Abstract
I-κB kinases (IKKs) are key regulators of NF-κB signaling. Three IKK isoforms – α, β and ε – have been linked to oncogenesis, yet the precise components of NF-κB signaling in ovarian cancer have not yet been dissected. We surveyed 120 ovarian cancer specimens for IKKε expression. Notably, cytoplasmic expression was elevated in metastatic lesions relative to primary tumors (p=0.03). Therefore, we hypothesized that IKKε drives ovarian cancer metastasis. IKKε was identified previously as a breast cancer oncogene and was associated with poor clinical outcome in ovarian cancer. We now define an ovarian cancer-specific IKKε-regulated gene expression signature using stably expressed shRNA targeting IKKε. Pathway analysis of the signature indicated that IKKε regulates expression of genes involved in cell motility and inflammation. We further showed that IKKε depletion in metastatic ovarian cancer cell lines decreased growth, adhesion, and invasion. Consistently, human xenografts depleted of IKKε in mice demonstrated decreased aggressiveness, while overexpression of IKKε in a less invasive ovarian cancer cell line increased metastasis in vivo. Taken together, these data provide evidence that IKKε is a key coordinator of invasion and metastasis programs in ovarian cancer. Inhibition of IKKε signaling thus emerges as a viable therapeutic strategy in women whose ovarian cancer demonstrates aberrant activation of this pathway.
Keywords: Ovarian cancer, NF-kappaB, metastasis, invasion, gene expression
INTRODUCTION
Ovarian cancer affects 20,000 women annually in the United States. The fractional death rate exceeds 50% due to late stage at diagnosis, and eventual resistance to chemotherapy (1). Goals in the field include improved screening and diagnostics, and improved therapy of advanced disease at the outset and for recurrence (2, 3). Recent molecular profiles of ovarian cancers reveal marked heterogeneity even within defined histological subtypes. Gene expression defined subsets of patients presenting aggressive disease which respond differently to standard surgery and chemotherapy treatment (4). Attempts were made to correlate molecular signatures with better survival. The Cancer Genome Atlas (TCGA) has rapidly and comprehensively advanced the molecular profiling of ovarian cancer through large-scale gene expression profiling, comparative genomic hybridization, single nucleotide polymorphism analysis, and gene exon sequencing (5). These efforts have made clear that ovarian cancer is an extremely heterogeneous disease. A single approach to chemotherapy is unlikely to achieve similar success across all patients. Therefore, there is pressing need to identify the molecular etiology driving defined subgroups of ovarian cancers, and to develop treatments targeting such pathways in order to improve specific patient survival.
The involvement of NF-κB in cancer dissemination further makes it a logical target. IKKα promotes metastasis in prostate cancer, due to its ability to suppress Maspin expression (6). Activated IKKα in prostate cancer cells’ nuclei increased with advancing stage of prostate cancers, and inversely correlated with Maspin expression. We recently identified the NF-κB signaling pathway as a potential driver of ovarian cancer growth (7). We identified autonomous NF-κB signaling in a defined subset of ovarian cancer, driven by IKKβ and susceptible to therapeutic targeting of this kinase (8). However, the mechanism through which the NF-κB pathway affects the initiation, propagation and dissemination of ovarian cancer is unknown. I-κB kinases (IKKs) are key regulators of NF-κB signaling. Three IKK isoforms – α, β and ε – have been linked to oncogenesis, yet the precise components of NF-κB signaling mechanisms in ovarian cancer have not yet been dissected.
A systems biology approach discovered that IKKε functioned as an oncogene in breast cancer (9). The IKKε locus (1q32) was found to be amplified in ~30% of human breast cancer specimens, and correlated with high expression of IKKε transcripts and protein. The oncogenic activity of IKKε was regulated through the transcription factor cREL, indicating that the NF-κB pathway is a key downstream mediator of IKKε-induced transformation. Further evidence that IKKε acts through the NF-κB pathway in oncogenesis was derived from a positional scanning peptide library assay to identify substrates of IKKε (10). The deubiquitinating enzyme CYLD, a tumor suppressor and negative regulator of the NF-κB pathway, was of particular interest. IKKε exerted oncogenic pressure by inhibiting CYLD activity through phosphorylation, resulting in the blockage of the deubiquitination of TRAF2 and NEMO, positive regulators of the classical NF-κB pathway.
Recent work associated expression of IKKε with poor outcome in women with ovarian cancer (11). IKKε was found to be over-expressed and activated in primary ovarian cancer specimens and cell lines. Its over-expression in tumors was associated with high grade, advanced stage disease. In cell lines, IKKε promoted resistance to platinum-based chemotherapy, a class of agents included in the initial therapy of advanced ovarian cancer. A role for IKKε in ovarian cancer was implicated by these results, without definition of its mechanism and function. We therefore sought, in the current study, to examine IKKε in ovarian cancer. Here, we have identified IKKε as a key coordinator of invasion and metastasis in ovarian cancer.
MATERIALS and METHODS
Patients and tissue microarray samples
Tissue microarrays (TMAs) contained 2-mm cores (n=270) from 120 ovarian carcinoma specimens (42 primary carcinomas, 78 solid metastases) operated at the Norwegian Radium Hospital. Metastases were to the omentum (n=46), peritoneum (n=15), intestine (n=12), lymph nodes (n=3) or other sites (n=2). Tumors were from 57 patients, of whom 38 had both primary carcinoma and one or more metastasis for evaluation, 4 had only primary carcinoma and 14 had only one or more metastasis. Tumors underwent microscopic confirmation of diagnosis, histological type and grade by a gynecopathologist (BD). Grading was according to the FIGO system. The Regional Committee for Medical Research Ethics in Norway approved the study. Antibodies and staining conditions were previously detailed (7).
Cell lines and culture conditions
Ovarian cancer cell lines CAOV3, A2780, MDA-Ovcar3, Skov3, A2780-1A9, and HeyA8 cells were a gift from Dr. Elise Kohn. Ovarian cancer cell lines IGROV1, Ovcar3, Ovcar4, Ovcar5, and Ovcar8 were obtained from the NCI-Frederick DCTD tumor/cell line repository (Frederick, MD). Low passage Caov3 cells were purchased from ATCC (Manassas, VA). All ovarian lines were cultured in RPMI plus 10% fetal bovine serum (Hyclone, Pittsburg, PA) and standard antibiotics. Cultures were maintained at 37°C in a 5% CO2 atmosphere. The Kohn cell lines were authenticated in July 2009 at the Johns Hopkins University Fragment Analysis Facility (Baltimore, MD) using Promega PowerPlex 1.2 System to test for STR markers (D16S539, D7S820, D13S317, D5S818, CSF1PO, TPOX, THO1, and vWA) and amelogenin for gender determination. Authenticity was confirmed against the ATCC database (12), CLIMA database (13) and NCI-60 database published data (14). ATCC lines and NCI-60 lines were authenticated at the source.
Expression and shRNA constructs have been described previously and are detailed in Supplementary Methods.
Cell growth and invasion assays have been described previously and are detailed in Supplementary Methods.
Western blot analysis
Protein was extracted from ovarian cancer cell lines using standard methods (8). Protein concentrations were estimated with BCA Protein Assay Kit (Thermo Scientific, Rockford, IL). SDS-Page and Western blot analysis were performed using the NuPage system (Invitrogen, Carlsbad, CA) and the Supersignal Chemiluminescent Substrate system (Thermo Scientific, Rockford, IL), respectively. Antibodies are listed in Supplementary Methods.
Mouse xenografts
Female Athymic Nu/Nu mice were obtained at 6–8 weeks of age from NCI-Frederick (Frederick, MD), and acclimated for 5–7 days. Care was provided in accordance to procedures in the Guide for the Care and Use of Laboratory Animals (NIH Publication No. 86-23, 1985). Experiments were conducted according to a protocol approved by the NCI Animal Care and Use Committee. Briefly, 1 million control shRNA or IKKε shRNA expressing ovarian cells were injected intraperitoneally into groups of 10 mice; whole body weights were obtained three times per week for 5 weeks post-injection. Ovcar5 and Ovcar8 cells were tested in two independent experiments. Caov3 (2 million cells) transduced with either empty vector, wild type IKKε or myristoylated IKKε, were injected intraperitoneally into groups of 6 mice or PBS into 4 mice; whole body weights were obtained three times per week for 4 weeks post-injection. Animals were sacrificed humanely and received complete necropsies, at which time wet organ weights, tissues and samples were obtained. Immunohistochemistry for GFP expression (Abcam, cat. No. ab290) and hematoxylin-eosin staining was performed on paraffin-embedded, formalin fixed sections, for evaluation of xenograft localization and morphology. Gross and microscopic pathology evaluation was performed by MA, and scored using the following definitions: Xenografts present in the peritoneal cavity from IP injected cell lines involving omental fat are coded as “primary”; xenografts in the same mouse in other abdominal organs (liver, diaphragm, pancreas, other sites) are coded as “secondary.” Xenografts were classified as “superficial” if they were implanted on the organ surface without disruption of the capsule, or “invasive” if they had disrupted the surface of the organ and were present within the organ parenchyma.
Gene expression analysis
Total RNA was isolated from 6 independent cultures of ovarian cells grown in 6-well plates using Trizol (Invitrogen, Carlsbad, CA) according to manufacturer protocol. Details of analysis are provided in Supplementary Methods. The following GEO datasests were analyzed: GSE9899, GSE18520 (15) the cancer genome atlas (TCGA) (16).
Quantitative PCR
Total RNA was isolated using RNeasy Mini kit (Qiagen). One microgram of total RNA was converted to cDNA using iScript cDNA Synthesis Kit (Bio-Rad). cDNA was diluted 1:5 in H2O for real-time PCR (QuantiTect SYBR Green PCR Kit, Qiagen), performed in triplicates, by 7900HT Fast Real-Time PCR System (Applied Biosystems). Each mRNA expression level was normalized by that of GAPDH. Primers are listed in Supplementary Methods.
RESULTS
IKKε is more highly expressed in metastatic ovarian cancers compared to primary tumors
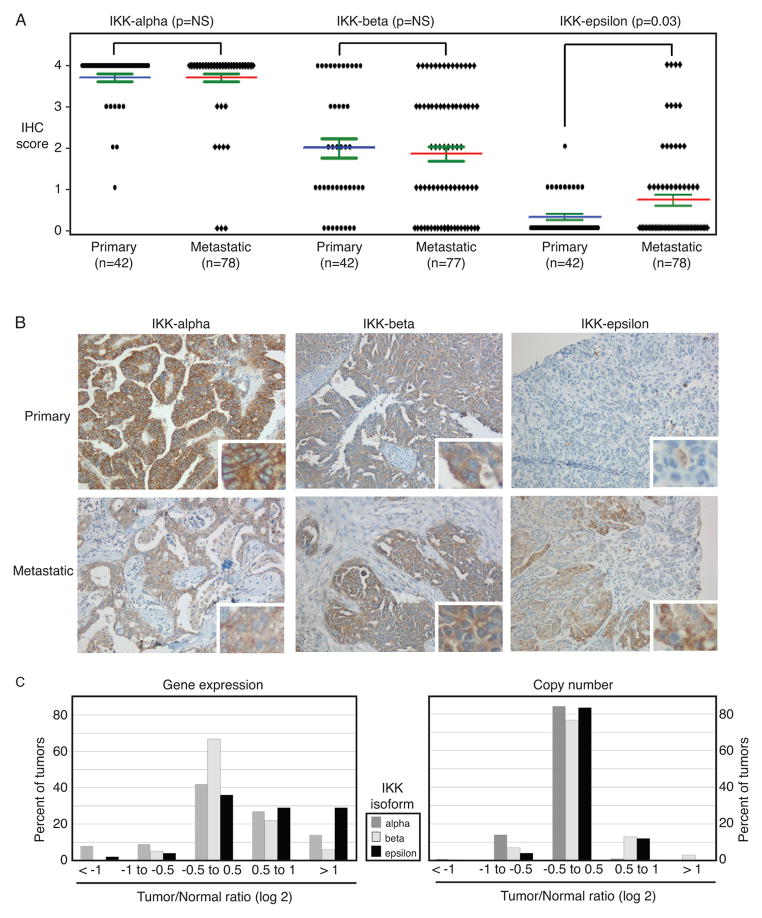
We surveyed 120 ovarian cancer specimens (42 primary carcinomas, 78 solid metastases) for expression of IKK proteins. Immunohistochemical analysis of three IKK proteins – IKKα, IKKβ, and IKKε – showed that IKKε was the only isoform that was differentially expressed between primary tumors and metastatic lesions. Importantly, IKKε was significantly (p=0.03) more frequently present in metastatic ovarian cancer cells relative to primary tumor sites (Figure 1A, 1B and Supplementary Table 2), whereas IKKα and IKKβ were evenly distributed between primary and metastatic tumors. TCGA measured gene expression and chromosomal amplifications or deletions in approximately 500 serous ovarian cancers that were stage IIIC at the time of initial diagnosis (5). These TCGA data show that at the time of initial diagnosis, IKKε is more frequently overexpressed than IKKα or IKKβ in advanced ovarian cancer (Figure 1C, left). Overexpression occurs at the transcriptional level, since there was no significant change in copy number in this dataset (Figure 1C, right). These findings prompted the hypothesis that IKKε promotes ovarian cancer metastasis.
Figure 1. IKKε is higher in metastatic ovarian cancers compared to primary tumors.
(A) Ovarian cancer tissue specimens were analyzed for relative expression of IKK isoforms – α, β, ε, - in primary and metastatic tumors. Expression (IHC score) is based on percent of cells showing expression of each protein (7). (B) Representative cases are shown at a 200 X magnification. (C) TCGA datasets were examined for expression of each IKK isoform (left) and copy number for the IKKα, IKKβ and IKKε loci (right) in ovarian cancer cases compared to non-cancer specimens. Relative expression is shown as log2 ratio of tumor to normal.
IKKε regulates expression of genes involved in cellular motility and inflammation
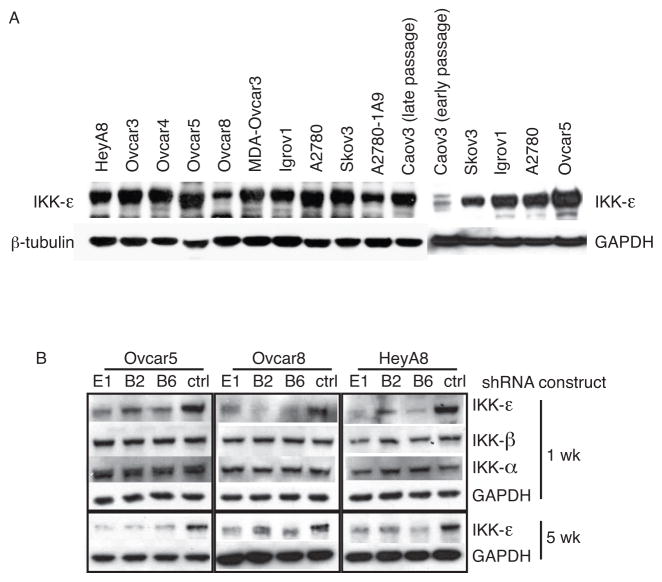
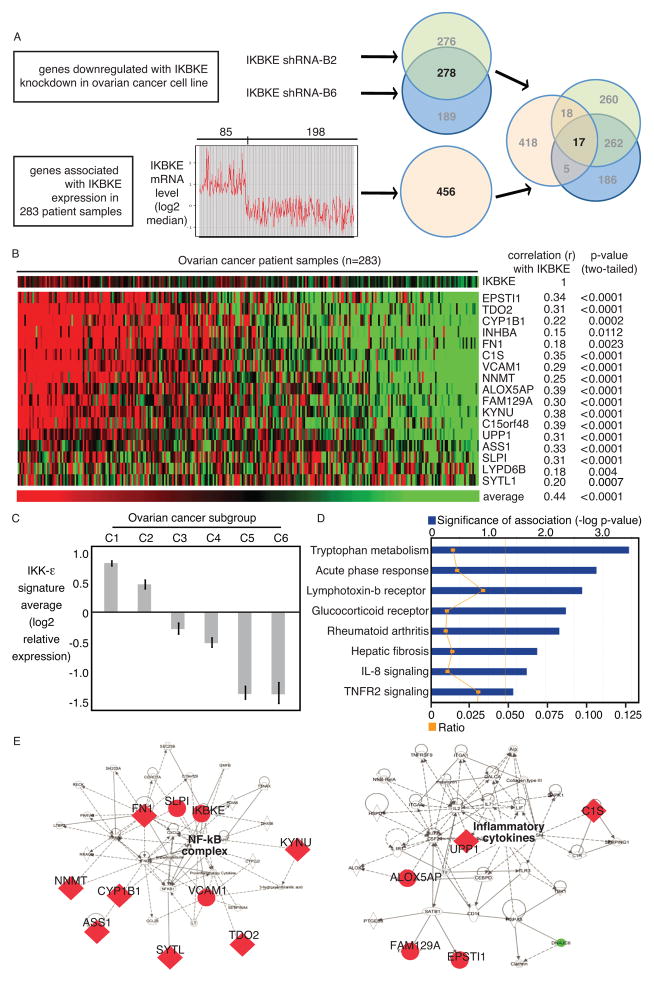
Ovarian cancer cell lines, all derived from advanced cancers, consistently expressed IKKε (Figure 2A). In order to dissect the functional role of IKKε in ovarian cancer, we screened 13 RNA interference constructs directed against IKKε, and selected the most effective shRNAs for further experiments (Supplementary Figure 1A, 1B). Three shRNA constructs effectively depleted IKKε, but not IKKβ or IKKα (Supplementary Figure 2A, 2B). Knockdown was confirmed in three ovarian cancer cell lines after initial selection, and the stability of knockdown was reconfirmed after the selected cells were maintained in the absence of selection agent (Figure 2B). Gene expression profiles of ovarian cancer cells depleted of IKKε identified molecules potentially causing the biological phenotype observed in human tumors. We identified 278 genes regulated by selective depletion of IKKε with two individual shRNAs in six biological replicates in Ovcar5 (Figure 3A). The signature was further refined by examining the expression of these genes in primary ovarian cancer specimens (GSE9899). Seventeen genes were both downregulated after IKKε depletion, and associated with elevated IKKε in primary ovarian cancers. These genes were associated with expression of IKKε itself, and were co-regulated across three independent datasets (Figure 3B, Supplementary figure 2C), suggesting a functional relationship in ovarian cancer. Strikingly, all 17 genes were overexpressed and/or amplified in the TCGA dataset (Supplementary figure 2D). Furthermore, the 17 IKKε signature genes were not equally represented among six previously defined molecular subgroups of ovarian cancer (17): the C1 and C2 subgroups had relatively high expression, while the C5 and C6 subgroups had low expression (Figure 3C). This previously published classification identified C1 tumors as having a “stromal” signature, and the C2 type expressed an “immune” signature. Consistent with our finding, the previous observation also showed an enrichment of metastatic tumors within the C1 subgroup, with high desmoplasia. Importantly, Ingenuity pathway analysis identified inflammatory signaling as the most significantly regulated pathway (Figure 3D). Accordingly, cellular movement and inflammation were the most significant networks affected by this set of genes (Figure 3E). Seven of the 17 genes are known to function in metastasis and invasion. We chose these seven genes to validate the microarray data. We verified their knockdown by qPCR in two ovarian cancer cell lines (Supplementary Figure 2E, 2F). Taken together, our analyses showed that IKKε modulates genes involved in cellular movement and inflammation, two functions critical for a metastatic phenotype.
Figure 2. IKKε shRNAs specifically knock down its expression in ovarian cancer cells.
(A) IKKε protein was highly expressed by Western blot in the majority of ovarian cancer cell lines. (B) Three shRNA constructs were tested for specificity towards IKKε, compared to IKKα or IKKβ, in cell lines Ovcar5, Ovcar8 and HeyA8. Knockdown of IKKε was measured after 1 week selection in puromycin, and 5 weeks after maintenance in medium without puromycin.
Figure 3. IKKε regulates expression of genes involved in cellular motility and inflammation.
(A) Schematic of workflow for determining the ovarian cancer-specific IKKε gene signature. First, 278 genes were experimentally determined by depleting IKKε with 2 different shRNA constructs in Ovcar5 cells. In parallel, 456 genes were found to be associated with IKKε over-expression in 283 publicly available ovarian cancer gene expression profiles (GSE9899). The intersection of these two sets identified 17 genes. (B) The relative expression of these 17 genes in 283 ovarian cancer patient specimens is ranked by average expression of the 17 genes. Correlation of each gene with IKKε expression is also noted. (C) The 17-gene expression average was calculated for each of the 6 previously published subgroups of ovarian cancer (12). Log2 average was median-centered for the entire dataset. (D, E) The 17 genes were analyzed by Ingenuity Pathway Analysis. The most significantly related pathway was a cell-motility network, centered on the NF-κB complex.
IKKε has a modest effect on ovarian cancer growth and invasion in vitro
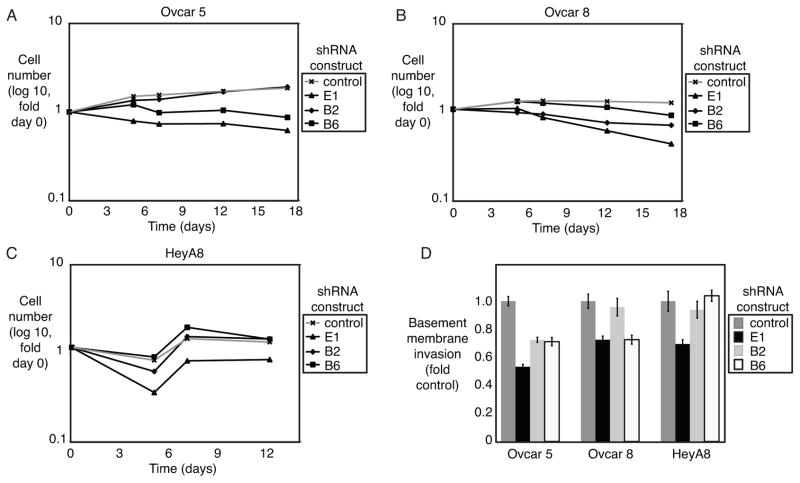
Depletion of IKKε by shRNA had a modest effect decreasing cell growth in vitro (Figure 4A–C, Supplementary Figure 3A). Interestingly, cell lines also showed a decrease in basement membrane invasion with IKKε depletion (Figure 4D, Supplementary Figure 3B). These results are consistent with the hypothesized role for IKKε-programmed events in regulating interactions of the tumor cell with its microenvironment. Therefore, we proceeded with in vivo xenograft experiments in order to assess the contribution of IKKε in a metastasis model.
Figure 4. IKKε modestly affects ovarian cancer growth and invasion in vitro.
(A–C) Ovarian cancer cell lines Ovcar 5, Ovcar8, and HeyA8 were quantified by flow cytometry for GFP expression (co-expressed with shRNA construct) for 12–17 days after selection for IKKε depletion. Measurements are normalized to day 0 for each cell line. (D) Invasion through basement membrane was measured after depletion of IKKε, and normalized to control shRNA for each cell line. Error bars represent S.E.
IKKε promotes tumor growth, invasion and metastasis in vivo
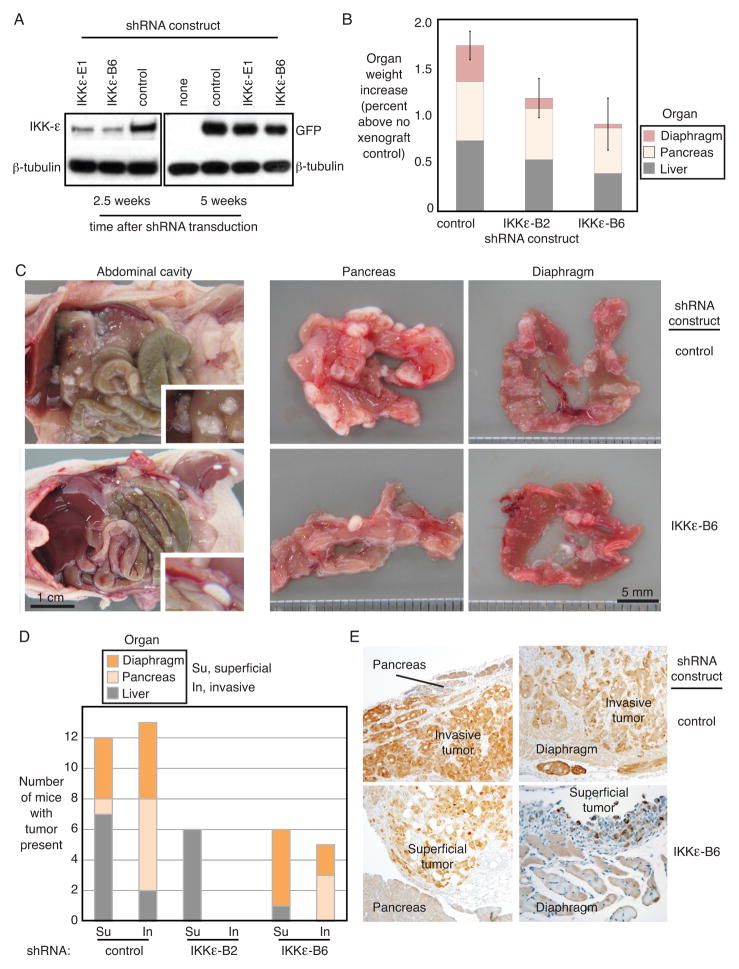
Ovarian cancer cell lines differed in their ability to form tumors in mice (Supplementary Figure 4A). HeyA8, Ovcar8 and Ovcar5, expressing high levels of IKKε, were among the most aggressive tumors, whereas low passage Caov3 that expressed low IKKε was less malignant. Ovarian cancer cell lines Ovcar8 and Ovcar5 were stably transduced with shRNA constructs targeting IKKε or control, and co-expressing GFP (Figure 5A). Persistent knockdown of IKKε was measured at the midpoint of the xenograft timecourse (2.5 weeks), and maintenance of the shRNA construct in the ovarian cancer cells persisted until the end of the experiment (5 weeks) as evidenced by the co-expressed GFP protein. Cohorts of Ovcar5 or Ovcar 8 cells were tested in two independent experiments. Animal weight gain tended to be lower in mice inoculated with ovarian cancer cells depleted of IKKε, compared to controls, without reaching statistical significance (Supplementary Figure 4B). Organ weights of liver, pancreas and diaphragm were measured at the time of necropsy. Organ weights of animals that had been injected with IKKε-depleted Ovcar5 xenografts were lower than control xenografts, indicating that IKKε-deficient tumor cells had undergone less metastasis and invasion (Figure 5B). IKKε-depleted xenografts appeared markedly different from control cells on gross anatomical examination (Figure 5C). Control cells were well-attached and adhered to abdominal organs, whereas IKKε-depleted Ovcar8 cells floated in ascites fluid and cascaded out of the abdomen when the cavity was opened. Ovcar5 cells did not float but adhered less well to the organs (data not shown). In addition, more tumor nodules were visible in the pancreas and diaphragm of the control xenograft animals compared to the IKKε-depleted xenografts of both cell lines. Quantification of metastatic xenograft invasion into solid organs showed decreased invasivness of Ovcar5 cells that had been depleted of IKKε by either of the shRNA constructs (Figure 5D). A similar pattern was observed in Ovcar8 cells (Supplementary Figure 4C). Consistently, microscopy of tissue sections supported this finding of decreased invasive capacity in the absence of IKKε expression (Figure 5E). Persistent genomic integration of the shRNA construct was confirmed by the presence of co-expressed GFP protein in the xenograft. Control xenografts with strong immunohistochemical staining of GFP protein showed severe invasion into the organ parenchyma, whereas the GFP positive IKKε-depleted xenografts remained in the omental fat layer, outside of solid organs. Of note, expression of the shRNA construct was maintained over the 5-week interval in vivo, since GFP protein was present in the tumor cells. Xenografts expressing control shRNA showed higher capacity to invade solid organs, as compared to the IKKε-deficient cells, which generally remained adjacent to the solid organ.
Figure 5. IKKε promotes tumor growth, invasion and metastasis in vivo.
(A) Persistent integration of the shRNA construct in Ovcar8 cells over the time course of mouse xenograft experiments was confirmed by Western blot of IKKε protein (2.5 weeks) and co-expressed GFP (5 weeks). β-tubulin was a loading control. (B) Organ weight was measured in mice after necropsy. Shown are the average organ weights for diaphragm, pancreas and liver (Ovcar5 xenografts, n=9 each). Organ weight is expressed as percent body weight, increased over organs from non-xenograft controls (PBS injected intraperitoneally). Error bars represent S.E. (C) Gross anatomical examination of the abdominal cavity showed decreased adhesion to external organ surfaces in IKKε-depleted Ovcar8 xenografts (left panels). Tumor adherence to pancreas and diaphragm also appeared decreased (right panels). (D) Quantification of xenograft invasion into secondary organs showed decreased invasiveness of Ovcar5 cells depleted of IKKε by either of the shRNA constructs. Shown is the number of animals with tumor present in each organ. (E) Immunohistochemical analysis of tissues exhibiting Ovcar5 xenografts, stained for GFP as a marker of shRNA construct integration. GFP protein is shown by the brown immunohistochemical stain, at 10X magnification.
Overexpression of IKKε promotes metastasis to secondary organ sites in vivo
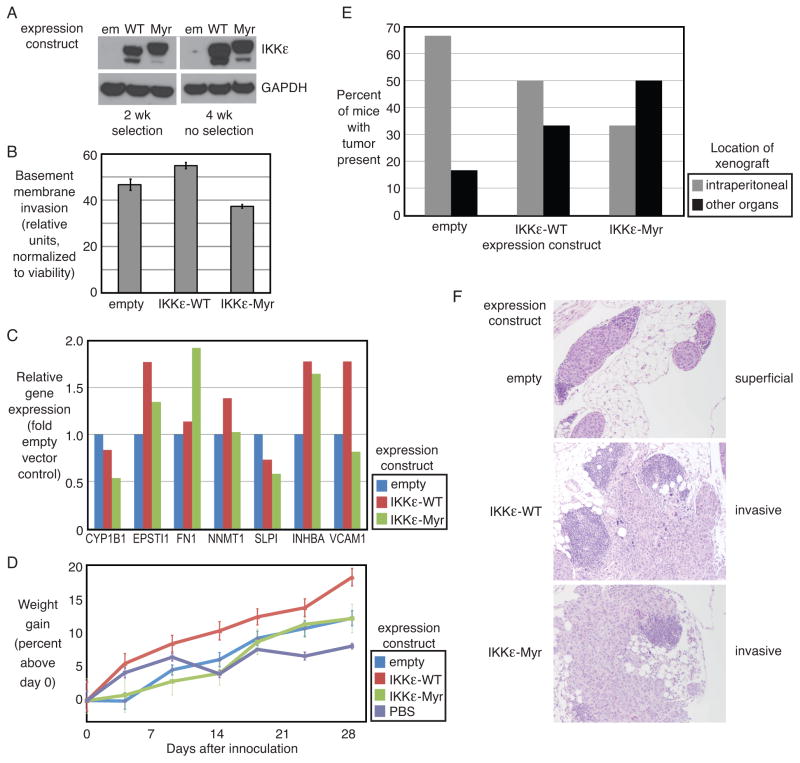
As a complement to the loss-of-function shRNA model, we developed a gain-of-function system by overexpressing IKKε in the less invasive ovarian cancer cell line Caov3, which expresses endogenously low IKKε. We over-expressed the wild type IKKε protein in order to mimic endogenous over-expression, and separately introduced the myristoylated version of IKKε, since this was previously determined to be oncogenic (9). Caov3 cells were stably transduced with either wild type or myristoylated IKKε (Figure 6A). Exogenous IKKε protein expression was confirmed after selection, and in cells cultured for 4 weeks without selection agent to verify maintenance of the transgene. Cells expressing wild type IKKε showed a slight increase in invasion through basement membrane in vitro (Figure 6B). Cells expressing myristoylated IKKε proliferated more; however, fewer of these cells invaded, when compared to cells transfected with empty vector or wild-type IKKε, even without normalization to viability (Supplementary Figure 4D). We queried by qPCR whether the seven IKKε-related metastasis genes were up-regulated upon introduction of IKKε (Figure 6C). Five of the 7 genes were upregulated, supporting their functional relationship downstream of IKKε. IKKε-overexpressing Caov3 cells were injected into mice to determine their behavior in vivo. Mice inoculated with the Caov3 cells overexpressing wild type IKKε gained weight at a significantly faster rate than the other groups (Figure 6D). Xenograft present in the peritoneal cavity and omental fat from IP injected cell lines was coded as primary. Xenografts in the same mouse in other abdominal organs and outside of the peritoneal cavity (liver, diaphragm, pancreas, other sites) were coded as secondary. Both wild type and myristoylated IKKε resulted in more xenografts in secondary organ locations as compared to empty vector control (Figure 6E). The IKKε-Myr acted differently in vivo, compared to in vitro invasion assays, suggesting that these assays are measuring different biologic processes and underscoring the importance of including both experiments. Tissue microscopy confirmed the increased ability of Caov3 cells containing the IKKε constructs to invade tissues (Figure 6F).
Figure 6. IKKε overexpression promotes ovarian cancer metastasis.
(A) Integration of the IKKε expression constructs was confirmed in Caov3 cells after 2 weeks in the presence of selection agent (G418) and after maintaining the culture without selection for 4 weeks (timecourse of xenograft experiments) as confirmed by Western blot of IKKε protein. GAPDH was a loading control. (B) Invasion through basement membrane was measured with overexpression of two IKKε constructs, and normalized to XTT viability for each cell line. (C) Shown is the relative expression of 7 IKKε-related metastasis-related genes. Expression was normalized to GAPDH and shown as fold change from cells transduced with empty vector. (D) Mice (n=6 each) were injected with Caov3 cell line engineered to express one of two different IKKε expression constructs or control. Weights were measured weekly. (E) Xenograft present in the peritoneal cavity or omental fat is denoted as “primary;” xenografts in other abdominal organs (liver, diaphragm, pancreas, other sites) are noted as “secondary.” Shown is the percent of mice with tumor in each of these locations. (F) Photomicrographs of representative tissue sections demonstrating superficial implantation of xenograft (top) or invasion into organ parenchyma (middle and bottom).
DISCUSSION
IKKε is an oncogene in breast cancer, and promotes chemoresistance in ovarian cancer (9, 11). Previous studies linked IKKε to poor prognosis in ovarian cancer patients (11). Its over-expression in ovarian cancer cell lines caused resistance to platinum chemotherapy, standard of care for initial treatment of advanced ovarian cancer. Our work presented herein suggests another mechanism for the poor prognosis conferred by high IKKε expression, namely an increased ability to disseminate. Our initial finding of IKKε overexpression in metastatic tumors compared to primary tumors was unexpected. Previous work in prostate cancer showed that IKKα was the differentially expressed isoform promoting metastasis (6). Therefore, we expected a similar mechanism of IKK-related metastasis in ovarian cancer. Instead, IKKα was equally expressed in most ovarian cancers examined. In contrast, IKKε was more highly expressed in metastatic ovarian cancers, and showed uniformly low expression in primary sites of ovarian cancer. Most metastatic ovarian cancers, however, also showed low expression of IKKε, suggesting that this is not the sole mechanism promoting ovarian cancer metastasis. This is consistent with our previous findings of NF-κB activity in ovarian cancer, where IKKβ was active in a subset of tumors, but was not a uniform target for all ovarian cancers (7, 8). Interestingly, our results suggest that IKKε differentially influences functions of proliferation versus invasion in individual cell lines. We found a similar phenomenon with IKKβ in ovarian cancer (8) where the pattern of functions mediated by IKKβ was not uniform across the cell lines tested, indicating a differential predominance of each effect among distinct ovarian cancers. These findings underscore the heterogeneous nature of ovarian cancer, supporting the hypothesis that there are multiple molecularly defined subtypes of this disease. Future clinical studies with NF-κB targeted agents should seek to identify biomarkers specific to IKK activity in order to appropriately allocate such therapies to patients whose tumors are more likely to respond.
We now show that IKKε coordinates genes promoting ovarian cancer metastasis. IKKε controlled genes involved in inflammation, cellular movement, and interaction with extracellular matrix, all functions linked to metastatic potential. IKKε had a moderate effect in maintaining cellular proliferation, but more prominently promoted invasive capacity of a subset of ovarian cancer cell lines.
The primary method of dissemination of epithelial ovarian cancer is through shedding from the primary tumor. Omental metastases occur in 70–80% of advanced ovarian cancers. In our mouse model, tumor was injected directly into the peritoneal cavity. Thus the model mimics ovarian cancer that has intitally shed. The omentum may play a role in trapping and destroying cancer cells (18). In a rat model, primary omentectomy followed by injection of tumor resulted in worse survival and more disseminated disease than in rats with an intact omentum (19). Since our xenografts are initially introduced into the peritoneal cavity of the mice, we coded our omental tumors the “primary” site and the extra-omental tumor deposits the “secondary.” Tumor that succeessfully moves past the omentum reflects its more aggressive biology. In mice, IKKε-deficient xenografts showed decreased tumor burden and invasion into secondary organs, supporting our hypothesis that IKKε promotes a program of metastasis in ovarian cancer. Conversely, exogenous over-expression of IKKε into a non-metastatic cell line increased the cells’ ability to invade tissues and disseminate to secondary abdominal organs.
Several genes comprising the 17-gene ovarian cancer-specific IKKε signature are known to be involved in cellular invasion and metastasis. For example, overexpression of SLPI increased tumor formation and dissemination in orthotopic mouse model of ovarian cancer, independent of its protease inhibition activity (20). NNMT induced cellular migration and invasion in renal carcinoma cells and bladder cancer (21, 22). FN1 significantly increased migration of melanoma cells combined with POSTN (23). INHBA was a top-ranked gene in a metastasis-associated expression signature from multi-cancer computational analysis (24). Consistently, a previous analysis of ovarian carcinoma also identified NNMT, FN1, and INHBA as metastasis-associated genes (25). CYP1B1 was overexpressed in both primary and metastatic ovarian cancers (26). In addition, EPSTI1 was highly up-regulated in invasive breast carcinoma compared with normal breast, suggesting its expression may be a crucial event in invasion and metastasis of cancer (27). The decreased metastatic potential in our IKKε-depleted xenografts are well explained by these genes’ function. The critical finding from our current analysis is that IKKε regulates these genes, as a metastasis-promoting cassette, in ovarian cancer. It is unlikely that any individual gene in the signature is responsible for the metastatic program induced by IKKε, but that coordinated expression of this gene cassette is required. However, the mechanism by which IKKε is activated in ovarian cancer is unknown, and is the subject of ongoing research.
Gene expression profiles previously defined six distinct subtypes of ovarian cancers (17). The ovarian cancer IKKε signature defined here was over-represented in the C1 and C2 subtypes, suggesting that IKKε could drive the phenotype of these cancers. The C1 subtype included cancers with high stromal response and desmoplasia. Notably, the patients with the C1-type tumors had the shortest progression-free and overall survival. It is possible that IKKε contributes to the poor-prognostic phenotype of the C1 subgroup by increasing the cancers’ metastatic potential, given our current findings in vitro and in vivo.
Conversely, 13 of the IKKε signature genes are under-expressed in the C5 subtype of ovarian cancers, which included mesenchymal-like tumors with a low immune signature. This inverse relationship with the C5 subgroup suggests that these genes may be required for an immune signature in ovarian cancer. Pathway analysis and networks corroborated this idea, since the genes are known to be instrumental in acute phase response, lymphotoxin beta receptor, IL-8 and TNFR2 signaling pathways, all of which contribute to an inflammatory response network.
IKKε may therefore provide a therapeutic target for C1-type ovarian cancers. IKKε-specific inhibitors are not currently under clinical development, but it may be possible to therapeutically interfere with IKKε signaling upstream or downstream of IKKε target pathways. For example, TNFR can stimulate the growth and survival of some ovarian cancers, likely through activation of canonical NF-κB signaling. The same cytokine promotes apoptosis in other contexts. cIAP1 acts as a critical switch to promote the pro-survival NF-κB pathway and prevent caspase activation (28). An early event in apoptosis is the release of second mitochondrial-derived activator of caspases (SMAC). In normal cells SMAC promotes IAP degradation, thereby activating caspases and tipping the balance from survival to apoptosis. In tumor cells, however, apoptosis is dysregulated due to insufficient amounts of SMAC (29). Thus, SMAC mimetics under clinical development could have selectively toxic activity against those molecularly defined IKKε-driven and/or C1-type ovarian cancers.
Understanding IKKε-regulated signaling in ovarian cancer will identify novel pathway interactions for context-specific therapeutics in the poor prognostic group of women whose ovarian cancers over-express IKKε. Further clarification of the ovarian cancer-specific IKKε pathway is likely to provide several potential therapeutic avenues for individualized treatment of women with this molecularly defined variant of ovarian cancer.
Supplementary Material
Acknowledgments
This work was supported by the Intramural Research Program, CCR, NCI (C.M.A.), NIH Grant K99CA151746 (C.M.A.), and the Norwegian Cancer Society and Health Region of South-Eastern Norway (B.D.), and with Federal funds from the NCI, NIH, under contract number HSN261200800001E.
Footnotes
There are no conflicts to disclose
References
- 1.Pignata S, Cannella L, Leopardo D, Pisano C, Bruni GS, Facchini G. Chemotherapy in epithelial ovarian cancer. Cancer Lett. 2011;303:73–83. doi: 10.1016/j.canlet.2011.01.026. [DOI] [PubMed] [Google Scholar]
- 2.Annunziata CM, Azad N, Dhamoon AS, Whiteley G, Kohn EC. Ovarian cancer in the proteomics era. Int J Gynecol Cancer. 2008;18 (Suppl 1):1–6. doi: 10.1111/j.1525-1438.2007.01096.x. [DOI] [PubMed] [Google Scholar]
- 3.Martin LP, Schilder RJ. Management of recurrent ovarian carcinoma: current status and future directions. Semin Oncol. 2009;36:112–125. doi: 10.1053/j.seminoncol.2008.12.003. [DOI] [PubMed] [Google Scholar]
- 4.Bonome T, Levine DA, Shih J, Randonovich M, Pise-Masison CA, Bogomolniy F, et al. A gene signature predicting for survival in suboptimally debulked patients with ovarian cancer. Cancer Res. 2008;68:5478–86. doi: 10.1158/0008-5472.CAN-07-6595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.TCGA. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–15. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Luo JL, Tan W, Ricono JM, Korchynskyi O, Zhang M, Gonias SL, et al. Nuclear cytokine-activated IKKalpha controls prostate cancer metastasis by repressing Maspin. Nature. 2007;446:690–4. doi: 10.1038/nature05656. [DOI] [PubMed] [Google Scholar]
- 7.Annunziata CM, Stavnes HT, Kleinberg L, Berner A, Hernandez LF, Birrer MJ, et al. Nuclear factor kappaB transcription factors are coexpressed and convey a poor outcome in ovarian cancer. Cancer. 2010;116:3276–84. doi: 10.1002/cncr.25190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hernandez L, Hsu SC, Davidson B, Birrer MJ, Kohn EC, Annunziata CM. Activation of NF-kappaB signaling by inhibitor of NF-kappaB kinase beta increases aggressiveness of ovarian cancer. Cancer Res. 2010;70:4005–14. doi: 10.1158/0008-5472.CAN-09-3912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boehm JS, Zhao JJ, Yao J, Kim SY, Firestein R, Dunn IF, et al. Integrative genomic approaches identify IKBKE as a breast cancer oncogene. Cell. 2007;129:1065–79. doi: 10.1016/j.cell.2007.03.052. [DOI] [PubMed] [Google Scholar]
- 10.Hutti JE, Shen RR, Abbott DW, Zhou AY, Sprott KM, Asara JM, et al. Phosphorylation of the tumor suppressor CYLD by the breast cancer oncogene IKKepsilon promotes cell transformation. Mol Cell. 2009;34:461–72. doi: 10.1016/j.molcel.2009.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guo JP, Shu SK, He L, Lee YC, Kruk PA, Grenman S, et al. Deregulation of IKBKE is associated with tumor progression, poor prognosis, and cisplatin resistance in ovarian cancer. Am J Pathol. 2009;175:324–33. doi: 10.2353/ajpath.2009.080767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.http://www.atcc.org/CulturesandProducts/CellBiology/STRProfileDatabase/tabid/174/Default.aspx
- 13.http://bioinformatics.istge.it/clima/
- 14.Lorenzi PL, Reinhold WC, Varma S, et al. DNA fingerprinting of theNCI-60 cell line panel. Mol Cancer Ther. 2009;8:713–24. doi: 10.1158/1535-7163.MCT-08-0921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.http://www.ncbi.nlm.nih.gov/gds
- 16.http://cancergenome.nih.gov/
- 17.Tothill RW, Tinker AV, George J, Brown R, Fox SB, Lade S, et al. Novel molecular subtypes of serous and endometrioid ovarian cancer linked to clinical outcome. Clin Cancer Res. 2008;14:5198–208. doi: 10.1158/1078-0432.CCR-08-0196. [DOI] [PubMed] [Google Scholar]
- 18.Koenen HJ, Smit MJ, et al. Effect of intraperitoneal administration of granulocyte/macrophage-colony-stimulating factor in rats on omental milky-spot composition and tumoricidal activity in vivo and in vitro. Cancer Immunol Immunother. 1996;42:310–316. doi: 10.1007/s002620050288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yokoyama Y, Hirakawa H, Wang H, Mizunuma H. Is omentectomy mandatory in the operation for ovarian cancer? Preliminary results in a rat study. Eur J Obstet Gynecol Reprod Biol. 2012 doi: 10.1016/j.ejogrb.2012.05.020. in press. [DOI] [PubMed] [Google Scholar]
- 20.Devoogdt N, Rasool N, Hoskins E, Simpkins F, Tchabo N, Kohn EC. Overexpression of protease inhibitor-dead secretory leukocyte protease inhibitor causes more aggressive ovarian cancer in vitro and in vivo. Cancer Sci. 2009;100:434–40. doi: 10.1111/j.1349-7006.2009.01076.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tang SW, Yang TC, Lin WC, Chang WH, Wang CC, Lai MK, Lin JY. Nicotinamide N-methyltransferase induces cellular invasion through activating matrix metalloproteinase-2 expression in clear cell renal cell carcinoma cells. Carcinogenesis. 2011;32:138–45. doi: 10.1093/carcin/bgq225. [DOI] [PubMed] [Google Scholar]
- 22.Wu Y, Siadaty MS, Berens ME, Hampton GM, Theodorescu D. Overlapping gene expression profiles of cell migration and tumor invasion in human bladder cancer identify metallothionein 1E and nicotinamide N-methyltransferase as novel regulators of cell migration. Oncogene. 2008;27:6679–89. doi: 10.1038/onc.2008.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Soikkeli J, Podlasz P, Yin M, Nummela P, Jahkola T, Virolainen S, et al. Metastatic outgrowth encompasses COL-I, FN1, and POSTN up-regulation and assembly to fibrillar networks regulating cell adhesion, migration, and growth. Am J Pathol. 2010;177:387–403. doi: 10.2353/ajpath.2010.090748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim H, Watkinson J, Varadan V, Anastassiou D. Multi-cancer computational analysis reveals invasion-associated variant of desmoplastic reaction involving INHBA, THBS2 and COL11A1. BMC Med Genomics. 2010;3:51. doi: 10.1186/1755-8794-3-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bignotti E, Tassi RA, Calza S, Ravaggi A, Bandiera E, Rossi E, et al. Gene expression profile of ovarian serous papillary carcinomas: identification of metastasis-associated genes. Am J Obstet Gynecol. 2007;196:245 e241–211. doi: 10.1016/j.ajog.2006.10.874. [DOI] [PubMed] [Google Scholar]
- 26.McFadyen MC, Cruickshank ME, Miller ID, McLeod HL, Melvin WT, Haites NE, et al. Cytochrome P450 CYP1B1 over-expression in primary and metastatic ovarian cancer. Br J Cancer. 2001;85:242–6. doi: 10.1054/bjoc.2001.1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nielsen HL, Ronnov-Jessen L, Villadsen R, Petersen OW. Identification of EPSTI1, a novel gene induced by epithelial-stromal interaction in human breast cancer. Genomics. 2002;79:703–10. doi: 10.1006/geno.2002.6755. [DOI] [PubMed] [Google Scholar]
- 28.Gyrd-Hansen M, Meier P. IAPs: from caspase inhibitors to modulators of NF-kappaB, inflammation and cancer. Nat Rev Cancer. 2010;10:561–574. doi: 10.1038/nrc2889. [DOI] [PubMed] [Google Scholar]
- 29.Vince JE, Wong WW, Khan N, Feltham R, Chau D, Ahmed AU, et al. IAP antagonists target cIAP1 to induce TNFalpha-dependent apoptosis. Cell. 2007;131:682–93. doi: 10.1016/j.cell.2007.10.037. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.