Background: The role of Cb1r signaling-mediated activation of Crebh in regulating lipid metabolism and insulin signaling is currently unknown.
Results: Cb1r signaling regulates Lipin1 gene transcription via Crebh and inhibits insulin receptor signaling.
Conclusion: Crebh-mediated activation of Lipin1 by Cb1r deregulates insulin receptor signaling.
Significance: Relieving higher hepatic DAG levels by targeting its upstream regulators might be beneficial to restore insulin signaling.
Keywords: Diacylglycerol, Endocannabinoids, Gene Regulation, Metabolic Regulation, Transcription Regulation
Abstract
Activation of hepatic cannabinoid 1 receptor (Cb1r) signaling has been implicated in the development of phenotypes associated with fatty liver, hypertriglyceridemia, and insulin resistance. In the current study, we have elucidated the critical role of endoplasmic reticulum-bound transcription factor cyclic AMP-response element-binding protein H (Crebh) in mediating activated Cb1r signaling in inducing phosphatidic acid phosphatase Lipin1 gene expression and subsequently deregulating hepatic insulin receptor signaling. Cb1r agonist (2-arachidonoylglycerol (2-AG)) treatment induced Lipin1 gene expression in a Crebh-dependent manner via recruiting CREBH to the endogenous Lipin1 gene promoter. Adenoviral overexpression of Crebh or 2-AG treatment in mice induced Lipin1 gene expression to increase the hepatic diacylglycerol (DAG) level and phosphorylation of protein kinase Cϵ (PKCϵ). This in turn inhibited hepatic insulin receptor signaling. Knockdown of Crebh or Cb1r antagonism attenuated 2-AG-mediated induction of Lipin1 gene expression and decreased DAG production in mouse liver and subsequently restored insulin receptor signaling. Similarly, knockdown of Lipin1 attenuated the 2-AG-induced increase in the DAG level and PKCϵ phosphorylation. Finally, shRNA-mediated knockdown of Crebh partially but significantly blunted Lipin1 expression and the DAG level in db/db mice. These results demonstrate a novel mechanism by which Cb1r signaling induces Lipin1 gene expression and increases DAG production by activating Crebh, thereby deregulating insulin receptor signaling pathway and lipid homeostasis.
Introduction
The endocannabinoid system comprises the cannabinoid receptors type 1 (Cb1r),4 which is expressed at high levels in the brain but is also present at much lower concentrations in peripheral tissues, and Cb2r, which is expressed predominantly in immune and hematopoietic cells (1). Arachidonoyl ethanolamide (anandamide) and 2-arachidonoylglycerol (2-AG) are endocannabinoids and lipid mediators that activate these receptors. Various studies in rodent models (2) and patients (3, 4) have provided evidence of hyperactivity of the endocannabinoid system and its correlation to obesity due to impaired energy balance. Mice deficient in Cb1r are resistant to diet-induced obesity and steatosis, whereas in wild-type mice, chronic treatment with Cb1r antagonists reverses these diet-induced effects (5). Both genetically and diet-induced obese animal models show elevated levels of endocannabinoids in the hypothalamus and peripheral tissues (1, 2, 5). Recently, using a liver-specific Cb1r knock-out mouse model, it was demonstrated that peripheral Cb1r could be selectively targeted for the treatment of fatty liver, impaired glucose homeostasis, and dyslipidemia to reduce the neuropsychiatric side effects of nonselective Cb1r signaling blockade in treatment of obesity-associated conditions (5). Overall, both clinical (3, 4, 6) and animal data regarding the Cb1r blockade (2, 5) overwhelmingly suggest the beneficial actions of Cb1r antagonism on lipid metabolism and insulin receptor signaling.
Endoplasmic reticulum-bound liver-specific transcription factor cyclic AMP-response element-binding protein H (CREBH) has been reported to be induced by the acute inflammatory response-inducing factor lipopolysaccharide and proinflammatory cytokines interleukin-6 and tumor necrosis factor α (7). Previously, we have reported that Crebh plays an important mediatory role in hormonal regulation of hepatic gluconeogenesis under fasting or insulin-resistant conditions in rodent liver and demonstrated that Crebh is dramatically induced and activated in diet-induced obese rodent models (8). In a recent study, we found that Cb1r signaling activates Crebh via the c-Jun N-terminal kinase pathway and elucidated the molecular mechanism involved in Cb1r signaling-mediated regulation of hepatic gluconeogenesis (9). Crebh has also been demonstrated to be involved in the regulation of hepatic iron (10) and triglyceride metabolism (11). Crebh gene expression is known to be regulated by nuclear receptors HNF4α and peroxisome proliferator-activated receptor α as well as fatty acids, thereby suggesting a possible role of Crebh in fatty acid metabolism (12, 13). Because of its liver-specific expression and stress sensor activation, Crebh is emerging as a key player in regulating various hepatic metabolic pathways.
Lipin1 plays a crucial role in lipid metabolism in adipose tissue, skeletal muscle, and liver. Lipin1 was identified as a cytosolic phosphatidic acid phosphatase that generates DAG in response to an increase in intracellular free fatty acid levels (14). Interestingly, Lipin1-deficient mice have a reduced adipose tissue mass, mild hyperglycemia, and insulin resistance, whereas enhanced Lipin1 expression in the adipose tissue or skeletal muscle of transgenic mice leads to obesity (15). Lipin1-deficienct mice display lipodystrophy-associated insulin resistance perhaps due to the lack of adipokine generation, showing the importance of adipose-specific Lipin1 function in lipid homeostasis and systemic insulin receptor signaling (15, 16). However, the role of Lipin1 in liver is complex as contradicting evidence suggests that LIPIN1 functions as a transcriptional coactivator for peroxisome proliferator-activated receptor α/PGC-1α to regulate fatty acid oxidation gene expression, and LIPIN1 has also been demonstrated to contribute to the regulation of triglyceride synthesis and VLDL secretion (17). It has been reported that the glucocorticoid receptor regulates the mouse Lipin1 promoter by the synthetic glucocorticoid dexamethasone (14) and that regulation of Lipin1 gene expression is mediated by sterol (18) and estrogen-related receptor γ (19), thereby suggesting the involvement of certain factor(s) affecting lipogenesis and adipogenesis by modulating Lipin1 gene expression.
Although Cb1r signaling has been known to affect lipid metabolism, the molecular mechanism behind this phenomenon is still unclear. Therefore, in the current study, we investigated the effect of Cb1r signaling pathway on lipid metabolism and insulin receptor signaling and the mediatory role of CREBH in this context. Our study delineates an unprecedented link by which Cb1r signaling pathway-induced activation of Crebh regulates Lipin1 gene expression and hepatic insulin receptor signaling.
EXPERIMENTAL PROCEDURES
Animals
Seven-week-old C57BL/6J (B6) and diabetic db/db mice (The Jackson Laboratory) were used. Mice were treated with a single dose of 2-AG (Tocris; 5 mg/kg intraperitoneally; 12 h) or pretreated with a single dose of AM251 (Tocris; 5 mg/kg intraperitoneally; 12 h) followed by 2-AG treatment. Delivery of recombinant adenoviruses (100 multiplicity of infection/virus) was performed via tail vein injection. For adenoviral overexpression, mice were sacrificed 72 h following viral delivery. For adenovirus-mediated knockdown experiments, mice were sacrificed 96 h after viral delivery. Following completion of experiments, liver tissues were collected for total RNA isolation or protein extraction and DAG measurement. All animal experiments were approved by the Institutional Animal Use and Care Committee of the Korea Research Institute of Bioscience and Biotechnology and performed in accordance with the United States National Institutes of Health Guidelines for Animal Experiments (NIH Publication No. 85-23, revised 1996).
Plasmids
Mouse Lipin1 gene promoter serial deletion-luciferase constructs and expression vectors (pcDNA3-FLAG) encoding CREBH-N, ATF6-N, and dominant negative CREBH have been described previously (8, 9, 20). CREBH response element mutant-luciferase constructs were cloned and confirmed by DNA sequencing.
Cell Culture, Isolation of Primary Rat Hepatocytes, and Adenoviral Infection
Transient transfection assays were performed in AML12 cells using Lipofectamine 2000 reagent (according to the manufacturer's protocol) with the indicated treatments as described previously (8, 9). Primary rat hepatocytes were prepared from 200–250-g Sprague-Dawley rats by a collagenase perfusion method as described previously (8, 9). For adenoviral infections, cells were washed with PBS and left for 2–3 h in serum-free medium containing the appropriate amount of viral particles (100 multiplicity of infection/virus). Medium was replaced with fresh growth medium for an additional 36–72 h before any treatment. All adenoviruses used in this study have been described previously (8, 20).
Semiquantitative PCR, Quantitative PCR (qPCR), and Western Blot Analysis
Total RNA from either primary hepatocytes or liver tissues was extracted using an easy-spin RNA extraction kit (Intron Biotechnology). cDNAs were generated by Superscript II enzyme (Invitrogen) and analyzed by semiquantitative PCR (1% agarose gel stained with EtBr) and qPCR using a SYBR Green PCR kit and Rotor Gene 6000 Real Time System (Corbett Life Science). All data were normalized to β-actin expression. For Western blot analysis, cell lysates were prepared from primary rat hepatocytes or liver tissues of experimental animals, and Western blotting was performed using the indicated antibodies.
Chromatin Immunoprecipitation (ChIP) Assay
The ChIP assay was performed according to the manufacturer's protocol (Upstate). Immunoprecipitation was performed using polyclonal anti-CREBH antibody (Orbigen) or IgG (as a negative control). After recovering DNA, qPCR was performed using primers encompassing the mouse Lipin1 promoter (−531/−256 and −1010/−846 bp) region.
Measurement of DAG Level
Liver tissues were processed for liquid chromatography/mass spectrometric determination of the DAG level as described previously (20).
Statistics
Values are expressed as mean ± S.E. Statistical significance was calculated using an unpaired Student's t test and one-way analysis of variance. Differences were considered significant at p ≤ 0.05.
RESULTS
Activated Cb1r Signaling and Crebh Induce Hepatic Lipin1 Gene Expression in Vivo
In a recent study, we demonstrated that activated Cb1r signaling induces Crebh gene expression and generates the active form of CREBH (9). We have also shown that Crebh plays an important role in the regulation of hepatic gluconeogenesis and is hyperactive under insulin resistance conditions (8). In previous studies, it has been shown that chronic alcohol exposure increases the hepatic endocannabinoid 2-AG but not the arachidonoyl ethanolamide level to activate Cb1r signaling, which subsequently leads to an aberrant increase in hepatic lipogenesis, resulting in fatty liver (5). Therefore, to dissect the role of Cb1r and Crebh in lipid metabolism, we initially treated mice with 2-AG or infected mice with adenovirus (Ad) CREBH-N (active form of CREBH) and analyzed the expression of key genes involved in lipid metabolism (Table 1). 2-AG treatment led to an increase in several genes involved in this pathway (Srebp1c, mtGPAT1, Cd36, Dgat1, Dgat2, and Lipin1) along with a significant decrease in genes involved in fatty acid oxidation (Cpt1 and Ucp2). Interestingly, Crebh overexpression showed significant up-regulation of Lipin1 and to some extent Dgat2 mRNA levels, whereas the expression of several other genes was not affected under this condition. This indicated a common role of activated Cb1r signaling and Crebh in regulating Lipin1 gene expression.
TABLE 1.
Key genes involved in lipid metabolism
Mice (n = 4–5/group) were treated with 2-AG for 12 h or were infected with the indicated adenoviruses (100 multiplicity of infection/virus) for 72 h. Following completion of the experiments, mice were sacrificed, and liver tissues were obtained for qPCR analyses of the indicated genes. Acc1, acetyl-coenzyme A carboxylase 1; Scd1, stearoyl-CoA desaturase; ChREBP, carbohydrate-response element-binding protein.
| Genes | mRNA level (-fold change) |
|
|---|---|---|
| 2-AG vs. control | Ad-CREBH vs. Ad-GFP | |
| Srebp1c | 3.28a | 0.83 |
| Acc1 | 1.34 | 0.79 |
| Scd1 | 0.87 | 1.13 |
| ChREBP | 1.11 | 1.03 |
| mtGPAT1 | 1.43a | 0.92 |
| Cpt1 | 0.66a | 0.79 |
| Ucp2 | 0.53a | 0.81 |
| Cd36 | 2.11a | 1.09 |
| Dgat1 | 1.76a | 1.12 |
| Dgat2 | 2.54a | 1.97a |
| Lipin1 | 5.71a | 4.43a |
| Lipin2 | 1.21 | 0.73 |
| Crebh | 5.91a | 19.28a |
a p < 0.05 versus control or Ad-GFP.
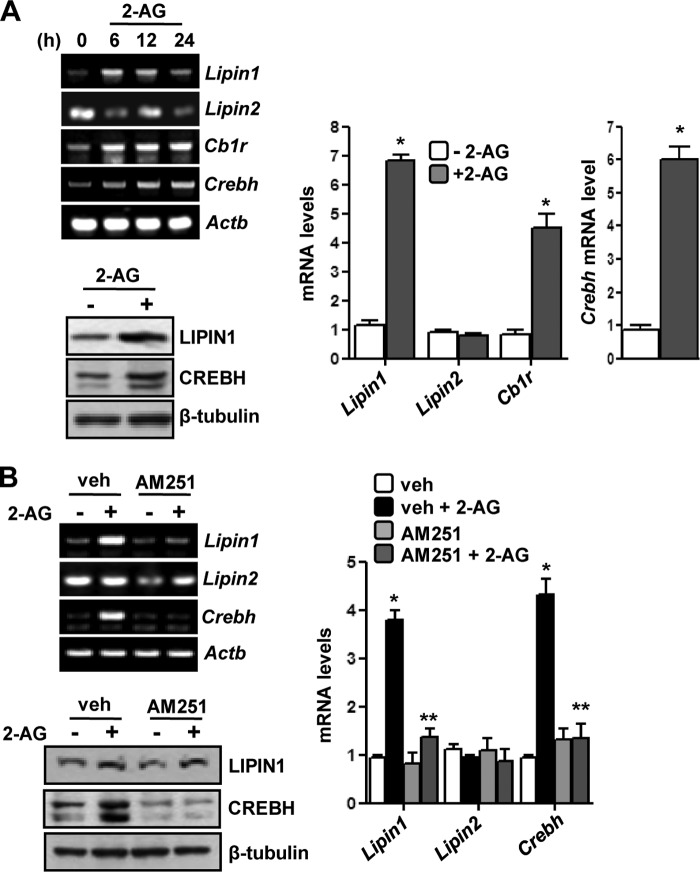
Next, we tried to systematically analyze the effect of 2-AG on Lipin1 gene regulation. A time course 2-AG treatment in mice showed significant induction of Lipin1 but not Lipin2 mRNA levels along with an increase of Cb1r and Crebh mRNA levels as observed from semiquantitative and qPCR analyses (Fig. 1A, left top and right). Similarly, 2-AG treatment also increased the LIPIN1 protein level and generated the active form of CREBH (Fig. 1A, left bottom). To confirm that 2-AG-induced activation of Cb1r signaling pathway induces Crebh and Lipin1 gene expression, we used Cb1r antagonist AM251 pretreatment in mice exposed to 2-AG. Pretreatment with AM251 significantly attenuated the 2-AG-mediated induction of Crebh as well Lipin1 mRNA and protein levels (Fig. 1B), and as expected, no changes were observed in the mRNA level of Lipin2 gene. Overall, these results indicate a connection between Cb1r signaling-mediated activation of Crebh and induction of Lipin1 gene expression in vivo.
FIGURE 1.
Activated Cb1r induces Lipin1 gene expression via Crebh. A, mice (n = 5 per group) were treated with 2-AG at the indicated times or for 12 h, and liver tissues were obtained for semiquantitative PCR (top left) or qPCR (right) analyses and for measuring protein levels (bottom left). *, p < 0.05 versus untreated control. B, mice (n = 5 per group) were treated with 2-AG for 12 h or treated with AM251 for 12 h preceding 2-AG treatment for a further 12 h, and liver tissues were obtained for semiquantitative PCR (top left) or qPCR (right) analyses and for measuring protein levels (bottom left).*, p < 0.05 versus control; **, p < 0.05 versus vehicle (veh) + 2-AG. All data represent mean ± S.E. (error bars).
CREBH Is a Transcriptional Regulator of Hepatic Lipin1
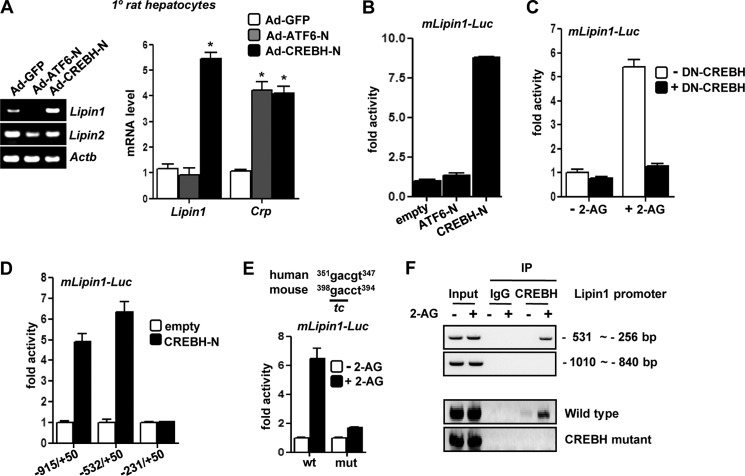
To elucidate the role of Crebh in 2-AG-mediated induction of Lipin1 gene expression, we sought to investigate whether Crebh directly regulates Lipin1 gene transcription. Initially, to confirm the specificity of Crebh-mediated activation of Lipin1 gene expression, we infected primary rat hepatocytes with adenoviruses encoding active forms of CREBH (CREBH-N) and ATF6 (ATF6-N), a member of the CREB/ATF family closely related to CREBH. Overexpression of Crebh significantly and specifically induced Lipin1 gene expression, whereas overexpression of Atf6 led to no significant change in the Lipin1 mRNA level (Fig. 2A). However, both Crebh and Atf6 overexpression significantly induced the C-reactive protein (Crp) mRNA level, a previously reported common target of both Crebh and Atf6. This indicates that the adenoviruses were in proper working conditions. Next, to verify the role of Crebh in regulating Lipin1 at the transcriptional level, we performed transient transfection using the Lipin1 promoter in AML12 cells. Similar to the induction of the Lipin1 mRNA level, overexpression of Crebh activated the Lipin1 gene promoter, whereas Atf6 overexpression failed to show any significant activation (Fig. 2B). Overall, these results suggest that Crebh specifically regulates Lipin1 gene transcription.
FIGURE 2.
Crebh is a transcriptional regulator of Lipin1 gene expression. A, primary (1°) rat hepatocytes were infected with adenoviruses encoding GFP, FLAG-ATF6-N, and FLAG-CREBH-N for 48 h. RNA was extracted for semiquantitative PCR (left) and qPCR analyses (right). *, p < 0.05 versus Ad-GFP. B–E, transient transfection assays were performed in AML12 cells with mLipin1-luc reporter constructs along with co-transfection of the indicated expression vectors (B–D) as indicated or treatment with 2-AG for 12 h (B and E) prior to luciferase assays. -Fold activity represents relative luciferase activity:β-gal activity. All transfections were performed in triplicates and are representative of three to four independent experiments. F, untransfected AML12 cells (top) or AML12 cells transfected with wild type or CREBH mutant constructs of mLipin1-Luc (bottom) were treated with vehicle or 2-AG for 12 h, and immunoprecipitation (IP) of AML12 chromatin from cells exposed to vehicle or 2-AG was performed with IgG or CREBH antibody. The percentage of DNA immunoprecipitated with CREBH antibody relative to input chromatin was demonstrated by semiquantitative PCR analysis. *, p < 0.05 versus control. All data represent mean ± S.E. (error bars) of at least three independent experiments. DN, dominant negative.
Next, we investigated the mediatory role of Crebh in 2-AG activation of the Lipin1 promoter. Co-transfection of the Lipin1 promoter with plasmid encoding Crebh cDNA lacking the transcriptional activation domain (dominant negative CREBH) was done in AML12 cells, and then cells were treated with 2-AG (Fig. 2C). 2-AG significantly activated the Lipin1 promoter under normal conditions but failed to activate the Lipin1 promoter in cells co-transfected with dominant negative CREBH, thereby suggesting that Crebh plays a major mediatory role in activating Lipin1 gene transcription by Cb1r signaling pathway.
Next, using several deletion constructs, we tried to map the CREBH-responsive region in the Lipin1 gene promoter. Deletion mapping results demonstrated that the putative CREBH-responsive region lies within −531 to −231 bp of the transcription start site of the Lipin1 promoter (Fig. 2D). We identified a putative binding region of CREBH (Fig. 2E). 2-AG treatment significantly activated the wild type (WT) Lipin1 promoter construct but failed to activate the CREBH mutant Lipin1 promoter. Similarly, 2-AG treatment led to the recruitment of endogenous CREBH to the endogenous Lipin1 gene promoter (Fig. 2F, top), and this increased occupancy of endogenous CREBH on the Lipin1 gene promoter upon 2-AG treatment was mitigated in the CREBH mutant Lipin1 promoter construct as demonstrated by ChIP assays (Fig. 2F, bottom). These results confirm that induction of Lipin1 by Cb1r signaling is mediated via Crebh.
Knockdown of Crebh Attenuates Lipin1 Induction and Decreases DAG Production by Hepatic Cb1r Signaling in Vivo
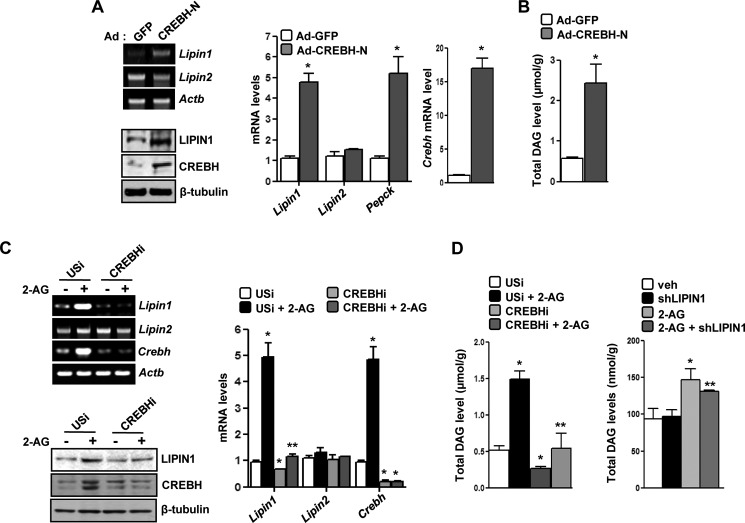
Previous reports have identified Lipin1 as a mammalian cytosolic phosphatidic acid phosphatase in various cell types and demonstrated that Lipin1 increases the hepatic DAG level (20). Adenoviral overexpression of CREBH in mice resulted in a significant increase of the Lipin1 mRNA and protein levels comparable with another Crebh target, phosphoenolpyruvate carboxykinase (Pepck) gene expression, in vivo (Fig. 3A) and also led to a significant increase in the DAG level (∼2.5-fold) (Fig. 3B). As expected, Lipin2 gene expression showed no significant change in this condition.
FIGURE 3.
Crebh-mediated induction of Lipin1 gene expression and increase in hepatic DAG level in vivo. A and B, mice (n = 5) were infected with the indicated adenoviruses for 72 h. Following completion of the experiments, mice were sacrificed, and liver tissues were obtained for semiquantitative PCR (top left) or qPCR (right) analyses and for measuring protein levels (bottom left). The DAG level was measured in Ad-GFP- and Ad-CREBH-N-infected mouse liver samples (B). *, p < 0.05 versus Ad-GFP. C, mice (n = 5) were infected with the indicated adenoviruses, and 96 h postinfection, mice were treated with 2-AG for a further 12 h. Liver tissues were obtained for semiquantitative PCR (top left) or qPCR (right) analyses and for measuring protein (bottom left). *, p < 0.05 versus USi; **, p < 0.05 versus USi + 2-AG. Data represent mean ± S.E. D, mice (n = 5; left) or AML12 cells (right) were infected with the indicated adenoviruses, and 96 h postinfection, mice were treated with 2-AG for a further 12 h. The DAG level was measured in adenovirus-infected mouse liver samples (left) and in AML12 cells (right). *, p < 0.05 versus USi/vehicle (veh); **, p < 0.05 versus USi/vehicle + 2-AG. All data represent mean ± S.E. (error bars).
Next, to ascertain the role of activated Cb1r signaling and the mediatory role of Crebh in this context, we overexpressed short hairpin RNA (shRNA) adenoviruses encoding unspecific RNAi (USi) and Crebh RNAi (CREBHi) in mice preceding 2-AG treatment. 2-AG treatment led to significant induction of Lipin1 and Crebh mRNA and protein levels in USi-infected livers but failed to substantially induce Lipin1 mRNA or protein in CREBHi-infected livers (Fig. 3C). Similarly, DAG production was significantly higher in 2-AG-treated mice compared with the vehicle-treated mice under the USi-infected condition but was dramatically reduced to the basal level under the CREBHi-infected condition (Fig. 3D, left). Indeed, CREBHi-infected mouse livers produced lower amounts of DAG compared with USi-infected littermates (Fig. 3D, left). To confirm the involvement of Lipin1 in Cb1r/Crebh-mediated regulation of DAG production, AML12 cells were infected with adenovirus encoding Lipin1 RNAi (shLipin1) preceding 2-AG treatment. 2-AG treatment led to an increase of the DAG level (∼60% compared with untreated cells). Similarly to CREBHi, Lipin1 knockdown led to a significant decrease in the total DAG level compared with 2-AG-treated cells (∼15% decrease from 2-AG-treated cells), thereby indicating the dependence of the Cb1r/Crebh pathway on Lipin1 to regulate DAG production (Fig. 3D, right). These observations indicate the crucial role played by Crebh in mediating the effect of Cb1r signaling on Lipin1 gene regulation and DAG production.
Cb1r Signaling Pathway Inhibits Hepatic Insulin Receptor Signaling via Crebh-mediated Induction of Lipin1
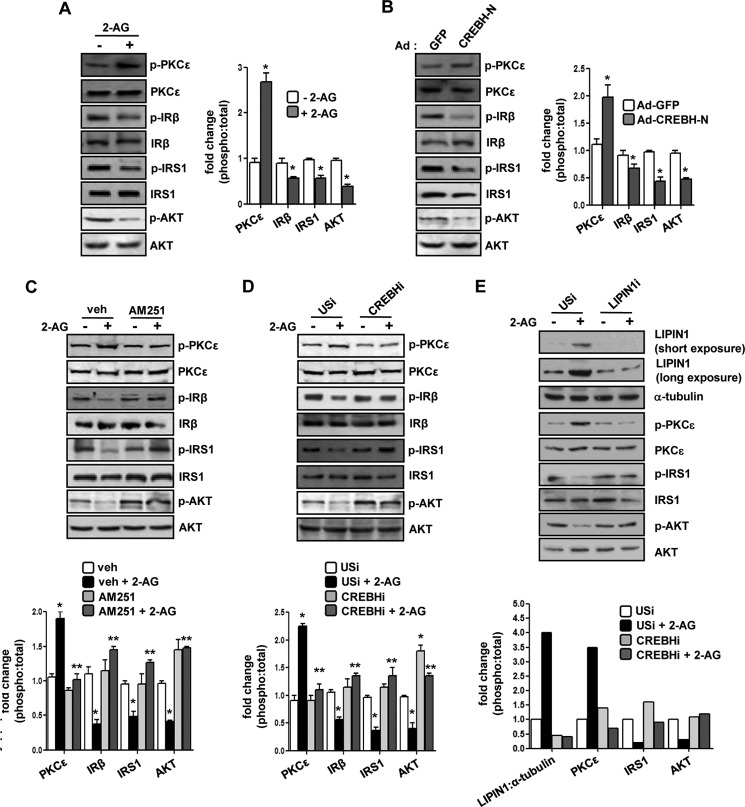
It has been demonstrated previously that increased production of DAG is linked to obesity-related insulin resistance in peripheral tissues (21, 22). Therefore, we sought to investigate the effect of Cb1r-mediated induction of Crebh and Lipin1 gene expression and increased DAG production on hepatic insulin receptor signaling in vivo. Both 2-AG treatment and Crebh overexpression in mice showed a remarkable increase in Ser-729 phosphorylation of PKCϵ, a major non-canonical isoform of PKC found in the liver. In addition, there was a significant reduction in the phosphorylation of insulin receptor β and its downstream signaling components, Tyr-989 phosphorylation of insulin receptor substrate 1 (IRS1) and Ser-473 phosphorylation of AKT, upon these treatments (Fig. 4, A and B). These results indicate that increased expression of Lipin1 in these conditions could promote deregulation of insulin signaling in the liver.
FIGURE 4.
Crebh-mediated induction of Lipin1 gene expression is linked to perturbation of hepatic insulin receptor signaling in vivo. A–E, experiments were performed as described in Fig. 1 (for A and C) or Fig. 3 (for B, D, and E), and liver tissues (A–D) or AML12 cell lysates (E) were obtained for Western blot analysis with the indicated antibodies. Protein levels were quantified by densitometry analysis (phospho:total form). *, p < 0.05 versus control and Ad-GFP; **, p < 0.05 versus vehicle (veh) + 2-AG and USi + 2-AG. All data represent mean ± S.E. (error bars). IRβ, insulin receptor β.
Therefore, we next assessed the phosphorylation status of these signaling components upon Cb1r antagonism or knockdown of Crebh (Fig. 4, C and D). Consistent with our earlier results that showed reduced gene expression of Lipin1 and a decrease in the DAG level under these conditions, we found that AM251 pretreatment or knockdown of Crebh significantly attenuated 2-AG-mediated activation of PKCϵ and led to a significant increase in insulin receptor β, IRS1, and AKT phosphorylation compared with 2-AG-treated mice (Fig. 4, C and D). Furthermore, to reconfirm that the 2-AG-induced deregulation of insulin receptor signaling pathway is mediated by Lipin1, the phosphorylation status of the above mentioned signaling kinases was assessed in AML12 cells upon knockdown of Lipin1 preceding 2-AG treatment (Fig. 4E). Similar to the effect of Crebh knockdown, Lipin1 knockdown significantly attenuated 2-AG-induced activation of PKCϵ and released the inhibitory effect of 2-AG on the IRS1 and AKT phosphorylation status, thereby suggesting that the 2-AG-induced deregulation of the insulin signaling pathway is mediated via Lipin1. Overall, these results indicate that the blockade of Crebh significantly attenuates Cb1r-mediated deregulation of hepatic insulin signaling via a reduction in Lipin1 gene expression and decreased DAG production in vivo.
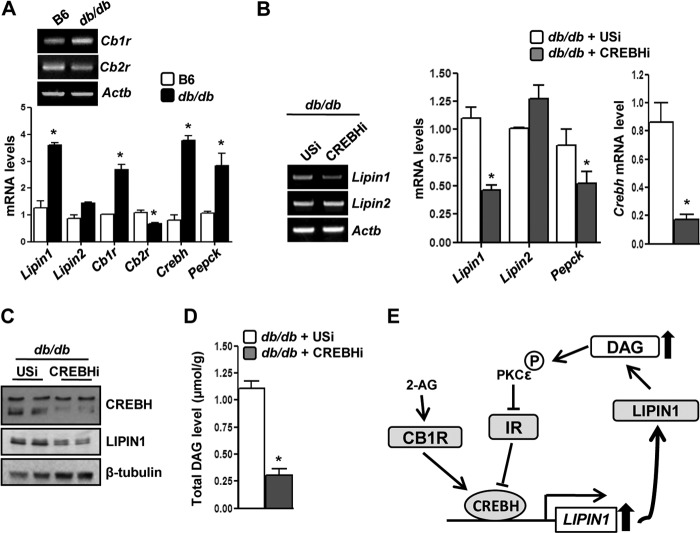
Knockdown of Crebh Attenuates Induction of Lipin1 Expression and Decreases DAG Production in Insulin Resistance Condition
Initially, we observed that Crebh and Lipin1 gene expression was significantly higher in db/db mice compared with normal mice (Fig. 5A). A recent report (5) demonstrated that Cb1r gene expression follows a pattern similar to that of Crebh and Lipin1 in the diet-induced obesity condition. Interestingly, we found that Cb1r gene expression was also significantly higher in db/db mice compared with normal mice, whereas Cb2r gene expression was significantly lower under similar conditions (Fig. 5A). These results indicate a constitutively activated status of the Cb1r signaling pathway in the insulin-resistant condition. Therefore, to ascertain the significance of Crebh-mediated induction of Lipin1 gene expression and the increase in DAG production from a physiological perspective, we overexpressed Crebh RNAi in db/db mice. Crebh knockdown resulted in a significant decrease in the Lipin1 mRNA and protein levels (Fig. 5, B and C), and as a consequence, there was a dramatic reduction in the DAG level of db/db mice under this condition (Fig. 5D). However, the phosphorylation status of the components of the insulin signaling pathway showed marginal changes (data not shown) upon Crebh knockdown. Overall, our results provide a novel link connecting hepatic Cb1r signaling with deregulation of the insulin receptor signaling pathway and identify Crebh as a crucial player in regulating lipid metabolism via Lipin1 gene induction in vivo.
FIGURE 5.
Knockdown of Crebh reverses perturbation of hepatic insulin receptor signaling in db/db mice. A, semiquantitative PCR (top) or qPCR (bottom) analyses of gene expression in B6 or db/db mice (n = 4–5). *, p < 0.05 versus B6. B–D, semiquantitative PCR (B, left) or qPCR (B, right) analyses, protein levels (C), and DAG levels (D) were measured in db/db mice (n = 5) infected with Ad-USi or Ad-CREBH RNAi. *, p < 0.05 versus USi. All data represent mean ± S.E. (error bars). E, proposed model depicting the effect of endocannabinoid-mediated activation of Cb1r in inducing and activating CREBH to disrupt hepatic insulin signaling via induction of Lipin1 gene expression and by increasing DAG production. IR, insulin receptor.
DISCUSSION
Crebh has been demonstrated to play an important role in hepatic gluconeogenesis, triglyceride metabolism, and iron metabolism by transcriptional regulation of key enzyme genes involved in those metabolic pathways (8–11). In our current study, we investigated and suggest a novel role of Crebh in regulating hepatic lipid metabolism by transcriptional activation of the Lipin1 gene. Cb1r-mediated activation of CREBH and Lipin1 induction lead to increased DAG production in vivo and phosphorylation of PKCϵ. Activation of PKCϵ by this signaling cascade imparts insulin resistance effects via disruption of the insulin receptor signaling pathway. Antagonism of Cb1r signaling repressed the activation of CREBH and its downstream target Lipin1, and Cb1r activation under Crebh-deficient conditions failed to induce the Lipin1-mediated increase in DAG production, ultimately leading to recovery of insulin receptor signaling component activity. Hepatic Cb1r, Crebh, and Lipin1 gene expression is higher in various models of insulin resistance (5, 8, 20). Conversely, in our previous study, we demonstrated that insulin treatment (or AKT co-transfection) leads to diminished Crebh mRNA levels (as well as promoter activity) (8). Knockdown of Crebh in an insulin-resistant rodent model (db/db mice) also showed significant lowering of the pre-existing high DAG level. Overall, our current study unravels a connection among Cb1r, Crebh, and Lipin1 and sheds light onto a novel molecular mechanism by which activation of CB1R leads to deregulation of the insulin signaling pathway in the liver (a schematic model is shown in Fig. 5E), indicating that targeted disruption of hepatic Cb1r signaling and CREBH activity might provide plausible therapeutic approaches to restore proper functioning of insulin receptor signaling.
The rate of de novo hepatic lipogenesis was increased by Cb1r agonists and decreased by Cb1r antagonists in rodents. Adipose tissue may be the source of liver fat as it has been demonstrated that activation of Cb1r in adipocytes promotes lipogenesis, and the released fatty acids are converted to triglycerides by the liver (1, 2, 4). Conversely, Cb1r blockade with antagonists leads to reduction of hepatic triglycerides with an increased rate of secretion of triglyceride-rich VLDL from the liver of insulin-resistant rodents. However, because of the deleterious side effects of Cb1r antagonists, it is essential to elucidate the Cb1r signaling pathway in detail to unravel downstream targets for possible therapeutic benefits. In this context, our study demonstrates the involvement of the endoplasmic reticulum-bound transcription factor Crebh in a mediatory role and may explain in part the molecular basis of Cb1r-mediated up-regulation of lipogenesis and DAG production that ultimately causes insulin resistance via deregulation of the insulin receptor signaling. Recently, it has been shown that chronic ethanol feeding increases the endocannabinoid 2-AG but not the arachidonoyl ethanolamide level in mouse liver, and this increase was followed by a robust change in the lipogenic gene program with a significant increase in gene expression of Srebp1c and its targets, whereas AMP-activated protein kinase and fatty acid oxidation were considerably attenuated under these conditions (5). In our previous study, we found that 2-AG treatment also led to increased CREBH activity (9), and in insulin resistance conditions, both are expressed at a higher level (5, 8). Therefore, we presumed that Crebh might mediate some of the effects associated with Cb1r activation by regulating the expression of some key genes involved in lipogenesis and the triglyceride synthesis pathway. However, our results (Table 1) demonstrated that Cb1r and Crebh have common targets further downstream in the lipogenic program and confirmed that Lipin1, a key gene involved in DAG production as well as perturbation of hepatic insulin signaling, is the common mediator of activated Cb1r and Crebh effects.
Recent reports have demonstrated the role of Lipin1 in promoting insulin resistance via enhanced DAG production and PKCϵ activation, triglyceride formation, and VLDL secretion in the liver (20). CREB/CRTC2 was identified as a regulator of Lipin1 gene transcription under fasting-refeeding and diet-induced obesity conditions to explain the role of Lipin1 in insulin resistance (20). Interestingly, ethanol is also known to increase the hepatic DAG level and promote insulin resistance, although the underlying molecular mechanism was not clearly established (23). In this context, an important observation was that chronic ethanol feeding increases the endocannabinoid 2-AG but not the arachidonoyl ethanolamide level in mouse liver (5). Thus, in accordance with our previous report (9), 2-AG-mediated activation of CREBH and CREBH-mediated transcriptional activation of Lipin1 gene may provide the missing link connecting alcohol injury to enhanced DAG production and insulin resistance. Previous studies have revealed the importance of DAG as a signaling molecule to activate PKCθ in muscle or PKCϵ in liver to target either insulin receptor substrates or insulin receptor, respectively (22, 24). This phenomenon was further confirmed by knockdown of Crebh in 2-AG-treated or db/db mice that showed a concomitant decrease in PKCϵ activity and Lipin1-mediated DAG production as well as considerable recovery in the activities of insulin receptor and its downstream signaling components (IRS1 and AKT). Our findings were further supported by a recent report in which a lowering of the hepatic triglyceride level in Crebh knock-out mouse models was observed (11).
Interestingly, our results demonstrated a moderate but significant decrease in the total DAG level upon Lipin1 knockdown in cells treated with 2-AG. This observation indicates the possibility of a Lipin1-independent effect of 2-AG in increasing the total DAG level and consequently the inhibiting insulin receptor signaling pathway. DAG is an interesting candidate responsible for lipid-induced insulin resistance and was found to be associated with insulin resistance after high fat feeding and/or Cb1r activation (5, 25). DAG can be derived from multiple sources. However, in the case of lipid-induced insulin resistance, the de novo synthesis of DAG via esterification of glycerol 3-phosphate is suggested to be the most important route. The Cb1r signaling pathway contributes to this source of DAG synthesis (1, 5). Other sources of DAG synthesis include the breakdown of phospholipids by the enzyme phospholipase C or phospholipase D-mediated hydrolysis of phosphatidylcholine (26). Especially in the phospholipase D pathway, both of the phosphatidic acid phosphatase family proteins diacylglycerol phosphate phosphatase and lipid-phosphate phosphatase contribute to the synthesis of DAG (27, 28). Alternatively, hydrolysis of triacylglycerol by the activity of lipases also results in increased DAG levels (29). Therefore, investigating the various factors involved in these multiple pathways that contribute to DAG synthesis in 2-AG-treated samples and analyzing the total DAG level upon knockdown or pharmacological inhibition of these factors (in the presence of 2-AG treatment) will provide additional information regarding the involvement of other contributory factors downstream of the Cb1r pathway and independent of Lipin1.
Overall, our study demonstrates the molecular link between Cb1r and insulin receptor signaling dysfunction in a lipogenic setting with crucial roles played by endoplasmic reticulum stress-activated transcription factor Crebh and Lipin1. These findings at least in part support the hypothesis that relieving higher hepatic DAG levels by targeting its upstream regulators might be beneficial to restore insulin signaling. However, further studies are necessary to assess the relative contribution of this signaling cascade that can be targeted for potential therapeutic purposes in insulin resistance conditions.
Acknowledgments
We thank our laboratory members and Dr. Seok-Yong Choi for cooperation and discussions related to this work.
This work was supported by the Future-based Technology Development Program (BIO Fields) through the Korea Research Foundation funded by the Ministry of Education, Science and Technology (Grant 2010-0019512) (to H.-S. C), a National Creative Research Initiatives grant funded by the Korean Ministry of Education, Science and Technology (Grant 20110018305) (to H.-S. C), and the Korea Research Institute of Bioscience and Biotechnology Research Initiative Program of Korea (to C.-H. L.).
- CB1R
- cannabinoid receptor type 1
- CREBH
- cAMP-response element binding protein H
- 2-AG
- 2-arachidonoyl glycerol
- DAG
- diacylglycerol
- USi
- unspecific RNAi
- Ad
- adenovirus
- CB2R
- cannabinoid receptor type 2
- qPCR
- quantitative PCR
- ATF
- activating transcription factor
- CREBHi
- Crebh RNAi
- IRS1
- insulin receptor substrate 1
- Srebp1c
- sterol response element-binding protein 1c
- GPAT
- acyl-CoA:glycerol-sn-3-phosphate acyltransferase
- Cpt1
- carnitine palmitoyltransferase 1
- Ucp2
- uncoupling protein 2
- Cd36
- cluster of differentiation 36
- Dgat
- acyl-CoA, diacylglycerol acyltransferase.
REFERENCES
- 1. Kunos G., Osei-Hyiaman D., Bátkai S., Sharkey K. A., Makriyannis A. (2009) Should peripheral CB1 cannabinoid receptors be selectively targeted for therapeutic gain? Trends Pharmacol. Sci. 30, 1–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Nogueiras R., Veyrat-Durebex C., Suchanek P. M., Klein M., Tschöp J., Caldwell C., Woods S. C., Wittmann G., Watanabe M., Liposits Z., Fekete C., Reizes O., Rohner-Jeanrenaud F., Tschöp M. H. (2008) Peripheral, but not central, CB1 antagonism provides food intake-independent metabolic benefits in diet-induced obese rats. Diabetes 57, 2977–2991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Monteleone P., Matias I., Martiadis V., De Petrocellis L., Maj M., Di Marzo V. (2005) Blood levels of the endocannabinoid anandamide are increased in anorexia nervosa and in binge-eating disorder, but not in bulimia nervosa. Neuropsychopharmacology 30, 1216–1221 [DOI] [PubMed] [Google Scholar]
- 4. Matias I., Gonthier M. P., Orlando P., Martiadis V., De Petrocellis L., Cervino C., Petrosino S., Hoareau L., Festy F., Pasquali R., Roche R., Maj M., Pagotto U., Monteleone P., Di Marzo V. (2006) Regulation, function, and dysregulation of endocannabinoids in models of adipose and β-pancreatic cells and in obesity and hyperglycemia. J. Clin. Endocrinol. Metab. 91, 3171–3180 [DOI] [PubMed] [Google Scholar]
- 5. Jeong W. I., Osei-Hyiaman D., Park O., Liu J., Bátkai S., Mukhopadhyay P., Horiguchi N., Harvey-White J., Marsicano G., Lutz B., Gao B., Kunos G. (2008) Paracrine activation of hepatic CB1 receptors by stellate cell-derived endocannabinoids mediates alcoholic fatty liver. Cell Metab. 7, 227–235 [DOI] [PubMed] [Google Scholar]
- 6. Després J. P., Golay A., Sjöström L.; Rimonabant in Obesity-Lipids Study Group (2005) Effects of rimonabant on metabolic risk factors in overweight patients with dyslipidemia. N. Engl. J. Med. 353, 2121–2134 [DOI] [PubMed] [Google Scholar]
- 7. Zhang K., Shen X., Wu J., Sakaki K., Saunders T., Rutkowski D. T., Back S. H., Kaufman R. J. (2006) Endoplasmic reticulum stress activates cleavage of CREBH to induce a systemic inflammatory response. Cell 124, 587–599 [DOI] [PubMed] [Google Scholar]
- 8. Lee M. W., Chanda D., Yang J., Oh H., Kim S. S., Yoon Y. S., Hong S., Park K. G., Lee I. K., Choi C. S., Hanson R. W., Choi H. S., Koo S. H. (2010) Regulation of hepatic gluconeogenesis by an ER-bound transcription factor, CREBH. Cell Metab. 11, 331–339 [DOI] [PubMed] [Google Scholar]
- 9. Chanda D., Kim D. K., Li T., Kim Y. H., Koo S. H., Lee C. H., Chiang J. Y., Choi H. S. (2011) Cannabinoid receptor type 1 (CB1R) signaling regulates hepatic gluconeogenesis via induction of endoplasmic reticulum-bound transcription factor cAMP-responsive element-binding protein H (CREBH) in primary hepatocytes. J. Biol. Chem. 286, 27971–27979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Vecchi C., Montosi G., Zhang K., Lamberti I., Duncan S. A., Kaufman R. J., Pietrangelo A. (2009) ER stress controls iron metabolism through induction of hepcidin. Science 325, 877–880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lee J. H., Giannikopoulos P., Duncan S. A., Wang J., Johansen C. T., Brown J. D., Plutzky J., Hegele R. A., Glimcher L. H., Lee A. H. (2011) The transcription factor cyclic AMP-responsive element-binding protein H regulates triglyceride metabolism. Nat. Med. 17, 812–815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Luebke-Wheeler J., Zhang K., Battle M., Si-Tayeb K., Garrison W., Chhinder S., Li J., Kaufman R. J., Duncan S. A. (2008) Hepatocyte nuclear factor 4α is implicated in endoplasmic reticulum stress-induced acute phase response by regulating expression of cyclic adenosine monophosphate responsive element binding protein H. Hepatology 48, 1242–1250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Danno H., Ishii K. A., Nakagawa Y., Mikami M., Yamamoto T., Yabe S., Furusawa M., Kumadaki S., Watanabe K., Shimizu H., Matsuzaka T., Kobayashi K., Takahashi A., Yatoh S., Suzuki H., Yamada N., Shimano H. (2010) The liver-enriched transcription factor CREBH is nutritionally regulated and activated by fatty acids and PPARα. Biochem. Biophys. Res. Commun. 391, 1222–1227 [DOI] [PubMed] [Google Scholar]
- 14. Reue K. (2009) The lipin family: mutations and metabolism. Curr. Opin. Lipidol. 20, 165–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Erion D. M., Shulman G. I. (2010) Diacylglycerol-mediated insulin resistance. Nat. Med. 16, 400–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Samuel V. T., Petersen K. F., Shulman G. I. (2010) Lipid-induced insulin resistance: unravelling the mechanism. Lancet 375, 2267–2277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Finck B. N., Gropler M. C., Chen Z., Leone T. C., Croce M. A., Harris T. E., Lawrence J. C., Jr., Kelly D. P. (2006) Lipin 1 is an inducible amplifier of the hepatic PGC-1α/PPARα regulatory pathway. Cell Metab. 4, 199–210 [DOI] [PubMed] [Google Scholar]
- 18. Ishimoto K., Nakamura H., Tachibana K., Yamasaki D., Ota A., Hirano K., Tanaka T., Hamakubo T., Sakai J., Kodama T., Doi T. (2009) Sterol-mediated regulation of human lipin 1 gene expression in hepatoblastoma cells. J. Biol. Chem. 284, 22195–22205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kim D. K., Kim J. R., Koh M., Kim Y. D., Lee J. M., Chanda D., Park S. B., Min J. J., Lee C. H., Park T. S., Choi H. S. (2011) Estrogen-related receptor γ (ERRγ) is a novel transcriptional regulator of phosphatidic acid phosphatase, LIPIN1, and inhibits hepatic insulin signaling. J. Biol. Chem. 286, 38035–38042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ryu D., Oh K. J., Jo H. Y., Hedrick S., Kim Y. N., Hwang Y. J., Park T. S., Han J. S., Choi C. S., Montminy M., Koo S. H. (2009) TORC2 regulates hepatic insulin signaling via a mammalian phosphatidic acid phosphatase, LIPIN1. Cell Metab. 9, 240–251 [DOI] [PubMed] [Google Scholar]
- 21. Samuel V. T., Liu Z. X., Qu X., Elder B. D., Bilz S., Befroy D., Romanelli A. J., Shulman G. I. (2004) Mechanism of hepatic insulin resistance in non-alcoholic fatty liver disease. J. Biol. Chem. 279, 32345–32353 [DOI] [PubMed] [Google Scholar]
- 22. Savage D. B., Petersen K. F., Shulman G. I. (2007) Disordered lipid metabolism and the pathogenesis of insulin resistance. Physiol. Rev. 87, 507–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kaiser J. P., Beier J. I., Zhang J., David Hoetker J., von Montfort C., Guo L., Zheng Y., Monia B. P., Bhatnagar A., Arteel G. E. (2009) PKCϵ plays a causal role in acute ethanol-induced steatosis. Arch. Biochem. Biophys. 482, 104–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Choi C. S., Savage D. B., Kulkarni A., Yu X. X., Liu Z. X., Morino K., Kim S., Distefano A., Samuel V. T., Neschen S., Zhang D., Wang A., Zhang X. M., Kahn M., Cline G. W., Pandey S. K., Geisler J. G., Bhanot S., Monia B. P., Shulman G. I. (2007) Suppression of diacylglycerol acyltransferase-2 (DGAT2), but not DGAT1, with antisense oligonucleotides reverses diet-induced hepatic steatosis and insulin resistance. J. Biol. Chem. 282, 22678–22688 [DOI] [PubMed] [Google Scholar]
- 25. Heydrick S. J., Ruderman N. B., Kurowski T. G., Adams H. B., Chen K. S. (1991) Enhanced stimulation of diacylglycerol and lipid synthesis by insulin in denervated muscle. Altered protein kinase C activity and possible link to insulin resistance. Diabetes 40, 1707–1711 [DOI] [PubMed] [Google Scholar]
- 26. Brose N., Betz A., Wegmeyer H. (2004) Divergent and convergent signaling by the diacylglycerol second messenger pathway in mammals. Curr. Opin. Neurobiol. 14, 328–340 [DOI] [PubMed] [Google Scholar]
- 27. Waggoner D. W., Xu J., Singh I., Jasinska R., Zhang Q. X., Brindley D. N. (1999) Structural organization of mammalian lipid phosphate phosphatases: implications for signal transduction. Biochim. Biophys. Acta 1439, 299–316 [DOI] [PubMed] [Google Scholar]
- 28. Brindley D. N., Pilquil C., Sariahmetoglu M., Reue K. (2009) Phosphatidate degradation: phosphatidate phosphatases (lipins) and lipid phosphate phosphatases. Biochim. Biophys. Acta 1791, 956–961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Carrasco S., Mérida I. (2007) Diacylglycerol, when simplicity becomes complex. Trends Biochem. Sci. 32, 27–36 [DOI] [PubMed] [Google Scholar]