Abstract
Controller (C) proteins regulate the expression of restriction–modification (RM) genes in a wide variety of RM systems. However, the RM system Esp1396I is of particular interest as the C protein regulates both the restriction endonuclease (R) gene and the methyltransferase (M) gene. The mechanism of this finely tuned genetic switch depends on differential binding affinities for the promoters controlling the R and M genes, which in turn depends on differential DNA sequence recognition and the ability to recognize dual symmetries. We report here the crystal structure of the C protein bound to the M promoter, and compare the binding affinities for each operator sequence by surface plasmon resonance. Comparison of the structure of the transcriptional repression complex at the M promoter with that of the transcriptional activation complex at the R promoter shows how subtle changes in protein–DNA interactions, underpinned by small conformational changes in the protein, can explain the molecular basis of differential regulation of gene expression.
INTRODUCTION
Restriction–modification (RM) systems protect bacteria from invasion by bacteriophage and may play a role in restricting the flow of genetic information in bacterial populations (1, 2). RM systems encode a restriction endonuclease (ENase) and a DNA methyltransferase (MTase) that recognize the same DNA sequence. The DNA MTase protects the host DNA from cleavage by the associated restriction enzyme, while digesting (restricting) foreign DNA (2). There are a variety of control mechanisms that ensure the correct temporal expression of RM genes, to ensure that the host DNA is methylated prior to exposure to the ENase.
The best known of these mechanisms employs a ‘controller’ (C) protein encoded by a gene downstream of its own promoter, and co-transcribed with the restriction endonuclease (R) gene as a single transcriptional unit (3–7). The C protein binds at various sites within the C/R promoter to regulate transcription of its own gene and the associated endonuclease gene (8). The time-dependence of the activity of this switch has been demonstrated in vitro, and ENase expression was shown to be delayed with respect to the MTase when the C protein is expressed in a new host in vivo (9,10).
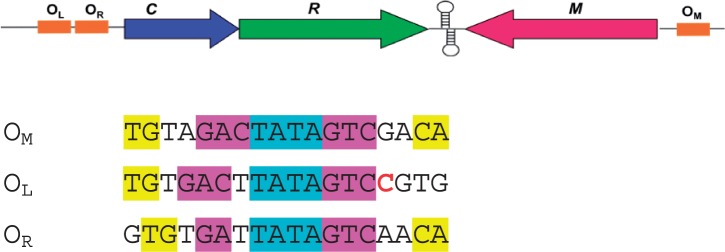
In typical C-protein systems, the operator sequence at the C/R promoter has two operator sites (denoted OL and OR) (11,12). OL is distal to the gene and has a high affinity for a C-protein dimer. When bound at this site, the σ subunit of RNA polymerase is recruited and both the C and R genes are switched on. OR is a much weaker binding site proximal to the gene; however, when a C-protein dimer is bound to OL then the affinity for OR is greatly increased and at high protein concentrations, this site is occupied and the gene is down-regulated (12–14). In the RM system Esp1396I, the C protein also represses the constitutively expressed methyltransferase (M) gene by binding as a dimer to the promoter that overlaps the transcriptional start site of this gene (15). The C/R genes and the M gene in this system are transcribed convergently from different promoters (See Figure 1).
Figure 1.
Regulation of restriction (R) and modification (M) genes by C.Esp1396I. The upper figure shows convergent gene organization and the location of the three operator sites: OM, OL and OR. The sequences of these sites are shown below, with the specific recognition motifs shown in magenta and yellow, and the central TATA in cyan. The C implicated in a possible interaction with D34 is indicated in red. Adapted from Bogdanova et al. (15).
Analysis of C-protein binding sites in a wide variety of RM systems suggested a repeating quasi-symmetrical consensus sequence consisting of two sets of inverted repeats or ‘C-boxes’ [GACT(N3)AGTC(N4)GACT(N3)AGTC] upstream of the C/R genes (6,8,12). However, the degree of sequence homology between species is moderate and the internal symmetry within and between ‘C-boxes’ is also weak in most C/R promoters (16). Moreover, the proposed 3-bp ‘spacers’ within the left and right operator sequences are also largely conserved between species, the consensus sequence being TAT. However, subsequent structural studies of C-protein–DNA complexes suggest that the binding site may be better described as a 4-bp alternating pyrimidine–purine spacer (e.g. TATA) separating two tri-nucleotide recognition sites, rather than a 3-bp spacer separating two 4-bp recognition sequences (11,17).
The first published structure of a C protein bound to DNA was that of C.Esp1396I bound as a tetramer, with two dimers bound adjacently on the 35-bp operator sequence (OL + OR) of the C/R promoter (11). The structure revealed the mechanism whereby cooperative binding of dimers to the DNA operator control the switch from activation to repression of the C and R genes. In the crystal structure of the complex (PDB code: 3CLC), two dimers are bound to the DNA, each centred on the pseudo-dyad located between the central A and T bases in the TATA sequence within each operator site, and interacting across the major groove at the centre of the DNA.
Subsequent high resolution crystallographic studies of the complex with the OL operator (17) showed more clearly the nature of the sequence specific contacts to the bases within the recognition site (‘direct readout’), as well as the non-specific interactions with the severely bent phosphodiester backbone (‘indirect readout’). We now report the structure of a dimer of C.Esp1396I bound to OM and investigate the affinities of the protein for its three natural promoters, OM, OR and OL, in order to understand the structural and mechanistic basis of differential DNA sequence recognition that underpins this elegant genetic switch.
MATERIALS AND METHODS
Purification
Large-scale cultures of Escherichia coli BL21(DE3) containing the plasmid pET–28b/esp1396IC were grown. Over-expressed C.Esp1396I containing an N-terminal hexa-histidine tag (C.Esp1396I-6His) was harvested by sonication and separated from the cell lysate using nickel affinity chromatography. The His-tag was removed by thrombin digestion but the purified protein retained a GSH tripeptide (C.Esp1396I-GSH). Size exclusion chromatography was performed on a 26/60 Sephacryl S-200 HR size exclusion column in order to separate C.Esp1396I-GSH from cleaved His-tag, uncleaved protein and thrombin. For structural studies and for biophysical analysis, the protein was concentrated using heparin affinity chromatography. The DNA oligonucleotides were purified as previously described and annealed to form a duplex, prior to complex formation (11).
Analytical ultracentrifugation
Sedimentation equilibrium experiments were performed at 20°C with a range of protein concentrations using an Optima XL-A analytical ultracentrifuge (Beckman–Coulter, Palo Alto, CA, USA). Preliminary studies were done at 28 000 r.p.m. covering the range 1–30 µM protein. Subsequent runs were carried out at rotor speeds of 15 000, 21 000 and 28 000 r.p.m. Scans were done at wavelengths of 225 and 280 nm with a radial step size of 0.01 mm after 21 h equilibration. The scans for 1, 5 and 10 µM protein were globally fitted to a self-association model using SEDPHAT to determine the dissociation constant for the dimer (Kdim). The values for partial specific volume and buffer density were calculated using SEDNTERP and the errors were estimated using F-statistics. The Kdim was used to calculate the dimer concentration [D] in a sample of known total protein concentration, PT, using the following relationship:
| (1) |
Surface plasmon resonance
5’ biotinylated synthetic oligonucleotides containing either the OM, OL, OR or both the OL and OR sequences (OL+R) were immobilized on the surface of a SA sensor chip on a Biacore T-100. C.Esp1396I-GSH was dialyzed against the running buffer (10 mM HEPES pH 7.4, 100 mM NaCl, 5 mM MgCl2, 5 mM CaCl2, 0.05% v/v Tween-20) before a range of concentrations were injected over the chip for 30 s at a flow rate of 30 µl/min. Kinetic analysis was performed using the 1:1 binding model (with mass-transfer correction enabled) provided in the BiaEval software (version 2.0.2). For the kinetic analysis, the protein concentration was adjusted to the actual dimer concentration using Equation (1). Equilibrium analysis was performed by fitting the data to either a one-site model:
| (2) |
or (for the OL+R data) a two-site model:
| (3) |
using GraFit version 5.0.11 (Erithacus Software Ltd.). R is the response generated on reaching equilibrium, Rmax is the maximum response that can be generated by saturating the binding sites on the immobilized ligand, KD is the dissociation constant for the interaction (with KD1 and KD2 denoting the dissociation constants for the interaction between OL and OR, respectively) and the dimer concentration, [D], is given by Equation (1).
Crystallization of complexes
The DNA containing OM was designed to promote the crystallization of the complex in a single orientation in a similar manner to the OL complex. The DNA consisted of an 18-bp duplex with 5’ overhangs of A on one strand and T on the other. C.Esp1396I was incubated with the DNA at varying ratios (1:1, 1.5:1, 2:1, 2.5:1, 3:1 and 4:1 protein monomer:DNA) prior to crystal screening. The protein–DNA complex was subjected to sparse matrix screening using the Honeybee robot (Digilabs) to set up sitting drops. Subsequent crystallizations of protein–DNA complex were done at a ratio of 2:1 (protein monomers:DNA) with a final DNA concentration of ∼20 µM. Sitting drops were set up using 2 µl complex and 2 µl of the well solution. The initial conditions were optimized by varying the pH from 7 to 8.5 (in 0.5 unit increments), while simultaneously varying the PEG 1500 concentration from 5 to 30% w/v (in 5% increments). The trays were incubated at 16°C and checked at regular intervals using polarizing light microscopy. Suitable crystals were cryoprotected in 30% v/v glycerol, cryocooled in liquid nitrogen and stored, prior to exposure to synchrotron radiation. The crystals that gave rise to the final OM structure formed in 0.1 M SPG (succinate/phosphate/glycine) buffer pH 8, 25% w/v PEG 1500 with spermidine at a final concentration of 10 µM in the drop.
Structure solution and refinement
Cryocooled crystals of the OM complex were exposed to synchrotron radiation on ID14-4 at the ESRF (Grenoble). A selection of crystals was screened using the automated sample changer and data sets were collected at 100 K using an ADSC 4Q CCD detector. The OM complex crystallized in space group P21 and 180 images were collected with an oscillation angle of 1°. The data were processed and scaled using MOSFLM/SCALA (18) as this provided better integration statistics than processing the data using XDS/XSCALE (19). The collection and refinement statistics are shown in Table 1.
Table 1.
Crystallographic parameters
| Data collection | |
| Space group | P21 |
| Unit-cell parameters (Å, °) | a = 47.5 |
| b = 147.1 | |
| c = 47.8 | |
| α = γ = 90 | |
| β = 93.7 | |
| Resolution limits (Å) | 45.36–2.7 (2.85–2.7) |
| Rmergea (%) | 6.6 (20.1) |
| I/σ(I) | 7.4 (3.8) |
| Completeness (%) | 98.9 (99.7) |
| Refinement parameters | |
| NCS | |
| Groups | 1 |
| Chains in group | A, B, E and F |
| Residue range | 5–75 |
| Restraint level | Tight |
| TLS | |
| Groups | 10 |
| Chains (residues) | A, C, D, F, G and H (1–79) |
| B and E (1–41,48–79) | |
| B and E (42–47) | |
| Refinement model statistics | |
| No. of reflections | 61 350 |
| Rcryst/Rfreeb (%) | 19.6/23.7 |
| No. of atoms | |
| Protein | 2496 |
| DNA | 1546 |
| Water | 12 |
| Average B factors (Å2) | |
| Protein | 31.8 |
| DNA | 35.6 |
| Water | 34.8 |
| RMS deviations from ideal | |
| Bond lengths (Å) | 0.015 |
| Angles (°) | 2.1 |
X-ray crystal data, refinement and model statistics for the OM complex structure. Values in parentheses are for the highest resolution shell.
aRmerge = ΣhklΣi|Ii(hkl)−«I(hkl)»|/ΣhklΣiIi(hkl), where «I(hkl)» is the mean intensity of reflection I(hkl) and Ii(hkl) is the intensity of an individual measurement of reflection I(hkl).
bRcryst = Σhkl│|Fobs|−|Fcalc|│/Σhkl|Fobs|, where Fobs is the observed structure factor amplitude and Fcalc is the calculated structure factor amplitude. Rfree is the same as Rcryst but for 5% of structure factor amplitudes that were set aside during refinement.
The scaled data were phased by molecular replacement using Phaser (20). Chains A and B along with 10 bp from the OL structure (chains C and D) were used as separate ensembles to search for a replacement solution. The OM structure contained two complexes (i.e. two dimers, each bound to a DNA duplex) in the asymmetric unit. The structure was refined to 2.7 Å using iterative cycles of REFMAC5 (21) and real-space refinement in COOT (22). Non-crystallographic symmetry (NCS) and TLS restraints were used in REFMAC5 and the missing bases were manually added into interpretable electron density using COOT. The restraints used in refinement are shown in Table 1. Solvent atoms were added manually in COOT. 5% of structure-factor amplitudes were set aside during refinement for Rfree calculations. The final structure refined with R/Rfree = 19.6/23.7% and contained all 76 DNA bases (38 per duplex) and the following amino acid residues: 2–77 (chain A), 2–78 (chain B), 1–77 (chain E) and 4–78 (chain D); 99.7% of amino acid residues were in the preferred region of the Ramachandran plot. The coordinates of the DNA–protein complex have been deposited in the protein databank (PDB code: 3UFD). Molecular structures were visualized with Pymol (23). Amino acid residue numbers refer to the native sequence; the tripeptide sequence remaining after removal of the affinity tag was not observed in the electron density map as, presumably, it is disordered.
RESULTS
The C protein C.Esp1396I was expressed and purified as described previously (24; see also ‘Materials and Methods’ section). Structural analysis of the interaction of the protein with the M operator was then undertaken by means of X-ray crystallography and the interaction was further characterized in solution by analytical ultracentrifugation (AUC). Surface plasmon resonance (SPR) was then performed to compare binding affinities of the protein for each of the three natural operator sites.
Crystallographic analysis of the DNA–protein complex
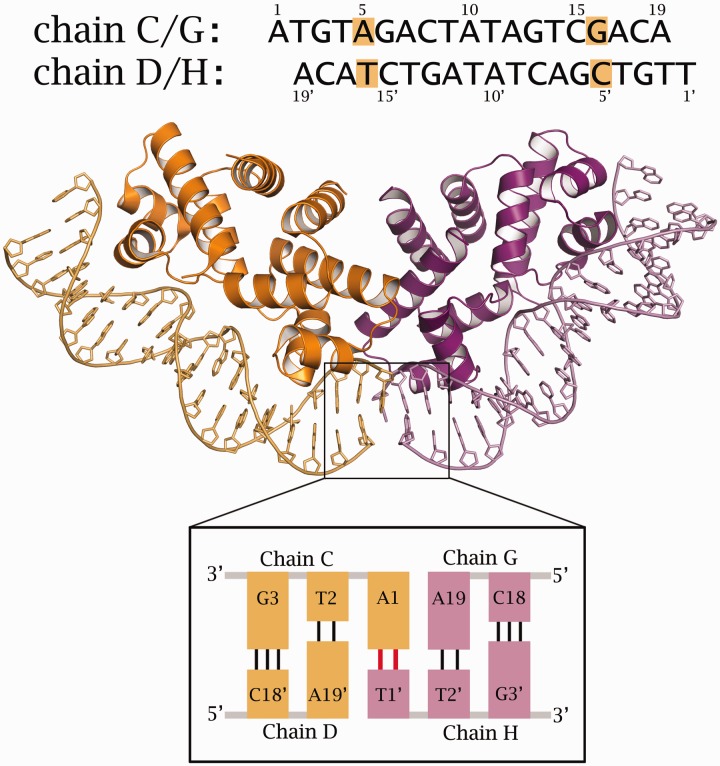
DNA–protein complexes were formed with an 18-bp DNA duplex consisting of two 19-bases oligonucleotides (thus forming 5’ A/T overhangs). This sequence contains the MTase gene operator sequence (OM) and was designed to aid the formation of pseudo-continuous DNA in a single orientation and thus overcome the symmetry-averaging problems encountered in the tetramer complex structure (11). Optimum crystallization conditions for the complex were determined from trials based on the PACT screen (Molecular Dimensions). X-ray diffraction data from suitable crystals were collected at 100 K at the ESRF (Grenoble). The space group was determined as P21, with two independent protein–DNA complexes in the asymmetric unit. The structure was solved by molecular replacement and refined by iterative cycles of reciprocal space refinement (REFMAC5) and real space refinement (COOT) to 2.7 Å resolution (see Table 1). Chains A–D comprise one DNA–protein complex, where A and B refer to the two subunits of a protein dimer, and C and D to the two strands of the DNA duplex (Figure 2). Chains E–H comprise the second DNA–protein complex in the asymmetric unit (E and F corresponding to the protein dimer, G and H to the DNA duplex).
Figure 2.
Structure of the two nucleoprotein complexes in the asymmetric unit of the C.Esp1396I/OM complex. Top: The sequence of the two DNA chains highlights the non-symmetric base pairs (AT and CG). Bottom: The two DNA duplexes in each complex are held together by an AT base pair formed from the 5’ overhanging bases.
The non-symmetric bases were identifiable during the building of the DNA duplex and thus the orientation of the DNA was defined. In particular, the purine/pyrimidine (A5/T16) and the pyrimidine/purine (C5′/G16′) base pairs could be distinguished (Figure 2). The terminal A and T bases could also be distinguished in the map, thus confirming the orientation of the DNA duplex. In contrast to the structure of the OL complex, where Hoogsteen base pairs are involved in the interaction between adjacent duplexes (17), the two terminal bases in the OM complex form Watson–Crick base pairs between duplexes, resulting in end-to-end packing of the DNA (Figure 2). In addition, R43 and K17 side-chains from adjacent complexes are involved in packing interactions that are mediated by an anion, most probably chloride (Supplementary Figure S1). Representative electron density in the map is illustrated in the vicinity of the dimer interface and around a region of the DNA (Supplementary Figure S2).
During the initial stages of refinement, the flexible loop regions (residues 43–46) were not subject to NCS restraints, since there are two stable conformations available for this loop in the free protein (24). However, after the initial refinement, the electron density maps were sufficiently clear to see that all four subunits had the flexible loop in the same conformation. Thus, subsequent rounds of refinement were carried out with tight NCS restraints also applied to the flexible loop region. Subsequent models therefore refer to the structure of a single complex (chains A–D). Although NCS restraints were not applied to the two DNA duplexes in the asymmetric unit, subsequent analysis shows that the two DNA helices have almost identical structures (see below).
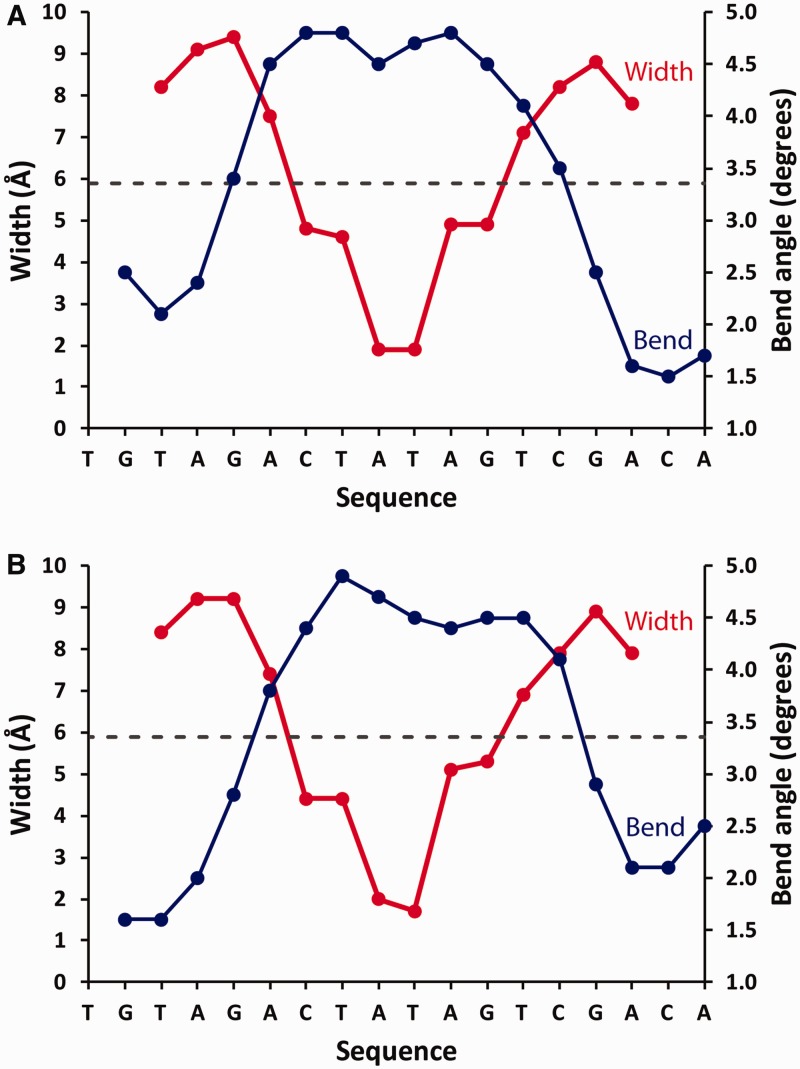
DNA structure in the complex
The DNA conformation in the nucleoprotein complex was analysed using the online CURVES server (25). The local DNA bend angle and the compression of the minor groove in the two complexes in the asymmetric unit is illustrated in Figure 3. The DNA helices in both complexes exhibit an overall bend angle of 56°. Additionally, both complexes show a very similar degree of local bending and minor groove compression at equivalent base pairs, despite the fact that no NCS restraints were applied to the DNA during refinement. The minor groove width varies from ∼10 Å to ∼2 Å, being most severely compressed at the TATA sequence. The compression of the minor groove is accompanied by an increased local bend angle.
Figure 3.
Analysis of DNA structure in the two DNA–protein complexes in the asymmetric unit, showing the local bend angle and groove width at each base pair.
The DNA in the OM complex has a higher overall bend angle (56°) than the DNA in the OL complex (41°), possibly reflecting the decreased spacing of the conserved elements in the sequence in OM. In the OL complex, the GAC/GTC sequences are separated by 5 bp and are positioned non-symmetrically relative to the TATA sequence (Figure 1). In the OM complex, the C-boxes are separated by 4 bp and are positioned symmetrically around the TATA sequence.
The compression of the minor groove is achieved through interactions between the phosphate backbone of the DNA and the amino acid residues of C.Esp1396I (Supplementary Figure S3). Residues D34, Y37, T49, S52 and N47 play a critical role in the compression of the phosphodiester backbone around the TATA sequence. Equivalent residues from each monomer interact with the backbone of a DNA strand either side of the TATA site and the distances between these residues in the two monomers determine the angle by which the DNA is bent. There are additional protein–DNA backbone contacts on the opposite strand that stabilize the complex, notably from amino acid residues R17, Q24, S39, R43 and N44 to the DNA phosphate backbone around the conserved TG nucleotides.
DNA sequence recognition
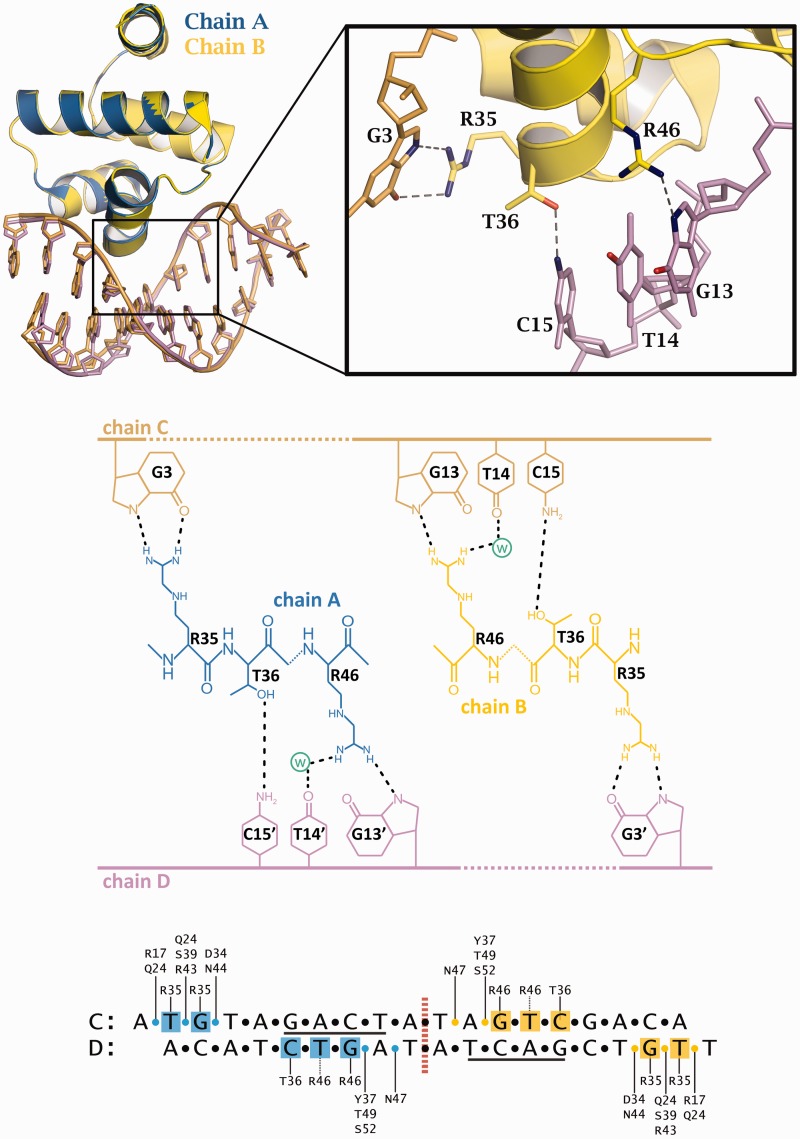
Direct readout of the OM operator DNA sequence is accomplished via the sidechains of the amino acid residues R35, T36 and R46 (Figure 4), which interact with the GAC/GTC motifs. In fact, all the contacts to this motif are made to the GTC bases on one strand. The γ-hydroxyl of the T36 sidechain interacts with the N4 amino group of cytosine C15. The R46 sidechain interacts with the N7 of G13. The interaction of the second NH of the guanidinium group with the carbonyl oxygen (O4) of G13 appears to be mediated through a water molecule. Likewise, there is a water molecule in a position to mediate the interaction of the R46 guanidinium group with the carbonyl oxygen of the thymine base, T14.
Figure 4.
DNA–protein contacts. Top: Rotation and superposition of the two subunits of the complex show symmetrical interactions to the DNA (inset: interactions of amino acids R35, T36 and R46 with bases G3 on one strand and G13, T14 and C15 on the other; the water atom is omitted for clarity). Middle: Schematic representation of the hydrogen bonding contacts. Bottom: Overview of specific base contacts and contacts to the DNA phosphates (yellow and blue circles).
The R35 sidechain is involved in both direct and indirect readout of the DNA sequence in the OM complex, as was found in both the OL complex and the tetrameric repression complex structures. The planar guanidinium head group of the arginine forms hydrogen bonds with the N7 and O6 of G3 but it is also involved in a π-stacking interaction with the adjacent base, T2 (Supplementary Figure S4). However, in contrast to the OL structure, the interaction of R35 with the TG motif is equivalent for both subunits, reflecting the symmetry of the DNA sequence at the M operator. It should be noted that the R35 from a given subunit (e.g. subunit A) interacts with different DNA strands when recognizing the GTC motif (on strand D) and the TG motif (on strand C). These interactions will further strengthen the integrity of the dimer in the nucleoprotein complex.
From comparison of C protein sequences and their cognate DNA binding sequences, it has been shown that there is a correlation between the identity of an amino acid residue in the recognition helix and the base sequence of the operator that it binds (26); specifically, it has been proposed that an aspartate at position 34 (or its equivalent in other C proteins) correlates with a cytosine base being present at the 3’ side of one of the GTC motifs, whereas a histidine at this site is most often found when there is a thymine at this site in the DNA sequence. C.Esp1396I belongs to the former category (i.e. possessing a DRTY rather than an HRTY motif in the recognition helix). We see no interaction of D34 with this base in the complex with the OM or the OL operator; instead, the D34 sidechain contacts the phosphodiester backbone of the DNA (see Supplementary Figure S3). There may conceivably be ‘indirect’ interactions to the base via a solvent molecule (although none is visible in the crystal structure).
Are there any other clues to a possible structural/biological role for D34? Somewhat surprisingly, the correlation observed only applies to the second of the four ‘C-boxes’ in the promoters studied [box 1B in the nomenclature of Mruk et al. (26)] and not to box 1A, where the symmetry related subunit of the dimer binds at the OL site (and nor does it apply to either site in OR). We also note that of the three DNA sequences that C.Esp1396I binds, each has a different base (G, C, A) at the site that has been proposed to interact with D34 (Figure 1). Indeed, the strongest binding site (OM) has a G at this site, a clear exception to the observations of Mruk et al. (26). Thus a direct role for D34 in binding to an isolated DNA operator site is unlikely.
However, we have previously shown that the C-protein subunit bound to box 1B of OL is involved in cooperative binding to the adjacent subunit bound to box 2A, at the interface between the two dimers in the tetrameric repression complex (11). Since the DRTY correlation with a cytosine base is only found at the second of the four repeating elements in the C/R promoter, we are tempted to speculate that D34 may play a role in repression at the promoter. The adjacent residue, R35, of this subunit interacts with E25 of the adjacent subunit via an ion pair mechanism in the tetrameric (repression) complex, and is a major contributor to the observed cooperativity between the two sites (11). The R35 of the adjacent subunit, however, binds to the G of the highly conserved central GT motif between OL and OR. It is possible that D34 may play an as yet unidentified role in that complex network of interactions, perhaps also involving the cytosine on the 3’ side of the GTC motif. If so, then presumably a histidine in the HRTY motif could make an equivalent interaction with a thymine at that site, to explain the observations of Mruk et al.(26).
Hydrodynamic analysis
The dissociation constant (Kdim) for the monomer–dimer equilibrium is an important parameter in the operation of the genetic switch, especially at low levels of expression of the C protein. Moreover, an accurate value of Kdim needs to be determined experimentally in order to obtain the relevant DNA binding constants. Thus, in order to obtain the DNA binding affinities of C.Esp1396I for its various operators, we first analysed the monomer–dimer equilibrium of the protein by sedimentation equilibrium in the AUC.
Since the protein has only one tyrosine, its extinction coefficient is too low to allow accurate determination of the Kdim, when low concentrations of protein are required. We therefore mutated the tyrosine residue Y29 into a tryptophan by site-directed mutagenesis. The mutation was confirmed by DNA sequencing of the gene, and the presence of a tryptophan could also be deduced from the fluorescence emission spectrum of the purified protein. Y29 is located far from the dimerization interface, and does not participate in DNA binding since it lies at the C-terminal end of helix 2. From dynamic light scattering, the hydrodynamic radius of the Y29W mutant protein (2.4 nm) was indistinguishable from that of the native protein, and its DNA binding properties were also found to be unchanged (data not shown).
The absorbance scans of the Y29W mutant of C.Esp1396I in the concentration range 1–30 µM were analysed using a single species model in SEDPHAT in order to determine the weight average molecular weight (Supplementary Figure S5). At low concentrations, the molecular weight was determined to be 8.8 ± 1.2 kDa, in agreement with the theoretical mass of a C.Esp1396I monomer (9.5 kDa). At higher concentrations (>10 µM), the molecular weight was found to be 19.4 ± 0.5 kDa, corresponding to the expected mass of a C.Esp1396I dimer. Thus, the Kdim for the monomer–dimer equilibrium is within the range of 1–10 µM.
A more accurate equilibrium constant was then determined by globally fitting the absorption scans measured at three different concentrations (1, 5 and 10 µM) and three different rotor speeds (15, 21 and 28 k.r.p.m.) to a self-associating species model (27) using SEDPHAT (see Supplementary Figure S6). This yielded a value for the Kdim of 1.6 µM corresponding to a free energy of dimerization (at 20°C) of −32.5 kJ/mol. The dimerization constant is of the same order of magnitude as that for C.AhdI (Kdim = 2.5 µM), consistent with the surface areas of their respective dimer interfaces (∼1900 Å2 versus ∼1400 Å2) and the similar H-bonding interactions between monomers in each case (14,24).
DNA binding analysis
(SPR experiments were conducted to investigate the DNA binding affinities of C.Esp1396I for the relevant promoter sites. Four different biotinylated duplexes containing either OM, OL, OR or the double site (OL+R) were each immobilized on separate streptavadin chips. For each experiment, the C.Esp1396I protein was injected using a range of concentrations, and the response measured as a function of time. The range of protein concentrations required to elicit a significant response for each DNA sequence varied greatly (up to 50-fold), reflecting the variation in DNA binding affinity at each site.
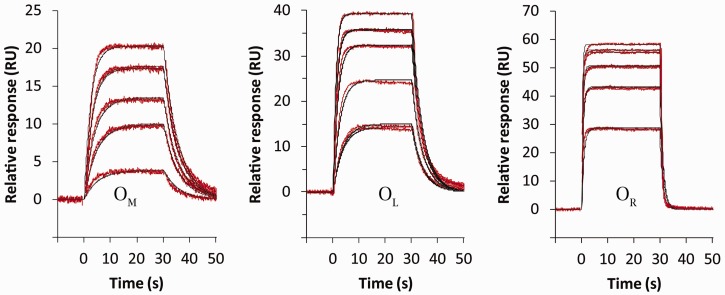
Following injection of the protein, the sensorgrams quickly reached their maximum response, and then remained constant throughout the 30 s injection (Figure 5). At this point the rates of binding and dissociation are equal and equilibrium has been attained. It is possible to obtain KD for the interaction by plotting the equilibrium response against protein concentration and fitting to the relevant binding equation. However, since C.Esp1396I binds as a dimer, the concentration of the active dimer must first be determined, since the total protein concentration is, in some cases, below the Kdim. This can be estimated using the dimer dissociation constant of 1.6 μM determined by AUC (see ‘Materials and Methods’ section). The analysis assumes that the monomer–dimer equilibrium is not affected by the small amount of protein dimer that binds to DNA during the experiment; for the relatively low loadings of DNA immobilized on the surface, this is likely to be a valid approximation.
Figure 5.
SPR kinetic analysis. For each operator site, the C protein was injected into the sample channel at five different protein concentrations (in duplicate), and the responses recorded after subtraction from the reference channel. Data were fitted to obtain the on- and off-rates for the interaction (see Table 1).
For the individual operator sites, the standard single-site binding equation was used to determine the dissociation constant of the dimer–DNA interaction, KD (Figure 6). For the duplex containing OL+R, a 2-site model was used with dissociation constants, KD1 and KD2, where KD1 describes binding to OL and KD2 describes binding to OR. By determining the affinity of C.Esp1396I for OL in isolation, KD1 can be fixed, which permits an accurate determination of the affinity for the second site (OR) when OL is already occupied.
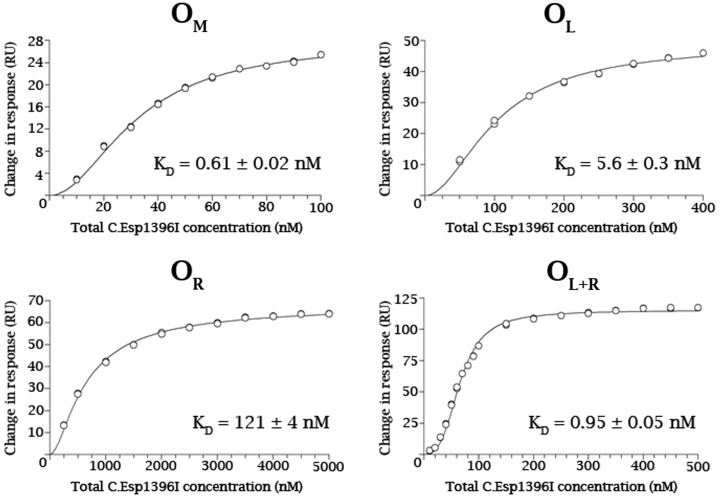
Figure 6.
Equilibrium binding analysis. Equilibrium binding at saturation was plotted against total protein concentration (expressed as monomer) from the SPR data shown in Figure 5. For OM, OL and OR, the curves were fitted to a single-site binding model to obtain the relevant dissociation constants, KD. For OL + OR, a two site binding model was employed; the Kd for binding to OL was fixed at the value obtained experimentally for the isolated operator site, thus permitting the determination of the affinity for the second site (OR) when OL is already occupied.
From the result of the equilibrium analysis, C.Esp1396I has the highest affinity for OM (KD = 0.61 nM), intermediate affinity for OL (KD = 5.6 nM) and the lowest affinity for OR (KD = 120 nM). Once the OL site has been occupied by a C.Esp1396I dimer, however, the affinity between C.Esp1396I and OR increases ∼130-fold (KD2 = 0.94 nM), indicating that there is a very high cooperativity of binding between the two operator sites.
The on- and off-rates of the interaction can also be measured by kinetic analysis of the sensorgrams (Figure 5), except for the case of two-site binding to OL+R, which cannot be described by any of the available binding models. From the ratio of the on- and off-rates, the binding constants for the three single operator sites can be obtained (See Table 2). The KD obtained by equilibrium measurements were generally higher than those obtained from kinetic analysis, but they were of the same magnitude, and were in approximately the same ratio (200:8:1 for OR:OL:OM). Thus, the SPR experiments show that the affinity for the OM site is around 8-fold higher than that for OL which, in turn, is 25-fold higher than that for OR.
Table 2.
Rate constants from kinetic analysis of the SPR data for C.Esp1396I binding to the three operator sites, OM, OL and OR
| ka (M−1.s−1) | kd (s−1) | KD (nM) | |
|---|---|---|---|
| OM | 1.61 ± 0.01 × 108 | 0.177 ± 0.001 | 1.10 ± 0.01 |
| OL | 2.99 ± 0.02 × 107 | 0.254 ± 0.001 | 8.5 ± 0.1 |
| OR | 3.88 ± 0.05 × 106 | 0.887 ± 0.004 | 229 ± 4 |
The equilibrium dissociation constants, KD, were in each case determined from the ratio of the off-rate (ka) to the on-rate (kd).
DISCUSSION
Overall, the structure of the C.Esp1396I/OM complex resembles that determined for the OL complex at the C/R promoter (17). Crucially, however, there are key structural differences that determine the differential DNA binding affinity for the endonuclease and M promoters. Figure 7 shows the superposition of the OM and OL complexes (RMSD of 0.36 Å). The majority of backbone and sidechain positions are essentially identical (Figure 7a), the exception being the conformation of the flexible loop (residues 43–46) of one of the two subunits of the dimer, which differs significantly between the two complexes (Figure 7b).
Figure 7.
Comparison of the structures of complexes of C.Esp1396I bound to the operators OL and OM (yellow and magenta, respectively) showing the displacement of the DNA bases. Although the sidechains of the alpha-helices in the two complexes are superimposed (a) the loop regions (b) are in quite different conformations, resulting in large displacements of amino acid side chains of N44 and S45, together with a smaller movement of R46.
The DNA bend in both complexes is centred on the alternating pyrimidine–purine sequence, TATA. Either side of this, in both complexes, the GAC motif (or more specifically, the complementary sequence GTC) is recognized by hydrogen bonding interactions with amino acid residues T36 and R46. In the OM complex, this motif is symmetrically disposed, 2 bp either side of the dyad axis within the central TATA, so that the centre of the GAC (=GTC) motifs are separated by 7 bp. However, in the OL complex, these motifs are asymmetrically arranged, 2 and 3 bp respectively from the pseudo-dyad axis, leading to an 8-bp separation between their centres (see Figure 1).
This additional separation of ca. 3.4 Å between these sites forces a conformational change in one of the subunits of the OL complex, in order to accommodate the displacement of the GTC motifs. It is notable that the overall position of the alpha-helices remains the same when compared with the OM complex; however, a localized conformational change in the flexible loop region (amino acid residues 43–46), leads to a significant rotation of the R46 sidechain that contacts the GTC motif.
The OM sequence is almost perfectly symmetrical, differing by only 1 bp (the A:T at position 5 is a C:G at the equivalent position on the other strand—see Figure 2). However, there are no contacts from the protein to the DNA at this position. The TG/CA and the GAC/GTC sequences are symmetrically arranged, and make specific hydrogen bonds to each subunit of the protein (including one via a water molecule). The TATA sequence does not make sequence-specific H-bonds, but instead makes numerous interactions with the protein via the phosphate groups of the DNA backbone to stabilize the highly deformed DNA helix at this point—a form of ‘indirect’ sequence read-out.
In comparison, the OL sequence lacks one of the TG motifs (see Figure 1), and thus loses a strong interaction with R35 (including two charged H-bonds and a base stacking interaction). Although OL has the GAC/GTC motif that is recognized by the protein, the extra base ‘insertion’ requires a conformational change in one of the protein subunits. Together, these changes in DNA sequence reduce the binding affinity by a factor of ∼8. We have previously shown that mutation of R35 to alanine abrogates binding to the OL+R operator, since in this case interactions with two TG motifs are lost (11).
There is no structure available for the OR complex, but such a complex would most likely lose the interaction with one of the TG motifs (Figure 1), unless the change in spacing can be accommodated by a conformational change, which itself would add an energy penalty. In addition, one of the GAC motifs becomes a GAT, thus losing the interaction with R46 on one subunit. Compared to OM, these two alterations to the DNA sequence, taken together, reduce the binding affinity by a factor of ∼200.
The order in which C.Esp1396I binds to its operator sequences is vital for the temporal regulation of the RM system. This is determined initially by the relative affinities of the C protein for the OL and OM binding sites, and subsequently by the cooperativity between OL and OR binding sites at the C/R promoter. Initially, C.Esp1396I is expressed at a low level from a weak C-independent promoter. The M gene (esp1396IM) is expressed constitutively, allowing the host genome to be methylated and thus protected from the action of the restriction enzyme. As the C.Esp1396I concentration slowly increases, protein dimers are formed. Initially, C-protein dimers bind to the highest affinity site OM and down-regulate the expression of esp1396IM, where the binding site overlaps the start of transcription (15). Subsequently, C-protein dimers bind to OL, up-regulating transcription from the C/R operon (esp1396IC/R) through cooperative recruitment of RNA polymerase, leading to a positive feedback loop. Thus the concentration of C.Esp1396I dimers will increase exponentially. At these higher concentrations, a further C-protein dimer binds cooperatively to the OR site to displace RNA polymerase, resulting in a negative feedback loop as the expression of esp1396IC/R is down-regulated. Ultimately, when both C/R and M promoters are repressed, the levels of C protein will fall, leading to de-repression of the M gene and thus enabling transcription of the M gene. Further regulation at the level of translation may also be involved, adding an additional level of fine tuning of the genetic switch.
The transcriptional regulation of the RM genes is ultimately dependent upon a localized conformational change in the C protein that is confined to a few amino acids residues in the loop region between helices 3 and 4. This conformational change is sufficient to allow variations in the spacing between specific DNA sequences of the ‘C-box’ motifs (specifically the trinucleotide sequence GAC/GTC) in relation to the TATA sequence that defines the centre of the bend in the DNA. There is, however, a free energy penalty to pay, as is evident from the 200-fold variation in DNA binding affinities between the three operator sites. In the OM promoter complex, there is almost perfect dyad symmetry within the C-protein dimer, matching a similar symmetry in the DNA sequence. In contrast, the shift in the pseudo-dyad axis relating the C-boxes in OL forces a conformational change in the loop of one subunit of the protein dimer, thus breaking the symmetry, and contributing to an almost 10-fold decrease in binding affinity, compared to the symmetrical binding site, OM.
This subtle change in the conformation of the protein underpins the differential affinity for the respective operator sites and controls the order in which the RM genes are switched on and off. The correct balance between methylation and restriction is thereby maintained, thus ensuring that the integrity of the bacterial genome is not compromised by premature expression of the endonuclease, while at the same time ensuring that DNA methyltransferase activity is kept in check.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1–6.
FUNDING
Biotechnology and Biological Sciences Research Council (BBSRC) [BB/E000878/1 to G.G.K. and BB/H00680X/1 to G.G.K. and J.E.M.]; Research Councils UK Academic Fellowship (to J.E.M.); University of Portsmouth IBBS PhD studentship (to N.J.B.). Funding for open access charge: BBSRC.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We are grateful to the ESRF (France) and Diamond Light Source (UK) and associated beam-line staff for provision of synchrotron radiation facilities.
REFERENCES
- 1.Jeltsch A. Maintenance of species identity and controlling speciation of bacteria: a new function for restriction/ modification systems? Gene. 2003;317:13–16. doi: 10.1016/s0378-1119(03)00652-8. [DOI] [PubMed] [Google Scholar]
- 2.Wilson GG, Murray NE. Restriction and modification systems. Ann. Rev. Genet. 1991;25:585–627. doi: 10.1146/annurev.ge.25.120191.003101. [DOI] [PubMed] [Google Scholar]
- 3.Tao T, Bourne JC, Blumenthal RM. A family of regulatory genes associated with type II restriction-modification systems. J. Bacteriol. 1991;173:1367–1375. doi: 10.1128/jb.173.4.1367-1375.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ives CL, Nathan PD, Brooks JE. Regulation of the BamHI restriction-modification system by a small intergenic open reading frame, bamHIC, in both Escherichia coli and Bacillus subtilis. J. Bacteriol. 1992;174:7194–7201. doi: 10.1128/jb.174.22.7194-7201.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rimseliene R, Vaisvila R, Janulaitis A. The eco72IC gene specifies a trans-acting factor which influences expression of both DNA methyltransferase and endonuclease from the Eco72I restriction-modification system. Gene. 1995;157:217–219. doi: 10.1016/0378-1119(94)00794-s. [DOI] [PubMed] [Google Scholar]
- 6.Vijesurier RM, Carlock L, Blumenthal RM, Dunbar JC. Role and mechanism of action of C·PvuII, a regulatory protein conserved among restriction-modification systems. J. Bacteriol. 2000;182:477–487. doi: 10.1128/jb.182.2.477-487.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cesnaviciene E, Mitkaite G, Stankevicius K, Janulaitis A, Lubys A. Esp1396I restriction-modification system: structural organization and mode of regulation. Nucleic Acids Res. 2003;31:743–749. doi: 10.1093/nar/gkg135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Knowle D, Lintner RE, Touma YM, Blumenthal RM. Nature of the promoter activated by C.PvuII, an unusual regulatory protein conserved among restriction-modification systems. J. Bacteriol. 2005;187:488–497. doi: 10.1128/JB.187.2.488-497.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bogdanova E, Djordjevic M, Papapanagiotou I, Heyduk T, Kneale G, Severinov K. Transcription regulation of the type II restriction-modification system AhdI. Nucleic Acids Res. 2008;36:1429–1442. doi: 10.1093/nar/gkm1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mruk I, Blumenthal RM. Real-time kinetics of restriction–modification gene expression after entry into a new host cell. Nucleic Acids Res. 2008;36:2581–2593. doi: 10.1093/nar/gkn097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McGeehan JE, Streeter SD, Thresh SJ, Ball N, Ravelli RB, Kneale GG. Structural analysis of the genetic switch that regulates the expression of restriction-modification genes. Nucleic Acids Res. 2008;36:4778–4787. doi: 10.1093/nar/gkn448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Streeter SD, Papapanagiotou I, McGeehan JE, Kneale GG. DNA footprinting and biophysical characterisation of the controller protein C.AhdI suggests the basis of a genetic switch. Nucleic Acids Res. 2004;32:6445–6453. doi: 10.1093/nar/gkh975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McGeehan JE, Papapanagiotou I, Streeter SD, Kneale GG. Cooperative binding of the C.AhdI controller protein to the C/R promoter and its role in endonuclease gene expression. J. Mol. Biol. 2006;358:523–531. doi: 10.1016/j.jmb.2006.02.003. [DOI] [PubMed] [Google Scholar]
- 14.McGeehan JE, Streeter S, Papapanagiotou I, Fox GC, Kneale GG. High-resolution crystal structure of the restriction-modification controller protein C.AhdI from Aeromonas hydrophila. J. Mol. Biol. 2005;346:689–701. doi: 10.1016/j.jmb.2004.12.025. [DOI] [PubMed] [Google Scholar]
- 15.Bogdanova E, Zakharova M, Streeter S, Taylor J, Heyduk T, Kneale GG, Severinov K. Transcription regulation of restriction-modification system Esp1396I. Nucleic Acids Res. 2009;37:3354–3366. doi: 10.1093/nar/gkp210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sorokin V, Severinov K, Gelfand MS. Systematic prediction of control proteins and their DNA binding sites. Nucleic Acids Res. 2009;37:441–451. doi: 10.1093/nar/gkn931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McGeehan J, Ball NJ, Streeter SD, Thresh S-J, Kneale GG. Recognition of dual symmetry by the controller protein C.Esp1396I based on the structure of the transcriptional activation complex. Nucleic. Acids Res. 2012;40:4158–4167. doi: 10.1093/nar/gkr1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leslie AGW. Recent changes to the MOSFLM package for processing film and image plate data. Joint CCP4 + ESF-EAMCB Newsletter on Protein Crystallography. 1992 No. 26. [Google Scholar]
- 19.Kabsch W. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J. Appl. Crystallogr. 1993;26:795–800. [Google Scholar]
- 20.McCoy AJ, Grosse-Kunstleve RW, Storoni LC, Read RJ. Likelihood-enhanced fast translation functions. Acta Crystallogr. D: Biol. Crystallogr. 2005;61:458–464. doi: 10.1107/S0907444905001617. [DOI] [PubMed] [Google Scholar]
- 21.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D: Biol. Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 22.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D: Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 23.Delano WL. The PyMOL Molecular Graphics System. San Carlos, CA, USA: DeLano Scientific; 2002. [Google Scholar]
- 24.Ball N, Streeter S, Kneale GG, McGeehan J. Structure of the restriction-modification controller protein C.Esp1396I. Acta Crystallogr. D Biol. Crystallogr. 2009;D65: 900–905. doi: 10.1107/S0907444909020514. [DOI] [PubMed] [Google Scholar]
- 25.Lavery R, Moakher M, Maddocks JH, Petkeviciute D, Zakrzewska K. Conformational analysis of nucleic acids revisited: Curves+ Nucleic Acids Res. 2009;37:5917–5929. doi: 10.1093/nar/gkp608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mruk I, Rajesh P, Blumenthal RM. Regulatory circuit based on autogenous activation-repression: roles of C-boxes and spacer sequences in control of the PvuII restriction-modification system. Nucleic Acids Res. 2007;35:6935–6952. doi: 10.1093/nar/gkm837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vistica J, Dam J, Balbo A, Yikilmaz E, Mariuzza RA, Rouault TA, Schuck P. Sedimentation equilibrium analysis of protein interactions with global implicit mass conservation constraints and systematic noise decomposition. Anal. Biochem. 2004;326:234–256. doi: 10.1016/j.ab.2003.12.014. [DOI] [PubMed] [Google Scholar]