Background: IL-10 inhibits TNFα expression in the anti-inflammatory response.
Results: IL-10 activates SHIP1 phosphatase to inhibit TNFα translation through polysome disassembly and Mnk1 activation.
Conclusion: SHIP1 is involved in the regulatory mechanism of IL-10 for controlling TNFα translation.
Significance: This suggests a STAT3-independent pathway for IL-10 in controlling inflammation through SHIP1.
Keywords: Inflammation, Macrophages, Signaling, Translation Regulation, Tumor Necrosis Factor (TNF), SHIP1 Inositol Phosphatase, Interleukin-10
Abstract
Production of the proinflammatory cytokine TNFα by activated macrophages is an important component of host defense. However, TNFα production must be tightly controlled to avoid pathological consequences. The anti-inflammatory cytokine IL-10 inhibits TNFα mRNA expression through activation of the STAT3 transcription factor pathway and subsequent expression of STAT3-dependent gene products. We hypothesized that IL-10 must also have more rapid mechanisms of action and show that IL-10 rapidly shifts existing TNFα mRNA from polyribosome-associated polysomes to monosomes. This translation suppression requires the presence of SHIP1 (SH2 domain-containing inositol 5′-phosphatase 1) and involves inhibition of Mnk1 (MAPK signal-integrating kinase 1). Furthermore, activating SHIP1 using a small-molecule agonist mimics the inhibitory effect of IL-10 on Mnk1 phosphorylation and TNFα translation. Our data support the existence of an alternative STAT3-independent pathway through SHIP1 for IL-10 to regulate TNFα translation during the anti-inflammatory response.
Introduction
The host defense against bacteria and other pathogens includes production of inflammatory cytokines such as TNFα by macrophages. Bacterial cell wall products such as LPS bind to receptors on the macrophage and initiate a signal transduction cascade, resulting in TNFα transcription (1). For translation to occur, p38 MAPK must be activated to relieve translational silencing that is mediated, in part, by proteins interacting with AU-rich elements (AREs)2 in the 3′-UTR of TNFα mRNA (2–6). One p38 MAPK substrate involved in regulating TNFα translation is Mnk1 (MAPK signal-integrating kinase 1) (7). Mnk1 can be activated by p38 MAPK or ERK1/2 and phosphorylates a number of ARE-binding proteins (5, 8) as well as eIF4E, a component of the translation initiation complex (9, 10).
Although TNFα is beneficial for potentiating the inflammatory response, the magnitude and duration of its production must be controlled and appropriately terminated to prevent tissue damage. The anti-inflammatory cytokine IL-10 is a key moderator of inflammation, and loss of normal levels of IL-10 or its receptor results in inflammatory disorders in both mouse and man (11–14). Studies on the mechanism by which IL-10 receptor signaling inhibits TNFα expression have concentrated on activation of the STAT3 transcription factor and induction of STAT3-regulated genes (15–19). The gene products implicated in inhibiting TNFα expression include Bcl-3, an inhibitor of TNFα transcription (20); tristetraprolin, which destabilizes TNFα mRNA (21); and ETV3 and SBNO2, which modify the chromatin structure of the TNFα gene (22). However, we reasoned that IL-10 must also have a more immediate mechanism of action because physiologically in a whole organism, IL-10 is not encountered by macrophages until they have already become activated. In fact, an activated macrophage itself is a major source of IL-10 through autocrine production, which intrinsically delays the exposure of the activated macrophage to IL-10. We thus investigated and found that IL-10 also has immediate effects on TNFα translation, which do not require de novo transcription of STAT3-dependent genes.
This STAT3-independent pathway involves SHIP1 (SH2 domain-containing inositol 5′-phosphatase 1). SHIP1 has already been implicated in negative regulation of immune and inflammatory cell activation by antagonizing the PI3K pathway (23, 24). We now show that IL-10 inhibits TNFα translation through SHIP1-dependent inhibition of Mnk1. This inhibitory effect appears to be modulated by phosphorylation of one of two of its upstream kinases, p38 MAPK. Consistent with this finding, activation of SHIP1 using a small-molecule SHIP1 agonist mimicked the effect of IL-10 by inhibiting Mnk1 activation and TNFα translation.
EXPERIMENTAL PROCEDURES
Cells and Reagents
RAW 264.7 cells were obtained from American Type Culture Collection and maintained in Dulbecco's modified Eagle's medium supplemented with 9% (v/v) Fetal calf serum (Thermo Fisher Scientific). Primary cells (peritoneal macrophages) from 6–20-week-old male and female BALB/c wild-type (SHIP1+/+) or SHIP1−/− mice (Dr. Gerald Krystal, BC Cancer Research Centre, Vancouver, British Columbia, Canada) were isolated by peritoneal lavage with 3–5 ml of sterile PBS (Thermo Fisher Scientific). Cells were collected and transferred to Iscove's modified Dulbecco's medium (Thermo Fisher Scientific) supplemented with 10% (v/v) fetal calf serum, 10 μm β-mercaptoethanol, 150 μm monothioglycolate, and 1 mm l-glutamine. All cells were cultured at 37 °C and at 5% CO2 and 95% humidity.
Antibodies against phospho-Mnk1 (Thr-197/Thr-202), phospho-ERK1/2 (p44/p42 MAPK; Thr-202/Tyr-204), phospho-p38 (Thr-180/Tyr-182), phospho-eIF4E (Ser-209), PI3K p85, and Akt (pan) were purchased from Cell Signaling Technologies. Anti-SHIP1 (P1C1) antibody was purchased from Santa Cruz Biotechnology. CGP57380 (Calbiochem) was dissolved in dimethyl sulfoxide, and AQX-MN100 (Aquinox Pharmaceuticals, Vancouver, British Columbia, Canada) was dissolved in ethanol. Recombinant mouse IL-10 was expressed and purified in-house.
Immunoblot Analysis
Peritoneal macrophages were plated at a density of 3 × 105 cells/well on a 24-well tissue culture plate and allowed to adhere for 1 h, followed by replacement with fresh medium for 3 h prior to stimulation with 1 ng/ml LPS (Escherichia coli serotype 0111:B4; Sigma) with and without 100 ng/ml IL-10 for 1 h. Cells were rinsed with cold PBS and lysed with 250 μl of hot 2× Laemmli solubilization buffer. Proteins were separated by 10% SDS-PAGE, followed by electroblotting onto PVDF membrane. Blots were developed with the appropriate Alexa Fluor® 660 or 800-conjugated anti-IgG antibodies (Invitrogen) and imaged using a LI-COR Odyssey imager.
SHIP1 and Scrambled Knockdowns
siRNA oligonucleotides targeting SHIP1 or a nonspecific scrambled sequence were designed using BLOCK-iTTM siRNA online design software (Invitrogen) and cloned into a microRNA expression vector (pTRIPZ, Thermo Fisher Scientific) under the control of a doxycycline/tetracycline-inducible promoter. Lentivirus produced from these vectors was used to transduce RAW 264.7 cells in the presence of 8 μg/ml protamine sulfate and selected for resistance in 3 μg/ml puromycin. Further selection was provided by induction of GFP expression by 2 μg/ml doxycycline (Sigma) and sorting for GFP fluorescence.
TNFα mRNA Fractionation and Quantification
siRNA lentivirus-transduced cells were treated with 2 μg/ml doxycycline 48 h prior to stimulation, when the cells were replated at a density of 8 × 106 cells in a 10-cm tissue culture plate at ∼32 h. Parental RAW 264.7 cells and peritoneal macrophages were plated as described above but without doxycycline treatment. For Mnk1 inhibitor treatment, cells were replaced with fresh medium containing vehicle (dimethyl sulfoxide) or 75 μm CGP57380 30 min prior to stimulation. Cells were stimulated with 1 ng/ml LPS for 45 min, followed by the addition of 100 ng/ml IL-10 or 10 μm AQX-MN100 for 15 min where indicated.
Cells were rinsed with 5 ml of cold PBS and then lifted in 500 μl of buffer containing 10 mm Tris-HCl (pH 7.4), 10 mm potassium chloride, 10 mm magnesium chloride, 20 mm dithiothreitol, 150 μg/ml cycloheximide, 0.5% Nonidet P-40, and 500 units of Protector RNase (Roche Diagnostics) with a cell scraper and lysed for 30 min at 4 °C. The soluble lysates were collected by centrifugation at 12,000 × g for 10 min, and sucrose was added to 500 μl of the lysates to 10% (w/v). A 125-μl aliquot was set aside to extract the total unfractionated RNA, and 450 μl was layered on top of a stepwise sucrose gradient containing 450 μl of 30% (w/v), 250 μl of 50% (w/v), and 200 μl of 70% (w/v) sucrose. All sucrose solutions were generated in the above lysis buffer lacking Nonidet P-40. Samples were separated by ultracentrifugation at 152,000 × g for 35 min, followed by collection of 125-μl fractions. Each fraction was supplemented with 150 ng of total human mRNA and 30 μg of GlycoBlue (Invitrogen) prior to extraction of RNA with TRIzol reagent (Invitrogen) following the manufacturer's protocol and then treated with DNase (Roche Diagnostics). cDNA was generated using a RevertAid H minus first strand cDNA synthesis kit (Roche Diagnostics) and analyzed by SYBR Green-based real-time quantitative PCR using mouse TNFα and human GAPDH gene-specific primers. The recovery of mouse TNFα mRNA in each fraction was normalized by comparison with the level of human GAPDH present in each fraction. The amount of mouse TNFα mRNA in each fraction is plotted as a percentage of the sum of mouse TNFα mRNAs in all fractions. The amount of mouse TNFα mRNA in the starting unfractionated sample was normalized to endogenous mouse GAPDH levels.
TNFα Production Measurements
Parental RAW 264.7 cells were seeded at a density of 2 × 105 cells in 24-well tissue culture plates overnight and then pre-activated with 1 ng/ml LPS for 45 min. The cells were rinsed with 1 ml of fresh medium and replaced with 1 ng/ml LPS with 0–100 ng/ml IL-10 for 15 min prior to collecting the media supernatant for ELISA analysis with a OptEIATM mouse TNF ELISA kit (BD Biosciences).
RESULTS
IL-10 Regulates TNFα Translation through SHIP1
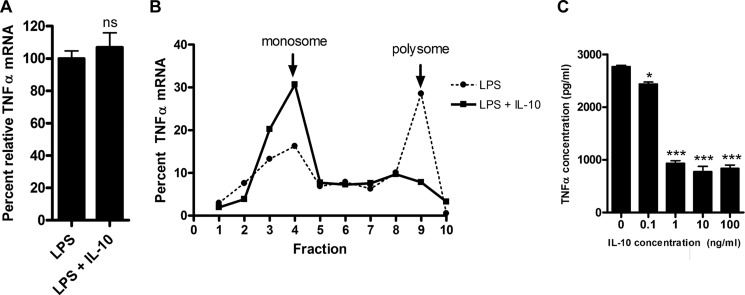
In a previous study, Kontoyiannis et al. (25) reported that the co-addition of IL-10 and LPS reduced the amount of TNFα produced, at least in part through inhibiting TNFα protein translation. Because certain populations of macrophages would be activated by LPS prior to exposure to IL-10 in many physiological settings, we examined whether IL-10 could inhibit TNFα translation if added to LPS-pre-activated macrophages. RAW 264.7 cells were stimulated for 45 min with LPS and then treated for 15 min with IL-10. RNA was isolated under nondenaturing conditions as described by Kontoyiannis et al. (25) and fractionated over a sucrose gradient to separate the heavier polyribosome-associated (polysome) mRNA from the lighter monoribosome-associated (monosome) mRNA. The relative amounts of TNFα mRNA associated with polysomes and monosomes were quantified to determine the amount of rapidly translated TNFα. As shown in Fig. 1A, the addition of IL-10 for 15 min did not change the total amount of TNFα mRNA present, but did shift the TNFα mRNA population completely from polysomes to monosomes (Fig. 1B). This decrease in polysome-associated TNFα mRNA is reflected in decreased amounts of TNFα protein detected in culture supernatants of LPS-pre-activated cells treated with 1–100 ng/ml IL-10 (Fig. 1C).
FIGURE 1.
IL-10 inhibits TNFα synthesis through polysome disassembly in pre-activated cells. A, relative total TNFα mRNA from RAW 264.7 cells pre-activated with 1 ng/ml LPS for 45 min and then treated with 100 ng/ml IL-10 for an additional 15 min. Relative TNFα mRNA was normalized against mouse GAPDH mRNA and then expressed as a percentage against cells that were not treated. Statistical significance between treatments was calculated by Student's unpaired two-tailed t test with 95% confidence. ns, not significant (p = 0.3077). B, TNFα mRNA distribution in total mRNA from A fractionated over a sucrose density gradient. Fraction 10 indicates the bottom of the gradient. mRNA levels were measured by real-time quantitative PCR in each fraction and expressed as a percentage of the sum of the TNFα mRNA signal from all 10 fractions. Results are representative of two independent experiments. C, TNFα protein production in cell supernatants was measured by ELISA from pre-activated cells from A except that cells were treated with a range of IL-10 concentrations. Results are representative of three experiments. Statistical significance between treatments was calculated by one-way analysis of variance with 95% confidence. *, p < 0.05; ***, p < 0.001.
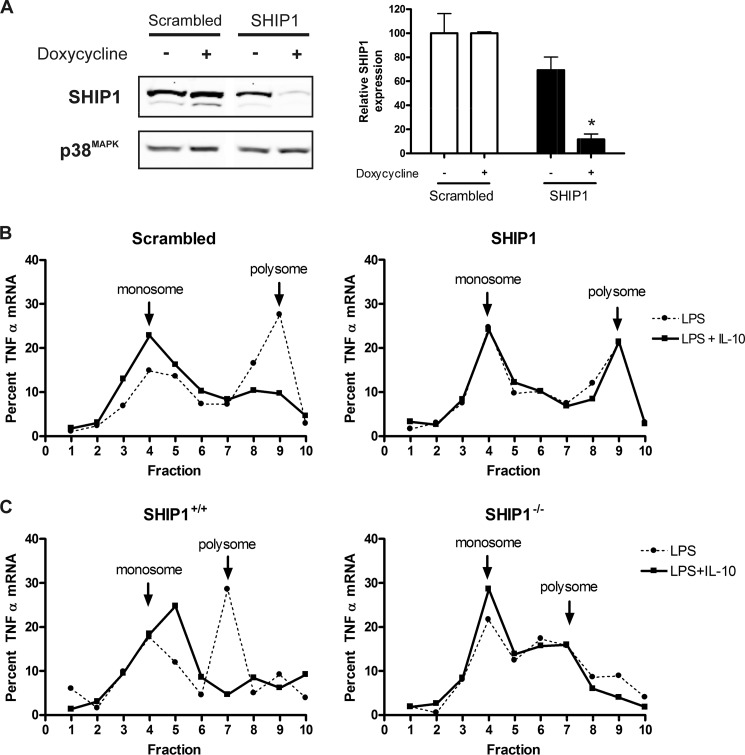
The IL-10 receptor signaling pathway is well known for activation of the STAT3 transcription factor (15–19). However, in other studies, we found that IL-10 also activates the SHIP1 inositol 5′-phosphatase.3 The action of the SHIP1 pathway is predicted to mediate the immediate responses to IL-10 because, unlike a STAT3 transcription factor-dependent pathway, it does not depend on de novo gene transcription (23, 24). We thus examined the ability of IL-10 to shift TNFα mRNA to monosomes in RAW 264.7 cells in which SHIP1 expression had been reduced by siRNA knockdown. siRNA sequences were cloned into the pTRIPZ vector, which expresses siRNA sequences in the context of a microRNA element and in a doxycycline-inducible manner. Fig. 2A shows SHIP1 expression in cells transduced with SHIP1 or scrambled siRNA of a nonspecific sequence and treated with or without doxycycline for 48 h. Densitometry of the SHIP1 bands suggested that <15% of SHIP1 expression remained when cells were treated with doxycycline.
FIGURE 2.
IL-10 regulates TNFα translation through SHIP1 in pre-activated cells. A, siRNA-inducible knockdown of SHIP1. Stably transduced cells carrying siRNA constructs of SHIP1 or a scrambled sequence were placed behind a doxycycline-inducible promoter, and the level of SHIP1 expression was assessed by immunoblotting and densitometric analysis after 48 h of induction. Protein levels were normalized against p38 MAPK, and relative SHIP1 levels were plotted as a percentage of SHIP1 from the scrambled cells. Statistical significance between treatments was calculated by Student's unpaired two-tailed t test with 95% confidence. *, p < 0.05. B, gradient fractionation of total mRNA from scrambled or SHIP1 siRNA knockdown pre-activated cells as described in the legend to Fig. 1. Results are representative of two experiments. C, gradient fractionation of total mRNA from wild-type (SHIP1+/+) or SHIP1 knock-out (SHIP1−/−) pre-activated peritoneal macrophages as described in the legend to Fig. 1. Results are representative of two experiments.
SHIP1 or scrambled siRNA cells were stimulated for 45 min with 1 ng/ml LPS and then treated with 100 ng/ml IL-10 for 15 min, and monosome/polysome-associated TNFα mRNA was analyzed as described above. As shown in Fig. 2B, IL-10 was unable to shift TNFα mRNA to the monosomes in SHIP1 knockdown cells. However, IL-10 shifted TNFα mRNA to the monosomes as efficiently in the scrambled cells as it did in the parental RAW 264.7 cells (Fig. 1). These studies were repeated in primary peritoneal macrophages freshly isolated from wild-type (SHIP1+/+) or SHIP1−/− mice with similar results (Fig. 2C).
IL-10 Inhibits TNFα Translation through SHIP1-mediated Inhibition of Mnk1
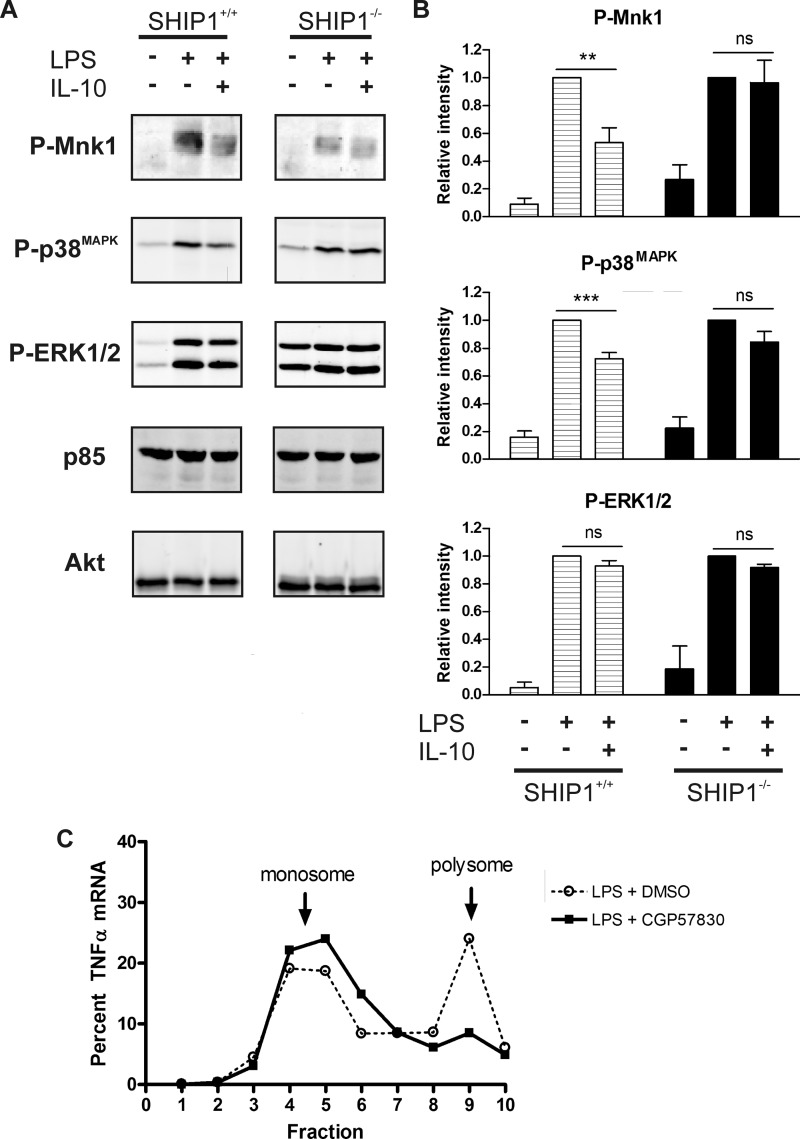
Several groups have shown that LPS induction of TNFα translation in macrophages involves Mnk1 (26–28). We thus tested whether IL-10 inhibits Mnk1 phosphorylation and whether this requires SHIP1. Peritoneal macrophages from wild-type (SHIP1+/+) and SHIP1−/− mice were stimulated with 1 ng/ml LPS with or without 100 ng/ml IL-10 and subjected to immunoblot analyses with antibody specific to phosphorylated Mnk1. As shown in Fig. 3A, IL-10 inhibited Mnk1 phosphorylation in SHIP1+/+ cells, but not in SHIP1−/− cells. Fig. 3B summarizes the densitometry of three to five independent experiments. We also examined the phosphorylation status of the p38 MAPK and ERK1/2 kinases because of their reported role as upstream activators of Mnk1 (29). We found that IL-10 also reduced the LPS-induced phosphorylation of p38 MAPK only in SHIP1+/+ cells (Fig. 3, A and B). ERK1/2 phosphorylation did not appear to be significantly inhibited by IL-10 regardless of the presence of SHIP1 (Fig. 3, A and B). Together, these results suggest that IL-10 regulates Mnk1 through SHIP1 by inhibiting the p38 MAPK pathway.
FIGURE 3.
IL-10 inhibits TNFα translation through SHIP1 via Mnk1. A, immunoblot analysis of cell lysates from peritoneal macrophages isolated from wild-type (SHIP1+/+) or SHIP1 knock-out (SHIP1−/−) mice stimulated with 1 ng/ml LPS in the absence or presence of 100 ng/ml IL-10 for 1 h. Results are representative of three to five experiments. B, densitometric analyses were done by normalizing the intensity to Akt or PI3K p85 protein levels. The average relative intensities from four experiments were then plotted relative to the LPS-only fractions. Statistical significance between LPS ± IL-10 samples was calculated by Student's paired two-tailed t test with 95% confidence. **, p < 0.01; ***, p < 0.001; ns, not significant (p = 0.8280 (phospho-Mnk1), p = 0.1059 (phospho-p38), and p = 0.4069 (phospho-ERK1/2) for SHIP+/+ mice and p = 0.1722 (phospho-ERK1/2) for SHIP−/− mice). C, gradient fractionation of total mRNA from RAW 264.7 cells pretreated with vehicle (dimethyl sulfoxide (DMSO)) or 75 μm Mnk1 inhibitor (CGP57380) as described in the legend to Fig. 1, except without IL-10 treatment.
Finally, we confirmed Mnk1 regulation of TNFα mRNA polysome assembly using the Mnk1 inhibitor CGP57380, which has been previously shown to decrease TNFα induced upon LPS stimulation (26). RAW 264.7 cells were pretreated with 75 μm CGP57380 or dimethyl sulfoxide vehicle for 30 min prior to stimulation with 1 ng/ml LPS for 45 min. mRNAs were then isolated and fractionated as described for Fig. 1. As shown in Fig. 3C, CGP57380 effectively abolished association of TNFα mRNA with polysomes.
SHIP1 Activation Is Sufficient to Inhibit Mnk1 Phosphorylation and TNFα Translation
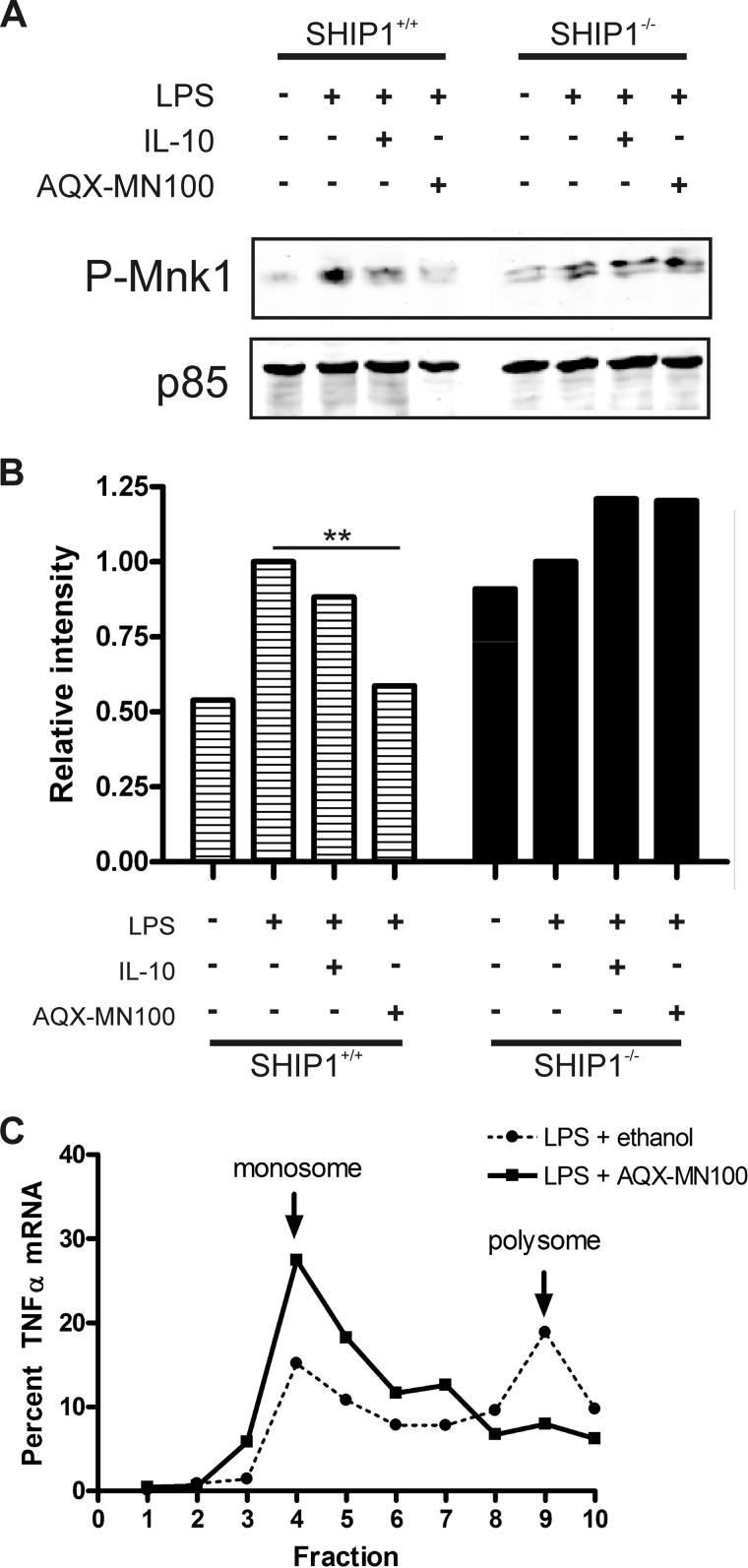
Our data suggest that IL-10 inhibits TNFα mRNA association with polysomes via activation of SHIP1. SHIP1 is an allosterically regulated enzyme, and we have previously described a small-molecule agonist of SHIP1 called AQX-MN100, which binds to the allosteric activation site of SHIP1 (30) and stimulates SHIP1 phosphatase activity. We thus used AQX-MN100 to determine whether SHIP1 activation alone would be sufficient to inhibit LPS-induced phosphorylation of Mnk1 and shift TNFα mRNA from polysomes to monosomes.
SHIP1+/+ and SHIP1−/− peritoneal macrophages were treated with LPS in the absence or presence of IL-10 or AQX-MN100, and cell lysates were analyzed for Mnk1 phosphorylation as described above. As shown in Fig. 4A, both IL-10 and AQX-MN100 reduced LPS-induced Mnk1 phosphorylation in SHIP1+/+ cells. In fact, the inhibitory effect of AQX-MN100 was ∼30% stronger than that of IL-10 (Fig. 4B). However, neither IL-10 nor AQX-MN100 was able to inhibit Mnk1 phosphorylation in SHIP−/− cells. This suggests that SHIP1 activation is required and is sufficient to inhibit Mnk1 phosphorylation. We then examined the effect of AQX-MN100 treatment on TNFα mRNA polysome assembly. Cells were treated with 1 ng/ml LPS for 45 min prior to the addition of 10 μm AQX-MN100 or ethanol vehicle as a control for 15 min. mRNA was isolated and subjected to sucrose density fractionation as described above. Fig. 4C shows that AQX-MN100 treatment shifted the TNFα mRNA from polysomes to monosomes, as observed with IL-10 treatment (Fig. 1).
FIGURE 4.
Small-molecule agonist-activated SHIP1 inhibits TNFα translation through Mnk1. A, immunoblot analysis of peritoneal macrophage cell lysates isolated from wild-type (SHIP1+/+) or SHIP1 knock-out (SHIP1−/−) mice stimulated with 10 ng/ml LPS in the absence or presence of 100 ng/ml IL-10 or 10 μm AQX-MN100 for 1 h. Results are representative of two experiments. B, densitometric analysis of phospho-Mnk1 levels in A normalized to the levels of PI3K p85 protein and plotted relative to the LPS-only samples. Statistical significance between treatments was calculated by one-way analysis of variance with 95% confidence. **, p < 0.01. C, gradient fractionation of total mRNA from pre-activated RAW 264.7 cells as described in the legend to Fig. 1, except with 10 μm AQX-MN100 treatment instead of IL-10 treatment.
DISCUSSION
IL-10 inhibition of TNFα production occurs at the level of transcription, mRNA stability, and translation (17, 21, 25). Studies on the mechanism by which IL-10 exerts its actions suggest that IL-10 activation of the STAT3 transcription factor and induction of STAT3-responsive gene products are sufficient for the anti-inflammatory response of IL-10 (15–19). However, we reasoned that, in a physiological setting, IL-10 target cells are usually already activated prior to exposure to IL-10, so it must also have more immediate mechanisms of action. We surveyed the occurrence of known IL-10 regulatory effects on TNFα mRNA expression when IL-10 is added after the cells are activated. As described previously by Denys et al. (31), IL-10 has only marginal effects on TNFα mRNA transcription if added to cells after LPS addition. However, we did find that IL-10 could efficiently shift the population of TNFα mRNA from polysomes to monosomes even when IL-10 was added to cells already activated by LPS.
Kontoyiannis et al. (25) were the first to show that when IL-10 is added simultaneously with LPS to macrophage cells, the proportion of TNFα mRNA in polysomes is significantly reduced compared with cells treated with LPS alone. Their data also suggest that IL-10 inhibition of p38 MAPK is responsible for the decreased polysome association, a conclusion consistent with the known requirement for p38 MAPK in TNFα translation (32). Subsequently, Mnk1 was discovered to be a p38 MAPK-activated kinase required for TNFα translation in T lymphocytes (5), prompting us to examine the effect of IL-10 on Mnk1. Our data show that IL-10 inhibits the activation of Mnk1 through SHIP1. Of the two known upstream activators of Mnk1, p38 MAPK and ERK1/2, our data suggest that IL-10 inhibition of Mnk1 occurs through SHIP1-dependent inhibition of p38 MAPK activation. The outcome of the SHIP1-dependent inhibition of p38 MAPK and Mnk1 kinases is an immediate decrease in translation of existing TNFα mRNA.
It is important to identify the Mnk1 substrate whose activity is attenuated by IL-10 treatment. The RNA-binding proteins eIF4E and heterogeneous nuclear ribonucleoprotein A1 (hnRNPA1) have been described as being Mnk1 substrates in macrophage or T cells and have been described to have a role in regulating TNFα translation (5, 26). The translation initiation factor eIF4E binds to the 5′-cap structure of mRNAs and the scaffold protein eIF4G to form a translation preinitiation complex along with the small ribosomal subunit (33). Although we and others (26, 27, 34) found that LPS induces Mnk1-dependent phosphorylation of eIF4E at Ser-209, we did not observe IL-10 inhibition of eIF4E phosphorylation (supplemental Fig. 1). However, the role of Ser-209 phosphorylation in eIF4E function is not clear (8, 35). It has been suggested that the phosphorylation of eIF4E is required for oncogenic transformation in cancer cells rather than regulating translation in normal cells, where it appears to be dispensable during normal development (36–38). eIF4E activity is also regulated through binding to the 4E-BP repressor protein to form an inactive complex (39). Phosphorylation of 4E-BP1 by Akt kinase causes release of eIF4E (40). LPS also activates the Akt pathway in macrophages (41), and although we observed increased 4E-BP1 phosphorylation upon LPS stimulation, no changes were observed with IL-10 treatment (supplemental Fig. 2), suggesting that IL-10 does not regulate eIF4E through its binding proteins. The hnRNPA1 protein binds to the TNFα mRNA ARE and has been shown to undergo phosphorylation in response to T cell receptor stimulation in the Jurkat T cell line (5). Phosphorylation reduces the affinity of binding to TNFα mRNA, but like eIF4E, the role of hnRNPA1 in translation is also still not clear (8). However, we did not observe changes in the amount of hnRNPA1 protein binding to the TNF ARE in an RNA pulldown assay of lysates from macrophages treated with LPS in the presence or absence of IL-10 (supplemental Fig. 3). Thus, we do not have evidence that hnRNPA1 is a substrate of Mnk1 in macrophages. Future studies will be directed toward identifying the Mnk1 substrate that mediates LPS-stimulated TNFα mRNA translation and whose action is inhibited by IL-10 signaling. Other potential Mnk1 substrates include ARE-binding proteins that are implicated in regulating translation. These include TIA-1 (42), TIAR (43), CUGBP2 (44), FXR1P (45), and tristetraprolin (46).
In conclusion, we have described a new mechanism by which IL-10 inhibits TNFα expression. This SHIP1-dependent inhibition of Mnk1 and TNFα translation occurs very rapidly and may represent the first response to IL-10. On the other hand, IL-10-induced, STAT3-regulated gene products mediate the subsequent long-term inhibitory effect of IL-10 on target cells. The implication that SHIP1 is involved in regulating the translation of proinflammatory genes such as TNFα also provides a new target for treating inflammatory diseases and disorders.
Supplementary Material
Acknowledgments
We thank Eva So and Sylvia Cheung for critical reading of the manuscript.
This work was supported by Research Grant MOP-84539 from the Canadian Institutes of Health Research (to A. L.-F. M.), and Canadian Cancer Society Research Institute Grant 017289 (to R. J. A.). A. M.-L. held CIHR Doctoral Research, and Michael Smith Foundation for Health Research (MSFHR) awards. The CIHR Transplantation Training Program provided graduate scholarships (to A. M. L., G. B. G., and C. S. C.).

This article contains supplemental “Experimental Procedures” and Figs. 1–3.
A. Ming-Lum, E. So, S. T. Cheung, C. S. Chan, E. MacCarrell, T. Chang, A. Ghanipour, P. Qasimi, L. M. Sly, S. W. Chung, R. J. Anderson, C. J. Ong, G. Krystal, and A. L.-F. Mui, unpublished data.
- ARE
- AU-rich element
- hnRNPA1
- heterogeneous nuclear ribonucleoprotein A1.
REFERENCES
- 1. Chow J. C., Young D. W., Golenbock D. T., Christ W. J., Gusovsky F. (1999) Toll-like receptor-4 mediates lipopolysaccharide-induced signal transduction. J. Biol. Chem. 274, 10689–10692 [DOI] [PubMed] [Google Scholar]
- 2. Kruys V., Kemmer K., Shakhov A., Jongeneel V., Beutler B. (1992) Constitutive activity of the tumor necrosis factor promoter is canceled by the 3′-untranslated region in nonmacrophage cell lines; a trans-dominant factor overcomes this suppressive effect. Proc. Natl. Acad. Sci. U.S.A. 89, 673–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lai W. S., Carballo E., Strum J. R., Kennington E. A., Phillips R. S., Blackshear P. J. (1999) Evidence that tristetraprolin binds to AU-rich elements and promotes the deadenylation and destabilization of tumor necrosis factor-α mRNA. Mol. Cell. Biol. 19, 4311–4323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Piecyk M., Wax S., Beck A. R., Kedersha N., Gupta M., Maritim B., Chen S., Gueydan C., Kruys V., Streuli M., Anderson P. (2000) TIA-1 is a translational silencer that selectively regulates the expression of TNFα. EMBO J. 19, 4154–4163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Buxadé M., Parra J. L., Rousseau S., Shpiro N., Marquez R., Morrice N., Bain J., Espel E., Proud C. G. (2005) The Mnks are novel components in the control of TNFα biosynthesis and phosphorylate and regulate hnRNP A1. Immunity 23, 177–189 [DOI] [PubMed] [Google Scholar]
- 6. Beisang D., Bohjanen P. R. (2012) Perspectives on the ARE as it turns 25 years old. Wiley Interdiscip. Rev. RNA 3, 719–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cargnello M., Roux P. P. (2011) Activation and function of the MAPKs and their substrates, the MAPK-activated protein kinases. Microbiol. Mol. Biol. Rev. 75, 50–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Buxade M., Parra-Palau J. L., Proud C. G. (2008) The Mnks: MAP kinase-interacting kinases (MAP kinase signal-integrating kinases). Front. Biosci. 13, 5359–5373 [DOI] [PubMed] [Google Scholar]
- 9. Flynn A., Proud C. G. (1995) Serine 209, not serine 53, is the major site of phosphorylation in initiation factor eIF4E in serum-treated Chinese hamster ovary cells. J. Biol. Chem. 270, 21684–21688 [DOI] [PubMed] [Google Scholar]
- 10. Joshi B., Cai A. L., Keiper B. D., Minich W. B., Mendez R., Beach C. M., Stepinski J., Stolarski R., Darzynkiewicz E., Rhoads R. E. (1995) Phosphorylation of eukaryotic protein synthesis initiation factor 4E at Ser-209. J. Biol. Chem. 270, 14597–14603 [DOI] [PubMed] [Google Scholar]
- 11. Kühn R., Löhler J., Rennick D., Rajewsky K., Müller W. (1993) Interleukin-10-deficient mice develop chronic enterocolitis. Cell 75, 263–274 [DOI] [PubMed] [Google Scholar]
- 12. Glocker E. O., Kotlarz D., Klein C., Shah N., Grimbacher B. (2012) IL-10 and IL-10 receptor defects in humans. Ann. N.Y. Acad. Sci. 1246, 102–107 [DOI] [PubMed] [Google Scholar]
- 13. Franke A., Balschun T., Karlsen T. H., Sventoraityte J., Nikolaus S., Mayr G., Domingues F. S., Albrecht M., Nothnagel M., Ellinghaus D., Sina C., Onnie C. M., Weersma R. K., Stokkers P. C., Wijmenga C., Gazouli M., Strachan D., McArdle W. L., Vermeire S., Rutgeerts P., Rosenstiel P., Krawczak M., Vatn M. H., IBSEN study group, Mathew C. G., Schreiber S. (2008) Sequence variants in IL10, ARPC2, and multiple other loci contribute to ulcerative colitis susceptibility. Nat. Genet. 40, 1319–1323 [DOI] [PubMed] [Google Scholar]
- 14. Spencer S. D., Di Marco F., Hooley J., Pitts-Meek S., Bauer M., Ryan A. M., Sordat B., Gibbs V. C., Aguet M. (1998) The orphan receptor CRF2-4 is an essential subunit of the interleukin-10 receptor. J. Exp. Med. 187, 571–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Murray P. J. (2006) Understanding and exploiting the endogenous interleukin-10/STAT3-mediated anti-inflammatory response. Curr. Opin. Pharmacol. 6, 379–386 [DOI] [PubMed] [Google Scholar]
- 16. El Kasmi K. C., Holst J., Coffre M., Mielke L., de Pauw A., Lhocine N., Smith A. M., Rutschman R., Kaushal D., Shen Y., Suda T., Donnelly R. P., Myers M. G., Jr., Alexander W., Vignali D. A., Watowich S. S., Ernst M., Hilton D. J., Murray P. J. (2006) General nature of the STAT3-activated anti-inflammatory response. J. Immunol. 177, 7880–7888 [DOI] [PubMed] [Google Scholar]
- 17. Murray P. J. (2005) The primary mechanism of the IL-10-regulated anti-inflammatory response is to selectively inhibit transcription. Proc. Natl. Acad. Sci. U.S.A. 102, 8686–8691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Murray P. J. (2006) STAT3-mediated anti-inflammatory signaling. Biochem. Soc. Trans. 34, 1028–1031 [DOI] [PubMed] [Google Scholar]
- 19. Hutchins A. P., Poulain S., Miranda-Saavedra D. (2012) Genome-wide analysis of STAT3 binding in vivo predicts effectors of the anti-inflammatory response in macrophages. Blood 119, e110–e119 [DOI] [PubMed] [Google Scholar]
- 20. Kuwata H., Watanabe Y., Miyoshi H., Yamamoto M., Kaisho T., Takeda K., Akira S. (2003) IL-10-inducible Bcl-3 negatively regulates LPS-induced TNFα production in macrophages. Blood 102, 4123–4129 [DOI] [PubMed] [Google Scholar]
- 21. Schaljo B., Kratochvill F., Gratz N., Sadzak I., Sauer I., Hammer M., Vogl C., Strobl B., Müller M., Blackshear P. J., Poli V., Lang R., Murray P. J., Kovarik P. (2009) Tristetraprolin is required for full anti-inflammatory response of murine macrophages to IL-10. J. Immunol. 183, 1197–1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. El Kasmi K. C., Smith A. M., Williams L., Neale G., Panopolous A., Watowich S. S., Häcker H., Foxwell B. M., Murray P. J. (2007) Cutting edge: a transcriptional repressor and corepressor induced by the STAT3-regulated anti-inflammatory signaling pathway. J. Immunol. 179, 7215–7219 [DOI] [PubMed] [Google Scholar]
- 23. Kerr W. G. (2011) Inhibitor and activator: dual functions for SHIP in immunity and cancer. Ann. N.Y. Acad. Sci. 1217, 1–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sly L. M., Ho V., Antignano F., Ruschmann J., Hamilton M., Lam V., Rauh M. J., Krystal G. (2007) The role of SHIP in macrophages. Front. Biosci. 12, 2836–2848 [DOI] [PubMed] [Google Scholar]
- 25. Kontoyiannis D., Kotlyarov A., Carballo E., Alexopoulou L., Blackshear P. J., Gaestel M., Davis R., Flavell R., Kollias G. (2001) Interleukin-10 targets p38 MAPK to modulate ARE-dependent TNF mRNA translation and limit intestinal pathology. EMBO J. 20, 3760–3770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Andersson K., Sundler R. (2006) Post-transcriptional regulation of TNFα expression via eukaryotic initiation factor 4E (eIF4E) phosphorylation in mouse macrophages. Cytokine 33, 52–57 [DOI] [PubMed] [Google Scholar]
- 27. Wan Y., Xiao H., Affolter J., Kim T. W., Bulek K., Chaudhuri S., Carlson D., Hamilton T., Mazumder B., Stark G. R., Thomas J., Li X. (2009) Interleukin-1 receptor-associated kinase 2 is critical for lipopolysaccharide-mediated post-transcriptional control. J. Biol. Chem. 284, 10367–10375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rowlett R. M., Chrestensen C. A., Schroeder M. J., Harp M. G., Pelo J. W., Shabanowitz J., DeRose R., Hunt D. F., Sturgill T. W., Worthington M. T. (2008) Inhibition of tristetraprolin deadenylation by poly(A)-binding protein. Am. J. Physiol. Gastrointest. Liver Physiol. 295, G421–G430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Stamou P., Kontoyiannis D. L. (2010) Post-transcriptional regulation of TNF mRNA: a paradigm of signal-dependent mRNA utilization and its relevance to pathology. Curr. Dir. Autoimmun. 11, 61–79 [DOI] [PubMed] [Google Scholar]
- 30. Ong C. J., Ming-Lum A., Nodwell M., Ghanipour A., Yang L., Williams D. E., Kim J., Demirjian L., Qasimi P., Ruschmann J., Cao L. P., Ma K., Chung S. W., Duronio V., Andersen R. J., Krystal G., Mui A. L. (2007) Small-molecule agonists of SHIP1 inhibit the phosphoinositide 3-kinase pathway in hematopoietic cells. Blood 110, 1942–1949 [DOI] [PubMed] [Google Scholar]
- 31. Denys A., Udalova I. A., Smith C., Williams L. M., Ciesielski C. J., Campbell J., Andrews C., Kwaitkowski D., Foxwell B. M. (2002) Evidence for a dual mechanism for IL-10 suppression of TNFα production that does not involve inhibition of p38 mitogen-activated protein kinase or NF-κB in primary human macrophages. J. Immunol. 168, 4837–4845 [DOI] [PubMed] [Google Scholar]
- 32. Ronkina N., Menon M. B., Schwermann J., Tiedje C., Hitti E., Kotlyarov A., Gaestel M. (2010) MAPKAP kinases MK2 and MK3 in inflammation: complex regulation of TNF biosynthesis via expression and phosphorylation of tristetraprolin. Biochem. Pharmacol. 80, 1915–1920 [DOI] [PubMed] [Google Scholar]
- 33. Polunovsky V. A., Bitterman P. B. (2006) The cap-dependent translation apparatus integrates and amplifies cancer pathways. RNA Biol. 3, 10–17 [DOI] [PubMed] [Google Scholar]
- 34. Rowlett R. M., Chrestensen C. A., Nyce M., Harp M. G., Pelo J. W., Cominelli F., Ernst P. B., Pizarro T. T., Sturgill T. W., Worthington M. T. (2008) MNK kinases regulate multiple TLR pathways and innate proinflammatory cytokines in macrophages. Am. J. Physiol. Gastrointest. Liver Physiol. 294, G452–G459 [DOI] [PubMed] [Google Scholar]
- 35. Scheper G. C., Proud C. G. (2002) Does phosphorylation of the cap-binding protein eIF4E play a role in translation initiation? Eur. J. Biochem. 269, 5350–5359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hay N. (2010) Mnk earmarks eIF4E for cancer therapy. Proc. Natl. Acad. Sci. U.S.A. 107, 13975–13976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hou J., Lam F., Proud C., Wang S. (2012) Targeting Mnks for cancer therapy. Oncotarget 3, 118–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ueda T., Watanabe-Fukunaga R., Fukuyama H., Nagata S., Fukunaga R. (2004) Mnk2 and Mnk1 are essential for constitutive and inducible phosphorylation of eukaryotic initiation factor 4E but not for cell growth or development. Mol. Cell. Biol. 24, 6539–6549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Yanagiya A., Suyama E., Adachi H., Svitkin Y. V., Aza-Blanc P., Imataka H., Mikami S., Martineau Y., Ronai Z. A., Sonenberg N. (2012) Translational homeostasis via the mRNA cap-binding protein, eIF4E. Mol. Cell 46, 847–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Gingras A. C., Kennedy S. G., O'Leary M. A., Sonenberg N., Hay N. (1998) 4E-BP1, a repressor of mRNA translation, is phosphorylated and inactivated by the Akt(PKB) signaling pathway. Genes Dev. 12, 502–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Monick M. M., Carter A. B., Robeff P. K., Flaherty D. M., Peterson M. W., Hunninghake G. W. (2001) Lipopolysaccharide activates Akt in human alveolar macrophages, resulting in nuclear accumulation and transcriptional activity of β-catenin. J. Immunol. 166, 4713–4720 [DOI] [PubMed] [Google Scholar]
- 42. Lopez de Silanes I., Galban S., Martindale J. L., Yang X., Mazan-Mamczarz K., Indig F. E., Falco G., Zhan M., Gorospe M. (2005) Identification and functional outcome of mRNAs associated with RNA-binding protein TIA-1. Mol. Cell. Biol. 25, 9520–9531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Gueydan C., Droogmans L., Chalon P., Huez G., Caput D., Kruys V. (1999) Identification of TIAR as a protein binding to the translational regulatory AU-rich element of tumor necrosis factor-α mRNA. J. Biol. Chem. 274, 2322–2326 [DOI] [PubMed] [Google Scholar]
- 44. Mukhopadhyay D., Houchen C. W., Kennedy S., Dieckgraefe B. K., Anant S. (2003) Coupled mRNA stabilization and translational silencing of cyclooxygenase-2 by a novel RNA-binding protein, CUGBP2. Mol. Cell 11, 113–126 [DOI] [PubMed] [Google Scholar]
- 45. Anderson P., Kedersha N. (2007) On again, off again: the SRC-3 transcriptional coactivator moonlights as a translational corepressor. Mol. Cell 25, 796–797 [DOI] [PubMed] [Google Scholar]
- 46. Qi M. Y., Wang Z. Z., Zhang Z., Shao Q., Zeng A., Li X. Q., Li W. Q., Wang C., Tian F. J., Li Q., Zou J., Qin Y. W., Brewer G., Huang S., Jing Q. (2012) AU-rich element-dependent translation repression requires the cooperation of tristetraprolin and RCK/P54. Mol. Cell. Biol. 32, 913–928 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.