Abstract
The heterogeneous nature of stem cells is an important issue in both research and therapeutic use in terms of directing cell lineage differentiation pathways, as well as self-renewal properties. Using flow cytometry we have identified two distinct subpopulations by size, large and small, within cultures of human embryonic stem (hES) cell lines. These two cell populations respond differentially to retinoic acid (RA) differentiation and several endocrine disruptor compounds (EDC). The large cell population responds to retinoic acid differentiation with a greater than a 50% reduction in cell number and loss of Oct-4 expression, whereas the number of the small cell population does not change and Oct-4 protein expression is maintained. In addition, four estrogenic compounds altered SSEA-3 expression differentially between the two cell subpopulations changing their ratios relative to each other. Both populations express stem cell markers Oct-4, Nanog, Tra-1-60, Tra-1-80 and SSEA-4, but express low levels of differentiation markers common to the three germ layers. Cloning studies indicate that both populations can revive the parental population. Furthermore, whole genome microarray identified approximately 400 genes with significantly different expression between the two populations (p<0.01). We propose the differential response to RA in these populations is due to differential gene expression of Notch signaling members, CoupTF1 and CoupTF2, chromatin remodeling and histone modifying genes that render the small population resistant to RA differentiation. The findings that hES cells exist as heterogeneous populations with distinct responses to differentiation signals and environmental stimuli will be relevant for their use for drug discovery and disease therapy.
Keywords: Human embryonic stem cell, Heterogeneous population, Retinoic acid differentiation, Endocrine disruptor compounds, Notch signaling, Chromatin remodeling
1. Introduction
Human embryonic stem cells (hESC) are derived from a small group of cells from the inner cell mass of human blastocysts. They are defined as being pluripotent and thus are able to self-renew and contain the capacity to develop into all three primordial germ cell layers (Thomson et al., 1998). They can be maintained in vitro as immortal pluripotent cells, but are responsive to various differentiation signals, such as growth factors and retinoic acid (RA) (Hu and Zhang, 2010; Niederreither and Dolle, 2008; Strickland and Mahdavi, 1978). Treatment with RA causes hESC cells to differentiate predominately into neural progenitors with the characteristics of Pax6-positive radial glial cells (Kayama et al., 2009). Human stem cell lines H1, H9 and BGN1 express stem cell markers characteristic of a puripotent stem cell (Oct-4, Nanog, SSea-4, Tra-1-60 and Tra-1-81), have a normal karyotype of either XY or XX and form teratomas with all three germ layers present when injected into Beige-Scid mice (Jiang et al., 2010; Lecina et al., 2010; Meng et al., 2010).
While many of the hundreds of hESC derived worldwide share many similarities, it is also clear that they exhibit multiple differences that may reflect their different genetic backgrounds, environmental exposures, methods of derivation and culture conditions (Allegrucci et al., 2007; Rao, 2008). Indeed, individual cell lines, while sharing many characteristic surface makers, including glycolipid antigens and keratin sulphate antigens as well as pluripotency factors, are not identical with respect to gene expression (Canham et al., 2010; Sharov et al., 2011; Tavakoli et al., 2009). As an example, a broad range of surface antigens including but not exclusive of CD133, SSEA1, CD117 and CD135 were observed on some but not all hESCs in culture, raising the possibility of subpopulations within individual cell lines (Nagano et al., 2008) Elegant FACS sorting experiments using CD133 and CD135 markers in combination with fluorescently tagged hESCs (King et al., 2009) provided more recently strong support for the view that existing stem cell lines are indeed heterogeneous. These studies demonstrated the existence of distinct subpopulations differentially expressing surface makers and consistent with the concept that not all the cells within a hESC line are pluripotent, but may have the propensity to differentiate towards a particular lineage depending on endogenous and exogenous signals (King et al., 2009).
There are clear and compelling molecular mechanisms by which chromatin architecture, microRNAs, DNA methylation, histone modifications, and ATP-dependent chromatin remodeling complexes may play a role in pluripotency and differentiation (Card et al., 2008; Hawkins et al., 2010; Meshorer and Misteli, 2006; Young, 2011). Indeed, it is now well established that the mammalian SWI/SNF complex is essential for embryonic stem cell self-renewal and pluripotency in mice (Gao et al., 2008; Ho et al., 2009; Yan et al., 2008). Similarly, differences in histone modifications, imprinted genes, and DNA methylation are providing insights into the fundamental characteristics of hESCs (Hawkins et al., 2010; Lengner et al., 2010; Sharov et al., 2011). Finally, it is clear that the context of stem cell derivation, the in vivo niche, and the subsequent culture conditions and attendant signaling programs are critical to understanding pluripotency (Harb et al., 2008; Stewart et al., 2008). The plethora of studies undertaken to characterize human ES cells assume that clonal hESCs are stable homogenous populations. We have examined the possibility that hESCs exist as heterogeneous populations of pluripotent stem cells distinguishable by physical characteristics and response to differentiation agents, which can alter their distribution.
We show that hESCs are heterogeneous populations that show distinct characteristics in size, cell cycle profile, gene expression, and responsiveness to retinoic acid differentiation and several endocrine disruptor compounds. The two populations express the nuclear stem cell markers, Oct-4 and nanog, and differentially express stem cell surface markers including Tra-1-60 and Tra-1-80. Furthermore, whole human genome microarray analysis indicates roughly 400 genes and ESTs that were significantly differentially expressed (p<0.01) between the two subpopulations. The two populations expressing different gene profile signatures may account for their differential response to RA and upon exposure to endocrine disrupting chemicals, a class of compounds important for human health and development. Finally, differential responses to these compounds is evident in the gene expression differences between the two populations for notch signaling, CoupTF1 and CoupTF2, components of the SWI/SNF complex and histone-modifying enzymes.
2. Material and Methods
2.1. Culture of hES Cells and In Vitro Differentiation and Endocrine Disruptor Treatment of hES Cells
H1 and H9 hES cells (WA01 and WA09, WiCell Research Institute, Madison, WI) and BGN1 hES cells (NIH BG01, hESBGN-01, BresaGen, Inc., Athens, GA) were cultured on gelatinized (0.2%) 6-well plates plated with irradiated (8000 rads) mouse embryonic feeder cells (CF-1, Chemicon International, Phillipsburg, NJ) in media containing DMEM F-12 (Invitrogen, Carlsbad, CA) supplemented with 20% Knockout Serum Replacement (Invitrogen), 1 mM l-glutamine (Invitrogen), 0.1 mM β-mercaptoethanol, 1% nonessential amino acids (Invitrogen), and 4 ng/mL human βFGF (Invitrogen) at 37°C and 5% CO2. Cells were maintained in fresh media every day and passaged harvesting with collagenase (Gibco, Grand Island, NY), split weekly 1:6 and replated in gelatinized (0.1%) 6 well plates. H1 and H9 hES cells were used between passage 39 and passage 49, and BGN1 hES cells between passage 35 and passage 45. H1 and BGN1 cells were also cultured on matrigel plates with mTeSR™1 media (STEMCELL Technologies, WiCell Research Institute, Madison, WI).
Retinoic acid (RA) a standard method for differentiating stem cells was used to differentiate both H1 and BGN1 hES cells. Three biological replicates of H1 and BGN1 hES cells were treated for 6 or 12 days with retinoic acid (1 μM, Sigma-Aldrich, St. Louis, MO) and then sorted to obtain subpopulations. Three biological replicates of H1 and H9 hES cells were also treated for 2 days with 4 endocrine disruptor compounds; 75 nM tamoxifen (Sigma-Aldrich), 75 nM kaempferol (Ivychem), 75 nM kepone (Cerilliant) or 37.5 nM apigenin (LKT Laboratories, Inc). Equivalent volumes or concentrations of DMSO (Caledon Laboratories, Georgetown, Canada), ETOH (Warner-Graham Company, Cockeysville, MD) or 10nM β-estradiol (Sigma-Aldrich) were used as control. After two days of treatment, cells were trypsinized, washed with 3% BSA/PBS, hybridized with FITC Rat anti-SSEA-3 antibody (BD Pharmingen) for 30 minutes, then washed twice with 3% BSA/PBS, filtered and separated on the flow cytometer as described below. All cells tested negative for mycoplasma.
2.2. Purification of Subpopulations of ES Cells by Flow Cytometry
Undifferentiated and RA treated ES cells were isolated by trypsin (Gibco). Cells were initially stained with propidium iodide (PI; 10 μg/ml, final) just prior to examination to remove any dead or dying cells from the analysis. Stained cells were examined on a FACSVantage SE flow cytometer equipped with digital electronics (Becton Dickinson, San Jose, CA). All samples were excited at 488 nm and cells were analyzed on the forward-scatter versus PI dot plot where gates were set to isolate the viable larger (high forward scatter) cells from the smaller (lower forward scatter) cells for further analysis. Sorted cells were collected by centrifugation at 3000 rpm for 25 minutes. Ten thousand cells were examined per sample for analysis using BD FACSDiVa software. Cells treated with endocrine disruptor compounds were gated for large and small populations and then examined for percent of positive FITC cells.
2.3. Cell Imaging by Flow Cytometry
H1 cells were stained with DRAQ5 (Biostatus Limited, Leicestershire, United Kingdom) at a final concentration of 5 μM for 30 minutes at room temperature. Propidium iodide (Sigma Aldrich, St. Louis, MO, USA) was added to a final concentration of 1 μg/ml immediately prior to analysis. Cells were analyzed using an ImageStreamX flow cytometer (Amnis Corporation, Seattle, WA, USA). Signals from PI and DRAQ5 were detected in channels 4 and 5, respectively, while bright field and side-scatter were detected in channels 1 and 6, respectively. Data was collected by INSPIRE acquisition software using 488 nm (PI) and 633 nm (DRAQ5) lasers with appropriate compensation controls and a 40-X objective with extended depth of field settings. Data from a minimum of 3,000 cells were collected and analyzed using IDEAS software. DRAQ5 positive, PI negative cells were focused, and then examined on a Brightfield Area versus Aspect Ratio dot plot. Gates were drawn around the small and large population of cells for image analysis.
2.4. Cell Size Analysis
Cell size was determined for the undifferentiated and RA treated ES cells (5000 cells per analysis) using a Cell Lab Quanta SC flow cytometer (Beckman Coulter, Miami, FL). The electronic volume (EV) parameter was calibrated using 488 excitable 6 μm beads. Cell Lab Quanta Flow methodology to measure cell size, as this uses electronic impedance or conductance (similar to a Coulter Counter) to size the cells, is an accepted and standard method for sizing cells (Bortner and Cidlowski, 2007). ES cells were initially stained with propidium iodide (PI) to a final concentration of 10 μg/ml final just prior to examination. All samples were triggered off FL-2 (575 nm) from the 488 nm laser and initially examined on an FL-3 (670 nm LP) histogram to gate the viable population of cells. Twenty thousand viable cells were recorded and examined on an EV versus FL-3 dot plot or an EV histogram. Mean cell volume (MCV) and cell diameter were determined using the Cell Lab Quanta SC software from gated regions for the large and small populations of cells. Images of sorted cells were acquired while in suspension after sorting to verify single cells and not aggregate cells were sized.
Annixen-V Staining for Apoptotic Cells
H1 hES cells were stained with 2 μl of Annexin-V-FITC (Trevigen, Gaithersburg, MD) and PI for 20 minutes, gated for large and small cell populations then examined for Annexin-V-FITC and PI fluorescence. Acquired events for each population were divided into four quadrants; Q1: FITC-PI+, cells which are necrotic and dead; Q2: FITC+PI+, apoptotic dead cells; Q3: FITC-PI−, viable cells; and finally Q4: FITC+PI−, cells which are potentially apoptotic but still alive.
2.5. Cell Cycle Analysis Using Propidium Iodide and DRAQ5
Cell cycle analysis was performed on the isolated ES cell populations by initially sorting 250,000 cells directly into 5 ml of cold 70% ethanol and stored at 20°C overnight. Cells were prepared for flow cytometry by washing in 3 ml of PBS, then staining them with a PI solution (20 μg/ml PI/10 Units/ml RNase One (Promega) for at least 20 min. at room temperature. Flow cytometry analysis was carried out using a BD Biosciences FACSort (San Jose, CA), gating on a PI area versus width dot plot to exclude cell debris and cell aggregates. Seven thousand five hundred cells were examined in this gate on a PI area (DNA content) histogram using CellQuest software. Additionally, the cell cycle was analyzed using ModFit software (Verity Software House, Topsham, ME) to determine the percent of cells in each phase of the cell cycle.
Cell cycle analysis was performed on the isolated ES cell populations in triplicate from two biological repeats using DRAQ5 (Biostatus, Ltd., Leicestershire, UK) at a final concentration of 25 μM to stain DNA. Cells stained with DRAQ5 were incubated for 5 min. at 37°C then immediately examined on a Becton Dickinson FACSort. Stained cells were initially examined on an FL-3 area versus width dot plot to gate out doublets and debris, than analyzed on an FL-3 (DNA content) histogram. Ten thousand cells were examined per sample for analysis using either CellQuest or ModFit software to determine the percent of cells in each phase of the cell cycle.
2.6. Tumor Initiation and Histology
A standard test for pluripotency is to inject hES cells into immuno-compromised mice to determine their ability to form a teratoma tumor with all three primordial germ layers present (Lee et al., 2005). To obtain relatively pure subpopulations of large and small BGN1 hES cells were harvested with trypsin (Invitrogen, Life tech, Grand Island, NY), washed and prepared for sorting in hES media. Unsorted cells for injection (4 × 106 per site) were harvested either by collagenase (Gibco) or trypsin Sterile flow sorted cells were collected by centrifugation and subcutaneously injected into Beige-Scid mice (Charles River Laboratory, Wilmington, MA) (4 × 106 cells per injection site, two sites per mouse). Sorted cells injected in the presence of 0.1% matrigel (BD Bioscience, San Jose, CA) in DMEM/F-12. Mice were palpated weekly to identify tumors. After tumors reached approximately 1 cm in diameter, they were removed and transferred into 10% formulin overnight and then to 10% ETOH before sectioning. Sections were stained H&E and examined for the presence of the three primary germ layers. Mice were observed up to one year for tumor formation.
2.7. Isolation of Cellular Protein and Western Blot Analysis
Total Protein was isolated from sorted cells using Buffer X (100 mM Tris-HCl [pH 8.5], 250 mM NaCl, 1% [vol/vol] NP-40, 1 mM EDTA, 1 mM phenylmethylsulfonyl fluoride, 1 μg of leupeptin/ml). Proteins were resolved by SDS-PAGE, transferred to a Nitrocellulose Membrane followed by western blotting using various antibodies. Antibodies against the stem cell markers Oct-4 (sc-5279), Tra-1-60 (sc-21705) Tra-1-81 (sc-21706) and SSEA-4 (sc-21704) were obtained from Santa Cruz Biotechnology whereas c-myc (9E10) antibody was from Abcam (Cambridge, MA) and SSEA-3-FITC from BD Pharmingen (San Jose, CA). The anti-actin antibody (A-2066) was from Sigma.
2.8. Microarray Analysis
RNA from sorted subpopulations was isolated using a midi-Rneasy Qiagen kit and treated with Dnase1 (Invitrogen). Four independent biological sort repeats for each subpopulation were hybridized to Affymetrix Human Genome U133 Plus 2.0 arrays (Affymetrix, Santa Clara, CA). Starting with 1 μg of total RNA, biotin-labeled cDNA was produced using the Affymetrix 3′ Amplification One-Cycle Target labeling kit according to manufacturer’s protocol. For each array, 15μg of amplified codas were fragmented and hybridized to the array for 16 hours in a rotating hybridization oven using the Affymetrix Eukaryotic Target Hybridization Controls and protocol. Slides were stained and washed as indicated in the Antibody Amplification Stain for Eukaryotic Targets protocol using the Affymetrix Fluidics Station FS450. Arrays were then scanned with an Affymetrix Scanner 3000. Data was obtained using the Genechip® Operating Software and imported into the Rosetta Resolver system (version 7.1)(Rosetta Inpharmatics LLC, Seattle, WA). Biological replicates were combined using an error-weighted average as described in Weng et al. (Weng et al., 2006). P-values and fold changes based on the ratio of small large were generated in Resolver and used as a threshold for selecting differentially expressed genes. RT-PCR, western blotting and flow cytometry validated significant changes in gene expression. Ingenuity Pathway Analysis was performed using all differentially expressed probes.
2.9. Quantitative RT-PCR Analysis
Total RNA was used to synthesize cDNA using the SuperScript First-Strand Synthesis System for RT-PCR (Invitrogen) as recommended by the manufacturer. QPCR was performed using Brilliant SYBR Green QPCR Reagent (Stratagene, La Jolla, CA). Relative expression levels were normalized to human glyceraldehydes-3-phosphate dehydrogenase (GAPDH). Primer sequences used for tissue-specific marker genes are as previously described (Lee et al., 2005). All primer sequences used in this study are available upon request.
3. Results
3.1. Characterization of Heterogeneous Sub-Populations within Embryonic Stem Cells
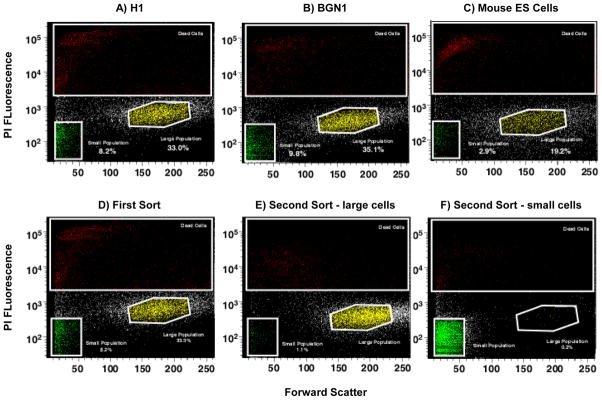
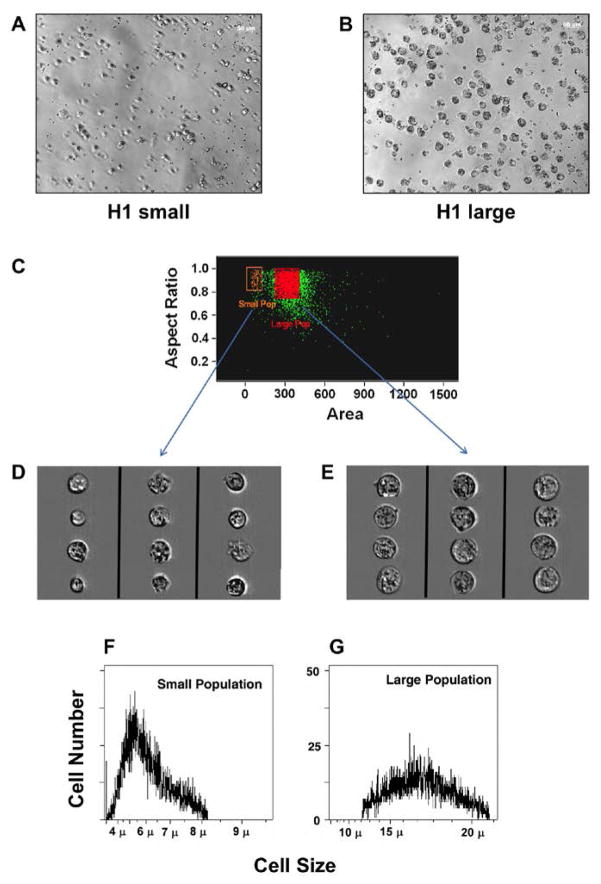
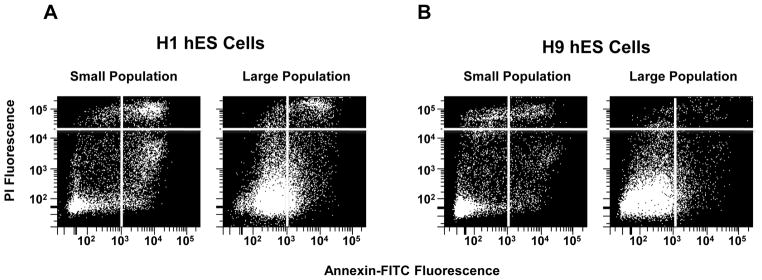
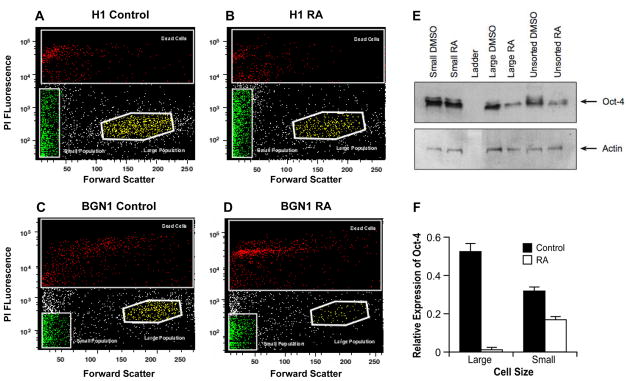
Two different subpopulations were identified from hESC lines (H1 and BGN1) and analyzed by flow cytometry using forward and side scatter light displaying two distinct subpopulations of embryonic stems cells (Figure 1A and 1B). The two populations were also identified in H9 hES cells and a human iPS cell line recently developed in our laboratory (data not shown). To confirm the existence of heterogeneous populations in other model systems, mouse embryonic stem cells were sorted in a similar manner. Mouse ES cells exhibit heterogeneous subpopulations similar to hESCs (Figure 1C). The populations of large and small cells were discrete and visually far apart after using tight gates for sorting. The subpopulations were sorted and then re-sorted to determine their purity. Re-analysis of the double-sorted populations confirmed 99% or greater purity in the isolated subsets (Figure 1D–F). Large cells re-sorted contained 1.1% small cells and the small cell population re-sorted contained 0.2% large cells. This is further emphasized by an enrichment of each population visually when either large or small previously sorted cells are re-sorted. H1 hES cells grown in the absence of a feeder layer of MEF cells on matrigel also demonstrated the presence of two subpopulations (Supplemental Figure 1). Both H1 and BGN1 cells were sorted and imaged under the microscope to be sure they were singular cells and not aggregate cells (Figure 2A–B) only H1 is shown. In addition, H1 hES cells were stained with PI and analyzed using an ImageStreamX flow cytometer to monitor viable cells using PI and DRAQ5. Images of viable cells (DRAQ5+/PI−) were collected for gated small and large populations (Figure 2C–E). 15–20% of gated small events were intact cells, corresponding to a similar percentage of DRAQ5+/PI− events in the small population, further indication of a large amount of debris in this size range. Using 488 excitable 6 μm beads the large population is approximately 2.5 fold larger in physical size than the small population. The viable population of the large and small cell populations were approximately 16 and 6.5 microns in size respectively, shown on the histogram (Figure 2F–G). To further establish the distinct identity of the two subpopulations and eliminate the possibility that the small population may be shrunken large cells going through apoptosis, H1 hES cells were stained with Annexin-V-FITC and PI, gated for large and small populations then examined for Annexin-V-FITC and PI fluorescence. Although there was evidence of some Annexin positive viable cells in both populations, the majority of the events were PI and Annexin negative indicating viable cells. Roughly 71% of events gated small and 89% of gated large events in 20,000 were negative for PI and Annexin, as viable non-apoptotic cells with the remaining percentage distributed between necrotic (PI positive, Annexin negative), dead (PI positive, Annexin positive) or apoptotic (PI negative, Annexin positive) (Figure 3).
Figure 1. Stem cells display heterogeneous populations.
Identification of two PI negative viable subpopulations of ES cells gated by size using flow cytometry with forward and side scatter. Human and mouse embryonic cell profile exhibiting two viable populations based on size indicated as large and small populations. A) H1 hES cells. B) BGN1 hES cells. C) Mouse ES cells. D) Whole population of H1 hES cells sorted and gated by size after PI treatment to remove dead cells. E) H1 Large population of gated re-sorted cells from figure 1D. F) Small population of gated re-sorted cells from figure 1D.
Figure 2. Two viable populations of hES have distinct sizes.
Isolation of cells from the two subpopulations using different methods, demonstrated viability and size differences. A) Microphotograph of H1 small cells sorted for size. B) Microphotograph of H1 large cells resorted for size. C) Image stream analysis of PI negative small and large populations stained with DRAQ5. D) Image stream analysis of small cells E) Image stream analysis of large cells. F) Cell Lab Quanta SC Analysis of cell size for small cells. G) Large cells.
Figure 3. Majority of small and large cell populations are viable.
H1 and H9 hES cells were stained for Annexin-V-FITC and PI and then sorted for large and small populations. Annexin-V-FITC vs IP fluorescence was determined and divided into four quadrants for analysis. A) H1 quadrant distributions of small and large cells showing the majority (60 – 80%) fall into quadrant 3 which represent viable non-apoptotic cells that are negative for Annexin-V and negative for PI B) Annexin-V-FITC and PI quadrant expression levels of H9 hES cell small and large cell populations.
3.2. Cell Cycle Analysis
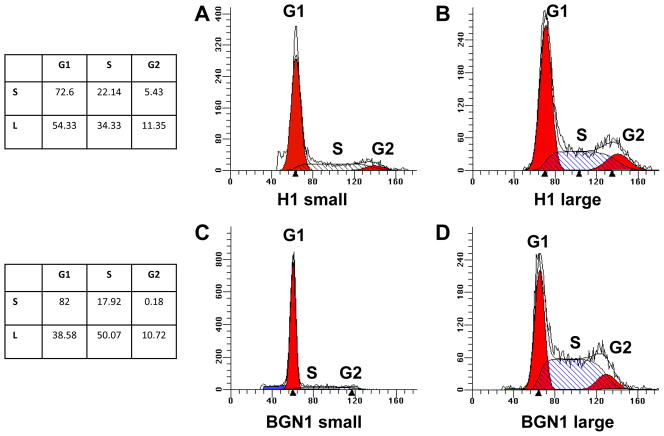
Cell cycle analysis of H1 and BGN1 hES cells in both populations showed that the two populations of cells exhibited distinct profiles (Figure 4). The large population was very similar in both lines, with the G0–G1 percentage of cells being approximately 40–55%, the G2–M percentage of cells being approximately 12% and the S – phase percentage of cells being 35–50%. The small cell population had a different distribution of cells in cell cycle. The G0 – G1 percentage was much higher at 70–80%. Few cells were represented in the G2–M cell cycle stage at 6% or less, and S-phase was 18–22% (Figure 4, A–D We have previously shown that whole cell population of BGN1 has a short G1 phase (21%) and a high proportion of cells in S-phase (50%) (Card et al., 2008)
Figure 4. Cell cycle profiles of large and small populations of hES cells.
H1 and BGN1 hES cell cycle profiles of large and small populations identifying percentage of cells in G0–G1, S-phase (hatch bars) and G2 using the PI method of staining and Mod-fit software A) H1 hES cell cycle profile of the small population B) H1 hES cell cycle profile of the large population C) BGN1 hES small population cell cycle profile D) BGN1 hES cell large population cell cycle profile.
3.3. Isolated Large and Small Clones Revert to Heterogeneous Populations
In the next series of experiments we double sorted the cells and attempted to maintain independent small and large populations of stem cells. However, at passage 2, a typical small cell clone contained 98% small cells, but by passage 5 it reverted to a parental population profile as shown in Figure 1, suggesting that the small population can repopulate the large population (Supplemental figure 2). Consequently, a greater expression of Oct-4 protein levels, similar to large hES cells, was also noted in the small cell clone at passage 5 compared to passage 2 (data not shown). Several large cell clones were isolated and at early establishment stages displayed distinct morphology from the small cell clones. The large population cells were flat and compact resembling the original population of parental stem cells. By passage 2, the resorted cells had a population distribution of small and large cells comparable to the parental population (Supplemental Figure 2).
3.4. In vivo Teratoma Assay
A standard test for pluripotency is to inject hES cells into immuno-compromised mice to determine their ability to form a teratoma tumor with all three primordial germ layers present. Unsorted BGN1 hES cells disassociated with collagenase treatment produced tumors in 70% of injection sites from 18 injections (Supplemental Table 1). However, only 33% of the hES cells disassociated with trypsin produced tumors when injected. The BGN1 large cell population produced only one teratoma tumor out of 12 injection sites and a mix of large plus the small cell population yielded one teratoma tumor in 2 sites. The BGN1 small population did not form a teratoma. The formation of all three germ layers gives support that the large cell population may represent a true population of embryonic stem cells with pluripotency. Histology of the teratoma tumors that formed showed that all three germ layers were present as identified by neural, gastrointestinal epithelial and smooth muscle cells (Supplemental Figure 3).
3.5. Differential Response to Retinoic Acid between Sub-Populations
The nuclear transcription factor Oct-4 is crucial in maintaining pluripotency in embryonic stem cells and is lost in mouse and human differentiated embryonic stem cells (Pan et al., 2002). RA predominantly induces neuronal differentiation of ES cells (Parsons et al.) and down regulates Oct-4 expression in ES cells (Schoorlemmer et al., 1994). H1 and BGN1 hES cells were treated with RA for 6 days and then sorted for large and small cell populations, The large cell population decreased by approximately 50% to 75% in cell number in both H1 and BGN1 hES cell lines after RA treatment, whereas the small population did not change in gated cell number (Figure 5A–D). Oct-4 protein and RNA levels were examined after RA treatment in both H1 and BGN1 stem cells, although data is only shown for H1 hES cells. After 6 days RA treatment, Oct-4 protein expression decreased significantly in the large population, whereas in the small population Oct-4 remained relatively unchanged (Figure 5E, lanes 2 and 5). After 12 days of treatment Oct-4 mRNA expression was significantly decreased in large cells which showed greater than 90% loss of Oct-4 expression while the small cell population retained up to 50% Oct-4 expression (Figure 5F).
Figure 5. Differential response of the two populations to retinoic acid treatment.
Flow cytometry forward vs. side scatter profiles of distribution of large and small H1 and BGN1 hES cells after 12-day treatment with RA or DMSO. A) Forward/Side scatter of H1 cells treated for 12 days with DMSO. B) H1 cell distribution of subpopulations treated for 12 days with RA. C) BGN1 cells treated with DMSO. D) BGN1 cells treated with RA. E) Oct-4 protein expression in H1 hES cells treated with DMSO or retinoic acid. Actin is used as a loading control. F) Oct-4 mRNA expression in H1 hES large and small population after 12 days of RA treatment.
3.6. Retinoic Acid Receptors are expressed in both Sub-Populations
In order to determine if the differential response to RA by the two populations was due to differential expression of retinoic acid receptors; six different RA receptors (RARA, RARB, RARG, RXRA, RXRB and RXRG) in three biological experiments of H1 hES were sorted for large and small cell populations after a 6-day treatment of RA and analyzed by qPCR. The expression levels of the different receptors varied between the two populations, however, both populations expressed all 6 receptors with RARB being the predominant receptor expressed in both populations (Supplemental Figure 4). All six receptors were induced with RA in the large cell population, four of which were statistically significant, RARA, RARB, RARG and RXRG, whereas only RARB and to a lesser degree RXRG, were induced in the small cell population. RARB could be induced in both populations by as much as 10-fold (Supplemental Figure 4).
3.7. Microarray Analysis of the Subpopulations Revealed Different Gene Expression Profiles
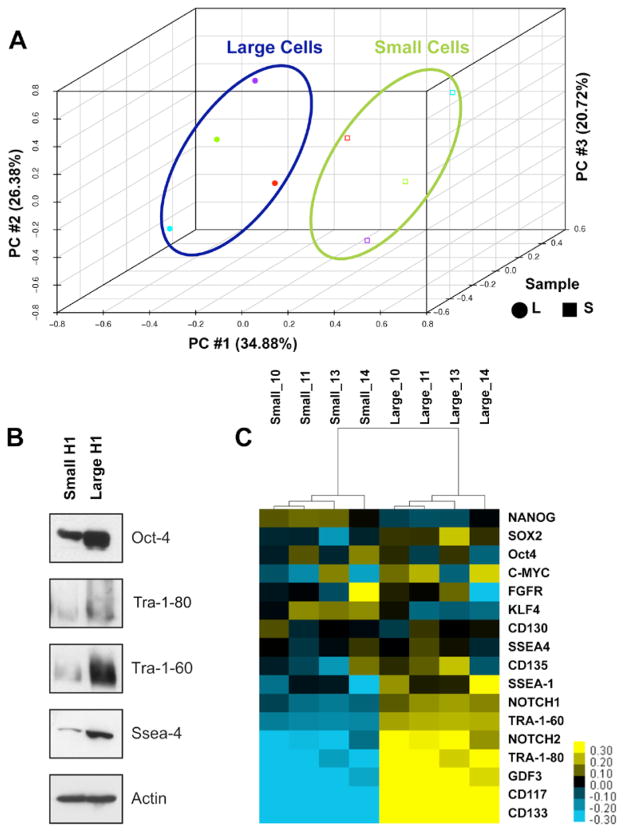
Microarray analysis was performed on four independently sorted biological samples of the H1 hES large and small cell subpopulations. Principal Component analysis demonstrated that the first three principal components captured more than 80% of the variability in the dataset and samples were clustered in two groups with a clear separation between the large and small cell populations (Figure 6A). The present/absent calls from the preliminary analysis of the microarray data indicated that transcripts corresponding to 34,131 probe sets were present in both populations with 16,145 absent (p<0.001). In at least one of the populations 1,496 of the probes were detectable and differentially expressed (p<0.01) of which 440 were greater than 2-fold. Converting the probe set to Unigene resulted in 399 genes with statistically significant differential expression (p<0.01). This set included 233 genes that were up regulated in the large cell population relative to the small population, while the remaining 166 genes were up regulated in the small cell population compared to the large (http://www.ncbi.nlm.nih.gov/geo/ accession number GSF24530).
Figure 6. Differential expression of stem cell markers in two subpopulations of H1 hES cells.
A) PCA analysis using the whole genome microarray data indicates clear separation between H1 hES Cell large and small subpopulations. B) A heat map depicting expression of stem cell marker and associated genes in 4 biological replicates of large and small subpopulation. Yellow indicates higher expression and blue indicates a lower expression of individual genes within the two populations. C) Protein expression of key stem cell markers, Tra-1-60, Tra-1-81, and SSEA-4 within the two populations. Actin is used as loading control.
Of the pathways determined to be significant (p<0.05), genes that were highly expressed in the large population were involved in several biosynthetic and metabolic pathways, including N- and O- glycan biosynthesis, sphingolipid, riboflavin, and starch and sucrose metabolism. Genes highly expressed in the small cell population were members of several signaling pathways, including estrogen receptor, glucocorticoid, and IGF-1/integrin. During development many cellular cues result from integrin signaling (Prowse et al.). For example, Integrin alpha-6, a transmembrane protein that mediates interactions between adhesion molecules on adjacent cells and/or the extracellular matrix (ECM) and important in myogenic stem cell differentiation (Wilschut et al.), is up regulated in the large population. This protein was expressed in 97% of the large cell population compared to only 20% in small cells (Supplemental Figure 5).
3.8. Differential Expression of Stem Cell Markers between Subpopulations
Although both subpopulations expressed traditional stem cell markers associated with pluripotent cells, the large cell population expressed in general a higher level of all stem cell markers than the small cell population (Figure 6B – C). As an example, H1 hES cell subpopulations showed that the large population consistently expressed high protein levels for the stem cell markers Tra-1-60, Tra-1-80 and SSEA-4 (Figure 6C).
Further microarray analysis identified stem cell markers associated with the pluripotency state (Figure 6C). Expression of transcripts encoding standard stem cell markers, including Nanog, Sox2 and Oct4, did not significantly vary between subpopulations. Examination of genes used in IPS studies (c-myc, KLF4 and FGFR) to revert differentiated fibroblast cells to a pluripotent state did not significantly differ between the two subpopulations, although KLF4 appears to be more highly expressed in the small cell population and c-Myc in the large population. However expression of certain cell surface markers associated with stem cell pluripotency, TRA-1-80, TRA-1-60, SSEA-1, and SSEA-4 appear elevated in the large compared to small sub-populations (Figure 6B & C). In addition, both microarray and FACS analysis showed higher SSEA-3 expression in large compared to small population (data not shown). In general, expression of differentiation markers was not significantly different between the small and large cell populations. However, the large cell population had higher expression levels of Cdx2 (trophectoderm), Gata6 and Gata4 (endoderm), enolase (mesoderm) and Pax6 (ectoderm). Trophectoderm maker hCG alpha and mesoderm marker β-hemoglobin were very similar in expression (Supplemental Figure 6). These data would lend support that both populations are potentially pluripotent and stem cell-like in nature but does not rule out that one population may be in an early differentiating state, which could limit or facilitate differentiation into diverse cell lineages (Figure 6B).
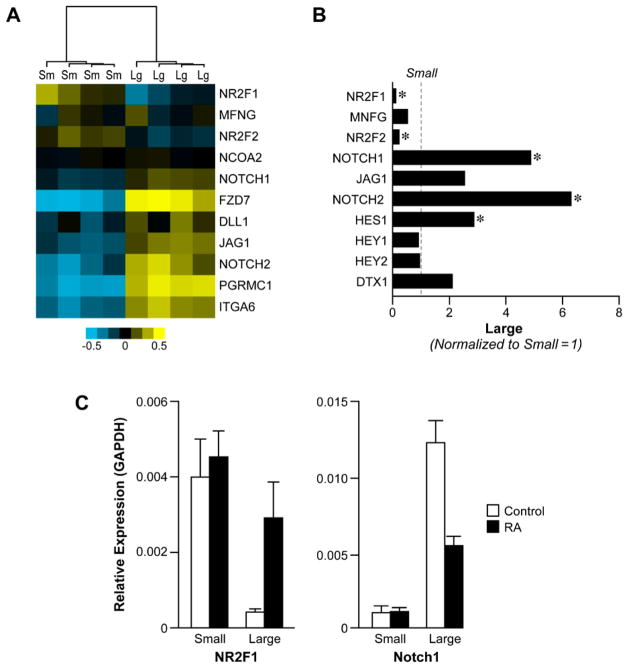
Next we explored the Notch proteins, which are known to regulate cell fate lineages during development and to be associated with self-renewal properties in various adult tissue stem cells (Artavanis-Tsakonas et al., 1995; Kumano et al., 2003; Mizutani et al., 2007). Members of the Notch signaling pathway, Notch1 & 2, JAG1 and Delta, had significantly higher expression in the H1 hES large cell population in the microarray analysis) and 2.5 to 5-fold by qPCR analysis (Figure 7A and B). Negative regulators of notch signaling CoupTF1 (NR2F1), CoupTF2 (NR2F2) and manic fringe showed significant higher gene expression in the H1 hES small cell population (Figure 6A–C). CoupTF1 (NR2F1) and CoupTF2 (NR2F2) have been shown to negatively regulate RA and Notch signaling ultimately repressing differentiation (Ben-Shushan et al., 1995; Studer et al., 2005; Tran et al., 1992; You et al., 2005). Apart from Notch signaling, which may suggest interdependency between the two populations, other signaling axis may be important in determining the small and large populations. Our microarray analysis indicates that expression of GDF3, a BMP inhibitor, is 2 fold higher in large cell population (Figure 6B). One might expect the expression of signaling receptors ALK7 and TDFG-1 to be more highly expressed in the small population, but in fact the two receptors were higher in the large cell population by 1.05 and 1.17, respectively (http://www.ncbi.nlm.nih.gov/geo/ accession number GSF24530).
Figure 7. Differential expression of notch signaling pathway genes and CoupTF1&2.
A) Heat map depicting expression of Notch signaling components and orphan receptors, CoupTF1 and CoupTF2 in H1 hES small and large cell population. B) mRNA expression of CoupTF1&2 and notch signaling pathway genes. mRNA expression is normalized to small cell population. C) Relative mRNA expression of CoupTF1 and Notch1 after treating the small and large cell population with RA for 6 days.
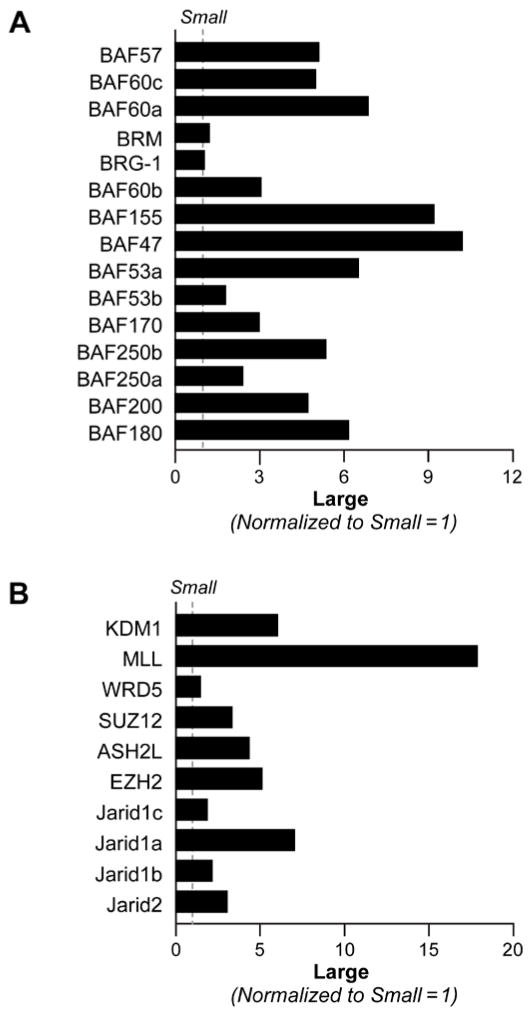
Given the substantial differences in gene expression between the large and small cells we next looked to differences in global regulators of transcription. Several studies have suggested a role of the SWI/SNF chromatin remodeling complex and coordinated regulation of epigenetic marks by histone modifying enzymes in the maintenance of stem cell state. Further microarray analysis showed that members of the BRG1 associated factors (BAF) within the SWI/SNF complex and histone-modifying enzymes were differentially expressed in the two populations. qPCR analysis showed that BAF proteins particularly BAF47, BAF155, BAF60a, BAF53a, BAF180 and BAF250b were expressed at much higher levels in the large cell population compared to small population, (Figure 8A). In contrast, BRG-1 and BRM expression were equal between the two populations (Figure 8A) Histone modifications have been reported to result in specific changes in local chromatin architecture which allow the ES cell genome to maintain pluripotency while poised for differentiation (Adamo et al., 2011b; Bertani et al., 2011; Foster et al., 2010; Fuentes et al., 2011; Lim et al., 2009). Analysis of a subset of the enzymes that maintain these marks showed that KDM1, MLL1 and JARID1A were significantly higher in large cell compared to small cell population (Figure 8B).
Figure 8. Differential mRNA expression of chromatin remodeling and histone modifying genes between small and large cell populations.
A) Quantitative RT-PCR analysis showing mRNA expression of BRG1 associated factors within the SWI/SNF complex in H1 hES small and large cell populations. mRNA expression is normalized to the levels expressed in small population. B) ) Quantitative RT-PCR analysis showing mRNA expression of histone modifying enzymes in H1 hES small and large cell populations. mRNA expression is normalized to the levels expressed in small population.
3.9. Effect of RA Treatment on Notch Signaling and Chromatin Modifying Enzymes
Since RA is widely used to differentiate ES cells we tested the effect of RA treatment on expression of those components differentially expressed in large cell compared to small cell population. CoupTF1&2, Notch signaling components, and Notch target genes mRNA expression was examined in H1 hES cells after 6 days of RA treatment. In the large cell population, expression of Notch1, Jag1 significantly decreased whereas that of Notch2 increased after RA treatment. In the small cell population expression of Notch2 significantly increased, whereas Notch1 and MNFG decreased (Supplemental Figure 7A – B). In addition, treatment with RA induced expression of Notch signaling target genes, Hes1, Hey1 and DTX-1 2–3-fold in the large population but not in the small cell population (Supplemental Figure 7A–B). There was a 5–7-fold increase in CoupTF1&2 expressions in the large cell population after a 6-day treatment of RA and relatively no change in the small cell population (Supplemental Figure 7A – B) Treatment with RA also affected the expression of a number of SWI/SNF components and histone modifying enzymes in H1 hES large and small cell populations. There was no significant change in expression of either group of genes in the small population cells treated with RA (Supplemental Figure 8A). In the large cell population treatment with RA increased BRM expression 3.5 fold with moderate increases in BRG1, BAF155 and BAF53b (Supplemental 8B). Similar to the SWI/SNF components there were no significant changes in the expression of histone modifying enzymes in the small cell population after RA treatment Supplemental Figure 9A). In the large population expression of KDM1 (LSD1), MLL1, EZH2, and JARID2 decreased by 60% or greater, whereas KDM5A (JARID1A) and KDM5B (JARID1B) both showed modest increase (Supplemental Figure 9B).
3.10. Differential Response of Subpopulations to Endocrine Disruptor Compounds
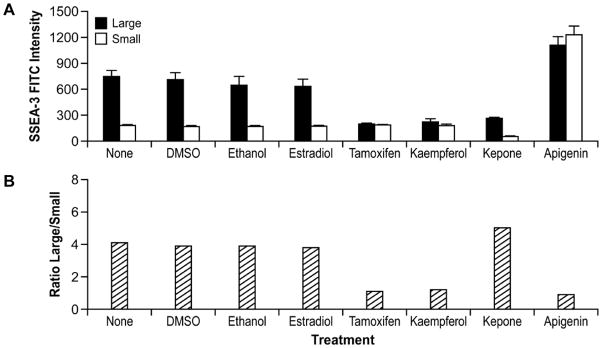
Given the differential response of the two populations to retinoic acid, we next examined if the two populations respond differentially to a limited number of estrogenic endocrine disrupting compounds (EDCs). We monitored response to EDCs by measuring the levels of the stem cell surface marker SSEA-3 and Oct-4 protein expression in H1 and H9 hES cells. Four EDC’s consistently changed SSEA-3 levels throughout the cell population, as well as differentially affected SSEA-3 expression in the two subpopulations. SSEA-3 expression was not significantly different among non-treated, vehicle controls or estrogen treated cells (Figure 9A and Supplemental Figure 10A). Treatment with three EDC compounds: tamoxifen, kaempferol, and kepone for 2 days decreased expression of SSEA-3 in both H1 and H9 hES cells. The fourth compound, apigenin, increased SSEA-3 expression in H1 and H9 hES cells (Supplemental Figure 10A). In addition tamoxifen and kaempferol decreased SSEA-3 expression by 70% in H1 and H9 hES large cell population, without substantially altering SSEA-3 expression in small cell populations (Figure 9A). This had the effect of changing the ratio of large to small cells in the population from 4:1 to 1:1 (Figure 9B). In contrast to the effects seen with kaempferol and tamoxifen, treatment with kepone decreased both large and small cells that expressed SSEA-3, such that the ratio of large to small SSEA-3 positive cells remained unchanged (Figure 9A–B). Finally apigenin treatment led to a third response such that expression of SSEA-3 increased in both populations. The small population SSEA-3 expression increased 2 to 4-fold, once again reducing the ratio of large and small cell to 1:1, as seen with tamoxifen and kaempferol (Figure 9A – B). We analyzed Oct4 protein expression after EDC treatment. Treatment with tamoxifen, kaempferol and to a lesser degree kepone and apigenin decreased Oct-4 protein expression in the cells (Supplemental Figure 10B). In addition Treatment with most of the EDC’s did not change cell morphology; however, treatment with kaempferol changed overall cell morphology resulting in enlarged cells (Supplemental Figure 10C).
Figure 9. Differential responses by subpopulations to treatment with endocrine disruptor compounds.
A) H9 hES cells were grown and exposed to EDC for 2 days and hybridized with SSEA-3-FITC and then sorted for large and small populations expressing SSEA-3. Similar results are shown in B) Ratio of SSEA-3 positive large cells to SSEA-3 positive small cells.
4. Discussion
The ability of hES cells to respond along distinct developmental pathways in response to different signaling cues is an area of intense investigation. Equally important is the ability, in the absence of external stimuli, to self-renew and retain their unique developmental potential. We show that well established hES cells are heterogeneous in nature with respect to cell size and that the two sub-populations vary in their response to retinoic acid (RA) differentiation and exposure to endocrine disrupting compounds (EDCs). In order to understand why these two subpopulations responded differentially to various stimulatory drugs, extensive characterization of gene expression using microarray analysis demonstrated the two populations vary in distinct gene expression profiles that may dictate specific cell lineages, such as integrin alpha6 and stem surface markers. Genes associated with self-renewal and pluripotency were expressed in both subpopulations albeit at varying levels. In general, metabolic and structural genes were increased in the large population, whereas genes encoding signaling pathways for example Notch were decreased in the small population perhaps suggesting a functional dependency between the two populations or ability of either population to respond to specific signaling cues leading to distinct cell states. For example, from microarray and qRTPCR analysis we find the expression of notch signaling components differs between the two cell populations (Figure 7). The large population expresses high levels of Notch1 and Notch 2 whereas the small population expresses high levels of the orphan receptors CoupTF1 and 2, which are negative regulators of Notch signaling. Retinoic acid is routinely used to induce neuronal differentiation of which the consequence is a decrease Oct-4 expression. The Oct-4 promoter harbors a RA-responsive element that can bind RAR/RXR and thus be down regulated directly by RA and induce neuronal differentiation (Schoorlemmer et al., 1995; Schoorlemmer et al., 1994; Zechel, 2005). Studies in P19 cells have shown that orphan receptors, CoupTF1 and 2 act as negative regulators of the RA response pathway by binding with high affinity to the responsive elements activated by RAR/RXR heterodimers (Ben-Shushan et al., 1995). Indeed we demonstrate that CoupTF1 and 2 expression is higher in the small population, and Oct-4 levels remain relatively unchanged after RA exposure, suggesting that differences in gene signature between the two populations can dictate different cell fate. The large cell population is more susceptible to RA differentiation signals than the small population. The differential response to RA may be important in understanding mechanisms by which heterogeneous hES cells can potentially dictate lineage specificity and additional facets of stem cell biology.
Our study also shows that the two hES populations differ in their expression of key subunits of the SWI/SNF ATP dependent chromatin remodeling complex and a significant number of histone modifying enzymes (Figure 8). Chromatin architecture and epigenetic mechanisms play important roles in determining ES cell fate transitions. Recent studies have implicated specific subunits of ATP-dependent chromatin remodeling complex (SWI/SNF) in regulating differentiation and self-renewal. The switching of the BAF subunits correlates with the transition from pluripotency to progenitor and eventually differentiated cells (Ho et al., 2009; Yoo and Crabtree, 2009; Yoo et al., 2009). Epigenetic mechanisms include modification of histones. Slight variations of epigenetic components can result in significant changes in chromatin environment within the hES cell. For example, recent studies have revealed a bivalent chromatin state may regulate the ‘ON’ and “OFF” states of key pluripotency factors to drive cell fate transitions (Adamo et al.; Bernstein et al., 2006; Meshorer and Misteli, 2006; Wu et al., 2009; Yoo and Crabtree, 2009). In addition, expression of polycomb and the MLL enzyme complexes that regulate this bivalent state are tightly controlled (Adamo et al.; Ahn et al.; Bertani et al.; Foster et al.; Fuentes et al.; Lim et al., 2009; Pasini et al.; Shen et al., 2009; Wang et al., 2007). Our study finds that members of the polycomb complex (EZH2, SUZ12) and MLL complex (MLL1, ASH2L, LSD1, JARID1A) are differentially expressed between the two populations (Figure 8) and these components respond to RA treatment only in the large population (Supplemental Figure 9). The importance of epigenetic control during neural development and maintenance of pluripotency through the regulation of bivalent domains has been identified in several studies with LSD1 (KDM1), JARID2, MLL1 (Adamo et al., 2011b; Bernstein et al., 2006; Chen et al., 2007; Das et al., 2007; Foster et al., 2010; Fuentes et al., 2011; Lim et al., 2009; Sun et al., 2010; Wang et al., 2007; Wu et al., 2009; Zhang et al., 2011). Our results indicate SWI/SNF chromatin remodeling and polycomb components differ between the two hES populations. We propose that different ratios of the BAF subunits of the SWI/SNF complex and the histone modifying enzymes may affect the ability of the small cell population to differentiate upon RA treatment as shown by the negligible loss of OCT-4 (Figure 5).
The importance of the environment to human health and disease has sparked intense interest in the consequences of early exposures. It is clear that environmental exposures can influence human development and potentially lead to carcinogenesis but little is known how stem cells respond to environmental exposures to various EDCs, whether they are pharmaceutical drugs, pesticides, herbicides or naturally occurring substances in food. We observed that four compounds, tamoxifen, kaempferol, kepone and apigenin altered SSEA-3 expression in the entire population as well as the two subpopulations of both H1 and H9 hES cells. We propose the differential response elicited by EDCs and differentiation stimuli may be due in part to different distinct gene expression profiles between the two subpopulations as discussed above. The four compounds are all estrogenic compounds, although they may function through ER dependent or independent pathways. ER expression in hES cells is not well characterized and a recent analysis of hES cells did not detect any ER transcript (Xie et al., 2009). Tamoxifen, a chemotherapeutic drug, is both an estrogen antagonist and agonist depending on the tissue (Ali et al., 2011; Frasor et al., 2006). Tamoxifen reduced SSEA-3 expression in the large cell population, but did not alter SSEA-3 expression in the small cell population. This response was similar using the compound kaempferol, a naturally occurring antioxidant flavonol found in many plant sources. Interestingly, Apigenin, a bioflavonoid with antioxidant properties thought to have many health benefits similar to kaempferol, elicits an increase in SSEA-3 expression in both small and large population of hES cells. Apigenin has been reported to improve the efficiency of iPS reprogramming of fibroblasts through the up regulation of e-cadherin very early in the reprogramming process and may be necessary for reprogramming to occur (Chen et al., 2010) Tamoxifen, keampferol and apigenin all changed the hES subpopulations to a 1:1 ratio, either by decreasing SSEA-3 in large cells or increasing expression in small hES cells. Kepone, a carcinogenic compound found in insecticides and herbicides, reduced SSEA-3 expression in both small and large populations, thereby maintaining a ratio of 4:1 between large and small cells similar to control hES cells. The altering of the ratios of large to small cells may potentially predispose the population in terms of directing cell lineage differentiation pathways, as well as self-renewal properties.
In summary, the two subpopulations we have identified based on size, are similar in their stem and cell surface marker expression as the subpopulations recently described in the literature (Hough et al., 2009; Laslett et al., 2007) where a model of multiple pluripotency states or a continuum of cell states between the pluripotent cell and the lineage committed hES cell were proposed. Using two cell surface proteoglycan markers, GCTM-2 and TG30, the authors showed significant heterogeneity within a single stem cell colony. Transcript analysis of the single cell colony showed that there was a gradient and a hierarchy of expression of pluripotency genes, OCT-4, NANOG, DNMT3B, the growth factor GDF3 and the nodal receptor TDGF-1 in the single cell population. In accordance to previous data, the current study supports heterogeneity of hES cells and shows distinct differences between two defined populations as defined by cell surface markers, TRA-1-60, TRA-1-80, SSEA3/4 in addition to notch signaling components. Similar to Hough et. al, we show that the large cell population expresses higher levels of GDF3, DNMT3B andTDGF-1(data not shown). Conversely, we show that NANOG and GTCM-2 are more highly expressed in the small cell population, suggesting that GTCM-2 expression does not correlate with higher GDF3 expression in our defined hES subpopulations, a finding that could be accounted for by differences in hES cell types used in the study. Other researchers have identified subpopulations of hES cells based on the expression levels of SSEA-3 (Stewart et al., 2006) or NANOG expression (Singh et al., 2007). Our observations would be consistent with the findings of both groups showing low SSEA-3 expression correlates with a less active cell cycle and low NANOG expression correlates with high GATA6 expression. Different hES cell lines have been shown to have varying differentiation capabilities (Tavakoli et al., 2009), and similar to our small cell population, very small embryonic-like stem cells (VSELs) have been discovered in cord blood and are pluripotent with maximum regenerative potential (Bhartiya et al.). Our in vivo studies confirm the pluripotency of the large cell population. However, injection of small cell population into beige-scid mice did not produce a teratoma. These observations suggest that there most likely exists a continuum in expression of pluripontency markers and subsets of subpopulations based on distinct expression of specific markers will continue to be identified. Eventually differences in expression of various hES heterogenenous subpopulations may give us more insight into the complex determination of cell fate and the interplay between transcription factors, chromatin remodeling factors, histone modifying enzymes and cell signaling pathways. Human stem cells are not a homogeneous population and display distinct characteristics that may impact their use for human therapeutic research as well as basic research. Finally, RA and EDC studies suggest that the two subpopulations have different sensitivities to signaling cues, which may dictate ES composition and therefore have consequences in human development and disease. These observations are broadly relevant as they suggest avenues for rapid identification of populations of stem cells that might be used more efficiently directed along specific developmental pathways for formation of particular progenitor cells.
Supplementary Material
Highlights.
Little is known about heterogeneity of human embryonic stem cell (hESCs) and how they functionally respond to signaling cues.
We show that hESCs have two heterogeneous populations with distinct characteristics including size, and gene expression signatures that dictate their responsiveness to retinoic acid differentiation and endocrine disruptor compounds.
Differential expression of key signaling and chromatin regulators in part accounts for the ability of subpopulations to differentially respond to signaling cues.
The presence of heterogeneous population in hESCs is important in directing cell lineage for regenerative medicine and understanding cellular development in response to endogenous signals and environmental exposures.
Acknowledgments
This research was supported by the Intramural Research Program of the National Institute of Environmental Health Sciences, NIH; project number Z01 ES071006-10.
We would like to thank the members of the Chromatin and Gene Expression Group of LMC for their help in preparation of this manuscript: Drs G. Hu, L. Lazarus, B. Rao and V. Davis for review of the manuscript. We thank Dhiral Phadke for preparing heat maps to represent the microarray data.
Footnotes
The microarray data discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/ and are accessible through GEO Series accession number GSE24530
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adamo A, Barrero MJ, Belmonte JC. LSD1 and pluripotency: a new player in the network. Cell Cycle. 2011a;10:3215–3216. doi: 10.4161/cc.10.19.17052. [DOI] [PubMed] [Google Scholar]
- Adamo A, Sese B, Boue S, Castano J, Paramonov I, Barrero MJ, Izpisua Belmonte JC. LSD1 regulates the balance between self-renewal and differentiation in human embryonic stem cells. Nature cell biology. 2011b;13:652–659. doi: 10.1038/ncb2246. [DOI] [PubMed] [Google Scholar]
- Ahn SE, Kim S, Park KH, Moon SH, Lee HJ, Kim GJ, Lee YJ, Cha KY, Chung HM. Primary bone-derived cells induce osteogenic differentiation without exogenous factors in human embryonic stem cells. Biochem Biophys Res Commun. 2006;340:403–408. doi: 10.1016/j.bbrc.2005.12.020. [DOI] [PubMed] [Google Scholar]
- Ali S, Buluwela L, Coombes RC. Antiestrogens and their therapeutic applications in breast cancer and other diseases. Annu Rev Med. 2011;62:217–232. doi: 10.1146/annurev-med-052209-100305. [DOI] [PubMed] [Google Scholar]
- Allegrucci C, Wu YZ, Thurston A, Denning CN, Priddle H, Mummery CL, Ward-van Oostwaard D, Andrews PW, Stojkovic M, Smith N, Parkin T, Jones ME, Warren G, Yu L, Brena RM, Plass C, Young LE. Restriction landmark genome scanning identifies culture-induced DNA methylation instability in the human embryonic stem cell epigenome. Hum Mol Genet. 2007;16:1253–1268. doi: 10.1093/hmg/ddm074. [DOI] [PubMed] [Google Scholar]
- Artavanis-Tsakonas S, Matsuno K, Fortini ME. Notch signaling. Science. 1995;268:225–232. doi: 10.1126/science.7716513. [DOI] [PubMed] [Google Scholar]
- Ben-Shushan E, Sharir H, Pikarsky E, Bergman Y. A dynamic balance between ARP-1/COUP-TFII, EAR-3/COUP-TFI, and retinoic acid receptor:retinoid X receptor heterodimers regulates Oct-3/4 expression in embryonal carcinoma cells. Mol Cell Biol. 1995;15:1034–1048. doi: 10.1128/mcb.15.2.1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, Jaenisch R, Wagschal A, Feil R, Schreiber SL, Lander ES. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Bertani S, Sauer S, Bolotin E, Sauer F. The noncoding RNA Mistral activates Hoxa6 and Hoxa7 expression and stem cell differentiation by recruiting MLL1 to chromatin. Molecular cell. 2011;43:1040–1046. doi: 10.1016/j.molcel.2011.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Bhartiya D, Shaikh A, Nagvenkar P, Kasiviswanathan S, Pethe P, Pawani H, Mohanty S, Rao SG, Zaveri K, Hinduja I. Very small embryonic-like stem cells with maximum regenerative potential get discarded during cord blood banking and bone marrow processing for autologous stem cell therapy. Stem cells and development. 2012;21:1–6. doi: 10.1089/scd.2011.0311. [DOI] [PubMed] [Google Scholar]
- Bortner CD, Cidlowski JA. Cell shrinkage and monovalent cation fluxes: role in apoptosis. Arch Biochem Biophys. 2007;462:176–188. doi: 10.1016/j.abb.2007.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canham MA, Sharov AA, Ko MS, Brickman JM. Functional heterogeneity of embryonic stem cells revealed through translational amplification of an early endodermal transcript. PLoS biology. 2010;8:e1000379. doi: 10.1371/journal.pbio.1000379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Card DA, Hebbar PB, Li L, Trotter KW, Komatsu Y, Mishina Y, Archer TK. Oct4/Sox2-regulated miR-302 targets cyclin D1 in human embryonic stem cells. Mol Cell Biol. 2008;28:6426–6438. doi: 10.1128/MCB.00359-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen PS, Wang CC, Bortner CD, Peng GS, Wu X, Pang H, Lu RB, Gean PW, Chuang DM, Hong JS. Valproic acid and other histone deacetylase inhibitors induce microglial apoptosis and attenuate lipopolysaccharide-induced dopaminergic neurotoxicity. Neuroscience. 2007;149:203–212. doi: 10.1016/j.neuroscience.2007.06.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T, Yuan D, Wei B, Jiang J, Kang J, Ling K, Gu Y, Li J, Xiao L, Pei G. E-cadherin-mediated cell-cell contact is critical for induced pluripotent stem cell generation. Stem Cells. 2010;28:1315–1325. doi: 10.1002/stem.456. [DOI] [PubMed] [Google Scholar]
- Das AV, James J, Bhattacharya S, Imbalzano AN, Antony ML, Hegde G, Zhao X, Mallya K, Ahmad F, Knudsen E, Ahmad I. SWI/SNF chromatin remodeling ATPase Brm regulates the differentiation of early retinal stem cells/progenitors by influencing Brn3b expression and Notch signaling. J Biol Chem. 2007;282:35187–35201. doi: 10.1074/jbc.M706742200. [DOI] [PubMed] [Google Scholar]
- Foster CT, Dovey OM, Lezina L, Luo JL, Gant TW, Barlev N, Bradley A, Cowley SM. Lysine-specific demethylase 1 regulates the embryonic transcriptome and CoREST stability. Molecular and cellular biology. 2010;30:4851–4863. doi: 10.1128/MCB.00521-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frasor J, Chang EC, Komm B, Lin CY, Vega VB, Liu ET, Miller LD, Smeds J, Bergh J, Katzenellenbogen BS. Gene expression preferentially regulated by tamoxifen in breast cancer cells and correlations with clinical outcome. Cancer Res. 2006;66:7334–7340. doi: 10.1158/0008-5472.CAN-05-4269. [DOI] [PubMed] [Google Scholar]
- Fuentes P, Canovas J, Berndt FA, Noctor SC, Kukuljan M. CoREST/LSD1 Control the Development of Pyramidal Cortical Neurons. Cerebral cortex. 2011 doi: 10.1093/cercor/bhr218. [DOI] [PubMed] [Google Scholar]
- Gao X, Tate P, Hu P, Tjian R, Skarnes WC, Wang Z. ES cell pluripotency and germ-layer formation require the SWI/SNF chromatin remodeling component BAF250a. Proc Natl Acad Sci U S A. 2008;105:6656–6661. doi: 10.1073/pnas.0801802105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harb N, Archer TK, Sato N. The Rho-Rock-Myosin signaling axis determines cell-cell integrity of self-renewing pluripotent stem cells. PLoS One. 2008;3:e3001. doi: 10.1371/journal.pone.0003001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins RD, Hon GC, Lee LK, Ngo Q, Lister R, Pelizzola M, Edsall LE, Kuan S, Luu Y, Klugman S, Antosiewicz-Bourget J, Ye Z, Espinoza C, Agarwahl S, Shen L, Ruotti V, Wang W, Stewart R, Thomson JA, Ecker JR, Ren B. Distinct epigenomic landscapes of pluripotent and lineage-committed human cells. Cell Stem Cell. 2010;6:479–491. doi: 10.1016/j.stem.2010.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho L, Jothi R, Ronan JL, Cui K, Zhao K, Crabtree GR. An embryonic stem cell chromatin remodeling complex, esBAF, is an essential component of the core pluripotency transcriptional network. Proc Natl Acad Sci U S A. 2009;106:5187–5191. doi: 10.1073/pnas.0812888106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hough SR, Laslett AL, Grimmond SB, Kolle G, Pera MF. A continuum of cell states spans pluripotency and lineage commitment in human embryonic stem cells. PLoS One. 2009;4:e7708. doi: 10.1371/journal.pone.0007708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu BY, Zhang SC. Directed differentiation of neural-stem cells and subtype-specific neurons from hESCs. Methods Mol Biol. 2010;636:123–137. doi: 10.1007/978-1-60761-691-7_8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, Lin X, Feng Y, Xie Y, Han J, Zhang Y, Wang ZZ, Chen T. Hemato-endothelial differentiation from lentiviral-transduced human embryonic stem cells retains durable reporter gene expression under the control of ubiquitin promoter. Cytotechnology. 2010;62:31–42. doi: 10.1007/s10616-010-9258-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayama M, Kurokawa MS, Ueda Y, Ueno H, Kumagai Y, Chiba S, Takada E, Ueno S, Tadokoro M, Suzuki N. Transfection with pax6 Gene of Mouse Embryonic Stem Cells and Subsequent Cell Cloning Induced Retinal Neuron Progenitors, Including Retinal Ganglion Cell-Like Cells, in vitro. Ophthalmic Res. 2009;43:79–91. doi: 10.1159/000247592. [DOI] [PubMed] [Google Scholar]
- King FW, Ritner C, Liszewski W, Kwan HC, Pedersen A, Leavitt AD, Bernstein HS. Subpopulations of human embryonic stem cells with distinct tissue-specific fates can be selected from pluripotent cultures. Stem Cells Dev. 2009;18:1441–1450. doi: 10.1089/scd.2009.0012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumano K, Chiba S, Kunisato A, Sata M, Saito T, Nakagami-Yamaguchi E, Yamaguchi T, Masuda S, Shimizu K, Takahashi T, Ogawa S, Hamada Y, Hirai H. Notch1 but not Notch2 is essential for generating hematopoietic stem cells from endothelial cells. Immunity. 2003;18:699–711. doi: 10.1016/s1074-7613(03)00117-1. [DOI] [PubMed] [Google Scholar]
- Laslett AL, Grimmond S, Gardiner B, Stamp L, Lin A, Hawes SM, Wormald S, Nikolic-Paterson D, Haylock D, Pera MF. Transcriptional analysis of early lineage commitment in human embryonic stem cells. BMC Dev Biol. 2007;7:12. doi: 10.1186/1471-213X-7-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecina M, Ting S, Choo A, Reuveny S, Oh S. Scalable Platform for hESC Differentiation to Cardiomyocytes in Suspended Microcarrier Cultures. Tissue Eng Part C Methods. 2010 doi: 10.1089/ten.TEC.2010.0104. [DOI] [PubMed] [Google Scholar]
- Lee JB, Kim JM, Kim SJ, Park JH, Hong SH, Roh SI, Kim MK, Yoon HS. Comparative characteristics of three human embryonic stem cell lines. Mol Cells. 2005;19:31–38. [PubMed] [Google Scholar]
- Lengner CJ, Gimelbrant AA, Erwin JA, Cheng AW, Guenther MG, Welstead GG, Alagappan R, Frampton GM, Xu P, Muffat J, Santagata S, Powers D, Barrett CB, Young RA, Lee JT, Jaenisch R, Mitalipova M. Derivation of pre-X inactivation human embryonic stem cells under physiological oxygen concentrations. Cell. 2010;141:872–883. doi: 10.1016/j.cell.2010.04.010. [DOI] [PubMed] [Google Scholar]
- Lim DA, Huang YC, Swigut T, Mirick AL, Garcia-Verdugo JM, Wysocka J, Ernst P, Alvarez-Buylla A. Chromatin remodelling factor Mll1 is essential for neurogenesis from postnatal neural stem cells. Nature. 2009;458:529–533. doi: 10.1038/nature07726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng Y, Eshghi S, Li YJ, Schmidt R, Schaffer DV, Healy KE. Characterization of integrin engagement during defined human embryonic stem cell culture. FASEB J. 2010;24:1056–1065. doi: 10.1096/fj.08-126821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meshorer E, Misteli T. Chromatin in pluripotent embryonic stem cells and differentiation. Nat Rev Mol Cell Biol. 2006;7:540–546. doi: 10.1038/nrm1938. [DOI] [PubMed] [Google Scholar]
- Mizutani K, Yoon K, Dang L, Tokunaga A, Gaiano N. Differential Notch signalling distinguishes neural stem cells from intermediate progenitors. Nature. 2007;449:351–355. doi: 10.1038/nature06090. [DOI] [PubMed] [Google Scholar]
- Nagano K, Yoshida Y, Isobe T. Cell surface biomarkers of embryonic stem cells. Proteomics. 2008;8:4025–4035. doi: 10.1002/pmic.200800073. [DOI] [PubMed] [Google Scholar]
- Niederreither K, Dolle P. Retinoic acid in development: towards an integrated view. Nat Rev Genet. 2008;9:541–553. doi: 10.1038/nrg2340. [DOI] [PubMed] [Google Scholar]
- Pan GJ, Chang ZY, Scholer HR, Pei D. Stem cell pluripotency and transcription factor Oct4. Cell Res. 2002;12:321–329. doi: 10.1038/sj.cr.7290134. [DOI] [PubMed] [Google Scholar]
- Parsons XH, Teng YD, Parsons JF, Snyder EY, Smotrich DB, Moore DA. Efficient derivation of human neuronal progenitors and neurons from pluripotent human embryonic stem cells with small molecule induction. Journal of visualized experiments : JoVE. 2011 doi: 10.3791/3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasini D, Cloos PA, Walfridsson J, Olsson L, Bukowski JP, Johansen JV, Bak M, Tommerup N, Rappsilber J, Helin K. JARID2 regulates binding of the Polycomb repressive complex 2 to target genes in ES cells. Nature. 2010;464:306–310. doi: 10.1038/nature08788. [DOI] [PubMed] [Google Scholar]
- Prowse AB, Chong F, Gray PP, Munro TP. Stem cell integrins: implications for ex-vivo culture and cellular therapies. Stem cell research. 2011;6:1–12. doi: 10.1016/j.scr.2010.09.005. [DOI] [PubMed] [Google Scholar]
- Rao M. Scalable human ES culture for therapeutic use: propagation, differentiation, genetic modification and regulatory issues. Gene Ther. 2008;15:82–88. doi: 10.1038/sj.gt.3303061. [DOI] [PubMed] [Google Scholar]
- Schoorlemmer J, Jonk L, Sanbing S, van Puijenbroek A, Feijen A, Kruijer W. Regulation of Oct-4 gene expression during differentiation of EC cells. Mol Biol Rep. 1995;21:129–140. doi: 10.1007/BF00997235. [DOI] [PubMed] [Google Scholar]
- Schoorlemmer J, van Puijenbroek A, van Den Eijnden M, Jonk L, Pals C, Kruijer W. Characterization of a negative retinoic acid response element in the murine Oct4 promoter. Mol Cell Biol. 1994;14:1122–1136. doi: 10.1128/mcb.14.2.1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharov AA, Nishiyama A, Piao Y, Correa-Cerro LS, Amano T, Thomas M, Mehta S, Ko MS. Responsiveness of genes to manipulation of transcription factors in ES cells is associated with histone modifications and tissue specificity. BMC Genomics. 2011;12:102. doi: 10.1186/1471-2164-12-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X, Kim W, Fujiwara Y, Simon MD, Liu Y, Mysliwiec MR, Yuan GC, Lee Y, Orkin SH. Jumonji modulates polycomb activity and self-renewal versus differentiation of stem cells. Cell. 2009;139:1303–1314. doi: 10.1016/j.cell.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh AM, Hamazaki T, Hankowski KE, Terada N. A heterogeneous expression pattern for Nanog in embryonic stem cells. Stem Cells. 2007;25:2534–2542. doi: 10.1634/stemcells.2007-0126. [DOI] [PubMed] [Google Scholar]
- Stewart MH, Bosse M, Chadwick K, Menendez P, Bendall SC, Bhatia M. Clonal isolation of hESCs reveals heterogeneity within the pluripotent stem cell compartment. Nat Methods. 2006;3:807–815. doi: 10.1038/nmeth939. [DOI] [PubMed] [Google Scholar]
- Stewart R, Yang C, Anyfantis G, Przyborski S, Hole N, Strachan T, Stojkovic M, Keith WN, Armstrong L, Lako M. Silencing of the expression of pluripotent driven-reporter genes stably transfected into human pluripotent cells. Regen Med. 2008;3:505–522. doi: 10.2217/17460751.3.4.505. [DOI] [PubMed] [Google Scholar]
- Strickland S, Mahdavi V. The induction of differentiation in teratocarcinoma stem cells by retinoic acid. Cell. 1978;15:393–403. doi: 10.1016/0092-8674(78)90008-9. [DOI] [PubMed] [Google Scholar]
- Studer M, Filosa A, Rubenstein JL. The nuclear receptor COUP-TFI represses differentiation of Cajal-Retzius cells. Brain Res Bull. 2005;66:394–401. doi: 10.1016/j.brainresbull.2004.11.027. [DOI] [PubMed] [Google Scholar]
- Sun G, Alzayady K, Stewart R, Ye P, Yang S, Li W, Shi Y. Histone demethylase LSD1 regulates neural stem cell proliferation. Molecular and cellular biology. 2010;30:1997–2005. doi: 10.1128/MCB.01116-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavakoli T, Xu X, Derby E, Serebryakova Y, Reid Y, Rao MS, Mattson MP, Ma W. Self-renewal and differentiation capabilities are variable between human embryonic stem cell lines I3, I6 and BG01V. BMC Cell Biol. 2009;10:44. doi: 10.1186/1471-2121-10-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, Jones JM. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- Tran P, Zhang XK, Salbert G, Hermann T, Lehmann JM, Pfahl M. COUP orphan receptors are negative regulators of retinoic acid response pathways. Mol Cell Biol. 1992;12:4666–4676. doi: 10.1128/mcb.12.10.4666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Scully K, Zhu X, Cai L, Zhang J, Prefontaine GG, Krones A, Ohgi KA, Zhu P, Garcia-Bassets I, Liu F, Taylor H, Lozach J, Jayes FL, Korach KS, Glass CK, Fu XD, Rosenfeld MG. Opposing LSD1 complexes function in developmental gene activation and repression programmes. Nature. 2007;446:882–887. doi: 10.1038/nature05671. [DOI] [PubMed] [Google Scholar]
- Weng L, Dai H, Zhan Y, He Y, Stepaniants SB, Bassett DE. Rosetta error model for gene expression analysis. Bioinformatics. 2006;22:1111–1121. doi: 10.1093/bioinformatics/btl045. [DOI] [PubMed] [Google Scholar]
- Wilschut KJ, van Tol HT, Arkesteijn GJ, Haagsman HP, Roelen BA. Alpha 6 integrin is important for myogenic stem cell differentiation. Stem cell research. 2011;7:112–123. doi: 10.1016/j.scr.2011.05.001. [DOI] [PubMed] [Google Scholar]
- Wu M, Zhang Y, Wu NH, Shen YF. Histone marks and chromatin remodelers on the regulation of neurogenin1 gene in RA induced neuronal differentiation of P19 cells. J Cell Biochem. 2009;107:264–271. doi: 10.1002/jcb.22122. [DOI] [PubMed] [Google Scholar]
- Xie CQ, Jeong Y, Fu M, Bookout AL, Garcia-Barrio MT, Sun T, Kim BH, Xie Y, Root S, Zhang J, Xu RH, Chen YE, Mangelsdorf DJ. Expression profiling of nuclear receptors in human and mouse embryonic stem cells. Molecular endocrinology. 2009;23:724–733. doi: 10.1210/me.2008-0465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Z, Wang Z, Sharova L, Sharov AA, Ling C, Piao Y, Aiba K, Matoba R, Wang W, Ko MS. BAF250B-associated SWI/SNF chromatin-remodeling complex is required to maintain undifferentiated mouse embryonic stem cells. Stem Cells. 2008;26:1155–1165. doi: 10.1634/stemcells.2007-0846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo AS, Crabtree GR. ATP-dependent chromatin remodeling in neural development. Curr Opin Neurobiol. 2009;19:120–126. doi: 10.1016/j.conb.2009.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo AS, Staahl BT, Chen L, Crabtree GR. MicroRNA-mediated switching of chromatin-remodelling complexes in neural development. Nature. 2009;460:642–646. doi: 10.1038/nature08139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You LR, Lin FJ, Lee CT, DeMayo FJ, Tsai MJ, Tsai SY. Suppression of Notch signalling by the COUP-TFII transcription factor regulates vein identity. Nature. 2005;435:98–104. doi: 10.1038/nature03511. [DOI] [PubMed] [Google Scholar]
- Young RA. Control of the embryonic stem cell state. Cell. 2011;144:940–954. doi: 10.1016/j.cell.2011.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zechel C. Requirement of retinoic acid receptor isotypes alpha, beta, and gamma during the initial steps of neural differentiation of PCC7 cells. Mol Endocrinol. 2005;19:1629–1645. doi: 10.1210/me.2004-0540. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Jones A, Sun CW, Li C, Chang CW, Joo HY, Dai Q, Mysliwiec MR, Wu LC, Guo Y, Yang W, Liu K, Pawlik KM, Erdjument-Bromage H, Tempst P, Lee Y, Min J, Townes TM, Wang H. PRC2 complexes with JARID2, MTF2, and esPRC2p48 in ES cells to modulate ES cell pluripotency and somatic cell reprogramming. Stem Cells. 2011;29:229–240. doi: 10.1002/stem.578. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.