Background: Matrix metalloprotease 11 induces tumorigenesis and promotes invasive tissue behavior. Its function is hardly understood.
Results: MMP-11 activatable cell-permeable FRET substrates were developed and tested in vitro, visualizing MMP-11 activity.
Conclusion: Rational design of FRET substrates yielded new imaging tools for MMP-11.
Significance: MMP-11 imaging can assist functional studies, investigating native substrates, or interaction partners of MMP-11.
Keywords: Cell Invasion, Extracellular Matrix Proteins, Fluorescence Resonance Energy Transfer (FRET), Imaging, Matrix Metalloproteinase (MMP), Cell Penetrating Peptides, Enzyme Substrate Design
Abstract
Matrix metalloprotease 11 (MMP-11), a protease associated with invasion and aggressiveness of cancerous tissue, was postulated as a prognostic marker for pancreatic, breast, and colon cancer patients. Expression analysis, however, did not reveal localization and regulation of this protease. Thus, cellular tools for the visualization of MMP-11 are highly desirable to monitor presence and activity and to elucidate the functional role of MMP-11. Therefore, fluorescein-Dabcyl-labeled Foerster resonance energy transfer (FRET) substrates were developed. The design focused on enhanced peptide binding to human MMP-11, employing an unusual amino acid for the specificity pocket P1′. The addition of several arginines resulted in a cell-permeable FRET substrate SM-P124 (Ac-GRRRK(Dabcyl)-GGAANC(MeOBn)RMGG-fluorescein). In vitro evaluation of SM-P124 with human MMP-11 showed a 25-fold increase of affinity (kcat/Km = 9.16 × 103 m−1 s−1, Km = 8 μm) compared with previously published substrates. Incubation of pancreatic adenocarcinoma cell line MIA PaCa-2 and mamma adenocarcinoma cell line MCF-7 with the substrate SM-P124 (5 μm) indicated intra- and extracellular MMP-11 activity. A negative control cell line (Jurkat) showed no fluorescent signal either intra- or extracellularly. Negative control FRET substrate SM-P123 produced only insignificant extracellular fluorescence without any intracellular fluorescence. SM-P124 therefore enabled intra- and extracellular tracking of MMP-11-overexpressing cancers such as pancreatic and breast adenocarcinoma and might contribute to the understanding of the activation pathways leading to MMP-11-mediated invasive processes.
Introduction
Matrix metalloproteases (MMPs)2 are calcium-containing, zinc-dependent endopeptidases secreted to the extracellular matrix (ECM), functioning as degradation/remodeling enzymes (1). MMPs constitute an important family of proteases, participating in ECM breakdown and enabling invasive behavior of cancerous tissue (2). Most MMPs such as MMP-1, MMP-3, MMP-10, MMP-11, and MMP-13 are not expressed in healthy and resting tissues (1). For instance, MMP-11−/−-deficient knock-out mice exhibited no phenotype and even impede tumorigenesis (3). MMP-11 expression is activated upon repair and remodeling processes and in diseased tissues only and is observed in 80–100% of invasive carcinomas of the breast, colon, pancreas, head and neck, lung, ovary, prostate, skin, and uterus (4–9). In breast, pancreatic, and colon cancer, where it is especially overexpressed (immunohistochemistry) and up-regulated (mRNA), its presence is a prognostic marker for survival and has been proposed as an antigen for cancer immunotherapy (10–14). MMP-11 is strongly overexpressed by established cancer cell lines such as MIA PaCa-2 and MCF-7 (15, 16). Interestingly, MMP-11 is not expressed by tumor cells themselves, but in tumor stroma by fibroblasts surrounding tumor cells, pointing toward a role in invasion (11, 12). 20–50% of the tumor mass in solid tumors consist of stromal cells with up to 90% in some specific cancer types (15).
MMP-11 differs strongly from other MMPs with respect to time and fashion of expression as well as to activation and substrate preference (17–19). MMP-11 hardly cleaves ECM components but rather natural substrates like serin protease inhibitors (serpins) such as α2-macroglobulin and α1-proteinase inhibitor, as well as insulin-like growth factor-binding protein 1 (20, 21). α2-Macroglobulin is believed to inhibit MMP-11 by a trapping mechanism (22). Degradation of insulin-like growth factor-binding protein 1 by MMP-11 might, however, result in inhibition of apoptosis and increased tumorigenesis (1, 2, 19). Additionally, degradation of serpins such as α1-proteinase inhibitor results in increased concentrations of serin proteases that focally break down the basement membrane, initiating early invasive processes (3, 4, 23).
Activation of MMP-11 differs, because of its furin cleavage site (18). The prodomain, responsible for autoinhibition of MMP-11 (proMMP-11), is cleaved intracellularly by the Golgi-associated protease furin, resulting in secretion of the α-isoform (48 kDa) to the extracellular space (17). Additionally, a nonsecreted, directly translated, fully active β-isoform (40 kDa), lacking the prodomain as well as 32 N-terminal amino acids was detected in cultured cells and placenta (24, 25). The function of this β-MMP-11, however, remains unknown. Detailed experimental results about the activity of both isoforms of MMP-11 have not been performed, and their entanglement in tumor progression, invasion, or apoptosis is still poorly understood (25).
Therefore, imaging probes for MMP-11 are important tools to further clarify activation, regulation, and functional significance of both extracellular α-MMP-11 and even more intracellularly active β-MMP-11.
For the visualization of MMP active tissues, intracellular imaging has distinct advantages, because extracellular fluorescent probes can diffuse away from their site of formation, delivering less precise spatial images of molecular target distribution. Moreover, intracellular imaging probes accumulate in the cell over time and increase the signal to noise ratio significantly. Addressing molecular targets such as MMP-11 in living organism calls for highly affine, specific probes that can be activated and produce a distinct, amplified signal to detect small intra- or extracellular concentrations of the enzyme (26).
Fluorescent optical imaging is rapidly expanding as a noninvasive method for disease and tumor progression evaluation, providing high sensitivity at nanomolar concentrations, use of nonradioactive materials, and safe detection, while employing readily available instruments at moderate cost (27, 28). Especially peptides susceptible for protease cleavage are used to specifically detect the presence and activity of enzymes in vitro and in vivo. Combination of these protease-sensitive peptides with fluorescent labels allows the generation of “smart” probes, developing a distinct fluorescent signal upon cleavage, whereas the inactivated probe remains invisible, because of FRET (29). Unfortunately, benchmark peptidic FRET substrates for MMPs such as Mca-PLG-LDap(Dnp)AR-NH2 (OmniMMPTM; Enzo Life Sciences, Inc.) are not cleaved by hMMP-11 (30). Commercially available substrates for MMP-11 (Mca-PLA-C(OMeBn)WARDap(Dnp)-NH2; Merck; kcat/Km = 3.6 × 104 m−1 s−1), however, have only been tested with murine MMP-11 (mMMP-11) and use fluorophores that strongly interfere with cellular autofluorescence (31). Additionally, autofluorescence of common aromatic building blocks used in screenings for potential binders can interfere with the assay readout, especially with substrates detected at shorter wavelengths. Therefore, substrates, readily available for cellular detection and monitoring of enzyme activities intra- and extracellularly, as well as for compound screenings, pose a strong need for further development. Here we describe the synthesis, in vitro evaluation, and live cell visualization of hMMP-11 activated, cell-penetrating peptides that can be used as imaging tools for the detection of tumorigenesis and invasion-related protease MMP-11.
EXPERIMENTAL PROCEDURES
Reagents and Resins
Reagents and resins were purchased from Sigma-Aldrich, Acros Organics, Novabiochem, and ChemPep Inc. and were used without further purification. All of the matrix metalloprotease catalytic domains were purchased from Enzo Life Sciences, Inc. (Lörrach, Germany). Purchased MMP-11 was produced in Escherichia coli. from human cDNA. A unit for the protease activity was not provided for MMP-11. The original buffer system of 50 mm Tris-HCl, 5 mm CaCl2, 300 mm NaCl, 20 μm ZnCl2, 0.5% Brij-35, and 30% glycerol at pH 7.5 was adjusted to a final concentration of 50% glycerol and 1% BSA to increase stability.
Synthesis
Fluorescein-Dabcyl-labeled substrates were synthesized via Fmoc-mediated solid phase synthesis on O-(methoxyethoxy-methyl)-5-amino-fluorescein 2-chlorotrityl resin that has been reported previously (32). First, Fmoc-protected glycine was added to a solution of 5 eq of HATU and 10 eq of 2,4,6-trimethylpyridine (collidine) in DMF/DCM (1:1, v/v) to the prepared O-(methoxy-ethoxymethyl)-5-amino-fluorescein 2-chlorotrityl resin for 3 h at room temperature, twice. Fmoc determination yielded 58% coupling efficiency. The remaining coupling positions were capped with MeOH/DIPEA/DCM (0.5:1:1, v/v/v). The Fmoc group was removed with 20% piperidine in DMF, twice, for 10 min. The following amino acids were coupled by the addition of 3 eq of Fmoc-protected amino acid, 3 eq of diisopropylcarbodiimide (DIC), 3 eq of hydroxy benzotriazole (HOBt), and 3 eq of diisopropylethylamine (DIPEA) in DMF for 3 h. Fmoc deblocking and coupling steps were repeated for additional amino acid couplings. Completeness of couplings was tested with a Kaiser test (33). The Dabcyl quencher was introduced by coupling 3 eq of Nα-Fmoc-Nϵ-4-[4-(dimethyl-amino)-phenylazo)benzoyl]-l-lysine with 3 eq of HOBt and 3 eq of DIC in DMF overnight at room temperature. All peptides were N-acetylated in the last step with 1 ml of acetic anhydride/DCM/DIPEA (1:1:1, v/v/v) for 10 min at room temperature. Cleavage cocktails were applied to the resins for 24 h at room temperature. DCM/TFA/H2O/Phenol/EDT/TIS (50:40:4:1:2.5:2.5, v/v/v/m/v/v) for substrates containing MeOBn protected cysteine, whereas DCM/TFA/H2O/Phenol/EDT/TIS (5:90:2:1:1:1, v/v/v/m/v/v) was used for leucine containing peptides. Substrates were precipitated in ether and were washed thrice with ether/MeCN (95:5) followed by drying in vacuo and isolation by reverse phase HPLC (Agilent 1100 Series, C18 ec, 100 Å, 7 μm, 21 × 250 mm column (Macherey-Nagel; Nucleodur®).
AMCA-labeled Substrate Synthesis
SM-P155 was synthesized employing AMCA that was Fmoc-protected as has been reported previously (34). Fmoc-protected Rink amide resin was deprotected with 20% piperidine in DMF, twice, for 10 min at room temperature. 3 eq of Fmoc-AMCA were activated with 3 eq of HOBt and 3 eq of DIC. The mixture was added to the Rink amide resin and was stirred for 3 h at room temperature. Loading efficiency (∼85%) was determined via Fmoc determination. Glycine was coupled using 5 eq of glycine, 5 eq of HATU, and 10 eq of collidine with Fmoc-deprotected AMCA-resin overnight at room temperature, twice. The following amino acid couplings afforded 3 eq of amino acid, 3 eq of HOBt, and 3 eq of DIC in DMF for 3 h at room temperature. Repeated Fmoc deblocking and coupling yielded the AMCA-labeled peptide sequence. The quencher was introduced by coupling of 3 eq of Fmoc-N-(2,4-dinitrophenyl)-l-lysine (Fmoc-K(Dnp)-OH) with equivalent amounts of HOBt and DIC in DMF for 3 h at room temperature. Completed sequences were acetylated in the last step with a 1:1:1, v/v/v mixture of acetic anhydride/DIPEA/DCM for 10 min at room temperature.
Absorption and Emission Spectra
All substrates were measured on a Tecan Saphire2 Reader on a Corning number 3544 microplate in MMP assay buffer at pH 7.5 at concentrations of 5 and 0.5 μm for absorption and emission spectra, respectively. The spectra were recorded at 483 ± 10 nm (excitation) and 525 ± 10 nm (emission).
In Vitro MMP Assay
50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 10 mm CaCl2, and 0.05% Brij-35 was used as the buffer system (35). All of the substrates were prepared as 10 mm stock solutions in Me2SO. A stock dilution series in Me2SO ranging from 400 to 20 μm, and 0 μm (Me2SO) as a negative control was prepared with a further dilution factor of 20. 1 μl of each stock concentration in three replicates was pipetted to the microplate and 19 μl of enzyme (12.5 nm for MMP-11 and 0.625 nm for MMP-14, determined by enzyme titration) were added to start kinetic measurements. Substrate in buffer without enzyme functioned as a negative control and as a fluorescence background measurement. 100% hydrolysis was measured in respect to these fluorescence standards: Ac-C(MeOBn)RMGG-Fluo, Ac-C(MeOBn)VRGG-Fluo, Ac-C(MeOBn)-WARGG-Fluo, Ac-LRMGG-Fluo, Ac-LVRGG-Fluo, and Ac-LWARGG-Fluo. Concentrations used correspond to 2–10% (final assay concentration, 1–1000 nm) of hydrolyzed substrate to avoid quenching effects in solution. MMP-11 cleavage was monitored at λ = 483 ± 10 nm (excitation) and λ = 525 ± 10 nm (emission). All plates were covered with a PCR-foil Ultra Clear room temperature-PCR (G060/UC-RT) from G. Kisker GbR to avoid evaporation during kinetic measurements at 28 °C. The results were analyzed by plotting and fitting the curves with Prism 5.0 (Graph Pad Software, Inc., San Diego, CA) to the Michaelis-Menten equation (Equation 1),
where Y is the substrate velocity in nm/min, and X is the substrate concentration in μm.
MMP-11 Inhibition Assay
Stock concentrations in Me2SO were prepared of GM6001 ranging from 3.125 mm to 31.25 μm (dilution factor to final concentration = 50, 1% Me2SO). 1 μl of each stock dilution of inhibitor was pipetted into three wells each and was mixed with 24 μl of 2 nm MMP-11 (final concentration, 1 nm) in assay buffer. The plate was incubated for 30 min at 37 °C, and 25 μl of 1.5 μm SM-P124 (final concentration, 0.75 μm) in assay buffer were added to all wells to start the reaction. Fluorescence intensity was recorded for 30 min at 37 °C. As a substrate activity control, several wells of MMP-11 with SM-P124 but without inhibitor were measured, as well as just SM-P124 in assay buffer serving as a fluorescence background measurement.
LC/MS-TOF Assay Validation
LC/MS-TOF measurements were performed on an Agilent Accurate-Mass 6220 TOF LC/MS station equipped with a diode array detector and a 1200 series LC. Liquid chromatography was performed employing Agilent's ZORBAX® Eclipse column XDB-C18 Rapid Resolution HT 50 × 4.6 mm with 1.8-μm particle size. High resolution masses of all compounds were determined with a deviation of <0.5.
Western Blot
The cells were lysed in lysis buffer (50 mm Tris-HCl, pH 7.5, 140 mm LiCl, 2 mm EDTA, pH 8, 0.5% Nonidet P-40, 1 Complete Mini EDTA-free Protease Inhibitor Mixture Tablet (Roche Applied Science)). Protein concentration of supernatants was determined with a Bradford Assay. Laemmli buffer was added (5:1), the samples were denatured at 95 °C for 5 min and were loaded to the gel (Criterion; Tris-glycine 4–20%). Bio-Rad Precision Plus Protein Standard Dual Color was used as a protein size reference. The gel was run at 200 V for 75 min in Tris-glycine buffer. The proteins were electroblotted to a PVDF membrane at 200 mA for 1 h. Washing steps in TBS-T buffer (3 × 10 min) were applied after blocking overnight with 5% BSA in TBS-T and after each incubation with antibodies. Anti-MMP-11 (Abcam, monoclonal mouse antibody, clone SL3.05 (ab85648)) was applied in blocking buffer (5% BSA in TBS-T buffer) at a 1:500 dilution for 1.5 h at room temperature. Goat anti-mouse HRP-labeled antibody from Dianova was diluted 1:10,000 and was applied for 1 h at room temperature. ECL Western blotting substrate (Pierce) was mixed 1:1 and applied to the membrane for 1.5 min at room temperature. The images were recorded on a Fuji LSA 4000 luminescent analyzer. The membrane was stripped (4 × 10 min 6 m guanidine HCl, 20 mm Tris-HCl, pH 7.5, 0.1 m β-mercaptoethanol, 0.2% Nonidet P-40). HRP-labeled anti-β-Actin antibody (Sigma-Aldrich; 1:20,000) was applied for 2 h at room temperature. Lanes from β-Actin were quantified with gel analyzing software (ImageJ; National Institutes of Health, Bethesda, MD). MMP-11 lanes were normalized against β-actin quantities and were evaluated graphically (GraphPad Prism 5.0). A t test was performed to test for significance of the displayed results.
Cellular Kinetic Assay
MIA PaCa-2 cells were a kind gift from Westfälische Wilhelms-Universität Münster, and MCF-7 were a kind gift from Universität Kiel, whereas Jurkat cells were taken from Leibniz-Institut für Molekulare Pharmakologie stock. All of the cells were tested for mycoplasma 2 weeks before the performed experiments (MycoAlertTM; Lonza). DMEM or RPMI 1640 was supplemented with 10% FBS and 1% penicillin/streptavidin (all from Invitrogen). The cell lines were cultured in Corning flasks, treated under a sterile hood, and incubated at 37 °C and 5% CO2 atmosphere. The cells were split and transferred at near confluence after treatment of 0.05% trypsin/EDTA in DMEM. 4000 cells/well were seeded 2 days prior to 384-well microplates (Corning number 3856 plate) in 50 μl of medium. Jurkat cells (RPMI 1640) were added to the microplate at the date of measurement in serum-free medium. 50 μl of 5 μm MMP-11 active substrate and negative control substrate dissolved in serum-free medium were added for measurement. Fluorescent background was recorded by the addition of Me2SO instead of substrate. Me2SO concentration in assay employed was 0.025%. The plates were covered with PCR-foil Ultra Clear RT-PCR to avoid evaporation. Fluorescence was measured with a Tecan Saphire2 reader at 483 ± 10 nm (excitation) and 525 ± 10 nm (emission). The results were analyzed by plotting the curves with Prism 5.0 (Graph Pad Software, Inc., San Diego, CA).
Confocal Laser Scanning Microscopy
Live cell images were recorded on a LSM 510 laser scanning confocal microscope from Zeiss equipped with a 63 × 1.2 W correction ring objective (C-Apochromat; Carl Zeiss Jena GmbH). A 200-milliwatt argon laser (488 nm, 5%) was used for excitation and a LP505 long pass filter for emission, respectively. 30,000 cells in 2 ml of DMEM medium (10% FBS, 1% penicillin/streptavidin) were seeded on a 30-mm round glass slide of 1-mm height (Thermo-Scientific) that were precoated with poly-l-lysine (Sigma Aldrich) 1 day prior in a 6-well Corning number 3331 plate. Cellular nuclei were stained with 10 μm Hoechst dye 33342 (Sigma Aldrich) right before live cell imaging. The glass slides were adjusted to a carrier and 500 μl of D-PBS were applied. Additional 500 μl of 10 μm imaging substrate in D-PBS were added to a final concentration of 5 μm substrate. A time series was recorded every 5 min for 30 min. Broad spectrum MMP inhibitor GM6001 (50 μm final, ilomastat, biomol, Ki(MMP-14) = 13.2 nm) was added 30 min prior to the addition of substrate (final concentration, 2.5 μm) and was incubated at 37 °C and 5% CO2.
RESULTS
Rational Design of FRET Substrates
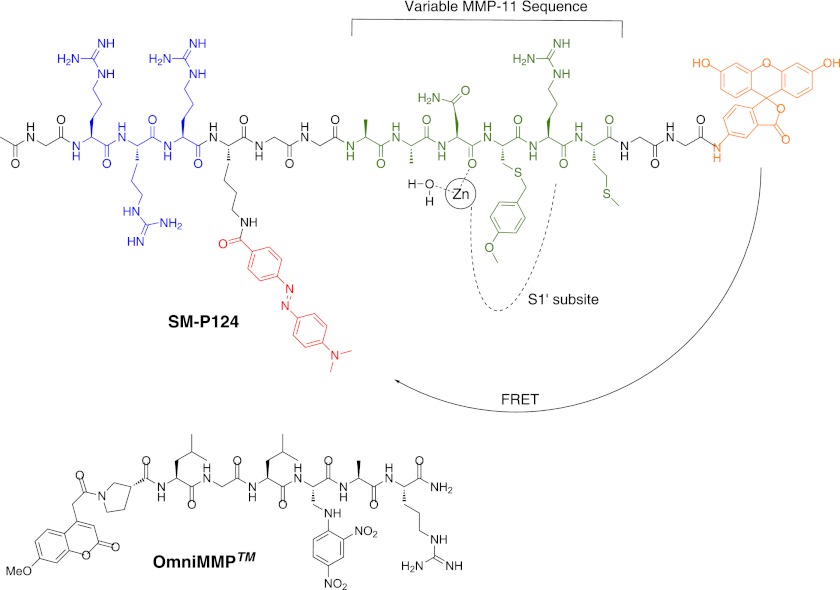
Substrate sequences for hMMP-11 discovered from phage display experiments (19) (AAN-LVR and YAE-LRM) were combined with sequences containing an unnatural amino acid residue in P1′ position (36), addressing the specificity pocket of MMP-11 (PLA-C(MeOBn)WAR). The latter sequence was previously designed for murine MMP-11. mMMP-11 is 10 times as active as its human isoform, because of a A235P mutation in the Met turn close to the binding cleft. The proline in mMMP-11 enables hydrogen bonding toward the binding substrate, stabilizing the substrate-enzyme complex, resulting in enhanced enzymatic activity of mMMP-11 (19). Introduction of the large, hydrophobic para-methoxybenzyl-protected cysteine residue C(MeOBn) in the P1′ site of substrates was reported to increase activity of mMMP-11 substrates 26-fold (37). Because the P1′ pocket is conserved from murine to human MMP-11 (19), employing this unnatural amino acid was anticipated to also increase substrate activity for hMMP-11. Further optimization of the recognition sequence for hMMP-11 was targeted by variation of the amino acids in the P3-P1 positions, employing the AAN, YAE, or PLA motif of the presented sequences. P2′ and P3′ were occupied either by the RM, VR, or WAR motif, respectively. The specificity pocket S1′ was addressed by leucine or 4-methoxybenzyl cysteine for affinity maturation. Furthermore, two glycine residues were added to each end of the recognition sequence as spacers to exclude interactions of the FRET pair with the enzyme. The FRET pair fluorescein and Dabcyl was selected, because longer excitation wavelengths (excitation, 483 nm; emission, 525 nm) proved to be preferable for screens and cellular measurements, avoiding correlation with autofluorescence (Fig. 1) (31). Moreover, Dabcyl acts as a dark quencher, absorbing the donor's emission and relaxing from its excited state via vibrational energy rather than light (Fig. 2, A and B). Thereby, Dabcyl efficiently reduces the fluorescence of fluorescein 35-fold, when in close proximity (Fig. 2C) (38). Cleavage of the constructed FRET-labeled peptide sequences by the catalytic domain of hMMP-11 therefore resulted in dequenching of fluorescein and the generation of a significant fluorescent signal.
FIGURE 1.
FRET substrate design. Orange, fluorescein as FRET donor; red, Dabcyl as FRET acceptor; green, MMP-11 recognition sequence; blue, cell penetrating unit/solubility adjustments. OmniMMPTM (standard MMP substrate) with leucine in P1′, N-terminal coumarin functions as a FRET donor, and dinitrophenyl in P2′ serving as FRET acceptor is not cleaved by hMMP-11.
FIGURE 2.
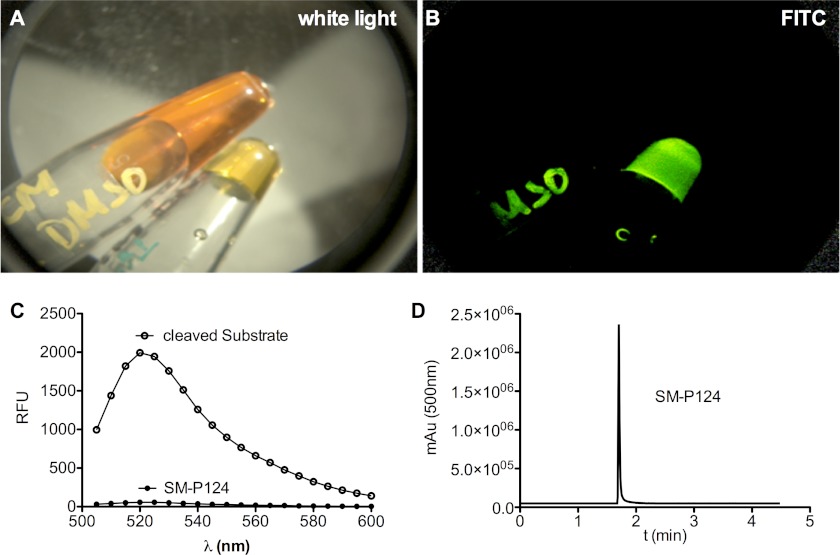
A, white light image. B, fluorescence filtered image. Intact SM-P124 in the upper vial shows the dark quenching effect, lacking fluorescence output of SM-P124. The lower vial represents completely hydrolyzed FRET substrate, demonstrating strong fluorescence. C, emission spectra of SM-P124 and its cleaved analog (including error bars). D, chromatogram of SM-P124 after cleavage from resin and reverse phase HPLC purification. RFU, relative fluorescence units.
Our preliminary investigations indicated low solubility (<10 μm in aqueous buffer) of the described FRET substrates. The addition of three arginines at the N terminus of the designed FRET substrates was found to enhance solubility sufficiently while leaving the energy transfer unaffected. Introduction of several arginines is known to increase cell permeability. Unfortunately, nona-arginine inserts also increase cell toxicity (39). Therefore, only three arginines were coupled to the N terminus of the prepared substrates. Consequently, a collection of 16 sequences (Table 1) engineered as cellular imaging tools by the addition of a FRET pair and inclusion of a solubility-enhancing RRR motif was designed.
TABLE 1.
Synthesized FRET substrates
Recognition sequences are represented in bold type.
| Entry | Sequence | Name | Mass |
|
|---|---|---|---|---|
| Calculated | Found | |||
| 1 | Ac-GRRRK(Dabcyl)GGYAENGG-Fluo (negative control) | SM-P123 | 943.4222 | 943.9315 (M+2)/2 |
| 2 | Ac-GRRRK(Dabcyl)GGAAN-C(MeOBn)RMGG-Fluo | SM-P124 | 2289.0341 | 2289.0422 |
| 3 | Ac-GRRRK(Dabcyl)GGNAA-C(MeOBn)RMGG-Fluo | SM-P125 | 1145.5171 | 1144.8886 (M+2)/2 |
| 4 | Ac-GRRRK(Dabcyl)GGYAE-C(MeOBn)RMGG-Fluo | SM-P127 | 2396.0600 | 2396.0668 |
| 5 | Ac-GRRRK(Dabcyl)GGAAN-LRMGG-Fluo | SM-P130 | 2179.0515 | 2179.0421 |
| 6 | Ac-GRRRK(Dabcyl)GGYAE-LRMGG-Fluo | SM-P133 | 2286.0774 | 2286.0633 |
| 7 | Ac-GRRRK(Dabcyl)GGAAN-C(MeOBn)VRGG-Fluo | SM-P148 | 1129.0347 | 1129.0343 (M+2)/2 |
| 8 | Ac-GRRRK(Dabcyl)GGYAE-C(MeOBn)VRGG-Fluo | SM-P151 | 2364.0880 | 2364.0724 |
| 9 | Ac-GRRRK(Dabcyl)GGAAN-LVRGG-Fluo | SM-P154 | 1074.0434 | 1074.0430 (M+2)/2 |
| 10 | Ac-GRRRK(Dabcyl)GGYAE-LVRGG-Fluo | SM-P157 | 2254.1053 | 2254.0928 |
| 11 | Ac-GRRRK(Dabcyl)GGPLA-C(MeOBn)WARGG-Fluo | SM-P166 | 1220.5871 | 1220.5859 (M+2)/2 |
| 12 | Ac-GRRRK(Dabcyl)GGAAN-C(MeOBn)WARGG-Fluo | SM-P169 | 1208.0587 | 1208.0588 (M+2)/2 |
| 13 | Ac-GRRRK(Dabcyl)GGYAE-C(MeOBn)WARGG-Fluo | SM-P172 | 2522.1360 | 2522.1140 |
| 14 | Ac-GRRRK(Dabcyl)GGPLA-LWARGG-Fluo | SM-P175 | 1165.5958 | 1165.5944 (M+2)/2 |
| 15 | Ac-GRRRK(Dabcyl)GGAAN-LWARGG-Fluo | SM-P178 | 1153.0674 | 1153.0669 (M+2)/2 |
| 16 | Ac-GRRRK(Dabcyl)GGYAE-LWARGG-Fluo | SM-P181 | 2412.1533 | 2412.1313 |
| 17 | Ac-RRRK(Dnp)GAAN-C(MeOBn)RMG-AMCA-NH2 | SM-P155 | 639.9545 | 640.297 (M+3)/3 |
| 18 | Mca-PLG-LDap(Dnp)AR-NH2a | OmniMMPTM | 1092.4989 | |
a Purchased from Enzo Life Sciences.
Synthesis of FRET Substrates
Synthesis of FRET substrates was performed on solid phase with C-terminally fluorescein labeled 2-chloro-trityl polystyrene resin. To all coupling solutions, 3 eq of DIPEA were added to prevent early cleavage of the attached fluorophore from the resin, because of acidification during the reaction. Additionally, methoxybenzyl-protected cysteine containing sequences afforded a refined cleavage mixture, because of cleavage condition interference of unnatural amino acid and the attached arginine motif with a total of up to four Pbf protecting groups in some sequences. The interference occurs because concentrations above 45% of TFA cleave the methoxybenzyl group off the cysteine in P1′ position. However, cleavage of the multiple Pbf protecting groups of the attached arginines affords 90% TFA as well as nucleophile scavengers such as triethylsilane (TIS) and water (40, 41). The cleavage mixture was therefore modified, whereas 40% TFA were employed with the addition of 1% phenol to capture cleaved Pbf groups, whereas cleavage was extended to 24 h at room temperature. EDT, TIS, and H2O were added as reducing agent and nucleophile scavengers, respectively. A representative HPLC spectrum is shown in Fig. 2D.
In Vitro Substrate Cleavage Evaluation
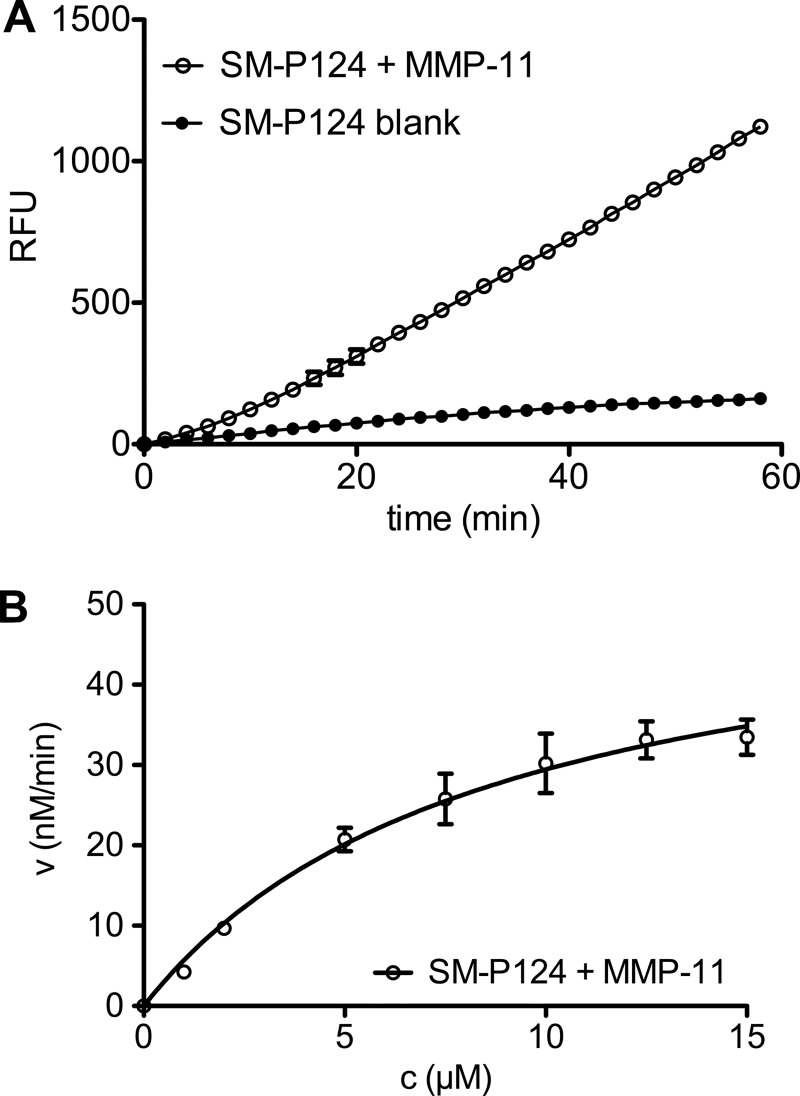
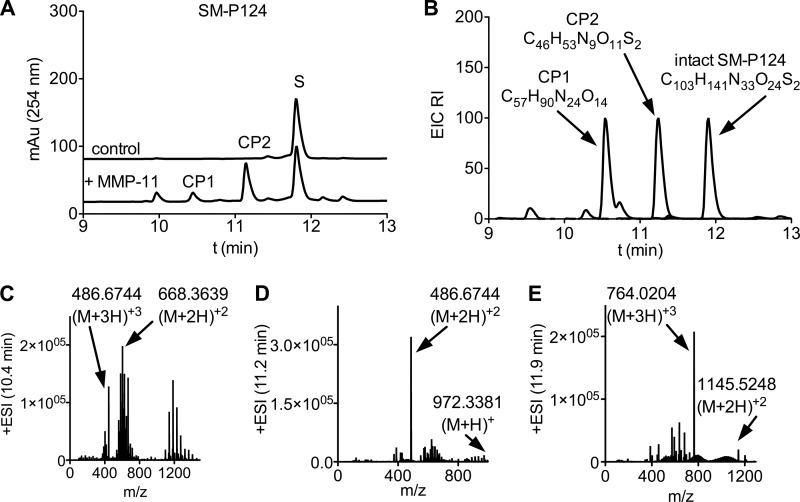
The synthesized substrates were evaluated in vitro with the catalytic domain of hMMP-11 by measuring increase of fluorescence over time (Fig. 3A). Maximal velocity of the enzymatic cleavage (Vmax) and rate of catalytic reaction (kcat) of this FRET assay were determined by a Michaelis-Menten fitting (Fig. 3B). Substrate cleavage was confirmed by identification of the cleavage products by high resolution mass spectrometry (LC/MS-TOF) (Fig. 4, A–E). Only four of the 16 prepared substrates were cleaved. Surprisingly, none of the literature-reported or commercially available sequences (Table 1, entries 6, 9, 11, and 18) were cleaved by hMMP-11 after derivatization as imaging tools.
FIGURE 3.
A, 12.5 nm MMP-11 incubated with 12. 5 μm FRET substrate SM-P124 and blank control (12.5 μm SM-P124 only). B, Michaelis-Menten plot of SM-P124 with 12.5 nm hMMP-11 catalytic domain. RFU, relative fluorescence units.
FIGURE 4.
A, chromatograms of 12. 5 μm SM-P124 (S). Top trace, blank control in assay buffer; bottom trace, 30 min after addition of 12.5 nm hMMP-11 cleavage product peaks (CP1 and CP2) are detected at 10.4 and 11.2 min. B, extracted ion chromatogram (EIC) overlay of CP1, CP2, and SM-P124. Composition formulas were determined by Agilent's LC/MS-TOF software and were in agreement with calculated formulas for cleaved fragments. C–E, mass spectra of extracted ion chromatograms. Cleaved fragments were identified by two or more corresponding mass peaks. ESI, electrospray ionization.
The four active sequences (SM-P124, SM-P125, SM-P130, and SM-P148) (Table 2) were also evaluated for their differential activity with hMMP-14 as selectivity is a challenging task, because the catalytic cleft of MMPs is highly conserved. MMP-14, similar to MMP-11, is also activated intracellulary and is secreted to the ECM (42).
TABLE 2.
In vitro evaluation of prepared FRET substrates
The values represent three independent measurements including three replicates. FRET substrates were all compared with the same batch of hMMP-11 cat domain.
| Entry | Name | hMMP-11 |
hMMP-14 |
||
|---|---|---|---|---|---|
| kcat/Km | Km | kcat/Km | Km | ||
| ×103m−1 s−1 | μm | ×103m−1 s−1 | μm | ||
| 1 | OmniMMPTM | Not cleaved | 75.23 | 9.93 ± 1.88 | |
| 2 | SM-P155 | 3.40 | 10.40 ± 4.11 | 51.70 | 39.27 ± 19.40 |
| 3 | SM-P130 | 1.27 | 5.30 ± 3.92 | 3.44 | 10.93 ± 4.51 |
| 4 | SM-P148 | 1.05 | 41.05 ± 181.1b | 9.65 | 25.75 ± 44.92b |
| 5 | SM-P124 | 2.40a | 4.96 ± 1.69 | 17.31 | 15.64 ± 3.25 |
| 6 | SM-P125 | 0.63 | 5.06 ± 3.15 | 6.76 | 23.44 ± 72.79 |
| 7 | SM-P123 (negative control) | Not cleaved | Not cleaved | ||
a The value differs from final value because of single tested enzyme batch.
b Approximate value because of high background fluorescence.
SM-P124 (Ac-GRRRK(Dabcyl)GGAAN-C(MeOBn)RMGG-Fluo) was validated as the most active fluorescein-Dabcyl labeled sequence for hMMP-11. Exchanging the P1 residue with the P3 residue resulted in reduced activity (SM-P125). Replacing the activity enhancing unnatural amino acid in P1′ by the usual leucine expectedly reduced the substrate activity for MMP-11. Interestingly, it enhanced selectivity by reducing activity for MMP-14 (SM-P130). SM-P148 was excluded for further analysis because it was not efficiently quenched and produced a strong background signal (data not shown).
To evaluate the impact of the selected FRET pair on activity and selectivity, the FRET pair of SM-P124 was exchanged against a low wavelength pair, AMCA-Dnp (SM-P155; excitation, 325 nm; emission, 393 nm). Indeed, introduction of the smaller, less space-demanding AMCA-Dnp pair resulted in a 1.5-fold activity increase, but surprisingly the affinity toward hMMP-11 dropped 2-fold. Additionally, activity for MMP-14 was strongly enforced, reducing selectivity.
Selectivity Evaluation of FRET Substrates
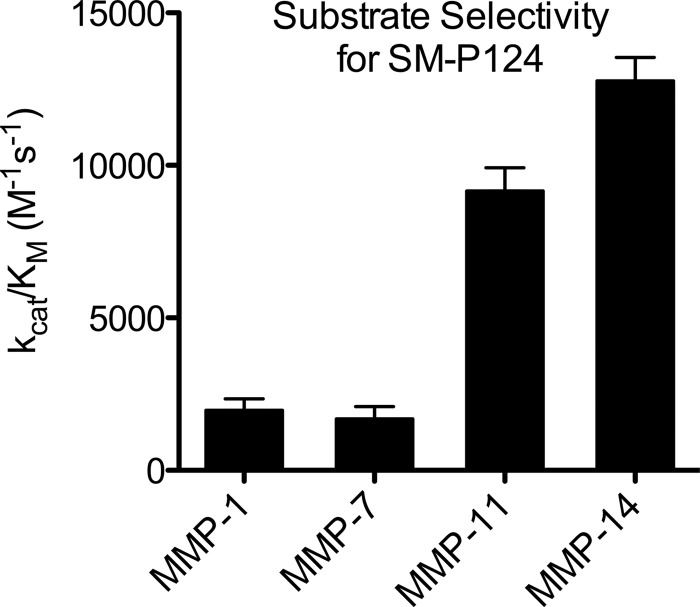
SM-P124 was furthermore tested with MMP-1, MMP-7, and MMP-14 (Table 3). Hardly any cleavage of the substrate bearing a methoxybenzyl-protected cysteine in P1′ position was observed for MMP-1 and MMP-7 in LC/MS-TOF measurements (data not shown). However, some fluorescence evolved over time, and a Michaelis-Menten plot was employed to determine the substrate activity. Fig. 5 compares the activities of SM-P124 with the activities of MMP-1, MMP-7, MMP-11, and MMP-14 and depicts the selectivity for MMP-11 and MMP-14.
TABLE 3.
Selectivity evaluation
The values represent three independent measurements, including three replicates each with a variation of hMMP-11 batches.
| Entry | Name | SM-P124 |
||
|---|---|---|---|---|
| kcat/Km | Km | kcat | ||
| ×103m−1 s−1 | μm | s−1 | ||
| 1 | MMP-1 | 1.49 | 5.46 ± 0.86 | 0.008 ± 0.001 |
| 2 | MMP-7 | 1.87 | 13.66 ± 5.08 | 0.025 ± 0.005 |
| 3 | MMP-11 | 9.18 | 8.23 ± 1.08 | 0.078 ± 0.004 |
| 4 | MMP-14 | 14.77 | 21.87 ± 6.55 | 0.310 ± 0.062 |
FIGURE 5.
kcat/Km values for SM-P124 with varying MMPs.
Comparison with Previously Published Substrates
Repetitive measurements of SM-P124 with different batches of hMMP-11 catalytic domain resulted in a final activity value of kcat/Km = 9.16 × 103 m−1 s−1, Km = 8 μm. Compared with the phage display-derived, unlabeled peptide MA-18 reported by Pan et al. (Ref. 19; kcat/Km = 2030 m−1 s−1, Km = 207 μm), SM-P124 was four times more active, and its affinity for hMMP-11 had increased 26-fold. At the same time, MA-18 preferred MMP-11 over MMP-14 by a factor of 10, whereas SM-P124, showing an affinity almost three times stronger for MMP-11 than for MMP-14, had 1.5-fold the activity for MMP-14 than for MMP-11 (kcat/Km(MMP-14) = 14820 m−1 s−1, Km = 22 μm).
Cellular Evaluation of FRET Substrates
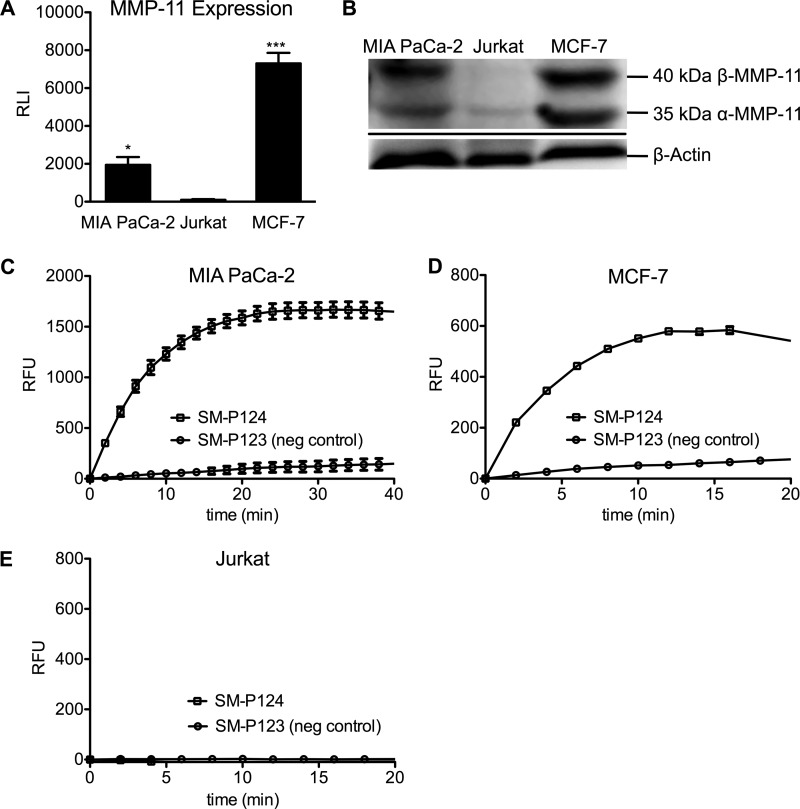
SM-P124 was evaluated in cellular assays with MIA PaCa-2, MCF-7, and Jurkat. MMP-11 expression of cell lines was validated via Western blot (Fig. 6, A and B) and showed strong expression of β-MMP-11 (37 kDa) as well as the inactivated α-MMP-11 (35 kDa) for MIA Paca-2 (p = 0.0002) and MCF-7 cells (p = 0.0114), whereas Jurkat cells were negative for MMP-11 expression. Fluorescence increase over time was monitored with all three cell lines after addition of FRET substrate SM-P124 and negative control SM-P123 (Ac-GRRRK(Dabcyl)-GYAENG-Fluo). Cleavage of SM-P124 was found in MMP-11 positive MIA PaCa-2 and MCF-7 cells with a maximum turnover after 20 min. SM-P124 turnover signal for MIA PaCa-2 cells increased 42-fold (SM-P123 was calculated as background signal) and 8-fold for MCF-7 cells (Fig. 6, C–E). This was surprising, because MMP-11 protein level was higher in MCF-7 than in MIA PaCa-2. Jurkat cells showed no fluorescent signal increase and therefore no substrate turnover. The negative control sequence SM-P123 yielded only insignificant increase in fluorescence over time when incubated with MIA PaCa-2 and MCF-7 cells, whereas no signal evolution was observed with Jurkat.
FIGURE 6.
A, MMP-11 expression normalized to β-actin. The asterisks represent a degree of significance to Jurkat expression. B, Western blot. C and D, 5 μm SM-P124 and SM-P123 with MIA PaCa-2 and MCF-7 cells confirming substrate cleavage for SM-P124. E, no signal increase was observed for both substrates with Jurkat cells. All data are presented with error bars. RLI, relative luminescence intensity; RFU, relative fluorescence units.
Laser Scanning Confocal Microscopy Substrate Cleavage Study
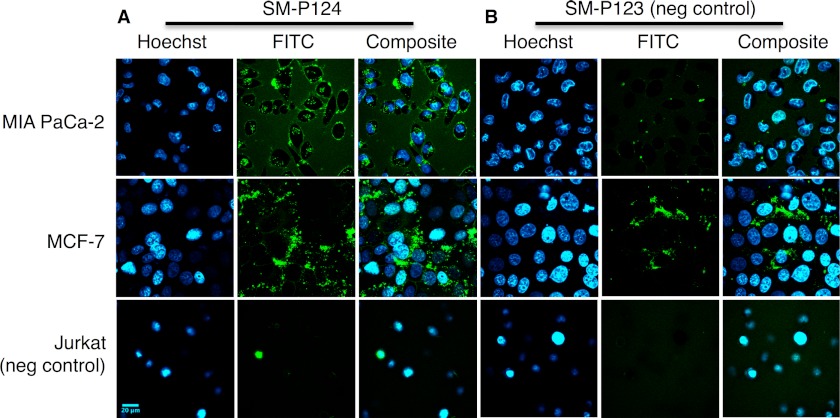
Laser scanning confocal microscopy images confirmed substrate cleavage of 5 μm SM-P124 (Fig. 7A) extracellularly and intracellularly in live MIA PaCa-2 and MCF-7 cells. The substrate was internalized immediately (t = 30 s) and stained the membrane and intracellular vesicles with both MMP-11-overexpressing cell lines. However, membrane staining of MIA PaCa-2 cells was not observed with negative control substrate SM-P123 (Fig. 7B). Only a faint fluorescent signal was detected intracellularly, proving internalization and hardly any unspecific cleavage of the negative control peptide. Additionally, SM-P124 produced a significant fluorescent signal in the surrounding medium of MIA PaCa-2 cells, whereas SM-P123 did not. MCF-7 cells, imaged in a monolayer that excluded all medium from the imaged plane, rather showed distinct membrane staining in between the cells in addition to the cytosolic distribution of intracellular substrate cleavage of SM-P124. Some signal evolution was also observed with SM-P123, pointing toward unspecific cleavage in cell monolayers. Still, the signal observed from SM-P124 cleavage is significantly stronger than that recorded of SM-P123. Images recorded with MMP-11 active SM-P124 and negative control peptide SM-P123 with Jurkat cells exhibited no fluorescence from the medium, nor membrane staining or intracellular cleavage.
FIGURE 7.
Images represent 146.25 μm × 146.25 μm slides. A, confocal images recorded 30 min after the addition of 5 μm SM-P124 to MIA PaCa-2, MCF-7, and Jurkat cells showing staining of medium, cell membrane, and intracellular vesicles with MMP-11 positive cells. B, 5 μm negative control SM-P123 yielded faint activity inside MIA PaCa-2 cells and some unspecific cleavage in MCF-7 monolayer. However, no signal evolution from Jurkat cells was observed.
Inhibition of MMPs Other than MMP-11
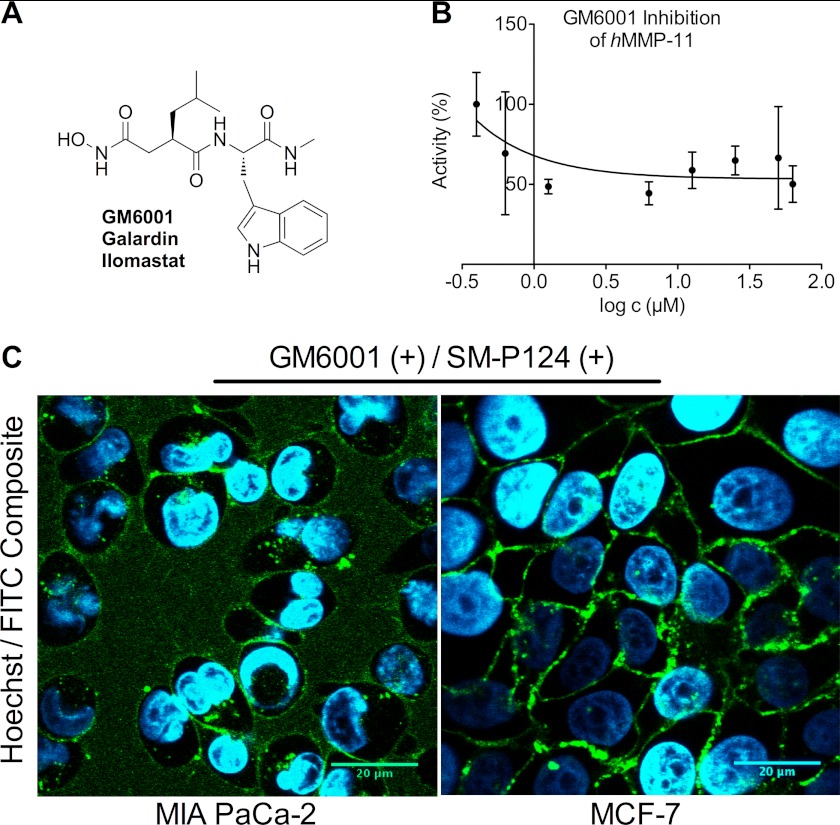
GM6001 (Fig. 8A) is a known broad spectrum inhibitor of, for example, MMP-1, -2, -3, -7, -8, -9, -10, -12, and -14 (43–45) that is able to also inhibit intracellular targets (46). The inhibitor was tested with the catalytic domain of hMMP-11, and GM6001 failed to inhibit substrate cleavage of SM-P124 (Fig. 8B). The substrate turnover rate was solely reduced to 50%. Therefore, live MIA PaCa-2 and MCF-7 cells were incubated with GM6001 to block MMP activity other than MMP-11. Confocal microscopy revealed the same specific cleavage pattern of SM-P124 (2.5 μm) on MIA PaCa-2 and MCF-7 cells (Fig. 8B). Again, cleavage was observed in the medium as well as on the membrane and in intracellular vesicles of MIA PaCa-2 cells. MCF-7 cells showed predominant extracellular membrane staining, because of accumulation of secreted protein to the extracellular space, which was limited in densely seeded cells.
FIGURE 8.
A, chemical structure of broad MMP inhibitor GM6001. B, inhibition of hMMP-11 catalytic domain with GM6001 resulted in a 50% decrease of the substrate turnover rate and therefore failed to diminish substrate cleavage of SM-P124. C, images represent 92.14-μm × 92.14-μm slides and were recorded 35 min after the addition of 2.5 μm SM-P124 to cells after prior incubation with 50 μm GM6001. MMP-11 specific cleavage of SM-P124 is observed especially intracellularly in MIA PaCa-2 and at the membrane of MCF-7 cells.
DISCUSSION
Considering the recorded data of FRET substrates for hMMP-11 several conclusions can be drawn. MMP-11 substrates require the unnatural, large, hydrophobic side chain of 4-methoxybenzyl-cysteine for significant activity. The most active fluorescein-Dabcyl-labeled substrate for hMMP-11, SM-P124 (Ac-GRRRK(Dabcyl)GGAAN-C(MeOBn)RMGG-Fluo) possessed arginine and methionine in P2′ and P3′, respectively, rather than valine and arginine as in SM-P148, which reduced FRET efficacy. The AAN motif was essential in positions P3-P1 because substrates harboring the YAE or PLA motif in this position were inactive. Alanine was essential in P3 because substrates containing proline or tyrosine in P3 were also inactive. Asparagine was tolerated in P3; however, the shorter amide side chain led to a 2-fold decrease in activity (SM-P125). C(MeOBn) binding to the S1′ pocket increased activity (SM-P124) but also increased activity for MMP-14. Because MMP-11 is the only metalloprotease presenting a glutamine in position 215 in the S1′ pocket, an interaction with this specific amino acid should strongly increase selectivity for MMP-11 (37, 47). This hypothesis could not be confirmed, in fact, leucine in P1′ increased the activity ratio of MMP-11 over MMP-14 6-fold (SM-P130).
In summary, the rational combination of native amino acid sequences with an unnatural side chain in P1′ and modification with a fluorescein-Dabcyl FRET pair yielded the first successfully evaluated, cellularly active FRET substrate for hMMP-11. SM-P124 displayed a 4-fold increased activity compared with nonlabeled substrates for hMMP-11 (19). It was selective for MMP-11 in comparison with MMP-1 and MMP-7. This is in agreement with structural data of the S1′ pocket of MMP-1 and MMP-7, harboring arginine and tyrosine, respectively, where MMP-11 holds a glutamine and where leucine is found in all other MMPs (37). Therefore, MMP-1 and MMP-7 form a shallow pocket, interfering with long, unnatural residues like Cys(MeOBn) and resulting in near inactivity of SM-P124 for these MMPs. On the contrary, SM-P124 was recognized as a substrate by MMP-14 with similar activity as for MMP-11.
The designed imaging probes were successfully evaluated at the cellular level. This is the first time that MMP-11 activity was visualized in living cells. Fluorescence that evolved from the medium and membrane was pointing toward cleavage from secreted MMP-11, whereas endosomal staining by SM-P124 in MIA PaCa-2 cells suggested the presence of MMP-11 activity within the vesicles. In agreement, the negative control peptide SM-P123 hardly displayed any intracellular fluorescence with MMP-11 positive cell lines and none at all with the negative control cell line Jurkat. These findings consolidated MMP-specific signal evolution of SM-P124 as well as cellular substrate stability, because of minimal unspecific cleavage of SM-P123. It proves that SM-P124 is cleaved through protease cleavage rather than lysosomal degradation. Furthermore, lysosomal degradation affords pH values of ∼4, at which fluorescein hardly emits any fluorescence and at which MMPs stop functioning. In MCF-7 cells in vitro fluorescence measurement over time indicated a less intensive signal evolution than for MIA PaCa-2 cells, even though the Western blot showed higher MMP-11 protein concentration in MCF-7 cells. These differences were only observed in the fluorescence plate reader. MIA PaCa-2 cells showed a strong fluorescence of the medium during confocal microscopy, resulting in a higher overall signal in the plate reader. Because of clustering of MCF-7 cells, less surface for secretion is available, and secreted α-MMP-11 will distribute in the connecting cell tissue, as observed in the confocal images. Minor unspecific cleavage was observed with SM-P123 in MCF-7 in the connective tissue. However, the intracellular signal detected was only observed with SM-P124.
Inhibition of the MMP family with the exception of MMP-11, with GM6001 resulted in substrate cleavage of SM-P124, caused by the ineffectiveness of ilomastat for hMMP-11. It was shown that selectivity concerns can therefore be addressed by blocking broad MMP activity while retaining specific MMP-11 activity. This creates novel opportunities also for in vivo applications and could be especially interesting for orthotopic model studies in terms of target distribution. The flexible design of the substrate allows an exchange of the FRET pair by nearly infrared fluorophores for noninvasive optical imaging. Proteolytic stability of the substrate for in vivo applications and its systemic retention can be increased by substrate coupling to a polymeric support. This enhances the pharmacokinetic properties of the substrate in vivo and enables MMP-11 imaging in animal models (48).3 Because conventional MMP substrates are not cleaved by MMP-11, this new tool will allow in vivo imaging with focus on pancreatic or breast adenocarcinoma animal models.
In addition, SM-P124 as an imaging substrate for MMP-11 serves as a potent chemical tool to further study its physiological role. It might help to explain the entanglement of MMP-11 in early invasion and the negative prognosis of MMP-11 positive cancers. Moreover, it can contribute to the development of targeted diagnostic approaches to monitor tumor progression or even increase understanding of the reported degradation of insulin-like growth factor-binding protein 1, because it results in the activation of insulin-like growth factor II and therefore in enhanced cell survival and proliferation of diseased, carcinogenic tissue (49, 50). Likewise, MMP-11 cleaves serine protease inhibitors, thereby influencing severe ECM turnover and was shown to cleave two intracellular proteins, namely heat shock protein 90β1 and galectin-1, the latter being associated with tumor cell migration, inflammation stimulation, and apoptosis of activated T cells (51, 52). Therefore, studies investigating the role of both isoforms, but especially nonsecreted β-MMP-11, clarifying its regulation or localization, are lacking. Optical imaging with SM-P124 might elucidate the role of intra- and extracellularly active MMP-11, because colocalization with potential substrates might reveal its regulation and mechanism of action.
Acknowledgments
We especially thank Christoph Bremer and Anke Hahnenkamp for the MIA PaCa-2 cell line, Günther Klöppel and Bence Sipos for providing the MCF-7 cell line, Jens-Peter von Kries for the cell culturing opportunity, and Burkhard Wiesner for the confocal microscope opportunity
This work was supported by MolDiag-PaCa, Sixth Framework Programme Priority Grant LSH.2004-1.2.2.1.
B. S. Meyer, K. Koschek, A. Hahnenkamp, C. Bremer, and J. Rademann, manuscript in preparation.
- MMP
- matrix metalloprotease
- AMCA
- 7-amino-4-methyl-coumarin acetamide
- C(MeOBn)
- 4-methoxybenzyl-protected cysteine
- Dabcyl
- 4-[4-(dimethyl-amino)-phenylazo)benzoyl]-l-lysine
- Dap
- diaminopropyl
- Dnp
- dinitrophenyl
- FRET
- Förster resonance energy transfer
- GM6001
- galardin/ilomastat
- HOBt
- hydroxy benzotriazole
- Mca
- 7-methoxycoumarin
- Pbf
- 2,2,4,6,7-pentamethyldihydrobenzofuran-5-sulfonyl
- ECM
- extracellular matrix
- h
- human
- m
- murine
- Fmoc
- N-(9-fluorenyl)methoxycarbonyl
- HATU
- O-(7-azabenzotriazol-1-yl)-N,N,N′,N′-tetramethyl-uronium hexafluorophosphate
- DIC
- diisopropylcarbodiimide
- DIPEA
- diisopropylethylamine
- TIS
- triethylsilane
- EDT
- ethane-1,2-dithiol
- DCM
- dichloromethane
- DMF
- dimethylformamide.
REFERENCES
- 1. Snoek-van Beurden P. A., Von den Hoff J. W. (2005) Zymographic techniques for the analysis of matrix metalloproteinases and their inhibitors. BioTechniques 38, 73–83 [DOI] [PubMed] [Google Scholar]
- 2. Andreini C., Banci L., Bertini I., Luchinat C., Rosato A. (2004) Bioinformatic comparison of structures and homology-models of matrix metalloproteinases. J. Proteome Res. 3, 21–31 [DOI] [PubMed] [Google Scholar]
- 3. Masson R., Lefebvre O., Noël A., Fahime M. E., Chenard M. P., Wendling C., Kebers F., LeMeur M., Dierich A., Foidart J. M., Basset P., Rio M. C. (1998) In vivo evidence that the stromelysin-3 metalloproteinase contributes in a paracrine manner to epithelial cell malignancy. J. Cell Biol. 140, 1535–1541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Arora S., Kaur J., Sharma C., Mathur M., Bahadur S., Shukla N. K., Deo S. V., Ralhan R. (2005) Stromelysin 3, Ets-1, and vascular endothelial growth factor expression in oral precancerous and cancerous lesions. Correlation with microvessel density, progression, and prognosis. Clin. Cancer Res. 11, 2272–2284 [DOI] [PubMed] [Google Scholar]
- 5. Baldini E., Toller M., Graziano F. M., Russo F. P., Pepe M., Biordi L., Marchioni E., Curcio F., Ulisse S., Ambesi-Impiombato F. S., D'Armiento M. (2004) Expression of matrix metalloproteinases and their specific inhibitors in normal and different human thyroid tumor cell lines. Thyroid 14, 881–888 [DOI] [PubMed] [Google Scholar]
- 6. Leivo I., Jee K., Heikinheimo K., Laine M., Ollila J., Nagy B., Knuutila S. (2005) Characterization of gene expression in major types of salivary gland carcinomas with epithelial differentiation. Cancer Gen. Cytogen. 156, 104–113 [DOI] [PubMed] [Google Scholar]
- 7. Denys H., De Wever O., Nusgens B., Kong Y., Sciot R., Le A. T., Van Dam K., Jadidizadeh A., Tejpar S., Mareel M., Alman B., Cassiman J. J. (2004) Invasion and MMP expression profile in desmoid tumours. Br. J. Cancer 90, 1443–1449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yamashita K., Tanaka Y., Mimori K., Inoue H., Mori M. (2004) Differential expression of MMP and uPA systems and prognostic relevance of their expression in esophageal squamous cell carcinoma. Int. J. Cancer 110, 201–207 [DOI] [PubMed] [Google Scholar]
- 9. Basset P., Bellocq J. P., Wolf C., Stoll I., Hutin P., Limacher J. M. (1990) A novel metalloproteinase gene specifically expressed in stromal cells of breast carcinomas. Nature 348, 699–704 [DOI] [PubMed] [Google Scholar]
- 10. Jones L. E., Humphreys M. J., Campbell F., Neoptolemos J. P., Boyd M. T. (2004) Comprehensive analysis of matrix metalloproteinase and tissue inhibitor expression in pancreatic cancer. Increased expression of matrix metalloproteinase-7 predicts poor survival. Clin. Cancer Res. 10, 2832–2845 [DOI] [PubMed] [Google Scholar]
- 11. Ahmad A., Hanby A., Dublin E., Poulsom R., Smith P., Barnes D., Rubens R., Anglard P., Hart I. (1998) Stromelysin 3. An independent prognostic factor for relapse-free survival in node-positive breast cancer and demonstration of novel breast carcinoma cell expression. Am. J. Pathol. 152, 721–728 [PMC free article] [PubMed] [Google Scholar]
- 12. Rouyer N., Wolf C., Chenard M. P., Rio M. C., Chambon P., Bellocq J. P., Basset P. (1994) Stromelysin-3 gene expression in human cancer. An overview. Invasion Metastasis 14, 269–275 [PubMed] [Google Scholar]
- 13. Thewes M., Pohlmann G., Atkinson M., Mueller J., Pütz B., Höfler H. (1996) Stromelysin-3 (ST-3) mRNA expression in colorectal carcinomas. Localization and clinicopathologic correlations. Diagn. Mol. Pathol. 5, 284–290 [DOI] [PubMed] [Google Scholar]
- 14. Peruzzi D., Mori F., Conforti A., Lazzaro D., De Rinaldis E., Ciliberto G., La Monica N., Aurisicchio L. (2009) MMP-11. A novel target antigen for cancer immunotherapy. Clin. Cancer Res. 15, 4104–4113 [DOI] [PubMed] [Google Scholar]
- 15. Laurell H., Bouisson M., Berthelemy P., Rochaix P., Dejean S., Besse P., Susini C., Pradayrol L., Vaysse N., Buscail L. (2006) Identification of biomarkers of human pancreatic adenocarcinomas by expression profiling and validation with gene expression analysis in endoscopic ultrasound-guided fine needle aspiration samples. World J. Gastroenterol., 12, 3344–3351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fromigué O., Louis K., Wu E., Belhacène N., Loubat A., Shipp M., Auberger P., Mari B. (2003) Active stromelysin-3 (MMP-11) increases MCF-7 survival in three-dimensional Matrigel culture via activation of p42/p44 MAP-kinase. Int. J. Cancer 106, 355–363 [DOI] [PubMed] [Google Scholar]
- 17. Pei D., Weiss S. J. (1995) Furin-dependent intracellular activation of the human stromelysin-3 zymogen. Nature 375, 244–247 [DOI] [PubMed] [Google Scholar]
- 18. Noël A., Santavicca M., Stoll I., L'Hoir C., Staub A., Murphy G., Rio M. C., Basset P. (1995) Identification of structural determinants controlling human and mouse stromelysin-3 proteolytic activities. J. Biol. Chem. 270, 22866–22872 [DOI] [PubMed] [Google Scholar]
- 19. Pan W., Arnone M., Kendall M., Grafstrom R. H., Seitz S. P., Wasserman Z. R., Albright C. F. (2003) Identification of peptide substrates for human MMP-11 (stromelysin-3) using phage display. J. Biol. Chem. 278, 27820–27827 [DOI] [PubMed] [Google Scholar]
- 20. Chaussain-Miller C., Fioretti F., Goldberg M., Menashi S. (2006) The role of matrix metalloproteinases (MMPs) in human caries. J. Dent. Res. 85, 22–32 [DOI] [PubMed] [Google Scholar]
- 21. Delain E., Pochon F., Barray M., Van Leuven F. (1992) Ultrastructure of α2-macroglobulins. Electron Microsc. Rev. 5, 231–281 [DOI] [PubMed] [Google Scholar]
- 22. del Mar Barbacid M., Fernández-Resa P., Buesa J. M., Márquez G., Aracil M., Quesadaand A. R., Mira E. (1998) Expression and purification of human stromelysin 1 and 3 from baculovirus-infected insect cells. Protein Expr. Purif. 13, 243–250 [DOI] [PubMed] [Google Scholar]
- 23. Schmitt M., Harbeck N., Thomssen C., Wilhelm O., Magdolen V., Reuning U., Ulm K., Höfler H., Jänicke F., Graeff H. (1997) Clinical impact of the plasminogen activation system in tumor invasion and metastasis. Prognostic relevance and target for therapy. Thromb. Haemost. 78, 285–296 [PubMed] [Google Scholar]
- 24. Luo D., Mari B., Stoll I., Anglard P. (2002) Alternative splicing and promoter usage generates an intracellular stromelysin 3 isoform directly translated as an active matrix metalloproteinase. J. Biol. Chem. 277, 25527–25536 [DOI] [PubMed] [Google Scholar]
- 25. Visse R., Nagase H. (2003) Matrix metalloproteinases and tissue inhibitors of metalloproteinases. Structure, function, and biochemistry. Circ. Res. 92, 827–839 [DOI] [PubMed] [Google Scholar]
- 26. Weissleder R., Mahmood U. (2001) Molecular imaging. Radiology 219, 316–333 [DOI] [PubMed] [Google Scholar]
- 27. Kovar J. L., Simpson M. A., Schutz-Geschwender A., Olive D. M. (2007) A systematic approach to the development of fluorescent contrast agents for optical imaging of mouse cancer models. Anal. Biochem. 367, 1–12 [DOI] [PubMed] [Google Scholar]
- 28. Lee S., Xie J., Chen X. (2010) Peptides and peptide hormones for molecular imaging and disease diagnosis. Chem. Rev. 110, 3087–3111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Förster T. (1948) Intermolecular energy transfer and fluorescence. Annals of Physics 6, 55–75 [Google Scholar]
- 30. Kannan R., Ruff M., Kochins J. G., Manly S. P., Stoll I., El Fahime M., Noël A., Foidart J. M., Rio M. C., Dive V., Basset P. (1999) Purification of active matrix metalloproteinase catalytic domains and its use for screening of specific stromelysin-3 inhibitors. Protein Expr. Purif. 16, 76–83 [DOI] [PubMed] [Google Scholar]
- 31. Monici M. (2005) Cell and tissue autofluorescence research and diagnostic applications. Biotechnol. Annu. Rev. 11, 227–256 [DOI] [PubMed] [Google Scholar]
- 32. Uryga-Polowy V., Kosslick D., Freund C., Rademann J. (2008) Resin-bound aminofluorescein for C-terminal labeling of peptides. High-affinity polarization probes binding to polyproline-specific GYF domains. ChemBioChem 9, 2452–2462 [DOI] [PubMed] [Google Scholar]
- 33. Coin I., Beyermann M., Bienert M. (2007) Solid-phase peptide synthesis. From standard procedures to the synthesis of difficult sequences. Nat. Protoc. 2, 3247–3256 [DOI] [PubMed] [Google Scholar]
- 34. Wood W. J., Patterson A. W., Tsuruoka H., Jain R. K., Ellman J. A. (2005) Substrate activity screening. A fragment-based method for the rapid identification of nonpeptidic protease inhibitors. J. Am. Chem. Soc. 127, 15521–15527 [DOI] [PubMed] [Google Scholar]
- 35. Nuti E., Panelli L., Casalini F., Avramova S. I., Orlandini E., Santamaria S., Nencetti S., Tuccinardi T., Martinelli A., Cercignani G., D'Amelio N., Maiocchi A., Uggeri F., Rossello A. (2009) Design, synthesis, biological evaluation, and NMR studies of a new series of arylsulfones as selective and potent matrix metalloproteinase-12 inhibitors. J. Med. Chem. 52, 6347–6361 [DOI] [PubMed] [Google Scholar]
- 36. Mucha A., Cuniasse P., Kannan R., Beau F., Yiotakis A., Basset P., Dive V. (1998) Membrane type-1 matrix metalloprotease and stromelysin-3 cleave more efficiently synthetic substrates containing unusual amino Acids in their P1′ positions. J. Biol. Chem. 273, 2763–2768 [DOI] [PubMed] [Google Scholar]
- 37. Holtz B., Cuniasse P., Boulay A., Kannan R., Mucha A., Beau F., Basset P., Dive V. (1999) Role of the S1′ subsite glutamine 215 in activity and specificity of stromelysin-3 by site-directed mutagenesis. Biochemistry 38, 12174–12179 [DOI] [PubMed] [Google Scholar]
- 38. Nazarenko I. A., Bhatnagar S. K., Hohman R. J. (1997) A closed tube format for amplification and detection of DNA based on energy transfer. Nucleic Acids Res. 25, 2516–2521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Fuchs S. M., Rutkoski T. J., Kung V. M., Groeschl R. T., Raines R. T. (2007) Increasing the potency of a cytotoxin with an arginine graft. Protein Eng. Des. Sel. 20, 505–509 [DOI] [PubMed] [Google Scholar]
- 40. Ramage R., Green J., Blake A. J. (1991) An acid labile arginine derivative for peptide synthesis. NG-2,2,5,7,8-pentamethylchroman-6-sulphonyl-l-arginine. Tetrahedron 47, 6353–6370 [Google Scholar]
- 41. Isidro-Llobet A., Alvarez M., Albericio F. (2009) Amino acid-protecting groups. Chem. Rev. 109, 2455–2504 [DOI] [PubMed] [Google Scholar]
- 42. Apte S. S., Fukai N., Beier D. R., Olsen B. R. (1997) The matrix metalloproteinase-14 (MMP-14) gene is structurally distinct from other MMP genes and is co-expressed with the TIMP-2 gene during mouse embryogenesis. J. Biol. Chem. 272, 25511–25517 [DOI] [PubMed] [Google Scholar]
- 43. Saghatelian A., Jessani N., Joseph A., Humphrey M., Cravatt B. F. (2004) Activity-based probes for the proteomic profiling of metalloproteases. Proc. Natl. Acad. Sci. U.S.A. 101, 10000–10005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Yang J., Zhang Z., Lin J., Lu J., Liu B. F., Zeng S., Luo Q. (2007) Detection of MMP activity in living cells by a genetically encoded surface-displayed FRET sensor. Biochim. Biophys. Acta 1773, 400–407 [DOI] [PubMed] [Google Scholar]
- 45. Ledour G., Moroy G., Rouffet M., Bourguet E., Guillaume D., Decarme M., Elmourabit H., Augé F., Alix A. J., Laronze J. Y., Bellon G., Hornebeck W., Sapi J. (2008) Introduction of the 4-(4-bromophenyl)benzenesulfonyl group to hydrazide analogs of ilomastat leads to potent gelatinase B (MMP-9) inhibitors with improved selectivity. Bioorg. Med. Chem. 16, 8745–8759 [DOI] [PubMed] [Google Scholar]
- 46. Balakrishnan A., Patel B., Sieber S. A., Chen D., Pachikara N., Zhong G., Cravatt B. F., Fan H. (2006) Metalloprotease inhibitors GM6001 and TAPI-0 inhibit the obligate intracellular human pathogen Chlamydia trachomatis by targeting peptide deformylase of the bacterium. J. Biol. Chem. 281, 16691–16699 [DOI] [PubMed] [Google Scholar]
- 47. Gall A. L., Ruff M., Kannan R., Cuniasse P., Yiotakis A., Dive V., Rio M. C., Basset P., Moras D. (2001) Crystal structure of the stromelysin-3 (MMP-11) catalytic domain complexed with a phosphinic inhibitor mimicking the transition-state. J. Mol. Biol. 301, 577–586 [DOI] [PubMed] [Google Scholar]
- 48. Bremer C., Bredow S., Mahmood U., Weissleder R., Tung C. H. (2001) Optical imaging of matrix metalloproteinase-2 activity in tumors. Feasibility study in a mouse model. Radiology 221, 523–529 [DOI] [PubMed] [Google Scholar]
- 49. Egeblad M., Werb Z. (2002) New functions for the matrix metalloproteinases in cancer progression. Nat. Rev. Cancer 2, 161–174 [DOI] [PubMed] [Google Scholar]
- 50. Hartmann W., Koch A., Brune H., Waha A., Schüller U., Dani I., Denkhaus D., Langmann W., Bode U., Wiestler O. D., Schilling K., Pietsch T. (2005) Insulin-like growth factor II is involved in the proliferation control of medulloblastoma and its cerebellar precursor cells. Am. J. Pathol. 166, 1153–1162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Mañes S., Mira E., Barbacid M. M., Ciprés A., Fernández-Resa P., Buesa J. M., Mérida I., Aracil M., Márquez G., Martínez-A C. (1997) Identification of insulin-like growth factor-binding protein-1 as a potential physiological substrate for human stromelysin-3. J. Biol. Chem. 272, 25706–25712 [DOI] [PubMed] [Google Scholar]
- 52. Kleifeld O., Doucet A., auf dem Keller U., Prudova A., Schilling O., Kainthan R. K., Starr A. E., Foster L. J., Kizhakkedathu J. N., Overall C. M. (2010) Isotopic labeling of terminal amines in complex samples identifies protein N-termini and protease cleavage products. Nat. Biotechnol. 28, 281–288 [DOI] [PubMed] [Google Scholar]