Background: Small molecule antagonists to the kinase domain induce ectodomain dimerization.
Results: EGF and kinase inhibitors that stabilize the asymmetric kinase domain dimer induce identical transmembrane domain interfaces as shown by cross-linking but distinct ectodomain conformations as shown by EM.
Conclusion: Transmembrane domain dimerization is not sufficient to induce the EGF-induced ectodomain conformation.
Significance: Outside-in and inside-out signaling differ in EGFR.
Keywords: Electron Microscopy (EM), Epidermal Growth Factor (EGF), Epidermal Growth Factor Receptor (EGFR), Immunology, Signaling
Abstract
We study a mechanism by which dimerization of the EGF receptor (EGFR) cytoplasmic domain is transmitted to the ectodomain. Therapeutic and other small molecule antagonists to the kinase domain that stabilize its active conformation, but not those that stabilize an inactive conformation, stabilize ectodomain dimerization. Inhibitor-induced dimerization requires an asymmetric kinase domain interface associated with activation. EGF and kinase inhibitors stimulate formation of identical dimer interfaces in the EGFR transmembrane domain, as shown by disulfide cross-linking. Disulfide cross-linking at an interface in domain IV in the ectodomain was also stimulated similarly; however, EGF but not inhibitors stimulated cross-linking in domain II. Inhibitors similarly induced noncovalent dimerization in nearly full-length, detergent-solubilized EGFR as shown by gel filtration. EGFR ectodomain deletion resulted in spontaneous dimerization, whereas deletion of exons 2–7, in which extracellular domains III and IV are retained, did not. In EM, kinase inhibitor-induced dimers lacked any well defined orientation between the ectodomain monomers. Fab of the therapeutic antibody cetuximab to domain III confirmed a variable position and orientation of this domain in inhibitor-induced dimers but suggested that the C termini of domain IV of the two monomers were in close proximity, consistent with dimerization in the transmembrane domains. The results provide insights into the relative energetics of intracellular and extracellular dimerization in EGFR and have significance for physiologic dimerization through the asymmetric kinase interface, bidirectional signal transmission in EGFR, and mechanism of action of therapeutics.
Introduction
The EGF receptor (EGFR)3 plays a critical role in cell survival and proliferation. Aberrant activation of this receptor tyrosine kinase is associated with a variety of human cancers (1). Small molecule kinase antagonists and monoclonal antibodies targeting EGFR are approved cancer therapeutics (1, 2).
Structural and biochemical studies have revealed how ligand binding promotes dimerization of the EGFR ectodomain, transmembrane (TM) domain, and kinase domain and activates its kinase activity (3–10). The EGFR ectodomain contains four domains: ligand-binding domains I and III and cysteine-rich domains II and IV. Domain II is masked in the tethered, unliganded receptor monomer. Binding of growth factor to both domains I and III induces their closer approach. Large conformational changes in the ectodomain free domain II and enable it to mediate dimerization (3, 11). In addition, dimerization in domain IV proximal to the membrane brings the C termini of two ectodomain monomers into close proximity, so that their single-pass TM domains can dimerize (9).
Ectodomain dimerization drives kinase activation by an allosteric mechanism in which two kinase domains form an asymmetric dimer. The C-lobe of one kinase domain interacts with the N-lobe of the second kinase domain and stabilizes the latter in the active conformation (10). A juxtamembrane region of 41 residues lies between the EGFR TM and kinase domains. The juxtamembrane region plays a role in kinase activation by stabilizing the asymmetric kinase dimer (8, 12). However, many aspects of how the conformation of the ectodomain couples to kinase activation remain unclear.
Previous studies with EGFR suggest a rather flexible linkage between the ectodomain and the intracellular domain. Cross-linking between the TM domains and between domains IV of intact EGFR on cell surfaces was found to be greatly stimulated by the addition of EGF; however, highly specific interfaces, including those in domain IV identified in ectodomain crystal structures and those in the TM domain identified by ligand-stimulated cross-linking, were shown by mutation not to be required for EGFR signaling (9). Furthermore, negative-stain EM with nearly full-length EGFR in detergent micelles showed that the ligand-bound ectodomain dimer can couple to multiple kinase domain arrangements (7). The EGF-dimerized ectodomain was found in association with rod-like asymmetric kinase dimers, as well as with symmetric kinase dimers and unassociated kinase monomers. Furthermore, small molecule kinase antagonists were found to shift the equilibrium between these three different states of association of the kinase domain. These results are consistent with loose linkage between the EGFR ectodomain and intracellular domain (9).
Because ectodomain dimerization can stimulate kinase domain dimerization and outside-in signaling, it might be expected that kinase domain dimerization could stimulate ectodomain dimerization and hence some type of inside-out signaling. Indeed, some studies have detected a certain extent of EGFR dimerization in the absence of exogenous ligand and found that such dimerization is dependent on an intact kinase domain (13, 14). Furthermore, the quinazoline-based kinase antagonists PD153035, AG-1478, and AG-1517 were reported to increase the formation of such ligand-independent EGFR dimers (15–17).
The mechanism(s) underlying kinase domain-driven dimerization remain poorly understood. Do the signals that are propagated to the cell membrane or ectodomain involve conformational transitions that are similar to those propagated by EGF in the opposite direction? Little is known, except that the binding of a conformation-dependent antibody to the ectodomain is increased by AG-1478 (17). Furthermore, is a specific type of kinase dimerization interface required for kinase-dependent TM and ectodomain dimerization? Recent studies have shown that association of kinase domains across a specific, asymmetric dimerization interface activates kinase activity and that different classes of kinase antagonists favor the active-like conformation seen with asymmetric dimer interface or an inactive-like conformation seen with distinct symmetric dimer interfaces in crystal lattices or a monomeric state seen in EM (7, 18–21). Each of these classes of antagonists includes quinazoline-based antagonists.
Here, we show that kinase-mediated EGFR dimerization is dependent on kinase domain conformation and requires formation of an asymmetric dimer. Kinase inhibitors that stabilize the active conformation of the kinase, but not those that stabilize inactive conformations, promote dimer formation. Furthermore, association occurs in the TM domain through interfaces similar to those found in ligand-induced dimerization. However, the ectodomain adopts a conformation distinct from the liganded dimer. This is shown both by a lack of cross-linking in domain II and by visualization of dimers in EM. Thus, one dimeric kinase domain conformation can couple to different ectodomain conformations, further demonstrating complexity in coupling between the ectodomain and intracellular domains in EGFR transmembrane signaling.
EXPERIMENTAL PROCEDURES
Reagents and Cell Lines
The kinase inhibitors gefitinib and erlotinib were from LC Laboratories (Woburn, MA); lapatinib was from Enzo Life Sciences (Farmingdale, NY); and PD168393 was from Calbiochem (Billerica, MA). HKI-272 was kindly provided by Dr. Janne Pasi (Dana-Farber Cancer Research Institute, Boston, MA). Recombinant human EGF was from PeproTech, Inc. (Rocky Hill, NJ). Antibodies to protein C and to phosphotyrosine (clone 4G10) were from Roche Applied Science and Millipore (Billerica, MA), respectively. Cetuximab Fab was kindly provided by Dr. Kathryn Ferguson, University of Pennsylvania (Philadelphia, PA).
I682Q and V924R mutations were generated in wild-type EGFR or the A623C mutant using QuikChange II XL from Agilent Technologies (Wilmington, DE). The full EGFR sequence was determined for each mutant. Stable Ba/F3 transfectants were generated as described (9).
Inhibitor and EGF Treatment of Cells, Lysate Preparation, and Western Blot
Inhibitor stocks (10 mm) in Me2SO were stored at −80 °C. Ba/F3 transfectants were serum- and IL-3-starved for 3 h and incubated with kinase inhibitors (final concentration, 10 μm) or Me2SO (0.1%) for 45 min at 37 °C in culture medium with 5% CO2. The cells were then treated with or without EGF at a final concentration of 100 nm for 5 min at 37 °C. Lysate preparation and Western blots were as described (9).
Disulfide and Chemical Cross-linking
Disulfide cross-linking of Ba/F3 stable cell lines that express EGFR cysteine substitutions was as described (9). Briefly, for mutants with cysteine substitutions in the extracellular domain, including the linker to the TM domain, the cells were lysed without permeabilization and oxidant treatment. For disulfide cross-linking of transmembrane cysteine mutants, the cells were pretreated with 2-bromopalmitate to inhibit palmitoylation of cysteines and permeabilized by two cycles of freezing and thawing. Permeabilized cells were treated with CuSO4/o-phenanthroline for 10 min at room temperature before detergent lysis. For extracellular region chemical cross-linking, the cells were washed twice with cold PBS and incubated with 1 mm bis(sulfosuccinimidyl) suberate (BS3) in PBS for 1 h at 4 °C. The reactions were quenched by addition of Tris, pH 7.4, at a final concentration of 50 mm for 15 min. After two washes in PBS, cells were lysed and subjected to Western blotting. EGFR was quantitated using antibody to the protein C tag as described previously (9).
Expression, Purification, and Gel Filtration of EGFR Proteins
The EGFR ΔectoΔ998 construct was prepared by deleting residues 1–612 from the previously described EGFR Δ998 construct using PCR amplification (7). The EGFR V924R Δ998 mutation was generated in EGFR Δ998 using QuikChange II XL. Expression of EGFR mutants in HEK293T cells, solubilization in Triton X-100, purification, and exchange into 0.2 mm dodecylmaltoside during the final gel filtration step were as described (7). For chromatographic analysis, 0.7 μm affinity-purified receptors, with or without 40 μm cetuximab Fab, were treated with 200 μm PD168393 or 200 μm HKI-272 for 5 min on ice before fractionation on Superose 6 HR equilibrated with 20 mm Tris, pH 8.0, 200 mm NaCl, 1 mm EDTA, 0.2 mm DTT, and 0.2 mm dodecylmaltoside (running buffer). Peak fractions corresponding to dimeric material were collected and immediately applied to EM grids for negative staining. Treatments were identical for EGFR ΔectoΔ998, except it was 0.2 μm in concentration.
Negative Staining, EM Image Collection, and Data Processing
Negative staining, EM image collection and data processing were as described (7). Briefly, proteins were applied to a glow-discharged, carbon-coated grid and stained with four drops of 0.75% (w/v) uranyl formate. EM images were collected in low dose mode with a Tecnai T12 microscope (FEI) at an acceleration voltage of 120 kV. The images were recorded at 67,000× magnification and a defocus of −1.5 μm. The pixel size at the specimen level was 4.5 Å. The particles were interactively picked using BOXER in EMAN (22) and processed with SPIDER (23). Particles for each sample were subjected to iterative K-means classification, multireference alignment, and averaging. For cross-correlation, evenly spaced projections from crystal structures were aligned with masked EM class averages; cross-correlation coefficients are reported for the most similar crystal structure projection.
RESULTS
Gefitinib Induces Dimerization of the Transmembrane Domain and Extracellular Linker Region of EGFR
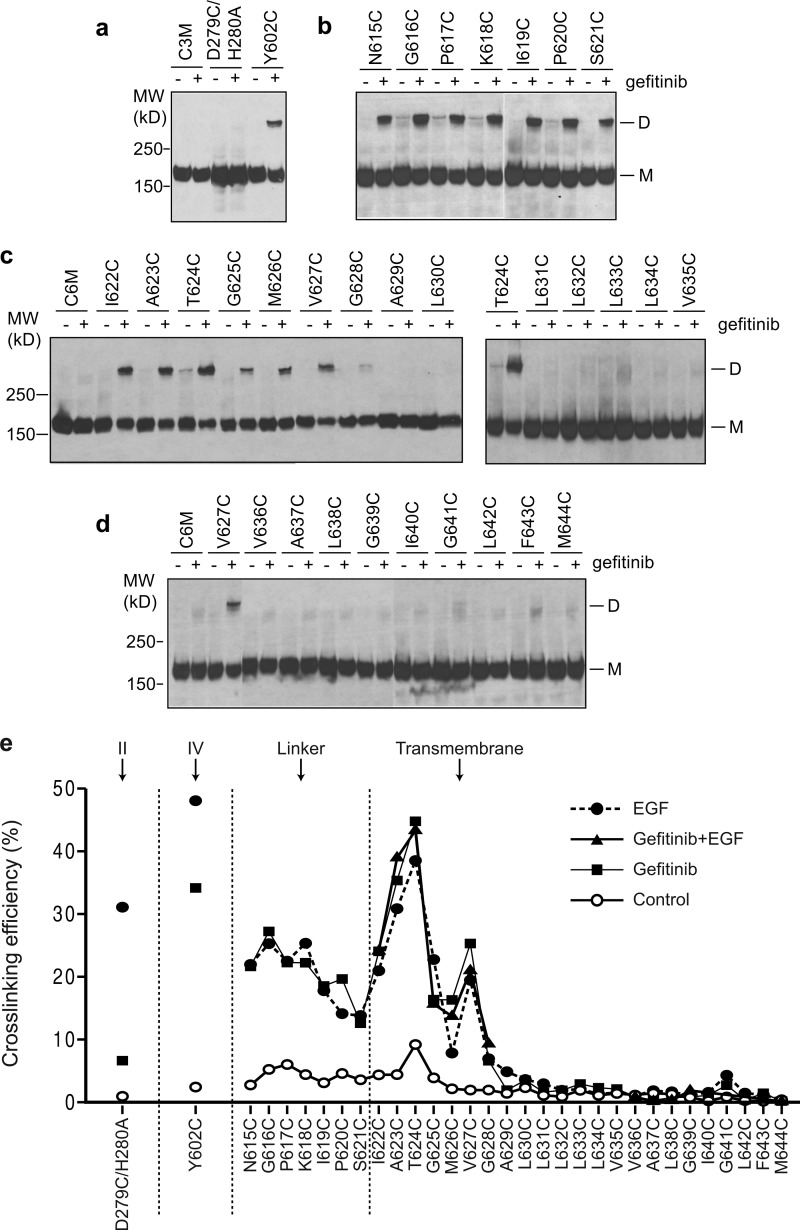
Ba/F3 stable cell lines that express EGFR with single cysteine substitutions in the extracellular linker region (residues 615–621) and TM domain (residues 622–644) were used to test the ability of gefitinib and, for comparison, EGF to induce EGFR dimerization. Disulfide bond formation between mutationally introduced cysteines was assayed by nonreducing SDS-PAGE of cell lysates and Western blotting (9) (Fig. 1). Compared with control, gefitinib markedly stimulated dimerization through cysteines in the extracellular linker region (Fig. 1b) and TM domain (Fig. 1c). A specific orientation between dimerized transmembrane α-helices was suggested by a major cross-linking peak at residues 623 and 624 and a smaller peak at residue 627 (Fig. 1e). The difference of 3.5 residues in peak position corresponds to the α-helical periodicity of 3.6 residues/turn. Remarkably, the pattern of disulfide cross-linking in the linker and TM regions induced by gefitinib was identical to that induced by EGF (9) and by the combination of gefitinib and EGF (Fig. 1e). These results suggest that gefitinib and EGF induce structurally similar dimerization interfaces in the EGFR linker and TM domains.
FIGURE 1.
Disulfide cross-linking of EGFR cysteine mutants in Ba/F3 transfectants treated with or without EGFR-specific kinase inhibitor, gefitinib. The cells were treated with 10 μm gefitinib or Me2SO control for 45 min. The lysates were subjected to nonreducing SDS 5% PAGE and Western blotting with protein C antibody to detect EGFR protein. a, cysteine substitutions in the domain II (D279C/H280A) and domain IV (Y602C) interfaces in the crystal structure of liganded dimer. b, linker residues. c and d, TM domain residues. e, comparison of disulfide cross-linking induced by gefitinib, EGF, and gefitinib in combination with EGF. EGF-induced cross-linking data were published previously (9). MW, molecular mass; D, dimeric; M, monomeric.
We also examined dimerization at two interfaces in the ectodomain. The D279C/H280A mutation assays dimerization in domain II, which is at the center of the dimerization interface. The Y602C mutation is at the membrane-proximal end of the highly elongated domain IV. Domain IV is more flexible than domains I–III in ectodomain dimers, as shown by its high temperature (B) factors in the crystal structure (9). Gefitinib enabled substantial cross-linking of Y602C at the domain IV–IV interface, inducing 70% as much as EGF (Fig. 1e). In contrast, cross-linking with gefitinib was substantially less at residue 279 in domain II, only 19% as much as with EGF (Fig. 1e). These results suggest that although the membrane-proximal ends of domain IV in gefitinib-induced dimers are nearby one another, there are large differences in the structure of the ectodomain in EGF-induced and gefitinib-induced dimers, particularly in the region of the dimerization arm in domain II.
The Effect of Kinase Inhibitors on Kinase Conformation Correlates with Effect on EGFR Dimerization
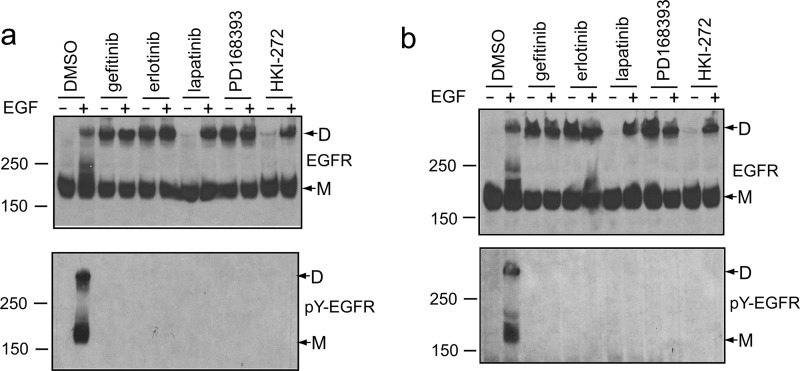
Although multiple kinase inhibitors bind to the ATP-binding pocket and inhibit kinase activity, co-crystal structures have shown that they differ in the kinase conformation that they stabilize and the types of kinase dimer interfaces that form in crystal lattices (10, 18–21). To test for differences among inhibitors, Ba/F3 cell transfectants were incubated with inhibitors or control, treated without or with EGF, and examined for cross-linking at TM residue 623 (Fig. 2a). In controls, EGF induced dimerization (Fig. 2a, upper panel) and phosphorylation on tyrosine (Fig. 2a, lower panel). In contrast, EGF-induced autophosphorylation was completely blocked in presence of kinase inhibitors (Fig. 2a, lower panel). Three kinase inhibitors that stabilize the active conformation of the kinase domain, gefitinib, erlotinib, and the covalent inhibitor PD168393 all induced EGFR TM domain dimerization (Fig. 2a, upper panel). Combination with EGF did not further increase cross-linking (Fig. 2a, upper panel). By contrast, two inhibitors that stabilize the inactive conformation of the kinase domain, lapatinib and the covalent inhibitor HKI-272, did not induce any dimerization (Fig. 2a, upper panel). However, these inhibitors had no effect on dimerization in the TM domains induced by EGF (Fig. 2a, upper panel). Essentially identical effects were observed with the N615C mutation, which assesses dimerization in the ectodomain-TM linker (Fig. 2b). These results suggest that the active kinase domain conformation is linked to the dimerization of EGFR at the TM domain and the extracellular linker, even in the absence of EGF.
FIGURE 2.
Two distinct classes of kinase inhibitors have different effect on EGFR dimerization. a and b, Ba/F3 stable transfectants expressing EGFR cysteine mutant A623C (a) or N615C (b) were incubated with 10 μm gefitinib, erlotinib, lapatinib, PD168393, HKI-272, or Me2SO (DMSO) for 45 min and treated with or without 100 nm EGF for 5 min. The lysates were subjected to nonreducing SDS 5% PAGE and Western blotting with protein C antibody (upper panel) or phosphotyrosine antibody 4G10 (lower panel). D, dimeric; M, monomeric.
The Asymmetric Kinase Domain Interface Is Required for Kinase Inhibitor-induced EGFR Dimerization
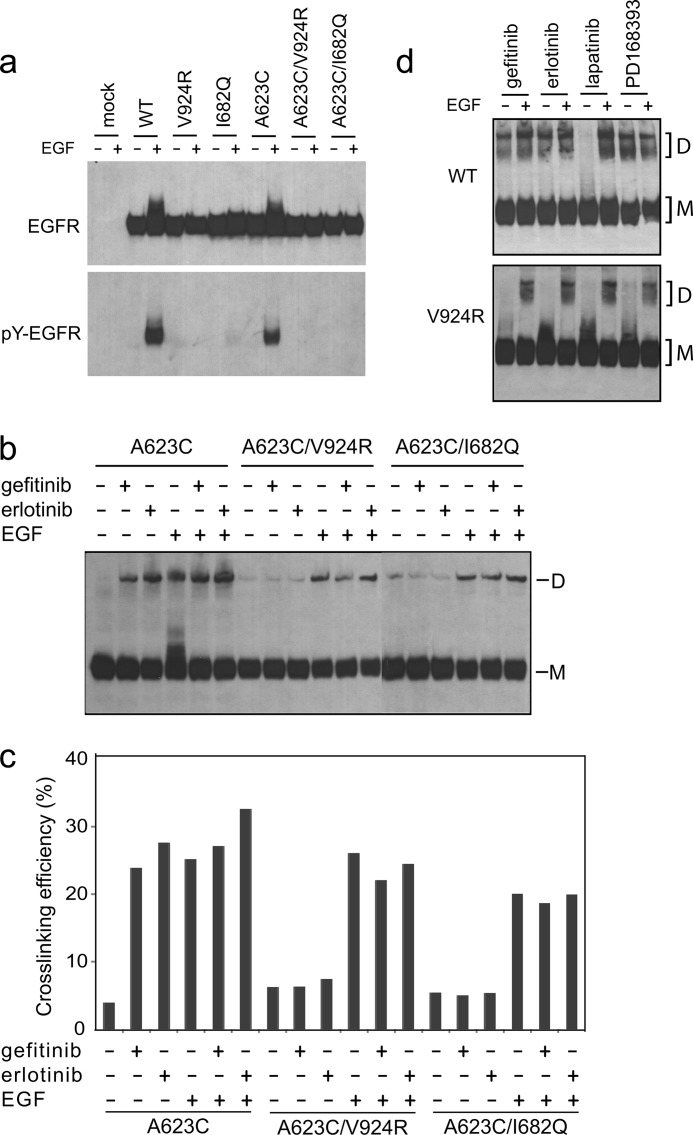
To test whether the specific asymmetric kinase domain dimerization interface identified in crystal structures is required for kinase inhibitor-induced dimerization, we introduced mutations known to disrupt this interface, V924R and I682Q (10). Consistent with previous reports in 293T cells (9), the V924R mutation in the kinase C-lobe and the I682Q mutation in the kinase N-lobe completely blocked EGF-stimulated EGFR autophosphorylation in Ba/F3 cells (Fig. 3a). As described above, the inhibitors gefitinib and erlotinib that stabilize the active kinase conformation each induced cross-linking of the A623C mutant in the TM domain (Fig. 3, b and c). However, combination with the I682Q or V924R mutations blocked or greatly diminished inhibitor-induced dimerization (Fig. 3, b and c). In contrast, the I682Q and V924R mutations had no effect on EGF-induced dimerization (Fig. 3, b and c), showing that EGF-induced ectodomain dimerization is capable of driving TM domain association even in the absence of the asymmetric kinase domain interface.
FIGURE 3.
Disruption of the asymmetric kinase domain interface by mutations abolishes kinase inhibitor-induced EGFR dimerization. a, Ba/F3 transfectants expressing WT or mutant EGFR were treated with or without EGF. The lysates were subjected to reducing SDS 7.5% PAGE and Western blotting with protein C antibody (upper panel) or phosphotyrosine antibody 4G10 (lower panel). b, cells were incubated with 10 μm gefitinib or erlotinib and treated with or without EGF. The lysates were subjected to nonreducing SDS 5% PAGE and Western blotting with protein C antibody. c, quantitation of cross-linking in b. d, Ba/F3 transfectants expressing WT (upper panel) or mutant V924R (lower panel) were treated with kinase inhibitors with or without EGF as in b. Surface cross-linking was carried by incubation with 1 mm BS3 for 1 h at 4 °C. The lysates were subjected to reducing SDS 5% PAGE and Western blotting with protein C antibody. D, dimeric; M, monomeric.
Cell surface chemical cross-linking further demonstrated the requirement of the asymmetric kinase domain interface for kinase inhibitor-mediated dimerization. Ba/F3 cells expressing wild-type EGFR or mutant V924R were incubated with kinase inhibitors, treated without or with EGF, and subjected to cross-linking using BS3 (Fig. 3d). Gefitinib, erlotinib, PD168393, and EGF induced dimers that were cross-linked by BS3; in contrast, lapatinib did not (Fig. 3d). Furthermore, the V924R mutation in the asymmetric interface abolished chemical cross-linking stimulated by gefitinib, erlotinib, and PD168393 (Fig. 3d). These results with BS3 are in complete agreement with the results of disulfide cross-linking in domain IV, the ectodomain linker, and TM domain.
To rule out the possibility that the V924R mutation might impair binding of inhibitors to the kinase domain, we measured inhibitor binding to wild-type and V924R mutant kinase domains (residues 672–998) using a fluorescence-quenching assay (20). The V924R mutation has no effect on the affinity of either gefitinib or erlotinib for the isolated kinase domain (Table 1 and supplemental Fig. S1). Furthermore, inhibitors were used at concentrations (10 μm for transfectants and 200 μm for purified receptors) more than 100-fold above their KD values.
TABLE 1.
Inhibitor binding to EGFR WT and mutant kinase domains
| Kinase domain |
KD |
|
|---|---|---|
| Gefitinib | Erlotinib | |
| nm | ||
| WT | 29.9 ± 3.0 | 99.8 ± 35 |
| V924R | 26.6 ± 0.3 | 80.6 ± 7.6 |
Biochemical Characterization of Kinase Inhibitor-induced EGFR Dimerization
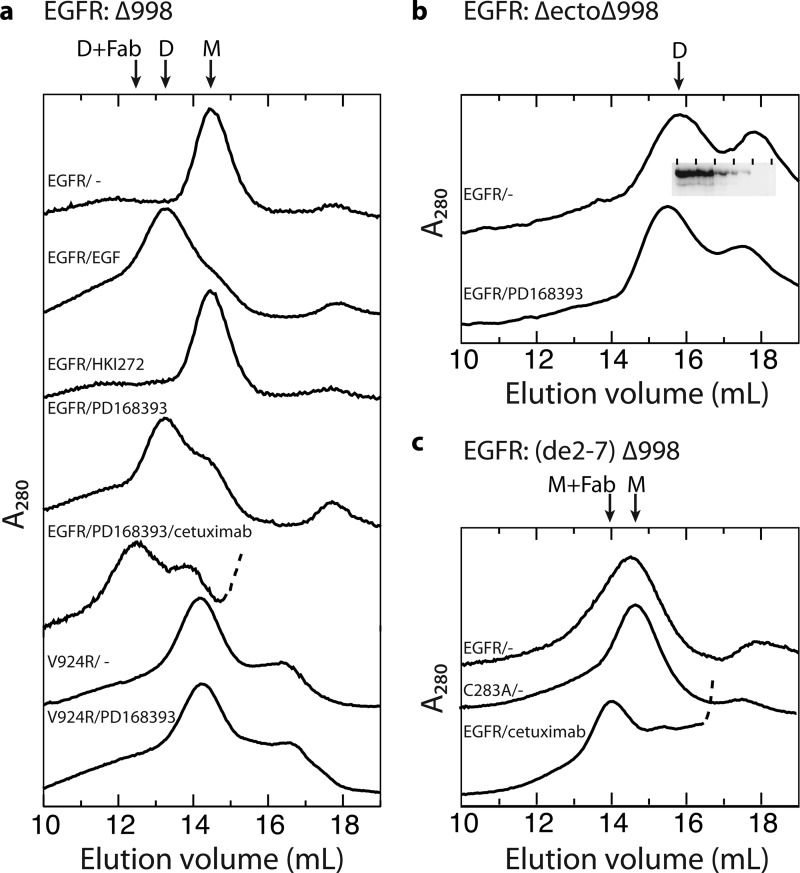
EGFR dimerization was further assessed in vitro, by treating purified wild-type or V924R receptor with EGF or inhibitors, followed by gel filtration. In the presence of dodecylmaltoside detergent, the EGFR Δ998 mutant that lacks the 999–1186 autophosphorylation tail elutes as monomers or dimers in absence and presence of EGF, respectively (Fig. 4a), as previously reported for full-length and Δ998 mutant EGFR (7, 24). After treatment with the covalent inhibitor HKI272 to stabilize the inactive kinase domain conformation, EGFR Δ998 remained monomeric (Fig. 4a). In contrast, after treatment with the covalent inhibitor PD168393 to stabilize the active kinase domain conformation, EGFR eluted largely as a dimer (Fig. 4a). However, PD168393 had no effect on the elution profile of the V924R EGFR mutant, which remained monomeric (Fig. 4a). These results show that both in cells and in vitro, kinase inhibitors that stabilize the active kinase domain conformation, but not those that stabilize the inactive conformation, promote EGFR dimerization through the kinase domain and that this dimerization requires the asymmetric N-lobe/C-lobe kinase domain interface.
FIGURE 4.
Gel filtration chromatograms of detergent-solubilized, purified EGFR proteins. The purified EGFR truncation mutants indicated in panels a–c, with any additional mutations shown above each trace (0.7 μm), were treated with or without kinase inhibitors (200 μm), EGF (20 μm), or cetuximab Fab (40 μm) for 5 min on ice for 5 min and subjected to gel filtration on a Superose 6 HR column equilibrated with running buffer containing 0.2 mm dodecylmaltoside and 0.2 mm DTT. The inset in b shows Western blots with protein C antibody of fractions from the upper trace, demonstrating that EGFR is present only in the dimer peak, and not in the second peak. The arrows mark positions of dimeric (D), monomeric (M), and Fab complex material (Fab). Traces of Fab complexes are truncated so that only the beginning, rising portion of the Fab peak is shown, which is dashed.
Formation of an Asymmetric Kinase Dimer with Ectodomain Conformation Distinct from EGF-liganded EGFR
Representative mutant or drug-treated EGFR Δ998 dimer or monomer gel filtration peak fractions, shown in Fig. 4, were subjected to negative-stain EM single-particle analysis. Particles were picked and class-averaged using multireference K-means classification. The numbers of particles, representative fields, and all class averages are shown in supplemental Fig. S2–S6. The studies were carried out exactly as described previously on EGFR Δ998 (7), and class averages from that study are shown for comparison in Fig. 5 for monomeric EGFR (Fig. 5a) and the dimeric EGFR complex with EGF (Fig. 5b).
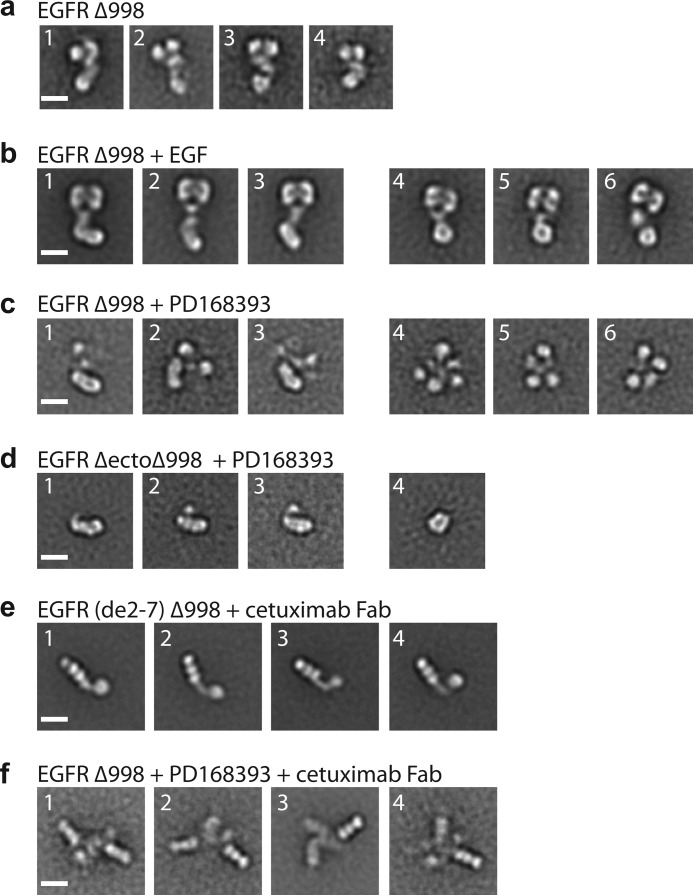
FIGURE 5.
Kinase inhibitor promotes formation of the asymmetric kinase dimer with ectodomain conformation distinct from liganded dimer. a, representative class averages of unliganded, monomeric EGFR Δ998. b, representative class averages of EGF-bound, dimeric EGFR Δ998, in which symmetric ectodomain dimers are linked to asymmetric kinase dimers (subpanels 1–3) or symmetric kinase dimers (subpanels 4–6). c, representative class averages of PD168393-bound EGFR Δ998 with asymmetric kinase dimer (subpanels 1–3) or less interpretable densities (subpanels 4–6). d, representative class averages of PD168393-bound EGFR ΔectoΔ998 with asymmetric kinase dimer (subpanels 1–3) or symmetric kinase dimer (subpanel 4). e, representative class averages of EGFR(de2–7)Δ998 in complex with cetuximab Fab. f, representative class averages of EGFR Δ998 with PD168393 in complex with cetuximab Fab. Scale bars, 10 nm.
In contrast to EGF-induced EGFR Δ998 dimers, PD168393-induced EGFR Δ998 dimers were substantially more heterogeneous in appearance (Fig. 5c). Not one of the 50 class averages showed the heart-shaped density characteristic of the liganded dimeric ectodomain (compare Fig. 5c and supplemental Fig. S3 with Fig. 5b and Ref. 7). However, the EGFR Δ998 + PD168393 particles shared enough characteristics to produce class averages with distinct features; furthermore, most class averages fell into one of two overall groups (Fig. 5c). One group of class averages contains an elongated, rod-shaped density (Fig. 5c, averages 1–3). This elongated, rod-shaped density is linked to two or more smaller, isolated densities. A second group of class averages contain four to five isolated densities tied together at the center (Fig. 5c, averages 4–6).
The elongated, rod-shaped density in Fig. 5c (averages 1–3) resembles the asymmetric kinase dimer found in ligand-bound EGF receptors (Fig. 5b, averages 1–3, and Ref. 7). Indeed, the masked rod-like densities cross-correlated well with the projections of the asymmetric kinase dimer crystal structure (Fig. 6a). By elimination, the smaller densities must correspond to ectodomain modules associated with the asymmetric kinase domain. However, because of variation in relative positions between the ectodomains and the kinase domains and between two ectodomain monomers or within the ectodomain monomers themselves, no assignment of ectodomain density could be made. The density for the ectodomain region was frequently weak in these class averages, another symptom of differing relative positions, which result in averaging out of domains. However, the class averages definitely showed that the ectodomain in the EGFR Δ998 +PD168393 preparations differed in conformation from that in EGF-liganded EGFR Δ998. In the second group of class averages (Fig. 5c, averages 4–6), it was not possible to distinguish between ectodomain and kinase domain densities.
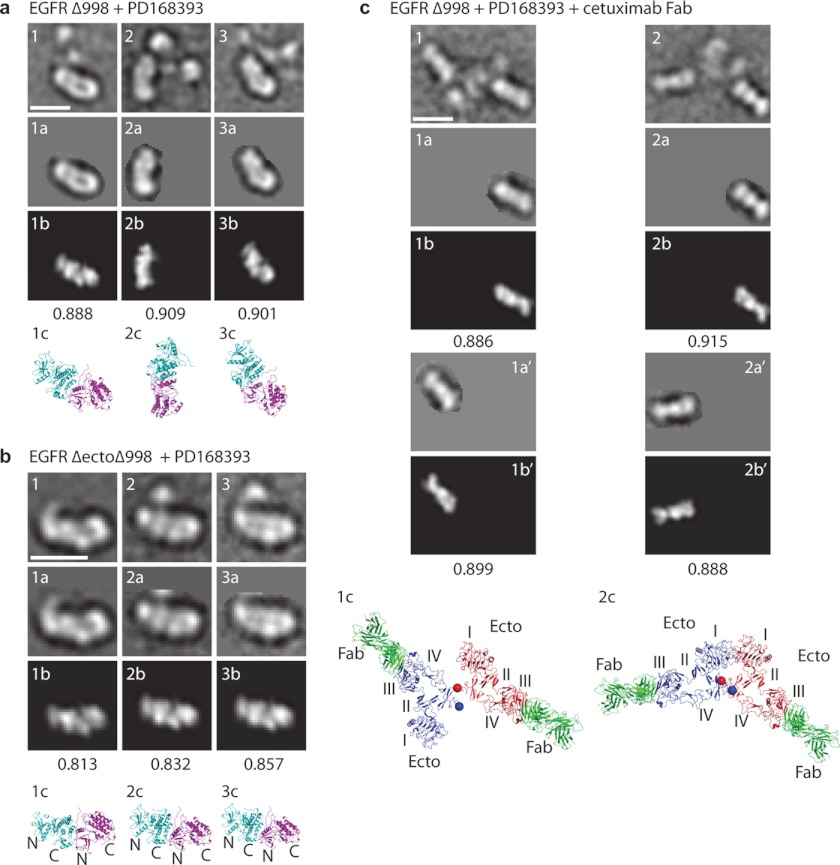
FIGURE 6.
Correlation of EM densities with crystal structures. Representative class averages are shown in the first row of each panel, with masked areas in the second row (labeled a), best correlating projections with crystal structures and cross-correlation scores in the third row (labeled b), and ribbon diagrams in the same orientation but at larger scale in the bottom row (labeled c). The cross-correlations in a and b are with the asymmetric kinase dimer from (10) (Protein Data Bank code 2GS6). Cross-correlations in c are with the Fab and EGFR domain III moieties (residues 311–503) from the crystal structure of cetuximab Fab bound to EGFR (25). In c, each monomeric unit was separately masked and cross-correlated. Using the orientation established for the Fab and domain III, the entire EGFR ectodomain Fab complex for each monomer is shown, preserving their relative spatial orientations, in the bottom panel of c. Scale bars, 10 nm.
To obtain more homogenous and better-resolved averages of the kinase moiety after treatment with PD168393, we used an EGFR construct lacking the ectodomain and the C-terminal autophosphorylation tail (EGFR ΔectoΔ998). This construct dimerized even in the absence of PD168393, and PD168393 addition caused earlier elution in gel filtration (Fig. 4b). EGFR ΔectoΔ998 in presence of PD168393 showed an elongated rod-shaped density composed of four smaller, linearly arranged globular densities (Fig. 5d, averages 1–3, and supplemental Fig. S3). These rod-shaped densities cross-correlated well with crystal structure projections of the asymmetrically associated kinase domain dimer (Fig. 6b). The four densities corresponded to the linear N-lobe:C-lobe/N-lobe:C-lobe arrangement in asymmetric dimers, with the larger globular densities corresponding to the larger C-lobes. A smaller density appeared to correspond to the TM/juxtamembrane region and was asymmetrically joined to the kinase dimer (Fig. 5d, averages 1–3), similarly to the asymmetric attachment of the EGFR/EGF ectodomain complex to the asymmetric kinase dimer (Fig. 5b, averages 1–3). In a few EGFR ΔectoΔ998 + PD168393 class averages, two kinases formed a ring-shaped density (Fig. 5d, panel 4), similar to the symmetric kinase dimer found in EGF-bound receptors (Fig. 5b, panels 4–6). In contrast, class averages of EGFR ΔectoΔ998 in absence of PD168393, although dimeric, are markedly more heterogeneous (supplemental Fig. S4).
An EGFR mutant with exons 2–7 deleted (de2-7, also known as vIII) is commonly overexpressed in glioblastoma (Ref. 17 and references therein). EGFR (de2-7) retains Cys-238, the disulfide-forming partner of which is removed in the de2-7 deletion. EGFR C283A (de2-7) Δ998 eluted as a monomer in gel filtration, whereas EGFR (de2-7) Δ998 eluted as a broader peak, suggesting the presence of some dimer (Fig. 4c). Complexes with cetuximab were isolated by gel filtration (Fig. 4c). Cetuximab binds to domain III of the EGFR ectodomain, as shown with a crystal structure of its Fab complexed with the ectodomain (25). The cetuximab Fab complex with EGFR (de2-7) Δ998 showed three linearly arranged densities corresponding to cetuximab Fab CH1 + CL, VH + VL, and EGFR domain III (Fig. 5e and supplemental Fig. S5). In addition, the monomeric complexes showed one or two densities corresponding to domain IV, the TM and juxtamembrane region, and the kinase domain (Fig. 5e).
To determine the orientation between ectodomain monomers in PD168393-induced EGFR Δ998 dimers, we prepared complexes with cetuximab Fab (Fig. 4a). Class averages showed two Fab fragments, each bound to domain III (Fig. 5f and supplemental Fig. S6). As seen in EGFR (de2-7) Δ998 monomers, each monomer in PD168393-induced EGFR Δ998 dimers contained three globular densities corresponding to EGFR domain III, bound to cetuximab VH + VL and CH1 + CL. These three linearly arranged units in each monomer were located distally in dimers. Density was often poorer in the central region of dimers, which may result from the collapse of the kinase dimer and ectodomain monomers in different orientations on top of one another or flexibility of domains I and II relative to domain III. The portion of the crystal structure corresponding to cetuximab Fab bound to domain III was separately cross-correlated with each masked monomer in the dimer class averages (Fig. 6c). Correlations were excellent. Maintaining the spatial relationships between the two monomers, the two-dimensional distances between the centers of two domain III modules in PD168393-EGFR dimers were 118 ± 25 Å (means ± S.D., n = 3). This is larger than the distances between domain III modules in EGF-EGFR dimers in EM (taken between ventricle-like densities in heart-shaped dimers) of 77 ± 7 Å, n = 26 measured from the class averages in Ref. 7 or in crystal structures of 70 Å (9).
The tethered (monomeric) structure of the EGFR ectodomain is little affected by cetuximab, which occludes the EGF-binding site on domain III (25). Using our domain III-Fab cross-correlations, we added back the remainder of the tethered EGFR monomer conformation (Fig. 6c, lower panel). Plausible models resulted for the ectodomain portions of EGFR-PD168393 dimers. The C termini of the domain IV modules in each monomer (Thr-614) were in close proximity, within 20–30 Å in 3D (Fig. 6c, panels 1c and 2c, spheres). This close proximity supports a model in which the EGFR TM domains are dimerized following PD168393-induced dimerization of the kinase domains. These results demonstrate that although inhibitors that stabilize the active kinase domain conformation promote formation of the asymmetric kinase domain dimer, they do not promote an EGF-complexed conformation of the ectodomain, and instead the ectodomain conformation is consistent with the presence of two closely associated ectodomain monomers, either in tethered or untethered conformations.
DISCUSSION
Communication between the EGFR extracellular and intracellular domains is known to be complex (7, 9, 26, 27). Ligand binding to the ectodomain induces receptor dimerization and kinase activation (28). However, quinazoline inhibitors of the kinase domain can also induce EGFR dimerization, and mutations in the cytoplasmic portion of EGFR can affect the monomer-dimer equilibrium and the affinity for EGF (2, 16, 17, 26, 27). We have shown selective induction of receptor dimerization by inhibitors that stabilize the active kinase conformation and demonstrated that receptors dimerized through the kinase domain differ from EGF-dimerized receptors in the structure of their ectodomain.
Previous work has shown that quinazoline class EGFR tyrosine kinase antagonists could induce dimerization of a subset of EGFR receptors as shown by cross-linking with the cell-impermeable reagent BS3 (2, 16, 17). However, we demonstrate that not all quinazoline class antagonists have this effect, because gefitinib, erlotinib, and PD168393, which induce dimerization, as well as lapatinib that does not induce dimerization, are quinazolines (29). Furthermore, our results suggest that inhibitor-induced EGFR dimerization is dependent on the conformational state of the kinase that is stabilized by the inhibitor. Gefitinib, erlotinib, and PD168393 are inhibitors that bind to the ATP-binding pocket and stabilize the kinase in active conformation (18, 20, 21). On the other hand, lapatinib and HKI-272 are inhibitors that stabilize the kinase in inactive conformation by pushing the kinase C-helix out of its active state (19, 30). We found that only inhibitors that stabilize the kinase in active conformation promoted ligand-independent EGFR dimerization on the cell surface, as detected by disulfide-cross-linking or chemical cross-linking. Moreover, gel filtration of purified, nearly full-length, detergent-solubilized receptors demonstrated that gefitinib and PD168393, but not lapatinib and HKI-272, induced EGFR dimerization.
We further showed that the asymmetric kinase interface is required for kinase inhibitor-induced receptor dimerization. Mutations on opposite sides of this interface, I682Q in the N-lobe and V924R in the C-lobe, each abolished dimerization in the EGFR TM domain induced by gefitinib and erlotinib. Affinity measurements on the isolated kinase domain ruled out the possibility that V924R mutation impairs the binding of inhibitors to the kinase domain. Moreover, the V924R mutation abolished the ability of PD168393 to induce noncovalent dimerization of detergent-solubilized EGFR as shown by gel filtration.
We previously demonstrated that EGF was sufficient to dimerize the same detergent-solubilized, nearly full-length receptor preparation as used here (7). The vast majority of such dimeric particles had the same heart-shaped ligand-bound ectodomain as seen in crystal structures, and ∼40% of these particles had asymmetrically associated kinase domains. PD168393 and gefitinib slightly enhanced the proportion of asymmetric kinase dimers to 50–60%, whereas lapatinib and HKI-272 abolished them (7). Here, we have extended these results by demonstrating that even in the absence of EGF, PD168393 could induce formation of asymmetric kinase dimers; however, there was no evidence for the heart-shaped, ligand-bound-like conformation of the ectodomain.
We also examined an analogous construct lacking the ectodomain, EGFR ΔectoΔ998. The detergent-solubilized EGFR ΔectoΔ998 preparation spontaneously dimerized in gel filtration. In the presence of PD168393, asymmetric kinase dimers were clearly evident. The absence of the ectodomain enabled higher weighting of the kinase domains in class averages. Enough detail could be seen in the kinase dimer to resolve the N- and C-lobes of the monomers and confirm the N-lobe/C-lobe asymmetric interface. Lack of formation of noncovalent dimers by EGFR Δ998 and EGFR (de2-7) Δ998 and formation of noncovalent dimers by EGFR ΔectoΔ998 preparations are consistent with a role for the ectodomain in sterically interfering with dimer formation. Kinase inhibitor-induced EGFR dimers and EGF-induced EGFR dimers appeared to associate identically in their TM domains. Cross-linking of cysteines introduced into the TM domain gave identical patterns of two peaks near the extracellular boundary. Furthermore, disulfide cross-linking in domain IV occurred almost as efficiently with PD168393 as with EGF, consistent with close proximity of domain IV in kinase inhibitor-induced dimers.
However, the structure of the EGFR ectodomain clearly differs in kinase inhibitor- and EGF-induced dimers. Disulfide cross-linking in domain II was induced to markedly lower levels by kinase inhibitors than by EGF. Furthermore, EM showed a complete lack of the characteristic EGF-induced heart-shaped ectodomain dimer in kinase inhibitor-induced dimers and a lack of a well defined orientation between the two ectodomain monomers. There also was no evidence for a second type of heart-shaped but less symmetric ectodomain dimer, which is a model for understanding negative cooperativity and may have either one or two bound ligands (31).
Complexes with cetuximab Fab enabled the location and orientation of EGFR domain III, to which cetuximab binds, to be defined within kinase inhibitor-induced dimers. Using the orientation between domains III and IV known from crystal structures of tethered EGFR ectodomain monomers and liganded EGFR dimers, the two monomers in inhibitor-induced dimers were found by cross-correlation between EM and crystal structures to be in close proximity near the C termini of domain IV. This finding agreed with findings from disulfide cross-linking of close proximity at domain IV and at the TM domain in dimers, but not at domain II.
The orientation between the two ectodomain monomers in inhibitor-induced dimers was variable, consistent with the lack of any well defined ectodomain dimer in kinase-induced dimers either in the presence or absence of cetuximab Fab. The kinase inhibitor-induced dimers in presence of cetuximab Fab did not show good density for EGFR domains I, II, and IV. This may in part be due to the collapse of the ectodomain and kinase domains on top of one another and the small sizes of domains II and IV. Another interesting source of heterogeneity is suggested by the partial exposure of an antibody epitope in domain II in kinase-induced dimers (17). The 806 epitope is exposed in EGFR (de2-7), but not in tethered, monomeric EGFR or liganded, dimeric EGFR (Ref. 17 and references therein). A crystal structure of 806 Fab bound to its target disulfide-linked module within domain II demonstrates that its epitope is buried in a module-module interface within domain II in tethered, monomeric EGFR and liganded, dimeric EGFR (32). Taken together, our EM results and the 806 Fab results suggest that when two ectodomain monomers are brought into close proximity by TM domain association in kinase inhibitor-induced dimers, the tethered ectodomain conformation is destabilized, and domain II becomes more flexible.
Our results have important implications for the relative energetics of EGFR ectodomain and cytoplasmic dimerization. Formation of the asymmetric kinase domain dimer is sufficient to drive dimerization of the TM domain and ectodomain linker, but not to form the heart-shaped, ectodomain dimer. In contrast, ligand binding not only drives ectodomain dimerization but also formation of the asymmetric kinase domain dimer. Therefore, ectodomain dimerization is more energetically costly than kinase domain dimerization. These results are consistent with the effects of kinase domain mutations on rate of dimer formation (26) and affinity for EGF (27). Conversely, the latter studies demonstrating an effect of the kinase domain on EGFR properties in the absence of kinase inhibitors suggest that our studies with kinase inhibitors are also relevant to the physiology of EGFR dimerization through the asymmetric interface in the absence of inhibitors. The relatively high energy of ectodomain dimerization compared with kinase domain association through the asymmetric interface is also consistent with evidence for negative cooperativity in EGF binding to EGFR and lateral propagation of signaling through EGFR (27).
Previously, a specific transmembrane dimer interface was found not to be required for ligand-induced kinase activation in cells (9). Our studies provide further support for the concept that the conformations of the EGFR extra- and intracellular domains are only loosely coupled through the membrane. Not only can one ligand-bound ectodomain dimer conformation couple to multiple kinase domain arrangements (7), but as shown here the asymmetric kinase dimer can also couple to two different ectodomain states. Our results have important implications for therapeutics directed to both the ectodomain and kinase domain of EGFR.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grant HL-103526. This work was also supported by the Howard Hughes Medical Institute (to T. W.).

This article contains supplemental text and Figs. S1–S6.
- EGFR
- EGF receptor
- TM
- transmembrane
- BS3
- bis(sulfosuccinimidyl) suberate.
REFERENCES
- 1. Hynes N. E., Lane H. A. (2005) ERBB receptors and cancer. The complexity of targeted inhibitors. Nat. Rev. Cancer 5, 341–354 [DOI] [PubMed] [Google Scholar]
- 2. Arteaga C. L. (2003) ErbB-targeted therapeutic approaches in human cancer. Exp. Cell Res. 284, 122–130 [DOI] [PubMed] [Google Scholar]
- 3. Ferguson K. M., Berger M. B., Mendrola J. M., Cho H. S., Leahy D. J., Lemmon M. A. (2003) EGF activates its receptor by removing interactions that autoinhibit ectodomain dimerization. Mol. Cell 11, 507–517 [DOI] [PubMed] [Google Scholar]
- 4. Ogiso H., Ishitani R., Nureki O., Fukai S., Yamanaka M., Kim J. H., Saito K., Sakamoto A., Inoue M., Shirouzu M., Yokoyama S. (2002) Crystal structure of the complex of human epidermal growth factor and receptor extracellular domains. Cell 110, 775–787 [DOI] [PubMed] [Google Scholar]
- 5. Garrett T. P., McKern N. M., Lou M., Elleman T. C., Adams T. E., Lovrecz G. O., Zhu H. J., Walker F., Frenkel M. J., Hoyne P. A., Jorissen R. N., Nice E. C., Burgess A. W., Ward C. W. (2002) Crystal structure of a truncated epidermal growth factor receptor extracellular domain bound to transforming growth factor α. Cell 110, 763–773 [DOI] [PubMed] [Google Scholar]
- 6. Cho H. S., Leahy D. J. (2002) Structure of the extracellular region of HER3 reveals an interdomain tether. Science 297, 1330–1333 [DOI] [PubMed] [Google Scholar]
- 7. Mi L. Z., Lu C., Li Z., Nishida N., Walz T., Springer T. A. (2011) Simultaneous visualization of the extracellular and cytoplasmic domains of the epidermal growth factor receptor. Nat. Struct. Mol. Biol. 18, 984–989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Jura N., Endres N. F., Engel K., Deindl S., Das R., Lamers M. H., Wemmer D. E., Zhang X., Kuriyan J. (2009) Mechanism for activation of the EGF receptor catalytic domain by the juxtamembrane segment. Cell 137, 1293–1307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lu C., Mi L. Z., Grey M. J., Zhu J., Graef E., Yokoyama S., Springer T. A. (2010) Structural evidence for loose linkage between ligand binding and kinase activation in the epidermal growth factor receptor. Mol. Cell Biol. 30, 5432–5443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhang X., Gureasko J., Shen K., Cole P. A., Kuriyan J. (2006) An allosteric mechanism for activation of the kinase domain of epidermal growth factor receptor. Cell 125, 1137–1149 [DOI] [PubMed] [Google Scholar]
- 11. Dawson J. P., Berger M. B., Lin C. C., Schlessinger J., Lemmon M. A., Ferguson K. M. (2005) Epidermal growth factor receptor dimerization and activation require ligand-induced conformational changes in the dimer interface. Mol. Cell Biol. 25, 7734–7742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Red Brewer M., Choi S. H., Alvarado D., Moravcevic K., Pozzi A., Lemmon M. A., Carpenter G. (2009) The juxtamembrane region of the EGF receptor functions as an activation domain. Mol. Cell 34, 641–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yu X., Sharma K. D., Takahashi T., Iwamoto R., Mekada E. (2002) Ligand-independent dimer formation of epidermal growth factor receptor (EGFR) is a step separable from ligand-induced EGFR signaling. Mol. Biol. Cell 13, 2547–2557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chantry A. (1995) The kinase domain and membrane localization determine intracellular interactions between epidermal growth factor receptors. J. Biol. Chem. 270, 3068–3073 [PubMed] [Google Scholar]
- 15. Arteaga C. L., Ramsey T. T., Shawver L. K., Guyer C. A. (1997) Unliganded epidermal growth factor receptor dimerization induced by direct interaction of quinazolines with the ATP binding site. J. Biol. Chem. 272, 23247–23254 [DOI] [PubMed] [Google Scholar]
- 16. Lichtner R. B., Menrad A., Sommer A., Klar U., Schneider M. R. (2001) Signaling-inactive epidermal growth factor receptor/ligand complexes in intact carcinoma cells by quinazoline tyrosine kinase inhibitors. Cancer Res. 61, 5790–5795 [PubMed] [Google Scholar]
- 17. Gan H. K., Walker F., Burgess A. W., Rigopoulos A., Scott A. M., Johns T. G. (2007) The epidermal growth factor receptor (EGFR) tyrosine kinase inhibitor AG1478 increases the formation of inactive untethered EGFR dimers. Implications for combination therapy with monoclonal antibody 806. J. Biol. Chem. 282, 2840–2850 [DOI] [PubMed] [Google Scholar]
- 18. Stamos J., Sliwkowski M. X., Eigenbrot C. (2002) Structure of the epidermal growth factor receptor kinase domain alone and in complex with a 4-anilinoquinazoline inhibitor. J. Biol. Chem. 277, 46265–46272 [DOI] [PubMed] [Google Scholar]
- 19. Wood E. R., Truesdale A. T., McDonald O. B., Yuan D., Hassell A., Dickerson S. H., Ellis B., Pennisi C., Horne E., Lackey K., Alligood K. J., Rusnak D. W., Gilmer T. M., Shewchuk L. (2004) A unique structure for epidermal growth factor receptor bound to GW572016 (Lapatinib). Relationships among protein conformation, inhibitor off-rate, and receptor activity in tumor cells. Cancer Res. 64, 6652–6659 [DOI] [PubMed] [Google Scholar]
- 20. Yun C. H., Boggon T. J., Li Y., Woo M. S., Greulich H., Meyerson M., Eck M. J. (2007) Structures of lung cancer-derived EGFR mutants and inhibitor complexes. Mechanism of activation and insights into differential inhibitor sensitivity. Cancer Cell 11, 217–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Blair J. A., Rauh D., Kung C., Yun C. H., Fan Q. W., Rode H., Zhang C., Eck M. J., Weiss W. A., Shokat K. M. (2007) Structure-guided development of affinity probes for tyrosine kinases using chemical genetics. Nat. Chem. Biol. 3, 229–238 [DOI] [PubMed] [Google Scholar]
- 22. Ludtke S. J., Baldwin P. R., Chiu W. (1999) EMAN. Semiautomated software for high-resolution single-particle reconstructions. J. Struct. Biol. 128, 82–97 [DOI] [PubMed] [Google Scholar]
- 23. Frank J., Radermacher M., Penczek P., Zhu J., Li Y., Ladjadj M., Leith A. (1996) SPIDER and WEB. Processing and visualization of images in 3D electron microscopy and related fields. J. Struct. Biol. 116, 190–199 [DOI] [PubMed] [Google Scholar]
- 24. Mi L. Z., Grey M. J., Nishida N., Walz T., Lu C., Springer T. A. (2008) Functional and structural stability of the epidermal growth factor receptor in detergent micelles and phospholipid nanodiscs. Biochemistry 47, 10314–10323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Li S., Schmitz K. R., Jeffrey P. D., Wiltzius J. J., Kussie P., Ferguson K. M. (2005) Structural basis for inhibition of the epidermal growth factor receptor by cetuximab. Cancer Cell 7, 301–311 [DOI] [PubMed] [Google Scholar]
- 26. Chung I., Akita R., Vandlen R., Toomre D., Schlessinger J., Mellman I. (2010) Spatial control of EGF receptor activation by reversible dimerization on living cells. Nature 464, 783–787 [DOI] [PubMed] [Google Scholar]
- 27. Macdonald J. L., Pike L. J. (2008) Heterogeneity in EGF-binding affinities arises from negative cooperativity in an aggregating system. Proc. Natl. Acad. Sci. U.S.A. 105, 112–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yarden Y., Schlessinger J. (1987) Self-phosphorylation of epidermal growth factor receptor. Evidence for a model of intermolecular allosteric activation. Biochemistry 26, 1434–1442 [DOI] [PubMed] [Google Scholar]
- 29. Eck M. J., Yun C. H. (2010) Structural and mechanistic underpinnings of the differential drug sensitivity of EGFR mutations in non-small cell lung cancer. Biochim. Biophys. Acta 1804, 559–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yun C. H., Mengwasser K. E., Toms A. V., Woo M. S., Greulich H., Wong K. K., Meyerson M., Eck M. J. (2008) The T790M mutation in EGFR kinase causes drug resistance by increasing the affinity for ATP. Proc. Natl. Acad. Sci. U.S.A. 105, 2070–2075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Alvarado D., Klein D. E., Lemmon M. A. (2009) ErbB2 resembles an autoinhibited invertebrate epidermal growth factor receptor. Nature 461, 287–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Garrett T. P., Burgess A. W., Gan H. K., Luwor R. B., Cartwright G., Walker F., Orchard S. G., Clayton A. H., Nice E. C., Rothacker J., Catimel B., Cavenee W. K., Old L. J., Stockert E., Ritter G., Adams T. E., Hoyne P. A., Wittrup D., Chao G., Cochran J. R., Luo C., Lou M., Huyton T., Xu Y., Fairlie W. D., Yao S., Scott A. M., Johns T. G. (2009) Antibodies specifically targeting a locally misfolded region of tumor associated EGFR. Proc. Natl. Acad. Sci. U.S.A. 106, 5082–5087 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.