Background: Malaria parasites detoxify copious amounts of heme during human infection, but enzyme involvement remains unclear.
Results: Parasite-infected and uninfected erythrocytes contain indistinguishable low levels of heme catabolites, and a heme oxygenase-like parasite protein binds but does not degrade heme.
Conclusion: P. falciparum parasites do not enzymatically degrade heme.
Significance: This study comprehensively answers a long standing question regarding heme metabolism in this important parasite.
Keywords: Biosensors, Heme, Heme Oxygenase, Malaria, Mass Spectrometry (MS), Parasite Metabolism, Parasitology
Abstract
Malaria parasites generate vast quantities of heme during blood stage infection via hemoglobin digestion and limited de novo biosynthesis, but it remains unclear if parasites metabolize heme for utilization or disposal. Recent in vitro experiments with a heme oxygenase (HO)-like protein from Plasmodium falciparum suggested that parasites may enzymatically degrade some heme to the canonical HO product, biliverdin (BV), or its downstream metabolite, bilirubin (BR). To directly test for BV and BR production by P. falciparum parasites, we DMSO-extracted equal numbers of infected and uninfected erythrocytes and developed a sensitive LC-MS/MS assay to quantify these tetrapyrroles. We found comparable low levels of BV and BR in both samples, suggesting the absence of HO activity in parasites. We further tested live parasites by targeted expression of a fluorescent BV-binding protein within the parasite cytosol, mitochondrion, and plant-like plastid. This probe could detect exogenously added BV but gave no signal indicative of endogenous BV production within parasites. Finally, we recombinantly expressed and tested the proposed heme degrading activity of the HO-like protein, PfHO. Although PfHO bound heme and protoporphyrin IX with modest affinity, it did not catalyze heme degradation in vivo within bacteria or in vitro in UV absorbance and HPLC assays. These observations are consistent with PfHO's lack of a heme-coordinating His residue and suggest an alternative function within parasites. We conclude that P. falciparum parasites lack a canonical HO pathway for heme degradation and thus rely fully on alternative mechanisms for heme detoxification and iron acquisition during blood stage infection.
Introduction
During intraerythrocytic infection, malaria parasites of the genus Plasmodium generate vast quantities of free heme within their acidic food vacuole during large scale digestion of host hemoglobin. Peak heme concentrations within this organelle have been estimated to reach several hundred millimolar (1). At the same time, parasites appear to maintain an active heme biosynthetic pathway that spans three subcellular compartments and culminates in de novo heme production within the mitochondrion (Fig. 1A) (2–4). Because parasites sequester the majority of host-derived heme as crystalline hemozoin within the digestive vacuole (5) and are thought to principally incorporate endogenous heme into mitochondrial cytochromes (2), it remains unclear whether any heme is enzymatically metabolized.
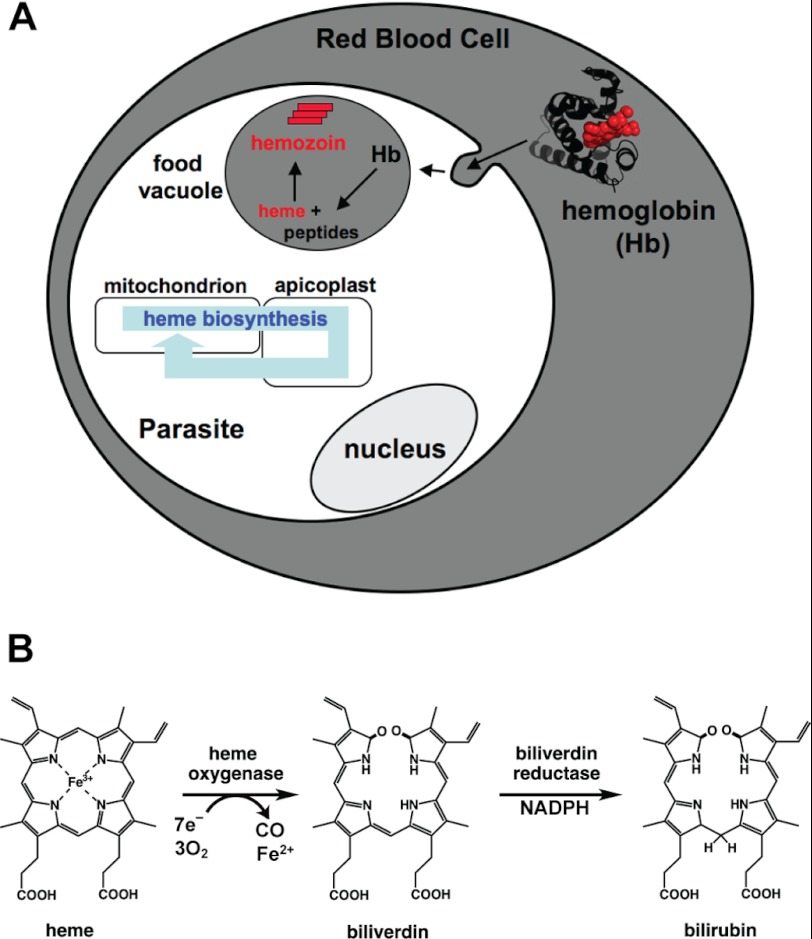
FIGURE 1.
Pathways for heme generation in parasites and enzymatic heme degradation. A, intraerythrocytic parasites generate heme by proteolytic degradation of host hemoglobin in the digestive food vacuole and by de novo biosynthesis coordinated between the mitochondrion, apicoplast, and cytosol. For simplicity, all membranes are shown as single layers and hemoglobin is depicted as a monomer. B, enzymatic heme degradation proceeds via cleavage of the porphyrin ring by a heme oxygenase to generate biliverdin IXα, which can then be reduced by a biliverdin reductase to form bilirubin IXα.
Heme oxygenases (HO)6 are conserved enzymes found in nearly all kingdoms of life that catalyze the oxidative cleavage of the heme macrocycle to release iron and initiate derivatization of the tetrapyrrole backbone for downstream metabolic usage or disposal. The vast majority of HO enzymes studied to date have a conserved α-helical fold, utilize cytochrome P450 reductase or reduced ferredoxin as an electron source, and cleave heme at the α-methine group to produce the canonical reaction product, biliverdin IXα (BV) (Fig. 1B) (6–8). BV can be further processed to downstream metabolites by additional enzymes, including biliverdin reductase (BVR) that utilizes NADPH to reduce BV at the γ-methine group to produce bilirubin IXα (BR) (Fig. 1B). The direct and downstream products of HO-catalyzed heme degradation can all serve useful metabolic roles within cells as follows. BV in its native or derivatized forms is incorporated into light-sensing phytochromes within plants and some bacteria (9); the BV-BR redox pair can serve as a potent buffer against oxidant stress (10, 11); BR can be further processed for excretion and disposal of the tetrapyrrole backbone (12, 13); BV, BR, and CO can serve as cellular signaling molecules (12, 13); and the iron released by heme cleavage can be scavenged for use in other iron-containing proteins (14).
Despite the abundance of heme encountered during intraerythrocytic infection and a need to scavenge host iron for use in parasite proteins, the human malaria parasite, Plasmodium falciparum, has been assumed to lack enzymes that process heme for metabolic utilization or disposal (1, 15–21). Nevertheless, recent in vitro studies of a recombinant HO-like protein (PF3D7_1011900, formerly PF10_0116) from P. falciparum (termed PfHO) showing sequence similarity to known HO enzymes have suggested that parasites may enzymatically degrade some heme to BV or BR, using an apicoplast-targeted ferredoxin as a reductant co-factor (22, 23). Because of conflicting suggestions and assumptions in the literature and because no direct tests of enzymatic heme degradation by parasites have been reported, confusion and uncertainty persist regarding the presence of a heme oxygenase pathway within blood stage P. falciparum parasites (2, 19, 24, 25).
To comprehensively test intraerythrocytic P. falciparum parasites for enzymatic heme degradation products, we developed a sensitive liquid chromatography-tandem mass spectrometry (LC-MS/MS) assay using 13C-labeled internal standards to directly quantify BV and BR levels in infected versus uninfected erythrocytes. We further tested live parasites for BV production by episomally expressing a fluorescent BV biosensor within the parasite cytosol, mitochondrion, and apicoplast. Finally, we recombinantly expressed and tested the heme binding and degrading properties of PfHO, including assays with purified P. falciparum ferredoxin. We found no evidence for BV or BR production either by parasites or by purified PfHO, strongly suggesting the absence of a canonical heme oxygenase pathway in parasites. The inability of PfHO to degrade heme is consistent with its lack of the conserved His residue used by all known HO enzymes to coordinate heme and suggests functional diversification to serve an alternative role within parasites.
EXPERIMENTAL PROCEDURES
Materials
All reagents were of the highest purity commercially available. Biliverdin IXα and bilirubin IXα were purchased from Frontier Scientific Inc. (Logan, UT); DMSO, reduced NADPH, spinach ferredoxin (Fd), spinach ferredoxin-NADP+ reductase (FNR), heme (hemin chloride), protoporphyrin IX, sodium ascorbate, bovine liver catalase, deferoxamine, BSA, equine heart myoglobin, and equine cytochrome c were purchased from Sigma; [13C]glycerol was purchased from Cambridge Isotope Laboratories Inc. (Andover, MA); and IPTG was purchased from Gold Biotechnology Inc. (St. Louis, MO).
Cloning and Site-directed Mutagenesis
HO1 from Synechocystis sp. 6803 (SynHO1) and the engineered infrared fluorescent protein 1.4 (IFP) were subcloned by PCR from the pBAD vector obtained from Roger Tsien (University of California, San Diego) into the NdeI/XhoI sites of pET22b (SynHO1) and the NcoI/XhoI sites of pET21d and pET28a (IFP) in-frame with a C-terminal hexa-His tag. Homo sapiens HO1 (HuHO1) was subcloned into the NdeI/SalI sites of pET22b (stop codon before optional hexa-His tag) by digesting the pBAce vector obtained from Paul Ortiz de Montellano (University of California, San Francisco) with NdeI/SalI to release the HuHO1 gene, followed by ligation into pET22b. The HO domains (following the chloroplast-targeting leader sequences) of Arabidopsis thaliana HO1 (AtHO1, residues 55–282) and AtHO2 (residues 57–299) were cloned by PCR from A. thaliana cDNA obtained from Barbara Kunkel (Washington University in St. Louis) into the NcoI/XhoI sites of pET28a, in-frame with the C-terminal hexa-His tag. A codon-optimized version (most abundant Escherichia coli codon for each amino acid) of the HO domain of AtHO2 was synthesized by end-overlap assembly PCR (26) and cloned into the NdeI/XhoI sites of pET28a, in-frame with the N-terminal hexa-His tag. The codon-optimized version of AtHO2 was used for all experiments based on its superior expression relative to the construct cloned from A. thaliana cDNA. Rattus norvegicus BVR was subcloned by PCR from a pET23 vector obtained from Paul Ortiz de Montellano (University of California, San Francisco) into the NcoI/XhoI sites of pET28a in-frame with the C-terminal hexa-His tag.
The HO-like domain of PfHO (PF3D7_1011900, formerly PF10_0116, residues 84–305) was cloned by PCR from P. falciparum (3D7) cDNA into the NcoI/XhoI sites of pET21d in-frame with the C-terminal hexa-His tag. A codon-optimized version of the PfHO construct (most abundant E. coli codon for each amino acid) was ordered from IDT, Inc. (Coralville, IA), and subcloned by PCR into the NcoI/XhoI sites of pET21d in-frame with the C-terminal hexa-His tag. The codon-optimized version of PfHO was used for all experiments based on its superior expression relative to the construct cloned from parasite cDNA. A codon-optimized version (most abundant E. coli codon for each amino acid) of the ferredoxin domain of PfFd (residues 97–194, following the apicoplast-targeting leader sequence) was ordered from IDT, Inc. (Coralville, IA), and subcloned by PCR into the NcoI/XhoI sites of pET28a in-frame with the C-terminal hexa-His tag. The P. falciparum proteins encoded by gene ID numbers PF3D7_0317800 (formerly PFC0785c), PF3D7_1002500 (formerly PF10_0029), and PF3D7_0511400 (formerly PFE0565w, amino acids 69–381, after the PEXEL motif) were cloned by PCR from P. falciparum (3D7) cDNA into the NcoI/XhoI sites of pET21d in-frame with the C-terminal hexa-His tag (PFC0785c) or into the NdeI/XhoI sites of pET28a in-frame with the N-terminal hexa-His tag (PF10_0029 and PFE0565w).
For IFP expression within P. falciparum parasites, IFP was subcloned by PCR into the XhoI/AvrII sites of an episomal overexpression (EOE) vector that contains the Hsp86 promoter sequence in the 5′-UTR and appends IFP with a C-terminal GFP tag (27). The EOE vector also contained a separate yeast dihydroorotate dehydrogenase resistance cassette for positive selection with DSM1 (28). To target expression of IFP to the parasite apicoplast or mitochondrion, the leader sequences of acyl carrier protein (residues 1–60) (29) or citrate synthase (residues 1–120) (30) were cloned by PCR from P. falciparum (3D7) cDNA into the XhoI site in-frame with the IFP gene previously cloned into the EOE vector.
The PfHO K114H and AtHO2 R88H mutants were prepared by QuikChange® II site-directed mutagenesis kit (Agilent Technologies; Santa Clara, CA) using the codon-optimized constructs described above. Sequences of all clones and mutants described above were confirmed by sequencing mini-prep DNA on Applied Biosciences 3130 and 3730 DNA Sequencers at the Protein and Nucleic Acid Chemistry Laboratory at the Washington University School of Medicine.
Production of 13C-Labeled Biliverdin and Bilirubin Internal Standards for Mass Spectrometry
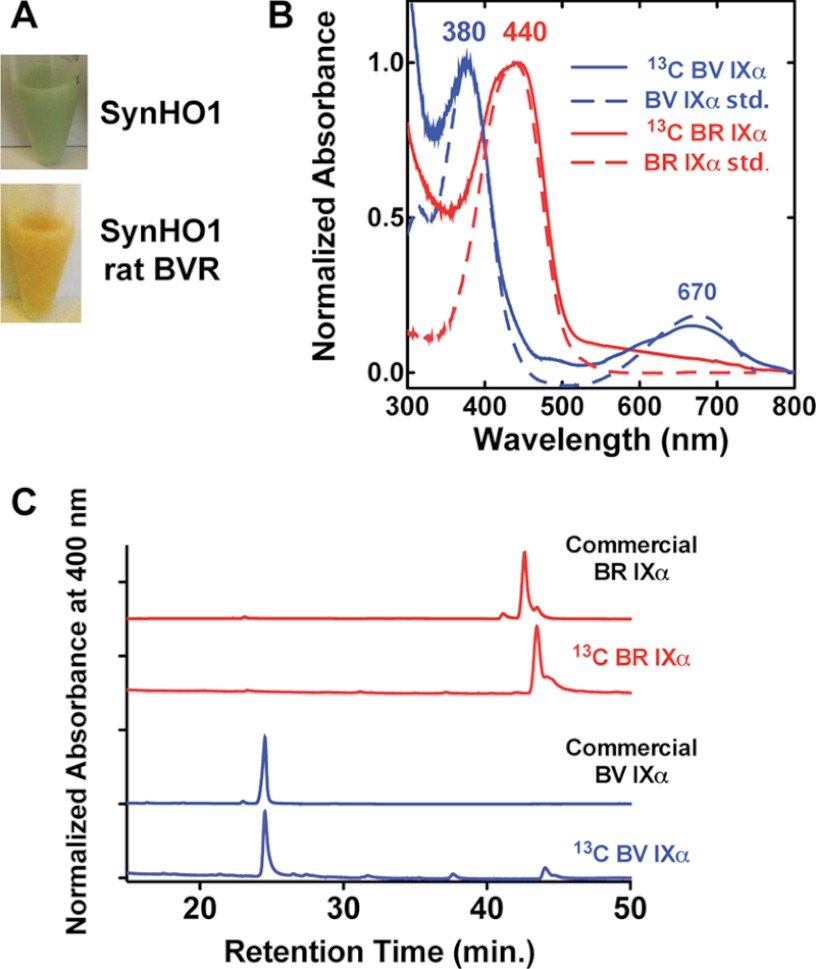
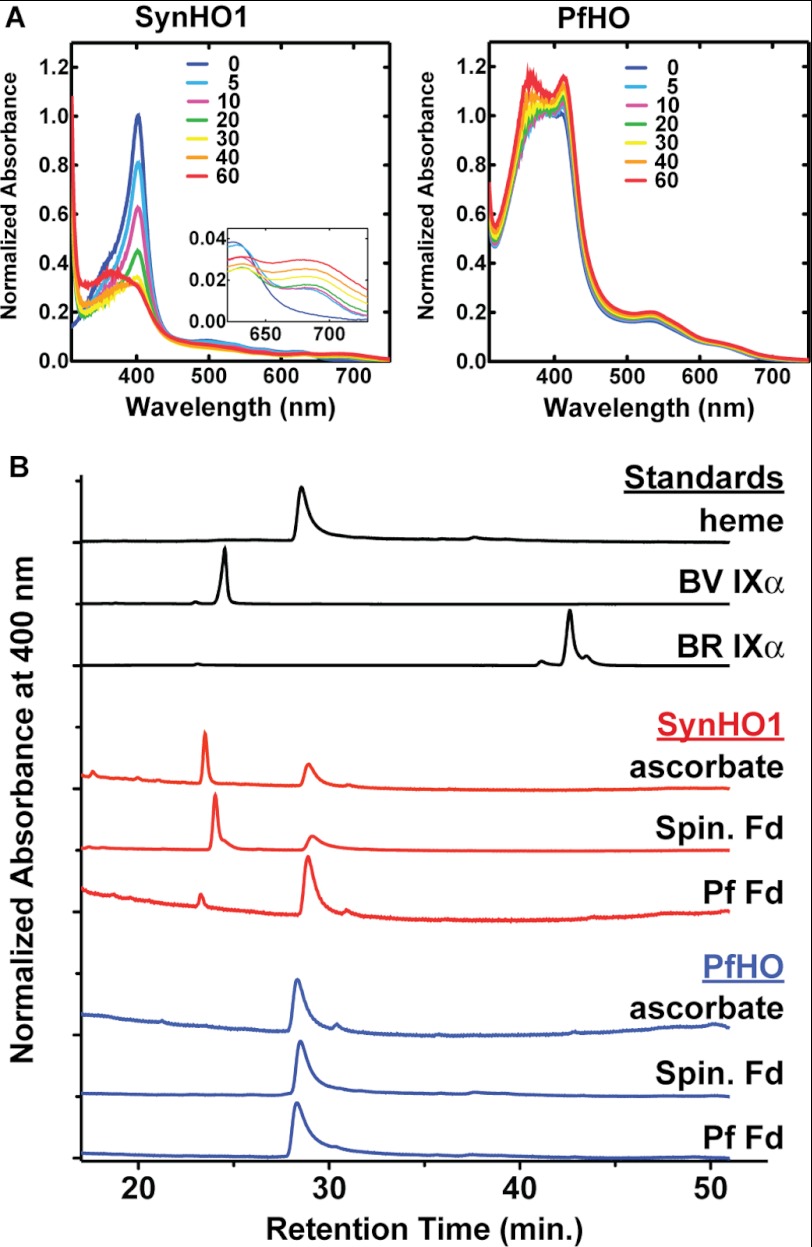
For in situ production of 13C-labeled BV and BR within E. coli, BL21/DE3 bacteria were transformed with SynHO1/pET22b (BV production) or SynHO1/pET22b and rat BVR/pET28a (BR production) and grown to saturation in an overnight LB culture supplemented with 50 μg/ml carbenicillin (pET22b) or 50 μg/ml carbenicillin and 30 μg/ml kanamycin (pET22b/pET28a). One ml of each overnight culture was diluted into 250 ml of M9 minimal medium (31) supplemented with 0.2% [13C]glycerol as the sole carbon source. Cells were grown at 37 °C for approximately 7 h to an A600 ∼0.5; protein expression was induced with 1 mm IPTG, and cells were grown a further 12–15 h at 37 °C prior to harvesting by centrifugation. Labeled BV and BR were extracted by resuspending pellets in 1 ml of DMSO, sonicating three times for 20 s (50% duty cycle, 50% power) on a Branson Ultrasonics (Danbury, CT) micro-tip sonicator, vortexing lysates three times for 1 min, isolating the DMSO-soluble supernatant by centrifugation, and passing it through a 0.45-μm syringe filter. The identity of the 13C-labeled BV and BR standards was confirmed by LC-MS/MS and by comparison of UV-visible absorbance and HPLC retention time on a reverse-phase C18 column to commercial BV and BR standards (Fig. 2). 13C labeling was >99% for both tetrapyrroles.
FIGURE 2.
Characterization of enzymatically produced 13C-labeled BV and BR mass spectrometry standards. 13C-Labeled BV IXα and BR IXα produced in E. coli by overexpression of SynHO1 or SynHO1 plus rat BVR turned bacteria the expected green or orange color due to accumulation of each tetrapyrrole (A). The DMSO-extracted tetrapyrroles had nearly identical UV-visible absorbance spectra (B) and reverse-phase C18 HPLC retention times (C) as commercial BV IXα and BR IXα standards.
LC-MS/MS Quantification of Biliverdin and Bilirubin in Infected Versus Uninfected Red Blood Cells
Human red blood cells that had been washed and separated to remove serum and retain only mature RBCs were obtained from the Blood Bank at Barnes-Jewish Hospital (St. Louis, MO) and used for analysis of uninfected RBCs and for parasite culturing. Uninfected RBC samples containing ∼1 × 109 human RBCs were prepared by pipetting 200 μl of a 50% hematocrit RBC solution in RPMI 1640 medium into a 15-ml conical tube, gently centrifuging, and aspirating off the supernatant. Infected RBC samples containing ∼1 × 109 parasitized RBCs were prepared by passing 6 × 12-ml plates of an asynchronous P. falciparum 3D7 culture at 2% hematocrit and ∼20% parasitemia over a MACS magnetic separation column (Miltenyi Biotec) to remove uninfected RBCs, eluting the retained trophozoite and schizont population into a 15-ml conical tube, centrifuging the sample 3 min at 1000 rcf, and aspirating off the supernatant. Positive control samples contained a 50-μl pellet of an equal mixture of BL21/DE3 E. coli bacteria transformed with SynHO1/pET22b or SynHO1/pET22b and rat BVR/pET28a for which protein expression had been induced (1 mm IPTG) for 4 h at 37 °C in LB medium with appropriate antibiotics. Each sample above was resuspended in 500 μl of DMSO and supplemented with 0.1 pmol of 13C-labeled BV and 1 nmol of 13C-labeled BR internal standards, giving a total sample volume of 700 μl. A negative control (blank) sample was prepared that contained only DMSO and internal standards. Each sample was vortexed three times for 1 min, sonicated on ice three times for 15 s (50% duty cycle, 50% power) on a Branson Ultrasonics (Danbury, CT) micro-tip sonicator, vortexed three times for 1 min, and centrifuged 10 min at 13,000 rpm (16,000 rcf), and the supernatant was transferred to clean tubes.
LC-MS/MS analyses were carried out using an AB Sciex API-4000 QTrap tandem mass spectrometer running Analyst version 1.5.1 (AB Sciex, Foster City, CA) coupled to a Shimadzu UFLC (Kyoto, Japan) and operated in the positive ion mode using the Turbo V ESI ion source, as described previously (32). Briefly, 1 μl of each sample was injected onto a Betasil C18 column (50 mm × 2.1 mm × 5 μm) with a flow rate of 0.4 ml/min. The mobile phase was held constant at 95% solvent A (0.1% aqueous formic acid) and 5% solvent B (100% acetonitrile containing 0.1% formic acid) for 1 min, increased via linear gradient from 5 to 98% B over 8 min, and then maintained at 98% B for 10 min. The column was allowed to equilibrate under starting conditions for 3 min before injecting the next sample. The ion spray voltage was set to 5000 V. The heater temperature was 400 °C. The declustering potential, nebulizer gas (G1), auxiliary gas (G2), and collision energy were set at 110, 35, 35, and 35 V, respectively. Fragmentation profiling of analytes was carried out using multiple reaction monitoring in the positive ion mode with precursor and product ions at their experimentally determined optimal values of m/z 584.5 and 299.2 for bilirubin, 617.7 and 316.4 for 13C-labeled bilirubin, 583.4 and 297.2 for biliverdin, and 616.5 and 314.2 for 13C-labeled biliverdin. Isolation widths for precursor and product ions were 0.7 and 1.1 m/z units, respectively. Collision energies were set at 30 for bilirubin and 35 for biliverdin, and analyte ion intensities were quantified by peak integration using the Analyst software and normalized to the peak area for the BV or BR internal standard in each sample. Absolute BV and BR concentrations in uninfected RBCs were estimated by multiplying the average normalized peak area detected for each analyte (0.066 for BR and 0.481 for BV) by the moles of 13C-labeled BV or 13C-labeled BR added to each sample (given above) and dividing by 1 × 109 RBCs and by 90 fl, the average volume of an RBC (33). We estimated the detection limit of our assay as 0.005 of the peak intensities for the BV and BR internal standards, which corresponds to absolute limits of detection of 10−18 mol of BV and 10−14 mol of BR.
Parasite Culturing and Transfection
Asynchronous P. falciparum 3D7 parasites were cultured in human red blood cells (described above) in RPMI 1640 medium with 0.5% Albumax as described previously (34, 35). For transfections, parasites were incubated with 100 μg of midi-prep DNA of each of the EOE constructs described above encoding IFP-GFP, ACPL-IFP-GFP, and CSL-IFP-GFP and transfected as described previously (36). Plasmid-containing parasites expressing the yeast dihydroorotate dehydrogenase marker were selected using 2 μm DSM1 (28).
Microscopy
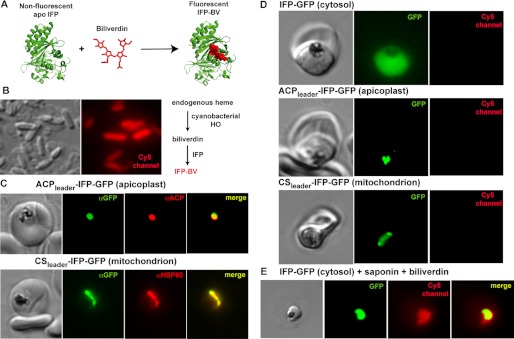
Images of live E. coli bacteria and live and fixed P. falciparum parasites were acquired on an Axio Imager.M1 epifluorescence microscope (Carl Zeiss Microimaging, Inc.) equipped with a Hamamatsu ORCA-ER digital CCD camera and running Axiovision 4.8 software, as described previously (37). Nuclei of live bacteria or parasites were stained by adding 5 μm Hoechst 3342 to culture aliquots immediately prior to mounting on a microscope slide. Immunofluorescence images of fixed parasites were acquired by fixing parasites as described previously (30, 38) and staining with a 1:500 dilution of polyclonal goat anti-GFP 5450 antibody (Abcam) and either a 1:5000 dilution of polyclonal rabbit anti-ACP antibody (38) or a 1:500 dilution of polyclonal rabbit anti-HSP60 antibody (38), followed by staining with 1:500 dilutions of Alexa Fluor 488-conjugated chicken anti-goat and Alexa Fluor 555-conjugated donkey anti-rabbit secondary antibodies (Molecular Probes). Fixed parasite nuclei were stained by mounting parasite-coated coverslips using Prolong Gold Antifade Reagent with DAPI (Molecular Probes, Inc.). As a positive control that episomally expressed IFP within parasites was competent to bind biliverdin and give detectable fluorescence on the Cy5 channel, RBCs infected with parasites expressing IFP-GFP were permeabilized by incubation for 10 min in 0.04% saponin supplemented with 100 μm biliverdin. After 10 min, parasites were mounted on coverslips for imaging. Sample images were acquired on the bright field, GFP, DsRed, and Cy5 channels and processed using the Axiovision 4.8 software and Adobe Photoshop CS4. Images on the GFP and Cy5 channels were processed with identical brightness and contrast settings.
Sequence Analysis, Sequence Alignment, and Structural Modeling of PfHO
BLAST and CS-BLAST analyses of PfHO were performed at their websites. The HO-like domain of PfHO (XP_001347401.2, residues 84–305) was aligned to SynHO1 (P72849), HuHO1 (NP_002124.1), HuHO2 (NP_002125.3), RnHO1 (NP_036712.1), CdHO (CAE50198.1), and the HO domains of AtHO1 (AEC07872.1, residues 55–282) and AtHO4 (AEE33533.1, residues 57–299) using ClustalW as implemented within the MegAlign module of Lasergene 10.0 (DNASTAR, Madison, WI). This alignment was used by MegAlign as input to generate the phylogenetic tree shown in Fig. 5B.
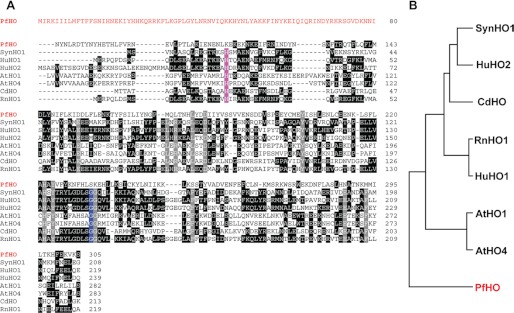
FIGURE 5.
Sequence analysis of PfHO relative to known heme oxygenases. A, ClustalW alignment of PfHO with known heme oxygenases from Synechocystis sp. 6803 (SynHO1), H. sapiens (HuHO1 and HuHO2), A. thaliana (AtHO1 and AtHO4), Corynebacterium diphtheriae (CdHO), and R. norvegicus (RnHO1). The amino acid leader sequence of PfHO is shown in red. Conserved residues in all seven known HOs are colored gray. The conserved proximal His residue that coordinates the bound heme and the conserved distal Gly residue are colored violet and blue, respectively. Identical residues in three or more proteins are in black. For simplicity, the chloroplast-targeting leader sequences of AtHO1 and AtHO4 and sequences of all proteins that extend beyond the C terminus of PfHO have been omitted. B, phylogenetic relationship of PfHO relative to the aligned heme oxygenases.
A structural model of the HO-like domain of PfHO (residues 84–305) was generated using the Rosetta Comparative and De Novo Modeling software as implemented on the Robetta Protein Structure Prediction Server (39) and aligned with the x-ray crystallographic structure of heme-bound SynHO1 (Protein Data Bank code 1WE1) (40) using MacPyMOL (41).
Bacterial Protein Expression
SynHO1, IFP, HuHO1, AtHO1, AtHO2, PfHO, and PfFd were expressed in BL21/DE3 E. coli bacteria by transformation with mini-prep plasmid DNA and overnight growth and selection at 37 °C in a 10-ml LB culture supplemented with either 50 μg/ml carbenicillin (pET21d and pET22b) or 30 μg/ml kanamycin (pET28a). For co-expression of SynHO1, HuHO1, AtHO1, AtHO2, and PfHO with IFP, bacteria were transformed with plasmid DNA encoding each desired protein but with orthogonal resistance cassettes (e.g. SynHO1/pET21d plus IFP/pET28a) and selected with both carbenicillin and kanamycin. Overnight cultures were diluted 1:100 into LB supplemented with appropriate antibiotics and grown at 37 °C to an A600 ∼0.5. Protein expression was induced by addition of 1 mm IPTG and further growth of 4 h at 37 °C or overnight at 20 °C. Bacterial pellets were harvested by centrifugation, resuspended in 50 mm Tris·HCl (pH 8), 100 mm NaCl, and lysed by sonication as described above. Lysates were clarified by centrifugation. Soluble protein expression was assessed by anti-poly-His Western blot using an HRP-conjugated or unconjugated monoclonal mouse anti-poly-His antibody (Sigma) and visualization with either ECL-Plus chemiluminescence reagent (PerkinElmer Life Sciences) or an IR800-conjugated chicken anti-mouse secondary antibody and the Odyssey Imaging System (LI-COR Biosciences), respectively.
Protein Purification
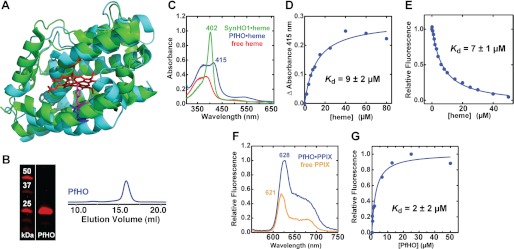
SynHO1, PfHO, and PfFd were expressed in 1.5-liter cultures of LB medium as described above. Bacterial pellets were isolated by centrifugation and resuspended in 25 ml of HisTrap buffer A (20 mm sodium phosphate (pH 7.5), 0.5 m NaCl, 5 mm imidazole). Bacteria were lysed by sonication 10 times for 30-s cycles using a Branson Ultrasonics cell disrupter equipped with a micro-tip and operated at 50% duty cycle and 50% power. Lysates were clarified by centrifugation for 30 min at 18,000 rpm (39,000 rcf) and passage through a 0.2-μm syringe filter. Clarified lysates were manually loaded over a 5-ml HisTrap HP metal chelation column (GE Healthcare) that had been preloaded with Ni2+ and equilibrated in HisTrap buffer A. The loaded column was connected to an Akta FPLC system and washed with 20 column volumes (CV) of buffer A using a flow rate of 5 ml/min while monitoring UV absorbance at 280 nm. The column was further washed with 5 CV of 5% HisTrap buffer B (buffer A supplemented with 0.5 m imidazole). The recombinant protein was eluted from the column using a linear gradient of 5–100% buffer B over 5 CV, followed by an isocratic flow at 100% B for 3 CV. Fractions with substantial absorbance at 280 nm were pooled and concentrated to ∼1 ml using an Amicon spin concentrator (Millipore) with a 10-kDa molecular mass cutoff. The concentrate was spin-filtered at 0.2 μm and injected over a 24-ml Superdex-200 gel filtration column that had been equilibrated in 50 mm Tris·HCl (pH 8), 250 mm NaCl. The protein was eluted using an isocratic flow at 0.5 ml/min, and fractions with substantial 280 nm absorbance were pooled and concentrated to ∼0.5 ml using Amicon spin concentrators. Typical yields were 10–15 mg of protein from a 1.5-liter culture. Protein concentrations were determined by absorbance at 280 nm using molar extinction coefficients of 21,890 m−1 cm−1 (SynHO1), 31,860 m−1 cm−1 (PfHO), and 8940 m−1cm−1 (PfFd) calculated from the ExPASy website (42). Protein purity was confirmed by the appearance of a single band by Coomassie-stained SDS-PAGE. Protein masses for SynHO1 and PfHO were confirmed by LC-MS. All three proteins migrated at the correct molecular weight by SDS-PAGE and had the expected properties for folded monomeric proteins.
UV-visible Absorbance and Fluorescence Spectroscopy
Fluorescence excitation and emission spectra were acquired for clarified bacterial lysates (excitation 630 nm, emission 730 nm) and protoporphyrin IX (excitation 400 nm, emission 620 nm) on a Cary Eclipse fluorescence spectrometer (Varian Inc.) using 10-nm slit widths and a PMT detector voltage setting of 800. UV-visible absorbance spectra were acquired on a DU 640 spectrophotometer (Beckman Coulter) using a 1-cm path length. For heme binding measurements, a 0.5-cm path length cuvette was used.
Measurement of Heme and Protoporphyrin IX Binding to PfHO
Heme binding to PfHO was measured in 50 mm Tris·HCl (pH 8) and 250 mm NaCl by acquiring UV-visible absorbance spectra (300–700 nm) of 0 to 80 μm heme in the presence or absence of 2 μm PfHO. Difference spectra were calculated by subtracting the free heme spectra from the heme + PfHO spectra. The absorbance difference at 415 nm was plotted as a function of heme concentration and fit with a quadratic binding equation using GraphPad Prism. Analysis of replicate experiments gave an average Kd of 9 ± 2 μm.
Heme binding was independently measured by monitoring fluorescence quenching (excitation 280 nm, emission 387 nm) of the endogenous Trp residues of 0.5 μm PfHO under identical solution conditions as above. The relative fluorescence was plotted as a function of heme concentration and fit with a quadratic binding equation in GraphPad Prism to determine an apparent Kd of 7 ± 1 μm.
Protoporphyrin IX (PPIX) binding to PfHO was measured in 50 mm Tris·HCl (pH 8) and 250 mm NaCl by acquiring fluorescence emission spectra (excitation 400 nm) of 100 nm PPIX in the presence of 0–50 μm PfHO. The fluorescence increase at 628 nm was plotted as a function of PfHO concentration and fit with a quadratic binding equation in GraphPad Prism. Analysis of replicate experiments gave an average Kd of 2 ± 2 μm.
In Vitro Heme Degradation Assays
The heme degradation activities of PfHO and SynHO1 were assessed using ascorbate, spinach ferredoxin (Spin. Fd), or recombinant P. falciparum ferredoxin (PfFd) as the reductant and monitored by UV-visible absorbance spectroscopy or analytical HPLC, as described previously with minor modifications (43, 44). For reactions using ascorbate, 100-μl samples contained 10 μm heme, 20 μm SynHO1 or 100 μm PfHO, 10 mm ascorbate, 1 μm catalase, 50 mm Tris·HCl (pH 8), and 100 mm NaCl. For reactions using ferredoxin, 100-μl samples contained 10 μm heme, 20 μm SynHO1 or 100 μm PfHO, 10 μm Spin. Fd or PfFd, 0.025 units/ml Spin. FNR, 100 μm NADPH, 1 μm catalase, 50 mm Tris·HCl (pH 8), and 100 mm NaCl. Reactions were also carried out with the addition of 1 mm deferoxamine, and identical results (i.e. no detectable heme degradation for PfHO) were observed in all cases. Reactions were initiated by adding the ascorbate or ferredoxin reductant to sample mixtures containing the other components. For UV-visible assays, the reaction mixture was transferred to a cuvette, and spectra were acquired at the indicated time points. For HPLC assays, reaction mixtures were incubated for 1–2 h at room temperature prior to injection.
HPLC Analysis of Heme Degradation Reaction Products
20 μl of each reaction mixture above or 10 μm solutions of commercial standards (heme, BV, and BR) were injected over a 150-mm Vydac analytical C18 reverse-phase HPLC column (5 μm bore size, 2.1 mm inner diameter) connected to a Waters HPLC system (626 Pump, 6002 Controller, and 486 Absorbance Detector) controlled by the Waters Empower2 software. A constant flow rate of 0.3 ml/min was used, and absorbance was monitored at 400 nm. Upon injection, the system was maintained at 80% buffer A (1% acetic acid in water) and 20% B buffer (50% acetone, 38% ethanol, 11% water, 1% acetic acid) for 2 min, followed by a linear gradient of 20–100% B over 40 min, isocratic flow at 100% B for 4 min, and a linear gradient of 100 to 20% B over 2 min. The system was equilibrated at 20% B for ≥10 min prior to each new injection.
RESULTS
Measurement of Biliverdin and Bilirubin Levels in Infected Versus Uninfected Erythrocytes by LC-MS/MS
Mature red blood cells do not contain the human HO1 or HO2 proteins found in other human cell types (45–47) and thus lack an enzymatic pathway for heme degradation. If blood stage P. falciparum parasites express enzymes that degrade heme to BV or BR, then we expected parasitized RBCs to contain increased levels of one or both of these metabolites relative to uninfected RBCs. To directly measure and compare BV and BR levels in parasite-infected versus uninfected RBCs, we developed a sensitive LC-MS/MS assay to specifically quantify these tetrapyrroles.
We first biosynthetically generated 13C-labeled BV and BR to serve as internal mass spectrometry standards by heterologous overexpression of the cyanobacterial HO1 from Synechocystis sp. PCC6803 (SynHO1) alone or in combination with rat BVR in E. coli grown in minimal media with [13C]glycerol as the sole carbon source. SynHO1 has previously been shown to be active when expressed within E. coli and to enzymatically produce BV from the endogenous heme produced by these bacteria, which ordinarily do not express an HO (43, 48, 49). Active production of BV and BR in E. coli upon expression of these enzymes was signaled by the green and orange color of the respective bacteria (Fig. 2A). The 13C-labeled BV and BR produced in this fashion were isolated by sonication and extraction of bacterial pellets in DMSO, and the clarified extracts were used without further purification. Analysis of the extracted tetrapyrroles by UV-visible absorbance, HPLC retention time, and LC-MS/MS confirmed uniform 13C labeling and identical properties to commercial BV IXα and BR IXα standards (Fig. 2, B and C).
To quantify BV and BR levels in complex cellular extracts, we used LC-MS/MS to specifically measure these tetrapyrroles based upon their characteristic collisionally induced dissociation ion fragmentation pathways. Each compound's product ion spectrum was determined using authentic BV and BR commercial standards, and the dominant peaks corresponded to previously reported values (50, 51). 13C-Labeled internal standards exhibited identical chromatographic retention times as commercial BV and BR, with precursor and product ions shifted by the m/z values expected following 13C enrichment. The dominant product ion for each standard, corresponding to dissociation of the aliphatic bridge between the two inner pyrrole moieties of BV and BR, was chosen to construct a targeted LC-multiple reaction monitoring analysis. Quantification of each analyte was then achieved by determining the ratio of its LC-multiple reaction monitoring peak area to that of its co-eluting 13C-labeled internal standard. To validate our detection of these analytes, we measured BV and BR levels in negative and positive control samples that consisted of 13C internal standards added to DMSO alone or to DMSO extracts of E. coli bacteria expressing SynHO1 or SynHO1/Rat BVR in rich media. Negligible amounts of unlabeled BV and BR were detected in the mock sample, and robust peaks were detected for BV and BR in extracts of bacteria expressing the active HO and HO/BVR enzymes (Fig. 3A).
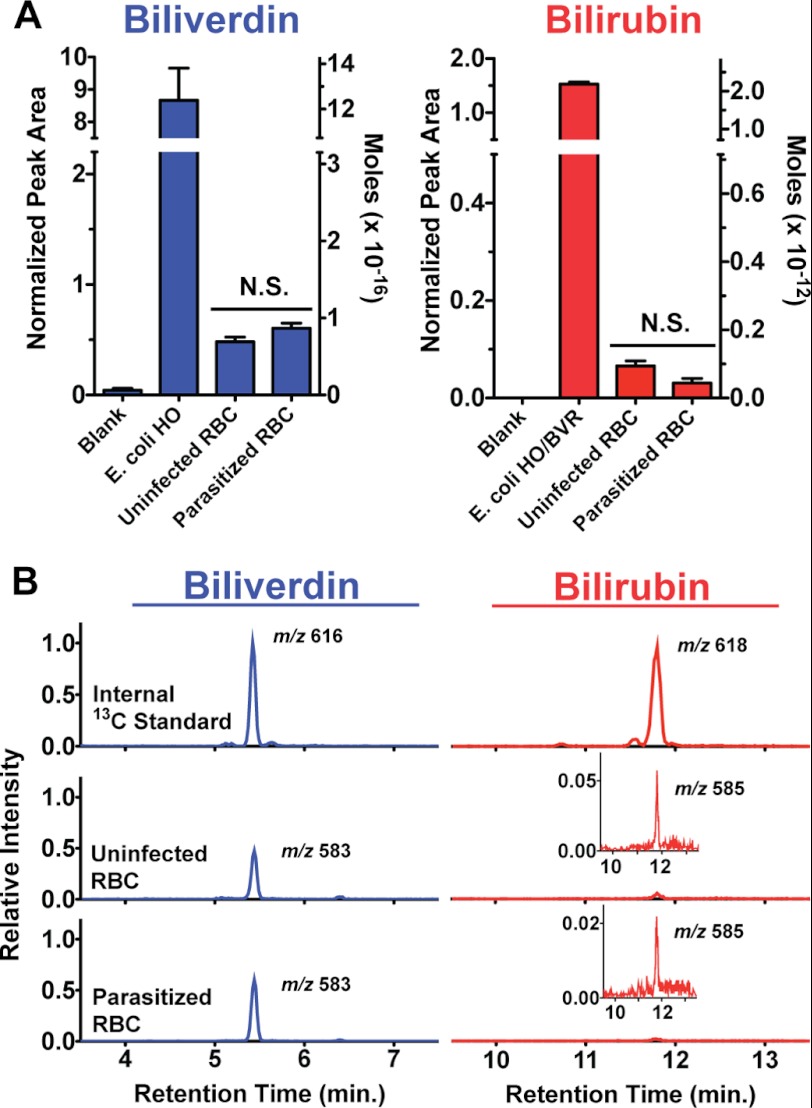
FIGURE 3.
LC-MS/MS measurement of biliverdin and bilirubin in parasite-infected versus uninfected red blood cells. Cells were extracted in DMSO, known amounts of 13C-labeled BV and BR internal standards were added, and samples were analyzed by LC-MS/MS. The integrated area of the BV and BR peak detected for each sample was normalized against the respective peak area for each internal standard. The average normalized peak area and standard error from four independently prepared and processed replicates are shown in A, and representative mass spectra with indicated [M + H]+ masses are shown in B. N.S. = not significantly different by unpaired t test (p = 0.11 [BV], 0.05 [BR]). The insets in B are expanded y axis views of the detected peaks. Estimated molar amounts of each analyte detected in injected samples have been included on the right y axis in A to facilitate comparison between BV and BR levels, as the normalized peak areas for the two analytes cannot be directly compared due to differing amounts of BV and BR internal standards added to samples (see “Experimental Procedures”).
To test whether P. falciparum-infected erythrocytes have altered BV or BR levels relative to uninfected erythrocytes, we prepared samples that contained ∼1 × 109 cells of each type. Infected RBC samples contained an asynchronous mixture of P. falciparum (3D7)-parasitized cells that had been passed over a magnetic column to remove uninfected RBCs and enrich to ≥95% parasitemia. A known amount of internal standards was added to each sample (four replicates of each), which were sonicated and extracted in DMSO, clarified by centrifugation, and analyzed by LC-MS/MS. DMSO was used for extractions based on literature reports (52, 53) and our observation of robust BV and BR solubility in this solvent.
Measurable amounts of both BV and BR were detected in uninfected RBC samples (Fig. 3, A and B). Based on the amount of each 13C-labeled standard added to samples, the number of RBCs in each sample, and an average RBC volume of 90 fl (33), our measurements can be used to estimate that a mature RBC contains 1 μm BR and 1 nm BV. The larger estimated RBC concentration for BR relative to BV is consistent with the absence of human HO1 or HO2 but the presence of human BVR in mature RBCs (45–47). These levels may remain from an earlier stage of erythrocytic development or reflect uptake from human serum, which contains ∼10 μm BR (54).
The parasitized RBC samples contained BV and BR levels that were indistinguishable from those of uninfected RBC samples (Fig. 3, A and B). On the basis of these results, we conclude that P. falciparum infection of erythrocytes results in insignificant changes in the levels of BV and BR detected within RBCs, as expected for the absence of enzymatic heme degradation within parasites.
Targeted Expression of a Fluorescent Biliverdin Biosensor in Live Parasites
To further test the conclusion that parasites do not degrade heme to BV and to extend our analysis to live parasites, we episomally expressed a GFP-tagged version of the fluorescent biliverdin-binding protein, IFP, within parasites. IFP is an engineered, monomeric version of the chromophore-binding domain of the bacteriophytochrome from Deinococcus radiodurans that has been mutationally altered to increase its fluorescence upon binding and covalent attachment of its cognate ligand, biliverdin IXα (Fig. 4A) (55, 56). The fluorescence excitation and emission maxima of 684 and 704 nm, respectively, for mature IFP-BV make it readily detectable on the Cy5 channel of a fluorescence microscope, and IFP remains competent to bind BV and fluoresce when fused to other proteins (55).
FIGURE 4.
Targeted episomal expression of a fluorescent biliverdin biosensor in P. falciparum parasites. A, unliganded and nonfluorescent IFP covalently binds free biliverdin to generate mature IFP-BV, whose fluorescence can be detected on the Cy5 channel. This scheme was constructed using the BV-bound x-ray structure of the D. radiodurans chromophore binding domain (Protein Data Bank code 3S7O), from which IFP is derived. B, heterologous co-expression of SynHO1 and IFP in E. coli enables in situ fluorescence detection of the BV product of SynHO1 within live bacteria imaged on the Cy5 channel of a fluorescence microscope. C, immunofluorescence microscopy of fixed parasites confirms apicoplast and mitochondrial targeting of episomally expressed IFP bearing a C-terminal GFP tag and the N-terminal leader sequence from either acyl carrier protein (ACP) or citrate synthase (CS), respectively. Parasites were stained with monoclonal αGFP and either a polyclonal αACP (apicoplast) or αHSP60 (mitochondrion) antibodies. The epitope recognized by the αACP antibody is distinct from the ACP leader sequence. D, fluorescence images of live parasites episomally expressing IFP within the parasite cytoplasm, apicoplast, or mitochondrion. E, fluorescence image of a live parasite expressing cytosolic IFP-GFP. The parasite was released from its host RBC via saponin treatment and incubated in exogenous biliverdin. Images on GFP and Cy5 channels were processed with identical brightness and contrast settings.
To validate our ability to use IFP as a fluorescent reporter of in situ BV production within live cells, we transformed E. coli bacteria with a pET28a plasmid encoding IFP alone or in combination with a pET21d plasmid encoding SynHO1 and induced protein expression by adding 1 mm IPTG. Bacteria expressing IFP alone gave no detectable fluorescence on the Cy5 channel, as expected for the absence of HO activity and BV production by most E. coli strains (57). In contrast, robust fluorescent signal for mature IFP-BV was readily detected on the Cy5 channel within bacteria co-expressing both IFP and SynHO1 (Fig. 4B). Although BV stably accumulates in SynHO1-expressing bacteria due to the absence of additional BV-processing enzymes, any BV produced by HO activity within parasites might not accumulate due to additional enzymatic conversion of BV into downstream metabolites such as BR. However, IFP has previously been shown to give detectable IFP-BV signal in human cell lines (55), where BV is transiently produced by human HO activity but does not accumulate due to processing by human BVR and other enzymes (13). Thus, we expected IFP to be sensitive to even transient levels of any BV that might be produced within parasites.
To test for BV production within live P. falciparum (3D7) parasites, we episomally expressed IFP within the parasite cytosol, mitochondrion, or apicoplast, subcellular compartments previously proposed as possible venues for HO activity within parasites (22), and we evaluated live parasites for detectable IFP-BV signal on the Cy5 channel. Targeted IFP expression was carried out by transfecting parasites with an EOE vector containing the Hsp86 promoter upstream of the IFP gene that had been cloned in-frame with a C-terminal GFP tag, which served as a reporter of protein expression and localization (27). For apicoplast and mitochondrial targeting, the leader sequence of either acyl carrier protein or citrate synthase, which had previously been shown to target episomal protein expression to the parasite apicoplast (29) or mitochondrion (30), respectively, was appended to the 5′ end of the IFP gene. The correct targeting of IFP to these two organelles was confirmed by fixing parasites and co-staining with anti-GFP and either anti-ACP (apicoplast marker) or anti-HSP60 (mitochondrial marker) antibodies (Fig. 4C).
Robust GFP fluorescence was detected in live parasites in the appropriate subcellular compartment for all three constructs, indicating protein expression in all three lines. However, no fluorescence for mature IFP-BV was detectable on the Cy5 channel in any parasite (Fig. 4D). To confirm that IFP was indeed expressed and competent to bind BV and give detectable fluorescence, we permeabilized infected RBCs with 0.04% saponin, a mild detergent, and added 100 μm BV to the culture medium. Under these conditions, we detected similar fluorescence signals on both the GFP and Cy5 channels for cytosolic IFP (Fig. 4E), as expected for IFP-BV formation and confirming proper functioning of the IFP reporter. Our results provide no evidence for BV production within live parasites and further support our conclusion based on the mass spectrometry results above that parasites do not enzymatically degrade heme.
Sequence Analysis of the HO-like P. falciparum Protein, PfHO
P. falciparum parasites have historically been assumed to lack HO enzymes (1, 15–21), based on the absence of parasite genes that show strong sequence identity to known HO enzymes from other organisms (58) and based on the recognition that most of the heme generated by hemoglobin catabolism is not degraded but incorporated into hemozoin (5). A recent study, however, identified a P. falciparum gene (PF3D7_1011900, formerly PF10_0116) encoding an HO-like protein (termed PfHO) that shows modest sequence similarity to a known HO from A. thaliana, AtHO4 (22). On the basis of in vitro assays performed with the recombinant HO-like domain of PfHO and recombinant P. falciparum ferredoxin, it was proposed that PfHO catalyzes the degradation of heme to bilirubin (22). A subsequent study of recombinant full-length PfHO (amino acids 1–305) by a different group concluded that PfHO catalyzes degradation of heme to biliverdin (23). To evaluate whether PfHO is indeed a heme-degrading enzyme, we first analyzed its sequence features and then tested the ability of recombinant PfHO to bind and degrade heme.
The PfHO gene is predicted to encode a 305-amino acid protein, and sequence analysis using BLAST (59) or CS-Blast (60) indicates substantial homology to hypothetical proteins from other Plasmodium species. However, only sequence beyond the first 100 amino acids shows low level homology to annotated HO proteins from more distantly related organisms. Further sequence analysis using the PATS and PlasmoAP tools on the PlasmoDB website suggests that residues 1–100, whose sequence is not HO-like and shares homology only with other Plasmodium spp. proteins, may constitute a leader sequence that confers subcellular targeting within the parasite (Fig. 5A). The PATS algorithm (61) identifies PfHO as a likely apicoplast-targeted protein, but PlasmoAP (62) returns an ambiguous verdict as follows: residues 20–100 have expected features of an apicoplast-targeting transit peptide, but the algorithm is uncertain whether residues 1–20 constitute a signal peptide that targets the nascent protein to the secretory system for routing to the apicoplast.
To compare the sequence of PfHO to known HO enzymes from diverse organisms and to assess whether PfHO possesses the conserved residues expected of an active HO, we used ClustalW to align the HO domain of PfHO and known bacterial, animal, and plant HO enzymes (6, 43, 63), including the previously aligned AtHO4 (Fig. 5A). Among these eight proteins, PfHO has the most divergent sequence and phylogenetically groups apart from the other seven (Fig. 5B). According to this alignment, there are 14 residues that are conserved between all seven of the known HO enzymes. These conserved residues include the proximal His side chain (Fig. 5A, violet) that is used by all known HO enzymes to coordinate the iron of the bound heme and a distal Gly residue (blue) that is positioned directly above the bound heme and that kinks the distal helix to position active site residues on this face and direct hydroxylation of the α-meso carbon of heme (6, 7, 64).
These two highly conserved residues are conspicuously absent in the PfHO sequence, which aligns with a Lys residue in place of the proximal His ligand and a Ser in place of the Gly. Indeed, a structural model of PfHO generated by the Rosetta Protein Structure Prediction software (39), when superimposed with the SynHO1 x-ray crystal structure, suggests that no His or other residue within the PfHO active site is appropriately positioned to serve as a proximal ligand to the iron of bound heme (Fig. 6A). On the basis of these sequence differences from known HO enzymes, we considered PfHO unlikely to be an active HO.
FIGURE 6.
Heme binding by PfHO versus SynHO1. A, structural alignment of the x-ray crystallographic model of SynHO1 containing bound heme (green, Protein Data Bank code 1WE1) with the Rosetta-derived model of the HO domain of PfHO (cyan) (root mean square deviation = 1.4 Å). The bound heme is shown in red; the distal Gly residue and the proximal His ligand of SynHO1 are shown in orange and violet, respectively, and the aligned Lys residue in PfHO is shown in blue. B, Coomassie-stained SDS-polyacrylamide gel and gel filtration chromatogram of purified PfHO. C, UV-visible absorbance spectrum of 10 μm heme free in solution (red) or bound to 20 μm SynHO1 (green) or 200 μm PfHO (blue). D, heme binding to 2 μm PfHO leads to a saturable absorbance increase at 415 nm. Plotting this increase as a function of heme concentration and fitting with a quadratic binding equation (R2 = 0.98) gives an apparent dissociation constant (Kd) of 9 ± 2 μm. E, heme binding to PfHO leads to quenching of endogenous Trp fluorescence at 387 nm. Fitting this fluorescence decrease with a quadratic binding equation (R2 = 0.99) gives an apparent affinity of 7 ± 1 μm. F, fluorescence emission spectrum (excitation 400 nm) of 10 μm PPIX free in solution (orange) or bound to 20 μm PfHO (blue). G, binding of 0.1 μm PPIX to PfHO leads to a saturable increase in PPIX fluorescence intensity. Plotting the fluorescence increase at 628 nm versus PfHO concentration and fitting with a quadratic binding equation (R2 = 0.97) yields an apparent Kd of 2 ± 2 μm.
Functional Tests of Heme Binding and Degradation by PfHO
To directly test the ability of PfHO to bind and degrade heme in vitro, we recombinantly expressed the HO domain of PfHO in bacteria and purified it to homogeneity using metal chelation and size-exclusion chromatography. We observed robust, soluble expression, and the protein eluted from a gel filtration column as a single symmetric peak at the expected retention time for a folded, monomeric 28-kDa protein (Fig. 6B). We simultaneously expressed and purified SynHO1 to serve as a positive control for heme binding and degradation by a known HO and the apicoplast-targeted P. falciparum ferredoxin (PfFd) to test its proposed ability to support HO-catalyzed heme degradation. Both proteins were purified to homogeneity and had the expected HO and Fd properties previously reported for these two recombinant proteins (43, 65).
All known HO enzymes bind heme using a conserved His-heme interaction that gives rise to an intense Soret peak in UV absorbance spectra similar to the 402-nm maximum observed for SynHO1 (Fig. 6C) (43, 44). Heme binding to PfHO, however, is signaled by a broad increase in UV-visible absorbance and a minor peak at 415 nm rather than a single intense Soret peak, consistent with PfHO's lack of a proximal His ligand and the suggestion based on modeling that the PfHO active site is not optimized to coordinate the iron of bound heme (Figs. 5A and 6A). Plotting the observed absorbance increase at 415 nm as a function of PfHO concentration and fitting with a quadratic binding equation gives an apparent dissociation constant (Kd) of 9 μm (Fig. 6D), indicating modest affinity and consistent with prior reports (22, 23). A nearly identical heme affinity was determined by measuring quenching of PfHO tryptophan fluorescence upon heme binding (Fig. 6E).
To further test the ligand-binding preferences of PfHO, we acquired fluorescence spectra of the heme precursor, PPIX, in the absence and presence of excess PfHO (Fig. 6F). Similar to prior studies of protein-bound PPIX (66), we observed an increase in fluorescence intensity and a shift in emission maximum from 621 to 628 nm upon PPIX binding to PfHO. Fitting this fluorescence increase as a function of PfHO concentration yielded an apparent affinity (Kd) of 2 μm for PPIX binding (Fig. 6G). The tighter observed binding of PPIX versus heme to PfHO further suggests that the binding pocket of PfHO is not optimized for coordination of heme.
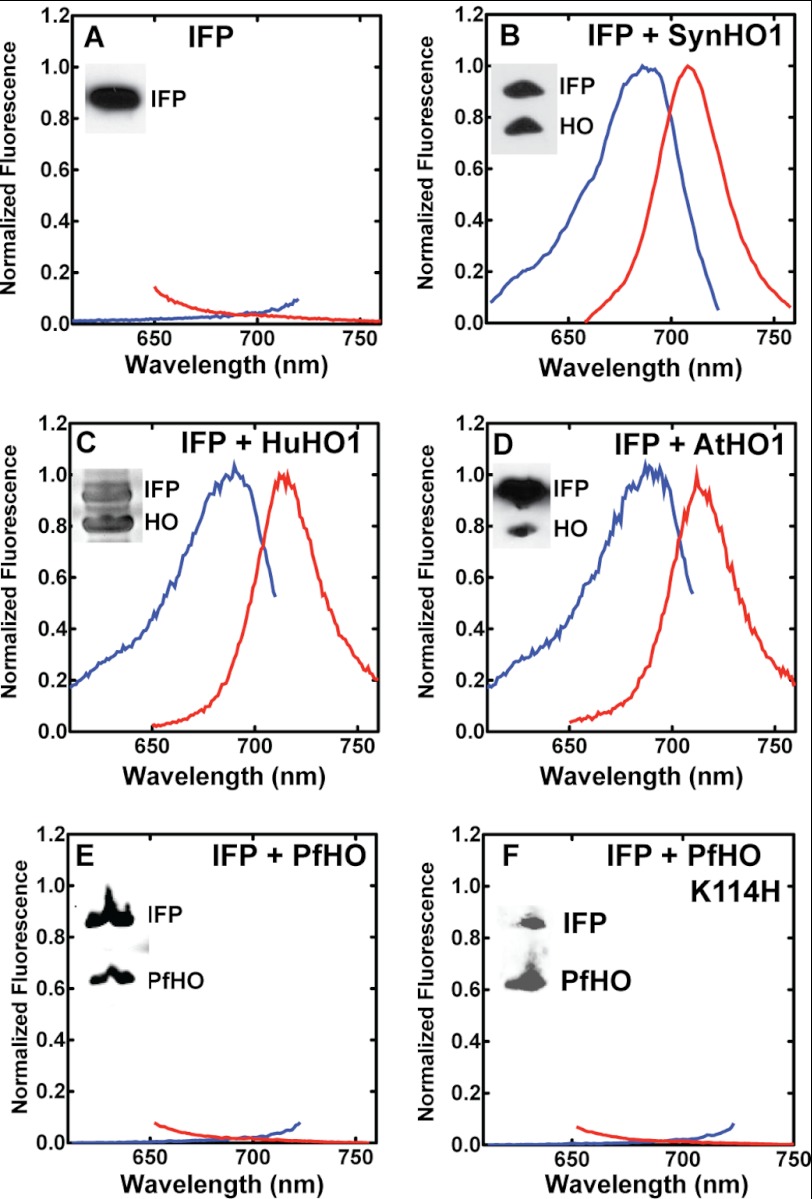
HO enzymes from many diverse organisms are enzymatically active when heterologously expressed within E. coli (48, 63, 67, 68), and co-expressions with the BV-binding domain of the D. radiodurans phytochrome (63) and its IFP derivative (55) have previously been used as sensitive reporters of recombinant HO activity in bacteria (shown schematically in Fig. 4B). As an initial test of the ability of PfHO to catalyze heme degradation in comparison with known HOs, we co-expressed IFP with PfHO, SynHO1, human HO1 (HuHO1), and A. thaliana HO1 (AtHO1) in E. coli and tested the clarified lysates for the expected fluorescence excitation and emission peaks of IFP-BV as a reporter of in situ conversion of endogenous heme to BV by these proteins. IFP expressed alone in bacteria gave no detectable fluorescence signal, but co-expression with SynHO1, HuHO1, and AtHO1 all gave the expected IFP-BV peaks for active BV production by these proteins (Fig. 7, A–D). In contrast, co-expression of PfHO and IFP gave no detectable fluorescence signal (Fig. 7E), indicating that PfHO does not catalyze BV production within E. coli and thus has divergent properties from these animal, plant, and bacterial HO enzymes.
FIGURE 7.
Testing HO activity in E. coli by co-expression with IFP. Fluorescence excitation (blue, emission at 730 nm) and emission (red, excitation at 630 nm) spectra of clarified bacterial lysates expressing IFP only (A), IFP and SynHO1 (B), IFP and HuHO1 (C), IFP and AtHO1 (D), IFP and PfHO (E), or IFP and PfHO K114H (F). The inset in each spectrum is an α-polyhistidine Western blot of lysate supernatants confirming soluble expression of the indicated proteins, except C, which is a Coomassie-stained gel of the clarified lysate as the HuHO1 was not His-tagged.
Although bacterial co-expression with IFP is a powerful assay of HO activity in vivo, it is limited by its exclusive sensitivity to BV production, the canonical product of HO-catalyzed heme degradation. PfHO, however, has previously been proposed by different groups to degrade heme to either BV (23) or BR (22). We note that the proposed conversion of heme to BR by PfHO is without biochemical precedent. Indeed, there is no known enzyme that can catalyze both an oxidative cleavage of the α-meso bridge and a reduction of the γ-meso bridge of the heme macrocycle (Fig. 1B), chemically distinct reactions that are canonically catalyzed by the discrete enzymes HO and BVR (7, 69). Because of these distinct product proposals for PfHO, we directly tested the heme-degrading properties of purified PfHO in vitro to assess formation of any PfHO-catalyzed breakdown products of heme, regardless of identity. These assays were performed in parallel with SynHO1 as a positive control for heme degradation by a known HO.
HO activity requires a reductant co-factor as an electron source, which is thought to be cytochrome P450 reductase for mammalian HOs and reduced ferredoxin for plant and bacterial HOs (7). In vitro, HOs appear to be highly promiscuous in the reductant co-factor they can utilize, including showing activity with ascorbate (43). Heme degradation by PfHO and SynHO1 was tested in parallel by UV-visible absorbance and analytical HPLC using ascorbate, spinach Fd, and the recombinant P. falciparum ferredoxin (PfFd) as co-factors, and 1 μm catalase was included in all reactions to prevent nonenzymatic coupled oxidation of heme (70).
Although the SynHO1·heme complex plus ascorbate showed the same time-dependent loss of the 402-nm Soret peak and rise in 670-nm absorbance previously reported for heme conversion to BV (43), we observed no time-dependent reductions in absorbance for the PfHO·heme complex over the same time scale (Fig. 8A). Reaction mixtures for each protein with each of the three reductant co-factors were analyzed by reverse-phase C18 HPLC and compared with commercial heme, BV, and BR standards for product identification (Fig. 8B). BV production by SynHO1 was detected with all three reductants, although only trace formation of BV was detected with PfFd. This lower activity with PfFd is consistent with its more positive redox potential (−266 mV) relative to spinach (−415 mV) and other plant ferredoxins (65, 71, 72) and suggests that PfFd is not optimized to support HO activity. In contrast to SynHO1, neither BV nor BR nor any other heme degradation products were detected for PfHO with any of the co-factors, supporting our prior conclusion that PfHO is not a heme-degrading enzyme.
FIGURE 8.
In vitro HO activity of purified SynHO1 versus PfHO. A, time-dependent (min) UV-visible absorbance changes of 10 μm heme bound to 20 μm SynHO1 or 100 μm PfHO. Samples contained 10 mm ascorbate, 1 μm catalase, 50 mm Tris·HCl (pH 8), and 100 mm NaCl. B, reverse-phase C18 HPLC analysis of tetrapyrrole standards or reaction products of 10 μm heme incubated with 20 μm SynHO1 or 100 μm PfHO in 50 mm Tris·HCl (pH 8), 100 mm NaCl, 1 μm catalase, and either (i) 10 mm ascorbate or (ii) 10 μm Spin. or P. falciparum (Pf) Fd, 0.025 units/ml spinach FNR, and 100 μm NADPH.
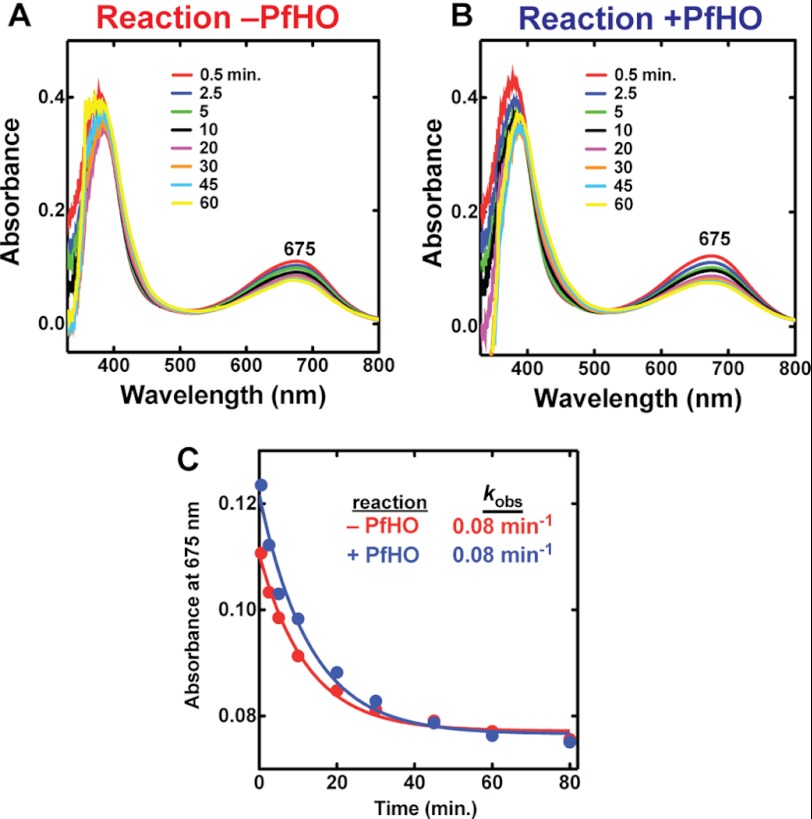
We also tested the previously proposed ability of PfHO to catalyze reduction of BV in the presence of PfFd (22). Although we observed the same time-dependent loss of BV absorbance at 675 nm as reported previously, this absorbance change occurred with an identical rate constant in the absence of PfHO, indicating that it was not catalyzed by PfHO (Fig. 9). To test whether the absence of a proximal His ligand in PfHO is alone responsible for the lack of detectable HO activity, we mutated Lys-114 to His and co-expressed this mutant with IFP in bacteria. As with WT PfHO, the K114H mutant had no detectable activity (Fig. 7F), indicating that additional sequence changes within the remainder of the protein also contribute to the lack of HO activity. In this regard, PfHO is similar to the HO2 protein from A. thaliana, which aligns with an Arg in place of the conserved proximal His in AtHO1 (73) and for which the WT and R88H proteins have no detectable HO activity (63).7
FIGURE 9.
UV-visible absorbance spectra of biliverdin in the presence or absence of PfHO. 10 μm BV was incubated with 10 μm PfFd, 0.025 units/ml Spin. FNR, 1 μm catalase, and 100 μm NADPH in 50 mm Tris·HCl (pH 8) and 100 mm NaCl in the absence (A) or presence (B) of 100 μm PfHO. C, the time-dependent loss (min) of BV absorbance at 675 nm observed in A and B was fit to an exponential decay equation (R2 = 0.98), giving an identical rate constant (kobs) independent of whether PfHO was present or absent.
Finally, we tested three additional P. falciparum proteins identified by BLAST search as showing low level sequence homology to AtHO4 or HuHO2 and retaining the conserved His at the expected sequence position of the proximal heme ligand. All three gave soluble expression in bacteria but displayed no detectable HO activity when co-expressed with IFP (supplemental Fig. 1). In summary, we find no evidence for enzymatic heme degradation by P. falciparum parasites.
DISCUSSION
Heme is a ubiquitous and essential biological co-factor that is required for diverse metabolic processes. Malaria parasites appear to require heme, but their cellular usage of it remains unclear (2). Indeed, the only known essential role for heme in parasites is incorporation into mitochondrial cytochromes to support electron transport, not for energy production but to recycle the ubiquinone co-enzyme of dihydroorotate dehydrogenase for pyrimidine biosynthesis (74). It remains unknown whether heme serves other essential roles within parasites, either as a metabolic end product itself, as a porphyrin precursor of downstream derivatives, or as a source or carrier of iron (2). Parasites require iron for multiple cellular processes, but it is unknown if they derive iron from host heme or scavenge it from other bioavailable iron sources in RBCs (75).
HO enzymes are broadly expressed by many organisms to degrade heme for disposal, to process it for metabolic utilization of the tetrapyrrole backbone, or to release and scavenge the protoporphyrin-bound iron (7, 13, 14). Considering the abundance of potentially cytotoxic heme liberated by hemoglobin digestion and the nutritional demand for iron from the host RBC, malaria parasites might have been expected to have evolved an HO pathway to degrade and detoxify heme. However, hemozoin formation, rather than heme degradation, has long been recognized to be the dominant mechanism of heme detoxification in malaria parasites. In this regard, Plasmodium parasites are similar to other hematophagous organisms, such as the blood fluke Schistosoma mansoni and the cow tick Boophilus microplus, which primarily detoxify excess dietary heme through sequestration rather than degradation (25, 76, 77).
Despite recognition of this central role for hemozoin formation, it has remained unclear and largely untested whether P. falciparum or other blood-feeding organisms also enzymatically process a small portion of the heme they encounter. Indeed, the kissing bug Rhodnius prolixus also detoxifies the prodigious heme encountered during human hemoglobin digestion by sequestering it as hemozoin (77), but recent work provides direct evidence that this insect also degrades some heme to biliverdin (51). Prior to our work, no direct tests of enzymatic heme degradation by Plasmodium spp. parasites had been reported in the literature. Our data from multiple experimental tests (Figs. 3 and 4) strongly suggest that P. falciparum parasites do not enzymatically degrade heme to biliverdin or bilirubin and thus lack a canonical HO pathway.
Recent studies have suggested that a fraction of host-derived heme escapes mineralization within the parasite's food vacuole and must be neutralized by alternative means. In the absence of an HO pathway, this trace heme may be oxidatively cleaved by nonenzymatic mechanisms proposed to operate within blood stage parasites (15, 16). Parasites do need a way to scavenge iron from the host, and many bacterial pathogens express HO enzymes to cleave and release heme-coordinated iron (14). Our data, however, suggest that iron acquisition by the parasite does not involve enzymatic degradation of host heme. Parasites may therefore satisfy their requirements by scavenging the iron released by trace nonenzymatic degradation of host heme or from other bioavailable sources within RBCs (75).
The absence of detectable metabolites from HO-catalyzed heme degradation in P. falciparum parasites is consistent with the dearth of clear HO genes in the parasite genome. Several genes identified by BLAST search show low level sequence similarity to known HO enzymes, but we found no detectable heme degradation by these parasite proteins. The most HO-like of these proteins, PfHO, was previously proposed to degrade heme based on limited in vitro data (22, 23). We carried out extensive tests of this proposed activity but found no evidence for PfHO catalysis of heme degradation (Figs. 7 and 8). The lack of catalytic activity by PfHO is consistent with the absence of the conserved His residue used by all known heme-degrading enzymes to coordinate the iron of bound heme, as well as the lack of a critical Gly residue that kinks and positions the distal helix above the bound heme and is required for activity (Figs. 5A and 6A) (7, 64).
These sequence differences relative to known HO enzymes and its lack of detectable heme degrading activity suggest that PfHO fulfills an alternative function within parasites. The protein does bind heme with modest affinity but without the canonical Soret peak expected for iron coordination by a protein optimized to bind heme (78), and PfHO appears to have a higher affinity for protoporphyrin IX (Fig. 6). These observations may suggest that heme is not the cognate ligand in vivo. The full-length protein contains a leader sequence upstream of the HO-like domain that shows features of an apicoplast-targeting motif, but the absence of a clear signal peptide leads to an ambiguous targeting prediction by the PATS and PlasmoAP algorithms (61, 62). Preliminary biochemical studies of parasites using a PfHO-specific antibody indicate that the protein is expressed during blood stage infection and has a complex distribution to multiple subcellular compartments.7
HO-like but catalytically inactive proteins have been identified in other organisms, but their cellular function remains unclear. The best studied example is the HO2 gene from A. thaliana. This protein shows strong sequence similarity to the AtHO1, AtHO3, and AtHO4 enzymes expressed by this plant but has an Arg in place of the conserved proximal His (73), has no detectable HO activity (63, 79), and like PfHO appears to bind protoporphyrin IX tighter than heme (79). Disruption of the AtHO2 gene in plants results in a modest developmental phenotype related to but more subtle than the observed phenotype from disruption of the AtHO1 gene involved in phytochrome chromophore biosynthesis (73), possibly reflecting a role for AtHO2 in porphyrin trafficking rather than degradation (79). PfHO may carry out an analogous trafficking role within parasites.
Given its substantial sequence similarity to the three other HO genes in A. thaliana, AtHO2 may have evolved via gene duplication and subsequent mutational drift to fulfill an alternative cellular role. The origin of PfHO is less obvious, as there is no HO homolog in the parasite genome to suggest that PfHO arose via recent gene duplication. It is possible that a distant evolutionary ancestor of P. falciparum contained an HO gene that was duplicated to give rise to PfHO and then subsequently lost. Indeed, clear PfHO orthologs are present within all Plasmodium species, suggesting functional conservation within all of these parasites. It remains an intriguing future challenge to unravel the cellular function of PfHO within parasites.
Supplementary Material
Acknowledgments
We thank Roger Tsien (University of California, San Diego) for plasmids encoding IFP and SynHO1, Paul Ortiz de Montellano (University of California, San Francisco) for plasmids encoding HuHO1 and rat BVR, Barbara Kunkel (Washington University in St. Louis) for providing A. thaliana cDNA, and members of the Goldberg laboratory for helpful comments and discussions.
This work was supported in part by National Institutes of Health Grants HD001459, HL101263, and DK064540 (to J. P. H.).

This article contains supplemental Fig. S1.
P. A. Sigala and D. E. Goldberg, unpublished results.
- HO
- heme oxygenase
- BV
- biliverdin IXα
- BR
- bilirubin IXα
- DMSO
- dimethyl sulfoxide
- IFP
- infrared fluorescent protein
- BVR
- biliverdin reductase
- ACP
- acyl carrier protein
- DHOD
- dihydroorotate dehydrogenase
- IPTG
- isopropyl 1-thio-β-d-galactopyranoside
- EOE
- episomal overexpression
- PPIX
- protoporphyrin IX
- Fd
- ferredoxin
- FNR
- ferredoxin-NADP+ reductase
- CV
- column volume
- Spin.
- spinach.
REFERENCES
- 1. Francis S. E., Sullivan D. J., Jr., Goldberg D. E. (1997) Hemoglobin metabolism in the malaria parasite Plasmodium falciparum. Annu. Rev. Microbiol. 51, 97–123 [DOI] [PubMed] [Google Scholar]
- 2. van Dooren G. G., Kennedy A. T., McFadden G. I. (2012) The use and abuse of heme in apicomplexan parasites. Antioxid. Redox Signal. 17, 634–656 [DOI] [PubMed] [Google Scholar]
- 3. Surolia N., Padmanaban G. (1992) De novo biosynthesis of heme offers a new chemotherapeutic target in the human malarial parasite. Biochem. Biophys. Res. Commun. 187, 744–750 [DOI] [PubMed] [Google Scholar]
- 4. Nagaraj V. A., Prasad D., Rangarajan P. N., Padmanaban G. (2009) Mitochondrial localization of functional ferrochelatase from Plasmodium falciparum. Mol. Biochem. Parasitol. 168, 109–112 [DOI] [PubMed] [Google Scholar]
- 5. Egan T. J., Combrinck J. M., Egan J., Hearne G. R., Marques H. M., Ntenteni S., Sewell B. T., Smith P. J., Taylor D., van Schalkwyk D. A., Walden J. C. (2002) Fate of heme iron in the malaria parasite Plasmodium falciparum. Biochem. J. 365, 343–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wilks A. (2002) Heme oxygenase. Evolution, structure, and mechanism. Antioxid. Redox Signal. 4, 603–614 [DOI] [PubMed] [Google Scholar]
- 7. Unno M., Matsui T., Ikeda-Saito M. (2007) Structure and catalytic mechanism of heme oxygenase. Nat. Prod. Rep. 24, 553–570 [DOI] [PubMed] [Google Scholar]
- 8. Ortiz de Montellano P. R. (2000) The mechanism of heme oxygenase. Curr. Opin. Chem. Biol. 4, 221–227 [DOI] [PubMed] [Google Scholar]
- 9. Beale S. I. (1993) Biosynthesis of phycobilins. Chem. Rev. 93, 785–802 [Google Scholar]
- 10. Stocker R., Yamamoto Y., McDonagh A. F., Glazer A. N., Ames B. N. (1987) Bilirubin is an antioxidant of possible physiological importance. Science 235, 1043–1046 [DOI] [PubMed] [Google Scholar]
- 11. Baranano D. E., Rao M., Ferris C. D., Snyder S. H. (2002) Biliverdin reductase. A major physiologic cytoprotectant. Proc. Natl. Acad. Sci. U.S.A. 99, 16093–16098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Vítek L., Ostrow J. D. (2009) Bilirubin chemistry and metabolism; harmful and protective aspects. Curr. Pharm. Des. 15, 2869–2883 [DOI] [PubMed] [Google Scholar]
- 13. Kirkby K. A., Adin C. A. (2006) Products of heme oxygenase and their potential therapeutic applications. Am. J. Physiol. Renal Physiol. 290, F563–F571 [DOI] [PubMed] [Google Scholar]
- 14. Wilks A., Burkhard K. A. (2007) Heme and virulence. How bacterial pathogens regulate, transport, and utilize heme. Nat. Prod. Rep. 24, 511–522 [DOI] [PubMed] [Google Scholar]
- 15. Loria P., Miller S., Foley M., Tilley L. (1999) Inhibition of the peroxidative degradation of heme as the basis of action of chloroquine and other quinoline antimalarials. Biochem. J. 339, 363–370 [PMC free article] [PubMed] [Google Scholar]
- 16. Zhang J., Krugliak M., Ginsburg H. (1999) The fate of ferriprotoporphyrin IX in malaria-infected erythrocytes in conjunction with the mode of action of antimalarial drugs. Mol. Biochem. Parasitol. 99, 129–141 [DOI] [PubMed] [Google Scholar]
- 17. Eckman J. R., Modler S., Eaton J. W., Berger E., Engel R. R. (1977) Host heme catabolism in drug-sensitive and drug-resistant malaria. J. Lab. Clin. Med. 90, 767–770 [PubMed] [Google Scholar]
- 18. Slater A. F., Swiggard W. J., Orton B. R., Flitter W. D., Goldberg D. E., Cerami A., Henderson G. B. (1991) An iron-carboxylate bond links the heme units of malaria pigment. Proc. Natl. Acad. Sci. U.S.A. 88, 325–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Weinberg E. D., Moon J. (2009) Malaria and iron. History and review. Drug Metab. Rev. 41, 644–662 [DOI] [PubMed] [Google Scholar]
- 20. Jani D., Nagarkatti R., Beatty W., Angel R., Slebodnick C., Andersen J., Kumar S., Rathore D. (2008) HDP. A novel heme detoxification protein from the malaria parasite. PLoS Pathog. 4, e1000053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ehlgen F., Pham J. S., de Koning-Ward T., Cowman A. F., Ralph S. A. (2012) Investigation of the Plasmodium falciparum food vacuole through inducible expression of the chloroquine resistance transporter (PfCRT). PLoS One 7, e38781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Okada K. (2009) The novel heme oxygenase-like protein from Plasmodium falciparum converts heme to bilirubin IXα in the apicoplast. FEBS Lett. 583, 313–319 [DOI] [PubMed] [Google Scholar]
- 23. Sartorello R., Budu A., Bagnaresi P., Fernandes C. A., Sato P. M., Bueno V. B., Fontes M. R., Oliveira P. L., Paiva-Silva G. O., Alves S. V., Netto L. E., Catalani L. H., Garcia C. R. (2010) In vivo uptake of a haem analogue Zn protoporphyrin IX by the human malaria parasite P. falciparum-infected red blood cells. Cell Biol. Int. 34, 859–865 [DOI] [PubMed] [Google Scholar]
- 24. Seeber F., Soldati-Favre D. (2010) Metabolic pathways in the apicoplast of apicomplexa. Int. Rev. Cell Mol. Biol. 281, 161–228 [DOI] [PubMed] [Google Scholar]
- 25. Toh S. Q., Glanfield A., Gobert G. N., Jones M. K. (2010) Heme and blood-feeding parasites. Friends or foes? Parasit. Vectors 3, 108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rydzanicz R., Zhao X. S., Johnson P. E. (2005) Assembly PCR oligo marker. A tool for designing oligodeoxynucleotides for constructing long DNA molecules for RNA production. Nucleic Acids Res. 33, W521–W525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Russo I., Oksman A., Goldberg D. E. (2009) Fatty acid acylation regulates trafficking of the unusual Plasmodium falciparum calpain to the nucleolus. Mol. Microbiol. 72, 229–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ganesan S. M., Morrisey J. M., Ke H., Painter H. J., Laroiya K., Phillips M. A., Rathod P. K., Mather M. W., Vaidya A. B. (2011) Yeast dihydroorotate dehydrogenase as a new selectable marker for Plasmodium falciparum transfection. Mol. Biochem. Parasitol. 177, 29–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Waller R. F., Reed M. B., Cowman A. F., McFadden G. I. (2000) Protein trafficking to the plastid of Plasmodium falciparum is via the secretory pathway. EMBO J. 19, 1794–1802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tonkin C. J., van Dooren G. G., Spurck T. P., Struck N. S., Good R. T., Handman E., Cowman A. F., McFadden G. I. (2004) Localization of organellar proteins in Plasmodium falciparum using a novel set of transfection vectors and a new immunofluorescence fixation method. Mol. Biochem. Parasitol. 137, 13–21 [DOI] [PubMed] [Google Scholar]
- 31. Marley J., Lu M., Bracken C. (2001) A method for efficient isotopic labeling of recombinant proteins. J. Biomol. NMR 20, 71–75 [DOI] [PubMed] [Google Scholar]
- 32. Chaturvedi K. S., Hung C. S., Crowley J. R., Stapleton A. E., Henderson J. P. (2012) The siderophore yersiniabactin binds copper to protect pathogens during infection. Nat. Chem. Biol. 8, 731–736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. McLaren C. E., Brittenham G. M., Hasselblad V. (1987) Statistical and graphical evaluation of erythrocyte volume distributions. Am. J. Physiol. 252, H857–H866 [DOI] [PubMed] [Google Scholar]
- 34. Drew M. E., Banerjee R., Uffman E. W., Gilbertson S., Rosenthal P. J., Goldberg D. E. (2008) Plasmodium food vacuole plasmepsins are activated by falcipains. J. Biol. Chem. 283, 12870–12876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Trager W., Jensen J. B. (1976) Human malaria parasites in continuous culture. Science 193, 673–675 [DOI] [PubMed] [Google Scholar]
- 36. Istvan E. S., Dharia N. V., Bopp S. E., Gluzman I., Winzeler E. A., Goldberg D. E. (2011) Validation of isoleucine utilization targets in Plasmodium falciparum. Proc. Natl. Acad. Sci. U.S.A. 108, 1627–1632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Klemba M., Beatty W., Gluzman I., Goldberg D. E. (2004) Trafficking of plasmepsin II to the food vacuole of the malaria parasite Plasmodium falciparum. J. Cell Biol. 164, 47–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ponpuak M., Klemba M., Park M., Gluzman I. Y., Lamppa G. K., Goldberg D. E. (2007) A role for falcilysin in transit peptide degradation in the Plasmodium falciparum apicoplast. Mol. Microbiol. 63, 314–334 [DOI] [PubMed] [Google Scholar]
- 39. Raman S., Vernon R., Thompson J., Tyka M., Sadreyev R., Pei J., Kim D., Kellogg E., DiMaio F., Lange O., Kinch L., Sheffler W., Kim B. H., Das R., Grishin N. V., Baker D. (2009) Structure prediction for CASP8 with all-atom refinement using Rosetta. Proteins 77, 89–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sugishima M., Migita C. T., Zhang X., Yoshida T., Fukuyama K. (2004) Crystal structure of heme oxygenase-1 from cyanobacterium. Synechocystis sp. PCC 6803 in complex with heme. Eur. J. Biochem. 271, 4517–4525 [DOI] [PubMed] [Google Scholar]
- 41. DeLano W. L. (2007) MacPyMOL. A PyMOL-based Molecular Graphics Application for MacOS X, DeLano Scientific LLC, Palo Alto, CA [Google Scholar]
- 42. Gill S. C., von Hippel P. H. (1989) Calculation of protein extinction coefficients from amino acid sequence data. Anal. Biochem. 182, 319–326 [DOI] [PubMed] [Google Scholar]
- 43. Migita C. T., Zhang X., Yoshida T. (2003) Expression and characterization of cyanobacterium heme oxygenase, a key enzyme in the phycobilin synthesis. Properties of the heme complex of recombinant active enzyme. Eur. J. Biochem. 270, 687–698 [DOI] [PubMed] [Google Scholar]
- 44. Muramoto T., Tsurui N., Terry M. J., Yokota A., Kohchi T. (2002) Expression and biochemical properties of a ferredoxin-dependent heme oxygenase required for phytochrome chromophore synthesis. Plant Physiol. 130, 1958–1966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Nagababu E., Rifkind J. M. (2004) Heme degradation by reactive oxygen species. Antioxid. Redox Signal. 6, 967–978 [DOI] [PubMed] [Google Scholar]
- 46. Pasini E. M., Kirkegaard M., Mortensen P., Lutz H. U., Thomas A. W., Mann M. (2006) In-depth analysis of the membrane and cytosolic proteome of red blood cells. Blood 108, 791–801 [DOI] [PubMed] [Google Scholar]
- 47. Roux-Dalvai F., Gonzalez de Peredo A., Simó C., Guerrier L., Bouyssié D., Zanella A., Citterio A., Burlet-Schiltz O., Boschetti E., Righetti P. G., Monsarrat B. (2008) Extensive analysis of the cytoplasmic proteome of human erythrocytes using the peptide ligand library technology and advanced mass spectrometry. Mol. Cell. Proteomics 7, 2254–2269 [DOI] [PubMed] [Google Scholar]
- 48. Cornejo J., Willows R. D., Beale S. I. (1998) Phytobilin biosynthesis. Cloning and expression of a gene encoding soluble ferredoxin-dependent heme oxygenase from Synechocystis sp. PCC 6803. Plant J. 15, 99–107 [DOI] [PubMed] [Google Scholar]
- 49. Gambetta G. A., Lagarias J. C. (2001) Genetic engineering of phytochrome biosynthesis in bacteria. Proc. Natl. Acad. Sci. U.S.A. 98, 10566–10571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Quinn K. D., Nguyen N. Q., Wach M. M., Wood T. D. (2012) Tandem mass spectrometry of bilin tetrapyrroles by electrospray ionization and collision-induced dissociation. Rapid Commun. Mass Spectrom. 26, 1767–1775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Paiva-Silva G. O., Cruz-Oliveira C., Nakayasu E. S., Maya-Monteiro C. M., Dunkov B. C., Masuda H., Almeida I. C., Oliveira P. L. (2006) A heme-degradation pathway in a blood-sucking insect. Proc. Natl. Acad. Sci. U.S.A. 103, 8030–8035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. McDonagh A. F., Palma L. A. (1980) Preparation and properties of crystalline biliverdin IXα. Simple methods for preparing isomerically homogeneous biliverdin and [14C]biliverdin by using 2,3-dichloro-5,6-dicyanobenzoquinone. Biochem. J. 189, 193–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Brodersen R. (1979) Bilirubin. Solubility and interaction with albumin and phospholipid. J. Biol. Chem. 254, 2364–2369 [PubMed] [Google Scholar]
- 54. Psychogios N., Hau D. D., Peng J., Guo A. C., Mandal R., Bouatra S., Sinelnikov I., Krishnamurthy R., Eisner R., Gautam B., Young N., Xia J., Knox C., Dong E., Huang P., Hollander Z., Pedersen T. L., Smith S. R., Bamforth F., Greiner R., McManus B., Newman J. W., Goodfriend T., Wishart D. S. (2011) The human serum metabolome. PLoS One 6, e16957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Shu X., Royant A., Lin M. Z., Aguilera T. A., Lev-Ram V., Steinbach P. A., Tsien R. Y. (2009) Mammalian expression of infrared fluorescent proteins engineered from a bacterial phytochrome. Science 324, 804–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Wagner J. R., Brunzelle J. S., Forest K. T., Vierstra R. D. (2005) A light-sensing knot revealed by the structure of the chromophore-binding domain of phytochrome. Nature 438, 325–331 [DOI] [PubMed] [Google Scholar]
- 57. Suits M. D., Pal G. P., Nakatsu K., Matte A., Cygler M., Jia Z. (2005) Identification of an Escherichia coli O157:H7 heme oxygenase with tandem functional repeats. Proc. Natl. Acad. Sci. U.S.A. 102, 16955–16960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Gardner M. J., Hall N., Fung E., White O., Berriman M., Hyman R. W., Carlton J. M., Pain A., Nelson K. E., Bowman S., Paulsen I. T., James K., Eisen J. A., Rutherford K., Salzberg S. L., Craig A., Kyes S., Chan M. S., Nene V., Shallom S. J., Suh B., Peterson J., Angiuoli S., Pertea M., Allen J., Selengut J., Haft D., Mather M. W., Vaidya A. B., Martin D. M., Fairlamb A. H., Fraunholz M. J., Roos D. S., Ralph S. A., McFadden G. I., Cummings L. M., Subramanian G. M., Mungall C., Venter J. C., Carucci D. J., Hoffman S. L., Newbold C., Davis R. W., Fraser C. M., Barrell B. (2002) Genome sequence of the human malaria parasite Plasmodium falciparum. Nature 419, 498–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Altschul S. F., Madden T. L., Schäffer A. A., Zhang J., Zhang Z., Miller W., Lipman D. J. (1997) Gapped BLAST and PSI-BLAST. A new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Biegert A., Söding J. (2009) Sequence context-specific profiles for homology searching. Proc. Natl. Acad. Sci. U.S.A. 106, 3770–3775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Zuegge J., Ralph S., Schmuker M., McFadden G. I., Schneider G. (2001) Deciphering apicoplast targeting signals–feature extraction from nuclear-encoded precursors of Plasmodium falciparum apicoplast proteins. Gene 280, 19–26 [DOI] [PubMed] [Google Scholar]
- 62. Foth B. J., Ralph S. A., Tonkin C. J., Struck N. S., Fraunholz M., Roos D. S., Cowman A. F., McFadden G. I. (2003) Dissecting apicoplast targeting in the malaria parasite Plasmodium falciparum. Science 299, 705–708 [DOI] [PubMed] [Google Scholar]
- 63. Emborg T. J., Walker J. M., Noh B., Vierstra R. D. (2006) Multiple heme oxygenase family members contribute to the biosynthesis of the phytochrome chromophore in Arabidopsis. Plant Physiol. 140, 856–868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Liu Y., Koenigs Lightning L., Huang H., Moënne-Loccoz P., Schuller D. J., Poulos T. L., Loehr T. M., Ortiz de Montellano P. R. (2000) Replacement of the distal glycine 139 transforms human heme oxygenase-1 into a peroxidase. J. Biol. Chem. 275, 34501–34507 [DOI] [PubMed] [Google Scholar]
- 65. Kimata-Ariga Y., Kurisu G., Kusunoki M., Aoki S., Sato D., Kobayashi T., Kita K., Horii T., Hase T. (2007) Cloning and characterization of ferredoxin and ferredoxin-NADP+ reductase from human malaria parasite. J. Biochem. 141, 421–428 [DOI] [PubMed] [Google Scholar]
- 66. Knobler E., Poh-Fitzpatrick M. B., Kravetz D., Vincent W. R., Muller-Eberhard U., Vincent S. H. (1989) Interaction of hemopexin, albumin, and liver fatty acid-binding protein with protoporphyrin. Hepatology 10, 995–997 [DOI] [PubMed] [Google Scholar]
- 67. Wilks A., Black S. M., Miller W. L., Ortiz de Montellano P. R. (1995) Expression and characterization of truncated human heme oxygenase (hHO-1) and a fusion protein of hHO-1 with human cytochrome P450 reductase. Biochemistry 34, 4421–4427 [DOI] [PubMed] [Google Scholar]
- 68. Wilks A., Schmitt M. P. (1998) Expression and characterization of a heme oxygenase (Hmu O) from Corynebacterium diphtheriae. Iron acquisition requires oxidative cleavage of the heme macrocycle. J. Biol. Chem. 273, 837–841 [DOI] [PubMed] [Google Scholar]
- 69. Maines M. D. (2005) New insights into biliverdin reductase functions. Linking heme metabolism to cell signaling. Physiology 20, 382–389 [DOI] [PubMed] [Google Scholar]
- 70. Sigman J. A., Wang X., Lu Y. (2001) Coupled oxidation of heme by myoglobin is mediated by exogenous peroxide. J. Am. Chem. Soc. 123, 6945–6946 [DOI] [PubMed] [Google Scholar]
- 71. Hanke G. T., Kimata-Ariga Y., Taniguchi I., Hase T. (2004) A post-genomic characterization of Arabidopsis ferredoxins. Plant Physiol. 134, 255–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Cammack R., Rao K. K., Bargeron C. P., Hutson K. G., Andrew P. W., Rogers L. J. (1977) Midpoint redox potentials of plant and algal ferredoxins. Biochem. J. 168, 205–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Davis S. J., Bhoo S. H., Durski A. M., Walker J. M., Vierstra R. D. (2001) The heme-oxygenase family required for phytochrome chromophore biosynthesis is necessary for proper photomorphogenesis in higher plants. Plant Physiol. 126, 656–669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Painter H. J., Morrisey J. M., Mather M. W., Vaidya A. B. (2007) Specific role of mitochondrial electron transport in blood stage Plasmodium falciparum. Nature 446, 88–91 [DOI] [PubMed] [Google Scholar]
- 75. Scholl P. F., Tripathi A. K., Sullivan D. J. (2005) Bioavailable iron and heme metabolism in Plasmodium falciparum. Curr. Top. Microbiol. Immunol. 295, 293–324 [DOI] [PubMed] [Google Scholar]
- 76. Lara F. A., Lins U., Paiva-Silva G., Almeida I. C., Braga C. M., Miguens F. C., Oliveira P. L., Dansa-Petretski M. (2003) A new intracellular pathway of heme detoxification in the midgut of the cattle tick Boophilus microplus. Aggregation inside a specialized organelle, the hemosome. J. Exp. Biol. 206, 1707–1715 [DOI] [PubMed] [Google Scholar]
- 77. Oliveira M. F., Kycia S. W., Gomez A., Kosar A. J., Bohle D. S., Hempelmann E., Menezes D., Vannier-Santos M. A., Oliveira P. L., Ferreira S. T. (2005) Structural and morphological characterization of hemozoin produced by Schistosoma mansoni and Rhodnius prolixus. FEBS Lett. 579, 6010–6016 [DOI] [PubMed] [Google Scholar]
- 78. Fufezan C., Zhang J., Gunner M. R. (2008) Ligand preference and orientation in b- and c-type heme-binding proteins. Proteins 73, 690–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Gisk B., Yasui Y., Kohchi T., Frankenberg-Dinkel N. (2010) Characterization of the heme oxygenase protein family in Arabidopsis thaliana reveals a diversity of functions. Biochem. J. 425, 425–434 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.