Abstract
Lung cancer is the leading cause of cancer deaths worldwide, and current therapies fail to treat this disease in the vast majority of cases. The RAS and p53 pathways are two of the most frequently altered pathways in lung cancers, with such alterations resulting in loss of responsiveness to current therapies and decreased patient survival. The microRNA-34 (mir-34) gene family members are downstream transcriptional targets of p53, and miR-34 expression is reduced in p53 mutant tumors; thus, we hypothesized that treating mutant Kras;p53 tumors with miR-34 would represent a powerful new therapeutic to suppress lung tumorigenesis. To this end we examined the therapeutically resistant KrasLSL-G12D/+;Trp53LSL-R172H/+ mouse lung cancer model. We characterized tumor progression in these mice following lung-specific transgene activation and found tumors as early as 10 weeks post-activation, and severe lung inflammation by 22 weeks. Tumors harvested from these lungs have elevated levels of oncogenic miRNAs miR-21 and miR-155; are deficient for p53-regulated miRNAs; and have heightened expression of miR-34 target genes, such as Met and Bcl-2. In the presence of exogenous miR-34, epithelial cells derived from these tumors show reduced proliferation and invasion. In vivo treatment with miR-34a prevented tumor formation and progression in KrasLSL-G12D/+;Trp53LSL-R172H/+ mice. Animals infected with mir-34a-expressing lentivirus at the same time as transgene activation had little to no evidence of tumorigenesis, and lentivirus-induced miR-34a also prevented further progression of pre-formed tumors. These data support the use of miR-34 as a lung tumor-preventative and tumor-static agent.
Keywords: miRNA, lung cancer, therapy, Kras, p53, in vivo
Introduction
Lung cancer causes an estimated 25% of all cancer-related deaths in the United States: more than breast, prostate and colon cancer combined(1). In 2007 approximately 160,000 people died from lung cancer(2) and although survival rates for individuals with other cancers have improved in the past few decades, lung cancer survival has remained unchanged, at or below 15% five-year survival for individuals with advanced lung cancer. Collectively these facts indicate that there is a pressing need for better lung cancer treatment options.
Acquired genetic aberrations contributing to lung cancer include amplification and/or mutational activation of BCL-2, c-MYC and the EGFR-RAS pathway(3). In particular, KRAS mutations occur in ~10–30% of non-small cell lung cancer (NSCLC) cases(4). These somatic alterations include missense mutations at codon 12, 13, or 61 that impair intrinsic KRAS GTP hydrolysis and render KRAS constitutively active. Activated KRAS subsequently signals downstream to activate the Raf/Mek/Erk, Ral-GDS and PI3-kinase pathways.
Conversely, tumor suppressor pathways such as those involving p53 and retinoblastoma/p16 are silenced due to inactivating coding-mutations, loss of heterozygosity (LOH), promoter silencing, or changes in protein stability. The TRP53 tumor suppressor that encodes p53 is mutated in more than 50% of NSCLC(3). Although multiple TRP53 lesions have been described, the mutations tend to cluster into two distinct subtypes: those that impair p53 from binding to its DNA element and those that alter the conformation of p53(5). Regardless, loss of p53 signaling leads to uncontrolled cellular division and apoptotic avoidance.
In an attempt to better understand the molecular events and pathophysiology involved in lung cancer development, several labs have generated genetically engineered mouse models that seek to recapitulate human lung carcinoma. The most common of these make use of oncogenic Kras. In these models mutant Kras is expressed from a somatic latent allele(6) or in a tissue specific manner using a lox-stop-lox cassette (LSL)(7). Similarly, several Trp53 mutant models have been reported that spontaneously(8, 9) develop lung cancer and perhaps more commonly sensitize mice to carcinogens(8–10). Although a double-transgenic model has been evaluated by Jacks and colleagues(11); their study focused on KrasLSL-G12D in combination with Trp53flx/flx. In this model, cells that have undergone Cre-induced recombination have activated Kras in a p53 null background. Because 80% of human tumors with atypical p53 harbor missense mutations at specific hot-spots including arginine-175 (R175), we sought to evaluate these two aberrantly regulated events in a single model of NSCLC that more accurately resembles the genetics of advanced human NSCLC. To this end we characterized tumor progression in KrasLSL-G12D/+;Trp53LSL-R172H/+ doubly transgenic animals (Trp53R172 is the mouse ortholog of human TRP53R175[(5)]), and evaluated molecular anomalies from dissected tumors including altered expression of p53-regulated microRNAs (miRNAs). As a transcription factor, p53 has been identified as a direct regulator of several miRNAs, including miR-34a(12–15), miR-34b/c(13, 16), miR-107(17), miR-145(18, 19), miR-192 and miR-215(20, 21).
The discovery of miRNAs has produced profound changes in our understanding of gene regulation during disease progression. With respect to cancer, oncogenic miRNAs (oncomiRs) function to suppress the expression of tumor suppressor genes(22, 23). Conversely, tumor suppressive miRNAs, such as let-7 and miR-34, have been found to repress the expression of oncogenes, such as KRAS, MET, BCL-2, and c-MYC(13, 24–26). Our group and others showed that let-7 or miR-34 can both prevent(27, 28) and reverse(29, 30) tumorigenesis in vivo in the KrasG12D autochthonous model of NSCLC. Indeed, therapeutic use of miRNAs is now being evaluated extensively(23).
The first family of miRNAs identified that is directly regulated by p53 includes miR-34a, miR-34b, and miR-34c. While all three are transcriptionally induced by p53 and therefore low in p53-mutated cells, additional mechanisms of miR-34 silencing exist. For example, all three family members are subject to methylation-dependent promoter silencing, while miR-34a also maps to a chromosomal region often deleted in lung cancer(31). In any case, restoration of miR-34 can induce cell-cycle arrest, apoptosis and/or cellular senescence(12–16). Therefore, perhaps restoring some of these p53 dependent miRNAs in tumor cells in which p53 is mutated or where these miRNAs are silenced or genetically lost will prove therapeutically effective against these cancers.
In this current study we identify miR-34 as a promising therapeutic in the powerful and relevant KrasG12D/+;Trp53R172H/+ autochthonous model of human lung cancer. Epithelial cells isolated from lung tumors from these mice show diminished proliferation and invasiveness in the presence of miR-34. Furthermore, we demonstrate a striking effect of miR-34 in preventing endogenously occurring tumor initiation and progression in this therapeutically resistant mouse model of human NSCLC. These therapeutic cell culture and in vivo delivery experiments support the pursuit of miR-34 replacement as a therapy for human lung cancer.
Methods
In vivo adenoviral infection and lenti-miRNA delivery to KrasLSL-G12D/+;Trp53LSL-R172H/+mice
KrasLSL-G12D(strain number 01XJ6) andTrp53LSL-R172H (strain number 01X13) mice were obtained from the NCI-Frederick Mouse Repository. Trp53LSL-R172H mice were bred into the KrasLSL-G12D background. For subsequent rounds of breeding double heterozygotes were bred to KrasLSL-G12D to avoid homozygosing the Trp53LSL-R172H allele, which in the absence of Cre-recombinase resembles a Trp53 null animal. Mice were housed in the Yale University Animal Facility under the guidelines held by the Institutional Animal Care and Use Committee.
Double heterozygous mice were intubated based on the protocol of Dupage et al(11). In brief, mice were anesthetized with a mixture of ketamine and xylazine until unresponsive to a toe pinch. Animals were then individually positioned on a mouse intubation platform with a fiber optic light aimed at their thoracic region to illuminate the trachea. Exel Safelet IV cathethers were gently inserted into the trachea, guided by the illumination. Once in position, 65 μl of virus (see below and main text for concentration) was administered into the catheter. Following successful delivery, the catheter was removed and mice were allowed to fully recover before returning to cages. The respective dose of Ad-Cre indicated in the main text was diluted in 1X Minimal Essential Medium (MEM) with 0.01M CaCl2. In all cases lentiviral particles were administered at 3 × 106 transducing units (TU)/animal in the presence of 100 μg/ml polybrene (Santa Cruz Biotechnology) resuspended in 1X MEM. Adenoviral and lentiviral particles were purchased from the Gene Transfer Vector Core Facility at the University of Iowa.
Tumor histology and immunohistochemistry
When required, animals were sacrificed, perfused through the right ventricle with 10 mls of phosphate buffered saline, tissues were harvested and fixed in 10% natural buffered formalin and paraffin embedded according to standard procedures. Sections were cut at 5 μm, stained by hematoxylin and eosin and evaluated by digital light microscope using a Zeiss dissection microscope, AxioCam MRc 5 camera, and AxioVision 4.7.1 imaging software. Tumor burden was scored using the tumor area and dividing by the total area of the lung. Areas were determined using ImageJ software (NIH).
Generation and characterization of KrasG12D/+;Trp53 lung epithelial cell lines
Lungs were harvested from KrasG12D/+;Trp53R172H/+ mice and tumors were dissected, minced and resuspended in RPMI supplemented with 1 mg/ml collagenase for 1 hour at 37°C. Cells in suspension were removed and washed 3 times with RPMI supplemented with 10% fetal bovine serum (FBS) followed by plating on collagen/fibronectin coated plates. Cells were passaged on coated plates for three passages at which point colonies with epithelial morphology were isolated and propagated. Although the cells morphologically resemble epithelial cells further molecular testing was not performed. Cells were maintained in RPMI supplemented with 10% FBS and 1X penicillin/streptomycin. The p53 wildtype cell line used in this study, mur-kr, was a gift from Katerina Politi, Yale University, and was initially generated by Tyler Jacks, MIT(32). All three cell-lines used were tested for p53 and Kras status to confirm the genetic identity at both loci prior to further evaluation.
To investigate relative levels of p53 and p53 target genes following p53 activation, cells were cultured in the presence of 40 μM cisplatin or serum free media for the indicated times. Protein or total RNA was isolated to evaluate p53 activation or p53 target induction respectively.
Protein isolation and immunoblotting
Cells lines were transfected with pre-miR-34a or control (Ambion, Austin TX) using dharmafect1 (Thermo Scientific), incubated for 48 hours and lysed in RIPA buffer containing protease and phosphatase inhibitors. Frozen tumor tissue was ground to a fine powder using a mortal and pestle followed by protein extraction in RIPA buffer. Total protein was determined using the BioRad Protein Assay and in all cases 75–100 μg of protein extract was size fractionated on 12% SDS/PAGE gels and transferred to PVDF membranes. Membranes were blocked in a solution of 1% non-fat milk in tris-buffered saline plus tween (TBST) followed by incubation with primary antibodies from Santa Cruz: Kras (1:200), Bcl-2 (1:500), Cdk4 (1:500) or Cell Signaling: c-Myc (1:500), Met (1:500), p53 (1:1000), p-p53 (1:1000) and subsequent incubation with respective secondary. Detection was achieved using West Dura (Pierce) and autoradiography.
Quantitative real-time PCR
Total RNA was isolated (miRNeasy, Qiagen) from either dissected tumors or KrasG12D/+;Trp53R172H/+ cell lines, which was then reverse transcribed using miScript II RT PCR kit (Qiagen). Reverse transcription reactions were diluted 10 fold and used in subsequent quantitative real-time PCR (qRT-PCR) reactions to detect mature miRNAs (miScript, Qiagen), mRNAs (Quantifast, Qiagen) and precursor miRNAs (Quantifast, Qiagen). In each case data were normalized to RNU6B (mature miRNAs) or Actin (precursor miRNAs and mRNAs). Probes for qRT-PCR were obtained from Qiagen.
Invasion assay
KrasG12D/+;Trp53R172H/+ lung epithelial cells were transduced with an equal amount (MOI=3) of lenti-control or lenti-miR-34 particles in the presence of 10 μg/ml polybrene. Transduction was verified via GFP expression: lentiviral particles coexpress the miRNA of interest and GFP. Forty-eight hours post transduction cells were replated at 1 × 105 cells/invasion chamber (BD Biosciences) in serum free media. Serum containing media placed outside of the chamber was used as a chemoattractant. Eighteen hours later cells were fixed and stained. Noninvading cells were removed from the inside of the chamber while cells on the exterior of the chamber were visualized and photographed. Invading cells from three fields of view for each transduction were counted from two biological replicates.
Proliferation, colony formation and wound healing assays
Cells were seeded in 6-well plates at 5 × 104 cells/well. Cells were transduced the following day, after approximately one round of doubling, based on 1 × 105 cells/well with a MOI of 5 TU of lenti-miR-34a or lenti-control in the presence of 10 μg/ml of polybrene. Four days following transduction, cells were replated in either 96-well plates at 4 × 103 cells/well (proliferation), 6-well plates at 500 cells/well (colony formation), or in 12-well plates at 1 × 105 cells/well (wound healing). For colony formation, cells were left undisturbed for 8 days at which point crystal violet was used to visualize colonies. For proliferation assays, cells were incubated an additional 4 days after plating. On the 4th day cells were fixed with tricholoracetic acid and stained with 0.4% (wt/vol) sulforhodamine B (SRB) dissolved in 1% acetic acid. Unbound dye was removed by four washes with 1% acetic acid, and protein-bound dye was extracted with 10 mM unbuffered Tris base for determination of absorbance. Colorimetric values were obtained and plotted with respect to control-infected cells. For wound healing experiments, a scratch was made with a pipet tip after the cells reached confluency (72 hours post plating). Cells were imaged immediately and again at 24 and 48 hours after generating the scratch.
Results
KrasG12D/+;Trp53R172H/+ mice develop lung adenocarcinoma and subsequent inflammation
In carcinoma of the lung, activated KRAS and mutant p53 are two of the most common anomalies. KRAS is activated in 10–30% of NSCLC while p53 mutations exist in ~50%(3). To better understand the contribution that these alterations make to carcinogenesis, orthologues of the most commonly occurring human mutations of KRAS (G12) and TRP53 (R175) have been generated in mouse models and evaluated individually and jointly in multiple tissues including the lung. We chose to evaluate one of the double mutant models, KrasLSL-G12D/+;Trp53LSL-R172H/+ because it closely resembles the combination of mutations commonly found in human lung cancer(5). To this end, we crossed the Trp53LSL-R172H/+ line with the KrasLSL-G12D/+ line generating a doubly transgenic mouse heterozygous for both Cre-inducible alleles. These transgenes were activated in the lung only, using adenoviral particles expressing Cre-recombinase (Ad-Cre) that were delivered intratracheally based on the methods of DuPage et al(11), which ensures that all the animals in the study received a near-equal number of viral particles.
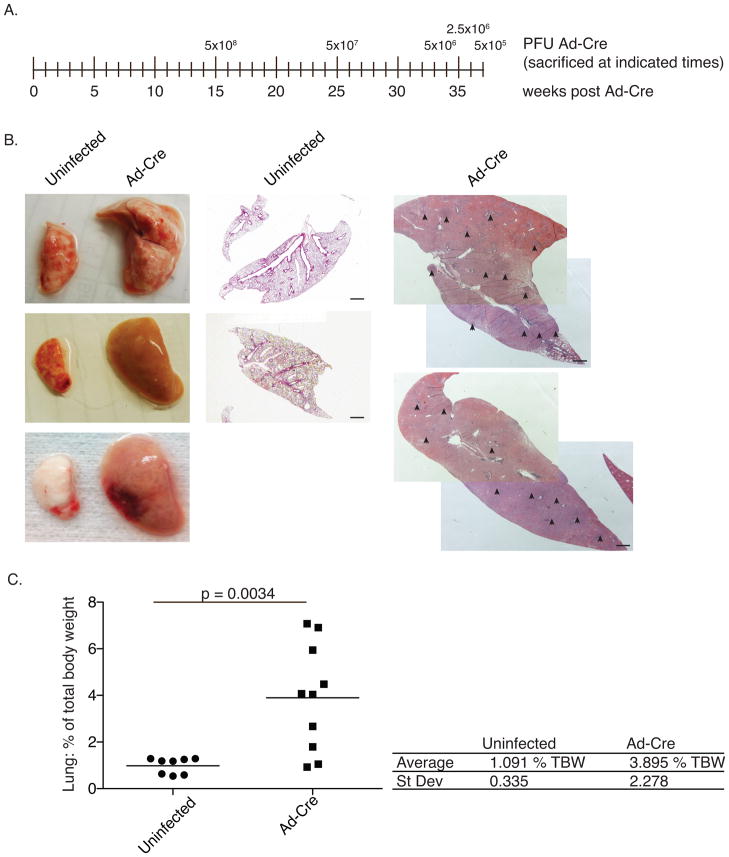
In an attempt to identify a viral dose that generated a modest number of lesions in the lung a dosing experiment was performed. Ad-Cre ranging from 5×105–5×108 plaque forming units (PFU) was intratracheally administered. At varying times following infection, animals developed phenotypes reminiscent of human lung cancer including labored breathing and decline in weight. Additionally their coat became scruffy, and in some severe cases the animals developed hunched posture. In accordance with regulatory guidelines, once clinical signs of cancer were apparent and quality of life diminished, the animals were sacrificed. As expected, animals infected with higher doses presented with clinical signs earlier (~15 weeks) than lower dosed animals (~37 weeks) (Fig. 1a). However, because animals were sacrificed when quality of life diminished many of the final pathologies were similar between the groups. For example, upon gross inspection, the majority of the animals (7/10) had excessively large lungs, taking up the bulk of the thoracic cavity. The lungs weighed an average of 0.92 grams (+/− 0.39 g), representing 3.9% (+/− 2.28%) of total body weight (TBW) (Fig. 1b and c). All uninfected control mice had lung weights of less than 0.3 grams (roughly 1% of TBW). This was specific to the lung as weights of the liver and kidneys remained similar between infected and control mice (Supplementary Fig. 1). Histologically, the lungs of the infected animals were largely tumor-filled and inflamed (Fig. 1b). Inflammation occurred in most animals with time, independent of Ad-Cre dose.
Figure 1. Tumors formed in KrasLSL-G12D/+;Trp53LSL-R172H/+mice.
Two animals were infected with each doses of Ad-Cre ranging from 5×105 PFU to 5×108 PFU. When quality of life was diminished animals were sacrificed. (a) Time-line indicating dose and time of sacrifice. (b) Representative lungs from control and Ad-Cre infected animals. Center and right panels are hematoxylin and eosin (H&E) stains of histological sections; note that the scale is the same for control and infected samples. Arrowheads point to tumors. Black bars = 1 mm (c) Ad-Cre infected lungs were grossly enlarged at time of sacrifice relative to uninfected control lungs. Following perfusion, lungs were harvested, weighed, and are graphed as a percentage of total body weight for each individual mouse. The black bar indicates the mean. p-values were determined by two-tailed t-test.
Because such inflammation has not been previously reported to follow Ad-Cre infection, it was essential to determine if the inflammation was caused by the adenoviral delivery or was a consequence of the tumorigenesis. Therefore, we setup a time-course using a relatively low dose of Ad-Cre, 5×106 PFU, sacrificing 2 or 3 animals every few weeks. In the previous experiment, clinical signs were not evident until at least 25–30 weeks at this dose (Fig. 1), and fewer tumors developed. Thus, with this dose we could afford the time to allow the few tumors to advance to higher-grade, more invasive adenocarcinomas.
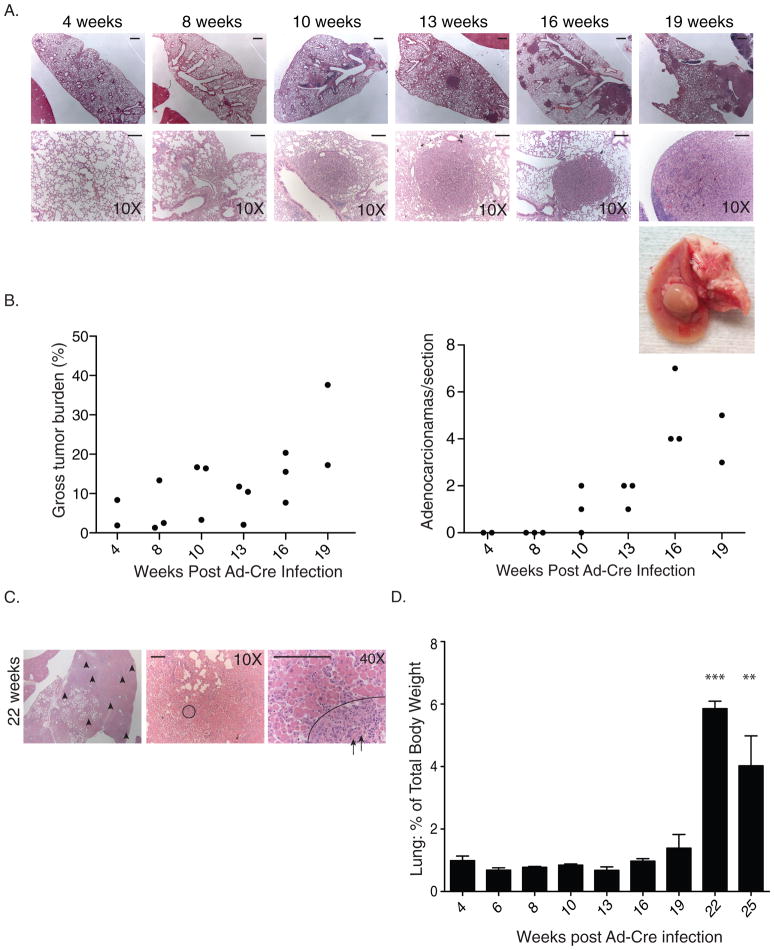
The results of the time course revealed two major findings. Firstly, nodules developed approximately 10 to 13 weeks following infection, with tumor burden increasing with time (Fig. 2a and b), and preceding any histologically visible inflammation. This suggested that the Ad-Cre viral delivery, per se, did not cause the inflammatory response in these animals, and instead that tumorigenesis was the likely driver of inflammation. At later time points we noticed a substantial change both histologically and at the gross morphological level. At 22 weeks following transgene activation, immune cell infiltration became evident (Fig. 2c). This coincided with a marked increase in lung weight that was sustained at 25 weeks (Fig. 2d). We hypothesize that the elevated lung weight was contributing to the hunched posture we observed in animals at later time-points.
Figure 2. Time course of tumor progression in KrasLSL-G12D/+;Trp53LSL-R172H/+ mice.
Animals were infected with 5×105 PFU of Ad-Cre. Three animals were sacrificed at each of the times indicated. Following perfusion, lungs were harvested, weighed, sectioned, and stained with H&E. (a) Representative images of KrasLSL-G12D/+;Trp53LSL-R172H/+ lungs. Top row: bars = 1 mm. Bottom row: bars = 0.125 mm (b) Tumor burden was scored by calculating the area of tumor tissue as a percentage of the total area of the lung for each animal. The number of adenocarcinomas from each mouse, averaged across three sections, is shown. (c) H&E stain of lungs at 22 weeks post Ad-Cre with evidence of infiltration. Tumors (in purple) are indicated with arrowheads in the first panel. The remainder of the lung has been infiltrated with immune cells (pink). The circle in the second panel indicates a well-differentiated tumor that is shown at higher magnification in the last panel. While the majority of the immune cells are external to the tumor, arrows point to immune cells that have infiltrated the tumor. Bars = 0.125 mm (d) The lung weight for each mouse is shown as a percentage of total body weight. Error bars indicate standard deviation. (n=3 for each time-point) ** p-value <0.01, ***p-value <0.001. p-values were determined by two-tailed t-test.
While the overall tumor burden in these animals increased with time, so did the occurrence of higher-grade adenocarcinomas (Fig. 2b), suggesting that many of these tumors may be acquiring the aggressive nature of later-stage human NSCLC. We hypothesized based on data from similar models(11) that these high-grade adenocarcinomas may acquire metastatic characteristics. In a single mouse that was sacrificed 22 weeks following transgene activation we detected a metastatic nodule in the liver supporting our initial hypothesis (data not shown). No further metastases to other organs or in any other mice were detected. This is expected since studies in the KrasG12D/+;p53flx/flx model found that only ~20% of animals had distant metastatic disease(11).
miR-34 levels are down regulated and certain target genes are up-regulated in KrasG12D/+;Trp53R172H/+ tumors
Because the majority of human tumors with heterozygous p53 inactivation go on to lose the remaining wildtype allele of p53, we sequenced the p53 locus from multiple dissected tumors, observing loss of heterozygosity in a subset of tumors (2/5) that were evaluated (data not shown).
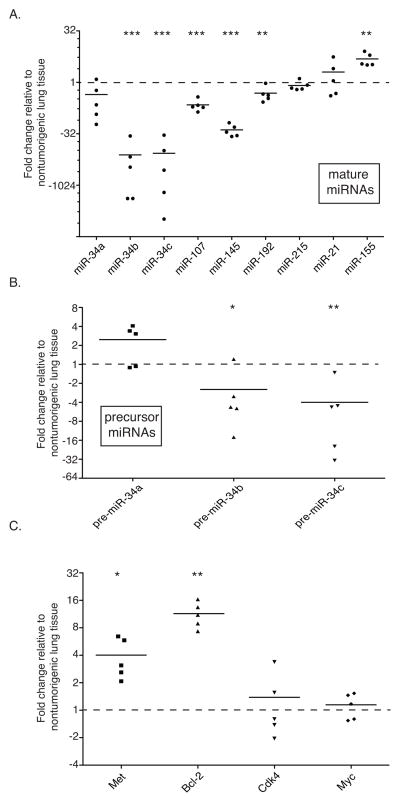
Due to the genomic loss and/or mutant signature of Trp53, we proposed that the known p53-regulated miRNAs (miR-34a, miR-34b/c, miR-107, miR-145, miR-192, miR-215) would be decreased in the tumor tissue as well. Individual tumors were dissected from KrasG12D/+;Trp53R172H/+ lungs and total RNA extracted. Following reverse transcription, quantitative real-time PCR (qRT-PCR) for each of the mature miRNAs (Fig 3a) and miR-34 precursors was performed (Fig. 3b). While all of the putative p53-regulated mature miRNAs were decreased in the tumor tissues relative to control, miR-34b and miR-34c were almost undetectable in the tumor samples analyzed. This decrease was evident at the precursor miRNA level as well (Fig. 3b). Interestingly, pre-miR-34a, which is transcribed independently of pre-miR-34b/c, was elevated in the tumors that retained one wild-type copy of p53. In tumors that had lost p53 heterozygosity, pre-miR-34a levels were markedly lower (Fig. 3b).
Figure 3. Loss of p53 regulated miRNAs and up-regulation of miR-34 target genes in KrasLSL-G12D/+;Trp53LSL-R172H/+ tumors.
Total RNA was extracted from individually dissected tumors or normal lung of uninfected control mice and transcript abundance was quantified. (a) Mature miRNA levels were measured by qRT-PCR, normalized to RNU6B, and graphed relative to normal lung tissue from 3 uninfected littermates. (b) Precursor miR-34 family members were evaluated by qRT-PCR, normalized to Actin, and graphed relative to normal uninfected lung tissue. The three points above the mean for pre-miR-34a represent tumors that were heterozygous for Trp53; the two below are hemizygous. (c) miR-34 target genes, Met, Bcl-2, Cdk-4, and Myc were evaluated by qRT-PCR, normalized to Actin, and graphed relative to normal uninfected lung tissue. In all three figures black bars indicate the mean. * p-value < 0.05, ** p-value <0.01, ***p-value <0.001. p-values were determined by two-tailed t-test.
Because the greatest and most consistent reductions were evident for two of the miR-34 family members, we proceeded to evaluate miR-34 target gene levels. As expected, the transcript level of two miR-34 targets, Bcl-2 and Met(33), were elevated in all of the tumors (Fig. 3c). Perhaps somewhat surprising, c-Myc and Cdk4, two additional miR-34 target genes, were unaffected at the transcriptional level, with mRNA levels similar to those in non-cancerous lung tissue (Fig. 3c). It remains possible that the levels of the c-Myc and Cdk4 proteins were elevated due to decreased miR-34 levels, as miRNA targeting can either cause mRNA degradation (detectible by qRT-PCR) or mRNA sequestration (not detectible)(reviewed in(34)). Due to minimal amounts of tissue obtained, it was not possible to measure protein levels directly.
Ectopic mir-34a reduces proliferation and invasion of KrasG12D/+;Trp53R172H/+ lung epithelial cells in culture
In an effort to determine if reexpressing mir-34a could repress miR-34 target genes and attenuate cellular proliferation and/or migration, lung epithelial cell lines were generated from the KrasG12D/+;Trp53R172H/+ tumors. These primary cell lines are stable and two lines are have remained in culture beyond 35 passages both of which support growth in nude mice. Each of the lines were sequenced and determined to be heterozygous at the p53 locus.
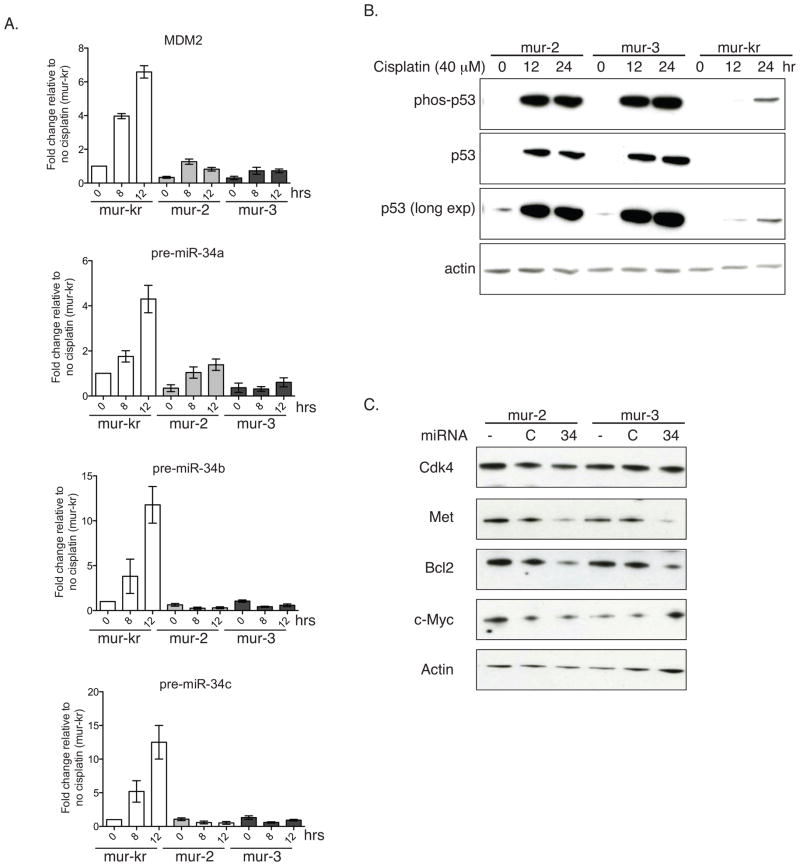
These KrasG12D/+;Trp53R172H/+ cells were first examined for the induction of p53-regulated miRNAs following either serum withdrawal or cisplatin treatment. Both result in up-regulation and activation of p53 leading to transcriptional elevations in p53-dependent target genes(35). For this evaluation, we included a murine cell line with mutant Kras but with wild-type p53 (mur-kr) to confirm induction of miRNAs in a p53 wild-type background. In this p53 wild-type cell line p53 protein levels and serine-15 phosphorylation were elevated in a time–dependent manner following cisplatin treatment (Fig. 4b). Interestingly, the phosphorylation and p53 accumulation in the mutant lines exceeded that of the p53 wild-type cell lines. It certainly is possible that the mutation enhances stability of p53. In fact, as expected, the p53 transcriptional target MDM2, which acts to enhance p53 degradation, was not induced in the mutant cell lines. This lack of MDM2 expression is likely contributing to the increase in p53 protein in these mutant lines. Similar to MDM2, pre-miR-34a/b/c were unaffected (miR-34b/c) or only minimally induced (miR-34-a) following cisplatin treatment. These data suggest that although upstream regulators of p53 are functioning to induce p53 expression and phosphorylation in the mutant cell lines, the mutant p53 is not capable of inducing transcriptional activation of its targets. We saw a similar trend when cells were cultured in the absence of serum (Sup Fig. 2), which also acts to stimulate p53(36). Serum withdrawal failed to induce miR-34 family members in KrasG12D/+;Trp53R172/+ cell lines to the extent that they were induced in KrasG12D/+;Trp53+/+ cells. Both of these assays show a trend similar to what we observed in the primary lung cancer specimens (Figs. 3a and b).
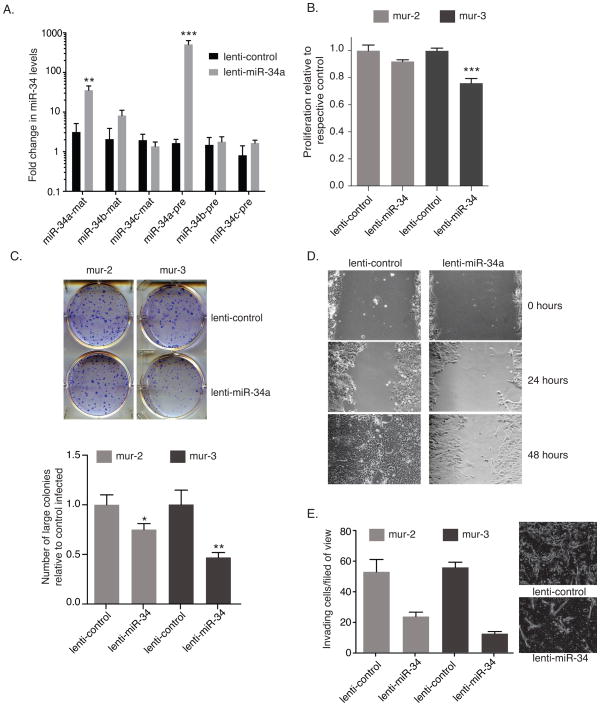
Figure 4. Ectopic mir-34a suppresses miR-34 target genes.
(a and b) KrasLSL-G12D/+;Trp53LSL-R172H/+ cell lines (mur-2, and mur-3) and a KrasLSL-G12D/+ cell line (mur-kr) were grown in the presence or absence of 40 μM cisplatin. (a) Total RNA was extracted and reverse transcribed, and levels of MDM2 and pre-miR-34a–c were measured by qRT-PCR. Data are normalized to Actin and shown relative to unstimulated mur-kr cells. (b) Protein was extracted, resolved via SDS-PAGE and immunoblotted for p-S15-p53 and p53 (c) Mutant lines were treated with exogenous pre-mir-34a and assayed for c-Myc, Met, Cdk4, and Bcl2 by immunoblot. Actin serves as a loading control in both b and c.
Based on these observations and on the tumor suppressive role of miR-34, we assessed the effects of ectopic miR-34a to our established cell lines. Following transfection with pre-miR-34a expression of miR-34 target genes was evaluated by immunoblotting. In p53 mutant cells, pre-miR-34a reduced Met and Bcl-2 (Fig. 4c), the same two targets that were transcriptionally elevated in the tumor tissue. Conversely, c-Myc and Cdk4 had varying responses depending on the cell line, suggesting that miR-34 may not be a predominant regulator of c-Myc and Cdk4 in these cells.
To assay for long-term functional effects of miR-34a on these cell lines, we utilized replication incompetent miR-34a expressing lentiviral particles, which upon integrating into the genome of transduced cells results in sustained high pre-miR-34a expression. Indeed, transduction with lenti-miR-34a resulted in a ~300 fold induction of pre-miR-34a, which translated into a ~33 fold increase in mature miR-34a (Fig. 5a). Importantly, proliferation decreased in cells transduced with lenti-miR-34a (Fig. 5b). The decrease was greatest for mur-3, at a 25% reduction. The reduced proliferation was recapitulated when analyzed by colony formation assay. Lenti-miR-34a transduced cells formed fewer and smaller colonies in these assays (Fig. 5c); in particular, the reduction in large colonies was dramatic and unmistakable. Cells transduced with lenti-miR-34a were also unable to recover in wound healing assays to the same degree as cells transduced with control virus (Fig. 5d).
Figure 5. Lentivirus-expressed miR-34a reduces proliferation and migration of KrasLSL-G12D/+;Trp53LSL-R172H/+ cells.
Murine epithelial cells were transduced with a lenti-miR-34a or lenti-control at a MOI of 5 TU. (a) Forty-eight hours following transduction, total RNA was isolated from mur-3 and assayed for precursor and mature miR-34 family members by qRT-PCR. Mature miRNA levels were normalized to RNU6B, and precursor miRNAs were normalized to β-actin. All data are plotted relative to untransduced cells. (b, c, and d) KrasLSL-G12D/+;Trp53LSL-R172H/+ cells were transduced with lenti-miR-34a or lenti-control at a MOI of 5 TU. Forty-eight hours following transduction, cells were reseeded for (b) proliferation (c) colony formation or (d) wound healing assays –mur-3. (b) Following reseeding, cells incubated for an additional forty-eight hours followed by staining with sulforhodamine B to analyze cell density. Data are plotted relative to each respective cell line transduced with lenti-control. (c) Five-hundred cells were seeded for colony formation. Cells proliferated undisturbed for 8 days followed by fixation and staining. The graph represents fold change in large colonies for each of the transduced lines. (d) Twenty-four hours after seeding cells were scratched and imaged. Additional images were acquired at 24 and 48 hours following the initial scratch. Error bars indicate standard deviation. (e) Cells were transduced at a MOI or 3. Fourth-eight hours later cells were reseeded in transwell chambers in SFM. Serum containing media was used as a chemoattractant. Invading cells were counted 16 hours after seeding. Data represent average of three fields of view from one biological replicate. Image insert shows a represented field of view from each treatment. p-values were determined by two-tailed t-test.
Because human NSCLC often metastasizes we evaluated the effect of exogenous miR-34a on invasion. We first compared these mouse cell lines to a genetically similar human cell line, H358. Like the murine lines, H358 harbors mutations in both KRAS and TRP53. Interestingly both of the murine cell lines were more invasive than H358 (data not shown). In fact H358 cells were relatively uninvasive. Importantly, invasion of the murine cells could be reduced if cells were first transduced with lenti-miR-34a before seeding in transwell migration chambers (Fig 5e). Collectively these cell-based in vitro studies suggest that miR-34 replacement is a viable option and might be effective in vivo to diminish the proliferation and/or invasion of cancer cells.
mir-34a prevents KrasG12D/+;Trp53R172H/+ lung adenocarcinoma
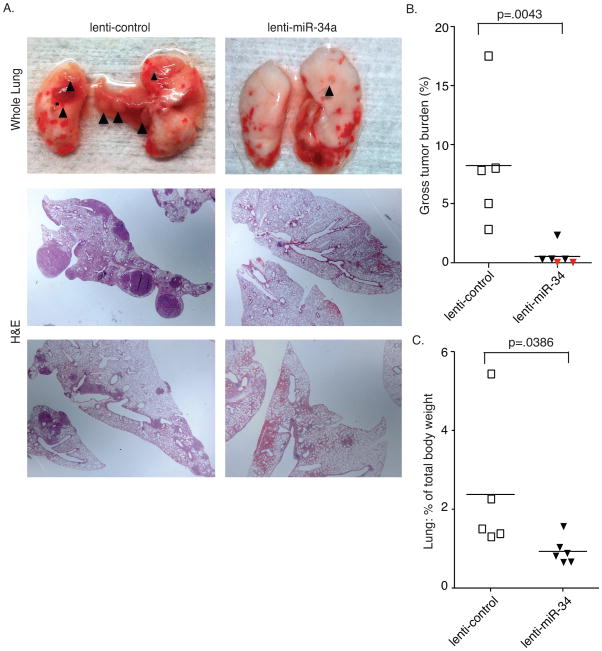
Attempts to prevent tumor formation in the KrasG12D/+;Trp53R172H/+ model have not yet been reported, presumably due to the aggressive nature of the tumors. Based on the promising results from the cell culture experiments we hypothesized that miR-34a overexpression might prevent KrasG12D/+;Trp53R172/+ driven lung tumors from forming. To test this, KrasG12D/+;Trp53R172/+ animals were infected with 5×106 PFU of Ad-Cre at the same time as 3×106 transducing units (TU) of lenti-miR-34a or lenti-control were delivered. Nineteen weeks following infection (a time that preceded substantial inflammation; Fig. 2a), animals were sacrificed and the tumor burden scored. Astonishingly, lenti-miR-34a almost completely abrogated tumor formation in this highly aggressive model system (Fig. 6 and Suppl. Fig 3). At both the gross level and histologically, lenti-miR-34a treated animals had minimal tumor formation (minimal hyperplasia) and in two out of six lenti-miR-34a treated mice there was no histological evidence of tumor formation (Fig. 6b, red diamonds and Suppl. Fig 3 –lenti-miR-34 treated animals 1 and 4). The volumes of the hyperplastic regions that were evident from the lenti-miR-34a treated mice were much smaller than the lenti-control treated tumors, suggesting that miR-34a treatment can not only suppress tumor initiation but may also play a role in preventing further progression. Through this analysis we also noted that the lenti-miR-34a treated lungs were smaller than the lenti-control treated animals (Fig. 6c), suggesting that exogenous miR-34a may prevent at least the early stages of immune cell infiltration.
Figure 6. lentiviral delivery of miR-34a impairs lung tumor initiation.
KrasLSL-G12D/+;Trp53LSL-R172H/+ mice were intubated and infected simultaneously with 5×106PFU of Ad-Cre and 3×106 TU of lenti-miR-34a or lenti-control. Animals were sacrificed 19 weeks following viral delivery. (a) top panel – whole lung analysis. black triangles indicate visible nodules. Bottom panels – H&E stain of representative sections from each treatment group. (b) Gross tumor burden was determined by quantifying the percentage of tumor tissue in the total lung. Each symbol represents the analysis of an individual animal; red diamonds represent animals with no discernable tumors in three sections following H&E evaluation. (c) The lungs of each mouse were graphed as a percentage of total body weight. Black bar indicates mean. p-values were determined by one-tailed t-test.
mir-34a treatment prevents progression of already established KrasG12D/+;Trp53R172H/+ lung tumors
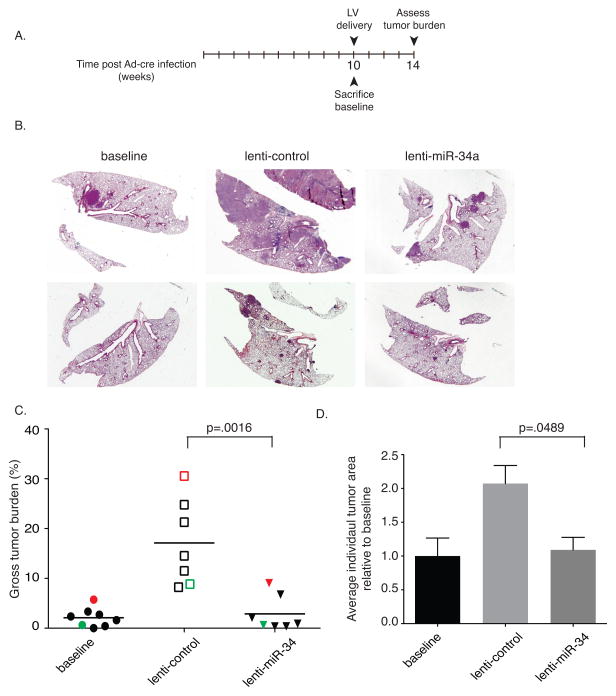
Following up on these promising results, we sought to evaluate the therapeutic potential of mir-34a in already established lung tumors. Tumors in KrasG12D/+;Trp53R172H/+ animals were allowed to preform for 10 weeks before lenti-miR-34a treatment began. At this time a sub-set of animals were sacrificed to determine baseline tumor levels before treatment began (Fig. 7a). The remaining animals were subdivided into two groups. One half was treated with lenti-miR-34a and the other with lenti-control. Four weeks following lentiviral delivery, animals were sacrificed and tumor burden was scored. Tumors in the lenti-control treated group continued to progress, in some cases to volumes that exceeded 40-fold of that of the average baseline volume (Fig. 7b–d). In contrast, tumors exposed to lenti-miR-34a did not increase in volume (compared to baseline samples), suggesting that tumor progression was inhibited by treatment with miR-34a.
Figure 7. lentiviral delivered miR-34a prevents tumor progression in KrasLSL-G12D/+;Trp53LSL-R172H/+ mice.
(a) Mice were intubated and infected with 5×106PFU of Ad-Cre followed by a 10-week incubation period that allowed for the formation of tumors. At 10 weeks baseline animals were sacrificed while the remaining animals were subdivided into two groups, receiving 3×106 TU lenti-control or lenti-mir-34a. After an additional four weeks, animals were sacrificed, and their lungs harvested, weighed, sectioned and stained. (b) H&E staining of representative lungs from each treatment arm. (c) Gross tumor burden was quantified based on percentage of tumor relative to total lung. Red symbols represent results obtained from the same animal as the upper H&E sections shown in (b), green symbols represent the lower H&E sections shown in (b). (d) Tumor area, averaged over three sections, from each treatment is shown relative to the average tumor area from the baseline animals. Error bars indicate standard error of the mean. p-values were determined by two-tailed t-test.
Discussion
Our work uses a model of aggressive lung cancer to evaluate the feasibility of translating miR-34 therapy into the clinic. Though labor intensive, use of this genetically engineered model has multiple advantages over more commonly used xenograft models. These tumors are autochthonous, that is, endogenous to the organism and cells are not manipulated in culture where they can acquire additional mutations. Further they grow in a native, immune-competent environment where they obtain signals and challenges from surrounding tissue as would occur in human cancer. Indeed, such models have proven more accurate for the evaluation of therapeutic efficiency(37).
Our pre-clinical study in this doubly transgenic autochthonous Kras;p53 lung cancer model is the first to show that miR-34a represents a potential treatment option for NSCLC. Based on our results, miR-34a delivered to human KRAS positive and p53 negative lung cancer patients would be predicted to delay tumor progression and potentially extend survival.
Therapeutically, miRNAs and their inhibitors, show a great deal of promise in pre-clinical models(23), possibly due to the pleotropic role of many miRNAs in affecting a wide range of target genes. In this respect, miR-34 is no exception: the miR-34 family has been reported to prevent the translation of genes involved in growth, proliferation, cell-cycle regulation, and anti-apoptotic signaling(33). In our study we show that at least two miR-34 target genes, Met and Bcl-2, whose expression has repeatedly been shown to correlate with multiple cancers, are elevated in KrasG12D/+;Trp53R172H/+ tumors and repressed in a mir-34a dependent manner in culture. Interestingly, some human non-small cell lung cancer cell lines are dependent on MET for growth and survival(38) suggesting that miR-34 replacement therapies could sensitize these otherwise resistant cells.
Although our study shows proof-of-concept in lung cancer, miR-34 replacement therapies could potentially treat more than just lung cancer. BCL-2, an inhibitor of the intrinsic apoptotic signaling cascade, is overexpressed in melanoma, chronic lymphocytic leukemia, and brain, breast, and prostate cancer(39). Similarly, MET, a receptor tyrosine kinase that is a driver of metastatic progression is often altered in lung, kidney, liver, stomach, breast, and brain tumors(40). Based on these and other findings MET and BCL-2 are attractive targets for therapeutic intervention. For example, antibodies are being evaluated as MET antagonists, as are MET kinase inhibitors(40). Our work suggests that miR-34 may be a promising therapeutic candidate to add to this list and may be more efficacious due to its coordinated ability to simultaneously reduce BCL-2. Due to the role of both MET and BCL-2 in chemotherapeutic resistance, miR-34 should be considered as a potential sensitizer for other therapeutic regimens(39, 41, 42).
Although p53 is the gene most often mutated in cancer, there are many cases of tumor profiling where the p53 gene is intact. Perhaps in these situations, p53-dependent regulation of target genes is lost via alternate mechanisms. This has been documented for both MDM2 and p14Arf, two important upstream regulators of p53 stability. MDM2, which is amplified in ~7% of NSCLC, reduces p53 levels through enhancing proteasome degradation of p53. Conversely, roughly 40% of NSCLC cases report loss of p14Arf, which inhibits MDM2, promoting p53 stabilization. In both of these cases, miR-34 therapy might still be fruitful, as miR-34 acts downstream of these genes. Finally, therapeutic miR-34 could also be utilized in cases where loss of miR-34 family members is evident, independent of p53 status. For example, mir-34b and c are located in a fragile region of the genome often lost in cancers(31), while mir-34a and mir-34b/c are both frequent victims of promoter methylation and subsequent silencing. Such changes may be present in a substantial fraction of tumors that do not harbor canonical p53 mutations, suggesting that these, too, may respond to exogenous miR-34 therapy.
Supplementary Material
Acknowledgments
Financial support: NIH CA131301
We thank Melissa Bonk, Kerry Card, and Jamie Graham (Yale Animal Resource Center) for animal care; Amos Brooks, Rebecca Melucci and Deidre Salemme (Yale’s Research Histology core) for embedding, sectioning, and staining lung tissue; Drs. Christopher Cheng and Zachary Pincus for critically reading this manuscript.
Grant Support: This work was supported by an NIH grant to FJS and Joanne Weidhaas (NCI R01 CA131301). AK was supported by a US National Institutes of Health (NIH) grant (1F32CA153885-01) and an American Cancer Society Postdoctoral Fellowship (120,766-PF-11-244-01-TBG).
Footnotes
Conflicts of interest: FS is a consultant to Mirna Therapeutics, which has licensed technology on miRNA therapeutics from Yale University. Mirna Therapeutics was not involved in any way with this study.
References
- 1.Jemal A, Thomas A, Murray T, Thun M. Cancer statistics, 2002. CA: a cancer journal for clinicians. 2002;52(1):23–47. doi: 10.3322/canjclin.52.1.23. Epub 2002/01/30. PubMed PMID: 11814064. [DOI] [PubMed] [Google Scholar]
- 2.U.S. Cancer Statistics Working Group. Atlanta, GA: U.S. Department of Health and Human Services, Centers for Disease Control and Prevention and National Cancer Institute; 2010. [cited 2010]. Available from: http://www.cdc.gov/uscs. [Google Scholar]
- 3.Herbst RS, Heymach JV, Lippman SM. Lung cancer. N Engl J Med. 2008;359(13):1367–80. doi: 10.1056/NEJMra0802714. Epub 2008/09/26 PubMed PMID: 18815398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pylayeva-Gupta Y, Grabocka E, Bar-Sagi D. RAS oncogenes: weaving a tumorigenic web. Nat Rev Cancer. 2011;11(11):761–74. doi: 10.1038/nrc3106. Epub 2011/10/14. PubMed PMID: 21993244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brosh R, Rotter V. When mutants gain new powers: news from the mutant p53 field. Nat Rev Cancer. 2009;9(10):701–13. doi: 10.1038/nrc2693. Epub 2009/08/21. nrc2693 [pii] [DOI] [PubMed] [Google Scholar]
- 6.Johnson L, Mercer K, Greenbaum D, Bronson RT, Crowley D, Tuveson DA, et al. Somatic activation of the K-ras oncogene causes early onset lung cancer in mice. Nature. 2001;410(6832):1111–6. doi: 10.1038/35074129. Epub 2001/04/27. [DOI] [PubMed] [Google Scholar]
- 7.Meuwissen R, Berns A. Mouse models for human lung cancer. Genes Dev. 2005;19(6):643–64. doi: 10.1101/gad.1284505. Epub 2005/03/17. [DOI] [PubMed] [Google Scholar]
- 8.Tchou-Wong KM, Jiang Y, Yee H, LaRosa J, Lee TC, Pellicer A, et al. Lung-specific expression of dominant-negative mutant p53 in transgenic mice increases spontaneous and benzo(a)pyrene-induced lung cancer. American journal of respiratory cell and molecular biology. 2002;27(2):186–93. doi: 10.1165/ajrcmb.27.2.4799. Epub 2002/08/02. PubMed PMID: 12151310. [DOI] [PubMed] [Google Scholar]
- 9.Wang Y, Zhang Z, Lubet R, You M. Tobacco smoke-induced lung tumorigenesis in mutant A/J mice with alterations in K-ras, p53, or Ink4a/Arf. Oncogene. 2005;24(18):3042–9. doi: 10.1038/sj.onc.1208390. Epub 2005/04/23. [DOI] [PubMed] [Google Scholar]
- 10.Wang Y, Zhang Z, Lubet RA, You M. A mouse model for tumor progression of lung cancer in ras and p53 transgenic mice. Oncogene. 2006;25(8):1277–80. doi: 10.1038/sj.onc.1209182. Epub 2005/10/26. [DOI] [PubMed] [Google Scholar]
- 11.DuPage M, Dooley AL, Jacks T. Conditional mouse lung cancer models using adenoviral or lentiviral delivery of Cre recombinase. Nature protocols. 2009;4(7):1064–72. doi: 10.1038/nprot.2009.95.. Epub 2009/06/30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chang TC, Wentzel EA, Kent OA, Ramachandran K, Mullendore M, Lee KH, et al. Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol Cell. 2007;26(5):745–52. doi: 10.1016/j.molcel.2007.05.010.. Epub 2007/06/02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.He L, He X, Lim LP, de Stanchina E, Xuan Z, Liang Y, et al. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447(7148):1130–4. doi: 10.1038/nature05939.. Epub 2007/06/08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Raver-Shapira N, Marciano E, Meiri E, Spector Y, Rosenfeld N, Moskovits N, et al. Transcriptional activation of miR-34a contributes to p53-mediated apoptosis. Mol Cell. 2007;26(5):731–43. doi: 10.1016/j.molcel.2007.05.017.. Epub 2007/06/02. [DOI] [PubMed] [Google Scholar]
- 15.Tarasov V, Jung P, Verdoodt B, Lodygin D, Epanchintsev A, Menssen A, et al. Differential regulation of microRNAs by p53 revealed by massively parallel sequencing: miR-34a is a p53 target that induces apoptosis and G1-arrest. Cell Cycle. 2007;6(13):1586–93. doi: 10.4161/cc.6.13.4436. Epub 2007/06/08. PubMed PMID: 17554199. [DOI] [PubMed] [Google Scholar]
- 16.Corney DC, Flesken-Nikitin A, Godwin AK, Wang W, Nikitin AY. MicroRNA-34b and MicroRNA-34c are targets of p53 and cooperate in control of cell proliferation and adhesion-independent growth. Cancer Res. 2007;67(18):8433–8. doi: 10.1158/0008-5472.CAN-07-1585.. Epub 2007/09/08. [DOI] [PubMed] [Google Scholar]
- 17.Yamakuchi M, Lotterman CD, Bao C, Hruban RH, Karim B, Mendell JT, et al. P53-induced microRNA-107 inhibits HIF-1 and tumor angiogenesis. Proc Natl Acad Sci U S A. 2010;107(14):6334–9. doi: 10.1073/pnas.0911082107.. Epub 2010/03/24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sachdeva M, Zhu S, Wu F, Wu H, Walia V, Kumar S, et al. p53 represses c-Myc through induction of the tumor suppressor miR-145. Proc Natl Acad Sci U S A. 2009;106(9):3207–12. doi: 10.1073/pnas.0808042106.. Epub 2009/02/10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Suh SO, Chen Y, Zaman MS, Hirata H, Yamamura S, Shahryari V, et al. MicroRNA-145 is regulated by DNA methylation and p53 gene mutation in prostate cancer. Carcinogenesis. 2011;32(5):772–8. doi: 10.1093/carcin/bgr036.. Epub 2011/02/26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Georges SA, Biery MC, Kim SY, Schelter JM, Guo J, Chang AN, et al. Coordinated regulation of cell cycle transcripts by p53-Inducible microRNAs, miR-192 and miR-215. Cancer Res. 2008;68(24):10105–12. doi: 10.1158/0008-5472.CAN-08-1846.. Epub 2008/12/17. [DOI] [PubMed] [Google Scholar]
- 21.Braun CJ, Zhang X, Savelyeva I, Wolff S, Moll UM, Schepeler T, et al. p53-Responsive micrornas 192 and 215 are capable of inducing cell cycle arrest. Cancer Res. 2008;68(24):10094–104. doi: 10.1158/0008-5472.CAN-08-1569.. Epub 2008/12/17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer. 2006;6(4):259–69. doi: 10.1038/nrc1840.. Epub 2006/03/25. nrc1840 [pii] [DOI] [PubMed] [Google Scholar]
- 23.Kasinski AL, Slack FJ. MicroRNAs en route to the clinic: progress in validating and targeting microRNAs for cancer therapy. Nat Rev Cancer. 2011;11(12):849–64. doi: 10.1038/nrc3166.. Epub 2011/11/25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, et al. RAS is regulated by the let-7 microRNA family. Cell. 2005;120(5):635–47. doi: 10.1016/j.cell.2005.01.014.. Epub 2005/03/16. S0092-8674(05)00088-7 [pii] [DOI] [PubMed] [Google Scholar]
- 25.Bommer GT, Gerin I, Feng Y, Kaczorowski AJ, Kuick R, Love RE, et al. p53-mediated activation of miRNA34 candidate tumor-suppressor genes. Curr Biol. 2007;17(15):1298–307. doi: 10.1016/j.cub.2007.06.068.. Epub 2007/07/28. [DOI] [PubMed] [Google Scholar]
- 26.Christoffersen NR, Shalgi R, Frankel LB, Leucci E, Lees M, Klausen M, et al. p53-independent upregulation of miR-34a during oncogene-induced senescence represses MYC. Cell Death Differ. 2010;17(2):236–45. doi: 10.1038/cdd.2009.109.. Epub 2009/08/22. [DOI] [PubMed] [Google Scholar]
- 27.Esquela-Kerscher A, Trang P, Wiggins JF, Patrawala L, Cheng A, Ford L, et al. The let-7 microRNA reduces tumor growth in mouse models of lung cancer. Cell Cycle. 2008;7(6):759–64. doi: 10.4161/cc.7.6.5834. Epub 2008/03/18. 5834 [pii]. PubMed PMID: 18344688. [DOI] [PubMed] [Google Scholar]
- 28.Kumar MS, Erkeland SJ, Pester RE, Chen CY, Ebert MS, Sharp PA, et al. Suppression of non-small cell lung tumor development by the let-7 microRNA family. Proc Natl Acad Sci U S A. 2008;105(10):3903–8. doi: 10.1073/pnas.0712321105.. Epub 2008/03/01. 0712321105 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Trang P, Medina PP, Wiggins JF, Ruffino L, Kelnar K, Omotola M, et al. Regression of murine lung tumors by the let-7 microRNA. Oncogene. 2009;29(11):1580–7. doi: 10.1038/onc.2009.445.. Epub 2009/12/08. onc2009445 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Trang P, Wiggins JF, Daige CL, Cho C, Omotola M, Brown D, et al. Systemic delivery of tumor suppressor microRNA mimics using a neutral lipid emulsion inhibits lung tumors in mice. Mol Ther. 2011;19(6):1116–22. doi: 10.1038/mt.2011.48.. Epub 2011/03/24. mt201148 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, Yendamuri S, et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci U S A. 2004;101(9):2999–3004. doi: 10.1073/pnas.0307323101 0307323101. Epub 2004/02/20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meylan E, Dooley AL, Feldser DM, Shen L, Turk E, Ouyang C, et al. Requirement for NF-kappaB signalling in a mouse model of lung adenocarcinoma. Nature. 2009;462(7269):104–7. doi: 10.1038/nature08462.. Epub 2009/10/23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hermeking H. The miR-34 family in cancer and apoptosis. Cell Death Differ. 2009;17(2):193–9. doi: 10.1038/cdd.2009.56.. Epub 2009/05/23. cdd200956 [pii] [DOI] [PubMed] [Google Scholar]
- 34.Valencia-Sanchez MA, Liu J, Hannon GJ, Parker R. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev. 2006;20(5):515–24. doi: 10.1101/gad.1399806.. Epub 2006/03/03. [DOI] [PubMed] [Google Scholar]
- 35.Pabla N, Huang S, Mi QS, Daniel R, Dong Z. ATR-Chk2 signaling in p53 activation and DNA damage response during cisplatin-induced apoptosis. J Biol Chem. 2008;283(10):6572–83. doi: 10.1074/jbc.M707568200.. Epub 2007/12/29. [DOI] [PubMed] [Google Scholar]
- 36.Itahana K, Dimri GP, Hara E, Itahana Y, Zou Y, Desprez PY, et al. A role for p53 in maintaining and establishing the quiescence growth arrest in human cells. J Biol Chem. 2002;277(20):18206–14. doi: 10.1074/jbc.M201028200.. Epub 2002/03/07. [DOI] [PubMed] [Google Scholar]
- 37.Becher OJ, Holland EC. Genetically engineered models have advantages over xenografts for preclinical studies. Cancer Res. 2006;66(7):3355–8. doi: 10.1158/0008-5472.CAN-05-3827.. discussion 8–9. Epub 2006/04/06. [DOI] [PubMed] [Google Scholar]
- 38.Lutterbach B, Zeng Q, Davis LJ, Hatch H, Hang G, Kohl NE, et al. Lung cancer cell lines harboring MET gene amplification are dependent on Met for growth and survival. Cancer Res. 2007;67(5):2081–8. doi: 10.1158/0008-5472.CAN-06-3495.. Epub 2007/03/03. [DOI] [PubMed] [Google Scholar]
- 39.Kelly PN, Strasser A. The role of Bcl-2 and its pro-survival relatives in tumourigenesis and cancer therapy. Cell Death Differ. 2011;18(9):1414–24. doi: 10.1038/cdd.2011.17.. Epub 2011/03/19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gherardi E, Birchmeier W, Birchmeier C, Vande Woude G. Targeting MET in cancer: rationale and progress. Nat Rev Cancer. 2012;12(2):89–103. doi: 10.1038/nrc3205.. Epub 2012/01/25. [DOI] [PubMed] [Google Scholar]
- 41.Engelman JA, Zejnullahu K, Mitsudomi T, Song Y, Hyland C, Park JO, et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science. 2007;316(5827):1039–43. doi: 10.1126/science.1141478.. Epub 2007/04/28. [DOI] [PubMed] [Google Scholar]
- 42.Garofalo M, Romano G, Di Leva G, Nuovo G, Jeon YJ, Ngankeu A, et al. EGFR and MET receptor tyrosine kinase-altered microRNA expression induces tumorigenesis and gefitinib resistance in lung cancers. Nat Med. 2012;18(1):74–82. doi: 10.1038/nm.2577.. Epub 2011/12/14. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.