Background: The eIF2α kinase Gcn2 aids cells to overcome amino acid starvation. For sensing starvation, Gcn2 must bind Gcn1, and both must contact the ribosome.
Results: Overexpression of translation elongation factor eEF3 inhibits Gcn1-mediated Gcn2 activation.
Conclusion: eEF3 keeps Gcn2 in its latent state under nutrient replete conditions.
Significance: We discovered a novel unanticipated role of a general translation factor in controlling Gcn2.
Keywords: Serine-Threonine Protein Kinase, Signaling, Stress Response, Translation Elongation Factors, Yeast, Gcn1, Gcn2, Amino Acid Starvation, eIF2alpha Kinase
Abstract
In eukaryotes, phosphorylation of translation initiation factor 2α (eIF2α) by the kinase Gcn2 (general control nonderepressible 2) is a key response to amino acid starvation. Sensing starvation requires that Gcn2 directly contacts its effector protein Gcn1, and both must contact the ribosome. We have proposed that Gcn2 is activated by uncharged tRNA bound to the ribosomal decoding (A) site, in a manner facilitated by ribosome-bound Gcn1. Protein synthesis requires cyclical association of eukaryotic elongation factors (eEFs) with the ribosome. Gcn1 and Gcn2 are large proteins, raising the question of whether translation and monitoring amino acid availability can occur on the same ribosome. Part of the ribosome-binding domain in Gcn1 has homology to one of the ribosome-binding domains in eEF3, suggesting that these proteins utilize overlapping binding sites on the ribosome and consequently cannot function simultaneously on the same ribosome. Supporting this idea, we found that eEF3 overexpression in Saccharomyces cerevisiae diminished growth on amino acid starvation medium (Gcn− phenotype) and decreased eIF2α phosphorylation, and that the growth defect associated with constitutively active Gcn2 was diminished by eEF3 overexpression. Overexpression of the eEF3 HEAT domain, or C terminus, was sufficient to confer a Gcn− phenotype, and both fragments have ribosome affinity. eEF3 overexpression did not significantly affect Gcn1-ribosome association, but it exacerbated the Gcn− phenotype of Gcn1-M7A that has reduced ribosome affinity. Together, this suggests that eEF3 blocks Gcn1 regulatory function on the ribosome. We propose that the Gcn1-Gcn2 complex only functions on ribosomes with A-site-bound uncharged tRNA, because eEF3 does not occupy these stalled complexes.
Introduction
Protein synthesis requires specific soluble factors that cycle on and off the ribosome in an orderly fashion. Eukaryotic translation elongation factor 1A (eEF1A)4 delivers amino acyl tRNA (aa-tRNA) to the ribosomal acceptor site (A-site) in a codon-specific manner (1). Elongation factor 2 (eEF2) is essential for translocating the ribosome along the mRNA by one triplet codon. The delivery of aa-tRNA to the A-site is coupled with the release of deacylated tRNA (tRNAdeacyl) from the E-site (1). In yeast it has been shown that this coupled mechanism is triggered by a soluble ATPase called elongation factor 3 (eEF3) (2–4). For nonfungal eukaryotes the factor equivalent to eEF3 has not been found yet. However, the high conservation of the translational process implies that nonfungal organisms must have a factor with a function equivalent to that of eEF3. Supporting this idea, it was found that metazoan ribosomes have an intrinsic ATPase activity that is absent in yeast ribosomes (5), suggesting that in metazoans a ribosome bound factor executes the eEF3-like function rather than a soluble factor. Interestingly, in Escherichia coli the protein RbbA (ribosome-bound ATPase) was identified that exhibits ATPase activity and is tightly associated with ribosomes (6, 7) and thus may have an eEF3-like function.
eEF3 consists of several domains. The N-terminal domain contains repeats that were first found in the Huntingtin protein, eEF3, protein phosphatase 2A, and TOR and were therefore called HEAT repeats (1). HEAT repeats are predicted to be interaction surfaces for other proteins and/or nucleic acids (8), and in fact an eEF3 fragment encompassing this domain was reported to bind 18 S rRNA, and structural analyses of ribosome bound eEF3 revealed that the eEF3 HEAT repeats interact with rRNA and ribosomal proteins of the small ribosomal subunit (9, 10). Adjacent to the HEAT repeat domain is a four-helix bundle of unknown function. The eEF3 C terminus contains two ATP-binding cassettes (ABC) (1). Unique to eEF3 is the insertion of a chromodomain within the C-terminal ABC cassette II (10). The chromodomain, as well as the ABCII portion N-terminal to this domain, contacts the small and large ribosomal subunit (10).
Constant and efficient protein synthesis is essential to life; hence a constant supply of amino acids must be maintained in the cell. For a timely response to amino acid shortages, cells need to constantly monitor amino acid availability, and studies in yeast suggested that this sensing occurs on the ribosome by the protein kinase Gcn2 (general control nonderepressible 2) and its effector protein Gcn1 (11–13). Gcn2 and Gcn1 are present in all eukaryotes, suggesting that they are universally essential for sensing and overcoming amino acid starvation. Upon sensing amino acid shortage, the Gcn2 kinase domain becomes activated, which subsequently phosphorylates the α subunit of translation initiation factor 2 (eIF2α) (14). eIF2 in its GTP-bound form is essential for initiating protein synthesis in that it delivers initiator methionyl tRNA (Met-tRNAiMet) to the ribosome. After completing translation initiation, eIF2 is released in its GDP-bound form, and it needs to be recycled to its GTP bound form by its guanine nucleotide exchange factor eIF2B. Phosphorylation of eIF2α by Gcn2 converts eIF2 to a competitive inhibitor of its own guanine nucleotide exchange factor, leading to reduced global protein synthesis and thus to reduced overall consumption of amino acids. Simultaneously, eIF2α phosphorylation evokes increased translation of specific genes containing unique upstream open reading frames in their mRNA. These genes encode for transcriptional activators, e.g., Gcn4 in yeast and ATF4 in mammals, that up-regulate the expression of many genes including those coding for key amino acid biosynthetic enzymes (14). Thus, increased Gcn4 or ATF4 protein levels lead to increased amino acid biosynthesis, thereby relieving cells from amino acid starvation or imbalance. Interestingly, starvation for any amino acid activates Gcn2 and stimulates the synthesis of all 20 amino acids; therefore the signal transduction pathway governing Gcn2 was called general amino acid control (GAAC) (14).
There are important gaps in our knowledge about the molecular mechanisms underlying the perception of the amino acid starvation signal. Amino acid starvation leads to increased cellular levels of tRNAdeacyl, which is the direct signal for amino acid starvation (14, 15). Yeast studies suggest that tRNAdeacyl is detected by the histidyl tRNA synthetase-like domain in Gcn2, which leads to the activation of its adjacent kinase domain and to subsequent phosphorylation of its substrate, eIF2α (14). The effector protein Gcn1 is essential for Gcn2 function in vivo. A gcn1Δ strain is unable to activate Gcn2 upon amino acid starvation (Gcn− phenotype); however, Gcn2 kinase activity can be detected in the whole cell extract of a gcn1Δ strain, suggesting that Gcn1 is not required for the kinase activity per se but for transferring the starvation signal to Gcn2 (14). We and others showed that for signal perception Gcn2 must directly bind to its effector protein Gcn1 and that both proteins must associate with translating ribosomes (13, 14). The binding domains in the Gcn1 and Gcn2 proteins required for Gcn1-Gcn2 interaction are physically distinct from the ribosome-binding domains in either protein, supporting the idea that Gcn1 and Gcn2 form a trimeric complex with the ribosome and that the starvation signal is detected on the ribosome.
The Gcn1 middle portion binds the N terminus of Gcn20, a protein required for full Gcn2 activation (16). Gcn20 does not bind to ribosomes, but studies suggest that it may affect or regulate Gcn1-ribosome association and thus the GAAC system; however, the mechanism remains elusive (17). Interestingly, the Gcn1 middle portion has HEAT repeats and has homology to the N-terminal HEAT repeat domain of eEF3, and the Gcn20 C terminus has homology to the eEF3 C terminus including the ABC cassettes (17). This suggests that the Gcn1/20 complex has a similar function as eEF3, just that it is involved in delivering or releasing tRNAdeacyl from the ribosomal A-site instead of the E-site (17). This idea is also based on findings in prokaryotes where under amino acid starvation tRNAdeacyl binds in the A-site in a codon-specific manner (18, 19). Interestingly, it was shown that also in eukaryotes tRNAdeacyl can enter the A-site in a codon-specific manner (20), suggesting that the mechanism of the starvation signal occurring in the A-site may be conserved from prokaryotes to eukaryotes. Although in prokaryotes tRNAdeacyl binding to the A-site then leads to the activation of the ppGpp synthetase RelA and the stringent response (18, 19), in eukaryotes this leads to Gcn1-dependent Gcn2 stimulation and the activation of the GAAC. We have proposed a model for Gcn1-mediated Gcn2 activation in which under amino acid starvation tRNAdeacyl enter the ribosomal A-site in a codon-specific manner. Gcn1 is directly involved in the transfer of the starvation signal to Gcn2, by one or more of the following mechanisms: (a) delivery of tRNAdeacyl to the ribosomal A-site, (b) transfer of tRNAdeacyl from the A-site to Gcn2, and/or (c) being a scaffold protein that positions Gcn2 and potential additional factors in such a way that Gcn2 can detect the starvation signal in the A-site (12, 17). Several lines of evidence support our idea that Gcn1 can access the A-site, directly or indirectly. We found that overexpression of Gcn1 leads to hypersensitivity to paromomycin, a drug binding in the A-site. Conversely, GCN1 deletion increased paromomycin resistance. Furthermore, internal deletions in Gcn1 affecting ribosome binding simultaneously increased paromomycin resistance, and so did deletion of GCN20 (12).
Interestingly, we have recently discovered that eEF1A directly binds to the Gcn2 C terminus (21). Under amino acid starvation, this interaction is lost in vivo, tRNAdeacyl prevents eEF1A-Gcn2 interaction in vitro, and eEF1A prevents eIF2α phosphorylation in vitro but not Gcn2 autophosphorylation. Together, this strongly suggested that eEF1A prevents full Gcn2 activation under replete conditions and that under amino acid starvation uncharged tRNAs channeled to Gcn2 remove eEF1A from Gcn2 and thereby allow Gcn2 activation and subsequent eIF2α phosphorylation (21).
Structural studies of the eEF3-ribosome complex revealed that eEF3 binds close to the E-site (10); however, considering that Gcn1 is a very large protein, that the N-terminal 2052 amino acids in Gcn1 are involved in ribosome binding, and that it has homology to eEF3, it is likely that on the ribosome some of the eEF3-binding sites overlap with a subset of those for Gcn1. If this is true, then eEF3 and Gcn1 cannot function on the same ribosome. Here we present several lines of evidence supporting this prediction. We found that overexpression of eEF3 impairs Gcn1-mediated Gcn2 activation. The eEF3 HEAT domain was sufficient for impairing Gcn1 function when overexpressed. Remarkably, we found that overexpressing the eEF3 C terminus (amino acids 910–1044) also was sufficient for impairing Gcn1 function and that this eEF3 segment also can bind to elongating ribosomes in vivo. Interestingly, eEF3 overexpression did not significantly affect Gcn1-ribosome or Gcn2-ribosome association; however, eEF3 overexpression exacerbated the Gcn− phenotype associated with Gcn1-M7A, a Gcn1 mutant protein with reduced ribosome affinity. Together this supports the idea that eEF3 overexpression does not simply remove Gcn1 from the ribosome but instead affects Gcn1 function on the ribosome. We propose that eEF3 blocks a productive Gcn1-ribosome association, supporting the idea that Gcn1-Gcn2 detects the amino acid starvation signal on stalled elongating ribosomes lacking eEF3. Under replete conditions eEF3 may work in concert with eEF1A to prevent Gcn2 activation
EXPERIMENTAL PROCEDURES
Yeast Strains and Plasmids
Yeast strains and plasmids used in this study are summarized in Tables 1 and 2, respectively. The vectors used were pRS316 (22), pEG(KT) (22), and pES128-9 (12).
TABLE 1.
Strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| Genetic background: H1511 | ||
| H1511 | MATα ura3-52 trp1-63 leu2-3,112 GAL2+ | Ref. 36 |
| H2556 | As H1511, and gcn1Δ | Vazquez de Aldana and Hinnebusch |
| Genetic background: H1402 | ||
| H1402 | MATα inol ura3-52 leu2-3,112 (HIS4-lacZ ura3-52) | Ref. 37 |
| H1613 | As H1402, and GCN2c-E601K-E1591K | Ref. 38 |
| Genetic background: H1515 | ||
| H1515 | MATa ura3-52 leu2-3 leu2-112 trp1-Δ63 | Cigan and Hinnebusch |
| H2544 | As H1515, and (GAL-CYC1-PKR LEU2) at leu2 | Romano and Hinnebusch |
| H1894 | As H1515, and gcn2Δ | Ref. 39 |
TABLE 2.
Plasmids used in this study
YEF3 codes for yeast eEF3, and E3L codes for vaccinia virus protein E3.
| Plasmid | Gene | Selectable marker (all AmpR) | Vector | Source |
|---|---|---|---|---|
| Yeast gene fusions, under GAL1-CYC1 promoter | ||||
| pGST-YEF3 | GST-YEF3a,b | URA3, leu2-d, 2 μ | pEG(KT) | Ref. 40 |
| pTKB705 | GST-YEF3b | URA3, leu2-d, 2 μ | pTKB544 | Ref. 27 |
| pTKB706 | GST-YEF3[1–775]b,c | URA3, leu2-d, 2 μ | pTKB544 | Ref. 27 |
| pTKB707 | GST-YEF3[100–367]b,c | URA3, leu2-d, 2 μ | pTKB544 | Ref. 27 |
| pTKB708 | GST-YEF3[775–910]b,c | URA3, leu2-d, 2 μ | pTKB544 | Ref. 27 |
| pTKB709 | GST-YEF3[910–1044]b,c | URA3, leu2-d, 2 μ | pTKB544 | Ref. 27 |
| pTKB710 | GST-YEF3[775–1044]b,c | URA3, leu2-d, 2 μ | pTKB544 | Ref. 27 |
| pES340-EF3-2-8 | GST-YEF3a,b | URA3, 2 μ | pES128-9 | This study |
| p2245 | HA-E3Lb | URA3, leu2-d, 2 μ | pEG(KT) | Ref. 28 |
| Yeast genes, under their own promoter | ||||
| p1832 | GCN1-mycd | LEU2, CEN6/ARSH4 | pRS316 | Ref. 16 |
| pES239-4-4a | gcn1-M7A-mycd | LEU2, CEN6/ARSH4 | pRS316 | This study |
a This construct lacks the first 10 N-terminal amino acids of eEF3.
b Epitope tag at the N terminus of the ORF.
c The numbers in brackets indicate amino acids encoded by the respective gene construct.
d Epitope tag at the C terminus of the ORF.
Semiquantitative Growth Assays
Yeast strains were grown to saturation in 4 ml of liquid medium, subjected to serial 10-fold dilutions, and then 5 μl of each dilution was transferred to solid medium containing substances to cover auxotrophies. If necessary, galactose was used as sole carbon source instead of glucose to induce the expression of genes under the galactose-inducible promoter. Amino acid analogues 3-amino-2,4-triazole (3AT) and sulfometuron methyl (SM) were added to autoclaved medium to trigger starvation for histidine or branched chain amino acids, respectively. The plates were incubated at 30 °C until colonies were visible.
Ribosome Co-sedimentation Assays
In vivo association of proteins with polyribosomes was determined as described previously (13). Briefly, yeast cells were grown to exponential phase in 300 ml of liquid medium to an A600 of ∼1 in 1-liter baffled flasks. The cultures were poured into a 400-ml centrifuge bottle containing 75 g of ice chips and 8.1 ml of formaldehyde (final concentration, 1%). The cells were then mixed and incubated on ice for 1 h with gentle mixing every 15 min. The cells were pelleted for 5 min at 4200 rpm (model J6 centrifuge; Beckman), washed with 5 ml of breaking buffer (20 mm Tris/HCl, pH 7.5, 50 mm KCl, 10 mm MgCl2, 1 mm dithiothreitol, 1 mm phenylmethylsulfonyl fluoride, 5 mm NaF, and 0.1 mg/ml each of pepstatin, aprotinin, and leupeptin), and resuspended in 200 μl of breaking buffer. Highly concentrated whole cell extract was generated as described in detail elsewhere (23), and 30 A260 units of extract were resolved on a 4.5–45% sucrose density gradient by centrifugation for 2 h at 39,000 rpm at 4 °C using the TH641 rotor (Sorvall). 1-ml fractions were collected while scanning continuously for A254 units using the Bio-Rad BioLogic fractionation system. If required, proteins were precipitated by adding 1 ml of isopropanol to each fraction, and the samples were then kept at −80 °C overnight; then the proteins were pelleted at 12,000 rpm for 30 min at 4 °C, the supernatant was removed, the pellet was dried under vacuum for 10–15 min, and finally the protein pellets were resuspended in 2× Laemmli loading dye.
Assaying for eIF2α Phosphorylation
Exponentially growing cells were subjected to amino acid starvation by adding either 3AT or SM and then subjected to formaldehyde cross-linking as done for polyribosome co-sedimentation assays. Whole cell extracts were generated as published elsewhere (23), and equal amounts of cell extract (5 μg) were subjected to SDS-PAGE and immunoblotting as described below.
Protein Techniques
Proteins were separated by SDS-PAGE using 4–12% gradient gels and transferred to PVDF membranes (Millipore) according to the manufacturer's protocol. Proteins were detected by the chemiluminescence detection system (Pierce) using antibodies against Gcn1 (HL1405, 1:1000 (16)), Gcn2 (1:1000 (21)), eIF2α (1:2000 (24)), eIF2α phosphorylated on Ser-51 (1:5000; Bio-Source International, Inc.), Pgk1 (1:5000; Life Technologies), RPL39 (1:5000 (25)), RPS22 (1:2000; from Dr. Jan van't Riet), or GST (1:2000; Santa Cruz). Immune complexes were visualized using horseradish peroxidase conjugated to donkey anti-rabbit antibodies (for the detection of Gcn1, GST, eIF2α, eIF2α-P, and RPS22 antibodies), goat anti-guinea pig (Gcn2) (Santa Cruz), or to sheep anti-mouse antibodies (Pgk1 and RPL39) (Pierce).
RESULTS
eEF3 Overexpression Elicits Sensitivity to 3-Amino-2,4-triazole (3ATs)
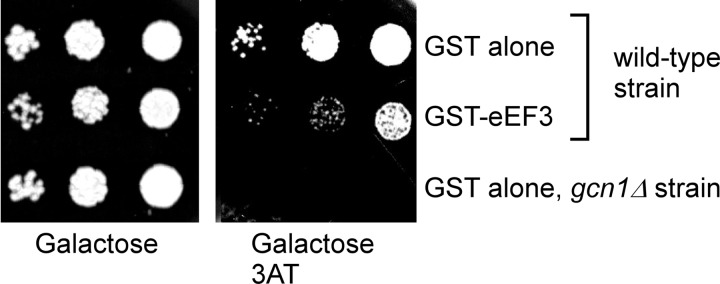
If Gcn1 binds to the same or overlapping regions of the ribosome as eEF3, then eEF3 overexpression should affect Gcn1 function and consequently Gcn2 activation. Strains with impaired Gcn2 activity can easily be scored by their inability to grow on medium containing 3AT, a drug causing histidine starvation by inhibiting the histidine biosynthetic enzyme encoded by HIS3 (26). To test our prediction that eEF3 affects Gcn1 function and thus impairs Gcn2 activation, we introduced into the yeast wild-type strain H1511 a plasmid expressing GST-tagged eEF3 from a galactose-inducible promoter or a plasmid harboring GST alone. The resulting transformants were subjected to semiquantitative growth assays using solid medium containing galactose as carbon source, and 3AT or no 3AT as control. As reported before (27), under replete conditions eEF3 overexpression did not lead to a significant growth defect (Fig. 1, left panel), indicating that eEF3 overexpression does not severely affect general protein synthesis or other essential processes in the cell. Via Western blot analysis, we verified that GST-eEF3 was overexpressed in the respective strains (data not shown). Interestingly, we found that eEF3 overexpression led to reduced growth in presence of 3AT (3AT sensitivity, 3ATs) as compared with the strain overexpressing GST alone (Fig. 1, compare first and second rows), suggesting that GAAC response is impaired by eEF3 overexpression.
FIGURE 1.
eEF3 overexpression causes sensitivity to the amino acid analogue 3AT. Saturated overnight cultures of wild-type strain H1511 harboring a plasmid for the galactose-inducible overexpression of GST tagged eEF3 (GST-eEF3) or GST alone (plasmids pGST-YEF3 and pEG(KT), respectively) or the isogenic gcn1Δ strain H2556 overexpressing GST alone (pEG(KT)) were subjected to 10-fold serial dilutions. 5 μl of each dilution was transferred to solid medium containing galactose as carbon source, and 120 mm 3AT if indicated, and incubated at 30 °C for 3–5 days.
The Growth Defect Associated with Gcn2c Is Partially Suppressed by eEF3 Overexpression
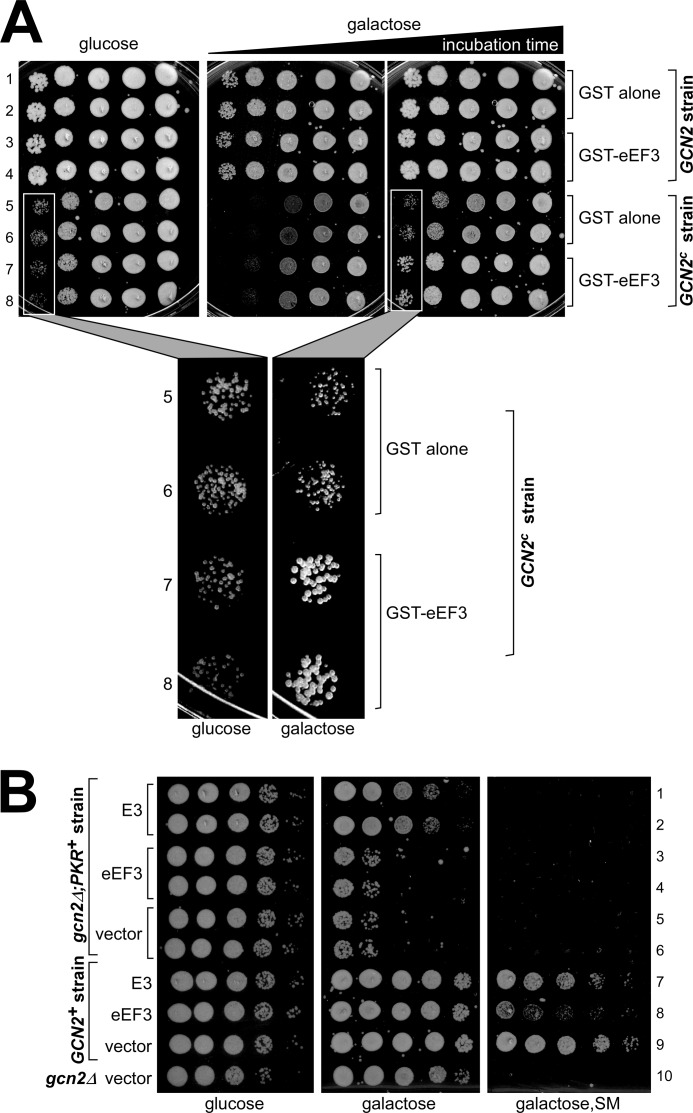
Having found that eEF3 overexpression leads to 3ATs, we wished to find additional genetic evidence that does not involve the usage of drugs that supports our idea that eEF3 overexpression impairs Gcn2 activation. Constitutively active Gcn2 (e.g., Gcn2c-E601K-E1591K, here abbreviated as Gcn2c) leads to increased levels of phosphorylated eIF2α (eIF2α-P) and to a general reduction in protein synthesis, and consequently to a slow growth (Slg−) phenotype (14). Gcn2c still requires Gcn1 for its activity, presumably because it still needs to receive tRNAdeacyl to become constitutively active. If overexpressed eEF3 affects Gcn1 function, then it should simultaneously diminish the constitutive kinase activity of Gcn2c, visible by a reversion of the Slg− phenotype conferred by Gcn2c. To test this, we introduced a plasmid expressing GST-tagged eEF3 under a galactose-inducible promoter or a plasmid harboring GST alone into isogenic strains harboring either the GCN2c allele or the GCN2 wild-type gene on its chromosome, and the resulting transformants were subjected to semiquantitive growth assays. As expected, on medium-containing glucose, i.e., no eEF3 overexpression, the strain harboring the GCN2c allele showed reduced growth compared with the wild-type control (Fig. 2A, left panel, rows 1–4 versus rows 5–8). On galactose, however, the GCN2c strain overexpressing eEF3 showed improved growth as compared with the GCN2c strain overexpressing GST alone (Fig. 2A, rows 7 and 8 versus rows 5 and 6), supporting the idea that GAAC response is impaired by eEF3 overexpression.
FIGURE 2.
eEF3 overexpression partially reverts the growth defect associated with constitutively active Gcn2. A, 2 ml of liquid medium was inoculated with one (GCN2 strain H1613) or four (GCN2c strain harboring allele GCN2c-E601K-E1591K) (to compensate for its slower growth rate) fresh transformants harboring plasmid borne GST alone or GST-eEF3 under a galactose-inducible promotor (pGST-YEF3 or pEG(KT), respectively). After 24 h of incubation at 30 °C, the saturated culture was subjected to 10-fold serial dilutions. 5 μl of each dilution was transferred to solid medium containing glucose or galactose as a carbon source and incubated at 30 °C until colonies were visible. B, eEF3 overexpression does not mitigate the growth defect associated with PKR expression. gcn2Δ strain H2544 harboring chromosomally integrated and galactose-inducible PKR and isogenic wild-type and gcn2Δ strains (H1515 and H1894) were transformed with high copy plasmid borne and galactose-inducible genes coding for GST-eEF3, GST alone, or E3, as indicated (pES340-EF3-2-8, pES128-9, and p2245). The resulting transformants were subjected to semiquantitative growth assays as outlined in Fig. 1.
Similarly to Gcn2c, expression of the mammalian double-stranded RNA-dependent eIF2α kinase PKR in yeast leads to a severe growth defect caused by high levels of eIF2α phosphorylation (28). In contrast to Gcn2, PKR does not require Gcn1 for function. If eEF3 impairs GAAC function by inhibiting Gcn1, and not by, for example, stimulating an eIF2α-P phosphatase or preventing eIF2α kinase domains from accessing their substrate, then, in contrast to Gcn2c, the Slg− phenotype elicited by PKR should not be diminished by eEF3 overexpression. To test this, we repeated the semiquantitative growth assay using a gcn2Δ strain overexpressing PKR from a chromosomally integrated PKR construct driven by a galactose-inducible promotor and also harboring a high copy plasmid containing a galactose-inducible gene coding for GST-eEF3, GST alone, or the PKR inhibitor from vaccinia virus, E3 (28). We found that, in contrast to E3, GST-eEF3 overexpression did not mitigate the growth defect elicited by PKR (Fig. 2B, galactose, rows 1 and 2 versus rows 3 and 4 versus rows 5 and 6). The fact that E3 did reduce the Slg− phenotype elicited by PKR indicated that a PKR inhibitor can successfully inhibit PKR activity even when its expression was induced at the same time as that of PKR. We verified that eEF3 overexpression inhibits GAAC in this strain background (Fig. 2B, galactose, SM, row 8 versus row 9). Taken together, our results support the idea that overexpressed eEF3 impairs Gcn1 function in mediating Gcn2 activation.
Overexpressed eEF3 Reduces eIF2α Phosphorylation Levels
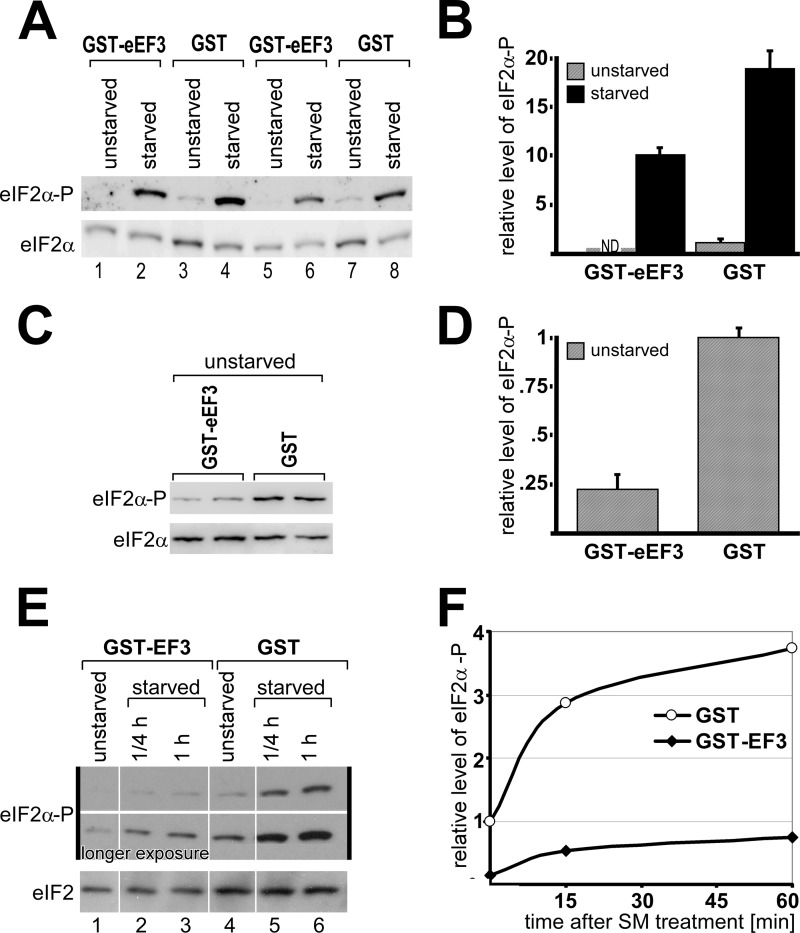
If it is true that eEF3 overexpression prevents Gcn2 activation and GAAC response, then this should be associated with reduced phosphorylation levels of eIF2α, the substrate of Gcn2. To test this, wild-type strain H1511 harboring a plasmid expressing GST-eEF3, or GST alone, from a galactose-inducible promotor, were grown to exponential phase in medium containing galactose as a carbon source. The cells were starved for histidine by adding 3AT and then were cross-linked with formaldehyde to prevent phosphorylation or de-phosphorylation events that may occur during the harvesting or processing of samples. Whole cell extracts were subjected to SDS-PAGE and immunoblotting analysis to determine the relative amount of eIF2α phosphorylation (Fig. 3). As expected, amino acid starvation led to increased eIF2α-P levels (Fig. 3, A, lanes 3 and 7 versus lanes 4 and 8, and B); however, when eEF3 was overexpressed in starved cells, the amount of eIF2α-P was reduced by ∼50% (Fig. 3, A, lanes 2 and 6 versus lanes 4 and 8, and B). Interestingly, we found that even under nutrient-replete conditions, eEF3 overexpression reduced eIF2α-P levels, by a factor of 4.5, to 22% of that of the wild-type control overexpressing GST alone (Fig. 3, C and D).
FIGURE 3.
eEF3 overexpression leads to reduced eIF2α phosphorylation. Yeast strains from Fig. 1 were grown to exponential phase and subjected to amino acid starvation by adding 3AT to a final concentration of 10 mm 20 min before harvesting (A and C) or by adding SM to a final concentration of 1.25 μm 15 min or 1 h before harvesting (E). As control, no 3AT or SM was added (unstarved). Whole cell extract was generated and subjected to SDS-PAGE and immunoblotting using antibodies against eIF2α and the phosphorylated form of eIF2α (eIF2α-P), respectively. The amount of eIF2α and eIF2α-P was quantified using the program ImageJ and plotted for 3AT starvation (B and D) and SM starvation (F), respectively, relative to the total amount of eIF2α and relative to the eIF2α-P/eIF2α ratio of the strain expressing GST under replete conditions.
Next we repeated the above experiment but instead of 3AT we used SM, a drug causing starvation for branched chain amino acids by inhibiting acetolactate synthase, the first common enzyme in the branched chain amino acid biosynthetic pathway (29). As expected, the addition of SM led to increased eIF2α-P levels (Fig. 3, E, lane 4 versus lanes 5 and 6, and F). As found above, under starvation conditions eEF3 overexpression significantly reduced the amount of eIF2α-P levels by a factor of about 5 (Fig. 3, E, lanes 2 and 3 versus lanes 5 and 6, and F). We again found that eEF3 overexpression reduced eIF2α-P levels even under replete conditions, by a factor of 6.7, to 15% of that of the wild-type control overexpressing GST alone (Fig. 3, E, lane 1 versus lane 4, and F). Taken together, our results strongly suggest that eEF3 overexpression inhibits Gcn2 activation in response to amino acid limitation but also under replete conditions. Thus, overexpressed eEF3 impairs the GAAC pathway (Gcn− phenotype, general control nonderepressible).
eEF3 Overexpression Does Not Simply Remove Gcn1 from the Ribosome
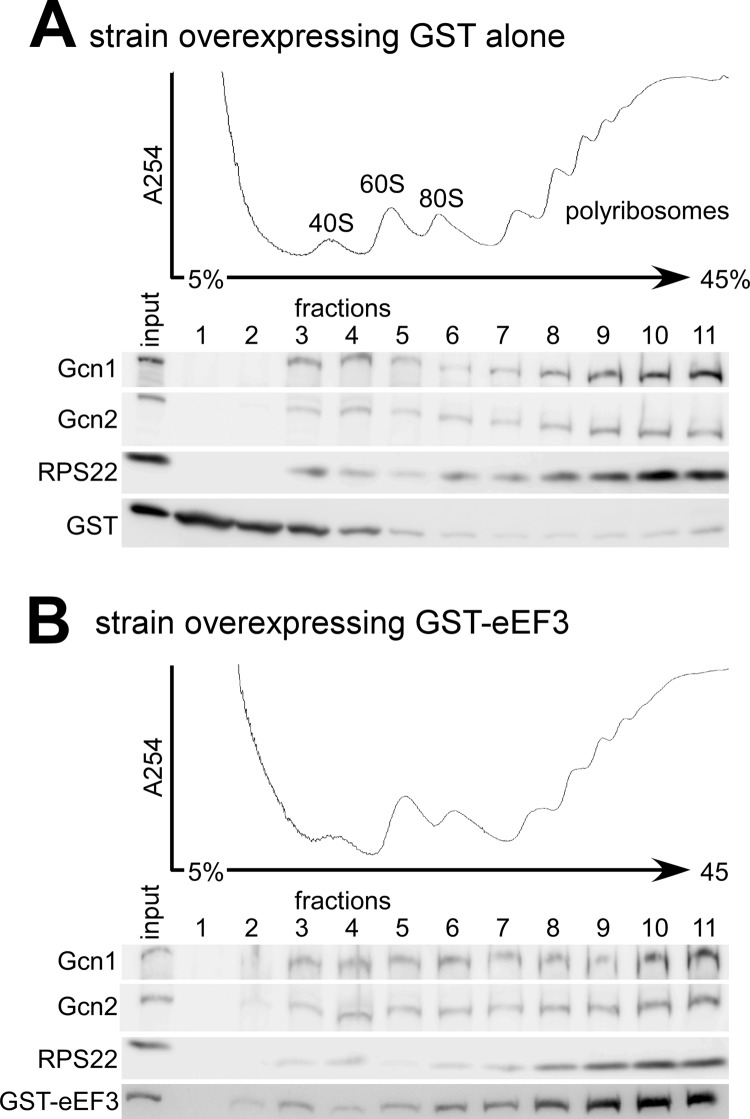
The above results support the idea that the Gcn− phenotype associated with eEF3 overexpression is due to eEF3 affecting Gcn1-ribosome interaction. This may be facilitated by eEF3 removing Gcn1 from the ribosome. Alternatively, considering that three-quarters of the 2672-amino acid-long Gcn1 is involved in ribosome binding, eEF3 may not remove Gcn1 from the ribosome but instead may affect only a subset of the Gcn1 ribosome contact points that are essential for Gcn1 function. To investigate whether eEF3 overexpression removes Gcn1 from the ribosome, we performed ribosome co-sedimentation assays under nearly physiological conditions as published previously (13), using the yeast wild-type strain H1511 harboring plasmid borne GST-eEF3 under a galactose-inducible promoter or GST alone. As published previously (13, 17, 30), in a strain overexpressing GST alone, Gcn1 and Gcn2 co-sedimented with polysomes, indicative of a Gcn1-ribosome and Gcn2-ribosome association (Fig. 4A). We found that even though overexpressed eEF3 co-sedimented with polyribosomes, this did not significantly affect polysome co-sedimentation of Gcn1 or Gcn2 (Fig. 4, A versus B). We did verify that the strains overexpressed eEF3 (data not shown). Considering the fact that on one hand eEF3 overexpression lead to a drastic—at least 4.5-fold—reduction of Gcn2 function in phosphorylating eIF2α (Fig. 3, C and D), but on the other hand eEF3 overexpression barely affected Gcn1-ribosome or Gcn2-ribosome interaction, this suggests that inhibition of Gcn2 function is not simply due to eEF3 removing Gcn1 from the ribosome. Taken together, this supports the idea that eEF3 may only affect a few of the many ribosome contact points in Gcn1 and that Gcn1 is still able to bind to the ribosome via the remaining unaffected ribosome contact points, but that the affected sites are crucial for Gcn1 function in Gcn2 activation.
FIGURE 4.
The effect of eEF3 overexpression on Gcn1-ribosome association. Wild-type strain H1511 overexpressing GST alone (plasmid pEG(KT)) (A) or GST-eEF3 (pGST-YEF3) (B), respectively, from a plasmid and a galactose-inducible promoter were grown to exponential phase to A600 = 1 and cross-linked with formaldehyde immediately before harvesting. Whole cell extracts were prepared and resolved by velocity sedimentation through 4.5–45% sucrose gradients. Fractions were collected while measuring A254 continuously to identify the positions of polyribosomes, 80 S ribosomes, and 40 S and 60 S ribosomal subunits. Equal amounts of each fraction were subjected to SDS-PAGE and immunoblot analysis using antibodies against Gcn1, Gcn2, GST, and the small ribosomal protein RPS22.
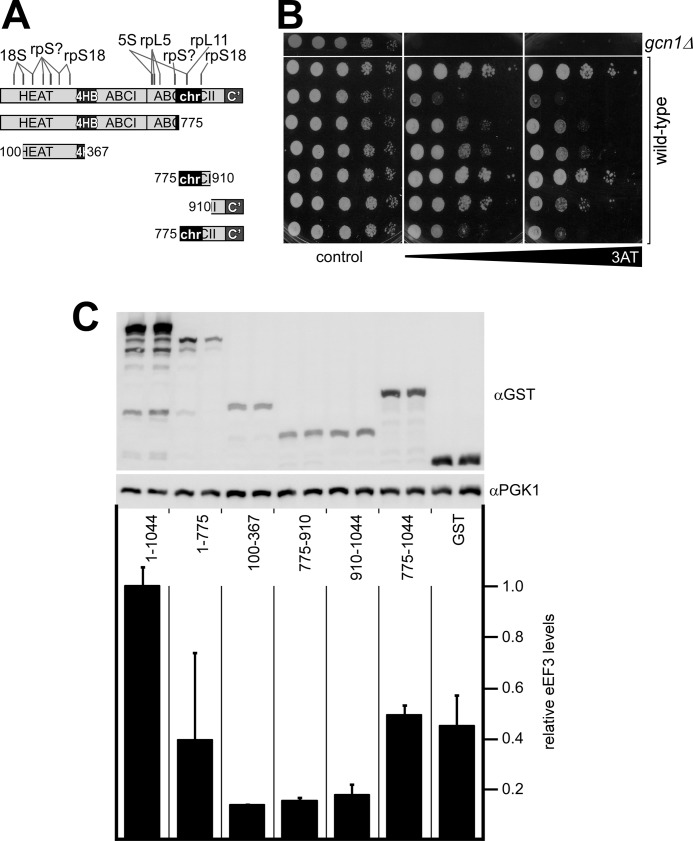
Overexpression of the eEF3 HEAT Domain, or the eEF3 C-terminal Domain (CTD), Is Sufficient for Impairing Gcn2 Activation
If eEF3 overexpression impairs Gcn1 function on the ribosome, then the ribosome binding activity of eEF3 must be essential for affecting Gcn1 function in activating Gcn2. To test this prediction, we aimed to map the domain in eEF3 responsible for affecting Gcn1 function and to compare it with the ribosome-binding domains in eEF3. For eEF3, several ribosome-binding sites have been mapped so far (9, 10) (Fig. 5A). One binding site is the chromodomain that is situated within the ABC cassette II in the eEF3 C terminus. The part of ABC cassette II N-terminal to the chromodomain also contacts the ribosome. Finally, the HEAT domain in the eEF3 N terminus has ribosome binding activities. To map the domain in eEF3 responsible for affecting Gcn1 function, various eEF3 fragments (Fig. 5A), and full-length eEF3 as control, fused to GST, were overexpressed in wild-type yeast cells, and the cells were scored for 3ATs as described above (Fig. 5B). In addition, we determined whether the eEF3 fragments were overexpressed as well as full-length GST-eEF3 (Fig. 5C). As reported above, in contrast to GST alone, overexpression of full-length eEF3 elicited a growth defect in presence of 3AT (Fig. 5B, compare second and third rows). We found that the eEF3 fragment encompassing amino acids 1–775, GST-eEF3[1–775], containing the HEAT domain, the ABC cassette I, and part of the ABC cassette II, conferred 3ATs. This phenotype was not as strong as that found for full-length eEF3, possibly because GST-eEF3[1–775] was not overexpressed as well as GST-eEF3 (Fig. 5C). Interestingly, a smaller fragment only harboring the HEAT domain, in GST-eEF3[100–367], conferred the same degree of 3ATs as GST-eEF3[1–775], suggesting that the HEAT domain was sufficient to elicit 3ATs. A fragment harboring only the chromodomain, in GST-eEF3[775–910], did not elicit 3AT sensitivity. This is most likely not due to its expression levels, because GST-eEF3[100–367] was expressed almost as well as GST-eEF3[775–910] and was able to cause 3ATs, suggesting that overexpressing the chromodomain is not sufficient to impair Gcn1 function. Remarkably, we found that overexpression of the eEF3 CTD encompassing amino acids 910–1044 was sufficient for causing a 3ATs phenotype (Fig. 5B). Because the expression levels of GST-eEF3[100–367] and GST-eEF3[910–1044] were similar and both fragments elicited 3ATs phenotypes of similar strength, this suggests that the HEAT domain and the CTD are equally important for affecting Gcn1 function. A fragment containing the CTD and the chromodomain, in GST-eEF3[775–1040], caused an even stronger phenotype than the CTD alone; in GST-eEF3[910–1040], however, we cannot exclude the possibility that this is due to its 2.5-fold higher expression level as compared with GST-eEF3[910–1040] (Fig. 5C).
FIGURE 5.
Overexpression of the eEF3 HEAT domain and CTD, respectively, is sufficient for causing a Gcn− phenotype. A, overview of full-length eEF3 and eEF3 fragments used in this study. The regions in eEF3 interacting with their respective ribosomal components are indicated. rpL, large ribosomal protein; rpS, small ribosomal protein, 18S, 18 S rRNAs; 5S, 5 S rRNAs. rpS? delineates an unassigned small ribosomal protein. Information was taken from Ref. 10. B, gcn1Δ strain H2556 and isogenic wild-type strain H1511, as indicated on the right, harboring plasmid borne and galactose-inducible GST alone (pEG(KT), top two rows), GST-eEF3 (pTKB705, third row), or GST-eEF3 fragments as indicated on the left in A (from top to bottom: pTKB706, pTKB707, pTKB708, pTKB709, and pTKB710), were subjected to growth assays as described in Fig. 1, using medium containing galactose (control) or galactose and 120 or 200 mm 3AT to induce histidine starvation. C, expression levels of the GST-eEF3 and GST-eEF3 fragments were determined from the strains in B by generating whole cell extracts from exponentially growing cells and subjecting them to SDS-PAGE and immunoblotting assays using antibodies against the GST epitope of eEF3 and the eEF3 fragments and against Pgk1 as control for equal loading. The relative expression levels of the GST-eEF3 proteins were determined from at least two independent transformants using the ImageJ software. The GST-eEF3 signals were normalized to that of Pgk1 for each sample, and the resulting values were averaged and plotted relative to the GST/Pgk1 value of full-length GST-eEF3.
The eEF3 C-terminal amino acids 775–1044, encompassing the CTD and part of the chromodomain, were reported previously to have ribosome binding activity in vitro, and it was proposed that the four distinct blocks of Arg and Lys residues found between amino acids 987 and 1044 mediate ribosome binding (31). In the eEF3-ribosome co-crystal structure and cryoEM studies, the chromodomain was found to contact the ribosome; however, no data for the C terminus were available because the eEF3 used in this study lacked the C-terminal charged amino acids (amino acids 981–1044) (10). Therefore, we next aimed to investigate whether the eEF3 CTD has ribosome binding activity by conducting ribosome co-sedimentation assays as described above. As expected, we found that GST-eEF3[775–1044] harboring some of the ribosome contact points co-sedimented with polyribosomes but GST alone did not, suggesting that it binds ribosomes in vivo as reported previously under in vitro conditions (31) (Fig. 6, top panel versus bottom panel). Interestingly, the eEF3 CTD encompassing only amino acids 910–1044 co-sedimented with polyribosomes as well, suggesting that this fragment does contain ribosome binding activity (Fig. 6, middle panel).
FIGURE 6.
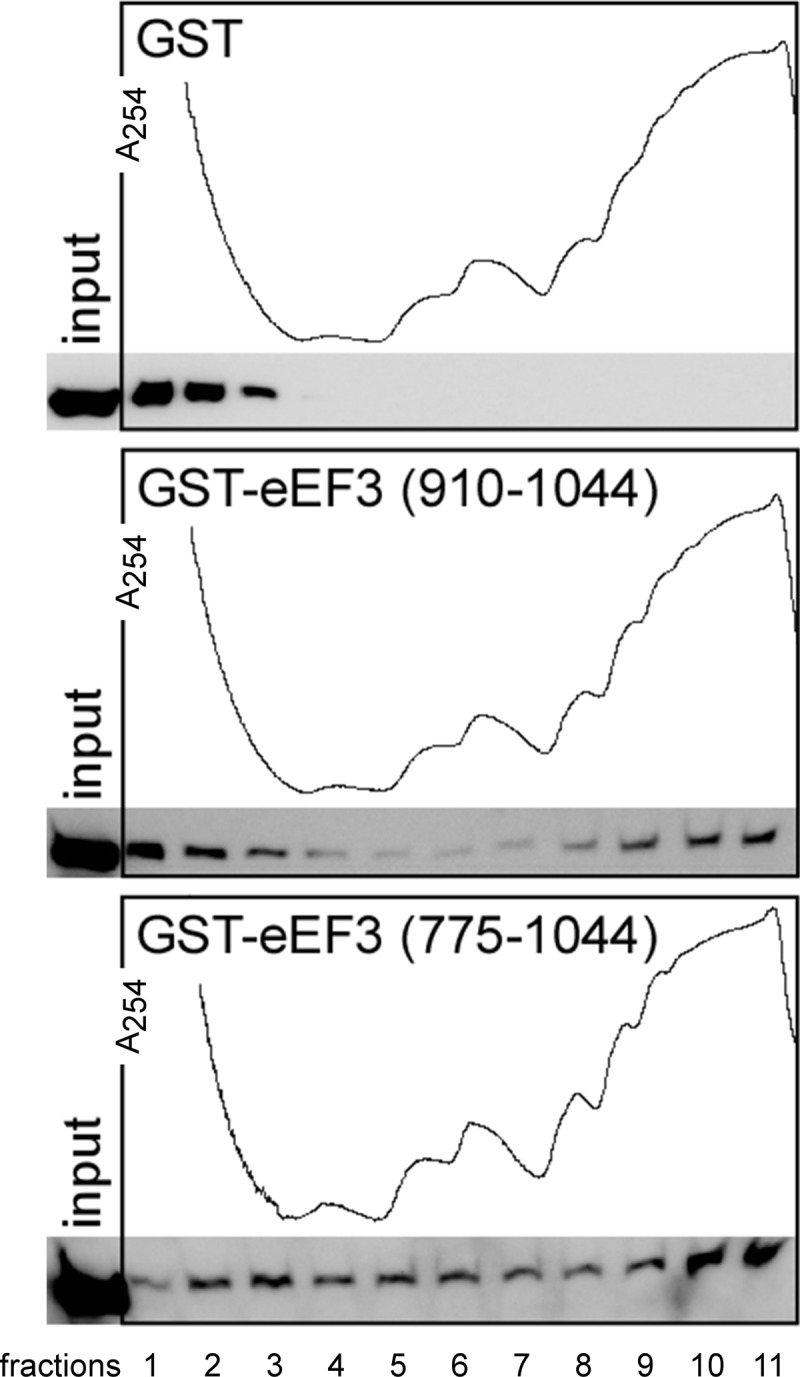
The eEF3 C-terminal amino acids 910–1044 are sufficient for co-sedimenting with polyribosomes. Yeast strain H1511 overexpressing GST alone (plasmid pEG(KT)) or the eEF3 C-terminal fragments encompassing amino acids 910–1044 or 775–1044 (pTKB709 or pTKB710) were subjected to ribosome co-sedimentation assays as outlined in Fig. 4.
Taken together, our results suggest that the eEF3 HEAT domain and eEF3-CTD, respectively, are sufficient for impairing Gcn2 function when overexpressed. Because both fragments have ribosome affinity, this supports the idea that Gcn1 function is impaired on the ribosome, thereby preventing Gcn2 activation.
The Gcn1 M7A Mutation Exacerbates the Gcn− Phenotype Associated with eEF3 Overexpression
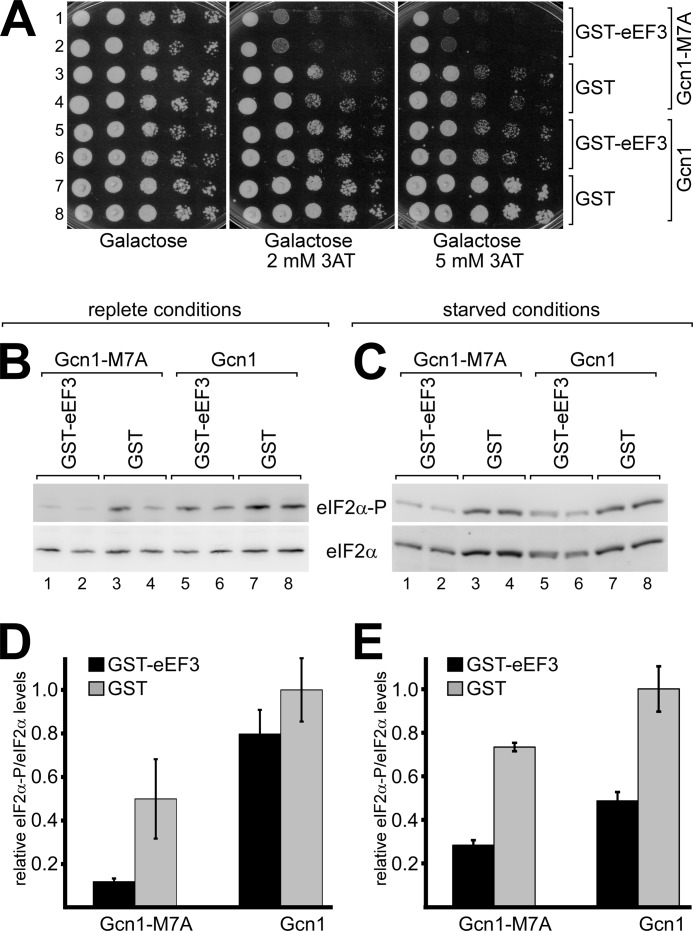
If eEF3 affects only a subset of the many ribosome contact points in Gcn1, then eEF3 should be more efficient in inhibiting a Gcn1 protein that has impaired ribosome binding activity. The Gcn1-M7A mutant protein is known to have reduced ribosome affinity, and this is associated with a reduced ability to activate Gcn2 visible by a slight 3ATs phenotype and reduced eIF2α phosphorylation (13). If eEF3 overexpression affects a subset of ribosome contact points in Gcn1, then eEF3 overexpression should exacerbate the 3ATs phenotype of Gcn1-M7A. To test this, we generated cells that expressed plasmid borne GCN1 or gcn1-M7A from its own promoter and over-expressed from a second plasmid GST alone or GST-eEF3 from a galactose-inducible promoter. The resulting transformants were then subjected to semiquantitative growth assays. As published previously, the gcn1-M7A strain is somewhat more sensitive to 3AT than the GCN1+ strain (Fig. 7A, rows 3 and 4 versus rows 7 and 8). As reported above, overexpression of GST-eEF3 in the GCN1+ strain elicited a 3ATs phenotype (Fig. 7A, rows 5 and 6 versus rows 7 and 8). However, we found that in a gcn1-M7A strain, eEF3 overexpression elicited a 3ATs that was much stronger than in a GCN1+ strain (Fig. 7A, rows 1 and 2 versus rows 3 and 4); in particular eEF3 overexpression led to a more than 10-fold reduction in growth as compared with overexpression of GST alone (Fig. 7A, 2 mm 3AT, rows 1 and 2 versus rows 3 and 4). Considering that the combined effect of the M7A mutation and eEF3 overexpression on cell growth in the presence of 3AT was clearly stronger than that of M7A or eEF3 overexpression alone, this indicates that the M7A mutation exacerbated the 3ATs phenotype associated with eEF3 overexpression, strongly supporting the idea that eEF3 overexpression impairs Gcn1 function on the ribosome.
FIGURE 7.
eEF3 overexpression exacerbates the 3ATs phenotype associated with the gcn1-M7A mutation. A, gcn1Δ strain H2556 overexpressing eEF3 fused to GST or GST alone (plasmid pES340-EF3-2-8 or pES128-9), respectively, from a galactose-inducible promoter, and carrying a plasmid expressing either Gcn1 or Gcn1-M7A from its native promoter (p1832, pES239-4-4a), were subjected to a growth assay as outlined in Fig. 1. B, strains in A were grown to exponential phase, and whole cell extracts were generated and subjected to SDS-PAGE and immunoblotting using antibodies against eIF2α and specifically against phosphorylated eIF2α (eIF2α-P). C, strains in A were grown to the exponential phase, and then 3AT was added to a final concentration of 10 mm. After 30 min the cells were harvested, and whole cell extracts were generated and subjected to SDS-PAGE and immunoblotting as in B. D and E, the amount of eIF2α phosphorylation in B and C was quantified relative to the respective total amount of eIF2α, and relative to the eIF2α-P/eIF2α ratio of the strain expressing GST and Gcn1, and the data are shown in bar graphs.
To verify that the exacerbation of the 3ATs phenotype was due to Gcn2 inhibition, we scored the levels of eIF2α phosphorylation. As shown above, eEF3 overexpression in a GCN1+ strain reduced the amount of eIF2α phosphorylation under replete as well as starved conditions (Fig. 7, B and C, lanes 5 and 6 versus lanes 7 and 8; D; and E), and as reported previously the M7A mutation reduced the levels of eIF2α phosphorylation (Fig. 7, B and C, lanes 3 and 4 versus lanes 7 and 8; D, and E) (13). Under starvation conditions, eEF3 overexpression in the GCN1+ strain reduced eIF2α phosphorylation 2.05-fold; however, in a gcn1-M7A strain, eIF2α phosphorylation was reduced even further, 2.58-fold as compared with the gcn1-M7A strain overexpressing GST alone (Fig. 7E), and this difference was even more pronounced under replete conditions (Fig. 7D). Together this supports the idea that eEF3 overexpression exacerbated the Gcn− phenotype of the Gcn1 M7A mutation. Considering that the M7A mutation affects Gcn1-ribosome association but no other known Gcn1 function, such as Gcn20 binding and Gcn2 binding, this strongly supports our idea that eEF3 overexpression affects Gcn1 function on the ribosome.
DISCUSSION
Continuous protein synthesis is essential to life, and so is a constant supply of amino acids. Eukaryotes possess the kinase Gcn2, which enables them to monitor amino acid availability and to cope with and overcome amino acid starvation. In our current working model, the starvation signal is uncharged tRNAs binding in the ribosomal A-site in a codon-specific manner (12, 17). The proteins Gcn1 and Gcn2 bind to ribosomes, and Gcn1 is involved in transfer of the starvation signal to the kinase Gcn2, which then becomes activated (14). Gcn20 binds to Gcn1 and is not essential for Gcn2 activation, and it was proposed that it may be required to regulate Gcn1-ribosome association under yet unknown conditions (16, 17). Gcn1 and Gcn2 are large proteins, and thus far it is unknown whether these proteins can function productively on ribosomes in all stages of the elongation cycle. Because eEF3 has homology to Gcn1, this question was of particular interest for elongating ribosomes that harbor eEF3.
In this study we show several lines of evidence supporting the idea that Gcn1 cannot function properly when eEF3 is overexpressed in the cell. eEF3 overexpression in an otherwise wild-type strain disabled growth on starvation medium (Gcn− phenotype), and this correlated with reduced phosphorylation of eIF2α, the substrate of Gcn2. Furthermore, eEF3 overexpression diminished the growth defect associated with constitutively active Gcn2 that still requires Gcn1 for function but did not suppress the growth defect associated with PKR expression.
Interestingly, eEF3 overexpression also reduced basal eIF2α phosphorylation levels. In line with our model, this would suggest that a low amount of uncharged tRNAs occurs in the A-site even under replete conditions, leading to a basal eIF2α phosphorylation level and thus basal levels of amino acid biosynthesis. This observation coincides with the fact that Gcn2c proteins still require Gcn1 for their hyperactive kinase function in nutrient-replete cells (14). Under replete conditions Gcn1 would transfer the rarely occurring uncharged tRNA to Gcn2c, which then becomes turned on inappropriately.
The ribosome-binding region in Gcn1 includes the middle portion of Gcn1 that has HEAT repeats, and these HEAT repeats have homology to the eEF3 HEAT repeat domain that also has ribosome binding activity. Thus, it was possible that these HEAT repeats utilize the same or overlapping binding sites on the ribosome and that eEF3 and Gcn1 compete for ribosome association. Supporting this idea, we found that overexpression of the eEF3 HEAT domain was sufficient for impairing GAAC. Remarkably, we found that overexpression of the eEF3 CTD, encompassing amino acids 910–1044, was also sufficient for impairing GAAC, and we demonstrated that this fragment can co-sediment with polysomes in vivo. Indeed, the eEF3 C terminus is Lys-rich, and it has been predicted previously that these amino acids may be involved in ribosome binding (31). Because eEF3 is not known to dimerize, the interaction of the eEF3 C terminus with the ribosome most likely is not mediated by endogenous eEF3. Thus, we propose that binding of the eEF3 HEAT domain and CTD to the ribosome are involved in its ability to impair Gcn1-mediated Gcn2 activation by tRNAdeacyl. The eEF3 HEAT domain or the eEF3 CTD were not overexpressed as strongly as full-length eEF3, and the eEF3 fragments did not impair the GAAC as strongly as full-length eEF3; therefore we could not determine whether the HEAT domain and CTD work in concert to inhibit GAAC or whether their function in inhibiting GAAC is redundant.
Gcn1-ribosome and Gcn2-ribosome interactions were not significantly affected by eEF3 overexpression, suggesting that on the ribosome the binding sites for Gcn1/Gcn2 and eEF3 are not identical. Indeed, the ribosome binding area in Gcn1 is very large, 2052 amino acids, suggesting that it has multiple ribosome-binding sites (13). Thus, eEF3 overexpression might compete with only one or a few ribosome interaction site(s) in Gcn1 and thereby impair Gcn1 function, whereas other ribosome-binding sites in Gcn1 would still support efficient Gcn1-ribosome interaction. The ribosome contact competed by eEF3 might be required for the ability of the Gcn1/20-Gcn2 to properly sense uncharged tRNAs in the A-site.
We have obtained evidence supporting the idea that eEF3 does in fact affect Gcn1 function on the ribosome and not via an indirect mechanism. The M7A substitution in Gcn1—alanine replacements of 12 basic amino acids located in a predicted amphipathic α helix (between amino acids 754 and 796)—is known to weaken Gcn1-ribosome association by 25% but not to affect the association with other known Gcn1 binding partners such as Gcn2 or Gcn20 (13). The weakened ribosome binding affinity of Gcn1-M7A is correlated with a weak Gcn− phenotype and a modest reduction in eIF2α phosphorylation, indicative of its reduced ability to activate Gcn2 (13). We found that eEF3 overexpression exacerbated the Gcn− phenotype of Gcn1-M7A and further reduced the eIF2α phosphorylation levels of a gcn1-M7A strain, supporting the idea that eEF3 inhibits Gcn1 function on the ribosome. It is known that overexpression of a protein may lead to unspecific effects; however, the facts that not all eEF3 fragments impaired Gcn2 function when overexpressed, and that eEF3 overexpression exacerbated the Gcn− phenotype associated with point mutations in gcn1-M7A that specifically affect Gcn1-ribosome association, support the idea that the effect of eEF3 overexpression on Gcn2 activation is specific.
If eEF3 association with translating ribosomes interferes with the ability of the Gcn1/20 complex to activate Gcn2, how then can Gcn2 be activated on elongating ribosomes? One possibility is that eEF3 and Gcn1 bind to different conformational states of the ribosome, allowing Gcn1/20-Gcn2 to access the conformational state that lacks eEF3. An attractive model would be that eEF3 does not bind with high affinity to an elongating ribosome containing tRNAdeacyl in the A-site, which is the relevant substrate for the Gcn1/20-Gcn2 complex. In fact, there is evidence that eEF3 interacts with eEF1A to help catalyze delivery of the eEF1A-GTP-amino acyl tRNA ternary complex (TC) to the ribosomal A-site (1, 27). Thus, eEF3 and Gcn1/20-Gcn2 would not normally occupy the same ribosomal complexes, whereas overexpressing eEF3 would force it to bind to the stalled complexes containing tRNAdeacyl in the A-site, by mass action, and thereby interfere with Gcn2 activation. In this view, the productive TC-eEF3 interaction at the A-site, in addition to the scarcity of tRNAsdeacyl entering the A-site, plays an important role in blocking activation of Gcn2 in nutrient-replete conditions. Interestingly, we have just recently found several lines of evidence strongly suggesting that eEF1A inhibits Gcn2 activation under amino acid replete conditions (21). eEF1A directly binds to the Gcn2 C terminus, and in vivo the eEF1A-Gcn2 interaction is lost when cells are starving for amino acids. In vitro, eEF1A inhibits Gcn2-mediated eIF2α phosphorylation but not Gcn2 autophosphorylation. Considering that eEF3 interacts with eEF1A to help catalyze delivery of the TC to the A-site, it is tempting to speculate that eEF1A and eEF3 work in concert to ensure that Gcn2 stays in its latent state under amino acid replete conditions.
Considering that Gcn1 and Gcn2 can directly interact with each other (12), it is likely that Gcn1 and Gcn2 form a permanent complex that shuttles on and off the ribosome. In view of the fact that ribosomes are 15 or 350 times more abundant in the cell than Gcn1 or Gcn2 (32), stable Gcn1-Gcn2 association would be necessary to ensure that Gcn1 and Gcn2 reside on the same ribosome at any given time to allow Gcn1-mediated transfer of tRNAdeacyl to Gcn2. Gcn20 binds strongly to Gcn1 and is unstable in gcn1Δ cells, strongly suggesting that Gcn20 remains associated with Gcn1 (16). Thus, it seems likely that the Gcn2-Gcn1/20 complex randomly probes ribosomes for the presence of tRNAdeacyl versus TC in the A-site and the attendant absence of eEF3.
We showed previously that Gcn1 overexpression leads to increased amounts of Gcn1 binding to polyribosomes, and this is associated with reduced cell growth, which could be due to interference with eEF3 or eEF1A function (12). Thus, the cellular Gcn1 levels are probably low to minimize its possible interference with elongation (12).
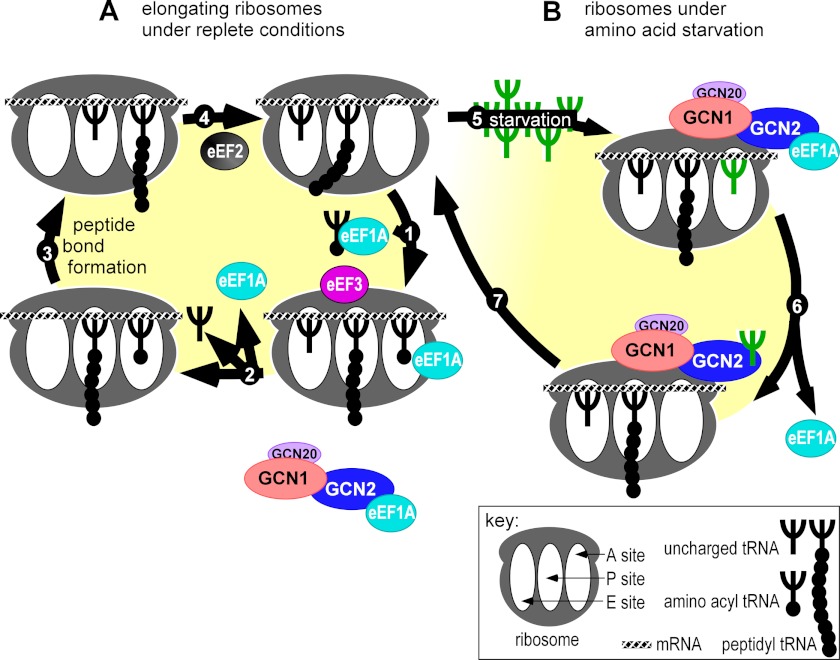
Our data led to a refined model for sensing amino acid starvation as shown in Fig. 8. When during each round of the elongation cycle the A-site is available for accommodating the next tRNA in a codon-specific manner, either under replete conditions aa-tRNA is delivered by eEF1A (Fig. 8A, arrow 1), or under starvation conditions tRNAdeacyl enters the A-site (Fig. 8B, arrow 5). If aa-tRNA is delivered, then eEF3 triggers the concomitant release of tRNAdeacyl from the E-site (Fig. 8B, arrow 2). Subsequently translation elongation can proceed to the next steps involving peptide bond formation and translocation (Fig. 8A, arrows 3 and 4). In such productive elongation complexes, the HEAT domain and C terminus of eEF3 would prevent productive Gcn1-ribosome interactions and thereby prevent Gcn2 from detecting tRNAsdeacyl, e.g., tRNAsdeacyl leaving the E-site. Furthermore, eEF1A binding to the Gcn2 C terminus would prevent Gcn2 from substrate phosphorylation (21). Because eEF1A is abundant in the cell, it remains to be determined whether this inhibition is mediated by the eEF1A delivering aa-tRNA to the A-site or by another eEF1A molecule. For simplicity, the nonfunctional Gcn1/20-Gcn2 complex was drawn separately from the ribosome and associated with an eEF1A molecule not involved in the translation elongation cycle (Fig. 8A, bottom part). As suggested above, eEF1A and eEF3 might cooperate to maintain Gcn2 latency under amino acid replete conditions. Although eEF3 would prevent Gcn1-mediated delivery of tRNAsdeacyl, for example the ones naturally occurring during the translation elongation cycle, to Gcn2, eEF1A would ensure that Gcn2 does not phosphorylate its substrate. If under amino acid starvation tRNAdeacyl enters the A-site (Fig. 8B, arrow 5), and in line with the model predicted for the E. coli stringent response, the E-site tRNAdeacyl does not leave the ribosome (33). The affinity of eEF3 for these stalled complexes would be lower, possibly because of the absence of eEF1A at the A-site, allowing Gcn1/20 to interact with the A-site, directly or indirectly, and mediate the transfer of tRNAdeacyl to Gcn2 for kinase activation (Fig. 8B, arrow 6). This tRNAdeacyl leads to eEF1A dissociating from Gcn2 and consequently allowing Gcn2-mediated eIF2α phosphorylation. It is possible that apart from triggering the GAAC response, the Gcn1/20-Gcn2 complex may be necessary to vacate the A-site to prevent premature peptide chain termination triggered by A-site bound tRNAdeacyl (34, 35) and to enable the “post-starved” ribosome to enter again the translation elongation cycle (Fig. 8B, arrow 7). It still remains to be determined how tRNAdeacyl enters the A-site, via diffusion or via delivery by another protein; however, the model can accommodate both mechanisms.
FIGURE 8.
Model for sensing amino acid starvation during the translation elongation cycle. A, translation elongation is a cyclic process where during each round one amino acid is added to the growing peptide chain, for more detail see main text. We propose that during the elongation cycle eEF3 and eEF1A cooperate in keeping Gcn2 in its latent state. eEF3 hinders Gcn1 from forming a productive complex with the ribosome, thereby preventing Gcn1-mediated transfer of tRNAsdeacyl to Gcn2, e.g., tRNAsdeacyl exiting the E-site. eEF1A associating with the Gcn2 C terminus impairs Gcn2 in phosphorylating its substrate eIF2α (21). It remains to be determined whether eEF1A delivering aa-tRNA to the A-site inhibits Gcn2 or whether a second eEF1A molecule executes this function. For simplicity, the nonfunctional Gcn1/20-Gcn2 complex was drawn separately from the ribosome and associated with an eEF1A molecule not involved in the translation elongation cycle (see bottom part of Fig. 7A). B, under starvation conditions the cellular levels of tRNAdeacyl increases. Arrow 5, tRNAdeacyl enters the A-site in a codon-dependent manner. Arrow 6, eEF3 not binding the ribosome allows Gcn1 to form a productive interaction with the ribosome, consequently mediating the removal of tRNAdeacyl from the A-site. Under starvation conditions eEF1A has dissociated from the Gcn2 C terminus, thus allowing Gcn2 to phosphorylate its substrate. The mechanism for eEF1A-Gcn2 dissociation still remains to be elucidated; however, our data suggest that tRNAdeacyl delivered to Gcn2 may remove eEF1A from Gcn2 (21). Arrow 7, the Gcn1/20-Gcn2 complex may be necessary to vacate the A-site to prevent premature peptide chain termination triggered by A-site bound tRNAdeacyl (34, 35) and to enable the post-starved ribosome to resume the translation elongation process. For simplicity, ribosomes and proteins are drawn as spheres. For more detail see main text.
Under replete conditions, if in a rare event tRNAdeacyl enters the A-site, one could envision that eEF3 makes way for the formation of a productive Gcn1-ribosome interaction, which then mediates the removal of tRNAdeacyl from the A-site and its delivery to Gcn2. At the same time eEF1A-mediated Gcn2 inhibition would cease and thereby allow eIF2α phosphorylation by Gcn2, accounting for the basal eIF2-P levels in unstarved yeast cells.
eEF3 is a fungal specific elongation factor, and it has been proposed that in nonfungal eukaryotes a factor permanently attached to the ribosome executes the function of eEF3 (5). We believe that our model may also be applicable to nonfungal eukaryotes. Gcn1 would be able to attach to a ribosome that harbors an integral eEF3-like protein. Conformational changes in the ribosome or of the integral eEF3-like protein then may allow or prevent Gcn1 function in transferring the starvation signal to Gcn2. Further studies will be necessary to test this model in nonfungal eukaryotes. Such studies may be of high interest in regards to finding specific anti-fungal agents.
We cannot exclude the possibility that eEF3 overexpression prevents Gcn2 function by a mechanism different to that proposed above. For example eEF3 could prevent the Gcn2 kinase domain from accessing and phosphorylating eIF2α, or eEF3 could promote eIF2α dephosphorylation by activating a phosphatase. However, these scenarios may be less likely considering that overexpression of eEF3 did not mitigate the toxicity of overexpressing the eIF2α kinase PKR when expressed in yeast instead of Gcn2. Taken together, our data provide strong evidence that eEF3 negatively regulates Gcn2 activation in a manner requiring the HEAT domain and/or requiring an accessory ribosome-binding domain at the eEF3 C terminus that is dispensable for eEF3 essential function in translation elongation (1, 27). These findings provide added support for the model that Gcn2 is activated by uncharged tRNAs on elongating ribosomes and reveal an unanticipated role for a general elongation factor, in addition to eEF1A, in controlling this process. Mapping the binding sites for Gcn1 and the eEF3 C terminus on the ribosome will be necessary to provide a full understanding of how eEF3 blocks Gcn2 activation. Together with our recent findings that eEF1A inhibits eIF2α phosphorylation by Gcn2, we propose that eEF3 works in concert with eEF1A to keep Gcn2 in its latent state when amino acids are plentiful.
Acknowledgments
We are grateful to Tracey Waller for technical support; Kalpana Chakraburtty and Terri Kinzy for plasmids; Thomas Dever, Maurice Swanson, and Jan van't Riet for antibodies; Kalpana Chakraburtty for helpful comments during the start-up of this project; and Terri Kinzy for helpful discussions.
This work was supported in part by National Institutes of Health Intramural Research Program (to A. G. H.). This work was also supported by Marsden Fund Council Grant MAU0607 administered by the Royal Society of New Zealand, a Massey University Technicians Award (to E. S. for supporting S. J. L.), the Massey University Research Fund (to E. S.), and a Massey University Ph.D. Scholarship (to J. V.).
- eEF
- eukaryotic translation elongation factor
- aa-tRNA
- amino acyl tRNA
- 3AT
- 3-amino-2,4-triazole
- GAAC
- general amino acid control
- PKR
- protein kinase R
- SM
- sulfometuron methyl
- tRNAdeacyl
- deacylated tRNA
- ABC
- ATP-binding cassette
- 3ATs
- 3AT sensitivity
- CTD
- C-terminal domain
- TC
- eEF1A·GTP·amino acyl tRNA ternary complex.
REFERENCES
- 1. Taylor D. R., Frank J., Kinzy T. G. (ed) (2006) Structure and Function of the Eukaryotic Ribosome and Elongation Factors, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 2. Dasmahapatra B., Chakraburtty K. (1981) Protein synthesis in yeast. I. Purification and properties of elongation factor 3 from Saccharomyces cerevisiae. J. Biol. Chem. 256, 9999–10004 [PubMed] [Google Scholar]
- 3. Skogerson L., Wakatama E. (1976) A ribosome-dependent GTPase from yeast distinct from elongation factor 2. Proc. Natl. Acad. Sci. U.S.A. 73, 73–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Triana-Alonso F. J., Chakraburtty K., Nierhaus K. H. (1995) The elongation factor 3 unique in higher fungi and essential for protein biosynthesis is an E site factor. J. Biol. Chem. 270, 20473–20478 [DOI] [PubMed] [Google Scholar]
- 5. El'skaya A. V., Ovcharenko G. V., Palchevskii S. S., Petrushenko Z. M., Triana-Alonso F. J., Nierhaus K. H. (1997) Three tRNA binding sites in rabbit liver ribosomes and role of the intrinsic ATPase in 80S ribosomes from higher eukaryotes. Biochemistry 36, 10492–10497 [DOI] [PubMed] [Google Scholar]
- 6. Kiel M. C., Aoki H., Ganoza M. C. (1999) Identification of a ribosomal ATPase in Escherichia coli cells. Biochimie 81, 1097–1108 [DOI] [PubMed] [Google Scholar]
- 7. Kiel M. C., Ganoza M. C. (2001) Functional interactions of an Escherichia coli ribosomal ATPase. Eur. J. Biochem. 268, 278–286 [DOI] [PubMed] [Google Scholar]
- 8. Andrade M. A., Petosa C., O'Donoghue S. I., Müller C. W., Bork P. (2001) Comparison of ARM and HEAT protein repeats. J. Mol. Biol. 309, 1–18 [DOI] [PubMed] [Google Scholar]
- 9. Gontarek R. R., Li H., Nurse K., Prescott C. D. (1998) The N terminus of eukaryotic translation elongation factor 3 interacts with 18 S rRNA and 80 S ribosomes. J. Biol. Chem. 273, 10249–10252 [DOI] [PubMed] [Google Scholar]
- 10. Andersen C. B., Becker T., Blau M., Anand M., Halic M., Balar B., Mielke T., Boesen T., Pedersen J. S., Spahn C. M., Kinzy T. G., Andersen G. R., Beckmann R. (2006) Structure of eEF3 and the mechanism of transfer RNA release from the E-site. Nature 443, 663–668 [DOI] [PubMed] [Google Scholar]
- 11. Marton M. J., Crouch D., Hinnebusch A. G. (1993) GCN1, a translational activator of GCN4 in Saccharomyces cerevisiae, is required for phosphorylation of eukaryotic translation initiation factor 2 by protein kinase GCN2. Mol. Cell Biol. 13, 3541–3556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sattlegger E., Hinnebusch A. G. (2000) Separate domains in GCN1 for binding protein kinase GCN2 and ribosomes are required for GCN2 activation in amino acid-starved cells. EMBO J. 19, 6622–6633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sattlegger E., Hinnebusch A. G. (2005) Polyribosome binding by GCN1 is required for full activation of eukaryotic translation initiation factor 2α kinase GCN2 during amino acid starvation. J. Biol. Chem. 280, 16514–16521 [DOI] [PubMed] [Google Scholar]
- 14. Hinnebusch A. G. (2005) Translational regulation of GCN4 and the general amino acid control of yeast. Annu. Rev. Microbiol. 59, 407–450 [DOI] [PubMed] [Google Scholar]
- 15. Zaborske J. M., Wu X., Wek R. C., Pan T. (2010) Selective control of amino acid metabolism by the GCN2 eIF2 kinase pathway in Saccharomyces cerevisiae. BMC Biochem. 11, 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Vazquez de Aldana C. R., Marton M. J., Hinnebusch A. G. (1995) GCN20, a novel ATP binding cassette protein, and GCN1 reside in a complex that mediates activation of the eIF-2 α kinase GCN2 in amino acid-starved cells. EMBO J. 14, 3184–3199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Marton M. J., Vazquez de Aldana C. R., Qiu H., Chakraburtty K., Hinnebusch A. G. (1997) Evidence that GCN1 and GCN20, translational regulators of GCN4, function on elongating ribosomes in activation of eIF2α kinase GCN2. Mol. Cell Biol. 17, 4474–4489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cashel M R. K. (1987) Eschericha coli and Salmonella typhimurium: Cellular and Molecular Biology, (Neidhardt F. C., Ingraham J. L., Magasanik B., Low K. B., Schaechter M., Umbarger H. E., eds), pp. 1410–1438, American Society for Microbiology, Washington, D.C [Google Scholar]
- 19. Goldman E., Jakubowski H. (1990) Uncharged tRNA, protein synthesis, and the bacterial stringent response. Mol. Microbiol. 4, 2035–2040 [DOI] [PubMed] [Google Scholar]
- 20. Murchie M. J., Leader D. P. (1978) Codon-specific interaction of uncharged transfer-RNA with eukaryotic ribosomes. Biochim. Biophys. Acta 520, 233–236 [DOI] [PubMed] [Google Scholar]
- 21. Visweswaraiah J., Lageix S., Castilho B. A., Izotova L., Kinzy T. G., Hinnebusch A. G., Sattlegger E. (2011) Evidence that eukaryotic translation elongation factor 1A (eEF1A) binds the Gcn2 protein C terminus and inhibits Gcn2 activity. J. Biol. Chem. 286, 36568–36579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sikorski R. S., Hieter P. (1989) A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122, 19–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Visweswaraiah J., Dautel M., Sattlegger E. (2011) Generating highly concentrated yeast whole cell extract using low-cost equipment. Nat. Prot. Exchange doi: 10.1038/protex. 2011.212 [DOI] [Google Scholar]
- 24. Dever T. E., Feng L., Wek R. C., Cigan A. M., Donahue T. F., Hinnebusch A. G. (1992) Phosphorylation of initiation factor 2α by protein kinase GCN2 mediates gene-specific translational control of GCN4 in yeast. Cell 68, 585–596 [DOI] [PubMed] [Google Scholar]
- 25. Anderson J. T., Paddy M. R., Swanson M. S. (1993) PUB1 is a major nuclear and cytoplasmic polyadenylated RNA-binding protein in Saccharomyces cerevisiae. Mol. Cell. Biol. 13, 6102–6113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hilton J. L., Kearney P. C., Ames B. N. (1965) Mode of action of the herbicide, 3-amino-1,2,4-triazole(amitrole): inhibition of an enzyme of histidine biosynthesis. Arch. Biochem. Biophys. 112, 544–547 [DOI] [PubMed] [Google Scholar]
- 27. Anand M., Balar B., Ulloque R., Gross S. R., Kinzy T. G. (2006) Domain and nucleotide dependence of the interaction between Saccharomyces cerevisiae translation elongation factors 3 and 1A. J. Biol. Chem. 281, 32318–32326 [DOI] [PubMed] [Google Scholar]
- 28. Romano P. R., Zhang F., Tan S. L., Garcia-Barrio M. T., Katze M. G., Dever T. E., Hinnebusch A. G. (1998) Inhibition of double-stranded RNA-dependent protein kinase PKR by vaccinia virus E3. Role of complex formation and the E3 N-terminal domain. Mol. Cell. Biol. 18, 7304–7316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. LaRossa R. A., Schloss J. V. (1984) The sulfonylurea herbicide sulfometuron methyl is an extremely potent and selective inhibitor of acetolactate synthase in Salmonella typhimurium. J. Biol. Chem. 259, 8753–8757 [PubMed] [Google Scholar]
- 30. Ramirez M., Wek R. C., Hinnebusch A. G. (1991) Ribosome association of GCN2 protein kinase, a translational activator of the GCN4 gene of Saccharomyces cerevisiae. Mol. Cell. Biol. 11, 3027–3036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kambampati R., Chakraburtty K. (1997) Functional subdomains of yeast elongation factor 3. Localization of ribosome-binding domain. J. Biol. Chem. 272, 6377–6381 [DOI] [PubMed] [Google Scholar]
- 32. Ghaemmaghami S., Huh W. K., Bower K., Howson R. W., Belle A., Dephoure N., O'Shea E. K., Weissman J. S. (2003) Global analysis of protein expression in yeast. Nature 425, 737–741 [DOI] [PubMed] [Google Scholar]
- 33. Wendrich T. M., Blaha G., Wilson D. N., Marahiel M. A., Nierhaus K. H. (2002) Dissection of the mechanism for the stringent factor RelA. Mol. Cell 10, 779–788 [DOI] [PubMed] [Google Scholar]
- 34. Caskey C. T., Beaudet A. L., Scolnick E. M., Rosman M. (1971) Hydrolysis of fMet-tRNA by peptidyl transferase. Proc. Natl. Acad. Sci. U.S.A. 68, 3163–3167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zavialov A. V., Mora L., Buckingham R. H., Ehrenberg M. (2002) Release of peptide promoted by the GGQ motif of class 1 release factors regulates the GTPase activity of RF3. Mol. Cell 10, 789–798 [DOI] [PubMed] [Google Scholar]
- 36. Foiani M., Cigan A. M., Paddon C. J., Harashima S., Hinnebusch A. G. (1991) GCD2, a translational repressor of the GCN4 gene, has a general function in the initiation of protein synthesis in Saccharomyces cerevisiae. Mol. Cell. Biol. 11, 3203–3216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hannig E. M., Williams N. P., Wek R. C., Hinnebusch A. G. (1990) The translational activator GCN3 functions downstream from GCN1 and GCN2 in the regulatory pathway that couples GCN4 expression to amino acid availability in Saccharomyces cerevisiae. Genetics 126, 549–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ramirez M., Wek R. C., Vazquez deAldana C. R., Jackson B. M., Freeman B., Hinnebusch A. G. (1992) Mutations activating the yeast eIF-2 α kinase GCN2. Isolation of alleles altering the domain related to histidyl-tRNA synthetases. Mol. Cell. Biol. 12, 5801–5815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kawagishi-Kobayashi M., Silverman J. B., Ung T. L., Dever T. E. (1997) Regulation of the protein kinase PKR by the vaccinia virus pseudosubstrate inhibitor K3L is dependent on residues conserved between the K3L protein and the PKR substrate eIF2α. Mol. Cell. Biol. 17, 4146–4158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kambampati R., Chakraburtty K. (1997) Overexpression and purification of elongation factor 3 from Saccharomyces cerevisiae. Protein Expr. Purif 10, 209–213 [DOI] [PubMed] [Google Scholar]