Background: Activated Hh signaling drives skin cancer development.
Results: Activated Hh signaling in mice increases IL-11, IL-11Ra, and STAT3 phosphorylation. STAT3 or IL-11Ra knockout reduces SmoM2-mediated carcinogenesis. The Smo agonist alone induces STAT3 phosphorylation in an IL11Ra-dependent manner.
Conclusion: The IL-11Ra/STAT3 signaling axis is required for Hh-mediated carcinogenesis.
Significance: This is the first report to link STAT3 signaling to Hh-mediated carcinogenesis.
Keywords: Carcinogenesis, Hedgehog, Signal Transduction, Skin, STAT Transcription Factor, STAT3, Stem Cells, BCC, IL-11, Smoothened
Abstract
Activation of the Hedgehog (Hh) pathway is known to drive development of basal cell carcinoma and medulloblastomas and to associate with many other types of cancer, but the exact molecular mechanisms underlying the carcinogenesis process remain elusive. We discovered that skin tumors derived from epidermal expression of oncogenic Smo, SmoM2, have elevated levels of IL-11, IL-11Rα, and STAT3 phosphorylation at Tyr705. The relevance of our data to human conditions was reflected by the fact that all human basal cell carcinomas examined have detectable STAT3 phosphorylation, mostly in keratinocytes. The functional relevance of STAT3 in Smo-mediated carcinogenesis was revealed by epidermal specific knockout of STAT3. We showed that removal of STAT3 from mouse epidermis dramatically reduced SmoM2-mediated cell proliferation, leading to a significant decrease in epidermal thickness and tumor development. We also observed a significant reduction of epidermal stem/progenitor cell population and cyclin D1 expression in mice with epidermis-specific knockout of STAT3. Our evidence indicates that STAT3 signaling activation may be mediated by the IL-11/IL-11Rα signaling axis. We showed that tumor development was reduced after induced expression of SmoM2 in IL-11Rα null mice. Similarly, neutralizing antibodies for IL-11 reduced the tumor size. In two Hh-responsive cell lines, ES14 and C3H10T1/2, we found that addition of Smo agonist purmorphamine is sufficient to induce STAT3 phosphorylation at Tyr705, but this effect was abolished after IL-11Rα down-regulation by shRNAs. Taken together, our results support an important role of the IL-11Rα/STAT3 signaling axis for Hh signaling-mediated signaling and carcinogenesis.
Introduction
The Hh pathway plays an important role in cell differentiation, tissue polarity, cell proliferation, and carcinogenesis (1–5). The seven transmembrane domain-containing protein smoothened (SMO)3 serves as the key player for signal transduction of this pathway, whose function is inhibited by another transmembrane protein, Patched (PTC), in the absence of Hh ligands. Binding of Hh to its receptor PTC releases this inhibition, allowing SMO to signal downstream, leading to formation of active forms of Gli transcription factors. Gli molecules can regulate target gene expression by direct association with a specific sequence located at the promoter region of the target genes (6).
Most BCCs and a subset of medulloblastomas have constitutive activation of Hh signaling because of genetic mutations of PTCH1, SMO, or Su(Fu) (7–16). Mice with tissue-specific expression of oncogenic SMO (6) or Ptch1+/− (17) develop spontaneous BCC-like tumors (11) and medulloblastomas. Thus, activation of Hh signaling can drive the development of these tumors. Furthermore, Hh signaling activation has been reported in a variety of human cancers, including gastrointestinal, lung, prostate, and breast cancers (reviewed in Refs. 1, 5, 18, 19).
Although the biological relevance of Hh signaling to human cancer is very clear, the molecular basis by which Hh signaling induces cell proliferation and carcinogenesis remains largely elusive. Although several downstream mediators for BCC development have been described, including BCL2 (20, 21), PDGFRα (22), CD95 (12, 23), and BMI1 (24), we do not know the actual contribution of these mediators to Hh-induced BCC carcinogenesis. Increasing evidence indicate the importance of signaling cross-talks between Hh signaling and other pathways during carcinogenesis, such as PI3K-AKT, p53, wnt, and epidermal growth factor receptor signaling (reviewed in Refs. 1, 5). It is not clear how these cross-talks contribute to tumor initiation or tumor progression.
To study the molecular mechanisms underlying BCC development, we compared gene expression between SmoM2-mediated tumors and normal skin keratinocytes and discovered a significant increase in expression of IL-11 and IL-11Rα, upstream stimulators of STAT3 signaling. As an important proto-oncogene, STAT3 is known to regulate hair follicle stem cell population and skin wound healing and is critical for skin carcinogenesis of SCCs. To understand the significance of STAT3 signaling for BCCs, we used tissue-specific gene knockout of STAT3 or down-regulation of its upstream activator IL-11Rα in the mouse model of BCCs. By examining the effects of these genetic deficiencies on tumor development, we revealed a critical role of the IL-11Rα/STAT3 signaling axis for Hh-mediated carcinogenesis.
EXPERIMENTAL PROCEDURES
Mice
K14-cre mice (25) were obtained from the Emice program. K14creER (26), IL11Ra knockout (27), and R26-SmoM2YFP mice (28) were purchased from The Jackson Laboratories (Bar Harbor, Maine). Mice were maintained and mated under pathogen-free husbandry conditions. Mice with conditional knockout of STAT3, generated as described previously, were generously provided by Dr. Xin-Yuan Fu (29). Gli1 null mice were provided by Alex Joyner (30). To obtain a Gli1 null background in K14cre+SmoM2+ mice, Gli1 null mice were mated with K14cre and R26-SmoM2YFP mice separately. The resulting Gli1+/− mice with K14cre+ or SmoM2+ were mated to obtain K14cre+SmoM2+Gli1−/− mice. To obtain IL-11Ra null mice with K14creER+ and SmoM2YFP+, we first generated IL11Ra+/−/K14creER+/SmoM2YFP+ mice, and these mice were then mated to each other to obtain the right genotypes (IL11Ra+/+/K14creER+/SmoM2YFP+, IL11Ra+/−/K14creER+/SmoM2YFP+, and IL11Ra−/−/K14creER+/SmoM2YFP+). The off-spring from the mating was screened using PCR to determine their transgenic status according to the instruction reported previously. SmoM2 expression in these mice was induced by oral administration of tamoxifen according to a protocol published previously (26). Neutralizing antibodies to IL-11 (purchased from R&D Systems) or the control IgG were injected subcutaneously into the site with tumors (5 μg in 10 μl of PBS) twice a week for 2 weeks. Five mice per group on average were used in this study. All animal studies were approved by Institutional Animal Care and Use Committee at The Indiana University.
Histology and Microscopic BCC Analysis
Skin tissues were collected after each experiment. Half of the tissue was frozen and stored at −80 °C. The other half was fixed in formalin overnight, paraffin-embedded, sectioned at 5 μm, and stained with H&E. Eight epidermal areas in each mouse were randomly chosen in each H&E-stained slide (12) to measure the ratio of tumor area over the total tissue area with ImagineJ software. The average number of mice in each group was five.
Epidermis Separation and Single Cell Isolation
Mouse skins were soaked in dispase® solution (5 mg/ml in PBS) for 3 h at 37 °C to remove the epidermis from the skin with forceps. One part of the epidermis was used for total RNA extraction with the Ambion RNA extract kit and the rest for single cell isolation. Epidermal single cells were obtained by digestion of epidermis with collagenase IV for 2 h at 37 °C. Expression of Hh target genes was examined by PCR, as reported previously (31).
Immunofluorescent and Immunohistochemistry Staining
Mouse skin tissues were embedded in paraffin (for paraffin section) or optimal cutting temperature. Tissue sections of 5-μm thickness were prepared for immunohistochemistry using a biotin-based staining kit from DAKO, as described previously (12). Specific antibodies to pSTAT3-Y70 and Cyclin D1 were purchased from Cell Signaling Technology, Ki67 from ABCam, and Keratin 14 from Covance. For immunofluorescent staining, slides were first fixed in 4% paraformaldehyde at room temperature for 30 min. After blocking with horse serum, tissue sections were incubated with specific antibodies for 3 h, followed by fluorescence-labeling secondary antibodies for 1 h at room temperature. Thorough washing was performed between antibody incubations to reduce nonspecific staining. DAPI staining was used to stain nucleus.
RT-PCR and Real-time PCR
Total RNA was isolated from the tissues using TRIzol reagent (Sigma) according to the instructions of the manufacturer. 1 μg of total RNA was reverse-transcribed into cDNAs using the first-strand synthesis kit (Roche). We performed real-time RT-PCR with a procedure reported previously (3).
EdU Labeling and Flow Cytometry
5-ethynyl-2′-deoxyuridine (EdU) labeling was carried out 14 h before animal sacrifice by intraperitoneal injection of EdU solution according to the instructions of the manufacturer (32). Single cells from epidermis were subjected to antibody staining to recognize CD11b, Gr1, CD34, and CD49F. Antibodies with fluorescent labeling were purchased from eBioscience and were incubated with cells for 30 min on ice (with 1:200 dilutions). Cells were further incubated with DAPI just before flow cytometry analysis. DAPI-negative cells (live cells) were gated for expression analysis of specific cell surface markers.
Cell Culture and ShRNA Expression
C3H10T1/2 and ES14 cells were cultured as described previously (33). ShRNAs to IL-11Rα were purchased from Sigma and were introduced into cells via lentivirus-mediated gene expression. Of the five ShRNAs tested, two independent ShRNAs were shown to reduce expression of IL-11Rα and were used in this study. To stimulate Hh signaling activation, the Smo agonist purmorphamine (at 5 μm) was added to the culture medium. Protocols for motor neuron differentiation and induction of alkaline phosphatase have been described previously (33).
Western Blot Analysis
Epidermis was first lysed with a protein loading buffer in ultrasound bath for 5 min. Specific antibodies to STAT3, pSTAT3-Y705, and cyclin D1 were purchased from Cell Signaling Technology, Inc. Proteins were detected according to a procedure reported previously (34).
Statistical Analyses
were performed using Student's t test (two-tailed) to compare the results, with p values of < 0.05 indicating a statistically significant difference.
RESULTS
Activation of STAT3 Signaling in SmoM2-mediated Skin Tumors
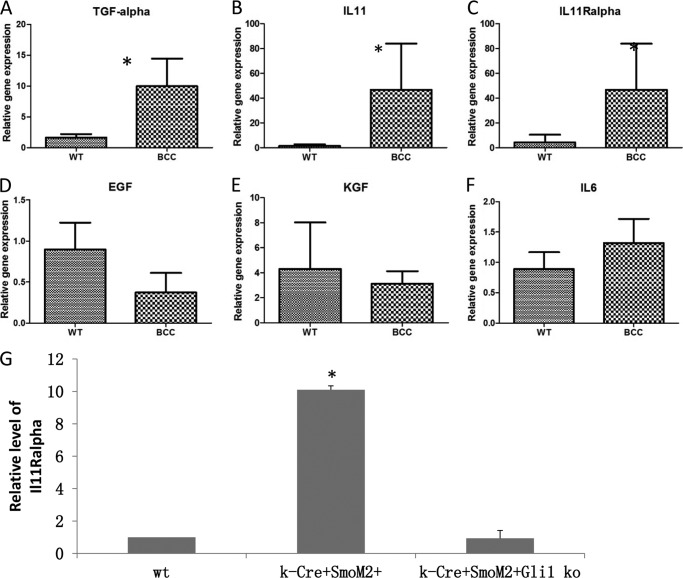
Almost all basal cell carcinomas contain activated Hh signaling, resulting frequently from inactivated mutation of tumor suppressor gene PTCH1 or gain-of-function mutation of proto-oncogene Smo. Skin-specific expression of mutant Smo, SmoM2, via mating of R26-SmoM2YFP mice (28) with K14-cre mice (25) exhibits phenotypes of BCCs (33) and is regarded as an important model for studying Hh-mediated carcinogenesis. To further understand molecular bases of Hh-mediated carcinogenesis, we performed gene expression profiling of SmoM2YFP-expressing keratinocytes using Affymetrix arrays and revealed changes in several cytokine molecules that are known to associate with STAT3 signaling (33). To confirm the data from the gene expression arrays, we first used real-time PCR to detect expression of STAT3 signaling activators in epidermis with or without SmoM2YFP expression. As expected, we detected activation of the Hh pathway, as indicated by elevated expression of the Hh target gene Gli1 in mice with SmoM2YFP expression (33) (supplemental Fig. 1). In addition, expression of several STAT3 signaling activators, TGFα, IL-11Rα, and IL-11, was elevated in epidermis with SmoM2YFP expression (Fig. 1, A–C). EGF, KGF, and IL-6, other putative STAT3 activators, were not significantly changed by SmoM2 expression (Fig. 1, D–F). The level of IL-11Rα expression, not IL-11 and TGFα, was dramatically reduced in keratinocytes after knockout of the downstream transcription factor Gli1 (Fig. 1G), suggesting that IL-11Rα may be regulated by Hh signaling. These results indicate that activation of Hh signaling in mouse epidermis mediates activation of STAT3 signaling.
FIGURE 1.
Expression of growth factors and cytokines in SmoM2-expressing skin tumors. Real-time PCR was used to detect the expression of cytokines and growth factors in skin tumors and normal epidermis. Expression of several STAT3 activators were examined, including TGFα (A), IL-11 (B), IL-11Rα (C), EGF (D), KGF (E), and IL-6 (F). Fold changes in SmoM2 expressing epidermis (BCC) in comparison with normal epidermis (WT, no SmoM2 expression, as 1) was shown. *, p < 0.05. Expression of TGFα, IL-11, and IL-11Rα was further examined in keratinocytes with Gli1 knockout to determine the role of Gli transcription factors in the gene expression regulation (G shows the data on IL-11Rα).
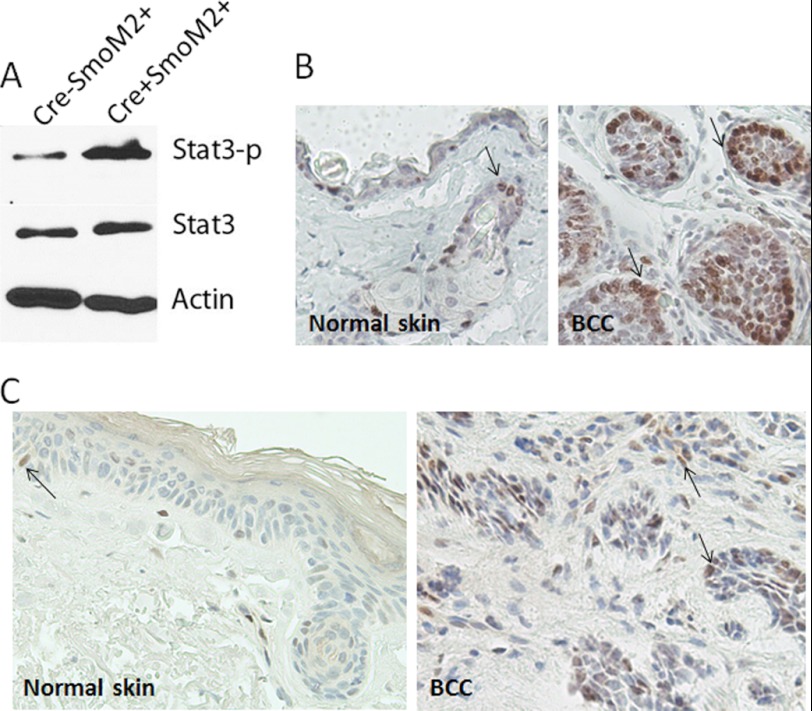
Phosphorylation at a critical tyrosine residue, Tyr705, induces STAT3 dimerization through phosphotyrosine-SH2 domain interaction, resulting in its nuclear localization and target gene expression (35). We examined whether the level of STAT3 phosphorylation is induced in epidermis by SmoM2 expression. Western blot analysis revealed a higher level of STAT3 phosphorylation in epidermis of K14cre+R26-SmoM2YFP+ mice, not in K14cre−R26-SmoM2YFP+ mice in which SmoM2YFP was not expressed (Fig. 2A). We did not observe a significant change in the total STAT3 protein, indicating that SmoM2 expression in epidermis can increase STAT3 phosphorylation. To confirm the Western blotting data, we detected STAT3 phosphorylation by immunohistochemistry. As shown in Fig. 2B, STAT3 phosphorylation at Tyr705 was seen mostly in the tumor compartment in mouse skin. We noticed that strong staining of STAT3 phosphorylation was often observed in the periphery of the tumor, not in the center of the tumor. In normal skin, some staining in hair follicles and basal cells were also observed (Fig. 2B, left panel). To test whether our results are relevant to human conditions, we performed immunohistochemistry in human BCCs and found STAT3 phosphorylation in all specimens examined (n = 25, Fig. 2C). The phospho-STAT3 positivity in human normal skin was not seen frequently. Taken together, our data indicate that activation of STAT3 signaling is commonly observed in Hh signaling-mediated BCCs of mice and humans.
FIGURE 2.
Elevated level of STAT3 phosphorylation in SmoM2-mediated BCCs of mice and humans. Specific antibodies to pSTAT3-Y705 were used to detect STAT3 activation in mouse epidermis by Western blotting (A) and immunohistochemistry (B and C) according to the protocol of the manufacturer. The epidermis was separated from the dermis by merging the skin in dispase® (5 mg/ml in PBS) for 2 h at 37 °C. The protein loading controls were endogenous STAT3 and β-actin. The negative control for immunohistochemistry was one section without primary antibodies. Positive staining is shown in brown (arrows). Tissues were counterstained with hematoxylin (blue). Positive staining of pSTAT3-Y705 was seen in the nucleus.
STAT3 Knockout in Epidermis Significantly Inhibits SmoM2-induced Skin Carcinogenesis
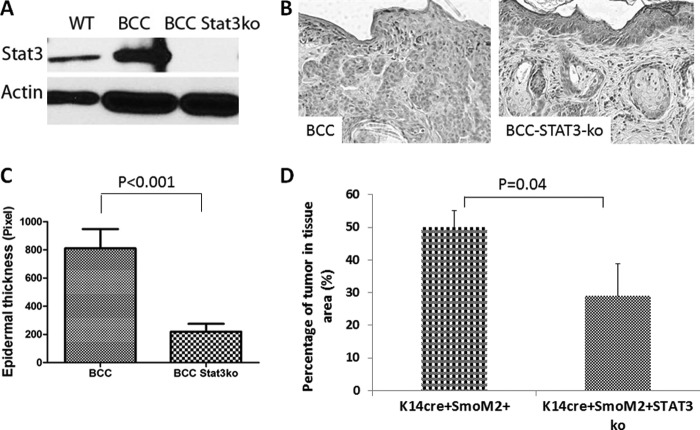
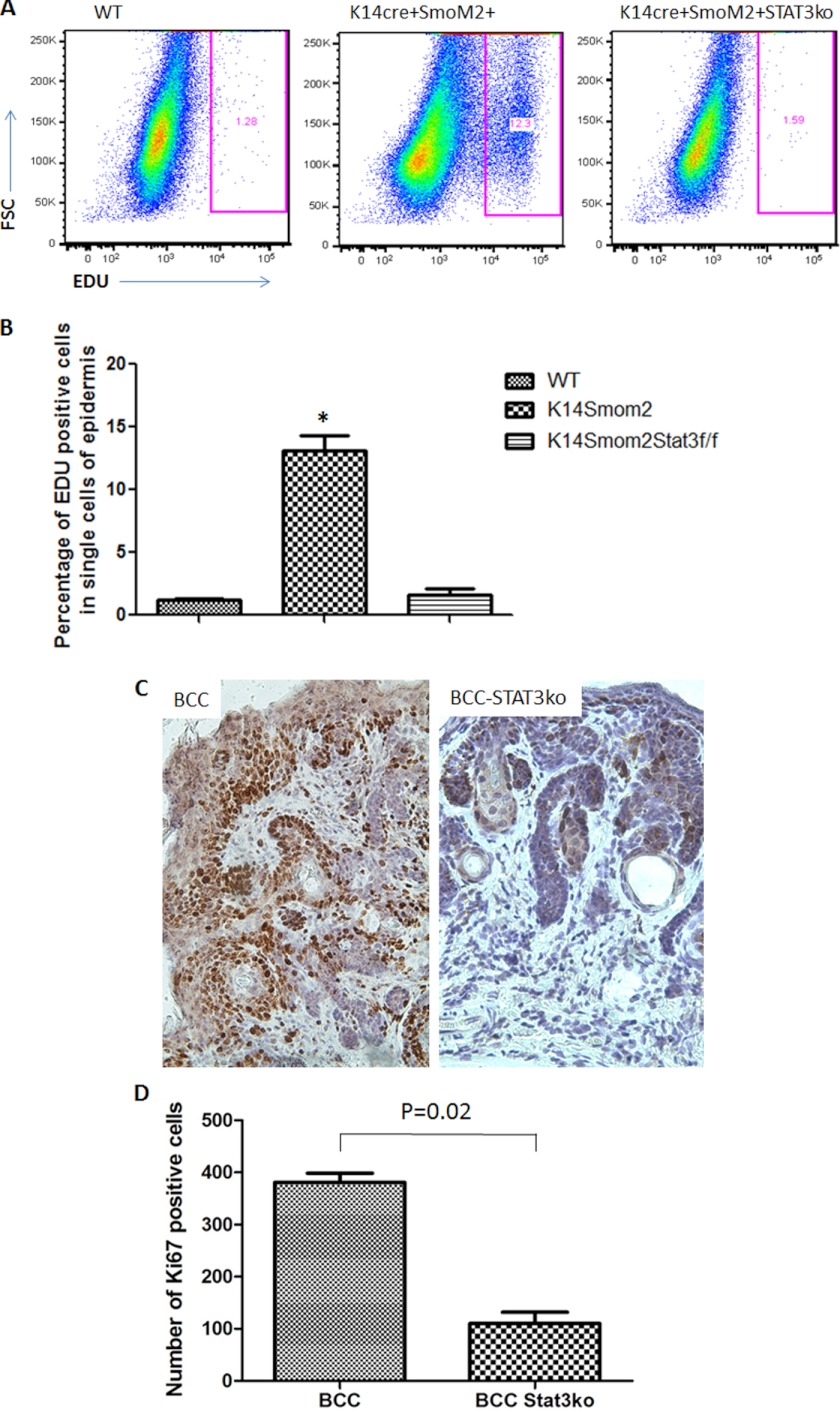
To demonstrate the significance of STAT3 signaling for Hh signaling-mediated tumors, we generated mice with keratin 14 promoter-driven STAT3 knockout in our mouse model of BCCs. We predict that if STAT3 signaling is important for SmoM2-mediated tumor development, deletion of STAT3 in the epidermis should reduce tumor formation in K14cre/R26-SmoM2YFP mice. Deletion of STAT3 was confirmed in the skin epidermis by Western blotting (Fig. 3A) and immunohistochemistry (supplemental Fig. 2). In the absence of SmoM2 expression, K14Cre-mediated STAT3 knockout had no visible phenotypes, suggesting STAT3 knockout does not affect the viability of mice, which was consistent with a previous study (36). Skin histology of mice with or without STAT3 in K14cre+SmoM2YFP+ mice was further examined. We found that SmoM2-expressing mice without STAT3 had a 70% reduction in skin thickness (Fig. 3, B and C). The percentage of tumor area in the epidermis was also reduced from 50% to 30% (Fig. 3D). To examine the cellular effects of STAT3 in mouse skin, we first examined cell proliferation following EDU labeling (32). EDU labeling was increased in K14cre+SmoM2YFP+ mice but was reduced to the basal level after STAT3 knockout (Fig. 4, A and B). Expression of Ki-67, another marker for cell proliferation, was also reduced in STAT3 knockout tumor specimens (Fig. 4, C and D). However, there were no significant changes in the level of active caspase 3, a marker for cell apoptosis in skin tissues without STAT3 (data not shown). These results indicate that STAT3 ablation in epidermis significantly reduces SmoM2-mediated cell proliferation and tumor development.
FIGURE 3.
Epidermal STAT3 knockout and its effects in SmoM2-mediated carcinogenesis. STAT3 knockout in epidermis was achieved through crossing K14cre+/R26-SmoM2YFP+ mice with STAT3f/f mice, followed by additional crossing among the F1 mice. Expression of STAT3 was examined by Western blotting (A) or immunohistochemistry (supplemental Fig. 2). The wild-type control in Western blotting was from K14cre−SmoM2YFP+STAT3f/f mice (A). B, typical H&E staining images from skin of K14cre+/SmoM2YFP+/STAT3wt (BCC) and K14cre+/SmoM2YFP+STAT3f/f (BCC-STAT3-ko) mice. C, data from the ImageJ analysis of epidermis thickness of the two genotypes described in B. D, the percentage of tumor areas in the skin tissue from mice with BCCs with or without epidermal STAT3 knockout. Five or more mice were used in each group. p < 0.05 was regarded as statistically significant.
FIGURE 4.
The effect of STAT3 knockout on cell proliferation in SmoM2-expressing epidermis. We compared K14cre+/SmoM2YFP+/STAT3wt mice with K14cre+/SmoM2YFP+STAT3f/f mice for cell proliferation in epidermis by EdU labeling and Ki-67 staining. EdU labeling was performed 14 h before animal sacrifice, and positive cells were analyzed by flow cytometry (A). B, data summary from all mice from each group. The percentage of EdU-positive cells in tumor-bearing K14cre+/SmoM2YFP+/STAT3wt mice (K14creSmom2) compared with that from K14cre+/SmoM2YFP+STAT3f/f mice (K14creSmom2STAT3f/f) (p < 0.0001). Immunohistochemistry was performed with specific antibodies to Ki-67 (C), and the number of Ki-67 positive cells/field (×200, one field = 0.9 mm2) was counted under the microscope. The number of positive cells shown in D was an average from five mice (eight fields per mouse). *, p < 0.05.
Alteration of Epidermal Stem/Progenitor Cell Population following STAT3 Knockout
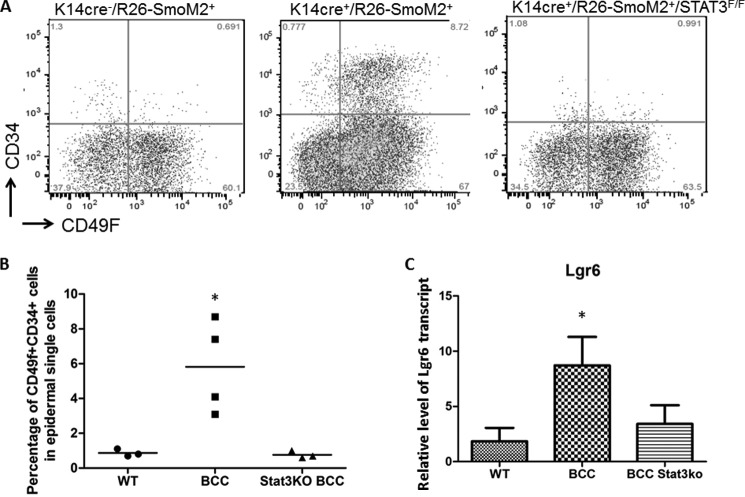
Like many types of cancer, skin cancer BCC is composed of many cell types: epidermal keratinocytes, stem/progenitor cells, fibroblasts, a variety of T cells, and myeloid-derived cells. Using flow cytometry, we detected a significant increase in the epidermal stem/progenitor cell population (indicated by expression of CD34/CD49F) (37, 38) as well as in myeloid-derived cells (CD11b+, Gr1+) in K14cre+SmoM2YFP+ mice (39) (Fig. 5, A and B, and supplemental Fig. 3). We did not observe significant changes in other cell populations. As indicated in Fig. 5, A and B, the population of CD34/CD49F-positive cells, the putative epidermal stem/progenitor cells, was increased significantly in SmoM2-expressing epidermis but was decreased in mice without epidermal STAT3. This effect appears to be STAT3-specific because we did not observe alteration of the Hh target gene Gli1 after STAT3 knockout (supplemental Fig. 1). Furthermore, expression of the Lgr6 transcript, another marker for BCC stem/progenitor cells (38), was increased in epidermis of K14cre+SmoM2YFP+ mice but reduced to the basal level in K14cre+R26-SmoM2YFP+/Stat3f/f mice, as detected by real-time PCR (Fig. 5C). Together, these results indicate that SmoM2 expression causes an increase of epidermal stem/progenitor cell population in a STAT3 signaling-dependent manner. In contrast, no changes in myeloid-derived suppressor cell population were observed in mice following STAT3 knockout (supplemental Fig. 3).
FIGURE 5.
The effect of STAT3 ablation on expression of epidermal stem/progenitor cell markers. Of the several markers for epidermal stem/progenitor cells, we obtained a significant increase in cell surface expression of CD34 and CD49F (α 6 integrin) and Lgr6 expression in SmoM2-derived tumor tissues. Single cells from epidermis were isolated to detect CD34 and CD49F expression. As indicated in A, cell surface expression of CD34 and CD49F was increased in epidermal single cells of Cre+SmoM2YFP+ mice. B, summary of our data on CD34 and CD49F expression. Each dot represents the result from one mouse. C, real-time PCR analysis of the Lgr6 transcript. STAT3 deficiency did not change the level of the Hh target gene Gli1 (supplemental Fig. 1). *, p < 0.05.
Furthermore, STAT3 knockout in K14cre+SmoM2YFP+ mice increased the lifespan of tumor-bearing mice. The lifespan of K14cre+SmoM2YFP+ mice is averaged at 12 weeks, whereas mice with K14cre+SmoM2YFP+STAT3f/f lived more than 20 weeks on average (p value< 0.001, supplemental Fig. 4).
Taken together, these results indicate an important role of STAT3 signaling in regulation of the epidermal stem/progenitor cell population during development of Hh signaling-mediated tumors.
STAT3 Regulates Expression of Cyclin D1 in SmoM2-induced Skin Tumors
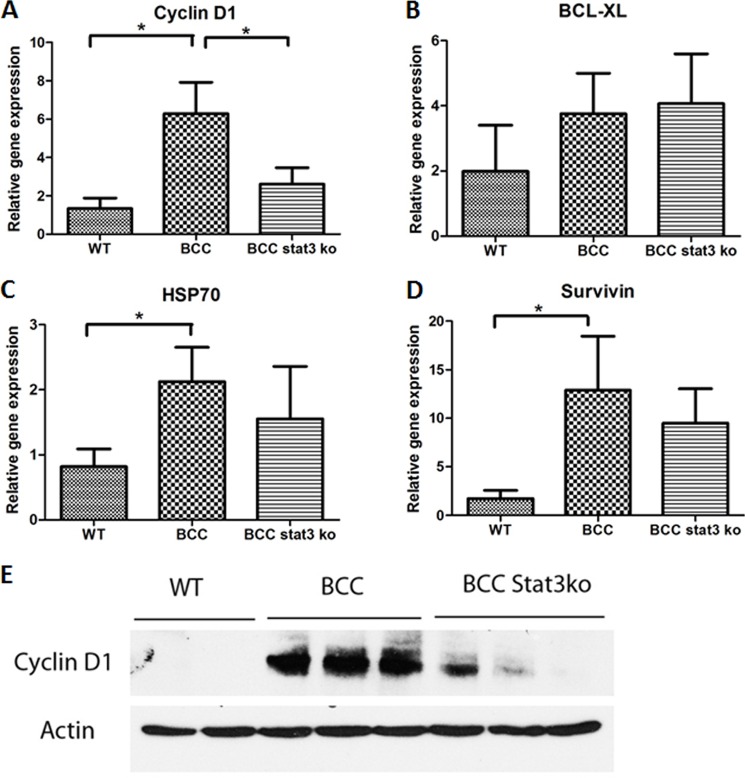
As a STAT3 target gene, cyclin D1 is known to mediate cell proliferation (40). We noticed that elevated expression of cyclin D1 is associated with EDU labeling and Ki-67 expression, suggesting that cyclin D1 may be an important factor driving cell proliferation in SmoM2-mediated carcinogenesis. We examined whether STAT3 signaling is responsible for cyclin D1 expression in epidermis. We assessed expression of several STAT3 target genes with real-time PCR. As indicated in Fig. 6A, cyclin D1 was induced in the tumor of K14cre+SmoM2YFP+ mice, as reported previously by others (41), but was down-regulated in skin without epidermal STAT3. The data from real-time PCR were further confirmed by Western blot analysis (Fig. 6E). Two other STAT3 target genes, survivin and HSP70, were also induced by SmoM2 expression. However, STAT3 knockout did not completely change their expression (Fig. 6, C and D), suggesting other mechanisms for regulating surviving and HSP70 expression. BCL-XL, another gene that can be regulated by STAT3, was not changed in epidermis by SmoM2 expression (Fig. 6B). This is not unexpected because expression of these genes is regulated by many other factors in addition to STAT3 signaling (42–44). To be sure that the effect of epidermal specific STAT3 knockout was not due to indirect regulation of Hh signaling, we compared skin tissues from K14cre+SmoM2+ STAT3+/+ mice with K14cre+ SmoM2+STAT3F/F mice on Hh target gene expression and found no significant changes in Gli1 (using the K14 transcript as the internal control (supplemental Fig. 1). These results indicate that cyclin D1-mediated cell proliferation in SmoM2-mediated carcinogenesis is regulated largely by STAT3 signaling.
FIGURE 6.
Regulation of Cyclin D1 expression by STAT3 signaling in SmoM2-mediated carcinogenesis. Expression of several putative STAT3 target genes in tumor-bearing epidermis was assessed by real-time PCR. These genes include cyclin D1 (A), BCL-XL (B), HSP70 (C), and Survivin (D). Following STAT3 ablation in epidermis, SmoM2-induced expression of cyclin D1 was examined at the protein level, which was detected by Western blotting (E). Please note that STAT3 ablation in epidermis did not completely wipe out all cyclin D1 expression. To rule out the possibility that the effect of STAT3 knockout was due to nonspecific regulation of Hh signaling, we detected Hh target gene expression from skin tissues of K14cre-Rosa26-SmoM2/STAT3+/+ mice or K14cre-Rosa26-SmoM2/STAT3flp/flp mice, with the K14 transcript as the internal control (supplemental Fig. 1), and found that Gli1 expression was not significantly altered by STAT3 depletion. *, p < 0.05.
Effects of IL-11Rα and IL-11 Down-regulation on SmoM2-mediated Tumor Formation
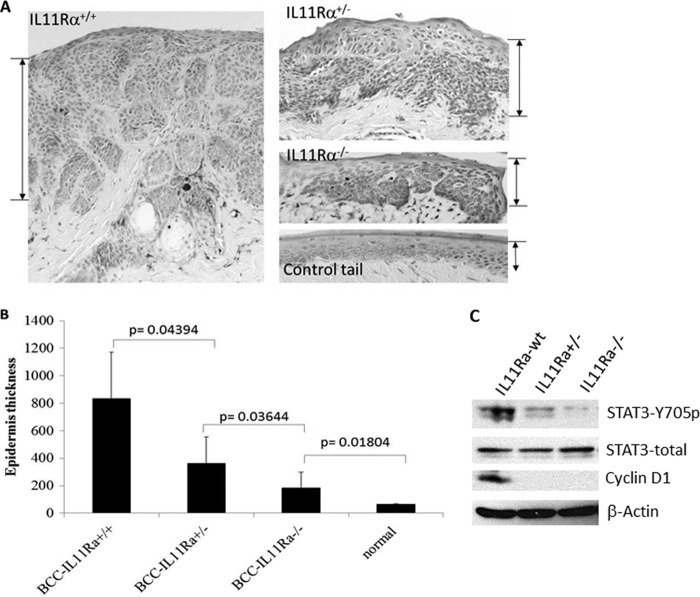
If the IL-11/IL-11Rα signaling axis is responsible for STAT3 activation during BCC development, down-regulation of IL-11Rα or IL-11 should reduce SmoM2-mediated tumor formation. We generated mice with epidermis-specific SmoM2 expression in IL-11Rα+/− or −/− background. As shown in Fig. 7, A and B, skin thickness was reduced over 50% in IL-11Rα+/− and over 70% in IL-11Rα−/− mice. The level of STAT3 phosphorylation was barely detectable in IL-11−/− mice (Fig. 7C). Even in IL-11+/− mice, the level of STAT3 phosphorylation was reduced significantly. Consistent with the data from epidermis-specific knockout of STAT3, cell proliferation, as indicated by Ki-67 staining, was reduced significantly in IL-11Rα−/− mice (supplemental Fig. 5, A and B). We also noticed a significant reduction of Lgr6 expression, a marker for epidermal stem cell marker, in IL-11Rα−/−/SmoM2+/K14creER+ mice (supplemental Fig. 5C). Cyclin D1 in tumors of IL-11Rα+/− or IL-11Rα−/− mice was also markedly reduced (Fig. 7C). These results indicate that IL-11Rα is a critical factor in mediating STAT3 activation and tumor development in this BCC model.
FIGURE 7.
Effects of the defective IL-11/IL-11Rα signaling axis on SmoM2-mediated carcinogenesis. A, typical H&E staining images of skin specimens from tumor-bearing mice with IL-11Rα+/+, IL-11Rα+/−, or IL-11Rα−/−, with the thickness shown by the bars. B, average epidermal thickness, a measure for BCC tumor size, from mice with different genotypes. p < 0.05 indicates statistically significant. C, expression of phosphorylated STAT3, cyclin D1, total STAT3, and β-actin in skin tissues from different genotypes.
IL-11Rα serves as the specific cytokine receptor for IL-11 by forming a hexamer complex with gp130β and IL-11 to activate the downstream effector STAT3 (45). If the IL-11/IL-11Rα signaling axis is the major contributor for STAT3 activation, we predicted that depleting IL-11 in the tumor microenvironment should reduce tumor size. To test this hypothesis, we treated skin lesions with neutralizing antibodies for IL-11 via subcutaneous injection (5 μg in 10 μl of PBS). Mouse IgG proteins were used as control. We found that although skin lesions with control IgG treatment had ∼50% tumor/tissue area, treatment with IL-11 neutralizing antibodies reduced the tumor to ∼35% (supplemental Fig. 6, A and B). Further analyses indicated that skin areas injected with IL-11 neutralizing antibodies had a reduced level of STAT3 phosphorylation (supplemental Fig. 6C). These results are consistent with our data from epidermis-specific knockout of STAT3 or IL-11Rα knockout. Thus, it appears that interruption of any part of the IL-11Rα/Stat3 signaling axis is sufficient to inhibit SmoM2-mediated carcinogenesis.
Reducing IL-11Rα Expression Prevents Smo-mediated STAT3 Phosphorylation
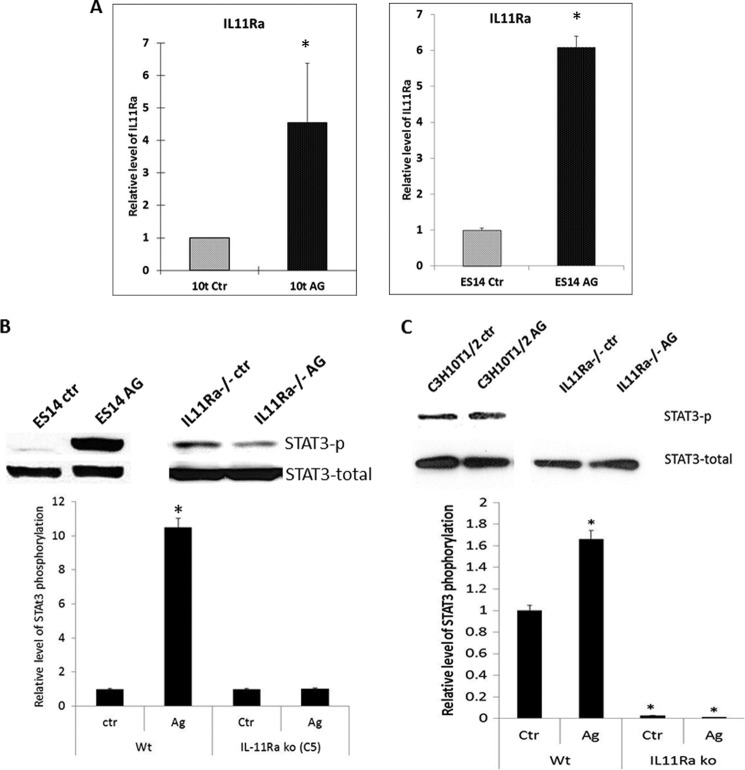
To demonstrate direct regulation of STAT3 activity by Hh signaling, we chose two Hh-responsive cell lines: mouse ES cell line ES14 and mesenchymal stem cell line C3H10T1/2. We showed previously that addition of Smo agonist purmorphamine in these two cell lines induces Hh target gene expression (33). As shown in Fig. 8A, we also observed elevated expression of IL-11Rα in response to purmorphamine treatment in both cell lines. Furthermore, we detected an increase in the level of STAT3 phosphorylation (Fig. 8, B and C, top left panels). To test whether IL-11Rα mediates Smo-induced STAT3 signaling activation, we used specific shRNAs to down-regulate IL-11Rα expression. After treatment of these cells with purmorphamine, we detected STAT3 phosphorylation. If IL-11Rα is the major factor for Hh-mediated STAT3 phosphorylation, down-regulation of IL-11Rα should be able to prevent purmorphamine-induced STAT3 phosphorylation. As shown in Fig. 8, B and C, top right panels, we found that STAT3 phosphorylation was not induced by purmorphamine in cells with down-regulation of IL-11Rα. The effect of purmorphamine was more dramatic in ES14 cells than C3H10T1/2 cells. In C3H10T1/2 cells the effect of purmorphamine was not as dramatic, but down-regulation of IL-11Rα significantly reduced the basal level of STAT3 phosphorylation. The high basal level of IL-11/IL-11Rα expression in C3H10T1/2 cells (data not shown) may be responsible for this effect. IL-11Rα down-regulation abolished Hh-mediated motor neuron differentiation and alkaline phosphatase expression (a marker for osteoblast differentiation (supplemental Fig. 7).
FIGURE 8.
Requirement of IL-11Rα for Hh signaling-induced STAT3 phosphorylation. A, expression of IL-11Rα in ES14 and C3H10T1/2 (10t) cells in presence of purmorphamine (AG). B, STAT3 phosphorylation in Hh-responsive mouse ES14 cells following treatment with Smo agonist AG. C, STAT3 phosphorylation in C3H10T1/2 cells in the presence or absence of AG. In B and C, the top panels show the Western blotting images, and the bottom panels show the analysis of the images (averaged from three independent experiments). *, p < 0.05. Note the low levels of STAT3 phosphorylation after IL-11Rα down-regulation (IL11Ra−/−).
In summary, we found that Hh signaling activation induces STAT3 phosphorylation through regulation of IL-11Rα in mouse models of BCC and in two Hh-responsive cell lines. We demonstrated that disruption of the IL-11Rα/STAT3 signaling axis either by epidermis-specific knockout of STAT3, IL-11Rα knockout, or subcutaneous injection of IL-11 neutralizing antibodies significantly reduced cell proliferation, skin stem/progenitor cell population, and tumor development. Using Hh-responsive ES cells, we showed that down-regulation of IL-11Rα alone is sufficient to abolish Hh-mediated STAT3 activation and cell differentiation processes. These data indicate that the IL-11Rα/STAT3 signaling axis plays a critical role in Hh-mediated biological processes.
DISCUSSION
As a key player in Hh signaling, activated mutations of smoothened are known to cause development of cancer. Tissue-specific expression of Smo mutant SmoM2 in mice greatly contributes to our understanding of Hh-mediated carcinogenesis (28, 33, 46–51). Currently, it is not very clear which specific molecules mediate Hh-signaling in cancer development. In this study, we discovered an important role of the IL-11Rα/STAT3 signaling axis in Hh-mediated cell functions and carcinogenesis. STAT3 signaling is a known pathway critical for development of many cancer types, including skin cancer (52). Through epidermis-specific gene knockout of STAT3, IL-11Rα knockout, and subcutaneous injection of IL-11-neutralizing antibodies, we demonstrated a critical role of the IL-11Rα/STAT3 signaling axis for regulation of cell proliferation, epidermal stem/progenitor cell expansion, and tumor development. Because of the fact that isolated BCC cells lose Hh signaling once culture in dishes, we chose two Hh-responsive cell lines to demonstrate that addition of Smo agonist purmorphamine induces STAT3 phosphorylation directly, and down-regulation of IL-11Rα abolishes this effect, supporting the critical role of IL-11Rα for Hh-induced STAT3 signaling. In contrast, IL-11 expression was induced during BCC formation but was not affected by Gli1 knockout. On the basis of these results, we believe that unlike IL-11Rα, IL-11 is indirectly regulated by Hh signaling. The effects of IL-11/IL-11Rα are mediated by a multimeric complex comprising IL-11, the ligand binding IL-11Rα, and the ubiquitously expressed gp130Rβ. As a result, STAT3 signaling is activated during SmoM2-mediated carcinogenesis (see supplemental Fig. 8 for the model).
Like IL-6, the cellular response of IL-11 is determined by the more restricted expression level of the specific α-receptor subunit IL-11Rα. There are two IL-11Rα isoforms, but only expression of IL-11Rα1 was altered in the BCC model (data not shown). As a member of IL-6 cytokine family, IL-11 can exert both immunomodulatory effects as well as epithelial effects (45). When STAT3 was knocked out in the epidermis, we did not observe any changes in myeloid-derived cell population (CD11b+Gr1+ cells), indicating that the immunomodulatory effects of IL-11 do not play a dominant role in the BCC model. In contrast, we observed significant changes in expression of cyclin D1 (Fig. 6), cell proliferation (Fig. 4), and stem/progenitor cell population (Fig. 5). Thus, our data strongly support a non-immunomodulatory effect of the IL-11Rα/STAT3 signaling axis in the BCC model.
In the last few years, elevated expression of IL-11Rα and STAT3 signaling has been reported in a number of cancer types, including osteosarcomas, prostate cancer, and gastric carcinomas (53–56). In a mouse model of gastric cancer, it is demonstrated that IL-11Rα knockout prevents tumor formation (57). It is not clear what regulates the abnormally high expression of IL-11Rα, and it will be interesting to examine whether Hh signaling plays a role in these cancers.
Our studies are consistent with the data from two-stage skin carcinogenesis studies in which keratinocyte-specific knockout of STAT3 prevents the 12-dimethylbenz(α)anthracene/12-O-tetradecanoylphorbol-13-acetate-induced development of SCC (58). Previous studies indicate that STAT3 can be activated by the epidermal growth factor receptor (59). Although SCC can be induced by the 12-dimethylbenz(α)anthracene/12-O-tetradecanoylphorbol-13-acetate, BCCs only develop after activation of the Hh signaling pathway via loss of function of Ptch1 or expression of oncogenic SmoM2. In our mouse model, we did observe significant changes in expression of epidermal growth factor receptor ligand TGFα but not EGF or KGF. In addition, we observed significant expression of IL-11 and IL-11Rα, suggesting that the triggers of STAT3 activation in our system may be different from that of the SCC model. Like other studies, our study only removed STAT3 signaling in the epidermis. Thus, the stromal effects of STAT3 cannot be concluded in our study. STAT3 signaling in tumor stroma is known to be responsible for regulation of myeloid-derived suppressor cells in a variety of cancers (39). Because of the lethality from STAT3 knockout, a better understanding of stromal STAT3 signaling in BCC can be examined using gene knockout of the upstream activators such as inducible knockout of IL-11Rα. Similarly, the downstream targets in our model are quite different from those of the SCC model. The mechanisms underlying the differences in STAT3 activators and downstream targets in different tumors are currently unknown. We suspect a few possibilities, and one of them is that cells of origin for SCCs and BCCs may be different (because of different tumor microenvironments).
In summary, our data provide evidence to support the following picture. SmoM2 induces expression of IL-11Rα in keratinocytes. Together with elevated expression of IL-11 in the tumor microenvironment, STAT3 signaling is activated. As a result of STAT3 activation, cyclin D1 expression is up-regulated, leading to increased cell proliferation, epidermal stem/progenitor cell accumulation, and development of BCCs (see details in supplemental Fig. 8 for our model).
Supplementary Material
Acknowledgments
We thank Drs. Xin-Yuan Fu and Weinian Shou for providing STAT3F/F mice and Dr. Alex Joyner for providing Gli1 null mice.
This work was supported by National Cancer Institute Grants R01CA155086 and R01CA94160, by the Wells Center for Pediatric Research, and by the Indiana University Simon Cancer Center.

This article contains supplemental Figs. 1–8.
- SMO
- smoothened
- PTC
- Patched
- Hh
- Hedgehog
- BCC
- basal cell carcinoma
- SCC
- squamous cell carcinoma.
REFERENCES
- 1. Yang L., Xie G., Fan Q., Xie J. (2010) Activation of the hedgehog signaling pathway in human cancer and the clinical implications. Oncogene 29, 469–481 [DOI] [PubMed] [Google Scholar]
- 2. Parkin C. A., Ingham P. W. (2008) The adventures of Sonic Hedgehog in development and repair. I. Hedgehog signaling in gastrointestinal development and disease. Am. J. Physiol. Gastrointest. Liver Physiol. 294, G363–367 [DOI] [PubMed] [Google Scholar]
- 3. Jiang J., Hui C. C. (2008) Hedgehog signaling in development and cancer. Dev. Cell 15, 801–812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. McMahon A. P., Ingham PW, Tabin CJ. (2003) Developmental roles and clinical significance of hedgehog signaling. Curr. Top. Dev. Biol. 53, 1–114 [DOI] [PubMed] [Google Scholar]
- 5. Epstein E. H. (2008) Basal cell carcinomas. Attack of the hedgehog. Nat. Rev. Cancer 8, 743–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kinzler K. W., Vogelstein B. (1990) The GLI gene encodes a nuclear protein which binds specific sequences in the human genome. Mol. Cell. Biol. 10, 634–642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Johnson R. L., Rothman A. L., Xie J., Goodrich L. V., Bare J. W., Bonifas J. M., Quinn A. G., Myers R. M., Cox D. R., Epstein E. H., Jr., Scott M. P. (1996) Human homolog of patched, a candidate gene for the basal cell nevus syndrome. Science 272, 1668–1671 [DOI] [PubMed] [Google Scholar]
- 8. Hahn H., Christiansen J., Wicking C., Zaphiropoulos P. G., Chidambaram A., Gerrard B., Vorechovsky I., Bale A. E., Toftgard R., Dean M., Wainwright B. (1996) A mammalian patched homolog is expressed in target tissues of sonic hedgehog and maps to a region associated with developmental abnormalities. J. Biol. Chem. 271, 12125–12128 [DOI] [PubMed] [Google Scholar]
- 9. Hahn H., Wicking C., Zaphiropoulous P. G., Gailani M. R., Shanley S., Chidambaram A., Vorechovsky I., Holmberg E., Unden A. B., Gillies S., Negus K., Smyth I., Pressman C., Leffell D. J., Gerrard B., Goldstein A. M., Dean M., Toftgard R., Chenevix-Trench G., Wainwright B., Bale A. E. (1996) Mutations of the human homolog of Drosophila patched in the nevoid basal cell carcinoma syndrome. Cell 85, 841–851 [DOI] [PubMed] [Google Scholar]
- 10. Hahn H., Wojnowski L., Zimmer A. M., Hall J., Miller G., Zimmer A. (1998) Rhabdomyosarcomas and radiation hypersensitivity in a mouse model of Gorlin syndrome. Nat Med 4, 619–622 [DOI] [PubMed] [Google Scholar]
- 11. Aszterbaum M., Epstein J., Oro A., Douglas V., LeBoit P. E., Scott M. P., Epstein E. H., Jr. (1999) Ultraviolet and ionizing radiation enhance the growth of BCCs and trichoblastomas in patched heterozygous knockout mice. Nat. Med. 5, 1285–1291 [DOI] [PubMed] [Google Scholar]
- 12. Athar M., Li C., Tang X., Chi S., Zhang X., Kim A. L., Tyring S. K., Kopelovich L., Hebert J., Epstein E. H., Jr., Bickers D. R., Xie J. (2004) Inhibition of smoothened signaling prevents ultraviolet B-induced basal cell carcinomas through regulation of Fas expression and apoptosis. Cancer Res. 64, 7545–7552 [DOI] [PubMed] [Google Scholar]
- 13. Xie J., Murone M., Luoh S. M., Ryan A., Gu Q., Zhang C., Bonifas J. M., Lam C. W., Hynes M., Goddard A., Rosenthal A., Epstein E. H., Jr., de Sauvage F. J. (1998) Activating Smoothened mutations in sporadic basal cell carcinoma. Nature 391, 90–92 [DOI] [PubMed] [Google Scholar]
- 14. Reifenberger J., Wolter M., Knobbe C. B., Köhler B., Schönicke A., Scharwächter C., Kumar K., Blaschke B., Ruzicka T., Reifenberger G. (2005) Somatic mutations in the PTCH, SMOH, SUFUH, and TP53 genes in sporadic basal cell carcinomas. Br. J. Dermatol. 152, 43–51 [DOI] [PubMed] [Google Scholar]
- 15. Reifenberger J., Wolter M., Weber R. G., Megahed M., Ruzicka T., Lichter P., Reifenberger G. (1998) Missense mutations in SMOH in sporadic basal cell carcinomas of the skin and primitive neuroectodermal tumors of the central nervous system. Cancer Res. 58, 1798–1803 [PubMed] [Google Scholar]
- 16. Svärd J., Heby-Henricson K., Henricson K. H., Persson-Lek M., Rozell B., Lauth M., Bergström A., Ericson J., Toftgård R., Teglund S. (2006) Genetic elimination of Suppressor of fused reveals an essential repressor function in the mammalian Hedgehog signaling pathway. Dev. Cell 10, 187–197 [DOI] [PubMed] [Google Scholar]
- 17. Goodrich L. V., Milenković L., Higgins K. M., Scott M. P. (1997) Altered neural cell fates and medulloblastoma in mouse patched mutants. Science 277, 1109–1113 [DOI] [PubMed] [Google Scholar]
- 18. Metcalfe C., de Sauvage F. J. (2011) Hedgehog fights back. Mechanisms of acquired resistance against Smoothened antagonists. Cancer Res. 71, 5057–5061 [DOI] [PubMed] [Google Scholar]
- 19. Ng J. M., Curran T. (2011) The Hedgehog's tale. Developing strategies for targeting cancer. Nat. Rev. Cancer 11, 493–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bigelow R. L., Chari N. S., Unden A. B., Spurgers K. B., Lee S., Roop D. R., Toftgard R., McDonnell T. J. (2004) Transcriptional regulation of bcl-2 mediated by the sonic hedgehog signaling pathway through gli-1. J. Biol. Chem. 279, 1197–1205 [DOI] [PubMed] [Google Scholar]
- 21. Regl G., Kasper M., Schnidar H., Eichberger T., Neill G. W., Philpott M. P., Esterbauer H., Hauser-Kronberger C., Frischauf A. M., Aberger F. (2004) Activation of the BCL2 promoter in response to Hedgehog/GLI signal transduction is predominantly mediated by GLI2. Cancer Res. 64, 7724–7731 [DOI] [PubMed] [Google Scholar]
- 22. Xie J., Aszterbaum M., Zhang X., Bonifas J. M., Zachary C., Epstein E., McCormick F. (2001) A role of PDGFRα in basal cell carcinoma proliferation. Proc. Natl. Acad. Sci. U.S.A. 98, 9255–9259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Li C., Chi S., He N., Zhang X., Guicherit O., Wagner R., Tyring S., Xie J. (2004) IFNα induces Fas expression and apoptosis in hedgehog pathway-activated BCC cells through inhibiting Ras-Erk signaling. Oncogene 23, 1608–1617 [DOI] [PubMed] [Google Scholar]
- 24. Leung C., Lingbeek M., Shakhova O., Liu J., Tanger E., Saremaslani P., Van Lohuizen M., Marino S. (2004) Bmi1 is essential for cerebellar development and is overexpressed in human medulloblastomas. Nature 428, 337–341 [DOI] [PubMed] [Google Scholar]
- 25. Jonkers J., Meuwissen R., van der Gulden H., Peterse H., van der Valk M., Berns A. (2001) Synergistic tumor suppressor activity of BRCA2 and p53 in a conditional mouse model for breast cancer. Nat. Genet. 29, 418–425 [DOI] [PubMed] [Google Scholar]
- 26. Vasioukhin V., Degenstein L., Wise B., Fuchs E. (1999) The magical touch. Genome targeting in epidermal stem cells induced by tamoxifen application to mouse skin. Proc. Natl. Acad. Sci. U.S.A. 96, 8551–8556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Nandurkar H. H., Robb L., Tarlinton D., Barnett L., Köntgen F., Begley C. G. (1997) Adult mice with targeted mutation of the interleukin-11 receptor (IL11Ra) display normal hematopoiesis. Blood 90, 2148–2159 [PubMed] [Google Scholar]
- 28. Mao J., Ligon K. L., Rakhlin E. Y., Thayer S. P., Bronson R. T., Rowitch D., McMahon A. P. (2006) A novel somatic mouse model to survey tumorigenic potential applied to the Hedgehog pathway. Cancer Res. 66, 10171–10178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kano A., Wolfgang M. J., Gao Q., Jacoby J., Chai G. X., Hansen W., Iwamoto Y., Pober J. S., Flavell R. A., Fu X. Y. (2003) Endothelial cells require STAT3 for protection against endotoxin-induced inflammation. J. Exp. Med. 198, 1517–1525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Park H. L., Bai C., Platt K. A., Matise M. P., Beeghly A., Hui C. C., Nakashima M., Joyner A. L. (2000) Mouse Gli1 mutants are viable but have defects in SHH signaling in combination with a Gli2 mutation. Development 127, 1593–1605 [DOI] [PubMed] [Google Scholar]
- 31. He J., Sheng T., Stelter A. A., Li C., Zhang X., Sinha M., Luxon B. A., Xie J. (2006) Suppressing Wnt signaling by the hedgehog pathway through sFRP-1. J. Biol. Chem. 281, 35598–35602 [DOI] [PubMed] [Google Scholar]
- 32. Diermeier-Daucher S., Clarke S. T., Hill D., Vollmann-Zwerenz A., Bradford J. A., Brockhoff G. (2009) Cell type specific applicability of 5-ethynyl-2′-deoxyuridine (EdU) for dynamic proliferation assessment in flow cytometry. Cytometry. 75, 535–546 [DOI] [PubMed] [Google Scholar]
- 33. Fan Q., He M., Sheng T., Zhang X., Sinha M., Luxon B., Zhao X., Xie J. (2010) Requirement of TGFβ signaling for SMO-mediated carcinogenesis. J. Biol. Chem. 285, 36570–36576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chi S., Xie G., Liu H., Chen K., Zhang X., Li C., Xie J. (2012) Cell. Signal. 24, 1222–1228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Yu H., Pardoll D., Jove R. (2009) STATs in cancer inflammation and immunity. A leading role for STAT3. Nat. Rev. Cancer 9, 798–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sano S., Itami S., Takeda K., Tarutani M., Yamaguchi Y., Miura H., Yoshikawa K., Akira S., Takeda J. (1999) Keratinocyte-specific ablation of Stat3 exhibits impaired skin remodeling but does not affect skin morphogenesis. EMBO J. 18, 4657–4668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Blanpain C., Lowry W. E., Geoghegan A., Polak L., Fuchs E. (2004) Self-renewal, multipotency, and the existence of two cell populations within an epithelial stem cell niche. Cell 118, 635–648 [DOI] [PubMed] [Google Scholar]
- 38. Snippert H. J., Haegebarth A., Kasper M., Jaks V., van Es J. H., Barker N., van de Wetering M., van den Born M., Begthel H., Vries R. G., Stange D. E., Toftgård R., Clevers H. (2010) Lgr6 marks stem cells in the hair follicle that generate all cell lineages of the skin. Science 327, 1385–1389 [DOI] [PubMed] [Google Scholar]
- 39. Gabrilovich D. I., Nagaraj S. (2009) Myeloid-derived suppressor cells as regulators of the immune system. Nat. Rev. Immunol. 9, 162–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sinibaldi D., Wharton W., Turkson J., Bowman T., Pledger W. J., Jove R. (2000) Induction of p21WAF1/CIP1 and cyclin D1 expression by the Src oncoprotein in mouse fibroblasts. Role of activated STAT3 signaling. Oncogene 19, 5419–5427 [DOI] [PubMed] [Google Scholar]
- 41. Dijkgraaf G. J., Alicke B., Weinmann L., Januario T., West K., Modrusan Z., Burdick D., Goldsmith R., Robarge K., Sutherlin D., Scales S. J., Gould S. E., Yauch R. L., de Sauvage F. J. (2011) Small molecule inhibition of GDC-0449 refractory smoothened mutants and downstream mechanisms of drug resistance. Cancer Res. 71, 435–444 [DOI] [PubMed] [Google Scholar]
- 42. Mita A. C., Mita M. M., Nawrocki S. T., Giles F. J. (2008) Survivin: key regulator of mitosis and apoptosis and novel target for cancer therapeutics. Clin. Cancer Res. 14, 5000–5005 [DOI] [PubMed] [Google Scholar]
- 43. Kiang J. G., Tsokos G. C. (1998) Heat shock protein 70 kDa. Molecular biology, biochemistry, and physiology. Pharmacol. Ther. 80, 183–201 [DOI] [PubMed] [Google Scholar]
- 44. Grad J. M., Zeng X. R., Boise L. H. (2000) Regulation of Bcl-xL. A little bit of this and a little bit of STAT. Curr. Opin. Oncol. 12, 543–549 [DOI] [PubMed] [Google Scholar]
- 45. Putoczki T., Ernst M. (2010) More than a sidekick. The IL-6 family cytokine IL-11 links inflammation to cancer. J. Leukocyte Biol. 88, 1109–1117 [DOI] [PubMed] [Google Scholar]
- 46. Hatanaka H., Takada S., Tsukui M., Choi Y. L., Kurashina K., Soda M., Yamashita Y., Haruta H., Hamada T., Tamada K., Hosoya Y., Sata N., Nagai H., Yasuda Y., Sugano K., Mano H. (2010) Identification of the transforming activity of Indian hedgehog by retroviral expression screening. Cancer Sci. 101, 60–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Youssef K. K., Van Keymeulen A., Lapouge G., Beck B., Michaux C., Achouri Y., Sotiropoulou P. A., Blanpain C. (2010) Identification of the cell lineage at the origin of basal cell carcinoma. Nat. Cell Biol. 12, 299–305 [DOI] [PubMed] [Google Scholar]
- 48. Grachtchouk M., Pero J., Yang S. H., Ermilov A. N., Michael L. E., Wang A., Wilbert D., Patel R. M., Ferris J., Diener J., Allen M., Lim S., Syu L. J., Verhaegen M., Dlugosz A. A. (2011) Basal cell carcinomas in mice arise from hair follicle stem cells and multiple epithelial progenitor populations. J. Clin. Invest. 121, 1768–1781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wang G. Y., Wang J., Mancianti M. L., Epstein E. H., Jr. (2011) Basal cell carcinomas arise from hair follicle stem cells in Ptch1(+/−) mice. Cancer Cell 19, 114–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Gao J., Graves S., Koch U., Liu S., Jankovic V., Buonamici S., El Andaloussi A., Nimer S. D., Kee B. L., Taichman R., Radtke F., Aifantis I. (2009) Hedgehog signaling is dispensable for adult hematopoietic stem cell function. Cell Stem Cell 4, 548–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hofmann I., Stover E. H., Cullen D. E., Mao J., Morgan K. J., Lee B. H., Kharas M. G., Miller P. G., Cornejo M. G., Okabe R., Armstrong S. A., Ghilardi N., Gould S., de Sauvage F. J., McMahon A. P., Gilliland D. G. (2009) Hedgehog signaling is dispensable for adult murine hematopoietic stem cell function and hematopoiesis. Cell Stem Cell 4, 559–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kim D. J., Chan K. S., Sano S., Digiovanni J. (2007) Signal transducer and activator of transcription 3 (Stat3) in epithelial carcinogenesis. Mol. Carcinog. 46, 725–731 [DOI] [PubMed] [Google Scholar]
- 53. Campbell C. L., Jiang Z., Savarese D. M., Savarese T. M. (2001) Increased expression of the interleukin-11 receptor and evidence of STAT3 activation in prostate carcinoma. Am J. Pathol. 158, 25–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Zurita A. J., Troncoso P., Cardó-Vila M., Logothetis C. J., Pasqualini R., Arap W. (2004) Combinatorial screenings in patients. The interleukin-11 receptor α as a candidate target in the progression of human prostate cancer. Cancer Res. 64, 435–439 [DOI] [PubMed] [Google Scholar]
- 55. Lewis V. O., Ozawa M. G., Deavers M. T., Wang G., Shintani T., Arap W., Pasqualini R. (2009) The interleukin-11 receptor α as a candidate ligand-directed target in osteosarcoma. Consistent data from cell lines, orthotopic models, and human tumor samples. Cancer Res. 69, 1995–1999 [DOI] [PubMed] [Google Scholar]
- 56. Nakayama T., Yoshizaki A., Izumida S., Suehiro T., Miura S., Uemura T., Yakata Y., Shichijo K., Yamashita S., Sekin I. (2007) Expression of interleukin-11 (IL-11) and IL-11 receptor α in human gastric carcinoma and IL-11 up-regulates the invasive activity of human gastric carcinoma cells. Int. J. Oncol. 30, 825–833 [PubMed] [Google Scholar]
- 57. Ernst M., Najdovska M., Grail D., Lundgren-May T., Buchert M., Tye H., Matthews V. B., Armes J., Bhathal P. S., Hughes N. R., Marcusson E. G., Karras J. G., Na S., Sedgwick J. D., Hertzog P. J., Jenkins B. J. (2008) STAT3 and STAT1 mediate IL-11-dependent and inflammation-associated gastric tumorigenesis in gp130 receptor mutant mice. J. Clin. Invest. 118, 1727–1738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Chan K. S., Sano S., Kiguchi K., Anders J., Komazawa N., Takeda J., DiGiovanni J. (2004) Disruption of Stat3 reveals a critical role in both the initiation and the promotion stages of epithelial carcinogenesis. J. Clin. Invest. 114, 720–728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Chan K. S., Carbajal S., Kiguchi K., Clifford J., Sano S., DiGiovanni J. (2004) Epidermal growth factor receptor-mediated activation of Stat3 during multistage skin carcinogenesis. Cancer Res. 64, 2382–2389 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.