Background: NF-κB is a key regulator of anti-apoptotic processes and plays a role in gonad formation in mammals.
Results: NF-κB activation leads to female-biased sex differentiation in zebrafish.
Conclusion: Anti-apoptotic signaling during the juvenile ovary stage is needed for the maintenance of oocytes in zebrafish.
Significance: Unraveling the regulation of apoptotic processes during gonadal transformation will facilitate understanding the molecular mechanism of zebrafish sex differentiation.
Keywords: Apoptosis, Development, Inflammation, Signal Transduction, Zebra Fish, Gonad, Gonad Transformation, Sex Determination, Sex Differentiation, Sex Reversal
Abstract
Testis differentiation in zebrafish involves juvenile ovary to testis transformation initiated by an apoptotic wave. The molecular regulation of this transformation process is not fully understood. NF-κB is activated at an early stage of development and has been shown to interact with steroidogenic factor-1 in mammals, leading to the suppression of anti-Müllerian hormone (Amh) gene expression. Because steroidogenic factor-1 and Amh are important for proper testis development, NF-κB-mediated induction of anti-apoptotic genes could, therefore, also play a role in zebrafish gonad differentiation. The aim of this study was to examine the potential role of NF-κB in zebrafish gonad differentiation. Exposure of juvenile zebrafish to heat-killed Escherichia coli activated the NF-κB pathways and resulted in an increased ratio of females from 30 to 85%. Microarray and quantitative real-time-PCR analysis of gonads showed elevated expression of NF-κB-regulated genes. To confirm the involvement of NF-κB-induced anti-apoptotic effects, zebrafish were treated with sodium deoxycholate, a known inducer of NF-κB or NF-κB activation inhibitor (NAI). Sodium deoxycholate treatment mimicked the effect of heat-killed bacteria and resulted in an increased proportion of females from 25 to 45%, whereas the inhibition of NF-κB using NAI resulted in a decrease in females from 45 to 20%. This study provides proof for an essential role of NF-κB in gonadal differentiation of zebrafish and represents an important step toward the complete understanding of the complicated process of sex differentiation in this species and possibly other cyprinid teleosts as well.
Introduction
The molecular mechanism of sex determination is unknown in zebrafish (Danio rerio). Earlier, neither cytogenetic studies (1, 2) nor comparative analysis of recombination rates between the two sexes (3) nor an array-based genome screen (4) have led to the identification of sex chromosomes. Recently, analysis of genetic linkage maps have revealed the presence of regions associated with sex determination on four chromosomes (5, 6), indicating polygenic sex determination in zebrafish (4–7).
On the other hand, a number of candidate genes with potential role in sexual development have been identified, including Sry-related HMG box gene 9 (sox9), anti-Müllerian hormone (amh), cytochrome P450, family 19, subfamily A, polypeptide 1a and b (cyp19a1a and b), nuclear receptor subfamily 5, group A (nr5a1 a and b), forkhead box protein L2 (foxl2), dead end (dnd), and factor in germ line α (figα) (7–10). Nr5a or steroidogenic factor-1 controls the expression of sox9, cyp19a1a, and amh (7, 10). In mammals, SOX9 is involved in the regulation of Amh4 (11). In mice, homozygous mutation of Sox9 binding site in Amh leads to lack of its transcription and development of pseudohermaphrodites (12), whereas mutations in the sox9 gene result in sex reversal in XY campomelic dysplasia patients (13).
Zebrafish gonadal differentiation starts with the formation of a juvenile ovarian structure that either matures into adult ovaries or transforms into testes (14, 15). The testis transformation process has been suggested to depend on apoptosis (16). This was further supported by a recent study where mutations in the Fanconi anemia, complementation group L (fancl) gene led to germ cell apoptosis and consequently activated the testis development pathway (17). Introducing mutations in the tumor protein p53 (tp53) gene counteracted the effect of the fancl mutations (17), suggesting a role for apoptosis in this process. In line with this, germ cell numbers are also important for ovarian development and complete absence of germ cells results in development of sterile males (9, 18).
Nuclear factor κ-light-chain-enhancer of activated B cells (NF-κB) is involved in regulation of inflammation, apoptosis, cell growth, and differentiation and can be activated by various physical and chemical factors (19). NF-κB is a protein complex composed of homo- or heterodimers of five members of the Rel family including NF-κB1 (p50), NF-κB2 (p52), Rel A (p65), Rel B, and c-Rel. These proteins are capable of binding one another with different binding specificities resulting in different DNA binding properties. The p65/p50 dimer is the most abundant heterodimeric form present in cells and is involved in transcription activation of a multitude of genes (19, 20). NF-κB subunits are generally sequestered in the cytoplasm by the inhibitor protein IκB. Numerous factors including UV irradiation, stress, cytokine, and free radicals can promote IκB degradation of the IκB/NF-κB complex, allowing the translocation of NF-κB to the nucleus resulting in subsequent induction of the transcription of its target genes (20–22).
NF-κB activation blocks apoptotic processes and promotes cell survival by interacting with the inhibitor of apoptosis protein (IAP) family of genes (23, 24). NF-κB is also known to interact with other nuclear receptors including the glucorticoid and androgen receptor as well as with other proteins to regulate gene expression (25, 26). NF-κB is highly expressed in mammalian Sertoli cells; it is involved in regulation of spermatogenesis (27) and down-regulates the Amh gene expression in mammalian testis. This is due to NF-κB interaction with steroidogenic factor-1, which in turn leads to the recruitment of histone deacetylase and suppression of Amh gene expression (28). NF-κB is also involved in the interleukin 1- and tumor necrosis factor-α (TNFα)-mediated down-regulation of Sox9 expression in mouse chondrocytic cells (29).
Zebrafish embryos and larvae are dependent on a functional innate immune system at an early stage of development (≥1 days post fertilization (dpf)). This includes macrophage and neutrophil differentiation (30), indicating that the NF-κB signaling system is present and active before testis transformation. In addition, a study on goldfish has shown that TNFα inhibits testicular testosterone production (31).
The aim of this study was to investigate the possible role of NF-κB in zebrafish sex differentiation. Induction of the NF-κB signaling pathway resulted in up-regulation of inflammatory and anti-apoptotic genes, and this correlated to female-biased sex ratio. In contrast, inhibition of NF-κB resulted in an increased proportion of males. This study demonstrates the involvement of NF-κB signaling in the maintenance of ovarian development and the inhibition of the transformation of juvenile ovaries to adult testis.
EXPERIMENTAL PROCEDURES
Breeding
Adult zebrafish were maintained in a recirculating system (Aquaneering) with a 14-h light/10-h dark cycle. The fish were fed twice a day with newly hatched Artemia salina nauplii and commercial flake food (Tetrarubin). The male and female brooders were kept in separate aquaria at 26–27 °C, and they were allowed to breed once a week. The fish handling procedures were approved by the Swedish Ethical Committee in Linköping (Permit 32-10).
Preparation of Heat-killed Bacteria
E. coli MG1655 was grown on Luria-Bertani (LB) agar and incubated at 37 °C overnight. One colony was inoculated into 10 ml of LB broth and incubated on a shaker (200 rpm) at 37 °C overnight. The bacteria were then centrifuged and washed with 5 ml of phosphate-buffered saline (Sigma). The bacterial pellet was resuspended in 2 ml of PBS and killed by heating to 70 °C for 1 h. To ensure 100% bacteria death, 10 μl of the heat-killed suspension was plated and incubated overnight at 37 °C.
NFκB-pGL4 Plasmid Construction
The pGL4 plasmid with promoter-less Luciferase gene and neomycin selection marker was purchased from Promega, and the NFκB cis-element insert was obtained from commercially available pNFκB-Luc plasmid (BD Biosciences). The pGL4 luciferase plasmid and the pNFκB-Luc plasmid were cut with HindIII and KpnI (Fermentas) and gel-purified by using Wizard SV Gel and PCR Clean-Up System (Promega). The NFκB cis-element and the pGL4 luciferase plasmid were ligated using T4 DNA ligase (Fermentas).
Cell Culture and Development of Stably Transfected Cells
ZFL cells were maintained at 28 °C and 3% CO2 in a humidified incubator in a complex media containing 50% L15 (Invitrogen), 35% DMEM-high glucose (PAA Laboratories), and 15% Ham's F-12 (Invitrogen) supplemented with 5% fetal bovine serum (Hyclone), 15 mm Hepes buffer (Lonza), 0.15 g/liter sodium bicarbonate (Biochrom AG), 50 ng/ml mouse EGF, and 0.01 mg/ml bovine insulin. For stable transfection, ZFL cells were plated in 24-well plates (BD Biosciences) (80,000 cells/well). The next day the cells were transfected with the NFκB-pGL4 plasmid (500 ng/well) using Lipofectamine 2000 (Invitrogen). After 48 h the cells were treated with 1 mg/ml G418 antibiotic (Duchefa Biochemie). Antibiotic-resistant colonies were selected, and the cells were screened for NFκB activity using the Luciferase assay kit (Promega).
ZFL Cell-based Experiments
Before exposure the stably transfected ZFL cells (nZFL) were plated in 24-well plates (80,000 cells/well) and incubated for 16–18 h at 28 °C and 3% CO2. Wild type ZFL cells were plated in 12-well plates (BD Biosciences) (2 × 105 cells/well) and incubated for 16–18 h at 28 °C and 3% CO2. The experiments were started by adding heat-killed bacteria at 5 × 106, 1 × 107, 5 × 107, or 1 × 108 colony forming units (cfu)/ml, sodium deoxycholate (DOC) at 200 or 300 μm (Sigma), or 40 nm NF-κB activation inhibitor (NAI) (Calbiochem) prepared in fresh media to the cells. The cells were incubated for 12 h (Luciferase assays) or 24 h (quantitative RT-PCR (qRT-PCR) assays) at 28 °C and 3% CO2. Detection of luciferase activity was performed using Luciferase assay kit, whereas cells used for qRT-PCR analysis were lysed with TRI-Reagent (Sigma).
Maintenance and Exposure of Juvenile Zebrafish
In the evening, pairs of adult zebrafish brooders were transferred to mating containers (Aquaneering). The next morning fertilized embryos were collected and divided in groups of 50 in 115-mm diameter crystallization dishes containing 100 ml of water and maintained at 28 °C. At 4 dpf, the juveniles were transferred to the circulating system. The water flow was adjusted to 20–30 drops/min, 14-h light/10-h dark cycle and fed twice with newly hatched A. salina nauplii and commercial flake food (Larval AP 100).
At the time of exposure, juvenile zebrafish were transferred back to crystallization dishes of 115-mm diameter with 100 ml of water (15 or 20 dpf) or 1-liter containers (180/100 mm) with 500 ml of water (41 or 70 dpf) with 20 individuals in each container. The juveniles were exposed to heat-killed E. coli (1× 106, 5 × 106, 1× 107, 5 × 107), DOC (200 μm), or NAI (20 nm). The fish were fed twice a day, and 50% of the water was changed every third day and heat-killed E. coli, DOC, or NAI was replenished to maintain the original concentrations. The water quality was monitored over the course of studies. The temperature was maintained at 25 ± 0.2 °C. Water pH averaged 7.3 ± 0.2, and salinity averaged 0.08 ± 0.01%. The nitrite level averaged 0.04 ± 0.05 mg/liter, whereas nitrate and ammonia level remained undetectable. Survival through the experimental exposures averaged 85 ± 5%. There was no effect of the different treatments on these parameters.
The exposures were terminated at different time points depending on the experiment and assay. For sample collection at 21 dpf, zebrafish juveniles were sacrificed by snap-freezing in liquid nitrogen and stored at −80 °C until further use. Juveniles of 35, 42, and 71 dpf of age were dissected under a stereomicroscope, and their gonads were isolated, snap-frozen in liquid nitrogen, and stored at −80 °C until further use. For sex ratio determination, the fishes were transferred back to the circulating system at the end of exposure (at 35 dpf), and the sex ratios were determined after dissection and microscopic observation of the gonads at 70 dpf.
RNA Extraction and qRT-PCR
Cells, juvenile zebrafish, and isolated gonads were homogenized in 200 μl Trizol Reagent (Sigma) and RNA was isolated according to manufacturer's instruction. cDNA synthesis was performed using qScript cDNA synthesis kit (Quanta Biosciences). Primers were designed for listed genes (supplemental Table S1). SYBR Green (Quanta) was used to determine the expression levels of all genes. Thermocycling conditions for SYBR Green consisted of a denaturation step for 5 min at 95 °C followed by 40 cycles of 95 °C for 2 s and 60 °C for 30 s. Data analysis was performed using standard curve method and ΔΔCt method (32).
Microarray-based Transcriptomics of Gonadal Samples
The gonads of 35 dpf juvenile zebrafish were isolated, and RNA was extracted from individual samples using TRIzol reagent according to manufacturer's instruction. Amplification and labeling of total RNA was performed as described previously (33) with the following modifications; 5 μg of antisense RNA was labeled with Alexa Fluor 647, whereas 20 μg of a common reference consisting of pooled antisense RNA from one individual in each treatment group (a total of 4 individuals) was labeled with Alexa Fluor 555. Microarray printing, hybridization, scanning, and data processing were performed as described previously (33). A total of 11 samples were analyzed: 6 control individuals, 3 individuals from the intermediate treatment, and 2 individuals from the highest treatment dose.
Analysis of Microarray Data
Raw expression values obtained from the GenePix Pro 6.0 software were background-corrected and normalized using R 2.9.0 (34) and the BioC limma 2.18.2 package (35, 36). Briefly, array intensity values were background corrected using the normexp method (37) followed by print-tip loess normalization within arrays and quantile normalization between arrays. Differential expression was calculated by fitting a linear model to a group means parameterization. Multiple testing was corrected by controlling the false discovery rate (38). Principal component analysis of the normalized expression values was conducted in R using the prcomp function. The gene symbol and putative function for each EST sequence was obtained based on the best zebrafish BLAST hit against the RefSeq RNA database (as of April 12, 2011) unless indicated otherwise. The full set of microarray expression data has been deposited in ArrayExpress under the accession number E-MEXP-3249.
Testis Culture
Adult zebrafish were euthanized, and their testes were isolated. The testes were sterilized with 0.5% v/v commercial bleach in PBS (Sigma) for 2 min and washed 3 times in PBS. The testes were transferred to 24-well plate (BD Biosciences) and maintained at 28 °C and 3% CO2 in a humidified incubator in ZFL cell media containing antibiotic-antimycotic solution (Invitrogen). The explants were cultured in parallel and exposed to heat-killed bacteria (5 × 107), DOC (200 μm), and NAI (20 nm) for 24 h.
Western Blot Analysis
The ovary samples from 71 dpf individuals were lysed in radioimmune precipitation assay buffer (150 mm NaCl, 1% Nonidet P-40, 0.5% DOC, 0.1% SDS, 50 mm Tris, pH 8) containing protease inhibitor mixture (Sigma). Protein content was quantified using Bradford reagent (Bio-Rad), and 30 μg of each sample was resolved by 12% SDS-PAGE. The proteins were transferred to a PVDF membrane (Amersham Biosciences). The membrane was incubated for 1 h in Tris-buffered saline containing 0.1% Tween to prevent nonspecific binding and probed with anti-caspase 3a antibody (AnaSpec) at 1:1000 dilution overnight. The HRP-conjugated anti rabbit IgG (Amersham Biosciences) was used at a 1:3000 dilution for 1 h, and the immunoreactive complex was detected by Super Signal West Pico chemiluminescent substrate (Thermo Scientific). The membrane was then stripped and probed for β-actin using mouse anti β-actin antibody (Sigma) at a 1:5000 dilution. The HRP-conjugated anti-mouse IgG (Amersham Biosciences) was used at 1:3000 dilution for 1 h. The bands were quantified using the ImageJ software (National Institutes of Health) and normalized with their respective β-actin level.
Statistical Analysis
Statistical significance were determined using a two-tailed non-paired Student's t test for two-group comparison or one-way analysis of variance followed by Dunnett post test for multiple group comparison, and differences were considered significant if the p values were <0.05 (*, p < 0.05; **, p < 0.01; ***, p < 0.001). Statistical analyses were performed using GraphPad Prism 5 software (GraphPad Software).
RESULTS
Heat-killed Escherichia coli Activate Zebrafish NF-κB Resulting in Female-biased Sex Ratios
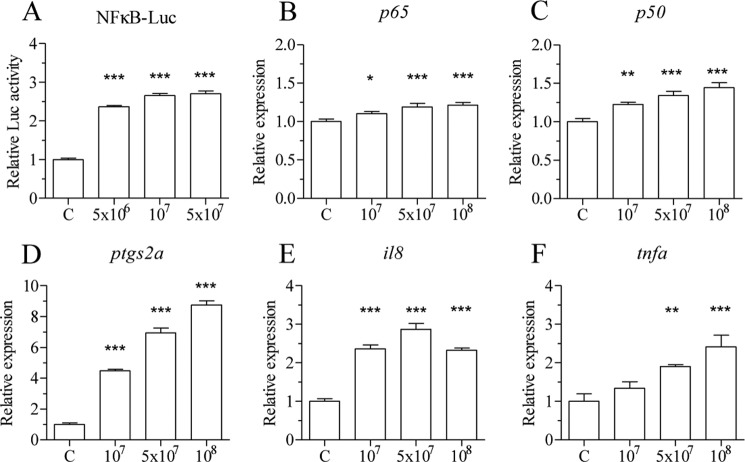
ZFL cells stably transfected with the NF-κB-pGL4 vector (nZFL) were exposed to heat-killed E. coli to determine whether this would result in activation of NF-κB. A dose-dependent activation of NF-κB was observed after exposure of nZFL cells to heat-killed E. coli (Fig. 1A). The qPCR analysis also showed up-regulation of inflammatory genes like p65, p50, prostaglandin-endoperoxide synthase 2a (ptgs2a), interleukin 8 (il8), and tumor necrosis factor a (tnfa) in response to heat-killed E. coli after exposure of ZFL cells (Fig. 1, B–F).
FIGURE 1.
Heat-killed bacteria activate NF-κB and inflammatory genes in vitro. nZFL cells were treated with heat-killed E. coli (5 × 106, 1× 107, and 5 × 107 cfu/ml), and the luciferase activity was determined after 12 h of exposure (A). ZFL cells were exposed to heat-killed E. coli (1× 107, 5 × 107, and 1× 108 cfu/ml) for 24 h, and qPCR analysis was performed to determine p65 (B), nfkb1/p50 (C), ptgs2a (D), il8 (E) and tnfa (F). One-way analysis of variance was performed to determine statistical significant difference (*, p < 0.05; **, p < 0.01; ***, p < 0.001). n = 4. Error bars represent the mean ± S.D.
Zebrafish juveniles are known to enter a juvenile ovary stage between 14 and 28 dpf (14, 15). This transition was indicated by the elevated levels of female-specific zona pellucida (zp2) gene at 19 dpf (Fig. 2A). Based on this information, all the exposures were started before or at the time of entry into the juvenile ovary stage.
FIGURE 2.
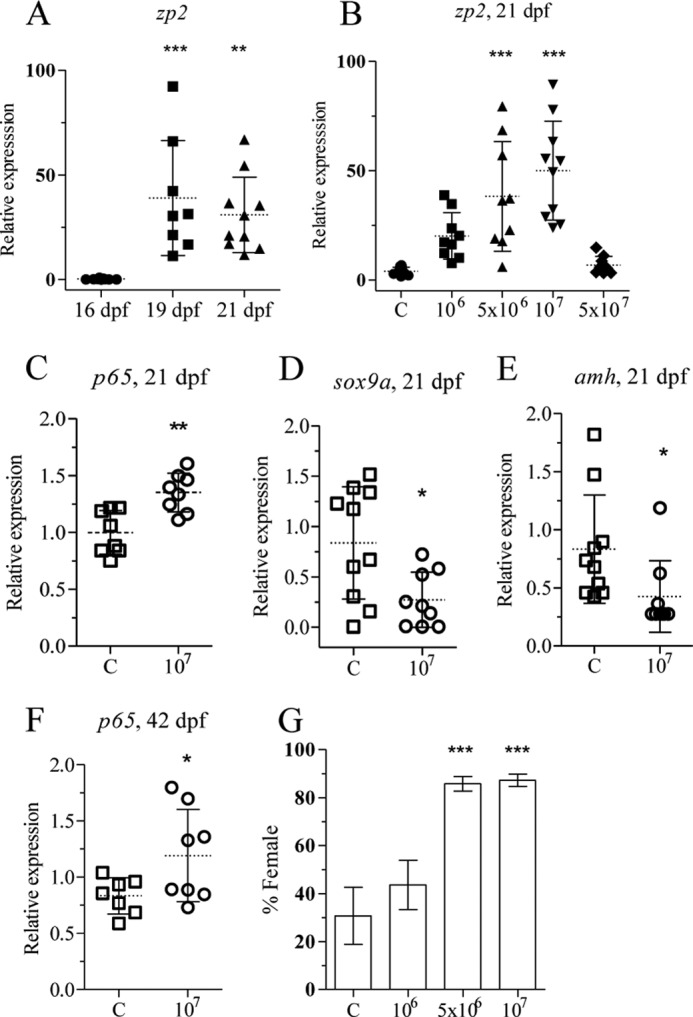
Heat-killed E. coli up-regulates p65 and zp2 and leads to a female-biased population. zp2 expression was determined by qRT-PCR from individuals (16, 19, and 21 dpf) using ef1a and 18 S rRNA as reference genes (A). Zebrafish juveniles were exposed to different concentrations of heat-killed bacteria, and zp2 expression was determined by qRT-PCR from individuals exposed from 15–21 dpf (B). Zebrafish juveniles at 20 and 41 dpf were exposed to heat-killed bacteria (107 cfu/ml) for 24 h. Analysis of p65 (C), sox9a (D), and amh (E) expression at 21 dpf juveniles and of p65 (F) expression at 42 dpf gonads was performed by qRT-PCR. Juveniles at 15 dpf were exposed to heat-killed E. coli for 20 days, and their sex was determined at 70 dpf. The mean and S.D. of three independent experiments is shown. Each group contained at least 20 individuals (G). Statistically significant differences between groups were determined using Student's t test for two groups and one-way analysis of variance test for more than two groups (*, p < 0.05; **, p < 0.01; ***, p < 0.001). Error bars represent the mean ± S.D.
Exposure to heat-killed bacteria from 15 to 21 dpf resulted in dose-dependent up-regulation of the zp2 gene expression and showed a bell-shaped curve with no effect at the highest dose (Fig. 2B). Next, zebrafish juveniles were exposed to 1 × 107 cfu/ml heat-killed bacteria for 24 h at 20 dpf to determine whether this would result in in vivo up-regulation of NF-κB. The transcript level of p65 was significantly up-regulated, whereas sox9a and amh expression were down-regulated (Fig. 2, C–E). This demonstrates that heat-killed bacteria induce the NF-κB pathway, similar to what is reported in mammals (28), NF-κB also down-regulates amh expression in zebrafish juveniles. To confirm if NF-κB activation occurred in the gonads, juveniles at 41 dpf were exposed to heat-killed bacteria for 24 h, and the gonads were excised. qRT-PCR analysis showed significant up-regulation of gonadal p65 expression after the treatment (Fig. 2F). Together, these results demonstrate that treatment with heat-killed E. coli led to activation of NF-κB and induction of female-specific gene expression.
To determine whether treatment with heat-killed bacteria would lead to alterations in sex ratio, zebrafish juveniles were exposed to three concentrations of heat killed bacteria (1 × 106, 5 × 106 or 1 × 107 cfu/ml) during the critical period of sex differentiation (15–35 dpf). The sex ratio was determined at 70 dpf and demonstrated an increase in the proportion of females from 30 to 85% (Fig. 2G). These results indicated that activation of NF-κB and the inflammatory signaling pathways interfere with testis formation in zebrafish.
Microarray-based Transcriptomic Analysis Indicate the Involvement of NF-κB Pathway in the Feminization Process
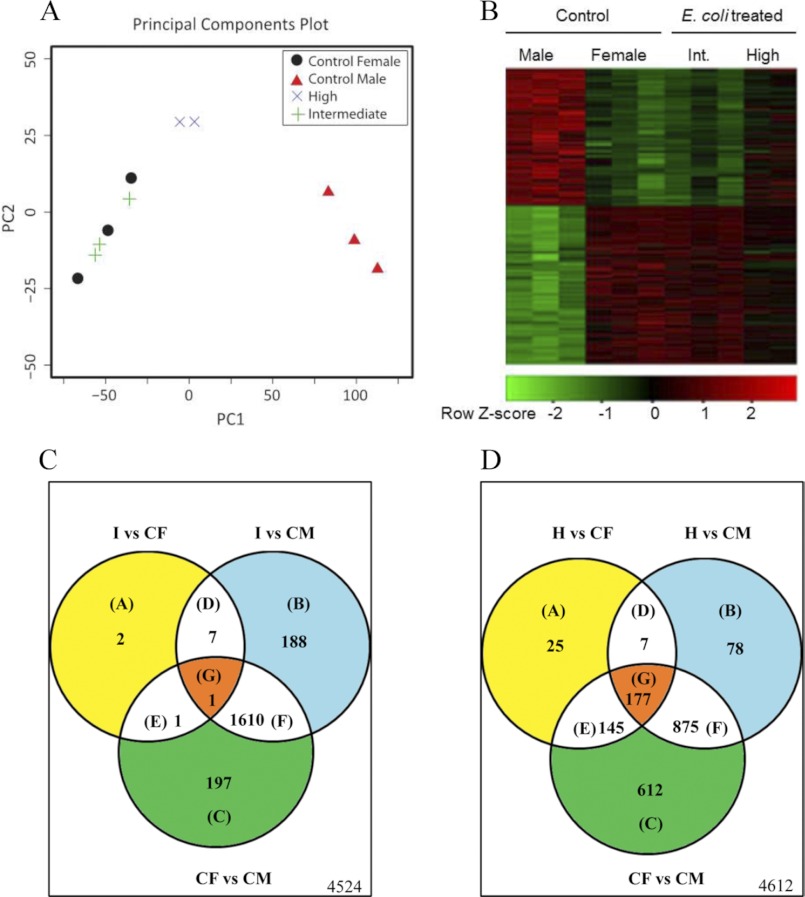
A custom-printed cDNA array comprising 6370 unique gonad-derived clones (33) was used to assess changes in the gene expression landscape of juvenile zebrafish gonads in response to two different concentrations of heat-killed E. coli (5 × 106 and 1 × 107 cfu/ml) between 15 and 35 dpf. Principal component analysis of the normalized array expression values illustrated that exposure to heat-killed bacteria defined the main treatment effect seen in the expression data (Fig. 3A). Each treatment group formed a tight cluster indicating no outliers among the individual transcriptomes analyzed. The results showed that individuals treated with an intermediate dose of heat-killed bacteria clustered more tightly with control females (and further apart from the control males) than the high dose group, and this may be due to the bell-shaped dose response curve observed with heat-killed bacteria (Fig. 2B). Sexually dimorphic expression of a selected set of genes between the control males and females is shown in the form of a heatmap (Fig. 3B; -fold change ≥; q value <0.05). Comparative analysis of the sex-specific gene expression in the treated groups revealed a striking similarity between the gonadal transcriptomic profiles of those treated with the intermediate dose and the control females, highlighting that treatment with heat-killed E. coli-enhanced female gonad differentiation and/or inhibited male gonad differentiation (supplemental Table S2).
FIGURE 3.
The gonadal transcriptomes of individuals treated with heat-killed E. coli are similar to those of control females. Differential gene expression was determined by a homemade gonadal cDNA microarray in the differentiating zebrafish gonads at 35 dpf after treatment with an intermediate (Int; 5 × 106 cfu/ml) and high dose (107 cfu/ml) of heat-killed E. coli. A, shown is a visualization of the main source of experimental variation using principal components analysis of all normalized array expression values. B, shown is a Heatmap visualization of 1812 array features differentially expressed between the control male and control female samples (-fold change ≥ 2-fold; q value < 0.05). Rows were scaled to have a mean of zero and S.D. of one. C and D, shown is a Venn analysis of genes differentially expressed (-fold change ≥ 2-fold; q value < 0.05) between the two sexes from the E. coli treatment groups and the control samples, respectively. 1) gene sets are differentially expressed after treatment with intermediate dose of heat-killed bacteria. 2) gene sets are differentially expressed after treatment with high dose of heat-killed bacteria. CF, control female; CM, control male; H, high dose (107 cfu/ml); I, intermediate dose (5 × 106 cfu/ml). The number of genes that were not differentially expressed is indicated in the lower right corner of each panel.
Venn analysis was performed on the microarray data to identify genes specifically expressed in treated fish in comparison to control males and females (Fig. 3, C and D). Comparison of the gonadal transcriptome profile of the group treated with an intermediate dose with that of control females (I versus CF) showed that only 11 genes differed in their expression pattern between these groups (Fig. 3C). Two of the genes that showed differential expression were dead (Asp-Glu-Ala-Asp) box polypeptide 3 (ddx3), a RNA helicase involved in spermatogenesis (39), and chordin (chd), which plays an important role in the specification of dorsal-ventral axis during zebrafish development (40). On the other hand, 1809 genes showed significant differential expression between the gonads of individuals treated with the intermediate dose and those of the control males (I versus CM) (Fig. 3C).
Similar results were observed with the individuals treated with high dose of heat-killed bacteria, as they have also demonstrated increased female bias (Fig. 2G). The expression level of 354 genes (supplemental Table S3) differed between individuals treated with the a high dose of heat-killed E. coli and the control females (H versus CF) compared with 1138 array features differentially expressed between the high dose group and control males (H versus CM; Fig. 3D). Taken together, this indicated that the gene expression pattern elicited by heat-killed E. coli treatment closely resembled that of the control females as opposed to control males.
Genes showing the largest differential expression levels between individuals treated with the intermediate dose of heat-killed E. coli and control males (I versus CM) are listed in Table 1. Several of these genes, including zp2, zp3, and zygote arrest 1 (zar1) as well as the anti-apoptotic gene baculoviral IAP repeat-containing 5B (birc5b) were shown to be expressed in a female-specific manner in the treated individuals. Analysis of the differences in gene expression between control females and males and between the group treated with intermediate dose of heat-killed E. coli and control males showed that the expression level of amh, cyp19a1a and b, and cytochrome P450, family 11, subfamily C, polypeptide 1 (cyp11c1) did not differ between these groups at 35 dpf (Table 2). However, zp2 showed higher expression in females and treated individuals than in males (Table 3), whereas nr5a1a showed higher expression in males (Table 2).
TABLE 1.
Genes displaying the largest differential expression in gonads from juveniles exposed to intermediate levels of heat-killed E. coli compared to control male samples from 15–35 dpf
| Gene symbol | Genes up-regulated in individuals treated with heat-killed bacteria |
|||
|---|---|---|---|---|
| I vs. CMa | q Valueb | Gene name | RefSeq ID | |
| sinup | 6.138972 | 3.8461E-13 | Siaz-interacting nuclear protein | NM_197937 |
| tpi1a | 5.778892 | 1.3865E-13 | Triosephosphate isomerase 1a | NM_153667 |
| stau2 | 5.729454 | 9.1741E-13 | Staufen, RNA-binding protein, homolog 2 (Drosophila) | NM_200925 |
| zp3 | 5.513024 | 3.5595E-12 | Zona pellucida glycoprotein 3 | NM_131331 |
| zp2.2 | 5.508029 | 3.0255E-11 | Zona pellucida glycoprotein 2.2 | NM_131827 |
| cldng | 5.502843 | 8.075E-12 | Claudin g | NM_180965 |
| snrpb | 5.379755 | 1.1409E-14 | Small nuclear ribonucleoprotein polypeptides B and B1 | NM_205667 |
| scg5 | 5.368851 | 2.5534E-12 | Secretogranin V | NM_200726 |
| mid1ip1 | 5.338822 | 1.3481E-14 | MID1-interacting protein 1 | NM_213439 |
| birc5b | 5.314876 | 5.4488E-12 | Baculoviral IAP repeat-containing 5B | NM_145195 |
| mt2 | 5.266296 | 3.5563E-12 | Metallothionein 2 | NM_001131053 |
| cyc1 | 5.202272 | 1.1492E-12 | Cytochrome c-1 | NM_001037393 |
| ccne | 5.179989 | 1.6367E-15 | Cyclin E | NM_130995 |
| slc16a3 | 5.150553 | 2.3088E-15 | Solute carrier family 16 member 3 | NM_212708 |
| retsatl | 5.095099 | 1.5198E-10 | Retinol saturase (all-trans-retinol 13,14-reductase)-like | NM_001040004 |
| ccna1 | 5.044201 | 2.5394E-13 | Cyclin A1 | NM_212818 |
| zp2 | 4.998241 | 9.6232E-12 | Zona pellucida glycoprotein 2 | NM_131330 |
| cldnd | 4.878126 | 1.2491E-12 | Claudin d | NM_180964 |
| thy1 | 4.843528 | 9.3008E-08 | Thy-1 cell surface antigen | NM_198065 |
| zp2l1 | 4.824493 | 1.5182E-12 | Zona pellucida glycoprotein 2,-like 1 | NM_001105104 |
| zp2.2 | 4.618628 | 4.1268E-10 | Zona pellucida glycoprotein 2.2 | NM_131827 |
| zar1 | 4.589525 | 6.0927E-11 | Zygote arrest 1 | NM_194381 |
| col1a2 | −6.30784 | 1.1828E-10 | Collagen, type I, α2 | NM_182968 |
| col1a3 | −5.40109 | 2.396E-11 | Collagen, type I, α1b | NM_201478 |
| cpa5 | −5.1099 | 0.0004497 | Carboxypeptidase A5 | NM_199271 |
| acta2 | −4.77678 | 1.1754E-08 | Actin, α2, smooth muscle, aorta | NM_212620 |
| rdh1l | −4.74052 | 3.8436E-08 | Dehydrogenase/reductase (SDR family) member 9 | NM_199609 |
| scp2 | −4.74032 | 1.7504E-09 | Sterol carrier protein 2 | NM_200865 |
| hbbe2 | −4.72664 | 6.9984E-08 | Hemoglobin β embryonic-2 | NM_212846 |
| rtn1a | −4.58485 | 1.8505E-08 | Reticulon 1a | NM_001029967 |
| gpx4a | −4.42594 | 6.1645E-06 | Glutathione peroxidase 4a | NM_001007282 |
| aldob | −4.3543 | 3.1299E-06 | Aldolase b, fructose-bisphosphate | NM_194367 |
| sparc | −4.2209 | 1.0631E-07 | Secreted acidic cysteine rich glycoprotein | NM_001001942 |
| hbae1 | −4.19334 | 1.0071E-08 | Hemoglobin α embryonic-1 | NM_182940 |
| cebpb | −3.81485 | 4.0853E-13 | CCAAT/enhancer binding protein (C/EBP), β | NM_131884 |
| try | −3.78429 | 0.00010216 | Trypsin | NM_131708 |
| acta2 | −3.71892 | 1.0572E-09 | Actin, α2, smooth muscle, aorta | NM_212620 |
| lpl | −3.69103 | 3.8436E-08 | Lipoprotein lipase | NM_131127 |
| btg1 | −3.69101 | 3.4084E-11 | B-cell translocation gene 1 | NM_200020 |
| fabp3 | −3.65859 | 9.0136E-08 | Fatty acid-binding protein 3, muscle and heart | NM_152961 |
| krt4 | −3.62196 | 7.8536E-07 | Keratin 4 | NM_131509 |
| rfc4 | −3.59129 | 1.0284E-07 | Replication factorC (activator 1) 4 | NM_214737 |
| gpx1a | −3.54498 | 4.1707E-07 | Glutathione peroxidase 1a | NM_001007281 |
| hbbe1 | −3.53021 | 0.00441842 | Hemoglobin β embryonic-2 | NM_131759 |
a -Fold change values between the intermediate and control male groups (I vs. CM) have been log2-transformed.
b The q value represents the false discovery rate corrected p-value derived from the linear fit to a group means parameterization.
TABLE 2.
Expression levels of genes with potential role in gonad differentiation were analyzed by microarrays using gonads from intermediate (I) doses of heat-killed E. coli from 15–35 dpf compared to those of control males (CM) or control females (CF)
The q value represents the false discovery rate-corrected p value derived from the linear fit to a group means paramaterization. -Fold change values between the control male and female (CF vs. CM) and the individuals treated with intermediate dose of heat-killed E. coli and control male (I vs. CM) have been log2-transformed.
| Gene | CF vs. CM | q value | I vs. CM | q value | Name |
|---|---|---|---|---|---|
| cyp19a1a | 0.116 | 0.675 | 0.146 | 0.594 | Cytochrome P450, family 19, subfamily A, polypeptide 1a |
| amh | 0.109 | 0.675 | 0.104 | 0.692 | Anti-Müllerian hormone |
| cyp19a1b | 0.063 | 0.856 | 0.028 | 0.939 | Cytochrome P450, family 19, subfamily A, polypeptide 1b |
| sox9a | 0.009 | 0.981 | 0.083 | 0.813 | SRY-box containing gene 9a |
| foxl2 | 0.009 | 0.981 | 0.022 | 0.956 | Forkhead box L2 |
| cyp11c1 | −0.391 | 0.136 | −0.357 | 0.174 | Cytochrome P450, family 11, subfamily C, polypeptide 1 |
| nr5a1b | −0.176 | 0.498 | 0.013 | 0.965 | Nuclear receptor subfamily 5, group A, member 1b |
| nr5a5 | −0.446 | 0.152 | −0.451 | 0.147 | Nuclear receptor subfamily 5, group A, member 5 |
| nr5a1a | −1.070 | 2.69E-06 | −1.180 | 6.42E-07 | Nuclear receptor subfamily 5, group A, member 1a |
TABLE 3.
Analysis of differential expression of seven genes by qRT-PCR to validate microarray data
CF, control female, n = 3; CM, control male, n = 2; I, intermediate (5 × 106 cfu/ml), n = 3; H, high (107 cfu/ml), n = 2; MA, microarray.
| Gene symbol | CF vs. CM |
I vs. CM |
H vs. CM |
|||
|---|---|---|---|---|---|---|
| MAa | qPCR | MA | qPCR | MA | qPCR | |
| bcl2 | −1.34 | 1.53 | −1.31 | 2.41b | 1.09 | 0.64 |
| dap3 | 1.57c | 13.04d | 1.67c | 21.00c | 1.17 | 5.49d |
| tp53 | 1.12 | 7.52c | 1.20 | 14.94d | 1.01 | 3.85d |
| pycard | −1.55b | −0.85 | −1.77b | −0.99 | 1.29 | −0.57 |
| casp3a | 1.4b | 42.84d | 1.42b | 89.61d | 1.05 | 10.82b |
| wt1a | −1.01 | 5.91 | −1.05 | 7.54 | 1.07 | 3.22 |
| zp2 | 40.59c | 388.56c | 31.96c | 450.17d | 17.50c | 102.64b |
a -Fold changes in gene expression for CF, I, and H compared to CM were calculated according to the formula, -fold change = ((CF, I, or H) − CM)/CM, using gonads from 35-dpf-old juveniles. Statistical significant difference from CM within a gene was determined using two-tailed Student́s t-test.
b p < 0.05.
c, p < 0.001.
d, p < 0.01.
Apoptotic Signaling Pathways Are Involved in the Survival of Oocytes
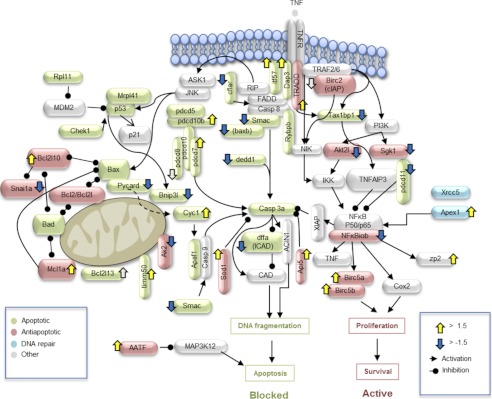
To perform a more detailed assessment of the potential role of apoptotic signaling pathways in zebrafish gonad differentiation, we analyzed the expression of those genes involved in these processes that were present on the microarray. The resulting gene expression pattern was consistent with the activation of anti-apoptotic pathways and the inhibition of apoptotic pathways in response to treatment with heat-killed E. coli (Fig. 4; Table 4). However, a number of genes involved in apoptotic pathway remained up-regulated in both female fish and those treated with heat-killed bacteria. Several crucial pro-apoptotic genes, including direct IAP-binding protein with low phosphatidylinositol/second mitochondria-derived activator of caspase (diablo/smac), CASP8 and FADD-like apoptosis regulator (cflar), death effector domain-containing 1 (dedd1), and PYD and CARD domain containing (pycard) were suppressed in treated individuals compared with controls, whereas genes involved in proliferation, including two birc5 genes (birc5a and -b), were up-regulated (Table 4). To confirm the microarray data, the expression patterns of a selected group of genes, including B-cell leukemia/lymphoma 2 (bcl2), death associated protein 3 (dap3), tp53, pycard, Wilms tumor 1a (wt1a), and zp2 were validated by qRT-PCR (Table 3). The obtained results confirmed the transcriptional activity profiles observed by the microarray for these genes.
FIGURE 4.
Alteration of apoptotic/anti-apoptotic signaling by heat-killed E. coli treatment. Exposure of juvenile zebrafish to an intermediate concentration (5 × 106 cfu/ml) of heat-killed E. coli was performed between 15 and 35 dpf. Gonads were dissected at 35 dpf. After RNA extraction, microarray was used to evaluate apoptotic and anti-apoptotic gene expression in individual gonads. Several critical apoptotic genes were suppressed (cflar, dedd1, pycard, bnip3l, and smac), whereas anti-apoptotic (bcl2l10, aatf, api5, and mcl1a) and NF-κB-target genes (birc5a and birc5b) were up-regulated.
TABLE 4.
Apoptotic and anti-apoptotic gene expression was analyzed by microarrays in gonads of zebrafish juveniles exposed to intermediate (I) and high (H) doses of heat-killed E. coli from 15–35 dpf compared to those of control males (CM) or control females (CF)
The q value represents the false discovery rate corrected p value derived from the linear fit to a group means paramaterization. -Fold change values between the treated groups and controls have been log2 transformed.
| Gene | CF vs. CM | q Value | I vs. CM | q value | Name |
|---|---|---|---|---|---|
| cyc1 | 5.545 | 4.22421E-13 | 5.202 | 1.14919E-12 | Cytochrome c1 |
| birc5b | 5.458 | 3.48241E-12 | 5.315 | 5.44875E-12 | Baculoviral IAP repeat-containing 5B |
| mcl1a | 2.387 | 6.10573E-11 | 2.294 | 1.19507E-10 | Myeloid cell leukemia sequence 1a |
| birc5a | 1.804 | 6.38669E-08 | 1.778 | 7.91592E-08 | Baculoviral IAP repeat-containing 5A |
| pdcd7 | 1.413 | 4.07714E-08 | 1.598 | 5.69641E-09 | Programmed cell death protein 7 homolog |
| sod1 | 1.080 | 6.32605E-05 | 1.150 | 2.98632E-05 | Superoxide dismutase |
| bcl2l13 | 0.982 | 0.244392481 | 0.323 | 0.732653483 | Bcl-2-like 13 protein |
| tradd | 0.945 | 1.72234E-06 | 1.006 | 6.88107E-07 | TNFR1-associated DEATH domain protein |
| ift57 | 0.942 | 2.5255E-06 | 0.828 | 1.45804E-05 | Intraflagellar transport protein 57 |
| timm50 | 0.938 | 0.000141313 | 0.663 | 0.003967945 | Import inner membrane translocase subunit TIM50 |
| bcl2l10 | 0.725 | 0.000281879 | 0.668 | 0.000662021 | Bcl-2-like 10 protein |
| pdcd10b | 0.722 | 2.70697E-05 | 0.824 | 4.76193E-06 | Programmed cell death protein 10B |
| dap3 | 0.651 | 0.000115272 | 0.741 | 2.46396E-05 | Death- associated protein 3 |
| aatf | 0.646 | 0.000488524 | 1.026 | 1.65076E-06 | Apoptosis-antagonizing transcription factor |
| apex1 | 0.640 | 0.002657661 | 0.905 | 8.01035E-05 | APEX nuclease |
| chek1 | 0.569 | 0.002904756 | 0.540 | 0.004424879 | Serine/threonine-protein kinase Chk1 |
| pdcd5 | 0.497 | 0.003502437 | 0.529 | 0.002112974 | Programmed cell death protein 5 |
| casp3a | 0.489 | 0.021534335 | 0.505 | 0.018007274 | Caspase 3 |
| api5 | 0.311 | 0.179674239 | 0.700 | 0.00309605 | Apoptosis inhibitor 5 |
| rybpb | 0.223 | 0.409033765 | 0.143 | 0.618438834 | Death effector domain-associated factor B |
| birc2 | 0.187 | 0.337374145 | −0.170 | 0.389206313 | Baculoviral IAP repeat-containing protein 2 |
| xrcc5 | 0.178 | 0.342167521 | 0.232 | 0.206242708 | X-ray repair cross-complementing protein 5 |
| tp53 | 0.166 | 0.547499145 | 0.267 | 0.308605818 | Tumor suppressor p53 |
| pdcd10a | −0.021 | 0.934394561 | −0.130 | 0.566305667 | Programmed cell death protein 10A |
| mrpl41 | −0.045 | 0.763067184 | −0.253 | 0.050968156 | Mitochondrial ribosomal protein L41 |
| bax | −0.076 | 0.813846548 | −0.169 | 0.581940727 | Bcl-2-associated X protein |
| bcl2l | −0.136 | 0.496224495 | −0.191 | 0.32022252 | Bcl-2-like 1 protein |
| apaf1 | −0.159 | 0.581732089 | −0.117 | 0.697171658 | Apoptotic protease-activating factor 1 |
| bad | −0.334 | 0.215793567 | −0.191 | 0.509176093 | Bcl2 antagonist of cell death |
| bcl2 | −0.418 | 0.151307689 | −0.389 | 0.183090613 | B-cell lymphoma 2 |
| dffa | −0.537 | 0.001465837 | −0.704 | 8.72749E-05 | DNA fragmentation factor subunit α |
| pdcd8 | −0.574 | 0.006444647 | −0.680 | 0.001647751 | Programmed cell death protein 8 |
| pycard | −0.637 | 0.04378308 | −0.820 | 0.010418878 | Apoptosis-associated speck-like prot. containing CARD |
| nfkbiab | −0.694 | 0.005897655 | −0.946 | 0.000374682 | NF-κB light polypeptide gene enhancer inhibitor αb |
| sgk1 | −1.002 | 0.000559778 | −0.694 | 0.011662921 | Serum/glucocorticoid-regulated kinase 1 |
| akt2l | −1.036 | 0.007551645 | −1.119 | 0.004243975 | V-akt murine thymoma viral oncogene homolog 2. like |
| rpl11 | −1.050 | 2.06218E-05 | −1.098 | 1.15201E-05 | 60 S ribosomal protein L11 |
| snai1a | −1.094 | 4.67951E-06 | −1.129 | 3.03591E-06 | Snail homolog 1a |
| ak2 | −1.246 | 7.86169E-06 | −1.452 | 9.0004E-07 | Adenylate kinase isoenzyme 2 |
| baxb | −1.343 | 1.49003E-07 | −1.381 | 9.54774E-08 | Bcl2-associated X protein, b |
| dedd1 | −1.635 | 7.21776E-08 | −1.574 | 1.29146E-07 | Death effector domain-containing 1 |
| smac | −1.695 | 0.000642094 | −1.522 | 0.001775805 | Second mitochondria-derived activator of caspase |
| tax1bp1 | −1.841 | 1.8459E-08 | −2.063 | 2.89485E-09 | Tax1-binding protein 1 |
| bnip3l | −1.941 | 4.97092E-09 | −1.902 | 7.09148E-09 | BCL2/adenovirus E1B 19 kDa-interacting protein 3-like |
| cflar | −2.585 | 5.34657E-11 | −2.610 | 4.51162E-11 | CASP8 and FADD-like apoptosis regulator precursor |
| pdcd11 | −3.346 | 6.17826E-08 | −2.841 | 7.32152E-07 | Programmed cell death protein 11 |
Treatment with Sodium Deoxycholate Induces NF-κB Pathway Resulting in Female-biased Sex Ratios
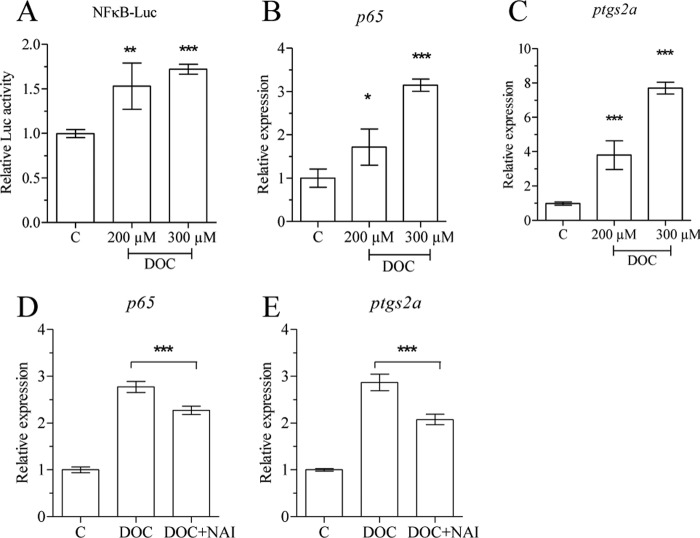
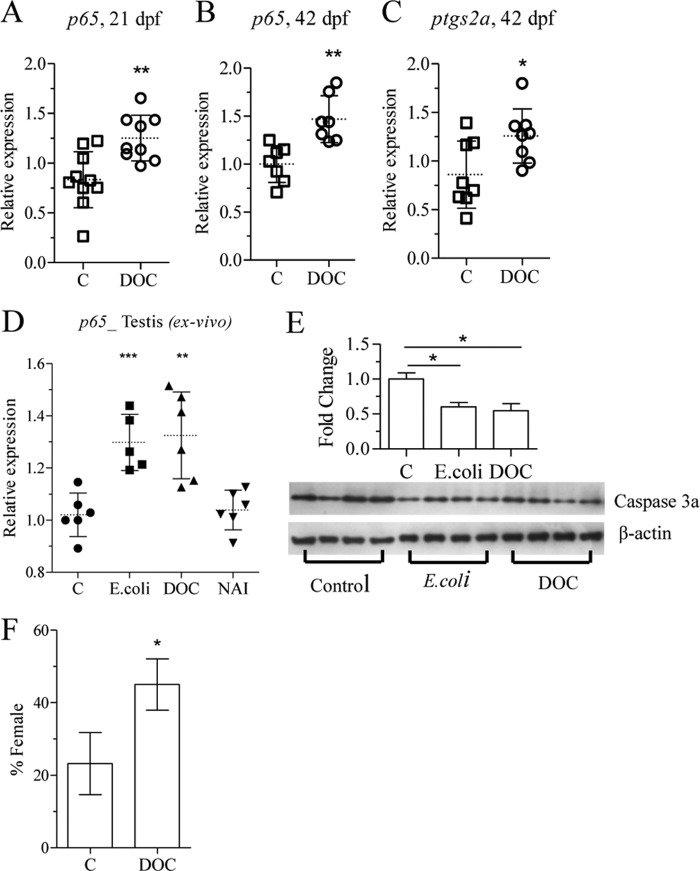
To confirm the involvement of NF-κB in gonad differentiation, we exposed zebrafish cells and juveniles to a known NF-κB activator, DOC (41). Exposure of nZFL and ZFL cells to DOC resulted in NF-κB activation (Fig. 5A) and up-regulation of both p65 and ptgs2a expression (Fig. 5, B and C). Exposure of zebrafish to 200 μm DOC from 15 to 21 dpf also caused an up-regulation of p65 expression (Fig. 6A), whereas the levels of sox9a, amh, and wt1a transcripts were not significantly reduced (data not shown), suggesting that DOC triggers a weaker response than heat-killed bacteria. However, significantly up-regulated p65 and ptgs2a expression (Fig. 6, B and C) was detected in the gonads of zebrafish juveniles exposed to DOC for 24 h at 41 dpf. The expression of p65 expression was also up-regulated in testis explants in response to heat-killed bacteria and DOC (Fig. 6D), whereas Western blot analysis of ovary samples showed a decrease in the level of caspase 3a protein (Fig. 6E), confirming that the treatment inhibited gonadal apoptosis. The sex ratio was determined at 70 dpf and revealed an increased percentage of females from 25 to 45% after exposure to 200 μm DOC from 15 to 35 dpf (Fig. 6F).
FIGURE 5.
DOC activates NFκB and up-regulates p65 and ptgs2a expression in vitro. nZFL were exposed to DOC (200 μm, 300 μm) for 12 h, and luciferase activity was analyzed to check NF-κB activation (A). ZFL cells were exposed to DOC (200 μm, 300 μm) alone and combination (200 μm DOC) with NAI (40 nm) for 24 h, and total RNA was extracted followed by qRT-PCR analysis of p65 (B and D) and ptgs2a (C and E) expression. The statistically significant difference between groups was determined using Student's t test (*, p < 0.05; **, p < 0.01; ***, p < 0.001). n = 4. Error bars represent the mean ± S.D.
FIGURE 6.
DOC treatment up-regulates the expression of inflammatory genes leading to female-biased population. Zebrafish juveniles were exposed to DOC (200 μm) and analyzed for changes in p65, ptgs2a, and sex ratio. Shown is analysis of p65 levels after 6 days of exposure at 15 dpf (A). 24 h of exposure of 41 dpf juveniles was followed by analysis of p65 (B) and ptgs2a (C) expression in the gonads. Testis explants were exposed to heat-killed bacteria (5 × 107), DOC (200 μm), and NAI (20 nm) for 24 h, and total RNA was extracted followed by qRT-PCR analysis of p65 (D). Zebrafish juveniles at 70 dpf were exposed to heat-killed bacteria (5 × 107) and DOC (300 μm) for 2 days, and gonads were isolated for Western blot analysis (E). Larvae at 15 dpf were exposed to DOC (200 μm) for 20 days, and sex ratio was determined at 70 dpf. The mean and S.D. of two independent experiments are shown. Each group contained at least 30 individuals (F). Statistically significant difference between groups was determined using Student's t test (*, p < 0.05; **, p < 0.01; ***, p < 0.001). Error bars represent the mean ± S.D.
NAI Exposure Down-regulates p65 and ptgs2a Expression Resulting in Male-biased Sex Ratios
NAI is a 6-amino-quinazoline-derived compound that is reported to inhibit NF-κB activation and TNF-α production (42). ZFL cells exposed to 200 μm DOC alone or in combination with 40 nm NAI were analyzed for p65 and ptgs2a expression. NAI treatment significantly down-regulated DOC-induced ptgs2a and p65 gene expression (Fig. 5, D and E). A significant down-regulation of p65 and up-regulation of caspase 8, apoptosis-related cysteine peptidase (casp8), cyclin-dependent kinase inhibitor 1A (p21), and smac (Fig. 7, A–D) expression has been observed in juveniles exposed to 20 nm NAI for 24 h at 20 dpf. Six days of NAI exposure of zebrafish juveniles (15–21 dpf) resulted in decreased expression of ptgs2a, X-linked inhibitor of apoptosis (xiap), and zp2 (Fig. 8, A–C). Exposure of juveniles to NAI for 24 h at 41 dpf resulted in reduced gonadal ptgs2a gene expression (Fig. 8D), confirming the effect of NF-κB inhibition on gonadal gene expression. Furthermore exposure of zebrafish juveniles to 20 nm NAI from 15 to 35 dpf resulted in a decreased percentage of females from 45 to 20% (Fig. 8E). These results further support a role of NF-κB in the maintenance of oocyte development during zebrafish sex differentiation.
FIGURE 7.
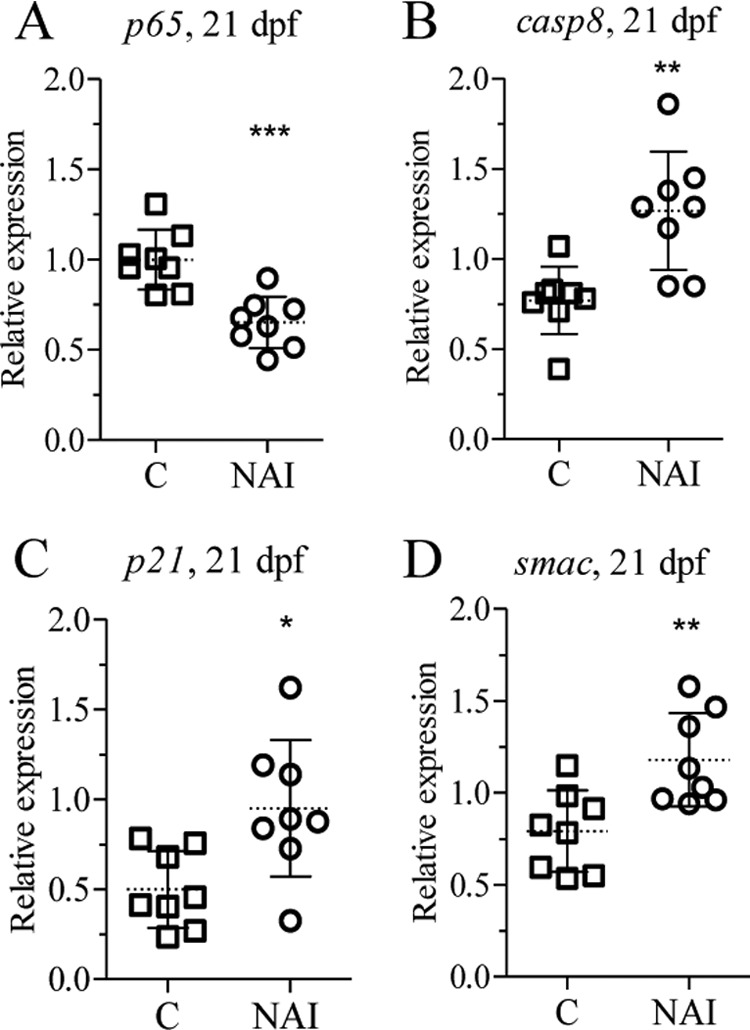
NAI exposure alters apoptotic and anti-apoptotic gene expression in vivo. Zebrafish juveniles at 20 dpf were exposed to NAI (20 nm) for 24 h and analyzed for p65 (A), casp8 (B), p21 (C), and smac (D) transcript levels. The statistically significance difference from the control was determined using Student's t test (*, p < 0.05; **, p < 0.01; ***, p < 0.001). n = 8. Error bars represent the mean ± S.D.
FIGURE 8.
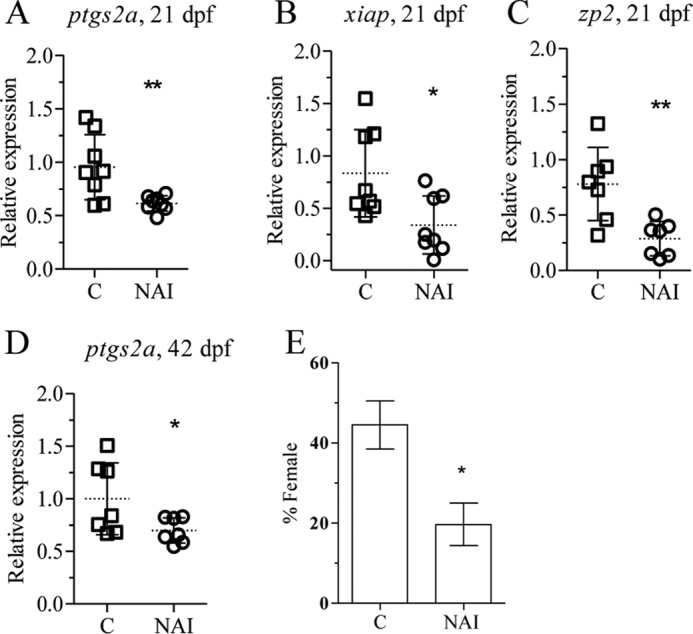
NAI treatment down-regulates the expression of inflammatory genes leading to male-biased population. Zebrafish juveniles were exposed to 20 nm NAI for different periods of time followed by qPCR analysis of gene expression and sex ratio. Exposure at 15 dpf for 6 days was followed by analysis of ptgs2a (A), xiap (B), and zp2 (C). Zebrafish juveniles at 41 dpf were exposed for 24 h followed by qPCR analysis of ptgs2a (D). Juveniles at 15 dpf were exposed for 20 days, and the sex ratio was determined at 70 dpf (E). The mean and S.D. of three independent experiments are shown. Each group contained at least 30 individuals. Statistically significance difference from the control was determined using Student's t test (* p < 0.05; **, p < 0.01). Error bars represent the mean ± S.D.
DISCUSSION
Although the molecular mechanism of zebrafish sex determination remains unknown, several genes, including sox9, amh, nr5a, and doublesex and mab-3 related transcription factor 1 (dmrt1), have been proposed to be involved in the initial stages of testis and ovary differentiation (7, 8) Testis differentiation in zebrafish starts with a juvenile ovary stage followed by a transformation involving apoptotic loss of oocytes leading to the eventual development of male sex organs (14, 16). Analysis of temporal expression profile of the zp2 gene suggests that the juvenile ovary stage is initiated before 19 dpf, indicating that all of our treatments were started before the initiation of the juvenile ovary stage. In this study we show that induction of gonadal inflammation, through treatment with heat-killed E. coli or DOC during the critical stage of gonad differentiation, activates NF-κB and anti-apoptotic signaling, resulting in maintenance of oocyte development and inhibition of testis formation. In contrast, inhibition of NF-κB activity by exposure to NAI results in an induction of testis differentiation. These results demonstrate that NF-κB is involved in the regulation of the gonadal differentiation process in zebrafish. Identifying the mechanisms of gonadal differentiation in zebrafish as well as interlinks and alterations by apoptotic and inflammatory (anti-apoptotic) responses are key factors in the understanding of possible endogenous and environmental influences during this delicate process.
NF-κB is a potent inducer of inflammatory responses as well as a modulator of apoptotic signaling (23). Several critical pro-apoptotic genes, including smac, pycard, BCL2/adenovirus E1B interacting protein-like b (bnip3l), and Tax1 (human T-cell leukemia virus type I)-binding protein 1b (tax1bp1) and cflar, were repressed by exposure to heat-killed E. coli. The decrease of caspase 3a protein levels in the ovaries confirms the anti-apoptotic effect of the treatment. The pro-apoptotic activity of these genes is well documented, and their increased expression leads to the activation of several caspases, including casp3, casp8, and casp9 (43). Their roles in the activation and translocation of proteins associated with the mitochondrial apoptotic pathway have also been demonstrated (44). Furthermore, the expression of several genes with anti-apoptotic functions associated with NF-κB signaling, including tnfrsf1a-associated via death domain (tradd), birc5a, birc5b, apoptosis inhibitor 5 (api5), apoptosis antagonizing transcription factor (aatf), and myeloid cell leukemia sequence 1a (mcl1a), was significantly up-regulated in response to induced inflammation. In addition, expression of the NF-κB inhibitory gene nuclear factor of κ light polypeptide gene enhancer in B-cells inhibitor, αb (nfkbiab) was suppressed. These genes have been reported to alter apoptotic signals transduced through the extrinsic TNF receptor (45) and the intrinsic (mitochondrial) (46) pathways. This is in agreement with the observed feminization after exposure to heat-killed bacteria and DOC. To confirm the involvement of NF-κB in this process, we used NAI to block NF-κB activation.
Short term exposure of larvae to NAI down-regulated p65, ptgs2a, and xiap expression, whereas the activity of apoptotic genes, smac, casp8, and p21 was significantly increased. The expression of female-specific zp2 gene was also down-regulated, and this correlated with male-biased populations after long term exposure. Down-regulation of zp2 and the male bias sex ratio, therefore, involves NF-κB inhibition and induction of apoptotic signals leading to oocyte apoptosis and initiation of testis differentiation. It was interesting to note that gonadal ptgs2a was up-regulated in DOC-treated individuals and down-regulated in NAI-treated individuals. At present, data linking prostaglandins to a direct role in sex differentiation in zebrafish are lacking. Therefore, further studies are clearly needed to elucidate the role of cyclooxygenase-2 and prostaglandins in these processes. There have been indications of the involvement of nr5a genes in zebrafish sex determination and gonad differentiation (8). The induction of Amh by Sox9, via steroidogenic factor-1, is well documented in mammals (47), but the molecular basis for gonad differentiation in zebrafish remains unclear. In this study only nr5a1a expression was found to be up-regulated in male fish at 35 dpf, whereas we did not observe any differential expression pattern for cyp19a1a, cyp19a1b, or cyp11c1 in either control fish or after exposure to heat-killed E. coli from 15 to 35 dpf. This suggests that altered 17β-estradiol and 11-ketotestosterone levels are a consequence of gonad differentiation. The recent discovery of sex-associated genomic regions on chromosomes 3, 4, 5, and 16 (5, 6) makes it unlikely that zebrafish would have a classical sex determination system. Instead, the interplay between different loci may be instrumental to the development of testis or ovary in the species (4, 7). As the identified regions account only for some of the variance, it is likely that other genes are also involved in the process.
Zebrafish gonad differentiation involves a juvenile ovary stage after which the gonad either receives signals to maintain differentiating oocytes or to induce oocyte apoptosis and trigger the transformation into a testis structure. The correlation between NF-κB activity and sex differentiation shown in this study is in good agreement with earlier studies suggesting the involvement of apoptosis and tp53 in testis development (16, 17). Further research is needed to elucidate the sex-specific signaling pathways leading to the regulation of NF-κB activity in zebrafish during the juvenile ovary stage, thereby controlling the further development into the ovary and testis.
Supplementary Material
This research was financed by the Swedish Research Council, the Knowledge Foundation, Sweden, and Örebro University (to P. E.-O.) and by the Agri-Food and Veterinary Authority as well as Temasek Life Sciences Laboratory, Singapore (to L. O.).

This article contains supplemental Tables 1–3.
The full set of microarray expression data has been deposited in ArrayExpress under the accession number E-MEXP-3249.
- AMH
- anti-Müllerian hormone
- IAP
- inhibitor of apoptosis protein
- DOC
- sodium deoxycholate
- NAI
- NF-κB activation inhibitor
- qRT-PCR
- quantitative RT-PCR
- I
- intermediate
- CM
- control male
- CF
- control females.
REFERENCES
- 1. Sola L., Gornung E. (2001) Classical and molecular cytogenetics of the zebrafish, Danio rerio (Cyprinidae, Cypriniformes). An overview. Genetica 111, 397–412 [DOI] [PubMed] [Google Scholar]
- 2. Wallace B. M., Wallace H. (2003) Synaptonemal complex karyotype of zebrafish. Heredity 90, 136–140 [DOI] [PubMed] [Google Scholar]
- 3. Singer A., Perlman H., Yan Y., Walker C., Corley-Smith G., Brandhorst B., Postlethwait J. (2002) Sex-specific recombination rates in zebrafish (Danio rerio). Genetics 160, 649–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Liew W. C., Bartfai R., Lim Z., Sreenivasan R., Siegfried K. R., Orban L. (2012) Polygenic sex determination system in zebrafish. PLoS One 7, e34397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bradley K. M., Breyer J.P., Melville D.B., Broman K.W., Knapik E.W., Smith J.R. (2011) An SNP-based linkage map for zebrafish reveals sex determination loci. G3 1, 3–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Anderson J. L., Rodríguez Marí A., Braasch I., Amores A., Hohenlohe P., Batzel P., Postlethwait J. H. (2012) Multiple sex-associated regions and a putative sex chromosome in zebrafish revealed by RAD mapping and population genomics. PLoS One 7, e40701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Orban L., Sreenivasan R., Olsson P. E. (2009) Long and winding roads. Testis differentiation in zebrafish. Mol. Cell. Endocrinol. 312, 35–41 [DOI] [PubMed] [Google Scholar]
- 8. von Hofsten J., Olsson P. E. (2005) Zebrafish sex determination and differentiation. Involvement of FTZ-F1 genes. Reprod. Biol. Endocrinol. 3, 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Siegfried K. R., Nüsslein-Volhard C. (2008) Germ line control of female sex determination in zebrafish. Dev. Biol. 324, 277–287 [DOI] [PubMed] [Google Scholar]
- 10. Rodríguez-Marí A., Yan Y. L., Bremiller R. A., Wilson C., Cañestro C., Postlethwait J. H. (2005) Characterization and expression pattern of zebrafish anti-Müllerian hormone (amh) relative to sox9a, sox9b, and cyp19a1a during gonad development. Gene Expression Patterns 5, 655–667 [DOI] [PubMed] [Google Scholar]
- 11. Lasala C., Carré-Eusèbe D., Picard J. Y., Rey R. (2004) Subcellular and molecular mechanisms regulating anti-Müllerian hormone gene expression in mammalian and nonmammalian species. DNA Cell Biol. 23, 572–585 [DOI] [PubMed] [Google Scholar]
- 12. Arango N. A., Lovell-Badge R., Behringer R. R. (1999) Targeted mutagenesis of the endogenous mouse Mis gene promoter. In vivo definition of genetic pathways of vertebrate sexual development. Cell 99, 409–419 [DOI] [PubMed] [Google Scholar]
- 13. Wagner T., Wirth J., Meyer J., Zabel B., Held M., Zimmer J., Pasantes J., Bricarelli F. D., Keutel J., Hustert E., Wolf U., Tommerup N., Schempp W., Scherer G. (1994). Cell 79, 1111–1120 [DOI] [PubMed] [Google Scholar]
- 14. Takahashi H. (1977) Juvenile Hermaphroditism in the Zebrafish, Brachydanio rerio. Bull. Fac. Fish. Hokkaido Univ. 28, 57–65 [Google Scholar]
- 15. Wang X. G., Bartfai R., Sleptsova-Freidrich I., Orban L. (2007) The timing and extent of “juvenile ovary” phase are highly variable during zebrafish testis differentiation. J. Fish Biol. 70, 33–44 [Google Scholar]
- 16. Uchida D., Yamashita M., Kitano T., Iguchi T. (2002) Oocyte apoptosis during the transition from ovary-like tissue to testes during sex differentiation of juvenile zebrafish. J. Exp. Biol. 205, 711–718 [DOI] [PubMed] [Google Scholar]
- 17. Rodríguez-Marí A., Cañestro C., Bremiller R. A., Nguyen-Johnson A., Asakawa K., Kawakami K., Postlethwait J. H. (2010) Sex reversal in zebrafish fancl mutants is caused by Tp53-mediated germ cell apoptosis. PLoS Genet. 6, e1001034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Slanchev K., Stebler J., de la Cueva-Méndez G., Raz E. (2005) Development without germ cells. The role of the germ line in zebrafish sex differentiation. Proc. Natl. Acad. Sci. U.S.A. 102, 4074–4079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Siebenlist U., Franzoso G., Brown K. (1994) Structure, regulation, and function of NF-κB. Annu. Rev. Cell Biol. 10, 405–455 [DOI] [PubMed] [Google Scholar]
- 20. Ghosh S., Hayden M. S. (2008) New regulators of NF-κB in inflammation. Nat. Rev. Immunol. 8, 837–848 [DOI] [PubMed] [Google Scholar]
- 21. Xiao W. (2004) Advances in NF-κB signaling transduction and transcription. Cell. Mol. Immunol. 1, 425–435 [PubMed] [Google Scholar]
- 22. Finco T. S., Baldwin A. S. (1995) Mechanistic aspects of NF-κB regulation. The emerging role of phosphorylation and proteolysis. Immunity 3, 263–272 [DOI] [PubMed] [Google Scholar]
- 23. Aggarwal B. B., Sethi G., Nair A., Ichikawa H. (2006) Nuclear factor-κB. A holy grail in cancer prevention and therapy. Curr. Signal Transduct. Ther. 1, 25–52 [Google Scholar]
- 24. Srinivasula S. M., Ashwell J. D. (2008) IAPs. What's in a name? Mol. Cell 30, 123–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rao N. A., McCalman M. T., Moulos P., Francoijs K. J., Chatziioannou A., Kolisis F. N., Alexis M. N., Mitsiou D. J., Stunnenberg H. G. (2011) Coactivation of GR and NF-κB alters the repertoire of their binding sites and target genes. Genome Res. 21, 1404–1416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Palvimo J. J., Reinikainen P., Ikonen T., Kallio P. J., Moilanen A., Jänne O. A. (1996) Mutual transcription interference between Rel A and androgen receptor. J. Biol. Chem. 271, 24151–24156 [DOI] [PubMed] [Google Scholar]
- 27. Delfino F., Walker W.H. (1998) Stage-specific nuclear expression of NF-κB in mammalian testis. Mol. Endocrinol. 12, 1696–1707 [DOI] [PubMed] [Google Scholar]
- 28. Hong C. Y., Park J. H., Seo K. H., Kim J. M., Im S. Y., Lee J. W., Choi H. S., Lee K. (2003) Expression of MIS in the testis is down-regulated by tumor necrosis factor α through the negative regulation of SF-1 transactivation by NF-κB. Mol. Cell. Biol. 23, 6000–6012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Murakami S., Lefebvre V., de Crombrugghe B. (2000) Potent inhibition of the master chondrogenic factor Sox9 gene by interleukin-1 and tumor necrosis factor-α. J. Biol. Chem. 275, 3687–3692 [DOI] [PubMed] [Google Scholar]
- 30. Meijer A. H., van der Sar A. M., Cunha C., Lamers G. E., Laplante M. A., Kikuta H., Bitter W., Becker T. S., Spaink H. P. (2008) Identification and real-time imaging of a myc-expressing neutrophil population involved in inflammation and mycobacterial granuloma formation in zebrafish. Dev. Comp. Immunol. 32, 36–49 [DOI] [PubMed] [Google Scholar]
- 31. Lister A., Van Der Kraak G. (2002) Modulation of goldfish testicular testosterone production in vitro by tumor necrosis factor a, interleukin-1β, and macrophage conditioned media. J. Exp. Zool. 292, 477–486 [DOI] [PubMed] [Google Scholar]
- 32. Schmittgen T. D., Livak K.J. (2008) Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 3, 1101–1108 [DOI] [PubMed] [Google Scholar]
- 33. Sreenivasan R., Cai M., Bartfai R., Wang X., Christoffels A., Orban L. (2008) Transcriptomic analyses reveal novel genes with sexually dimorphic expression in the zebrafish gonad and brain. PLoS One 3, e1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ihaka R., Gentleman R. (1996) R: A language for data analysis and graphics. J. Comput. Graph. Stat. 5, 299–314 [Google Scholar]
- 35. Smyth G. K. (2004) Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 3, 3. [DOI] [PubMed] [Google Scholar]
- 36. Smyth G. K., Speed T. (2003) Normalization of cDNA microarray data. Methods 31, 265–273 [DOI] [PubMed] [Google Scholar]
- 37. Ritchie M. E., Silver J., Oshlack A., Holmes M., Diyagama D., Holloway A., Smyth G. K. (2007) A comparison of background correction methods for two-color microarrays. Bioinformatics 23, 2700–2707 [DOI] [PubMed] [Google Scholar]
- 38. Benjamini Y., Hochberg Y. (1995) Controlling the false discovery rate. A practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57, 289–300 [Google Scholar]
- 39. Sekiguchi T., Iida H., Fukumura J., Nishimoto T. (2004) Human DDX3Y, the Y-encoded isoform of RNA helicase DDX3, rescues a hamster temperature-sensitive ET24 mutant cell line with a DDX3X mutation. Exp. Cell Res. 300, 213–222 [DOI] [PubMed] [Google Scholar]
- 40. Gilardelli C. N., Pozzoli O., Sordino P., Matassi G., Cotelli F. (2004) Functional and hierarchical interactions among zebrafish vox/vent homeobox genes. Dev. Dyn. 230, 494–508 [DOI] [PubMed] [Google Scholar]
- 41. Payne C. M., Weber C., Crowley-Skillicorn C., Dvorak K., Bernstein H., Bernstein C., Holubec H., Dvorakova B., Garewal H. (2007) Deoxycholate induces mitochondrial oxidative stress and activates NF-κB through multiple mechanisms in HCT-116 colon epithelial cells. Carcinogenesis 28, 215–222 [DOI] [PubMed] [Google Scholar]
- 42. Tobe M., Isobe Y., Tomizawa H., Nagasaki T., Takahashi H., Hayashi H. (2003) A novel structural class of potent inhibitors of NF-κB activation. Structure-activity relationships and biological effects of 6-aminoquinazoline derivatives. Bioorg. Med. Chem. 11, 3869–3878 [DOI] [PubMed] [Google Scholar]
- 43. Shiozaki E. N., Shi Y. (2004) Caspases, IAPs, and Smac/DIABLO: Mechanisms from structural biology. Trends Biochem. Sci. 39, 486–494 [DOI] [PubMed] [Google Scholar]
- 44. Ohtsuka T., Ryu H., Minamishima Y. A., Macip S., Sagara J., Nakayama K. I., Aaronson S. A., Lee S. W. (2004) ASC is a Bax adaptor and regulates the p53-Bax mitochondrial apoptosis pathway. Nat. Cell Biol. 6, 121–128 [DOI] [PubMed] [Google Scholar]
- 45. Hsu H., Xiong J., Goeddel D.V. (1995) The TNF receptor 1-associated protein TRADD signals cell death and NF-κB activation. Cell 81, 495–504 [DOI] [PubMed] [Google Scholar]
- 46. Rigou P., Piddubnyak V., Faye A., Rain J. C., Michel L., Calvo F., Poyet J. L. (2009) The antiapoptotic protein AAC-11 interacts with and regulates Acinus-mediated DNA fragmentation. EMBO J. 28, 1576–1588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lasala C., Schteingart H. F., Arouche N., Bedecarrás P., Grinspon R. P., Picard J.Y., Josso N., di Clemente N., Rey R. A. (2011) SOX9 and SF1 are involved in cyclic AMP-mediated up-regulation of anti-Müllerian gene expression in the testicular prepubertal Sertoli cell line SMAT1. Am. J. Physiol. Endocrinol. Metab. 301, E539–E547 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.