Background: Malaria parasite egress from host erythrocytes requires cysteine protease activity, but the identity of key parasite proteases is unknown.
Results: SERA6 is an essential parasite cysteine protease that resides in the parasitophorous vacuole and is activated by SUB1, a parasite serine protease.
Conclusion: SERA6 may play a role in egress.
Significance: SERA6 is a potential new antimalarial drug target.
Keywords: Cysteine Protease, Malaria, Microbial Pathogenesis, Plasmodium, Serine Protease, SERA, SUB1, Egress
Abstract
The malaria parasite replicates within an intraerythrocytic parasitophorous vacuole (PV). The PV and host cell membranes eventually rupture, releasing merozoites in a process called egress. Certain inhibitors of serine and cysteine proteases block egress, indicating a crucial role for proteases. The Plasmodium falciparum genome encodes nine serine-repeat antigens (SERAs), each of which contains a central domain homologous to the papain-like (clan CA, family C1) protease family. SERA5 and SERA6 are indispensable in blood-stage parasites, but the function of neither is known. Here we show that SERA6 localizes to the PV where it is precisely cleaved just prior to egress by an essential serine protease called PfSUB1. Mutations that replace the predicted catalytic Cys of SERA6, or that block SERA6 processing by PfSUB1, could not be stably introduced into the parasite genomic sera6 locus, indicating that SERA6 is an essential enzyme and that processing is important for its function. We demonstrate that cleavage of SERA6 by PfSUB1 converts it to an active cysteine protease. Our observations reveal a proteolytic activation step in the malarial PV that may be required for release of the parasite from its host erythrocyte.
Introduction
Some of the worlds most important parasitic diseases, including malaria, toxoplasmosis, and cryptosporidiosis, are caused by protozoan pathogens of the phylum Apicomplexa. A unifying feature of this group of parasites is their requirement to replicate within the confines of a host cell. Blood-stage forms of the malaria parasite, Plasmodium spp., invade erythrocytes where they undergo multiple rounds of nuclear division within a membrane-bound parasitophorous vacuole (PV).6 Daughter merozoites eventually bud from the mature intracellular schizont then the enclosing PV membrane (PVM) and host cell membrane rupture in a process called egress to release the new generation of parasites. Numerous reports have shown that egress can be efficiently blocked by inhibitors of serine and cysteine proteases. As an example of the latter, treatment of mature Plasmodium falciparum schizonts with the epoxide trans-epoxysuccinyl-l-leucylamido(4-guanidino)butane (E64), an inhibitor of cysteine proteases of the papain-like superfamily (MEROPS clan CA, family C1), results in “sacks” of mature merozoites that remain trapped within at least one bounding membrane (1–4). Mechanical rupture of these stalled segmented schizonts releases fully invasive parasites (1, 5), demonstrating that E64 does not ablate merozoite viability per se. Despite these and other similar observations implicating a critical role for proteases, only one molecularly defined parasite protease, an essential subtilisin-like serine protease called SUB1, has been experimentally directly implicated in blood-stage egress. P. falciparum SUB1 (PfSUB1: PlasmodDB 3D7 ID PF3D7_0507500, MEROPS ID S08.012) is expressed during schizont maturation and is initially stored in a set of parasite secretory organelles called exonemes (6). Just prior to egress, PfSUB1 is discharged into the PV lumen where it precisely cleaves several merozoite surface and PV proteins (6–8). These PfSUB1 substrates include an abundant, soluble parasite PV protein called SERA5 (PlasmoDB 3D7 ID PF3D7_0207600), which is cleaved by PfSUB1 to release a central papain-like domain. SERA5 is encoded by a member of a large family of SERA genes, orthologues of which are found in the genomes of all Plasmodium species examined (9). The number of SERA family members varies between Plasmodium species; for example, there are 9 SERA genes in P. falciparum, and 5 in the rodent malarial species Plasmodium berghei. Importantly, in most SERA orthologues there is clear conservation of the SUB1 processing sites (9–11), suggesting that they too may be substrates for SUB1. In support of this, earlier data from this laboratory (6) indicate that in P. falciparum both SERA4 and SERA6 may be similarly cleaved by PfSUB1. Although most SERA genes are expressed in the asexual blood-stage life cycle (12, 13), one (SERA8 in P. falciparum, or its orthologue SERA5 in P. berghei) is expressed exclusively during mosquito stages of the parasite life cycle, where it has been shown to be required for egress of midgut sporozoites from oocysts (14). This finding has prompted speculation that the blood-stage SERAs may play an analogous role in merozoite egress from erythrocytes. In view of the temporal association between SERA5 processing and egress in P. falciparum (15–17), plus the observation that selective pharmacological inhibition of PfSUB1 inhibits egress (6, 18), and the fact that proteases are often activated by proteolysis, we have previously suggested (19) that processing of SERA5, and also perhaps of other blood-stage SERAs, by PfSUB1 may activate them in some way to play a proteolytic role in the events accompanying egress. However, there is no direct evidence that SUB1-mediated processing of any SERA family member is important for maintenance of the parasite life cycle.
Of the nine P. falciparum SERA family members, SERA6, -7, and -8 possess a canonical Cys residue at the position of the active site nucleophile in their papain-like central domains. In the remaining P. falciparum SERAs this residue is replaced with a Ser. Importantly, gene disruption studies in P. falciparum have shown that only SERA5 (“Ser”-type) and the much less abundant SERA6 (“Cys”-type: P. falciparum 3D7 PlasmoDB ID PF3D7_0207500) are essential in asexual blood stages (12, 20). Experimental Cys to Ser substitution at the active site of a number of cysteine proteases, including papain (21), streptopain (22), and cathepsin L (23), abolishes or profoundly reduces the proteolytic activity of those enzymes, so the absence of a catalytic Cys in SERA5 has led to suggestions that it and the other Ser-type SERAs are unlikely to be proteases. In support of this, only very limited protease activity was found associated with a recombinant form of the SERA5 papain-like domain (24), and x-ray crystallographic studies of the same protein have revealed aberrant structural features that cast doubt on its capacity to interact in a canonical manner with polypeptide substrates (25). In contrast, the papain-like domain of P. falciparum SERA6 possesses a canonical active-site Cys (Cys-644), and both homology modeling (24) and phylogenetic analyses (9, 20) have suggested that SERA6 likely plays a functionally distinct role from that of the Ser-type SERAs. Despite this, there is no experimental evidence that SERA6 can mediate protease activity, and its physiological function remains unknown.
Here we examine the importance and function of SERA6 and its proteolytic processing in the asexual blood-stage life cycle of the parasite. We first confirm that SERA6 localizes to the PV in P. falciparum and show that it is an authentic endogenous PfSUB1 substrate, being precisely cleaved by PfSUB1 at 3 positions at or around the time of egress. Using targeted homologous recombination, we demonstrate that the predicted catalytic Cys of SERA6 is required for asexual blood-stage P. falciparum viability, suggesting that SERA6 is an essential enzyme. We similarly show that mutations in SERA6 that block processing by PfSUB1 are not tolerated in the parasite, suggesting that the processing is important for SERA6 function. Finally, we demonstrate that cleavage by PfSUB1 of the P. berghei orthologue of SERA6 converts it to a form with protease activity. Our observations suggest that SERA6 may participate in an essential proteolytic pathway that is triggered by discharge of PfSUB1 into the PV just prior to egress.
EXPERIMENTAL PROCEDURES
Recombinant Expression and Purification of PfSUB1, SERA6, and PbSERA3, and Production of Antibodies
Recombinant PfSUB1 (rPfSUB1) was produced, purified, and quantified as described previously (10, 26). One unit of rPfSUB1 is defined as the amount of protease that hydrolyzes 1 pmol of the fluorogenic peptide substrate SERA4st1F-6R12 (Ac-CKITAQDDEESC labeled on both cysteine side chains with iodoacetamido tetramethylrhodamine) (10), in 1 min at a substrate concentration of 0.1 μm in digestion buffer (25 mm Tris-HCl, pH 8.2, 12 mm CaCl2, 25 mm CHAPS) at 21 °C. Three regions of the P. falciparum SERA6 sequence were expressed as recombinant proteins in Escherichia coli SHuffle® T7 Express lysY competent cells (New England Biolabs) using a synthetic SERA6 gene (sera6synth; produced by Geneart AG), codon-optimized for expression in Pichia pastoris and based on the predicted P. falciparum FCBR SERA6 sequence reported by Knapp et al. (27) (PFASERP H, GenBankTM accession number M55428). This differs from the predicted 3D7 SERA6 primary sequence (PlasmoDB ID PF3D7_0207500) by just two substitutions, at P894T and N929D (the last residue in each case is the FCBR one). The sera6synth gene was provided as an insert with terminal AscI and PacI sites in pGA14. Polypeptides comprising SERA6 residues Asp371–Gln927 (called S6C1) and residues Ser928–Val1031 (called S6 C-term) were expressed with N-terminal hexahistidine (His6) and S-tags using the pEt-30Xa/LIC system (Invitrogen). S6C1 accumulated in insoluble inclusion bodies that were extensively washed, solubilized into 8 m urea in 20 mm Tris-HCl, pH 8.2, and buffer exchanged into phosphate-buffered saline (PBS). S6 C-term was expressed in soluble form and was purified by nickel-chelate chromatography using nickel-nitrilotriacetic acid Superflow resin (Qiagen). The resin was washed in 300 mm NaCl, 50 mm arginine in 50 mm NaH2PO4, pH 8, then bound proteins were eluted using the same buffer containing 1 m arginine. Eluted fractions were dialyzed overnight against PBS at 4 °C. A third recombinant protein corresponding to SERA6 residues Gly35–Lys66 (called S6 N-term) was expressed with an N-terminal glutathione S-transferase (GST)-tag using expression vector pGEX-SERA6 (a kind gift of Brendan Crabb, Burnet Institute, Melbourne, Australia) and was affinity purified using glutathione-agarose (Sigma) as described previously (12). Yields of the purified recombinant proteins were ∼20 mg/liter of E. coli culture (S6C1), ∼5 (S6 C-term) and ∼2 mg/liter (S6 N-term). The three purified polypeptides were used to immunize mice, and S6C1 was also used to immunize rabbits (performed by Harlan Laboratories, UK). Before use, all sera were adsorbed against E. coli acetone powder (40 mg/ml serum) to deplete antibodies against bacterial proteins.
The synthetic sera6synth gene was also used to express a recombinant form of full-length P. falciparum SERA6 (minus its secretory signal peptide) called rSERA6wt (residues Glu23–Val1031), plus three processing site mutants in which residues Ala369–Gln370 (called rSERA6mutS1), or Gly926–Gln927 (rSERA6mutS2), or all four residues (rSERA6mutS1S2), were substituted with Leu by site-directed mutagenesis using the QuikChange II kit (Agilent Technologies) (see supplemental Table S1 for all oligonucleotide primers used in this study). All four recombinant proteins were expressed with N-terminal His and S-tags in the pEt-30Xa/LIC system (Invitrogen) in E. coli BL21-Gold DE3 cells (Stratagene). Purification and use of these proteins is described below.
A central region of P. berghei ANKA PbSERA3 (PlasmodDB ID PBANKA_030490), the orthologue of P. falciparum SERA6 (9), was expressed in E. coli using a synthetic pbsera3 gene (pbsera3synth; Geneart AG) codon-optimized for expression in Spodoptera frugiperda (Sf9, Invitrogen) cells. For this, PbSERA3 residues Ser425–Gln935 (called PbS3C1) were expressed with N-terminal His and S-tags using the pEt-30Xa/LIC system. PbS3C1 accumulated in insoluble inclusion bodies, which were washed, solubilized in 8 m urea, 20 mm Tris-HCl, pH 8.2, and purified by nickel-nitrilotriacetic acid chromatography, eluting with 8 m urea, 250 mm imidazole in 100 mm NaH2PO4, 10 mm Tris-HCl, pH 4. Yields of purified protein were ∼5 mg/liter of E. coli culture. The protein was buffer-exchanged into PBS and used to immunize mice. Prior to use, the resulting sera (anti-PbS3C1) were adsorbed against E. coli acetone powder to remove antibodies to bacterial contaminants.
Full-length PbSERA3 and a PbSERA3 C641A mutant (called rPbSERA3 and rPbSERA3C641A) were expressed as secreted proteins in insect cells, using the pbsera3synth gene. Substitution of the predicted catalytic Cys641 codon in pbsera3synth with an Ala codon was performed by site-directed mutagenesis (QuikChange II, Agilent Technologies) using oligonucleotide primer PbSERA3_C/A_fwd (supplemental Table S1) and its complementary reverse primer. The pbsera3synth and pbsera3synthC641A coding sequences were introduced into the pFastBacTM vector (Invitrogen), and recombinant bacmid was prepared by site-directed transposition according to the manufacturer's instructions for the Bac-to-Bac® Baculovirus Expression System (Invitrogen). Recombinant virus was generated by transfecting Sf9 cells with bacmid DNA. For recombinant protein production, High FiveTM cells (Invitrogen) grown in ESF 921 medium (Invitrogen) containing tunicamycin (Sigma; 0.5 μg ml−1) were infected with recombinant virus as described previously (26). Accumulation of secreted recombinant protein was monitored by Western blot using the mouse anti-PbS3C1 antibodies or polyclonal mouse antibodies against PbSERA3 (kindly provided by Volker Heussler, University of Bern, Switzerland). Clarified culture supernatants were adjusted to 50 mm Tris-HCl, pH 8.2, applied to a HiPrep Q XL 16/10 anion exchange column (GE Healthcare), and bound proteins were eluted with a gradient of 0–400 mm NaCl in 50 mm Tris-HCl, pH 8.2. The peak containing rPbSERA3 or rPbSERA3C641A (which showed identical elution profiles) was pooled, concentrated, and further purified by gel filtration chromatography on a Superdex 200 HR 10/30 column equilibrated in 25 mm HEPES, pH 7.4, 150 mm NaCl, 12 mm CaCl2. Yields of purified protein were very low (∼2–10 μg/L insect cell supernatant). Purified protein was stored in aliquots at −70 °C until required.
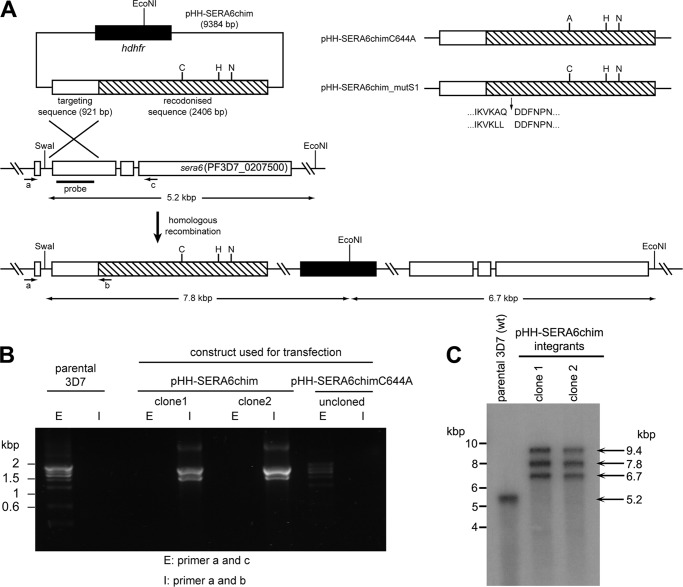
Construction of Homologous Integration Construct pHH-SERA6chim and Derivatives
Plasmid constructs for modification of the P. falciparum sera6 locus by single crossover targeted homologous recombination were based on plasmid pHH-SERA6synth, which was in turn derived from the backbone of pHH1-ΔSERA4 (a kind gift of Brendan Crabb, Burnet Institute, Melbourne, Australia). To generate pHH-SERA6synth, the promoter and 5′ coding region of the genomic sera6 gene (from 979 bp upstream of the translation start site to the middle of exon 2, 474 bp downstream of the translational start site) were amplified with primers SERA6_prom_fwd and SERA6_prom_rev (supplemental Table S1), digested with BglII and XhoI and ligated into similarly digested pHH1-ΔSERA4 in place of the SERA4 sequence. This placed it directly upstream of the P. berghei dihydrofolate reductase-thymidylate synthase (dhfrts) 3′ UTR, which regulates correct transcription termination and polyadenylation of the transcript, as described previously (6, 28). The remainder of the SERA6 coding region (2406 bp) was then amplified from the recodonized sera6synth sequence using primers SERA6synth_fwd and SERA6synth_rev (supplemental Table S1), digested with XhoI at both ends, and ligated in-frame with the authentic sera6 sequence to generate a vector (pHH-SERA6synth) that encoded the complete sera6 sequence. The promoter and first 624 bp of the sera6synth sequence were then excised from pHH-SERA6synth using restriction enzymes BamHI and AleI, and replaced with a 921-bp region of genomic sera6 DNA sequence amplified with primers SERA6gen_homology_fwd and SERA6gen_homology_rev (supplemental Table S1). This extends from intron 1 to the region of genomic sequence corresponding to the AleI site in the sera6synth sequence. The resulting construct (pHH-SERA6chim) therefore contained a region of sera6 genomic DNA in-frame with the sera6synth sequence, but lacked a promoter and the first sera6 exon. Homologous recombination by single crossover integration of pHH-SERA6chim at the sera6 locus would therefore be predicted to generate a full-length sera6 chimera, and a downstream truncated gene with no promoter. Mutant derivative pHH-SERA6chimC644A, in which the putative catalytic Cys codon Cys644 was replaced with an Ala codon, as well as derivative pHH-SERA6chim_mutS1, in which the codons encoding residues Ala369 and Gln370 were converted to Leu codons, were produced by QuikChange II site-directed mutagenesis, using primers SERA6_C/A_fwd and SERA6st1_mut_fwd, respectively, plus their complementary reverse primers (supplemental Table S1). All final constructs were fully sequenced on both strands.
Parasite Cultures and Transfections
In vitro cultures of the asexual blood stages of P. falciparum clone 3D7 were maintained and synchronized according to standard procedures (6, 29) in RPMI 1640 medium containing the serum substitute Albumax (Invitrogen). Ring stage parasites at 5–10% parasitaemia were transfected by electroporation with 70 μg of plasmid DNA and selection for lines carrying the plasmid was performed by culture in medium containing 10 nm WR99210 (Jacobus Pharmaceuticals, NJ). Selection for integration of transfected DNA was promoted by cycles of culture in the absence and presence of drug, as described previously (28). Where integration was detected by diagnostic PCR, clones were obtained by limiting dilution. Mature P. falciparum schizonts were enriched from highly synchronous cultures using Percoll (GE Healthcare) as described previously (28).
SDS-PAGE, Western Blot, and Saponin and Streptolysin O (SLO) Treatment
Percoll-enriched schizonts were pelleted in aliquots of 4 × 108 cells, then resuspended into either 10 volumes of PBS containing 4 hemolytic units of activated SLO (Sigma), or 10 volumes of 0.15% (w/v) saponin (Sigma) in PBS. Following incubation for 6 min at room temperature, parasite-containing and soluble fractions were separated by centrifugation at 12,400 × g for 10 min at 4 °C. Soluble and parasite fractions were solubilized into SDS sample buffer and subjected to SDS-PAGE under reducing conditions followed by transfer to Hybond-C Extra nitrocellulose membranes (GE Healthcare). Membranes were probed as described previously (30). Most antibodies used were produced in this study, as described above. Other antibodies were the SERA5-specific monoclonal antibody 24C6.1F1 (15), a kind gift of Jean-Francois Dubremetz (University of Montpellier 2, France), the MSP1-specific human mAb X509 (31), and polyclonal antibodies against aldolase of Arabidopsis thaliana (Agrisera, Sweden), which show cross-reactivity with P. falciparum aldolase.
Indirect Immunofluorescence Analysis (IFA)
Thin blood films were prepared from synchronous P. falciparum cultures enriched in mature schizonts. Slides were air-dried, fixed with 4% (w/v) paraformaldehyde for 30 min, permeabilized with 0.1% Triton X-100 for 10 min, and blocked overnight in 3% (w/v) bovine serum albumin (BSA) in PBS. Slides were probed with polyclonal anti-S6C1 to detect SERA6, and mAb 24C6.1F1 or mAb 89.1 to detect SERA5 or MSP1, respectively, as described previously (28). Primary antibodies were detected using Alexa Fluor 488 or Alexa Fluor 594-conjugated anti-mouse, anti-rabbit, or anti-human secondary antibodies (Invitrogen, Merck Chemicals Ltd.), then slides were stained with 4,6-diamidino-2-phenylindole (DAPI), mounted in Citifluor (Citifluor Ltd., Canterbury, United Kingdom), and images were captured using AxioVision 3.1 software on an Axioplan 2 Imaging system (Zeiss) using a Plan-APOCHROMAT 1006/1.4 oil immersion objective.
Peptide Substrates and rPfSUB1 Digestion Assays
N-Acetylated synthetic peptide PbSERA3st2 (Ac-DVSGQSENHQ-OH) was obtained from BIOMATIK (USA), as was peptide Ac-CVTAQSDEDC-OH. The latter peptide was labeled on both cysteine side chains with 6-iodoacetamido tetramethylrhodamine, and purified as described previously (32) to produce fluorogenic peptide PbSERA3st1F-6R. For digestion with rPfSUB1, peptides were diluted from stock solutions in dimethyl sulfoxide into digestion buffer supplemented with rPfSUB1 (∼1 unit/ml) and incubated at 21 °C. To monitor cleavage of PbSERA3st1F-6R, fluorescence increase of a 0.1 μm solution was continuously monitored with time as described previously (32), using a Cary Eclipse fluorescence spectrophotometer (Varian, UK) fitted with a 96-well microplate reader accessory. To confirm the specificity of peptide substrate cleavage, samples of partially digested peptides were fractionated by RP-HPLC on a Vydac 4.6 mm × 25-cm C18 column, eluting at 1 ml/min with a 0–45% (v/v) gradient of acetonitrile in 0.1% (v/v) TFA over 25 min, using either fluorescence (excitation at 552 nm, emission at 580 nm), or absorbance at 220 nm to detect the products of cleavage. Digestion products were identified by electrospray mass spectrometry as described previously (6, 32). For examining cleavage of native SERA6, predominantly full-length parasite-derived SERA6 was purified from hypotonic extracts of P. falciparum schizonts as described previously (6). Parasite protein or recombinant proteins were buffer-exchanged into 25 mm HEPES, 12 mm CaCl2, pH 7.4, and supplemented with ∼1 unit/ml of purified rPfSUB1 (10, 26) plus in some experiments purified recombinant PfSUB1 prodomain (PD; 85 nm final concentration), a selective nanomolar inhibitor of PfSUB1 (30), then incubated at 37 °C and sampled at intervals. Samples were examined by Western blot using the anti-SERA6 or anti-PbSERA3 antibodies as described above.
Mapping of PfSUB1 Cleavage Sites in SERA6 by In-gel Digestion
Recombinant full-length P. falciparum SERA6 proteins rSERA6wt and processing site mutants rSERA6mutS1, rSERA6mutS2, and rSERA6mutS1S2, accumulated as insoluble inclusion bodies when expressed in E. coli BL21-Gold DE3 cells. Isolated inclusion bodies were washed twice in PBS, 0.5% (v/v) Triton X-100 followed by sequential washes in 2, 4, and 6 m urea in 20 mm Tris-HCl, pH 8.2, 50 mm DTT. Washed inclusion bodies were then solubilized directly into SDS sample buffer containing 0.1 m DTT, and subjected to SDS-PAGE on 10 or 15% gels. The major product, corresponding to full-length protein, was identified by staining with Coomassie Blue, then excised from the gel to separate it from breakdown products (yields of the purified recombinant proteins were ∼5 mg/liter of E. coli culture). Excised protein bands were washed four times for 30 min each in 20 mm Tris-HCl, pH 8.2, 2.5% (v/v) Triton X-100 followed by a 20-min wash in 25 mm HEPES, pH 7.4, 150 mm NaCl, 12 mm CaCl2, then suspended in a minimal volume of the same buffer and subjected to in-gel digestion with rPfSUB1 (or mock digestion). Gel pieces were then heated in SDS sample buffer to solubilize the protein fragments, and again subjected to SDS-PAGE, followed by transfer to PVDF membranes and staining with Coomassie Blue. N-terminal sequencing was performed by the Protein and Nucleic Acid facility at the Department of Biochemistry, University of Cambridge, UK. Phenylhydantoin yields in the first five cycles of Edman degradation ranged from 1 to ∼4 pmol.
Southern Blot and PCR Analysis of Transgenic P. falciparum Lines
Parasite genomic DNA was isolated from cultured parasites using the DNeasy blood and tissue kit (Qiagen), according to the manufacturer's protocol. Diagnostic PCR to screen for integration of transfection constructs was performed using forward primer A (5′-AAAAGTAAAAGACCAAATGATA-3′), designed to hybridize to the 5′ UTR of the sera6 gene, and reverse primer B (5′-AAGTAGGAGTCGGACTTAGAA-3′) designed to anneal to the recodonized sera6synth sequence used in the pHH-SERA6chim construct. The unmodified endogenous sera6 gene was detected using the same forward primer combined with a different reverse primer (primer C; 5′-GATTCACTAGGTAATGTTAGACCTC-3′) designed to anneal to the endogenous sera6 coding sequence but not to the recodonized sera6synth sequence. PCR was conducted with an initial denaturation step at 94 °C for 2 min followed by 35 cycles of denaturation at 94 °C, annealing at 55 °C and extension at 72 °C, and concluded with a final extension of 72 °C for 6 min. For Southern blot, genomic DNA from wild type parental clone 3D7 parasites or cloned transgenic parasites was digested with restriction enzymes SwaI and EcoNI, fractionated by agarose gel electrophoresis on 0.7% agarose gels, transferred to Hybond-N nylon membranes (GE Healthcare), then probed with a 440-bp 32P-labeled PCR product amplified using primers SERA6int_probe_fwd and SERA6int_probe_rev (supplemental Table S1) from the 3′-proximal coding region of the genomic 3D7 sera6 gene.
Immunoelectron Microscopy
Mature P. falciparum schizonts (∼3 × 108) were fixed on ice by suspending in 0.075% (v/v) glutaraldehyde, 4% (w/v) paraformaldehyde in PBS, pH 7.2, for 15 min, then washed in ice-cold PBS, dehydrated at 4 °C through a graded ethanol series to 75% (v/v), infiltrated with LR White resin (Agar Scientific, UK), and polymerized by incubating at 55 °C for 24 h. Sections were mounted onto pioloform-treated 300 mesh nickel grids (thin/thick bars). Sections were then blocked for 45 min at room temperature with 1% (w/v) BSA, 0.01% (v/v) Tween 20 in PBS (BSA/T/PBS) followed by incubation with anti-S6C1 antiserum diluted 1:150 in BSA/T/PBS for 2 h at room temperature. Sections were then washed 5 times for 5 min in BSA/T/PBS followed by a 90-min incubation at room temperature with goat anti-rabbit IgG antibody (BB International, UK) conjugated to 10-nm gold particles, diluted 1:60 in BSA/T/PBS. Control samples were similarly probed with the corresponding preimmune serum. Sections were finally counterstained for 2 min with 2% (w/v) aqueous uranyl acetate and images were captured digitally with a Jeol 1200 EX transmission electron microscope.
RESULTS
SERA6 Is a PV Protein That May Be Partially Membrane Associated
The PVM forms an interface between the intravacuolar parasite and the host erythrocyte cytosol. A feature of soluble P. falciparum PV proteins is that they are retained within the PV following treatment of erythrocytic schizonts with SLO, which selectively permeabilizes the host erythrocyte membrane without affecting the integrity of the PVM (33, 34). In contrast, soluble PV proteins are efficiently released from schizonts by the nonionic glycoside detergent saponin, which at low concentrations permeabilizes both the erythrocyte membrane and the PVM without disrupting the parasite plasma membrane (35). Whereas P. falciparum SERA5 is a well established abundant PV protein (e.g. 36), SERA6 is much less well characterized, and no previous study has examined its subcellular localization using the above approach.
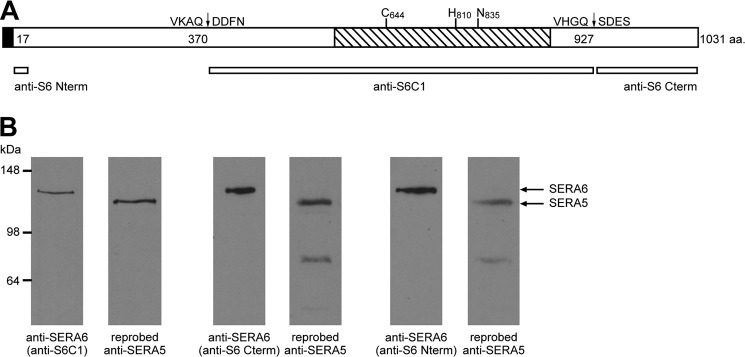
To aid in characterizing SERA6 localization and processing, we first raised antibodies specific for recombinant SERA6 polypeptides representing two of the proteolytic fragments predicted to be generated by PfSUB1 processing (6). In addition, a further polyclonal antibody and a mAb were raised against a previously described recombinant protein corresponding to a short region very close to the N terminus of SERA6 (Fig. 1A). All four antibodies recognized a single ∼135 kDa species in Western blot analysis of extracts of P. falciparum mid-stage (∼40 h) schizonts (Fig. 1B), likely corresponding to full-length SERA6. In support of this, reprobing the same blots with the SERA5-specific mAb 24C6.1F1 detected full-length SERA5 as a distinct, dominant ∼120-kDa band as well as traces of the well described 73- and 50-kDa SERA5 processing products (6, 37). This result is in agreement with previous indications (12, 13) that full-length SERA6 migrates more slowly on SDS-PAGE than full-length SERA5, and suggested that the new anti-SERA6 antibodies exhibit no significant cross-reactivity with other members of the blood-stage expressed SERA family. The new antibodies were therefore considered suitable reagents for examination of the localization and fate of SERA6.
FIGURE 1.
A, schematic of SERA6 primary structure and recombinant SERA6 proteins used for antibody production. The papain-like domain (residues Ser606–Lys880 in both the FCBR and 3D7 SERA6 sequence, PlasmodDB ID PF3D7_0207500 (25)) is shown hatched, with relative positions of predicted catalytic triad residues (Cys644, His810, and Asn835) indicated. Predicted positions of PfSUB1 cleavage called site 1 (367VKAQ↓DDFN374) and site 2 (924VHGQ↓SDES931 in the FCBR sequence as shown, or 924VHGQ↓SNES931 in the 3D7 sequence) are also indicated, based on alignments with known cleavage sites in SERA5 (6). The predicted secretory signal peptide is indicated by a black box. Antibodies were raised against recombinant proteins (narrow bars) corresponding to residues Gly35–Lys66 (anti-S6 N-term), Asp371–Gln927 (anti-S6C1), and the C-terminal residues Ser928–Val1031 (anti-S6 C-term). B, anti-SERA6 antibodies recognize a single major species in Western blot analysis of schizonts extracts, and do not cross-react with SERA5, the most abundant SERA family member. Extracts (∼5 μg total protein loaded per lane) were probed with the indicated anti-SERA6 antibodies, then membranes were stripped and re-probed with the anti-SERA5 mAb 24C6.1F1 to confirm that the proteins recognized are of different masses. Signals corresponding to full-length proteins are indicated with arrows. Faint bands at lower molecular masses are processing products of SERA5.
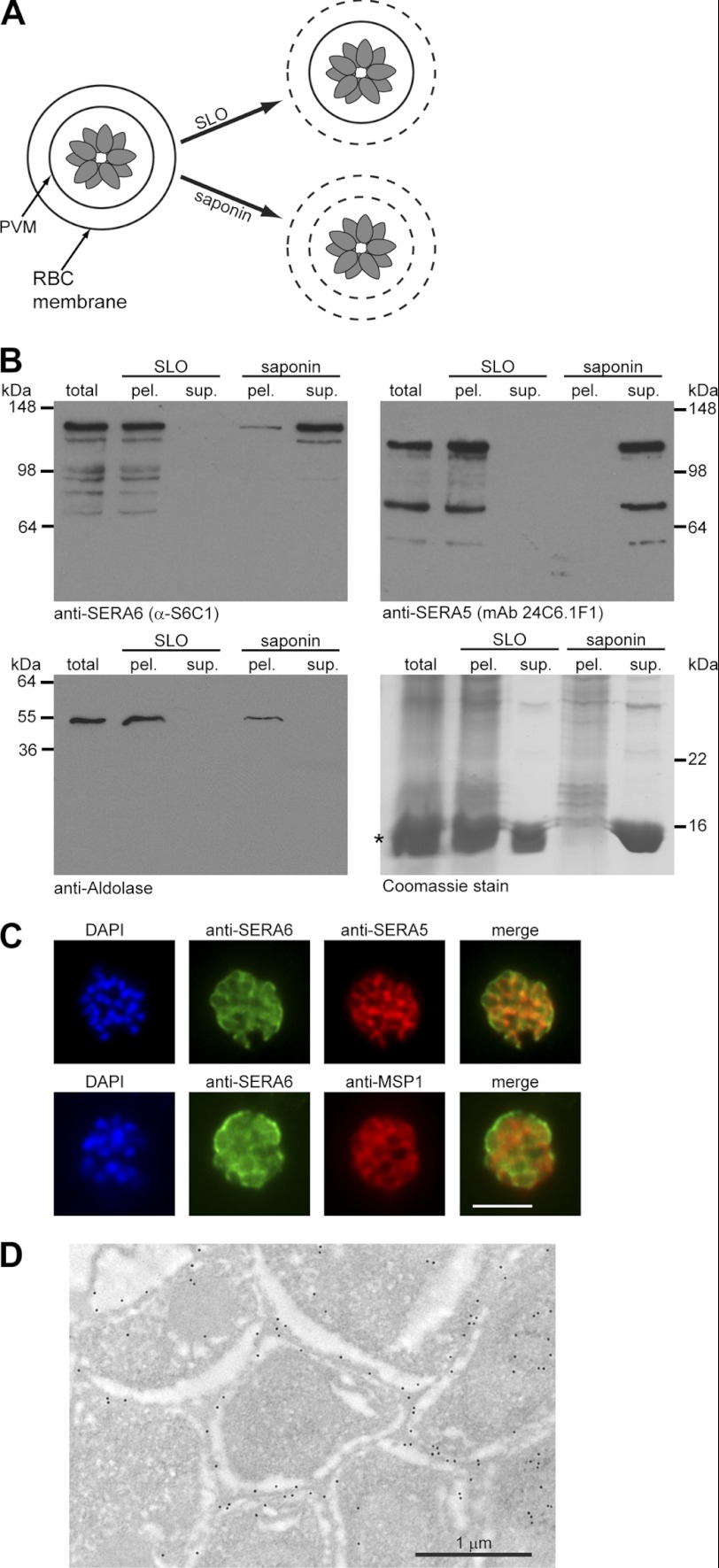
Asexual blood stages of the 3D7 clone of P. falciparum have an in vitro life cycle of ∼45–46 h. Preparations of highly synchronous mature schizonts (44–46 h old) were treated with SLO to selectively permeabilize the host erythrocyte membrane, or alternatively treated with saponin to rupture both the erythrocyte membrane and the PVM (Fig. 2A). Released soluble PV and erythrocyte proteins were separated from membrane-associated components by centrifugation and the various fractions were examined by Western blot. As shown in Fig. 2B, the solubility profile of SERA6 was virtually identical to that of SERA5; whereas neither protein was significantly released from schizonts by SLO treatment, saponin effectively released both proteins into the soluble fraction. Interestingly, saponin-mediated release of SERA6 was reproducibly (in n = 6 independent experiments) slightly less efficient than that of SERA5. Indirect IFA of fixed and permeabilized schizonts using the SERA6-specific antibodies revealed a signal with the “wagon-wheel” pattern characteristic of PV proteins. This IFA signal largely co-localized with that of SERA5 (Fig. 2C, upper panel). However, in a substantial proportion of parasites (71% of 106 parasites examined in detail) the SERA6 antibodies were observed to stain the outer confines of the intracellular parasite more intensely than the anti-SERA5 antibodies. This well delineated peripheral signal was clearly external to the parasite plasma membrane, as defined using antibodies to the major merozoite surface protein MSP1 (Fig. 2C, lower panel), and so it likely corresponds to the PVM. To obtain higher resolution localization of SERA6, the anti-S6C1 antibodies were used in immunoelectron microscopic (immuno-EM) analysis of fixed segmented schizonts. This confirmed that SERA6 predominantly localizes to the space surrounding individual segmented merozoites (Fig. 2D), and again provided evidence for some association with the PVM (supplemental Fig. S1). Collectively, these results suggest that SERA6 accumulates as a predominantly soluble protein in the same subcellular compartment as SERA5, the PV, but has a somewhat different distribution in this compartment, perhaps due to partial association with the PVM.
FIGURE 2.
SERA6 is a soluble PV protein that may partially interact with the PVM. A, schematic indicating protocol for selective permeabilization of schizonts. SLO permeabilizes just the erythrocyte plasma membrane, allowing release only of host cell cytosolic components, whereas saponin disrupts both the erythrocyte membrane and PVM. B, schizonts were treated with either SLO or saponin, supernatant (sup.) or parasite-containing (pel.) fractions were separated by centrifugation, and the fractions (∼10 μg of protein loaded per lane) were analyzed by Western blot in comparison with a total schizont extract (total) produced by extracting intact schizonts directly into SDS. SERA6 is present in the pellet fraction after SLO treatment, but predominantly in the supernatant after saponin treatment, suggesting that SERA6 is a soluble PV protein. SERA5, an established PV protein, served as a positive control. Aldolase, a cytosolic parasite protein, served as control for parasite lysis. Coomassie Blue staining of the various fractions (bottom right) was used to assess the efficiency of fractionation. The band marked with the asterisk indicates the position of hemoglobin, a soluble component of the erythrocyte cytosol. C, IFA demonstrates co-localization of the anti-SERA6 and anti-SERA5 signal in mature schizonts, except that the SERA6 signal additionally shows an association with the outer confines of the intracellular parasite, probably corresponding to the PVM. Scale bar, 5 μm. D, immunoelectron microscopic localization of SERA6 in a P. falciparum schizont, using the anti-S6C1 antibodies labeled with 10 nm immunogold. The majority of the signal is clearly associated with the peripheral space between intracellular merozoites, consistent with a PV localization. Magnification, ×15,000.
SERA6 Is an Endogenous Substrate for PfSUB1
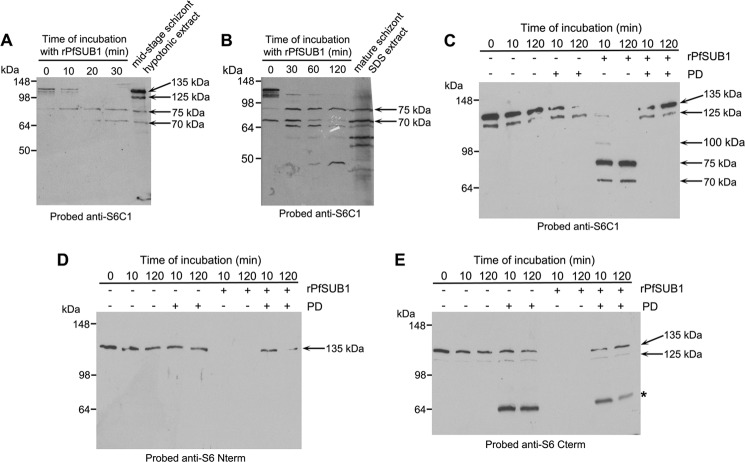
Cleavage of SERA5 by PfSUB1 occurs at two internal motifs referred to as site 1 and site 2, which flank the central papain-like domain, as well as at a third more N-terminal site that is present in only some allelic forms of SERA5 (6, 38). Sites 1 and 2 share substantial conservation across most members of the SERA family (6, 9, 10). We have previously shown (6) that synthetic decapeptides based on the putative P. falciparum SERA6 sites 1 and 2 (see Fig. 1A) are cleaved as predicted by recombinant PfSUB1 (rPfSUB1), supporting the notion that SERA6 may be an authentic PfSUB1 substrate. However, whether SERA6 in the parasite is processed at these or other positions has not been experimentally demonstrated. In the fractionation experiments shown in Fig. 2B, it was noticed that SERA6 in extracts of mature schizonts (some of which are often close to the point of egress due to imperfect synchrony of the cultures) was often detected in a partially fragmented form, suggestive of proteolytic processing. To examine this in more detail, parasite SERA6 was released from predominantly mid-stage (∼40 h) schizonts, using hypotonic lysis rather than saponin to avoid potential confounding effects of the presence of detergent (which can activate or inhibit proteases). The full-length form of the protein was then enriched from these extracts using a combination of gel filtration and ion exchange chromatography as previously described (6). Incubation in vitro of this partially purified, predominantly full-length SERA6 with rPfSUB1 resulted in its rapid conversion to a set of up to 5 major anti-S6C1-reactive fragments that migrated on SDS-PAGE with a profile that was similar to that of the original hypotonic extract before purification (Fig. 3A), or of SERA6 released by SDS extraction of highly mature segmented, ∼46 h schizonts (Fig. 3B). Importantly, the partially purified full-length parasite SERA6 was stable in vitro in the absence of added rPfSUB1, and the cleavage observed upon addition of rPfSUB1 was completely prevented by prior addition of recombinant PfSUB1 PD, a potent (∼5 nm) selective inhibitor of PfSUB1 (30), showing that all the processing observed required PfSUB1 activity (Fig. 3C). Further analysis (Fig. 3, C–E) showed that whereas the full-length protein, as well as minor 125- and 100-kDa forms and the dominant 75- and 70-kDa digestion products were all recognized by the anti-S6C1 antibodies, only the full-length protein was recognized by the anti-S6 N-term antibodies, whereas only the full-length and 125-kDa forms were recognized by the anti-S6 C-term antibodies. It is conceivable that some of the polypeptides detected in the mature schizont extracts could represent cross-reaction of the anti-S6C1 antibodies with other SERA isoforms, so it could not be definitely concluded that all were derived from SERA6. Collectively, however, these observations suggested that SERA6 processing observed in very mature schizonts is likely mediated by the endogenous parasite PfSUB1. Furthermore, the limited number of SERA6 fragments detected in late P. falciparum schizont extracts suggested that PfSUB1 cleaves SERA6 at just a few discrete sites. Importantly, the observation that extracts of highly mature segmented schizonts (i.e. following budding of mature merozoites from the mature schizont) contained virtually no full-length SERA6 (Fig. 3B, right-hand lane) suggests that endogenous processing of SERA6 may go to completion very late in schizont maturation. This is consistent with it being triggered by discharge of PfSUB1 into the PV just prior to egress, as in the case of SERA5 processing. However, in contrast to the situation with SERA5, processing products of which are easily detected in parasite culture supernatants following egress, analysis of culture supernatants with the anti-SERA6 antibodies failed to reproducibly detect any released SERA6 processing products (data not shown). This may be due to the relatively low abundance of SERA6 and/or instability of the SERA6 processing products following schizont rupture.
FIGURE 3.
SERA6 is an endogenous PfSUB1 substrate. A, Western blot analysis of processing in vitro of parasite-derived SERA6 by rPfSUB1. Time course of processing, showing conversion of predominantly full-length SERA6 to fragments of 75 and 70 kDa that co-migrate with similar species present in a hypotonic extract of mid-stage schizonts. B, similar to A, but showing a comparison of the in vitro processing products with SERA6-derived species present in an SDS extract of mature (∼46 h) schizonts. Note the absence of full-length SERA6 in the mature schizont extracts. C and D, parasite-derived SERA6 is stable in vitro in the absence of added rPfSUB1, and all observed processing requires PfSUB1 activity. Western blots of in vitro incubations of parasite-derived SERA6 with (+) or without (−) rPfSUB1 or recombinant PfSUB1 PD, a potent and selective inhibitor of PfSUB1. Identical digests were probed with anti-S6C1 (C), anti-S6 N-term (D), or anti-S6 C-term antibodies (E). The estimated molecular mass of major antibody-reactive species is indicated throughout, and ∼5 μg of total protein was loaded per lane. The band marked with the asterisk in E indicates cross-reactivity of the anti-S6 C-term antibodies with the PD, perhaps due to the presence of a His6 sequence on both the PD and the recombinant antigen used to make the anti-S6 C-term antibodies (see “Experimental Procedures”).
Mapping of the PfSUB1 Processing Sites in SERA6
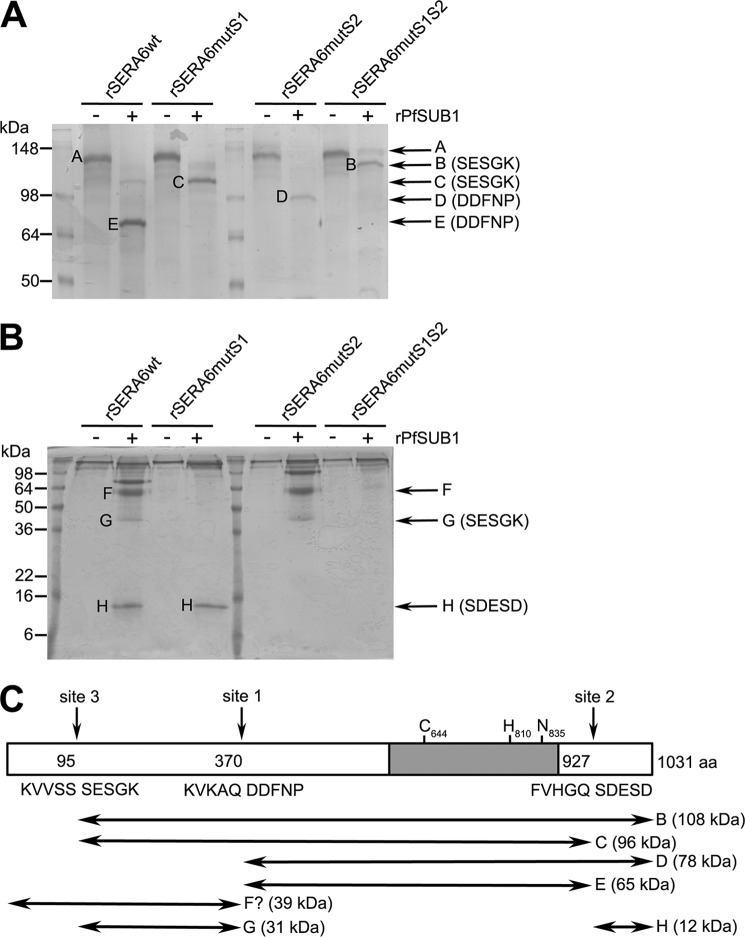
The low abundance of SERA6 in the parasite made it unfeasible to attempt N-terminal sequence analysis of parasite-derived protein (as previously achieved for SERA5) (6). Previous work has indicated that PfSUB1 efficiently and correctly cleaves short synthetic peptides based on cleavage sites in a number of known substrates (6–8, 26, 39), indicating that cleavage specificity is based primarily on the amino acid sequence flanking the site rather than higher-order structural features. Accordingly, to attempt to precisely map the sites of cleavage in P. falciparum SERA6 we expressed the full-length protein (minus its secretory signal peptide) in recombinant form in E. coli, then examined the consequences of digesting the purified protein (referred to as rSERA6wt) with rPfSUB1 in vitro. The rSERA6wt was expressed in inclusion bodies as an insoluble protein, rendering it difficult to manipulate, so we devised a protocol that involved prior denaturation and fractionation of the protein by SDS-PAGE, followed by “in-gel” digestion with rPfSUB1 of isolated full-length protein in excised gel slices. The digestion products were visualized by a second round of SDS-PAGE and Coomassie Blue staining (Fig. 4, A and B). In initial experiments, the ∼135 kDa full-length rSERA6wt (labeled fragment A) was rapidly converted by rPfSUB1 to four stable products of 75 (fragment E), 60 (fragment F), 40 (fragment G), and 12 kDa (fragment H) via short-lived intermediates of ∼125 and 100 kDa (not shown). Analysis by Edman degradation of fragments E and H revealed N-terminal sequences DDFNP and SDESD, respectively, consistent with cleavage of the precursor at the predicted sites 1 and 2 (Fig. 4C, also see Fig. 1A). N-terminal sequencing of fragment G produced the sequence SESGK, which corresponds to Ser96–Lys100 of the SERA6 sequence. This lies directly downstream of the sequence 91KVVSS95, indicating an additional cleavage by PfSUB1 at the motif 91KVVSS↓SESGK100 (the downward-pointing arrow indicates the position of cleavage) within the N-terminal region of SERA6. By analogy with a positionally similar, allele-specific third cleavage site in SERA5 (6, 38), this is referred to as site 3 (Fig. 4C). To test this interpretation and to further examine the derivation of the short-lived processing intermediates, three mutant forms of rSERA6wt were produced, possessing Leu substitutions of the P1 and P2 positions at either sites 1 or 2, or at both sites. Previous work (7, 26) has shown that PfSUB1 cannot cleave C-terminal to a Leu residue, and that it has a strict requirement for small residues at P2, so these di-leucine substitutions were predicted to block PfSUB1 processing. Digestion of rSERA6mutS1 with rPfSUB1 resulted again in the formation of fragment H as expected, plus, instead of fragment E, a stable 110-kDa species (fragment C), both via a short-lived intermediate processing product of ∼125 kDa. N-terminal sequencing of fragment C produced the sequence SESGK, identical to that of fragment G. Digestion of mutant rSERA6mutS2 resulted as expected in formation of a novel stable species (fragment D) intermediate in size between fragments C and E, but with the same N-terminal sequence as fragment E. No fragment H was observed in this digest, consistent with efficient blockade of cleavage at site 2. Digestion of the double mutant rSERA6mutS1S2 produced just a single detectable digestion product of 125 kDa (fragment B), with the N-terminal sequence SESGK, consistent with cleavage only at site 3. The short-lived 125- and 100-kDa processing intermediates observed in the processed rSERA6wt preparations presumably result from cleavage at site 3; they would therefore likely be identical to fragments B and C (Fig. 4C), although this could not be confirmed due to their low abundance. N-terminal sequencing of fragment A, the undigested full-length rSERA6wt, gave no signal, presumably due to blockade of the N-terminal residue. A similar result was obtained with fragment F, consistent with this possessing the same N terminus as the full-length protein.
FIGURE 4.
SERA6 is cleaved by PfSUB1 at three positions. A and B, Coomassie-stained SDS-PAGE gels showing the results of in vitro digestion of recombinant SERA6 with rPfSUB1. Digests (∼4 μg of purified protein loaded per lane) were fractionated on either 10 (A) or 15% (B) gels to obtain optimal resolution of precursor proteins and both large and small digestion products. Well resolved digestion products and their positions of migration (arrows) are labeled B-H, and the first 5 amino acid residues identified in Edman degradation analysis of each band are indicated on the right in parentheses. No sequence was obtained for bands A (precursor protein) and F. C, schematic indicating identified sites of cleavage in relationship to the full-length P. falciparum FCBR SERA6 sequence and the internal papain-like domain (shaded) containing the predicted catalytic triad residues Cys644, His810, and Asn835. The double-headed arrows indicate the likely identity of the products of digestion with rPfSUB1 of rSERA6wt, rSERA6mutS1, rSERA6mutS2, and rSERA6mutS1S2. Also indicated (in parentheses) is the predicted molecular mass of each species based on its amino acid composition. The provenance of digestion product F is uncertain (indicated by a question mark), but likely corresponds to that shown. Cleavage sites 1 and 2 are between Gln370–Asp371 and Gln927–Ser928, respectively, as previously predicted based on similarity with processing sites in SERA5 (6). Site 3 (Ser95–Ser96) has all the features of the previously determined PfSUB1 consensus motif (6, 8, 10), except for the presence of a Ser residue at P2 rather than the commonly observed Ala or Gly.
Collectively, these data suggest that SERA6 can be cleaved by PfSUB1 at just 3 discrete positions (Fig. 4C). Two of these (sites 1 and 2) are identical to those previously predicted on the basis of sequence alignments with SERA5 and experimental cleavage of short synthetic peptides (Ac-KVKAQ↓DDFNP and Ac-FVHGQ↓SNESD (6); sites of cleavage are indicated by downward-pointing arrows) based on sequences flanking these sites. The third (site 3) lies just 95 residues away from the N terminus of the SERA6 precursor. Based on a collective comparison of the rSERA6wt-derived and parasite-derived SERA6 digestion products and their reactivity with the various anti-SERA6 antibodies (Figs. 3 and 4), it can be tentatively proposed that the 125-kDa endogenous parasite SERA6 product (which is not recognized by anti-S6 N-term antibodies; Fig. 3D), likely corresponds to fragment B in Fig. 4C. The low abundance 100-kDa parasite intermediate product likely corresponds to fragment C; this would be expected to be recognized by the anti-S6 C-term antibodies, but the lack of a signal at 100 kDa in Fig. 3E may be explained by the relatively low titer of these antibodies (compare the overall intensity of the Western blot signals in Fig. 3, C and E). The major 75-kDa parasite-derived SERA6 product likely corresponds to fragment E, the central segment of SERA6 produced by cleavage at both sites 1 and 2. The possible provenance of the 70-kDa and smaller parasite-derived species is discussed below. These results are consistent with a model in which SERA6 is subjected to processing by PfSUB1 in the latter stages of schizont maturation, presumably following discharge of PfSUB1 into the PV. Importantly, as with SERA5, processing of SERA6 by PfSUB1 would be expected to release the papain-like central domain, which lies between sites 1 and 2.
The Predicted Active Site Cys of SERA6, and SERA6 Processing by PfSUB1, Is Required for Parasite Viability
Comprehensive earlier studies from Crabb and colleagues (12, 20) showed that, of the nine P. falciparum SERA genes, only those encoding SERA5 and SERA6 could not be truncated or deleted in asexual blood-stage parasites. This suggests an indispensable role for both genes in that part of the life cycle. However, the consequences of more subtle gene modifications designed to specifically modify any putative enzyme activity were not addressed. To do this, we adapted a previously used strategy (6, 39, 40) in which we introduced into the parasite circular DNA constructs designed to integrate by single crossover homologous recombination into the sera6 locus. Each construct contained a targeting sequence corresponding to 921 bp of the internal authentic sera6 sequence, fused in-frame to synthetic “recodonized” sequence comprising 2406 bp of the 3′-proximal SERA6 open reading frame. The synthetic sequence (called sera6synth) shares only ∼74% identity at the nucleotide level with the authentic sera6 gene, reducing the likelihood of recombination downstream of the junction between the authentic and synthetic sequence. Correct integration of the control parental construct, pHH-SERA6chim (Fig. 5A), was expected to produce a chimeric sera6 gene, still under the control of its endogenous promoter, and encoding the reconstituted authentic SERA6 primary sequence. Integration of pHH-SERA6chimC644A, in contrast, was predicted to replace the codon for the nucleophilic active site Cys644 of SERA6 with an Ala codon. This would be expected to ablate any enzymatic activity in the gene product. Aside from this substitution of 2 bp at the Cys644 codon, the two DNA constructs were otherwise identical. Both carried the human dhfr gene, which confers resistance to the antifolate drug WR99210.
FIGURE 5.
Modification of the P. falciparum sera6 locus by homologous recombination. A, schematic showing the major elements incorporated into transfection construct pHH-SERA6chim, and the expected mode of single crossover integration into the endogenous sera6 locus. Targeting sequence derived from the authentic sera6 gene was fused in-frame to the recodonized sequence (hatched) derived from the sera6synth gene, to select for recombination upstream of the junction between the two. Relative positions of the codons encoding the putative catalytic Cys, His, and Asn residues are shown, as is the single EcoNI restriction site within the hdhfr gene (black), which confers resistance to the antifolate drug WR99210. For clarity, the P. berghei dhfr 3′ UTR, which lies downstream of the sera6synth sequence in the plasmid is not shown. The endogenous sera6 locus, which contains 3 introns (27), is shown as is the expected result of homologous integration of the entire plasmid and the architecture of the modified locus relative to flanking SwaI and EcoNI sites. Note that episomal plasmids are harbored as concatamers in P. falciparum so integration of more than one copy often occurs. However, because the plasmid contains only a partial sequence not preceded by a promoter, the only copy to be transcribed is the modified chimeric gene directly downstream of the endogenous promoter. Construct pHH-SERA6chim was designed to simply reconstitute the SERA6 coding sequence (as a control), pHH-SERA6chimC644A was designed to replace the catalytic Cys with an Ala, whereas construct pHH-SERA6chim_mutS1 was designed to incorporate mutations into site 1 that render it refractory to cleavage by PfSUB1. B, diagnostic PCR detects homologous integration of construct pHH-SERA6chim but not of pHH-SERA6chimC644A. Primer pair a and c were designed to amplify a product only from the unmodified sera6 locus, whereas primers a and b amplify a product that can only arise from the predicted integration event. Shown is agarose gel electrophoresis of PCR amplicons obtained using as template genomic DNA from parental wild-type 3D7 parasites, two clones obtained by limiting dilution from a line transfected with pHH-SERA6chim (similar results were obtained from the noncloned line; not shown), or from a line transfected with pHH-SERA6chimC644A and maintained for 5 drug cycles. No integration of pHH-SERA6chimC644A was detected. Note that in each case, the major amplicons obtained by PCR were confirmed as the expected product by sequence analysis (not shown). The additional minor amplicons were not identified but are probably the result of mispriming, often observed when amplifying from the highly AT-rich P. falciparum genomic DNA. C, Southern blot of genomic DNA from the parental 3D7 clone and the two clones derived from parasites transfected with pHH-SERA6chim, showing that the plasmid had integrated as predicted into the sera6 locus. A region of the targeting sequence was used as probe (panel A). The endogenous sera6 locus is modified in the transfected clones and integration bands of the predicted size appear. The uppermost, ∼9.4-kbp signal observed in the integrants is the same size as the input plasmid and the same intensity as the other bands, suggesting that each clone had integrated 2 copies of the plasmid.
The pHH-SERA6chim and pHH-SERA6chimC644A plasmid constructs were separately transfected into 3D7 parasites. For each transfection, WR99210-resistant parasites harboring the plasmids were readily selected within 2–3 weeks of transfection. The cultures were then subjected to cycles of culture with and without WR99210 to select for integrant parasites (41). After 2–3 cycles of drug treatment, diagnostic PCR analysis indicated the presence of homologous recombination events in each of 7 parasite lines transfected with pHH-SERA6chim (not shown). Homologous integration in the expected manner was confirmed by PCR analysis of the uncloned pHH-SERA6chim lines (not shown), as well as PCR and Southern blot analysis of 2 clones derived by limiting dilution (Fig. 5, B and C). SERA6 expression from these clones was normal, as expected, and they exhibited no phenotype in comparison with the parental 3D7 clone (not shown). In contrast, no integration of the input plasmid was detected in any of the 7 lines transfected with pHH-SERA6chimC644A, either by diagnostic PCR (Fig. 5B) or by Southern blot (not shown), even after up to 6 cycles of drug treatment. Because constructs pHH-SERA6chim and pHH-SERA6chimC644A differ by just 2 bp within the recodonized sequence, our inability to detect parasites harboring integration of the latter is very unlikely to be due to technical issues or differences in the recombination frequency of the constructs. These results strongly suggest that replacement of the catalytic Cys644 of SERA6 with an Ala residue is deleterious or lethal for the parasite, consistent with the notion that SERA6 has catalytic activity that is important for parasite survival.
To explore the importance of proteolytic processing of SERA6 by PfSUB1, we took a similar gene targeting approach, exploiting the evidence described above (Fig. 4) that di-leucine substitutions of the P1 and P2 residues at the processing sites can efficiently block cleavage by PfSUB1. For this we produced the construct pHH-SERA6chim_mutS1 (Fig. 5A), which again was identical to the control construct pHH-SERA6chim except for the presence of a 5-bp substitution that converted the Ala369 and Gln370 codons adjacent to the site 1 processing site to Leu codons. Correct integration of this construct was predicted to produce a mutant sera6 gene product that would be refractory to PfSUB1-mediated cleavage at site 1. No integration of this construct could ever be detected following transfection, even following up to 6 cycles of drug treatment (data not shown). This result strongly suggests that PfSUB1-mediated processing of SERA6 is important for maintenance of the P. falciparum asexual blood-stage life cycle in vitro.
Processing by PfSUB1 of the P. berghei Orthologue of SERA6 Converts It to an Active Cysteine Protease
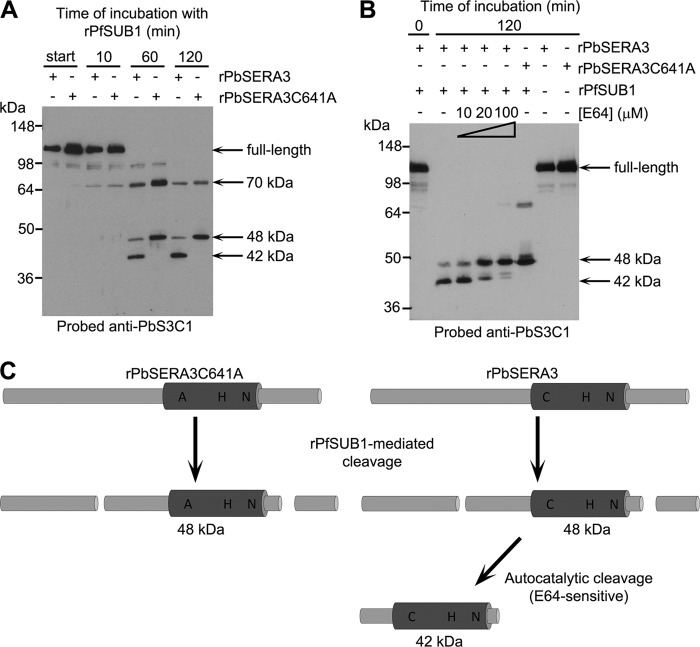
In the light of our genetic evidence that the putative active site Cys of SERA6 is functionally important, and that PfSUB1-mediated processing of SERA6 is critical for parasite viability, we hypothesized that processing by PfSUB1 might activate SERA6 to play a proteolytic role in the parasite life cycle. To seek direct evidence for this, we attempted to obtain recombinant SERA6 in a soluble, correctly folded form. Exhaustive attempts to refold the recombinant S6C1 and rSERA6wt bacterial products described above were unsuccessful, and attempts to express the recodonized sera6synth gene in P. pastoris or in baculovirus-infected insect cells resulted in no detectable protein expression (not shown). Because small modifications in amino acid sequence can significantly modify solubility and/or expression levels, we turned to the P. berghei orthologue of SERA6, PbSERA3. In this case, low levels of apparently full-length secreted soluble recombinant PbSERA3 (rPbSERA3) could be obtained upon expression in recombinant baculovirus of a synthetic pbsera3 gene (pbsera3synth), codon-optimized for expression in Sf9 insect cells. Initial experiments indicated that the presence of a C-terminal His6 tag sequence interfered with expression of this protein (data not shown), so all subsequent work used a construct lacking a tag. To provide a control for studying the potential protease activity of rPbSERA3, we produced a mutant form of the protein (called rPbSERA3C641A) in which the predicted catalytic Cys641 residue was substituted with Ala. Expression and purification of the two recombinant proteins under identical conditions resulted in levels of protein that were very similar, as ascertained by Western blot using antibodies specific for the papain-like central domain of PbSERA3 (supplemental Fig. S2). In initial experiments, the partially purified rPbSERA3 and rPbSERA3C641A were examined for protease activity in a range of assay systems, including gelatin gel zymograms and incubation with a panel of commercially obtainable fluorogenic peptide substrates. No protease activity was detected (not shown). We therefore decided to study the consequences of cleavage of the protein with PfSUB1. Previous modeling studies of several SUB1 orthologues (10) have indicated that the substrate specificity of SUB1 orthologues from P. falciparum and P. berghei are similar. In agreement with this, initial experiments (supplemental Fig. S3 and Table S2) showed that rPfSUB1 can efficiently cleave short synthetic peptides based on the two predicted PbSUB1 sites within PbSERA3. This suggested that rPfSUB1 could be used as a surrogate for PbSUB1 in in vitro digestion experiments. As shown in Fig. 6A, incubation of both rPbSERA3 and rPbSERA3C641A with rPfSUB1 resulted in their time-dependent conversion to products of 70 and 48 kDa reactive with the anti-PbS3C1 antibodies. Intriguingly, however, we noticed that whereas the PfSUB1-digested rPbSERA3C641A mutant accumulated predominantly in the 48-kDa form, digested rPbSERA3 underwent further cleavage to a 42-kDa form, despite the fact that it differs from rPbSERA3C641A only by the presence of the Cys641 residue, which lies some distance from either predicted processing site in the PbSERA3 sequence (supplemental Fig. S3A). To examine this phenomenon in more detail, the digestion experiments were repeated in the presence of E64, a peptidyl epoxide that selectively inhibits many cysteine proteases but does not affect PfSUB1 activity (26). These experiments (Fig. 6B) showed that, whereas cleavage of rPbSERA3 to the 48-kDa form was unaffected by E64 (as expected if this is directly mediated by PfSUB1), further conversion to the 42-kDa form was selectively sensitive to E64 in a dose-responsive manner. Importantly, both rPbSERA3 and rPbSERA3C641A were completely stable in vitro during purification (supplemental Fig. S2) or upon incubation in the absence of added PfSUB1 (Fig. 6B, two right-hand lanes), and cleavage of both required PfSUB1 activity because it was completely inhibited by addition of PD (not shown). These results indicate that cleavage of rPbSERA3 by PfSUB1 results in activation of an rPbSERA3-associated autoprocessing activity that mediates its further cleavage to the 42-kDa form (Fig. 6C). The autoprocessing activity of rPbSERA3 requires the presence of its putative active site Cys641 residue, and is sensitive to E64. Extensive efforts to identify the site(s) of cleavage involved in conversion to the 42-kDa form, or to demonstrate activity of the 42-kDa form in trans against a range of fluorogenic peptide substrates were unsuccessful, probably due to the very low yields of recombinant PbSERA3 obtained in the baculovirus expression system. Nonetheless, these results provide the first evidence that a SERA6 orthologue possesses protease activity and, significantly, suggest that this activity is enhanced or activated by SUB1-mediated cleavage.
FIGURE 6.
Cleavage of PbSERA3 by PfSUB1 activates an autoprocessing activity. A, Western blot showing the results of digesting rPbSERA3 and rPbSERA3C641A with rPfSUB1. Both substrates are converted in a time-dependent manner via a 70-kDa intermediate to a 48-kDa product, but only rPbSERA3 undergoes further conversion to a 42-kDa species. Identical results were obtained with three different independently purified batches of rPbSERA3 and rPbSERA3C641A (not shown). B, Western blot showing the effects of E64 on the conversion of rPbSERA3 to the 42-kDa form. E64 was present in the digestion reactions at concentrations of 10, 20, or 100 μm. Incubation of rPbSERA3 or rPbSERA3C641A under identical conditions without added rPfSUB1 resulted in no conversion. All products detected likely include the papain-like catalytic domain of PbSERA3 because this is the region against which the anti-PbS3C1 antibodies were raised. Approximately 0.5 μg of purified protein was loaded per lane. C, schematic depicting a model for digestion of rPbSERA3 and rPbSERA3C641A with rPfSUB1. The papain-like domain of each protein is indicated (dark gray cylinder), with approximate positions of the catalytic triad residues (or the Cys-to-Ala substitution in rPbSERA3C641A) shown. Cleavage by rPfSUB1 is depicted as taking place at two positions equivalent to the predicted sites 1 and 2 in PbSERA3 (supplemental Fig. S3), but this was not confirmed. Similarly, the precise positions at which further autocatalytic conversion of the rPbSERA3 48-kDa form to its 42-kDa product could not be identified.
DISCUSSION
Despite compelling evidence that egress of the blood-stage malaria merozoite requires protease activity and is sensitive to selective inhibitors of cysteine proteases, little is known about the effector enzymes involved. We have previously shown that P. falciparum egress is associated with PfSUB1-mediated processing of several merozoite surface and PV proteins, all triggered by the regulated discharge of PfSUB1 into the PV. In the present study, by demonstrating: 1) that SERA6 is a PV-resident PfSUB1 substrate; 2) that SERA6 plays an essential catalytic role in the parasite; 3) that processing of SERA6 by PfSUB1 is important for parasite viability; and 4) that a SERA6 orthologue possesses protease activity that is activated by PfSUB1 cleavage, we provide the first evidence linking these two phenomena and raise the possibility that SERA6 may have a critical proteolytic function at egress.
Only three previous studies (12, 13, 27) have sought to determine the subcellular location of P. falciparum SERA6. These used either light microscopy (IFA), which suffers from the limitations imposed by the very small dimensions of the parasite, or immuno-EM. Here, by combining both approaches with selective permeabilization of the parasitized cell we have confirmed that SERA6 is predominantly a soluble PV protein, but also provided suggestive evidence that it is partly associated with the PVM. It is worth noting that our permeabilization data are consistent with previous results from Nyalwidhe and Lingelbach (36) who used SLO treatment of schizonts followed by treatment with sulfo-NHS-LC-biotin to selectively biotinylate PV proteins (the PVM is thought to contain nonselective pores that allow access of the biotinylation reagent to the PV lumen). SERA6 was among the set of parasite proteins labeled in that study, in accord with our localization data. Similarly, IFA studies on PbSERA3 in liver stage schizonts of P. berghei has shown that PbSERA3 leaks into the cytosol of the infected hepatocyte upon rupture of the PVM, again consistent with a PV localization for that orthologue (11).
Using full-length, native parasite SERA6 isolated from mid-stage schizonts, we next showed that the protein could be digested in vitro with rPfSUB1 to produce a fragmentation pattern that shared clear similarities with the endogenous pattern of SERA6 processing detected in mature schizonts. This implicates SERA6 as an authentic PfSUB1 substrate in vivo. Importantly, extracts of highly mature schizonts contained little or no full-length SERA6, consistent with processing taking place at around the time of PfSUB1 discharge into the PV just prior to egress. Mapping of the cleavage sites in SERA6 using N-terminal sequencing of recombinant digestion products confirmed the identity of two cleavage sites (sites 1 and 2) previously predicted on the basis of similarity with those in SERA5 (6), and identified a third site close to the N terminus of the precursor. This site (site 3) had not been predicted based on previous data, primarily because all previously identified PfSUB1 cleavage sites possess either Ala or Gly at the P2 position (8, 10); SERA6 site 3 is therefore unusual in that it comprises the first example of an authentic PfSUB1 cleavage site possessing a P2 Ser residue, an observation that suggests that other small residues may also be acceptable at P2. It is worth noting that a third cleavage site is also present in SERA5, within an allele-specific polymorphic region close to its N terminus (6, 38). The site 3 cleavage sites might be functionally analogous.
The low abundance of SERA6 in the parasite precluded structural analysis of native protein by mass spectrometry or N-terminal sequencing, so we were unable to unambiguously assign the identities of the various native cleavage products detectable in schizont extracts. However, on the basis of the reactivity of the native processing fragments with our antibody reagents, together with our analysis of in vitro rPfSUB1-digested native and recombinant SERA6, we propose that the 125-kDa native SERA6 processing fragment, which was recognized by the anti-S6C1 and anti-S6 C-term, but not by the anti-S6 N-term antibodies, represents the full-length protein following removal of the extreme N terminus by cleavage at site 3. In contrast, the 75- and 70-kDa processing products were recognized by the anti-S6C1 antibodies but by neither the anti-S6 N-term nor the anti-S6 C-term antibodies. Previous studies on SERA5 have shown that, following cleavage by PfSUB1 at sites 1 and 2, the resulting P56 fragment is subject to further “trimming” at its C terminus by a distinct leupeptin-sensitive cysteine protease (“protease X”) (19), converting it to a P50 terminal form. If SERA6 undergoes processing in a similar manner, the 75-kDa species likely results from cleavage at both sites 1 and 2, whereas the 70-kDa species may represent a C terminally truncated form of the 75-kDa fragment. The fact that the 70-kDa form appeared in in vitro PfSUB1 digests of native SERA6 (Fig. 3, A–C) may be due to co-purification of protease X with the parasite-derived SERA6. The smaller SERA6 processing fragments observed in mature schizont extracts (Fig. 3B, right-hand lane) may be due to autocatalytic processing activity of the proteolytically active SERA6 (see below).
Previous work from Crabb and colleagues (12) found that the P. falciparum sera6 gene was refractory to disruption by homologous recombination, suggesting an essential function in asexual blood-stage growth. Attempts to disrupt the orthologous P. berghei pbsera3 gene have also been unsuccessful (42). To address the possibility that SERA6 may have essential catalytic activity, we adopted a more targeted approach specifically designed to assess the importance of the predicted nucleophilic active-site Cys644 residue. Using single crossover recombination, we were readily able to modify the genomic sera6 locus to generate transgenic parasites expressing wild-type protein from a chimeric sera6 gene under the control of its endogenous promoter. However, introduction into the parasite of a virtually identical targeting construct designed to substitute the Cys644 codon with an Ala codon consistently failed to produce integrant parasites, strongly suggesting that integration of that construct produced nonviable or poorly replicating parasites (which would be rapidly overgrown in culture). Similar negative results with the pHH-SERA6chim_mutS1 construct, designed to introduce a di-leucine substitution known to render site 1 refractory to PfSUB1 cleavage, suggested that processing at that position is important for parasite viability. In the absence of robust conditional gene knock-out or allelic replacement technologies suitable for use in Plasmodium blood stages, these results provide the best available evidence that SERA6 performs a critical catalytic role, and that processing by PfSUB1 is required for that or another essential function.
To directly address whether SERA6 possesses protease activity, we attempted to produce correctly folded recombinant protein using a range of expression systems. This failed, so we turned to PbSERA3, expressing it successfully, albeit only at very low levels, in a full-length secreted form in insect cells. The rPbSERA3 was soluble and apparently stable during expression and purification, as was a rPbSERA3C641A mutant. We reasoned that one approach to exploring the potential protease activity of rPbSERA3 would be to examine the consequences of it being cleaved in a manner similar to that predicted to occur upon exposure to PbSUB1, the P. berghei orthologue of PfSUB1. Previous homology modeling work (10) has shown that the substrate specificity of PfSUB1 and PbSUB1 are similar (though not identical), and this was confirmed by the demonstration here that PfSUB1 correctly cleaves two synthetic peptide substrates based on sequence flanking the predicted sites 1 and 2 in PbSERA3. We therefore digested rPbSERA3 with rPfSUB1, anticipating that this should mimic cleavage of PbSERA3 by PbSUB1. Although both proteins were efficiently digested to a 48-kDa form, rPbSER3A, but not rPbSERA3C641A, underwent almost quantitative further conversion to a smaller 42-kDa product. The conversion displayed by rPbSERA3 is highly unlikely to be directly mediated by PfSUB1 or a contaminating protease activity for the following four reasons. First, it was absent from the rPbSERA3C641A mutant protein, despite the fact that both proteins were expressed and purified under identical conditions. Second, it was ablated by a single point mutation of the predicted catalytic Cys641, which lies some distance away from any predicted PfSUB1 cleavage site in the primary sequence of PbSERA3. Third, the activity was sensitive to the cysteine protease inhibitor E64, to which PfSUB1 is completely insensitive (as indicated by the lack of any effect of E64 on digestion to the 48-kDa form). Finally, the conversion activity required processing by PfSUB1, as the unprocessed rPbSER3A displayed complete stability in the absence of PfSUB1.
We conclude from these results that processing by PfSUB1 activates the protease activity of PbSERA3. By analogy, we propose that PfSUB1 processing likely similarly activates P. falciparum SERA6. It may be significant that processing of PbSERA3 has previously been reported to take place in liver stage schizonts, where expression of the protein is up-regulated shortly before egress of liver stage merozoites (11). Many proteases, including clan CA papain-like proteases, are activated by proteolysis, either through the action of a distinct protease (43), or autocatalytically either in cis by an intramolecular (unimolecular) cleavage event or in trans through autoproteolysis (22, 44–46). Whether autoprocessing is a requirement for SERA6/PbSERA3 function is unknown, but certainly it may be responsible for some of the smaller SERA6 fragments detected in mature schizont extracts that are unlikely to be direct products of PfSUB1 cleavage. Unfortunately, the low yields of rPbSERA3 produced in our insect cell expression system precluded further analysis of the activity or substrate specificity of the “activated” PbSERA3. Future work will focus on increasing expression yields or exploring alternative expression systems to obtain useful amounts of recombinant protease.
Perhaps the most convincing experimental evidence for a role of cysteine proteases in asexual blood stage egress in P. falciparum has derived from the effects of E64, which are evident at concentrations as low as 10 μm. Could the potent inhibitory effect of E64 on egress be a consequence of its inhibition of SERA6 activity? The relative insensitivity to E64 of the autocatalytic activity associated with PfSUB1-cleaved PbSERA3 suggests to us that this is unlikely (although it is possible that SERA6 may be more sensitive to E64 than its PbSERA3 orthologue). Previous work from Greenbaum and colleagues (47) has suggested that host erythrocyte calpain-1 may play a role in rupture of the host cell membrane at egress. Because this is the bounding membrane in which merozoites appear to remain trapped following blockade of egress by E64, we consider it more likely that calpain-1 is the primary target of E64. SUB1-mediated processing of SERA6/PbSERA3 at egress may enable it to perform a distinct essential proteolytic function in the asexual blood-stage life cycle. This could include a direct role in PVM rupture.
Supplementary Material
Acknowledgments
We are grateful to Steve Howell (National Institute of Medical Research) for mass spectrometry analysis and Mike Weldon (Protein and Nucleic Acid facility) for N-terminal sequencing.
This work was supported in part by United Kingdom Medical Research Council Grant U117532063.

This article contains supplemental Figs. S1–S3 and Tables S1 and S2.
- PV
- parasitophorous vacuole
- PVM
- parasitophorous vacuole membrane
- SLO
- streptolysin O
- IFA
- immunofluorescence analysis
- PD
- prodomain.
REFERENCES
- 1. Salmon B. L., Oksman A., Goldberg D. E. (2001) Malaria parasite exit from the host erythrocyte. A two-step process requiring extraerythrocytic proteolysis. Proc. Natl. Acad. Sci. U.S.A. 98, 271–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wickham M. E., Culvenor J. G., Cowman A. F. (2003) Selective inhibition of a two-step egress of malaria parasites from the host erythrocyte. J. Biol. Chem. 278, 37658–37663 [DOI] [PubMed] [Google Scholar]
- 3. Soni S., Dhawan S., Rosen K. M., Chafel M., Chishti A. H., Hanspal M. (2005) Characterization of events preceding the release of malaria parasite from the host red blood cell. Blood Cells Mol. Dis. 35, 201–211 [DOI] [PubMed] [Google Scholar]
- 4. Glushakova S., Mazar J., Hohmann-Marriott M. F., Hama E., Zimmerberg J. (2009) Irreversible effect of cysteine protease inhibitors on the release of malaria parasites from infected erythrocytes. Cell Microbiol. 11, 95–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Boyle M. J., Wilson D. W., Richards J. S., Riglar D. T., Tetteh K. K., Conway D. J., Ralph S. A., Baum J., Beeson J. G. (2010) Isolation of viable Plasmodium falciparum merozoites to define erythrocyte invasion events and advance vaccine and drug development. Proc. Natl. Acad. Sci. U.S.A. 107, 14378–14383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Yeoh S., O'Donnell R. A., Koussis K., Dluzewski A. R., Ansell K. H., Osborne S. A., Hackett F., Withers-Martinez C., Mitchell G. H., Bannister L. H., Bryans J. S., Kettleborough C. A., Blackman M. J. (2007) Subcellular discharge of a serine protease mediates release of invasive malaria parasites from host erythrocytes. Cell 131, 1072–1083 [DOI] [PubMed] [Google Scholar]
- 7. Koussis K., Withers-Martinez C., Yeoh S., Child M., Hackett F., Knuepfer E., Juliano L., Woehlbier U., Bujard H., Blackman M. J. (2009) A multifunctional serine protease primes the malaria parasite for red blood cell invasion. EMBO J. 28, 725–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Silmon de Monerri N. C., Flynn H. R., Campos M. G., Hackett F., Koussis K., Withers-Martinez C., Skehel J. M., Blackman M. J. (2011) Global identification of multiple substrates for Plasmodium falciparum SUB1, an essential malarial processing protease. Infect. Immun. 79, 1086–1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Arisue N., Kawai S., Hirai M., Palacpac N. M., Jia M., Kaneko A., Tanabe K., Horii T. (2011) Clues to evolution of the SERA multigene family in 18 Plasmodium species. PLoS One 6, e17775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Withers-Martinez C., Suarez C., Fulle S., Kher S., Penzo M., Ebejer J. P., Koussis K., Hackett F., Jirgensons A., Finn P., Blackman M. J. (2012) Plasmodium subtilisin-like protease 1 (SUB1). Insights into the active-site structure, specificity and function of a pan-malaria drug target. Int. J. Parasitol. 42, 597–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Schmidt-Christensen A., Sturm A., Horstmann S., Heussler V. T. (2008) Expression and processing of Plasmodium berghei SERA3 during liver stages. Cell Microbiol. 10, 1723–1734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Miller S. K., Good R. T., Drew D. R., Delorenzi M., Sanders P. R., Hodder A. N., Speed T. P., Cowman A. F., de Koning-Ward T. F., Crabb B. S. (2002) A subset of Plasmodium falciparum SERA genes are expressed and appear to play an important role in the erythrocytic cycle. J. Biol. Chem. 277, 47524–47532 [DOI] [PubMed] [Google Scholar]
- 13. Aoki S., Li J., Itagaki S., Okech B. A., Egwang T. G., Matsuoka H., Palacpac N. M., Mitamura T., Horii T. (2002) Serine repeat antigen (SERA5) is predominantly expressed among the SERA multigene family of Plasmodium falciparum, and the acquired antibody titers correlate with serum inhibition of the parasite growth. J. Biol. Chem. 277, 47533–47540 [DOI] [PubMed] [Google Scholar]
- 14. Aly A. S., Matuschewski K. (2005) A malarial cysteine protease is necessary for Plasmodium sporozoite egress from oocysts. J. Exp. Med. 202, 225–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Delplace P., Dubremetz J. F., Fortier B., Vernes A. (1985) A 50-kDa exoantigen specific to the merozoite release-reinvasion stage of Plasmodium falciparum. Mol. Biochem. Parasitol. 17, 239–251 [DOI] [PubMed] [Google Scholar]
- 16. Delplace P., Bhatia A., Cagnard M., Camus D., Colombet G., Debrabant A., Dubremetz J. F., Dubreuil N., Prensier G., Fortier B. (1988) Protein p126. A parasitophorous vacuole antigen associated with the release of Plasmodium falciparum merozoites. Biol. Cell 64, 215–221 [DOI] [PubMed] [Google Scholar]
- 17. Delplace P., Fortier B., Tronchin G., Dubremetz J. F., Vernes A. (1987) Localization, biosynthesis, processing and isolation of a major 126-kDa antigen of the parasitophorous vacuole of Plasmodium falciparum. Mol. Biochem. Parasitol. 23, 193–201 [DOI] [PubMed] [Google Scholar]
- 18. Arastu-Kapur S., Ponder E. L., Fonović U. P., Yeoh S., Yuan F., Fonović M., Grainger M., Phillips C. I., Powers J. C., Bogyo M. (2008) Identification of proteases that regulate erythrocyte rupture by the malaria parasite Plasmodium falciparum. Nat. Chem. Biol. 4, 203–213 [DOI] [PubMed] [Google Scholar]
- 19. Blackman M. J. (2008) Malarial proteases and host cell egress: an “emerging” cascade. Cell. Microbiol. 10, 1925–1934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. McCoubrie J. E., Miller S. K., Sargeant T., Good R. T., Hodder A. N., Speed T. P., de Koning-Ward T. F., Crabb B. S. (2007) Evidence for a common role for the serine-type Plasmodium falciparum serine repeat antigen proteases. Implications for vaccine and drug design. Infect. Immun. 75, 5565–5574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Clark P. I., Lowe G. (1978) Conversion of the active-site cysteine residue of papain into a dehydroserine, a serine and a glycine residue. Eur. J. Biochem. 84, 293–299 [DOI] [PubMed] [Google Scholar]
- 22. Doran J. D., Nomizu M., Takebe S., Ménard R., Griffith D., Ziomek E. (1999) Autocatalytic processing of the streptococcal cysteine protease zymogen. Processing mechanism and characterization of the autoproteolytic cleavage sites. Eur. J. Biochem. 263, 145–151 [DOI] [PubMed] [Google Scholar]
- 23. Coulombe R., Grochulski P., Sivaraman J., Ménard R., Mort J. S., Cygler M. (1996) Structure of human procathepsin L reveals the molecular basis of inhibition by the prosegment. EMBO J. 15, 5492–5503 [PMC free article] [PubMed] [Google Scholar]
- 24. Hodder A. N., Drew D. R., Epa V. C., Delorenzi M., Bourgon R., Miller S. K., Moritz R. L., Frecklington D. F., Simpson R. J., Speed T. P., Pike R. N., Crabb B. S. (2003) Enzymic, phylogenetic, and structural characterization of the unusual papain-like protease domain of Plasmodium falciparum SERA5. J. Biol. Chem. 278, 48169–48177 [DOI] [PubMed] [Google Scholar]
- 25. Hodder A. N., Malby R. L., Clarke O. B., Fairlie W. D., Colman P. M., Crabb B. S., Smith B. J. (2009) Structural insights into the protease-like antigen Plasmodium falciparum SERA5 and its noncanonical active-site serine. J. Mol. Biol. 392, 154–165 [DOI] [PubMed] [Google Scholar]
- 26. Withers-Martinez C., Saldanha J. W., Ely B., Hackett F., O'Connor T., Blackman M. J. (2002) Expression of recombinant Plasmodium falciparum subtilisin-like protease-1 in insect cells. Characterization, comparison with the parasite protease, and homology modeling. J. Biol. Chem. 277, 29698–29709 [DOI] [PubMed] [Google Scholar]
- 27. Knapp B., Nau U., Hundt E., Küpper H. A. (1991) A new blood stage antigen of Plasmodium falciparum highly homologous to the serine-stretch protein SERP. Mol. Biochem. Parasitol. 44, 1–13 [DOI] [PubMed] [Google Scholar]
- 28. Harris P. K., Yeoh S., Dluzewski A. R., O'Donnell R. A., Withers-Martinez C., Hackett F., Bannister L. H., Mitchell G. H., Blackman M. J. (2005) Molecular identification of a malaria merozoite surface sheddase. PLoS Pathog. 1, 241–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Blackman M. J. (1994) Purification of Plasmodium falciparum merozoites for analysis of the processing of merozoite surface protein-1. Methods Cell Biol. 45, 213–220 [DOI] [PubMed] [Google Scholar]
- 30. Jean L., Hackett F., Martin S. R., Blackman M. J. (2003) Functional characterization of the propeptide of Plasmodium falciparum subtilisin-like protease-1. J. Biol. Chem. 278, 28572–28579 [DOI] [PubMed] [Google Scholar]
- 31. Blackman M. J., Whittle H., Holder A. A. (1991) Processing of the Plasmodium falciparum major merozoite surface protein-1. Identification of a 33-kilodalton secondary processing product which is shed prior to erythrocyte invasion. Mol. Biochem. Parasitol. 49, 35–44 [DOI] [PubMed] [Google Scholar]
- 32. Blackman M. J., Corrie J. E., Croney J. C., Kelly G., Eccleston J. F., Jameson D. M. (2002) Structural and biochemical characterization of a fluorogenic rhodamine-labeled malarial protease substrate. Biochemistry 41, 12244–12252 [DOI] [PubMed] [Google Scholar]
- 33. Ansorge I., Paprotka K., Bhakdi S., Lingelbach K. (1997) Permeabilization of the erythrocyte membrane with streptolysin O allows access to the vacuolar membrane of Plasmodium falciparum and a molecular analysis of membrane topology. Mol. Biochem. Parasitol. 84, 259–261 [DOI] [PubMed] [Google Scholar]
- 34. Ansorge I., Benting J., Bhakdi S., Lingelbach K. (1996) Protein sorting in Plasmodium falciparum-infected red blood cells permeabilized with the pore-forming protein streptolysin O. Biochem. J. 315, 307–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Burghaus P. A., Lingelbach K. (2001) Luciferase, when fused to an N-terminal signal peptide, is secreted from transfected Plasmodium falciparum and transported to the cytosol of infected erythrocytes. J. Biol. Chem. 276, 26838–26845 [DOI] [PubMed] [Google Scholar]
- 36. Nyalwidhe J., Lingelbach K. (2006) Proteases and chaperones are the most abundant proteins in the parasitophorous vacuole of Plasmodium falciparum-infected erythrocytes. Proteomics 6, 1563–1573 [DOI] [PubMed] [Google Scholar]
- 37. Debrabant A., Maes P., Delplace P., Dubremetz J. F., Tartar A., Camus D. (1992) Intramolecular mapping of Plasmodium falciparum P126 proteolytic fragments by N-terminal amino acid sequencing. Mol. Biochem. Parasitol. 53, 89–95 [DOI] [PubMed] [Google Scholar]
- 38. Li J., Mitamura T., Fox B. A., Bzik D. J., Horii T. (2002) Differential localization of processed fragments of Plasmodium falciparum serine repeat antigen and further processing of its N-terminal 47-kDa fragment. Parasitol. Int. 51, 343–352 [DOI] [PubMed] [Google Scholar]
- 39. Child M. A., Epp C., Bujard H., Blackman M. J. (2010) Regulated maturation of malaria merozoite surface protein-1 is essential for parasite growth. Mol. Microbiol. 78, 187–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Olivieri A., Collins C. R., Hackett F., Withers-Martinez C., Marshall J., Flynn H. R., Skehel J. M., Blackman M. J. (2011) Juxtamembrane shedding of Plasmodium falciparum AMA1 is sequence independent and essential, and helps evade invasion-inhibitory antibodies. PLoS Pathog. 7, e1002448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Crabb B. S., Rug M., Gilberger T. W., Thompson J. K., Triglia T., Maier A. G., Cowman A. F. (2004)Transfection of the human malaria parasite Plasmodium falciparum. Methods Mol. Biol. 270, 263–276 [DOI] [PubMed] [Google Scholar]
- 42. Putrianti E. D., Schmidt-Christensen A., Arnold I., Heussler V. T., Matuschewski K., Silvie O. (2010) The Plasmodium serine-type SERA proteases display distinct expression patterns and nonessential in vivo roles during life cycle progression of the malaria parasite. Cell. Microbiol. 12, 725–739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sajid M., McKerrow J. H., Hansell E., Mathieu M. A., Lucas K. D., Hsieh I., Greenbaum D., Bogyo M., Salter J. P., Lim K. C., Franklin C., Kim J. H., Caffrey C. R. (2003) Functional expression and characterization of Schistosoma mansoni cathepsin B and its trans-activation by an endogenous asparaginyl endopeptidase. Mol. Biochem. Parasitol. 131, 65–75 [DOI] [PubMed] [Google Scholar]
- 44. Pungercar J. R., Caglic D., Sajid M., Dolinar M., Vasiljeva O., Pozgan U., Turk D., Bogyo M., Turk V., Turk B. (2009) Autocatalytic processing of procathepsin B is triggered by proenzyme activity. FEBS J. 276, 660–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Quraishi O., Storer A. C. (2001) Identification of internal autoproteolytic cleavage sites within the prosegments of recombinant procathepsin B and procathepsin S. Contribution of a plausible unimolecular autoproteolytic event for the processing of zymogens belonging to the papain family. J. Biol. Chem. 276, 8118–8124 [DOI] [PubMed] [Google Scholar]
- 46. Ménard R., Carmona E., Takebe S., Dufour E., Plouffe C., Mason P., Mort J. S. (1998) Autocatalytic processing of recombinant human procathepsin L. Contribution of both intermolecular and unimolecular events in the processing of procathepsin L in vitro. J. Biol. Chem. 273, 4478–4484 [DOI] [PubMed] [Google Scholar]
- 47. Chandramohanadas R., Davis P. H., Beiting D. P., Harbut M. B., Darling C., Velmourougane G., Lee M. Y., Greer P. A., Roos D. S., Greenbaum D. C. (2009) Apicomplexan parasites co-opt host calpains to facilitate their escape from infected cells. Science 324, 794–797 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.