Abstract
We performed a detailed genomic investigation of the chimpanzee locus syntenic to human chromosome 4q35.2, associated to the facioscapulohumeral dystrophy. Two contigs of approximately 150 kb and 200 kb were derived from PTR chromosomes 4q35 and 3p12, respectively: both regions showed a very similar sequence organization, including D4Z4 and Beta satellite linked clusters. Starting from these findings, we derived a hypothetical evolutionary history of human 4q35, 10q26 and 3p12 chromosome regions focusing on the D4Z4–Beta satellite linked organization. The D4Z4 unit showed an open reading frame (DUX4) at both PTR 4q35 and 3p12 regions; furthermore some subregions of the Beta satellite unit showed a high degree of conservation between chimpanzee and humans. In conclusion, this paper provides evidence that at the 4q subtelomere the linkage between D4Z4 and Beta satellite arrays is a feature that appeared late during evolution and is conserved between chimpanzee and humans.
Keywords: Primate evolution, Chimpanzee, Beta satellite, D4Z4, 4q35, FSHD
Highlights
► A detailed genomic analysis of the PTR locus syntenic to human chromosome 4q35.2. ► PTR 4q35 and 3p12 regions carried a very similar D4Z4 and Beta satellite linked clusters. ► We derived a presumable evolutionary history of human 4q35, 10q26 and 3p12 regions. ► PTR and HSA subregions of the Beta satellite showed a high degree of conservation. ► 4q D4Z4–Beta satellite linked arrays appeared very late during evolution.
1. Introduction
The human genome contains a large amount (> 50%) of repetitive sequences, which were, for a long time, regarded as parasitic or “junk” DNA with no phenotypic impact [1,2]. However, recent data suggest that retroelements as well as tandem repeats (i.e. satellite DNA) are involved in mammalian gene regulation [3,4]. In fact, transcripts derived from satellite DNAs seem to participate in the epigenetic process(es) of chromatin remodeling and heterochromatin formation, which are crucial for genome stability [5,6]. Furthermore, some satellite DNAs show the conservation of particular structural motifs; these selective constraints are probably related to their interaction with proteins involved in heterochromatin formation [7]. An example of the importance of clustered repeats for human cell physiology can be derived from the contraction of the 3.3 kb (D4Z4) megasatellite located at chromosome 4q35.2 which is associated to FSHD manifestation [8–10]. FSHD is an autosomal dominant disease showing at the subtelomere of chromosome 4q 1–10 copies of D4Z4, whereas in the normal population the number of repeats is highly polymorphic ranging from 11 to 150. The 4q subtelomere comes in two genomic variants [11], alleles 4qA and 4qB, which are equally common in the general population and show an equal variation in size and somatic instability [12]. Nonetheless, FSHD alleles are always of the 4qA type [13], which differs from 4qB due to the presence, downstream to the D4Z4 array, of the pLAM sequence (260 bp) followed by the Beta satellite cluster of 6.2 kb in length. In normal individuals the FSHD region is methylated and displays the typical feature of unexpressed euchromatin [14,15], whereas in FSHD patients hypomethylation of the contracted D4Z4 alleles has been reported [16–18]. Furthermore, DNA hypermethylation is associated with the recruitment of various methylated DNA binding proteins as well as histone-modifying deacetylases and methyltransferases, all involved in chromatin condensation [19]. Thus, in FSHD hypomethylation of the contracted 4q D4Z4 alleles supports the relevance of these epigenetic changes able to induce a more euchromatic-like D4Z4 array, a fact that seems directly associated with FSHD expression [20]. Both histone modifications and DNA hypomethylation of D4Z4 are likely to be disease-related, affecting in some way the functionality of the 4qter region, most probably through the up-regulation of D4Z4-associated genes. Accordingly, it has been observed that the number of residual D4Z4 copies is inversely correlated to the age of onset and severity of the disease [21]. Nevertheless, patients showing the same extent of the deletion may differ in disease progression [22], and monozygotic twins show discordant or extremely variable phenotypes. Furthermore, a small percentage of FSHD patients (phenotypic FSHD or FSHD-2) do not carry the D4Z4 contraction at 4q35.2, whereas some individuals are asymptomatic carriers of the D4Z4 contraction [23,24]. These observations strongly suggest that FSHD manifestation might rely on epigenetic modifications other than those attributable to D4Z4.
To address this question, after having defined the molecular features of the 4q region in the gorilla [25], we decided to investigate the sequence composition of the orthologous region in chimpanzee (PTR). Since most biological phenomena can be better understood if studied in an evolutionary frame, the identification of conserved and/or lost structural differences between human and primate 4q subtelomeres might be relevant to understand the etiology of the disease. This paper reports that at the 4q subtelomere the linkage between D4Z4 and Beta satellite arrays is a feature that seems to be appeared late during evolution and is conserved between chimpanzee and humans, and that Beta satellite sequences may play a role in the epigenetic regulation of the 4q35.2 region.
2. Materials and methods
2.1. Genomic library screening
The genomic library was obtained from BAC/PAC Resources, Children's Hospital, Oakland Research Institute (CHORI-251 segments 3 and 4: Chimpanzee BAC Library). High-density arrayed BAC filters were hybridized according to the instructions provided by BAC/PAC Resources [BACPAC Resources Center [http://bacpac.chori.org/] with a 32P-labeled D4Z4 probe (77 M12-KpnI D4Z4 probe) obtained by subcloning a KpnI fragment of 3.3 kb from a previously isolated human genomic BAC (77 M12) from the human shared BAC library CHORI-16 (BACPAC Resources Center).
2.2. DNA isolation and Southern blot hybridization
BAC clones were grown in LB medium supplemented with chloramphenicol. BAC DNA was purified using a Sigma PhasePrep BAC kit (Sigma Aldrich, USA), digested with restriction enzymes (Biolabs, USA), fractionated by means of agarose gel electrophoresis, blotted onto nylon filters (Hybond N+, Amersham, UK), and hybridized with a 32P-labeled probe representative of D4Z4 (77 M12-KpnI D4Z4) and Beta satellite sequences (A17a plasmid) [26].
Molecular hybridization was performed in 2 × SSC at 60 °C overnight, and the filters were washed at 60 °C in 1 × SSC twice for 20 min. The hybridization signals were quantified by means of phosphoimaging (Typhoon 9200, Amersham).
2.3. DNA sequencing and in silico analysis
T7/Sp6 BAC ends were sequenced using the Big Dye terminator 3.1 system as described by BAC/PAC Resources [BACPAC Resources Center [http://bacpac.chori.org/]. All fluorescent traces were analyzed using the Applied Biosystem Model 3100 DNA Sequencing System (Applied Biosystem). The newly derived nucleotide sequences are available at NCBI [http://www.ncbi.nlm.nih.gov/] with accession numbers: JM170483 (584M21-Sp6), JM170484 (584M21-T7), JM170481 (578I19-Sp6), JM170482 (578I19-T7), JM170493 (709I17-Sp6), JM170494 (709I17-T7), JM170491 (705I9-Sp6), JM170492 (705I9-T7), JM170497 (784B11-Sp6), JM170498 (784B11-T7), JM170501 (802O1-Sp6), JM170502 (802O1-T7), JM170499 (790M3-Sp6), JM170500 (790M3-T7), JM170495 (773B17-Sp6), JM170496 (773B17-T7), JM170485 (590D11-Sp6), JM170486 (590D11-T7), JM170487 (618G2-Sp6), JM170488 (618G2-T7), JS886734 (678H18-Sp6), JS886735 (678H18-T7), JM170489 (700D23-Sp6), JM170490 (700D23-T7), JM170503 (809J1-Sp6), and JM170504 (809J1-T7). Internal BAC sequencing was carried out at BMR Genomics (Padua, Italy) by the ROCHE-GS-FLX 454 platform. A total of four BACs were sequenced and the newly derived nucleotide sequences are available at NCBI with accession numbers: JF900598 (BAC 584M21), JF900596 (BAC 709I17), JF900597 (BAC 705I9), and JF900595 (BAC 809J1). DNA sequence analysis was performed using DNASTAR software and NCBI and UCSC Genome Bioinformatics Sites facilities [http://www.ncbi.nlm.nih.gov/; http://genome.cse.ucsc.edu/].
2.4. Fluorescent in situ hybridization (FISH) on metaphase chromosome spreads
The metaphase chromosome spreads were obtained by standard methods from Pan troglodytes (PTR) lymphoblastoid or fibroblast cell lines. The primate cell lines were provided by M. Rocchi [27]. Probes were labeled with digoxigenin-dUTP or biotin-dUTP (Roche Diagnostic) using a nick translation kit (Roche Diagnostic), and detected either using fluorescein-streptavidin (SIGMA) for the biotin-labeled probes, or CY3-anti-digoxigenin conjugated antibodies (Jackson ImmunoResearch Laboratories) for the digoxigenin-labeled probes. The chromosomes were counterstained with propidium iodide and DAPI in antifade (Vectashild), and then visualized using a Leitz DM-RB microscope equipped with DAPI and FITC/TRITC epifluorescence optics. Hybridizations were performed in 50% formamide (v/v), 10% dextran sulfate, 2 × SSC at 37 °C, in the presence of human Cot1 DNA (Gibco-BRL). Post-hybridization washing included 50% formamide, 2 × SSC at 42 °C, followed by three washes in 1 × SSC at 60 °C (HSA), or 50% formamide, 1 × SSC at 37 °C, followed by three washes in 1 × SSC at 42 °C (GGO and PTR). The chromosomes were counterstained with DAPI (4′,6-diamidino-2-phenylindole), and digital images were captured using a Leica DMRXA epifluorescence microscope equipped with a cooled CCD camera (Princeton Instruments). The fluorescence signals were recorded separately as gray-scale images. The images were pseudocolored and merged using Adobe Photoshop software.
In all the paper great apes' chromosomes are identified as the homologous human chromosomes.
3. Results
3.1. Isolation and characterization by Southern blot hybridization of D4Z4-positive PTR genomic clones
The PTR genomic library CHORI-251 (four genome equivalents) was screened with a human D4Z4 probe allowing the isolation of a total of thirteen BACs. The PTR clones were analyzed for the presence of D4Z4 and Beta satellite clusters by digestion with different restriction enzymes and hybridization with the corresponding human probes (Fig. S1). Four clones (584M21, 578I19, 709I17 and 705I9) were D4Z4 +/Beta satellite + (Fig. S1), and showed D4Z4 and Beta satellite restriction enzyme variants (Fig. S1 and Table S1). In particular, the clones 584M21/578I19 and 709I17/705I9 contained Kpn +/BlnI − and Kpn −/BlnI + D4Z4 units, respectively. Furthermore, the clones 584M21/578I19 and 709I17/705I9 carried a Beta satellite cluster originating a comparable ladder of 68 bp mers when digested by Sau3A, but a distinct PstI restriction pattern (Fig. S1 and Supplementary Table 1). These results indicate that the two pairs of BACs 584M21/578I19 and 709I17/705I9 carry distinct D4Z4 and Beta satellite clusters, suggesting their derivation from different chimpanzee chromosome regions. Among the remaining nine clones, six (773B17, 790M3, 802O1, 809J1, 590D11, and 618G2) showed Kpn +/BlnI − D4Z4 units and three (784B11, 700D3 and 678H18) Kpn −/BlnI + D4Z4 units (Fig. S1, Table S1 and not shown).
3.2. Terminal sequencing and databank analysis of D4Z4-positive PTR genomic clones
The isolated PTR clones were then characterized by end sequencing, and the derived sequences compared with the human genome databank (hg19). Considering D4Z4 +/Beta satellite + positive BACs, 584M21-Sp6 and 578I19-T7 showed a high percentage of identity with 4q35.2 (98.6% and 96.8%, respectively), whereas 584M21-T7 and 578I19-Sp6 with 3q29 (97.2% and 98.6%, respectively) (Table S2 and Fig. S2). Human genome databank analysis also evidenced that chromosome 3q29 contains duplicated sequences present in several subtelomeres, such as 1p36.33, 5q35.3 and 19q13.3 (not shown). Conversely, 709I17-T7 and 705I9-T7 showed identity with the 3p12.3 chromosome region, whereas the opposite ends aligned with 4q35.2 (709I17-Sp6) and with D4Z4 (705I9-Sp6) (Table S2 and Fig. S2). From these data it can be derived that BAC 578I19 is comprised within BAC 584 M21 (single asterisk in Fig. S2), and that 709I17 spans from the FRG2C gene on 3p12.3 to subtelomeric sequences located downstream to D4Z4/Beta satellite clusters on both 4q35 and 10q26 chromosomes (double asterisk in Fig. S2 and Table S2). Furthermore BAC 705I9 is delimited by D4Z4 and 3p12.3 sequences (triple asterisk in Fig. S2 and not shown). Since analyses by Southern blot hybridization showed the occurrence within these BACs of both D4Z4 and Beta satellite clusters, the results strongly indicate that in chimpanzee a very similar repetitive sequence block is located on both 4q35 and 3p12. Of the remaining nine clones, five showed at both ends D4Z4 sequences (809J1, 590D11, 618G2, 700D3 and 678H18), three D4Z4 and 4q35 sequences (773B17, 790M3 and 802O1), and one D4Z4 and 3p12 (784B11) sequences. Chromosome coordinates and identity, and GenBank accession numbers of the BAC ends are reported in Table S2 and in Fig. S2.
3.3. Chromosome location by FISH of D4Z4/Beta satellite positive PTR genomic clones
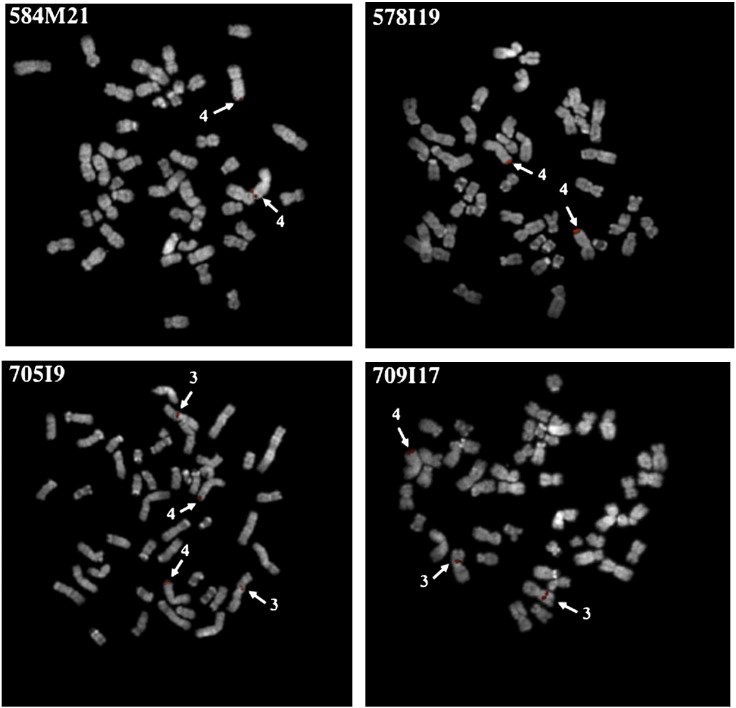
Total DNA or inter-Alu sequences from BACs 584M21, 578I19, 709I17 and 705I9 were used as probes on PTR metaphase chromosome spreads. As reported in Table S3 and in Fig. 1, the BACs can be subdivided into two classes, those essentially mapping to 4q35 (584M21 and 578I19) and those also highlighting the 3p12 chromosome region (709I17 and 705I9). These results provide additional evidence that, in chimpanzee, the 3p12 and 4q35 chromosome regions share a large duplication including sequences other than D4Z4 and Beta satellite.
Fig. 1.
Chromosomal location by FISH on chimpanzee metaphase chromosome spreads of unique and low repetitive sequences contained in PTR BACs 584M21, 578I19, 705I9 and 709I17. In all FISH experiments inter-Alu from the corresponding BAC was used as a probe. The roman numbers refer to the homologous human chromosome.
3.4. Sequencing of D4Z4/Beta satellite positive PTR genomic clones
To define the sequence content and organization of 4q35 and 3p12 chromosomes in chimpanzee, BACs 584M21, 709I17 and 705I9 were sequenced. The sequencing project also included BAC 809J1 as representative of DNA inserts flanked at both ends by D4Z4 repeats. BAC inserts were not completely sequenced, and thus they contain some gaps of unknown length. The alignment with the human genome databank (hg19) evidenced for BAC 584M21 a high percentage of identity (> 98%) with both 4q35.2 (subsequences 12, 17 and 18; total length of 52.888 kb) and 3q29 (subsequences 14 and 13; total length of 33.898 kb) (Table S4). Furthermore, in BAC 584M21, a fragment of D4Z4 linked to a cluster of Beta satellite of about 6 kb (subsequence 15 asterisked in Table S4; 11.488 kb in length) showed an identity of 96.2% with 10q26.3. Interestingly, this DNA region also showed a similar percentage of identity (> 97%) with 18p11.32. Also the BAC 809J1 belonged to the 4q35 chromosome; in fact it aligned (> 98% of identity) with the block of sequences that in human chromosome 4q35.2 are comprised between DUX4c and the D4Z4 cluster (Table S4). Regarding the sequence organization of the 3p12 chromosome, it is noteworthy that BACs 709I17 (subsequence 35; 11.360 in length) and 705I9 (subsequence 69; 8.122 kb in length) (double asterisks in Table S4) overlapped with 99.8% of identity (not shown) and hence they were considered as a contig. The overlapping region showed the presence of a D4Z4–Beta satellite sequence organization very similar to that found in 4q35. The sequence upstream to the D4Z4/Beta satellite region (subsequences 36 and 37 of BAC 709I17; total length of 44.098 kb) displayed identity with both 3p12.3 and 4q35.2. In particular, 4q35.2-like sequences included the FRG2 gene and the polymorphic marker SSLP, whereas 3p12.3-like sequences a short cluster of divergent Beta satellite (approximately 2 kb in length) showing a high percentage of identity (98.4%) with the human orthologous region (not shown). Conversely, the sequence downstream to the D4Z4/Beta satellite region, (subsequences 67, 61 and 62 of BAC 705I9: total length of 46.066 kb) aligned only with 3p12.3 (identity index of approximately 97%). Coordinates on different chromosomes, identities with human genome databank (hg19) and accession numbers of the derived BAC sequences are listed in Table S4. A graphical representation of the alignment of subsequences from BACs 584M21, 709I17 and 705I9 on human chromosomes obtained by BLAT is reported in Figs. S3 and S4.
In summary, BAC 584M21 spans from upstream the FRG2 gene to subtelomeric sequences located downstream to the linked clusters of D4Z4 and Beta satellite on chromosomes 4q35.2/10q26.3. Towards the telomere, this block of sequences is followed by other subtelomeric sequences duplicated on several chromosomes (Fig. S3). Overlapping BACs 709I17 and 705I9 span from the FRG2C gene to subtelomeric sequences downstream to the clusters of D4Z4 and Beta satellite on chromosomes 4q35.2/10q26.3, where they are linked to approximately 110 kb of 3p12.3 sequences. The approximate length of D4Z4 clusters was derived from the estimated length of the analyzed BACs obtained by PFGE (not shown): all the sequenced BACs seem to carry a D4Z4 cluster of 15–20 repetitive units.
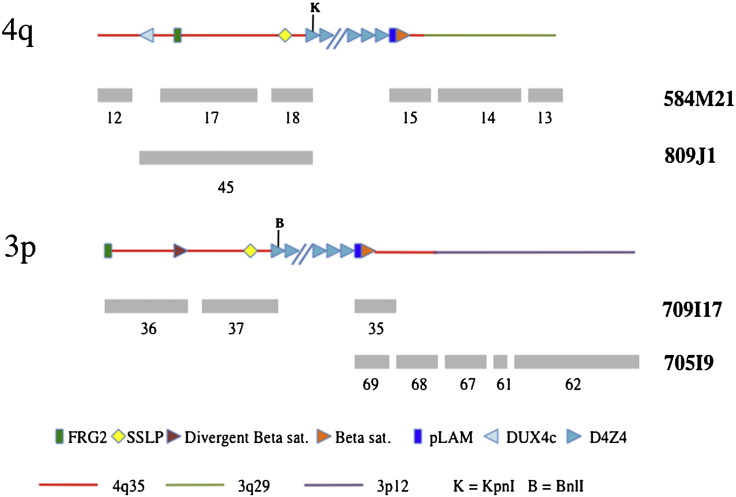
The organization of analyzed BACs is depicted in Fig. 2: BACs 584M21 and 809J1 span approximately 150 kb of the chimpanzee 4qter region, whereas approximately 200 kb of the chimpanzee 3p12 chromosome are present in BACs 709I17 and 705I9. Thus, in chimpanzee 4q35 and 3p12 chromosomes carry a very similar repetitive sequence organization.
Fig. 2.
Schematic sequence organization of PTR BACs 584M21, 809J1, 705I9 and 709I17. Gray rectangles and numbers beneath the schematic organization identify the sequenced subregions (see Supplementary Table 4). The internal organization of each BAC has been derived by sequence alignment to human genome databank (hg19), and by the reciprocal sequence identity as for the pairs of BACs 584M21/809J1, and 1705I9/709I17. Chromosome location at 4q and 3p of the sequenced BACs was derived by FISH analysis and by nucleotide sequence identity. In this regard, within BAC 584M21, 4q35-specific subtelomeric sequences (in red) are linked to PTR-specific subtelomeric sequences (light green), present in the human genome on several subtelomeres as 3q29, 1p36, 5q35, and 19p13, but not at 4q35. In addition, BAC 705I9, which overlaps with BAC 709I17, shows a linkage of 4q35 subtelomeric sequences with interstitial 3p12 sequences (in violet). // indicates interruption in the D4Z4 array that for these BACs has been estimated to be of approximately 15 copies of the repetitive unit. (For interpretation of the references to color in this figure legend, the reader is referred to the web of this article.)
Schematic sequence organization of PTR BACs 584M21, 809J1, 705I9 and 709I17. Gray rectangles and numbers beneath the schematic organization identify the sequenced subregions (see Supplementary Table 4). The internal organization of each BAC has been derived by sequence alignment to human genome databank (hg19), and by the reciprocal sequence identity as for the pairs of BACs 584M21/809J1, and 1705I9/709I17. Chromosome location at 4q and 3p of the sequenced BACs was derived by FISH analysis and by nucleotide sequence identity. In this regard, within BAC 584M21, 4q35-specific subtelomeric sequences (in red) are linked to PTR-specific subtelomeric sequences (light green), present in the human genome on several subtelomeres as 3q29, 1p36, 5q35, and 19p13, but not at 4q35. In addition, BAC 705I9, which overlaps with BAC 709I17, shows a linkage of 4q35 subtelomeric sequences with interstitial 3p12 sequences (in violet). // indicates interruption in the D4Z4 array that for these BACs has been estimated to be of approximately 15 copies of the repetitive unit.
3.5. Comparison of D4Z4 and Beta satellite repeats from PTR and HSA 4q35 chromosome
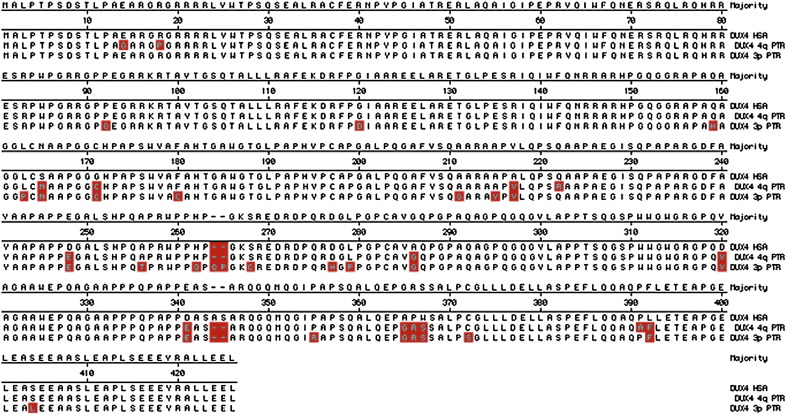
The employed sequencing approach did not allow to obtain full length D4Z4 units; this was probably due to difficulties in assembling the sequences of this tandem array, and/or to underrepresentation of D4Z4 sequences during the amplification step needed for the sequencing process. Nevertheless, by aligning several D4Z4 sequenced fragments from both 3p12 and 4q35 chromosome regions we derived two almost complete D4Z4 repeats labeled as D4Z4 4q (2992 bp) and D4Z4 3p (3263 bp) (accession numbers JN851147 and JN851148, respectively). These D4Z4 sequences (4q and 3p) contain an ORF of 1266 bp or 1272 bp, giving putative DUX4 protein of 422 (DUX4 4q) and 424 (DUX4 3p) aminoacids, respectively. Both protein sequences show a high percentage of identity (100% and 94.8% for DUX 4q and DUX 3p, respectively) with a previously reported putative DUX4 protein from Pan troglodytes (accession number CAL41938). Furthermore, PTR DUX 4q and 3p showed an identity of 95.8% and 92.5%, with a human DUX protein (accession number ADK24688). The alignment of chimpanzee and human DUX4 proteins is reported in Fig. 3.
Fig. 3.
Comparison of human and chimpanzee sequence features of D4Z4. Alignment of HSA and PTR 4q and 3p DUX4 amino-acid sequences. The three proteins show a length of 424 aa (HSA and 3p PTR) and 422 aa (4q PTR). The DUX 4q PTR protein showed a 100% identity with a previously reported chimpanzee DUX protein (accession number CAL41938).
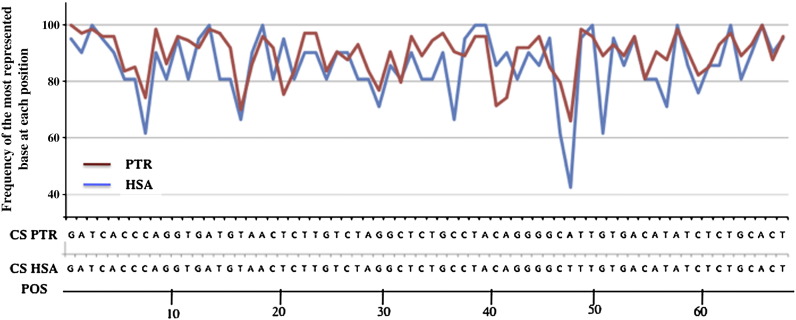
The nucleotide consensus sequences of PTR and HSA Beta satellites were then derived from 21 and 77 repeat units present, respectively, in the cluster contained within human clone U774496 (4qA) and chimpanzee BAC 584M21(4q) (Fig. 4). The two consensus showed a high degree of identity (98.5%), as also derived from the frequency profiles of the most represented base at each position (Fig. 4). We then compared two other PTR and HSA sequences, namely pLAM and SSLP, which are considered important markers of the human 4q35 region [9,10]. pLAM sequences present in chimpanzee 709I17 and 584M21 BACs exhibited an identity of 68% and 93%, respectively, when compared with the human pLAM (accession number Z252821) (not shown); furthermore both PTR pLAMs do not contain any poly(A) signal (not shown). As regard the SSLP sequence located upstream to the D4Z4 array, 709I17 and 584 M21 BACs showed a nucleotide core of respectively 160 and 173 bases (not shown).
Fig. 4.
Frequency of the most represented base at each position showed by Beta satellite DNA from PTR (red line) and HSA (blue line) as derived by the alignment of 74 repetitive units present in the cluster contained within PTR BAC 584M21-4q and of 21 repetitive units within the human clone U774496-4qA. Alignment of the consensus sequence of Beta satellite from PTR (CS PTR) and from HSA (CS HSA) derived from the values shown in the upper graphic. The alignment of two consensus showed a high degree of conservation (identity of 98.5%). POS = position of nucleotides within the repetitive unit.
4. Discussion
The data here presented, not only provide a detailed analysis of the chimpanzee genome, but also add new insights into the evolutionary history of the human 4q35 chromosome region, and open the possibility that other repetitive sequences, in addition to the D4Z4 array, play a role in the functionality of this genomic region. The analysis of a PTR genomic library for clones highly positive for the D4Z4 repeat yielded a total of thirteen genomic sectors that were subsequently grouped into different classes: clones containing a linkage of 4q35 sequences with sequences located in different human subtelomeres (i.e. 1p36.33 and 19p13.3), clones with a linkage of 4q35-like sequences and 3p12.3 sequences, and finally clones completely defined at both extremities by D4Z4 repeats. Furthermore, the PTR genomic clones exhibited in the D4Z4 unit a different restriction enzyme periodicity (KpnI or BlnI), and most importantly four of them, namely 584M21, 578I19, 709I17 and 705I9, carried a Beta satellite cluster linked to the D4Z4 array. Contrary to what is found in PTR and humans, in GGO this organization was not present on chromosome 4q but probably only on chromosome 4p. This conclusion was derived by the molecular analysis of the gorilla genome in which 4qter-derived genomic clones did not show the presence of Beta satellite sequences [25], and by FISH that showed the co-localization of D4Z4 and Beta satellite only on an internal location of chromosome 4p [28,25]. Furthermore, no Beta satellite sequences were detected by the screening of a rhesus monkey genomic library, and gibbon and orangutan Beta satellite sequences do not show a clustered organization [29]. Thus we suggest that, at 4qter, D4Z4 and Beta satellite linkage has recently appeared in the evolution, since this organization is only detected starting from the chimpanzee. This hypothesis is also supported by previous data reporting that the chimpanzee 4qter region is characterized by a sequence organization very similar to that found in humans [30]. Furthermore, in both studies only one type of 4qter organization was detected, and this was structurally similar to human 4qA up to a point just distal of the Beta satellite array, and included 4q specific subtelomeric sequences. Since this result was obtained by the screening of two independent PTR genomic libraries, it is reasonable to propose that PTR possesses only one type of 4qter organization belonging to the 4qA allele. A very similar sequence organization was detected on PTR chromosome 3p, where D4Z4 and Beta satellite linked arrays were embedded in 4q35-like sequences, spanning from the proximal FRG2C sequence to the distal subtelomeric sequences. Differences between the two chromosome regions included the occurrence on 3p of a divergent array of Beta satellite located upstream to the D4Z4 array, the restriction enzyme periodicity of D4Z4 (BlnI), and the number of PstI sites present in the Beta satellite linked to D4Z4. Starting from these findings in the PTR and from the organization of human syntenic regions, we derived a hypothetical evolutionary history of 4q35 and 3p12.2 that occurred after the divergence of chimpanzee from the human lineage (Fig. 5). An assumption is that a sequence organization composed of D4Z4 and Beta satellite linked arrays (4qA-like) was present at 4qter and 3p in a common ancestor of PTR and HSA. In HSA lineage, one of the two 4qA-like sequence organizations underwent the deletion of a region including one D4Z4, the pLAM and the Beta satellite cluster, originating the 4qB allele. Due to the identity of 97.8% between this PTR 4q region and human chromosome 18p11.32, it is possible to hypothesize that the deleted region has been transposed into the subtelomere of chromosome 18p. This hypothesis, in agreement with previous findings derived by the comparison of subtelomeric regions from human chromosomes 4qA and 4qB [11], suggests that the 4qA allele is evolutionary older than 4qB. A duplication event in PTR chromosome 4q interesting a region spanning from the FRG2 gene to the subtelomeric sequences, and including D4Z4 and Beta satellite linked arrays, might have originated human chromosome 10q. Human chromosomes 3p12.3 and 4q35.2 show sequence identity in a region of approximately 35–40 kb comprised between one inverted D4Z4 (probably a copy of DUX4c) upstream to FRG2, and sequences just proximal to the D4Z4 array. Hence, human chromosome 3p12 might have originated by the total deletion of D4Z4 and Beta satellite linked arrays and of 4q35 subtelomeric sequences (Fig. 5). Alternatively, D4Z4 and Beta satellite linked arrays were not present at 3p in a common ancestor of PTR and HSA, and thus this repetitive sequence organization appeared in the PTR lineage. Thus, after human/chimpanzee divergence D4Z4 and Beta satellite linked arrays underwent an extensive genomic remodeling leaving this sequence organization at the subtelomeric location of human chromosomes 4qA and 10q26. In this regard, search in human reference sequence (hg19) and in human unplaced contigs and clones for D4Z4 and Beta satellite positivity, strongly indicates that the linking of the two families of repeats is a peculiar feature of chromosomes 4q and 10q. In fact, as previously reported [31] particularly for the p arm of acrocentric chromosomes and for the Y chromosome, arrays of Beta satellite are intermingled with single or double D4Z4 repeats.
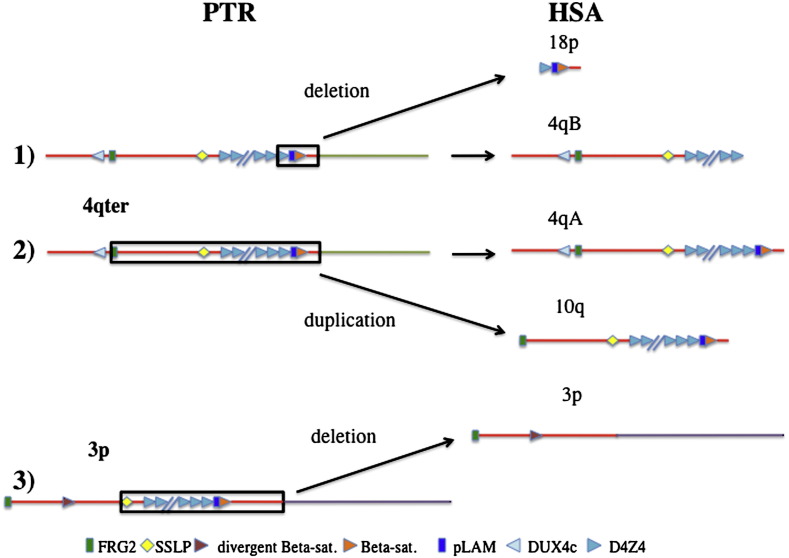
Fig. 5.
Schematic representation of the hypothetical evolutionary events occurring in the human lineage after the separation of chimpanzee. The PTR 4qter chromosome region is represented with two copies of the 4qA-like allele, whereas the interstitial 3p region with only one copy. (1) One 4qA-like allele underwent to an event of deletion and transposition to chromosome 18pter of a region comprising one D4Z4, pLAM, the Beta satellite array and subtelomeric sequences (boxed region on the upper 4qA-like copy). This deletion originated the human 4qB allele, whereas the other 4qter allele became allele 4qA. (2) One 4qA-like allele was used as template for the duplication of a region spanning from FRG2 to subtelomeric sequences, and including the full D4Z4 and Beta satellite linked arrays; this duplication was translocated to the qter region of chromosome 10. (3) The interstitial D4Z4 and Beta satellite linked arrays at PTR 3p12 was deleted (boxed region) on both chromosomes leaving upstream sequences just to the DUX4c and including the FRG2C gene and a short array of divergent Beta satellite.
A different evolutionary history characterizes the two families of repeats and their reciprocal sequence organization typical of human chromosomes 4q and 10q. D4Z4 repeats were not confined to primates, but found in several placental mammals thus indicating an ancient origin [32]. All the analyzed species showed the maintenance of the DUX ORF and an array organization containing at least 10 repeated units. Furthermore a survey of non-mammalian genomes identified a likely ancestor of the mammalian DUX gene [33]. Conversely, the appearance of clustered Beta satellite sequences can be traced back to the gorilla, where the bulk of the repeats were mapped interstitially to 4p and to the p arm of chromosome Y [28,29]. That the gorilla 4qter region contains clustered D4Z4 units but not Beta satellite linked array was confirmed by genome analysis [25]. Hence, D4Z4 and Beta satellite linked arrays at 4qter represents a recent evolutionary event occurring in the PTR after the divergence of the gorilla lineage. However, during human evolution these repetitive regions underwent further genomic remodeling that caused the loss of the Beta satellite cluster from one 4q allele and the appearance of D4Z4–Beta satellite linked organization at 10q. Nevertheless, as in PTR 4q and 3p, human D4Z4 units maintain a coding capacity at 4q as well as at 10q. The importance of this repetitive sequence organization for human cell physiology can be derived from the observation that D4Z4 array contraction of the 4qA allele causes a type of muscle dystrophy (FSHD) [8–10]. There is a general consensus in the field that D4Z4 deletion leads to epigenetic alterations that affect the expression profiles of in cis candidate genes (for review see [9,10,34]). Potential FSHD candidate genes are ANT1 (SLC25A4), FRG1, FRG2, DUX4 and DUX4c and in this regard in the last years many papers have attempted to define the “true” candidate gene(s) without conclusive results. Although DUX4 and FRG1 seem to be the main candidate genes, the heterogeneity of disease manifestations could probably underline a heterogeneity of gene expression due to different epigenetic alterations of in cis candidate genes. It has been previously demonstrated that both contracted and non contracted 4q35 alleles associate with the heterochromatic nuclear compartment (nuclear rim and nucleolar periphery) [35,36]. These studies also indicated that other chromosomal loci bearing the D4Z4 repeats, such as 10q and the p arm of acrocentric chromosomes showed a common affinity for these heterochromatic regions. Furthermore, a common feature of these D4Z4 chromosome regions is that they reside adjacent to blocks of Beta satellite DNA. Indeed, D4Z4 does not seem to be responsible for the localization to the heterochromatic compartment, but rather for the transcriptional repression of the region via heterochromatinization. In this regard, it has been demonstrated that in normal cells D4Z4 repeats are methylated and show histone marks of repression (H3K27me3), and that both features are lost in FSHD patients [16,17,37]. Furthermore a repressor complex composed of YY1, HMGB2 and nucleolin is specifically bound to D4Z4 [38,39]. For this reason a possible role of Beta satellite in augmenting the observed heterochromatic repressive signal at the 4q35 region by acting synergistically with the D4Z4 array in recruiting chromatin modifiers can be hypothesized. At present no data are available either on the polymorphism of 4q35 Beta satellite array within the human population, or whether the array might undergo contraction like the D4Z4 array. However, differently from the 4q35 D4Z4 array showing in the human population a great length polymorphism (ranging from 11 to 150 copies) [40,41], the few available information from the databank suggest a conservation in PTR and HSA of the length of 4q35 Beta satellite array, accounting for approximately 90 copies (6 kb). Moreover, we found that some subregions of the Beta satellite unit seem to be highly conserved between PTR and HSA. Although further computational and functional experiments are certainly required to confirm the conservation of Beta satellite sequence, our results suggest a possible biological role for this class of repeats. Interestingly, the obtained results are very similar to those previously reported by us [42]. In this regard, the observed variability of D4Z4-contracted FSHD manifestation, as well as FSHD manifestation in non-contracted individuals (phenotypic FSHD) and the absence of disease manifestation in carrier of D4Z4 contraction might be explained in term of Beta satellite DNA. In conclusion, these results have delineated the evolutionary history of linked D4Z4 and Beta satellite arrays, raising the possibility that Beta satellite repeats might be involved in the regulation of the FSHD region.
The following are the supplementary data related to this article.
A) Southern blot hybridization with a human D4Z4 probe (77M12-KpnI) of KpnI and BlnI digested DNA from D4Z4-positive PTR BACs isolated from the PTR genomic library CHORI-251, after fractionation by 1% agarose gel electrophoresis. Bands with length of 3.3 kb are indicated. M = molecular weight standard. B) Southern blot hybridization with a human Beta satellite probe (clone A17A; Agresti et al., 1987) of Sau3A and PstI digested DNA from D4Z4- and Beta satellite positive PTR BACs isolated from the PTR genomic library CHORI-251, after fractionation by 1% agarose gel electrophoresis. M = molecular weight standard. 1 ×, 2 ×, 3 × = monomer, dimer and trimer of the Beta satellite family of DNA repeats.
Restriction enzyme periodicity of D4Z4 unit and number of restriction enzyme sites within Beta satellite cluster of different PTR genomic clones.
Identity of PTR BAC ends with human genome databank (hg19).
Graphic representation of sequence identity (blat search at http://genome.cse.ucsc.edu/) of isolated PTR BAC ends to human genome databank (hg19). The analyzed ends did not include those showing similarities with D4Z4 (for the full list see Supplementary Table 2). From top to bottom: sequence identity with chromosomes 4q35.2, 10q26.3, 3q29 and 3p12.3. Sp6 and T7, after the BAC symbols identify the different ends of each BAC.
Chromosomal location by FISH of PTR genomic clones on PTR metaphases (major locations are in bold).
Identity of PTR BAC sequences with human genome databank (hg19).
Graphic representation of sequence identity (blat search at http://genome.cse.ucsc.edu/) of subsequences from PTR BAC 584M21 to human genome databank (hg19) (see also Supplementary Table 4). The different subsequences are identified by numbers (12, 17, 18, 15, 14 and 13) after the BAC identification number (584). From the top: sequence identity with chromosomes 4q35.2, 10q26.3, 3q29. Beta sat/tel: clone containing the linkage between the beta satellite array and subtelomeric sequences; subtel. sequences: block of subtelomeric sequences of PTR chromosome homologous to human chromosome 4; cen: centromere; tel: telomere.
Graphic representation of sequence identity (blat search at http://genome.cse.ucsc.edu/) of subsequences from PTR BACs 705I9 and 709I17 to human genome databank (hg19) (see also Supplementary Table 4). The different subsequences are identified by numbers (36, 37 and 35 for BAC 709I17, and 69, 68, 67, 61 and 62 for BAC 705I9) after the BAC identification number (709 or 705). From the top: sequence identity with chromosomes 10q26.3 and 3q12.3. Beta sat/tel: subsequences containing the linkage between the Beta satellite array and subtelomeric sequences; Beta sat 10q/4q and D4Z4: location, respectively, of the Beta satellite and D4Z4 arrays on human chromosomes 4q35.2 (allele 4qA) and 10q26.3; Beta sat 3p and DUX4c: location on human chromosome 3p12.3 of an array of divergent Beta satellite repeats and of DUX4c. cen: centromere; tel: telomere.
Acknowledgments
This work was supported by grants from Telethon to EG (GGP07078), Association Française contre les Myopathies (AFM) to EG (13160 and 14464) and PRIN 2008 to EG and RM.
Footnotes
Sequence data from this article have been deposited with the GenBank Data Library under accession numbers JM170481–JM170504, JS886734, JS886735, and JF900595–JF900598.
Contributor Information
Marta Giussani, Email: marta.giussani@unimi.it.
Maria Francesca Cardone, Email: mariafrancesca.cardone@entecra.it.
Beatrice Bodega, Email: BBodega@Dti.Telethon.it.
Enrico Ginelli, Email: enrico.ginelli@unimi.it.
Raffaella Meneveri, Email: raffaella.meneveri@unimib.it.
References
- 1.Doolittle W.F., Sapienza C. Selfish genes, the phenotype paradigm and genome evolution. Nature. 1980;284:601–603. doi: 10.1038/284601a0. [DOI] [PubMed] [Google Scholar]
- 2.Orgel L.E., Crick F.H. Selfish DNA: the ultimate parasite. Nature. 1980;284:604–607. doi: 10.1038/284604a0. [DOI] [PubMed] [Google Scholar]
- 3.Tomilin N.V. Regulation of mammalian gene expression by retroelements and non-coding tandem repeats. Bioessays. 2008;30:338–348. doi: 10.1002/bies.20741. [DOI] [PubMed] [Google Scholar]
- 4.Ugrakovic D. Functional elements residing within satellite DNAs. EMBO Rep. 2005;6:1035–1039. doi: 10.1038/sj.embor.7400558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guetg C., Lienemann P., Sirri V., Grummt I., Hernandez-Verdun D., Hottiger M.O., Fussenegger M., Santoro R. The NoRC complex mediates the heterochromatin formation and stability of silent rRNA genes and centromeric repeats. EMBO J. 2010;29:2135–2146. doi: 10.1038/emboj.2010.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Iotti G., Longobardi E., Masella S., Dardaei L., De Santis F., Micali N., Blasi F. Homeodomain transcription factor and tumor suppressor Prep1 is required to maintain genomic stability. Proc. Natl. Acad. Sci. U. S. A. 2011;108:E314–E322. doi: 10.1073/pnas.1105216108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mravinac B., Plohl M., Ugarkovic D. Preservation and high sequence conservation of satellite DNAs suggest functional constraints. J. Mol. Evol. 2005;61:542–550. doi: 10.1007/s00239-004-0342-y. [DOI] [PubMed] [Google Scholar]
- 8.Statland J.M., Tawil R. Facioscapulohumeral muscular dystrophy: molecular pathological advances and future directions. Curr. Opin. Neurol. 2011;24:423–428. doi: 10.1097/WCO.0b013e32834959af. [DOI] [PubMed] [Google Scholar]
- 9.van der Maarel S.M., Tawil R., Tapscott S.J. Facioscapulohumeral muscular dystrophy and DUX4: breaking the silence. Trends Mol. Med. 2011;17:252–258. doi: 10.1016/j.molmed.2011.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cabianca D.S., Gabellini D. The cell biology of disease: FSHD: copy number variations on the theme of muscular dystrophy. J. Cell Biol. 2010;191:1049–1060. doi: 10.1083/jcb.201007028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van Geel M., Dickson M.C., Beck A.F., Bolland D.J., Frants R.R., van der Maarel S.M., de Jong P.J., Hewitt J.E. Genomic analysis of human chromosome 10q and 4q telomeres suggests a common origin. Genomics. 2002;79:210–217. doi: 10.1006/geno.2002.6690. [DOI] [PubMed] [Google Scholar]
- 12.Buzhov B.T., Lemmers R.J., Tournev I., van der Wielen M.J., Ishpekova B., Petkov R., Petrova J., Frants R.R., Padberg G.W., van der Maarel S.M. Recurrent somatic mosaicism for D4Z4 contractions in a family with facioscapulohumeral muscular dystrophy. Neuromuscul. Disord. 2005;15:471–475. doi: 10.1016/j.nmd.2005.03.005. [DOI] [PubMed] [Google Scholar]
- 13.Lemmers R.J., de Kievit P., Sandkuijl L., Padberg G.W., van Ommen G.J., Frants R.R., van der Maarel S.M. Facioscapulohumeral muscular dystrophy is uniquely associated with one of the two variants of the 4q subtelomere. Nat. Genet. 2002;32:235–236. doi: 10.1038/ng999. [DOI] [PubMed] [Google Scholar]
- 14.van der Maarel S.M., Frants R.R., Padberg G.W. Facioscapulohumeral muscular dystrophy. Biochim. Biophys. Acta. 2007;1772:186–194. doi: 10.1016/j.bbadis.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 15.Jiang G., Yang F., van Overveld P.G., Vedanarayanan V., van der Maarel S., Ehrlich M. Testing the position-effect variegation hypothesis for facioscapulohumeral muscular dystrophy by analysis of histone modification and gene expression in subtelomeric 4q. Hum. Mol. Genet. 2003;12:2909–2921. doi: 10.1093/hmg/ddg323. [DOI] [PubMed] [Google Scholar]
- 16.van Overveld P.G., Lemmers R.J., Sandkuijl L.A., Enthoven L., Winokur S.T., Bakels F., Padberg G.W., van Ommen G.J., Frants R.R., van der Maarel S.M. Hypomethylation of D4Z4 in 4q-linked and non-4q-linked facioscapulohumeral muscular dystrophy. Nat. Genet. 2003;35:315–317. doi: 10.1038/ng1262. [DOI] [PubMed] [Google Scholar]
- 17.de Greef J.C., Lemmers R.J., van Engelen B.G., Sacconi S., Venance S.L., Frants R.R., Tawil R., van der Maarel S.M. Common epigenetic changes of D4Z4 in contraction-dependent and contraction-independent FSHD. Hum. Mutat. 2009;3:1449–1459. doi: 10.1002/humu.21091. [DOI] [PubMed] [Google Scholar]
- 18.de Greef J.C., Lemmers R.J., Camano P., Day J.W., Sacconi S., Dunand M., van Engelen B.G., Kiuru-Enari S., Padberg G.W., Rosa A.L., Desnuelle C., Spuler S., Tarnopolsky M., Venance S.L., Frants R.R., van der Maarel S.M., Tawil R. Clinical features of facioscapulohumeral muscular dystrophy 2. Neurology. 2010;75:1548–1554. doi: 10.1212/WNL.0b013e3181f96175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ballestar E., Wolffe A.P. Methyl-CpG-binding proteins. Targeting specific gene repression. Eur. J. Biochem. 2001;268:1–6. doi: 10.1046/j.1432-1327.2001.01869.x. [DOI] [PubMed] [Google Scholar]
- 20.de Greef J.C., Wohlgemuth M., Chan O.A., Hansson K.B., Smeets D., Frants R.R., Weemaes C.M., Padberg G.W., van der Maarel S.M. Hypomethylation is restricted to the D4Z4 repeat array in phenotypic FSHD. Neurology. 2007;69:1018–1026. doi: 10.1212/01.wnl.0000271391.44352.fe. [DOI] [PubMed] [Google Scholar]
- 21.Pandya S., King W., Tawil R. Facioscapulohumeral dystrophy. Phys. Ther. 2008;88:105–113. doi: 10.2522/ptj.20070104. [DOI] [PubMed] [Google Scholar]
- 22.Tawil R., van der Maarel S.M. Facioscapulohumeral dystrophy. Muscle Nerve. 2008;34:1–15. doi: 10.1002/mus.20522. [DOI] [PubMed] [Google Scholar]
- 23.Bastress K.L., Stajich J.M., Speer M.C., Gilbert J.R. The genes encoding for D4Z4-binding proteins HMGB2, YY1, NCL and MYOD are excluded as candidate genes for FSDH1B. Neuromuscul. Disord. 2005;15:316–320. doi: 10.1016/j.nmd.2004.12.006. [DOI] [PubMed] [Google Scholar]
- 24.Tonini M.M., Passos-Bueno M.R., Cerqueira A., Matioli S.R., Pavanello R., Zatz M. Asymptomatic carriers and gender differences in facioscapulohumeral muscular dystrophy (FSHD) Neuromuscul. Disord. 2004;14:33–38. doi: 10.1016/j.nmd.2003.07.001. [DOI] [PubMed] [Google Scholar]
- 25.Bodega B., Cardone M.F., Muller S., Neusser M., Orzan F., Rossi E., Battaglioli E., Marozzi A., Riva P., Rocchi M., Meneveri R., Ginelli E. Evolutionary genomic remodelling of the human 4q subtelomere (4q35.2) BMC Evol. Biol. 2007;7:39–51. doi: 10.1186/1471-2148-7-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meneveri R., Agresti A., Della Valle G., Talarico D., Siccardi A.G., Ginelli E. Identification of a human clustered G+C-rich DNA family of repeats (Sau3A family) J. Mol. Biol. 1985;186:483–489. doi: 10.1016/0022-2836(85)90123-8. [DOI] [PubMed] [Google Scholar]
- 27.Primate Cytogenetics. http://www.biologia.uniba.it/primates/cell_lines.html
- 28.Hirai H., Taguchi T., Godwin A.K. Genomic differentiation of 18S ribosomal DNA and Beta satellite DNA in the hominoid and its evolutionary aspects. Chromosome Res. 1999;7:531–540. doi: 10.1023/a:1009237412155. [DOI] [PubMed] [Google Scholar]
- 29.Cardone M.F., Ballarati L., Ventura M., Rocchi M., Marozzi A., Ginelli E., Meneveri R. Evolution of beta satellite DNA sequences: evidence for duplication-mediated repeat amplification and spreading. Mol. Biol. Evol. 2004;21:1792–1799. doi: 10.1093/molbev/msh190. [DOI] [PubMed] [Google Scholar]
- 30.Rudd M.K., Endicott R.M., Friedman C., Walker M., Young J.M., Osoegawa K., NISC Comparative Sequencing Program. de Jong P.J., Green E.D., Trask B.J. Comparative sequence analysis of primate subtelomeres originating from a chromosome fission event. Genome Res. 2009;19:33–41. doi: 10.1101/gr.083170.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Winokur S.T., Bengtsson U., Vargas J.C., Wasmuth J.J., Altherr M.R., Weiffenbach B., Jacobsen S.J. The evolutionary distribution and structural organization of the homeobox-containing repeat D4Z4 indicates a functional role for the ancestral copy in the FSHD region. Hum. Mol. Genet. 1996;5:1567–1755. doi: 10.1093/hmg/5.10.1567. [DOI] [PubMed] [Google Scholar]
- 32.Clapp J., Mitchell L.M., Bolland D.J., Fantes J., Corcoran A.E., Scotting P.J., Armour J.A., Hewitt J.E. Evolutionary conservation of a coding function for D4Z4, the tandem DNA repeat mutated in facioscapulohumeral muscular dystrophy. Am. J. Hum. Genet. 2007;81:264–279. doi: 10.1086/519311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Leidenroth A., Hewitt J.E. A family history of DUX4: phylogenetic analysis of DUXA, B, C and Duxbl reveals the ancestral DUX gene. BMC Evol. Biol. 2010;10:364–376. doi: 10.1186/1471-2148-10-364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Neguembor M.V., Gabellini D. In junk we trust: repetitive DNA, epigenetics and facioscapulohumeral muscular dystrophy. Epigenomics. 2010;2:271–287. doi: 10.2217/epi.10.8. [DOI] [PubMed] [Google Scholar]
- 35.Tam R., Smith K.P., Lawrence J.B. The 4q subtelomere harboring the FSHD locus is specifically anchored with peripheral heterochromatin unlike most human telomeres. J. Cell Biol. 2004;167:269–279. doi: 10.1083/jcb.200403128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Masny P.S., Bengtsson U., Chung S.A., Martin J.H., van Engelen B., van der Maarel S.M., Winokur S.T. Localization of 4q35.2 to the nuclear periphery: is FSHD a nuclear envelope disease? Hum. Mol. Genet. 2004;13:1857–1871. doi: 10.1093/hmg/ddh205. [DOI] [PubMed] [Google Scholar]
- 37.Zeng W., de Greef J.C., Chen Y.Y., Chien R., Kong X., Gregson H.C., Winokur S.T., Pyle A., Robertson K.D., Schmiesing J.A., Kimonis V.E., Balog J., Frants R.R., Jr Ball A.R., Lock L.F., Donovan P.J., van der Maarel S.M., Yokomori K. Specific loss of histone H3 lysine 9 trimethylation and HP1gamma/cohesin binding at D4Z4 repeats is associated with facioscapulohumeral dystrophy (FSHD) PLoS Genet. 2009;5:e1000559. doi: 10.1371/journal.pgen.1000559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gabellini D., Green M.R., Tupler R. Inappropriate gene activation in FSHD: a repressor complex binds a chromosomal repeat deleted in dystrophic muscle. Cell. 2002;110:339–348. doi: 10.1016/s0092-8674(02)00826-7. [DOI] [PubMed] [Google Scholar]
- 39.Bodega B., Ramirez G.D., Grasser F., Cheli S., Brunelli S., Mora M., Meneveri R., Marozzi A., Mueller S., Battaglioli E., Ginelli E. Remodeling of the chromatin structure of the facioscapulohumeral muscular dystrophy (FSHD) locus and upregulation of FSHD-related gene 1 (FRG1) expression during human myogenic differentiation. BMC Biol. 2009;7:41. doi: 10.1186/1741-7007-7-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wijmenga C., Hewitt J.E., Sandkuijl L.A., Clark L.N., Wright T.J., Dauwerse H.G., Gruter A.M., Hofker M.H., Moerer P., Williamson R., van Ommen G.J., Padberg G.W., Frants R.R. Chromosome 4q DNA rearrangements associated with facioscapulohumeral muscular dystrophy. Nat. Genet. 1992;2:26–30. doi: 10.1038/ng0992-26. [DOI] [PubMed] [Google Scholar]
- 41.van Deutekom J.C., Wijmenga C., van Tienhoven E.A., Gruter A.M., Hewitt J.E., Padberg G.W., van Ommen G.J., Hofker M.H., Frants R.R. FSHD associated DNA rearrangements are due to deletions of integral copies of a 3.2 kb tandemly repeated unit. Hum. Mol. Genet. 1993;2:2037–2042. doi: 10.1093/hmg/2.12.2037. [DOI] [PubMed] [Google Scholar]
- 42.Agresti A., Meneveri R., Siccardi A.G., Marozzi A., Corneo G., Gaudi S., Ginelli E. Linkage in human heterochromatin between highly divergent Sau3A repeats and a new family of repeated DNA sequences (HaeIII family) J. Mol. Biol. 1989;205:625–631. doi: 10.1016/0022-2836(89)90308-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A) Southern blot hybridization with a human D4Z4 probe (77M12-KpnI) of KpnI and BlnI digested DNA from D4Z4-positive PTR BACs isolated from the PTR genomic library CHORI-251, after fractionation by 1% agarose gel electrophoresis. Bands with length of 3.3 kb are indicated. M = molecular weight standard. B) Southern blot hybridization with a human Beta satellite probe (clone A17A; Agresti et al., 1987) of Sau3A and PstI digested DNA from D4Z4- and Beta satellite positive PTR BACs isolated from the PTR genomic library CHORI-251, after fractionation by 1% agarose gel electrophoresis. M = molecular weight standard. 1 ×, 2 ×, 3 × = monomer, dimer and trimer of the Beta satellite family of DNA repeats.
Restriction enzyme periodicity of D4Z4 unit and number of restriction enzyme sites within Beta satellite cluster of different PTR genomic clones.
Identity of PTR BAC ends with human genome databank (hg19).
Graphic representation of sequence identity (blat search at http://genome.cse.ucsc.edu/) of isolated PTR BAC ends to human genome databank (hg19). The analyzed ends did not include those showing similarities with D4Z4 (for the full list see Supplementary Table 2). From top to bottom: sequence identity with chromosomes 4q35.2, 10q26.3, 3q29 and 3p12.3. Sp6 and T7, after the BAC symbols identify the different ends of each BAC.
Chromosomal location by FISH of PTR genomic clones on PTR metaphases (major locations are in bold).
Identity of PTR BAC sequences with human genome databank (hg19).
Graphic representation of sequence identity (blat search at http://genome.cse.ucsc.edu/) of subsequences from PTR BAC 584M21 to human genome databank (hg19) (see also Supplementary Table 4). The different subsequences are identified by numbers (12, 17, 18, 15, 14 and 13) after the BAC identification number (584). From the top: sequence identity with chromosomes 4q35.2, 10q26.3, 3q29. Beta sat/tel: clone containing the linkage between the beta satellite array and subtelomeric sequences; subtel. sequences: block of subtelomeric sequences of PTR chromosome homologous to human chromosome 4; cen: centromere; tel: telomere.
Graphic representation of sequence identity (blat search at http://genome.cse.ucsc.edu/) of subsequences from PTR BACs 705I9 and 709I17 to human genome databank (hg19) (see also Supplementary Table 4). The different subsequences are identified by numbers (36, 37 and 35 for BAC 709I17, and 69, 68, 67, 61 and 62 for BAC 705I9) after the BAC identification number (709 or 705). From the top: sequence identity with chromosomes 10q26.3 and 3q12.3. Beta sat/tel: subsequences containing the linkage between the Beta satellite array and subtelomeric sequences; Beta sat 10q/4q and D4Z4: location, respectively, of the Beta satellite and D4Z4 arrays on human chromosomes 4q35.2 (allele 4qA) and 10q26.3; Beta sat 3p and DUX4c: location on human chromosome 3p12.3 of an array of divergent Beta satellite repeats and of DUX4c. cen: centromere; tel: telomere.