Abstract
This paper describes a possible application of luminescent Escherichia coli activated by blood serum for high-sensitivity and high-specificity assays of antibiotics in solutions. Antibiotics inhibited luminescence of a genetically engineered E. coli strain; the system sensitivity to some antibiotics grew notably after the cells had been preactivated by blood serum. The highest level of sensitivity (2.8 ± 0.6 ng/ml) of luminescent cells was obtained for aminoglycoside antibiotics (gentamicin and streptomycin). It is feasible to create the specific biosensor for antibiotics on the basis of bioluminescent E. coli strains by applying sera containing antibodies against the antibiotic under assay. The presence of antibodies specific for gentamicin in serum affects inhibition of luminescent cells by gentamicin but not inhibition by other antibiotics.
The widespread use of antibiotics in medicine, veterinary, biotechnology, and agriculture necessitates the application of high-sensitivity, specific, and simple methods for their detection, with the lower detectable concentrations ranging from 1 to 50 ng/ml. Antibiotics are assayed chromatographically, immunochemically, microbiologically, and by other methods (17). Bioluminescence procedures are among the most speedy and sensitive detection assays (1, 4, 12, 17, 18). The use of recombinant Escherichia coli strains (with genes incorporated from a bacterial luminescent system derived from marine bacteria) notably simplified the experimental assay using luminescent bacteria. Luminescent E. coli strains and some luminescent strains of other bacteria were used to assay the antibacterial effect of many antimicrobial agents (3, 6, 14, 15, 18, 20). The increase in the light response seen with a luxCDABE-based bioluminescence bacterium after exposure to some organic solvents was due to changed fatty acid synthesis patterns that affected the aldehyde supply for the bioluminescence reaction (7). The membranolytic effect of blood serum was studied by the use of E. coli with an insect luc gene (19). Although luminescent bacteria have been applied in tests of antibiotics, the problem of assay specificity has always been a concern (12, 17). Previous publications described a recombinant E. coli strain capable of luminescence only in the presence of tetracycline-type antibiotics (9-11, 13).
Earlier, we explored the effect of antibiotics and various samples of blood serum on luminescence of a recombinant E. coli strain (2). Streptomycin was used to exemplify a notable sensitization of serum-activated cells to the antibiotics. The objective of this research was to provide a high level of assay specificity for antibiotics by the use of recombinant E. coli strains. We propose the application of the specific immune sera as an antibiotic detector. The luminescence of cells treated by antibiotics should be less inhibited in the presence of antibodies, since antibodies trap a portion of the antibiotic. In this research, gentamicin was used to demonstrate antibiotic-caused inhibition effects on recombinant an E. coli strain.
Bacterial strains.
A luminescent E. coli strain was constructed using standard techniques (16). The bacterium E. coli K12 TG1 was used as a recipient of hybrid plasmids with the insertion of luxCDABE of Photobacterium leiognathi 54D10 (from the collection of M. V. Lomonosov Moscow State University) (5). The luxCDABE genes were isolated by the use of the vector plasmid pUC19. The luminescent E. coli TG1 clones were grown at 28°C up to the mid-logarithmic growth phase under submerged conditions in culture medium containing (in grams/liter) NaCl (5), Na2HPO4 · 12H2O (5.3), KH2PO4 · 2H2O (2.1), (NH4)2HPO4 (0.5), MgSO4 · 7 H2O (0.1), peptone (10), yeast extract (1), and glycerol (3 ml/liter) and supplemented with 100 μg ampicillin/ml and were then centrifuged at 400 × g for 20 min. The precipitated bacteria were resuspended in the cooled protective medium containing gelatin (1%), sodium glutamate (1%), and saccharose (10%) (pH 7.5 to 7.8) (5). The cell density was adjusted to 7 × 1010 to 2 × 1011 cells/ml. The cell aliquots were placed in glass weighing bottles, slowly frozen by liquid nitrogen, and lyophilized with a New Brunswick instrument (St. Albans, Hertfordshire, United Kingdom) for 20 h.
Sample preparation.
Lyophilized cells were rehydrated with cold distilled water and allowed to stay at 4°C for 0.5 h. The cells were than diluted with 5 mM HEPES-Na buffer (pH 7.2) to a concentration of (2.5 ± 1.0) × 107 cells/ml. Solutions of antibiotics (1 to 2 mg/ml) were prepared in the buffer or methanol and diluted with 5 mM HEPES-Na buffer to the desired concentration before being added to cells. For cell activation by serum, the serum aliquot (20 μl/ml) was added to E. coli suspension and incubated for 20 min; 0.05% Tween 20 was also added to the cells. Samples of normal sera as well as of antisera were from Immunotek (Moscow, Russia). The aliquots (0.8 ml) of serum-activated cells were placed in the luminometer measurement cuvettes and supplemented with 0.2 ml of antibiotic solution; the control sample of cells was supplemented with buffer. Finally, the cell concentration in the measurement cuvette was (2 ± 1) × 107 cells/ml, the quantity of serum amounted to 16 μl/ml (with no serum used in experiments with native cells), and the antibiotic concentration was adjusted.
Luminescence measurements.
The luminescence was recorded at 23°C with an LKB-1250 luminometer (LKB-Wallac, Turku, Finland). In every experiment up to eight samples were measured, with replicate measurements of identical sample preparations (usually n = 3); one of the samples served as a control. The luminescence intensity (I) was measured every 10 min after the antibiotic had been added, and the mean (± standard deviation [SD]) for the three tests of the sample was calculated. In our experiments, the measurement time corresponded to the time of incubation of cells with antibiotics. The effect of antibiotics on the luminescence of recombinant E. coli was characterized by the determination of the relative luminescence parameter Irel (the percentage of luminescence for the antibiotic-containing cell suspension versus the control value at measurement moment t). Further discussion will largely concern t = 60 min. The experimental error of Irel was less than 5%, which permitted us to determine the sensitivity of cells to antibiotics as the concentration at which the luminescence inhibition occurred was 10% (Irel = 90%).
Effects of the presence of antibiotics and sera on the luminescence of recombinant E. coli.
Recombinant E. coli cells are characterized by steady-state luminescence lasting many hours. The addition of antibiotics to cell suspensions inhibits E. coli luminescence (depending on the antibiotic concentration and the incubation time). The sensitivity of cells to some antibiotics after 60 min of incubation is shown in Table 1.
TABLE 1.
Sensitivity of luminescent E. coli (native and activated by normal serum) to some antibioticsa
| Antibiotic | Results (ng/ml)
|
|
|---|---|---|
| Native cells | Serum-activated cells | |
| Streptomycin | 78 ± 17 | 2.2 ± 0.5 |
| Gentamicin | 92 ± 20 | 3.3 ± 0.6 |
| Neomycin | 200 ± 35 | 15 ± 3.5 |
| Monomycin | 210 ± 45 | 16 ± 3.5 |
| Kanamycin | (3.8 ± 0.8) · 103 | 25 ± 5.5 |
| Oxytetracycline | 270 ± 30 | 45 ± 9 |
| Chloramphenicol | (2.2 ± 0.5) · 103 | (0.42 ± 0.05) · 103 |
| Erythromycin | (5.7 ± 1) · 103 | (2.5 ± 0.6) · 103 |
Cells were incubated with antibiotic for 60 min. The serum was added (at 20 μl/ml of cell suspension) 20 min before addition of the antibiotic. The cell suspension consisted of 5 mM HEPES-Na (pH 7.2), 0.05% Tween 20, and (2 ± 1) · 107 cells/ml and was used at 23°C. All values represent the means (± SD) of three independent experiments.
Addition of serum to genetically engineered E. coli cells activated their luminescence (2, 19). We examined the effects of the presence of human and rabbit sera, as well as that of the immune rabbit sera, on the cell luminescence. The serum samples tested influenced the cells in similar ways: E. coli incubation with serum activated cell luminescence; after the maximum level had been reached, luminescence slowly decreased. The incubation of cells with serum (20 μl of serum/ml of cell suspension) for 60 min intensified cell luminescence 8- to 10-fold.
Effect of antibiotics on luminescent E. coli activated by normal serum.
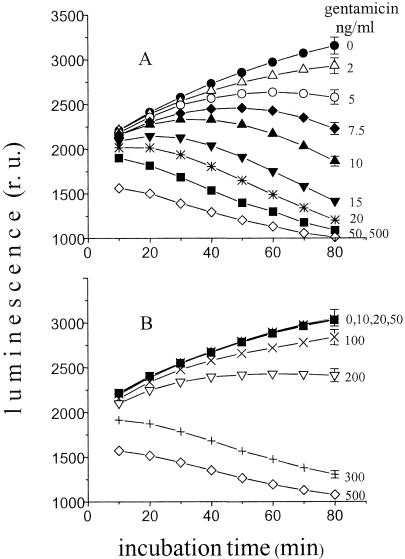
Fig. 1A shows the kinetics of luminescence of recombinant E. coli activated by the presence of normal serum at various gentamicin concentrations. The serum was added 20 min before the antibiotic. When added at that time, the serum activated the cell luminescence about sevenfold (data not shown). Then, gentamicin was added to the cells at t = 0 (Fig. 1A). The upper curve of the figure shows the kinetics of luminescence increase for the control suspension supplemented with buffer. The presence of gentamicin in other samples decreased E. coli luminescence intensity compared to the levels seen with the control. The effect depended on the gentamicin concentration in the cell suspension and on the measurement time point. Table 1 lists the levels of sensitivity of the biosensor to some antibiotics. Analysis of the tabulated results shows substantial sensitization of cells (activated by normal serum) to some antibiotics. Besides those indicated in Table 1, other antibiotics were tested. These antibiotics (bacitracin, benzylpenicillin, cyclosporine, cyclophosphamide, oxacillin, and rifamycin) showed a poor effect on genetically engineered E. coli luminescence levels in our experimental scheme (test sensitivity > 5 μg/ml).
FIG. 1.
Time dependencies of the luminescence of recombinant E. coli (activated by serum) at various gentamicin concentrations. (A) Normal serum; (B) gentamicin-specific immune serum. An 0.8-ml cell suspension was supplemented with 0.2 ml of antibiotic solution at t = 0. The serum was added (at 20 μl of serum/ml of cell suspension) to a cell suspension consisting of 5 mM HEPES-Na (pH 7.2), 0.05% Tween 20, and 2 × 107 cells/ml (at 23°C) 20 min before addition of the antibiotic. r.u., relative units.
The inhibition of bacterial luminescence by antibiotics in the presence of immune serum compared to the results seen with normal serum.
The experiment (in similarity to that described for Fig. 1A) was performed with recombinant E. coli activated by immune serum containing antibodies specific for gentamicin. In this case, the resultant luminescence kinetic curves for the samples (containing 2 to 50 ng of antibiotic/ml) coincided with those for the control sample (0 ng/ml). Inhibition of cell luminescence became notable at gentamicin concentrations of about 100 ng/ml and increased with the gentamicin concentration (Fig. 1B).
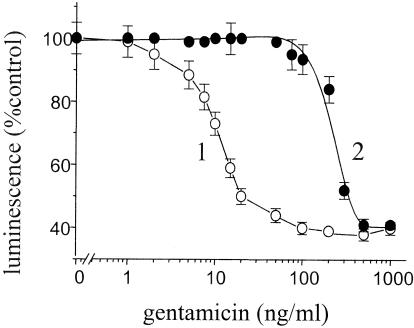
A comparison of the effects of various gentamicin concentrations on the relative luminescence levels of E. coli (activated by normal serum or specific immune serum) is presented in Fig. 2. In cases in which cells were activated by the presence of normal serum, the relative E. coli luminescence level decreased by 27% when the sample contained 10 ng of gentamicin/ml (Irel = 73% [curve 1]). In the presence of immune serum, inhibition of luminescence by the antibiotic was not observed at concentrations of gentamicin up to 50 ng/ml (Irel = 100% [curve 2]) and was less than 10% at 100 ng of gentamicin/ml (Irel = 94% [curve 2]). The absence of luminescence inhibition in the mixture containing antibodies specific for gentamicin is explicable by the formation of immune gentamicin-antibody complexes incapable of inhibiting E. coli luminescence. When the solution contained 200 ng of gentamicin/ml, the quantity of antibodies was not large enough to trap the total amount of the antibiotic in the solution and a portion of the antibiotic in the solution remained free; the level of inhibition in the presence of immune serum was 16%, whereas that measured in the presence of normal serum was 60%. Gentamicin (500 ng/ml) inhibited luminescence in the two cell suspensions equally, since in this case the antigen-antibody complex concentration was much less than that of the antibiotic. The effect of the presence of the immune serum may be dependent on both its quantity and its antibody content. Similar experiments were performed with other antibiotics for which we had immune sera; these antibiotics were streptomycin, tetracycline, and chloramphenicol (data not shown). The results for streptomycin with respect to the dependence of relative luminescence levels on antibiotic concentration were similar to those presented in Fig. 2. The maximum discrepancies for gentamicin and streptomycin in the Irel parameter in curves 1 and 2 were 50 to 60%. For chloramphenicol and tetracycline, Irel discrepancies were 15 to 30%. In these cases, when the test sensitivity is tens or hundreds of nanograms per milliliter it may be useful to increase the discrepancies by one of the two ways: either by enriching the antiserum with an immunoglobulin G fraction of immune serum or by adding more serum (up to 100 μl).
FIG. 2.
Relative luminescence levels of recombinant E. coli (activated by the presence of normal serum [○] or of gentamicin-specific immune serum [•]) as a function of gentamicin concentration. The duration of incubation of cells with antibiotic was 60 min. The serum was added (at 20 μl/ml of cell suspension) to a cell suspension consisting of 5 mM HEPES-Na (pH 7.2), 0.05% Tween 20, and 1.8 × 107 cells/ml (at 23°C) 20 min before addition of the antibiotic. Data represent the means of three independent measurements (± SD). The results are typical of four independent experiments.
Test specificity.
The specific action of the antibodies was demonstrated by a comparison of the inhibitory effect of the presence of streptomycin on the luminescence of cells activated by normal serum to the effect seen with gentamicin-specific immune serum. Here, the time dependencies of luminescence were identical for normal and immune sera. Gentamicin-specific serum formed no immune complexes with streptomycin and, hence, had no effect on the capacity of streptomycin to inhibit luminescence. Identical experiments were also conducted for the following combinations: antiserum against streptomycin plus gentamicin and antiserum against chloramphenicol plus streptomycin. In all cases, the presence of antibodies had no detectable effect on luminescence inhibition by antibiotics.
Conclusions. In this research, we used well-known published data on the inhibition of bacterial bioluminescence by antibiotics (3, 6, 12, 14, 15, 18) and on the use of immune sera to assay antibiotics (8, 17). The combination of these approaches with our biosensor afforded results that served as an experimental ground for the assay of antibiotics by the use of genetically engineered E. coli and immune sera. These results included (i) the inhibition of recombinant E. coli luminescence when minor doses of some antibiotics were added to cell suspensions (Table 1), (ii) a notable increase in cell sensitivity to some antibiotics when cells were activated by blood serum (Fig. 1 and Table 1) (the minor experimental error provides the high sensitivity of the method), and (iii) the discrepancies in antibiotic-caused luminescence inhibition results for E. coli activated by normal blood serum or by a specific immune serum (helping to identify the antibiotic) (Fig. 2). Our experimental approach is very simple, and (including all preliminary preparations for the procedure) the assay lasts no more than 3 h.
For quantitation of antibiotics by the use of luminescent E. coli, it is necessary to construct the calibration curves. Construction of the curves largely depends on the choice of incubation time for the cell suspension and antibiotic (2, 10, 12). The levels of cell luminescence can be changed by the presence of a number of organic substances; therefore, the assay requires adequate experimental controls for the interpretation of sensing data (7). The influence of biological fluids on E. coli luminescence needs to be taken into account when assays of antibiotics in various biological fluids are conducted; one method is that of adding a similar antibiotic-free fluid to the control sample. It may be useful to inactivate biological fluids by heating (6, 11, 12, 19). It is to be noted that the above-described experimental scheme is modifiable. For the efficient assay of other antimicrobial agents, it will be useful to alter the conditions of activation by serum and of cell incubation with antibiotics as well as to use other luminescent bacterium strains. Likewise, the experimental approach on offer is applicable for obtaining information about the presence (in serum) of antibodies against a definite antibiotic, which is important when determining allergic states.
The results of this research show that it is feasible to elaborate a new assay—simple, speedy, and providing high levels of sensitivity and specificity—for antimicrobial agents on the basis of genetically engineered E. coli strains and immune sera.
Acknowledgments
We express our appreciation to Z. V. Samsonova for providing the immune sera.
REFERENCES
- 1.Anko, M. L., J. Kurittu, and M. Karp. 2002. An Escherichia coli biosensor strain for amplified and high throughput detection of antimicrobial agents. J. Biomol. Screen. 7:119-125. [DOI] [PubMed] [Google Scholar]
- 2.Asrieli, T. V., I. I. Vlasova, E. M. Gavrilova, and V. S. Danilov. 2002. Effect of antibiotics on the luminescence of recombinant Escherichia coli strain activated by blood serum. Biotekhnologiya (Russia) 2:85-93. [Google Scholar]
- 3.Beard, S. J., V. Salisbury, R. J. Levis, J. A. Sharpe, and A. P. MacGowan. 2002. Expression of lux genes in a clinical isolate of Streptococcus pneumoniae: using bioluminescence to monitor gemifloxacin activity. Antimicrob. Agents Chemother. 46:538-542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Champiat, D., A. Roux, O. Lhomme, and G. Nosenzo. 1994. Biochemiluminescence and biomedical application. Cell Biol. Toxicol. 10:345-351. [DOI] [PubMed] [Google Scholar]
- 5.Danilov, V. S., A. P. Zarubina, G. E. Erochnikov, L. N. Solov'eva, F. V. Kartashev, and G. B. Zavil'gel'skii. 2002. Sensory bioluminescence systems based on lux-operons of various-type luminescent bacteria. Vestnik of Moscow University. Biologiya 3:20-24. [Google Scholar]
- 6.Hancock, J. T., V. Salisbury, M. C. Ovejero-Boglione, R. Cherry, C. Hoare, R. Eisenthal, and R. Harrison. 2002. Antimicrobial properties of milk: dependence on presence of xanthine oxidase and nitrite. Antimicrob. Agents Chemother. 46:3308-3310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heitzer, A., B. Applegate, S. Kehrmeyer, H. Pinkart, O. F. Webb, T. J. Phelps, D. C. White, and G. S. Sayler. 1998. Physiological considerations of environmental applications of lux reporter fusion. J. Microbiol. Methods 33:45-57. [Google Scholar]
- 8.Kolosova, A. Yu., A. N. Blintsov, Zh. V. Samsonova, and A. M. Egorov. 1998. Designing of solid-phase enzyme immunoassay for detection of gentamicin in human sera. Antibiot. Khimioter. (Russia) 43:9-13. [PubMed] [Google Scholar]
- 9.Korpela, M. T., J. S. Kurittu, J. T. Karvinen, and M. T. Karp. 1998. A recombinant Escherichia coli sensor strain for the detection of tetracyclines. Anal. Chem. 70:4457-4462. [DOI] [PubMed] [Google Scholar]
- 10.Kurittu, J., M. Karp, and M. Korpela. 2002. Detection of tetacyclines with luminescent bacterial strain. Luminescence 15:291-297. [DOI] [PubMed] [Google Scholar]
- 11.Kurittu, J., S. Lonnberg, M. Virta, and M. Karp. 2000. A group-specific microbiological test for the detection of tetracycline resides in raw milk. J. Agric. Food Chem. 48:3372-3377. [DOI] [PubMed] [Google Scholar]
- 12.Naveh, A., I. Potasman, H. Bassan, and S. Ulitzur. 1984. New rapid and sensitive bioluminescence assay for antibiotics that inhibit protein synthesis. J. Appl. Bacteriol. 56:457-463. [DOI] [PubMed] [Google Scholar]
- 13.Pillineu, T., G. Bylund, M. Virta, A. Niemi, and M. Karp. 2002. Detection of traces of tetracyclines from fish with a bioluminescent sensor strain incorporating bacterial luciferase reporter genes. J. Agric. Food Chem. 50:4812-4815. [DOI] [PubMed] [Google Scholar]
- 14.Rocchetta, H. L., C. J. Boylan, J. W. Foley, P. W. Iversen, D. L. LeTourneau, C. L. McMillian, P. R. Contag, D. E. Jenkins, and T. R. Parr, Jr. 2001. Validation of a noninvasive, real-time imaging technology using bioluminescent Escherichia coli in the neutropenic mouse thing model of infection. Antimicrob. Agents Chemother. 45:129-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Salisbury, V., A. Pfoestl, H. Wiesinger-Mayr, R. Lewis, K. E. Bowker, and A. P. MacGowan. 1999. Use of clinical Escherichia coli isolate expressing lux genes to study the antimicrobial pharmacodynamics of moxifloxacin. J. Antimicrob. Chemother. 43:829-832. [DOI] [PubMed] [Google Scholar]
- 16.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 17.Stead, D. A. 2000. Current methodologies for the analysis of aminoglycoside. J. Chromatogr. B 747:69-93. [DOI] [PubMed] [Google Scholar]
- 18.Ulitzur, S. 1986. Determination of antibiotic activities with the aid of luminous bacteria. Methods Enzymol. 133:275-284. [DOI] [PubMed] [Google Scholar]
- 19.Virta, M., M. Karp, S. Ronnemaa, and E.-M. Lilius. 1997. Kinetic measurement of the membranolytic activity of serum complement using bioluminescent bacteria. J. Immunol. Methods 201:215-221. [DOI] [PubMed] [Google Scholar]
- 20.Wahlström, G., and P. E. J. Saris. 1999. A nisin bioassay based on bioluminescence. Appl. Environ. Microbiol. 65:3742-3745. [DOI] [PMC free article] [PubMed] [Google Scholar]