Abstract
The developmental origin of adipose tissue and what controls its distribution is poorly understood. By linage tracing and gene expression analysis in mice, we provide evidence that mesenchymal precursors expressing Myf5—which are thought to give rise only to brown adipocytes and skeletal muscle—also give rise to a subset of white adipocytes. Furthermore, individual brown and white fats contain a mixture of adipocyte progenitor cells derived from Myf5+ and Myf5neg lineages, the number of which varies with depot location. Subsets of white adipocytes originating from both Myf5+ and Myf5neg precursors respond to β3-adrenoreceptor stimulation suggesting brite adipocytes may also have multiple origins. We additionally find that deleting PTEN with myf5-cre causes lipomatosis and partial lipodystrophy by selectively expanding the Myf5+ adipocyte lineages. Thus, the spectrum of adipocytes arising from Myf5+ precursors is broader than previously thought and differences in PI3K activity between adipocyte lineages alters body fat distribution.
Keywords: PTEN, PI3K, adipose, brown fat, white fat, brite fat, adipocyte progenitor cell, lipomatosis
INTRODUCTION
The obesity epidemic has intensified efforts to understand adipose tissue biology because obesity is a major risk factor for type 2 diabetes, dyslipidemias, cardiovascular disease, cancer, and other conditions [Reviewed in (Gesta et al., 2007)]. Adipose tissue is classified into two types: white adipose tissue (WAT)—the primary site of energy storage, and brown adipose tissue (BAT)—an energy expending tissue that regulates thermogenesis. The amount and distribution of white and brown adipose can vary considerably between individuals, significantly impacting the risk for developing metabolic complications [Reviewed in (Frontini and Cinti, 2010; Gesta et al., 2007; Tseng et al., 2010)]. However, what determines adipose tissue distribution in mammals is poorly understood.
It is becoming clear that different WAT depots are heterogeneous. For instance, in mammals individual WAT depots appear at different times in development and have unique functional characteristics [Reviewed in (Billon and Dani, 2011; Cristancho and Lazar, 2011; Gesta et al., 2007)]. Moreover, obesity characterized by increased visceral fat associates with high risk of metabolic disease, while the risk associated with increased subcutaneous fat is low. The distinction between subcutaneous and visceral fat may be over-simplified because evidence exists suggesting that metabolic properties also vary between some visceral fat depots (Edens et al., 1993; Fried et al., 1993; Tchkonia et al., 2005). Studies in mice and with human adipose tissue further indicates that individual adipocytes within a depot have different gene expression signatures and growth characteristics, indicating heterogeneity even exists within a single fat depot (Bluher et al., 2002; de Souza et al., 2001; Fortier et al., 2005; Jernas et al., 2006). These findings have led to a hypothesis that individual white adipocytes have different developmental origins. However, distinct white adipose tissue lineages have not been clearly identified.
Mounting evidence indicates that BAT exists in variable amounts in adult humans and positively influences human metabolism (Cypess et al., 2009; van Marken Lichtenbelt et al., 2009; Virtanen et al., 2009)[Reviewed in (Enerback, 2010; Nedergaard and Cannon, 2010)] and that therapeutically increasing brown fat energy expenditure could treat obesity [Reviewed in (Tseng et al., 2010)]. To this end, increasing attention is being given to brown adipose-like cells that can be induced to form within white fat by β3-adrenergic receptor stimulation. These “brite” (brown in white) or “recruitable” brown adipocytes, may potentially be targets for anti-obesity therapies aimed at increasing cellular bioenergetics [Reviewed in (Gesta et al., 2007; Tseng et al., 2010)].
One emerging view is that brown fat is developmentally more similar to skeletal muscle than to white fat [Reviewed in (Tseng et al., 2010)]. For example, gene expression analysis indicates primary brown but not white preadipocytes exhibit a myogenic-like transcriptional profile (Timmons et al., 2007). In vivo fate mapping experiments further suggest that brown fat originates from a mesenchymal precursor cell that expresses the myogenic transcription factor Myf5 (Atit et al., 2006; Seale et al., 2008). This has led to a widely accepted model in which brown fat and skeletal muscle share a common Myf5+ precursor cell—explaining the favorable metabolic properties of brown fat—and that an undefined Myf5neg precursor gives rise to white fat and to the inducible brite adipocytes [Reviewed in (Cristancho and Lazar, 2011; Enerback, 2009; Tseng et al., 2010)]. Recent progress has been made in identifying populations of cells within adipose tissues that contain adipocyte progenitor cells (APCs) (Rodeheffer et al., 2008; Tang et al., 2008). To what extent the Myf5+ lineage contributes to the BAT APC pool, and the identity of distinct lineages giving rise to WAT APCs and mature adipocytes is unclear.
The signals that regulate brown fat differentiation are poorly defined. In vitro, preadipocyte differentiation assays are supplemented with insulin, which activates the phosphatidylinositol 3-kinase (PI3K) signaling pathway and stimulates adipogenesis [Reviewed in (Cannon and Nedergaard, 2004; Rosen and MacDougald, 2006)]. PI3K activity is negatively regulated by PTEN (Phosphatase and tensin homolog). In this study, we genetically activated PI3K signaling (by conditionally deleting PTEN) specifically in the Myf5+ lineage to begin investigating how nutrient and growth factor sensing pathways regulate brown fat development in vivo. Strikingly, losing PTEN in the Myf5+ lineage alters whole body adipose tissue distribution resulting in combined lipomatosis and partial lipodystrophy. Unexpectedly, this results from the expansion of a subset of white and brown adipocytes that arise from Myf5-Cre+ precursors. Lineage tracing analysis further reveals that each fat depot contains adipocyte progenitor cells arising from Myf5+ precursors, the number of which varies depending upon depot location. Our results suggest that the spectrum of adipocytes originating from Myf5+ precursors is broader than previously thought and that activating PI3K in Myf5+ precursors redistributes body fat by selectively expanding the Myf5+ adipocyte lineages.
RESULTS
Deleting PTEN in Myf5+ precursors causes combined lipomatosis and partial lipodystrophy
To better understand the signaling mechanisms controlling brown fat development in vivo, we activated the PI3K signaling pathway in brown fat precursors by crossing PTENfl/fl mice to the myf5-cre knock-in line (Seale et al., 2008; Tallquist et al., 2000). Immediately from birth the myf5-cre;ptenfl/fl conditional knockout (PTENmyf5cKO) mice are distinguishable from their littermates by a horse-collar like growth and torpedo shape [Figure 1A and 1B]. Fewer than expected PTENmyf5cKO mice survive to weaning (25% expected; 15% actual), although mutant embryos are present in late development at the expected frequency, indicating that some PTENmyf5cKO mice die during or shortly after birth [Table S1]. PTENmyf5cKO mice die sporadically with a maximum lifespan of around 6 months [Figure S1A]. At 6–12 weeks of age PTENmyf5cKO mutants show no significant differences in total weight [Figure S1B]; however, they have slightly increased fat mass [Figure S1C].
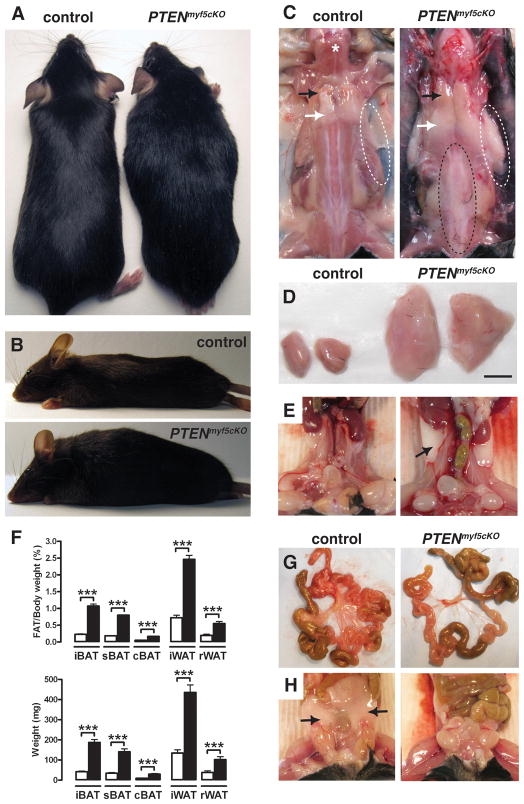
Figure 1. Deleting PTEN in the Myf5+ lineage causes severe combined lipomatosis and partial lipodystrophy.
(A) Anatomy of a 6-week-old PTENmyf5cKO mutant (right) and a littermate control (left). PTENmyf5cKO mice have a horse-collar-like growth and overall torpedo shape.
(B) Lateral view of a PTENmyf5cKO mouse (bottom panel) and a control (top panel).
(C) Macroscopic images of control and PTENmyf5cKO mouse. Black arrow indicates iBAT region; white arrow indicates iWAT. White dashed circles show axillary WAT (top panels). Vertebral WAT is indicated with a black dashed circle. A star indicates the trapezius muscle.
(D) Macroscopic images of iBAT (scale bar = 5mm)
(E) Macroscopic images of rWAT (black arrow).
(F) Fat mass relative to body weight (top panel) and total fat mass (bottom panel) for the indicated tissues in 6-week-old PTENmyf5cKO mice (black bars) and controls (white bars) (n=13; Bars represent mean± SEM; T-test; ***, p<0.001).
(G) Representative images of mesenteric fat in control (left panels) and PTENmyf5cKO mouse (right panels).
(H) Representative images of perigonal WAT (black arrow). See also Figure S1.
Despite their overall normal weight, individual tissue mass analysis indicates that the PTENmyf5cKO mutants have a severe adipose tissue distribution disorder. Most striking is their greatly enlarged interscapular, subscapular, and cervical brown fat (iBAT, sBAT, and cBAT), interscapular WAT (iWAT), retroperitoneal WAT (rWAT), and vertebral WAT depots [Figure 1C–F and S1D–F]. The mutant iBAT and iWAT show the greatest growth differential, increasing to a tissue to body weight ratio that is 4.7 and 3.4 times larger than litter-matched control tissues by 6 weeks of age [Figure 1D and 1F]. The oversized BAT is readily apparent in neonates, when its function is critical for thermogenesis, while the oversized WATs rapidly grow within a few weeks of birth [not shown]. Remarkably, other WAT depots such as the mesenteric, perigonadal (pgWAT), inguinal (ingWAT), gluteal, and posterior subcutaneous WATs are absent [Figure 1G, 1H, and S1F]. In contrast, the limb skeletal muscles (e.g. gastrocnemius, quadriceps, and triceps) are reduced in mass as are several other lean tissues including the heart, liver, kidney, lung, testes, and brain [Figure S1G]. Thus, PTENmyf5cKO mice cannot regulate adipose tissue distribution and develop severe combined lipomatosis, lipoatrophy, and lean tissue wasting.
Adipose tissues in PTENmyf5cKO mice have larger adipocytes and more cells
Hematoxylin & Eosin (H&E) staining indicates that the PTENmyf5cKO BAT lipid droplets maintain their typical multilocular appearance; however, the mutant tissues have visibly larger lipid droplets and fewer nuclei per mm2 [Figure 2A, 2B, S2A, and S2B], indicating individual mutant brown adipocytes are larger. Similar to the mutant BATs, the unilocular lipid droplets characteristic of white adipocytes are also larger in the PTENmyf5cKO rWAT and iWAT depots [Figure 2A]. The average lipid droplet diameter in the mutant WATs is nearly twice the diameter of lipid droplets in control white adipocytes [Figure 2C and S2C–E], indicating the increased size of PTENmyf5cKO fat depots is caused in part by hypertrophy. In contrast to the PTENmyf5cKO fats, the size of individual skeletal muscle fibers is decreased [Figure 2A, S2A, S2F, and S2G].
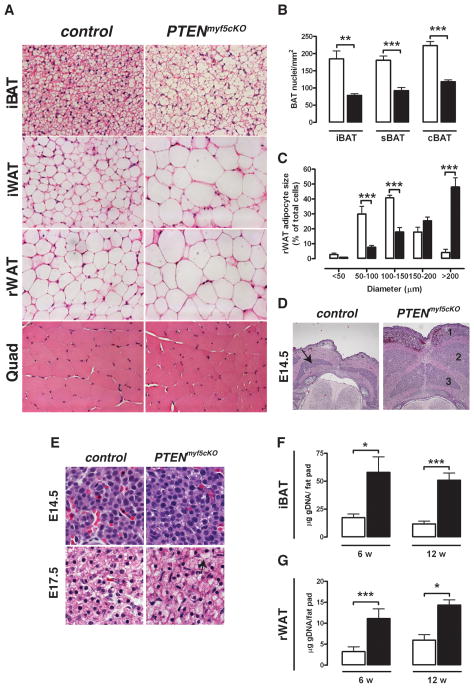
Figure 2. PTEN-deficient fats have large adipocytes and more total cells.
(A) H&E images of iBAT, iWAT, rWAT and quadriceps from a control and PTENmyf5cKO mouse (40×).
(B) Nuclei density per mm2 of iBAT, sBAT and cBAT (n=5, 6 w old).
(C) rWAT adipocytes cell diameter from 6 week old mice (n=7)
(D) Images of H&E stained E14.5 control and PTENmyf5cKO embryo sections (10×). Brown fat is marked with an arrow in control. In the mutant, iBAT (1), sBAT (2) and cBAT (3) precursor pools are enlarged.
(E) Detail of embryonic BAT precursors at E14.5 and E17.5. Lipid droplets can be seen forming prematurely in the mutants by E17.5 (arrow).
(F) Total genomic DNA purified iBAT in 6 and 12 week old control (white bars) and PTENmyf5cKO (black bars) (n=8).
(G) Total genomic DNA from rWAT (n=8). Bars represent mean± SEM. T-test; *, p<0.05, **, p< 0.01, ***, p<0.001). See also Figure S2.
Brown adipose precursor cell pools are easily visible by H&E staining of sectioned embryos. Deleting PTEN with myf5-cre increases the size of brown fat precursor cell pools in PTENmyf5cKO E14.5 embryos, consistent with a role for PTEN in regulating precursor cell number [Figure 2D][Reviewed in (Hill and Wu, 2009)]. At high magnification E14.5 PTENmyf5cKO precursors appear qualitatively more similar to controls, but by E17.5 they prematurely begin accumulating lipid [Figure 2E] suggesting aberrant lipid accumulation is secondary to a primary defect in cell number regulation. By 6 weeks of age the mutant iBAT contains 3.5-fold more total genomic DNA compared to control tissues indicating that the total cell number of the developed mutant brown fats is also increased [Figure 2F]. Similarly, 6 week mutant rWAT contains more total genomic DNA (a 3.6-fold increase over control) [Figure 2G]. Notably, the total DNA content of both depots between 6 and 12 weeks of age minimally changes indicating that the difference in total cell number between control and mutant fats may result in part from the expansion of embryonic/neonatal progenitor cells.
PTENmyf5cKO mice completely lack PTEN in both the white and brown fats
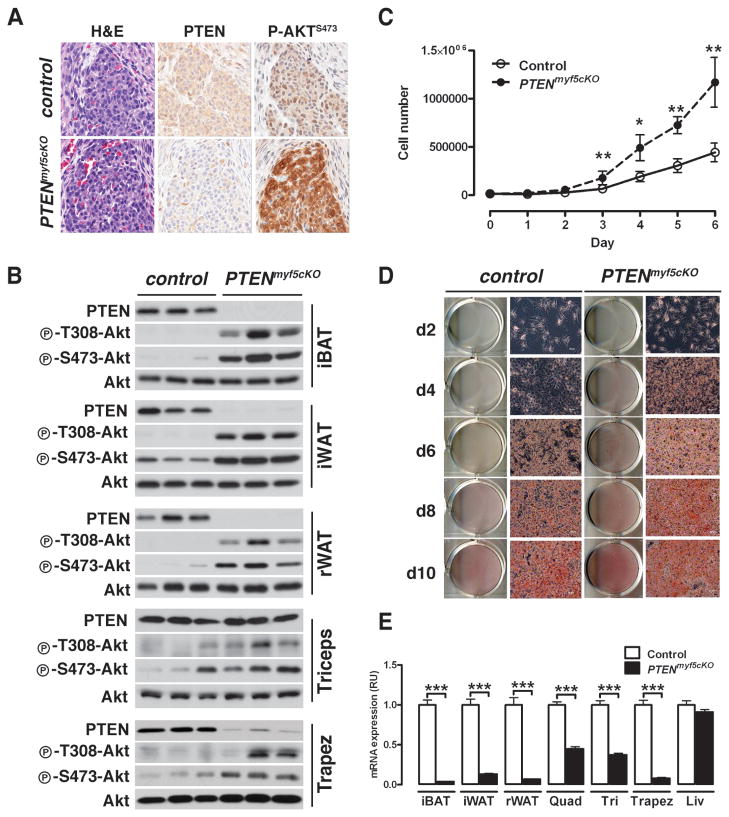
Immunohistochemical analysis reveals robust PTEN loss and strong PI3K pathway activation (by increased AKTS473 phosphorylation) in the embryonic brown fat precursor cell pools [Figure 3A], indicating myf5-cre deletes PTENfl/fl early in brown fat development and in most brown fat precursors. Immunoblotting of tissue lysates prepared from 6-week-old PTENmyf5cKO mice reveals robust PTEN ablation and AKT activation in the adult iBAT, sBAT, and cBAT depots [Figure 3B and S3A], further validating that myf5-cre targets PTENfl/fl deletion to the brown fat lineage. The myf5-cre also efficiently deletes PTEN in primary BAT preadipocytes, which we purified from PTENmyf5cKO neonates [Figure S3B]. Consistent with our in vivo findings, PTEN-deficient BAT preadipocytes proliferate faster than control preadipocytes in culture [Figure 3C], and when induced to differentiate prematurely accumulate lipid [Figure 3D].
Figure 3. PTENmyf5cKO mice completely lack PTEN in WAT and BAT.
(A) IHC and H&E stains on serial sections of E14.5 BAT precursor for PTEN and phospho-Akt-S473 in control and PTENmyf5cKO mice.
(B) PTEN, phospho-AktT308, and phospho-AktS473 levels in lysates from iBAT, iWAT, rWAT, triceps, and trapezius. See also Figure S3.
(C) Proliferation curve of primary BAT preadipocytes (n=3;. Points represent mean± SEM. T-test; *, p< 0.05; **, p<0.01).
(D) Primary BAT preadipocytes were differentiates and stained with ORO at different stages.
(E) Tissue mRNA expression of PTEN (n=5–6; Bars represent mean± SEM. T-test; ***, p<0.001).
Interestingly, we are unable to detect significant loss of PTEN in the limb muscles by immunoblot [Figure 3B and S3A], despite detecting recombination at the PTENfl/fl locus by PCR [not shown], and tracing the Myf5 lineage to all limb skeletal muscle fibers [Figure 4A, 4E and S3D]. We noted evidence in the literature suggesting that redundant Myf5+ and Myf5neg lineages converge to generate skeletal muscles (Gensch et al., 2008; Haldar et al., 2008). In fact, one report elegantly demonstrates that a greater fraction of Myf5+ precursors contributes to the epaxial lineages (e.g. vertebrae, rib, back, and neck muscles) compared to the hypaxial lineages (e.g. diaphragm, abdominal muscles, and limb muscles) (Haldar et al., 2008). Therefore, we reasoned that myf5-cre might inefficiently delete PTEN in the myotubes of limb muscles because a Myf5neg myoblast (expressing PTEN) fusing to a Myf5+ myoblast (deleted for PTEN) could partially compensate for the PTEN deficiency. If this is true, then one might expect PTEN loss to be more readily detectable in the neck musculature, which arises from a greater contribution of Myf5+ precursors compared to the limb musculature. This is indeed the case for the PTENmyf5cKO trapezius muscle, in which partial loss of PTEN and increased AKT phosphorylation is detectable by immunoblotting [Figure 3B]. Although PTEN protein levels are unchanged in limb muscles, PTEN mRNA levels are reduced [Figure 3E], suggesting that the limb muscles can also regulate PTEN expression post-translation (Wang and Jiang, 2008). Regardless, we conclude that in the PTENmyf5cKO mice, a systemic effect rather than intrinsic loss of PTEN causes the decrease in limb muscle fiber size and total mass.
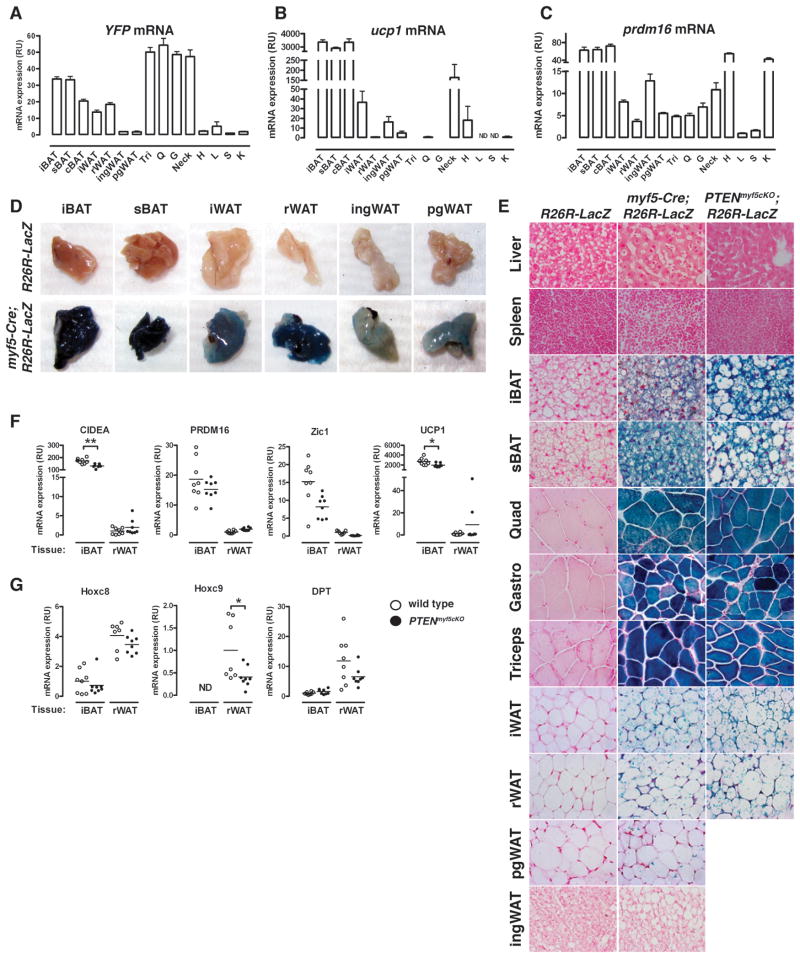
Figure 4. A subset of white adipocytes trace to the Myf5 lineage.
(A–C) YFP (A), Ucp1 (B), and Prdm16 (C) mRNA expression in tissues (n=5) prepared from myf5-cre;R26R-YFP mice (Tri:triceps; Q: quadriceps; G: gastrocnemius; H: heart; L: liver; S:spleen; K: kidney). In (A) YFP level detected in each tissue from mice lacking Cre was subtracted. Bars represent mean± SEM.
(D) Images of the indicated depots from a R26R-LacZ and a myf5-cre;R26R-LacZ mouse after X-Gal staining.
(E) β-galactosidase activity in tissues from negative control (R26R-LacZ) and from myf5-cre;R26R-LacZ or PTENmyf5cKO-LacZ mice. Counterstained with NFR. Liver and spleen at 40X; others at 63X. See also Figure S3.
(F) qRT-PCR analysis of Cidea, Prdm16, Zic1, Ucp1, HoxC8, HoxC9 and DPT in whole iBAT and rWAT. (n=8; Horizontal line indicates the mean. *, p<0.05; **, p< 0.01).
White fat is not predicted to arise from a Myf5+ precursor; therefore, the dramatic WAT phenotype of PTENmyf5cKO mice is unexpected. Surprisingly, immunoblotting reveals that myf5-cre robustly deletes PTENfl/fl in iWAT and rWAT to the same extent as it does in mutant BAT [Figure 3B]. Moreover, recombination at the PTEN locus is detected by PCR in the mutant WAT [not shown]; and PTEN mRNA expression is decreased in the mutant iWAT and rWAT to the same extent as it is in mutant iBAT [Figure 3E]. This contradicts the hypotheses that brown adipose is the only fat that arises from Myf5+ precursors and suggests some white adipocytes are also derived from a Myf5+ expressing progenitor cell.
A subset of white adipocytes arise from Myf5+ precursors
To explore the possibility that some white adipocytes arise from Myf5+ precursors, we generated myf5-cre;R26R-EYFP mice (which express YFP only in cells that have previously expressed the myf5-cre knock-in allele) to trace the Myf5+ lineage. As expected, the myf5-cre;R26R-EYFP mice express high levels of YFP mRNA in the iBAT, sBAT, and cBAT, and in skeletal muscles (triceps, quadriceps, gastrocnemius, and trapezius), but not in the heart, liver, spleen, or kidney [Figure 4A], which is consistent with brown fat and skeletal muscle arising from a Myf5+ precursor (Seale et al., 2008). Moreover, YFP expression is also low in both ingWAT and pgWAT. However, myf5-cre;R26R-EYFP mice express 15–20 times more YFP mRNA in the iWAT and rWAT compared to ingWAT and pgWAT, and at levels comparable to YFP expression in BAT. Notably, YFP mRNA expression in iWAT and rWAT does not correlate with ucp1 or prdm16 mRNA expression (brown fat markers) [Figure 4A–C]. For example, ucp1 and prdm16 express more highly in ingWAT [Figure 4B and 4C], which has low YFP expression [Figure 4A]. Thus, myf5-cre lineage tracing labels iWAT and rWAT in addition to BAT and skeletal muscle.
We also generated myf5-cre;R26R-LacZ mice to confirm by an alternate approach that iWAT and rWAT trace to the Myf5+ lineage. Consistent with YFP mRNA expression, myf5-cre;R26R-LacZ mice strongly express LacZ in whole BAT [Figure 4D]. Similarly, the rWAT and iWAT depots strongly stain LacZ positive. In contrast, the pgWAT and ingWAT faintly stain positive, while liver stains negative. Upon sectioning, strong LacZ expression is visible in nearly all iBAT and sBAT adipocytes, while many unilocular adipocytes in rWAT and iWAT also stain positive [Figure 4E and S3C]. Although difficult to find, a few LacZ positive cells are detectable in the pgWAT and to a lesser extent in the ingWAT; however, the adipocytes in these depots are predominantly LacZ negative [Figure 4E and S3C]. Notably, many adipocytes within ingWAT contain multilocular lipid droplets indicative of inducible brown fat cells (also called brite fat) [Figure 4E], which are thought to arise from a Myf5neg lineage (Seale et al., 2008). Consistent with this notion, none of these cells are LacZ positive.
We next profiled the gene expression pattern of iBAT and rWAT with well-established brown and white fat markers to ask if the WAT tracing to the Myf5+ lineage has a BAT signature (Schulz et al., 2011; Seale et al., 2008; Walden et al., 2011). By quantitative RT-PCR, we find as expected that whole iBAT more highly expresses the brown fat markers cidea, prdm16, zic1, and ucp1 compared to rWAT [Figure 4F (wild type denoting PTEN positive)]. Moreover, rWAT more highly expresses the white fat markers HoxC8, HoxC9, and Dpt compared to iBAT [Figure 4G], indicating that despite its strong labeling with myf5-cre, rWAT does not have a brown fat signature. These data suggest that Myf5+ precursors give rise to a subset of white adipocytes in addition to brown adipocytes.
The number of adipocyte progenitor cells arising from Myf5+ precursors varies with depot location
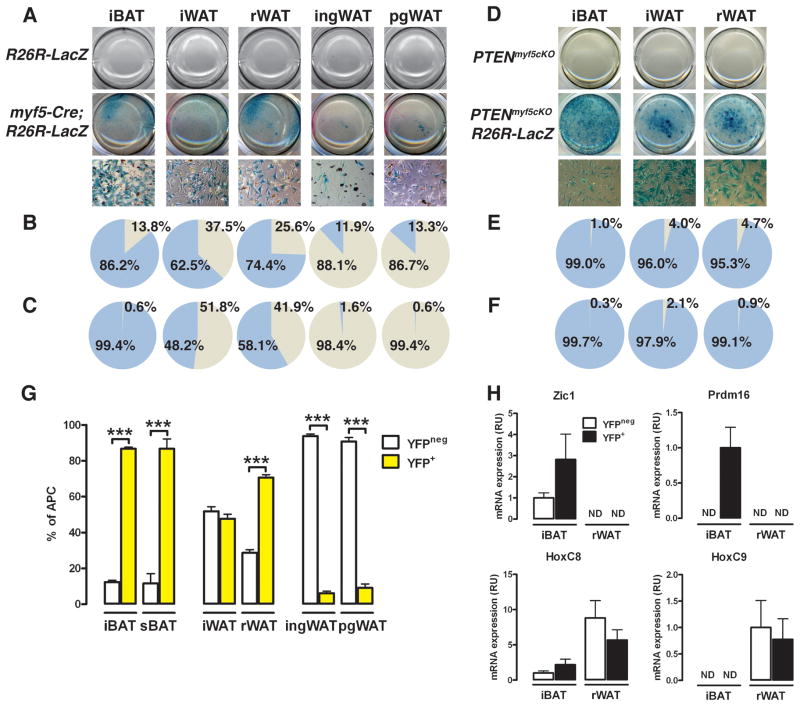
To more closely examine the Myf5-lineage contribution to each adipose tissue depot, we isolated individual depots from myf5-cre;R26R-LacZ mice, prepared their stromal vascular fractions (SVF), and examined cellular LacZ expression. Compared to cre-lacking R26R-LacZ controls, the plated SVF of myf5-cre;R26R-LacZ iBAT, iWAT, and rWAT strongly stain positive for LacZ [Figure 5A]. The iBAT SVF contains the highest percentage of LacZ positive cells at 86.2%, while 62.5 % and 74.4% respectively of the iWAT and rWAT SVF cells label LacZ positive [Figure 5B]. In contrast, only 11.9% of the ingWAT and 13.3% of the pgWAT SVF cells are LacZ positive. Nevertheless, the SVF of each depot contains cells originating from a Myf5+ precursor.
Figure 5. The number of Myf5-lineage derived adipocyte progenitor cells in each depot varies with its anatomical location.
(A) Macroscopic (top) and microscopic (40×-bottom) images of SVF cultures from R26R-LacZ and myf5-cre;R26R-LacZ mice from the indicated depots and stained with X-Gal.
(B) Positive and negative β-gal cell quantification shown in (A).
(C) Positive and negative β-gal activity in only the individual lipid containing cells (see Figure S5A).
(D) Macroscopic (top) and microscopic (40×-bottom) images of SVF cultures from PTENmyf5cKO and PTENmyf5cKO-LacZ mice stained with X-Gal.
(E) Positive and negative β-gal cell quantification shown in (D).
(F) Positive and negative β-gal activity in only the individual lipid containing cells (see Figure S5B).
(G) APCs were purified from the myf5-cre;R26R-YFP (wild type) fat depots. Yellow bars: YFP+ APCs; white bars: YFPneg. (n=9).
(H) qRT-PCR of Zic1, prdm16, HoxC8 and HoxC9 mRNA in YFP+ and YFPneg APCs purified from the iBAT and rWAT (n=3) of myf5-Cre;R26R-YFP mice. ND: not detectable. Bars represent mean± SEM; ***, p<0.001. See also Figure S4.
To determine the adipogenic potential of the LacZ-positive SVF cell populations, we differentiated the SVF cells purified from myf5-cre;R26R-LacZ mice and stained them for LacZ expression [Figure S4A]. Of the lipid-containing cells in the differentiated fractions, 99.4% of the iBAT-derived cells are LacZ-positive [Figure 5C], indicating the adipogenic cells isolated from BAT are predominantly of Myf5+ precursor origin. In iWAT and rWAT, LacZ labels 48.2% and 58.1% of the lipid-containing cells respectively, indicating that nearly half of the adipogenic cells in the SVFs of these depots also originate from a Myf5+ precursor. In contrast, only a few (<2%) of the lipid-containing cells in the ingWAT and pgWAT differentiated fractions are LacZ positive. In these depots, it appears that the majority of Myf5+ lineage SVF cells are non-adipogenic under these conditions.
The SVF contains blood, vascular, nerve, and matrix cells in addition to adipocyte progenitor cells (APCs). To specifically examine the Myf5-lineage contribution to the APC population in each depot, we generated myf5-cre;R26R-EYFP mice, purified the APCs from each depot and quantified how many of them express YFP by flow cytometry. We first separated the adipocyte pools from the SVF in each depot and examined YFP expression using co-expression of leptin and pref1 respectively to monitor fraction purity [Figure S4C and S4D]. Consistent with the LacZ lineage-tracing analysis, YFP expresses highly in both the adipocyte fraction and SVF of iBAT, sBAT, iWAT, and rWAT [Figure S4C and S4D]. The APCs are contained within a CD31−:CD45−:Ter119−:CD29+:CD34+:Sca1+ cell population (Rodeheffer et al., 2008). Approximately 85% of the APCs purified from the iBAT or sBAT SVFs express YFP protein [Figure 5G] consistent with brown fat progenitor cells being largely derived from Myf5+ precursors. In the iWAT and rWAT, 48.9% and 69.4% of the APCs respectively are also YFP positive, correlating well with the percentage of LacZ-positive adipogenic cells in their SVFs [compare to Figure 5C]. Also consistent with the LacZ lineage tracing, most APCs purified from ingWAT or pgWAT are YFPneg although a small number of APCs in each depot (6.1% and 9.1% respectively) do express YFP [Figure 5G]. This also correlates with the LacZ tracing in their SVFs [Figure 5B]. However, only 1.6% and 0.6% of the Myf5 lineage SVF cells in these depots are adipogenic in vitro [compare to Figure 5C]. Collectively, these data indicate all depots examined (BATs and WATs) contain varying amounts of adipocyte progenitors that arise from Myf5+ precursors.
Because Myf5+ precursors are thought to give rise only to BAT, we asked whether the Myf5-lineage APCs residing in WAT have a brown preadipocyte signature. Importantly, YFP mRNA expression is only detectable in the YFP+ APC populations, validating the sorting purity [Figure S4E, S7]. Both the YFP+ and YFPneg APCs purified from the iBAT of myf5-cre;R26R-EYFP mice express the brown fat marker Zic1, while only the YFP+ iBAT APC pool expresses prdm16 [Figure 5H]. Neither marker is detectable in the YFP+ APCs purified from rWAT indicating the Myf5+ white APCs do not express classic brown fat markers. Moreover, both the YFP+ and YFPneg APCs purified from rWAT more highly express the white fat markers HoxC8 and HoxC9, which express at low or undetectable levels respectively in the APCs purified from iBAT [Figure 5H]. Thus, Myf5-lineage APCs residing in WAT have a distinct gene signature and are not equivalent to BAT progenitors.
Prolonged exposure to a β3-adrenoreceptor agonist does not selectively target Myf5+ adipocyte lineages
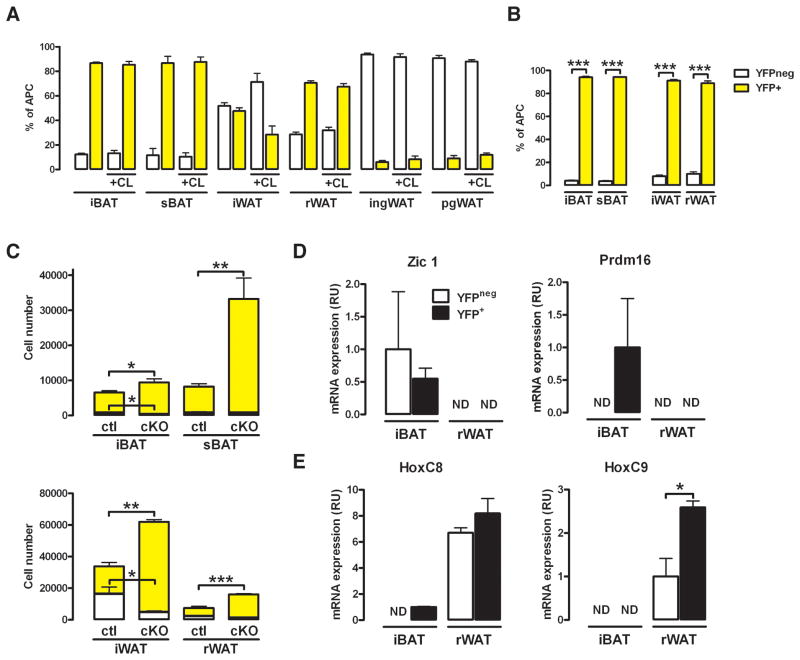
We next considered the possibility that the Myf5-lineage in WAT could be a source of a distinct population of brite fat cells different from the Myf5neg brite fat cells residing in inguinal WAT. It is reported for some strains that rWAT briefly expresses UCP1 in early post-natal development and might transiently function like classical brown fat, possibly regaining that function in adult mice when stimulated (Xue et al., 2007). We compared ucp1 expression at 1, 2, 3 and 7 weeks of age between the rWAT, ingWAT, and pgWAT. There is indeed a small transient increase in ucp1 expression in rWAT between 1 and 2 weeks of age (2.1 fold) [Figure S6A]; however, ingWAT transiently increases ucp1 expression to a much higher degree (33.3 fold). Thus, if rWAT transiently contributes to thermogenesis in these mice, that contribution is likely overshadowed by the ingWAT. To further test whether the Myf5-lineage adipocytes in WAT are the inducible brown fat cells, we treated myf5-cre;R26R-LacZ mice for 1 week with CL316,243 (a β3-adrenoreceptor agonist) and examined the overlap between multilocular cells and LacZ expression. Consistent with un-stimulated ingWAT [Figure 4E and S3C], none of the multilocular cells in either ingWAT or pgWAT following prolonged CL316,243 exposure are LacZ positive [Figure S5B]. In contrast, most of the CL316,243 induced multilocular cells in rWAT are LacZ positive [Figure S5B], consistent with the high level of Myf5-lineage tracing to this depot. Both ingWAT and rWAT strongly induce ucp1 expression relative to pgWAT upon CL316,243 treatment [Figure S5C]. Thus, β3-adrenoreceptor-stimulation induces brite adipocytes arising from both Myf5+ and Myf5neg lineages.
Next, we asked if prolonged exposure to CL316,243 selectively stimulates expansion of Myf5-lineage APCs. In myf5-cre;R26R-EYFP mice, CL316,243 has little effect on YFP+ APC composition in BAT, which is not surprising because brown adipocytes are already present and fully differentiated in classical BAT depots [Figure 6A]. Interestingly, CL316,243 increases the percentage of YFPneg APCs in the iWAT [Figure 6A], as well as total APC number [Figure S5D]. In contrast, CL316,243 has no effect on YFP+ APC composition in rWAT [Figure 6A]. CL316,243 also has little effect on the overall APC composition of ingWAT or pgWAT [Figure 6A], although the total number of YFPneg and YFP+ APCs in these depots increases [Figure S5D]. Thus, CL316,243 does not selectively stimulate expansion of the APCs arising from Myf5+ precursors.
Figure 6. Deleting PTEN with myf5-cre, but not prolonged CL316,243 treatment, selectively expands the Myf5+ adipocyte lineages.
(A) APCs were purified from myf5-cre;R26R-YFP mice treated with PBS or CL316,243 for a week (n=3). Yellow bars: YFP+ APCs; white bars: YFPneg.
(B) APCs were purified by FACS from the PTENmyf5cKO-YFP fat depots (n=4). Yellow bars represent YFP+ APCs; white bars are YFPneg. (C) Total number of YFP+ (yellow bars, top) and YFPneg cells (white bars, bottom) in each depot of myf5-cre;R26R-YFP and PTENmyf5cKO-YFP (cKO) mice (n=4).
(D) qRT-PCR of Zic1 and prdm16 mRNA in YFP+ and YFPneg APCs purified from the iBAT and rWAT (n=3) of PTENmyf5cKO-YFP mice. ND indicates not detectable.
(E) qRT-PCR of HoxC8 and HoxC9 mRNA as in (D). Bars represent mean± SEM; T-test; *, p<0.05, **, p<0.01, ***, p<0.001. See also Figure S5.
Deleting PTEN with myf5-cre selectively expands adipocyte lineages arising from Myf5+ precursors
Lineage tracing clearly indicates that iWAT and rWAT contain a mixture of APCs arising from both Myf5+ and Myf5neg precursors; however, sizing individual iWAT and rWAT adipocytes in the PTENmyf5cKO mice does not partition them into two distinct populations [Figure 2C]. Moreover, immunoblotting and RT-PCR analysis shows that myf5-cre completely deletes PTEN in iWAT and rWAT [Figure 3B and 3E]. This contradicts the lineage tracing data that shows the Myf5-lineage partially contributes to the adipocyte population in these depots and therefore predicts only partial PTEN deletion [Figure 5G]. To begin unraveling the mechanism by which PTEN loss in Myf5+ precursors redistributes body fat resulting in depots completely lacking PTEN, we first examined the Myf5 lineage in myf5-cre;PTENfl/fl;R26R-LacZ (PTENmyf5cKO-LacZ) mice. The PTENmyf5cKO-LacZ mice show strong uniform β-galactosidase staining in the iBAT, sBAT, iWAT, and rWAT depots compared to controls [Figure 4E]. Most SVF cells (>95%) purified from PTENmyf5cKO-LacZ iBAT, iWAT and rWAT stain positive for LacZ compared to 0% in control cells which lack the reporter (Figure 5D and 5E). After inducing differentiation, almost all of the lipid containing cells derived from iWAT (97.9%) and rWAT (99.1%) stain positive for LacZ (Figure 5F and S4B) indicating most PTENmyf5cKO-LacZ white adipocyte progenitors and mature adipocytes arise from Myf5+ precursors.
Next we generated myf5-cre;PTENfl/fl;R26R-EYFP (PTENmyf5cKO-YFP) mice to ask how deleting PTEN affects the Myf5 lineage contribution to the APC pools in each depot. Importantly, PTEN mRNA is barely detectable in the YFP+ APCs isolated from PTENmyf5cKO-YFP mice confirming high deletion efficiency [Figure S5E]. PTEN-deficient YFP+ APCs are also larger than control YFP+ APCs consistent with PTEN regulating cell size [Figure S6F]. A slight increase in the percentage of YFP+ APCs residing in BAT is detectable in PTENmyf5cKO-YFP mice (up from 90 to 95%) [Figure 6B]. Most importantly, compared to the Myf5 lineage contribution to the APC pools in PTEN positive mice [Figure 5G], the percentage of YFP+ APCs residing in iWAT and rWAT in PTENmyf5cKO-YFP mice increases to 91% and 89% respectively [Figure 6B]. Thus, deleting PTEN with myf5-cre increases the Myf5-lineage contribution to the APC pool in iWAT and rWAT to levels comparable with BAT.
Consistent with PTENmyf5cKO whole fat pads having more total cells [Figure 2D–G], the total number of YFP+ APCs residing in each PTENmyf5cKO-YFP fat depot also increases [Figure 6C]. To confirm that PTEN-deficient APCs have the same adipogenic potential as PTEN positive control APCs we performed adipogenic colony forming assays. Single Myf5-lineage APCs from control and PTEN-deficient BATs or WATs were sorted, clonally expanded, and differentiated (see Supplemental Methods). After induction more that 79% of the BAT and 73% of the WAT APC colonies contained cells staining positive for neutral lipids (Figure S5G and S5H). These results are consistent with recent reports (Joe et al., 2010; Lee et al., 2012). Adipogenic colony forming potential is as efficient if not higher when PTEN is deleted (86.16% for BAT and 79.49% for WAT) (Figure S5G and S5H) indicating hyperplasia of PTENmyf5cKO fats probably results from increased numbers of adipocyte progenitors.
Similar to what we observe in wild type tissues, the brown fat markers cidea, prdm16, zic1, and ucp1 remain highly expressed in PTENmyf5cKO whole iBAT compared to rWAT, with PTEN-deletion causing only a negligible drop in cidea and ucp1 expression in the iBAT [Figure 4F, PTENmyf5cKO]. This indicates PTEN loss does not affect BAT marker gene expression. Similarly, PTEN loss does not affect Hoxc8 and Dpt expression in rWAT, while Hoxc9 expression is slightly reduced [Figure 4G, PTENmyf5cKO]. Thus, white fat identity is also largely unperturbed by PTEN loss. Finally, we profiled the molecular identity of the YFP+ and YFPneg APCs purified from the PTENmyf5cKO-YFP iBAT and rWAT depots. Similar to wild type YFP+ APCs [Figure 5G and 5H], the PTEN-deficient BAT APCs express Zic1 and Prdm16, which are undetectable in PTENmyf5cKO-YFP rWAT APCs [Figure 6D]. Moreover, the white fat markers Hoxc8 and Hoxc9 express at low or undetectable levels respectively in PTENmyf5cKO-YFP iBAT APCs but continue to express highly in PTENmyf5cKO-YFP rWAT APCs [Figure 6E]. We conclude that deleting PTEN with myf5-cre does not affect APC identity or adipogenic colony forming potential, but expands the Myf5 adipocyte lineages such that all fat depots (brown and white) in PTENmyf5cKO mice are exclusively derived from Myf5+ precursors.
DISCUSSION
Little is known about the developmental origins of adipose tissue, what controls adipose tissue heterogeneity, and what determines the variable body fat distribution patterns seen in humans. Understanding this is clinically important because some fats (e.g. brown fat and subcutaneous white fat) show favorable metabolic characteristics, while other fats (e.g. visceral white fat) are linked to metabolic disorders. Moreover, some partial lipodystrophies, which in general are poorly understood, often manifest as adipose tissue distribution disorders. In this study we made the surprising finding that a subset of unilocular white adipocytes are derived from a precursor cell that expresses the myf5-cre knock-in allele, and that the contribution of this lineage to each adipose tissue varies with depot location.
Different fat depots develop at different stages of postnatal life and have unique characteristics [Reviewed in (Billon and Dani, 2011; Gesta et al., 2007)] This has led to the hypothesis that different white adipose depots originate from distinct mesodermal locations. However, a specific mesoderm-derived white adipose lineage had not been described. Our discovery that a subset of unilocular white adipocytes originates from Myf5-Cre+ precursors supports this hypothesis and identifies at least one WAT lineage arising from the mesoderm. There is also accumulating evidence that the adipocyte population within certain depots is heterogeneous (Bluher et al., 2002; Fortier et al., 2005). Moreover, two distinct subtypes of preadipocytes have been characterized in human fat, the proportions of which vary among depot location (Tchkonia et al., 2005). Our finding that WAT contains a mixed population of Myf5-Cre+ and Myf5-Creneg derived progenitors supports this.
We do not yet know if being of Myf5+ or Myf5neg origin has metabolic significance or if the Myf5 lineage contribution to each depot varies with strain background. Differences in these characteristics could affect body fat pattering or sensitivity to obesity. It is alternatively possible that despite distinct origins, Myf5+ and Myf5neg adipocytes are functionally identical. This would argue that while myf5-cre expression distinguishes the origins of adipocytes, it should not be used to delineate them functionally. Regardless, mutations affecting distinct developmental lineages could be the pathological basis of some human fat disorders (discussed below). Human adipose-derived stem cells (ASCs) are abundantly available and obtainable by minimally invasive procedures. Thus, understanding adipocyte lineage variations could have important therapeutic implications [Reviewed in (Tran and Kahn, 2010)]. For example, there may be favorable characteristics associated with a particular developmental origin and selecting for the optimum ASC population could offer distinct advantages.
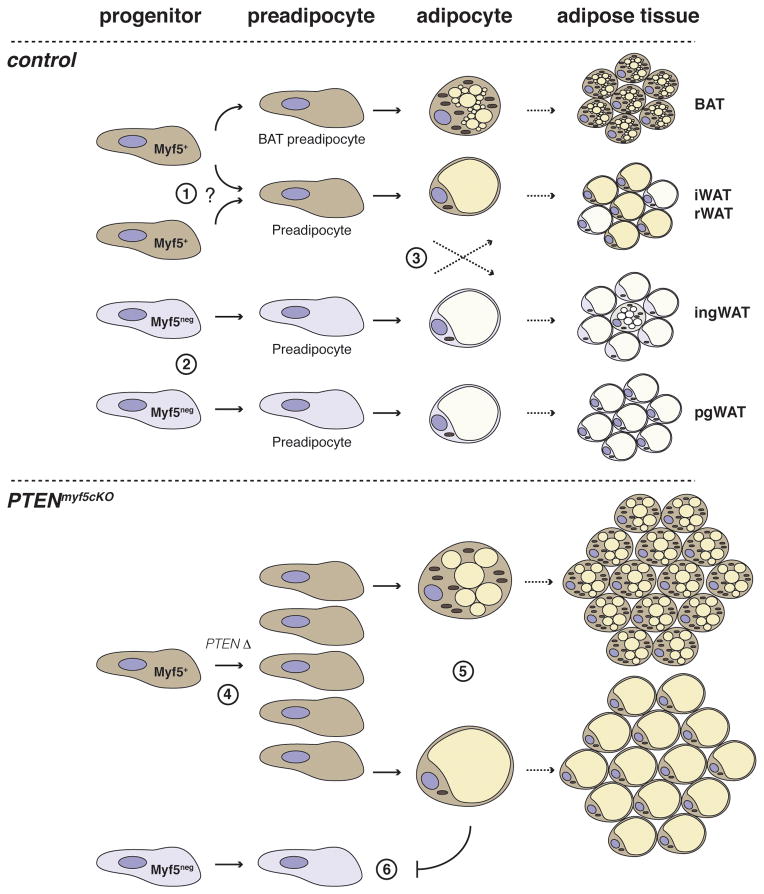
Previous work using the same Myf5-cre knock-in line suggested that classical brown fat adipocytes, but not the inducible brite fat cells, arise from Myf5+ precursors, leading to a widely accepted model in which brown fat and skeletal muscle share a common precursor (Seale et al., 2008)[Reviewed in (Billon and Dani, 2011; Cristancho and Lazar, 2011; Fruhbeck et al., 2009; Kajimura et al., 2010; Seale et al., 2009; Tseng et al., 2010)]. It is hypothesized that the common origin of brown adipocytes and muscle explains the favorable metabolic properties of brown fat. All white adipocytes and brite adipocytes are thought to arise from a unique undefined Myf5neg mesenchymal lineage(s). Our Myf5-Cre lineage tracing studies are consistent with brown adipocytes largely originating from a Myf5+ precursor. However, our work further suggests white fat depots contain a mixture of adipocytes and resident APCs derived from both Myf5+ and Myf5neg lineages, with the Myf5+ lineage being more highly represented in some fats (e.g. iWAT and rWAT) compared to others (e.g. ingWAT and pgWAT). We also find that the brite adipocytes in the inguinal WAT do not label with Myf5-Cre, which is also consistent with previous work. Although the same Myf5-Cre knock-in line is used in this study and in Seale et al, the PTEN-deletion phenotype described here led us to examine other white fat depots in addition to the largely Myf5neg inguinal and peri-gonadal WATs examined by Seale et al. The only discrepancy is that Seale et al used indirect immunofluorescence to show that the interscapular WAT depot is YFP negative, (Seale et al., 2008). We also cannot detect YFP signal by indirect immunofluorescence in interscapular WAT (or any other WAT) using this approach, likely reflecting a technical limitation. However, we additionally employed the ROSA26-LacZ reporter, RT-PCR, and FACS analysis to clearly show Myf5-Cre lineage tracing to the interscapular WAT. Based on these insights, which were uncovered by deleting PTEN with Myf5-Cre and confirmed through lineage tracing, we suggest an alternative model of adipose tissue development [Figure 7].
Figure 7. Model of adipose tissue development based on lineage analysis and conditional PTEN deletion with Myf5-Cre.
(1) Myf5-expressing progenitor cells give rise to a classical brown adipocytes as well as a subset of white adipocytes in iWAT and rWAT. Alternatively, Myf5-Cre expressing progenitor cells, distinct from those which give rise to classical BAT, could give rise to adipocytes in iWAT and rWAT– indicated by the question mark. (2) Myf5neg progenitors of unknown origin give rise to ingWAT and pgWAT. (3) Many adipose depots contain a mixed population of adipocyte progenitor cells arising from both Myf5-Cre+ and Myf5-Creneg precursors. The Myf5-lineage significantly contributes to the adipocyte population in iWAT and rWAT, while in ingWAT and pgWAT the significance of the resident Myf5+ cells is unknown. Notably, the multilocular “brite fat” cells residing in ingWAT at ambient temperature, or induced after prolonged exposure to CL316,243 (not shown in figure), do not trace to the Myf5-Cre lineage. (4) PTEN loss in Myf5+ precursors expands the Myf5+ preadipocyte pool. (5) Upon differentiation, the PTEN-deficient adipocyte lineages accumulate excess lipid, resulting in overgrowth. (6) The growing Myf5+ adipocyte lineages restrict development of Myf5neg adipocyte lineages, resulting in the selective expansion of fats exclusively derived from Myf5+ precursors.
We also considered the possibility that the Myf5-Cre marked adipocytes in the iWAT and rWAT could represent an inducible brown-adipocyte like lineage distinct from the Myf5neg brite fat cells in the inguinal WAT. However, in both the iWAT and rWAT, only a subset of the adipocytes arising from Myf5-Cre+ precursors become multilocular following prolonged CL316,243 treatment. We have adopted the perspective for now that the developmental origins of brown, brite, and at least a subset of white adipocytes cannot be delineated simply based on Myf5-Cre+ expression. Moreover, the heterogeneity within and between depots, particularly with respect to ucp1 expression dynamics, leads us to conclude that the iWAT, rWAT, ingWAT, and pgWAT depots should be considered as distinct types of white fat.
Recent studies of intramuscular adipose tissue find that these particular adipocytes also do not originate from a Myf5-Cre expressing precursor (Joe et al., 2010; Liu et al., 2012; Schulz et al., 2011). Notably, the study by Liu et al finds that Pax3-Cre+ precursors, like Myf5-Cre+ precursors, give rise to brown fat and importantly for our study, roughly half of the adipocytes in the iWAT (Liu et al., 2012). However, it is also possible that Pax3-Cre and Myf5-Cre could be transiently and independently expressed in distinct precursor pools, different from those that give rise to skeletal muscle or to brown fat. This requires further investigation. We currently favor the idea that Myf5-expressing precursors may be more akin to the multi-potential Engrailed-1-expressing precursors in the central dermomyotome, which are reported to become BAT, skeletal muscle, and dermis (Atit et al., 2006).
Insulin through PI3K regulates adipogenesis in both brown and white fat (Cannon and Nedergaard, 2004; Rosen and MacDougald, 2006). Therefore, it is perhaps not surprising that deleting PTEN with Myf5-Cre increases the lipid content of both brown and white adipocytes. However, a previous study targeting PTEN deletion to adipocytes with the aP2-Cre did not detect any changes in adipose tissue mass or in lipid droplet size (Kurlawalla-Martinez et al., 2005). Thus, for reasons unclear it appears that increasing PI3K signaling in brown or white preadipocytes can cause excessive lipid accumulation, but once an adipocyte fully differentiates, the ability to further increase lipid content through PI3K activation is lost. That PTEN regulates brown fat is supported by a recent paper describing a transgenic mouse model of PTEN, in which PTEN is overexpressed in all tissues (Ortega-Molina et al., 2012). Although systemic effects cannot be separated from tissue-specific effects in this model, PTEN overexpressing mice have reduced lipid content in their interscapular brown fat pads. Interestingly, globally overexpressing PTEN appears to increase BAT activity as well as increase the number of multilocular adipocytes in inguinal WAT, leading the authors to conclude that PTEN positively regulates energy expenditure; although the possibility that overexpressing PTEN also suppresses adipogenesis is not considered. Exactly how overexpressing PTEN increases energy expenditure and its physiological significance to normal regulation of thermogenesis remains unclear.
The fact that deleting PTEN with Myf5-Cre redistributes body fat raises the interesting possibility that variations in metabolic regulation between distinct adipocyte lineages could be a factor in controlling body fat distribution. For example, if a particular lineage is more insulin sensitive, that lineage may preferentially expand. Although PTEN loss affects many downstream pathways, the PTENmyf5cKO mice have low circulating glucose levels, and are highly glucose tolerant and insulin sensitive suggesting they are hyper sensitive to insulin (J.S-G. and D.A.G., unpublished). Thus, one factor contributing to the selective expansion of PTEN-deficient fats may be that adipocytes lacking PTEN preferentially clear and store glucose (as lipid) leaving no excess circulating energy available for storage in the Myf5neg adipocyte lineages. In this model, the Myf5neg lineages would never receive the adipogenic signals to differentiate. Although PTEN deletion is a genetically engineered and extreme example of insulin/PI3K pathway activation, more subtle and natural variations in metabolic regulation between distinct lineages could partly explain the variable body fat pattering seen in humans.
Interestingly, the PTENmyf5cKO phenotype is remarkably similar to a mysterious human disorder called Multiple Symmetric Lipomatosis (MSL) (also called Benign Symmetric Lipomatosis or Madelung’s disease) (Herbst, 2012). MSL is characterized by adipose tissue overgrowth in the upper back, neck, and shoulders and is often accompanied by decreased visceral adiposity and muscle atrophy. Patients suffer from disfiguration, neuropathy and are at risk for sudden death (Enzi et al., 2002; Guastella et al., 2002; Ramos et al., 2010). Although poorly characterized, the location, histological features, and metabolic properties of the diseased fat led to the proposal that MSL may be of brown fat origin (Cinti et al., 1983; Nisoli et al., 2002; Zancanaro et al., 1990). Here we describe a model of partial lipodystrophy and suggest one possible explanation for the pathology associated with these conditions is that a certain adipocyte lineage analogous to the Myf5 lineage may be selectively targeted resulting in altered body fat distribution.
In this report, we provide evidence that Myf5+ precursors give rise to both brown and white adipocytes, and show that activating PI3K signaling in the Myf5+ lineage redistributes body fat. Future studies aimed at defining the significance of adipocyte ancestry, the lineage composition of different fat depots, and to understanding how genetic variation or mutations affect distinct adipocyte precursor pools will significantly impact the understanding and treatment of common (e.g. obesity, metabolic syndrome, diabetes) and rare (e.g. lipomatosis, lipodystrophy) human diseases of adipose tissue imbalance.
EXPERIMENTAL PROCEDURES
Materials
PTEN, S473-Akt, T308-Akt, and pan Akt antibodies were from Cell Signaling Technologies. 7AAD (BD Biosciences), calcein blue (Invitrogen), CD29-Biotin, Streptavidin-PE-Cy7, CD31-PE, CD45-PE, Ter119-PE, CD34-Alexa700, Sca1-APC (all from eBiosciences). CL316243 was from Tocris. All other reagents were from Sigma-Aldrich.
Mice
PTEN floxed mice (Jackson laboratories) were crossed to mice expressing the Cre recombinase transgene from the Myf5 promoter (Myf5-CRE) (Tallquist et al., 2000). Rosa26-LSL-YFP and Rosa26–LSL-LacZ were from Jackson laboratories. Mice were kept on a daily 12 h light/dark cycle and fed a normal chow diet (Prolab® Isopro® RMH 3000) from LabDiet ad libitum. All animal experiments were approved by the University of Massachusetts Medical school animal care and use committee.
Tissue Harvest
Adipose tissue depots described in (Walden et al., 2011) were carefully dissected to avoid contamination from surrounding tissue. Interscapular WAT refers to the tissues named bsWAT and asWAT in (Walden et al., 2011).
Gene expression analysis
Total RNA was isolated from cells or tissues using Qiazol (Invitrogen) and RNeasy kit (Invitrogen). Equal amounts of RNA were retro-transcribed to cDNA using High capacity cDNA reverse transcription kit (#4368813, Applied Biosystems). qRT-PCR was run in 10 μL reactions in a StepOnePlus real-time PCR system machine from Applied Biosystems using SYBR Green PCR master mix (#4309156, Applied Biosystems) accordingly to manufacturer instructions. Standard and melting curves were run in every plate for every gene to ensure efficiency and specificity of the reaction. Primer sequences are listed in the Extended Experimental Procedures (Table S2).
Histology and image analysis
ImageJ (NIH) was used to calculate cell area, diameter and nuclei number in formalin fixed tissues sectioned at 5 μm of thickness. Embedding and sectioning was done by the UMass Morphology Core.
Primary cell isolation and FACS analysis
Primary brown preadipocytes were isolated and immortalized according to (Fasshauer et al., 2001). SVF from all adipose depots were isolated using the same protocol. Adipose precursor cells (APCs) were isolated from SVF according to previously established markers (Rodeheffer et al., 2008; Tang et al., 2008) using a FACS Aria II cell sorter equipped with FASCDiva software in the Umass Flow cytometry core facility.
Adipogenic colony formation assay
Single cell colony formation assay was done as previously reported (Lee et al., 2012) with minor modifications. Also see supplementary experimental procedures.
LacZ staining
LacZ staining was done as in (Tang et al., 2008) with minor modifications. Also see supplementary experimental procedures.
Statistical analysis
Unless otherwise stated, data are presented as the mean ± SEM. A T-test was used to determine statistical significance; *(<0.05); **(<0.01);***(<0.001).
Supplementary Material
HIGHLIGHTS.
BAT and WAT contain progenitor cells derived from both Myf5+ and Myf5neg lineages
The number of progenitors arising from Myf5+ precursors varies with depot location
Deleting PTEN in Myf5+ precursors selectively expands the Myf5+ adipocyte lineages
Deleting PTEN in Myf5+ precursors dramatically redistributes body fat
Acknowledgments
This work is supported by grants from the National Institutes of Health (R00CA129613 and R21CA161121) and awards from the Pew Charitable Trusts and Charles Hood Foundation to D.A.G. D.A.G. is a member of the UMass DERC (DK32520). J.S-G. is supported by the Beatriu de Pinós program from the Generalitat de Catalunya.
Footnotes
The authors declare no conflict of interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Atit R, Sgaier SK, Mohamed OA, Taketo MM, Dufort D, Joyner AL, Niswander L, Conlon RA. Beta-catenin activation is necessary and sufficient to specify the dorsal dermal fate in the mouse. Dev Biol. 2006;296:164–176. doi: 10.1016/j.ydbio.2006.04.449. [DOI] [PubMed] [Google Scholar]
- Billon N, Dani C. Developmental Origins of the Adipocyte Lineage: New Insights from Genetics and Genomics Studies. Stem Cell Rev. 2011 doi: 10.1007/s12015-011-9242-x. [DOI] [PubMed] [Google Scholar]
- Bluher M, Michael MD, Peroni OD, Ueki K, Carter N, Kahn BB, Kahn CR. Adipose tissue selective insulin receptor knockout protects against obesity and obesity-related glucose intolerance. Dev Cell. 2002;3:25–38. doi: 10.1016/s1534-5807(02)00199-5. [DOI] [PubMed] [Google Scholar]
- Cannon B, Nedergaard J. Brown adipose tissue: function and physiological significance. Physiol Rev. 2004;84:277–359. doi: 10.1152/physrev.00015.2003. [DOI] [PubMed] [Google Scholar]
- Cinti S, Enzi G, Cigolini M, Bosello O. Ultrastructural features of cultured mature adipocyte precursors from adipose tissue in multiple symmetric lipomatosis. Ultrastruct Pathol. 1983;5:145–152. doi: 10.3109/01913128309141834. [DOI] [PubMed] [Google Scholar]
- Cristancho AG, Lazar MA. Forming functional fat: a growing understanding of adipocyte differentiation. Nat Rev Mol Cell Biol. 2011;12:722–734. doi: 10.1038/nrm3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cypess AM, Lehman S, Williams G, Tal I, Rodman D, Goldfine AB, Kuo FC, Palmer EL, Tseng YH, Doria A, et al. Identification and importance of brown adipose tissue in adult humans. N Engl J Med. 2009;360:1509–1517. doi: 10.1056/NEJMoa0810780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Souza CJ, Eckhardt M, Gagen K, Dong M, Chen W, Laurent D, Burkey BF. Effects of pioglitazone on adipose tissue remodeling within the setting of obesity and insulin resistance. Diabetes. 2001;50:1863–1871. doi: 10.2337/diabetes.50.8.1863. [DOI] [PubMed] [Google Scholar]
- Edens NK, Fried SK, Kral JG, Hirsch J, Leibel RL. In vitro lipid synthesis in human adipose tissue from three abdominal sites. Am J Physiol. 1993;265:E374–379. doi: 10.1152/ajpendo.1993.265.3.E374. [DOI] [PubMed] [Google Scholar]
- Enerback S. The origins of brown adipose tissue. N Engl J Med. 2009;360:2021–2023. doi: 10.1056/NEJMcibr0809610. [DOI] [PubMed] [Google Scholar]
- Enerback S. Human brown adipose tissue. Cell Metab. 2010;11:248–252. doi: 10.1016/j.cmet.2010.03.008. [DOI] [PubMed] [Google Scholar]
- Enzi G, Busetto L, Ceschin E, Coin A, Digito M, Pigozzo S. Multiple symmetric lipomatosis: clinical aspects and outcome in a long-term longitudinal study. Int J Obes Relat Metab Disord. 2002;26:253–261. doi: 10.1038/sj.ijo.0801867. [DOI] [PubMed] [Google Scholar]
- Fasshauer M, Klein J, Kriauciunas KM, Ueki K, Benito M, Kahn CR. Essential role of insulin receptor substrate 1 in differentiation of brown adipocytes. Mol Cell Biol. 2001;21:319–329. doi: 10.1128/MCB.21.1.319-329.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortier M, Soni K, Laurin N, Wang SP, Mauriege P, Jirik FR, Mitchell GA. Human hormone-sensitive lipase (HSL): expression in white fat corrects the white adipose phenotype of HSL-deficient mice. J Lipid Res. 2005;46:1860–1867. doi: 10.1194/jlr.M500081-JLR200. [DOI] [PubMed] [Google Scholar]
- Fried SK, Leibel RL, Edens NK, Kral JG. Lipolysis in intraabdominal adipose tissues of obese women and men. Obes Res. 1993;1:443–448. doi: 10.1002/j.1550-8528.1993.tb00026.x. [DOI] [PubMed] [Google Scholar]
- Frontini A, Cinti S. Distribution and development of brown adipocytes in the murine and human adipose organ. Cell Metab. 2010;11:253–256. doi: 10.1016/j.cmet.2010.03.004. [DOI] [PubMed] [Google Scholar]
- Fruhbeck G, Becerril S, Sainz N, Garrastachu P, Garcia-Velloso MJ. BAT: a new target for human obesity? Trends Pharmacol Sci. 2009;30:387–396. doi: 10.1016/j.tips.2009.05.003. [DOI] [PubMed] [Google Scholar]
- Gensch N, Borchardt T, Schneider A, Riethmacher D, Braun T. Different autonomous myogenic cell populations revealed by ablation of Myf5-expressing cells during mouse embryogenesis. Development. 2008;135:1597–1604. doi: 10.1242/dev.019331. [DOI] [PubMed] [Google Scholar]
- Gesta S, Tseng YH, Kahn CR. Developmental origin of fat: tracking obesity to its source. Cell. 2007;131:242–256. doi: 10.1016/j.cell.2007.10.004. [DOI] [PubMed] [Google Scholar]
- Guastella C, Borsi C, Gibelli S, Della Berta LG. Madelung’s lipomatosis associated with head and neck malignant neoplasia: a study of 2 cases. Otolaryngol Head Neck Surg. 2002;126:191–192. doi: 10.1067/mhn.2002.122004. [DOI] [PubMed] [Google Scholar]
- Haldar M, Karan G, Tvrdik P, Capecchi MR. Two cell lineages, myf5 and myf5-independent, participate in mouse skeletal myogenesis. Dev Cell. 2008;14:437–445. doi: 10.1016/j.devcel.2008.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbst KL. Rare adipose disorders (RADs) masquerading as obesity. Acta Pharmacol Sin. 2012;33:155–172. doi: 10.1038/aps.2011.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill R, Wu H. PTEN, stem cells, and cancer stem cells. J Biol Chem. 2009;284:11755–11759. doi: 10.1074/jbc.R800071200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jernas M, Palming J, Sjoholm K, Jennische E, Svensson PA, Gabrielsson BG, Levin M, Sjogren A, Rudemo M, Lystig TC, et al. Separation of human adipocytes by size: hypertrophic fat cells display distinct gene expression. FASEB J. 2006;20:1540–1542. doi: 10.1096/fj.05-5678fje. [DOI] [PubMed] [Google Scholar]
- Joe AW, Yi L, Natarajan A, Le Grand F, So L, Wang J, Rudnicki MA, Rossi FM. Muscle injury activates resident fibro/adipogenic progenitors that facilitate myogenesis. Nat Cell Biol. 2010;12:153–163. doi: 10.1038/ncb2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajimura S, Seale P, Spiegelman BM. Transcriptional control of brown fat development. Cell Metab. 2010;11:257–262. doi: 10.1016/j.cmet.2010.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurlawalla-Martinez C, Stiles B, Wang Y, Devaskar SU, Kahn BB, Wu H. Insulin hypersensitivity and resistance to streptozotocin-induced diabetes in mice lacking PTEN in adipose tissue. Mol Cell Biol. 2005;25:2498–2510. doi: 10.1128/MCB.25.6.2498-2510.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YH, Petkova AP, Mottillo EP, Granneman JG. In vivo identification of bipotential adipocyte progenitors recruited by beta3-adrenoceptor activation and high-fat feeding. Cell Metab. 2012;15:480–491. doi: 10.1016/j.cmet.2012.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Liu Y, Lai X, Kuang S. Intramuscular adipose is derived from a non-Pax3 lineage and required for efficient regeneration of skeletal muscles. Dev Biol. 2012;361:27–38. doi: 10.1016/j.ydbio.2011.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nedergaard J, Cannon B. The changed metabolic world with human brown adipose tissue: therapeutic visions. Cell Metab. 2010;11:268–272. doi: 10.1016/j.cmet.2010.03.007. [DOI] [PubMed] [Google Scholar]
- Nisoli E, Regianini L, Briscini L, Bulbarelli A, Busetto L, Coin A, Enzi G, Carruba MO. Multiple symmetric lipomatosis may be the consequence of defective noradrenergic modulation of proliferation and differentiation of brown fat cells. J Pathol. 2002;198:378–387. doi: 10.1002/path.1212. [DOI] [PubMed] [Google Scholar]
- Ortega-Molina A, Efeyan A, Lopez-Guadamillas E, Munoz-Martin M, Gomez-Lopez G, Canamero M, Mulero F, Pastor J, Martinez S, Romanos E, et al. Pten positively regulates brown adipose function, energy expenditure, and longevity. Cell Metab. 2012;15:382–394. doi: 10.1016/j.cmet.2012.02.001. [DOI] [PubMed] [Google Scholar]
- Ramos S, Pinheiro S, Diogo C, Cabral L, Cruzeiro C. Madelung disease: a not-so-rare disorder. Ann Plast Surg. 2010;64:122–124. doi: 10.1097/SAP.0b013e31819bd729. [DOI] [PubMed] [Google Scholar]
- Rodeheffer MS, Birsoy K, Friedman JM. Identification of white adipocyte progenitor cells in vivo. Cell. 2008;135:240–249. doi: 10.1016/j.cell.2008.09.036. [DOI] [PubMed] [Google Scholar]
- Rosen ED, MacDougald OA. Adipocyte differentiation from the inside out. Nat Rev Mol Cell Biol. 2006;7:885–896. doi: 10.1038/nrm2066. [DOI] [PubMed] [Google Scholar]
- Schulz TJ, Huang TL, Tran TT, Zhang H, Townsend KL, Shadrach JL, Cerletti M, McDougall LE, Giorgadze N, Tchkonia T, et al. Identification of inducible brown adipocyte progenitors residing in skeletal muscle and white fat. Proc Natl Acad Sci U S A. 2011;108:143–148. doi: 10.1073/pnas.1010929108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seale P, Bjork B, Yang W, Kajimura S, Chin S, Kuang S, Scime A, Devarakonda S, Conroe HM, Erdjument-Bromage H, et al. PRDM16 controls a brown fat/skeletal muscle switch. Nature. 2008;454:961–967. doi: 10.1038/nature07182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seale P, Kajimura S, Spiegelman BM. Transcriptional control of brown adipocyte development and physiological function--of mice and men. Genes Dev. 2009;23:788–797. doi: 10.1101/gad.1779209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tallquist MD, Weismann KE, Hellstrom M, Soriano P. Early myotome specification regulates PDGFA expression and axial skeleton development. Development. 2000;127:5059–5070. doi: 10.1242/dev.127.23.5059. [DOI] [PubMed] [Google Scholar]
- Tang W, Zeve D, Suh JM, Bosnakovski D, Kyba M, Hammer RE, Tallquist MD, Graff JM. White fat progenitor cells reside in the adipose vasculature. Science. 2008;322:583–586. doi: 10.1126/science.1156232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tchkonia T, Tchoukalova YD, Giorgadze N, Pirtskhalava T, Karagiannides I, Forse RA, Koo A, Stevenson M, Chinnappan D, Cartwright A, et al. Abundance of two human preadipocyte subtypes with distinct capacities for replication, adipogenesis, and apoptosis varies among fat depots. Am J Physiol Endocrinol Metab. 2005;288:E267–277. doi: 10.1152/ajpendo.00265.2004. [DOI] [PubMed] [Google Scholar]
- Timmons JA, Wennmalm K, Larsson O, Walden TB, Lassmann T, Petrovic N, Hamilton DL, Gimeno RE, Wahlestedt C, Baar K, et al. Myogenic gene expression signature establishes that brown and white adipocytes originate from distinct cell lineages. Proc Natl Acad Sci U S A. 2007;104:4401–4406. doi: 10.1073/pnas.0610615104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran TT, Kahn CR. Transplantation of adipose tissue and stem cells: role in metabolism and disease. Nat Rev Endocrinol. 2010;6:195–213. doi: 10.1038/nrendo.2010.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng YH, Cypess AM, Kahn CR. Cellular bioenergetics as a target for obesity therapy. Nat Rev Drug Discov. 2010;9:465–482. doi: 10.1038/nrd3138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Marken Lichtenbelt WD, Vanhommerig JW, Smulders NM, Drossaerts JM, Kemerink GJ, Bouvy ND, Schrauwen P, Teule GJ. Cold-activated brown adipose tissue in healthy men. N Engl J Med. 2009;360:1500–1508. doi: 10.1056/NEJMoa0808718. [DOI] [PubMed] [Google Scholar]
- Virtanen KA, Lidell ME, Orava J, Heglind M, Westergren R, Niemi T, Taittonen M, Laine J, Savisto NJ, Enerback S, et al. Functional brown adipose tissue in healthy adults. N Engl J Med. 2009;360:1518–1525. doi: 10.1056/NEJMoa0808949. [DOI] [PubMed] [Google Scholar]
- Walden TB, Hansen IR, Timmons JA, Cannon B, Nedergaard J. Recruited versus nonrecruited molecular signatures of brown, “brite” and white adipose tissues. Am J Physiol Endocrinol Metab. 2011;302:E19–E31. doi: 10.1152/ajpendo.00249.2011. [DOI] [PubMed] [Google Scholar]
- Wang X, Jiang X. Post-translational regulation of PTEN. Oncogene. 2008;27:5454–5463. doi: 10.1038/onc.2008.242. [DOI] [PubMed] [Google Scholar]
- Xue B, Rim JS, Hogan JC, Coulter AA, Koza RA, Kozak LP. Genetic variability affects the development of brown adipocytes in white fat but not in interscapular brown fat. J Lipid Res. 2007;48:41–51. doi: 10.1194/jlr.M600287-JLR200. [DOI] [PubMed] [Google Scholar]
- Zancanaro C, Sbarbati A, Morroni M, Carraro R, Cigolini M, Enzi G, Cinti S. Multiple symmetric lipomatosis. Ultrastructural investigation of the tissue and preadipocytes in primary culture. Lab Invest. 1990;63:253–258. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.