Background: CD3-T cell receptor co-stimulation through SLAMF3/SLAMF6 promotes IL-17A production.
Results: SLAMF3/SLAMF6 and CD28 co-stimulation induces NFAT recruitment to IL17A. SLAMF3/SLAMF6 signaling increases nuclear RORγt abundance and recruitment to the IL17A promoter.
Conclusion: SLAMF3/SLAMF6 promote Th17 subsets in a RORγt-dependent fashion.
Significance: Deciphering the molecular mechanisms that control receptor-specific cytokine expression will help to understand immune regulation in health and disease.
Keywords: Interleukin, NFAT Transcription Factor, Signal Transduction, T Cell, T Cell Receptor, IL-17, RORgammat, SAP, SLAMF
Abstract
Th17 lymphocytes play a key role during immune responses against bacteria and fungi and are involved in the pathophysiology of multiple autoimmune disorders. The co-stimulatory molecules SLAMF3 and SLAMF6 have been implicated in the formation of Th17 phenotypes and IL-17A expression. Increased surface expression of SLAMF3 and SLAMF6 has been linked with disease activity in systemic lupus erythematosus. Here we demonstrate that in human total T lymphocytes the canonical CD28 and the non-canonical SLAMF3/SLAMF6 co-stimulatory pathways cooperate in the recruitment of the transcription factor NFAT1 to the IL17A promoter. Furthermore, the dominance of the SLAMF3/SLAMF6 pathway in inducing IL-17A production can be attributed to an increased nuclear abundance and recruitment of RORγt to the IL17A promoter. Thus, we have identified an additional mechanism that may be central for the specific control of IL17A gene regulation in systemic lupus erythematosus T lymphocytes.
Introduction
T lymphocytes play a central role during immune responses against pathogens. T lymphocyte activation is achieved through the recognition of the Ag-MHC complex by the T cell receptor complex and additional signals that are mediated through co-stimulatory pathways (1, 2). Co-stimulation through the canonical CD28 pathway is best characterized during T lymphocyte activation (3). A growing body of literature provides evidence for additional co-stimulatory molecules, including the family of signaling lymphocyte activation molecules (SLAMF)3 (4–6, 6, 7). SLAMs are trans-membrane receptors that have been demonstrated to mediate regulatory signals between immune cells by entertaining homophilic and/or heterophilic interactions. SLAMF receptors play a key role in the regulation of both the innate and adaptive immune system, and defective SLAM signaling has been linked with severe NK (natural killer), T, and B cell anomalies and reduced antibody production (6, 6–8).
The differentiation of CD4+ T helper (Th) cells into distinct effector populations is a key mechanism during adaptive immune responses. Recent reports suggest that co-stimulation of T helper populations through SLAMs is involved in T cell priming and lineage commitment, especially during germinal center formation (9–12). The recently discovered and highly specialized Th17 subset is characterized by abundant production of the proinflammatory cytokines IL-17A, IL-17F, IL-21, and IL-22. Th17 lymphocytes play a central role during immune responses to bacteria and fungi and have been implicated in the development of autoimmune disorders, including multiple sclerosis, rheumatoid arthritis, inflammatory bowel disease, and systemic lupus erythematosus (SLE) (10, 13–17).
We recently demonstrated that T lymphocytes from SLE patients exhibit increased SLAMF3 and SLAMF6 surface expression, the levels of which even reflect disease activity in these individuals (10). Co-activation of T lymphocytes with anti-SLAMF3 or anti-SLAMF6 antibodies along with CD3-T cell receptor stimulation induces IL-17A production and contributes to Th17 generation. However, the molecular downstream mechanisms following SLAM receptor activation and the involved transcription factor repertoires are not yet elucidated.
The calcium-dependent transcription factor family that is referred to as nuclear factor of activated T cells (NFAT) plays a central role during T helper lymphocyte differentiation. During this process, NFATs control the expression of genes that are essential for lineage determination and commitment, including growth factors, cytokines, and cell-cell interaction proteins (18–22). NFAT1 (also known as NFATc2) centrally contributes to the cytokine profile that is essential for Th17 generation. NFAT1 is involved in the induction of the lineage-determining cytokines IL-17A, IL-17F, IL-21, and IL-22 (19, 23, 24). Another central role for the differentiation of Th17 subsets has been documented for the two transcription factors RORα and RORγt. Both are required for the differentiation of Th17 lymphocytes from naive CD4+ T cells (25–28). SR1001 is a specific suppressor of RORα and RORγt activity. The application of SR1001 to in vitro T lymphocyte cultures results in reduced IL17A promoter activities and inhibits Th17 differentiation. In the Th17-dependent experimental autoimmune encephalopathy model, application of SR1001 results in an improvement of clinical scores and reduced expression of the proinflammatory cytokines IL-17A, IL-21, and IL-22 (27).
In this report, we demonstrate that both transcription factors, NFAT1 and RORγt, together are required in order to optimally induce IL17A in response to co-stimulation with CD28 and SLAMF3/SLAMF6. Differences in IL-17A expression levels in response to co-stimulation through the canonical CD28 pathway and the non-canonical SLAMF3/SLAMF6 co-stimulatory pathways can be explained by increased nuclear abundance of RORγt in response to SLAMF3/SLAMF6 signaling, resulting in enhanced recruitment to a RORγt(−183) binding site within the IL17A promoter and increased trans-activation.
MATERIALS AND METHODS
Study Subjects and T Cell Isolation
Blood samples were obtained from healthy platelet donors at the Kraft Family Blood Donor Center (Dana-Farber Cancer Institute, Boston, MA). Primary total T cells were isolated from peripheral venous blood by negative selection as described previously (15). All primary T cells were kept in RPMI medium supplemented with 10% fetal bovine serum.
Plasmids and Generation of Luciferase Reporter Constructs
Reporter constructs spanning the proximal 465 and 195 bp of the human IL17A promoter were PCR-amplified and cloned into luciferase vector pGL3-Basic (Promega) as reported previously (15) using primers with attached restriction sites for MluI and BglII. All plasmid DNA preparations were carried out with DNA purification kits (Qiagen) and sequence-verified (Genewiz, Cambridge, MA). Site-directed mutagenesis at the NFAT sites (−212 and −170) and the RORγt site (−183) within both reporter constructs IL17Ap(−465)_luc and IL17Ap(−195)_luc was performed using the PfuTurbo® DNA polymerase (Stratagene) according to the manufacturer's instructions.
Luciferase Assays in Primary Human T Cells
Three million primary human T cells were transfected with a total amount of 3 μg of plasmid DNA by the Amaxa transfection system (Lonza). Each reporter experiment included 10 ng of Renilla luciferase construct as an internal control. Eighteen hours after transfection, cells were collected and lysed, and luciferase activity was quantified using the Promega Dual-Luciferase assay system (Promega) according to the manufacturer's instructions. Luciferase experiments were repeated at least four times, and values in the bar diagrams are given as mean and S.E.
T Lymphocyte Culture and Th17 Differentiation Assays
Cell culture plates were precoated overnight with 0.5 μg/ml monoclonal anti-CD3 (BioXcell, clone OKT3), 0.5 μg/ml anti-CD28 (Biolegend), 0.5 μg/ml anti-SLAMF6 (Genentech, clone 24D8.1H5.1F5), or 0.5 μg/ml anti-SLAMF3 antibodies (Biolegend, clone HLy-9.1.25) as indicated. For transcription factor inhibition experiments and transcription factor immunoblotting assays, naive CD4+ T cells were differentiated into Th17 cells in serum-free X-VIVO10 medium (BioWhittaker) by the addition of IL-6 (25 ng/ml), TGF-β1 (5 ng/ml), IL-1β (12.5 ng/ml), IL-21 (25 ng/ml), and IL-23 (25 ng/ml) for the indicated time periods. IL-6, IL-1β, IL-23, and TGF-β1 were obtained from R&D Systems, and IL-21 was purchased from Cell Sciences. Supernatants were collected at different time points and tested for IL-17A (eBioscience) by ELISA. T lymphocytes were collected at different time points and tested for 1) the nuclear abundance of NFAT1 or RORγt at 72 or 120 h or 2) used for ChIP analysis (NFAT1 or RORγt recruitment) at 120 h as indicated.
T Cell Stimulation, NFAT and RORγt Inhibitors, and ELISAs
In order to assess the influence of NFAT and RORγt effects on IL-17A expression, both transcription factors were antagonized by applying specific inhibitors. NFAT was inhibited with 0.5 nm FK506/tacrolimus (Sigma) or 10 nm cyclosporin A (Sigma) as indicated, and RORγt was antagonized by 10 nm SR1001 (Cayman Chemical). Vehicle controls (DMSO for FK506/tacrolimus and CSA; 96% ethanol for SR1001) were included in order to exclude effects of the solvent on IL-17A expression. Supernatants were collected after 120 h and tested for IL-17A by ELISA (eBioscience).
NFAT Knockdown
In order to assess the effects of NFAT on IL-17A expression in response to co-stimulation with anti-CD28, SLAMF3, or SLAMF6 antibodies under polarizing conditions (10, 13–17), we knocked down NFAT1 with trivalent siRNAs (OriGene). In brief, naive CD4+ T cells were isolated through magnetic bead separation (Miltenyi). Two million naive CD4+ T cells were transfected with 80 μm NFAT1-specific siRNA (OriGene) using 40 μl/ml Lipofectamine 2000 (Invitrogen). Prior to these experiments, experimental conditions were optimized using Cy-3-labeled control siRNA (OriGene). Transfection efficiency for siRNAs was >70%. Cells were collected after 6 h, washed and resuspended in RPMI 1640 medium with 20% FBS, and cultured in the presence of Th17 priming cytokines for 72 h as published previously (10, 13–17). Cells and supernatants were harvested in order to assess IL-17A expression by ELISA (eBioscience) and quantitative RT-PCR. NFAT1 mRNA knockdown efficiency was ∼60–70% as assessed by quantitative RT-PCR with NFAT1-specific primers.
NFAT and RORγt Immunoblotting
For nuclear protein extraction, cells were handled on ice and lysed in 200 μl of lysis buffer (10 mm HEPES, pH 7.9), 10 mm KCl, 0.1 mm EDTA, 0.1 mm EGTA, supplemented with freshly added 1 mm DTT, 0.5 mm PMSF, 2 mm aprotinin, 1 mm leupeptin, 10 mm NaF, and 2 mm Na3VO4 for 15 min. Subsequently, Nonidet P-40 was added to the reaction mixture at a concentration of 0.6%. Nuclei were pelleted, resuspended, and washed. Nuclear extracts were resolved on 4–12% BisTris gels and transferred to a polyvinylidene difluoride membrane. Membranes were blocked with 5% nonfat milk in Tris-buffered saline with Tween 20 (TBS-T) for 1 h, incubated with primary antibody (1:1,000) overnight (NFATc2 and RORγt, both from Santa Cruz Biotechnology, Inc.), washed three times with TBS-T, incubated with horseradish peroxidase-conjugated secondary antibody (1:2,000) for 1 h, washed three times with TBS-T, developed with ECL reagents (GE Healthcare), and visualized on a Fujifilm LAS-4000 imager (GE Healthcare).
NFAT and RORγt ChIP and Reporter-ChIP Assays
Anti-NFAT1 antibodies were from Abcam, and anti-RORγt was from Santa Cruz Biotechnology, Inc. Nonspecific normal rabbit and normal mouse IgG were obtained from Upstate (Millipore). ChIP grade Protein A/G Plus-agarose was purchased from Pierce (Thermo Scientific). The ChIP assay was carried out according to the manufacturer's instructions (Upstate Biotechnology/Millipore). Briefly, human T cells were cross-linked with 1% formaldehyde, washed with cold phosphate-buffered saline, and lysed in buffer containing protease inhibitors (Roche Applied Science). Cell lysates were sonicated to shear DNA and sedimented, and diluted supernatants were immunoprecipitated with the indicated antibodies. A proportion (20%) of the diluted supernatants was kept as “input” (input represents PCR amplification of the total sample). Protein-DNA complexes were eluted in 1% SDS, 0.1 m NaHCO3 and reverse-cross-linked at 65 °C. DNA was recovered using the QIAamp DNA minikit (Qiagen) and subjected to PCR analysis on an ABI OneStepPlus real-time PCR system. The amount of immunoprecipitated DNA was subtracted by the amplified DNA that was bound by the nonspecific normal IgG and subsequently calculated as relative to the respective input DNA.
Reporter-ChIP was performed from T lymphocytes transfected with wild type IL17Ap(−195)_wild type, IL17Ap(−195)_mNFAT(−170), or IL17Ap(−195)_RORc(−183) plasmids. Subsequently, cells were lysed, DNA was sheared by sonication, and ChIP was carried out according to the manufacturer's instructions (Upstate Biotechnology/Millipore). Immunoprecipitated DNA was quantified by quantitative RT-PCR with sequence-specific primers.
Statistical Analyses
The paired two-tailed Student's t test was used for statistical analyses.
RESULTS
NFAT Regulates IL17A Promoter Activity in Response to Co-stimulation through CD28, SLAMF3, or SLAMF6
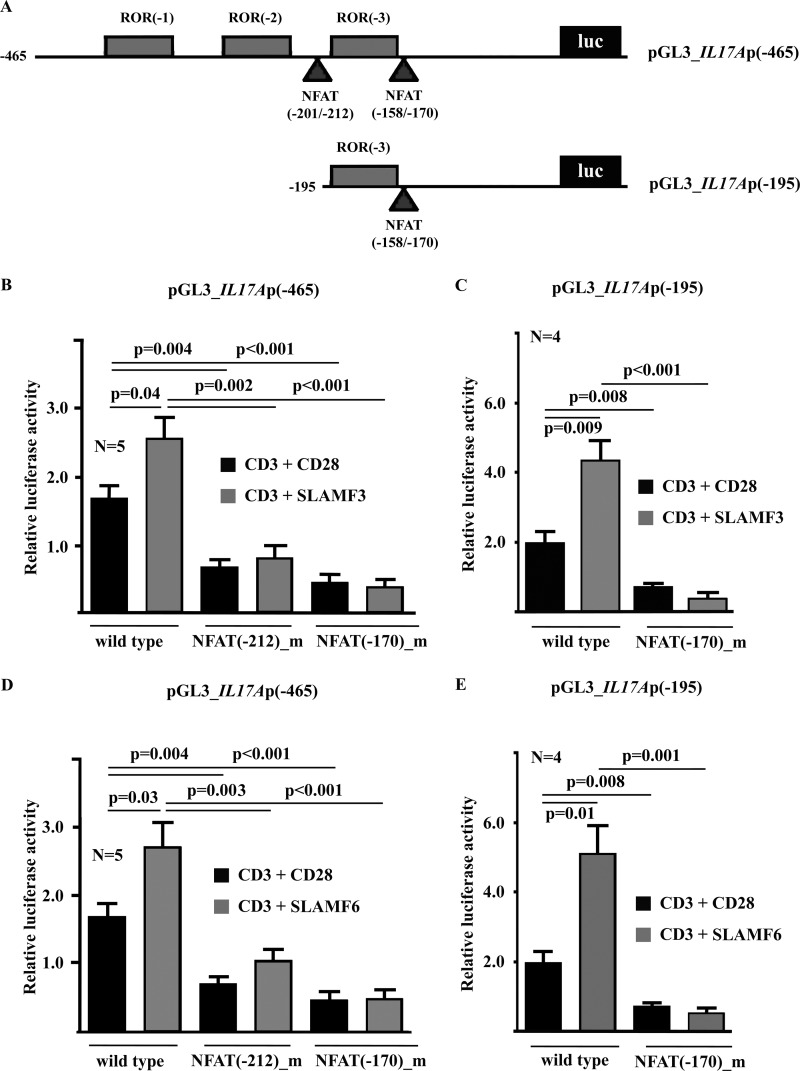
A thorough analysis of the human IL17A promoter yielded three putative RORγt sites and two potential NFAT sites (Fig. 1A). To investigate whether NFAT and RORγt regulate IL-17A expression in response to canonical co-stimulation through CD28 and/or co-stimulation through SLAMF3 or SLAMF6 at the transcriptional level, we performed luciferase assays using reporter constructs spanning the proximal 465 or 195 bp of the human IL17A promoter (Fig. 1A). Indeed, stimulation of T lymphocytes with anti-CD3 antibodies plus anti-CD28, anti-SLAMF3 (Fig. 1, B and C), or anti-SLAMF6 (Fig. 1, D and E) antibodies resulted in significant up-regulation of IL17A promoter activities in both constructs as compared with pGL3 empty vector. Next, we deleted either NFAT site (−212 or −170) within the 465 bp-spanning construct (Fig. 1, B and D) or the sole NFAT site (−170) within the 195 bp-spanning IL17A reporter construct (Fig. 1, C and E) and noted that the activity of both constructs lacking NFAT sites was significantly reduced (p < 0.001 under all conditions in the 465-bp construct and p < 0.01 in the 192-bp construct). These results suggest that both NFAT sites are crucial for IL17A transcription in human T cells.
FIGURE 1.
IL17A promoter activity in response to CD28, SLAMF3, or SLAMF6 co-stimulation requires intact NFAT binding sites. A, the IL17A promoter harbors multiple putative transcription factor binding sites, including three consensus binding sequences for RORγt and two consensus sequences for NFAT. In order to investigate IL17A promoter activity and the effects of transcription factors in response to co-stimulation with CD28, SLAMF3, or SLAMF6, two reporter constructs (−465 and −195 bp) were used. The effects of T lymphocyte stimulation with anti-CD3 antibodies and co-stimulation with anti-CD28, anti-SLAMF3 (B and C), or anti-SLAMF6 (D and E) antibodies on the −465 bp (B and D) and the −195 bp (C and E) IL17A promoter construct were tested as indicated. The consensus NFAT sites in both IL17A reporter constructs were subsequently deleted in order to assess their contribution to promoter activity. Error bars, S.E.
RORγt Regulates IL17A Promoter Activity in Response to Co-stimulation through CD28, SLAMF3, or SLAMF6
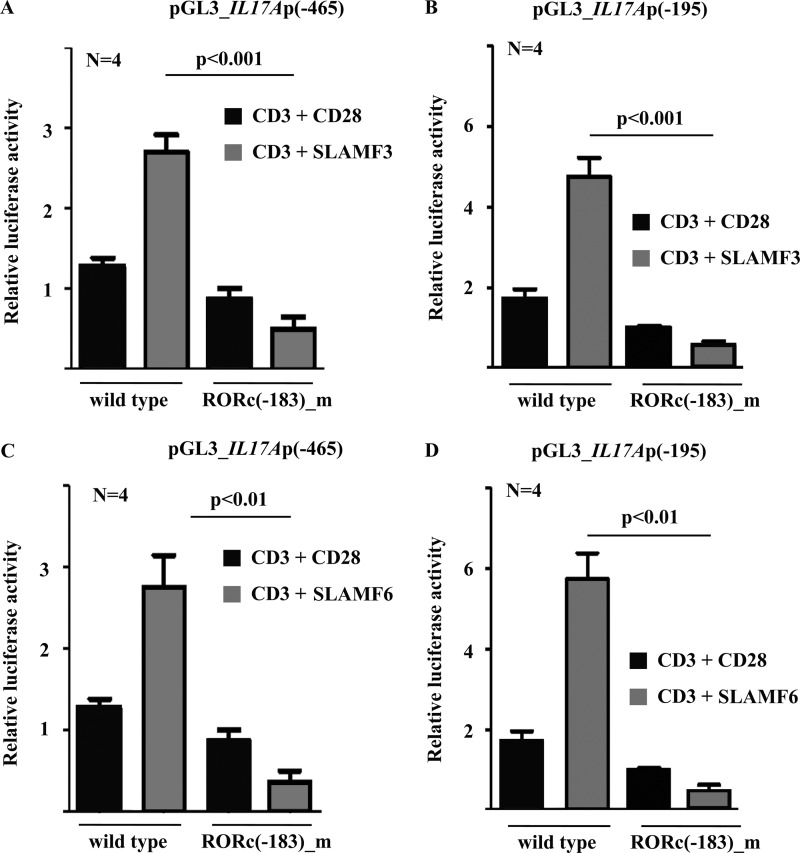
To investigate whether RORγt regulates IL17A transcription in response to canonical co-stimulation through CD28 and/or co-stimulation through SLAMF3 or SLAMF6, we performed luciferase assays using the aforementioned reporter constructs spanning the proximal 465 or 195 bp of the human IL17A promoter (Fig. 1A). Stimulation of T lymphocytes with anti-CD3 antibodies plus anti-CD28, anti-SLAMF3 (Fig. 1B), or anti-SLAMF6 (Fig. 1C) antibodies resulted in a significant induction of IL17A promoter activities in both constructs as compared with pGL3 empty vector. However, co-stimulation with anti-SLAMF3 or anti-SLAMF6 antibodies was superior to co-stimulation with anti-CD28 antibodies. Next, we deleted the RORγt site (−183) within the 195 bp-spanning IL17A reporter construct and noted that promoter activity was significantly reduced (p < 0.01). These results suggest that the RORγt(−183) site is crucial for RORγt-mediated IL17A transcription in human T cells in response to CD28 and SLAMF co-stimulation (Fig. 2, B and D).
FIGURE 2.
IL17A promoter activity in response to co-stimulation with anti-SLAMF3 and anti-SLAMF6 antibodies requires an intact RORγt binding site. The effects of T lymphocyte stimulation with anti-CD3 plus anti-CD28, anti-SLAMF3 (A and B), or anti-SLAMF6 (C and D) antibodies on the −465 bp (A and C) and the −195 bp (B and D) IL17A promoter construct were tested as indicated. One consensus RORγt site, −183 bp upstream from the transcriptional start codon, was subsequently deleted in both IL17A reporter constructs in order to assess its contribution to promoter activity. Error bars, S.E.
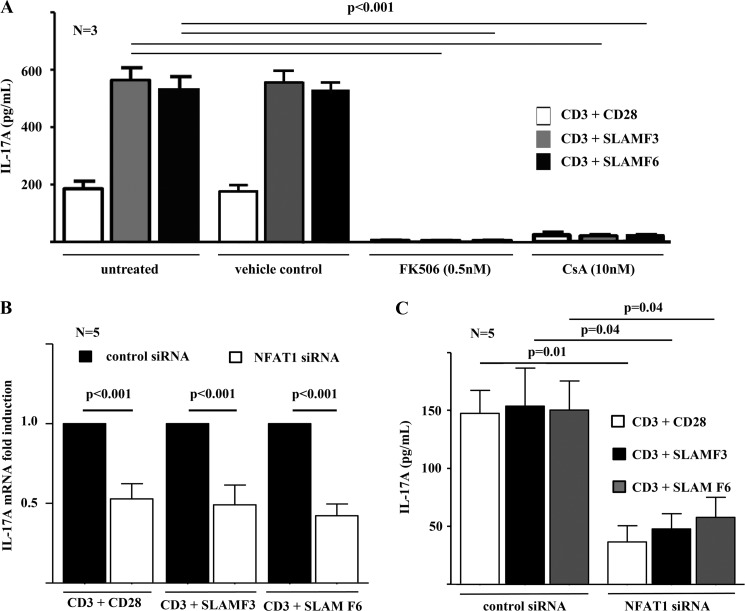
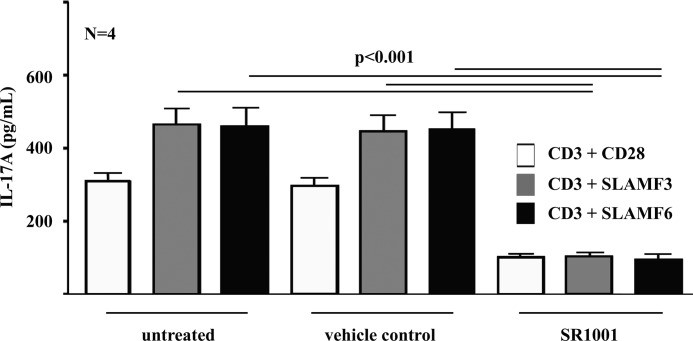
NFAT Inhibition Disrupts IL-17A Expression
In order to further investigate the involvement of NFAT and RORγt signaling in the induction of IL-17A expression and Th17 differentiation in response to either canonical CD28 or SLAMF3/SLAMF6 co-stimulation, we applied inhibitors for NFAT (FK506/tacrolimus and cyclosporin A) (29, 30) and RORγt (SR1001) (27) in naive CD4+ T lymphocyte cultures under Th17-polarizing conditions as indicated under “Materials and Methods.” Effects of the chemical solvents used were excluded by the inclusion of vehicle controls as indicated under “Materials and Methods.” Supernatants were harvested after 5 days of culture. Co-stimulation through SLAMF3/SLAMF6 resulted in significantly increased IL-17A expression when compared with canonical co-stimulation through CD28 (Figs. 3A and 4). Both NFAT inhibitors, FK506/tacrolimus and cyclosporin A, almost completely blocked IL-17A expression (p < 0.001) (Fig. 3A). The RORγt antagonist SR1001 significantly reduced IL-17A expression into the supernatant. However, IL-17A expression was not completely abrogated, suggesting additional effects of RORγt in addition to “preexisting” NFAT signaling that seems to be necessary for the induction of IL-17A expression (Fig. 3B).
FIGURE 3.
IL-17A expression depends on NFAT. A, the effects of NFAT signaling on IL-17A expression by T lymphocytes in response to stimulation with anti-CD3 plus anti-CD28, anti-SLAMF3 or anti-SLAMF6 antibodies were investigated, using the NFAT inhibitor FKK506/tacrolimus or cyclosporin A. Vehicle controls were included in order to exclude effects of the applied chemical solvent DMSO. NFAT1 was furthermore knocked down, applying specific NFAT1 siRNAs as compared with scrambled control siRNAs. The impacts of NFAT1 knockdown on IL-17A of mRNA (B) and protein (C) expression are given. Error bars, S.E.
FIGURE 4.
IL-17A expression depends on RORγt. The effects of RORγt signaling on IL-17A expression by T lymphocytes in response to stimulation with anti-CD3 plus anti-CD28, anti-SLAMF3, or anti-SLAMF6 antibodies were investigated, using the RORγt inhibitor SR1001 as indicated. Vehicle controls were included in order to exclude effects of the applied chemical solvent ethanol. Error bars, S.E.
NFAT Knockdown
Because calcineurin inhibition though FK506/tacrolimus or cyclosporin A is not very specific for NFAT, we specifically knocked down NFAT1 though trivalent siRNAs. Knockdown of NFAT1 in naive CD4+ T cells in the presence of Th17 polarizing cytokines and in response to co-stimulation though CD28, SLAMF3, or SLAMF6 resulted in equally reduced mRNA and protein expression of IL-17A (Fig. 3, B and C).
Co-stimulation through SLAMF3 or SLAMF6 Mediates Increased Nuclear Levels of RORγt
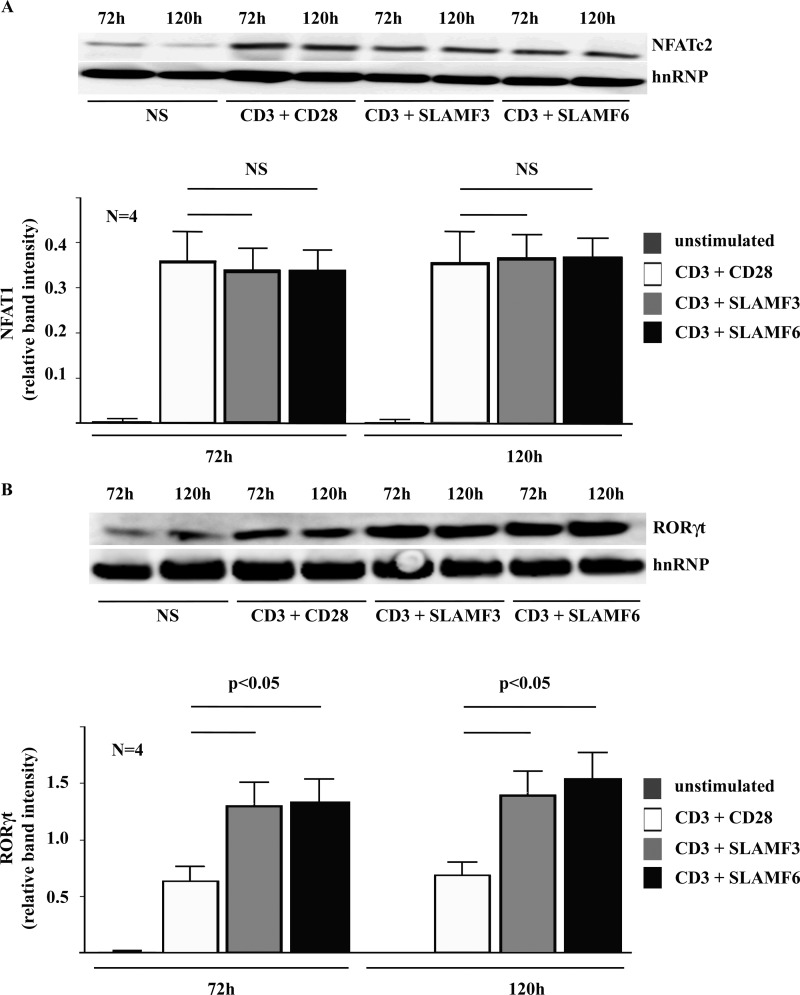
To determine whether varying nuclear levels of NFAT1 and RORγt may be responsible for differences in IL-17A induction and Th17 generation as a result of canonical co-stimulation through CD28 versus non-canonical co-stimulation through SLAMF3 or SLAMF6, we performed immunoblot analyses in nuclear lysates from T lymphocytes under Th17-polarizing conditions after 72 or 120 h as indicated.
NFAT1 was induced to comparable levels in response to stimulation with anti-CD3 plus anti-CD28 antibodies and anti-CD3 plus anti-SLAMF3 or anti-SLAMF6 antibodies (Fig. 5A). Interestingly, nuclear RORγt levels were significantly higher in response to T lymphocyte stimulation with anti-CD3 plus anti-SLAMF3 or anti-SLAMF6 antibodies when compared with co-stimulation with anti-CD28 antibodies (p < 0.05) (Fig. 5B), suggesting that differences in IL-17A expression levels in response to SLAMF3/SLAMF6 versus CD28 co-stimulation may be the result of variable RORγt induction.
FIGURE 5.
Differences in the IL-17A response to co-stimulation with anti-CD28 versus anti-SLAMF3 or anti-SLAMF6 antibodies are mediated by RORγt but not NFAT. Differences in the nuclear abundance of NFAT (A) and RORγt (B) in response to co-stimulation with anti-CD28 versus anti-SLAMF3 or anti-SLAMF6 antibodies were investigated using Western blot techniques. Error bars, S.E.
Co-Stimulation with SLAMF3 or SLAMF6 Results in Increased RORγt Recruitment to the IL17A Promoter
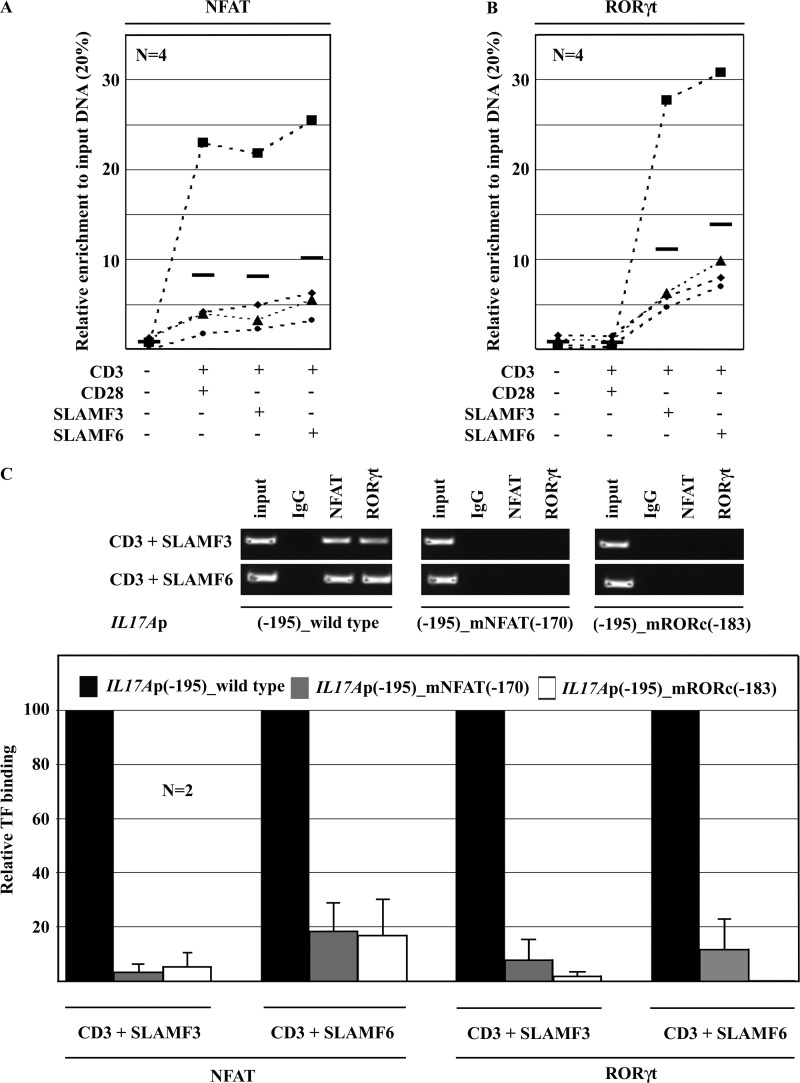
In order to investigate whether the increased nuclear abundance of RORγt in response to co-stimulation with SLAMF3 or SLAMF6 translates into increased RORγt recruitment to the IL17A promoter, we performed ChIP assays with NFAT and RORγt antibodies. In agreement with the aforementioned reporter studies and Western blot analyses, co-stimulation with anti-CD28 antibodies or anti-SLAMF3/SLAMF6 antibodies under Th17-polarizing conditions for 120 h resulted in an increased NFAT recruitment to the NFAT(−170) site. However, canonical and non-canonical co-stimuli did not cause differences in the level of NFAT recruitment (Fig. 6A, left). Co-stimulation with anti-SLAMF3/SLAMF6 antibodies resulted in markedly increased RORγt recruitment to the RORγt(−183) site in the IL17A promoter as compared with anti-CD28 antibodies (Fig. 5A, right).
FIGURE 6.
Differences in the IL-17A response to co-stimulation with anti-CD28 versus anti-SLAMF3 or anti-SLAMF6 antibodies are mediated by increased binding of RORγt but not NFAT to the IL-17A promoter. Differences in the recruitment of NFAT (A) and RORγt (B) to the IL17A promoter in response to co-stimulation with anti-CD28 versus anti-SLAMF3 or anti-SLAMF6 antibodies were investigated using ChIP. C, in order to assess the importance of co-recruitment of NFAT and RORγt to the proximal IL17A promoter (RORγt(−183) site and NFAT(−170) site), we deleted either site in the −195 bp IL17A reported construct and determined the recruitment of NFAT and RORγt in response to co-stimulation with anti-SLAMF3 or anti-SLAMF6 antibodies using ChIP. One representative experiment is given in the top panel, and averages and S.D. (error bars) values from two independent experiments are given in the bottom panel.
Both the NFAT(−170) and the RORγt(−183) Site Are Required for the Recruitment of Either Transcription Factor
The NFAT(−170) and the RORγt(−183) site are in close proximity within the IL17A promoter. Furthermore, the deletion of the NFAT(−170) site abrogated SLAMF3/SLAMF6 effects that strongly induce RORγt signaling. Thus, we asked whether the presence of either transcription factor binding site is required for the recruitment of the other factor. Applying reporter-ChIP techniques, we determined strong recruitment of NFAT and RORγt to the IL17A promoter construct in response to co-stimulation with SLAMF3 or SLAMF6 (Fig. 6B). Deletion of either the NFAT(−170) or the RORγt(−183) site within the 195 bp-spanning IL17A reporter construct resulted in significantly impaired binding of both NFAT and RORγt. Thus, the presence of both intact transcription factor binding sites is required for the recruitment of NFAT and/or RORγt.
DISCUSSION
Th17 cells are a highly specialized T cell subset and play a central role during immune responses to bacteria and fungi (26, 31, 32). We recently demonstrated that Th17 polarization is enhanced by non-canonical co-stimulation of T lymphocytes through SLAMF3 or SLAMF6. Furthermore, T lymphocytes from SLE patients express significantly more SLAMF3 and SLAMF6 on their surface when compared with T cells from healthy controls (10). Co-stimulation through SLAMF3 or SLAM6 follows different kinetics when compared with CD28. IL-17A production in response to CD28 co-stimulation peaks at day 3 and may relate to a normal immune response elicited by the antigen involving the CD3-T cell receptor complex. However, engagement of SLAMF3 or SLAMF6 initiates a prolonged inflammatory response, which may cause organ damage (10). Thus, we hypothesized that overexpression of SLAMF3/SLAMF6 on T lymphocytes may be a pathophysiological step that contributes to the pathogenesis of SLE (10).
It has been documented that SLAM molecules associate with intracellular adaptor proteins (e.g. those of the SLAM-associated protein (SAP) family) (10, 33–35). SAP proteins contribute to SLAM receptor activation as they mediate dimer formation between SLAM receptors. The downstream molecular mechanisms that are involved in SLAMF3/SLAMF6-mediated IL-17A induction remained to be elucidated. Here, we provide evidence for the involvement of both transcription factors, NFAT1 and RORγt, in the trans-activation of IL17A in response to co-stimulation with CD28, SLAMF3, or SLAMF6. Superior trans-activating effects in response to induction of the SLAMF3/SLAMF6 pathway could be explained by increased nuclear abundance of RORγt, resulting in enhanced recruitment to the RORγt(−183) site to the IL17A promoter.
The NFAT family of transcription factors, including NFAT1, is centrally involved in the transcriptional control of IL17A (19). Stimulation of the CD3-T cell receptor results in Ca2+ flux from the extracellular space and Ca2+ release from the endoplasmic reticulum (19, 36). In turn, NFAT controls the expression of target genes that are essential during lineage determination and commitment of Th17 cells, including IL-17A, IL-17F, IL-21, and IL-22 (19, 23, 24). Thus, we aimed to investigate whether differences in the induction of IL17A and Th17 polarization in response to co-stimulation through SLAMF3/SLAMF6 could be mediated by differential induction and/or recruitment of NFAT1. Applying luciferase reporter constructs, we determined superior induction of promoter activity in response to co-stimulation with SLAMF3/SLAMF6 when compared with CD28. Deleting either binding site, we demonstrated that NFAT recruitment to both the NFAT(−212) and the NFAT(−170) element are involved and essential for trans-activation of IL17A. In agreement with these findings, the suppression of NFAT activation with the calcineurin inhibitor FK506/tacrolimus or cyclosporin A in T cell cultures under Th17-polarizing conditions resulted in a complete abrogation of IL-17A protein expression under all co-stimulation conditions: CD28, SLAMF3, or SLAMF6. The same results were achieved by specific NFAT1 knockdown through siRNAs. However, no differences in the nuclear abundance of NFAT1 were detected in response to canonical (CD28) and non-canonical co-stimulation of T lymphocytes. Furthermore, NFAT recruitment to the IL17A promoter was comparable under all co-stimulatory conditions (CD28, SLAMF3, or SLAMF6). Taken together, these findings strongly suggest a key role for NFAT1 during the induction of IL-17A and Th17 generation. However, NFAT1 does not appear to be responsible for the variable kinetics and expression levels of IL-17A expression in response to canonical or non-canonical co-stimulation (10).
Members of the ROR transcription factor family, particularly RORα and RORγt, are centrally involved in the generation of Th17 phenotypes and IL-17A expression (25–28). Thus, we aimed to investigate the involvement of RORγt in the increased IL-17A expression in response to SLAMF3/SLAMF6 co-stimulation. Applying the aforementioned reported constructs, we determined reduced IL17A promoter activity in response to co-stimulation through CD28 and SLAMF3/SLAMF3 after deletion of the RORγt(−183) site. Suppression of RORα and RORγt by the specific inhibitor SR1001 (27) under Th17-polarizing conditions resulted in significantly reduced IL17A promoter activity in response to both canonical (CD28) and non-canonical (SLAMF3/SLAMF6) co-stimulation. In contrast to our findings using calcineurin inhibitors, SR1001 did not completely abrogate IL-17A protein expression in T cell cultures in Th17-polarizing conditions. SR1001 rather reduced IL-17 protein expression to the same levels in all groups (CD28 and SLAMF3/SLAMF6). This suggests that differences in the IL-17A expression in response to canonical and non-canonical co-stimulation are at least partially be due to variable effects of RORγt on IL17A transcription. Thus, we aimed to determine differences in the nuclear abundance of RORγt. Indeed, RORγt was detectable at higher levels in response to co-stimulation through SLAMF3/SLAMF6 when compared with CD28 co-stimulation under Th17-polarizing conditions. In agreement with this, RORγt recruitment to the IL17A promoter was markedly increased in response to co-stimulation with SLAMF3/SLAMF6.
Because NFAT family transcription factors, including NFAT1, have been demonstrated to interact with other transcription factors in order to differentially affect T lymphocyte functional outcomes (19), we asked whether the binding of NFAT may also affect RORγt recruitment to IL17A and vice versa. Indeed, deletion of the NFAT(−170) site in our −195 bp-spanning IL17A promoter construct strongly affected RORγt binding to its RORγt(−183) element, as assessed by reporter-ChIP, and deletion of the RORγt(−183) site resulted in impaired NFAT recruitment to NFAT(−170). This suggests interplay between the two transcription factors and points to the importance of both binding elements for IL-17A expression.
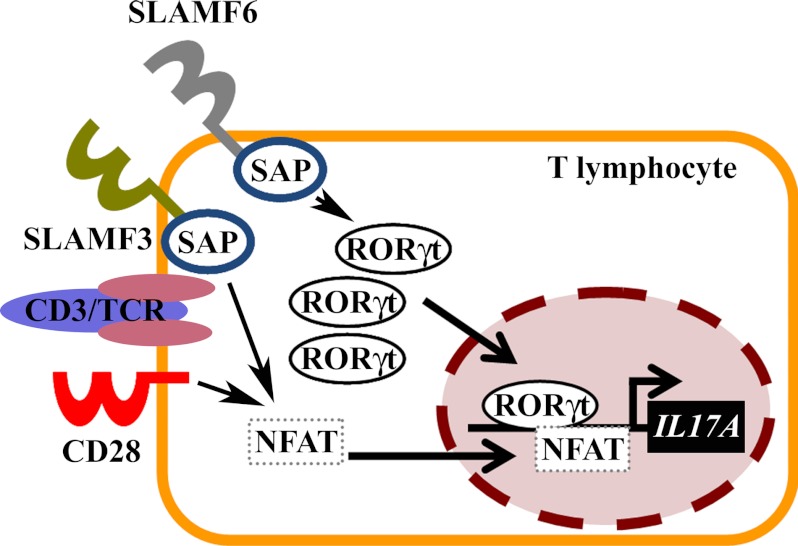
At this point, it remains to be answered whether additional transcription factors that play a role in the induction of IL-17A, such as Stat3 or IRF4, etc., are affected by SLAMF3/SLAMF6 (31). Furthermore, the molecular mechanisms downstream of SLAMF3/SLAMF6 and the interacting SAP proteins and upstream of NFAT/RORγt remain to be elucidated in order to link SLAM stimulation with the increased RORγt recruitment to IL17A (10, 33–35) (Fig. 7).
FIGURE 7.
Model of the effects of canonical co-stimulation through CD28 versus co-stimulation with SLAMF3 or SLAMF6 on IL-17 expression in T lymphocytes. Co-stimulation of T lymphocytes through the canonical CD28 pathway results in massively increased NFAT abundance in the nucleus but only slightly increased RORγt shuttling as compared with resting conditions. Co-stimulation with SLAMF3 or SLAMF6 results in a massive increase of NFAT and RORγt in the nucleus, resulting in recruitment of both to the IL17A promoter and strong trans-activation.
CONCLUSIONS
Co-stimulation of T lymphocytes through the SLAMF3/SLAMF6 pathways mediates more potent effects on IL-17A expression when compared with the canonical CD28 pathway. Although the transcription factor NFAT is necessary for the expression of IL-17A, it is not responsible for increased IL-17A expression in response to SLAM signaling. SLAMF3/SLAMF6 signaling mediates increased nuclear abundance and recruitment of RORγt to the proximal IL17A promoter, resulting in increased trans-activation and gene expression (Fig. 7).
This work was supported, in whole or in part, by National Institutes of Health Grants R01 AI49954 and P01 AI065687. This work was also supported by Deutsche Forschungsgemeinschaft Grant RA1927-1/1 (to T. R.).
- SLAMF
- signaling lymphocyte activation molecule family
- SLAM
- signaling lymphocyte activation molecule
- SLE
- systemic lupus erythematosus
- NFAT
- nuclear factor of activated T cells
- SAP
- SLAM-associated protein
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
REFERENCES
- 1. Collins M., Ling V., Carreno B. M. (2005) The B7 family of immune-regulatory ligands. Genome Biol. 6, 223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Sharpe A. H., Freeman G. J. (2002) The B7-CD28 superfamily. Nat. Rev. Immunol. 2, 116–126 [DOI] [PubMed] [Google Scholar]
- 3. Veillette A., Latour S., Davidson D. (2002) Negative regulation of immunoreceptor signaling. Annu. Rev. Immunol. 20, 669–707 [DOI] [PubMed] [Google Scholar]
- 4. Lucas P. J., Negishi I., Nakayama K., Fields L. E., Loh D. Y. (1995) Naive CD28-deficient T cells can initiate but not sustain an in vitro antigen-specific immune response. J. Immunol. 154, 5757–5768 [PubMed] [Google Scholar]
- 5. Shahinian A., Pfeffer K., Lee K. P., Kündig T. M., Kishihara K., Wakeham A., Kawai K., Ohashi P. S., Thompson C. B., Mak T. W. (1993) Differential T cell costimulatory requirements in CD28-deficient mice. Science 261, 609–612 [DOI] [PubMed] [Google Scholar]
- 6. Calpe S., Wang N., Romero X., Berger S. B., Lanyi A., Engel P., Terhorst C. (2008) The SLAM and SAP gene families control innate and adaptive immune responses. Adv. Immunol. 97, 177–250 [DOI] [PubMed] [Google Scholar]
- 7. Detre C., Keszei M., Romero X., Tsokos G. C., Terhorst C. (2010) SLAM family receptors and the SLAM-associated protein (SAP) modulate T cell functions. Semin. Immunopathol. 32, 157–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Veillette A., Dong Z., Latour S. (2007) Consequence of the SLAM-SAP signaling pathway in innate-like and conventional lymphocytes. Immunity 27, 698–710 [DOI] [PubMed] [Google Scholar]
- 9. Chatterjee M., Kis-Toth K., Thai T. H., Terhorst C., Tsokos G. C. (2011) SLAMF6-driven co-stimulation of human peripheral T cells is defective in SLE T cells. Autoimmunity 44, 211–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chatterjee M., Rauen T., Kis-Toth K., Kyttaris V. C., Hedrich C. M., Terhorst C., Tsokos G. C. (2012) Increased expression of SLAM receptors SLAMF3 and SLAMF6 in systemic lupus erythematosus T lymphocytes promotes Th17 differentiation. J. Immunol. 188, 1206–1212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Graham D. B., Bell M. P., McCausland M. M., Huntoon C. J., van Deursen J., Faubion W. A., Crotty S., McKean D. J. (2006) Ly9 (CD229)-deficient mice exhibit T cell defects yet do not share several phenotypic characteristics associated with SLAM- and SAP-deficient mice. J. Immunol. 176, 291–300 [DOI] [PubMed] [Google Scholar]
- 12. Valdez P. A., Wang H., Seshasayee D., van Lookeren Campagne M., Gurney A., Lee W. P., Grewal I. S. (2004) NTB-A, a new activating receptor in T cells that regulates autoimmune disease. J. Biol. Chem. 279, 18662–18669 [DOI] [PubMed] [Google Scholar]
- 13. Crispín J. C., Oukka M., Bayliss G., Cohen R. A., Van Beek C. A., Stillman I. E., Kyttaris V. C., Juang Y. T., Tsokos G. C. (2008) Expanded double negative T cells in patients with systemic lupus erythematosus produce IL-17 and infiltrate the kidneys. J. Immunol. 181, 8761–8766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Garrett-Sinha L. A., John S., Gaffen S. L. (2008) IL-17 and the Th17 lineage in systemic lupus erythematosus. Curr. Opin. Rheumatol. 20, 519–525 [DOI] [PubMed] [Google Scholar]
- 15. Rauen T., Hedrich C. M., Juang Y. T., Tenbrock K., Tsokos G. C. (2011) cAMP-responsive element modulator (CREM)α protein induces interleukin 17A expression and mediates epigenetic alterations at the interleukin-17A gene locus in patients with systemic lupus erythematosus. J. Biol. Chem. 286, 43437–43446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rauen T., Juang Y. T., Hedrich C. M., Kis-Toth K., Tsokos G. C. (2012) A novel isoform of the orphan receptor RORγt suppresses IL-17 production in human T cells. Genes Immun. 13, 346–350 [DOI] [PubMed] [Google Scholar]
- 17. Tsokos G. C. (2011) Systemic lupus erythematosus. N. Engl. J. Med. 365, 2110–2121 [DOI] [PubMed] [Google Scholar]
- 18. Graef I. A., Chen F., Crabtree G. R. (2001) NFAT signaling in vertebrate development. Curr. Opin. Genet. Dev. 11, 505–512 [DOI] [PubMed] [Google Scholar]
- 19. Hermann-Kleiter N., Baier G. (2010) NFAT pulls the strings during CD4+ T helper cell effector functions. Blood 115, 2989–2997 [DOI] [PubMed] [Google Scholar]
- 20. Macian F. (2005) NFAT proteins. Key regulators of T-cell development and function. Nat. Rev. Immunol. 5, 472–484 [DOI] [PubMed] [Google Scholar]
- 21. Rudensky A. Y., Gavin M., Zheng Y. (2006) FOXP3 and NFAT. Partners in tolerance. Cell 126, 253–256 [DOI] [PubMed] [Google Scholar]
- 22. Sundrud M. S., Rao A. (2007) New twists of T cell fate. Control of T cell activation and tolerance by TGF-β and NFAT. Curr. Opin. Immunol. 19, 287–293 [DOI] [PubMed] [Google Scholar]
- 23. Shen F., Hu Z., Goswami J., Gaffen S. L. (2006) Identification of common transcriptional regulatory elements in interleukin-17 target genes. J. Biol. Chem. 281, 24138–24148 [DOI] [PubMed] [Google Scholar]
- 24. Sun Z., Unutmaz D., Zou Y. R., Sunshine M. J., Pierani A., Brenner-Morton S., Mebius R. E., Littman D. R. (2000) Requirement for RORγ in thymocyte survival and lymphoid organ development. Science 288, 2369–2373 [DOI] [PubMed] [Google Scholar]
- 25. Bettelli E., Korn T., Oukka M., Kuchroo V. K. (2008) Induction and effector functions of TH17 cells. Nature 453, 1051–1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Littman D. R., Rudensky A. Y. (2010) Th17 and regulatory T cells in mediating and restraining inflammation. Cell 140, 845–858 [DOI] [PubMed] [Google Scholar]
- 27. Solt L. A., Kumar N., Nuhant P., Wang Y., Lauer J. L., Liu J., Istrate M. A., Kamenecka T. M., Roush W. R., Vidović D., Schürer S. C., Xu J., Wagoner G., Drew P. D., Griffin P. R., Burris T. P. (2011) Suppression of TH17 differentiation and autoimmunity by a synthetic ROR ligand. Nature 472, 491–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yang X. O., Pappu B. P., Nurieva R., Akimzhanov A., Kang H. S., Chung Y., Ma L., Shah B., Panopoulos A. D., Schluns K. S., Watowich S. S., Tian Q., Jetten A. M., Dong C. (2008) T helper 17 lineage differentiation is programmed by orphan nuclear receptors RORα and RORγ. Immunity. 28, 29–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Cippà P. E., Kraus A. K., Lindenmeyer M. T., Chen J., Guimezanes A., Bardwell P. D., Wekerle T., Wüthrich R. P., Fehr T. (2012) Resistance to ABT-737 in activated T lymphocytes. Molecular mechanisms and reversibility by inhibition of the calcineurin-NFAT pathway. Cell Death. Dis. 3, e299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kayama H., Koga R., Atarashi K., Okuyama M., Kimura T., Mak T. W., Uematsu S., Akira S., Takayanagi H., Honda K., Yamamoto M., Takeda K. (2009) NFATc1 mediates Toll-like receptor-independent innate immune responses during Trypanosoma cruzi infection. PLoS Pathog. 5, e1000514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Dong C. (2011) Genetic controls of Th17 cell differentiation and plasticity. Exp. Mol. Med. 43, 1–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wilke C. M., Bishop K., Fox D., Zou W. (2011) Deciphering the role of Th17 cells in human disease. Trends Immunol. 32, 603–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Henning G., Kraft M. S., Derfuss T., Pirzer R., de Saint-Basile G., Aversa G., Fleckenstein B., Meinl E. (2001) Signaling lymphocytic activation molecule (SLAM) regulates T cellular cytotoxicity. Eur. J. Immunol. 31, 2741–2750 [DOI] [PubMed] [Google Scholar]
- 34. Howie D., Okamoto S., Rietdijk S., Clarke K., Wang N., Gullo C., Bruggeman J. P., Manning S., Coyle A. J., Greenfield E., Kuchroo V., Terhorst C. (2002) The role of SAP in murine CD150 (SLAM)-mediated T-cell proliferation and interferon γ production. Blood 100, 2899–2907 [DOI] [PubMed] [Google Scholar]
- 35. Howie D., Laroux F. S., Morra M., Satoskar A. R., Rosas L. E., Faubion W. A., Julien A., Rietdijk S., Coyle A. J., Fraser C., Terhorst C. (2005) Cutting edge. The SLAM family receptor Ly108 controls T cell and neutrophil functions. J. Immunol. 174, 5931–5935 [DOI] [PubMed] [Google Scholar]
- 36. Li H., Rao A., Hogan P. G. (2011) Interaction of calcineurin with substrates and targeting proteins. Trends Cell Biol. 21, 91–103 [DOI] [PMC free article] [PubMed] [Google Scholar]