Background: GAT-2 is physiologically and pharmacologically important for regulating peripheral GABAergic mechanisms.
Results: We identify GAT-2 ligands, including drugs, metabolites, and fragments, using comparative modeling, virtual screening, and experiments.
Conclusion: GAT-2 is a high selectivity/low affinity transporter that is resistant to inhibition by typical GABAergic inhibitors.
Significance: Our results explain pharmacological and physiological effects of GAT-2 ligands and identify specificity determinants in the SLC6 family.
Keywords: Drug Discovery, Homology Modeling, Membrane Proteins, Membrane Transport, Protein Structure, Comparative Modeling, GABA Transporter, Protein Function Prediction, Protein Structure Prediction, Structure-based Ligand Discovery
Abstract
The solute carrier 6 (SLC6) is a family of ion-dependent transporters that mediate uptake into the cell of osmolytes such as neurotransmitters and amino acids. Four SLC6 members transport GABA, a key neurotransmitter that triggers inhibitory signaling pathways via various receptors (e.g., GABAA). The GABA transporters (GATs) regulate the concentration of GABA available for signaling and are thus targeted by a variety of anticonvulsant and relaxant drugs. Here, we characterize GAT-2, a transporter that plays a role in peripheral GABAergic mechanisms, by constructing comparative structural models based on crystallographic structures of the leucine transporter LeuT. Models of GAT-2 in two different conformations were constructed and experimentally validated, using site-directed mutagenesis. Computational screening of 594,166 compounds including drugs, metabolites, and fragment-like molecules from the ZINC database revealed distinct ligands for the two GAT-2 models. 31 small molecules, including high scoring compounds and molecules chemically related to known and predicted GAT-2 ligands, were experimentally tested in inhibition assays. Twelve ligands were found, six of which were chemically novel (e.g., homotaurine). Our results suggest that GAT-2 is a high selectivity/low affinity transporter that is resistant to inhibition by typical GABAergic inhibitors. Finally, we compared the binding site of GAT-2 with those of other SLC6 members, including the norepinephrine transporter and other GATs, to identify ligand specificity determinants for this family. Our combined approach may be useful for characterizing interactions between small molecules and other membrane proteins, as well as for describing substrate specificities in other protein families.
Introduction
The solute carrier 6 family (SLC6)5 consists of 20 Na+- and Cl−-dependent membrane transporters that regulate a variety of biological activities such as neurotransmission and metabolism (1). This transporter family can be classified into four groups based on their amino acid sequences: monoamine transporters, GABA transporters, amino acid transporters, and “orphan” transporters that may also transport amino acids (1, 2). Mutations in SLC6 members are associated with a wide spectrum of disorders such as obsessive compulsive disorder (serotonin transporter, SLC6A4), obesity (SLC6A14), and orthostatic hypotension (NET, SLC6A2) (2). SLC6 members are therefore targets for many prescription drugs, including antidepressants (e.g., venlafaxine (Effexor®)) and stimulants (e.g., methylphenidate (Ritalin®)) that often act on more than one transporter (3).
GABAergic Signaling and the GABA Transporters
GABA is a key inhibitory neurotransmitter in the mammalian brain and acts by binding to the GABAergic receptors in inhibitory neurons (e.g., GABAA) in the CNS (4). GABAergic mechanisms have also been found in the stomach, pancreas, intestine, testis, ovary, uterus, liver, urinary bladder, and kidney (5). Malfunctions of the GABAergic system have been associated with mucous overproduction in asthma (6), as well as with protective and regenerative effects on islet β cells in diabetes (7). The GABA transporter (GAT) family consists of four transporters that regulate the concentration of GABA available for signaling via the GABAergic receptors (4, 8). Therefore, the GATs are emerging drug targets for a variety of disorders, primarily those that have been associated with neurosignaling (4). For example, GAT-1 (SLC6A1) and GAT-3 (SLC6A11) are key targets for anticonvulsants and relaxants (e.g., tiagabine (Gabitril®)). These drugs increase the concentration of GABA in the synaptic cleft by inhibiting GAT-1- or GAT-3-mediated GABA reuptake. The GABA transporter 2 (GAT-2, SLC6A13) is primarily expressed in the liver, kidney, and other peripheral tissues such as the testis, retina, and lungs (4, 8, 9). Thus, GAT-2 might be physiologically important for regulating key peripheral GABAergic mechanisms such as those associated with asthma and diabetes (6, 7). Furthermore, GAT-2 might also play a pharmacological role in disposition and metabolism of GABAergic drugs in the liver and kidney or be a drug target itself (e.g., in asthma therapy).
Structure and Mechanism of GAT-2
GAT-2 contains one large domain with 12 predicted membrane-spanning helices (10). No structures of human SLC6 members, including GAT-2, have been determined at atomic resolution; however, x-ray structures of a bacterial homolog, the leucine transporter LeuT, have been determined in four different conformations that were proposed to represent different snapshots of the transport cycle (11–13). Additionally, LeuT complex structures with various substrates and inhibitors suggested a competitive inhibition mechanism in which larger ligands (e.g., tryptophan) stabilize an inhibited outward facing conformation (12). Interestingly, it was also shown that an additional substrate-binding site (S2) is located on the surface of LeuT (14–16) and that various inhibitors stabilize different conformations in LeuT via sites overlapping with the S2 site (11, 17). These observations are in agreement with the notion that LeuT and its human homologs transport ligands across the cell membrane via the “alternating access” transport mechanism (13, 19–25).
Here, we characterize the function of GAT-2 using an integrated computational and experimental approach. We constructed structural models for GAT-2 in two different conformations (i.e., occluded and outward facing states) in complex with GABA and experimentally validated the models using site-directed mutagenesis. We then performed virtual ligand screening against the modeled binding site to predict small molecules, including metabolites, prescription drugs, and fragment-like compounds, that interact with GAT-2. The predicted hits and additional molecules were then validated, using inhibition of radiolabeled substrate uptake experiments. We also compared the predicted GAT-2-binding site with those of other SLC6 members, including the norepinephrine transporter (NET), and the other human GATs. Finally, we discuss the application of these results to describe the specificity determinants in the SLC6 family, as well as the utility of our approach to identify residues important for function and chemically novel ligands for GAT-2 and other transporters.
EXPERIMENTAL PROCEDURES
Comparative Model Construction
GAT-2 was modeled based on x-ray structures of LeuT from Aquifex aeolicus in the occluded/outward facing (“occluded”) and the outward facing conformations (Protein Data Bank codes 2A65 (26) and 3F3A (12), respectively), using MODELLER-10v8 (see Fig. 1 and supplemental materials). For each conformation, 100 models were generated based on the GAT-2-LeuT alignment (supplemental Fig. S1), using the standard “automodel” class of MODELLER-10v8 (27). The models were assessed using Z-DOPE, a normalized atomic distance-dependent statistical potential based on known protein structures (see Table 1) (28). Moreover, the occluded GAT-2 conformation was modeled with nonprotein atoms, including the leucine molecules, ions (chloride and sodium), and other heteroatoms (i.e., the detergents), based on their corresponding coordinates in the template structure; the outward facing GAT-2 conformation was modeled similarly with nonprotein atoms, including the tryptophan molecules, sodium ions, and detergents that were used for crystallization. However, because the S1-binding site in the outward facing structure of LeuT is partially occupied by the detergent molecules B-octylgucoside and tetradecane, we removed the atoms of these molecules from the template structure. Finally, the models were refined by repacking the side chains on a fixed backbone with SCWRL4 (29), as well as by being subjected to 10,000 steps of conjugate gradient minimization under the Amber99SB-ILDN force field (Refs. 30 and 31 and supplemental materials).
FIGURE 1.
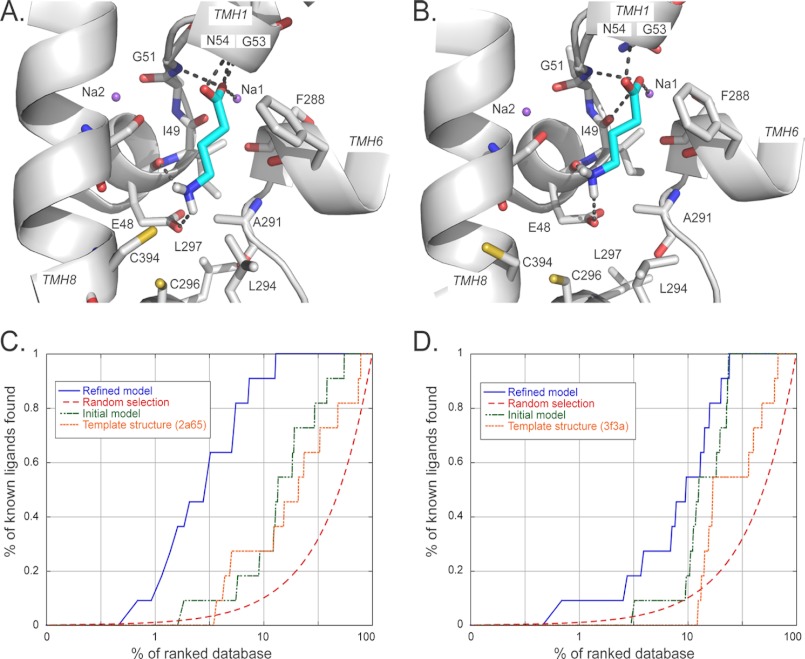
GAT-2-GABA models and their validation by ligand enrichment. A and B, predicted structures of the GAT-2-GABA complex in the occluded (A) and the outward facing (B) conformations. GABA is colored in cyan, with oxygen, nitrogen, and hydrogen atoms in red, blue, and white, respectively. The sodium ions Na1 and Na2 are visualized as purple spheres. The transmembrane helices of GAT-2 are depicted as white ribbons. Key residues are displayed as sticks. The hydrogen bonds between GABA and GAT-2 are shown as dotted gray lines; they involve the residues Glu-48, Gly-51, and Gly-53, as well as the sodium ion Na1 for both conformations models. GABA forms polar interactions with Asn-54 only in the occluded conformation model. C and D, enrichment plots for different structures of the occluded (C) and the outward facing (D) models: the refined GAT-2 models (blue), random selection (red), the initial GAT-2 models (green), and the LeuT template structures (orange).
TABLE 1.
Assessment of the GAT-2 models
| Modela | Z-DOPEb | logAUCc | EF1d |
|---|---|---|---|
| Occluded conformation | |||
| Template Structure | −2.5 | 25.9 | 0 |
| Initial model | −0.93 | 29.5 | 0 |
| Refined model | −0.36 | 53.4 | 17.0 |
| Outward facing conformation | |||
| Template Structure | −2.39 | 19.7 | 0 |
| Initial model | −1.13 | 29.1 | 0 |
| Refined model | −1 | 37.4 | 7.9 |
a Model marks the model used for the assessment. Template structure corresponds to the structures of the leucine transporter LeuT in the occluded (Protein Data Bank code 2A65) and the outward facing (Protein Data Bank code 3F3A) conformations. Initial model represents the initial models by MODELLER that achieved the best Z-DOPE (28) score, without refinement. Refined model marks our final refined models.
b Z-DOPE provides the score of the models using Z-DOPE, a normalized atomic distance-dependent statistical potential based on known protein structures (28). Per residue Z-DOPE score of the initial score was also compared with that of the template structure.
c logAUC marks the area under the logarithmic scale of the enrichment curve.
d EF1 represents the enrichment factor at 1% of the ranked database.
Ligand Docking and Virtual Screening
Virtual screening against the GAT-2 models was performed using a semiautomatic docking procedure (32–36). All of the docking calculations were performed with DOCK 3.5.54 (37, 38). The docking poses of database molecules were ranked by DOCK score consisting of van der Waals, Poisson-Boltzmann electrostatic, and ligand desolvation penalty terms. Importantly, binding affinity cannot yet be predicted accurately by docking (35, 39). Thus, poses of the 200–500 highest ranked compounds from each one of the computational screens were analyzed manually (36, 40).
The receptor structure was prepared by removing all nonprotein atoms, except for the sodium ions. Binding site residues were identified as residues with at least one atom within 10 Å of any heavy atoms of the ligand leucine from the initial model, using the program FILT (from the DOCK3.5 distribution). The solvent-accessible molecular surface of the protein-binding site was then calculated with the program DMS (41) using a probe radius of 1.4 Å. Receptor-derived spheres were calculated using the program SPHGEN (42) (part of the UCSF DOCK suite), whereas the ligand-derived spheres were generated from the positions of the heavy atoms of the crystallographic ligand, if available. In total, 45 matching spheres were used to orient ligands in the binding site.
Binding Site Assessment
The final models that were used for virtual screening were selected based on their ability to discriminate known ligands from decoys using docking. In particular, we calculated the enrichment for the known ligands among the top scoring decoy compounds, generated by the Directory of Useful Decoys protocol (32, 43). 11 GAT-2 ligands were collected from the literature (4, 9), as well as the UniProt (44) and Kyoto Encyclopedia of Genes and Genomes (KEGG) (45) databases. For each known ligand and 36 Directory of Useful Decoys-generated decoys, the best docking pose was computed. The corresponding docking scores were used to calculate EF1 (supplemental materials, Equation 1) and logAUC (supplemental materials, Equation 2), where final models were selected based on their logAUC score. For example, a random selection of known ligands from a database consisting of known ligands and decoys yields a logAUC of 14.5. Finally, the generated models were evaluated using enrichment calculations that included the ligands discovered in this study. The most enriching models were virtually identical to the models used for our original virtual screening.
Data Sets for Virtual Screening
We used the following three compound data sets for virtual screening. First, the KEGG DRUG database is a comprehensive information resource for approved drugs in Japan, the United States, and Europe. It includes all the marketed drugs in Japan, including prescription drugs and over the counter drugs. We used a filtered version that included 6,436 molecules suitable for docking (36). Second, the KEGG Ligand Compound database includes metabolites, biopolymers, and other chemical substances that are related to biological systems. We also used a filtered version of KEGG Ligand Compound that included 12,730 molecules. Third, the ZINC fragment-like set includes 575,000 purchasable organic molecules with fragment-like physicochemical properties, including a molecular mass of 250 Dalton or lower, five or fewer rotatable bonds, and an xlogP value of 3.5 or less, where xlogP is the octanol/water partition coefficient (logP) calculated by an atom additive method (43, 46).
Chemical Novelty Evaluation
For assessing chemical similarity between two compounds, we computed the Daylight 1024 hashed fingerprints (Daylight Chemical Information Systems, Inc., Laguna Niguel, CA). Tanimoto coefficients (Tc) were calculated between each docking discovered ligand and 14 annotated GAT-2 ligands in the ChEMBL database (48), using the program Pipeline Pilot. Tc values of <0.5 suggest that the molecule is a chemically novel GAT-2 ligand.
Subcloning and Transient Transfection of GAT-2 into HEK Cells
The full-length human GAT-2 cDNA clone was purchased from ATCC and subcloned into the pcDNA5/FRT expression vector (Invitrogen) according to the manufacturer's protocol. For directional subcloning, the full-length GAT-2 cDNA was excised from the host vector using the restriction enzymes XhoI and HindIII (New England Biolabs, Ipswich, MA). The expression vector was similarly cleaved with the same restriction enzymes, and the two DNA fragments were ligated using T4 DNA ligase (Invitrogen) according to the manufacturer's protocol. The ligated DNA was transformed into DH5a cells (Invitrogen) according to the manufacturer's protocol and plated on agar plates (TEKnova, Hollister, CA) overnight at 37 °C. The colonies were selected the following day and amplified in 5 ml of ampicillin-containing LB broth (TEKnova, Hilden, Germany) overnight. The cultures were then pelleted by centrifugation, and DNA was isolated and purified from the pellets using a Qiagen DNA extraction kit. The DNA was then sequenced (Quintara Biosciences, Albany, CA) to determine the validity of the pcDNA-5-GAT-2 clones. Clones with 100% identity to the GenBankTM reference human GAT-2 sequence were selected.
For transfection, HEK-Flpin cells were seeded in DMEM (Cell Culture Facility, University of California, San Francisco) supplemented with 10% FBS in poly-d-lysine-coated 24-well plates at a density of 600,000 cells/ml. Approximately 24 h later, the cells were transfected with Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol. In brief, for each plate 48 μl of Lipofectamine 2000 were added to 1152 μl of OptiMEM-medium (Cell Culture Facility, University of California, San Francisco), and the solution was incubated at room temperature for 5 min. In the meantime, OptiMEM solutions for each plate containing 19.2 μg of either pcDNA5/FRT (empty vector) or vector containing the DNA of interest (e.g., GAT-2- or site-directed mutant sequences) in a total volume of 1200 μl were prepared. After the 5-min incubation period, the two solutions were combined and incubated for 20 min at room temperature. During this time, the medium in the 24-well plates was changed to 0.5 ml/well prewarmed OptiMEM. Finally, 100 μl of the Lipofectamine-DNA solution were added to each well, and the plates were put back into the incubator. 12 h later the medium was changed to DMEM (Cell Culture Facility, University of California, San Francisco) substituted with 10% FBS, and uptake experiments were performed ∼48 h after transfection.
Uptake Experiments
The cells were washed with 0.5 ml of Hanks' buffered saline solution (HBSS; Cell Culture Facility, University of California, San Francisco) per well and then incubated for 10 min in 0.5 ml of HBSS/well at 37 °C. Then the buffer was removed and replaced with 0.5 ml of prewarmed HBSS containing 20 nm 3H-labeled (PerkinElmer Life Sciences) and 1 μm unlabeled GABA (Sigma-Aldrich) with or without the compound of interest at 50, 500, or 5000 μm. After incubating the plate at 37 °C for 2 min, the uptake was stopped by removing the medium and washing twice with 1 ml of ice-cold HBSS/well. The cells were lysed in 700 μl of lysis buffer (0.1 n NaOH and 0.1% SDS in bidistilled water) by shaking for 2.5 h. 600 μl of the lysate were added to 3 ml of EcoLite scintillation fluid (MP Bio, Solon, OH) and counted on a LS6500 Scintillation Counter (Beckman Coulter, Pasadena, CA). The counts were corrected for protein concentration, and the uptake was expressed as a percentage of control (labeled and unlabeled GABA). Protein concentrations were determined with a BCA assay kit (Thermo Scientific, Rockford, IL).
Site-directed Mutagenesis
Site-directed mutagenesis was performed using the QuikChange kit (Agilent Technologies, Santa Clara, CA) according to the manufacturer's protocol. In brief, 5 μl of 10× reaction buffer were mixed with 1 μl of DNA solution at 5 ng/μl, 1.25 μl of primer solution (containing 100 ng/μl of both forward and reverse primers each), 1 μl of dNTP mix, and 41.75 μl of double distilled H2O. Then 1 μl of PfuTurbo® DNA polymerase was added, and the PCR was started as described in the manufacturer's protocol with 16 cycles and 15 min at 68 °C for the elongation step. PCR products were digested with DpnI at 37 °C for 1 h followed by transfection into Epicurian Coli XL1-Blue supercompetent bacteria. The bacteria were plated, and the clones were selected and screened for the correct sequence.
Primers for site-directed mutagenesis were designed with the PrimerX web-based program using the following parameters: melting temperature between 78 and 85 °C, GC content between 40 and 60%, length between 25 and 45 bp, and both 5′- and 3′-flanking regions 11–25 bp in length with the primers terminating in G or C and the mutation site at the center of the primer.
Construction and Visualization of Chemical Similarity Networks
The graphs representing the chemical similarity networks (see Fig. 3) were constructed and visualized using Cytoscape 2.8.1 (49), relying on the ChemViz (v.1.1) and clusterMaker (v.1.9) plugins. In particular, we calculated the Tc values between all pairs of the 376 small molecules, using the default parameters in ChemViz, which calculates molecular descriptors for the compounds using the Chemistry Development Kit open source library (50). The layout of the final network was obtained using the edge-weighted spring-embedded algorithm in Cytoscape, using the calculated Tc values as weights (see Fig. 3A). The edges indicate similarities between molecules with Tc of at least 0.30. We also clustered the molecules using the Markov clustering algorithm (51) of the clusterMaker plugin, using the default parameters (see Fig. 3B).
FIGURE 3.
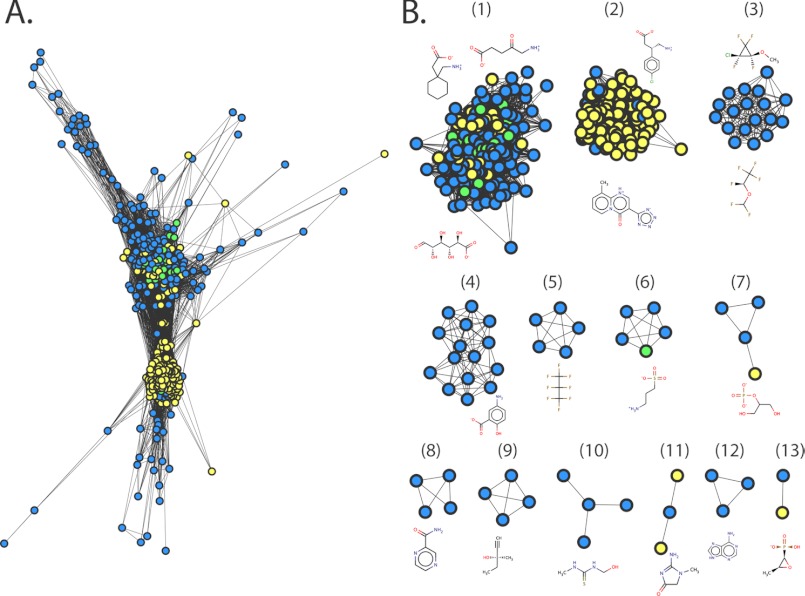
Chemical similarity network of predicted ligands. The relationships among the top ranked small molecule drugs from KEGG DRUG are visualized using Cytoscape 2.8.1. The nodes represent the small molecules predicted to bind GAT-2, using the occluded model (blue), the outward facing model (yellow), or both models (green). Each edge represents pairwise chemical similarity with Tc of at least 0.3. A, a similarity network using the edge-weighted spring-embedded layout algorithm in Cytoscape, which preserves all the relationships among the small molecules (49). B, a network with the 13 clusters of the small molecule drugs. The molecules were clustered using the Markov clustering algorithm (51) in Cytoscape. Representative small molecules structures of the clusters are visualized using MarvinView 5.4.1.1.
RESULTS
GAT-2 Models and Their Assessment
GAT-2 was modeled based on the structures of the leucine transporter LeuT from A. aeolicus in the occluded/outward facing (“occluded”) (26) and the outward facing conformations (12) (Fig. 1, A and B). The comparative models contain the whole transmembrane domain of the protein, including the 12 transmembrane helices and the S1-binding site residues. The refined models were assessed based on their ability to discriminate between known ligands and likely nonbinders (“decoys”), using “enrichment curves” derived from ligand docking calculations (Table 1; Fig. 1, C and D; and supplemental materials) (36). The final refined GAT-2 models in the occluded and outward facing conformations obtained logAUC scores of 53.4 and 37.4, respectively (Fig. 1 and Table 1). This result suggests that the occluded model is more accurate than the outward facing model and that both models are suitable for selecting ligands for experimental testing (32, 36). The logAUC values for the refined models are substantially better than those calculated for the template structures (25.9 and 19.7 for the occluded and outward facing models, respectively) and the initial models (29.5 and 29.1), as well as for random selection of ligands (14.5) (Table 1). We also assessed the models using the enrichment factor (EF1), which is the fraction of the annotated ligands among the 1% top scoring docking hits compared with their fraction in the entire docking database (supplemental materials) (32, 33, 43). The EF1 values for the occluded and outward facing models (17.0 and 7.9, respectively) also indicate that the models can potentially discriminate between known ligands and nonbinders.
Mode of GAT-2 Interaction with GABA
The occluded and outward facing GAT-2 models are structurally similar and include minor backbone and side chain rearrangements, similarly to the corresponding LeuT template structures (12). For example, the residues forming the extracellular gate (i.e., Tyr-129 and Phe-288) are 2 Å further away from each other in the outward facing conformation than in the occluded conformation, making additional volume accessible to a ligand (supplemental Fig. S2). The corresponding space of the outward facing LeuT structure (i.e., between Tyr-108 and Phe-253) is also partially occupied by a detergent that was used for crystallization (12). In addition, the majority of the key polar interactions between GABA and GAT-2 are conserved in both the occluded and the outward facing models; for example, the carboxyl group of GABA forms polar interactions with the sodium ion Na1, Gly-51, and Gly-53 in the models of both conformations, as well as with Asn-54 only in the occluded model and with Ile-49 only in the outward facing conformation. Furthermore, the amine group of GABA forms a hydrogen bond with the main chain oxygen of Glu-48, as well as polar interaction with the negatively charged side chain of the same residue. During refinement, the conformation of the Glu-48 side chain correlated with the enrichment scores for the models (i.e., models with buried Glu-48 obtained the worst enrichment scores and vice versa (supplemental Fig. S3)), suggesting the importance of this residue in ligand recognition. Thus, our combined model refinement/ligand docking approach was used to predict Glu-48 as a key residue for transport.
Model Validation Using Site-directed Mutagenesis
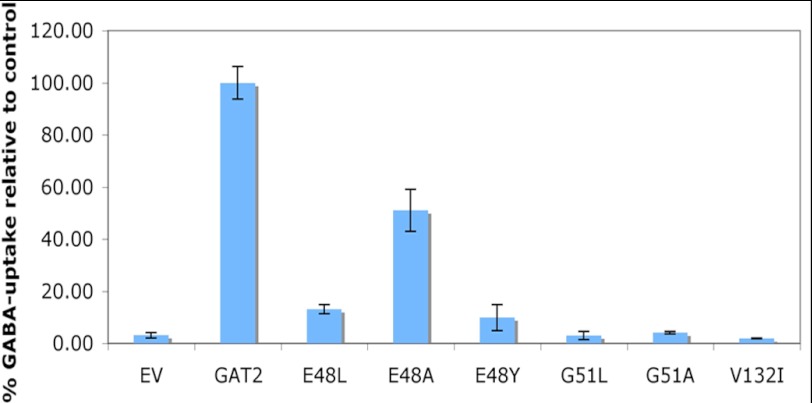
To validate the GAT-2 models, we mutated residues predicted to be involved in GABA binding or to be in close proximity to the binding site (Fig. 1). In particular, we looked at their effect on the uptake of the radiolabeled substrate 3H-GABA into transiently transfected HEK293 cells (“Experimental Procedures”) (Fig. 2). Among the most significant mutations were the G51L and G51A that completely abolished transport (Fig. 2). This result confirms the importance of Gly-51 in mediating the network of polar interactions between the residues in the GAT-2-binding site, the sodium ion Na1, and the GAT-2 ligands. This network is likely to be conserved among SLC6 members that transport amino acids, including the GATs (e.g., Gly-63 in GAT-1 (52)), but not in monoamine transporters, which have aspartate in the corresponding position (e.g., Asp-75 in NET (36)). In addition, mutations of Glu-48 hampered transport but did not prevent substrate uptake completely. A reduction of 3H-GABA uptake by ∼50% was observed for E48A and by ∼90% for E48L and E48Y. The position of Glu-48 in GAT-1 corresponds to a tyrosine in GAT-1 (Tyr-60); thus, the latter mutation (E48Y) suggests that GAT-2 and GAT-1 achieve specificity for GABA via different amino acid residues (52). Interestingly, although Val-132 does not directly interact with the ligand, the V132I mutation abolished transport completely, suggesting that it might have an indirect effect. Importantly, Val-132 corresponds to Ile-111 in LeuT in the S2 substrate-binding site (11), which has not yet been confirmed to exist on the surface of the human SLC6 members. Immunohistochemistry with a GAT antibody confirmed that all of the mutant proteins localized to the membrane similarly to the wild-type protein (data not shown). Finally, the location of the binding site is in agreement with those of the other GATs (20, 53). For example, the mouse GAT-3 (i.e., GAT-4) binding site is almost identical to that of our GAT-2 model (54).
FIGURE 2.
Validating predicted binding site residues by mutagenesis. Influence of site-directed mutations on 3H-GABA transport as compared with the wild-type sequence (GAT-2) and empty vector (EV). These results were obtained using HEK293 cells transiently transfected with the reference and mutated pcDNA5/FRT-GAT-2 or with the empty vector pcDNA5/FRT.
Virtual Screening of Small Molecule Libraries against the GAT-2 Models
We computationally screened filtered libraries of 6,436, 12,730, and 575,000 small molecules from the KEGG DRUG, KEGG Ligand Compound (45), and ZINC fragment-like (43) databases, respectively, against the refined models of the occluded and the outward facing conformations (Experimental Procedures). The KEGG DRUG set includes over the counter and prescription drugs that are marketed in Europe, Japan, and the United States; the KEGG Ligand Compound library consists of a variety of small molecules, biopolymers, and other chemical substances that are found in biological systems; the ZINC fragment-like data set includes purchasable small molecule ligands with fragment-like chemical properties (43, 46). Several known GAT-2 ligands were ranked highly in our screen, which increases our confidence in the models. For example, GABA was ranked 3 and 36 in the KEGG DRUG screens against the occluded and outward facing models, respectively.
Chemical Similarity of the Predicted Ligands
Relationships among predicted small molecules are illustrated using chemical similarity networks. We calculated the chemical similarity among the 200 top scoring hits of the KEGG DRUG screens against the outward facing and the occluded models. Except for four drugs, all 376 molecules were related to each other, including 24 molecules that were common to both lists (“Experimental Procedures”). This result indicates that the hits occupy a continuous area in the chemical space, even though there was no significant filtering bias in the virtual screens (Fig. 3A). These 376 molecules were grouped into 13 distinct clusters based on the similarity among their chemical structures (Fig. 3B). For example, cluster 9 includes small drugs containing an alkyne group (e.g., the hypnotic/sedative drug methylpentynol). Interestingly, molecules covering particular areas of the similarity network are predicted using screens against particular models. For example, molecules predicted only in the outward facing model screen are localized in the center of the similarity network (Fig. 3A) corresponding to cluster 2 (Fig. 3B). The majority of the molecules in this cluster are too large to fit into the binding site of the occluded model (e.g., pemirolast contains three aromatic rings) (Fig. 3B). Some hits are predicted to bind GAT-2 based on both models (e.g., homotaurine) (Fig. 4).
FIGURE 4.
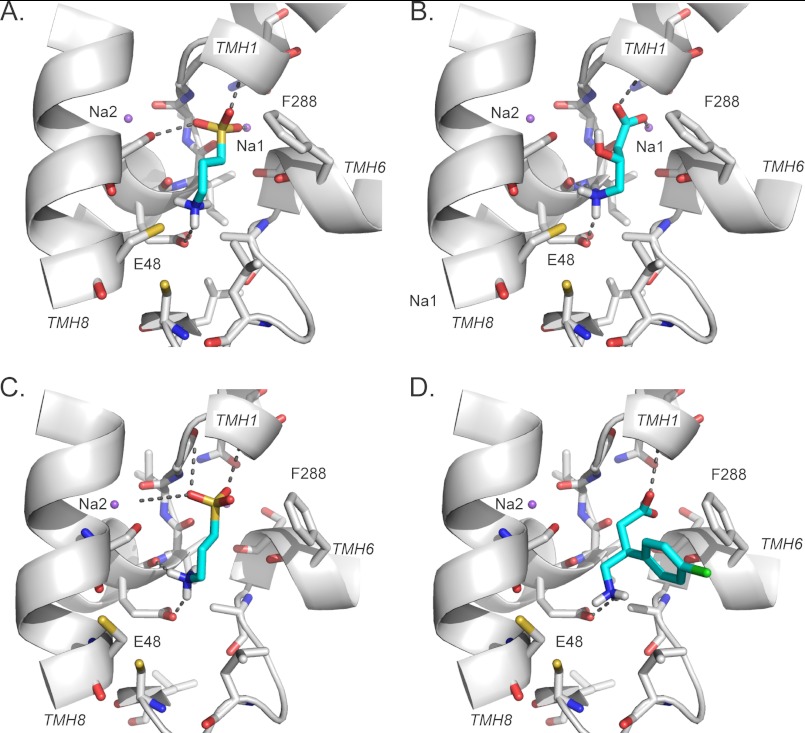
Predicted binding modes for GAT-2 validated ligands. A–D, predicted binding modes of newly identified GAT-2 ligands with GAT-2 models in the occluded (A and B) and the outward facing (C and D) conformations. Ligands are colored in cyan, with sulfur, chloride, oxygen, nitrogen, and hydrogen atoms in yellow, green, red, blue, and white, respectively. The sodium ions Na1 and Na2 are visualized as purple spheres. The transmembrane helices of GAT-2 are depicted as white ribbons. Key residues are displayed as sticks; the hydrogen bonds between ligands and key GAT-2 residues (e.g., Glu-48) are shown as dotted gray lines. The representative previously unknown ligands are homotaurine (A and C), GABOB (B), and baclofen (D).
Rationale for Selecting Molecules
The top 200–500 highest ranked hits in each of the six computational screens (i.e., three data sets against two models) were examined manually. In particular, we analyzed similarities of the predicted poses of these ligands to those in the predicted complexes of GAT-2 with known ligands, as well as frequent scaffolds and common pharmacological function (35, 36, 39, 40). Some of our selected compounds were similar to known GAT-2 ligands in their structure, function, or predicted mode of interaction with GAT-2. For example, the anticonvulsant γ-amino-β-hydroxybutyric acid (GABOB) is a derivative of GABA that is predicted to interact with the key residues Glu-48 and Gly-51, as well as with the sodium ion Na1, similarly to GABA (Figs. 1A and 4B).
Other molecules, however, were different from known GAT-2 ligands in structure, function, or predicted mode of interaction. For instance, homotaurine contains a sulfonic acid group, making it dissimilar to known GAT-2 ligands (Tc of 0.30) (supplemental Table S1). Nevertheless, our confidence in the homotaurine prediction was increased for two reasons. First, homotaurine was predicted as GAT-2 ligand by virtual screens against GAT-2 models in both the occluded and outward facing conformations. Second, the poses of homotaurine in the predicted complexes were similar to the poses of GABA in the corresponding GAT-2-GABA complexes (Fig. 4, A and C). In total, 31 of the predicted compounds were tested using a cis-inhibition assay that does not distinguish between inhibitors and substrates.
Experimental Validation of the Top Hits
To experimentally validate GAT-2 inhibition by the predicted compounds, we developed a cell-based cis-inhibition assay. This assay is based on the uptake of 3H-GABA in HEK293 cells transiently transfected with GAT-2 and on the capacity of inhibitors and substrates to reduce intracellular accumulation of the probe substrate. To choose an appropriate GABA concentration and a suitable uptake time, we performed time course and uptake kinetics experiments (supplemental Fig. S4). The functionality of the assay was demonstrated by comparing kinetic parameters with those previously reported for GAT-2 in published data (4, 8, 9) and by competitive inhibition of 3H-GABA uptake with unlabeled GABA in addition to the known GAT-2 inhibitors β-alanine and desipramine.
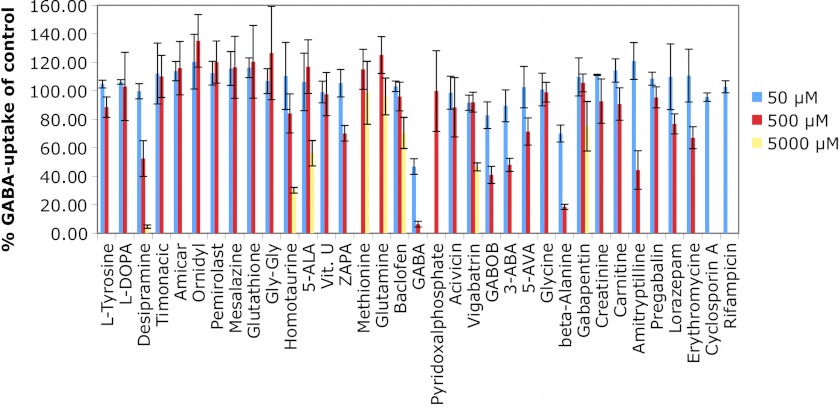
Twelve of the predicted molecules inhibited 3H-GABA uptake by 20% or more at concentrations ranging from 500 μm to 5 mm (Fig. 5 and supplemental Figs. S5 and S6). These values are in agreement with previous uptake inhibition data for GABAergic molecules (4, 9). Six of the identified ligands (homotaurine, baclofen, gabapentin, lorazepam, pyridoxal phosphate, and ZAPA) are chemically novel GAT-2 ligands (Fig. 5 and supplemental Table S1). Homotaurine, gabapentin, and ZAPA were predicted using the models in both conformations, whereas the larger molecules baclofen, lorazepam, and pyridoxal phosphate were predicted using only the outward facing model, demonstrating that virtual screening against different conformations can cover different parts of the chemical space (Fig. 4D). However, the affinity of the three ligands binding to the outward conformation is not as strong as that of the occluded model ligands. For example, the relaxant drug baclofen, a GABAB receptor agonist (55) that was recently approved for treating alcoholism (56), contains a chlorophenyl group, making it a chemically novel GAT-2 ligand (Tc of 0.36) (supplemental Table S1). In addition, the anticonvulsants GABOB and vigabatrin were also identified as GAT-2 ligands. However, although these molecules are highly similar in structure to GABA (i.e., Tc of 0.93 and 0.75, respectively), they are much weaker inhibitors of GAT-2 than GABA itself. For example, GABOB and vigabatrin inhibited 3H-GABA uptake by 59% at 500 μm and by 53% at 5 mm, respectively.
FIGURE 5.
cis-Inhibition studies of predicted GAT-2 inhibitors. Uptake of 3H-GABA in transiently transfected GAT-2-expressing HEK293 cells in the presence of various small molecule compounds is shown. The tested concentrations were 50 and 500 μm. A concentration of 5000 μm was only used where solubility and toxicity allowed. All of the data are shown with bars representing the S.E.
Similarly, other GABA analogs including pregabalin and amicar, as well as various amino acids, did not exert any effect on 3H-GABA uptake (Fig. 5 and supplemental Table S1). Moreover, classical inhibitors of transporters exhibit no (i.e., rifampicine and cyclosporine) or only weak (erythromycin) inhibitory effects on GAT-2 transport further demonstrating the high tolerance of GAT-2 against inhibition. We also tested compounds that are structurally related to known and predicted GAT-2 ligands (supplemental Table S1). However, of these predicted molecules only the chemically novel hit pyridoxal phosphate inhibited 3H-GABA uptake (Fig. 5). In summary, our results suggest that GAT-2 is more selective for inhibitors than the other GATs.
Finally, based on the observation that GAT-2 is highly selective for small GABA-like compounds, we computationally screened the ZINC fragment-like small molecule set (575,000 molecules) against the GAT-2 model. We experimentally validated three hits. 3-Aminobutanoic acid reduced 3H-GABA uptake by at least 50% at 500 μm, and 5-aminovaleric acid exhibited a 30% inhibition at 500 μm (Fig. 5). Furthermore, 5-aminolevulinic acid (5-ALA) inhibits 3H-GABA uptake by 44% at 5 mm (Fig. 5) and completely abolished transport at 20 mm (supplemental Fig. S6). The relatively weak inhibition by these previously unknown GABA-like ligands again indicates that GAT-2 is a highly selective transporter.
Comparison between the GAT-2 and the NET Models
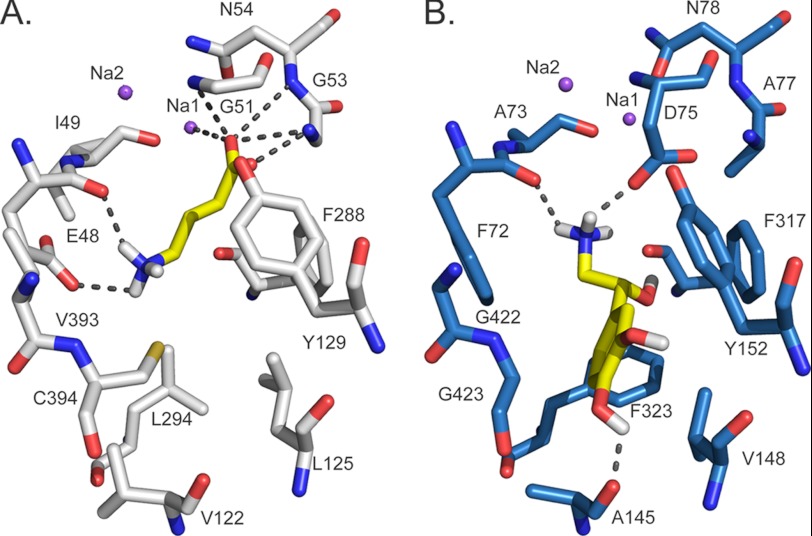
We compared the binding site of GAT-2 to that of NET (36), which shares about 45% sequence identity with GAT-2 (1, 2), to rationalize their variation in substrate specificity (i.e., GAT-2 and NET transport small GABA-like zwitterions and monoamines, respectively) (Fig. 6). The NET model, which was also constructed based the LeuT structure, is highly similar to models of the other SLC6 monoamine transporters serotonin transporter (57) and dopamine transporter (58). The predicted GAT-2 and NET structures are similar (root mean square deviation of 0.1 Å), sharing several features, including the location of the sodium ions and the S1-binding site, as well as the arrangement of several key binding site residues (i.e., Tyr-129, Phe-288, and Ser-289 in GAT-2 correspond to Tyr-152, Phe-317, and Ser-318 in NET).
FIGURE 6.
Comparison of GAT-2 and NET predicted binding sites. The final model of GAT-2 (A, white) and the model of NET (36) (B, blue) in the occluded conformation are shown with their corresponding substrates (i.e., GABA and norepinephrine, respectively) (yellow). Atoms are illustrated by sticks, with oxygen, nitrogen, and hydrogen atoms in red, blue, and white, respectively. The sodium ions Na1 and Na2 are visualized as purple spheres. GABA and norepinephrine are depicted in orange sticks, and their hydrogen bonds with GAT-2 (involving Glu-48, Gly-51, Gly-53, Asn-54, and Na1) and NET (involving Ala-145, Phe-72, and Asp-75) are shown as dotted gray lines.
However, the structural models of NET and GAT-2 include the following main differences that may rationalize their substrate specificities: First, Phe-72 and Phe-323 in NET are substituted for Glu-48 and Leu-294 in GAT-2. As a result, (i) the GAT-2-binding site consists of fewer aromatic residues than that of NET, in which the additional phenylalanine residues can make π-π interactions with ligands such as norepinephrine (36) and (ii) the corresponding region of the GAT-2-binding site is more acidic than that of NET; thus, it is capable of forming key polar interactions with the amine groups of the GAT-2 ligands, such as GABA (Fig. 6A, gray dotted lines). Second, the negatively charged Asp-75 in NET is replaced by Gly-51 in GAT-2, which is predicted to adopt a similar conformation to that of Gly-24 in the LeuT x-ray structure. Consequently, (i) the volume occupied by the aspartate side chain in NET is accessible for ligands in GAT-2, and (ii) the negative charge of the aspartate side chain is removed, and the positive charge of the sodium ion Na1 becomes accessible for ligands, changing the corresponding accessible surface of the binding site from acidic (NET) to basic (GAT-2). Third, Ala-145, Val-148, Gly-422, and Gly-423 in NET are replaced by the larger Val-122, Leu-125, Val-393, and Cys-394 in GAT-2, making the volume previously occupied by the aromatic rings of NET ligands inaccessible in GAT-2 (Fig. 6).
DISCUSSION
The function of transporters is determined by their structure, dynamics, and localization (12, 14, 17, 20–25). For example, the shape and physicochemical properties of the transporter binding site (i.e., specificity determinants) govern the molecules that bind to the transporter (i.e., binding specificity), which may help determine the molecules that get transported (i.e., substrate specificity). The mechanism of transport describes relationships between specificity determinants to binding specificity and substrate specificity. A key step toward describing the mechanisms of transport by solute carriers includes the characterization of transporter structures in different conformational states in complex with their ligands, through computation and/or experiment.
Four key findings are presented in this study. First and importantly, distinct ligands were identified using GAT-2 models in two different conformations: the occluded and the outward facing (Fig. 3). This finding highlights the importance of characterizing the dynamics of the transport process for GAT-2 and other transporters to improve our ability to find unknown ligands with novel scaffolds. Second, by applying our combined modeling/docking approach to GAT-2, a membrane protein that shares only about 23% sequence identity with its template structure (Fig. 1 and supplemental Fig. S1), we correctly predicted key residues for function (Figs. 1 and 2 and supplemental Fig. S3) and selectivity of chemically novel GAT-2 ligands (Fig. 5 and supplemental Table S1). This suggests that our combined experimental and computational structure-based approach can be useful for identifying functionally important residues in other proteins, as well as unknown interactions between these proteins and chemically novel small molecule ligands. Third, several drugs (e.g., baclofen) and metabolites (e.g., GABOB) that target proteins other than transporters (e.g., the GABA receptor GABAB) are also ligands of GAT-2 (Figs. 4 and 5, Table 1, and supplemental Table S1). GAT-2 inhibition by these molecules might contribute to their pharmacological (i.e., efficacy and/or side effects) and physiological functions, which is an example of polypharmacology, a phenomenon in which a drug binds multiple targets (59, 60). Fourth, a comparison of the GAT-2 and NET binding sites confirms previously proposed specificity determinants of the human SLC6 family (20, 21) and identifies unknown factors important for substrate specificity for this key transporter family (Fig. 6). We now discuss each of the four points in turn.
Distinct Ligands Are Identified Using Two Different GAT-2 Conformations
An important step toward a description of the transport mechanism for the SLC6 family includes the computational or experimental characterization of their structures in various conformations. It has been suggested that a LeuT substrate needs to both bind to the S1-binding site of the outward facing conformation and fit within the binding cavity of the occluded transporter state (12). Tryptophan, which is much larger than leucine, does not fit into the cavity in the occluded state and consequently traps LeuT in the outward facing conformation, thereby acting as a competitive inhibitor (12). Whether the model approximates an active or inhibited conformation of GAT-2 is expected to determine the type of ligands that are predicted by virtual screening against the model.
Our virtual screening, chemical similarity network of the hits (Fig. 3), structural comparison of the predicted complexes (Fig. 4), and uptake kinetic experiments (Fig. 5) indicate that small molecule ligands predicted using comparative models of the two different conformations are indeed chemically different. Molecules predicted to interact with GAT-2 using the outward facing model (e.g., baclofen) are larger and more hydrophobic than those predicted to bind to the occluded model (e.g., homotaurine); however, our experiments show that the larger molecules are not stronger inhibitors (Fig. 5). This observation suggests that the differences between substrates and competitive inhibitors of GAT-2 includes more features than just size and hydrophobicity and that characterization of additional conformations of SLC6 members in complex with their ligands is needed. Importantly, although the outward facing model is less accurate than the occluded model (Table 1), it is useful for identifying chemically novel ligands (supplemental Table S1). Recent studies using experimental techniques such as single molecule FRET (15, 16) and electron paramagnetic resonance (17) in combination with MD simulations (14), revealed that different inhibitors stabilize additional LeuT conformations. Furthermore, additional crystallographic structures of LeuT (13) and other proteins with the LeuT-like fold, such as the sodium-hydantoin transporter Mhp1 (61, 62), the amino acid antiporter AdiC (63), and the sodium/galactose transporter vSGLT (64–66), have revealed additional conformations that GAT-2 might adopt during transport or inhibition. Taken together with our results, future studies should screen against GAT-2 models in additional conformations to identify new classes of GAT-2 ligands including substrates and inhibitors.
Structure-based Ligand Discovery for Membrane Transporters
In virtual screening, large libraries of organic molecules are docked computationally against experimentally determined atomic structures of target proteins. For proteins with unknown structure, comparative or homology modeling can be applied when the target sequences are detectably related to an experimentally determined protein structure. Recent advances and automation in molecular docking and comparative modeling enabled the application of structure-based approaches to ligand discovery. For example, such protocols were applied to identify prescription drugs that interact with the NET (36), as well as potent and novel inhibitors for the Dopamine receptor D3 (40). In this study, we apply comparative modeling and virtual screening to characterize a relatively unstudied transporter, GAT-2, which is distantly related to its template structure LeuT. By constructing a large number of models and selecting the final models based on their enrichment scores, we predicted key residues for transport by GAT-2 (Fig. 1 and supplemental Fig. S3). These residues were validated experimentally via site-directed mutagenesis and kinetic measurements (Fig. 2). We also identified unknown ligands, including endogenous metabolites and prescription drugs that interact with GAT-2, to further characterize its physiological and pharmacological roles (Figs. 4 and 5 and supplemental Table S1). Recent structures of homologs of varied human SLCs (66) increase our ability to discover ligands for biomedically important transporters as well as other proteins.
Physiological and Pharmacological Implications of GAT-2 Inhibitors
Of the four GABA transporters, GAT-2 is the least studied. Its localization in the liver, kidney, pancreas, retina, and lung suggests that it plays an important role in GABAergic signaling in peripheral tissues. Identifying structure-function relationships of the transporter will enhance our understanding of the functional role of the transporter. Although six of the GAT-2 ligands identified in our screen contain novel scaffolds (e.g., baclofen), most of the identified ligands were chemically similar to GABA (supplemental Table S1). Strikingly, even ligands that are within one heavy atom of GABA, such as GABOB (supplemental Table S1) and 2,4-diaminobutyric acid (4, 9), have significantly lower affinity to GAT-2 than GABA itself. Therefore, we hypothesize that a potential inhibitor needs to fulfill specific structural requirements (e.g., size and configuration of charges) to bind GAT-2, unlike inhibitors of other SLC6 monoamine transporters (2, 67–69) and other GATs (4, 8, 9).
Because of the blood-brain barrier (BBB), peripherally expressed transporters such as GAT-2 are exposed to higher systemic concentrations of xenobiotics compared with transporters within the CNS. Although speculative, it is possible that GAT-2 may therefore have evolved to be more resistant to chemical inhibition. In particular, resistance to xenobiotic inhibitors, which would otherwise result in a reduction in its function, might be important. Notably, general transporter inhibitors such as cyclosporine were not able to inhibit GAT-2 even at high concentrations, and because of specific structural requirements, it seems unlikely that GAT-2 is inhibited by many commonly used drugs at their pharmacological concentrations. Nevertheless, several pharmacological agents may still be substrates of GAT-2. In particular, the reduction of GAT-2-mediated 3H-GABA uptake by several of the above mentioned compounds (e.g., vigabatrin) might indicate that they are substrates of this transporter, which would have important toxicological and pharmacological consequences. For example, the heme precursor 5-ALA is involved in the development of the neurological symptoms of porphyria (70, 71). Medical uses of 5-ALA include the photodynamic detection of various tumors (especially in the CNS) and its use as a photosensitizer for photodynamic therapy of many diseases (11, 72, 73). Nevertheless, to date it is not fully understood how 5-ALA crosses the BBB. However, it has been shown that GABA and 5-ALA share a common facilitator in Saccharomyces cerevisiae (47). In addition, based on mRNA expression data from our laboratory (data not shown), GAT-2 is enriched at the BBB and might thus be a candidate transporter for the translocation of 5-ALA through the BBB.
Toward a Description of Substrate Specificity in the SLC6 Family
Identification of structural relationships in transporter-ligand complexes among members of the SLC6 family and their correlation with experimental ligand binding results facilitates the description of specificity determinants within this transporter family. Although GAT-2 and NET are highly related in sequence (sequence identity of ∼45%), their substrates are chemically different (small linear amino acids and monoamines for GAT-2 and NET, respectively). The differences in the physicochemical properties of the small molecule substrates of these two SLC6 members are reflected in the following key differences in their corresponding binding sites (Fig. 6): (i) the number of aromatic residues (two in GAT-2 and four in NET), (ii) the number and location of the charged groups (Glu-48 and Na1 in GAT-2 and Asp-75 in NET), and (iii) the size and shape of the binding site (i.e., Ala-145, Val-148, Gly-422, and Gly-423 in NET are replaced by the larger Val-122, Leu-125, Val-393, and Cys-394 in GAT-2). Thus, our comparison between the models of GAT-2 and NET, representatives of two groups within the SLC6 family (i.e., the GABA transporters and the monoamine transporters, respectively), in complex with their ligands highlights key features used by SLC6 members to achieve substrate specificity (Fig. 6).
In addition, our results further support the finding that even though four SLC6 members transport GABA in humans, they achieve their specificities using different mechanisms. For example, the side chain of Glu-48 in GAT-2 is predicted to make key polar interactions with GABA, whereas this position in GAT-1 is occupied by a tyrosine (Tyr-60). Interestingly, the E48Y mutation in GAT-2 (Fig. 2) and the Y60E mutation in GAT-1 (52) significantly affected the functions of these transporters. Similarly, the mutation E61Y in the mouse homolog of GAT-3 (i.e., GAT-4) resulted in a negative effect on its transport activity (54). Furthermore, despite exhibiting high sequence similarity, the transporters GAT-2, GAT-3, and the betaine/GABA transporter (BGT-1) have considerable differences in affinity to GABA-like molecules (4, 9). Particularly, the residues in close proximity to the S1-binding site in GAT-2 and GAT-3 are highly similar (the only difference is Val-132 in GAT-2, which corresponds to Ile-150 in GAT-3) (supplemental Table S2). Surprisingly, the V132I GAT-2 mutant, which mimics GAT-3, almost completely lost its transport capability (Fig. 2). Although the corresponding residue in LeuT (Ile-111) is found in the S2 binding site (11), it has not been shown that human SLC6 members contain an additional high affinity substrate-binding site (i.e., S2); however, it is plausible that Val-132 is a part of a lower affinity binding site that is coupled allosterically to S1.
In summary, through modeling GAT-2 in two different conformations, our study revealed distinct ligands of GAT-2 and suggested that GAT-2, a peripheral transporter, selectively restricts binding of inhibitors to a greater degree than other GABA transporters. Finally, our results have broad implications for the characterization of SLC6 structures. In particular, the interactions of multiple conformations of transporters with ligands are needed to describe at higher resolution the specificity and mechanisms of transport. Furthermore, our approach is generally useful for describing substrate specificities in protein families other than the SLC6 family, including other transporters, receptors, and enzymes.
Supplementary Material
Acknowledgments
We thank Sook Wah Yee and Ethan Geier for helpful discussions, as well as Ursula Pieper, Ben Webb, and Elina Tjioe for technical assistance and maintenance of the computational resources required for this study.
This work was supported, in whole or in part, by National Institutes of Health Grants R01 GM54762, U54 GM093342, and P01 GM71790 (to A. S.); U54 GM074929 and U01 GM61390 (to A. S. and K. M. G.); R25 CA112355 (to A. D.); and F32 GM088991 (to A. Sc.). This work was also supported by Swiss National Science Foundation Grant for Prospective Researchers PBBSP3–133384 (to M. B. W.). We also acknowledge funding for computing hardware from Hewlett Packard, IBM, NetApps, Intel, Ron Conway, and Mike Homer.

This article contains supplemental text, Tables S1 and S2, and Figs. S1–S6.
- SLC
- solute carrier
- GAT
- GABA transporter
- NET
- norepinephrine transporter
- KEGG
- Kyoto Encyclopedia of Genes and Genomes
- Tc
- Tanimoto coefficient
- HBSS
- Hanks' buffered saline solution
- GABOB
- γ-amino-β-hydroxybutyric acid
- 5-ALA
- 5-aminolevulinic acid
- BBB
- blood-brain barrier.
REFERENCES
- 1. Chen N. H., Reith M. E., Quick M. W. (2004) Synaptic uptake and beyond. The sodium- and chloride-dependent neurotransmitter transporter family SLC6. Pflugers Arch. 447, 519–531 [DOI] [PubMed] [Google Scholar]
- 2. Hahn M. K., Blakely R. D. (2007) The functional impact of SLC6 transporter genetic variation. Annu. Rev. Pharmacol. Toxicol. 47, 401–441 [DOI] [PubMed] [Google Scholar]
- 3. Wishart D. S., Knox C., Guo A. C., Shrivastava S., Hassanali M., Stothard P., Chang Z., Woolsey J. (2006) DrugBank. A comprehensive resource for in silico drug discovery and exploration. Nucleic Acids Res. 34, D668–672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Madsen K. K., White H. S., Schousboe A. (2010) Neuronal and non-neuronal GABA transporters as targets for antiepileptic drugs. Pharmacol. Ther. 125, 394–401 [DOI] [PubMed] [Google Scholar]
- 5. Erdö S. L., Wolff J. R. (1990) γ-Aminobutyric acid outside the mammalian brain. J. Neurochem. 54, 363–372 [DOI] [PubMed] [Google Scholar]
- 6. Xiang Y. Y., Wang S., Liu M., Hirota J. A., Li J., Ju W., Fan Y., Kelly M. M., Ye B., Orser B., O'Byrne P. M., Inman M. D., Yang X., Lu W. Y. (2007) A GABAergic system in airway epithelium is essential for mucus overproduction in asthma. Nat. Med. 13, 862–867 [DOI] [PubMed] [Google Scholar]
- 7. Soltani N., Qiu H., Aleksic M., Glinka Y., Zhao F., Liu R., Li Y., Zhang N., Chakrabarti R., Ng T., Jin T., Zhang H., Lu W. Y., Feng Z. P., Prud'homme G. J., Wang Q. (2011) GABA exerts protective and regenerative effects on islet β cells and reverses diabetes. Proc. Natl. Acad. Sci. U.S.A. 108, 11692–11697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Nakashita M., Sasaki K., Sakai N., Saito N. (1997) Effects of tricyclic and tetracyclic antidepressants on the three subtypes of GABA transporter. Neurosci. Res. 29, 87–91 [DOI] [PubMed] [Google Scholar]
- 9. Christiansen B., Meinild A. K., Jensen A. A., Braüner-Osborne H. (2007) Cloning and characterization of a functional human γ-aminobutyric acid (GABA) transporter, human GAT-2. J. Biol. Chem. 282, 19331–19341 [DOI] [PubMed] [Google Scholar]
- 10. Pacholczyk T., Blakely R. D., Amara S. G. (1991) Expression cloning of a cocaine- and antidepressant-sensitive human noradrenaline transporter. Nature 350, 350–354 [DOI] [PubMed] [Google Scholar]
- 11. Nyola A., Karpowich N. K., Zhen J., Marden J., Reith M. E., Wang D. N. (2010) Substrate and drug binding sites in LeuT. Curr. Opin. Struct. Biol. 20, 415–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Singh S. K., Piscitelli C. L., Yamashita A., Gouaux E. (2008) A competitive inhibitor traps LeuT in an open-to-out conformation. Science 322, 1655–1661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Krishnamurthy H., Gouaux E. (2012) X-ray structures of LeuT in substrate-free outward-open and apo inward-open states. Nature 481, 469–474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Shi L., Quick M., Zhao Y., Weinstein H., Javitch J. A. (2008) The mechanism of a neurotransmitter:sodium symporter. Inward release of Na+ and substrate is triggered by substrate in a second binding site. Mol. Cell 30, 667–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhao Y., Terry D., Shi L., Weinstein H., Blanchard S. C., Javitch J. A. (2010) Single-molecule dynamics of gating in a neurotransmitter transporter homologue. Nature 465, 188–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhao Y., Terry D. S., Shi L., Quick M., Weinstein H., Blanchard S. C., Javitch J. A. (2011) Substrate-modulated gating dynamics in a Na+-coupled neurotransmitter transporter homologue. Nature 474, 109–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Claxton D. P., Quick M., Shi L., de Carvalho F. D., Weinstein H., Javitch J. A., McHaourab H. S. (2010) Ion/substrate-dependent conformational dynamics of a bacterial homolog of neurotransmitter:sodium symporters. Nat. Struct. Mol. Biol. 17, 822–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fukuda H., Casas A., Batlle A. (2005) Aminolevulinic acid. From its unique biological function to its star role in photodynamic therapy. Int. J. Biochem. Cell Biol. 37, 272–276 [DOI] [PubMed] [Google Scholar]
- 19. Forrest L. R., Rudnick G. (2009) The rocking bundle. A mechanism for ion-coupled solute flux by symmetrical transporters. Physiology 24, 377–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kanner B. I., Zomot E. (2008) Sodium-coupled neurotransmitter transporters. Chem Rev 108, 1654–1668 [DOI] [PubMed] [Google Scholar]
- 21. Krishnamurthy H., Piscitelli C. L., Gouaux E. (2009) Unlocking the molecular secrets of sodium-coupled transporters. Nature 459, 347–355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Forrest L. R., Zhang Y. W., Jacobs M. T., Gesmonde J., Xie L., Honig B. H., Rudnick G. (2008) Mechanism for alternating access in neurotransmitter transporters. Proc. Natl. Acad. Sci. U.S.A. 105, 10338–10343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jardetzky O. (1966) Simple allosteric model for membrane pumps. Nature 211, 969–970 [DOI] [PubMed] [Google Scholar]
- 24. Guan L., Kaback H. R. (2006) Lessons from lactose permease. Annu. Rev. Biophys. Biomol. Struct. 35, 67–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Abramson J., Wright E. M. (2009) Structure and function of Na+-symporters with inverted repeats. Curr. Opin. Struct. Biol. 19, 425–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yamashita A., Singh S. K., Kawate T., Jin Y., Gouaux E. (2005) Crystal structure of a bacterial homologue of Na+/Cl−-dependent neurotransmitter transporters. Nature 437, 215–223 [DOI] [PubMed] [Google Scholar]
- 27. Sali A., Blundell T. L. (1993) Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 234, 779–815 [DOI] [PubMed] [Google Scholar]
- 28. Shen M. Y., Sali A. (2006) Statistical potential for assessment and prediction of protein structures. Protein Sci. 15, 2507–2524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Krivov G. G., Shapovalov M. V., Dunbrack R. L., Jr. (2009) Improved prediction of protein side-chain conformations with SCWRL4. Proteins 77, 778–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lindorff-Larsen K., Piana S., Palmo K., Maragakis P., Klepeis J. L., Dror R. O., Shaw D. E. (2010) Improved side-chain torsion potentials for the Amber ff99SB protein force field. Proteins 78, 1950–1958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Shaw D. E., Maragakis P., Lindorff-Larsen K., Piana S., Dror R. O., Eastwood M. P., Bank J. A., Jumper J. M., Salmon J. K., Shan Y., Wriggers W. (2010) Atomic-level characterization of the structural dynamics of proteins. Science 330, 341–346 [DOI] [PubMed] [Google Scholar]
- 32. Huang N., Shoichet B. K., Irwin J. J. (2006) Benchmarking sets for molecular docking. J Med Chem 49, 6789–6801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Fan H., Irwin J. J., Webb B. M., Klebe G., Shoichet B. K., Sali A. (2009) Molecular docking screens using comparative models of proteins. J. Chem. Inf. Model. 49, 2512–2527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Irwin J. J., Shoichet B. K., Mysinger M. M., Huang N., Colizzi F., Wassam P., Cao Y. (2009) Automated docking screens. A feasibility study. J. Med. Chem. 52, 5712–5720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Shoichet B. K. (2004) Virtual screening of chemical libraries. Nature 432, 862–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Schlessinger A., Geier E., Fan H., Irwin J. J., Shoichet B. K., Giacomini K. M., Sali A. (2011) Structure-based discovery of prescription drugs that interact with the norepinephrine transporter, NET. Proc. Natl. Acad. Sci. U.S.A. 108, 15810–15815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lorber D. M., Shoichet B. K. (2005) Hierarchical docking of databases of multiple ligand conformations. Curr. Top. Med. Chem. 5, 739–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Mysinger M. M., Shoichet B. K. (2010) Rapid context-dependent ligand desolvation in molecular docking. J. Chem. Inf. Model. 50, 1561–1573 [DOI] [PubMed] [Google Scholar]
- 39. Kuntz I. D. (1992) Structure-based strategies for drug design and discovery. Science 257, 1078–1082 [DOI] [PubMed] [Google Scholar]
- 40. Carlsson J., Coleman R. G., Setola V., Irwin J. J., Fan H., Schlessinger A., Sali A., Roth B. L., Shoichet B. K. (2011) Ligand discovery from a dopamine D3 receptor homology model and crystal structure. Nat. Chem. Biol. 7, 769–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ferrin T. E., Huang C. C., Jarvis L. E., Langridge R. (1988) The midas display system. J. Mol. Graphics 6, 13–27 [Google Scholar]
- 42. Kuntz I. D., Blaney J. M., Oatley S. J., Langridge R., Ferrin T. E. (1982) A geometric approach to macromolecule-ligand interactions. J. Mol. Biol. 161, 269–288 [DOI] [PubMed] [Google Scholar]
- 43. Irwin J. J., Shoichet B. K. (2005) ZINC. A free database of commercially available compounds for virtual screening. J. Chem. Inf. Model. 45, 177–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. UniProt Consortium (2008) The universal protein resource (UniProt). Nucleic Acids Res. 36, D190–D195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Okuda S., Yamada T., Hamajima M., Itoh M., Katayama T., Bork P., Goto S., Kanehisa M. (2008) KEGG Atlas mapping for global analysis of metabolic pathways. Nucleic Acids Res. 36, W423–W426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Carr R. A., Congreve M., Murray C. W., Rees D. C. (2005) Fragment-based lead discovery. Leads by design. Drug Discov. Today 10, 987–992 [DOI] [PubMed] [Google Scholar]
- 47. Bermúdez Moretti M., Correa García S. R., Chianelli M. S., Ramos E. H., Mattoon J. R., Batlle A. (1995) Evidence that 4-aminobutyric acid and 5-aminolevulinic acid share a common transport system into Saccharomyces cerevisiae. Int. J. Biochem. Cell Biol. 27, 169–173 [DOI] [PubMed] [Google Scholar]
- 48. Overington J. (2009) ChEMBL. An interview with John Overington, team leader, chemogenomics at the European Bioinformatics Institute Outstation of the European Molecular Biology Laboratory (EMBL-EBI). Interview by Wendy A. Warr. J. Comput. Aided Mol. Des. 23, 195–198 [DOI] [PubMed] [Google Scholar]
- 49. Shannon P., Markiel A., Ozier O., Baliga N. S., Wang J. T., Ramage D., Amin N., Schwikowski B., Ideker T. (2003) Cytoscape. A software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Steinbeck C., Hoppe C., Kuhn S., Floris M., Guha R., Willighagen E. L. (2006) Recent developments of the chemistry development kit (CDK). An open-source Java library for chemo- and bioinformatics. Curr. Pharm. Des. 12, 2111–2120 [DOI] [PubMed] [Google Scholar]
- 51. Enright A. J., Van Dongen S., Ouzounis C. A. (2002) An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res. 30, 1575–1584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kanner B. I. (2003) Transmembrane domain I of the γ-aminobutyric acid transporter GAT-1 plays a crucial role in the transition between cation leak and transport modes. J. Biol. Chem. 278, 3705–3712 [DOI] [PubMed] [Google Scholar]
- 53. Beuming T., Shi L., Javitch J. A., Weinstein H. (2006) A comprehensive structure-based alignment of prokaryotic and eukaryotic neurotransmitter/Na+ symporters (NSS) aids in the use of the LeuT structure to probe NSS structure and function. Mol. Pharmacol. 70, 1630–1642 [DOI] [PubMed] [Google Scholar]
- 54. Melamed N., Kanner B. I. (2004) Transmembrane domains I and II of the γ-aminobutyric acid transporter GAT-4 contain molecular determinants of substrate specificity. Mol. Pharmacol. 65, 1452–1461 [DOI] [PubMed] [Google Scholar]
- 55. Carter L. P., Koek W., France C. P. (2009) Behavioral analyses of GHB. Receptor mechanisms. Pharmacol. Ther. 121, 100–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Garbutt J. C., Kampov-Polevoy A. B., Gallop R., Kalka-Juhl L., Flannery B. A. (2010) Efficacy and safety of baclofen for alcohol dependence. A randomized, double-blind, placebo-controlled trial. Alcohol Clin. Exp. Res. 34, 1849–1857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Celik L., Sinning S., Severinsen K., Hansen C. G., Møller M. S., Bols M., Wiborg O., Schiøtt B. (2008) Binding of serotonin to the human serotonin transporter. Molecular modeling and experimental validation. J. Am. Chem. Soc. 130, 3853–3865 [DOI] [PubMed] [Google Scholar]
- 58. Beuming T., Kniazeff J., Bergmann M. L., Shi L., Gracia L., Raniszewska K., Newman A. H., Javitch J. A., Weinstein H., Gether U., Loland C. J. (2008) The binding sites for cocaine and dopamine in the dopamine transporter overlap. Nat. Neurosci. 11, 780–789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Keiser M. J., Setola V., Irwin J. J., Laggner C., Abbas A. I., Hufeisen S. J., Jensen N. H., Kuijer M. B., Matos R. C., Tran T. B., Whaley R., Glennon R. A., Hert J., Thomas K. L., Edwards D. D., Shoichet B. K., Roth B. L. (2009) Predicting new molecular targets for known drugs. Nature 462, 175–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Roth B. L., Sheffler D. J., Kroeze W. K. (2004) Magic shotguns versus magic bullets. Selectively non-selective drugs for mood disorders and schizophrenia. Nat. Rev. Drug Discov. 3, 353–359 [DOI] [PubMed] [Google Scholar]
- 61. Weyand S., Shimamura T., Yajima S., Suzuki S., Mirza O., Krusong K., Carpenter E. P., Rutherford N. G., Hadden J. M., O'Reilly J., Ma P., Saidijam M., Patching S. G., Hope R. J., Norbertczak H. T., Roach P. C., Iwata S., Henderson P. J., Cameron A. D. (2008) Structure and molecular mechanism of a nucleobase-cation-symport-1 family transporter. Science 322, 709–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Shimamura T., Weyand S., Beckstein O., Rutherford N. G., Hadden J. M., Sharples D., Sansom M. S., Iwata S., Henderson P. J., Cameron A. D. (2010) Molecular basis of alternating access membrane transport by the sodium-hydantoin transporter Mhp1. Science 328, 470–473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Gao X., Zhou L., Jiao X., Lu F., Yan C., Zeng X., Wang J., Shi Y. (2010) Mechanism of substrate recognition and transport by an amino acid antiporter. Nature 463, 828–832 [DOI] [PubMed] [Google Scholar]
- 64. Faham S., Watanabe A., Besserer G. M., Cascio D., Specht A., Hirayama B. A., Wright E. M., Abramson J. (2008) The crystal structure of a sodium galactose transporter reveals mechanistic insights into Na+/sugar symport. Science 321, 810–814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Watanabe A., Choe S., Chaptal V., Rosenberg J. M., Wright E. M., Grabe M., Abramson J. (2010) The mechanism of sodium and substrate release from the binding pocket of vSGLT. Nature 468, 988–991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Forrest L. R., Krämer R., Ziegler C. (2011) The structural basis of secondary active transport mechanisms. Biochim. Biophys. Acta 1807, 167–188 [DOI] [PubMed] [Google Scholar]
- 67. Bednarczyk D. (2010) Fluorescence-based assays for the assessment of drug interaction with the human transporters OATP1B1 and OATP1B3. Anal. Biochem. 405, 50–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Andersen J., Kristensen A. S., Bang-Andersen B., Stromgaard K. (2009) Recent advances in the understanding of the interaction of antidepressant drugs with serotonin and norepinephrine transporters. Chem. Commun. (Camb.) 3677–3692 [DOI] [PubMed] [Google Scholar]
- 69. Kristensen A. S., Andersen J., Jørgensen T. N., Sørensen L., Eriksen J., Loland C. J., Strømgaard K., Gether U. (2011) SLC6 neurotransmitter transporters. Structure, function, and regulation. Pharmacol. Rev. 63, 585–640 [DOI] [PubMed] [Google Scholar]
- 70. Albers J. W., Fink J. K. (2004) Porphyric neuropathy. Muscle Nerve 30, 410–422 [DOI] [PubMed] [Google Scholar]
- 71. Lindberg R. L., Martini R., Baumgartner M., Erne B., Borg J., Zielasek J., Ricker K., Steck A., Toyka K. V., Meyer U. A. (1999) Motor neuropathy in porphobilinogen deaminase-deficient mice imitates the peripheral neuropathy of human acute porphyria. J. Clin. Invest. 103, 1127–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Musiol R., Serda M., Polanski J. (2011) Prodrugs in photodynamic anticancer therapy. Curr. Pharm. Des. 17, 3548–3559 [DOI] [PubMed] [Google Scholar]
- 73. Stummer W., Pichlmeier U., Meinel T., Wiestler O. D., Zanella F., Reulen H. J. (2006) Fluorescence-guided surgery with 5-aminolevulinic acid for resection of malignant glioma. A randomised controlled multicentre phase III trial. Lancet Oncol. 7, 392–401 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.