Background: Epf is a multidomain cell surface protein from Streptococcus pyogenes.
Results: Epf mediates adhesion to human epithelial cells through an N-terminal domain comprising two common binding modules.
Conclusion: Epf is an adhesin with a novel binding-domain supported on a stalk built from tandem helical repeat domains.
Significance: Adhesins such as Epf are important for colonization and infection by Streptococci.
Keywords: Bacterial Adhesion, Bacterial Pathogenesis, Cell Surface Protein, Crystal Structure, Streptococcus pyogenes, Carbohydrate-binding Module
Abstract
Streptococcus pyogenes is an exclusively human pathogen. Streptococcal attachment to and entry into epithelial cells is a prerequisite for a successful infection of the human host and requires adhesins. Here, we demonstrate that the multidomain protein Epf from S. pyogenes serotype M49 is a streptococcal adhesin. An epf-deficient mutant showed significantly decreased adhesion to and internalization into human keratinocytes. Cell adhesion is mediated by the N-terminal domain of Epf (EpfN) and increased by the human plasma protein plasminogen. The crystal structure of EpfN, solved at 1.6 Å resolution, shows that it consists of two subdomains: a carbohydrate-binding module and a fibronectin type III domain. Both fold types commonly participate in ligand receptor and protein-protein interactions. EpfN is followed by 18 repeats of a domain classified as DUF1542 (domain of unknown function 1542) and a C-terminal cell wall sorting signal. The DUF1542 repeats are not involved in adhesion, but biophysical studies show they are predominantly α-helical and form a fiber-like stalk of tandem DUF1542 domains. Epf thus conforms with the widespread family of adhesins known as MSCRAMMs (microbial surface components recognizing adhesive matrix molecules), in which a cell wall-attached stalk enables long range interactions via its adhesive N-terminal domain.
Introduction
The Gram-positive bacterium Streptococcus pyogenes (Group A streptococcus (GAS))6 is one of the most widespread human pathogens. GAS commonly colonizes the human skin and throat epithelia, causing mild infections such as pharyngitis and cellulitis. In some cases, it also invades underlying tissues and cells, leading to acute diseases such as necrotizing fasciitis. GAS infections can also result in autoimmune sequelae such as rheumatic heart disease, which is a major issue in developing countries (1).
Numerous studies have demonstrated that GAS adheres to a broad range of cell types and tissues (2) and can then be internalized into epithelial cells (3, 4). Both adherence and internalization are of clinical relevance for S. pyogenes infections (5, 6). An essential step for the contact between GAS and host cells involves binding to the human extracellular matrix component fibronectin (3, 7). The interaction of GAS with plasma proteins, e.g., plasminogen (8), also promotes adherence to and internalization into human epithelial cells (9, 10).
To be able to adhere to and invade host epithelia, GAS elaborates a plethora of virulence factors. The expression of these virulence factors is fine-tuned by two-component signal transduction systems and stand-alone transcriptional regulators. One of those regulators, Ralp3, was recently identified in a transcriptomics study of a Δnra mutant of GAS serotype M49 (11). Ralp3 was found to be encoded together with the two virulence factors, Eno (streptococcal surface enolase) and SagA (streptolysin S), and with Epf (extracellular protein factor) in the eno ralp3 epf sagA (ERES) genomic region. Of these genes, the eno sagA block is present in all GAS serotypes, whereas ralp3 and epf were shown to be restricted to serotypes M1, M4, M12, M28, and M49 (11). Interestingly, the novel protein Epf shows many hallmarks of a potential adhesin.
Like many members of the diverse family of bacterial adhesins called MSCRAMMs (microbial surface components recognizing adhesive matrix molecules) (12, 13), the 205-kDa protein Epf has a number of conserved C-terminal repeat domains and a unique N-terminal domain (see Fig. 1). It also has an N-terminal signal sequence and a C-terminal LPXTG-type sortase recognition motif, which indicates that Epf is attached to the streptococcal cell wall (11). In Epf from serotype M49, 18 C-terminal repeat domains, classified as DUF1542s (domains of unknown function 1542; PFAM accession number PF07564, amino acids 387–1841), were identified. The N-terminal domain has no homology with any protein of known structure or function (11).
FIGURE 1.
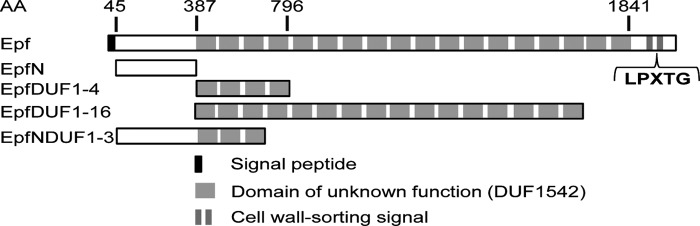
Domain structure of Epf from group A streptococcus serotype M49. The sequence range of each of the principal constructs used in this analysis, EpfN, EpfDUF1-4, EpfDUF1-16, and EpfNDUF1-3, is also shown.
An Epf orthologue from GAS serotype M1 has also been described and called LSA (14). LSA has an N-terminal domain similar to Epf and 20 DUF1542 repeat domains. Although no function has been attributed to either Epf or LSA, it was shown that LSA is essential for GAS virulence in a mouse model of infection (14). Other streptococcal species such as Streptococcus suis also produce proteins with a domain structure similar to that of Epf (15), including a unique N-terminal domain and C-terminal DUF1542 repeats.
Here, we investigated the potential function of Epf as an adhesin, analyzed the respective role of the Epf domains, solved the crystal structure of the N-terminal domain of Epf, and characterized the DUF1542 repeats. To our knowledge, this is the first study providing structural insight into the widespread group of proteins containing DUF1542 repeats.
EXPERIMENTAL PROCEDURES
Bacterial Strains, Eukaryotic Cells, and Culture Conditions
GAS serotype M49 strain 591 was isolated from a patient with a skin infection and was provided by R. Lütticken (Aachen, Germany). The construction of the epf− mutant strain was described previously (11). The GAS wild type strain was cultured in Todd-Hewitt broth (Invitrogen) supplemented with 0.5% (w/v) yeast extract (THY; Invitrogen) at 37 °C under a 5% CO2, 20% O2 atmosphere.
The human epithelial cell lines HaCaT (Deutsches Krebsforschungszentrum, Heidelberg, Germany), HEp-2 (ATCC CCL23), Detroit 562 (ATCC CCL-138), and Ca9-22 (TKG 0485) were used for standard adherence and internalization assays. HaCaT cells were maintained in DMEM (Invitrogen) GlutaMAXTM-I, 10% (v/v) FBS (Invitrogen). HEp-2 and Ca9-22 cells were maintained in DMEM, 2 mm l-glutamine, 10% (v/v) FBS, and Detroit 562 cells were maintained in DMEM/F-12, 2 mm l-glutamine, 10% (v/v) FBS.
Protein Production
Constructs EpfN, EpfDUF1-4, and EpfDUF1-16 were cloned from the gene epf (NCBI accession number ZP_00366506) from S. pyogenes strain 591 (serotype M49) into the expression vector pASK-IBA2 (IBA GmbH) and expressed in Escherichia coli BL21 (DE3) cells, as described elsewhere (16). These constructs were purified by affinity chromatography using Strep-Tactin Superflow High Capacity beads (IBA GmbH) (16). Two further constructs EpfN_b (Epf residues 58–349) and EpfN-DUF1-3 (residues 58–612) were also generated. These were cloned into the expression vector pProEx, expressed in E. coli BL21 (DE3) cells, and purified by immobilized metal affinity chromatography and size exclusion chromatography in buffer C (10 mm Tris-HCl, pH 7.4, 137 mm NaCl, 2.7 mm KCl, 0.3 mm NaN3). All constructs are shown in Fig. 1.
Eukaryotic Cell Adherence and Internalization
Adherence to and internalization into epithelial cells was quantified using the antibiotic protection assay (17). 24-well plates were inoculated with 2.5 × 105 cells/well in DMEM without antibiotics. The cells were allowed to grow to confluence. For the assay, the cells were washed with DMEM and infected with GAS M49 wild type and mutant strains at a multiplicity of infection of 1:10 in DMEM. Two hours after infection, the cells were washed extensively with PBS, detached from the wells by trypsin treatment, and lysed with sterile distilled water. The viable counts of GAS (colony-forming units) released from the lysed cells were determined by serial dilution in PBS and plating on THY agar. For the assessment of bacterial internalization, 2 h after infection, the cells were washed with PBS and incubated with DMEM supplemented with penicillin (50 units/ml) and streptomycin (5 mg/ml) for an additional 2 h. Subsequently, the cells were washed and lysed, and the GAS viable counts were determined as described above.
To measure the direct interaction of EpfN with epithelial cell lines, latex beads (carboxylate-modified, polystyrene, fluorescent yellow-green; Sigma) were used in adherence assays. Briefly, 108 yellow fluorescent latex beads (1 μm) were incubated with 50 μl of purified proteins (100 μg/ml) in PBS overnight at 4 °C. After washing steps, free binding sites on the bead surface were blocked by incubation in 200 μl of bovine serum albumin (10 mg/ml) for 1 h at room temperature. The beads were then washed again and suspended in DMEM without any supplementation. Beads exclusively treated with bovine serum albumin were used in all experiments as negative control. Cell lines were infected for 2 h under 5% CO2 atmosphere with the seeding strategy of 35 beads/cell. After incubation the cells were washed with PBS, and the number of adherent beads and cells were counted and related to each other. To elucidate the effect of plasminogen (Plg) on Epf adherence, cell lines were also pretreated with human Plg (2 μg ml−1) for 30 min. Unbound Plg was removed by washing the cells with PBS, and the cells were infected with beads as described above.
Purification of EpfN for Crystallization
For crystallization, EpfN was purified by affinity chromatography using Strep-Tactin Superflow High Capacity beads (IBA GmbH) followed by size exclusion chromatography in buffer C (16). Selenomethionine-substituted EpfN (SeMet-EpfN) was produced in a similar manner to native EpfN, with the exception that 15 min before induction the following amino acids were added to the expression culture: 60 mg/liter l-selenomethionine, 100 mg/liter l-lysine, 100 mg/liter l-phenylalanine, 100 mg/liter l-threonine, 80 mg/liter l-isoleucine, 80 mg/liter l-leucine, 80 mg/liter l-valine, as previously described (18). SeMet-EpfN was purified in the same way as EpfN, with the addition of β-mercaptoethanol to the initial purification buffers and of 1 mm Tris(2-carboxy-ethyl)phosphine hydrochloride to buffer C.
Crystallization of EpfN
EpfN was crystallized using an in situ proteolysis protocol, in which EpfN mixed with chymotrypsin at a weight ratio of 1000:1 was subjected to hanging drop vapor diffusion against 20–25% (w/v) PEG3350 and 100–400 mm KCH3COO or KCl (16). Crystals of SeMet-EpfN were obtained using a similar mixture of SeMet-EpfN with chymotrypsin in 20–25% (w/v) PEG3350 and 100–400 mm KCH3COO, which was streak-seeded with native EpfN crystals using a cat whisker. In all crystallization experiments, 1 μl of EpfN (19.5–20 mg/ml in buffer C) was mixed with an equal volume of the reservoir solution and equilibrated at 18 °C. For flash-cooling in liquid nitrogen, the crystals were cryo-protected with buffer C containing 30% (v/v) glycerol, 17.5% (w/v) PEG3350, and 140 mm KCH3COO (KCH3COO condition) or a mixture of 70% (v/v) ParatoneN and 30% (v/v) paraffin (KCl condition).
Structure Determination and Refinement
All of the data sets (Table 1) were collected at Beamline MX2 (03ID1) of the Australian Synchrotron (Victoria, Australia) and processed as described previously (16). The program suite AutoSHARP (19) was used to solve the structure of EpfN in a multiple-wavelength anomalous dispersion experiment with two data sets from a single SeMet-EpfN crystal (Selenium Remote and Selenium Inflection in Table 1). All four expected selenium sites were found (correlation coefficient of 0.289) using the SHELXC/D package (20), and phase information was successfully derived using SHARP (21) with a figure of merit of 0.33/0.13 (acentric/centric). The resulting electron density map was modified using SOLOMON (22). Using Arp/Warp (23), two EpfN chains were automatically built to 94% completeness. The EpfN model was refined using diffraction data from a crystal of native EpfN grown in KCH3COO. Refinement was performed in iterative cycles of manual building with COOT (24) and maximum-likelihood refinement in autoBUSTER (25) with 2-fold noncrystallographic symmetry constraints. Water molecules were picked manually and automatically using autoBUSTER. The final EpfN model consists of two chains A (residues 56–357) and B (57–353). Chain A was then used as a search model for molecular replacement with PHASER (26) to solve the structure of EpfN-KCl, which has 4 molecules per asymmetric unit. The EpfN-KCl model was refined as above, using 4-fold noncrystallographic symmetry constraints. Full details of both refinements are included in Table 1. Model geometry was assessed using MolProbity (27). The figures were created using PyMol (28).
TABLE 1.
Crystal data and statistics for data collection, structure determination, and refinement
| Data set | Selenium |
EpfN-KCH3COO | EpfN-KCl | |
|---|---|---|---|---|
| Remote | Inflection | |||
| Crystal data | ||||
| Growth conditions | 22% (w/v) PEG3350, 200 mm KCH3COO | 22% (w/v) PEG3350, 200 mm KCH3COO | 25% (w/v) PEG3350, 400 mm KCH3COO | 23% (w/v) PEG3350, 200 mm KCl |
| Space group | P212121 | P212121 | P212121 | P21 |
| Unit cell parameters (Å, °) | a = 60.46, b = 86.14, c = 118.23, α = β = γ = 90 | a = 60.55, b = 86.21, c = 118.47, α = β = γ = 90 | a = 60.27, b = 85.62, c = 117.78, α = β = γ = 90 | a = 71.08, b = 117.13, c = 73.04, α = 90, β = 106.46, γ = 90 |
| Molecules per a.u. | 2 | 2 | 2 | 4 |
| Data collection | ||||
| Wavelength (Å) | 0.95369 | 0.97941 | 0.97942 | 0.97942 |
| Resolution rangea | 86.17–1.80 (1.90–1.80) | 86.17–2.20 (2.32–2.20) | 19.88–1.58 (1.66–1.58) | 19.73–2.00 (2.11–2.00) |
| Total observationsa | 1,649,177 (219,367) | 921,369 (133,936) | 1,699,412 (137,780) | 283,329 (41,558) |
| Unique reflectionsa | 57,988 (8,357) | 32,146 (4,629) | 83,319 (11,097) | 76,117 (11,229) |
| Redundancya | 28.4 (26.2) | 28.7 (28.9) | 20.4 (12.4) | 3.7 (3.7) |
| Completeness (%)a | 99.9 (100) | 100 (100) | 98.7 (91.6) | 98.4 (99.2) |
| Mean I/σ(I)a | 25.3 (6.6) | 26.1 (9.6) | 35.1 (5.6) | 6.4 (2.2) |
| Rmerge (%)a | 13.3 (57.5) | 15.6 (49.8) | 7.0 (45.7) | 11.3 (44.1) |
| Rp.i.m. (%)a | 2.6 (11.5) | 3.0 (9.6) | 1.6 (12.9) | 7.6 (30.8) |
| Phasing | ||||
| Selenium sites | 4 | 4 | ||
| RCullis (%) | 84.2 | 83.6 | ||
| Phasing power | 0.95 | 0.95 | ||
| Refinement | ||||
| Resolution rangea | 19.80–1.58 (1.62–1.58) | 19.73–2.00 (2.05–2.00) | ||
| Number of reflectionsa | 83,232 (5,261) | 76,089 (5,651) | ||
| R/Rfree (%)a | 15.0/18.1 (19.5/23.4) | 17.2/21.5 (20.7/25.0) | ||
| Cutoff criterion (σF) | 0 | 0 | ||
| Number of atoms | ||||
| Protein | 4,980 | 9,528 | ||
| Water | 978 | 1035 | ||
| Root mean square deviations from standard values | ||||
| Bond lengths (Å) | 0.010 | 0.010 | ||
| Bond angles (°) | 1.07 | 1.13 | ||
| Wilson B (Å2) | 14.7 | 28.0 | ||
| Mean B, protein (Å2) | 14.7 | 30.8 | ||
| Mean B, solvent (Å2) | 29.7 | 34.1 | ||
| Ramachandran statistics | ||||
| Favored (%) | 98.8 | 98.0 | ||
| Allowed (%) | 1.2 | 2.0 | ||
| Outliers (%) | 0 | 0 | ||
| Poor rotamers (%) | 0.75 | 1.2 | ||
a The values in parentheses are for the outermost resolution shell.
Structure and Sequence Analysis
The Protein Data Bank (PDB) was searched for homologous structures using SSM (29) and DALI (30). The crystal packing was analyzed using PISA (31).
CD Spectroscopy
For CD analysis, the construct DUF1-4 was dialyzed extensively against 10 mm phosphate buffer, pH 7.7. CD spectra were recorded at a DUF1-4 concentration of 1 μm in a 1-mm quartz cuvette at 20 °C. The final spectrum is the average of 10 measurements; the spectrum for the buffer was deducted. The secondary structure composition was estimated using the SOMCD algorithm (32).
Electron Microscopy
EpfDUF1-16 (5 μl of a 0.1 mg/ml solution in buffer C) was adsorbed on to continuous carbon grids for 60 s. Sample solutions were blotted with Whatman #1 filter paper (Amersham Biosciences, GE Healthcare) and stained for 60 s with 20 μl uranyl acetate solution (1% w/v; Electron Microscopy Sciences, Hatfield, PA) followed by final blotting. Low dose EM was performed using a Tecnai 12 electron microscope (FEI, Hillsboro, Oregon), and images were recorded at a nominal magnification of 52,000× using an UltraScan 2k × 2k CCD camera (Gatan, Pleasanton, CA).
Small Angle X-ray Scattering (SAXS)
SAXS analysis was performed at the SAXS/WAXS Beamline of the Australian Synchrotron. The construct EpfN-DUF1-3 in buffer C was analyzed over a concentration range of 0.25–2.0 mg ml−1 at 4 °C. The data were recorded using a Pilatus 1M detector at a distance of 3.5 m. For absolute scaling, intensity data were normalized to water as standard. After buffer subtraction, the scattering curves were analyzed using PRIMUS (33) and AutoRG (34). The distance distribution function was calculated using GNOM (35). Sixteen dummy atom models were reconstructed using DAMMIF (36), of which fifteen were averaged with DAMAVER (37) and superimposed with the EpfN crystal structure using SUPCOMB13 (38). Further experimental details are listed in Table 2.
TABLE 2.
SAXS data collection and scattering derived parameters for construct EpfNDUF1-3
| Data collection parameters | |
| X-ray source | Australian Synchrotron |
| Wavelength (Å) | 1.127 |
| q range (Å−1) | 0.004–0.25 |
| Sample flow rate (4 μl s−1) | 4 |
| Exposure time/frame (s) | 1 |
| Concentration range (mg ml−1) | 0.25–2.0 |
| Structural parameters | |
| I(0) (cm−1) (from Guinier plot) | |
| 2 mg ml−1 | 0.108 ± 0.0001 |
| 1 mg ml−1 | 0.051 ± 0.0001 |
| 0.5 mg ml−1 | 0.025 ± 0.0001 |
| 0.25 mg ml−1 | 0.012 ± 0.0001 |
| Rg (Å) (from Guinier plot) | |
| 2 mg ml−1 | 58.2 ± 0.4 |
| 1 mg ml−1 | 57.6 ± 0.7 |
| 0.5 mg ml−1 | 56.9 ± 1.2 |
| 0.25 mg ml−1 | 57.3 ± 1.7 |
| I(0) (cm−1) (from P(r))a | 0.112 ± 0.001 |
| Rg (Å) (from P(r))a | 63.1 ± 0.4 |
| Dmax (Å)a | 215 |
| Molecular mass Mr (from I(0))a | 74,150 |
| Calculated monomeric mass Mr from sequence | 63,184 |
| Normalized spatial discrepancy of averaged dummy atom modelsa | 0.62 ± 0.07 |
a Reported for 2 mg ml−1 measurement, which was used for model building.
EpfN Binding to Immobilized Plasminogen
Native Plg was purified from human plasma as previously described (39). For the pulldown assay, purified Plg was prebound to lysine-Sepharose (GE Healthcare) in buffer D (5 mm Tris-HCl, pH 8.0, 100 mm phosphate); EpfN_b was then added to Plg at 5-fold molar excess and incubated at 4 °C in the same buffer overnight with mixing. Unbound material was removed by washing with buffer D, and bound material was eluted with 25 mm ϵ-amino caproic acid. Plg alone and EpfN_b alone were included as controls.
Surface Plasmon Resonance
The interactions between Plg and recombinant Epf were analyzed with a BIAcore3000 system (Biosensor, La Jolla, CA) using CM5 sensor chips as described before (40). Briefly, the ligand EpfN was immobilized on the flow cell surfaces of the chip to densities up to 1500 response units using standard amine-coupling chemistry and the software tool “Application Wizard-Surface Preparation” (BIA-core 3000 Instrument Handbook). Each analyte-ligand complex was allowed to associate and dissociate for 3 and 5 min, respectively, with background subtraction using an unmodified flow cell as reference surface. To collect binding data, the analytes, i.e. plasminogen, fibronectin, fibrinogen, and collagen-I dissolved in 10 mm HEPES, 150 mm NaCl, 3 mm EDTA, 0.005% P20, pH 7.4, were flowed over the ligand and reference surfaces at concentrations of 125 nm and at a flow rate of 30 μl min−1. For concentration series, plasminogen was tested at 50, 125, 250, 500, 1000, and 2000 nm. The ligand surface was regenerated with a 15-s injection of 0.1% SDS at the end of each binding cycle. Three independent replicate experiments were carried out to obtain meaningful and reliable results. The data from the BIAcore sensorgrams were fitted globally, using the one-step biomolecular association reaction model (1:1 Langmuir binding with drifting base line), which resulted in optimum mathematical fits, reflected by low χ2 values (<5).
RESULTS
Requirement of Epf for GAS Adherence and Internalization
Adherence to and internalization in human epithelial cells are essential steps for successful infection of the human host by GAS. We assayed the importance of Epf for these processes by infecting four different human epithelial cell lines for 2 h with wild type GAS serotype M49 and its epf-deficient mutant (epf−). The epf− mutant showed significantly reduced adherence to human skin keratinocytes (HaCaT cells), by ∼50% (Fig. 2A). GAS expresses a number of other adhesins, and this experiment shows that even on this background, Epf plays a significant role in adhesion. Similarly, the epf− mutant showed decreased internalization rates for both HaCaT cells and the gingival epithelial cell line Ca9-22 (Fig. 2B). The internalization rate for HaCaT cells was low under the conditions used but is well in the range of previous studies (41). Both processes, adherence and internalization, were unaffected in the case of Detroit562 (pharyngeal) and HEp-2 (laryngeal) cells.
FIGURE 2.
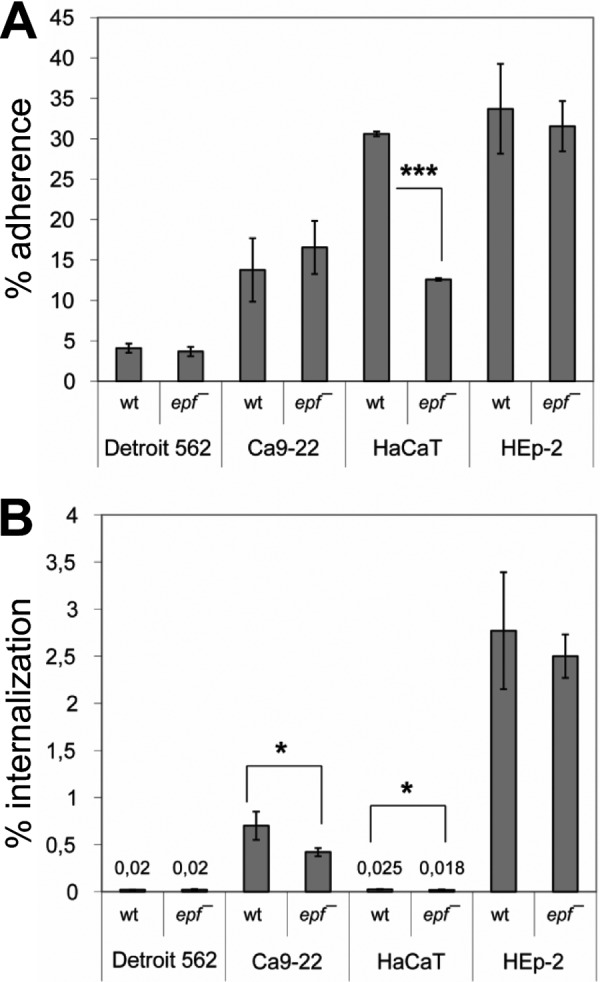
The impact of inactivation of epf on GAS M49 adherence to (A) and internalization into (B) human epithelial cells. The data for wild type (wt) and epf-deficient mutant (epf−) represent the mean values ± standard deviation from five independent experiments. Only significant differences are indicated (U test). *, p < 0.05; ***, p < 0.001).
Adherence of Epf Domains to Epithelial Cell Lines
To identify the domain(s) conferring the adhesin activity on Epf, the constructs EpfN, EpfDUF1-4, and EpfDUF1-16 (Fig. 1) were used to coat yellow fluorescent latex beads, which were incubated with human epithelial cell lines for 2 h. Uncoated beads were used as control and did not display any cell binding. In additional experiments, the cells were treated with Plg before infection with the Epf constructs. After the infection the cells were stained with DAPI (Roche Applied Science) and inspected by fluorescence microscopy. The results are summarized in Table 3.
TABLE 3.
Adherence of latex beads coated with the Epf constructs EpfN, EpfDU1-4, and EpfDUF1-16 to human epithelial cell lines
The p values were calculated using the U test. Plg, pretreatment of cells with plasminogen.
| Cell line | Beads coating | Cell treatment | No. of beads/cell | p |
|---|---|---|---|---|
| Ca9–22 | 0.050 ± 0.002 | |||
| EpfN | 0.216 ± 0.004 | 0.006 | ||
| EpfN | Plg | 0.229 ± 0.080 | 0.068 | |
| Detroit562 | 0.004 ± 0.001 | |||
| EpfN | 0.012 ± 0.002 | 0.058 | ||
| EpfN | Plg | 0.011 ± 0.011 | 0.076 | |
| HaCaT | 0.035 ± 0.025 | |||
| EpfN | 0.014 ± 0.001 | 0.211 | ||
| EpfN | Plg | 0.440 ± 0.035 | 0.002 | |
| HEp-2 | 0.070 ± 0.047 | |||
| EpfN | 0.350 ± 0.082 | 0.006 | ||
| EpfN | Plg | 0.650 ± 0.276 | 0.011 | |
| 0.051 ± 0.002 | ||||
| EpfDUF1-4 | 0.008 ± 0.006 | 0.076 | ||
| EpfDUF1-4 | Plg | 0.002 ± 0.081 | 0.068 | |
| 0.051 ± 0.002 | ||||
| EpfDUF1-16 | 0.040 ± 0.038 | 0.712 | ||
| EpfDUF1-16 | Plg | 0.022 ± 0.221 | 0.076 |
EpfN-coated beads were found to bind to both Ca9-22 and HEp-2 cells directly, whereas beads coated with the constructs EpfDUF1-4 and EpfDUF1-16, which lack EpfN, showed only background levels of adherence to any of the four tested cell lines (data only shown for HEp-2 cells). Pretreatment of the tested cell lines with Plg enabled EpfN-coated beads to adhere to HaCaT cells, enhanced their adherence to HEp-2 cells, and had no influence on their adherence to Ca9-22 and Detroit562 cells.
Structure of the N-terminal Domain of Epf
The N-terminal domain of Epf (EpfN) yielded diffracting crystals in the presence of the protease chymotrypsin, which trimmed the original EpfN construct (residues 45–386) to residues 52–357 in situ (16). Two different crystal forms were obtained depending on the presence of KCH3COO or KCl. They were in space groups P212121 and P21, with two and four molecules/asymmetric unit, respectively (Table 1). The structure of EpfN was solved using multiple-wavelength anomalous dispersion phases from a single crystal of SeMet-EpfN in the orthorhombic space group P212121. The initial model was then refined against data for both crystal forms of native EpfN. In all chains, in both crystal forms, there is clear electron density for residues 57–352, whereas residues 52–56 and 353–357 appear to be subject to varying degrees of disorder. Otherwise, no significant difference can be observed for the EpfN structures from these two space groups; the two structures show root mean square (rms) differences between the six chains of 0.3–0.7 Å for all 296 residues. In the following, we use as the reference structure EpfN chain A of the P212121 crystal form (EpfN-KCH3COO in Table 1), which was refined at a resolution of 1.6 Å (R/Rfree = 15.0/18.2%). This molecule is almost complete, comprising residues 56–357.
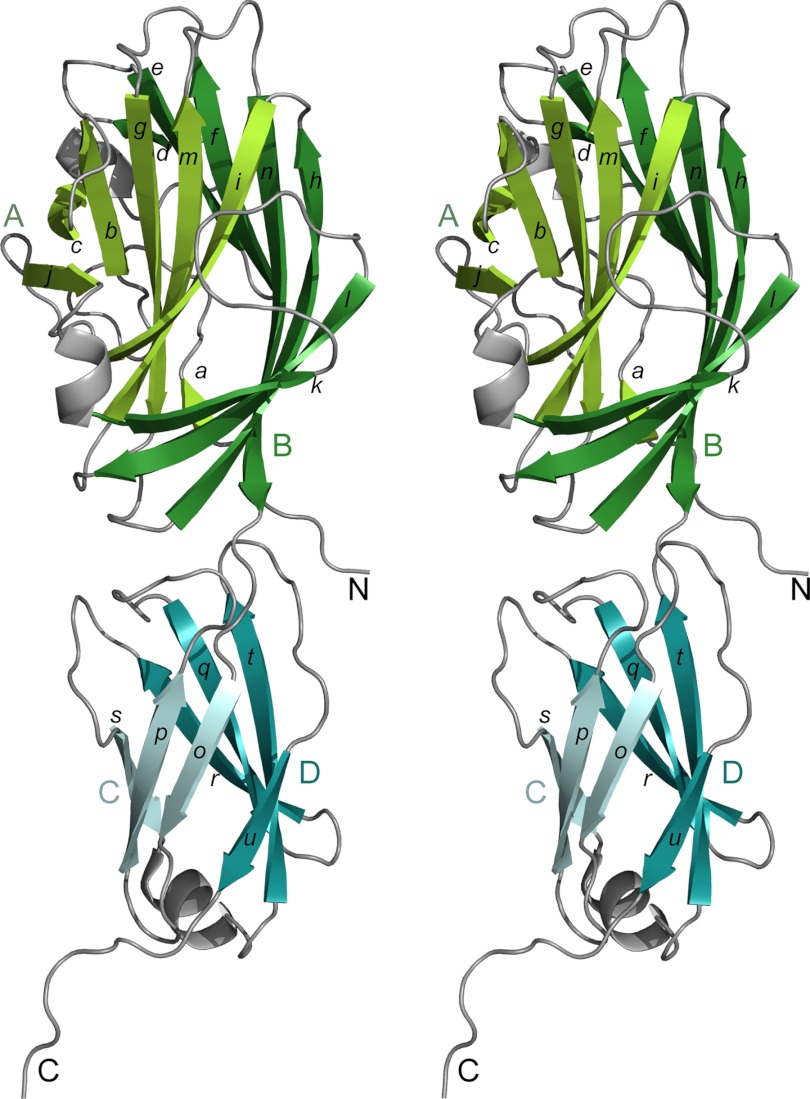
The EpfN molecule (Fig. 3) comprises two subdomains, EpfN1 (residues 56–251) and EpfN2 (residues 252–357), which together form an elongated shape with a length of ∼85 Å. Subdomain EpfN1 is folded into two twisted antiparallel β-sheets, A and B, with seven β-strands each. β-Sheet B (β-strands d, e, f, h, k, l, and n) forms half a barrel, which wraps around the almost complete barrel of β-sheet A (β-strands a, b, c, g, i, j, and m). Subdomain EpfN2 is folded as a β-sandwich of two antiparallel β-sheets, C and D, with three and four β-strands respectively (C: β-strands o, p, and s, D: β-strands q, r, t, and u). Three short α-helices decorate loops between the β-strands in both domains. The interface between the two domains EpfN1 and EpfN2 appears to be stabilized by an extensive hydrogen bond network that centers on the residues His-149, Arg-279, and Gln-334. Several ligands derived from the crystallization buffers were observed in the two EpfN structures, although most binding sites are occupied only in a single chain, e.g., by a potassium ion, glycerol, and an acetate ion.
FIGURE 3.
Structure of the N-terminal domain of Epf (EpfN), shown in stereo. The β-sheets of the EpfN1 subdomain are in green (sheet A lighter, sheet B darker), and those of the EpfN2 subdomain are in blue (sheet C lighter, sheet D darker). Lowercase letters indicate β-strands, and uppercase letters indicate the β-sheets and the N/C chain termini.
Recombinant EpfN is monomeric in solution, as judged by its retention volume in size exclusion chromatography (data not shown). The program PISA (31), which analyzes crystal packing contacts, gives a score of 0 for the likelihood that the crystal packing contacts imply oligomer formation.
Structural Relationships of the N-terminal Domain of Epf
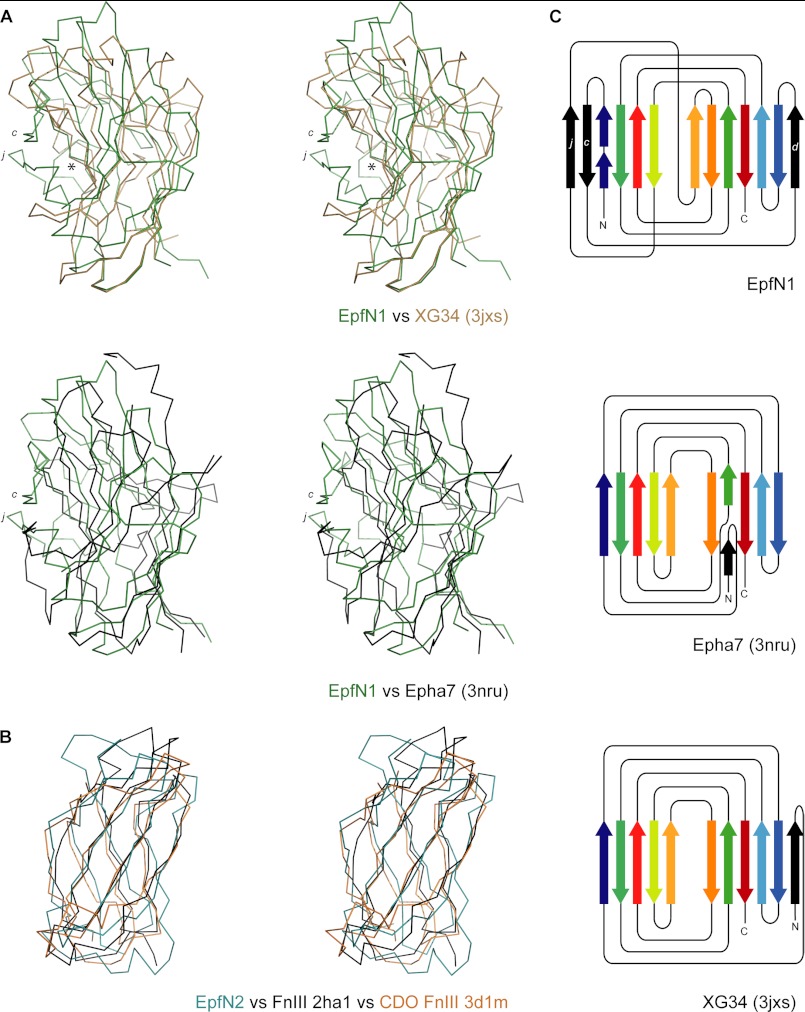
A search of the PDB for structural homologues of the full EpfN structure, using the programs SSM (29) and DALI (30), yielded no significant hits. The use of each EpfN subdomain in isolation, however, led to the identification of a number of proteins with similar folds, despite no significant sequence identity (Table 4 and Fig. 4).
TABLE 4.
Structural homologues of the two EpfN domains EpfN1 and EpfN2
The Protein Data Bank (PDB) was searched using SSM (29), and the most closely related structures are listed here with their ligand (if reported). rmsd, root mean square deviation; Nalg, number of aligned Cα positions; SI, sequence identity; NA, not applicable.
| Protein | Ligand | Organism | rmsd | Nalg | SI | PDB | Reference |
|---|---|---|---|---|---|---|---|
| Å | % | ||||||
| EpfN1 | |||||||
| Epha7 ligand-binding domain | Ephrin | Homo sapiens | 2.76 | 125 | <10 | 3NRU:K | NA |
| XG34 CBM | Xyloglucan | Rhodotermus marinus | 3.09 | 134 | <10 | 3JXS:B | Ref. 49 |
| Xylanase CBM4-2 | Xylan | Rhodothermus marinus | 2.89 | 128 | <10 | 1K45:A | Ref. 51 |
| Laminarinase CBM4 | Laminarin | Thermotoga marina | 2.71 | 118 | <10 | 1GUI:A | Ref. 52 |
| EpfN2 | |||||||
| Fibronectin type III domain | Homo sapiens | 2.42 | 76 | <10 | 2HA1:A | Ref. 43 | |
| FnIII domain of CDO | Desert hedgehog protein | Homo sapiens | 2.85 | 86 | <10 | 3D1M:D | Ref. 50 |
| FnIII domain NCAM1 | Homo sapiens | 2.68 | 86 | <10 | 2E3V:B | NA | |
| CHIR-AB1 | IgY | Gallus gallus | 2.46 | 80 | <10 | 2VSD:A | Ref. 53 |
FIGURE 4.
Structural relationships of the EpfN subdomains EpfN1 and EpfN2. A, structural superposition of EpfN1, in green, on to the carbohydrate-binding module of XG34 (PDB code 3jxs) (49), in brown, and the ligand-binding domain of the human ephrin receptor Epha7 (PDB code 3nru), in black; c and j designate the additional β-strands in EpfN1, and the asterisk indicates the carbohydrate-binding site in XG34. B, structural superposition of EpfN2, in green, on to the fibronection type III domains of fibronectin (2ha1) (43), in black, and human CDO (3d1m) (50), in brown. In each case, the fold is represented as a Cα trace in stereo; structures were aligned using SSM (31). C, topology diagrams of EpfN1, Epha7, and XG34. Aligned β-strands are marked in the same colors, and additional β-strands are colored black. For clarity, decorating α-helices, which are present in all three structures, were omitted, and the lengths of β-strands are not to scale.
Subdomain EpfN1 is structurally similar to a number of carbohydrate-binding modules (CBMs) with rms differences of 2.7–3.4 Å over 110–134 aligned Cα positions (Table 4). In general, CBMs are noncatalytic domains of carbohydrate-modifying enzymes (42). EpfN1 is also similar to the ligand-binding domain of the human ephrin receptor (PDB entry 3NRU). Although sharing the same basic fold, EpfN1 does, however, have distinct differences from the β-sandwich fold CBMs and the ephrin receptor because of the presence in EpfN1 of additional loops and β-strands, such as β-strands c, d, and j and their associated loops. Strands c and j protrude from β-sheet A, which usually forms a flat or concave ligand-binding site in a typical CBM (Fig. 4, A and C).
The fold of the EpfN2 subdomain conforms to that of fibronectin type III domains. EpfN2 can be superimposed on the first type III domain of fibronectin (PDB 2HA1) (43) with an rms difference of 2.42 Å over 76 aligned Cα positions (Table 4 and Fig. 4B). Superpositions of the structures of fibronectin type III-type domains from other proteins such as the signaling receptor CDO or the neural cell adhesion molecule NCAM yield rms differences in a similar range (Table 4).
Structure of the DUF1542 Repeats
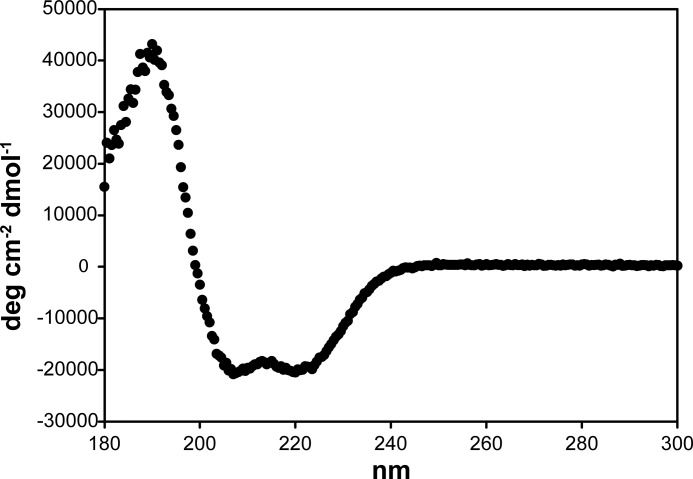
Although the EpfDUF1-4 construct, which comprises four DUF1542 repeats, could be expressed in soluble form, it has not yet been possible to obtain crystals. We therefore turned to CD spectroscopy. The CD spectrum of DUF1-4 proved to be typical of an α-helical protein (Fig. 5). Using the SOMCD algorithm (32), this construct is estimated to consist of more than 90% α-helical structure. In analogy to adhesins such as the M protein, we find increasing conservation of the DUF1542 repeats toward the C terminus of Epf. In contrast to the M protein, however, which forms a coiled-coil dimer, size exclusion chromatography-mutliple-angle laser light scattering analysis showed that the EpfDUF1-4 construct is monomeric (data not shown).
FIGURE 5.
CD spectrum of the Epf DUF1542 repeats. Construct EpfDUF1-4 was analyzed at a concentration of 1 μm in 10 mm phosphate buffer, pH 7.7; the final spectrum is the average of 10 measurements.
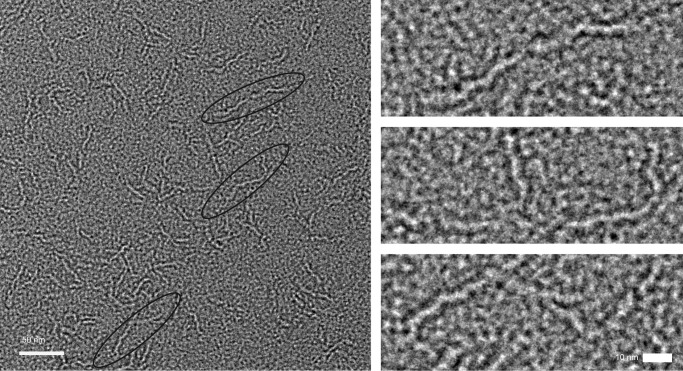
To gain further structural information on the C-terminal DUF1542 repeat region, we used EM and SAXS analysis. The construct EpfDUF1-16, comprising 16 DUF1542 repeats, was analyzed by negative staining EM, which revealed long, flexible, very thin fibrous structures (Fig. 6). Their apparent dimensions are 50–60 nm in length and 6 nm in thickness. Within a fiber, globular domains are recognizable. Thus, the DUF1542 repeats seem to form a series of small globular domains, arranged in tandem, to give a fiber-like stalk.
FIGURE 6.
Electron microscopy of Epf. Shown is an electron micrograph of the EpfDUF1-16 construct, comprising the C-terminal 16 DUF1542 repeats, at 52,000 magnification. Left panel, field of view showing EpfDUF1-16 particles (black ovals). The scale bar is 50 nm. Right panel, the three representative EpfDUF1-16 particles circled in the left panel are shown at higher magnification. The scale bar is 10 nm.
The construct EpfN-DUF1-3, which encompasses the EpfN domain and the first three DUF1542 repeats, was analyzed using SAXS (Table 2 and Fig. 7). The distance distribution function (Fig. 7C) is typical of extended multidomain proteins. An averaged dummy atom model was reconstructed for EpfN-DUF1-3 and shows an extended structure of ∼200 Å (Fig. 7D). Although this structure is mostly featureless, it is notable that there is a thicker part (∼90 Å long) at one end. The EpfN crystal structure could be superimposed on this thick part of the model (Fig. 7D), leaving ∼100–120 Å for the three DUF1542 repeats. Thus, by combining the SAXS model of EpfN-DUF1-3 with the EM data for DUF1-16, Epf can be visualized as a fiber-like structure with the adhesion domain EpfN at one end.
FIGURE 7.

Small-angle x-ray scattering analysis of an N-terminal construct of Epf. Small angle x-ray scattering analysis of the construct EpfNDUF1-3, comprising the Epf N-terminal domain and the first three DUF1542 domains, is shown. A, scattering profiles for this construct at four concentrations. Overlaid is the fit for a representative dummy atom model used for the averaged model shown in D (χ2 of 0.47). B, the Guinier plots corresponding to the profiles in A. C, the distance distribution function P(r) for EpfNDUF1-3 at 2 mg/ml. D, an average of 15 dummy atom models for EpfN-DUF1-3, with the EpfN crystal structure superimposed. The scale bar is 100 Å.
Binding of EpfN to Plasminogen
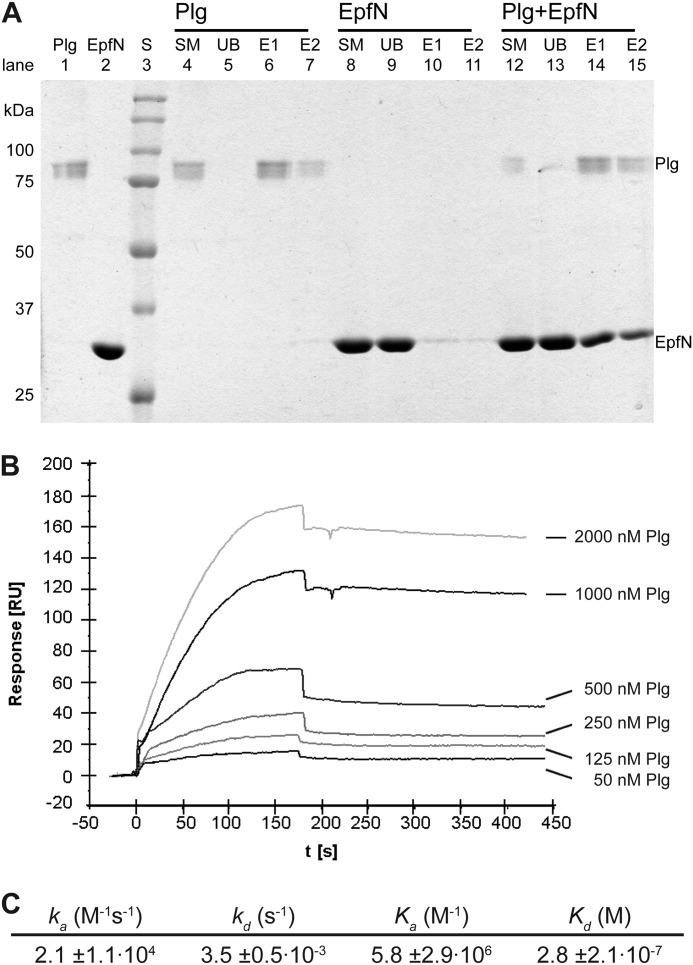
Following previous reports that Epf might bind to the human plasma protein Plg (11), we further assessed this potential interaction using pulldown assays. Plg can be immobilized on lysine-Sepharose through its kringle domains and eluted with the lysine analog ϵ-amino caproic acid (Fig. 8A). EpfN alone did not bind to lysine-Sepharose. However, when EpfN was incubated with Plg that was immobilized on lysine-Sepharose, both proteins then co-eluted after the addition of ϵ-amino caproic acid (Fig. 8A). These results indicate that immobilized plasminogen is able to bind EpfN.
FIGURE 8.
Interaction between EpfN and human plasminogen. A, binding of EpfN to immobilized plasminogen in a pulldown assay. Plg was immobilized on lysine-Sepharose and could be eluted with ϵ-amino caproic acid (lanes 4–7). EpfN alone did not bind to lysine-Sepharose (lanes 8–11) but co-eluted with plasminogen (lanes 12–15). Lanes 1 and 2 show pure plasminogen and EpfN samples. S, protein standards; SM, starting material; UB, unbound material after incubation with lysine-Sepharose; E1 and E2, fractions eluted with ϵ-amino caproic acid. B and C, quantification of the interaction of Plg (analyte) with immobilized EpfN (ligand) using surface plasmon resonance measurements. B, representative profile of the relative surface plasmon resonance responses for the association and dissociation of different analyte concentrations (50–2000 nm). Injection of the analyte started at time 0 and ended after 180 s, whereupon the dissociation phase was documented for at least 270 s. Shown is one representative concentration series out of three independent experiments. C, the association and dissociation data of the interaction were fitted globally using the one step biomolecular reaction model (1:1 Langmuir binding with drifting base-line model: A+B ⇄ AB), which resulted in optimum mathematical fits reflected by the lowest χ2 values (0.3–5). The values for association rate (ka), dissociation rate (kd), association constant (Ka), and dissociation constant (Kd) were calculated from the binding data with the BIAevaluation software. The mean values from three independent experiments are presented in the table ± standard deviation.
Quantification of the EpfN-Plasminogen Interaction
The binding strength between EpfN and Plg was quantified by surface plasmon resonance measurements employing the BIAcore system. For this real time biospecific interaction analysis, Plg was used as soluble analyte and EpfN as immobilized ligand. Increasing concentrations of the analyte were allowed to associate with EpfN, immobilized as the ligand on the CM5 chip. The sensorgram of this interaction was recorded (Fig. 8B), and the data obtained were used to calculate the dissociation constant (Kd; see also Fig. 8C). Given that typical Kd values for biologically significant interactions are in the nanomolar to low millimolar range, the determined Kd value of 0.28 μm for the EpfN-Plg interaction implies a biologically relevant interaction. Other proteins such as fibronectin, fibrinogen, and collagen-I did not bind to EpfN in the BIAcore system (data not shown).
DISCUSSION
One of the earliest events of the infection process is the attachment of bacteria to epithelial cells, mediated by surface proteins that serve as adhesins. Group A streptococci are known to produce a plethora of adhesins, which vary among different serotypes. Examples include the M-protein, which is the major cell wall-associated virulence factor (2), adhesive pili that are assembled by the action of sortase enzymes (44), and the fibronectin-binding protein FbaB (40). These are all anchored covalently to the cell wall by the action of sortases that recognize a characteristic LPXTG sorting motif. Understanding their roles and mechanisms of action is essential for understanding disease but is made more difficult by the exposure of these proteins to human immune surveillance. This typically results in wide sequence variation that makes it difficult to recognize structural and functional relationships from sequence alone.
Here, we have examined the structure and function of the putative streptococcal adhesin Epf. Epf and its homologues are found in serotypes M1, M4, M12, M28, and M49 and share several features that suggest they are adhesins. They are encoded in the so-called ERES pathogenicity island, adjacent to the transcriptional regulator ralp3 (11), they have an LPXTG sequence motif near the C terminus that implies covalent attachment to the cell wall, and they have a domain structure consisting of a unique N-terminal domain followed by a large number of conserved repeat domains, classified as DUF1542 repeats. This domain arrangement is analogous to that of many adhesins (45).
We have shown here that Epf is indeed an adhesin that is important for adhesion of GAS serotype M49 (strain 591) to human skin keratinocytes; an epf knock-out leads to a ∼50% decrease in adherence to these cells (Fig. 2A). Epf also appears to promote internalization into human skin keratinocytes and into the gingival cell line Ca9-22 (Fig. 2B). Interestingly, adhesion to other epithelial cell lines of pharyngeal or laryngeal origin was unaffected in the epf knock-out. The GAS strain 591 was originally isolated from a patient with a GAS skin infection, and in this clinical context, Epf on serotype M49 GAS may play a specific role in tissue tropism by conferring adherence to human skin.
Dissection of Epf into its N-terminal domain (EpfN) and constructs comprising varying numbers of the C-terminal DUF1542 repeats identifies EpfN as the adhesin domain. Only EpfN adhered to the human cell lines tested, namely to Ca9-22 (gingival) and HEp-2 (laryngeal) cells. In contrast, the DUF1542 repeats show no evidence of any involvement in adhesion. They appear to have a structural role, and the data from EM and SAXS analysis show that they form a fiber-like structure that supports the EpfN domain at its distal end.
This observation, that Epf consists of a ligand-binding N-terminal domain supported on a stalk formed by the C-terminal DUF1542 repeats, confirms the proposed similarities in overall architecture between Epf and other cell surface adhesins. Examples include the multidomain adhesin Cna from Staphylococcus aureus, which has a collagen-binding A domain near the N terminus, supported by a series of repetitive B domains (12, 13), and the pili expressed by many Gram-positive pathogens, which have their adhesin domains at their tip, supported by a polymeric shaft formed from multiple Ig-like domains (46).
The crystal structure of the EpfN domain reveals striking structural homologies that were not apparent from its sequence. EpfN is folded into two subdomains, EpfN1 and EpfN2, which, in contrast to the DUF1542 repeats, are primarily formed from β-strands. The EpfN1 subdomain shows a close structural similarity to the β-sandwich class CBMs (Table 4 and Fig. 4A), which are usually found as binding domains associated with carbohydrate-modifying enzymes such as cellulases (42). The EpfN2 subdomain has the fibronectin type III fold, found in many human proteins that mediate protein-protein interactions including fibronectin itself and the neural cell adhesion molecule (Table 4 and Fig. 4B). Thus, both EpfN1 and EpfN2 display structural homology to domains that commonly participate in receptor-ligand interactions, in full support of the role EpfN evidently plays in GAS adhesion.
How might EpfN adhere to cells? The similarity of the EpfN1 subdomain to CBM domains suggested that it could bind extracellular carbohydrates on the host cell surface. However, a glycan array screen performed by the Functional Glycomics Consortium yielded no potential ligands (data not shown). It is possible that the chosen experimental conditions prevented the identification of a ligand or that this ligand was not included in the array. We note, however, that most classic CBM domains bind carbohydrate ligands in the concave face of β-sheet A (42). In the case of EpfN1, however, this face seems to be occluded by the additional β-strands c and j from β-sheet A (Fig. 4A). Moreover, the carbohydrate binding sites of CBMs almost always include an associated metal ion (42), whereas EpfN1 contains no bound metal. Some CBMs do, however, bind ligands at the edge of β-sheets A and B, as observed for the ephrin receptor (47) and some classical CBMs (48). A potential candidate for such a binding site could be the edge of β-sheet B, where the surface-exposed residues Trp-104, Tyr-114, and Arg-125 are located; aromatic side chains are strongly associated with carbohydrate binding (42).
It is also possible that EpfN recognizes a protein ligand. Protein binding by an EpfN1-like domain has been observed for the ephrin receptor A7 (Epha7) ligand-binding domain (47), and fibronectin type III domains like EpfN2 are well known for their ability to bind protein ligands. An ability to bind protein ligands is supported by the demonstrated ability of Epf to bind to the human plasma protein Plg.
Pretreatment with Plg increases adherence of GAS to, and invasion into, HaCaT keratinocytes, through integrin-mediated pathways (10). In our present studies with recombinant EpfN, we observed that the pretreatment of human epithelial cells with Plg enhanced binding to HEp-2 cells and enabled binding to HaCaT cells by recombinant EpfN (Table 3). We further showed that lysine-immobilized Plg bound to EpfN in a pulldown assay (Fig. 8). The interaction between Epf and Plg is of medium strength with a Kd of 0.28 μm. However, this interaction was not strong enough to be detected using analytical size exclusion chromatography (no co-elution; data not shown). It seems possible that the significant effect of Plg on the adherence of recombinant EpfN to epithelial cells involves the activation of other receptors on the host cells or requires additional factors.
In conclusion, we have shown that Epf is an adhesin of group A streptococcus. Adhesion is mediated by an N-terminal domain EpfN that is built from two subdomains, both of which have folds that are typically involved in receptor-ligand interactions. These adhesin domains are projected away from the streptococcal cell wall toward the host receptor by a stalk formed from α-helical DUF1542 repeats, which form a long thin fiber-like structure. This would enable long range interactions with host receptors and ensure protrusion of the adhesin domain beyond the extracellular carbohydrate. DUF1542 repeats are found in many surface-exposed proteins from Gram-positive bacteria, and it is likely that other proteins from a variety of Gram-positive bacteria may also have an N-terminal domain similar to EpfN. However, structural data may be needed to establish their homology because of the high sequence variation typical for N-terminal domains of cell wall-attached proteins.
Acknowledgments
We thank A. J. Quek and D. Jeevarajah for technical assistance, M. Cumming and C. Day for help with SEC-MALLS analysis, J. Busby and L. O'Ryan for SAXS data collection, and D. C. Goldstone for helpful discussion.
This work was supported by the Medical Faculty of the University of Rostock in the framework of the FORUN program, the German Federal Ministry of Education and Research (BMBF) in the framework of the ERANet “PathoGenoMics” I and II program, the Health Research Council of New Zealand, the Tertiary Education Commission of New Zealand through funding of the Maurice Wilkins Centre for Molecular Biodiscovery and the National Health and Medical Research Council of Australia. Access to the Australian Synchrotron was supported by the New Zealand Synchrotron Group Ltd.
The atomic coordinates and structure factors (codes 4es8 and 4es9) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- GAS
- group A streptococcus
- CBM
- carbohydrate-binding module
- EpfN
- N-terminal domain of Epf
- PDB
- Protein Data Bank
- Plg
- plasminogen
- rms
- root mean square
- SAXS
- small angle x-ray scattering
- SeMet-EpfN
- selenomethionine-substituted EpfN.
REFERENCES
- 1. Cunningham M. W. (2000) Pathogenesis of group A streptococcal infections. Clin. Microbiol. Rev. 13, 470–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Courtney H. S., Hasty D. L., Dale J. B. (2002) Molecular mechanisms of adhesion, colonization, and invasion of group A streptococci. Ann. Med. 34, 77–87 [DOI] [PubMed] [Google Scholar]
- 3. Kreikemeyer B., Klenk M., Podbielski A. (2004) The intracellular status of Streptococcus pyogenes. Role of extracellular matrix-binding proteins and their regulation. Int. J. Med. Microbiol. 294, 177–188 [DOI] [PubMed] [Google Scholar]
- 4. LaPenta D., Rubens C., Chi E., Cleary P. P. (1994) Group A streptococci efficiently invade human respiratory epithelial cells. Proc. Natl. Acad. Sci. U.S.A. 91, 12115–12119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Osterlund A., Engstrand L. (1997) An intracellular sanctuary for Streptococcus pyogenes in human tonsillar epithelium. Studies of asymptomatic carriers and in vitro cultured biopsies. Acta Otolaryngol. 117, 883–888 [DOI] [PubMed] [Google Scholar]
- 6. Podbielski A., Kreikemeyer B. (2001) Persistence of group A streptococci in eukaryotic cells. A safe place? Lancet 358, 3–4 [DOI] [PubMed] [Google Scholar]
- 7. Cue D. R., Dombeck P. E., Lam H., Cleary P. P. (1998) Streptococcus pyogenes serotype M1 encodes multiple pathways for entry into human epithelial cells. Infect. Immun. 66, 4593–4601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Walker M. J., McArthur J. D., McKay F., Ranson M. (2005) Is plasminogen deployed as a Streptococcus pyogenes virulence factor? Trends Microbiol. 13, 308–313 [DOI] [PubMed] [Google Scholar]
- 9. Sanderson-Smith M. L., Dinkla K., Cole J. N., Cork A. J., Maamary P. G., McArthur J. D., Chhatwal G. S., Walker M. J. (2008) M protein-mediated plasminogen binding is essential for the virulence of an invasive Streptococcus pyogenes isolate. FASEB J. 22, 2715–2722 [DOI] [PubMed] [Google Scholar]
- 10. Siemens N., Patenge N., Otto J., Fiedler T., Kreikemeyer B. (2011) Streptococcus pyogenes M49 plasminogen/plasmin binding facilitates keratinocyte invasion via integrin-integrin-linked kinase (ILK) pathways and protects from macrophage killing. J. Biol. Chem. 286, 21612–21622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kreikemeyer B., Nakata M., Köller T., Hildisch H., Kourakos V., Standar K., Kawabata S., Glocker M. O., Podbielski A. (2007) The Streptococcus pyogenes serotype M49 Nra-Ralp3 transcriptional regulatory network and its control of virulence factor expression from the novel eno ralp3 epf sagA pathogenicity region. Infect. Immun. 75, 5698–5710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Joh D., Wann E. R., Kreikemeyer B., Speziale P., Höök M. (1999) Role of fibronectin-binding MSCRAMMs in bacterial adherence and entry into mammalian cells. Matrix Biol. 18, 211–223 [DOI] [PubMed] [Google Scholar]
- 13. Patti J. M., Jonsson H., Guss B., Switalski L. M., Wiberg K., Lindberg M., Höök M. (1992) Molecular characterization and expression of a gene encoding a Staphylococcus aureus collagen adhesin. J. Biol. Chem. 267, 4766–4772 [PubMed] [Google Scholar]
- 14. Kwinn L. A., Khosravi A., Aziz R. K., Timmer A. M., Doran K. S., Kotb M., Nizet V. (2007) Genetic characterization and virulence role of the RALP3/LSA locus upstream of the streptolysin S operon in invasive M1T1 group A Streptococcus. J. Bacteriol. 189, 1322–1329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Smith H. E., Reek F. H., Vecht U., Gielkens A. L., Smits M. A. (1993) Repeats in an extracellular protein of weakly pathogenic strains of Streptococcus suis type 2 are absent in pathogenic strains. Infect. Immun. 61, 3318–3326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Linke C., Siemens N., Middleditch M. J., Kreikemeyer B., Baker E. N. (2012) Purification, crystallization and preliminary crystallographic analysis of the adhesion domain of Epf from Streptococcus pyogenes. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 68, 793–797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ozeri V., Rosenshine I., Mosher D. F., Fässler R., Hanski E. (1998) Roles of integrins and fibronectin in the entry of Streptococcus pyogenes into cells via protein F1. Mol. Microbiol. 30, 625–637 [DOI] [PubMed] [Google Scholar]
- 18. Van Duyne G. D., Standaert R. F., Karplus P. A., Schreiber S. L., Clardy J. (1993) Atomic structures of the human immunophilin FKBP-12 complexes with FK506 and rapamycin. J. Mol. Biol. 229, 105–124 [DOI] [PubMed] [Google Scholar]
- 19. Vonrhein C., Blanc E., Roversi P., Bricogne G. (2007) Automated structure solution with autoSHARP. Methods Mol. Biol. 364, 215–230 [DOI] [PubMed] [Google Scholar]
- 20. Sheldrick G. M. (2010) Experimental phasing with SHELXC/D/E. Combining chain tracing with density modification. Acta Crystallogr. D Biol. Crystallogr. 66, 479–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bricogne G., Vonrhein C., Flensburg C., Schiltz M., Paciorek W. (2003) Generation, representation and flow of phase information in structure determination. Recent developments in and around SHARP 2.0. Acta Crystallogr. D Biol. Crystallogr. 59, 2023–2030 [DOI] [PubMed] [Google Scholar]
- 22. Abrahams J. P., Leslie A. G. (1996) Methods used in the structure determination of bovine mitochondrial F1 ATPase. Acta Crystallogr. D Biol. Crystallogr. 52, 30–42 [DOI] [PubMed] [Google Scholar]
- 23. Langer G., Cohen S. X., Lamzin V. S., Perrakis A. (2008) Automated macromolecular model building for x-ray crystallography using ARP/wARP version 7. Nat. Protoc. 3, 1171–1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Emsley P., Lohkamp B., Scott W. G., Cowtan K. (2010) Features and development of COOT. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Blanc E., Roversi P., Vonrhein C., Flensburg C., Lea S. M., Bricogne G. (2004) Refinement of severely incomplete structures with maximum likelihood in BUSTER-TNT. Acta Crystallogr. D Biol. Crystallogr. 60, 2210–2221 [DOI] [PubMed] [Google Scholar]
- 26. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chen V. B., Arendall W. B., 3rd, Headd J. J., Keedy D. A., Immormino R. M., Kapral G. J., Murray L. W., Richardson J. S., Richardson D. C. (2010) MolProbity. All-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. DeLano W. L. (2002) PyMOL molecular graphics system, DeLano Scientific, Palo Alto, CA [Google Scholar]
- 29. Krissinel E., Henrick K. (2004) Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr. D Biol. Crystallogr. 60, 2256–2268 [DOI] [PubMed] [Google Scholar]
- 30. Holm L., Rosenström P. (2010) DALI server. Conservation mapping in 3D. Nucleic Acids Res. 38, W545–W549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Krissinel E., Henrick K. (2007) Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 [DOI] [PubMed] [Google Scholar]
- 32. Unneberg P., Merelo J. J., Chacón P., Morán F. (2001) SOMCD. Method for evaluating protein secondary structure from UV circular dichroism spectra. Proteins 42, 460–470 [DOI] [PubMed] [Google Scholar]
- 33. Konarev P. V., Volkov V. V., Sokolova A. V., Koch M. H., Svergun D. I. (2003) PRIMUS. A Windows PC-based system for small-angle scattering data analysis. J. Appl. Crystallogr. 36, 1277–1282 [Google Scholar]
- 34. Petoukhov M. V., Konarev P. V., Kikhney A. G., Svergun D. I. (2007) ATSAS 2.1. Towards automated and web-supported small-angle scattering data analysis. J. Appl. Crystallogr. 40, s223–s228 [Google Scholar]
- 35. Semenyuk A. V., Svergun D. I. (1991) GNOM. A program package for small-angle scattering data processing. J. Appl. Crystallogr. 24, 537–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Franke D., Svergun D. I. (2009) DAMMIF, a program for rapid ab-initio shape determination in small-angle scattering. J. Appl. Crystallogr. 42, 342–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Volkov V. V., Svergun D. I. (2003) Uniqueness of ab initio shape determination in small-angle scattering. J. Appl. Crystallogr. 36, 860–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kozin M. B., Svergun D. I. (2001) Automated matching of high- and low-resolution structural models. J. Appl. Crystallogr. 34, 33–41 [Google Scholar]
- 39. Deutsch D. G., Mertz E. T. (1970) Plasminogen. Purification from human plasma by affinity chromatography. Science 170, 1095–1096 [DOI] [PubMed] [Google Scholar]
- 40. Kreikemeyer B., Oehmcke S., Nakata M., Hoffrogge R., Podbielski A. (2004) Streptococcus pyogenes fibronectin-binding protein F2. Expression profile, binding characteristics, and impact on eukaryotic cell interactions. J. Biol. Chem. 279, 15850–15859 [DOI] [PubMed] [Google Scholar]
- 41. Siemens N., Fiedler T., Normann J., Klein J., Münch R., Patenge N., Kreikemeyer B. (2012) Effects of the ERES pathogenicity region regulator Ralp3 on Streptococcus pyogenes serotype M49 virulence factor expression. J. Bacteriol. 194, 3618–3626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Boraston A. B., Bolam D. N., Gilbert H. J., Davies G. J. (2004) Carbohydrate-binding modules. Fine-tuning polysaccharide recognition. Biochem. J. 382, 769–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Vakonakis I., Staunton D., Rooney L. M., Campbell I. D. (2007) Interdomain association in fibronectin. Insight into cryptic sites and fibrillogenesis. EMBO J. 26, 2575–2583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Abbot E. L., Smith W. D., Siou G. P., Chiriboga C., Smith R. J., Wilson J. A., Hirst B. H., Kehoe M. A. (2007) Pili mediate specific adhesion of Streptococcus pyogenes to human tonsil and skin. Cell Microbiol. 9, 1822–1833 [DOI] [PubMed] [Google Scholar]
- 45. Vengadesan K., Narayana S. V. (2011) Structural biology of Gram-positive bacterial adhesins. Protein Sci. 20, 759–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kang H. J., Baker E. N. (2012) Structure and assembly of Gram-positive bacterial pili. unique covalent polymers. Curr. Opin. Struct. Biol. 22, 200–207 [DOI] [PubMed] [Google Scholar]
- 47. Himanen J. P., Goldgur Y., Miao H., Myshkin E., Guo H., Buck M., Nguyen M., Rajashankar K. R., Wang B., Nikolov D. B. (2009) Ligand recognition by A-class Eph receptors. crystal structures of the EphA2 ligand-binding domain and the EphA2/ephrin-A1 complex. EMBO Rep. 10, 722–728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Boraston A. B., Notenboom V., Warren R. A., Kilburn D. G., Rose D. R., Davies G. (2003) Structure and ligand binding of carbohydrate-binding module CsCBM6–3 reveals similarities with fucose-specific lectins and “galactose-binding” domains. J. Mol. Biol. 327, 659–669 [DOI] [PubMed] [Google Scholar]
- 49. Gullfot F., Tan T. C., von Schantz L., Karlsson E. N., Ohlin M., Brumer H., Divne C. (2010) The crystal structure of XG-34, an evolved xyloglucan-specific carbonyarate-binding module. Proteins 78, 785–789 [DOI] [PubMed] [Google Scholar]
- 50. McLellan J. S., Zheng X., Hauk G., Ghirlando R., Beachy P. A., Leahy D. J. (2008) The mode of Hedgehog binding to Ihog homologues is not conserved across different phyla. Nature 455, 979–983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Simpson P. J., Jamieson S. J., Abou-Hachem M., Karlsson E. N., Gilbert H. J., Holst O., Williamson M. P. (2002) The solution structure of the CBM4–2 carbohydrate binding module from a thermostable Rhodothermus marinus xylanase. Biochemistry 41, 5712–5719 [DOI] [PubMed] [Google Scholar]
- 52. Boraston A. B., Nurizzo D., Notenboom V., Ducros V., Rose D. R., Kilburn D. G., Davies G. J. (2002) Differential oligosaccharide recognition by evolutionarily-related β-1,4 and β-1,3 glucan-binding modules. J. Mol. Biol. 319, 1143–1156 [DOI] [PubMed] [Google Scholar]
- 53. Arnon T. I., Kaiser J. T., West A. P., Jr., Olson R., Diskin R., Viertlboeck B. C., Göbel T. W., Bjorkman P. J. (2008) The crystal structure of CHIR-AB1. A primordial avian classical Fc receptor. J. Mol. Biol. 381, 1012–1024 [DOI] [PMC free article] [PubMed] [Google Scholar]