Background: Autophagy is an essential physiological process involving dynamic membrane rearrangements.
Results: Pik1 and Mss4 are PtdIns 4- and PtdIns4P 5-kinases that are needed for selective and/or nonselective autophagy.
Conclusion: PtdIns4P is important in the mechanism of autophagy and affects Atg9 trafficking.
Significance: We show that PtdIns4P, similar to PtdIns3P, regulates autophagy.
Keywords: Autophagy, Membrane Trafficking, Protein Targeting, Stress, Yeast, Vacuole
Abstract
Macroautophagy (hereafter autophagy) is a degradative cellular pathway that protects eukaryotic cells from stress, starvation, and microbial infection. This process must be tightly controlled because too little or too much autophagy can be deleterious to cellular physiology. The phosphatidylinositol (PtdIns) 3-kinase Vps34 is a lipid kinase that regulates autophagy, but the role of other PtdIns kinases has not been examined. Here we demonstrate a role for PtdIns 4-kinases and PtdIns4P 5-kinases in selective and nonselective types of autophagy in yeast. The PtdIns 4-kinase Pik1 is involved in Atg9 trafficking through the Golgi and is involved in both nonselective and selective types of autophagy, whereas the PtdIns4P 5-kinase Mss4 is specifically involved in mitophagy but not nonselective autophagy. Our data indicate that phosphoinositide kinases have multiple roles in the regulation of autophagic pathways.
Introduction
Selective and nonselective autophagy play essential physiological roles in all eukaryotic cells by controlling the rapid degradation of long-lived proteins, unnecessary or damaged organelles, and/or bulk cytoplasm in response to various types of stress in their intracellular and extracellular environment (1). As our knowledge of the core machinery of autophagy expands (2), we find that there is much more to explore regarding autophagy regulation. Phosphoinositides are important regulators in cellular membrane dynamics (3). In yeast, cellular phosphatidylinositol can be phosphorylated by the PtdIns3 3-kinase Vps34 to generate phosphatidylinositol 3-phosphate (PtdIns3P), and further phosphorylation by the PtdIns3P 5-kinase Fab1 converts PtdIns3P into PtdIns(3,5)P2. The other route for PtdIns metabolism is phosphorylation by PtdIns 4-kinases to generate PtdIns4P, and further phosphorylation by Mss4 generates PtdIns(4,5)P2 (4). The PtdIns3P signaling pathway is important for autophagy. Yeast Vps34 forms an autophagy-specific complex containing Atg14, Vps30/Atg6, and Vps15 and is important for the organization of the phagophore assembly site (PAS) (5). The corresponding complex that generates PtdIns3P in mammals is also critical for autophagy. Furthermore, PtdIns(3,5)P2 plays a role in autophagy in mammalian cells (6). However, the potential roles of the second PtdIns metabolic route involving PtdIns 4-kinases in the autophagic pathway remain largely unexplored.
The Saccharomyces cerevisiae genome contains three PtdIns 4-kinases: the Golgi- and nucleus-localized Pik1, the plasma membrane-localized Stt4, and the nonessential Lsb6 (7–9). Pik1 and Stt4 are responsible for the synthesis of the majority of cellular PtdIns4P. Whereas the function of the nuclear population of Pik1 remains unclear, the Golgi-localized Pik1 is essential for secretory vesicle exit from the trans-Golgi network. Plasma membrane-localized Stt4 is involved in the Slt2 MAP kinase cascade (7); the PtdIns4P generated by Stt4 is converted into PtdIns(4,5)P2 by Mss4, which acts as an upstream regulator of this signaling pathway.
A previous study reported that PtdIns4P is required for peroxisome degradation by selective autophagy in the yeast Pichia pastoris (10). In this regard, PtdIns4P is generated by PpPik1 and is thought to participate in the formation of a pexophagy-specific membrane structure, termed the micropexophagic apparatus. The key autophagy-related (Atg) protein that binds this phosphoinositide in P. pastoris is PpAtg26; however, the homologous protein in Saccharomyces cerevisiae does not have a role in autophagy (11). These findings raise several important questions. For example, is PtdIns4P involved in other types of autophagy aside from pexophagy? P. pastoris, which is a methylotrophic yeast, has a Golgi complex that, in contrast to that of S. cerevisiae, is morphologically organized in a manner similar to the Golgi complex of mammalian cells. Accordingly, it is not clear whether PtdIns 4-kinases would play an equivalent role in budding yeast. In this report we analyzed the role of PtdIns 4-kinases in both selective and non-selective autophagic pathways and examined their mechanistic roles. Our data suggest that PtdIns 4-kinases have a conserved function in autophagy regulation. In yeast, defects in PtdIns 4-kinase activity affect trafficking of Atg9 and cause blocks both in nonselective autophagy and mitophagy.
EXPERIMENTAL PROCEDURES
Media and Growth Conditions
Yeast cells were grown in YPD (rich medium; 1% yeast extract, 2% peptone, 2% glucose), SMD (synthetic minimal medium; 0.67% yeast nitrogen base without amino acids, 2% glucose, supplemented with vitamins and auxotrophic amino acids as needed), YPL (1% yeast extract, 2% peptone, and 2% lactate), or SML (0.67% yeast nitrogen base, 2% lactate, and auxotrophic amino acids and vitamins as needed). Starvation experiments were performed in SD-N (synthetic medium lacking nitrogen; 0.17% yeast nitrogen base without ammonium sulfate or amino acids and 2% glucose). For temperature-sensitive mutants, cells were grown at 24 °C to midlog phase and shifted to 37 °C for 30 min to inactivate the mutants. If not otherwise indicated, cells were grown at 30 °C.
Strains and Plasmids
Plasmids used in this study are listed in supplemental S1. pCu416 and pCuGFP-Aut7(416) have been reported previously (12, 13). To construct pCu3HA-Pik1(416) and pCuGFP-Pik1(416), 3HA or green fluorescent protein (GFP) was first cloned into the SpeI/XmaI sites of pCu416 to generate pCu3HA(416) or pCuGFP(416), and then the open reading frame (ORF) of PIK1 was amplified from genomic DNA and cloned into the XmaI/SalI sites of pCu3HA(416) or pCuGFP(416). To make pCu3HA-Pik1(405), the Cu3HA-Pik1 coding sequence from pCu3HA-Pik1(416) was amplified and cloned into the SacII/BamHI sites of pRS405. pCuGFP-pik1–11(416) and pCuGFP-pik1–104(416) were generated by amplifying pik1–11, or pik1–104 from genomic DNA of the corresponding strains and then cloning into the XmaI/SalI sites of pCuGFP(416). To construct pCuRFP-Atg8(405), RFP was first cloned into the SpeI/XmaI sites of pCu416 to generate pCuRFP(416) followed by cloning of ATG8 into the XmaI/SalI sites of pCuRFP(416), and then the CuRFP-Atg8 coding sequence from pCuRFP-Atg8(416) was amplified and cloned into the SacII/BamHI sites of pRS405.
Yeast strains used in this study are listed in supplemental S2. Gene disruption and C-terminal tagging were carried out using a PCR-based method as described previously (14, 15). For integration of Cu3HA-Pik1, pCu3HA-Pik1(405) was linearized with BglII and integrated into the PIK1 locus. To integrate Atg9–3GFP, the pAtg9–3GFP(306) plasmid was linearized with BglII and integrated into the ATG9 genomic locus (16). RFP-Ape1 was incorporated into the chromosome by integrating AvrII-digested pRFP-Ape1(305) into the APE1 locus (17). To integrate GFP-Atg8 and RFP-Atg8, pGFP-Atg8(405) and pRFP-Atg8(405) were linearized with AflII and integrated into the LEU2 locus. The integration of GFP-Osh2-PH has been described previously (18).
To generate the pik1–11, and pik1–104 strains, the endogenous copy of PIK1 was replaced with mutant alleles by homologous recombination. First, the 333-base pair genomic region downstream of the PIK1 ORF including its terminator was cloned and ligated into the SacI/SacII sites of pFA6a-KAN or pFA6a-TRP1, which are downstream of the KAN or TRP1 genes. Next, the pik1–11 and pik1–104 sequences plus the CYC1 terminator were amplified by PCR from pCuGFP-pik1–11(416) and pCuGFP-pik1–104(416), respectively, and inserted into the XmaI/BglII sites of pFA6a-KAN or pFA6a-TRP1, which are upstream of the KAN or TRP1 genes, to complete the pFA6a-pik1–11-KAN, pFA6a-pik1–104-KAN, pFA6a-pik1–11-TRP1, and pFA6a-pik1–104-TRP1 plasmids. Then these plasmids were digested with XmaI/SacII, and the resulting fragments were transformed into yeast cells to replace the endogenous copy of PIK1. Incorporation of the mutations was confirmed by DNA sequencing.
Fluorescence Microscopy
Cells were cultured in YPD or SMD selective media to mid-log phase and then shifted to SD-N medium as indicated in the corresponding figure legends. Cells were pelleted by centrifugation and resuspended in fresh SMD to reflect growing conditions or in SD-N for starvation conditions. Cells were visualized with a microscope (DeltaVision Spectris; Applied Precision, Issaquah, WA) fitted with differential interference contrast optics and an Olympus camera IX-HLSH100. For each microscopy picture, 10–12 Z-section images were captured. The distance between two neighboring sections was 0.2–0.5 μm, and the total depth of each stack was 5.5 μm, which is the approximate diameter of a normal yeast cell. The images were deconvolved using softWoRx software (Applied Precision).
Autophagy Assays
To monitor bulk autophagy, the Pho8Δ60 assay and the GFP-Atg8 processing assay were performed as previously described (19, 20).
Mitophagy Assays
Om45-GFP processing and mitoPho8Δ60 assays were performed as described previously (21).
RESULTS
Golgi-localized Pik1 Is Required for Autophagy
To determine the role of the PtdIns kinases in autophagy, we first examined the autophagic phenotype of a pik1 mutant using a well established procedure, the GFP-Atg8 processing assay. The ubiquitin-like protein Atg8 is conjugated to phosphatidylethanolamine and remains associated with the completed autophagosome. When transported into the vacuole during autophagy, GFP-tagged Atg8 is hydrolyzed to yield free GFP. Thus, the relatively stable GFP moiety reflects the level of autophagy (20).
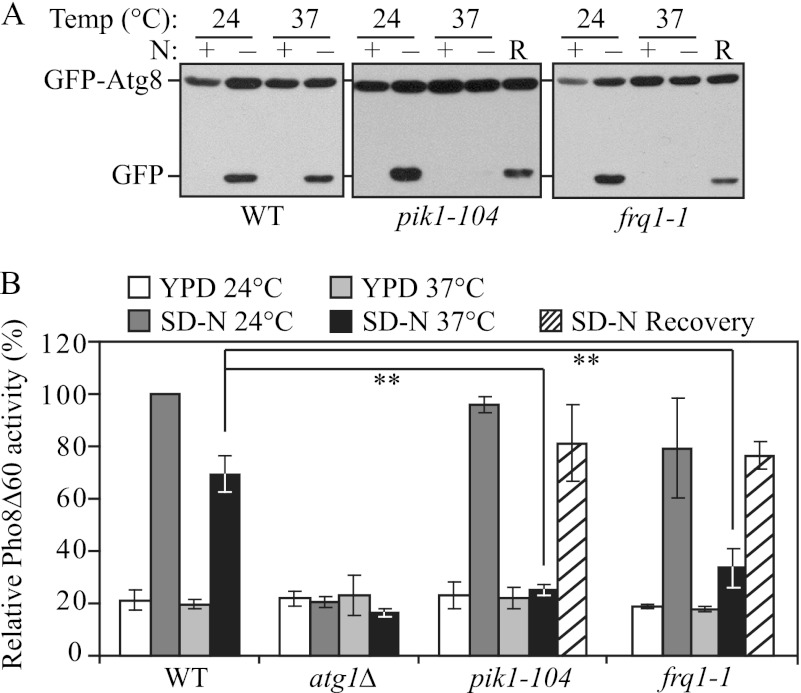
Using this method, we observed a strong autophagic defect in the temperature sensitive pik1–104 mutant (Fig. 1A). In nutrient-rich conditions at both the permissive temperature (PT) and non-permissive temperature (NPT), there was essentially no detectable free GFP as expected. After a 2-h nitrogen starvation at 24 °C there was a clear appearance of the free GFP band in wild-type and pik1 mutant cells. However, at 37 °C, significantly reduced free GFP was detected in the pik1–104 strain compared with the wild-type cells (Fig. 1A). To rule out the possibility that the observed autophagic defect might be due to cell death caused by inactivation of Pik1, which is an essential protein, we shifted the cells back to the PT for another 2-h starvation (recovery period (R)); we observed a clear induction of GFP-Atg8 processing during the recovery period, demonstrating that autophagy induction was restored and that the cells were still viable (Fig. 1A). The autophagy defect was rescued by a plasmid expressing the PIK1 gene chromosomally integrated into the pik1 mutant, suggesting the autophagic defect was indeed due to the functional impairment of Pik1 (data not shown).
FIGURE 1.
Pik1 is required for autophagy. A, GFP-Atg8 processing is blocked in the pik1–104 and frq1-1 mutants. Wild type (WT; BY4742) and pik1–104 (BY4742) and frq1-1 (BY4742) strains transformed with a plasmid expressing GFP-Atg8 under the control of the CUP1 promoter were grown in SMD-Ura medium at 24 °C to mid-log phase. For each strain, the culture was divided into two aliquots. One aliquot was preincubated at 37 °C for 30 min and shifted to SD-N for 2 h at 37 °C, then (for the mutants) were shifted back to 24 °C for another 2 h (recovery (R)). The other aliquot was kept at 24 °C for 30 min and then shifted to SD-N for 2 h at 24 °C. For each aliquot, samples were taken before (N+) and after (N−) starvation and at the recovery stage. Immunoblotting was done with anti-YFP antibody, and the positions of full-length GFP-Atg8 and free GFP are indicated. B, nonspecific autophagy is blocked in the pik1–104 and frq1-1 mutants. WT (YTS158) and atg1Δ (TYY127), pik1–104 (ZFY281), and frq1-1 (ZFY280) strains were cultured as in A, but the incubation time in SD-N was increased to 4 h; recovery was carried out as in A. The Pho8Δ60 activity was measured as described under “Experimental Procedures.” Error bars represent S.D., obtained from at least three independent repeats. **, p < 0.005.
To further confirm our observations, we used the Pho8Δ60 assay to quantitatively analyze the autophagic defect in the pik1 mutant. PHO8 encodes a vacuolar alkaline phosphatase, and its delivery into the vacuole through the secretory pathway is dependent on its N-terminal transmembrane domain (22). Pho8Δ60 is a truncated form that lacks the 60 N-terminal amino acid residues including the transmembrane domain; this altered protein is unable to enter the endoplasmic reticulum, remains in the cytosol, and is delivered into the vacuole only through bulk autophagy (19). In this way, the magnitude of bulk autophagy is quantified by measuring the activity of alkaline phosphatase in nitrogen starvation conditions. After a 4-h nitrogen starvation, wild-type cells showed a strong increase in Pho8Δ60 activity at both 24 and 37 °C, whereas the atg1Δ strain that is defective in autophagy showed only background levels of activity (Fig. 1B). In pik1–104, although the induction of the Pho8Δ60 activity was normal at the PT, there was essentially no increase at the NPT. Similar to the results from the GFP-Atg8 processing assay, the Pho8Δ60 activity of pik1–104 was partially recovered when the cells were shifted back to the PT (recovery) or upon the introduction of plasmid-expressed PIK1 (Fig. 1B and data not shown).
Consistent with a previous publication (23), our sequence analysis showed that the temperature-sensitive pik1–104 allele contains mutations (S412P, A455T, Y517H, D529G, Q731R, R802G) that are located within the C-terminal catalytic kinase domain of Pik1. This suggests that the PtdIns 4-kinase activity of Pik1 could be essential for autophagy. However, we also considered the possibility that these point mutations may affect the stability of the Pik1 protein at NPT. To test this, we examined Pik1 in temperature-sensitive mutant strains. When shifted to the NPT for 2 h, three Pik1 temperature-sensitive alleles, pik1–11, pik1–83, and pik1–104 all showed a decreased protein level (data not shown), suggesting that these point mutations destabilized Pik1 at the NPT.
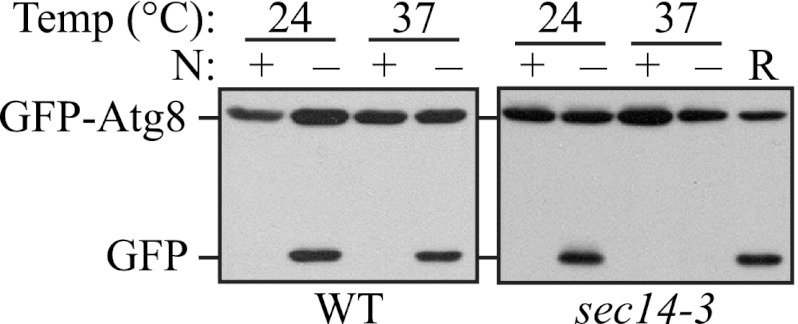
To extend our analysis on the requirement of Pik1 activity (i.e. the generation of PtdIns4P) for autophagy, we decided to examine an additional mutant strain that affects Pik1 function and the levels of PtdIns4P at the Golgi complex. The phosphatidylcholine/phosphatidylinositol transfer protein Sec14 is required for the transport of vesicles from the trans-Golgi (24, 25), and the sec14-3 mutant displays a major reduction in PtdIns4P, generating ∼50% of the wild-type level (26). Therefore, we examined whether autophagy is affected in the sec14-3 mutant using the GFP-Atg8 processing assay. The sec14-3 mutant cells displayed a clear absence of free GFP after 2 h of starvation at the NPT, indicating an autophagic defect, and this defect was recovered after the cells were shifted back to the PT (Fig. 2), indicating that the generation of PtdIns4P, dependent on Sec14 and Pik1, is essential for autophagy.
FIGURE 2.
Sec14 is required for autophagy. GFP-Atg8 processing is blocked in the sec14-3 mutant. WT (BY4742) and sec14-3 (BY4742) strains transformed with a plasmid expressing GFP-Atg8 under the control of the CUP1 promoter were cultured as in Fig. 1A. Samples were taken before (N+) and after (N−) starvation and at the recovery stage (R). Immunoblotting was done with anti-YFP antibody, and the positions of full-length GFP-Atg8 and free GFP are indicated.
Pik1 localizes to both the Golgi and the nucleus (8). To test which population of Pik1 is involved in autophagy, we analyzed a regulatory subunit of Pik1, Frq1, which is targeted to the Golgi and is required for the recruitment of Pik1 to this organelle (8, 23). As shown by the GFP-Atg8 processing assay, the frq1-1 mutant displayed a clear temperature-sensitive autophagy phenotype (Fig. 1A). At the NPT, the absence of free GFP after 2-h starvation indicated a defect of autophagy induction, and this defect was partially recovered after the cells were shifted back to the PT. As with the pik1–104 mutant, the autophagy impairment in the frq1-1 mutant was also observed using the Pho8Δ60 assay (Fig. 1B). This result suggested that the Golgi localization of Pik1 was essential for autophagy induction.
Pik1 Is Involved in Mitophagy
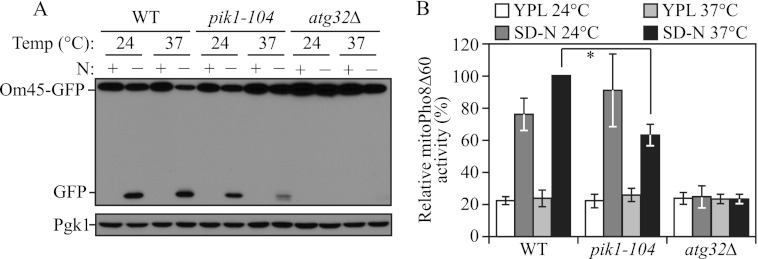
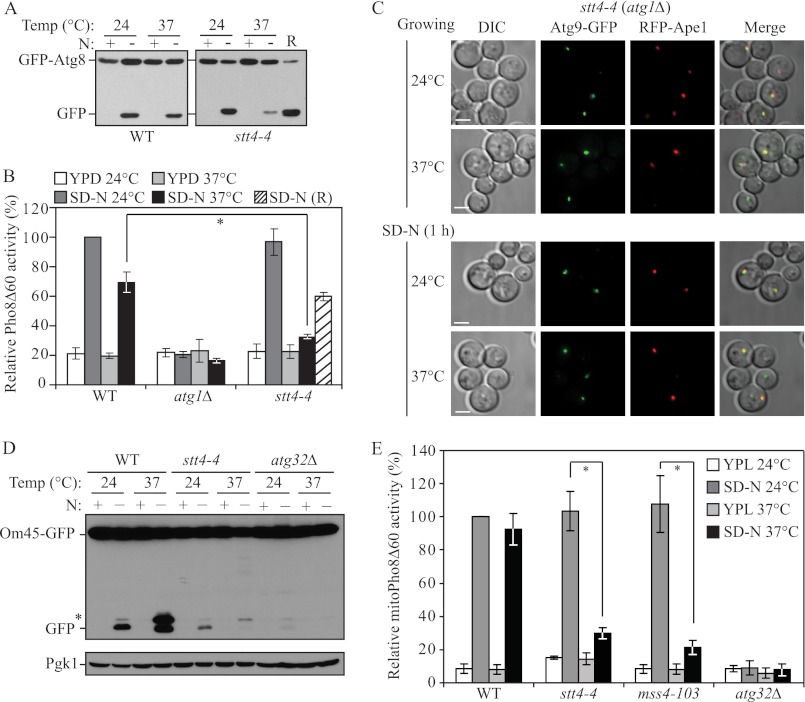
PpPik1 was previously reported to play a role in pexophagy in P. pastoris (10). Thus, we speculated that Pik1 may also play roles in selective autophagy in S. cerevisiae. Therefore, we extended our analysis of Pik1 to examine its role in mitophagy, the selective autophagic degradation of mitochondria. To detect mitophagy, we first utilized the Om45-GFP processing assay. OM45 encodes a mitochondrial outer membrane protein, and a chromosomally tagged version with the green fluorescent protein (GFP) at the C terminus is correctly localized on this organelle. Similar to the GFP-Atg8 processing assay, when mitophagy is induced, Om45-GFP, as part of the mitochondria, is delivered into the vacuole for degradation. Om45 is proteolytically removed or degraded, whereas the GFP moiety is relatively stable and accumulates in the vacuole. Therefore, mitophagy can be monitored based on the appearance of free GFP by immunoblot (27). When growing in rich medium containing lactate as the sole carbon source plus nitrogen (YPL), no free GFP band was detected at either 24 or 37 °C (Fig. 3A). After nitrogen starvation at 24 °C there was a clear appearance of the free GFP band in wild-type and pik1–104 mutant cells. At 37 °C, significantly reduced free GFP was detected in pik1–104 compared with the wild type (Fig. 3A), indicating a strong mitophagy block in the pik1–104 mutant. An atg32Δ mutant, which is completely defective in mitophagy, showed a complete block in Om45-GFP processing at either temperature as expected (28).
FIGURE 3.
Mitophagy is defective in the absence of Pik1 activity. A, Om45-GFP processing is blocked in the pik1–104 mutant. OM45 was chromosomally tagged with GFP in the wild-type (WT; TKYM107) and atg32Δ (TKYM137) and pik1–104 (KWY81) strains. Cells were cultured in YPL to mid-log phase at 24 °C, and then the culture was divided into two aliquots. One aliquot was kept at 24 °C for 30 min and then shifted to SD-N for 6 h at 24 °C. The other aliquot was preincubated at 37 °C for 30 min and then shifted to SD-N for 6 h at 37 °C. For each aliquot samples were taken before (N+) and after (N−) starvation. Protein extracts were probed with anti-YFP antibody and anti-Pgk1 antibody (the latter as a loading control). B, the mitoPho8Δ60 activity is reduced in the pik1–104 mutant. WT (KWY20) and atg32Δ (KWY22) and pik1–104 (KWY79) strains were cultured as in A. The mitoPho8Δ60 activity was measured as described under “Experimental Procedures”. Error bars correspond to the S.D. from at least three independent repeats. *, p < 0.01.
We also used the mitoPho8Δ60 assay to quantitatively examine the mitophagy defect in the pik1 mutant (21). The mitoPho8Δ60 construct is a modified version of Pho8Δ60 that allows the quantitative measurement of mitophagy activity. If PHO8Δ60 is fused with a mitochondrial targeting sequence, the encoded protein is specifically localized to mitochondria, and the ensuing alkaline phosphatase activity becomes an indicator of mitophagy (21). When mitophagy was induced by nitrogen starvation, wild-type cells showed a dramatic increase in mitoPho8Δ60 activity at both 24 and 37 °C (Fig. 3B). In the pik1–104 mutant, mitophagy induction at the PT was comparable with wild-type cells. In contrast, at the NPT there was a clear decrease of mitoPho8Δ60 activity (Fig. 3B), although not as strong as that seen with the Om45-GFP assay (Fig. 3A). This difference may reflect the different sensitivities of these two assays. Nonetheless, together, these results demonstrate that Pik1 is also involved in selective autophagy, in particular mitophagy.
Pik1 Affects the Anterograde Movement of Atg9
To further extend our analysis, we explored the mechanism through which Pik1 participates in the regulation of selective and nonselective autophagy. Inactivation of Sec7, which is indispensable for post-Golgi transport, affects Atg9 trafficking to the PAS (29). Atg9 is the only known conserved transmembrane protein of the yeast autophagy core machinery. Atg9 cycles between the PAS and other peripheral cytoplasmic sites (termed Atg9 reservoirs) and is important for the recruitment of additional Atg proteins (30). Because Pik1 is also critical for secretion at the Golgi, we hypothesized that the anterograde movement of Atg9 to the PAS would also be affected in the pik1 mutant.
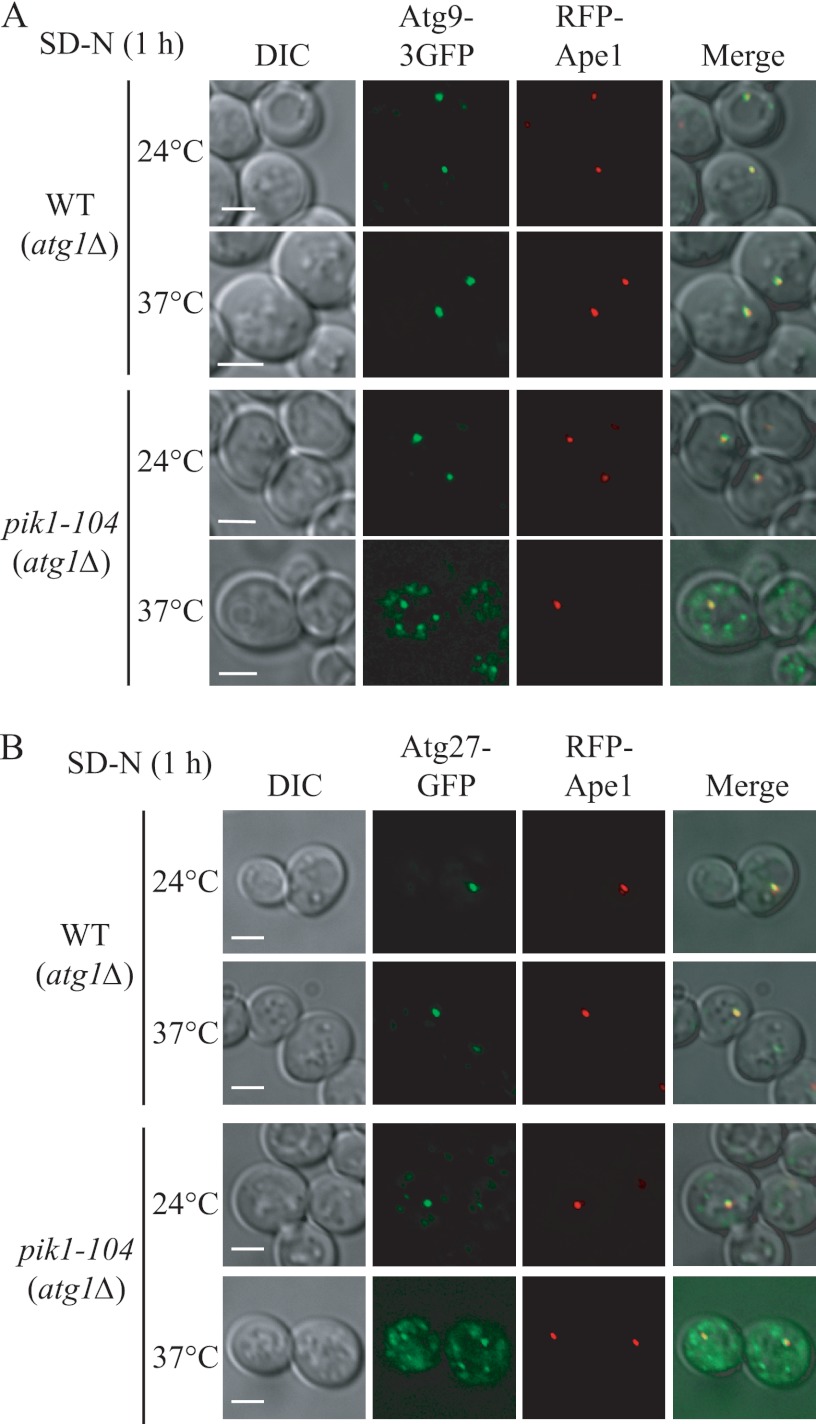
To test this, we used the TAKA (transport of Atg9 after knocking out ATG1) assay (20, 31). In brief, this assay is based on previous findings that Atg9 is retrieved from the PAS back to peripheral sites, and this retrieval process requires the Atg1-Atg13 complex (32); atg1Δ cells accumulate Atg9 almost exclusively at the PAS as a single dot unless the introduction of a second mutation causes its impaired anterograde transport. To check Atg9 localization, we made a functional C-terminal triple GFP fusion protein (Atg9–3GFP); this triple GFP chimera appears stable and does not interfere with the normal function and proper localization of Atg9 (16, 33). As reported previously, in atg1Δ cells, under both growing (data not shown) and starvation conditions, Atg9–3GFP displayed exclusively one single dot at the PAS (marked by RFP-Ape1) irrespective of the temperature (Fig. 4A). At 24 °C, atg1Δ pik1–104 cells showed the same pattern as the atg1Δ cells, whereas at the NPT, Atg9 was present in multiple puncta under both growing (data not shown) and starvation conditions (Fig. 4A). In some cells one of these dots colocalized with the PAS. These data indicated that there was a substantial reduction, but not a complete block, in the anterograde movement of Atg9 to the PAS in the pik1–104 mutant. A similar trafficking defect was also observed for Atg27, which is another transmembrane protein that also shuttles among the PAS, Golgi complex, and other peripheral sites, in the pik1–104 (Fig. 4B) and pik1–11 (data not shown) mutants.
FIGURE 4.
Atg9 and Atg27 trafficking is defective in the pik1 mutant. A, Atg9 anterograde movement to the PAS is defective in the pik1 mutant. WT (ZFY270) and pik1–104 and (ZFY273) strains in an atg1Δ background were cultured as in Fig. 1 and shifted to SD-N for 1 h. The localization of Atg9–3GFP and RFP-Ape1 was observed for samples cultured at the indicated temperature. B, Atg27 trafficking to the PAS is defective in the pik1 mutant. WT (ZFY070) and pik1–104 (ZFY301) strains were cultured as in A. The localization of Atg27-GFP and RFP-Ape1 was observed for samples cultured at the indicated temperature. Representative pictures from single Z-section images are shown. DIC, differential interference contrast. Scale bars, 5 μm.
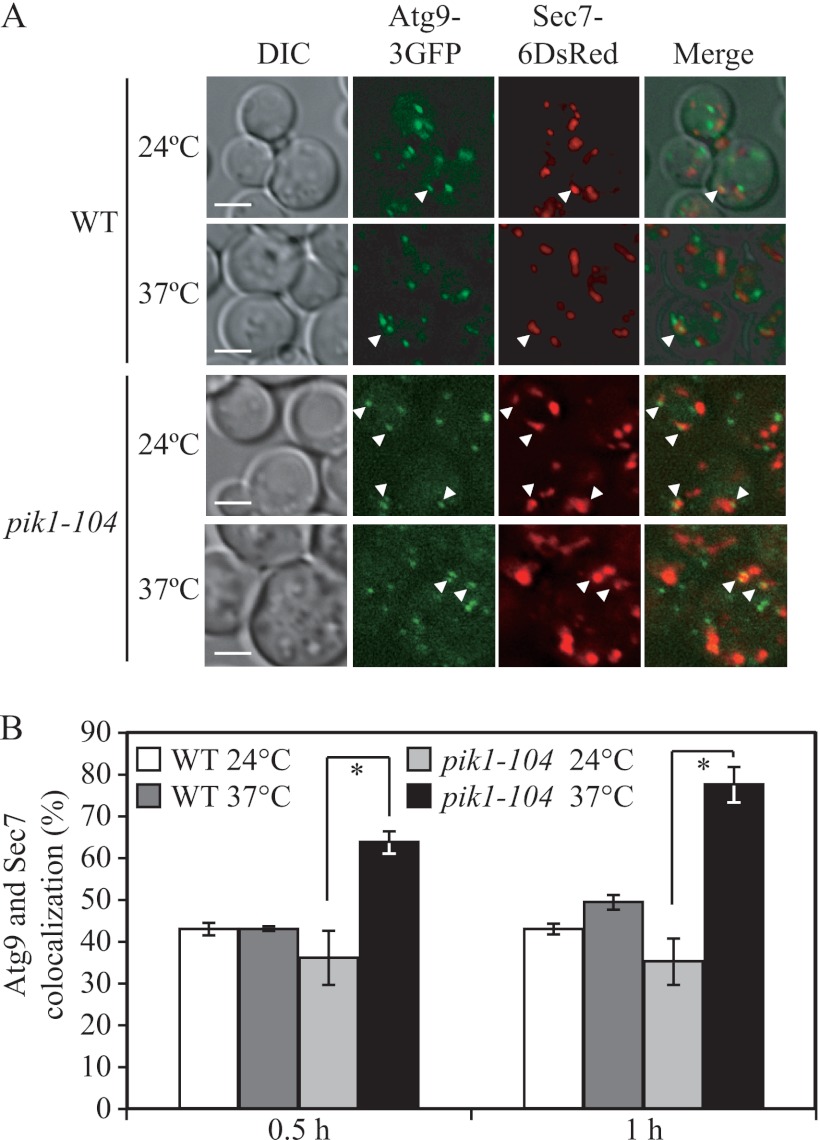
Because Pik1 functions at the late Golgi, we hypothesized that the defect in Atg9–3GFP localization to the PAS detected in the TAKA assay reflected accumulation of this protein in the trans-Golgi network. Accordingly, we further investigated the subcellular localization of Atg9–3GFP in the pik1 mutant. Newly synthesized Atg9 is transported into the ER and reaches its destination at the Atg9 reservoirs after passing through the Golgi complex, based on the observation that there is a partial colocalization between Atg9 and the late Golgi protein marker Sec7 (29). Therefore, we examined the colocalization of Atg9–3GFP with Sec7–6DsRed in a strain having the mutant pik1–104 allele. In wild-type cells, Atg9–3GFP was present in multiple puncta, partially colocalized with Sec7. When cultured at the PT and NPT for 30 min, the frequencies of colocalization (quantified by the percentage of cells that showed at least one colocalizing dot) in wild-type cells were 42.9 ± 1.7 and 42.9 ± 0.5% at 24 and 37 °C, respectively (Fig. 5A and B, data not shown). In the pik1–104 mutant, the frequency of Atg9 and Sec7 colocalization was 36.2 ± 6.3% at the PT. In contrast, at the NPT when Pik1 was inactivated, the frequency increased to 64.0 ± 2.5% (Fig. 5B). An additional 30-min incubation at the NPT increased the frequency of colocalization in the pik1–104 mutant to 77.5 ± 4.3% (Fig. 5B), whereas the wild-type strain showed only a minor increase in colocalization of 49.6 ± 1.5%. These data underscore the requirement of Pik1 in the exit of Atg9 from the Golgi complex or a post-Golgi compartment and, hence, provide an explanation for the failure of additional Atg9 to reach the PAS in the pik1 mutant. Taken together, our results indicate that Golgi-localized Pik1 is required for normal Atg9 trafficking through the secretory pathway and transport to the PAS, and this defect in Atg9 movement may be the cause of the autophagy defect seen in the pik1 mutants.
FIGURE 5.
Atg9 exit from the Golgi is defective in the pik1 mutant. A, WT (ZFY326) and pik1–104 (ZFY328) strains were cultured in SMD medium at 24 °C to mid-log phase and then divided into two aliquots. One aliquot was kept at 24 °C, and the other aliquot was shifted to 37 °C for 0.5 h. The localization of Atg9–3GFP and Sec7–6DsRed was observed for samples cultured at the indicated temperature. Representative pictures from single Z-section images are shown. DIC, differential interference contrast. Scale bars, 5 μm. B, quantification of the colocalization of Atg9–3GFP and Sec7–6DsRed from the data in A and after an additional 30 min at the indicated temperatures. Error bars represent the S.D. from at least three independent repeats. *, p < 0.05.
The Plasma Membrane-localized Stt4 Is Required for Autophagy and Mitophagy
Next, we explored the role of the plasma membrane-localized PtdIns 4-kinase Stt4 in autophagy using the GFP-Atg8 processing assay. Similar to the pik1 mutant, significantly reduced free GFP was detected in a stt4-4 mutant at 37 °C compared with wild-type cells, suggesting a strong defect in autophagy (Fig. 6A). Again, a subsequent 2-h starvation at 24 °C (recovery (R)) restored the autophagy phenotype. This result was also confirmed by the Pho8Δ60 assay (Fig. 6B), although in this case the recovery was clearly less robust; the stt4-4 mutant showed a quite severe growth phenotype at the NPT that may have compromised the ability of the mutant strain to recover when shifted back to the PT. To test whether a similar mechanism may apply for Stt4, we also examined Atg9 movement in the stt4-4 mutant by the TAKA assay; however, normal Atg9 trafficking to the PAS was observed in the stt4 mutant, as Atg9 accumulated as a single dot in the atg1Δ stt4-4 strain in growing conditions and after starvation at both PT and NPT (Fig. 6C). Therefore, the plasma membrane-localized Stt4 is required for autophagy by a mechanism distinct from that of Pik1.
FIGURE 6.
Stt4 is required for autophagy. A, GFP-Atg8 processing is blocked in the stt4-4 mutant. WT (BY4742) and stt4-4 (BY4741) strains transformed with a plasmid expressing GFP-Atg8 under the control of the CUP1 promoter were cultured as in Fig. 1A. Samples were taken before (N+) and after (N−) starvation and at the recovery stage (R). Immunoblotting was done with anti-YFP antibody, and the positions of full-length GFP-Atg8 and free GFP are indicated. B, Pho8Δ60 activity is blocked in the stt4-4 mutant. WT (YTS158), atg1Δ (TYY127) and stt4-4 (ZFY282) strains were cultured as in Fig. 1B. The Pho8Δ60 activity was measured as described under “Experimental Procedures.” Error bars represent S.D., which was obtained from at least three independent repeats. C, Atg9 anterograde movement to the PAS is normal in the stt4-4 mutant. The stt4-4 strain was cultured as in Fig. 4A. The localization of Atg9-GFP and RFP-Ape1 was observed for samples cultured at the indicated temperature in growing or starvation conditions. Representative pictures from single Z-section images are shown. DIC, differential interference contrast. Scale bars, 5 μm. D, Om45-GFP processing is blocked in the stt4-4 mutant. OM45 was chromosomally tagged with GFP in the WT (TKYM107), atg32Δ (TKYM137) and stt4-4 (KWY80) strains. Cells were cultured as in Fig. 3A. Samples were taken before (N+) and after (N−) starvation. Protein extracts were probed with anti-YFP antibody and anti-Pgk1 antibody (the latter as a loading control). The asterisk marks a nonspecific band. E, the mitoPho8Δ60 activity is reduced in the stt4-4 and mss4–103 mutants. Wild type (KWY84), atg32Δ (KWY87), stt4-4 (KWY85), and mss4–103 (KWY86) strains were cultured as in D. The mitoPho8Δ60 activity was measured as described under “Experimental Procedures.” Error bars correspond to the S.D. from at least three independent repeats. *, p < 0.01.
Next we tested the role of Stt4 in mitophagy using both the Om45-GFP processing and mitoPho8Δ60 assays. After a 6-h starvation at 37 °C in the stt4-4 mutant, a significantly reduced level of free GFP or mitoPho8Δ60 activity was observed compared with the wild type, indicating a strong mitophagy block in this mutant (Fig. 6, D and E). Taken together, these results demonstrated that Stt4 is required for both selective and nonselective autophagy.
The PtdIns4P 5-Kinase Mss4 Is Required Primarily for Selective Autophagy
Stt4 has been previously reported to function upstream of the Pkc1-mediated MAP kinase cascade (34). Mss4, the only identified PtdIns4P 5-kinase in yeast, converts PtdIns4P generated by Stt4 at the plasma membrane into PtdIns(4,5)P2, which is required for efficient Pkc1-dependent signaling in the cell wall integrity pathway (7). Pkc1 is involved in nonselective autophagy regulation (35), whereas the downstream MAP kinase of this pathway, Slt2, is involved only in selective types of autophagy (36, 37). Therefore, we tested the role of Mss4 in both autophagy and mitophagy.
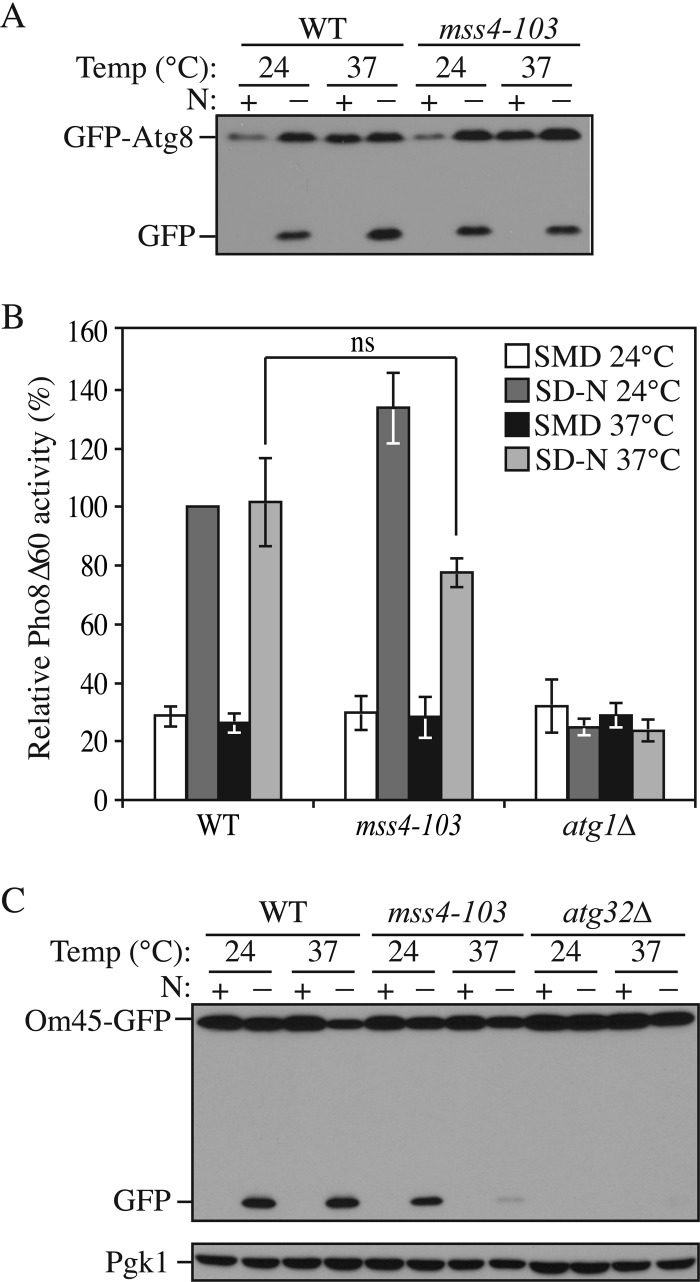
First we used the GFP-Atg8 processing assay to test the autophagy phenotype in a temperature-sensitive mss4–103 mutant strain. A comparable free GFP band was observed after a 2-h starvation at 37 °C in this mutant relative to the wild type, suggesting that there was not a major autophagy defect in the mss4–103 mutant (Fig. 7A). We further confirmed this result by the quantitative Pho8Δ60 assay; a slightly decreased but still comparable Pho8Δ60 activity was observed in the mss4–103 mutant at 37 °C (Fig. 7B). Therefore, these results demonstrated that Mss4 was not required for nonselective autophagy. This finding agrees with the observation that Stt4 appears to have an Mss4-independent function in the regulation of cell wall maintenance, as the stt4-4 mutant cannot be rescued by overexpression of Mss4 (38).
FIGURE 7.
Mss4 is required for mitophagy but not nonselective autophagy. A, GFP-Atg8 processing is normal in the mss4–103 mutant. WT (BY4742) and mss4–103 (BY4741) strains transformed with a plasmid expressing GFP-Atg8 under the control of the CUP1 promoter were cultured as in Fig. 1A but without the recovery stage. Samples were taken before (N+) and after (N−) starvation. Immunoblotting was done with anti-YFP antibody, and the positions of full-length GFP-Atg8 and free GFP are indicated. B, the Pho8Δ60 activity is normal in the mss4–103 mutant. WT (YTS158) and atg1Δ (TYY127) and mss4–103 (KWY83) strains were cultured as in Fig. 1B. The Pho8Δ60 activity was measured as described under “Experimental Procedures.” Error bars represent S.D., which was obtained from at least three independent repeats. C, Om45-GFP processing is blocked in the mss4–103 mutant. Wild-type (TKYM107), atg32Δ (TKYM137), and mss4–103 (KWY82) strains were cultured as in Fig. 3A. Samples were taken before (N+) and after (N−) starvation. Protein extracts were probed with anti-YFP antibody and anti-Pgk1 antibody (the latter as a loading control). ns, not significant. p > 0.05.
Next, we tested the role of Mss4 in mitophagy by both the Om45-GFP processing and mitoPho8Δ60 assays. After a 6-h starvation at 37 °C in the mss4–103 mutant, a significantly reduced level of free GFP or mitoPho8Δ60 activity was observed compared with the wild type, indicating a strong mitophagy block in this mutant almost as severe as seen in the atg32Δ strain (Fig. 7C and Fig. 6E). Taken together, these results demonstrated that Mss4 is specifically involved in selective but not nonselective autophagy.
DISCUSSION
The importance of phosphoinositides in autophagy regulation has been demonstrated by the study of the Vps34-containing PtdIns 3-kinase complex in yeast (5) and both the PtdIns 3-kinase and PtdIns3P 5-kinase in mammalian cells (39, 40). However, relatively little attention has been paid to the role of PtdIns 4-kinases in autophagy (10), and to our knowledge there is no information regarding the function of such enzymes in autophagy in S. cerevisiae. Here we reported the involvement of two other enzymes, a PtdIns 4-kinase and PtdIns4P 5-kinase, in selective and nonselective autophagy. We demonstrated that two major yeast PtdIns 4-kinases, Pik1 and Stt4, are both required for autophagy. In addition, we also showed that the PtdIns4P 5-kinase Mss4 is specifically involved in mitophagy.
Although the mechanism through which plasma membrane-localized Stt4 affects autophagy is still under study, we showed that Golgi-localized Pik1, which plays an essential role in vesicle exit from this organelle, may be involved in autophagy by affecting normal Atg9 trafficking to the PAS. This conclusion is consistent with several recent studies that demonstrate Atg9 traffics through part of the secretory pathway to the peripheral Atg9 reservoirs (41, 42), and exit from the Golgi complex is required for normal Atg9 trafficking (29, 44). However, little is known about Atg9 trafficking beyond Golgi exit. Atg9 does not colocalize with any known organelle markers after leaving the Golgi, and electron microscopy data suggest that Atg9 becomes part of a structure called the tubulovesicular cluster (41, 42). Atg27 and Atg23 are two other proteins found at the tubulovesicular cluster; however, except for these morphological studies, we do not know anything more about this structure. As Pik1-generated PtdIns4P is also important for vesicle transport after exit from the Golgi, we speculated that Pik1 might play a role(s) in directing Atg9-containing vesicles from the Golgi to their next destination. This function could be achieved by the recruitment of a PtdIns4P effector protein(s) that is involved in autophagy. Unfortunately, even though we tested several potential PtdIns4P effectors (Mdr1, Ysp2, Yfl042C, Yhr080C, and Ylr072W) that have a GRAM domain with the capacity to bind PtdIns4P (10), we did not observe an autophagy defect (data not shown). Therefore, a still unknown effector(s) might participate in PtdIns4P-mediated Atg9 trafficking to the tubulovesicular cluster. Identifying the potential effector(s) may provide critical insight into the formation of the tubulovesicular cluster and thus would be helpful in adding to our understanding of the organization of the PAS.
We also reported that Pik1 plays an important role in mitophagy. The observation that PtdIns4P participates in the formation of the micropexophagic apparatus provided the initial insight into the connection between PtdIns4P and selective autophagy (10). Our results that Pik1 is required for mitophagy extended this finding. For further exploration, we also tested possible colocalization of a PtdIns4P marker (GFP-Osh2-PH) with the mitophagy marker protein Atg32, but we did not observe a significant level of colocalization (data not shown). This result could reflect the inability of this PtdIns4P probe to mark all cellular PtdIns4P or the possibility that the population of PtdIns4P that participates in mitophagy may not be sufficient to be detectably marked by GFP-Osh2-PH.
In the meantime, it remains unclear as to how the plasma membrane-localized Stt4 participates in autophagy. Our results excluded the possibility that Stt4 acts solely as an upstream factor of Mss4 for autophagy because the PtdIns4P 5-kinase Mss4 that functions at the plasma membrane is dispensable for nonselective autophagy, although Mss4 is required for mitophagy. PtdIns(4,5)P2 is important for the organization of the actin cytoskeleton (45), which is involved in selective autophagy (43). The actin cytoskeleton may facilitate cargo transport to the PAS. However, we found that Atg32 trafficking to the mitochondria-specific PAS is essentially normal in the mss4–103 mutant (data not shown). Further study is thus required to understand the role of Mss4 in mitophagy.
In summary, our study established a role of PtdIns 4-kinases and PtdIns4P 5-kinases in selective and nonselective autophagy pathways and provides the first information on the mechanism by which the PtdIns 4-kinase may regulate these processes. Continued studies should provide additional insight into how these enzymes control autophagy.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grant GM053396 (to D. J. K.).

This article contains supplemental Tables S1 and S2.
- PtdIns
- phosphatidylinositol
- Atg
- autophagy-related
- PAS
- phagophore assembly site
- PT
- permissive temperature
- NPT
- nonpermissive temperature
- TAKA
- transport of Atg9 after knocking out ATG1.
REFERENCES
- 1. Levine B., Klionsky D. J. (2004) Development by self-digestion. Molecular mechanisms and biological functions of autophagy. Dev. Cell 6, 463–477 [DOI] [PubMed] [Google Scholar]
- 2. Xie Z., Klionsky D. J. (2007) Autophagosome formation. Core machinery and adaptations. Nat. Cell Biol. 9, 1102–1109 [DOI] [PubMed] [Google Scholar]
- 3. Di Paolo G., De Camilli P. (2006) Phosphoinositides in cell regulation and membrane dynamics. Nature 443, 651–657 [DOI] [PubMed] [Google Scholar]
- 4. Strahl T., Thorner J. (2007) Synthesis and function of membrane phosphoinositides in budding yeast, Saccharomyces cerevisiae. Biochim. Biophys. Acta 1771, 353–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kihara A., Noda T., Ishihara N., Ohsumi Y. (2001) Two distinct Vps34 phosphatidylinositol 3-kinase complexes function in autophagy and carboxypeptidase Y sorting in Saccharomyces cerevisiae. J. Cell Biol. 152, 519–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ferguson C. J., Lenk G. M., Meisler M. H. (2009) Defective autophagy in neurons and astrocytes from mice deficient in PI(3,5)P2. Hum. Mol. Genet. 18, 4868–4878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Audhya A., Emr S. D. (2002) Stt4 PI 4-kinase localizes to the plasma membrane and functions in the Pkc1-mediated MAP kinase cascade. Dev. Cell 2, 593–605 [DOI] [PubMed] [Google Scholar]
- 8. Strahl T., Hama H., DeWald D. B., Thorner J. (2005) Yeast phosphatidylinositol 4-kinase, Pik1, has essential roles at the Golgi and in the nucleus. J. Cell Biol. 171, 967–979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Han G. S., Audhya A., Markley D. J., Emr S. D., Carman G. M. (2002) The Saccharomyces cerevisiae LSB6 gene encodes phosphatidylinositol 4-kinase activity. J. Biol. Chem. 277, 47709–47718 [DOI] [PubMed] [Google Scholar]
- 10. Yamashita S., Oku M., Wasada Y., Ano Y., Sakai Y. (2006) PI4P-signaling pathway for the synthesis of a nascent membrane structure in selective autophagy. J. Cell Biol. 173, 709–717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cao Y., Klionsky D. J. (2007) Atg26 is not involved in autophagy-related pathways in Saccharomyces cerevisiae. Autophagy 3, 17–20 [DOI] [PubMed] [Google Scholar]
- 12. Labbé S., Thiele D. J. (1999) Copper ion inducible and repressible promoter systems in yeast. Methods Enzymol. 306, 145–153 [DOI] [PubMed] [Google Scholar]
- 13. Kim J., Huang W.-P., Klionsky D. J. (2001) Membrane recruitment of Aut7p in the autophagy and cytoplasm to vacuole targeting pathways requires Aut1p, Aut2p, and the autophagy conjugation complex. J. Cell Biol. 152, 51–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Longtine M. S., McKenzie A., III, Demarini D. J., Shah N. G., Wach A., Brachat A., Philippsen P., Pringle J. R. (1998) Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14, 953–961 [DOI] [PubMed] [Google Scholar]
- 15. Gueldener U., Heinisch J., Koehler G. J., Voss D., Hegemann J. H. (2002) A second set of loxP marker cassettes for Cre-mediated multiple gene knockouts in budding yeast. Nucleic Acids Res. 30, e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Monastyrska I., He C., Geng J., Hoppe A. D., Li Z., Klionsky D. J. (2008) Arp2 links autophagic machinery with the actin cytoskeleton. Mol. Biol. Cell 19, 1962–1975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Strømhaug P. E., Reggiori F., Guan J., Wang C.-W., Klionsky D. J. (2004) Atg21 is a phosphoinositide-binding protein required for efficient lipidation and localization of Atg8 during uptake of aminopeptidase I by selective autophagy. Mol. Biol. Cell 15, 3553–3566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Roy A., Levine T. P. (2004) Multiple pools of phosphatidylinositol 4-phosphate detected using the pleckstrin homology domain of Osh2p. J. Biol. Chem. 279, 44683–44689 [DOI] [PubMed] [Google Scholar]
- 19. Noda T., Matsuura A., Wada Y., Ohsumi Y. (1995) Novel system for monitoring autophagy in the yeast Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 210, 126–132 [DOI] [PubMed] [Google Scholar]
- 20. Shintani T., Klionsky D. J. (2004) Cargo proteins facilitate the formation of transport vesicles in the cytoplasm to vacuole targeting pathway. J. Biol. Chem. 279, 29889–29894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kanki T., Wang K., Klionsky D. J. (2010) A genomic screen for yeast mutants defective in mitophagy. Autophagy 6, 278–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Klionsky D. J., Emr S. D. (1989) Membrane protein sorting. Biosynthesis, transport, and processing of yeast vacuolar alkaline phosphatase. EMBO J. 8, 2241–2250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hendricks K. B., Wang B. Q., Schnieders E. A., Thorner J. (1999) Yeast homologue of neuronal frequenin is a regulator of phosphatidylinositol-4-OH kinase. Nat. Cell Biol. 1, 234–241 [DOI] [PubMed] [Google Scholar]
- 24. Bankaitis V. A., Aitken J. R., Cleves A. E., Dowhan W. (1990) An essential role for a phospholipid transfer protein in yeast Golgi function. Nature 347, 561–562 [DOI] [PubMed] [Google Scholar]
- 25. Bankaitis V. A., Malehorn D. E., Emr S. D., Greene R. (1989) The Saccharomyces cerevisiae SEC14 gene encodes a cytosolic factor that is required for transport of secretory proteins from the yeast Golgi complex. J. Cell Biol. 108, 1271–1281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hama H., Schnieders E. A., Thorner J., Takemoto J. Y., DeWald D. B. (1999) Direct involvement of phosphatidylinositol 4-phosphate in secretion in the yeast Saccharomyces cerevisiae. J. Biol. Chem. 274, 34294–34300 [DOI] [PubMed] [Google Scholar]
- 27. Kanki T., Klionsky D. J. (2008) Mitophagy in yeast occurs through a selective mechanism. J. Biol. Chem. 283, 32386–32393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kanki T., Wang K., Cao Y., Baba M., Klionsky D. J. (2009) Atg32 is a mitochondrial protein that confers selectivity during mitophagy. Dev. Cell 17, 98–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. van der Vaart A., Griffith J., Reggiori F. (2010) Exit from the Golgi is required for the expansion of the autophagosomal phagophore in yeast Saccharomyces cerevisiae. Mol. Biol. Cell 21, 2270–2284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. He C., Klionsky D. J. (2007) Atg9 trafficking in autophagy-related pathways. Autophagy 3, 271–274 [DOI] [PubMed] [Google Scholar]
- 31. Klionsky D. J., Baehrecke E. H., Brumell J. H., Chu C. T., Codogno P., Cuervo A. M., Debnath J., Deretic V., Elazar Z., Eskelinen E. L., Finkbeiner S., Fueyo-Margareto J., Gewirtz D., Jäättelä M., Kroemer G., Levine B., Melia T. J., Mizushima N., Rubinsztein D. C., Simonsen A., Thorburn A., Thumm M., Tooze S. A. (2011) A comprehensive glossary of autophagy-related molecules and processes (2nd edition). Autophagy 7, 1273–1294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Reggiori F., Tucker K. A., Stromhaug P. E., Klionsky D. J. (2004) The Atg1-Atg13 complex regulates Atg9 and Atg23 retrieval transport from the pre-autophagosomal structure. Dev. Cell 6, 79–90 [DOI] [PubMed] [Google Scholar]
- 33. He C., Baba M., Cao Y., Klionsky D. J. (2008) Self-interaction is critical for Atg9 transport and function at the phagophore assembly site during autophagy. Mol. Biol. Cell 19, 5506–5516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Yoshida S., Ohya Y., Goebl M., Nakano A., Anraku Y. (1994) A novel gene, STT4, encodes a phosphatidylinositol 4-kinase in the PKC1 protein kinase pathway of Saccharomyces cerevisiae. J. Biol. Chem. 269, 1166–1172 [PubMed] [Google Scholar]
- 35. Shahnazari S., Yen W.-L., Birmingham C. L., Shiu J., Namolovan A., Zheng Y. T., Nakayama K., Klionsky D. J., Brumell J. H. (2010) A diacylglycerol-dependent signaling pathway contributes to regulation of antibacterial autophagy. Cell Host Microbe 8, 137–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Manjithaya R., Jain S., Farré J. C., Subramani S. (2010) A yeast MAPK cascade regulates pexophagy but not other autophagy pathways. J. Cell Biol. 189, 303–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Mao K., Wang K., Zhao M., Xu T., Klionsky D. J. (2011) Two MAPK-signaling pathways are required for mitophagy in Saccharomyces cerevisiae. J. Cell Biol. 193, 755–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Audhya A., Foti M., Emr S. D. (2000) Distinct roles for the yeast phosphatidylinositol 4-kinases, Stt4p and Pik1p, in secretion, cell growth, and organelle membrane dynamics. Mol. Biol. Cell 11, 2673–2689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ho C. Y., Alghamdi T. A., Botelho R. J. (2012) Phosphatidylinositol-3,5-bisphosphate. No Longer the Poor PIP(2). Traffic 13, 1–8 [DOI] [PubMed] [Google Scholar]
- 40. Lindmo K., Stenmark H. (2006) Regulation of membrane traffic by phosphoinositide 3-kinases. J. Cell Sci. 119, 605–614 [DOI] [PubMed] [Google Scholar]
- 41. Mari M., Griffith J., Rieter E., Krishnappa L., Klionsky D. J., Reggiori F. (2010) An Atg9-containing compartment that functions in the early steps of autophagosome biogenesis. J. Cell Biol. 190, 1005–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Nair U., Jotwani A., Geng J., Gammoh N., Richerson D., Yen W. L., Griffith J., Nag S., Wang K., Moss T., Baba M., McNew J. A., Jiang X., Reggiori F., Melia T. J., Klionsky D. J. (2011) SNARE proteins are required for macroautophagy. Cell 146, 290–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Reggiori F., Monastyrska I., Shintani T., Klionsky D. J. (2005) The actin cytoskeleton is required for selective types of autophagy, but not nonspecific autophagy, in the yeast Saccharomyces cerevisiae. Mol. Biol. Cell 16, 5843–5856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Geng J., Nair U., Yasumura-Yorimitsu K., Klionsky D. J. (2010) Post-Golgi Sec proteins are required for autophagy in Saccharomyces cerevisiae. Mol. Biol. Cell 21, 2257–2269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Desrivières S., Cooke F. T., Parker P. J., Hall M. N. (1998) MSS4, a phosphatidylinositol-4-phosphate 5-kinase required for organization of the actin cytoskeleton in Saccharomyces cerevisiae. J. Biol. Chem. 273, 15787–15793 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.