Abstract
Physiological and molecular phylogenetic approaches were used to investigate variation among 12 cyanobacterial strains in their tolerance of sulfide, an inhibitor of oxygenic photosynthesis. Cyanobacteria from sulfidic habitats were found to be phylogenetically diverse and exhibited an approximately 50-fold variation in photosystem II performance in the presence of sulfide. Whereas the degree of tolerance was positively correlated with sulfide levels in the environment, a strain's phenotype could not be predicted from the tolerance of its closest relatives. These observations suggest that sulfide tolerance is a dynamic trait primarily shaped by environmental variation. Despite differences in absolute tolerance, similarities among strains in the effects of sulfide on chlorophyll fluorescence induction indicated a common mode of toxicity. Based on similarities with treatments known to disrupt the oxygen-evolving complex, it was concluded that sulfide toxicity resulted from inhibition of the donor side of photosystem II.
Understanding how evolutionary processes, environmental variation, and geography shape the distribution of genetic and ecological diversity of microorganisms is a fundamental challenge in microbial ecology. Cyanobacteria provide an excellent natural system for investigating the evolution of ecological differences, because these oxygenic phototrophs have diversified to exploit a wide variety of aquatic and terrestrial environments, including hot springs, polar ponds, hot and cold deserts, and hypersaline lagoons (38). By combining physiological experiments with molecular phylogenetics, it has become possible to study the origins of this ecological variation within an evolutionary framework. This integration of approaches has been used to identify several examples of the physiological adaptation of unique cyanobacterial lineages in response to major environmental variables, including salinity (14), light (25), and temperature (23).
Another key variable that can vary greatly across cyanobacterial habitats is sulfide, an inhibitor of oxygenic photosynthesis (27). While it is absent from stably oxygenated environments, sulfide is either permanently or periodically present in many ecosystems where cyanobacteria are found, as a result of its presence in source water and/or biological sulfate reduction. These include benthic microbial mat communities that are commonly found in hot springs and both hypersaline and marine sediments (29). Cyanobacteria likewise vary in sulfide tolerance, and strains that typically are not exposed to sulfide in nature, including most planktonic and heterocystous cyanobacteria, are extremely sensitive to and are irreversibly poisoned by this toxin (6, 10, 15, 27). In contrast, those from sulfidic habitats exhibit one or more adaptations for maintaining their photoautotrophic metabolism under such conditions. These include the maintenance of oxygenic photosynthesis by the resistance of photosystem (PS) II to sulfide (6, 10) and the ability to perform PS II-independent, anoxygenic photosynthesis with sulfide as an electron donor to PS I (9).
Many questions still remain about both the origins of the ecological variation among cyanobacteria in sulfide tolerance and the nature of sulfide toxicity. It has been proposed that sulfide-tolerant cyanobacteria may represent a unique radiation within an otherwise sensitive group (10). Indeed, many strains that have been isolated from sulfidic environments are morphologically similar members of the genus Oscillatoria. On the other hand, it has already been established that this genus is polyphyletic (e.g., reference 37), and the evolutionary relatedness of most of the strains used in previous studies of sulfide tolerance is not known. A better understanding of the genetic diversity of cyanobacteria from sulfidic environments would not only clarify the above but would also provide a context for interpreting the conflicting conclusions reached by previous investigations regarding the toxic effects of sulfide. Sulfide is known to bind metalloproteins (12) and could conceivably interfere with one or more components involved in photosynthetic electron transport. These include the manganese-containing, water-splitting complex on the donor side of PS II as well as cytochromes on the acceptor side. Whereas some data suggest that sulfide inhibits oxygenic photosynthesis by blocking electron flow from the donor side of PS II (11, 27), other results have implicated the acceptor side as the target (10). Because different studies have used not only different approaches (although all are based on chlorophyll fluorescence signals) but also different cyanobacterial strains of unknown relatedness, it is not clear whether the mode of sulfide toxicity truly differs across strains and, if so, whether toxicity varies in a phylogeny-dependent manner.
In order to address these issues, we sampled cyanobacteria that had been isolated from a variety of sulfidic habitats and were therefore expected to span the breadth of genetic diversity among sulfide-tolerant cyanobacteria. We characterized the evolutionary relationships of the strains by reconstructing phylogenies with 16S rRNA gene sequence data to test whether cyanobacteria from sulfidic habitats primarily represent an evolutionarily coherent assemblage that has diverged as a result of habitat diversification. We also assayed the resistance of PS II to sulfide by pulse-amplitude-modulated fluorometry (30) to investigate both whether the mode of sulfide toxicity varies among strains and the amount of genetic variation among strains with regard to resistance. Finally, using statistical models that take into account the potential phylogenetic dependence of the physiological data, we both estimated the correlation between sulfide tolerance and environmental sulfide and tested whether the kinetics of photosynthetic electron transport under optimal conditions provide any information about a strain's tolerance of this toxin.
MATERIALS AND METHODS
Sample collection and sulfide measurement.
Benthic microbial mat samples were collected from habitats that were either permanently sulfidic (due to the presence of primary sulfide in the system) or became sulfidic at night in the absence of oxygenic photosynthesis (due to biological sulfate reduction). The former category included the following sites: the Pond 2 evaporation pond of Exportadora de Sal SA, a salt works in Baja California Sur; Wilbur Hot Springs in northern California; and Keane Mine Spring, an ambient-temperature spring in Death Valley National Park. The latter category included the Pond 4 evaporation pond in Baja California Sur. In addition, we collected material from Boiling River Hot Spring in Yellowstone National Park, where Mastigocladus sp. grows as streamers in continuously high-flow, oxygenated water and is not exposed to sulfide (6). Mat material was sampled with a sterile 10-ml syringe and transferred to a 20-ml scintillation vial, where it was stored in the dark at room temperature until return to NASA-Ames Research Center. We used Pachmayr colorimetric assays (36) to estimate the total sulfide concentration in the water columns of our sites. For the Pond 4 site, we used an amperometric sulfide microelectrode (Unisense, Aarhus, Denmark) to estimate the in situ concentration of H2S in the cyanobacterial (Microcoleus sp.) layer of the mat (at a depth of approximately 600 to 800 μm below the mat surface). Electrode signal was calibrated with respect to sulfide concentration by monitoring the electrode current in nitrogen-sparged 50 mM KH2PO4 (pH 8) buffer to which different volumes of 100 mM Na2S-phosphate buffer master stock had been added. The relationship between current and sulfide concentration was linear over the range of sulfide concentrations observed in situ.
Strain isolation and maintenance.
Laboratory strains isolated for this study are included in Table 1. All are filamentous. To obtain clones, we took a direct isolation approach in which collections were streaked on agar plates (media for strains are given in Table 1) and an agar plug containing a single trichome was transferred with forceps to liquid medium. When sufficient biomass had accumulated, the process was repeated at least one additional time, depending on the presence of contaminating bacteria. Many of the clones exhibited motility, thereby facilitating culture purification. Each isolate was routinely maintained on a 12-h light-dark cycle, at or near the temperature of the collection from which it was isolated (Table 1). Strains have been deposited in the University of Oregon's Culture Collection of Microorganisms from Extreme Environments.
TABLE 1.
Strains used in this study
| Strainc | Origind | Culture temp (°C) | Mediuma | Referenceb |
|---|---|---|---|---|
| Geitlerinema strain B-33*c | Pond 2 evaporation pond, ESSA, Baja California Sur | 23 | NIOBG11 | This study |
| Mastigocladus strain Y-HR-9 | Boiling River, Yellowstone NP | 45 | BG11 | This study |
| Oscillatoria strain U-Stink* | Stinky Hot Springs, Utah | 35 | IOBG11 | |
| Oscillatoria strain DV-00-5* | Keane Mine Spring, Death Valley NP | 30 | BG11 | This study |
| Oscillatoria strain DV-00-7* | Keane Mine Spring, Death Valley NP | 30 | BG11 | This study |
| Oscillatoria strain 13-1* | Cirque Hot Spring, New Zealand | 45 | DTN | 13 |
| Oscillatoria strain PE* | New Zealand hot spring | 45 | DTN | 13 |
| Oscillatoria strain WHS-4* | Wilbur Hot Springs, Calif. | 37 | BG11 + 10% IO | This study |
| Oscillatoria strain NZ-TLS* | Lakeside Hot Spring, New Zealand | 35 | BG11 | 5 |
| Pseudanabaena strain CCMEE 5435 | Salton Sea, Calif. | 23 | IOBG11 | 39 |
| Microcoleus strain B-00-8 | Pond 4 evaporation pond, ESSA, Baja California Sur | 23 | IOBG11 | This study |
| Spirulina strain B-99-8* | Pond 2 evaporation pond, ESSA, Baja California Sur | 23 | NIOBG11 | This study |
Recipes for media per reference 7. NIOBG11, IOBG11 plus 0.5 g of ammonium sulfate per liter.
Strains that were not isolated in this study were obtained from the University of Oregon's Culture Collection of Microorganisms from Extreme Environments.
*, collection site permanently sulfidic (see text).
Abbreviations: ESSA, Exportadora de Sal SA; NP, National Park.
16S rRNA gene amplification and sequencing.
An approximately 1-kb fragment of the 16S rRNA gene spanning Escherichia coli nucleotide positions 359 to 1334 was amplified from genomic DNA of each laboratory strain as previously described (23). Amplified fragments were cleaned with the QIAquick PCR purification kit (Qiagen), and 5 μl of the purified product was added to a sequencing reaction mixture containing 10 pmol of sequencing primer and 4 μl of BigDye mix diluted 1:1 with buffer. Cycle sequencing conditions were 94°C for 10 s, 50°C for 5 s, and 60°C for 4 min for 25 cycles. Sequencing reactions were cleaned with a DyeEx 96 kit (Qiagen) and sequenced on an ABI 3700 apparatus. Both strands of the fragment were sequenced.
Phylogenetic reconstruction.
Nucleotide sequences were aligned as described by Miller and Castenholz (23). Cyanobacterial phylogenies were reconstructed using PAUP* version 4.0 (D. L. Swofford, Sinauer Associates, Sunderland, Mass.) by maximum likelihood, maximum parsimony, and neighbor-joining methods. Parameters in the likelihood model were estimated according to a general time-reversible model of nucleotide substitution with substitution rate heterogeneity among sites given by a discrete approximation of a gamma distribution. For both likelihood and parsimony methods, a heuristic search was performed using the TBR branch-swapping algorithm following obtainment of the starting tree by random stepwise sequence addition. The distance matrix used in the neighbor-joining method was estimated with a general time-reversible model of substitution. Phylogenies were bootstrapped with either 100 (likelihood) or 1,000 (parsimony and neighbor-joining) pseudoreplicates.
In vivo assay of PS II performance in the presence of sulfide.
The chlorophyll concentration of an exponentially growing culture was determined spectrophotometrically using the absorption at 665 nm of a methanol extract (made from a filtered 2-ml culture subsample) and an extinction coefficient of 0.075 ml μg−1. The remainder of the culture was then filtered and resuspended in fresh growth medium augmented with 25 mM HEPES buffer (pH 7.2) to a final concentration of 2.7 μg of chlorophyll a ml−1. For each data point, a sacrificial 3-ml aliquot of the suspension was delivered to a temperature-controlled cuvette chamber and dark adapted for 5 min at the strain's optimal temperature before addition, under weak illumination (<10 μmol of photons m−2 s−1) with a red safelight, of 100 μl from one of a series of working sulfide stock solutions. These working solutions were prepared by aliquoting known volumes of a Na2S-aqueous NaOH master stock (pH ∼12, with concentrations ranging between 90 and 100 mM) to serum vials containing nitrogen-sparged, HEPES-buffered growth medium. At the pH of these working solutions (pH 7.2), approximately 35% of the sulfide is expected to be in the form of H2S, which is a gas and therefore permeable across the cell membrane (17). Following 5 min of additional dark adaptation, chlorophyll fluorescence induction data were collected at 10-ms intervals with a pulse-amplitude-modulated fluorometer (DIVING-PAM; Walz) from which the fluorescence minimum FO and peak fluorescence FP were determined. Chlorophyll fluorescence was detected with a photodiode equipped with a 710-nm long-pass filter. The fluorescence minimum was measured under LED illumination with 0.15 μmol of photons m−2 s−1 of red light (peak emission at 650 nm) modulated at 0.6 kHz. Peak fluorescence was measured following illumination with an approximately 1-s pulse of saturating white light (ca. 4,000 μmol of photons m−2 s−1). After fluorescence induction, a 1-ml sample of the suspension was fixed in a scintillation vial containing 2 ml of 2% zinc acetate solution, and the sulfide concentration of the treatment was subsequently determined spectroscopically by the Pachmayr colorimetric assay (36).
PS II function at each sulfide concentration was estimated as the ratio of PS II variable fluorescence to the maximum fluorescence of the treatment, (FP − FO)/FP. As is frequently observed for dosage-mortality curves, relative PS II function (expressed as the percent reduction relative to the sulfide-free control) at increasing sulfide concentrations could generally be described by a cumulative normal distribution. Therefore, in order to estimate PS II performance in the presence of sulfide, relative PS II function was probit transformed (into standardized normal equivalent deviates) and linearly regressed on the logarithmically transformed sulfide concentration (SPSS version 8.0). The exception to this approach was the data set for Oscillatoria amphigranulata strain PE, the most sulfide-tolerant strain. In this case, a better model was obtained by linearly regressing untransformed PS II data on sulfide concentration. The means and confidence intervals of the sulfide concentration reducing photochemical efficiency by 50% were estimated by inverse prediction. Data for each strain were pooled from assays of at least two independent laboratory cultures.
Phylogenetic comparative methods.
Computer simulation and experimental evolution studies consistently indicate the need to consider the statistical nonindependence of physiological data collected from organisms that are related to each other by a branching phylogeny (22, 26). For correlation analyses, generalized least-squares models that take into account the phylogenetic structure of the data (22) were developed using the PGLS Relationships program in the software package compare version 4.4 (distributed by E. P. Martins; http://compare.bio.indiana.edu). To specify the elements in the variance-covariance matrix of the error term, branch length data from a 16S rRNA phylogeny were transformed into units of expected phenotypic similarity by modeling phenotypic evolution along the phylogeny as an Ornstein-Uhlenbeck process. In this model, the decrease in expected similarity as a function of evolutionary distance depends on the strength of the restraining force, alpha. When alpha is equal to zero, the relationship is linear. As alpha increases, phenotypic similarity between organisms decays exponentially more rapidly, so that at very high values the data can be considered independent of phylogeny (i.e., the use of standard least-squares regression is appropriate). Virtually any model of the microevolutionary process can be described by either the linear or the exponential model (16). For each pair of variables analyzed, regressions were run for different values of alpha, and the generalized least-square (GLS) model was optimized at the value of alpha that maximized the likelihood of the model.
The degree of phylogenetic constraint on trait evolution was estimated using the restricted maximum likelihood form of Lynch's phylogenetic mixed model (20) implemented in compare. This method partitions the phenotypic variance in a data set into phylogenetically heritable and ahistorical components, respectively. A high phylogenetic heritability, or resemblance among relatives, is indicative of constraints on phenotypic evolution. In contrast, a lack of constraint suggests that phenotypes are free to change in response to other factors that are not strictly inherited, such as environmental variation. The expected phenotypic similarity among strains due to phylogenetic heritability was modeled as a Brownian motion process by which change in the mean phenotype over time is independent and normally distributed with variance proportional to the branch lengths of a phylogeny in units of molecular sequence divergence. The parameter estimates reported were the averages obtained from three searches along the likelihood surface with different starting conditions.
Nucleotide sequence accession numbers.
Sequence data have been deposited in GenBank under accession numbers AY426541 to AY426549.
RESULTS AND DISCUSSION
Strain diversity.
Our sample (Table 1) consisted of several strains that had been isolated for this study from chemically, physically, and geographically diverse sulfidic microbial mat habitats as well as strains obtained from the Culture Collection of Microorganisms from Extreme Environments at the University of Oregon. It included strains of O. amphigranulata (strains 13-1 and PE), which had been reported to be inhibited by sulfide on the donor side of PS II (11). It also contained isolates from the same site as strains for which the acceptor side had previously been inferred to be the target of sulfide toxicity (Stinky Hot Springs, Utah, and Wilbur Hot Springs, Calif. [10]).
It appears that the water column overlaying most collection sites was permanently sulfidic. Previously reported estimates of sulfide include 40 μM (pH 8.5) at Cirque Hot Springs (13), 1,200 μM (pH 6.1) at Stinky Hot Springs (10), and 2,200 μM (pH 7.1) at Wilbur Hot Springs (10). We estimated the sulfide concentration (mean ± standard error) in the water columns of our site at the Pond 2 evaporation pond of a Mexican salt works to be 73 ± 6.5 μM (pH 9.3). At Keane Mine Spring in Death Valley National Park, sulfide was estimated to be 145 ± 2.3 μM (pH 7.0) at the site from which Oscillatoria strain DV-00-7 was isolated and 87 ± 3.1 μM (pH 7.0) at the location of the collection from which Oscillatoria strain DV-00-5 was obtained. By contrast, the water column of the Pond 4 evaporation pond, from which Microcoleus strain B-00-8 was isolated, was not permanently sulfidic. Instead, the Microcoleus mat layer (approximately 600 to 800 μm below the mat surface) experienced extreme diel fluctuations in sulfide exposure. This was due to movement of the vertical position of the sulfide-oxygen interface within the mat as a result of light-driven changes in the relative rates of photosynthetic oxygen generation and sulfate reduction over the course of a day. We estimated the mean in situ concentration of H2S in this layer to be 10 ± 2.9 μM for an entire diel cycle (unpublished data). With an average pH of 7.2 ± 0.09 during the period in which sulfide accumulates in the Microcoleus layer, this corresponds to a total mean soluble sulfide concentration of 29 μM. Finally, sulfide was not detected in water samples from Boiling River (our data and reference 6), a fast-flowing hot spring from which we isolated a strain of the heterocystous cyanobacterium Mastigocladus sp.
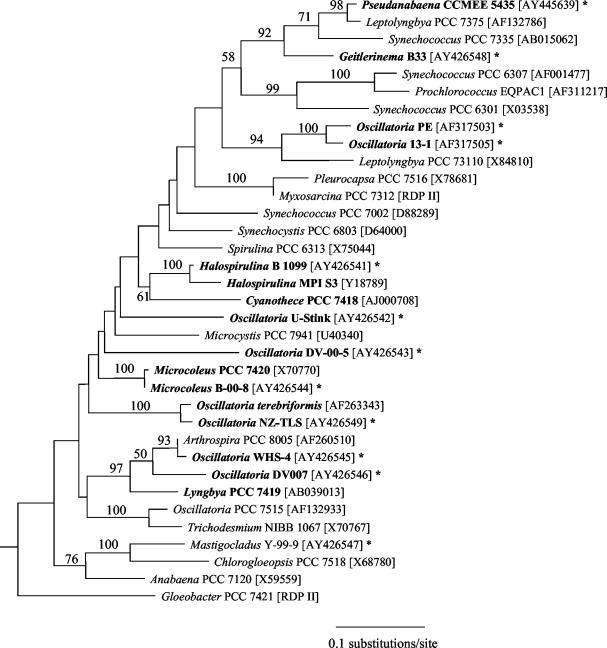
Cyanobacterial molecular phylogenies were reconstructed in order to evaluate whether strains from sulfidic habitats tend to cluster together phylogenetically or, alternatively, whether representatives from many lineages are capable of living in the presence of sulfide. Of particular interest was whether morphologically similar strains from extremely sulfide-rich habitats (e.g., Oscillatoria strains WHS-4 and U-Stink) were closely related. An approximately 1-kb fragment of the 16S rRNA gene was amplified and subsequently sequenced from the genomic DNA of each strain. These sequence data were aligned with additional cyanobacterial sequences, and phylogenies were reconstructed using maximum likelihood (Fig. 1), maximum parsimony, and neighbor-joining methods. Strains were dispersed throughout the phylogeny regardless of the method used, and strains U-Stink and WHS-4 were not closely related. However, there were also particular cases of phylogenetic clustering. For example, Oscillatoria strain WHS-4 and Oscillatoria strain DV-00-7 formed a strongly supported clade along with Arthrospira strain PCC 8005 and Lyngbya strain PCC 7419 in all phylogenies. In addition, Pseudanabaena strain CCMEE 5435 and Geitlerinema strain B33 clustered with Leptolyngbya strain PCC 7375 and Synechococcus strain PCC 7335 in all trees. Also, strains from New Zealand hot springs corresponding to the botanical species O. amphigranulata (strains 13-1 and PE) are sister taxa, as has been previously reported (24). Nevertheless, the general conclusion to be drawn from this phylogenetic framework is that many different evolutionary lineages within the cyanobacteria are capable of living in sulfidic habitats.
FIG. 1.
Maximum likelihood phylogeny for cyanobacteria inferred from an alignment of ca. 1 kb of the 16S rRNA gene. Strains known to have been isolated from habitats that are either periodically or permanently sulfidic are in bold. Strains assayed for PS II sulfide tolerance are indicated by an asterisk. Values at internodes indicate the number of times that the taxa to the right of the node formed a clade out of 100 bootstrap pseudoreplicated data sets. GenBank accession numbers and sequences retrieved from the Ribosomal Database Project II (RDP II) are indicated in brackets.
Effects of sulfide on PS II fluorescence.
In a dark-adapted sample in the absence of sulfide, chlorophyll-variable fluorescence induction following a brief pulse of saturating light reflects photosynthetic electron transport from a fluorescence minimum (FO), when all PS II reaction centers are open, to a maximum (FP), when most PS II are closed and most QA molecules, the primary electron acceptors in PS II, are reduced (4, 19). The addition of sulfide to dark-adapted suspensions of exponentially growing cells had a characteristic effect on all strains, i.e., the quenching of variable fluorescence (FV = FP − FO) due to a decrease in peak fluorescence FP and a small increase in FO at higher sulfide concentrations (Fig. 2). However, the rate at which the variable fluorescence was quenched varied greatly among strains and was indicative of differences in PS II efficiency in the presence of sulfide. In order to quantify these differences, we compared FV/FP as a function of sulfide among strains. In plants this ratio is a well-established index of PS II quantum efficiency (1). Because a large fraction of FO in cyanobacteria is contributed by phycobiliproteins rather than PS II, this ratio is not strictly equivalent to photochemical efficiency in these organisms. Still, it has been repeatedly shown to be a reliable indicator of PS II function in cyanobacteria (reviewed in reference 4). For each strain we determined the effect of sulfide concentration on FV/FP relative to that of its sulfide-free control (Fig. 3). Following transformation of these data, we used linear regression models and inverse prediction to estimate the hydrogen sulfide concentrations required to inhibit PS II activity by 50% (Table 2). There was an approximately 50-fold difference in PS II resistance between the least and most sensitive strains tested.
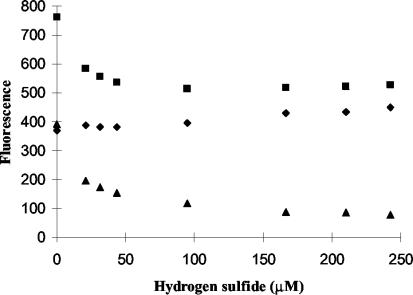
FIG. 2.
Typical profile of effects of hydrogen sulfide on chlorophyll fluorescence of dark-adapted cells following a saturating pulse of light. Sulfide primarily reduces PS II variable fluorescence (triangles) by lowering peak fluorescence (squares), although there is also a rise in the fluorescence minimum FO (diamonds) at higher sulfide concentrations. Data shown are for Oscillatoria strain U-Stink.
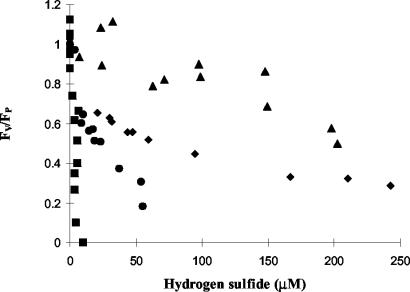
FIG. 3.
Cyanobacterial PS II function (FV/FP) relative to that of the sulfide-free control as a function of hydrogen sulfide dose. The range in response is illustrated with profiles for four strains: Oscillatoria strain 13-1 (squares), Oscillatoria strain DV-00-7 (circles), Oscillatoria strain U-Stink (diamonds), and Oscillatoria strain PE (triangles).
TABLE 2.
Hydrogen sulfide dose required to reduce PS II function by 50%
| Strain | [H2S] (μM) producing 50% inhibition [mean (95% CI)a] |
|---|---|
| Geitlerinema sp. strain B-33 | 35.4 (30.6, 40.6) |
| Mastigocladus sp. strain Y-HR-9 | 4.6 (1.1, 14.8) |
| Oscillatoria sp. strain U-Stink | 64.9 (54.7, 77.4) |
| Oscillatoria sp. strain DV-00-5 | 42.0 (33.3, 56.6) |
| Oscillatoria sp. strain DV-00-7 | 20.2 (15.6, 26.7) |
| Oscillatoria sp. strain 13-1 | 3.7 (2.1, 5.6) |
| Oscillatoria sp. strain PE | 204.5 (148.3, 260.7) |
| Oscillatoria sp. strain WHS-4 | 107.3 (65.4, 197.7) |
| Oscillatoria boryana NZ-TLS | 30.6 (23.5, 40.0) |
| Pseudanabaena sp. strain CCMEE 5435 | 18.8 (2.1, 97.3) |
| Microcoleus strain B-00-8 | 31.9 (17.4, 57.0) |
| Spirulina sp. strain B-99-8 | 67.0 (42.7, 113.8) |
CI, confidence interval.
The above observation that sulfide quenches variable fluorescence in all strains by a substantial decrease in FP and a smaller increase in FO (Fig. 2) suggests that oxygenic photosynthesis is poisoned by a similar mechanism regardless of either a strain's evolutionary history or its degree of sulfide tolerance (Fig. 3; Table 2). This pattern of variable fluorescence quenching has been observed in some, but not all, previous investigations of the effects of sulfide on chlorophyll fluorescence (11, 27). It has also been observed after treatment with either a low (<10 mM) concentration of hydroxylamine (11, 27, 31) or short-term heat shock (31, 33), both of which are known to disrupt electron transfer from the water-splitting complex to the PS II reaction center (8, 18). In all three cases the decrease in peak fluorescence upon illumination can be interpreted as the reduction in the proportion of reduced QA due to electron limitation from the donor side of PS II.
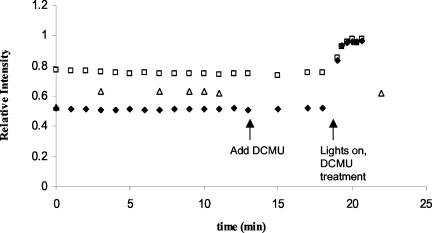
If sulfide does indeed inhibit PS II on the donor side and consequently reduces electron flow to the primary acceptor QA, then the fluorescence induction curves of sulfide-treated cells should exhibit other similarities with those following treatment with heat and hydroxylamine. For example, the rate of the fluorescence rise to FP should slow with increasing sulfide concentrations (31, 33), which was the case (Fig. 4). In addition, if there is electron limitation from the donor side, the fluorescence yield should recover the FP level of the sulfide-free control upon addition of dichlorophenyldimethyl urea (DCMU), which blocks photosynthetic electron flow between QA and QB and thereby leads to the accumulation of reduced QA and closed PS II reaction centers (35). We demonstrated this to be the case for exponentially growing cells of Oscillatoria strain U-Stink with our in vivo fluorescence assay protocol. Following addition of hydrogen sulfide to a final concentration of approximately 20 μM, peak fluorescence of dark-adapted cells was approximately 75% of FP in the sulfide-free control (Fig. 5). As expected, this loss of fluorescence was recovered following DCMU addition (to a final concentration of 7 μM) under low white light illumination (approximately 10 μmol of photons m−2 s−1). This recovery was not as rapid as was observed in the absence of sulfide (27) and can be explained by electron limitation from the donor side. In a parallel assay, sulfide and low light in the absence of DCMU were not sufficient to recover FP (Fig. 5). Rather, photosynthetic electron transport closes a fraction of, but not all, PS II reaction centers under these conditions, producing a steady-state fluorescence that is higher than the fluorescence minimum in the dark but lower than FP. It therefore appears likely that sulfide inhibits oxygenic photosynthesis at the donor side of PS II and that the mode of toxicity possibly involves the water-splitting complex. Demonstration of the existence of a K step (33) during fluorescence induction, which would require time resolution on the order of 10 μs, would definitively confirm the latter.
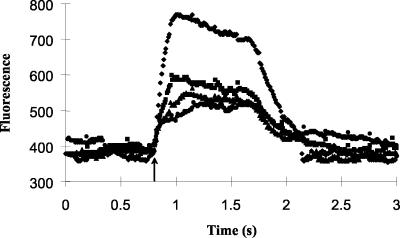
FIG. 4.
Effect of hydrogen sulfide on fluorescence induction following exposure of dark-adapted cells of Oscillatoria strain U-Stink to an approximately 1-s saturating pulse of light (arrow) at a hydrogen sulfide concentration of 0 (diamonds), 21 (squares), 43 (triangles), or 94 μM (circles).
FIG. 5.
Recovery of fluorescence yield in the light by the addition of DCMU. Peak fluorescence of dark-adapted cells of Oscillatoria strain U-Stink treated with 18 μM hydrogen sulfide (squares) decreased relative to FP of the sulfide-free control (with FP of the control defined as having a relative intensity equal to 1). After DCMU addition and under white light, all PS II reaction centers were closed, leading to a recovery of maximal fluorescence yield under low light (diamonds). This recovery was not observed under steady-state illumination if cells were assayed in the presence of sulfide but the absence of DCMU (triangles).
Degree of tolerance is correlated with environmental sulfide levels.
We developed generalized least-squares models to evaluate whether the substantial variation in sulfide tolerance observed among strains could be explained in part by differences in sulfide concentration across environments. A positive correlation would be evidence that the cyanobacterial photosynthetic apparatus has some capacity to track environmental sulfide levels. In order to take into account the potential statistical nonindependence of the physiological data due to the underlying phylogenetic structure, the elements in the variance-covariance matrix of the error term of the models were specified using branch length information from the 16S rRNA gene maximum likelihood phylogeny presented above (Fig. 1). In order to convert this phylogenetic information into the expected phenotypic similarity between two strains (i.e., their covariance, or statistical dependence, in the matrix), phenotypic evolution along the phylogeny was modeled as an Ornstein-Uhlenbeck process with the program compare (16). In this model, the shape of the curve describing the decay of phenotypic similarity as a function of sequence divergence depends on the value of the restraining force parameter alpha, which can be estimated from the data by maximum likelihood. When alpha is equal to zero, phenotypes evolve by a Brownian motion process, and similarity decreases linearly with increasing evolutionary distance (i.e., the expected phenotypic covariance between strains is simply the sum of the length of the branches on the phylogeny that they shared prior to their divergence from a common ancestor). As alpha gets larger, phenotypic similarity decays at an exponentially faster rate until the expected phenotypic covariance between two strains approaches zero, at which point the model becomes equivalent to standard least-squares regression.
Estimates of environmental sulfide were available for the collection sites of 9 of the 12 strains tested (see above). The dose required to reduce PS II activity by 50% (Table 2) was regressed on both total environmental sulfide and H2S concentration. The latter is thought to be largely responsible for sulfide toxicity, because it is the species that is permeable across the cell membrane (17). We calculated H2S concentration using pH estimates for each site and the acid-base equilibrium equation of Broderius and Smith (2). Both models were statistically significant, as indicated by their positive regression slopes (Table 3), and they also indicated a positive correlation between sulfide tolerance and environmental sulfide. Both total sulfide and H2S explained a substantial fraction of the variation in tolerance among strains (percent R2 equal to 69 and 51%, respectively).
TABLE 3.
Summary statistics for GLS models
| Model | Regression slopea (β1 ± SE) | Correlation (r) | Alpha |
|---|---|---|---|
| [H2S]50% on [S2−]env (μM) | 0.10 ± 0.026* | 0.83 | 8.2 |
| [H2S]50% on [H2S]env (μM) | 0.16 ± 0.060* | 0.72 | 15.5 |
| [H2S]50% on FI rise (ΔF s−1) | 0.02 ± 0.188 | 0.03 | 15.5 |
| [H2S]50% on FI time (s) | −41.1 ± 382.83 | −0.04 | 15.5 |
*, slope was significantly different from zero.
The correlation coefficients of the GLS models were close to those obtained by standard least-squares regression (0.82 for the total sulfide model, 0.71 for the H2S model), as expected given the high estimated values of alpha (Table 3). This observation that phylogenetic correlations are quickly erased from the phenotypic data suggests that the evolution of the cyanobacterial photosynthetic apparatus in response to changes in environmental sulfide is not constrained by prior evolutionary history. That is, tolerance may be gained or lost relatively rapidly in response to environmental variation in sulfide levels. This lack of constraint may help explain the dispersed pattern of sulfide tolerance throughout the cyanobacterial phylogeny. To further test that the degree of phylogenetic constraint on the evolution of PS II tolerance was low, we analyzed the tolerance data for all 12 strains with the restricted maximum likelihood form of the phylogenetic mixed model (20) implemented in compare. This application of quantitative genetic theory to the study of interspecific phenotypic evolution along a phylogeny partitions the phenotypic variance in a data set into phylogenetically heritable and ahistorical components, respectively. The former is the proportion of the total phenotypic variance that can be explained by phylogeny (i.e., by resemblance of relatives) and therefore takes on values between 0 and 1. Phylogenetic heritability was estimated to be 0 for each of three searches along the likelihood surface (log likelihood values ranged from 262.99 to 263.04). Low values of phylogenetic heritability suggest that phenotypes are not constrained by phylogeny and are free to change in response to other factors that are not strictly inherited, such as environmental variation.
Fluorescence induction kinetics are phylogenetically constrained.
Strasser et al. (34) were the first to demonstrate heterogeneity in fluorescence induction kinetics among cyanobacteria. In particular, they reported variation among strains in the time required to attain maximal fluorescence. Given that sulfide slows the fluorescence rise (Fig. 3), we used GLS models to test whether inherent differences in photosynthetic electron transport under optimal conditions (i.e., in sulfide-free controls) are associated with variation among strains in sulfide tolerance. The sulfide dose required to reduce PS II activity by 50% was regressed on two variables describing the fluorescence induction curve: the initial slope of the fluorescence rise (FI rise) and the time required to reach peak fluorescence (FI time). Neither was significantly associated with sulfide tolerance (Table 3). A phylogenetic mixed model further suggested that fluorescence induction time is primarily a product of phylogenetic, rather than ecological, history. Estimates of its phylogenetic heritability were very high, ranging between 0.859 and 0.862. This evidence for phylogenetic constraints on the evolution of the induction curve is best exemplified by Oscillatoria strains 13-1 and PE. Although they represent the extremes in sulfide tolerance observed in this study, the fluorescence induction times (0.68 and 0.44 s, respectively) of both strains are slower than those of all other strains (which range between 0.17 and 0.33 s).
Evolutionary and ecological implications.
Although sulfide is lethal in small doses for many strains (6, 10, 15), cyanobacteria are often found together with anoxygenic phototrophic bacteria in diverse environments where light and sulfide cooccur (29). Several lines of evidence support the conclusion that sulfide tolerance is a dynamic trait that can be gained (or lost) in this group in response to changes in environmental sulfide. First, strains from sulfidic environments exhibit great variation in PS II tolerance (Table 2) that we propose can be explained in part by the adaptive tracking of sulfide levels in the environment (Table 3). Additionally, the undetectable phylogenetic heritability of sulfide tolerance indicates that a strain's phenotype provides no information about that of its relatives, as expected for a trait that evolves in response to environmental change without being constrained by prior evolutionary and ecological history (20). By comparison, fluorescence induction, a sensitive indicator of photosynthetic electron transport, is under great phylogenetic constraint. Finally, strains from sulfidic habitats would not necessarily be expected to cluster together phylogenetically if changes in tolerance across different lineages to a large degree reflect relatively recent differences in the strength of environmental selection. It is therefore not surprising that sulfide-tolerant cyanobacteria are a genetically diverse group that has adapted to geographically distant and otherwise physically and chemically distinct sulfidic habitats (Fig. 1; Table 1).
While a substantial fraction of the phenotypic variation among strains in PS II tolerance may be explained by differences in environmental sulfide concentration, a lack of association in individual cases is potentially indicative of the presence of an alternative tolerance mechanism. In particular, several cyanobacteria from sulfidic habitats have the facultative capacity for PS I-driven anoxygenic photosynthesis with sulfide as an electron donor. Although many of these also have sulfide-tolerant PS II, Cohen et al. (10) have shown, for example, that Oscillatoria limnetica from sulfide-rich Solar Lake is tolerant of sulfide in spite of a sensitive PS II. This is because anoxygenic photosynthesis is induced at very low sulfide concentrations.
A qualitative survey of the phylogenetic distribution of anoxygenic photosynthesis suggests that it, like PS II tolerance, is distributed throughout the cyanobacterial phylogeny (Fig. 1). For example, Oscillatoria strains PE, 13-1, NZ-TLS, and U-Stink all are capable of anoxygenic photosynthesis. Cyanothece strain PCC 7418 is as well, but apparently only as a detoxification mechanism, as it cannot grow in the presence of sulfide (3, 15). The ability to utilize sulfide as an electron donor in photosynthesis also appears to be able to evolve rapidly. For example, Oscillatoria terebriformis from Hunter's Hot Springs, Oreg., from which the strain used for sequence analysis was isolated (Fig. 1), differed markedly from its close relative, strain NZ-TLS, in that it was unable to perform anoxygenic photosynthesis (6). In addition, O. terebriformis from a nearby spring containing moderate levels of primary sulfide did show some capacity for anoxygenic photosynthesis. This process depends upon the activity of sulfide-quinone reductase (SQR), a flavoprotein that transfers electrons from sulfide to the quinone pool on the donor side of PS I (32). SQR may play a role in other functions, such as anaerobic respiration (28), and some cyanobacteria that cannot perform anoxygenic photosynthesis, like Anabaena strain PCC 7120, have copies of sqr (3). Among other sequenced cyanobacterial genomes, BLAST searches revealed that only the hot spring cyanobacterium Synechococcus strain BP-1has a copy of this gene, while the planktonic Prochlorococcus strains MED-4 and MIT-9313, Synechococcus strain WH8102, Trichodesmium strain IMS101, and Nostoc strain PCC 73102 do not (data not shown). Synechocystis strain PCC 6803 has a distant homolog with unknown function. This observation raises the question of whether an ancestral sqr has been selectively maintained in cyanobacterial lineages with histories of sulfide exposure (and lost in others) or, alternatively, whether the ability to perform anoxygenic photosynthesis has been acquired in genetically divergent lineages with shared ecologies by horizontal gene transfer. One could discriminate between these possibilities by testing whether a genealogy reconstructed with a larger sqr data set than is currently available exhibits significant deviation from topological congruence with a phylogeny inferred with the 16S rRNA gene.
Our conclusion that the target of sulfide toxicity is the donor side of PS II agrees with two previous investigations (11, 27) and draws additional support from the similarities between sulfide exposure and treatment with either hydroxylamine or heat, both known to disrupt the oxygen-evolving complex (31, 33). It differs with another study (10), however, which had inferred that sulfide inhibited oxygenic photosynthesis at the acceptor side. This was based on observed sulfide-induced increases in variable fluorescence analogous to the effects of DCMU. Two of the strains in our study, Oscillatoria strains WHS-4 and U-Stink, were cloned by direct isolation from the same sites as two of the strains used by Cohen et al. (10). Like every other strain assayed, both showed decreases, rather than increases, in variable fluorescence following sulfide exposure. Although the explanation for the anomalous experimental results presented in reference 10 is not clear, the balance of evidence supports the donor side of PS II as the site of sulfide toxicity.
Acknowledgments
We thank two anonymous reviewers for their comments and suggestions.
S.R.M. was supported by a National Research Council research associateship award.
REFERENCES
- 1.Björkman, O., and B. Demmig. 1987. Photon yield of O2 evolution and chlorophyll fluorescence characteristics at 77K among vascular plants of diverse origins. Planta 170:489-504. [DOI] [PubMed] [Google Scholar]
- 2.Broderius, S. J., and L. L. Smith. 1977. Direct determination and calculation of aqueous hydrogen sulfide. Anal. Chem. 49:424-428. [Google Scholar]
- 3.Bronstein, M., M. Schütz, G. Hauska, E. Padan, and Y. Shahak. 2000. Cyanobacterial sulfide-quinone reductase: cloning and heterologous expression. J. Bacteriol. 182:3336-3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Campbell, D., V. Hurry, A. K. Clarke, P. Gustafsson, and G. Öquist. 1998. Chlorophyll fluorescence analysis of cyanobacterial photosynthesis and acclimation. Microbiol. Mol. Biol. Rev. 62:667-683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Castenholz, R. W. 1976. The effect of sulfide on the blue-green algae of hot springs. I. New Zealand and Iceland. J. Phycol. 12:54-68. [Google Scholar]
- 6.Castenholz, R. W. 1977. The effect of sulfide on the blue-green algae of hot springs. II. Yellowstone National Park. Microb. Ecol. 3:79-105. [DOI] [PubMed] [Google Scholar]
- 7.Castenholz, R. W. 1988. Culturing methods for cyanobacteria. Methods Enzymol. 167:68-93. [Google Scholar]
- 8.Cheniae, G. M., and I. F. Martin. 1970. Sites of function of manganese within photosystem II: roles in O2 evolution and system II. Biochim. Biophys. Acta 357:127-143. [DOI] [PubMed] [Google Scholar]
- 9.Cohen, Y., B. B. Jørgensen, E. Padan, and M. Shilo. 1975. Sulfide-dependent anoxygenic photosynthesis in the cyanobacterium Oscillatoria limnetica. Nature 257:489-491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cohen, Y., B. B. Jørgensen, N. P. Revsbech, and R. Poplawski. 1986. Adaptation to hydrogen sulfide of oxygenic and anoxygenic photosynthesis among cyanobacteria. Appl. Environ. Microbiol. 51:398-407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dodds, W. K., and R. W. Castenholz. 1990. Sulfide and pH effects on variable fluorescence of photosystem II in two strains of the cyanobacterium Oscillatoria amphigranulata. Photosynth. Res. 24:265-271. [DOI] [PubMed] [Google Scholar]
- 12.Evans, C. L. 1967. The toxicity of hydrogen sulphide and other sulphides. Q. J. Exp. Physiol. 52:231-248. [DOI] [PubMed] [Google Scholar]
- 13.Garcia-Pichel, F., and R. W. Castenholz. 1990. Comparative anoxygenic photosynthetic capacity in 7 strains of a thermophilic cyanobacterium. Arch. Microbiol. 153:344-351. [Google Scholar]
- 14.Garcia-Pichel, F., U. Nübel, and G. Muyzer. 1998. The phylogeny of unicellular, extremely halotolerant cyanobacteria. Arch. Microbiol. 169:469-482. [DOI] [PubMed] [Google Scholar]
- 15.Garlick, S., A. Oren, and E. Padan. 1977. Occurrence of facultative anoxygenic photosynthesis among filamentous and unicellular cyanobacteria. J. Bacteriol. 129:623-629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hansen, T. F., and E. P. Martins. 1996. Translating between microevolutionary process and macroevolutionary patterns: the correlation structure of interspecific data. Evolution 50:1404-1417. [DOI] [PubMed] [Google Scholar]
- 17.Howsley, R., and H. W. Pearson. 1979. pH dependent sulfide toxicity to oxygenic photosynthesis in cyanobacteria. FEMS Microbiol. 6:287-292. [Google Scholar]
- 18.Katoh, S., I. Ikegami, and A. Takamiya. 1970. Effects of hydroxylamine on electron transport system in chloroplasts. Arch. Biochem. Biophys. 141:207-218. [DOI] [PubMed] [Google Scholar]
- 19.Lazár, D. 1999. Chlorophyll a fluorescence induction. Biochim. Biophys. Acta 1412:1-28. [DOI] [PubMed] [Google Scholar]
- 20.Lynch, M. 1991. Methods for the analysis of comparative data in evolutionary biology. Evolution 45:1065-1080. [DOI] [PubMed] [Google Scholar]
- 21.Martins, E. P., and T. F. Hansen. 1997. Phylogenies and the comparative method: a general approach to incorporating phylogenetic information into the analysis of interspecific data. Am. Nat. 149:646-667. [Google Scholar]
- 22.Martins, E. P., A. F. Diniz-Filho, and E. A. Housworth. 2002. Adaptive constraints and the phylogenetic comparative method: a computer simulation test. Evolution 56:1-13. [PubMed] [Google Scholar]
- 23.Miller, S. R., and R. W. Castenholz. 2000. Evolution of thermotolerance in hot spring cyanobacteria of the genus Synechococcus. Appl. Environ. Microbiol. 66:4222-4229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miller, S. R., and R. W. Castenholz. 2001. Ecological physiology of Synechococcus sp. strain SH-94-5, a naturally occurring cyanobacterium deficient in nitrate assimilation. Appl. Environ. Microbiol. 67:3002-3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moore, L., G. Rocap, and S. Chisholm. 1998. Physiology and molecular phylogeny of coexisting Prochlorococcus ecotypes. Nature 393:464-467. [DOI] [PubMed] [Google Scholar]
- 26.Oakley, T. H., and C. W. Cunningham. 2000. Independent contrasts succeed where ancestor reconstruction fails in a known bacteriophage phylogeny. Evolution 54:397-405. [DOI] [PubMed] [Google Scholar]
- 27.Oren, A., E. Padan, and S. Malkin. 1979. Sulfide inhibition of photosystem II in cyanobacteria (blue-green algae) and tobacco chloroplasts. Biochim. Biophys. Acta 546:270-279. [DOI] [PubMed] [Google Scholar]
- 28.Oren, A., and M. Shilo. 1979. Anaerobic heterotrophic dark metabolism in the cyanobacterium Oscillatoria limnetica: sulfur respiration and lactate fermentation. Arch. Microbiol. 122:77-84. [Google Scholar]
- 29.Padan, E. 1979. Impact of facultative anaerobic photoautotrophic metabolism on ecology of cyanobacteria (blue-green algae). Adv. Microb. Ecol. 3:1-48. [Google Scholar]
- 30.Schreiber, U. 1986. Detection of rapid induction kinetics with a new type of high-frequency modulated chlorophyll fluorometer. Photosynth. Res. 9:261-272. [DOI] [PubMed] [Google Scholar]
- 31.Schreiber, U., and C. Neubauer. 1987. The polyphasic rise of chlorophyll fluorescence upon onset of strong continuous illumination. II. Partial control by the photosystem II donor side and possible ways of interpretation. Z. Naturforsch. Teil C 42:1255-1264. [Google Scholar]
- 32.Shahak, Y., B. Arieli, B. Binder, and E. Padan. 1987. Sulfide-dependent photosynthetic electron flow coupled to proton translocation in thylakoids of the cyanobacterium Oscillatoria limnetica. Arch. Biochem. Biophys. 259:605-615. [DOI] [PubMed] [Google Scholar]
- 33.Srivastava, A., B. Guisse, H. Greppin, and R. J. Strasser. 1997. Regulation of antenna structure and electron transport in photosystem II of Pisum sativum under elevated temperature probed by the fast polyphasic chlorophyll a fluorescence transient: OKJIP. Biochim. Biophys. Acta 1320:95-106. [Google Scholar]
- 34.Strasser, R. J., A. Srivastava, and Govindjee. 1995. Polyphasic chlorophyll a fluorescence transient in plants and cyanobacteria. Photochem. Photobiol. 61:32-42. [Google Scholar]
- 35.Trebst, A. 1980. Inhibitors in electron flow: tools for the functional and structural localization of carriers and energy conservation sites. Methods Enzymol. 69:675-715. [Google Scholar]
- 36.Trüper, H. G., and H. G. Schlegel. 1964. Sulfur metabolism in Thiorhodaceae. I. Qualitative measurements in growing cultures of Chromatium okenii. Antonie Leeuwenhoek 30:225-238. [DOI] [PubMed] [Google Scholar]
- 37.Turner, S. 1997. Molecular systematics of oxygenic photosynthetic bacteria. Plant Syst. E vol. Suppl. 11:13-52. [Google Scholar]
- 38.Whitton, B. A., and M. Potts (ed.). 2000. The ecology of cyanobacteria: their diversity in time and space. Kluwer, Dordrecht, The Netherlands.
- 39.Wood, A. M., S. R. Miller, W. K. W. Li, and R. W. Castenholz. 2002. Preliminary studies of cyanobacteria, picoplankton, and virioplankton in the Salton Sea with special attention to phylogenetic diversity among eight strains of filamentous cyanobacteria. Hydrobiologia 473:77-92. [Google Scholar]