Background: Self-renewal properties are attributed to critical amounts of the OCT4A transcription factor, and little is known about its post-translational regulation.
Results: OCT4A interacts with ERK1/2 and is phosphorylated at Ser-111, increasing its ubiquitination and degradation.
Discussion: These results suggest an increase in OCT4A degradation downstream of MEK1 activation and FGF2 treatment.
Significance: Controlling the mechanism by which cells balance self-renewal would advance our knowledge of stem cells.
Keywords: Embryonic Stem Cell, Protein Degradation, Signal Transduction, Stem Cells, Ubiquitination, OCT4A
Abstract
Embryonic stem cell self-renewal properties are attributed to critical amounts of OCT4A, but little is known about its post-translational regulation. Sequence analysis revealed that OCT4A contains five putative ERK1/2 phosphorylation sites. Consistent with the hypothesis that OCT4A is a putative ERK1/2 substrate, we demonstrate that OCT4A interacts with ERK1/2 by using both in vitro GST pulldown and in vivo co-immunoprecipitation assays. MS analysis identified phosphorylation of OCT4A at Ser-111. To investigate the possibility that ERK1/2 activation can enhance OCT4A degradation, we analyzed endogenous ubiquitination in cells transfected with FLAG-OCT4A alone or with constitutively active MEK1 (MEK1CA), and we observed that the extent of OCT4 ubiquitination was clearly increased when MEK1CA was coexpressed and that this increase was more evident after MG132 treatment. These results suggest an increase in OCT4A ubiquitination downstream of MEK1 activation, and this could account for the protein loss observed after FGF2 treatment and MEK1CA transfection. Understanding and controlling the mechanism by which stem cells balance self-renewal would substantially advance our knowledge of stem cells.
Introduction
The pluripotency and self-renewal of embryonic stem (ES)3 cells are maintained by several key transcription factors (1). OCT4 (also called OCT3, encoded by Pou5f1) is a member of the POU (Pit/OCT/Unc) family of transcription factors and is expressed in pluripotent cells, including early embryos, ES cells, embryonal carcinoma cells, and embryonic germ cells (2–5).
Recent analysis of the human ES cell phosphoproteome indicates that OCT4 is phosphorylated (6), but the functional consequences of these modifications have not been determined. In addition, limited data are available on the mechanisms regulating the nuclear entry of OCT4 and how its nuclear localization is linked to signaling pathways.
In this study, we demonstrate for the first time that the serine/threonine kinase ERK interacts with and phosphorylates OCT4. Moreover, we evaluated the mechanism by which ERK controls OCT4 stability as well as OCT4 subcellular localization, identifying Ser-111 as a novel important target for such control. Two isoforms of OCT4 have been identified, OCT4A and OCT4B (7), but only isoform A seems to be related to stemness, so our work was focused on OCT4A.
The function and regulation of OCT4 expression have been intensively studied in mice. Knocking out the Oct4 gene in mice results in death due to the lack of inner cell mass formation, suggesting that the gene is essential for establishment of pluripotent cell lineages in early mammalian development (8). In more recent works surrounding somatic cell reprogramming, OCT4 was shown to be one of four necessary factors for the derivation of induced pluripotent stem (iPS) cells (9). Subsequent studies showed that substitutions could be made for some or all of the other three factors (SOX2, KLF4, and c-Myc) (10), but the constant requirement of OCT4 reactivation and the known need for regulated OCT4 expression in the embryo and ES cells suggest a central role for OCT4 in the maintenance of pluripotency. Increasing data indicate that the activity of iPS factors is regulated through post-translational modifications. For example, phosphorylation of human SOX2 at Ser-249, Ser-250, and Ser-251 results in the inhibition of SOX2 DNA-binding activity (11), whereas acetylation of mouse SOX2 at Lys-75 by p300/CBP (cAMP-responsive element-binding protein-binding protein) family proteins enhances nuclear export and degradation of SOX2 through a ubiquitin-mediated degradation pathway (12). Additionally, phosphorylation of human OCT4 at Ser-229 by protein kinase A might partially regulate OCT4 transactivation activity (13). OCT4 is also a target for SUMO1 (small ubiquitin-related modifier 1) modification that occurs at a single lysine, Lys-118, located at the end of the N-terminal transactivation domain and next to the DNA-binding domain (14). These findings indicate that post-translational modifications of iPS factors are probably involved in the regulation of their activity, which could result in modulation of ES cell self-renewal activity.
It has been shown that a critical amount of OCT4 is required to maintain self-renewal of ES cells (15), but the mechanisms that regulate OCT4 expression and maintain it within a certain range are not well understood. The ubiquitin-proteasome pathway plays a crucial role in degradation of the proteins involved in many cellular processes (16–19), so this is an important mechanism that may be involved in OCT4A level regulation.
EXPERIMENTAL PROCEDURES
Reagents and Plasmids
The MEK1 inhibitor PD98059 was purchased from Cell Signaling. Cycloheximide (CHX), MG132, and leptomycin B were purchased from Sigma. FGF2 was purchased from PeproTech.
To construct human OCT4A, total RNA was prepared from NTera2 cells, and cDNA was synthesized using the SuperScriptTM first-strand synthesis system (Invitrogen). Full-length human OCT4A cDNA was subcloned into the pCMV-FLAG vector (Sigma) to create the FLAG-tagged human OCT4A construct and fused in-frame with the GST protein into pGEX4T1 (Sigma) to create the pGEX4T1-human OCT4A construct. pUSE-constitutively active MEK1 (MEK1CA) was from Upstate Biotechnology. To generate the OCT4A S111A and S111D mutants, the QuikChange II site-directed mutagenesis kit (Stratagene) was used according to the manufacturer's instructions.
Immunoprecipitation and Immunoblot Analysis
Cells were washed with ice-cold phosphate-buffered saline and lysed in radioimmune precipitation assay lysis buffer (20 mm Tris-HCl (pH 7.5) 150 mm NaCl, 1 mm Na2EDTA, 1 mm EGTA, 1% Nonidet P-40, 1% sodium deoxycholate, 2.5 mm sodium pyrophosphate, 1 mm β-glycerophosphate, 1 mm Na3VO4, and 1 μg/ml leupeptin) supplemented with a mixture of protease inhibitors and phosphatase inhibitors (Sigma). The NE-PER kit (Pierce) was used to perform cellular fractionation according to the manufacturer's instructions. Total cell lysates containing 1 mg of protein or nuclear and cytoplasmic fractions were immunoprecipitated with immobilized FLAG no agarose beads (Sigma). The agarose beads were washed, and bead-bound proteins were separated by 10% SDS-PAGE. Gel proteins were transferred onto nitrocellulose membranes (Millipore). After blocking with 5% BSA in TBS containing 0.1% Tween 20, the blots were probed with the following antibodies: anti-OCT4A and anti-poly(ADP-ribose) polymerase (Santa Cruz Biotechnology); anti-ERK and anti-phospho-ERK (Cell Signaling); anti-Nanog, anti-SOX2, anti-REX1, anti-BMP4, anti-DLX5, and anti-GATA6 (Abcam); anti-phosphoserine, anti-α-tubulin, and anti-FLAG (Sigma); and anti-ubiquitin (Covance).
GST Pulldown Assays
For GST pulldown assays, GST-OCT4A protein (10 μg) was incubated overnight at 4 °C with 5 mg of NTera2 cell extract, and the GST-tagged proteins were recovered by incubating the reaction at 4 °C for 3 h with 20 μl of glutathione-Sepharose 4B beads. The bead pellet was washed three times with 1 ml of buffer containing 20 mm Tris-HCl (pH 7.5), 150 mm NaCl, 10% glycerol, 1% Triton X-100, and 2 mm EDTA. Boiled samples were then subjected to 10% SDS-PAGE.
Real-time PCR
Total RNA was extracted from NTera2 cells using TRIzol. Quantitative PCR was conducted using SYBR Green on a 96-well plate using LightCycler 480 (Roche Applied Bioscience) after DNase I treatment. The total volume (20 μl) of each PCR contained SYBR Green PCR Master Mix, 10 ng of cDNA, and 0.4 μm each of the forward and reverse primers. Real-time PCR (n = 4) was performed using the following primer sequences: Nanog, 5′-ATGCCTCACACGGAGACTGT and 5′-AGGGCTGTCCTGAATAAGCA (66 bp, 55 °C, NM_024865); SOX2, 5′-ATGGGTTCGGTGGTCAAGT and 5′-CCTGTGGTTACCTCTTCCTCC (60 bp, 56 °C, NM_003106); REX1, 5′-TCTGAGTACATGACAGGCAAGAA and 5′-TCTGATAGGTCAATGCCAGGT (62 bp, 45 °C, NM_174900); BMP4, 5′-TCCACAGCACTGGTCTTGAG and 5′-GGGATGTTCTCCAGATGTTCTT (94 bp, 54 °C, NM_130851.2); DLX5, 5′-CTACAACCGCGTCCCAAG and 5′-GCCATTCACCATTCTCACCT (76 bp, 55 °C, NM_005221.5); GATA6, 5′-AATACTTCCCCCACAACACAA and 5′-CTCTCCCGCACCAGTCAT (69 bp, 55 °C, NM_005257.3); and RNA polymerase II 5′-CAAGTTCAACCAAGCCATTG and 5′-GTGGCAGGTTCTCCAAGG (78 bp, 56 °C, NM_000937.4). The transcript amount of each gene was normalized with RNA polymerase II. The relative -fold change in expression was calculated using the ΔΔCT method (CT values of <30) with respect to untreated NTera2 cells.
RNA Interference
We used SignalSilence® pool p44/42 (ERK1/2) MAPK siRNA (Cell Signaling Technology) to specifically inhibit p44/42 MAPK expression by RNA interference, a method whereby gene expression can be selectively silenced through the delivery of double-stranded RNA molecules to the cell. OCT4A endogenous expression was blocked using a construct that was previously shown to specifically knock down OCT4A mRNA (20).
Mass Spectrometry Analysis
Lysates of HeLa cells transfected with OCT4A and MEK1CA were subjected to immunoprecipitation with FLAG-conjugated agarose beads and analyzed by MS for phosphopeptide identification. The sample was concentrated to half the volume of the original sample (SpeedVac), reduced with DTT, and alkylated with iodoacetamide. A small amount of the sample was digested with trypsin for protein identification, and the remaining sample was digested with Glu-C for phosphopeptide analysis.
The trypsin-digested sample was desalted and purified using C18 ZipTips (Millipore), and the eluate was added onto an AnchorChip target for analysis on a Bruker Autoflex III MALDI TOF/TOF instrument. The peptide mixture was analyzed in positive reflector mode for accurate peptide mass determination. MALDI-MS/MS was performed, and peptides were selected for partial peptide sequencing. The MS and MS/MS spectra were combined and used for database searching using Mascot software.
Phosphopeptide enrichment was performed by TiO2 purification. The eluate from the TiO2 column was purified on a Poros R3 microcolumn and eluted for detection by MS. The peptide mixture was analyzed in positive reflector mode for accurate peptide mass determination. Peptides matching the OCT4A sequence were selected for partial peptide sequencing (MS/MS analysis). The MS and MS/MS spectra were combined and used for database searching using Mascot software.
Calf Intestinal Phosphatase (CIP) Treatment
The cell lysate was harvested, followed by incubation with 1 μl of 10 units/μl CIP (Sigma) in 50 μl of CIP buffer for 1 h at 37 °C. Reactions were stopped by two washes in CIP buffer and boiling in gel loading buffer.
Indirect Immunofluorescence Microscopy
Cells grown on coverslips were fixed in 4% paraformaldehyde and permeabilized with 0.2% Triton X-100. The cells were stained overnight with monoclonal anti-FLAG, polyclonal anti-HA (Sigma), and monoclonal anti-OCT4A antibodies in blocking buffer (3% BSA/PBS) and then rinsed and incubated with Alexa Fluor 594-conjugated anti-mouse and Alexa Fluor 488-conjugated anti-rabbit secondary antibodies (Invitrogen) for 1 h. Cells were rinsed with PBS, stained with DAPI, and mounted. The slides were examined using a fluorescence microscope (Eclipse TE300, Nikon) and digital image analysis software (IPLab, Scanalytics).
Protein Degradation Analysis
Cells were transfected with a plasmid encoding a FLAG-tagged version of the protein of interest. Where indicated, MEK1CA constructs was cotransfected. For half-life studies, CHX (20 μg/ml) was added to the media 40 h post-transfection. At various time points thereafter, cells were lysed, and protein amounts were measured by immunoblot analysis.
Cell Culture
NTera2 cells were maintained in high glucose DMEM containing 10% fetal bovine serum and 4 mm l-glutamine at 37 °C in 5% CO2. HeLa cells were maintained in high glucose DMEM containing 10% fetal bovine serum and 2 mm l-glutamine at 37 °C in 5% CO2. The concentrations and times used for each chemical treatment were as follows: PD98059, 20 μm for 4 h; MG132, 10 μm for 6 h; CHX, 1 μg/ml; and FGF2, 100 ng/ml.
Statistical Analysis
The data are expressed as means ± S.D. from an appropriate number of experiments as indicated in the figure legends. Statistical analysis was done using Student's t test, and p < 0.05 was considered significant.
RESULTS
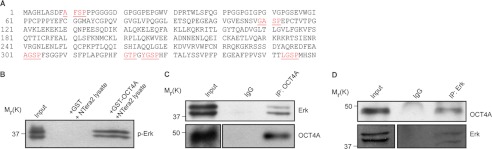
Although the transcriptional targets of the transcription factor OCT4A have been extensively studied, very little is known about how the protein itself is regulated, especially at the post-translational level. On the basis of recent studies describing post-translational modifications of OCT4 (6), we hypothesized that OCT4 activity might be regulated by post-translational modifications, specifically phosphorylation. OCT4A sequence analysis conducted with the database PhosphoMotif Finder (21) revealed that human OCT4A contains six putative ERK phosphorylation sites (Fig. 1A), which conforms to the optimal ERK motif XX(S/T)P. We therefore reasoned that OCT4A is an ERK substrate. Consistent with the hypothesis that OCT4A is a putative ERK substrate, using both in vitro GST pulldown (Fig. 1B) and in vivo co-immunoprecipitation (Fig. 1, C and D) assays, we were able to show that OCT4A specifically interacts with ERK.
FIGURE 1.
ERK interacts in vivo and in vitro with OCT4A. A, OCT4A protein sequence in which potential ERK phosphorylation sites are shown. B, GST pulldown assay was carried out using immobilized GST or GST-OCT4A fusion protein on agarose beads, followed by incubation with extracts prepared from NTera2 cells. The in vitro interaction of ERK with OCT4A was assessed by immunoblotting with an ERK-specific antibody. C and D, lysates of NTera2 cells were subjected to immunoprecipitation (IP) with anti-OCT4A (C) or anti-ERK antibody (D). The endogenous interaction of OCT4A with ERK was evaluated by immunoblotting using anti-ERK or anti-OCT4A antibody, respectively.
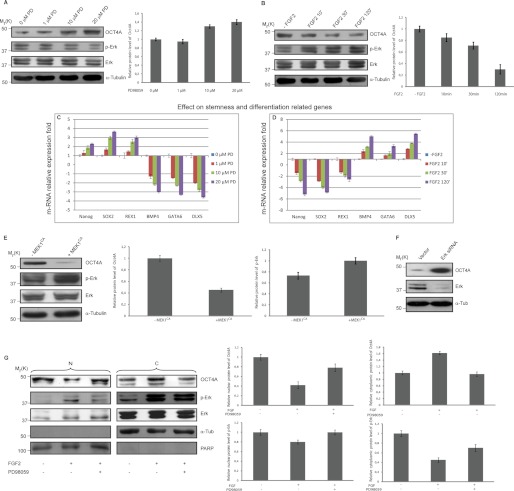
Next, to determine whether ERK signaling is involved in OCT4A expression regulation, we investigated the effects of activation and inhibition of this pathway on NTera2 cells, a human embryonal carcinoma cell line considered as a malignant counterpart of human ES cells. They closely resemble human ES cells and have often been used in studies of human embryogenesis (22). For this purpose, we treated NTera2 cells with the MEK inhibitor PD98059 and observed a dose-dependent increase in OCT4A protein levels concomitant with a robust inhibition of ERK activity as revealed by the loss of phospho-ERK (Fig. 2A). We then used FGF2, which potently activates the MAPK pathway, and observed a time-dependent decrease in OCT4A protein levels also concomitant with enhanced ERK phosphorylation (Fig. 2B). We also verified the expression patterns of differentially expressed genes that were previously evidenced to be downstream targets of OCT4, including pluripotency-related (Nanog, SOX2, and REX1) and differentiation-related (BMP4, GATA6, and DLX5) markers (23–26). Real-time PCR of samples prepared from both untreated and treated NTera2 cells demonstrated that Nanog, SOX2, and REX1 were up-regulated and that BMP4, GATA6, and DLX5 were down-regulated after MEK1-ERK1/2 inhibition due to PD98059 treatment (Fig. 2C). In contrast, Nanog, SOX2, and REX1 were down-regulated and BMP4, GATA6, and DLX5 were up-regulated after OCT4A-decreased expression due to FGF2 treatment (Fig. 2D). In addition, when MEK1CA that activates ERK was transfected into NTera2 cells, the total OCT4A protein was reduced (Fig. 2E). Similarly, using ERK siRNA to knock down the ERK protein expression level in NTera2 cells (Fig. 2F) led to an increase in OCT4A protein expression.
FIGURE 2.
ERK mediates the down-regulation of OCT4A and induces its nuclear exclusion. A and B, NTera2 cells were treated with increasing doses of PD98059 (PD; A) or with 100 ng/ml FGF2 for the indicated times (B), and the relative protein levels of OCT4A and phospho-ERK were quantified. Lysates were analyzed by immunoblotting with the indicated antibodies. C, real-time PCR results confirmed the up-regulation of the key pluripotency-controlling genes Nanog, SOX2, and REX1 and the down-regulation of the differentiation-related genes BMP4, GATA6, and DLX5 after PD98059 treatment. D, real-time PCR results confirmed the down-regulation of the key pluripotency-controlling genes Nanog, SOX2, and REX1 and the up-regulation of the differentiation-related genes BMP4, GATA6, and DLX5 after FGF2 treatment. E, lysates of NTera2 cells transfected with control or MEK1CA plasmids were analyzed by immunoblotting using the indicated antibodies, and the relative protein levels of OCT4A and phospho-ERK were quantified. F, lysates of NTera2 cells were subjected to immunoblotting with the indicated antibodies after being transfected with a control vector or ERK1 and ERK2 siRNAs. G, NTera2 cells were serum-starved and then treated with FGF2 (100 ng/ml) for 30 min in the presence (+) or absence (−) of PD98059. Nuclear (N) and cytoplasmic (C) fractions were analyzed by immunoblotting with the indicated antibodies. The relative protein levels of OCT4A (normalized to α-tubulin (α-Tub) or poly(ADP-ribose) polymerase (PARP)) and phospho-ERK (normalized to ERK) were calculated. Graphs show the mean value of the representative results from three experiments (n = 3) with S.D. conducted in duplicate for each. An equal amount of lysates was loaded in each lane.
It is known that the nuclear-cytoplasmic shuttling of many transcription factors is regulated through direct phosphorylation by specific kinases (27–31), so we asked whether ERK activation could also lead to a variation in OCT4A subcellular localization. To this end, nuclear and cytoplasmic fractions were obtained from NTera2 cells after ERK activation induced by FGF2 treatment for 30 min in either the presence or absence of MEK inhibitor PD98059, and we observed that OCT4A was reduced in the nucleus with FGF2 treatment (Fig. 2G), suggesting that ERK activation induces OCT4A nuclear exclusion.
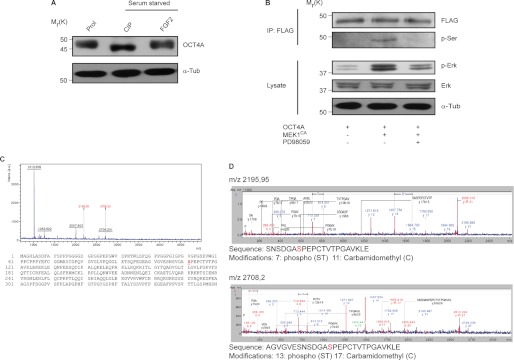
To explore the mechanism of ERK-mediated OCT4A down-regulation and nuclear exclusion, we investigated the possibility of a direct phosphorylation by ERK. Consistent with this hypothesis, immunoblot probing of OCT4A revealed a band shift (∼2 kDa) after treatment with CIP in NTera2 cells compared with proliferation conditions, which then shifted back after FGF2 stimulation (Fig. 3A). In addition, when lysates of HeLa cells transfected with FLAG-OCT4A alone or with MEK1CA were subjected to immunoprecipitation with anti-FLAG antibody and immunoblotting with anti-phosphoserine antibody, there were no visible serine phosphorylation levels in lysates that were not cotransfected with MEK1CA or treated with PD98059 compared with the cotransfected cells (Fig. 3B). Taken together, the data indicate that OCT4A is phosphorylated at a serine residue and support the hypothesis that it could be an ERK substrate.
FIGURE 3.
OCT4A is phosphorylated at Ser-111. A, lysates from NTera2 cells in proliferation (Prol) or serum-starved overnight and treated with CIP or stimulated with FGF2 for 30 min were subjected to immunoblotting. B, HeLa cells transfected with FLAG-OCT4A and control or MEK1CA plasmids were subjected to immunoprecipitation (IP) using immobilized FLAG on agarose beads and immunoblotting with either anti-phosphoserine or anti-FLAG antibody or direct immunoblotting with anti-phospho-ERK, anti-ERK, and anti-α-tubulin (α-Tub) antibodies. Where indicated, cells were treated with PD98059 for 4 h before the lysates were extracted. C, lysates from HeLa cells that had been cotransfected with OCT4A and MEK1CA were subjected to immunoprecipitation with anti-FLAG beads, and the isolated OCT4A was subjected to MS. The spectrum of the TiO2-purified sample and the position of the phosphoserine identified are reported. a.u., arbitrary units. D, annotated MS/MS spectra and phosphorylation sites.
To determine whether ERK can directly phosphorylate OCT4A, we performed MS analysis on extracts of HeLa cells transfected with FLAG-OCT4A and MEK1CA subjected to immunoprecipitation, and we identified phosphorylation of Ser-111 (Fig. 3, C and D), one of the putative ERK phosphorylation sites revealed by sequence analysis conducted with the database PhosphoMotif Finder.
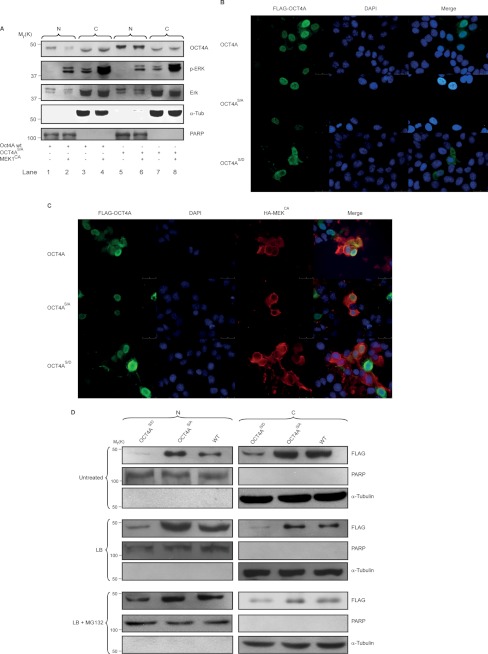
To verify the contribution of the identified Ser-111 in the regulation of OCT4A subcellular localization, this residue was mutated to alanine (S111A) to mimic constitutive dephosphorylation. The mutant was expressed in HeLa cells as a fusion protein with FLAG, and the subcellular localization was analyzed by immunoblotting. The generated S111A mutant accumulated more in the nucleus (Fig. 4A, lane 5) than in the cytosol (lane 7) compared with the wild type (lanes 1 and 3, respectively). In addition, when MEK1CA was cotransfected, the nuclear localization of the WT was dramatically reduced (Fig. 4A, lane 2), whereas the localization of the S111A mutant was not affected (lane 6). To gain further insight into the mechanism responsible for the increased nuclear accumulation of the S111A mutant, Ser-111 was mutated to aspartic acid (S111D) and used to transfect HeLa cells to mimic constitutive phosphorylation. We then compared its subcellular localization with respect to the WT and S111A mutant and by immunofluorescence in either the absence or presence of MEK1CA (Fig. 4, B and C). As shown, the S111A mutant accumulated more in the nucleus compared with the WT and S111D mutant. In contrast, the S111D mutant was localized more in the cytoplasm compared with the WT and S111A mutant (Fig. 4B). In addition, when MEK1CA was cotransfected, the WT cytoplasmic localization increased, the S111A mutant localization was not affected, and the S111D mutant showed a distribution similar to the cells not cotransfected with MEK1CA (Fig. 4C).
FIGURE 4.
OCT4A Ser-111 phosphorylation regulates OCT4A subcellular localization. A, NTera2 cells were serum-starved and then treated with FGF2 (100 ng/ml) for 30 min in the presence (+) or absence (−) of PD98059. Nuclear (N) and cytoplasmic (C) fractions were analyzed by immunoblotting with the indicated antibodies. An equal amount of lysates was loaded in each lane. α-Tub, α-tubulin; PARP, poly(ADP-ribose) polymerase. B, immunofluorescence of HeLa cells transfected with FLAG-OCT4A, FLAG-OCTA4(S111A) (OCTA4S/A), and FLAG-OCT4A(S111D) (OCTA4S/D). The results show that the generated S111A mutant accumulated more in the nucleus than in the cytosol compared with the WT and S111D mutant. C, immunofluorescence of HeLa cells transfected with FLAG-OCT4A, FLAG-OCTA4(S111A), and FLAG-OCT4A(S111D) (green) together with HA-MEK1CA (red). When MEK1CA was cotransfected, the nuclear localization of the WT was dramatically reduced, whereas that of the S111A and S111D mutants was not affected. Scale bars = 25 μm. Nuclei were counterstained with DAPI. D, nuclear and cytoplasmic fractions of HeLa cells treated with leptomycin B (LB) or with a combination of leptomycin B and MG132 (LB + MG132) or left untreated and transfected with WT OCT4A or the S111A or S111D mutant were subjected to immunoblotting.
We also compared the subcellular distribution of the WT and mutants after treatment of the cells with leptomycin B, an inhibitor of CRM1-mediated nuclear export, or a combination of MG132 and leptomycin B (Fig. 4D). As expected, the S111A mutant accumulated more in the nucleus than the S111D mutant and WT in untreated cells (Fig. 4D, upper panels). In addition, blocking nuclear export failed to clearly increase the S111D mutant nuclear accumulation, but there was an increase in the WT nuclear localization compared with the untreated cells (Fig. 4D, middle panels). A possible explanation may be that the S111D mutant is more sensible to degradation. In fact, the addition of leptomycin B to cells pretreated with MG132 (Fig. 4D, lower panels) allowed a more obvious nuclear accumulation of the S111D mutant. These results indicate that the S111D mutant can be imported into the nucleus, where it becomes susceptible to ubiquitin-dependent proteolysis. In addition, nuclear accumulation of the S111A mutant after leptomycin B treatment confirms that phosphorylation of Ser-111 plays an important role in OCT4A degradation and subcellular distribution.
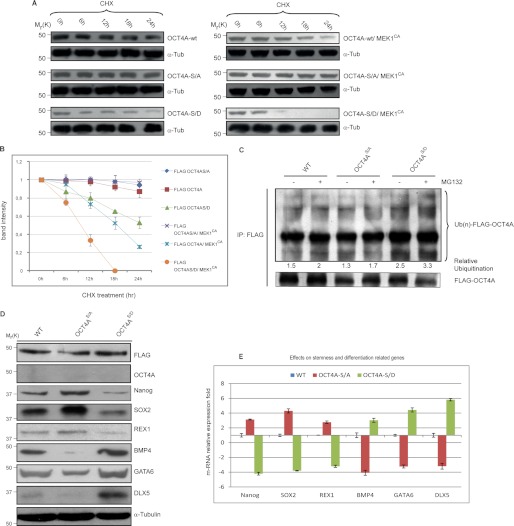
To investigate whether phosphorylation of Ser-111 is implicated in OCT4A stability and degradation, we analyzed OCT4A turnover by treating transfected HeLa cells with the protein synthesis inhibitor CHX. In the presence of MEK1CA, the turnover of FLAG-tagged WT OCT4A was substantially enhanced. In contrast, the FLAG-S111A mutant protein level remained relatively stable over the period of CHX treatment in the presence of MEK1CA, whereas the FLAG-S111D mutant was degraded significantly more rapidly than the WT (Fig. 5, A and B). It is important to underline that the rate of turnover of OCT4A(S111D) increased upon coexpression of MEK1CA, probably because activation of the ERK pathway may be implicated in other mechanisms that are also involved in OCT4A turnover and necessitate the previous phosphorylation of Ser-111.
FIGURE 5.
Ser-111 phosphorylation enhances OCT4A proteasome-dependent degradation. A, HeLa cells were transfected with indicated plasmids. At 24–48 h after transfection, cells were treated with 15 μg/ml CHX. At the indicated time points, whole cell lysates were prepared, and immunoblots were probed with the indicated antibodies. OCT4A-S/A, OCT4A(S111A); OCT4A-S/D, OCT4A(S111D). B, FLAG-OCT4A band intensity was normalized to α-tubulin (α-Tub) and then normalized to the t = 0 controls. Results are shown as means ± S.D. for three independent sets of experiments. C, HeLa cells transfected with the WT or the S111A or S111D mutant were lysed and immunoprecipitated (IP) using anti-FLAG beads. Immunoblotting was performed using anti-ubiquitin (Ub) and anti-FLAG antibodies. D, OCT4A-silenced NTera2 cells transfected with the WT or the S111A or S111D mutant were reacted with anti-FLAG, anti-OCT4A, anti-Nanog, anti-SOX2, anti-REX1, anti-BMP4, anti-GATA6, anti-DLX5, and anti-α-tubulin antibodies. E, the same stemness (Nanog, SOX2, and REX1) and differentiation (BMP4, GATA6, and DLX5) markers were tested by real-time PCR. Data were normalized to RNA polymerase II and are shown as means ± S.D. for three independent sets of experiments.
In addition, when we analyzed endogenous ubiquitination, the extent of S111D mutant ubiquitination was increased compared with the WT and S111A mutant (Fig. 5C). These results clearly show that phosphorylation of Ser-111 promotes degradation of OCT4A, implying a dominant-negative function of Ser-111 phosphorylation in the regulation of the OCT4A protein level. To verify the effect of OCT4A Ser-111 phosphorylation on stem cell maintenance, we silenced endogenous OCT4A using RNAi (Fig. 5D) (20), followed by transfection of both phosphorylated mutant forms and WT OCT4A in NTera2 cells, in which OCT4A function was demonstrated to be relevant (22). Consequently, we analyzed the expression of the stemness-related markers Nanog, SOX2, and REX1 and the differentiation markers BMP4, GATA6, and DLX5 (Fig. 5D), known to be downstream targets of OCT4A (23–26), by immunoblotting. The results show that OCT4A(S111D) mutant expression was associated with a decrease in Nanog, SOX2, and REX1 and an increase in BMP4, GATA6, and DLX5 compared with the WT. In contrast, OCT4A(S111A) mutant transfection led to an increase in Nanog, SOX2, and REX1 and a decrease in BMP4, GATA6, and DLX5, demonstrating that this phosphorylation is important for the maintenance of stemness. Similar results were obtained in real-time PCR experiments (Fig. 5E).
DISCUSSION
Our understanding of the core transcription factors that control ES cell regulatory processes as well as their involvement in generating iPS cells from somatic cells has expanded greatly in the past few years. OCT4A is one of the four critical factors involved in generating iPS cells (32). Although transcriptional networks controlled by OCT4 have been delineated (23, 25, 26) and several protein interactors of OCT4 have been identified (33, 34), the mechanisms through which the OCT4 protein itself is regulated have remained largely unexplored.
Export of proteins from the nucleus has been implicated in the control of several nuclear processes, primarily gene transcription. Clearly, nuclear exclusion of a transcription factor represents a potent mechanism to negatively affect its activity. We propose that this mechanism of regulation also applies to OCT4A. Our findings indicate that nuclear-cytoplasmic shuttling of OCT4A is regulated through a mechanism involving ERK activation, providing evidence for a role of the MEK-ERK signaling pathway in regulating the cellular localization and biological activity of this transcription factor.
We have demonstrated for the first time that OCT4A interacts in vivo and in vitro with ERK. In addition, our data suggest that ERK directly phosphorylates OCT4A at Ser-111 and that the S111D mutation decreases its nuclear accumulation.
We have shown that phosphorylation of Ser-111 is an important determinant of OCT4A stability and is involved in stemness regulation, as evidenced by the decreased expression of stemness-related markers and by the increased expression of differentiation-related markers. In particular, mutation of Ser-111 to aspartic acid, which mimics constitutive phosphorylation, causes OCT4A instability. Our data also suggest a broader role for Ser-111 phosphorylation in the regulation of OCT4A via ubiquitination and proteasomal degradation. We showed that the S111D mutant is more ubiquitinated than the WT and S111A mutant. In addition, when the proteasome was inhibited and leptomycin B was added, the S111D mutant accumulated in the nucleus. This result indicates that OCT4A degradation takes place in the nucleus (35). Xu et al. (35) demonstrated that OCT4A interacts specifically with a HECT domain-containing E3 ubiquitin-protein ligase (WWP2) that promotes OCT4A protein ubiquitination and degradation through the 26 S proteasome. In this work, we have demonstrated that OCT4A ubiquitination and degradation are signal-regulated processes, adding another piece to the complexity of stemness regulation. We hypothesize that phosphorylation of this serine residue increases not only OCT4A nuclear exclusion but also the susceptibility of OCT4A to interaction with and degradation by WWP2. Further studies are necessary to verify this hypothesis. It would be interesting to investigate whether the increase in OCT4A ubiquitination after ERK activation can be explained by ERK-mediated WWP2 up-regulation, given that the RAS-ERK pathway is known to up-regulate MDM2 protein expression (36).
To our knowledge, this is the first demonstration that human OCT4A is phosphorylated by ERK, promoting a decrease in its nuclear accumulation as well as its degradation. These findings identify a new mechanism by which the OCT4A protein level and functions are precisely controlled, extending the current knowledge regarding post-translational modification of OCT4A and providing evidence for negative OCT4A regulation by phosphorylation. Further investigation is required to determine to what extent these mechanisms will translate to the human ES cell system. In conclusion, our studies provide insight into how specific post-translational modifications influence OCT4A regulation, which can potentially be modulated to enhance stemness maintenance and iPS technology.
Footnotes
- ES
- embryonic stem
- iPS
- induced pluripotent stem
- CHX
- cycloheximide
- MEK1CA
- constitutively active MEK1
- CIP
- calf intestinal phosphatase.
REFERENCES
- 1. Niwa H. (2007) How is pluripotency determined and maintained? How is pluripotency determined and maintained? Development. 134, 635–646 [DOI] [PubMed] [Google Scholar]
- 2. Schöler H. R., Dressler G. R., Balling R., Rohdewohld H., Gruss P. (1990) Oct-4: a germ line-specific transcription factor mapping to the mouse t-complex. EMBO J. 9, 2185–2195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Rosner M. H., Vigano M. A., Ozato K., Timmons P. M., Poirier F., Rigby P. W., Staudt L. M. (1990) A POU domain transcription factor in early stem cells and germ cells of the mammalian embryo. Nature 345, 686–692 [DOI] [PubMed] [Google Scholar]
- 4. Okamoto K., Okazawa H., Okuda A., Sakai M., Muramatsu M., Hamada H. (1990) A novel octamer binding transcription factor is differentially expressed in mouse embryonic cells. Cell 60, 461–472 [DOI] [PubMed] [Google Scholar]
- 5. Hansis C., Grifo J. A., Krey L.C. (2000) Oct-4 expression in inner cell mass and trophectoderm of human blastocysts. Mol. Hum. Reprod. 6, 999–1004 [DOI] [PubMed] [Google Scholar]
- 6. Swaney D. L., Wenger C. D., Thomson J. A., Coon J. J. (2009) Human embryonic stem cell phosphoproteome revealed by electron transfer dissociation tandem mass spectrometry. Proc. Natl. Acad. Sci. U.S.A. 106, 995–1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lee J., Kim H. K., Rho J. Y., Han Y. M., Kim J. (2006) The human OCT4 isoforms differ in their ability to confer self-renewal. J. Biol. Chem. 281, 33554–33565 [DOI] [PubMed] [Google Scholar]
- 8. Nichols J., Zevnik B., Anastassiadis K., Niwa H., Klewe-Nebenius D., Chambers I., Schöler H., Smith A. (1998) Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell 95, 379–391 [DOI] [PubMed] [Google Scholar]
- 9. Takahashi K., Yamanaka S. (2006) Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663–676 [DOI] [PubMed] [Google Scholar]
- 10. Yu J., Vodyanik M. A., Smuga-Otto K., Antosiewicz-Bourget J., Frane J. L., Tian S., Nie J., Jonsdottir G. A., Ruotti V., Stewart R., Slukvin I. I., Thomson J. A. (2007) Induced pluripotent stem cell lines derived from human somatic cells. Science 318, 1917–1920 [DOI] [PubMed] [Google Scholar]
- 11. Tsuruzoe S., Ishihara K., Uchimura Y., Watanabe S., Sekita Y., Aoto T., Saitoh H., Yuasa Y., Niwa H., Kawasuji M., Baba H., Nakao M. (2006) Inhibition of DNA binding of Sox2 by the SUMO conjugation. Biochem. Biophys. Res. Commun. 351, 920–926 [DOI] [PubMed] [Google Scholar]
- 12. Baltus G. A., Kowalski M. P., Zhai H., Tutter A. V., Quinn D., Wall D., Kadam S. (2009) Acetylation of Sox2 induces its nuclear export in embryonic stem cells. Stem Cells 27, 2175–2184 [DOI] [PubMed] [Google Scholar]
- 13. Saxe J. P., Tomilin A., Schöler H. R., Plath K., Huang J. (2009) Post-translational regulation of Oct4 transcriptional activity. PLoS ONE 4, e4467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wei F., Schöler H. R., Atchison M. L. (2007) Sumoylation of Oct4 enhances its stability, DNA binding, and transactivation. J. Biol. Chem. 282, 21551–21560 [DOI] [PubMed] [Google Scholar]
- 15. Niwa H., Miyazaki J., Smith A. G. (2000) Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation, or self-renewal of ES cells. Nat. Genet. 24, 372–376 [DOI] [PubMed] [Google Scholar]
- 16. Conaway R. C., Brower C. S., Conaway J. W. (2002) Emerging roles of ubiquitin in transcription regulation. Science 296, 1254–1258 [DOI] [PubMed] [Google Scholar]
- 17. Ciechanover A. (1998) The ubiquitin-proteasome pathway: on protein death and cell life. EMBO J. 17, 7151–7160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Naujokat C., Hoffmann S. (2002) Role and function of the 26S proteasome in proliferation and apoptosis. Lab. Invest. 82, 965–980 [DOI] [PubMed] [Google Scholar]
- 19. Mayer R. J. (2000) The meteoric rise of regulated intracellular proteolysis. Nat. Rev. Mol. Cell Biol. 1, 145–148 [DOI] [PubMed] [Google Scholar]
- 20. Chew J. L., Loh Y. H., Zhang W., Chen X., Tam W. L., Yeap L. S., Li P., Ang Y. S., Lim B., Robson P., Ng H. H. (2005) Reciprocal transcriptional regulation of Pou5f1 and Sox2 via the Oct4/Sox2 complex in embryonic stem cells. Mol. Cell. Biol. 25, 6031–6046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Amanchy R., Periaswamy B., Mathivanan S., Reddy R., Tattikota S. G., Pandey A. (2007) A curated compendium of phosphorylation motifs. Nat. Biotechnol. 25, 285–286 [DOI] [PubMed] [Google Scholar]
- 22. Pal R., Ravindran G. (2006) Assessment of pluripotency and multilineage differentiation potential of NTERA-2 cells as a model for studying human embryonic stem cells. Cell Prolif. 39, 585–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Babaie Y., Herwig R., Greber B., Brink T. C., Wruck W., Groth D., Lehrach H., Burdon T., Adjaye J. (2007) Analysis of Oct4-Dependent transcriptional networks regulating self-renewal and pluripotency in human embryonic stem Cells. Stem Cells 25, 500–510 [DOI] [PubMed] [Google Scholar]
- 24. Rodda D. J., Chew J. L., Lim L. H., Loh Y. H., Wang B., Ng H. H., Robson P. (2005) Transcriptional regulation of Nanog by OCT4 and SOX2. J. Biol. Chem. 280, 24731–24737 [DOI] [PubMed] [Google Scholar]
- 25. Boyer L. A., Lee T. I., Cole M. F., Johnstone S. E., Levine S. S., Zucker J. P., Guenther M. G., Kumar R. M., Murray H. L., Jenner R. G., Gifford D. K., Melton D. A., Jaenisch R., Young R. A. (2005) Core transcriptional regulatory circuitry in human embryonic stem cells. Cell 122, 947–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Loh Y. H., Wu Q., Chew J. L., Vega V. B., Zhang W., Chen X., Bourque G., George J., Leong B., Liu J., Wong K. Y., Sung K. W., Lee C. W., Zhao X. D., Chiu K. P., Lipovich L., Kuznetsov V. A., Robson P., Stanton L. W., Wei C. L., Ruan Y., Lim B., Ng H. H. (2006) The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat. Genet. 38, 431–440 [DOI] [PubMed] [Google Scholar]
- 27. Chen R. H., Sarnecki C., Blenis J. (1992) Nuclear localization and regulation of erk- and rsk-encoded protein kinases. Mol. Cell. Biol. 12, 915–927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lenormand P., Sardet C., Pagès G., L'Allemain G., Brunet A., Pouysségur J. (1993) Growth factors induce nuclear translocation of MAP kinases (p42mapk and p44mapk) but not of their activator MAP kinase kinase (p45mapkk) in fibroblasts. J. Cell Biol. 122, 1079–1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Karin M., Hunter T. (1995) Transcriptional control by protein phosphorylation: signal transmission from the cell surface to the nucleus. Curr. Biol. 5, 747–757 [DOI] [PubMed] [Google Scholar]
- 30. Hill C. S., Treisman R. (1995) Transcriptional regulation by extracellular signals: mechanisms and specificity. Cell 80, 199–211 [DOI] [PubMed] [Google Scholar]
- 31. Hunter T., Karin M. (1992) The regulation of transcription by phosphorylation. Cell 70, 375–387 [DOI] [PubMed] [Google Scholar]
- 32. Yamanaka S. (2007) Strategies and new developments in the generation of patient-specific pluripotent stem cells. Cell Stem Cell 1, 39–49 [DOI] [PubMed] [Google Scholar]
- 33. Xu H. M., Liao B., Zhang Q. J., Wang B. B., Li H., Zhong X. M., Sheng H. Z., Zhao Y. X., Zhao Y. M., Jin Y. (2004) Wwp2, an E3 ubiquitin ligase that targets transcription factor Oct-4 for ubiquitination. J. Biol. Chem. 279, 23495–23503 [DOI] [PubMed] [Google Scholar]
- 34. Wang J., Rao S., Chu J., Shen X., Levasseur D. N., Theunissen T. W., Orkin S. H. (2006) A protein interaction network for pluripotency of embryonic stem cells. Nature 444, 364–368 [DOI] [PubMed] [Google Scholar]
- 35. Xu H., Wang W., Li C., Yu H., Yang A., Wang B., Jin Y. (2009) WWP2 promotes degradation of transcription factor OCT4 in human embryonic stem cells. Cell Res. 19, 561–573 [DOI] [PubMed] [Google Scholar]
- 36. Ries S., Biederer C., Woods D., Shifman O., Shirasawa S., Sasazuki T., McMahon M., Oren M., McCormick F. (2000) Opposing effects of Ras on p53: transcriptional activation of mdm2 and induction of p19ARF. Cell 103, 321–330 [DOI] [PubMed] [Google Scholar]