Background: CaBP7 is an EF-hand-containing transmembrane protein that inhibits PI4KIIIβ activity.
Results: PI4KIIIβ interacts with CaBP7 NTD, which exhibits an expansive hydrophobic pocket.
Conclusion: The structure of CaBP7 NTD is similar to that of CaM NTD but has a more expansive hydrophobic pocket containing fewer methionine residues.
Significance: Regulation of PI4P synthesis is essential for vesicle trafficking and secretory pathway function.
Keywords: Calcium, Calcium-binding Proteins, Calmodulin, Membrane Trafficking, Nuclear Magnetic Resonance, Structural Biology, CaBP7, CaBP8, Calneurons, PI4KIIIbeta, EF-hand
Abstract
Calcium-binding protein 7 (CaBP7) is a member of the calmodulin (CaM) superfamily that harbors two high affinity EF-hand motifs and a C-terminal transmembrane domain. CaBP7 has been previously shown to interact with and modulate phosphatidylinositol 4-kinase III-β (PI4KIIIβ) activity in in vitro assays and affects vesicle transport in neurons when overexpressed. Here we show that the N-terminal domain (NTD) of CaBP7 is sufficient to mediate the interaction of CaBP7 with PI4KIIIβ. CaBP7 NTD encompasses the two high affinity Ca2+ binding sites, and structural characterization through multiangle light scattering, circular dichroism, and NMR reveals unique properties for this domain. CaBP7 NTD binds specifically to Ca2+ but not Mg2+ and undergoes significant conformational changes in both secondary and tertiary structure upon Ca2+ binding. The Ca2+-bound form of CaBP7 NTD is monomeric and exhibits an open conformation similar to that of CaM. Ca2+-bound CaBP7 NTD has a solvent-exposed hydrophobic surface that is more expansive than observed in CaM or CaBP1. Within this hydrophobic pocket, there is a significant reduction in the number of methionine residues that are conserved in CaM and CaBP1 and shown to be important for target recognition. In CaBP7 NTD, these residues are replaced with isoleucine and leucine residues with branched side chains that are intrinsically more rigid than the flexible methionine side chain. We propose that these differences in surface hydrophobicity, charge, and methionine content may be important in determining highly specific interactions of CaBP7 with target proteins, such as PI4KIIIβ.
Introduction
The Ca2+-binding proteins (CaBPs)5 are a subfamily of the calmodulin (CaM)-related superfamily of EF-hand-containing calcium sensors (1–5). The CaBP family proteins are vertebrate-specific and comprise seven proteins (CaBP1 to -5 and CaBP7 and -8), which have emerged as important regulators of ion channel function and intracellular trafficking enzymes (6, 7). All members of the CaBP family have four EF-hand motifs with a domain organization similar to that of CaM; however, fundamental differences in EF-hand activity and subcellular targeting mechanisms have led to a further subdivision within this family (1, 8). CaBP1 to -5 have one inactive EF-hand motif (EF-2), which is unable to bind divalent cations, and are either cytosolic or targeted to membranes via post-translational attachment of an N-linked myristoyl chain (2). CaBP7 and CaBP8 (also known as calneuron II and calneuron I, respectively), however, have only two active EF-hand motifs and possess a single transmembrane domain (TMD) close to their C termini that allows them to associate with membranes of the trans-Golgi network and intracellular vesicles (5, 8–10). Evolutionary analysis of this family of calcium sensors suggests that CaBP7 and CaBP8 evolved independently of the rest of the CaBPs and represent a completely separate family of calcium sensors in their own right (1, 6).
CaBP7 and CaBP8 share a maximum of 24% overall homology with other CaBP members but are 63% homologous to each other (1). Both proteins are highly evolutionarily conserved, suggesting they might each perform important, non-redundant functions. The most highly conserved regions of CaBP7 and CaBP8 encode the two high affinity Ca2+ binding sites within the N-terminal half of the protein and the extreme C terminus encoding the TMD. The extreme N terminus and the connecting region between the EF-hands and the TMD are more variable.
CaBP1 to -5 have been shown to interact with a number of target proteins common to those of CaM, such as inositol trisphosphate receptors (11–14), L-type Ca2+ channels (15–21), and P/Q type Ca2+ channels (22–24). In contrast, only two CaBP7/8-interacting proteins have been described (10, 25). A direct interaction between CaBP8 and TRC40 (transmembrane domain recognition complex 40) has been demonstrated using co-immunoprecipitation, proximity ligation assays, and bioluminescence resonance energy transfer (10). TRC40 is important for the post-translational membrane insertion of tail-anchored proteins, such as CaBP7 and CaBP8, and is therefore unlikely to be a target of physiological regulation by these proteins (9, 10). Despite this, definitive proof that TRC40 is required for their membrane insertion has not yet been shown.
A unique role for CaBP7 and CaBP8 in the regulation of PI4KIIIβ has been more thoroughly characterized using both binding assays and in vitro functional assays (25). CaBP7 and CaBP8 are thought to act in conjunction with the more distantly related calcium sensor, neuronal calcium sensor 1 (NCS-1), which has been previously shown to promote PI4KIIIβ activity (25–29). Conversely, CaBP7 and CaBP8 act to inhibit phosphatidylinositol 4-phosphate production by PI4KIIIβ. The opposing physiological actions of CaBP7/8 and NCS-1 are suggested to provide a molecular switch regulating PI4KIIIβ function. At low Ca2+ levels, PI4KIIIβ is thought to preferentially bind to CaBP7 or CaBP8, placing a block on PI4KIIIβ activity, whereas at elevated Ca2+ levels, NCS-1 is able to compete with and displace CaBP7 and CaBP8. This in turn relieves kinase inhibition, and direct binding of NCS-1 to PI4KIIIβ further augments PI4KIIIβ activity, increasing phosphatidylinositol 4-phosphate production and stimulating trans-Golgi network to plasma membrane trafficking (25).
In this study, we show that the N-terminal domain (NTD) but not the C-terminal domain (CTD) of CaBP7 is able to interact independently with PI4KIIIβ. The NTD is also the portion of CaBP7 that displays the highest degree of homology with other CaBP family members. Importantly, caldendrin, an isoform of CaBP1 with an extended N terminus, has been shown to be unable to regulate PI4KIIIβ activity in vitro (25). It was therefore important to analyze the structure of CaBP7 NTD to discover how differences between EF-hand-containing calcium sensors may determine their unique and non-redundant interactions. Through biophysical and NMR spectroscopy analyses, this study examines how the three-dimensional structure of CaBP7 NTD compares with that of other similar EF-hand-containing Ca2+ sensors and what properties might determine the unique interaction of CaBP7 NTD with PI4KIIIβ. We show that the NTD of CaBP7 is monomeric, contains two functional EF-hand motifs that bind specifically to Ca2+, and has an unstructured region at its extreme N terminus. The overall structure is very similar to the C terminus of CaM but displays different surface properties and a unique unstructured N-terminal extension.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
CaBP7 NTD (residues 1–100) and CaBP7 CTD (residues 88–188) were subcloned from a synthetic gene (Integrated DNA Technologies, Leuven, Belgium) encoding human CaBP7 (NP_872333.1) codon optimized for expression in Escherichia coli and inserted into the pE-SUMOpro Kan vector (tebu-bio, Peterborough, UK). Expression of soluble His-SUMO-CaBP7 NTD, His-SUMO-CaBP7 CTD, or His-SUMO alone was induced in E. coli BL21 StarTM (DE3) (Invitrogen) using 1 mm isopropyl-1-thio-β-d-galactopyranoside at 18 °C for 16 h. Cells were harvested by centrifugation and resuspended in lysis buffer containing 50 mm sodium phosphate, pH 7.0, 300 mm NaCl plus protease inhibitors (Complete Mini protease inhibitor mixture tablets, Roche Applied Science). After cell lysis by one-shot cell disruption at 27 KPSI (Constant Sytems Ltd., Daventry, UK), soluble proteins were recovered by ultracentrifugation. The supernatant was applied to a charged HisTrap FF 5-ml affinity column and washed with 50 mm sodium phosphate buffer, pH 7.0, 300 mm NaCl, 25 mm imidazole, and the recombinant protein was eluted in 50 mm sodium phosphate, pH 7.0, 300 mm NaCl with a linear imidazole gradient from 25 to 500 mm. After buffer exchange into 20 mm HEPES, pH 6.5, 150 mm NaCl, the eluted protein was cleaved with recombinant SUMO protease and reapplied to a metal affinity column to remove the cleaved His-SUMO tag, and the flow-through containing CaBP7 NTD was collected. Based on SDS-PAGE analysis, SUMO tag cleavage was 90–95% complete under these conditions. CaBP7 NTD was further purified by gel filtration on a Superdex 75 column (GE Healthcare).
Uniformly isotope-labeled CaBP7 NTD was expressed in M9 minimal medium supplemented with 1 mm MgSO4, 50 μm CaCl2, 23 mm NH4Cl, 22 mm glucose, 100 μl of PTM1 salts. Isotopically labeled 15NH4Cl and [13C]glucose were the sole nitrogen and carbon sources, respectively. Single-labeled 15N, but not double-labeled protein preparations, were supplemented with 0.5 g of 15N-labeled ISOGRO® (ISOTEC®, Sigma) per 1 liter of M9 minimal medium.
Apo CaBP7 NTD was prepared for NMR and CD analysis by the addition of 5 mm EGTA and 5 mm EDTA to the sample followed by buffer exchange into 20 mm HEPES, pH 6.5, 150 mm NaCl, 30 mm n-octyl-β-d-glucopyranoside using a PD-10 column.
PI4KIIIβ Pull-down Assay
Bovine brain cytosolic protein extracts were prepared from whole homogenized tissue (FirstLink, Birmingham, UK) as described previously (30). Briefly, after initial extraction into cytosol buffer (10 mm HEPES, pH 7.8, 100 mm KCl, 2 mm MgCl2, 1 mm DTT), the cytosolic fraction was dialyzed overnight against PI4KIIIβ binding buffer (20 mm HEPES, pH 7.4, 100 mm KCl, 1 mm DTT, 100 μm ATP, 100 μm MgCl2, 1 mm CaCl2). The dialyzed extract was further clarified by centrifugation (15 000 × g, 15 min, 4 °C) prior to use.
Recombinant His-SUMO, His-SUMO-CaBP7 NTD, and His-SUMO-CaBP7 CTD expression was induced as described above. After soluble protein had been recovered by ultracentrifugation, the supernatant was incubated with 2 ml of loose metal affinity resin charged with cobalt (TalonTM resin, Takara Bio) equilibrated in lysis buffer to purify recombinant protein. After incubation for 1 h at 4 °C, the resin was washed with 40 volumes of lysis buffer and equilibrated with 30 volumes of PI4KIIIβ binding buffer. Each recombinant His-SUMO fusion protein (5–10 mg) was then incubated with clarified bovine brain cytosol for 2 h at 4 °C with constant agitation. After incubation with bovine brain cytosol, each column was washed with 40 volumes of PI4KIIIβ binding buffer. Specific Ca2+-dependent binding proteins were eluted by incubation with 2 ml of the PI4KIIIβ binding buffer supplemented with 5 mm EGTA for 10 min at room temperature. An additional high salt elution step isolated all other binding partners using PI4KIIIβ binding buffer supplemented with 1 m NaCl.
Eluted proteins were precipitated using the acetone/trichloroacetic acid (TCA) method. Briefly, 10 volumes of cold 10% TCA in acetone was added to each sample and incubated at 20 °C overnight. Samples were then centrifuged (15,000 × g, 10 min, 4 °C), and the supernatant was discarded. Each pellet was then washed in 5 volumes of cold acetone and incubated for 30 min at 20 °C before centrifugation (15,000 × g, 5 min, 4 °C). The supernatant was discarded, and the pellets were resuspended in 100 μl of SDS dissociation buffer (125 mm HEPES, pH 6.8, 10% sucrose, 10% glycerol, 4% SDS, 1% β-mercaptoethanol, 2 mm EDTA). Samples were boiled for 5 min and analyzed by SDS-PAGE (NuPAGE® NovexTM 12% BisTris gels, Invitrogen).
Western Blotting Protocol
Protein samples were transferred from SDS-polyacrylamide gels by transverse electrophoresis onto nitrocellulose membranes for detection by Western blotting. Filters were probed with anti-PI4KIIIβ antibody (1:500, Merck). Immunoreactivity was detected after application of an anti-rabbit secondary antibody conjugated to horseradish peroxidase and visualized using ECL regents. The same loading volumes and dilutions were used for all eluates to allow comparability between His-SUMO fusion proteins.
Native Polyacrylamide Gel Mobility Shift Assay
Protein samples (either control SUMO protein or CaBP7 NTD) were prepared in SDS-free loading buffer containing 50 mm Tris, pH 7.5, 10% glycerol, 10% sucrose, 2 mm EDTA, 2 mm EGTA. To investigate Ca2+ and Mg2+ binding, either 10 mm CaCl2, 10 mm MgCl2, or 10 mm CaCl2 and 10 mm MgCl2 were added to the loading buffer. Protein samples were not boiled and were resolved on native polyacrylamide gels containing 18% acrylamide. Gels were stained with SimplyBlueTM SafeStain (Invitrogen).
Circular Dichroism
A Jasco-J810 spectropolarimeter was used for CD measurements in the far UV region, from 190 to 260 nm. Spectra were recorded at protein concentrations of ∼0.25 mg/ml in a cuvette of 1-mm path length in a temperature-controlled cell holder at 25 °C. Spectra were monitored with a 0.2-nm step and 10 averages. Samples were prepared in 20 mm HEPES, pH 6.5, 150 mm NaCl, 30 mm n-octyl-β-d-glucopyranoside, and all sample spectra were base line-corrected by subtraction of the spectra for buffer alone.
Size Exclusion Chromatography-Multiangle Laser Light Scattering (SECMALLS) Analysis
CaBP7 NTD was run on a Superose 12 10/300GL gel filtration column (GE Healthcare) equilibrated in 20 mm HEPES, pH 6.5, 150 mm NaCl at 0.75 ml/min. Elution was monitored by a Wyatt EOS 18-angle laser photometer (Wyatt Technology, Santa Barbara, CA), an Optilab rEX refractive index detector, and a Jasco UV-2077 Plus UV-visible spectrophotometer (Jasco, Easton, MD); these were coupled to a quasi-elastic light scattering detector for simultaneous measurement of hydrodynamic radius. Molar mass measurements were performed using both Astra version 5.3.2.16 software (Wyatt Technology) and the “three-detector method.” Values of mass and hydrodynamic radius are expressed as means ± S.E.
NMR Spectroscopy
All NMR spectra were acquired at 303 K on Bruker AVANCE II+ 600- and 800-MHz spectrometers equipped with triple resonance cryoprobes. Samples for NMR were concentrated to 0.2–1 mm in 20 mm HEPES, pH 6.5, 150 mm NaCl. To prevent protein aggregation, 30 mm n-octyl-β-d-glucopyranoside was added. All samples were prepared in 90% H2O, 10% D2O unless otherwise stated. Sequence-specific backbone resonance assignment was obtained using HNCACB, CBCA(CO)NH, HNCO, and HN(CA)CO triple-resonance experiments. Side chain assignment was obtained using a three-dimensional HCCH-total correlation spectroscopy experiment (10.8 ms mixing time). NOEs were derived from three-dimensional 15N- and 13C-edited NOESY-HSQC experiments with 130-ms mixing times. One-bond 1H-15N residual dipolar couplings were obtained from 1H-15N IPAP-HSQC spectra collected in Pf1 bacteriophage (20 mg/ml). The final values of the magnitude and rhombic components in the refinement stage were 3.864 and 0.647, respectively. Hydrogen bond restraints were derived from 1H-15N HSQC spectra in 100% D2O. All spectra were processed with Bruker Topspin and analyzed using the CCPN Analysis package (31).
15N longitudinal and transverse relaxation rates were obtained on Bruker AVANCE II+ 800-MHz spectrometer using two-dimensional 1H-15N correlation pulse sequences. The 15N R1 and R2 values at 800 MHz were obtained using 12 delays (20, 50, 200, 400, 800, 1200, 1400, 1600, 1800, and 2000 ms for R1; 4.8, 9.6, 19.2, 28.8, 48, 72, 96, 120, 144, and 168 ms for R2).
Structure Calculation and Validation
Structures of CaBP7 NTD were calculated in a two-stage process using the ARIA (32) and CYANA (33) packages. CYANA was used to generate lists based on automated NOE assignment. The generated restraint lists were then used as input for final structure calculation in ARIA, with the inclusion of DANGLE-derived dihedral restraints (34), residual dipolar coupling, and distance restraints between Ca2+ and residues in positions 1, 3, 5, 7, and 12 in the EF-hand loops (1 and 2). Nine iterations were performed, generating 200 structures. After refinement, the 20 lowest energy structures were selected and analyzed. The final structures were assessed using PROCHECK-NMR (35) and validated by comparing the correlation between experimentally measured RDCs and back-calculated RDCs from the final structure using the PALES program (36). The NMR-derived structure and NMR assignments of CaBP7 NTD were deposited in the Protein Data Bank (PDB code 2LV7) and the Biological Magnetic Resonance Bank (code 18557).
RESULTS
CaBP7 N Terminus but Not C Terminus Binds PI4KIIIβ in a Ca2+-dependent Manner
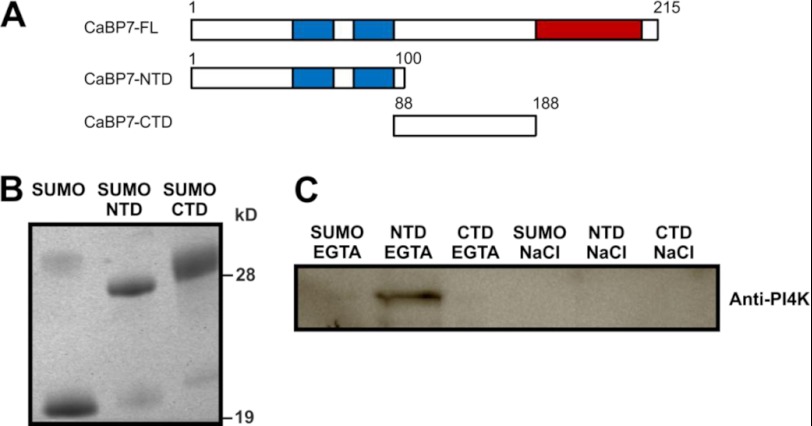
Attempts to express and purify full-length CaBP7 gave only a very low yield of soluble protein. The protein was found mainly in the insoluble fraction, which could be explained by the presence of the highly hydrophobic C-terminal TMD, causing aggregation of the protein (10). The structure and function of related calcium sensors, CaBP1 and CaM, have often been studied by splitting these proteins into independent N- or C-terminal lobes (37–41). A similar approach was adopted here in order to identify the region of CaBP7 that interacts with PI4KIIIβ. Affinity pull-down assays were performed using either the NTD (residues 1–100) or CTD (residues 88–188) of CaBP7 (Fig. 1A). The C-terminal TMD, which would normally be buried within intracellular membranes, was excluded from these experiments. Affinity purification columns were prepared with either immobilized His-SUMO, His-SUMO-CaBP7 NTD, or His-SUMO-CaBP7 CTD (Fig. 1, A and B). Each affinity column was incubated with bovine brain cytosol extract in the presence of Ca2+ to pull out specific binding partners. Ca2+-dependent interacting proteins were eluted using an EGTA wash followed by a high salt wash to elute any Ca2+-independent partners. Eluted samples were analyzed by Western blot by probing with a polyclonal PI4KIIIβ antibody. A PI4KIIIβ-reactive protein of the correct molecular mass was found to bind specifically to CaBP7 NTD and not CaBP7 CTD or the His-SUMO tag alone (Fig. 1C). PI4KIIIβ was eluted with the EGTA wash, indicating that this is a Ca2+-dependent interaction.
FIGURE 1.
Affinity pull-down assay of PI4KIIIβ from bovine brain cytosol fraction using either the NTD or CTD of CaBP7 immobilized through a His6 tag on cobalt-charged metal affinity resin. A, schematic diagram of CaBP7. Blue boxes indicate active EF-hand domains. Red boxes indicate the C-terminal TMD. B, Coomassie-stained SDS-polyacrylamide gel showing equivalent expression levels of SUMO, SUMO-CaBP7 NTD, and SUMO-CaBP7 CTD used in pull-down assays. C, Western blot showing an immunoreactive band for PI4KIIIβ, which is only extracted by SUMO-CaBP7 NTD and is eluted specifically by an EGTA wash. Bound proteins were eluted from resin using a 5 mm EGTA wash (first three lanes) followed by a 1 m NaCl wash (last three lanes).
CaBP7 NTD Binds Specifically to Ca2+ and Not Mg2+
Many EF-hand containing proteins have been demonstrated to bind to and undergo both Mg2+- and Ca2+-induced conformational changes (38, 42–44). Free Mg2+ levels in the resting cell are estimated at between 0.5 and 5 mm (45), and it is thought that for these proteins, the conformation in the presence of Mg2+ is more physiologically relevant than the apo form. The specificity of Ca2+ binding to CaBP7 NTD was investigated using native PAGE to monitor the electrophoretic mobility of CaBP7 NTD in the presence or absence of Ca2+ or Mg2+ (Fig. 2). The control protein, SUMO, exhibited no change in electrophoretic mobility under any of the conditions tested. An excess of Mg2+ failed to influence the migration of CaBP7 NTD when compared with Ca2+-free/Mg2+-free conditions. An excess of Ca2+, however, induced a significant retardation in the migration rate of CaBP7 NTD, which was unaffected by the presence of equimolar Mg2+. This significant Ca2+-dependent shift in electrophoretic mobility is consistent with specific binding of Ca2+ to CaBP7 NTD and is in agreement with previously published data demonstrating that full-length CaBP7 does not bind Mg2+ (25).
FIGURE 2.
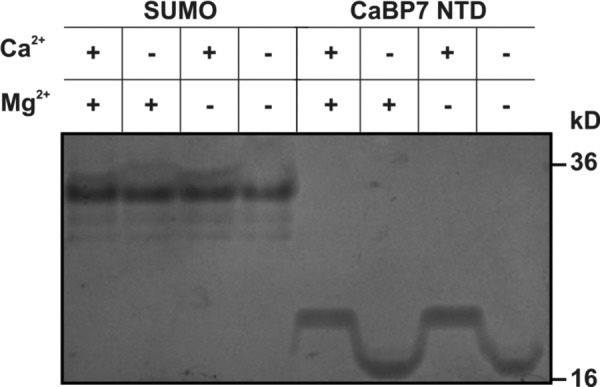
Coomassie-stained native polyacrylamide gel showing the electrophoretic gel mobility of SUMO and CaBP7 NTD in the presence or absence of an excess of Ca2+ or Mg2+. Samples were resolved on an 18% acrylamide gel in loading buffer containing 2 mm EGTA and 2 mm EDTA with either 10 mm CaCl2 or 10 mm MgCl2. The presence or absence of Ca2+ or Mg2+ is indicated by a plus or minus sign above each lane of the gel image.
Ca2+ Binding Induces a Change in the Helical Content of CaBP7 NTD
Circular dichroism analysis scanning between 190 and 260 nm demonstrated that both Ca2+-bound and apo-CaBP7 NTD adopt a high degree of helical content (Fig. 3). The Ca2+-bound form exhibited an increase in the negative value of ellipticity (2–7 millidegrees) between 200 and 230 nm compared with the apo form, demonstrating a significant increase in helical content. This suggests that the protein undergoes Ca2+-induced alterations in secondary structure.
FIGURE 3.
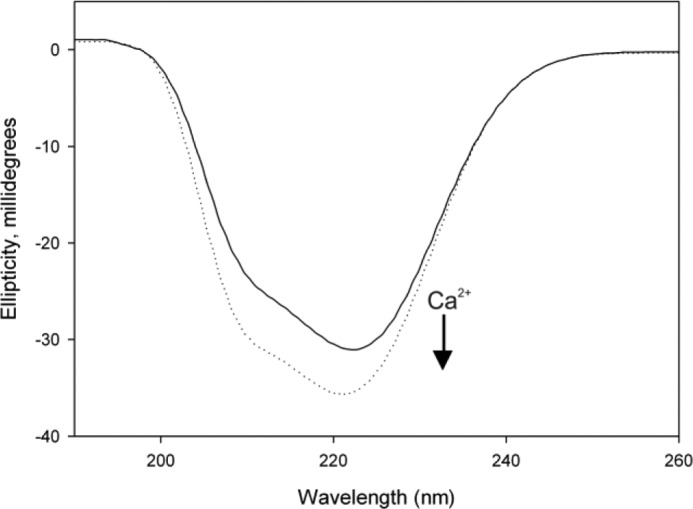
The effect of Ca2+ on far-UV CD of apo- and Ca2+-bound CaBP7 NTD. Changes to CaBP7 NTD secondary structure were monitored by far-UV CD spectroscopy. The solid line represents the trace for apo-CaBP7 NTD, and the dashed line represents the trace for Ca2+-bound CaBP7 NTD. To prepare apo-CaBP7 NTD, 5 mm EGTA and 5 mm EDTA were added to the sample and then removed using a desalting column. Final samples were analyzed in 20 mm HEPES, pH 6.5, 150 mm NaCl, 30 mm n-Octyl-β-d-glucopyranoside at 25 °C. Each spectrum is representative of 10 averaged scans and is normalized to the spectrum of buffer alone.
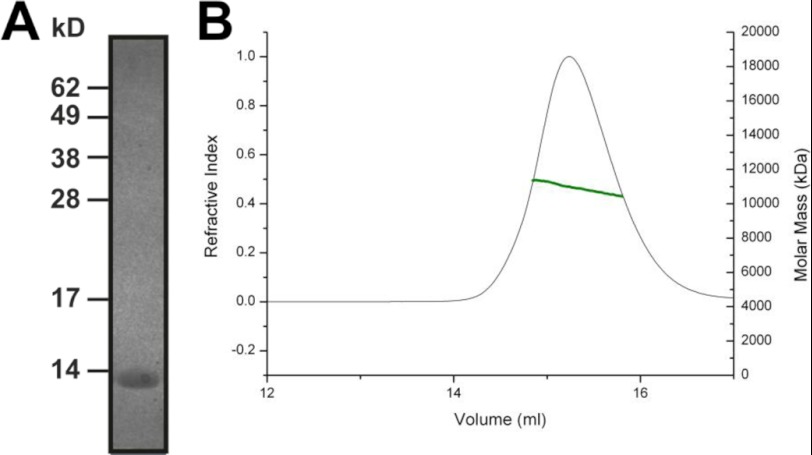
Ca2+-bound CaBP7 NTD Is Monomeric
A previous study has reported that full-length CaBP7 forms soluble dimers both in vitro and in vivo in a Ca2+-independent manner. To investigate whether Ca2+-bound CaBP7 NTD is monomeric or dimeric, SECMALLS and NMR analyses were performed. The theoretical molecular mass of CaBP7 NTD is 11.4 kDa (Fig. 4A). SECMALLS analysis showed that CaBP7 NTD has an apparent molecular mass of 11.1 ± 0.1% kDa (Fig. 4B), which is consistent with CaBP7 NTD existing in monomeric form under the conditions tested. In agreement with this, NMR spectroscopy also suggested that the protein was monomeric. The average rotational correlation time of CaBP7 NTD was calculated to be 7.34 ns, consistent with the apparent molecular mass calculated by SECMALLS.
FIGURE 4.
CaBP7 NTD is monomeric. A, Coomassie-stained SDS-polyacrylamide gel of CaBP7 NTD after purification and cleavage of a His-SUMO tag. The protein migrates at a rate consistent with the theoretical molecular mass of 11.4 kDa. B, SECMALLS characterization of Ca2+-bound CaBP7 NTD. The refractive index of the sample is plotted against elution volume. The molar mass distribution is indicated by the green line. The data illustrate that CaBP7 NTD forms a stable monomer in 20 mm HEPES, pH 6.5, 150 mm NaCl at 20 °C. There is a single peak with an average molar mass of 11.1 ± 0.11 kDa.
Ca2+ Binding Induces Changes in the Tertiary Structure of CaBP7 NTD
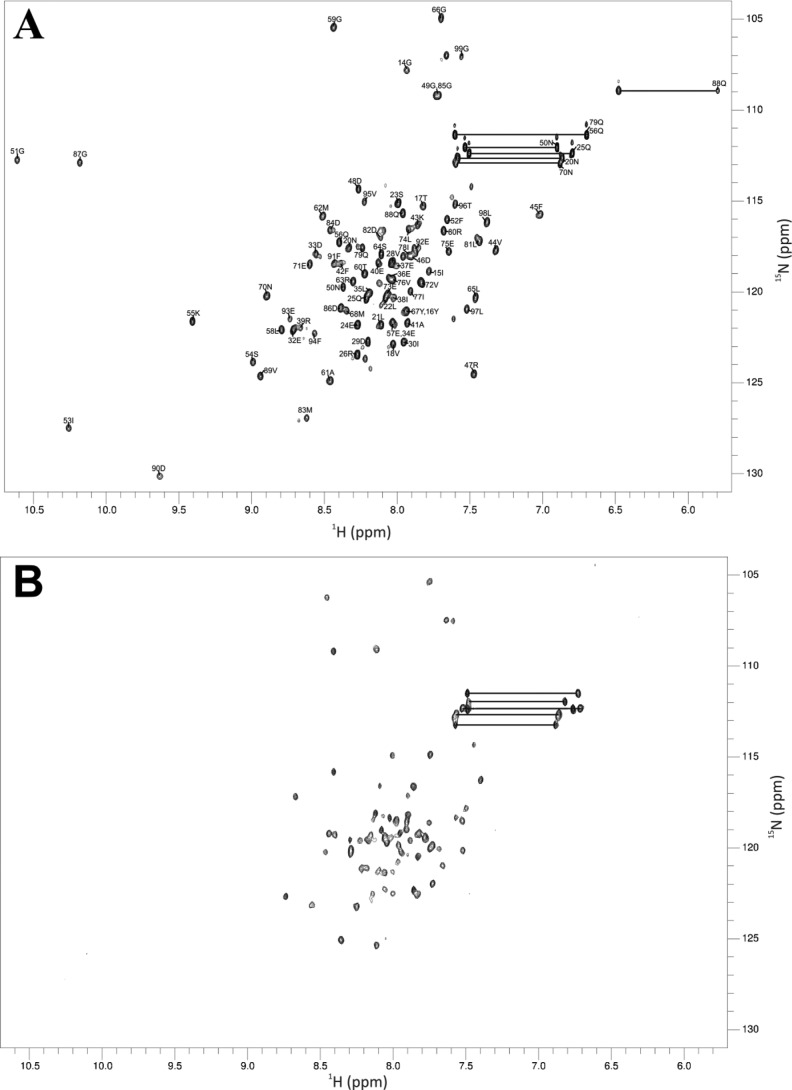
NMR spectroscopy was used to probe Ca2+-dependent conformational changes to CaBP7 NTD tertiary structure. The peaks in the 1H-15N HSQC NMR spectra represent main chain and side chain amide groups, providing a residue-specific fingerprint of the protein conformation. After purification, the protein was found to be in the Ca2+-bound form without the addition of external Ca2+. This could be discerned by the presence of two downfield shifted peaks (1H 10.5 and 10.2 ppm) that are characteristic of conserved glycine residues at the 6-position of divalent ion-occupied EF-hands (Fig. 5A). Treatment of the sample with divalent metal ion chelators, EGTA and EDTA, to generate apo-CaBP7 NTD caused a dramatic change in the 1H-15N HSQC spectrum (Fig. 5B). Notably, the two characteristic downfield shifted peaks at 1H 10.5 and 10.2 ppm were no longer visible. The HSQC spectrum for apo-CaBP7 NTD exhibited good peak dispersion with well resolved non-overlapping peaks, suggesting that the protein is folded in the absence of Ca2+. 107 peaks rather than the expected 92 backbone amide peaks (100 less the 7 prolines and the N terminus methionine residue) were observed, however, and a mixture of peak intensities was also apparent. The NMR data therefore suggest the presence of some conformational heterogeneity under the conditions used.
FIGURE 5.
1H-15N HSQC spectra of CaBP7 NTD. A, assigned 1H-15N HSQC spectrum of Ca2+-bound CaBP7 NTD; B, 1H-15N HSQC spectrum of apo-CaBP7 NTD. Sample heterogeneity of apo-CaBP7 NTD is seen from the unequal distribution of peak intensities. Asn and Gln side chain NH2 groups are indicated by peaks joined with a solid black line. Ca2+-bound CaBP7 NTD was prepared in 20 mm HEPES, pH 6.5, 150 mm NaCl, 30 mm n-octyl-β-d-glucopyranoside. Apo CaBP7 NTD was prepared by the addition of 5 mm EGTA and 5 mm EDTA to the sample followed by buffer exchange into 20 mm HEPES, pH 6.5, 150 mm NaCl, 30 mm n-octyl-β-d-glucopyranoside. Spectra were acquired at 303 K.
Under solution NMR conditions, the addition of Mg2+ to CaBP7 NTD induced protein precipitation, a documented effect of nonspecific binding of divalent cations to proteins with a net negative charge (46–49). The addition of Ca2+ to apo-CaBP7 NTD did not have the same effect, and a shift was seen from the apo form to the Ca2+-bound holo form. Significant chemical shift dispersion (1H 5.8 to 10.6 ppm) and uniform peak intensities were observed in the 1H-15N HSQC spectrum for Ca2+-bound CaBP7 NTD, suggesting that the protein was stably folded (Fig. 5A). In this spectrum, 99 peaks were observed compared with the expected 92, again suggesting a small degree of conformational heterogeneity. Using sequential connectivities from standard triple resonance spectra, 86.0% of the CaBP7 NTD backbone and 79.4% of the amino acid side chains were assigned (Fig. 5B). The first 13 amino acids from the N terminus exhibited weak NMR signals that could not be assigned. The two downfield shifted peaks at 1H 10.5 and 10.2 ppm were assigned to the glycines at the 6-position of each of the two EF-hands (Gly-51 and Gly-87) as expected and consistent with Ca2+ bound at both EF-1 and EF-2. Other dispersed peaks unique to the Ca2+-bound spectrum include Ile-53, Lys-55, Ser-54, Met-83, Val-89, Asp-90, and the side chain amides of Gln-88. Along with Gly-51 and Gly-87, these residues also reside within the Ca2+ binding loops of the two EF-hands. The spectra of the apo- and Ca2+-bound protein were too dissimilar to enable transfer of any backbone assignments between the two.
NMR-derived Structure of Ca2+-bound CaBP7 NTD
On the basis of the NMR assignments, the structure of CaBP7 NTD was calculated using 1965 interproton distance restraints obtained from 1H-15N and 1H-13C NOESY-HSQC spectra, 37 RDC restraints measured in Pf1 bacteriophage, 165 DANGLE-derived backbone torsion angles, and 32 hydrogen bonds (Table 1). The 20 calculated structures with the lowest total free energy have a backbone RMSD of 0.43 Å for the structured regions (Table 1). The final calculated structures were validated using PROCHECK NMR, which shows that 81.1% of the residues belong to the most favorable region of the Ramachandran plot. The correlation between the measured and back-calculated RDCs to the lowest energy structure in the ensemble shows a good fit between the two values, with an R correlation value of 0.996. For additional validation, the structure of CaBP7 NTD was calculated with the 37 experimentally derived RDC restraints removed, and the lowest energy structure was used to calculate the correlation between measured and back-calculated RDCs. In this case, the R correlation value was 0.899, showing a close correlation between the final structure and experimentally derived RDC values.
TABLE 1.
Structural statistics for the ensemble structures of CaBP7 NTD
| NOE restraints | 1965 |
| Ambiguous | 536 |
| Unambiguous | 1429 |
| Intra (|i − j| = 0) | 310 |
| Medium (0 < |i − j| < 5) | 830 |
| Long (|i − j| ≥ 5) | 289 |
| Hydrogen bond restraints | 32 |
| Dihedral angle restraints (Φ, Ψ) | 165 |
| Residual dipolar coupling restraints | 37 |
| PALES validation of RDCs | |
| R value | 0.996 |
| Q value | 0.091 |
| Root mean square deviation from ideal geometry | |
| Bond length (Å) | 0.0015 ± 0.00009 |
| Bond angle (degrees) | 0.315 ± 0.008 |
| Root mean square deviation from average structure (Å) | |
| Secondary structure (backbone) | 0.43 |
| Secondary structure (heavy) | 0.92 |
| Ramachandran plot (%) | |
| Most favored region | 81.1 |
| Allowed region | 18.1 |
| Disallowed region | 0.8 |
| Average energy (kcal/mol) | 606.16 |
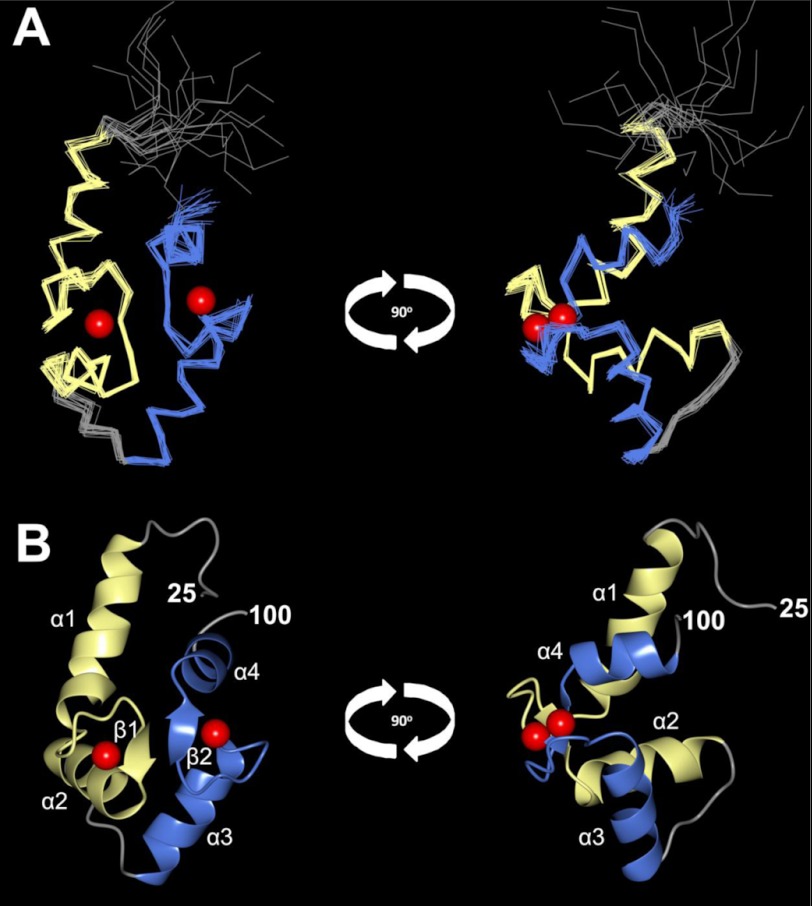
The final NMR-derived structure of Ca2+-bound CaBP7 NTD is illustrated in Fig. 6. The structure contains four α-helices and two β-strands: α1 (residues 32–45), α2 (residues 55–65), α3 (residues 72–81), α4 (residues 91–98), β1 (residues 53–54), β2 (residues 89–90). CaBP7 NTD contains two EF-hand domains: EF1 (Fig. 6, yellow, residues 32–65) and EF2 (Fig. 6, blue, residues 72–100) with a linker domain joining the two. The first 30 amino acids are unstructured.
FIGURE 6.
Main chain structures of Ca2+-bound CaBP7 NTD determined by solution NMR (PDB code 2LV7). Shown are superposition of the 20 lowest energy structures (A) and a ribbon representation of the lowest energy structure (B) seen from the front (left) and side (right). N-terminal residues (residues 1–25) are unstructured and are not shown. EF-hand 1 is shown in yellow, and EF-hand 2 is shown in blue. Red spheres represent bound Ca2+.
The geometry of covalent bonds chelating Ca2+ in each of the two EF-hands was modeled using structural constraints derived from the x-ray crystal structure of Ca2+-bound CaM, which has been shown to closely resemble the binding site geometry conserved in other EF-hand proteins (50). This approach has been used previously to model the Ca2+ binding geometry of CaBP1 (37).
Comparison of Ca2+-bound CaBP7 NTD with Ca2+-bound CaM and CaBP1
Comparison of the structure of CaBP7 NTD with the established structures of related EF-hand containing calcium sensors is important to further the understanding of how calcium sensors with similar sequence and structure are able to regulate distinct protein targets. Bioinformatic analyses have shown previously that CaBP7 exhibits a low degree of homology to CaM and other members of the CaBP family (1). The structure of CaM has been extensively studied both through x-ray crystallography and NMR analysis (50, 51). More recently, solution NMR and x-ray structures of CaBP1 have been solved (37, 39, 52), and it has been shown to closely resemble CaM in both its domain structure and its ability to regulate similar targets. Importantly, both proteins have been shown to possess two independent domains, the first encompassing EF-1 and EF-2 and the second encompassing EF-3 and EF-4.
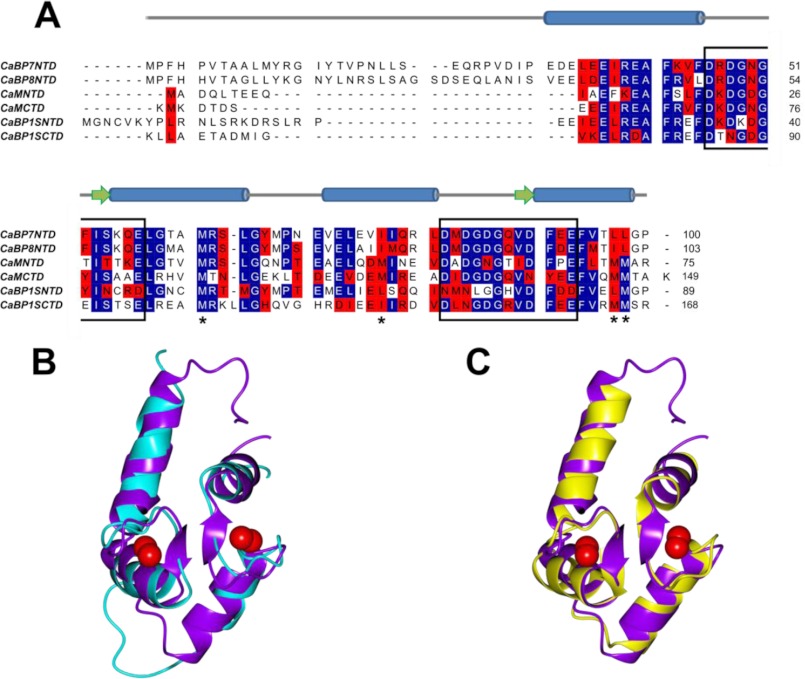
Fig. 7A shows a multiple-sequence alignment of CaBP7 NTD with the N- and C-terminal domains of CaM and CaBP1. CaBP8 NTD is also included for comparison. CaBP7 NTD has a similar level of sequence identity with both N- and C-lobes of CaM and CaBP1 when the non-homologous N termini and linker regions are excluded from alignments (Table 2). Comparison of the known structures of these proteins shows that the CTD of both CaM and CaBP1 are the most structurally homologous to CaBP7 NTD, with RMSD values of 1.35 and 2.12, respectively. Fig. 7, B and C, shows the NMR structure of CaBP7 NTD superposed with either CaM CTD or CaBP1 CTD, respectively. This figure illustrates the high degree of similarity in the overall tertiary structure of these Ca2+-sensing proteins. Table 3 shows the interhelical angles calculated between the helices of each EF-hand in Ca2+-bound CaBP7 NTD and Ca2+-bound or unbound CaM and CaBP1 (calculated using Helixang from the CCP4 suite of programs (53)). The interhelical angles for EF1 and EF2 in CaBP7 NTD are 116 and 87°, respectively, and reflect the Ca2+-bound open conformation that is typical of CaM-related Ca2+ sensors.
FIGURE 7.
Comparison of Ca2+-bound CaBP7 NTD, CaM CTD, and CaBP1 CTD sequence and main chain structure. A, sequence alignment of CaBP7 NTD with CaBP8 NTD and the N- and C-terminal domains of CaM and CaBP1. Identical amino acids are highlighted in blue, and similar amino acids are highlighted in red. Black boxes indicate the 12-amino acid Ca2+-binding loop of each EF-hand. Asterisks below the sequences indicate methionine residues that are conserved in CaM NTD and CTD. These are Met-36, -51, -71, and -72 in the N-terminal domain and Met-110, -125, -145, and -146 in the C-terminal domain. The NMR-derived secondary structure elements of CaBP7 NTD are indicated above the alignment. Cylinders, α-helices; arrows, β-sheets. B and C, ribbon representation of Ca2+-bound CaBP7 NTD (PDB code 2LV7, residues 30–100) lowest energy conformer (purple) superposed with Ca2+-bound CaBP1 CTD (cyan, PDB code 2LAP) (B) and Ca2+-bound CaM CTD (yellow, PDB code 1CLL, residues 80–147) (C). Red spheres represent bound Ca2+.
TABLE 2.
Percentage sequence identity and comparative RMSD values for Ca2+-bound CaBP1 (PDB codes 2LAN and 2LAP) and CaM (PDB code 1CLL) compared to CaBP7 NTD (PDB code 2LV7)
Sequence identity was calculated based on alignment of CaBP7, CaBP1, and CaM excluding non-homologous regions in the N termini or linker regions of the proteins. The comparative RMSD was calculated using PDBeFold (73).
| Sequence identity | RMSD | |
|---|---|---|
| % | Å | |
| CaM NTD | 47.8 | 1.66 |
| CaM CTD | 47.2 | 1.35 |
| CaBP1 NTD | 47.8 | 3.54 |
| CaBP1 CTD | 43 | 2.12 |
TABLE 3.
Interhelical angles of the EF-hands in CaM, CaBP1, and CaBP7 NTD
The PDB codes were as follows: apo-CaM, 1CFD; Ca2+-bound CaM, 1CLL; Mg2+-bound CaBP1, 2K7B and 2K7C; Ca2+-bound CaBP1, 2LAN and 2LAP; Ca2+-bound CaBP7 NTD, 2LV7. Angles were calculated using Helixang from the CCP4 suite of programs (53).
| Helix pair | Interhelical angles |
||||
|---|---|---|---|---|---|
| Apo-CaM | Ca2+-bound CaM | Mg2+-bound CaBP1 | Ca2+-bound CaBP1 | Ca2+-bound CaBP7 NTD | |
| degrees | |||||
| α1-α2 | 135 | 86 | 133 | 129 | 116 |
| α3-α4 | 134 | 94 | 139 | 138 | 87 |
| α5-α6 | 126 | 98 | 132 | 116 | |
| α7-α8 | 136 | 90 | 123 | 87 | |
Surface Properties of Ca2+-bound CaBP7 NTD Compared with Ca2+-bound CaM and CaBP1 CTDs
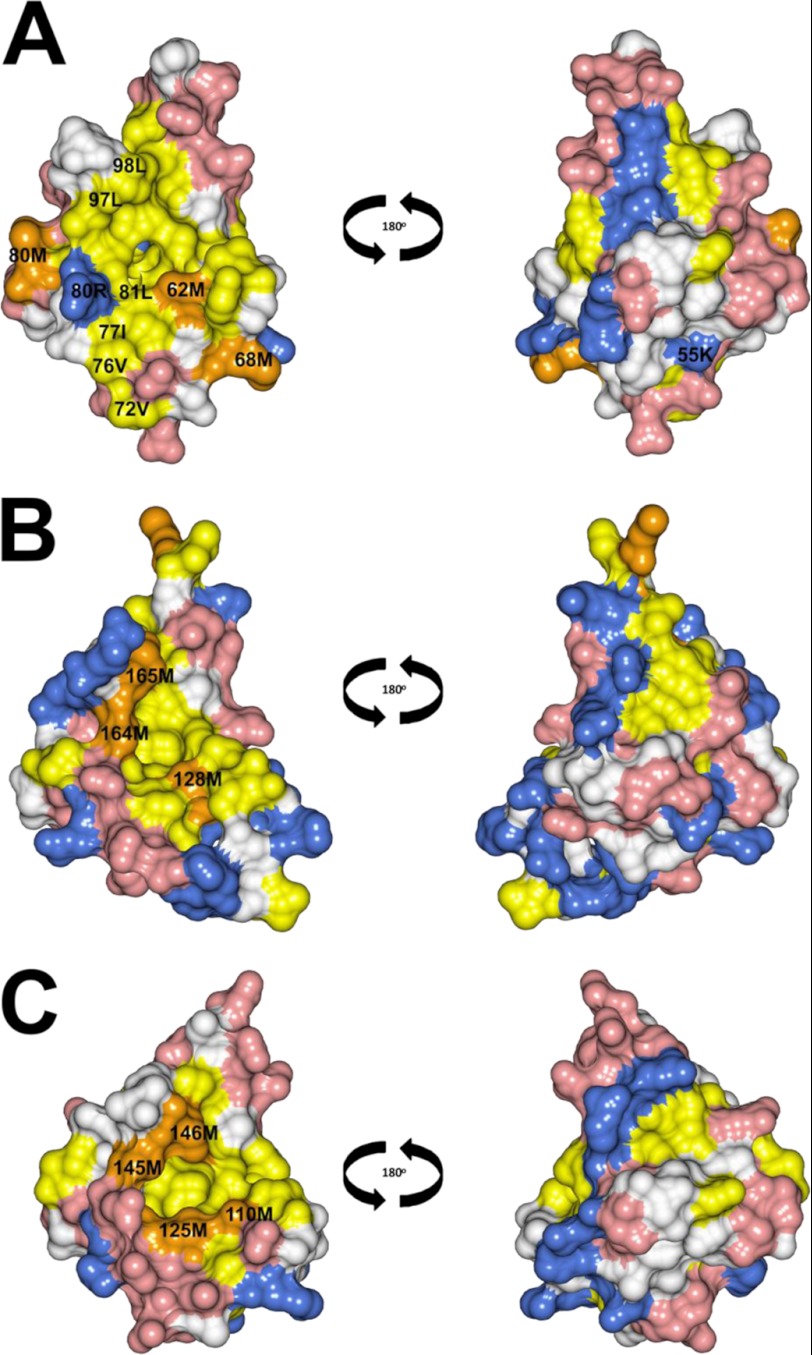
Space-filling representations of Ca2+-bound CaBP7 NTD, CaM CTD, and CaBP1 CTD are illustrated in Fig. 8. The back face of Ca2+-bound CaBP7 NTD displays few obvious deviations from the surface properties of CaM and CaBP1 CTDs. The most notable difference is a non-conserved basic residue (Lys-55), which replaces an uncharged amino acid in CaM and CaBP1.
FIGURE 8.
Space-filling representations of Ca2+-bound CaBP7 NTD (PDB code 2LV7, residues 30–100) (A), Ca2+-bound CaBP1 (PDB code 2LAP) (B), and Ca2+-bound CaM (PDB code 1CLL, residues 80–147) (C). The front face is shown on the left, and the back face is shown on the right. Acidic residues (Asp and Glu) and basic residues (Arg, His, and Lys) are shown in red and blue, respectively. Hydrophobic residues (Ile, Leu, Phe, Trp, Val, and Tyr) are shown in yellow with the exception of Met residues, which are highlighted in orange.
Analysis of the per residue solvent accessibility of CaBP7 NTD, CaBP1 CTD, and CaM CTD (PDBePISA (54)) revealed that the proportion of hydrophobic residues that contribute to the accessible surface area was 31.6, 28.6, and 18.8%, respectively. In agreement with these values, CaBP7 NTD exhibits a striking solvent-exposed hydrophobic surface (front face, shown in orange and yellow in Fig. 8A) that is more expansive than seen in either CaM or CaBP1. Notably, within this hydrophobic region, non-conserved hydrophobic residues can be observed in the interlinking loop between EF-1 and EF-2 (Met-68) and in the first helix of EF-2 (Val-72 and Val-76), which are either polar or uncharged at the corresponding positions in CaM and CaBP1. This results in a distinctive loss of charged residues surrounding the hydrophobic pocket; in CaM and CaBP1, these charged residues are thought to confer specific electrostatic contacts with target-interacting proteins (37). In addition, an acidic residue that is conserved in CaM and CaBP1 is replaced by a positively charged residue (Arg-80) in CaBP7.
Another notable difference between the three structures is the lack of methionine residues within the hydrophobic pocket of CaBP7 NTD. Methionine residues are shown in orange in Fig. 8 and can be observed outlining the hydrophobic pocket in CaM and CaBP1. There are four methionine residues in the CaM CTD that are also conserved within the CaM NTD (Fig. 7A). These residues have been shown to be of particular importance for determining interactions with target-binding proteins (55). Three of these methionine residues are conserved in CaBP1 CTD; however, only one is conserved in CaBP7 (Met-62). Ile-77, Leu-97, and Leu-98 in CaBP7 replace methionine residues at the equivalent positions in CaM (Fig. 7A). Additional non-conserved methionine residues, Met-68 and Met-83, are found outside of the hydrophobic pocket of CaBP7 (Fig. 8A).
DISCUSSION
This study describes the interaction of CaBP7 NTD with PI4KIIIβ and the subsequent determination of the NMR solution structure of Ca2+-bound CaBP7 NTD. In vitro binding data show that the CaBP7 NTD, encompassing its two active EF-hands, is able to bind to PI4KIIIβ from bovine brain cytosol, whereas CaBP7 CTD is not. PI4KIIIβ was found in the EGTA elution fraction for CaBP7 NTD only, suggesting that this is a Ca2+-dependent interaction. Full-length CaBP7 and CaBP8 have been previously demonstrated to interact with and inhibit PI4KIIIβ (25). Contradictory to the results presented here, this interaction was reported to be Ca2+-independent in in vitro assays. The differences in the Ca2+ dependence observed in these two studies is probably due to differences in assay conditions; however, in agreement with our findings, Mikhaylova et al. (25) reported that regulation of PI4KIIIβ by CaBP8 was Ca2+-dependent because inhibition was augmented by an elevation in Ca2+ levels in in vitro kinase assays.
CaBP7 and CaBP8 act in an opposing fashion to NCS-1 in the regulation of PI4KIIIβ activity. Despite both the CaBP and NCS families of proteins belonging to the CaM superfamily of EF-hand calcium sensors, they are known to possess numerous features that distinguish them from one another. Sequence analysis shows that these families are only distantly related, with less that 20% homology between members of the two families. This is reflected in structural studies showing that members of the NCS family typically possess a globular structure with their four EF-hands arranged in a tandem array (56–62). In contrast, the structure of CaBP1 has been shown to more closely resemble the characteristic dumbbell-like conformation of CaM, which possesses two independent domains that are able to differentially regulate target-binding proteins (37, 39, 52).
The structure of NCS-1 homologs from Schizosaccharomyces pombe (Ncs1) (63) and Saccharomyces cerevisiae (frequenin) (64) in complex with an interacting peptide from the yeast PI4KIIIβ homolog, Pik1, have been studied previously and have provided important insights into how NCS-1 may regulate PI4KIIIβ activity. In both systems, the Pik1 peptide forms two helices separated by a 20-residue linking loop. Upon Ca2+ binding to NCS-1, a hydrophobic pocket is exposed, allowing binding of the two Pik1 helices in an antiparallel manner. This forces the peptide into a U-turn conformation, and it is suggested that this could promote structural interactions between the lipid kinase unique and catalytic domains of Pik1, leading to optimal activation of lipid kinase activity (63, 64).
Regulation of PI4KIIIβ by NCS-1 is an evolutionarily conserved mechanism, and null mutations, which lack the NCS-1 orthologue, frequenin, or the PI4KIIIβ orthologue, Pik1, are lethal in yeast (65). Despite this, changes in NCS-1 regulation of PI4KIIIβ can be observed in higher organisms. The localization of NCS-1 in mammalian cells is not restricted to Golgi membranes as it is in yeast (28), and it has been found that the interaction of NCS-1 with PI4KIIIβ is of lower affinity than the interaction between yeast frequenin and Pik1 (66). Throughout evolution, there has been a significant expansion in the number of EF-hand containing Ca2+ sensors both in the neuronal calcium sensor family and in other vertebrate-specific families, such as the CaBPs (1, 67). This has led to increased complexity in the regulation of many processes, including ion channel function and intracellular trafficking. This may have led to alterations in the mechanisms underlying the modulation of PI4KIIIβ activity, and the discovery of an interaction of CaBP7 and CaBP8 with PI4KIIIβ supports this theory.
CaBP7 and CaBP8 form a separate subfamily within the CaBP family due to their unique pattern of EF-hand activity and their ability to target to cellular membranes via a C-terminal transmembrane domain (1, 8–10). Unlike other members of the CaBP family, which have three active EF-hand motifs and a redundant second EF-hand, CaBP7 and CaBP8 possess only two active EF-hands, which are both located in their N-terminal domain (6). Due to numerous amino acid deletions and substitutions, there is little sequence conservation within their C-terminal EF hand motifs. These differences are likely to be fundamental for determining distinct, non-overlapping target interactors for these proteins compared with the other CaBP proteins.
A previous study has reported a propensity of full-length CaBP7 and CaBP8 to dimerize in a Ca2+-independent manner both in vitro and in vivo (10). Hradsky et al. (10) suggested that CaBP7 and CaBP8 may have two dimerization interfaces: the hydrophobic TMD and the cytosolic domain. Here we show through SECMALLS and NMR analyses that Ca2+-bound CaBP7 NTD is homogeneously folded and monomeric in nature under the conditions tested. This suggests that the previously reported dimerization interface most likely requires residues present in CaBP7 CTD.
In agreement with previous studies, we show that CaBP7 NTD binds specifically to Ca2+ and not Mg2+ (25). Whereas the addition of Mg2+ to the protein resulted in precipitation, the addition of Ca2+ resulted in a significant conformational change as evidenced by comparison of 1H-15N HSQC NMR spectra of apo- and Ca2+-bound CaBP7 NTD. EF-hand domains in CaM-related calcium sensors typically adopt an open conformation upon binding to Ca2+, which exposes a highly hydrophobic pocket that can interact with target-binding proteins. The calculated interhelical angles within the Ca2+-bound NMR structure of CaBP7 NTD were in agreement with the protein adopting a similar open conformation, and far-UV CD analysis revealed an increase in helical content upon Ca2+ binding similar to that observed in CaM in previous studies (68). For NMR analyses, we could only obtain high quality spectra in the presence of n-octyl-β-d-glucopyranoside. Previous structural studies of small Ca2+-binding proteins have also employed this detergent (58, 61, 69). Significantly, our analyses show that Ca2+-bound CaBP7 NTD adopts a structural conformation that is very similar to that of CaM and CaBP1 CTDs. Based on this high degree of tertiary similarity, we believe that the presence of octyl glucoside has not significantly distorted or influenced the CaBP7 NTD structure. This conclusion is validated by comparison of CaBP7 NTD 1H-15N HSQC spectra in the absence and presence of 30 mm octyl glucoside (supplemental Fig. 1). No significant alterations were observed in key EF-hand residue chemical shifts (the same that undergo extensive changes between apo- and Ca2+-bound forms of CaBP7 NTD), although peak intensity and the number of visible peaks both increased in the presence of octyl glucoside. Collectively, these data indicate that 30 mm octyl glucoside did not alter the tertiary structure of CaBP7-NTD. CaBP7-NTD, CaM CTD, and CaBP1 CTD all contain four α-helices and two β-sheets of similar length and arrangement. In particular, the RMSD upon comparison of CaM and CaBP7 NTD was 1.36 Å, further demonstrating a tight correlation between the two structures.
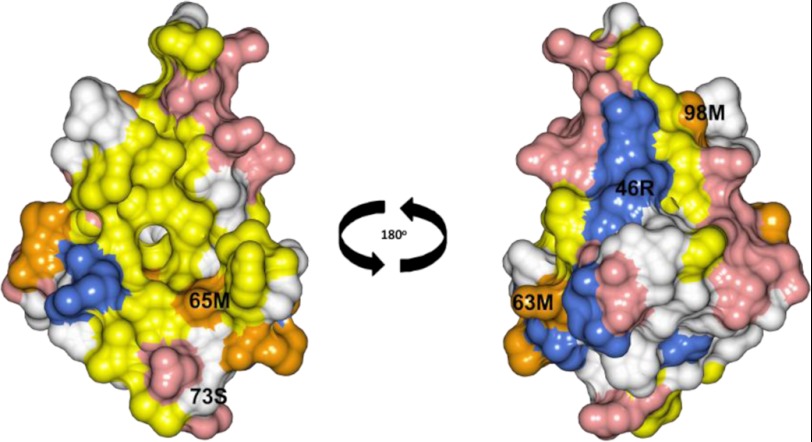
Despite the close similarities in the overall tertiary arrangement of the three proteins, analysis of their surface properties revealed more significant differences. CaBP7 NTD displays a more expansive hydrophobic pocket when compared with CaBP1 and CaM. Hydrophobic residues were found in place of charged residues at the corresponding positions in CaBP1 and CaM, resulting in the loss of an electrostatic patch. Interestingly, in CaM and CaBP1, the hydrophobic pocket forms the binding surface for target proteins. Charged residues may be important for determining the specificity of interactions, and the change in electrostatic potential may be significant for defining the ability of CaBP7 to regulate unique target proteins. Interestingly, comparison of the structured portion of CaBP7 NTD with CaBP8 NTD revealed 81% sequence identity and 95.6% sequence similarity between the two proteins with only conservative amino acid changes within the hydrophobic pocket. Modeling of the CaBP8 NTD structure based on the structured region of Ca2+-bound CaBP7 NTD suggests that the structures of the two proteins are probably very similar (Fig. 9), and this may explain their ability to interact with and regulate the same target protein, PI4KIIIβ.
FIGURE 9.
Space-filling representation of CaBP8 homology model. The front face is shown on the left, and the back face is shown on the right. Acidic residues (Asp and Glu) and basic residues (Arg, His, and Lys) are shown in red and blue, respectively. Hydrophobic residues (Ile, Leu, Phe, Trp, Val, and Tyr) are shown in yellow with the exception of Met residues, which are highlighted in orange. Solvent-exposed amino acids that are significantly different from the corresponding residues in CaBP7 are labeled. The predicted surface of CaBP8 is very similar to CaBP7 (Fig. 8A). The homology model was predicted using the Modeler software package (74) and residues 30–100 of the calculated Ca2+-bound CaBP7 NTD structure as a template.
The difference in the distribution of methionine residues on the surface of CaBP7 NTD is also conserved in CaBP8. CaM possesses nine methionine residues, which contribute ∼46% of the surface hydrophobicity in the N- and C-terminal domains (70). This high proportion of methionine residues is suggested to be the main reason that CaM is capable of binding such a diverse array of target proteins (70). The alignment in Fig. 7A demonstrates that there are four methionines in each of the N- and C-terminal EF-hand pairs of CaM, which reside at corresponding positions within the two sequences. CaBP1 CTD has methionines conserved at three of these positions, but CaBP7 and CaBP8 NTDs have only one. This supports previous bioinformatic analyses indicating that CaBP7 and CaBP8 evolved independently of other CaBPs (1, 6). In place of the three methionine residues, either isoleucine (Ile-77) or leucine (Leu-97 and -98) residues are found at these positions in CaBP7. Interestingly, conservative mutations of methionines to leucines in CaM, although not significantly altering the overall structure of the protein, significantly alter the ability of CaM to stimulate cAMP phosphodiesterase (71). Furthermore, structural studies have demonstrated a vital role for these methionine residues in interactions with numerous CaM substrates (55, 71).
The importance of methionine residues for determining protein interaction interfaces can be explained by considering the enhanced flexibility of their side chains when compared with the more rigid branched side chains of isoleucine and leucine (70, 71). Methionine side chains can adopt numerous configurations within proteins (72), meaning that a methionine-rich surface on a protein is potentially more malleable than a surface rich in isoleucine and leucine residues. The increased number of leucine and isoleucine residues in the exposed hydrophobic surface of CaBP7 NTD compared with CaM and CaBP1 may therefore make it less adaptable to binding partners of varying dimensions (71). This property may be important for conferring selectivity of binding to unique interactors, such as PI4KIIIβ.
The structural study presented here provides insights into how the structure of the EF-hand-containing NTD of CaBP7 compares with other related calcium sensors and how differences may confer unique regulatory activities. We have shown that this domain is sufficient for binding to the only known physiological target of CaBP7, PI4KIIIβ, whereas the CTD is not. Difficulties expressing high levels of mammalian PI4KIIIβ have precluded structural characterization of this protein, and although binding peptides important for interactions with the yeast orthologue of NCS-1 have been identified in Pik1, there is little sequence homology between this peptide and mammalian PI4KIIIβ. Further structural analysis of mammalian PI4KIIIβ will therefore be required in order to deduce the exact nature of its interaction with CaBP7 and how this might confer an inhibitory action opposite to the effects of NCS-1.
Supplementary Material
Acknowledgments
We thank Marjorie Howard and Dr. Thomas Jowitt (Faculty of Life Sciences, University of Manchester) for help with SECMALLS and circular dichroism analyses.
This work was supported by a Wellcome Trust prize Ph.D. studentship (to H. V. M.).
The atomic coordinates and structure factors (code 2LV7) have been deposited in the Protein Data Bank (http://wwpdb.org/).

This article contains supplemental Fig. 1.
- CaBP
- calcium-binding protein
- CaM
- calmodulin
- NTD
- N-terminal domain
- CTD
- C-terminal domain
- TMD
- transmembrane domain
- PI4KIIIβ
- phosphatidylinositol 4-kinase IIIβ
- SECMALLS
- size exclusion chromatography-multiangle laser light scattering
- NCS
- neuronal calcium sensor
- HSQC
- heteronuclear single quantum coherence
- RDC
- residual dipolar coupling
- RMSD
- root mean square deviation
- SUMO
- small ubiquitin-like modifier
- PDB
- Protein Data Bank
- BisTris
- bis(2-hydroxyethyl)iminotris(hydroxymethyl)methane.
REFERENCES
- 1. McCue H. V., Haynes L. P., Burgoyne R. D. (2010) Bioinformatic analysis of CaBP/calneuron proteins reveals a family of highly conserved vertebrate Ca2+-binding proteins. BMC Res. Notes 3, 118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Haeseleer F., Sokal I., Verlinde C. L., Erdjument-Bromage H., Tempst P., Pronin A. N., Benovic J. L., Fariss R. N., Palczewski K. (2000) Five members of a novel Ca2+-binding protein (CABP) subfamily with similarity to calmodulin. J. Biol. Chem. 275, 1247–1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Seidenbecher C. I., Langnaese K., Sanmartí-Vila L., Boeckers T. M., Smalla K. H., Sabel B. A., Garner C. C., Gundelfinger E. D., Kreutz M. R. (1998) Caldendrin, a novel neuronal calcium-binding protein confined to the somato-dendritic compartment. J. Biol. Chem. 273, 21324–21331 [DOI] [PubMed] [Google Scholar]
- 4. Wu Y. Q., Lin X., Liu C. M., Jamrich M., Shaffer L. G. (2001) Identification of a human brain-specific gene, calneuron 1, a new member of the calmodulin superfamily. Mol. Genet. Metab. 72, 343–350 [DOI] [PubMed] [Google Scholar]
- 5. Mikhaylova M., Sharma Y., Reissner C., Nagel F., Aravind P., Rajini B., Smalla K. H., Gundelfinger E. D., Kreutz M. R. (2006) Neuronal Ca2+ signaling via caldendrin and calneurons. Biochim. Biophys. Acta 1763, 1229–1237 [DOI] [PubMed] [Google Scholar]
- 6. Haynes L. P., McCue H. V., Burgoyne R. D. (2012) Evolution and functional diversity of the calcium-binding proteins (CaBPs). Front. Mol. Neurosci. 5, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. McCue H. V., Haynes L. P., Burgoyne R. D. (2010) The diversity of calcium sensor proteins in the regulation of neuronal function. Cold Spring Harb. Perspect. Biol. 2, a004085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. McCue H. V., Burgoyne R. D., Haynes L. P. (2009) Membrane targeting of the EF-hand containing calcium-sensing proteins CaBP7 and CaBP8. Biochem. Biophys. Res. Commun. 380, 825–831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. McCue H. V., Burgoyne R. D., Haynes L. P. (2011) Determination of the membrane topology of the small EF-hand Ca2+-sensing proteins CaBP7 and CaBP8. PLoS One 6, e17853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hradsky J., Raghuram V., Reddy P. P., Navarro G., Hupe M., Casado V., McCormick P. J., Sharma Y., Kreutz M. R., Mikhaylova M. (2011) Post-translational membrane insertion of tail-anchored transmembrane EF-hand Ca2+ sensor calneurons requires the TRC40/Asna1 protein chaperone. J. Biol. Chem. 286, 36762–36776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Haynes L. P., Tepikin A. V., Burgoyne R. D. (2004) Calcium-binding protein 1 is an inhibitor of agonist-evoked, inositol 1,4,5-trisphosphate-mediated calcium signaling. J. Biol. Chem. 279, 547–555 [DOI] [PubMed] [Google Scholar]
- 12. Yang J., McBride S., Mak D. O., Vardi N., Palczewski K., Haeseleer F., Foskett J. K. (2002) Identification of a family of calcium sensors as protein ligands of inositol trisphosphate receptor Ca2+ release channels. Proc. Natl. Acad. Sci. U.S.A. 99, 7711–7716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kasri N. N., Bultynck G., Smyth J., Szlufcik K., Parys J. B., Callewaert G., Missiaen L., Fissore R. A., Mikoshiba K., de Smedt H. (2004) The N-terminal Ca2+-independent calmodulin-binding site on the inositol 1,4,5-trisphosphate receptor is responsible for calmodulin inhibition, even though this inhibition requires Ca2+. Mol. Pharmacol. 66, 276–284 [DOI] [PubMed] [Google Scholar]
- 14. Cardy T. J., Taylor C. W. (1998) A novel role for calmodulin. Ca2+-independent inhibition of type-1 inositol trisphosphate receptors. Biochem. J. 334, 447–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhou H., Kim S. A., Kirk E. A., Tippens A. L., Sun H., Haeseleer F., Lee A. (2004) Ca2+-binding protein-1 facilitates and forms a postsynaptic complex with CaV1.2 (L-type) Ca2+ channels. J. Neurosci. 24, 4698–4708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhou H., Yu K., McCoy K. L., Lee A. (2005) Molecular mechanism for divergent regulation of CaV1.2 Ca2+ channels by calmodulin and Ca2+-binding protein-1. J. Biol. Chem. 280, 29612–29619 [DOI] [PubMed] [Google Scholar]
- 17. Tippens A. L., Lee A. (2007) Caldendrin, a neuron-specific modulator of CaV/1.2 (L-type) Ca2+ channels. J. Biol. Chem. 282, 8464–8473 [DOI] [PubMed] [Google Scholar]
- 18. Asmara H., Minobe E., Saud Z. A., Kameyama M. (2010) Interactions of calmodulin with the multiple binding sites of CaV1.2 Ca2+ channels. J. Pharmacol. Sci. 112, 397–404 [DOI] [PubMed] [Google Scholar]
- 19. Cui G., Meyer A. C., Calin-Jageman I., Neef J., Haeseleer F., Moser T., Lee A. (2007) Ca2+-binding proteins tune Ca2+ feedback to CaV1.3 channels in mouse auditory hair cells. J. Physiol. 585, 791–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Haeseleer F., Imanishi Y., Maeda T., Possin D. E., Maeda A., Lee A., Rieke F., Palczewski K. (2004) Essential role of Ca2+-binding protein 4, a CaV1.4 channel regulator, in photoreceptor synaptic function. Nat. Neurosci. 7, 1079–1087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yang P. S., Alseikhan B. A., Hiel H., Grant L., Mori M. X., Yang W., Fuchs P. A., Yue D. T. (2006) Switching of Ca2+-dependent inactivation of CaV1.3 channels by calcium-binding proteins of auditory hair cells. J. Neurosci. 26, 10677–10689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Few A. P., Lautermilch N. J., Westenbroek R. E., Scheuer T., Catterall W. A. (2005) Differential regulation of CaV2.1 channels by calcium-binding protein 1 and visinin-like protein-2 requires N-terminal myristoylation. J. Neurosci. 25, 7071–7080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lee A., Westenbroek R. E., Haeseleer F., Palczewski K., Scheuer T., Catterall W. A. (2002) Differential modulation of CaV2.1 channels by calmodulin and Ca2+-binding protein 1. Nat. Neurosci. 5, 210–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lee A., Wong S. T., Gallagher D., Li B., Storm D. R., Scheuer T., Catterall W. A. (1999) Ca2+/calmodulin binds to and modulates P/Q-type calcium channels. Nature 399, 155–159 [DOI] [PubMed] [Google Scholar]
- 25. Mikhaylova M., Reddy P. P., Munsch T., Landgraf P., Suman S. K., Smalla K. H., Gundelfinger E. D., Sharma Y., Kreutz M. R. (2009) Calneurons provide a calcium threshold for trans-Golgi network to plasma membrane trafficking. Proc. Natl. Acad. Sci. U.S.A. 106, 9093–9098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. de Barry J., Janoshazi A., Dupont J. L., Procksch O., Chasserot-Golaz S., Jeromin A., Vitale N. (2006) Functional implication of neuronal calcium sensor-1 and phosphoinositol 4-kinase-β interaction in regulated exocytosis of PC12 cells. J. Biol. Chem. 281, 18098–18111 [DOI] [PubMed] [Google Scholar]
- 27. Zheng Q., Bobich J. A., Vidugiriene J., McFadden S. C., Thomas F., Roder J., Jeromin A. (2005) Neuronal calcium sensor-1 facilitates neuronal exocytosis through phosphatidylinositol 4-kinase. J. Neurochem. 92, 442–451 [DOI] [PubMed] [Google Scholar]
- 28. Taverna E., Francolini M., Jeromin A., Hilfiker S., Roder J., Rosa P. (2002) Neuronal calcium sensor 1 and phosphatidylinositol 4-OH kinase β interact in neuronal cells and are translocated to membranes during nucleotide-evoked exocytosis. J. Cell Sci. 115, 3909–3922 [DOI] [PubMed] [Google Scholar]
- 29. Haynes L. P., Thomas G. M., Burgoyne R. D. (2005) Interaction of neuronal calcium sensor-1 and ADP-ribosylation factor 1 allows bidirectional control of phosphatidylinositol 4-kinase β and trans-Golgi network-plasma membrane traffic. J. Biol. Chem. 280, 6047–6054 [DOI] [PubMed] [Google Scholar]
- 30. Haynes L. P., Fitzgerald D. J., Wareing B., O'Callaghan D. W., Morgan A., Burgoyne R. D. (2006) Analysis of the interacting partners of the neuronal calcium-binding proteins L-CaBP1, hippocalcin, NCS-1, and neurocalcin δ. Proteomics 6, 1822–1832 [DOI] [PubMed] [Google Scholar]
- 31. Vranken W. F., Boucher W., Stevens T. J., Fogh R. H., Pajon A., Llinas M., Ulrich E. L., Markley J. L., Ionides J., Laue E. D. (2005) The CCPN data model for NMR spectroscopy. Development of a software pipeline. Proteins 59, 687–696 [DOI] [PubMed] [Google Scholar]
- 32. Linge J. P., Habeck M., Rieping W., Nilges M. (2003) ARIA. Automated NOE assignment and NMR structure calculation. Bioinformatics 19, 315–316 [DOI] [PubMed] [Google Scholar]
- 33. Güntert P. (2004) Automated NMR structure calculation with CYANA. Methods Mol. Biol. 278, 353–378 [DOI] [PubMed] [Google Scholar]
- 34. Cheung M. S., Maguire M. L., Stevens T. J., Broadhurst R. W. (2010) DANGLE. A Bayesian inferential method for predicting protein backbone dihedral angles and secondary structure. J. Magn. Reson. 202, 223–233 [DOI] [PubMed] [Google Scholar]
- 35. Laskowski R. A., Rullmannn J. A., MacArthur M. W., Kaptein R., Thornton J. M. (1996) AQUA and PROCHECK-NMR. Programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR 8, 477–486 [DOI] [PubMed] [Google Scholar]
- 36. Zweckstetter M. (2008) NMR. Prediction of molecular alignment from structure using the PALES software. Nature protocols 3, 679–690 [DOI] [PubMed] [Google Scholar]
- 37. Li C., Chan J., Haeseleer F., Mikoshiba K., Palczewski K., Ikura M., Ames J. B. (2009) Structural insights into Ca2+-dependent regulation of inositol 1,4,5-trisphosphate receptors by CaBP1. J. Biol. Chem. 284, 2472–2481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wingard J. N., Chan J., Bosanac I., Haeseleer F., Palczewski K., Ikura M., Ames J. B. (2005) Structural analysis of Mg2+ and Ca2+ binding to CaBP1, a neuron-specific regulator of calcium channels. J. Biol. Chem. 280, 37461–37470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Findeisen F., Minor D. L., Jr. (2010) Structural basis for the differential effects of CaBP1 and calmodulin on CaV1.2 calcium-dependent inactivation. Structure 18, 1617–1631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Liu Z., Vogel H. J. (2012) Structural basis for the regulation of L-type voltage-gated calcium channels. Interactions between the N-terminal cytoplasmic domain and Ca2+-calmodulin. Front. Mol. Neurosci. 5, 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kim E. Y., Rumpf C. H., Fujiwara Y., Cooley E. S., Van Petegem F., Minor D. L., Jr. (2008) Structures of CaV2 Ca2+/CaM-IQ domain complexes reveal binding modes that underlie calcium-dependent inactivation and facilitation. Structure 16, 1455–1467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Aravind P., Chandra K., Reddy P. P., Jeromin A., Chary K. V., Sharma Y. (2008) Regulatory and structural EF-hand motifs of neuronal calcium sensor-1. Mg2+ modulates Ca2+ binding, Ca2+-induced conformational changes, and equilibrium unfolding transitions. J. Mol. Biol. 376, 1100–1115 [DOI] [PubMed] [Google Scholar]
- 43. Lim S., Peshenko I., Dizhoor A., Ames J. B. (2009) Effects of Ca2+, Mg2+, and myristoylation on guanylyl cyclase-activating protein 1 structure and stability. Biochemistry 48, 850–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Osawa M., Dace A., Tong K. I., Valiveti A., Ikura M., Ames J. B. (2005) Mg2+ and Ca2+ differentially regulate DNA binding and dimerization of DREAM. J. Biol. Chem. 280, 18008–18014 [DOI] [PubMed] [Google Scholar]
- 45. Romani A., Scarpa A. (1992) Regulation of cell magnesium. Arch. Biochem. Biophys. 298, 1–12 [DOI] [PubMed] [Google Scholar]
- 46. Takeuchi T., Sarashina I., Iijima M., Endo K. (2008) In vitro regulation of CaCO3 crystal polymorphism by the highly acidic molluscan shell protein Aspein. FEBS Lett. 582, 591–596 [DOI] [PubMed] [Google Scholar]
- 47. Palmiter R. D. (1974) Magnesium precipitation of ribonucleoprotein complexes. Expedient techniques for the isolation of undergraded polysomes and messenger ribonucleic acid. Biochemistry 13, 3606–3615 [DOI] [PubMed] [Google Scholar]
- 48. Weisenberg R. C., Timasheff S. N. (1970) Aggregation of microtubule subunit protein. Effects of divalent cations, colchicine and vinblastine. Biochemistry 9, 4110–4116 [DOI] [PubMed] [Google Scholar]
- 49. Feric M., Zhao B., Hoffert J. D., Pisitkun T., Knepper M. A. (2011) Large-scale phosphoproteomic analysis of membrane proteins in renal proximal and distal tubule. Am. J. Physiol. Cell Physiol. 300, C755–C770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Chattopadhyaya R., Meador W. E., Means A. R., Quiocho F. A. (1992) Calmodulin structure refined at 1.7 Å resolution. J. Mol. Biol. 228, 1177–1192 [DOI] [PubMed] [Google Scholar]
- 51. Kuboniwa H., Tjandra N., Grzesiek S., Ren H., Klee C. B., Bax A. (1995) Solution structure of calcium-free calmodulin. Nat. Struct. Biol. 2, 768–776 [DOI] [PubMed] [Google Scholar]
- 52. Park S., Li C., Ames J. B. (2011) Protein Sci., DOI: 10.1002/pro.662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. McNicholas S., Potterton E., Wilson K. S., Noble M. E. (2011) Presenting your structures. The CCP4mg molecular graphics software. Acta Crystallogr. D 67, 386–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Krissinel E., Henrick K. (2007) Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 [DOI] [PubMed] [Google Scholar]
- 55. Ikura M., Ames J. B. (2006) Genetic polymorphism and protein conformational plasticity in the calmodulin superfamily. Two ways to promote multifunctionality. Proc. Natl. Acad. Sci. U.S.A. 103, 1159–1164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Flaherty K. M., Zozulya S., Stryer L., McKay D. B. (1993) Three-dimensional structure of recoverin, a calcium sensor in vision. Cell 75, 709–716 [DOI] [PubMed] [Google Scholar]
- 57. Stephen R., Palczewski K., Sousa M. C. (2006) The crystal structure of GCAP3 suggests molecular mechanism of GCAP-linked cone dystrophies. J. Mol. Biol. 359, 266–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Lusin J. D., Vanarotti M., Li C., Valiveti A., Ames J. B. (2008) NMR structure of DREAM. Implications for Ca2+-dependent DNA binding and protein dimerization. Biochemistry 47, 2252–2264 [DOI] [PubMed] [Google Scholar]
- 59. Bourne Y., Dannenberg J., Pollmann V., Marchot P., Pongs O. (2001) Immunocytochemical localization and crystal structure of human frequenin (neuronal calcium sensor 1). J. Biol. Chem. 276, 11949–11955 [DOI] [PubMed] [Google Scholar]
- 60. Vijay-Kumar S., Kumar V. D. (1999) Crystal structure of recombinant bovine neurocalcin. Nat. Struct. Biol. 6, 80–88 [DOI] [PubMed] [Google Scholar]
- 61. Ames J. B., Dizhoor A. M., Ikura M., Palczewski K., Stryer L. (1999) Three-dimensional structure of guanylyl cyclase-activating protein-2, a calcium-sensitive modulator of photoreceptor guanylyl cyclases. J. Biol. Chem. 274, 19329–19337 [DOI] [PubMed] [Google Scholar]
- 62. Ames J. B., Lim S. (2012) Molecular structure and target recognition of neuronal calcium sensor proteins. Biochim. Biophys. Acta 1820, 1205–1213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Lim S., Strahl T., Thorner J., Ames J. B. (2011) Structure of a Ca2+-myristoyl switch protein that controls activation of a phosphatidylinositol 4-kinase in fission yeast. J. Biol. Chem. 286, 12565–12577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Strahl T., Huttner I. G., Lusin J. D., Osawa M., King D., Thorner J., Ames J. B. (2007) Structural insights into activation of phosphatidylinositol 4-kinase (Pik1) by yeast frequenin (Frq1). J. Biol. Chem. 282, 30949–30959 [DOI] [PubMed] [Google Scholar]
- 65. Balla A., Balla T. (2006) Phosphatidylinositol 4-kinases. Old enzymes with emerging functions. Trends Cell Biol. 16, 351–361 [DOI] [PubMed] [Google Scholar]
- 66. Zhao X., Várnai P., Tuymetova G., Balla A., Tóth Z. E., Oker-Blom C., Roder J., Jeromin A., Balla T. (2001) Interaction of neuronal calcium sensor-1 (NCS-1) with phosphatidylinositol 4-kinase β stimulates lipid kinase activity and affects membrane trafficking in COS-7 cells. J. Biol. Chem. 276, 40183–40189 [DOI] [PubMed] [Google Scholar]
- 67. Burgoyne R. D., Weiss J. L. (2001) The neuronal calcium sensor family of Ca2+-binding proteins. Biochem. J. 353, 1–12 [PMC free article] [PubMed] [Google Scholar]
- 68. Dieter P., Cox J. A., Marmé D. (1985) Calcium-binding and its effect on circular dichroism of plant calmodulin. Planta 166, 216–218 [DOI] [PubMed] [Google Scholar]
- 69. Schwenk J., Zolles G., Kandias N. G., Neubauer I., Kalbacher H., Covarrubias M., Fakler B., Bentrop D. (2008) NMR analysis of KChIP4a reveals structural basis for control of surface expression of Kv4 channel complexes. J. Biol. Chem. 283, 18937–18946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. O'Neil K. T., DeGrado W. F. (1990) How calmodulin binds its targets. Sequence-independent recognition of amphiphilic α-helices. Trends Biochem. Sci. 15, 59–64 [DOI] [PubMed] [Google Scholar]
- 71. Zhang M., Li M., Wang J. H., Vogel H. J. (1994) The effect of Met → Leu mutations on calmodulin's ability to activate cyclic nucleotide phosphodiesterase. J. Biol. Chem. 269, 15546–15552 [PubMed] [Google Scholar]
- 72. Gellman S. H. (1991) On the role of methionine residues in the sequence-independent recognition of nonpolar protein surfaces. Biochemistry 30, 6633–6636 [DOI] [PubMed] [Google Scholar]
- 73. Krissinel E., Henrick K. (2004) Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr. D 60, 2256–2268 [DOI] [PubMed] [Google Scholar]
- 74. Sali A., Blundell T. L. (1993) Comparative protein modeling by satisfaction of spatial restraints. J. Mol. Biol. 234, 779–815 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.