terminal extracellular domains for all three splice variants remained available in the mutants to bind cadherin 23 in formation of the stereociliary tip-links.
Curiously, hair cell mechanotransduction in the CD2 and CD3 mutants was apparently unaltered compared with controls at the early post-natal age (P7/P8) used for measurements of outer hair cell function (39). The finding that the protocadherin 15 carboxyl-terminal deletion mutants did not have obvious mechanotransduction deficits represents a significant deviation from theory. The protocadherin 15 tip-link proteins are thought to be localized at the tops of shorter stereocilia, a site of calcium influx and the presumed site of a mechanotransduction channel or complex (5, 30). The protocadherin 15 carboxyl terminus, the only cytoplasmic domain in this protein, is hypothesized to couple to the channel, and its absence should impinge hair cell mechanotransduction.
However, another interpretation exists. The interesting question for knock-outs/mutants, in general, is which other proteins/channels may be up- and down-modulated. An up-expression of the GABAA chloride channel (38) and/or down modulation of IKL (37) have been reported for the HCN1 mutant. The change of IKL presumably occurs to maintain balance between inward and outward potassium flux regulating the membrane potential. Also, in our experiments, quantitative PCR indicated probable compensation, with a 9-fold increased expression of the mutant form of the HCN1 transcript relative to the HCN1 wild-type form in controls. In the study by Webb et al. (39), quantitative PCR indicated, very interestingly, that not only was the expression of CD3 splice transcript specifically eliminated for the protocadherin 15 CD3 knock-out but the expression of CD2 transcript was elevated, and by 20-fold. Given the identity/homology of the carboxyl-terminal sequence in protocadherin 15 CD2 and CD3, the question is whether protocadherin 15 CD2 can substitute for protocadherin 15 CD3 in binding to the amino terminus of HCN1.
HCN Channels in Inner Ear Hair Cells
There is little doubt about the existence of Ih in vestibular and lateral line hair cells (40–42), with sequencing of full-length HCN1 transcript and in situ hybridization-localization in saccular hair cells (1) and immunolocalization of HCN1 protein to both the basolateral membrane and stereocilia of hair cells in this vestibular hair cell model (2). Furthermore, Ih has been detected in a cochlear hair cell precursor line of the Immortomouse (43), and a cAMP-sensitive, nonselective cation conductance has been observed in both inner and outer hair cells of the young adult guinea pig cochlea (44). However, in contrast, possible involvement of HCN channels in MET is controversial. On the one hand, the specific inhibitor of Ih and Iinst, ZD7288, blocked mechanotransduction of mammalian cochlear outer hair cells and type II vestibular hair cells (35). Furthermore, ZD7288 and another “specific” inhibitor of Ih, DK-AH269, blocked HCN channels in hair cells of the zebrafish lateral line (42), accompanied by a partial block of MET. On the other hand, cochlear outer hair cells and type II vestibular hair cells from the HCN1 mutant did not reveal obvious deficits in the initial rapid inward component of MET currents elicited by mechanical stimuli, representing a test for MET (35). However, this latter finding by itself is identical to the lack of change in Iinst reported for the HCN1 mutant HCN−/−. Whether MET was influenced in cochlear inner hair cells or in type I vestibular hair cells of the HCN1 mutant or whether there were differences in MET adaptation for any hair cell type was not evaluated. However, in any case, there are differences in both overall conductance and Ca2+ permeability between HCN channels and MET. A question that emerges is whether MET comprises two channels (45), with the second channel, unlike the HCN channel, characterized by high Ca2+ permeability.
Other Ion Channels in Hair Cell Stereocilia
There is molecular and electrophysiological evidence that a number of ion channels are expressed in cochlear hair cell stereocilia. TRPML3 (46), polycystin-1 (47), amiloride-sensitive sodium channels (48), TRPA1 (49, 50), CFTR (51), TMC1 (52–54), CLIC5 (55), ATP (56, 57), TRPV1 (58), TRPV4 (59), HCN1 (2), CNG channels, CNGA2 and CNGA4 (60), and CNGA3 (61) have all been implicated as either being present and/or candidates for mechanotransduction channels. The multitude of channels would by itself point to the possibility of ion channel complexes and the possibility of separate modulation of initial rapid inward current and adaptation. Interpretations of the role of each channel expressed in hair cell stereocilia has often been dependent upon the use of mutants/knock-outs of the channel with the potential difficulty of the existence of more than one variable, i.e. indirect effects due to associated, possibly compensatory changes in other ion channels that would affect the overall interpretation. Furthermore, if genetic manipulation of an inward current such as Ih can affect an outward current, IKL (37), the possibility exists that genetic manipulation of an outward basolateral current may also affect an inward current.
Conclusions
Our analysis of singly isolated cochlear hair cells that has indicated transcript expression of HCN1 together with confocal and immunogold localization of HCN1 protein to cochlear hair cell stereocilia and subcuticular plate sites, as well as SPR results, are all consistent with the molecular resolution afforded in the determinations of calcium-dependent protein-protein interaction between HCN1 and protocadherin 15 CD3 (2). To the complex of HCN1 and protocadherin 15 CD3, we can now add F-actin-binding filamin A, known to interact with HCN1 (25), representing a possible mechanism to link a stereociliary HCN1 channel to the actin-cytoskeleton in stereocilia and mechanotransduction (26). Immunolocalizations of HCN2 to tip-link sites in this study would be consistent with the additional participation of this second HCN isoform in the formation of stereociliary HCN channels in cochlear hair cells. Curiously, although HCN2 immunoprecipitated HCN1, presumably via the amino-terminal sequence conserved across HCN isoforms, it was not present in the filamin A complex with HCN1 and protocadherin 15 CD3. Protocadherin 15 CD3 was not present in the complex immunoprecipitated by HCN2 that did include HCN1. HCN2 itself is immunoprecipitated by F-actin-binding fascin-2, and this complex includes HCN1 but not protocadherin 15 CD3. These results are consistent with separation of molecular function; HCN1 appears either to bind protocadherin 15 CD3 and filamin A or to bind HCN2. We previously showed a similar separation of molecular binding by alteration in [Ca2+]; protocadherin 15 CD3 binding to HCN1 was enhanced at a concentration of Ca2+ (associated with adaptation of mechanotransduction) that was higher than required for binding of HCN1 to HCN1 (as occurs in channel formation) (2). Interestingly, in all of the descriptions and characterizations of ion channels present in hair cell stereocilia, only HCN1, to date, has been found to bind to a stereociliary tip-link protein.
Supplementary Material
Acknowledgments
We thank Joyce Benjamins and Jeffrey Loeb for their input in quantitative PCR determinations.
This work was supported, in whole or in part, by National Institutes of Health Grants R01 DC004076 (to M. J. D.) and R01 DC000156 (to D. G. D.).

This article contains supplemental Figs. 1–4.
- HCN1
- hyperpolarization-activated, cyclic nucleotide-gated nonselective cation channel isoform 1
- HCN2
- hyperpolarization-activated, cyclic nucleotide-gated nonselective cation channel isoform 2
- PPI
- protein-protein interaction
- SPR
- surface plasmon resonance
- OHC
- outer hair cell
- IHC
- inner hair cell
- PIP2
- phosphatidylinositol (4,5)-bisphosphate
- PIP3
- phosphatidylinositol (3,4,5)-trisphosphate
- aa
- amino acid
- DAB
- 3,3′-diaminobenzidine
- MET
- mechanotransduction channel
- Ih
- hyperpolarization-activated, inwardly rectifying cation current
- Iinst
- instantaneous current coded for by HCN isoforms.
REFERENCES
- 1. Cho W. J., Drescher M. J., Hatfield J. S., Bessert D. A., Skoff R. P., Drescher D. G. (2003) Hyperpolarization-activated, cyclic AMP-gated, HCN1-like cation channel. The primary, full-length HCN isoform expressed in a saccular hair-cell layer. Neuroscience 118, 525–534 [DOI] [PubMed] [Google Scholar]
- 2. Ramakrishnan N. A., Drescher M. J., Barretto R. L., Beisel K. W., Hatfield J. S., Drescher D. G. (2009) Calcium-dependent binding of HCN1 channel protein to hair cell stereociliary tip link protein protocadherin 15 CD3. J. Biol. Chem. 284, 3227–3238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Seiler C., Finger-Baier K. C., Rinner O., Makhankov Y. V., Schwarz H., Neuhauss S. C., Nicolson T. (2005) Duplicated genes with split functions. Independent roles of protocadherin15 orthologues in zebrafish hearing and vision. Development 132, 615–623 [DOI] [PubMed] [Google Scholar]
- 4. Kazmierczak P., Sakaguchi H., Tokita J., Wilson-Kubalek E. M., Milligan R. A., Müller U., Kachar B. (2007) Cadherin 23 and protocadherin 15 interact to form tip-link filaments in sensory hair cells. Nature 449, 87–91 [DOI] [PubMed] [Google Scholar]
- 5. Grati M., Kachar B. (2011) Myosin VIIa and sans localization at stereocilia upper tip-link density implicates these Usher syndrome proteins in mechanotransduction. Proc. Natl. Acad. Sci. U.S.A. 108, 11476–11481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Oh C. K., Drescher M. J., Hatfield J. S., Drescher D. G. (1999) Selective expression of serotonin receptor transcripts in the mammalian cochlea and its subdivisions. Brain Res. Mol. Brain Res. 70, 135–140 [DOI] [PubMed] [Google Scholar]
- 7. Drescher M. J., Cho W. J., Folbe A. J., Selvakumar D., Kewson D. T., Abu-Hamdan M. D., Oh C. K., Ramakrishnan N. A., Hatfield J. S., Khan K. M., Anne S., Harpool E. C., Drescher D. G. (2010) An adenylyl cyclase signaling pathway predicts direct dopaminergic input to vestibular hair cells. Neuroscience 171, 1054–1074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Nolan M. F., Malleret G., Lee K. H., Gibbs E., Dudman J. T., Santoro B., Yin D., Thompson R. F., Siegelbaum S. A., Kandel E. R., Morozov A. (2003) The hyperpolarization-activated HCN1 channel is important for motor learning and neuronal integration by cerebellar Purkinje cells. Cell 115, 551–564 [DOI] [PubMed] [Google Scholar]
- 9. Knop G. C., Seeliger M. W., Thiel F., Mataruga A., Kaupp U. B., Friedburg C., Tanimoto N., Müller F. (2008) Light responses in the mouse retina are prolonged upon targeted deletion of the HCN1 channel gene. Eur. J. Neurosci. 28, 2221–2230 [DOI] [PubMed] [Google Scholar]
- 10. Much B., Wahl-Schott C., Zong X., Schneider A., Baumann L., Moosmang S., Ludwig A., Biel M. (2003) Role of subunit heteromerization and N-linked glycosylation in the formation of functional hyperpolarization-activated cyclic nucleotide-gated channels. J. Biol. Chem. 278, 43781–43786 [DOI] [PubMed] [Google Scholar]
- 11. Tran N., Proenza C., Macri V., Petigara F., Sloan E., Samler S., Accili E. A. (2002) A conserved domain in the NH2 terminus important for assembly and functional expression of pacemaker channels. J. Biol. Chem. 277, 43588–43592 [DOI] [PubMed] [Google Scholar]
- 12. Biel M., Wahl-Schott C., Michalakis S., Zong X. (2009) Hyperpolarization-activated cation channels. From genes to function. Physiol. Rev. 89, 847–885 [DOI] [PubMed] [Google Scholar]
- 13. Proenza C., Angoli D., Agranovich E., Macri V., Accili E. A. (2002) Pacemaker channels produce an instantaneous current. J. Biol. Chem. 277, 5101–5109 [DOI] [PubMed] [Google Scholar]
- 14. Gauss R., Seifert R., Kaupp U. B. (1998) Molecular identification of a hyperpolarization-activated channel in sea urchin sperm. Nature 393, 583–587 [DOI] [PubMed] [Google Scholar]
- 15. Ishii T. M., Takano M., Xie L. H., Noma A., Ohmori H. (1999) Molecular characterization of the hyperpolarization-activated cation channel in rabbit heart sinoatrial node. J. Biol. Chem. 274, 12835–12839 [DOI] [PubMed] [Google Scholar]
- 16. Seifert R., Scholten A., Gauss R., Mincheva A., Lichter P., Kaupp U. B. (1999) Molecular characterization of a slowly gating human hyperpolarization-activated channel predominantly expressed in thalamus, heart, and testis. Proc. Natl. Acad. Sci. U.S.A. 96, 9391–9396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Decher N., Bundis F., Vajna R., Steinmeyer K. (2003) KCNE2 modulates current amplitudes and activation kinetics of HCN. Influence of KCNE family members on HCN4 currents. Pflugers Arch. 446, 633–640 [DOI] [PubMed] [Google Scholar]
- 18. Macri V., Accili E. A. (2004) Structural elements of instantaneous and slow gating in hyperpolarization-activated cyclic nucleotide-gated channels. J. Biol. Chem. 279, 16832–16846 [DOI] [PubMed] [Google Scholar]
- 19. Azene E. M., Xue T., Marbán E., Tomaselli G. F., Li R. A. (2005) Nonequilibrium behavior of HCN channels. Insights into the role of HCN channels in native and engineered pacemakers. Cardiovasc. Res. 67, 263–273 [DOI] [PubMed] [Google Scholar]
- 20. Mistrík P., Pfeifer A., Biel M. (2006) The enhancement of HCN channel instantaneous current facilitated by slow deactivation is regulated by intracellular chloride concentration. Pflugers Arch. 452, 718–727 [DOI] [PubMed] [Google Scholar]
- 21. Ludwig A., Zong X., Jeglitsch M., Hofmann F., Biel M. (1998) A family of hyperpolarization-activated mammalian cation channels. Nature 393, 587–591 [DOI] [PubMed] [Google Scholar]
- 22. Macri V., Proenza C., Agranovich E., Angoli D., Accili E. A. (2002) Separable gating mechanisms in a mammalian pacemaker channel. J. Biol. Chem. 277, 35939–35946 [DOI] [PubMed] [Google Scholar]
- 23. Proenza C., Yellen G. (2006) Distinct populations of HCN pacemaker channels produce voltage-dependent and voltage-independent currents. J. Gen. Physiol. 127, 183–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lin W., Laitko U., Juranka P. F., Morris C. E. (2007) Dual stretch responses of mHCN2 pacemaker channels. Accelerated activation, accelerated deactivation. Biophys. J. 92, 1559–1572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gravante B., Barbuti A., Milanesi R., Zappi I., Viscomi C., DiFrancesco D. (2004) Interaction of the pacemaker channel HCN1 with filamin A. J. Biol. Chem. 279, 43847–43853 [DOI] [PubMed] [Google Scholar]
- 26. Ehrlicher A. J., Nakamura F., Hartwig J. H., Weitz D. A., Stossel T. P. (2011) Mechanical strain in actin networks regulates FilGAP and integrin binding to filamin A. Nature 478, 260–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Calloe K., Elmedyb P., Olesen S. P., Jorgensen N. K., Grunnet M. (2005) Hypoosmotic cell swelling as a novel mechanism for modulation of cloned HCN2 channels. Biophys. J. 89, 2159–2169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Emery E. C., Young G. T., Berrocoso E. M., Chen L., McNaughton P. A. (2011) HCN2 ion channels play a central role in inflammatory and neuropathic pain. Science 333, 1462–1466 [DOI] [PubMed] [Google Scholar]
- 29. Barrow A. J., Wu S. M. (2009) Low conductance HCN1 ion channels augment the frequency response of rod and cone photoreceptors. J. Neurosci. 29, 5841–5853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Farris H. E., Wells G. B., Ricci A. J. (2006) Steady-state adaptation of mechanotransduction modulates the resting potential of auditory hair cells, providing an assay for endolymph [Ca2+]. J. Neurosci. 26, 12526–12536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pian P., Bucchi A., Decostanzo A., Robinson R. B., Siegelbaum S. A. (2007) Modulation of cyclic nucleotide-regulated HCN channels by PIP2 and receptors coupled to phospholipase C. Pflugers Arch. 455, 125–145 [DOI] [PubMed] [Google Scholar]
- 32. Hirono M., Denis C. S., Richardson G. P., Gillespie P. G. (2004) Hair cells require phosphatidylinositol 4,5-bisphosphate for mechanical transduction and adaptation. Neuron 44, 309–320 [DOI] [PubMed] [Google Scholar]
- 33. Ahmed Z. M., Riazuddin S., Ahmad J., Bernstein S. L., Guo Y., Sabar M. F., Sieving P., Riazuddin S., Griffith A. J., Friedman T. B., Belyantseva I. A., Wilcox E. R. (2003) PCDH15 is expressed in the neurosensory epithelium of the eye and ear and mutant alleles are responsible for both USH1F and DFNB23. Hum. Mol. Genet. 12, 3215–3223 [DOI] [PubMed] [Google Scholar]
- 34. Yi E., Roux I., Glowatzki E. (2010) Dendritic HCN channels shape excitatory postsynaptic potentials at the inner hair cell afferent synapse in the mammalian cochlea. J. Neurophysiol. 103, 2532–2543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Horwitz G. C., Lelli A., Géléoc G. S., Holt J. R. (2010) HCN channels are not required for mechanotransduction in sensory hair cells of the mouse inner ear. PLoS One 5, e8627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ye B., Nerbonne J. M. (2009) Proteolytic processing of HCN2 and co-assembly with HCN4 in the generation of cardiac pacemaker channels. J. Biol. Chem. 284, 25553–25559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Cao X. J., Oertel D. (2011) The magnitudes of hyperpolarization-activated and low voltage-activated potassium currents co-vary in neurons of the ventral cochlear nucleus. J. Neurophysiol. 106, 630–640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Chen X., Shu S., Schwartz L. C., Sun C., Kapur J., Bayliss D. A. (2010) Homeostatic regulation of synaptic excitability. Tonic GABAA receptor currents replace Ih in cortical pyramidal neurons of HCN1 knock-out mice. J. Neurosci. 30, 2611–2622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Webb S. W., Grillet N., Andrade L. R., Xiong W., Swarthout L., Della Santina C. C., Kachar B., Müller U. (2011) Regulation of PCDH15 function in mechanosensory hair cells by alternative splicing of the cytoplasmic domain. Development 138, 1607–1617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Holt J. R., Eatock R. A. (1995) Inwardly rectifying currents of saccular hair cells from the leopard frog. J. Neurophysiol. 73, 1484–1502 [DOI] [PubMed] [Google Scholar]
- 41. Sugihara I., Furukawa T. (1996) Inwardly rectifying currents in hair cells and supporting cells in the goldfish sacculus. J. Physiol. 495, 665–679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Trapani J. G., Nicolson T. (2011) Mechanism of spontaneous activity in afferent neurons of the zebrafish lateral-line organ. J. Neurosci. 31, 1614–1623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Jagger D. J., Holley M. C., Ashmore J. F. (1999) Ionic currents expressed in a cell line derived from the organ of Corti of the Immortomouse. Pflugers Arch. 438, 8–14 [DOI] [PubMed] [Google Scholar]
- 44. Kolesnikov S. S., Rebrik T. I., Zhainazarov A. B., Tavartkiladze G. A., Kalamkarov G. R. (1991) A cyclic AMP-gated conductance in cochlear hair cells. FEBS Lett. 290, 167–170 [DOI] [PubMed] [Google Scholar]
- 45. van Netten S. M., Meulenberg C. J., Lennan G. W., Kros C. J. (2009) Pairwise coupling of hair cell transducer channels links auditory sensitivity and dynamic range. Pflugers Arch. 458, 273–281 [DOI] [PubMed] [Google Scholar]
- 46. van Aken A. F., Atiba-Davies M., Marcotti W., Goodyear R. J., Bryant J. E., Richardson G. P., Noben-Trauth K., Kros C. J. (2008) TRPML3 mutations cause impaired mechano-electrical transduction and depolarization by an inward-rectifier cation current in auditory hair cells of varitint-waddler mice. J. Physiol. 586, 5403–5418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Steigelman K. A., Lelli A., Wu X., Gao J., Lin S., Piontek K., Wodarczyk C., Boletta A., Kim H., Qian F., Germino G., Géléoc G. S., Holt J. R., Zuo J. (2011) Polycystin-1 is required for stereocilia structure but not for mechanotransduction in inner ear hair cells. J. Neurosci. 31, 12241–12250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hackney C. M., Furness D. N., Benos D. J., Woodley J. F., Barratt J. (1992) Putative immunolocalization of the mechanoelectrical transduction channels in mammalian cochlear hair cells. Proc. Biol. Sci. 248, 215–221 [DOI] [PubMed] [Google Scholar]
- 49. Corey D. P., García-Añoveros J., Holt J. R., Kwan K. Y., Lin S. Y., Vollrath M. A., Amalfitano A., Cheung E. L., Derfler B. H., Duggan A., Géléoc G. S., Gray P. A., Hoffman M. P., Rehm H. L., Tamasauskas D., Zhang D. S. (2004) TRPA1 is a candidate for the mechanosensitive transduction channel of vertebrate hair cells. Nature 432, 723–730 [DOI] [PubMed] [Google Scholar]
- 50. Stepanyan R. S., Indzhykulian A. A., Vélez-Ortega A. C., Boger E. T., Steyger P. S., Friedman T. B., Frolenkov G. I. (2011) TRPA1-mediated accumulation of aminoglycosides in mouse cochlear outer hair cells. J. Assoc. Res. Otolaryngol. 12, 729–740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Homma K., Miller K. K., Anderson C. T., Sengupta S., Du G. G., Aguiñaga S., Cheatham M., Dallos P., Zheng J. (2010) Interaction between CFTR and prestin (SLC26A5). Biochim. Biophys. Acta 1798, 1029–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Marcotti W., Kros C. J. (1999) Developmental expression of the potassium current IK,n contributes to maturation of mouse outer hair cells. J. Physiol. 520, 653–660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Marcotti W., Erven A., Johnson S. L., Steel K. P., Kros C. J. (2006) Tmc1 is necessary for normal functional maturation and survival of inner and outer hair cells in the mouse cochlea. J. Physiol. 574, 677–698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Kawashima Y., Géléoc G. S., Kurima K., Labay V., Lelli A., Asai Y., Makishima T., Wu D. K., Della Santina C. C., Holt J. R., Griffith A. J. (2011) Mechanotransduction in mouse inner ear hair cells requires transmembrane channel-like genes. J. Clin. Invest. 121, 4796–4809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Gagnon L. H., Longo-Guess C. M., Berryman M., Shin J. B., Saylor K. W., Yu H., Gillespie P. G., Johnson K. R., (2006) The chloride intracellular channel protein CLIC5 is expressed at high levels in hair cell stereocilia and is essential for normal inner ear function. J. Neurosci. 26, 10188–10198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Mockett B. G., Housley G. D., Thorne P. R. (1994) Fluorescence imaging of extracellular purinergic receptor sites and putative ecto-ATPase sites on isolated cochlear hair cells. J. Neurosci. 14, 6992–7007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Crumling M. A., Tong M., Aschenbach K. L., Liu L. Q., Pipitone C. M., Duncan R. K. (2009) P2X antagonists inhibit styryl dye entry into hair cells. Neuroscience 161, 1144–1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Mukherjea D., Jajoo S., Whitworth C., Bunch J. R., Turner J. G., Rybak L. P., Ramkumar V. (2008) Short interfering RNA against transient receptor potential vanilloid 1 attenuates cisplatin-induced hearing loss in the rat. J. Neurosci. 28, 13056–13065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Karasawa T., Wang Q., Fu Y., Cohen D. M., Steyger P. S. (2008) TRPV4 enhances the cellular uptake of aminoglycoside antibiotics. J. Cell Sci. 121, 2871–2879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Drescher M. J., Barretto R. L., Chaturvedi D., Beisel K. W., Hatfield J. S., Khan K. M., Drescher D. G. (2002) Expression of subunits for the cAMP-sensitive “olfactory” cyclic nucleotide-gated ion channel in the cochlea. Implications for signal transduction. Brain Res. Mol. Brain Res. 98, 1–14 [DOI] [PubMed] [Google Scholar]
- 61. Selvakumar D., Drescher M. J., Dowdall J. R., Khan K. M., Hatfield J. S., Ramakrishnan N. A., Drescher D. G. (2012) CNGA3 is expressed in inner ear hair cells and binds to an intracellular C-terminal domain of EMILIN1. Biochem. J. 443, 463–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Drescher D. G., Ramakrishnan N. A., Drescher M. J. (2009) Surface plasmon resonance (SPR) analysis of binding interactions of proteins in inner-ear sensory epithelia. Methods Mol. Biol. 493, 323–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.