Background: Toluene dioxygenase reductase converts the redox equivalents of NADH under oxic conditions.
Results: Reduced reductase forms a stable complex with NAD+ that shows slower electron transfer to dioxygen.
Conclusion: Charge transfer complex formation regulates the reactivity of the reductase.
Significance: Charge transfer complexes between flavins and nicotinamides are abundant and likely to have diverse mechanistic functions.
Keywords: Bioenergetics/Electron Transfer Complex, Electron Transfer, FAD, Flavin, Flavoproteins, Iron-Sulfur Protein
Abstract
The three-component toluene dioxygenase system consists of an FAD-containing reductase, a Rieske-type [2Fe-2S] ferredoxin, and a Rieske-type dioxygenase. The task of the FAD-containing reductase is to shuttle electrons from NADH to the ferredoxin, a reaction the enzyme has to catalyze in the presence of dioxygen. We investigated the kinetics of the reductase in the reductive and oxidative half-reaction and detected a stable charge transfer complex between the reduced reductase and NAD+ at the end of the reductive half-reaction, which is substantially less reactive toward dioxygen than the reduced reductase in the absence of NAD+. A plausible reason for the low reactivity toward dioxygen is revealed by the crystal structure of the complex between NAD+ and reduced reductase, which shows that the nicotinamide ring and the protein matrix shield the reactive C4a position of the isoalloxazine ring and force the tricycle into an atypical planar conformation, both factors disfavoring the reaction of the reduced flavin with dioxygen. A rapid electron transfer from the charge transfer complex to electron acceptors further reduces the risk of unwanted side reactions, and the crystal structure of a complex between the reductase and its cognate ferredoxin shows a short distance between the electron-donating and -accepting cofactors. Attraction between the two proteins is likely mediated by opposite charges at one large patch of the complex interface. The stability, specificity, and reactivity of the observed charge transfer and electron transfer complexes are thought to prevent the reaction of reductaseTOL with dioxygen and thus present a solution toward conflicting requirements.
Introduction
Toluene dioxygenase (EC 1.14.12.11) catalyzes the dihydroxylation of toluene to yield cis-(1R,2S)-dihydroxy-3-methylcyclohexa-3,5-diene (cis-toluene dihydrodiol). This is the first step in the bacterial degradation of toluene and allows Pseudomonas putida F1 to utilize toluene as the sole source of carbon and energy (1). Toluene dioxygenase is a member of the Rieske-type oxygenase family, enzymes that typically consist of two or three components to mediate electron transfer from NADH to the active site of a Rieske-type oxygenase component where dioxygen is activated and the substrate is turned over (2). Recently, the crystal structures of all three individual components of toluene dioxygenase have been reported, and the structure of the reductase component was found to belong to the glutathione reductase family (3, 4). In toluene dioxygenase, the flavin adenine dinucleotide (FAD/FADH2) redox center of reductaseTOL acts as a transformer by accepting two electrons from soluble nicotinamide adenine dinucleotide (NAD+/NADH) and donating them in two separate one-electron transfer steps to a Rieske-type ferredoxin, termed ferredoxinTOL (5). From ferredoxinTOL, the electrons are transferred to oxygenaseTOL.
ReductaseTOL belongs to the large family of bacterial oxygenase-coupled, NADH-dependent ferredoxin reductases. Oxygenase-coupled, NADH-dependent ferredoxin reductases are found in conjunction with Rieske-type oxygenases or cytochrome P450-dependent monooxygenases to which the structurally characterized ferredoxin reductase components of biphenyl dioxygenase (BphA4)3 (6) and the putidaredoxin reductase (7) belong. All of these proteins share similar active site architectures with a conserved Lys-Glu ion pair, which is in hydrogen-bonding distance to the N5 atom of the flavin cofactor. The Lys-Glu pair has also been identified in the crystal structure of reductaseTOL (4).
Reduced flavins are typically oxidized by dioxygen, producing reduced dioxygen species like peroxide and superoxide. We became interested in how this unwanted side reaction may be suppressed in flavoenzymes, which have to function in an oxic environment. We selected the toluene dioxygenase system for which our combined kinetic and structural analysis revealed how the reactivity of the reduced reductase with dioxygen may be suppressed by formation of a stable charge transfer complex and by rapid electron transfer to physiological and artificial electron acceptors.
EXPERIMENTAL PROCEDURES
Reagents
All chemicals used were of analytical grade. All chromatography materials were from Amersham Biosciences.
Cloning and Expression of ReductaseTOL and FerredoxinTOL
The nucleotide sequences of todA and todB, encoding for reductaseTOL and ferredoxinTOL, from P. putida F1 were obtained from GenBank (accession number J04996.1). The open reading frame of P. putida F1 todA and todB was amplified by polymerase chain reaction (PCR) using P. putida F1 genomic DNA as template and cloned into pET-15b or pET-11a (Novagen), respectively. The identity of the constructs was verified by DNA sequencing. ReductaseTOL and ferredoxinTOL were expressed under aerobic conditions in Escherichia coli CodonPlus (DE3)-RIL (Stratagene) using dYT medium (8) supplemented with 50 μg/ml carbenicillin and 34 μg/ml chloramphenicol at 22 °C. Expression was induced with 0.5 mm isopropyl β-d-galactopyranoside at an A600 nm of 0.5. Cells were harvested by centrifugation after 20 or 18 h in the case of reductaseTOL or ferredoxinTOL, respectively; frozen in liquid nitrogen; and stored at −30 °C until further use.
Purification of ReductaseTOL and FerredoxinTOL
Frozen cells containing the overexpressed reductaseTOL were resuspended in 50 mm Tris-HCl, pH 8.0, 20 mm NaCl, 20 mm imidazole, and 0.1 mm phenylmethylsulfonyl fluoride (PMSF; Carl Roth GmbH). Crude cell extract prepared by sonication and centrifugation was applied to a nickel-Sepharose high performance column, and the His-tagged protein was eluted with increasing imidazole concentration from 20 to 250 mm. ReductaseTOL-containing fractions were pooled, loaded on a Q-Sepharose Fast Flow column equilibrated with 50 mm Tris-HCl, pH 8.0 and eluted with a linear gradient of 0–500 mm NaCl. The fractions with the highest purity were pooled, and the buffer was exchanged to 50 mm Tris-HCl, pH 7.2 and 100 mm NaCl using a Sephadex G-25 column. Finally, the protein was concentrated to 23 mg ml−1, frozen in liquid nitrogen, and stored at −80 °C.
Frozen cells containing the overexpressed ferredoxinTOL were resuspended in 50 mm Tris-HCl, pH 6.9, 1 mm DTT, and 1 mm PMSF. Supernatant obtained after sonication and centrifugation was applied on a DEAE-Sepharose Fast Flow column. Elution was performed with increasing concentration from 0 to 500 mm NaCl with ferredoxinTOL eluting at around 270 mm NaCl. The pooled fractions were loaded on a CHTTM ceramic hydroxyapatite column equilibrated with 5 mm KH2PO4, pH 6.9 and 1 mm DTT. FerredoxinTOL does not bind to the resin and is found in the flow-through. Subsequently, the buffer was exchanged to 50 mm Tris-HCl, pH 6.9, 1 mm DTT, and 150 mm NaCl using a Sephadex G-25 column. Finally, the protein was concentrated to 10 mg ml−1, shock frozen in liquid nitrogen, and stored at −80 °C.
Enzyme Assays and Rapid Reaction Kinetics
Specific activities were determined by monitoring the NADH-dependent reduction of 2,6-dichlorophenolindophenol using the assay described by Subramanian et al. (9).
Stopped-flow measurements were performed under anoxic conditions in 50 mm MOPS/NaOH, pH 7.2 and 150 mm NaCl at 10 °C using an Applied Photophysics SX-20MV spectrophotometer with a 1-cm observation path length coupled to either a diode array detector or a photomultiplier. Enzyme solutions were made anoxic in a glass tonometer by repeated cycles of evacuation and equilibration with nitrogen gas. Apparent first-order rate constants (kobs) were derived from the kinetic traces using the Pro-Data software (Applied Photophysics, UK). All quoted concentrations are final concentrations after 1:1 (v/v) mixing.
Reductive Half-reaction
Spectral changes attributed to the reduction of reductaseTOL were monitored for the reaction of 15.3 μm reductaseTOL with 30 μm NADH using diode array detection. To estimate the limiting rate constant for the flavin reduction (klim) and the dissociation constant for the transient enzyme-substrate complex (KD), 2.4 μm reductaseTOL was mixed 1:1 with a solution containing 20–500 μm NADH. Each experiment was repeated at least four times for each substrate concentration. Hyperbolic plots of observed rate constants versus substrate concentration were fitted using Equation 1,
assuming a fast binding step followed by a slower quasi-irreversible electron transfer.
Oxidative Half-reaction
To obtain fully reduced reductaseTOL, the anoxic enzyme was reduced by addition of either stoichiometric amounts of NADH or sodium dithionite, respectively. The reoxidation by potassium hexacyanoferrate(III) or oxygen as oxidative substrate were monitored with a diode array detector.
Crystallization, Data Collection, Structure Solution, and Refinement
Crystals of the NAD+-reductaseTOL charge transfer complex were obtained by the hanging drop technique. ReductaseTOL was prepared with 3 times the concentration of NADH and crystallized in 2.2 m sodium malonate, pH 6.5. Crystals of the reductaseTOL-ferredoxinTOL complex were obtained by the sitting drop technique. ReductaseTOL and ferredoxinTOL had equimolar concentrations in the crystallization drop. Crystals grew above a reservoir solution of 0.1 m Bis-Tris, pH 6.5 and 20% (w/v) polyethylene glycol monomethyl ether 5000.
Diffraction data were collected at −180 °C at beam line BL14.2 (Berliner Elektronenspeicherring-Gesellschaft für Synchrotronstrahlung (BESSY), Berlin, Germany) (Table 1). Diffraction images were processed, and data were scaled using XDS (10).
TABLE 1.
Statistics on diffraction data and structure refinement
Numbers in parentheses correspond to the highest resolution shell. refl., reflections; ML, machine learning; r.m.s., root mean square.
| Charge transfer complex NAD+-reductaseTOL | ReductaseTOL-ferredoxinTOL complex | |
|---|---|---|
| Wavelength (Å) | 0.91841 | 0.91841 |
| Space group | P41212 | P65 |
| Cell constants (Å) | 78.4, 78.4, 158.7 | 120.3, 120.3, 60.4 |
| Total/unique refl. | 316,338/41,735 | 84,325/19,644 |
| Rsa (%) | 5.3 (59.3) | 15.2 (58.1) |
| Resolution (Å) | 20–1.80 (1.85–1.80) | 20–2.40 (2.46–2.40) |
| Completeness (%) | 90.1 (99) | 99.9 (100) |
| (I)/(σI) | 20.9 (2.8) | 7.9 (2.9) |
| Model R/Rfree factor (%)b | 18.4/21.5 | 16.7/22.8 |
| r.m.s. deviation from ideal geometry | ||
| Bonds (Å) | 0.011 | 0.004 |
| Angles (°) | 1.779 | 0.78 |
| ML-based coordinate error (Å) | 0.24 | 0.32 |
a Rs = ΣhΣi|Ii(h) − I(h)|/ΣhΣiIi(h) where i represents the independent observations of reflection h.
b The Rfree factor was calculated from 5% of the data, which were removed at random before the refinement was carried out. The R factor was calculated from the reflections of the working set and test set.
The structure was solved using Patterson search techniques with the crystal structures of ferredoxinTOL (Protein Data Bank code 3DQY) and reductaseTOL (Protein Data Bank code 3EF6) as homologous search models (4) using Phaser (11). The model was constructed using Coot (12). Positional, temperature factor, and TLS refinements (13) were carried out with PHENIX (14). The final refinement statistics are shown in Table 1. All figures were prepared using the PyMOL Molecular Graphics System, Version 1.5.0.1, Schrödinger, LLC (15).
Model Calculations
Electrostatic potential calculations were carried out using APBS (16) using the non-linear Poisson-Boltzmann equation with a solvent radius of 1.4 Å. The structures were prepared using PDB2PQR (17) and the Schrödinger 2010 suite (Protein Preparation Wizard, Epik, Version 2.1; Impact, Version 5.6; and Prime, Version 2.2). The partial charges of FAD used within Poisson-Boltzmann calculations were determined using semiempirical methods (PM3) as implemented in Gaussian03 (18). Partial charges for the [2Fe-2S] cluster were gained from a model containing the [2Fe-2S] cluster core, methylimidazoles, and methylthiols optimized using hybrid density functional theory (B3LYP) as implemented in Gaussian03 (18).
Data Bank Accession Codes
Coordinates and structure factor amplitudes have been deposited in the Protein Data Bank under codes 4EMI (NAD+-reductaseTOLCT) and 4EMJ (ferredoxinTOL-reductaseTOL).
RESULTS AND DISCUSSION
Reaction of ReductaseTOL with NADH and Charge Transfer Complex Formation
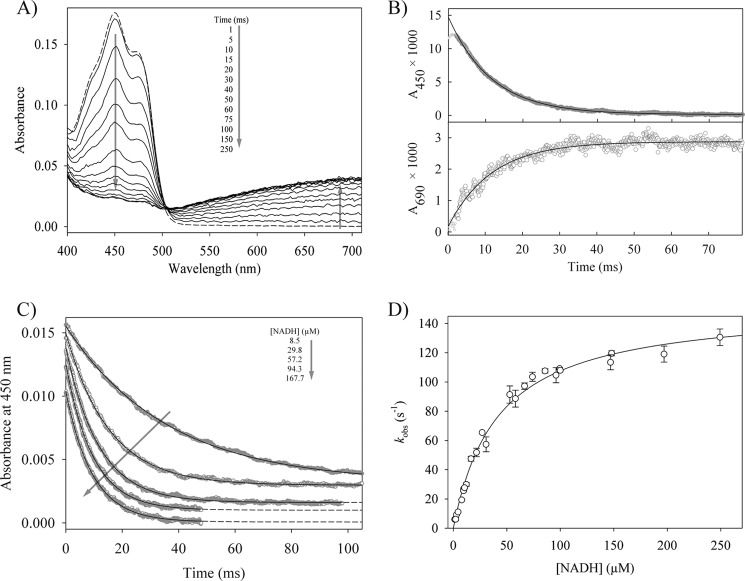
The reductive half-reaction of reductaseTOL was studied by multiple wavelength stopped-flow spectrophotometry, which was used to follow the reaction of oxidized reductaseTOL with NADH after rapid mixing under anoxic conditions. The first spectrum after mixing of the two components is similar to the spectrum of oxidized reductaseTOL and does not show any characteristic signal for a charge transfer (CT) complex between NADH and oxidized reductaseTOL (Fig. 1A). The NADH-dependent reduction of reductaseTOL was followed by recording the time-dependent absorption decrease at 450 nm, whereas the concomitant formation of a CT complex was indicated by an increase in absorption at 520–720 nm where the long wavelength detection is limited by the diode array detector (Fig. 1A). The observed transients can be described by a single exponential function (Fig. 1, B and C). Using the rapid equilibrium approximation for a two-step reaction (Equation 1), the dissociation constant (KD) for the reductaseTOL-NADH complex has been determined as 41 ± 4 μm with a limiting rate of reduction (kred) of 152 ± 4 s−1 (Fig. 1D). The increased broad absorption band between 520 and 720 nm is typical for CT complexes involving reduced flavins (19). The kinetic trace at 690 nm monitors the formation of the CT complex, and the agreement of the observed rate constants at 450 and 690 nm (Fig. 1B) suggests that the reduction of the flavin is immediately followed by the formation of the CT complex. The reaction sequence thus indicates that the CT complex formed involves the reduced FAD and the oxidized pyridine nucleotide (NAD+-reductaseTOLCT). The amplitude of the phase at 690 nm was used to calculate an extinction coefficient at 690 nm (ϵ690) of 2,082 m−1 cm−1, assuming the practically complete formation of the NAD+-reductaseTOLCT complex at high NADH concentrations. The derived ϵ690 value is in good agreement with the extinction coefficient of 2,000 m−1 cm−1 observed for a CT intermediate of melilotate hydroxylase (20).
FIGURE 1.
The reductive half-reaction of reductaseTOL. A, time-dependent spectral changes of 15.3 μm oxidized reductaseTOL reacting with 30 μm NADH. The dashed line represents the spectrum of oxidized reductaseTOL, and arrows indicate the direction of absorbance changes. B, time-dependent absorbance change at 450 and 690 nm for the reaction of 1.2 μm reductaseTOL with 57 μm NADH. The solid lines show single exponential fits to the data, yielding observed rate constants of 86.6 ± 0.2 (450 nm) and 85.3 ± 1.6 s−1 (690 nm), respectively. C, stopped-flow traces for the reaction of 1.2 μm reductaseTOL with different NADH concentrations. The traces have been shifted vertically for clarity. The solid lines are single exponential fits of the data. D, dependence of the observed rate constants at 450 nm on the concentration of NADH. The solid line shows the fit to Equation 1, yielding a dissociation constant for the Michaelis complex (reductaseTOL with NADH) of 41 ± 4 μm and a limiting rate constant of 152 ± 4 s−1 for the reduction of reductaseTOL. Error bars denote the S.D. of observed rate constants of at least four measurements per NADH concentration.
Reactivity of NAD+-ReductaseTOLCT
As the formed NAD+-reductaseTOLCT complex was observed to be stable in the stopped-flow experiment, it may also act as the physiological electron donor for ferredoxinTOL. Formation of CT complexes between flavoproteins and ligands in either oxidation state of the flavin can modulate the reactivity of the enzymes (19), and we were therefore interested in the mechanistic significance of the observed NAD+-reductaseTOLCT complex.
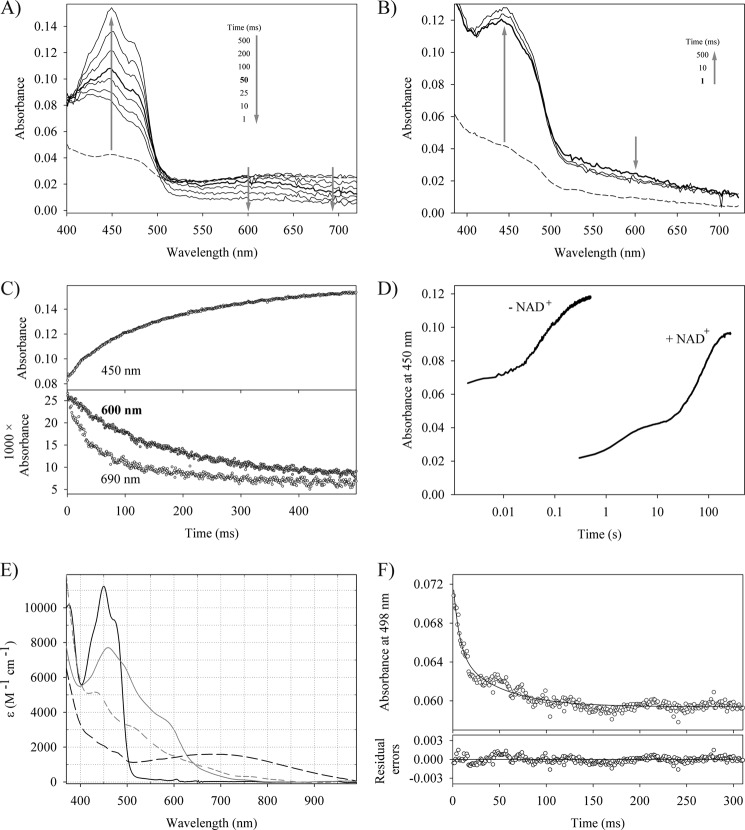
By titrating oxidized reductaseTOL with NADH under anoxic conditions, we were able to produce a dominant population of NAD+-reductaseTOLCT with some additional NAD+ and reduced NAD+-free reductaseTOL present in the assay. Although the electron transfer to ferredoxinTOL would correspond to the physiological reaction, we chose the artificial electron acceptor ferricyanide instead because the limited amounts available of ferredoxinTOL together with its spectral overlap with reductaseTOL precluded a comprehensive investigation. Single turnover kinetics initiated by mixing NAD+-reductaseTOLCT/reductaseTOL with potassium ferricyanide (potassium hexacyanoferrate(III)) revealed rapid electron transfers. The first spectrum taken about 1–2 ms after mixing shows a strongly increased absorption at 400–500 nm with a maximum at 430 nm compared with a mixture of NAD+-reductaseTOLCT with buffer only (Fig. 2A). To resolve the contributions of NAD+-reductaseTOLCT and NAD+-free reductaseTOL, we also followed the reoxidation of sodium dithionite-reduced reductaseTOL with ferricyanide (Fig. 2B). Sodium dithionite-reduced reductaseTOL is almost completely reoxidized within the dead time of the instrument by ferricyanide, and the shapes of the spectra show contributions from ferricyanide and indicate that the sodium dithionite-reduced enzyme tends to aggregate. We thus attribute the reoxidation of the mixture of NAD+-reductaseTOLCT and NAD+-free reductaseTOL within the dead time of the measurement to NAD+-free reductaseTOL alone. The NAD+-reductaseTOLCT complex in contrast was not oxidized in the dead time of the measurement as the characteristic CT band around 650–720 nm was not decreased (Fig. 2A). The subsequent decrease of the CT band (monitored at 690 nm) indicates the sequential oxidation of the NAD+-reductaseTOLCT complex via a neutral (blue) semiquinone species to the fully oxidized quinone state (Fig. 2C). Whereas the decay of the semiquinone species (monitored at 600 nm) and the formation of the quinone state occurred with the same rate, which is consistent with the second one-electron transfer step, the decay of the CT complex is faster (Fig. 2C). Analysis of the oxidative half-reaction thus indicates that NAD+-free enzyme is rapidly reoxidized by the electron acceptor (kobs > 300 s−1), whereas reoxidation of the NAD+-reductaseTOLCT is slower and results in the intermediate population of the blue semiquinone species. The separation of the phases corresponding to the two sequential electron transfers is only visible at low concentrations of ferricyanide. A tentative and limited analysis of the electron transfer between NAD+-reductaseTOLCT and ferredoxinTOL indicates comparable electron transfer rates as found for the artificial electron acceptor ferricyanide (Fig. 2, E and F).
FIGURE 2.
Oxidative half-reaction of reductaseTOL with various electron acceptors. Time-dependent spectral changes of 15 μm NADH-reduced reductaseTOL reacting with 30 μm potassium ferricyanide (A) and 8 μm dithionite-reduced reductaseTOL reacting with 20 μm potassium ferricyanide (B). The dashed line represents the spectrum of reduced reductaseTOL, and arrows indicate the direction of absorbance changes. C, time-dependent absorbance change at 450, 600, and 690 nm for the reaction shown in A. D, time-dependent change in absorption at 450 nm when reduced reductaseTOL is mixed with an air-saturated solution. The trace labeled − NAD+ shows the reoxidation of dithionite-reduced reductaseTOL, whereas that labeled + NAD+ shows the recorded trace for NADH-reduced reductaseTOL where the NAD+-reductaseTOLCT is found in solution. E, absorption spectra of oxidized reductaseTOL (solid black line), oxidized ferredoxinTOL (solid gray line), NAD+-reductaseTOLCT (dashed black line), and sodium dithionite-reduced ferredoxinTOL (dashed gray line). F, stopped-flow trace at 498 nm for the reaction of 19.8 μm reductaseTOL with 10 μm ferredoxinTOL. At this wavelength, the kinetics of the electron transfer to the iron-sulfur cluster of ferredoxinTOL is observed without significant contributions from the flavin species. The solid line shows a double exponential fit to the data, yielding observed rate constants of 153.8 and 21.4 s−1 and amplitudes of 0.0075 and 0.0056, respectively.
ReductaseTOL is part of a dioxygen-dependent multicomponent enzyme and thus has to selectively shuttle electrons to its physiological electron donor and not to the abundant dioxygen molecules. We therefore followed the oxidation of NAD+-reductaseTOLCT and compared it with the reoxidation of dithionite-reduced reductaseTOL after rapid mixing with an air-saturated buffer (Fig. 2D). An accumulation of intermediates like semiquinone species during oxidation was not observed. The reaction of dioxygen with both reduced reductaseTOL preparations was complex and cannot be interpreted in terms of a simple one- or two-step mechanism (Fig. 2D). Despite this complexity, it is obvious that dithionite-reduced reductaseTOL reacts with dioxygen about 2 orders of magnitude faster than NAD+-reductaseTOLCT (Fig. 2D).
Formation of CT complexes has been observed for several flavoproteins and is considered for most enzymes only as a catalysis-accompanying phenomenon, which is neither necessary nor beneficial for the reaction (19). In the apoptosis-inducing factor (AIF) protein, NADH binding and the subsequently slow reduction of the flavin result in the formation of an air-stable CT complex and a dimerization of the protein (21). The CT complex of AIF is ineffective in electron transfer, and CT complex formation probably enables AIF to act as a redox signaling molecule in apoptosis. For lactate monooxygenase, a change in reactivity with dioxygen upon CT complex formation has been observed (22); however, the CT complex was observed to react more, not less, rapidly with dioxygen. Being an oxygenase, lactate monooxygenase uses dioxygen as a substrate, and a more rapid reaction with dioxygen may thus contribute to catalysis. ReductaseTOL is part of an oxygenase system and thus has to function in the presence of dioxygen, but a reaction of reductaseTOL with dioxygen would waste reducing equivalents and produce superoxide or peroxide anions. The substantially reduced reactivity of NAD+-reductaseTOLCT with dioxygen compared with reduced reductaseTOL without NAD+ thus indicates that formation of the CT complex helps to control the reactivity of the reduced enzyme. In contrast to the related stable NAD+-AIF CT complex, which is ineffective in electron donation, NAD+-reductaseTOLCT still transfers its electrons to electron acceptors. Thus, formation of a stable CT complex with NAD+ likely serves under physiological conditions to reduce the risk of unwanted side reactions of the reduced reductase with dioxygen.
Structure of the NAD+-reductaseTOLCT Complex
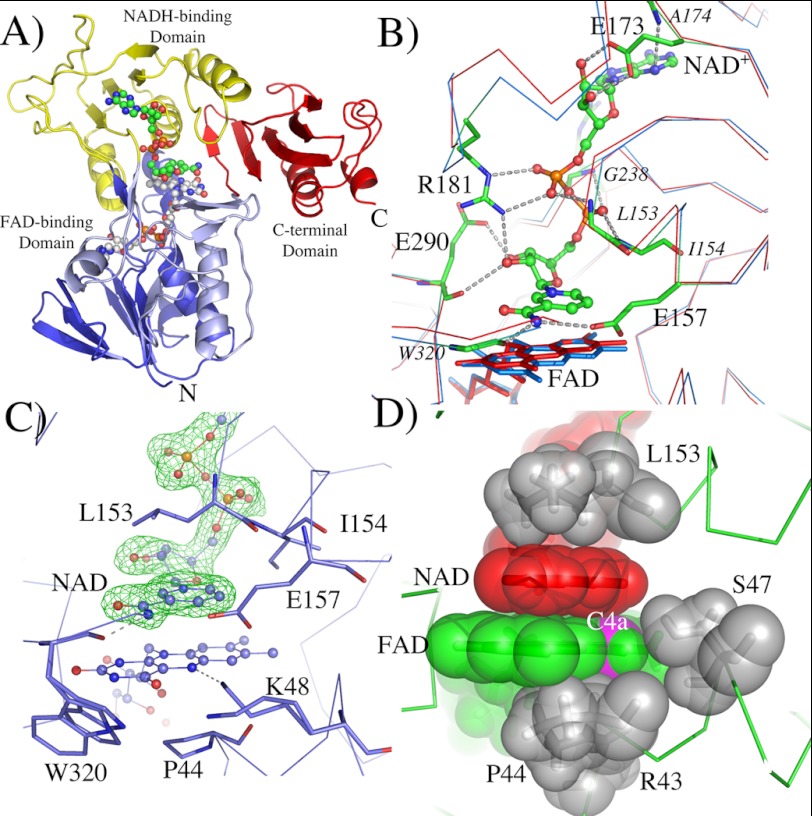
To understand how the CT complex modulates the reactivity of reductaseTOL with dioxygen, we crystallized reductaseTOL under anoxic conditions in the presence of NADH. Addition of NADH changed the color of the reductaseTOL solution used for crystallization from yellow to a faint blue, demonstrating the formation of NAD+-reductaseTOLCT with its broad absorption band at 500–720 nm (Fig. 1A). In principle, the blue color could also indicate the presence of the neutral semiquinone state of reductaseTOL; however, this would require the presence of electron acceptors, and the semiquinone state proved to be reactive during our transient kinetics. We therefore assumed that the blue color is due to the formation of the stable NAD+-reductaseTOLCT with the blue color originating from the CT band. Crystals appearing after 1 week had the same blue color as the concentrated NAD+-reductaseTOLCT and were frozen in liquid nitrogen inside the glove box in which they were grown previously. One of the blue crystals was used to collect a complete data set at synchrotron beamline BL14.2 (BESSY, Berlin, Germany). The structure was solved using Patterson search techniques with the structure of oxidized reductaseTOL as a homologous search model (Protein Data Bank code 3EF6 (4)) and was refined to 1.8-Å resolution (Table 1). The crystal structure clearly reveals the complete protein structure of reductaseTOL with its overall fold and arrangement of domains being very similar to oxidized reductaseTOL (4). As expected by the color of the crystal, additional electron density in the active site was observed, clearly showing the presence of NAD+. The NAD+ molecule is well defined in the electron density map (Fig. 3).
FIGURE 3.
Structure of the NAD+-reductaseTOL charge transfer complex. A, ribbon plot representation of NAD+ reductaseTOLCT. The N-terminal FAD-binding domain (residues 3–111) is colored in blue, the NADH-binding domain (residues 112–238) is shown in yellow, the C-terminal FAD-binding domain (residues 239–317) is depicted in light blue, and the C-terminal domain (residues 318–404) is colored in red. FAD is shown as sticks with carbon atoms in white, nitrogen atoms in blue, and oxygen atoms in red. NAD+ is also shown in stick presentation with carbon atoms in white. B, interaction of NAD+ with the protein matrix. Superimposition of oxidized reductaseTOL (red) with NAD+ reductaseTOLCT (blue) is shown. Main and side chains interacting with NAD+ are shown as sticks, and likely hydrogen bond and salt bridge interactions are indicated by dotted lines. Residues coordinating NAD+ with main chain atoms only are labeled in italic; all others are in regular font. C, NAD+ bound in the active site of reductaseTOL. An Fobs − Fcalc omit map for the bound NAD+ is shown at a contour level of 5 σ (green mesh). D, the coplanar arrangement of the nicotinamide (red) and isoalloxazine (green) ring together with residues of the protein matrix (gray) blocks access for dioxygen to the C4a carbon atom (magenta). All molecules are shown with transparent surfaces indicating their van der Waals radii.
NAD+ is bound in an extended conformation, and its nicotinamide ring is coplanar to the isoalloxazine ring and is lying above the re face of the flavin. The carboxamide group of NAD+ is near the pyrimidine ring of the flavin, whereas the pyridine ring is situated directly over the reactive “enediamine” subfunction of the flavin. The C4 atom of NAD+ is 3.2 Å from the N5 atom of the isoalloxazine and the N10-N5-C4 angle is 95°; both values are within the range typically found for flavoprotein-substrate complexes (23). The orientation of the nicotinamide in this product complex is such that in the substrate complex (NADH + oxidized reductaseTOL) the pro-S hydrogen of NADH would be directly above the N5 of the isoalloxazine. Thus, both the transient kinetics and the crystal structure indicate that the CT complex is formed immediately after hydride transfer from NADH to FAD, both reactions occur with the same rate constants (Fig. 1B), and conformational changes appear not to be necessary to form the CT complex after the hydride transfer occurs (Fig. 3).
Binding of NAD+ is accompanied by a few small conformational changes of the protein, and oxidized reductaseTOL and reduced NAD+-reductaseTOLCT can be superimposed with a root mean square deviation of Cα atoms of 0.66 Å. Although most conformational changes seem to originate from the interactions between the protein and the ADP part of NAD+ that are remote from the active site, the nicotinamide perturbs the structure in the direct vicinity of the isoalloxazine ring (Fig. 3B). Binding of nicotinamide pushed the complete isoalloxazine ring down, tilting it by 10° compared with the oxidized enzyme (Fig. 3B). The N5 atom of flavin moves 0.5 Å away from the C4 atom of nicotinamide, and the carbonyl oxygen O4 shifts by 0.7 Å to move away from the carboxamide group of NAD+. The carboxamide group of the nicotinamide is in hydrogen-bonding distance to the carboxylate group of Glu-157, thereby positioning the carboxylate 3.4 Å from the hydride donor/acceptor atom C4.
Conformational changes directly related to the reduction of the FAD are not discernible. The side chain of Lys-48 adopts two conformations of which the main conformer is in hydrogen-bonding distance to the N5 atom of FAD as has been observed in the oxidized state of reductaseTOL (4). More surprisingly, the isoalloxazine ring is completely planar in the reduced NAD+-reductaseTOLCT, which is unanticipated as reduced flavin cofactors typically show a butterfly-bent shape along the N10-N5 axis (Fig. 3, C and D) (24). The unexpected planarity of the isoalloxazine ring is most likely due to the stabilization of the π-π donor-acceptor interaction in the CT complex (19, 25).
The structure of NAD+-reductaseTOLCT is similar to that of related flavoprotein-NAD+ complexes like glutathione reductase (26, 27), the reductase component BphA4 of biphenyl dioxygenase (6, 28), and others. Notably, in the AIF, only small conformational changes occur upon reduction: a nearly perfect coplanar arrangement of nicotinamide and isoalloxazine ring and a resulting stable CT complex have been observed (21, 29). Thus, despite the different functions of AIF and reductaseTOL, their structure-reactivity relation appears to be similar. A surprisingly large number of differences are found when we compare our structure with that of the reduced (hydroquinone) state of BphA4 reported by Senda et al. (28). Despite an amino acid sequence identity of 34%, conserved active site architecture, and closely related functions in the hydroxylation of aromatic substrates, the structures of reduced BphA4 and reductaseTOL in complex with NAD+ differ considerably. Despite our co-crystallization approach, which in contrast to soaking experiments would allow for large conformational changes in solution before the crystal lattice is formed, we did not observe the redox-dependent conformational changes observed for BphA4: neither the bending of the isoalloxazine ring nor the flip of the ribityl chain of FAD, nor a conformational change of Lys-48 (corresponding to Lys-54 in BphA4), nor movement of a subdomain, termed the backrest domain, in the hydroquinone state (28).
Structure of the ReductaseTOL-FerredoxinTOL Complex
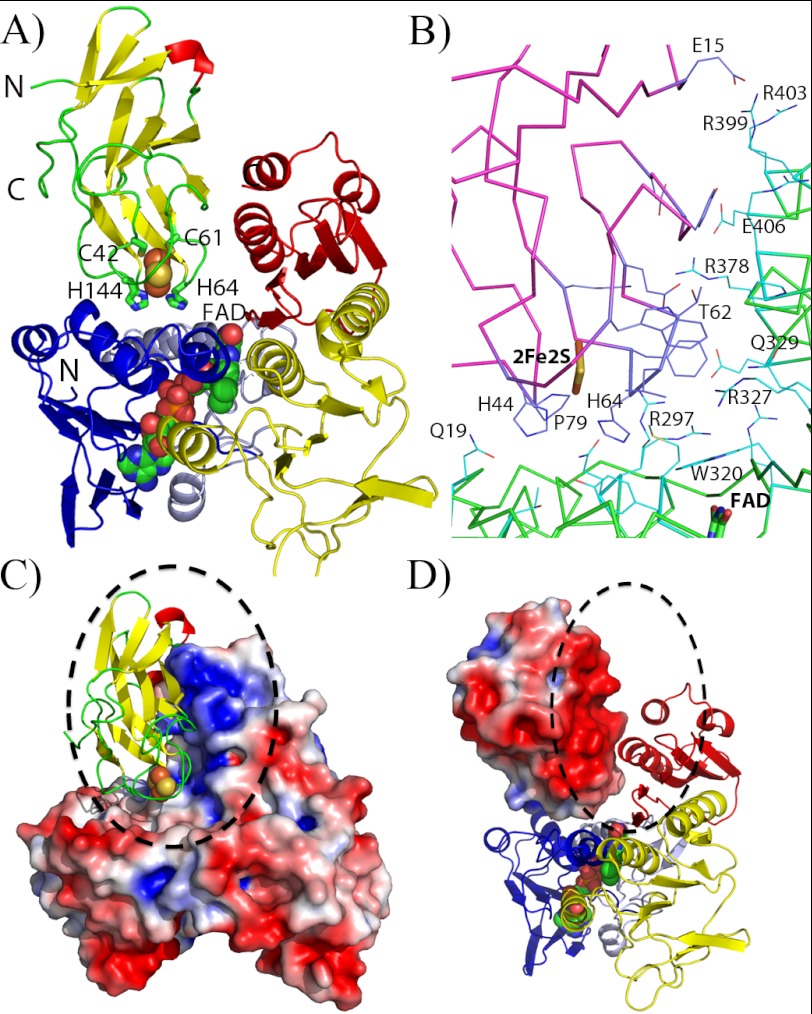
To determine the direction and distance between donor and acceptor in the physiological electron transfer of reductaseTOL, we determined the crystal structure of the complex between reductaseTOL and ferredoxinTOL. Prereduced reductaseTOL was incubated with oxidized ferredoxinTOL, and the solution was used for crystallization. Complex formation and crystallization were carried out under anoxic conditions. Orange crystals appeared after 2–3 weeks, indicating that reductaseTOL and ferredoxinTOL were both present in the crystal but in their oxidized states. Crystals diffracted x-rays to 2.4-Å resolution, and the structure of the reductaseTOL-ferredoxinTOL complex was determined by molecular replacement using the crystal structure of reductaseTOL as the homologous search model (Table 1). The presence of ferredoxinTOL in the crystal was immediately indicated after the replacement search by loose packing interactions and strong peaks in the Fobs − Fcalc difference maps, which revealed the position of the Rieske-type [2Fe-2S] cluster and its coordinating protein matrix. FerredoxinTOL was positioned in the electron density by Patterson search techniques.
The co-crystal structure of reductaseTOL-ferredoxinTOL reveals that ferredoxinTOL binds opposite to the substrate channel (Fig. 4A). The FAD-binding domain and the C-terminal domain create two faces with perpendicular orientation that are used to interact with two sides of ferredoxinTOL (Fig. 4A). Only 14% of the solvent-accessible surface area of ferredoxinTOL (759 from a total of 5,453 Å2) and 4.4% of reductaseTOL (769 from a total of 17,522 Å2) are covered upon complex formation. Salt bridge formation by Arg-378 with ferredoxinTOL is accompanied by a change of its side-chain conformation and an increased order of the C-terminal helix of reductaseTOL where Arg-378 is situated. Residues 403–406 of the C-terminal helix of reductaseTOL are not visible in the structure of oxidized reductaseTOL (Protein Data Bank code 3EF6) (4) but are clearly defined in the complex despite a lack of crystalline contacts in this area and the lower resolution of the complex structure compared with the structure of reductaseTOL (4). This small disorder-order transition is probably facilitated by the intramolecular salt bridge formed between Arg-378 and Glu-406 enabled by the conformational change of Arg-378 following complex formation (Fig. 4B).
FIGURE 4.
Structure of the reductaseTOL-ferredoxinTOL complex. A, overall structure of the reductaseTOL-ferredoxinTOL complex. The schematic representing ferredoxinTOL is colored according to secondary structure with β-sheets in yellow, α-helices in red, and loop regions in green. Residues coordinating the Rieske-type [2Fe-2S] cluster are shown as sticks and are labeled. ReductaseTOL is color-coded as in Fig. 3A with the FAD-binding domain in blue and light blue, the NADH-binding domain in yellow, and the C-terminal domain in red. B, binding interface between ferredoxinTOL(magenta ribbons; blue carbon atoms) and reductaseTOL(green ribbons; light blue carbon atoms). C, electrostatic potential mapped on the solvent-accessible surface of reductaseTOL with ferredoxinTOL displayed as in A. D, electrostatic potential mapped on the solvent-accessible surface of ferredoxinTOL with a schematic representation of reductaseTOL as in A.
Intra- as well as interprotein electron tunneling follows the same exponential rate dependence on distance (30), and short distances between electron donors and acceptors are needed or electron transfer may become rate-limiting. The shortest connection between the two cofactors is between the histidine-coordinated iron ion and the N3 atom of the isoalloxazine ring and at 11.7 Å is within a range allowing fast electron transfer between the two centers (31).
The electrostatic surface potentials of reductaseTOL and ferredoxinTOL reveal that the complex is most likely stabilized by Coulomb attraction between both proteins. The interface between the two proteins is dominated by large surface areas with opposing charges, positively charged on the side of reductaseTOL and negatively charged at the opposing side of ferredoxinTOL (Fig. 4, C and D). The Coulomb interaction between the large positively charged patch at the side of the C-terminal domain of reductaseTOL and the negatively charged patch of ferredoxinTOL is likely facilitating a long range electrostatic attraction between both proteins in the cell, allowing a rough preorientation of the electron-donating and -accepting cofactor. Small electron carrier proteins, like ferredoxinTOL, are responsible for electron shuttling between donor and acceptor sites by diffusional encounter and the formation of transient protein complexes (for reviews, see Refs. 32 and 33). It has been suggested that diffusional encounter of two electron transfer partners would drastically slow down complex formation and electron transfer if not for long range interactions and if attractive docking sites did not also allow the redox centers to be brought in close proximity (31). In the case of reductaseTOL and ferredoxinTOL, electrostatic attraction between both partners likely allows the formation of nonspecific complexes with a variety of different orientations in the ensemble as suggested to occur for several different electron transfer systems (34).
ReductaseTOL belongs to a large family of flavoproteins, which have two dinucleotide-binding domains in common (35). A shared feature of this enzyme family is a flow of electrons from a reduced pyridine cofactor via FAD by a reaction occurring on the re side of the isoalloxazine ring to a redox-active group, like disulfides or cysteine sulfenic acids, heme groups, or iron-sulfur, positioned more closely to the si side of FAD. Comparison of structures that include the electron acceptor reveals largely superimposable positions for the electron donor (FADH2) and electron acceptors both in intra- and intermolecular electron transfer, arguing for a rather strict conservation of the electron transfer pathway and its directionality in evolution (35). We observed the same directionality of electron flow in complexes of reductaseTOL with NAD+ and ferredoxinTOL.
Conclusions
The physiological role of reductaseTOL is to selectively transfer 2 reducing eq from NADH to ferredoxinTOL, resulting in the conversion of a hydride ion into a proton and two electrons. To achieve this goal, reductaseTOL needs to (a) bind NADH selectively (we determined a KD for the Michaelis complex between oxidized reductaseTOL and NADH of 41 ± 4 μm), (b) rapidly oxidize NADH (we determined a limiting rate constant for this reaction of 152 ± 4 s−1), (c) attract ferredoxinTOL to form a complex (crystal structure of ferredoxinTOL-reductaseTOL), and finally (d) transfer the electrons sequentially to two molecules of ferredoxinTOL. The last step in this sequence was investigated by us using the artificial electron acceptor ferricyanide, which allowed us to detect the formation of a neutral semiquinone state and the decay of the NAD+-reductaseTOLCT complex. Our combined kinetic and structural investigation uncovered that reductaseTOL utilizes at least two different strategies to overcome the problem of a reaction with dioxygen. 1) Reaction with dioxygen is disfavored because of the formation of a stable NAD+-reductaseTOL CT complex in which the nicotinamide ring shields the reactive C4a position of the flavin and forces the isoalloxazine ring into a planar, less reactive conformation. 2) A rapid reoxidation of the reduced flavin by electron transfer to an acceptor substantially decreases the risk of unwanted side reactions by decreasing the half-life of the reactive species. Rapid electron transfers are likely enhanced by a long range electrostatic attraction between reductaseTOL and ferredoxinTOL and short distances between the electron-donating and -accepting cofactors in the complex of the two proteins.
Acknowledgment
The staff at beamline BL14.2, BESSY (Helmholtz-Gesellschaft, Berlin, Germany) is acknowledged for help during data collection.
This work was supported by Deutsche Forschungsgemeinschaft Grant DO 785/2-2 (to H. D.).
The atomic coordinates and structure factors (codes 4EMI and 4EMJ) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- BphA4
- ferredoxin reductase component of biphenyl dioxygenase
- AIF
- apoptosis-inducing factor
- bis-Tris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- CT
- charge transfer
- TOL
- toluene.
REFERENCES
- 1. Gibson D. T., Koch J. R., Kallio R. E. (1968) Oxidative degradation of aromatic hydrocarbons by microorganisms. I. Enzymatic formation of catechol from benzene. Biochemistry 7, 2653–2662 [DOI] [PubMed] [Google Scholar]
- 2. Ferraro D. J., Gakhar L., Ramaswamy S. (2005) Rieske business: structure-function of Rieske non-heme oxygenases. Biochem. Biophys. Res. Commun. 338, 175–190 [DOI] [PubMed] [Google Scholar]
- 3. Lee K., Friemann R., Parales J. V., Gibson D. T., Ramaswamy S. (2005) Purification, crystallization and preliminary x-ray diffraction studies of the three components of the toluene 2,3-dioxygenase enzyme system. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 61, 669–672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Friemann R., Lee K., Brown E. N., Gibson D. T., Eklund H., Ramaswamy S. (2009) Structures of the multicomponent Rieske non-heme iron toluene 2,3-dioxygenase enzyme system. Acta Crystallogr. D Biol. Crystallogr. 65, 24–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Subramanian V., Liu T. N., Yeh W. K., Narro M., Gibson D. T. (1981) Purification and properties of NADH-ferredoxinTOL reductase. A component of toluene dioxygenase from Pseudomonas putida. J. Biol. Chem. 256, 2723–2730 [PubMed] [Google Scholar]
- 6. Senda T., Yamada T., Sakurai N., Kubota M., Nishizaki T., Masai E., Fukuda M., Mitsui Y. (2000) Crystal structure of NADH-dependent ferredoxin reductase component in biphenyl dioxygenase. J. Mol. Biol. 304, 397–410 [DOI] [PubMed] [Google Scholar]
- 7. Sevrioukova I. F., Li H., Poulos T. L. (2004) Crystal structure of putidaredoxin reductase from Pseudomonas putida, the final structural component of the cytochrome P450cam monooxygenase. J. Mol. Biol. 336, 889–902 [DOI] [PubMed] [Google Scholar]
- 8. Sambrook J., Russel D. (2001) Molecular Cloning: A Laboratory Manual, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 9. Subramanian V., Liu T. N., Yeh W. K., Serdar C. M., Wackett L. P., Gibson D. T. (1985) Purification and properties of ferredoxinTOL. A component of toluene dioxygenase from Pseudomonas putida F1. J. Biol. Chem. 260, 2355–2363 [PubMed] [Google Scholar]
- 10. Kabsch W. (1988) Evaluation of single-crystal x-ray diffraction data from a position-sensitive detector. J. Appl. Crystallogr. 21, 67–71 [Google Scholar]
- 11. McCoy A. J. (2007) Solving structures of protein complexes by molecular replacement with Phaser. Acta Crystallogr. D Biol. Crystallogr. 63, 32–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Emsley P., Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 13. Painter J., Merritt E. A. (2006) Optimal description of a protein structure in terms of multiple groups undergoing TLS motion. Acta Crystallogr. D Biol. Crystallogr. 62, 439–450 [DOI] [PubMed] [Google Scholar]
- 14. Adams P. D., Grosse-Kunstleve R. W., Hung L. W., Ioerger T. R., McCoy A. J., Moriarty N. W., Read R. J., Sacchettini J. C., Sauter N. K., Terwilliger T. C. (2002) PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr. 58, 1948–1954 [DOI] [PubMed] [Google Scholar]
- 15. DeLano W. L. (2002) The PyMOL Molecular Graphics System, Schrödinger, LLC, New York [Google Scholar]
- 16. Baker N. A., Sept D., Joseph S., Holst M. J., McCammon J. A. (2001) Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl. Acad. Sci. U.S.A. 98, 10037–10041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Dolinsky T. J., Nielsen J. E., McCammon J. A., Baker N. A. (2004) PDB2PQR: an automated pipeline for the setup of Poisson-Boltzmann electrostatics calculations. Nucleic Acids Res. 32, W665–W667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Frisch M. J., Trucks G. W., Schlegel H. B., Scuseria G. E., Robb M. A., Cheeseman J. R., Montgomery J. A., Jr., Vreven T., Kudin K. N., Burant J. C., Millam J. M., Iyengar S. S., Tomasi J., Barone V., Mennucci B., Cossi M., Scalmani G., Rega N., Petersson G. A., Nakatsuji H., Hada M., Ehara M., Toyota K., Fukuda R., Hasegawa J., Ishida M., Nakajima T., Honda Y., Kitao O., Nakai H., Klene M., Li X., Knox J. E., Hratchian H. P., Cross J. B., Adamo C., Jaramillo J., Gomperts R., Stratmann R. E., Yazyev O., Austin A. J., Cammi R., Pomelli C., Ochterski J. W., Ayala P. Y., Morokuma K., Voth G. A., Salvador P., Dannenberg J. J., Zakrzewski V. G., Dapprich S., Daniels A. D., Strain M. C., Farkas O., Malick D. K., Rabuck A. D., Raghavachari K., Foresman J. B., Ortiz J. V., Cui Q., Baboul A. G., Clifford S., Cioslowski J., Stefanov B. B., Liu G., Liashenko A., Piskorz P., Komaromi I., Martin R. L., Fox D. J., Keith T., Al-Laham M. A., Peng C. Y., Nanayakkara A., Challacombe M., Gill P. M. W., Johnson B., Chen W., Wong M. W., Gonzalez C., Pople J. A. (2003) Gaussian03, Gaussian, Inc., Pittsburgh, PA [Google Scholar]
- 19. Massey V., Ghisla S. (1974) Role of charge-transfer interactions in flavoprotein catalysis. Ann. N.Y. Acad. Sci. 227, 446–465 [Google Scholar]
- 20. Strickland S., Massey V. (1973) The mechanism of action of the flavoprotein melilotate hydroxylase. J. Biol. Chem. 248, 2953–2962 [PubMed] [Google Scholar]
- 21. Churbanova I. Y., Sevrioukova I. F. (2008) Redox-dependent changes in molecular properties of mitochondrial apoptosis-inducing factor. J. Biol. Chem. 283, 5622–5631 [DOI] [PubMed] [Google Scholar]
- 22. Lockridge O., Massey V., Sullivan P. A. (1972) Mechanism of action of the flavoenzyme lactate oxidase. J. Biol. Chem. 247, 8097–8106 [PubMed] [Google Scholar]
- 23. Fraaije M. W., Mattevi A. (2000) Flavoenzymes: diverse catalysts with recurrent features. Trends Biochem. Sci. 25, 126–132 [DOI] [PubMed] [Google Scholar]
- 24. Senda T., Senda M., Kimura S., Ishida T. (2009) Redox control of protein conformation in flavoproteins. Antioxid. Redox Signal. 11, 1741–1766 [DOI] [PubMed] [Google Scholar]
- 25. Sakurai T., Hosoya H. (1966) Charge-transfer complexes of nicotinamide-adenine dinucleotide analogues and flavin mononucleotide. Biochim. Biophys. Acta 112, 459–468 [DOI] [PubMed] [Google Scholar]
- 26. Karplus P. A., Schulz G. E. (1989) Substrate binding and catalysis by glutathione reductase as derived from refined enzyme: substrate crystal structures at 2 Å resolution. J. Mol. Biol. 210, 163–180 [DOI] [PubMed] [Google Scholar]
- 27. Karplus P. A., Pai E. F., Schulz G. E. (1989) A crystallographic study of the glutathione binding site of glutathione reductase at 0.3-nm resolution. Eur. J. Biochem. 178, 693–703 [DOI] [PubMed] [Google Scholar]
- 28. Senda M., Kishigami S., Kimura S., Fukuda M., Ishida T., Senda T. (2007) Molecular mechanism of the redox-dependent interaction between NADH-dependent ferredoxin reductase and Rieske-type [2Fe-2S] ferredoxin. J. Mol. Biol. 373, 382–400 [DOI] [PubMed] [Google Scholar]
- 29. Sevrioukova I. F. (2009) Redox-linked conformational dynamics in apoptosis-inducing factor. J. Mol. Biol. 390, 924–938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tezcan F. A., Crane B. R., Winkler J. R., Gray H. B. (2001) Electron tunneling in protein crystals. Proc. Natl. Acad. Sci. U.S.A. 98, 5002–5006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Page C. C., Moser C. C., Chen X., Dutton P. L. (1999) Natural engineering principles of electron tunnelling in biological oxidation-reduction. Nature 402, 47–52 [DOI] [PubMed] [Google Scholar]
- 32. Crowley P. B., Ubbink M. (2003) Close encounters of the transient kind: protein interactions in the photosynthetic redox chain investigated by NMR spectroscopy. Acc. Chem. Res. 36, 723–730 [DOI] [PubMed] [Google Scholar]
- 33. Crowley P. B., Carrondo M. A. (2004) The architecture of the binding site in redox protein complexes: implications for fast dissociation. Proteins 55, 603–612 [DOI] [PubMed] [Google Scholar]
- 34. Prudêncio M., Ubbink M. (2004) Transient complexes of redox proteins: structural and dynamic details from NMR studies. J. Mol. Recognit. 17, 524–539 [DOI] [PubMed] [Google Scholar]
- 35. Ojha S., Meng E. C., Babbitt P. C. (2007) Evolution of function in the “two dinucleotide binding domains” flavoproteins. PLoS Comput. Biol. 3, e121. [DOI] [PMC free article] [PubMed] [Google Scholar]