Background: Mycobacterium tuberculosis is an intracellular pathogen that survives under oxidative stress for which none of the common bacterial oxidation sensors have been described.
Results: A new oxidation sensor of M. tuberculosis is described.
Conclusion: MosR is a new thiol-based oxidation-sensing regulator of the MarR family.
Significance: M. tuberculosis must adapt to oxidative environments; therefore, targeting MosR or MosR-regulated proteins may provide ways to fight this bacterium in the future.
Keywords: Crystal Structure, Disulfide, Helix-Loop-Helix Transcription Factors, Mycobacterium tuberculosis, Reactive Oxygen Species (ROS)
Abstract
Mycobacterium tuberculosis thrives in oxidative environments such as the macrophage. To survive, the bacterium must sense and adapt to the oxidative conditions. Several antioxidant defenses including a thick cell wall, millimolar concentrations of small molecule thiols, and protective enzymes are known to help the bacterium withstand the oxidative stress. However, oxidation-sensing regulators that control these defenses have remained elusive. In this study, we report a new oxidation-sensing regulator, Rv1049 or MosR (M. tuberculosis oxidation-sensing regulator). MosR is a transcriptional repressor of the MarR family, which, similarly to Bacillus subtilis OhrR and Staphylococcus aureus MgrA, dissociates from DNA in the presence of oxidants, enabling transcription. MosR senses oxidation through a pair of cysteines near the N terminus (Cys-10 and Cys-12) that upon oxidation forms a disulfide bond. Disulfide formation rearranges a network of hydrogen bonds, which leads to a large conformational change of the protein and dissociation from DNA. MosR has been shown previously to play an important role in survival of the bacterium in the macrophage. In this study, we show that the main role of MosR is to up-regulate expression of rv1050 (a putative exported oxidoreductase that has not yet been characterized) in response to oxidants and propose that it is through this role that MosR contributes to the bacterium survival in the macrophage.
Introduction
Thanks to impressive international efforts, the rate of Mycobacterium tuberculosis infections is beginning to decrease (1). However, with an estimated 8.8 million cases of tuberculosis and 1.1 million deaths reported in 2010, there is still plenty to do before we can foresee the possible eradication of the disease. In addition, the demographics of the disease (it mostly occurs in developing countries), the vast reservoir for potential reactivation (up to one-third of the world population is latently infected), and the emergence of drug-resistant strains (650,000 estimated prevalent cases in 2010) make accomplishing its eradication a formidable task (1). Although current therapies are effective in killing the bacterium, they require multiple months of therapy, which reduces the compliance of patients and increases the risk of drug resistance. To develop superior therapies and vaccines, we need to gain better understanding of the physiology of the bacterium.
A poorly understood aspect of M. tuberculosis is the exact mechanism of how it detects and evades the host immune system. To survive inside macrophages, M. tuberculosis must withstand reactive oxygen species (ROS)2 produced by phagocyte oxidase (NOX2/gp91phox) and reactive nitrogen species (RNS) produced by inducible nitric-oxide synthase in the macrophage (2). The bacterium is aided by several antioxidant defenses including a thick cell wall rich in lipoarabinomannan, cyclopropanated mycolic acid, and phenolic glycolipid I (PGL-1) (3), millimolar concentration of mycothiol (4, 5), protective enzymes such as catalase (KatG) (6), superoxide dismutases (SodA and SodC) (7, 8), peroxidase and peroxynitrite reductase complex (AhpC, AhpD, SucB, and Lpd) (9, 10), and DNA-binding proteins such as Lrs2 (11), as well as an arsenal of exported proteins (12). However, little is known of transcriptional regulators sensitive to oxidation in M. tuberculosis that control these cellular defenses.
Transcriptional regulators responsive to oxidative stress are crucial for many pathogenic bacteria (13–15). For example, Staphylococcus aureus MgrA controls the transcriptional regulation of ∼350 genes in S. aureus and is a major virulence factor (16–18). In Pseudomonas aeruginosa, OxyR and SoxR control the expression of antioxidant genes and DNA repair enzymes that confer resistance to oxidative stress, antibiotics, heavy metals, and macrophage killing (19–21). Unlike reductive stress regulators such as WhiB3, which have been well characterized in recent years (22, 23), no oxidative stress regulators homologous to SoxR, OxyR, and OhrR have been described in M. tuberculosis to date. OxyR, although present in related species such as Mycobacterium leprae, has acquired several nonsense mutations in M. tuberculosis, and it is nonfunctional; SoxR, also present in the related species Mycobacterium smegmatis, does not have a homolog in M. tuberculosis; and so far, no MgrA/OhrR homologs have been described in M. tuberculosis.
Recently, Voskuil et al. (24) examined the transcriptional response of M. tuberculosis to ROS and RNS. They found that under low concentrations of H2O2 (0.05 and 0.5 mm), only furA, the regulator of catalase peroxidase (KatG), was induced. Concentrations of 50 mm or higher were lethal to the bacteria and gave relatively few transcriptional changes. Intermediate concentrations between 5 and 10 mm caused a number of genes and regulators to be induced including those involved in iron response, iron-sulfur cluster repair, ROS/RNS damage repair, and DNA repair. They concluded that besides FurA (25), IdeR (26), SigH (27), and SigE (28), which had been previously implicated in ROS resistance, there are 14 other unstudied regulators that can be induced upon ROS. In this study, we describe a new oxidation-sensitive transcriptional regulator in M. tuberculosis, a homolog of Bacillus subtilis OhrR and S. aureus MgrA that we named MosR (M. tuberculosis oxidation-sensing regulator) and propose a molecular mechanism for MosR in the context of infection.
EXPERIMENTAL PROCEDURES
All procedures involving live M. tuberculosis were carried out in a biosafety level III laboratory. Additional details for these and other experimental procedures can be found in the supplemental Experimental Procedures.
Protein Expression and Purification
All the MosR protein variants were expressed in Escherichia coli using the codon-optimized sequence on a modified pET28a vector containing a thrombin-cleavable C-terminal His tag. After overnight expression at 16 °C, the protein was purified by nickel-nitrilotriacetic acid affinity chromatography. Plasmids and primers used for cloning and expression are shown in supplemental Tables S1 and S2.
Protein Crystallization, X-ray Data Collection, Structure Determination, and Refinement
For crystallization purposes, a construct containing a four-amino acid truncation on the N terminus (amino acids 2–5) was used. Crystallization experiments were carried out using hanging-drop vapor diffusion at room temperature and 16 °C. Datasets on frozen crystals were collected at beamline 24-ID of the Argonne National Laboratory. All data were indexed, integrated, and scaled using HKL2000 (29). The structure of MosR-DNA complex was solved by molecular replacement in Phaser (30) using a model derived from the structure of OhrR with DNA (Protein Data Bank (PDB) ID 1Z9C) (31). To solve apo-MosR, we used a model based on the newly obtained structure of MosR-DNA. The structures were refined using PHENIX (32) and Coot (33). Figures were prepared using PyMOL (34). Full data collection and refinement statistics can be found in supplemental Table S3.
Electrophoretic Mobility Shift Assays
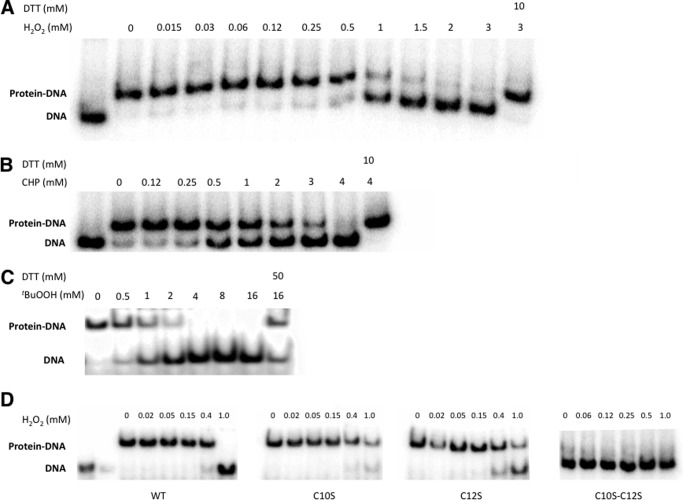
Purified recombinant MosR (50 nm) was combined with a 32P-labeled DNA (5 nm) corresponding to the 100–300-bp region upstream of the gene under investigation in the presence of excess salmon-sperm DNA and different concentration of oxidants as specified in Fig. 3. The mixture was incubated at room temperature for 30 min. DTT was added to the specified samples 15 min after the addition of the oxidant. Samples were run on 4% polyacrylamide native gels, and afterward the gels were dried and exposed to a phosphor screen overnight, and images acquired using a Bio-Rad Pharos FX molecular imager.
FIGURE 3.
MosR binds to DNA in an oxidation-dependent manner. A–C, electrophoretic mobility shift assays of MosR (25 nm) with DNA (5 nm) upon increasing concentrations of oxidants: H2O2 (A); cumene hydroperoxide (CHP) (B); and tert-butyl hydroperoxide (tBuOOH) (C). D, electrophoretic mobility shift assay for MosR, MosR_C10S, MosR_C12S, and MosR_C10SC12S upon increasing concentrations of H2O2. In this assay, complete dissociation is seen for the WT protein but not for the Cys-to-Ser substituted proteins. Double-substituted MosR_C10SC12S shows decreased DNA binding even under reducing conditions.
DNase I Footprinting Assay and Sequencing Reactions
DNase I footprinting assay was carried out using the protocol from Leblanc and Moss (35). Briefly, 50 nm DNA corresponding to the promoter region labeled on one end only was combined with 500 nm protein in the presence of 1 mm DTT. 0.5 units of DNase I were added to the mix and allowed to react for 4 min. Then the reaction was stopped by the addition of STOP solution. Sequencing reactions were performed using the Sequenase kit from Affymetrix. Both samples were run side-by-side on a 4% native gel. The images were acquired as in the electrophoretic mobility shift assays.
Construction of M. tuberculosis Mutant Strains
ΔmosR strain was constructed using the pGOAL/p2NIL system developed by Parish and Stoker (36). Complementation strains were prepared by transforming ΔmosR strain with the appropriate plasmid. A list with the plasmids used in this study can be found in supplemental Table S1.
RNA Isolation and Microarray
RNA isolation and microarray were performed as reported previously (24, 37).
RT-PCR
RT-PCR was performed using SuperScript one-step RT-PCR system (Invitrogen). Primers used are shown in supplemental Table S2.
RESULTS
MosR Is an OhrR/MgrA Homolog
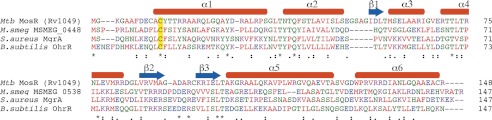
Intrigued by the lack of obvious oxidation-sensing regulators in M. tuberculosis, we decided to look for homologs of B. subtilis OhrR and S. aureus MgrA in M. tuberculosis H37Rv proteome. A direct BLAST-P search for homologs yielded no hits in M. tuberculosis, but found one candidate in the closely related species M. smegmatis that belongs to the MarR family, MSMEG_0448 (43% identical over 145 amino acids to OhrR and 35% identical over 143 amino acids to MgrA). MSMEG_0448 as an OhrR homolog is further supported by the recent discovery that adjacent protein MSMEG_0447, is a homolog of organic hydroperoxide resistance protein (Ohr) (38). A subsequent search for homologs of MSMEG_0448 found one potential candidate in M. tuberculosis: Rv1049 (26% identical over 147 amino acids) (Fig. 1, supplemental Fig. S1) that we named MosR. Although MosR is only 28% identical over 103 amino acids to OhrR and 28% identical over 106 amino acids to MgrA, we thought it might share an oxidation-sensing mechanism as it contains the cysteine presumed to serve for oxidation sensing in a conserved position (Cys-12) and an additional adjacent cysteine (Cys-10) not conserved in OhrR/MgrA. In addition, in these oxidation-sensing regulators, the conserved cysteine is typically surrounded by aromatic amino acids (39), which are also conserved in MosR (Tyr-13, Tyr-26, and Phe-40).
FIGURE 1.
MosR (Rv1049) is an OhrR/MgrA homolog. ClustalW sequence alignment of M. tuberculosis MosR (Mtb MosR), M. smegmatis MSMEG_0448, S. aureus MgrA, and B. subtilis OhrR. Conserved Cys is highlighted in yellow. Amino acids are colored according to physicochemical properties. Asterisks indicate conserved residues; colons indicate highly similar residues; periods indicate less similar residues.
MosR Contains a Reversible Disulfide Bond between Cys-10 and Cys-12
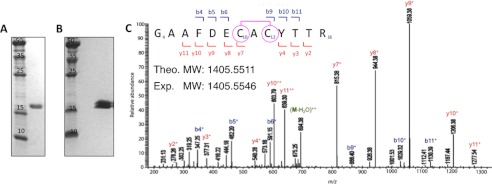
After failed attempts to express MosR in E. coli from the native DNA sequence, we turned to a synthetic variant containing the codon-optimized sequence for E. coli. This construct expressed well, and we were able to purify the protein to reasonable yields (0.1 mg/liter) and high purity using a nickel-nitrilotriacetic acid column (Fig. 2A). We noticed that if the protein was purified in the absence of reducing agents, a new band with retarded mobility appeared on a nonreducing SDS-PAGE gel, suggesting partial oxidation and formation of an intramolecular disulfide bond that may induce a conformational change of the protein (Fig. 2B). We then sought to identify and confirm the disulfide bond using mass spectrometry.
FIGURE 2.
MosR forms a reversible disulfide bond upon oxidation. A, SDS-PAGE gel showing purified reduced MosR. B, double band on nonreducing SDS-PAGE indicates partially oxidized protein. C, LC-MS/MS spectrum of peptide fragment 4GAAFDECACYTTR16 (observed m/z 1405.5546 corresponding to peptide theoretical mass of 1407.5321 Da minus two hydrogen atoms (−2 Da)) obtained after trypsin digestion of oxidized MosR. The fragment ions y7+ and b9+ corresponding to 4GAAFDECAC12 and 10CACYTTR16, respectively (observed m/z 815.38 and 886.40) indicate the presence of disulfide bond between Cys-10 and Cys-12. Fragment ion arising from the neutral loss of water (−18 Da) is marked as M-H2O. Theo. MW, theoretical molecular weight; Exp. MW, experimental molecular weight.
For this experiment, we subjected MosR to three different conditions (1 mm DTT, 1 h of air oxidation, and 5 mm H2O2) followed by alkylation, trypsin digestion, and LC-MS/MS analysis. In the presence of 1 mm DTT, we found no evidence for the presence of disulfide bonds in the protein. Under air oxidation, which mimics the conditions in which we observe a double band on SDS-PAGE gel, we found one disulfide bond between Cys-10 and Cys-12 (Fig. 2C). Under 5 mm H2O2, we found evidence for two disulfide bonds with one between Cys-10 and Cys-12 and another between Cys-96 and Cys-147 (supplemental Fig. S2). These data show that formation of disulfide between Cys-10 and Cys-12 is more facile and likely represents the primary biological mechanism used by MosR to sense oxidation.
Reduced MosR Binds to DNA, whereas Oxidized MosR Does Not
To test whether MosR functions as an oxidation-sensing transcriptional regulator, we tested its ability to bind to DNA in the presence of oxidants. We speculated that similar to other MarR proteins, MosR would be autoregulated, and therefore we chose a 300-bp fragment of DNA directly upstream of the mosR gene to test for DNA binding. The protein bound to the amplified DNA even in the presence of 10-fold excess of nonspecific DNA with a Kd of ∼15 nm (supplemental Fig. S3A). Subsequently, we tested the ability of the protein to bind DNA in the presence of increasing concentrations of oxidants including H2O2, cumene hydroperoxide and tert-butyl hydroperoxide, and the NO precursor spermine-NONOate (Fig. 3, A–C, supplemental Fig. S3B). In all cases, except with the NO precursor, we saw dissociation of the protein from DNA upon the addition of 0.5–1 mm of the respective oxidant. This dissociation was reversible upon the addition of the reducing agent, DTT (Fig. 3, A–C). Spermine-NONOate did not cause dissociation of the protein from the DNA up to 1 mm, and concentrations of 10 mm or higher led to protein aggregation due to the tendency of spermine to precipitate proteins.
We then assayed the role of the cysteines in oxidation-dependent DNA binding. MosR contains four cysteines: Cys-10, Cys-12, Cys-96, and Cys-147. Only Cys-12 is conserved among oxidation-sensing OhrR/MgrA proteins, and thus we focused our attention on this cysteine and the proximal Cys-10. Substitution of Cys-10 or Cys-12 by serine results in proteins with lower sensitivity to oxidation as shown in Fig. 3D. Under the assayed conditions, treatment with 1 mm H2O2 for 1 h is sufficient to completely dissociate the wild-type MosR but not the Cys-to-Ser substituted proteins. Double mutation of Cys-10 and Cys-12 to serine results in a protein with decreased DNA binding affinity even under reducing conditions. In our experience, substitution of Cys to Ser in this family of proteins often affects the stability of the protein. The partial dissociation of MosR-C10S and MosR-C12S with >1 mm H2O2 may be due to nonspecific oxidation of the protein or additional oxidation-sensing mechanisms. Nonetheless, the lower sensitivity of the substituted proteins with Cys-10 and Cys-12 replaced by Ser suggests that these residues play important roles in the oxidation-sensing mechanism, which is consistent with disulfide bond formation between these two residues. This type of behavior in which substitution from Cys-12 to Ser results in a protein less responsive to oxidation is reminiscent of OhrR and MgrA (16, 40).
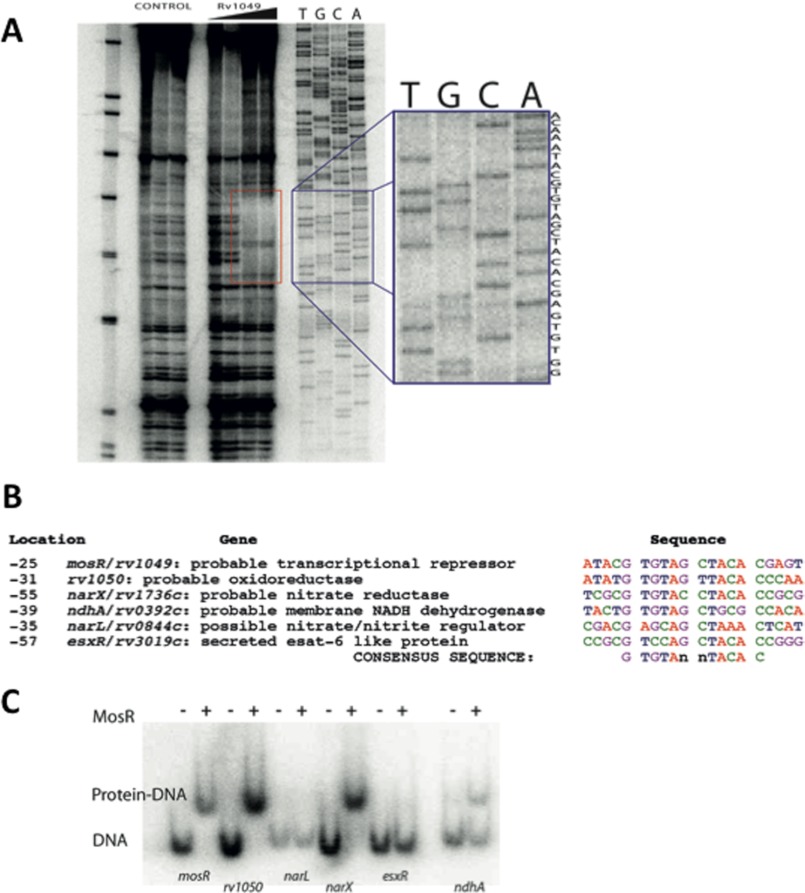
MosR Binds to a Specific DNA Sequence and Controls the Expression of Redox-related Genes
We then performed a DNase I footprinting assay to determine the specific sequence that MosR binds. A 26-base-long quasi-palindromic motif starting 31 bases upstream of mosR (rv1049) starting codon was revealed as the binding site of MosR (Fig. 4A). A pattern search for similar sequences in the genome of M. tuberculosis unveiled similar sequences in the promoter regions of rv1050, ndhA, narL, narX, ndhA, and esxR (Fig. 4B). We then tested for binding to those promoters and confirmed that in addition to binding to its own promoter, MosR can bind to the promoters of rv1050 and narX, and to a lower extent, ndhA (Fig. 4C). These experiments allowed us to determine the sequence GTGTAn nTACAC (n = A, C, G, or T), as the consensus recognition motif of MosR. This motif is strictly conserved in the promoters of mosR (or rv1049), rv1050, and narX (or rv1736c), but it contains two mismatches in the promoters of ndhA and esxR and three in narL promoter, which explains why MosR does not bind to these promoters or why it does so with reduced affinity. Interestingly, Rv1050 and NarX are thought to be involved in reduction processes, which is consistent with being controlled by an oxidation-sensitive transcriptional regulator. Rv1050 is an exported protein (41) predicted to function as an oxidoreductase, and NarX is a predicted nitrite reductase, part of the dosR regulon, known to be induced by NO and hypoxia (42, 43).
FIGURE 4.
MosR binds to a specific DNA sequence and controls expression of redox-related genes. A, DNase I footprinting assay. The red rectangle indicates the region where the protein binds. The blue rectangle shows the sequence for that region: ACAA ATACG TGTAG | CTACA CGAGT CTGG. B, similar sequences are found in the promoter regions of mosR, rv1050, narX, ndhA, narL, and esxR. The location number indicates bases from the start codon. C, electrophoretic mobility shift assay showing that MosR binds to the promoter regions of mosR, rv1050, narX, and ndhA.
MosR Structure Reveals a New Oxidation-sensing Mechanism
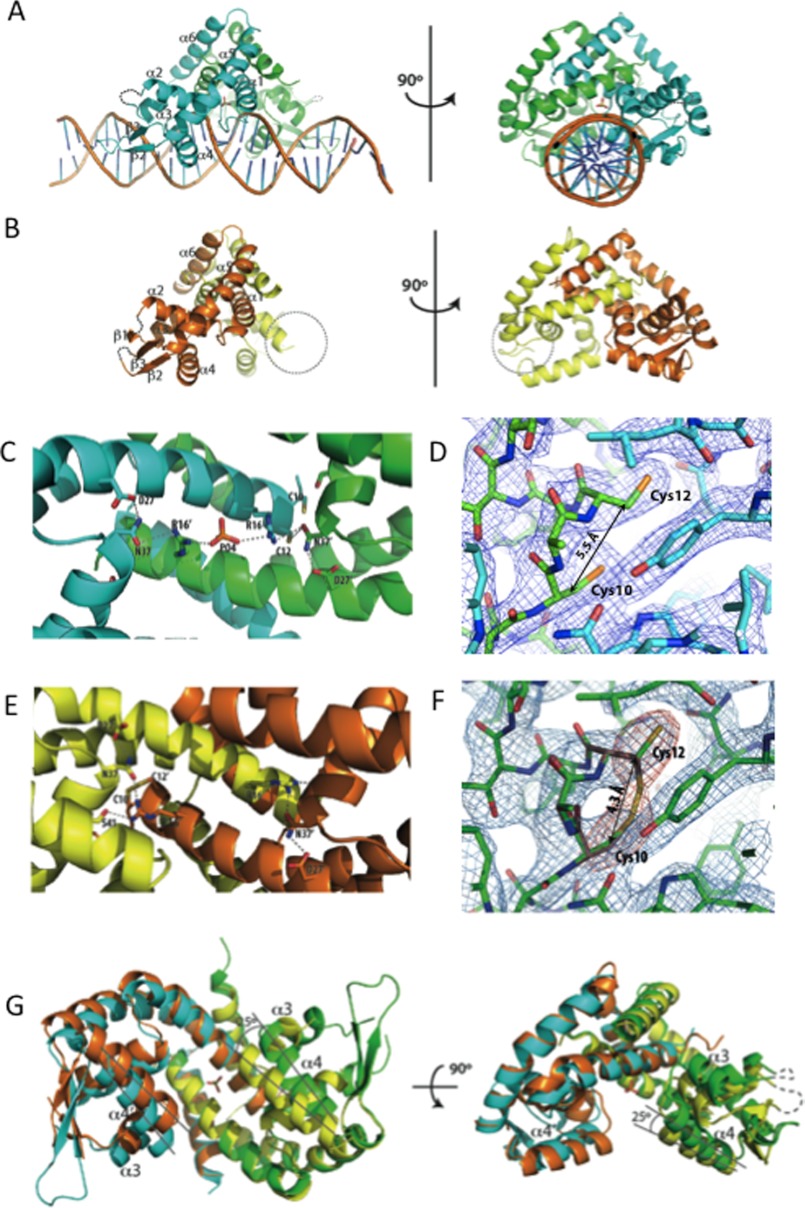
We next crystallized and solved the structures of MosR bound to a 28-bp duplex DNA and the apo form (Fig. 5, A and B). The MosR-DNA complex crystallized in the R3 space group, and the structure was solved to 3.1 Å resolution. Apo-MosR crystallized in the P212121 space group and was solved to 2.7 Å resolution. In both structures, MosR forms a dimer of triangular shape with the two monomers related by noncrystallographic symmetry. These two structures resemble the structures of OhrR and SlyA with and without DNA (31, 44) (supplemental Fig. S4, A and B). Each monomer contains six α-helices and three antiparallel β-strands forming a β-sheet (there are only two β-strands in the DNA-bound structure as β1 becomes disordered). Strand β1 is longer than in related structures due to the insertion of five amino acids (Gly-51–Ile-55) between α2 and α3. Helices α1, α5, and α6 form the dimerization domain with α6 going over α1 helix of the adjacent monomer. Helices α3 and α4 and the connecting loop form the characteristic helix-turn-helix DNA-binding motif.
FIGURE 5.
The structure of MosR reveals oxidation-sensing mechanism. A, reduced MosR-DNA structure. B, oxidized apo-MosR structure. The dotted circle indicates the wing region of chain B that had poor electron density and was not built. C, hydrogen-bonding interactions in the MosR-DNA (reduced) structure. D, detail of Cys-10–Cys-12 site in reduced MosR-DNA (reduced) structure. Electron density map corresponds to 2Fo - Fc map contoured at 1.5 σ. E, hydrogen-bonding interactions in the apo-MosR (oxidized) structure. F, detail of Cys-10–Cys-12 site in apo-MosR (oxidized) structure. In red, Fo - Fc simulated annealing omit maps for the sulfur atoms of Cys-10 and Cys-12 showing two possible conformations for Cys-12. Blue maps correspond 2Fo - Fc maps at 1.5 σ. G, superposition of the two structures seen from below and side (239 atoms, root mean square deviation = 1.83 Å2). Cyan and green, MosR-DNA structure. Orange and yellow, apo-MosR. Lines indicate movement of α4.
On the MosR-DNA complex structure, helices α1 and α1′ (prime refers to the opposing monomer) sit atop the DNA forming extensive contacts with the phosphate backbone on the minor groove of the DNA. α4 and α4′ fit into two consecutive major grooves recognizing the palindromic sequence GTGTAn nTACAC mainly through the side chains of Arg-70, Thr-71, Thr-72, and Asn-76 (supplemental Fig. S4, C and D). Between α4 and α5, there is a stretch of 21 amino acids forming two antiparallel β-strands (β2 and β3) that extend over the minor grooves making extensive contacts with phosphates, sugar oxygens, and the O2 position of a thymidine base three bases away from the end of the palindrome. On the base of the triangle, the side chains of α1, α2, α1′, and α2′ form an extended array of hydrogen bonds parallel to the helices. Starting on one end, Cys-12 (α1) interacts with Asn-37′ (α2′) from the other monomer, which further interacts with Arg-16 (α1) that is hydrogen-bonded to a phosphate ion situated in the center of the protein. Symmetrically, the phosphate interacts with Arg-16′ (α1′) from the other monomer, then Asn-37 (α2) and, finally, Cys-12′ (α1′) (Fig. 5C). In this structure, the Cβ atoms of Cys-10 and Cys-12 are positioned 5.5 Å away from each other, and both thiol groups point toward the C terminus of the helix, representing clearly the reduced form of the protein (Fig. 5D).
The apo-MosR structure represents the oxidized conformation of the protein. The Cβ atom of Cys-10 is positioned only 4.3 Å away from the Cβ of Cys-12 (when compared with 5.5 Å in the reduced form), and Cys-12 can fit into the electron density in two alternate conformations, pointing up (or reduced) and pointing down forming disulfide (or oxidized) (Fig. 5F). Although there is electron density for the both the oxidized and the reduced conformations of Cys-12, the overall conformation of the protein is consistent with the oxidized state. The fact that the Cys-12 is partially reduced is indicative of the reversible nature of the disulfide bond, and we believe it to be due to the reducing power of electrons from the x-ray beam (45) because the overall conformation is very different from the reduced DNA-bound form.
From these structures, we can infer that when the disulfide bond is formed, there is rearrangement of hydrogen bonds and, consequently, a large change in the conformation of the protein. Inversion in the orientation of Cys-12 (α1) breaks the hydrogen bond to Asn-37′ (α2′) and brings Arg-16 (α1) closer, which forms a new hydrogen bond to Ser-41′ (α2′) (Fig. 5E). This new arrangement of hydrogen bonds results in the translation of α2 ∼1.2 Å relative to α1. This movement in α2 pushes α3 ∼4.5 Å toward α4, which then rotates ∼25° inward to accommodate α3 (Fig. 5G). Rotation of α4 and α4′ prevents them from fitting into consecutive major grooves and thus impairs DNA binding.
With the structure at hand, we examined the possible roles of Cys-96 and Cys-147 in oxidation sensing. As mentioned above, by mass spectrometry, we found evidence of disulfide bond formation between these two cysteines. These cysteines are located on the surface of the protein and are more than 30 Å apart, which is incompatible with an intramolecular or interchain disulfide. This disulfide may arise from reaction between two dimers in solution. Formation of this second disulfide bond may be due to the strong oxidizing conditions employed experimentally, and it may or may not reflect a secondary oxidation-sensing mechanism of MosR that can be subjected to future studies.
Formation of an intramolecular disulfide bond between Cys-10 and Cys-12 is novel in this family of proteins. OhrR proteins can be classified as 1-Cys or 2-Cys proteins (46). 1-Cys proteins, typified by B. subtilis OhrR, have only one cysteine that can be oxidized to sulfenic acid or mixed disulfides (47), and this cysteine has been shown structurally in SarZ, another oxidation-sensitive transcriptional repressor from S. aureus (48). On the contrary, 2-Cys proteins, typified by Xanthomonas campestris OhrR, form intermolecular disulfide bond between the peroxidatic cysteine in the N terminus and a cysteine residue located on the C terminus of the other monomer (40, 49). To our knowledge, formation of a disulfide between two nearby cysteines from the same chain is unique to MosR and represents a new variant in oxidation sensing.
MosR Deletion Was Generated by Double-crossover Homologous Recombination
Following the protocol by Parish and Stoker (36), we generated an unmarked in-frame deletion strain of mosR in H37Rv background (ΔmosR). This strain harbors a 441-bp deletion inside mosR (rv1049) gene corresponding to amino acids 2–148. This deletion was confirmed by PCR and Southern blotting (supplemental Fig. S5, A and B). We also generated a complementation strain (ΔmosR::pUC-mosR HygR) by cloning mosR plus 318 bp upstream into the integrative plasmid pUC-GM-INT (50) and transforming ΔmosR cells with this plasmid. This strain was generated to ensure that any observed phenotypes were only due to the absence of mosR and not from inadvertent mutations introduced during cloning. We chose to include the promoter region to maintain similar expression levels as in wild-type strain. Additionally, this allowed us to perform site-directed mutagenesis in the complementation plasmid to generate a complementation strain expressing MosR-C12S to study the effect of this cysteine in vivo. Notably, mutation of mosR does not affect growth (supplemental Fig. S6), sensitivity to H2O2 and diamide, or sensitivity to the antituberculosis drugs isoniazid, rifampin, and ethambutol (supplemental Table S4).
Oxidation-responsive Transcriptional Regulation Was Confirmed in Vivo by RT-PCR
To verify that oxidation sensing is in fact occurring in vivo, we tested the mRNA levels of rv1050 by RT-PCR using sigA as control. We extracted RNA of M. tuberculosis cultures grown to early log phase treated with 0, 5, and 10 mm H2O2 for 30 min. As shown in Fig. 6, without H2O2 treatment, the levels of rv1050 mRNA on the Δrv1050 strain were 39-fold higher than in the wild-type strain, whereas the MosR and MosR-C12S complementation strains had levels similar to wild type (WT). Upon H2O2 treatment with 5 and 10 mm H2O2, the rv1050 levels in WT went up by 5.6- and 27-fold, respectively, reaching levels comparable with ΔmosR. The ΔmosR strain had very high levels of rv1050 transcript in the absence of oxidants. The addition of H2O2 caused a slight decrease in the rv1050 level, but it remained high when compared with uninduced WT. These results indicate that rv1050 is fully up-regulated in the mutant and that MosR is the sole regulator of rv1050. In the MosR-C12S complementation strain, there was a moderate 2.5-fold increase in rv1050 message upon 5 mm treatment (when compared with 5.6-fold increase in WT, p = 0.12) and 30-fold increase upon 10 mm treatment (when compared with 27 in WT). This result shows that MosR-C12S is less sensitive to moderate concentrations of oxidants, which agrees with the trends observed by electromobility shift assays (Fig. 3). The complementary strain behaved similarly to WT strain in all the conditions. Similar trends were observed when probed by Northern blotting (supplemental Fig. S7).
FIGURE 6.
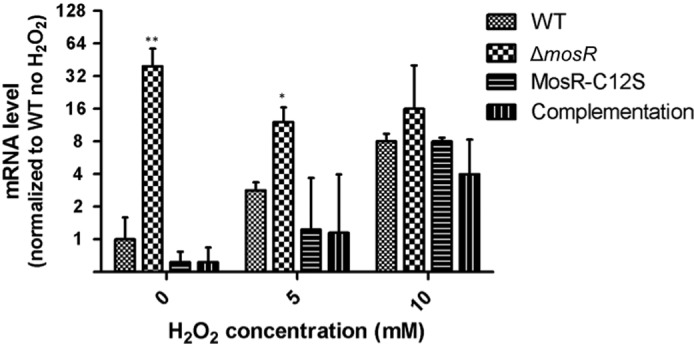
MosR controls rv1050 in response to oxidants. RT-PCR analysis of rv1050 expression in the wild-type, ΔmosR, MosR-C12S complementation, and MosR complementation strains during H2O2 treatment. Values are relative to wild-type strain under no H2O2 treatment. Results represent mean ± S.D. of three independent biological replicates and were normalized using sigA levels. (*, p < 0.05; **, p < 0.01 when compared with WT under the same treatment.).
The remarkably high levels of expression of rv1050 upon H2O2 treatment suggest that this protein of unknown function plays an important role in adaptation to oxidative stress. Increase in mosR and rv1050 expression upon the addition of oxidants has also been observed in previous microarray experiments with H2O2 (24) and diamide (51). Polyunsaturated fatty acids such as arachidonic acid and linoleic acid, which can readily undergo lipid peroxidation to generate ROS, also up-regulate the expression of rv1050, whereas saturated fatty acids such as oleic acid and palmitic acid down-regulate its expression.3
Global Gene Expression Profiling Shows Discrete Regulation by MosR
To determine the regulatory scope of MosR, we performed global gene expression profiling using RNA microarrays comparing the ΔmosR strain with H37Rv wild-type strain with and without H2O2 (supplemental Table S5). In the mutant versus WT under no H2O2 stress, we found only four genes that were differentially regulated: rv0350, rv1050, rv1361c, and rv3478. Out of these genes, rv1050 showed the most significant difference (on average 352-fold higher in ΔmosR than wild type), whereas the other genes displayed modest induction (rv0350, rv1361c, and rv2478, range: −9.1- to 2.5-fold). Under 10 mm H2O2 treatment, there was no difference between the wild-type and ΔmosR strains in rv1050 levels, which agrees with the RT-PCR results, and there were a total of nine genes, which showed 2–5-fold change (fadA2, rpsS, rplP, rpsQ, rv0839, rv1884c, rv2558, rv3614c, and rv3615c). None of these genes contain the consensus sequence in their promoter regions, and we attribute these changes to pleiotropic effects or to the perturbation of cellular pathways by the dramatic overexpression of rv1050.
Still, we were surprised not to find narX among the genes up-regulated in the ΔmosR strain. However, this can be explained by a second layer of regulation for this gene. It is known that narX is under the control of DosR and is only activated under hypoxia, nitric oxide, or carbon monoxide (43). It is possible that narX is under control of multiple regulators that need to work synergistically. In sum, this microarray experiment confirmed that MosR regulates rv1050, but unlike in S. aureus, where MgrA is a global regulator (17), M. tuberculosis MosR has narrow regulatory scope.
DISCUSSION
In this study, we presented a new oxidation-sensing transcriptional repressor of M. tuberculosis that belongs to the MarR family, and its plausible mechanism (Fig. 7). We described the identification of MosR using a bioinformatics approach, the biochemical characterization of this protein showing that DNA binding is dependent on the oxidation state of the protein and that it is fully reversible. Based on x-ray crystallography and mass spectrometry, we proposed that oxidation sensing is facilitated by formation of a reversible disulfide bond between Cys-10 and Cys-12, a new mechanism among MarR proteins. We also identified the DNA-binding sequence for this regulator by DNase I footprinting and showed that similar sequences are present in the promoter regions of itself, rv1050, narX, and ndhA. To validate the function in vivo, we constructed a mosR deletion strain and demonstrated that MosR controls the expression of rv1050 in response to oxidants using microarray and RT-PCR.
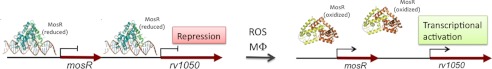
FIGURE 7.
Proposed model for MosR. MosR is an oxidation-sensing transcriptional regulator of itself and rv1050. In the absence of oxidative stress, MosR binds to DNA, repressing transcription. Upon oxidative stress (as in the macrophage, Mø) MosR dissociates from DNA, enabling transcription of itself and rv1050.
Interestingly, it had been shown previously that rv1050 is up-regulated 2-fold during infection in INF-γ-activated macrophages (53). In light of our results, we can now explain that this up-regulation is likely due to MosR sensing ROS produced by the macrophage.
Another notable aspect about MosR comes from a study by Rengarajan et al. (54) that analyzed the genes required for survival in macrophages. This study found that a transposon insertion in rv1049 (mosR) had the second largest increase in resistance to macrophage killing (5.6-fold increase). The authors note that the genes that give increased survival of the bacterium may pose a transcriptional burden under normal laboratory conditions. Now, based on our results, we can propose that mutation of mosR results in enhanced survival probably due to constitutively high expression of rv1050. Although we do not know the exact function of Rv1050, it likely has a protective effect in the bacterium. Unfortunately, that genome-wide study had no information on the effect of a transposon mutation directly on rv1050 probably because the experiment was carried out using a heterogeneous culture (the TraSH library), and exported proteins such as Rv1050 could be produced by a neighboring bacterium masking its effect. It will be very interesting to test whether mutation of rv1050 results in decreased intracellular survival.
Rv1050 is an exported protein predicted to function as an oxidoreductase. Given that arachidonic acid and linoleic acid are also among the signals that induce rv1050 up-regulation, we suspect that Rv1050 may affect fatty acid metabolism in the macrophage. It is known that M. tuberculosis-infected macrophages exhibit an altered arachidonic acid metabolism in favor of lipoxin A4 over prostaglandin E2 (PGE2), which promotes necrosis over apoptosis (52, 55). Although it is unknown how M. tuberculosis affects this process, it is reasonable to conjecture that exported proteins are used to manipulate the metabolism of the macrophage. Furthermore, if Rv1050 is indeed important for the survival of M. tuberculosis in the macrophage, one could envision targeting this exported protein as a new avenue for therapy. Targeting exported proteins could be particularly interesting as it circumvents the challenge of the drugs getting into the bacterium.
Supplementary Material
Acknowledgments
We thank Prof. T. Parish for the mycobacterial plasmids, Prof. Linda Adams for helpful discussions, Dr. L. Zhang, Dr. C. B. Poor, and the beamline staff for help collecting the x-ray diffraction data, S. Alvarez and L. Hicks for assistance with the mass spectrometry experiments, H. Gutka and M. Rutter for assistance with M. tuberculosis cultures and media, and Dr A. Tevar for proofreading this manuscript. The use of the Advanced Photon Source (beamline 24-ID) at Argonne National Laboratory was supported by the United States Department of Energy.
This work was supported, in whole or in part, by National Institutes of Health Grant AI074658 (to C. H.) from the NIAID. This work was also supported by the Chicago Biomedical Consortium with support from The Searle Funds at the Chicago Community Trust (to C. H. and S. G. F.), a Burroughs Wellcome Fund Investigator in the Pathogenesis of Infectious Disease Award (to C. H.), National Institutes of Health Grant R01 AI061505 (to M. I. V.), and a Graduate Fellowship from Fundación Caja Madrid (to P. B.).

This article contains supplemental Figs. S1–S7, Tables S1–S5, and Experimental Procedures.
The atomic coordinates and structure factors (codes 4FX0 and 4FX4) have been deposited in the Protein Data Bank (http://wwpdb.org/).
Y. Liu and G. Schoolnik, TB Database.
- ROS
- reactive oxygen species
- RNS
- reactive nitrogen species
- MosR
- M. tuberculosis oxidation-sensing regulator
- Ohr
- organic hydroperoxide resistance protein.
REFERENCES
- 1. World Health Organization (2011) Global Tuberculosis Control, WHO Report 2011, pp. 1–27, World Health Organization, Geneva, Switzerland [Google Scholar]
- 2. Ehrt S., Schnappinger D. (2009) Mycobacterial survival strategies in the phagosome: defense against host stresses. Cell. Microbiol. 11, 1170–1178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Flynn J. L., Chan J. (2001) Tuberculosis: latency and reactivation. Infect. Immun. 69, 4195–4201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Buchmeier N., Fahey R. C. (2006) The mshA gene encoding the glycosyltransferase of mycothiol biosynthesis is essential in Mycobacterium tuberculosis Erdman. FEMS Microbiol. Lett. 264, 74–79 [DOI] [PubMed] [Google Scholar]
- 5. Buchmeier N. A., Newton G. L., Fahey R. C. (2006) A mycothiol synthase mutant of Mycobacterium tuberculosis has an altered thiol-disulfide content and limited tolerance to stress. J. Bacteriol. 188, 6245–6252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Manca C., Paul S., Barry C. E., 3rd, Freedman V. H., Kaplan G. (1999) Mycobacterium tuberculosis catalase and peroxidase activities and resistance to oxidative killing in human monocytes in vitro. Infect. Immun. 67, 74–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Jackett P. S., Aber V. R., Lowrie D. B. (1978) Virulence and resistance to superoxide, low pH, and hydrogen peroxide among strains of Mycobacterium tuberculosis. J. Gen. Microbiol. 104, 37–45 [DOI] [PubMed] [Google Scholar]
- 8. Piddington D. L., Fang F. C., Laessig T., Cooper A. M., Orme I. M., Buchmeier N. A. (2001) Cu,Zn-superoxide dismutase of Mycobacterium tuberculosis contributes to survival in activated macrophages that are generating an oxidative burst. Infect. Immun. 69, 4980–4987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bryk R., Griffin P., Nathan C. (2000) Peroxynitrite reductase activity of bacterial peroxiredoxins. Nature 407, 211–215 [DOI] [PubMed] [Google Scholar]
- 10. Bryk R., Lima C. D., Erdjument-Bromage H., Tempst P., Nathan C. (2002) Metabolic enzymes of mycobacteria linked to antioxidant defense by a thioredoxin-like protein. Science 295, 1073–1077 [DOI] [PubMed] [Google Scholar]
- 11. Colangeli R., Haq A., Arcus V. L., Summers E., Magliozzo R. S., McBride A., Mitra A. K., Radjainia M., Khajo A., Jacobs W. R., Jr., Salgame P., Alland D. (2009) The multifunctional histone-like protein Lsr2 protects mycobacteria against reactive oxygen intermediates. Proc. Natl. Acad. Sci. U.S.A. 106, 4414–4418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. McCann J. R., McDonough J. A., Sullivan J. T., Feltcher M. E., Braunstein M. (2011) Genome-wide identification of Mycobacterium tuberculosis exported proteins with roles in intracellular growth. J. Bacteriol. 193, 854–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Green J., Paget M. S. (2004) Bacterial redox sensors. Nat. Rev. Microbiol. 2, 954–966 [DOI] [PubMed] [Google Scholar]
- 14. den Hengst C. D., Buttner M. J. (2008) Redox control in actinobacteria. Biochim. Biophys. Acta 1780, 1201–1216 [DOI] [PubMed] [Google Scholar]
- 15. Chen P. R., Brugarolas P., He C. (2011) Redox signaling in human pathogens. Antioxid. Redox Signal. 14, 1107–1118 [DOI] [PubMed] [Google Scholar]
- 16. Chen P. R., Bae T., Williams W. A., Duguid E. M., Rice P. A., Schneewind O., He C. (2006) An oxidation-sensing mechanism is used by the global regulator MgrA in Staphylococcus aureus. Nat. Chem. Biol. 2, 591–595 [DOI] [PubMed] [Google Scholar]
- 17. Luong T. T., Dunman P. M., Murphy E., Projan S. J., Lee C. Y. (2006) Transcription profiling of the mgrA Regulon in Staphylococcus aureus. J. Bacteriol. 188, 1899–1910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Luong T. T., Newell S. W., Lee C. Y. (2003) Mgr, a novel global regulator in Staphylococcus aureus. J. Bacteriol. 185, 3703–3710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ha U., Jin S. (1999) Expression of the soxR gene of Pseudomonas aeruginosa is inducible during infection of burn wounds in mice and is required to cause efficient bacteremia. Infect. Immun. 67, 5324–5331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kobayashi K., Tagawa S. (2004) Activation of SoxR-dependent transcription in Pseudomonas aeruginosa. J. Biochem. 136, 607–615 [DOI] [PubMed] [Google Scholar]
- 21. Ochsner U. A., Vasil M. L., Alsabbagh E., Parvatiyar K., Hassett D. J. (2000) Role of the Pseudomonas aeruginosa oxyR-recG operon in oxidative stress defense and DNA repair: OxyR-dependent regulation of katB-ankB, ahpB, and ahpC-ahpF. J. Bacteriol. 182, 4533–4544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Singh A., Crossman D. K., Mai D., Guidry L., Voskuil M. I., Renfrow M. B., Steyn A. J. (2009) Mycobacterium tuberculosis WhiB3 maintains redox homeostasis by regulating virulence lipid anabolism to modulate macrophage response. PLoS Pathog. 5, e1000545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Singh A., Guidry L., Narasimhulu K. V., Mai D., Trombley J., Redding K. E., Giles G. I., Lancaster J. R., Jr., Steyn A. J. (2007) Mycobacterium tuberculosis WhiB3 responds to O2 and nitric oxide via its [4Fe-4S] cluster and is essential for nutrient starvation survival. Proc. Natl. Acad. Sci. U.S.A. 104, 11562–11567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Voskuil M. I., Bartek I. L., Visconti K., Schoolnik G. K. (2011) The response of Mycobacterium tuberculosis to reactive oxygen and nitrogen species. Front. Microbiol. 2, 105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zahrt T. C., Song J., Siple J., Deretic V. (2001) Mycobacterial FurA is a negative regulator of catalase-peroxidase gene katG. Mol. Microbiol. 39, 1174–1185 [DOI] [PubMed] [Google Scholar]
- 26. Rodriguez G. M., Voskuil M. I., Gold B., Schoolnik G. K., Smith I. (2002) ideR, An essential gene in Mycobacterium tuberculosis: role of IdeR in iron-dependent gene expression, iron metabolism, and oxidative stress response. Infect. Immun. 70, 3371–3381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Fernandes N. D., Wu Q. L., Kong D., Puyang X., Garg S., Husson R. N. (1999) A mycobacterial extracytoplasmic sigma factor involved in survival following heat shock and oxidative stress. J. Bacteriol. 181, 4266–4274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wu Q. L., Kong D., Lam K., Husson R. N. (1997) A mycobacterial extracytoplasmic function sigma factor involved in survival following stress. J. Bacteriol. 179, 2922–2929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Otwinowski Z., Minor W. (1997) Processing of X-ray diffraction data collected in oscillation mode Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 30. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hong M., Fuangthong M., Helmann J. D., Brennan R. G. (2005) Structure of an OhrR-ohrA operator complex reveals the DNA binding mechanism of the MarR family. Mol. cell 20, 131–141 [DOI] [PubMed] [Google Scholar]
- 32. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Emsley P., Lohkamp B., Scott W. G., Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. DeLano W. L. (2010) The PyMOL Molecular Graphics System, version 1.3r1, Schrödinger, LLC, New York [Google Scholar]
- 35. Leblanc B., Moss T. (1994) DNase I footprinting. Methods Mol. Biol. 30, 1–10 [DOI] [PubMed] [Google Scholar]
- 36. Parish T., Stoker N. G. (2000) Use of a flexible cassette method to generate a double unmarked Mycobacterium tuberculosis tlyA plcABC mutant by gene replacement. Microbiology 146, 1969–1975 [DOI] [PubMed] [Google Scholar]
- 37. Wilson M., Voskuil M., Schnappinger D., Schoolnik G. K. (2001) Functional genomics of Mycobacterium tuberculosis using DNA microarrays. Methods Mol. Med. 54, 335–357 [DOI] [PubMed] [Google Scholar]
- 38. Ta P., Buchmeier N., Newton G. L., Rawat M., Fahey R. C. (2011) Organic hydroperoxide resistance protein and ergothioneine compensate for loss of mycothiol in Mycobacterium smegmatis mutants. J. Bacteriol. 193, 1981–1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Poole L. B., Nelson K. J. (2008) Discovering mechanisms of signaling-mediated cysteine oxidation. Curr. Opin. Chem. Biol. 12, 18–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Panmanee W., Vattanaviboon P., Poole L. B., Mongkolsuk S. (2006) Novel organic hydroperoxide-sensing and responding mechanisms for OhrR, a major bacterial sensor and regulator of organic hydroperoxide stress. J. Bacteriol. 188, 1389–1395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Målen H., Berven F. S., Fladmark K. E., Wiker H. G. (2007) Comprehensive analysis of exported proteins from Mycobacterium tuberculosis H37Rv. Proteomics 7, 1702–1718 [DOI] [PubMed] [Google Scholar]
- 42. Sherman D. R., Voskuil M., Schnappinger D., Liao R., Harrell M. I., Schoolnik G. K. (2001) Regulation of the Mycobacterium tuberculosis hypoxic response gene encoding α-crystallin. Proc. Natl. Acad. Sci. U.S.A. 98, 7534–7539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Voskuil M. I., Schnappinger D., Visconti K. C., Harrell M. I., Dolganov G. M., Sherman D. R., Schoolnik G. K. (2003) Inhibition of respiration by nitric oxide induces a Mycobacterium tuberculosis dormancy program. J. Exp. Med. 198, 705–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Dolan K. T., Duguid E. M., He C. (2011) Crystal structures of SlyA protein, a master virulence regulator of Salmonella, in free and DNA-bound states. J. Biol. Chem. 286, 22178–22185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Petrova T., Ginell S., Mitschler A., Kim Y., Lunin V. Y., Joachimiak G., Cousido-Siah A., Hazemann I., Podjarny A., Lazarski K., Joachimiak A. (2010) X-ray-induced deterioration of disulfide bridges at atomic resolution. Acta Crystallogr. D Biol. Crystallogr. 66, 1075–1091 [DOI] [PubMed] [Google Scholar]
- 46. Antelmann H., Helmann J. D. (2011) Thiol-based redox switches and gene regulation. Antioxid. Redox Signal 14, 1049–1063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lee J. W., Soonsanga S., Helmann J. D. (2007) A complex thiolate switch regulates the Bacillus subtilis organic peroxide sensor OhrR. Proc. Natl. Acad. Sci. U.S.A. 104, 8743–8748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Poor C. B., Chen P. R., Duguid E., Rice P. A., He C. (2009) Crystal structures of the reduced, sulfenic acid, and mixed disulfide forms of SarZ, a redox active global regulator in Staphylococcus aureus. J. Biol. Chem. 284, 23517–23524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Newberry K. J., Fuangthong M., Panmanee W., Mongkolsuk S., Brennan R. G. (2007) Structural mechanism of organic hydroperoxide induction of the transcription regulator OhrR. Mol. Cell 28, 652–664 [DOI] [PubMed] [Google Scholar]
- 50. Mahenthiralingam E., Marklund B. I., Brooks L. A., Smith D. A., Bancroft G. J., Stokes R. W. (1998) Site-directed mutagenesis of the 19-kilodalton lipoprotein antigen reveals no essential role for the protein in the growth and virulence of Mycobacterium intracellulare. Infect Immun. 66, 3626–3634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Fontán P. A., Voskuil M. I., Gomez M., Tan D., Pardini M., Manganelli R., Fattorini L., Schoolnik G. K., Smith I. (2009) The Mycobacterium tuberculosis sigma factor σB is required for full response to cell envelope stress and hypoxia in vitro, but it is dispensable for in vivo growth. J. Bacteriol. 191, 5628–5633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Divangahi M., Desjardins D., Nunes-Alves C., Remold H. G., Behar S. M. (2010) Eicosanoid pathways regulate adaptive immunity to Mycobacterium tuberculosis. Nat. Immunol. 11, 751–758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Schnappinger D., Ehrt S., Voskuil M. I., Liu Y., Mangan J. A., Monahan I. M., Dolganov G., Efron B., Butcher P. D., Nathan C., Schoolnik G. K. (2003) Transcriptional adaptation of Mycobacterium tuberculosis within macrophages: insights into the phagosomal environment. J. Exp. Med. 198, 693–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Rengarajan J., Bloom B. R., Rubin E. J. (2005) Genome-wide requirements for Mycobacterium tuberculosis adaptation and survival in macrophages. Proc. Natl. Acad. Sci. U.S.A. 102, 8327–8332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Chen M., Divangahi M., Gan H., Shin D. S., Hong S., Lee D. M., Serhan C. N., Behar S. M., Remold H. G. (2008) Lipid mediators in innate immunity against tuberculosis: opposing roles of PGE2 and LXA4 in the induction of macrophage death. J. Exp. Med. 205, 2791–2801 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.