Abstract
RNase P is an RNA-based enzyme primarily responsible for 5′-end pre-tRNA processing. A structure of the bacterial RNase P holoenzyme in complex with tRNAPhe revealed the structural basis for substrate recognition, identified the active site location, and showed how the protein component increases functionality. The active site includes at least two metal ions, a universal uridine (U52), and P RNA backbone moieties, but it is unclear whether an adjacent, bacterially conserved protein loop (residues 52–57) participates in catalysis. Here, mutagenesis combined with single-turnover reaction kinetics demonstrate that point mutations in this loop have either no or modest effects on catalytic efficiency. Similarly, amino acid changes in the ‘RNR’ region, which represent the most conserved region of bacterial RNase P proteins, exhibit negligible changes in catalytic efficiency. However, U52 and two bacterially conserved protein residues (F17 and R89) are essential for efficient Thermotoga maritima RNase P activity. The U52 nucleotide binds a metal ion at the active site, whereas F17 and R89 are positioned >20 Å from the cleavage site, probably making contacts with N−4 and N−5 nucleotides of the pre-tRNA 5′-leader. This suggests a synergistic coupling between transition state formation and substrate positioning via interactions with the leader.
INTRODUCTION
The conversion of precursor tRNA (pre-tRNA) into functional tRNA requires an RNA-based catalyst, ribonuclease (RNase) P, to remove the leader sequence on the 5′ end (1). This ribonucleoprotein (RNP) complex is composed of one essential RNA subunit and one or more protein subunits, which collectively enable substrate recognition and catalysis. RNase P recognizes its substrate in trans, is a metal-dependent multiple turnover enzyme, and accurately cleaves pre-tRNA transcripts throughout phylogeny, yet can also exhibit broad specificity and act on various non-tRNA substrates, such as mRNA, rRNA, transfer messenger (tm) RNA and riboswitches [see reviews (2–4)].
In bacteria, a single RNA (P RNA, ∼400 nucleotide long) and a small protein (P protein, ∼120 amino acids) comprise the enzyme. The RNA component can support catalysis in vitro in the absence of the protein component, but both subunits are essential in vivo, emphasizing the critical role of both components (1,5,6). Previous biochemical and structural information of the bacterial RNase P holoenzyme with and without pre-tRNA have addressed several key questions about the mechanism of this RNP and how it recognizes its RNA target. P RNA-tRNA recognition involves intermolecular base stacking, A-minor, and base-pairing interactions, whereas the P protein contacts the pre-tRNA leader region 5′ to the cleavage site and provides stabilization of conserved P RNA regions (Figure 1A and B) (1,7–21). In addition, divalent metal ions contribute to both substrate binding and catalysis (9,22–26), with magnesium hydroxide serving as the likely catalytic species required for phosphodiester cleavage (27,28). The location of the enzyme active site is inferred from biochemical studies (8,29–38) and the structure of the RNase P-tRNA complex (Figure 1C) (9). In the structure, the tRNA 5′ end sits within the core of enzyme, between the major groove of the P4 helix, a highly conserved loop region (termed conserved region V), a universal uridine nucleobase (U52) that is unstacked from the P4 helix, and two metal ions. Metal dependent catalysis by RNase P has been extensively studied with data generally supportive of 2–3 metal ions at the site of cleavage depending on monovalent conditions (23–26,30,39). Interestingly, both time-resolved Förster resonance energy transfer (trFRET) (22) and structural studies (9) suggest that the catalytically important metal ions possess differential binding affinity, where a high-affinity metal ion might position the scissile phosphate oxygen atoms of the substrate (prior to cleavage) and activate the nucleophillic hydroxyl ion (M1), and a weaker metal ion might enhance the cleavage step by stabilizing the transition state and mediating proton transfer during product release (M2). However, whereas the combined biochemical and structural knowledge offers a more consistent mechanistic hypothesis for this RNP enzyme, questions regarding specific interactions at the active site and between the P protein and pre-tRNA leader remain. Specifically, it is unclear whether a bacterially conserved loop region of the protein (β3α2 loop), positioned ∼8–10 Å from the active site, participates directly in catalysis. This region of the protein also resides adjacent to the bacterially conserved and positively charged 'RNR' amino acid motif that comprises a stretch of seven residues (R60-N61-K62-L64-K64-R65-W66 in Thermotoga maritima) (40,41). Within the β3α2 loop region, two protein residues (R52 and K56) are positioned ∼4.5 Å from the catalytically important M2 metal ion in the structure of the complex. In addition, though several studies have examined biochemically putative protein-pre-tRNA contacts located 5′ to the cleavage site (12,13,15,37,39,40,42,43), it is unclear how the amino acids lining the substrate binding track of the T. maritima protein contribute to enzymatic activity and which specific residues and atoms within the holoenzyme are involved in substrate alignment and are essential for efficient catalysis.
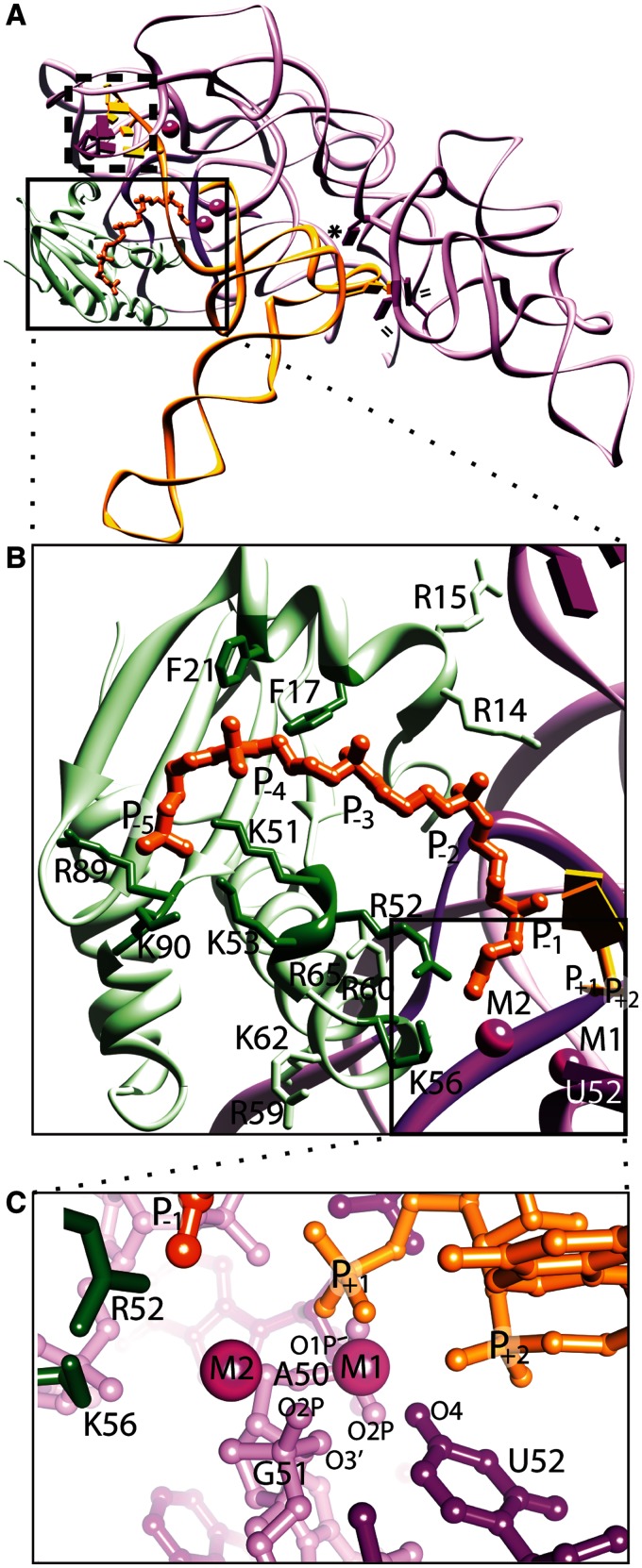
Figure 1.
Structure of bacterial RNase P holoenzyme in complex with tRNA and a short oligonucleotide leader (rcsb: 3Q1R). The T. maritima holoenzyme consists of a large P RNA (purple), a small protein (light green), and critical metal ions (magenta spheres), and makes several contacts with the tRNA (orange) and a short leader oligonucleotide (dark orange). (A) Ribbon representation emphasizing the tertiary P RNA-tRNA elements crucial for recognition, where double lines (=) represent hydrophobic base stacking, an asterisk (*) represents an A-minor interaction, and a dashed box represents intermolecular base pairs. In the structure of the complex, the protein sits atop and stabilizes conserved, non-helical regions of the P RNA (dark purple). The boxed region highlights the area where the P protein, P RNA and tRNA converge, including the active site. (B) Expanded view of the boxed region in (A), showing key protein residues that contact the P RNA (light green, including R14, R15, R59, R60, K62, R65), or residues that potentially contact the pre-tRNA leader region (dark green, including F17, F21, K51, R52, K53, K56, R89, K90). The phosphates of the pre-tRNA leader (labelled P−1 – P−5, dark orange) were observed in a structure of a holoenzyme-tRNA complex with a short leader soaked into the crystal, and the active site region (boxed) was inferred from the location of the +1 phosphate of the tRNA and the presence of two catalytically important metal ions (M1 and M2). (C) An expanded view of the active site of the bacterial RNase P enzyme. Specific P RNA oxygen atoms, including: G51 (O2P and O3′), A50 (O1P and O2P) and the O4 atom of a universal uridine nucleobase (U52), are likely to make contacts with the metal ions. Protein residues K56 and R52 are located ∼4.5 Å from the nearest metal ion (M2) and ∼7–8 Å from the phosphorus atom of the mature tRNA (+1 nucleotide). All structural figures were prepared using Chimera (61).
Here, we combine site-directed mutagenesis with single-turnover enzyme kinetics to assess the functional contributions of several protein residues within the pre-tRNA leader binding region, as well as protein residues that make structural contacts with the P RNA (Figure 1B). In addition, a U52C P RNA mutant holoenzyme, representing a single carboxyl to amine substitution, was examined. Based on the structure of the complex, the O4 atom of this bulged and universally conserved nucleotide makes first coordination sphere contacts with a catalytically important metal ion (M1) that also makes direct contacts with the reactive phosphate oxygen atoms (Figure 1C) (9). Based on results of single-turnover kinetic studies, we show that the U52C RNase P holoenzyme mutant results in severe catalytic defects. In addition, mutation of two amino acid in the P protein (F17A and R89A), which are positioned far from the active site and make putative contacts with nucleotides N−4 and N−5 of the pre-tRNA leader, also result in a significant loss of catalytic efficiency. Interestingly, point mutations of bacterially conserved amino acids closest to the active site (R52, K56) and those within the conserved ‘RNR’ region (R59–R65) have no or modest effects on catalytic efficiency. Comparative analysis of point mutants near the active site and along the path of the pre-tRNA leader identifies the location of critical binding contacts involved in substrate positioning and functionally confirms the location of the enzyme active site, in excellent agreement with the structural data.
MATERIALS AND METHODS
Preparation of RNase P, RNA substrate and point mutants
Wild-type T. maritima P RNA, U52C P RNA and the pre-tRNAPhe substrate were prepared and purified as previously described (9) with minor modifications. Modified RNAs (U52C P RNA and pre-tRNA substrate, which contains the leader sequence 5′-G−9 G−8 A−7 G−6 G−5 A−4 G−3 G−2 U−1-tRNA), were prepared from previous pUC19 plasmids where the T. maritima P RNA or tRNAPhe genes were inserted at FokI and BmsAI restriction sites, respectively (9,18). P RNA and pre-tRNA samples were purified by 6% and 8% denaturing polyacrylamide gel electrophoresis (PAGE), respectively, identified by ultraviolet absorbance, recovered by diffusion into 50 mM potassium acetate (pH 7) and 0.2 M potassium chloride, and ethanol precipitated. Centrifugation of the RNA (∼8000g) yielded a pellet, which was washed twice (80% ethanol), dried using centrifugal evaporation or lyophilization, and resuspended in diethyl pyrocarbonate (DEPC)-treated double distilled water (ddH2O). RNA samples are >95% pure, based on denaturing 8% PAGE analysis and staining with toluidine blue.
All mutants were made using the QuikChange (Stratagene) method and confirmed by sequencing the DNA covering the protein coding region. A pGEX4Ta expression vector containing the T. maritima wild-type gene (rnpA) was a gift from the Pace laboratory (University of Colorado–Boulder) and mutant RNase P proteins were expressed as GST-fusion peptides and purified as described previously (44) with the following modifications. Protein expression was done in Escherichia coli (BL21(DE3)pLysS cells; cell cultures were grown to an OD595 of 0.5–0.8 at 37°C, induced by the addition of 1 mM IPTG, and were subsequently incubated for 6–12 h at 30°C. Cells expressing each protein were harvested by centrifugation and snap frozen in liquid nitrogen until use. Cell pellets were re-suspended in lysis buffer (50 mM Tris HCl (pH 7.5), 4 mM EDTA, 10% glycerol, 0.1% (v/v) NP-40 and one-fourth of a tablet containing complete protease inhibitors (Roche). After cells were fully lysed by sonication (10–15 min. (30 s. on, 40 s. off)), 600 NIH units of thrombin were added and the lysate was incubated for 12–14 h at room temperature. The lysate was centrifuged (∼55 000g) for 30 min at 10°C, filtered (0.22 µM), and diluted to a final lysate solution of 5 M urea, 50 mM Tris (pH 7.5), 2 mM EDTA, 5% glycerol. Denatured RNase P protein was initially separated from the filtered lysate with a gradient elution buffer (50 mM Tris (pH 7.5), 0.2 mM EDTA, 2 M NaCl and 5 M urea) using cation exchange chromatography (Source™ 15 S, GE Healthcare). All wild-type and mutant RNase P proteins eluted between 0.7 M and 1.0 M NaCl under denaturing conditions. Dialysis in refolding buffer (50 mM Tris (pH 7.5), 0.2 mM EDTA, 1 M NaCl) for 1–2 days was used to remove urea. This solution was subsequently diluted to a final concentration of 0.2 M NaCl using 50 mM Tris (pH 7.5), 0.2 mM EDTA (buffer A) and re-applied to a 15 S column pre-equilibrated in buffer A. A shallow gradient elution using 50 mM Tris (pH 7.5), 0.2 mM EDTA, 2 M NaCl buffer was performed and all wild-type and mutant RNase P proteins eluted between 0.9 M and 1.2 M NaCl. This second column purification contained the fully re-folded P protein and was required to remove any partially positively charged polypeptides that may have co-eluted with the denatured P protein. Purified protein was dialyzed against 10 mM Tris pH 7.5, 0.1 mM EDTA, concentrated using a pre-equilibrated Amicon Ultra 3K centrifugation filter (Millipore), identified by SDS-PAGE (Supplementary Figure S1), confirmed using electrospray ionization mass spectrometry (ESI-MS) (data not shown), and stored at 4°C. All wild-type and mutant proteins did not degrade for several months, based on SDS-PAGE. All protein samples were always freshly prepared prior to RNase P holoenzyme activity assays.
CD spectroscopy of T. maritima RNase P protein and point mutants
Circular Dichroism (CD) measurements were obtained with a Jasco J-815 spectropolarimeter equipped with a Peltier device and routinely calibrated with d-10-camphorsulfonic acid (Keck Facility, Northwestern University). Wavelength scans between 180 nm and 260 nm were carried out at 20°C at a final protein concentration of 10 µM/l in 10 mM sodium phosphate, pH 8.0, 0.14 M NaF (0.1 mm path length demountable cuvette). The data pitch was 1 nm, the scan speed 20 nm/min, the response time 8 s, and the band width 1 nm. Data for wavelength scans are presented in units of molar ellipticity ([θ]), deg cm2 dmol−1 residue−1. The wild-type and RNase P protein mutants show that all mutants are well folded with no differences in their secondary structure (Supplementary Figure S2).
Thermotoga maritima RNase P activity assays and determination of kinetic parameters
To form the T. maritima RNase P holoenzyme, the purified, folded protein component and unfolded P RNA were mixed at a 1:1 molar ratio in 33 mM Tris, 66 mM HEPES, (pH 7.4), 0.1 mM EDTA (1X THE) and 100 mM ammonium acetate, as previously described (9,10). The solution was heated to 90°C (2 min), set aside at room temperature (3 min), MgCl2 added to a final 10 mM concentration and incubated for 10 min at 50°C, and subsequently incubated at 37°C until use. For assays containing only the RNase P RNA component, the identical folding protocol was performed except monovalent and divalent ions were added simultaneously 3 min after the short 90°C incubation period at concentrations of 1 M ammonium acetate and 0.1 M MgCl2 for high salt conditions (hs) and 0.6 M ammonium acetate and 50 mM MgCl2 for elevated salt conditions (Table 1). Holoenzyme or ribozyme were prepared and subsequently diluted to various concentrations (25–12 000 nM) for each cleavage reaction condition.
Table 1.
Single-turnover kinetic parameters measuring pre-tRNA cleavage by wild-type and mutant T. maritima RNase P
| Holoenzyme (protein/RNA) | Amino acid conservation (%)a | kcat / KM (106 M−1 min−1)b | n | Relative reactivityc |
|---|---|---|---|---|
| WT / WT | 10.8 ± 1.70 | 6 | 1 | |
| R14A / WT | 9.7 (13.5) | 3.90 ± 0.06 | 1 | 0.4 |
| R15A / WT | 16.6 (34.6) | 3.25 ± 0.15 | 2 | 0.3 |
| F17A / WT | 84.3 (97.7) | 0.50 ± 0.25 | 2 | 0.05 |
| F21A / WT | 58.1 (88.6) | 1.7 ± 0.10 | 2 | 0.2 |
| K51A / WT | 2.0 (2.65) | 4.2 ± 1.3 | 3 | 0.4 |
| R52A / WT | 9.0 (98.2) | 4.0 ± 1.0 | 2 | 0.4 |
| K53A / WT | 93.9 (99.0) | 2.4 ± 0.8 | 2 | 0.2 |
| K56A / WT | 26.4 (48.3) | 10.9 ± 1.9 | 2 | 1.1 |
| R59A / WT | 3.3 (7.7) | 11.9 ± 1.3 | 2 | 1.2 |
| R60A / WT | 99.8 (100) | 7.2 ± 2.6 | 2 | 0.7 |
| K62A / WT | 10.2 (79.2) | 10.2 ± 1.2 | 2 | 1 |
| R65A / WT | 100 | 15.7 ± 3.6 | 2 | 1.5 |
| R89A / WT | 70.8 (95.3) | 0.60 ± 0.30 | 3 | 0.06 |
| K90A / WT | 43.5 (48.8) | 2.65 ± 1.50 | 2 | 0.3 |
| WT / U52C | 0.8 ± 0.4 | 2 | 0.08 | |
| R89A / U52C | 0.050 ± 0.011 | 3 | 0.005 | |
| F17 / U52C | 0.006 ± 0.003 | 2 | 0.0006 | |
| - / WT (hs)d | 0.50 ± 0.03 | 1 | 0.05 | |
| - / WT (salt)e | 0.030 ± 0.028 | 1 | 0.003 |
aFrequency of occurrence of amino acid in a large group of bacterial RNase P proteins (n = 491). In parenthesis is shown the frequency of occurrence of R + K for either R or K, and F + Y for F. This reflects better the overall conservation of this amino acid.
bkcat/KM were determined from the observed reaction rates (kobs) at various concentrations and assuming Michaelis–Menten kinetics. The enzyme activity was obtained from the average of n complete time course reaction assays performed. For n > 1, the estimated error represents the propagated standard error from all time-dependent kinetic measurements. For n = 1, the estimated error was propagated from individual time-dependent kinetic measurements as determined by Kaleidagraph 4 (Synergy Software).
cApproximate relative enzyme activity using the ratio: kcat/KM(modified RNase P)/kcat/KM(wild-type RNase P).
dRibozyme (P RNA alone) under high salt (hs) conditions (100 mM MgCl2, 1.0 M NH4OAc).
eRibozyme (P RNA alone) under elevated salt conditions (50 mM MgCl2, 0.6 M NH4OAc).
To prepare the 32P-labelled pre-tRNA substrate, RNA was dephosphorylated for 30 min at 37°C using calf intestine alkaline phosphatase (NEB), followed by phenol/chloroform extraction and multiple ethanol precipitation washes. Incubation of dephosphorylated RNA and 25 µM 32P-ATP (6000 Ci/mmol, 10 mCi/ml, Perkin Elmer) with T4 polynucleotide kinase (NEB) for 30 min at 37°C, followed by passage over multiple spin columns (P6, Bio-Rad) resulted in a highly purified 32P-labelled substrate probe with nominal background of free 32P-ATP (<0.02%). Addition of 50 mM potassium acetate/200 mM potassium chloride followed by ethanol precipitation and subsequent 80% ethanol precipitation washes were performed and the dried RNA substrate was stored at −20°C until use. Prior to each cleavage reaction, the 32P-labelled RNA substrate was re-suspended in 1X THE, heated to 85–90°C for 2 min, set aside at room temperature (3 min), MgCl2 and ammonium acetate added to a final 10 mM and 0.1 M concentration, respectively, and the solution was incubated at 37°C until use.
Cleavage reactions were started by mixing enzyme and substrate (< ∼1 nM) at 37°C. Reactions were performed at linear enzyme concentration ranges, spanning 25–12 000 nM depending upon kobs (Supplementary Figure S3). An equal volume (typically 3 µl) was extracted from the reaction and immediately quenched with an 8 M urea/50 mM EDTA/bromophenol blue/xylene cyanol/5% glycerol mixture at various time points. Pre-equilibrated 10% denaturing PAGE was performed on all reaction mixtures, which enabled the clear separation of substrate from product (Figure 2). After exposure to a phosphorimaging screen (ranging from 8–15 min) and scanning [Pharos FX Plus (BioRad) or Storm 860 (GE Healthcare)], all reaction profiles were quantified using Quantity One 4.6.9 (BioRad) or ImageQuant™ TL software packages. A plot of the percentage of product formation over time provided the rate constant for cleavage at each concentration. Single-turnover conditions assuming pseudo first-order kinetics follow the equation P = P∞ (1 − e-kobst), where P is the fraction of pre-tRNA cleaved, P∞ is the fraction of uncleaved pre-tRNA at the end of the reaction and kobs is the observed reaction rate constant. Assuming Michaelis–Menten kinetics, plots of kobs versus enzyme concentration are linear. By measuring kobs at different concentrations within the linear range of the experiment, it is possible to obtain kcat/KM from the slope of the regression line. Standard error from the pseudo-first order kinetic curve fits for all data were generated by Kaleidagraph 4 (Synergy Software), were propagated throughout the kobs versus enzyme concentration regression analysis, and represent the reported estimated error (Table 1). Nearly all wild-type and mutant RNase P holoenzyme activity experiments were performed in duplicate, allowing for an accurate and comparative activity profile.
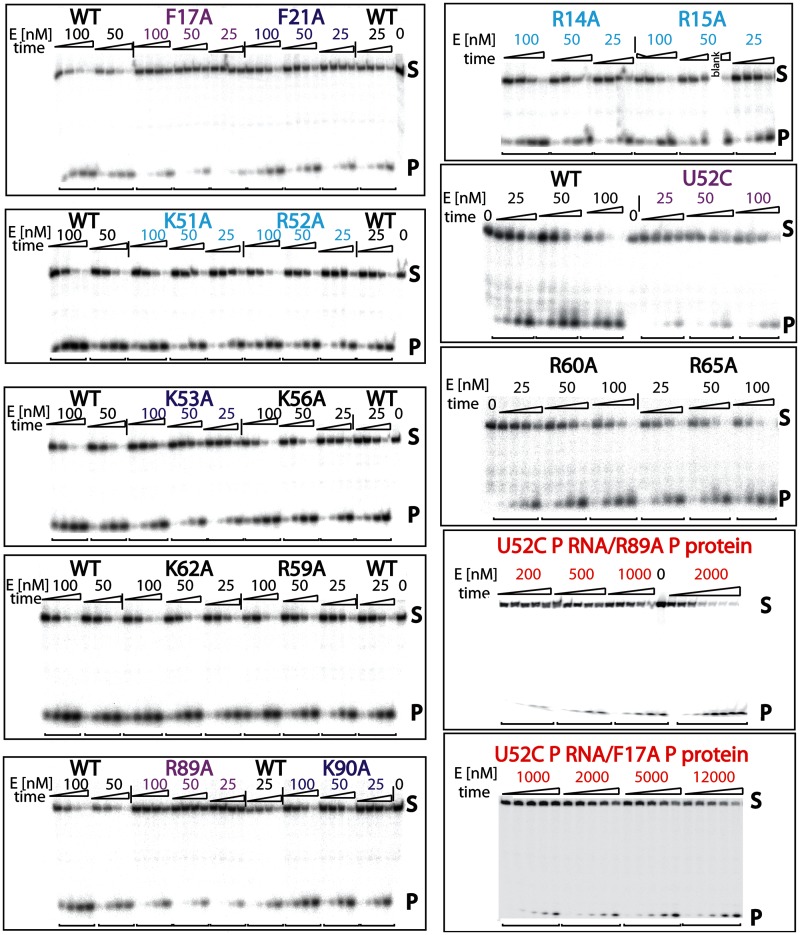
Figure 2.
Representative cleavage kinetics of pre-tRNA catalyzed by the wild-type and mutant T. maritima RNase P holoenzymes. Multiple time course reactions (time points: 0.25, 1, 4 and 16 min) at different enzyme concentrations enabled the quantitative measurement of single-turnover reaction kinetic parameters (kcat/KM) (Table I). In most cases, enzyme concentrations (25, 50, 100 and 200 nM) fell within the linear range, enabling extraction of kcat/KM from a plot of [E] versus kobs (see ‘Materials and Methods’ section and Supplementary Figure S3). In other cases, such as the double mutants (dm) U52C RNA/F17A protein or U52C RNA/R89A P protein (red), high holoenzyme concentrations (200–12000 nM) were required to observe the formation of any product. In addition, the P RNA ribozyme by itself (under high and medium divalent/monovalent conditions), as well as R89A, F17A and U52C P RNA mutants, were tested at higher enzyme concentrations (vide supra and Supplementary Figure S3). Compared with the wild-type T. maritima RNase P holoenzyme (black), point mutants range from showing no or little effect (black (K56A, R59A, R60A, K62A and R65A)), to modest (light blue (R14A, R15A, K51A, R52A)), substantial (dark blue (F21A, K53A, R90A)), severe (magenta (U52C P RNA, R89A and F17A)) and deleterious (red, dm U52C RNA/F17A protein or U52C RNA/R89A P protein) effects on catalytic activity.
RESULTS AND DISCUSSION
The structure of the T. maritima RNase P in complex with mature tRNA revealed the presence of two metal ions (M1 and M2) at the enzyme active site. M1 is located in a pocket formed by the P RNA and tRNA (∼3.0 Å from the 5′ phosphorus atom) and M2 is fully occupied in the presence of the 5′ leader (∼4.3 Å from the 5′ phosphorus atom). The M1 metal ion, putatively Mg2+, is coordinated by the O4 oxygen of a universally conserved uridine (U52) and phosphate oxygens from both the tRNA and the P RNA, possibly through inner sphere coordination. This metal-RNA environment comprises the putative RNase P enzyme active site and it has been proposed that the active site is the same in all organisms that contain an RNA-based RNase P (9). Indeed, the structural topology and metal-binding properties of the bacterial A- and B-type P RNAs appear to be highly similar in this region (9,19,45,46), although the conserved uridine in the B-type structure is in a different, probably artefactual, conformation as it is involved in crystallographic contacts and the structure does not include any ligands (19).
The central role of this uridine in the reaction had been surmised before from biochemical studies (31–33,47–49), but its location and role in the reaction were not completely clear. To test the effect of this uridine in the context of the holoenzyme, and not only the isolated RNA component (33,47), the uridine was mutated to a cytidine. This is the most parsimonious change possible within the four naturally occurring RNA nucleotides and involves only the change of the O4 oxygen to an amine with the two nucleobases of approximately the same size, therefore avoiding any possible deleterious stereochemical effects. In addition, previous studies with the E. coli ribozyme have shown that a U69C (U52C) mutation retains the ability to bind substrate and has no detectable change in Tb3+ cleavage patterns, suggesting that effects of this mutation do not result from alternative pairing interactions within the P4 helix (47). As expected, mutation of the uridine to a cytidine leads to a severe loss of catalytic activity; the cytidine mutant is over one order of magnitude less active than the wild-type holoenzyme (Table 1). This is similar to what was observed in the context of the bacterial RNA component alone (∼20–100-fold less active depending on metal ion concentrations (33,47) and also in a self-cleaving (cis-acting) archaeal RNase P ribozyme-substrate system (∼80-fold less active), where significant loss of activity was also observed when the universally conserved uridine was deleted or mutated. Further, this result is nearly identical to what was observed in the archaeal mutant holoenzyme, where deletion of the conserved uridine reduces activity by 13-fold compared with the WT holoenzyme under high divalent metal ion concentrations (100 mM Mg2+) (50). All these experiments are consistent and reveal that the U52C mutant dramatically decreases activity, presumably by abolishing efficient metal ion binding to the catalytically important M1 site. This strongly suggests that even though U52 is strictly conserved and binds to the catalytic metal ion, it is not irreplaceable. Clearly, coordination through the phosphate backbone oxygen-rich environment is sufficient to orient the catalytic metal ion correctly, but optimal catalysis may be obtained when the O4 oxygen is part of the coordinating ligands. This is supported by ribozyme experiments that show metal ion specificity changes in the absence of this specific uridine (33,47). Here, a mutation, deletion or a change in the helical register (positioning) of the uridine can dramatically alter metal ion affinity and resulted in large catalytic defects, reflected by a significant decrease in apparent cooperativity of rescuing metal binding at the reactive phosphate. The previous studies, coupled with the structure of the RNase P holoenzyme-tRNA complex and functional data presented herein, strongly point to a critical but not essential role of this universally conserved, bulged uridine in both metal ion coordination and substrate positioning.
The function of the protein component of bacterial RNase P has been studied previously, but a precise role has yet to be established. The protein does have a significant effect on catalysis and under similar ionic conditions the holoenzyme is well over 100 times more active than the RNA component alone. The structure of the RNase P/tRNA complex suggests that the primary roles of the protein component are to bind the pre-tRNA leader and facilitate a stable P RNA active site architecture, consistent with several prior biochemical and crosslinking studies (8,12,13,15,29,31–40,42,43). In addition, the P protein has also been shown to enhance the affinity of metal ion binding sites, helping to maintain a rigid RNA active site architecture (22).
The structure of the complex indicates that although the protein component is positioned adjacent to the active site, it does not participate directly in catalysis. Within the pre-tRNA leader track located 5′ to the cleavage site, it is likely that distinct P RNA-N−1 and P protein-N−4 interactions occur and are important for catalysis (9,13,15,22,25,51–55). Interestingly, it seems that for efficient cleavage there may also be some intrinsic sequence preference for binding pre-tRNAs containing a uracil and an adenosine at the N−1 and N−4, respectively (13,15,22). Based on these observations, a series of point mutants selected on the basis of high sequence conservation across bacteria and/or the structure of the complex were prepared (Figure 1B) (9). The P protein residues targeted comprise the leader binding groove observed in the crystal structure, the β3α2 loop facing the active site, and a pair of arginine residues that make direct contacts with the P15/L15 region of the P RNA. In some instances, similar mutations have been studied previously (12,13,15,37,39,40,42,43), but many of the mutants herein represent residues whose putative role was inferred from the RNase P/tRNA complex structure. In all mutants, the amino acids were replaced with alanine to avoid introducing steric side chain effects. More extensive mutagenesis studies were not done as the main goal was to probe the essentiality of the amino acids targeted and their possible role in catalysis or leader recognition. Many of the mutants targeted positively charged amino acids (lysines and arginines) as these are expected to interact directly with the RNA phosphate backbone. In two instances, F17 and F21, the mutation eliminated large aromatic groups that may intercalate in between the RNA bases, as previously proposed (22,37,56). Lastly, R89 and K90 are positively charged residues that had not been biochemically targeted previously, yet reside along the path of the pre-tRNA binding track in the structure of the RNase P-tRNA complex (9). CD measurements show that all mutant constructs exhibit a consistent molar ellipticity pattern, indicative of a uniform, well-folded protein domain with negligible secondary structure differences (Supplementary Figure S2). The list of the mutants is shown in Table I, their location is illustrated in Figure 1.
All the mutants were purified to homogeneity, enabling reconstitution of the holoenzyme. Quantification of the wild-type and mutant RNase P reaction profiles (Figure 2) enabled the accurate measurement of the kinetics of substrate cleavage (kcat/KM) under single-turnover conditions. Data from the cleavage reaction allow for a comparative kinetic analysis of all the RNase P protein mutants and the results are summarized in Table I and Supplementary Figure S3. The effect of alanine point mutations on RNase P holoenzyme activity varied greatly depending upon the specific location of the protein residue. For comparative purposes, the effect on enzyme activity by each mutation is classified into four groups: those exhibiting wt-like activity, or impairing activity either modestly, substantially or severely (Figure 3). This classification allows for easier comparison of the results.
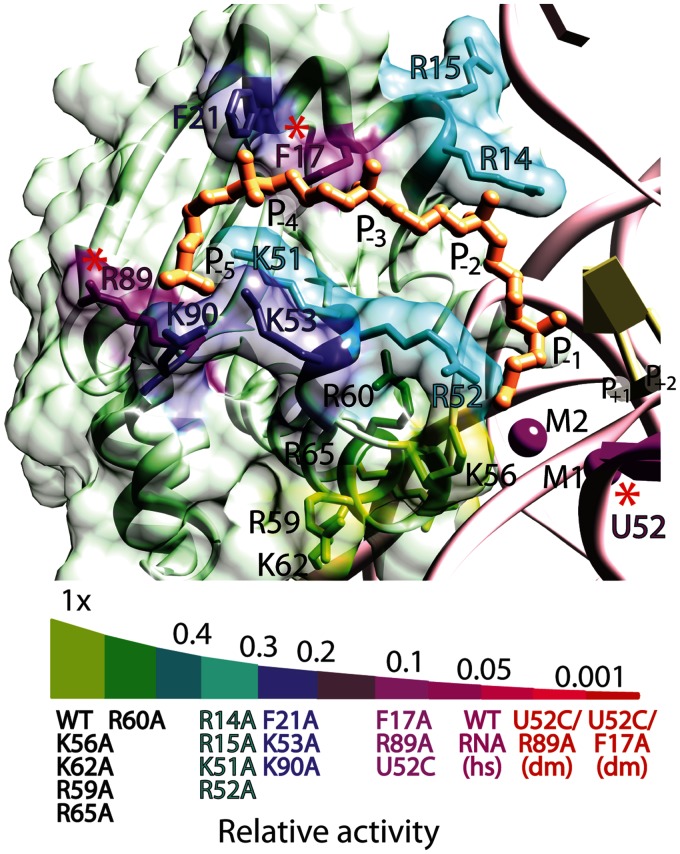
Figure 3.
Effects of mutations in the P protein on T. maritima RNase P activity. P protein variants with single-alanine exchanges were examined under single-turnover enzyme conditions and classified according to their effect on RNase P activity: wt-like (green), modest (cyan), substantial (blue) and severe (dark magenta) activity defects, as detailed in Figure 2. The results indicate that the protein residues showing the most severe reduction in activity are located far from the active site and contact the N−4 and N−5 phosphates of the leader. Single-nucleotide and amino acid exchanges that comprise the dm U52C RNA/F17A protein or U52C RNA/R89A protein RNase P holoenzymes are depicted by red asterisks. A relative activity scale classifies each mutant (as previously colour coded) into the various subgroups based on single-turnover enzyme kinetics. The dm RNase P holoenzyme mutants (red) exhibits a >210-fold decrease in activity compared with the T. maritima holoenzyme and are significantly less active than the wild-type P RNA ribozyme under high salt (magenta, hs) conditions.
Mutants displaying wt-like RNase P activity, such as K56A, R59A, R60A, K62A or R65A, are positioned within the (β3α2) loop or are part of the conserved ‘RNR’ motif of the protein. Based on the structure, these amino acids make important contacts with conserved regions of the P RNA, but do not interact with the pre-tRNA substrate directly. The lack of a marked effect of the mutants on activity is somewhat surprising as all of them contain positively charged side chains. In addition, a previous study of the Bacillus subtilis RNase P holoenzyme showed that R60A and R62A mutants reduced activity at pH 8.0 by 2 - and 5-fold, respectively, and further proposed that these residues may help to stabilize a metal dependent conformational change (43). Although it is clear from our results that these regions do not participate in catalysis, it is possible that some of the residues (such as R60, K62, and R65) support holoenzyme assembly and may indirectly enhance substrate affinity, as previously proposed (43). It is also possible that the positively charged tertiary amine folding buffer (Tris/HEPES) in our study helps to stabilize the holoenzyme (P RNA—‘RNR’) interface and masks minor holoenzyme destabilization effects that may be caused by the mutants. This may explain the discrepancy between the present R60A and K62A results and previous studies performed in MES pH 8.0 (43), where ‘RNR’-alanine mutations were shown to have marked effects on catalytic efficiency. Interestingly, mutating R65, which represents the only conserved and invariant amino acid across all bacteria, has no effect on catalytic efficiency, suggesting that R65 could serve a role in assembly and stabilization of the holoenzyme structure (43). In the case of the K56A mutant, which is the closest amino acid to the catalytically important M2 metal ion (∼4.2 Å) and is one of three protein residues within 10 Å of the reactive phosphate in the structure, no effect on catalysis was observed. This suggests that K56 does not assist in the formation of the M2 binding site, but may rather play a minor non-specific role in neutralizing the P RNA or pre-tRNA phosphate backbone. Taken together, these results suggest that the likely function for many of these amino acids is to stabilize the holoenzyme complex through protein/RNA interactions; the absence of these individual interactions is not sufficient to disrupt the complex and alter its activity.
A second class of mutants (R14A, R15A, K51A or R52A) exhibit a modest ∼2.5-fold decrease in activity compared with wild-type RNase P holoenzyme. Residues R14 and R15 make direct contacts with the P15/L15 region of the P RNA, are located far (>18 Å) from the active site, and appear to serve purely structural roles, though it is entirely possible that these P RNA-P protein interactions are functionally important during product release (9,57). Thus, mutations R14A and R15A could display diminished enzymatic activity by destabilizing the pseudoknot helical arm of the P RNA, potentially destabilizing the intermolecular P RNA (L15)—tRNA (3′-CCA) base pairs, and indirectly influencing the stability of the enzyme active site. In contrast, residues K51 and R52 are located very close to the active site (7–15 Å from the reactive phosphate) and are the most obvious residues within the pre-tRNA binding track to interact with nucleotides in the active site. Despite this location and the relatively high sequence conservation of R52 across bacteria (9), it appears that K51 and R52 serve minor roles in substrate positioning and are not directly involved in formation of the active site. The modest changes in catalytic activity displayed by these mutants is especially intriguing as R52 resides ∼5 Å from the M2 metal ion location and is uniquely poised to interact with the N−1 pre-tRNA nucleotide in the structure of the complex, whereas K51 could potentially interact with the N−3 or N−4 phosphate backbone. These residues may play electrostatic roles in stabilizing RNA phosphate backbone oxygens of the pre-tRNA leader, modestly enhancing substrate affinity and positioning, yet exhibit minimal effects on catalysis and are very unlikely to interact directly with the catalytically important M2 ion.
By comparison, point mutants F21A, K53A or K90A substantially reduce the catalytic efficiency of the RNase P holoenzyme (3–5-fold decrease). These residues are positioned within the pre-tRNA binding region of the protein and far from the active site (∼17–28 Å away from the reactive phosphate). Based on the structure of the complex, these residues likely make contacts or are nearby the N−4 and N−5 nucleotides of the pre-tRNA leader, where K53 and K90 are positioned on the phosphate backbone edge of the pre-tRNA leader and F21 is potentially poised to make a nucleobase interaction. Specifically, position 21 is occupied mostly by an F or a Y and its corresponding location in B. subtilis has been shown to make sequence-specific interactions with the N−4 nucleotide of the 5′ pre-tRNA leader, likely through hydrophobic base stacking (22). The results of that study are consistent with prior results (32,37,56,58) as well as the structure of the complex, all of which identify key interactions between the protein and the pre-tRNA substrate. Analysis of single-turnover kinetics of point mutants (F21A, K53A and K90A) further underscores the importance of the protein-N−4 and N−5 binding interactions and reveals that an extensive network of protein-pre-tRNA contacts far from the active site plays an integral role in both substrate recognition and catalytic efficiency.
Lastly, two point mutations in the protein (F17A, R89A) as well as the aforementioned U52C P RNA mutation caused severe catalytic defects. Both F17 and R89 are highly conserved in character. Position 17 is mostly an F or a Y whereas position 89 is almost always a positively charged residue (R or K). These mutations decrease activity by 13–20-fold, and are located either at the RNase P active site (U52C) or at regions far (>∼20 Å) from the reactive phosphate (R89 and F17). The comparative activity profile across the pre-tRNA binding track emphasizes critical RNase P protein-substrate binding at the N−4 and N−5 positions, as previously observed (12,13,15,37,39,40,42,43). Based on the structure of the complex, R89 is poised to make contacts with the phosphate backbone at the N−5 nucleotide and is adjacent to K90 and K53, whereas F17 is positioned to make contacts with the N−4 nucleobase of the pre-tRNA leader and is also located ∼4.5–5.0 Å from F21. Collectively, these interactions may participate in a specific hydrophobic base stacking and hydrogen bonding network that intricately links substrate binding to catalysis in the active site located over 20 Å away.
A specific RNase P interaction at the N−4 and N−5 pre-tRNA interface is consistent with multiple observations: an apparent sequence preference for adenosine likely occurs at the N−4 position, pre-tRNAs with <5 residues at their 5′ ends bind the RNase P holoenzyme with dramatically reduced affinity, transcribed tRNA genes vary in length but typically contain a 17–18 nucleotide leader region, and the length of the pre-tRNA 5′ leader can substantially alter the level of mature tRNA in vivo (22,42,59,60). These generalities combined with specific biochemical, genetic and structural studies of the RNase P-pre-tRNA complex (9,12,13,15,37,39,40,42,43,56), and the detrimental effects of the F17A and R89A mutants suggest that interactions at positions N−4 and N−5 are essential for accurate substrate binding and positioning.
Interestingly, although the N−1 and N−2 pre-tRNA nucleotides have been shown to make putative contacts with specific P RNA residues (9) and also clearly influence the binding and catalytic efficiency of RNase P (51–53), no convincing data have shown that specific protein residues make functionally important interactions with the N−1, N−2 or N−3 pre-tRNA region. A crosslinking study (with an approximate resolution of ∼12–20 Å) identified possible interactions between the pre-tRNA N−2 and N−3 regions and specific protein residues. However, it is not clear if these specific interactions directly contribute to binding or to catalysis, or if they may be attributed to changes during holoenzyme formation/assembly (56). Although it is certainly plausible that protein residues make transient contacts with N−1, N−2 and N−3 leader nucleotides, it is likely that these interactions serve only non-specific electrostatic roles, helping to stabilize binding of the leader phosphate backbone. A lack of specific protein-pre-tRNA contacts at N−1–N−3 immediately adjacent of the active site may enable RNase P to recognize a wide variety of naturally occurring non-canonical pre-tRNA substrates, allowing a certain degree of plasticity and broad specificity inherent to the enzyme. This protein/RNA interaction appears to be evolutionarily conserved across bacteria as the interactions between the pre-tRNA leader and the protein involve highly conserved regions, suggesting that the bacterial P protein has a defined, signature binding mode of interaction with all pre-tRNAs. By analogy, the highly conserved RNA–RNA tertiary interactions observed far away from the active site, involving the tRNA TΨC/D loop and Conserved Regions II and III in the P RNA (CRII/CRIII) and that serve as a ‘ruler’ to help establish that the correct acceptor stem lengths (∼7 bp long) are processed, also act as a key determinant in substrate recognition. It is clear that detailed structural information on the RNase P protein interactions with the pre-tRNA leader region will help to address how specific protein-N−4 and N−5 interactions might influence substrate affinity, positioning and sequence selection.
In addition to assessing the effect of single-point mutations on activity, two double mutants (U52C P RNA/R89A protein and U52C P RNA/F17A) RNase P holoenzymes were prepared and exhibited >210–1750-fold decreases in activity compared with the wild-type holoenzyme, respectively, and were found to be at least one order of magnitude less active than the wild-type ribozyme in the absence of the P protein (Table 1). These double mutants emphasize the synergistic coupling that likely occurs between accurate pre-tRNA substrate positioning and properly maintaining the active site geometry. It appears that each component individually affects catalytic efficiency by about the same amount, but combining such P RNA and protein variants has a substantially larger effect. Thus, our findings not only underscore the function of the universally conserved and bulged uridine nucleotide, which is to bind the catalytically important metal ion (M1) and accurately position the pre-tRNA substrate at the enzyme active site (9,33,47,50), but also helps identify the specific protein residues that are likely involved in providing critical binding interactions with the N−4 and N−5 nucleotides of the pre-tRNA substrate.
SUMMARY
Comparative kinetic analysis shows clearly that the bacterially conserved regions closest to the active site (R52, K56) and those within the conserved ‘RNR’ region (R59–R65) have only a modest effect on catalytic efficiency. This is consistent with the intermolecular contacts observed in the structure of the complex and indicates that these protein residues likely participate in hydrogen bonding, stacking interactions and electrostatic stabilization to conserved regions of the P RNA, but do not participate directly in catalysis. In contrast, bacterially conserved P protein locations F17 or R89, located within the substrate binding path but far (>20 Å) from the enzyme active site, have severe effects on RNase P holoenzyme activity emphasizing the important role that leader recognition and binding play in tRNA processing activity. In addition, a point mutation of a universally conserved P RNA residue (U52) located at the proposed active site of the enzyme also results in dramatically reduced enzyme activity compared with the wild-type T. maritima RNase P holoenzyme. Combined with previous biochemical studies and knowledge of the structure of the RNase P holoenzyme in complex with tRNA, these studies identify specific regions of RNase P that are critical to accurately align the substrate and assist in the catalytic reaction. Future experiments probing both the enzyme-substrate (E•S) and enzyme-product (E•P) interface will be essential to unravel the catalytic mechanism of pre-tRNA by RNase P.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1–3.
FUNDING
N.J.R. was an National Institutes of Health National Service Award postdoctoral fellow [5F32GM087055]. National Institutes of Health (NIH) [5R01GM058443 to A.M.]. Funding for open access charge: NIH.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Tao Pan for help and advice throughout the project and the reviewers for valuable comments and suggestions on the manuscript. We thank Clarence Chan for critical reading of the manuscript, Bhaskar Chetnani and Alfredo Torres-Larios for help and suggestions, Taylor Poor and Robert Lamb for mass spectrometry, Mai Yasunaga for help with CD measurements, Rakhi Rajan, Dan Grilley, Eliza Small, Georgette Moyle Heyrman and Linlin Zhao for technical advice, and F. Peter Guengerich for help and advice. We acknowledge staff and instrumentation support from the Keck Biophysics Facility.
REFERENCES
- 1.Guerrier-Takada C, Gardiner K, Marsh T, Pace N, Altman S. The RNA moiety of ribonuclease P is the catalytic subunit of the enzyme. Cell. 1983;35:849–857. doi: 10.1016/0092-8674(83)90117-4. [DOI] [PubMed] [Google Scholar]
- 2.Liu F, Altman S. Ribonuclease P. New York: Springer Science+Business Media, LLC; 2010. [Google Scholar]
- 3.Kazantsev AV, Pace NR. Bacterial RNase P: a new view of an ancient enzyme. Nat. Rev. Microbiol. 2006;4:729–740. doi: 10.1038/nrmicro1491. [DOI] [PubMed] [Google Scholar]
- 4.Hartmann RK, Gossringer M, Spath B, Fischer S, Marchfelder A. The making of tRNAs and more – RNase P and tRNase Z. Prog. Mol. Biol. Transl. Sci. 2009;85:319–368. doi: 10.1016/S0079-6603(08)00808-8. [DOI] [PubMed] [Google Scholar]
- 5.Apirion D. Genetic mapping and some characterization of the rnpA49 mutation of Escherichia coli that affects the RNA-processing enzyme ribonuclease P. Genetics. 1980;94:291–299. doi: 10.1093/genetics/94.2.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schedl P, Primakoff P. Mutants of Escherichia coli thermosensitive for the synthesis of transfer RNA. Proc. Natl Acad. Sci. USA. 1973;70:2091–2095. doi: 10.1073/pnas.70.7.2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pan T, Loria A, Zhong K. Probing of tertiary interactions in RNA: 2′-hydroxyl-base contacts between the RNase P RNA and pre-tRNA. Proc. Natl Acad. Sci. USA. 1995;92:12510–12514. doi: 10.1073/pnas.92.26.12510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kirsebom LA, Svard SG. Base pairing between Escherichia coli RNase P RNA and its substrate. EMBO J. 1994;13:4870–4876. doi: 10.1002/j.1460-2075.1994.tb06814.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reiter NJ, Osterman A, Torres-Larios A, Swinger KK, Pan T, Mondragon A. Structure of a bacterial ribonuclease P holoenzyme in complex with tRNA. Nature. 2010;468:784–789. doi: 10.1038/nature09516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Buck AH, Dalby AB, Poole AW, Kazantsev AV, Pace NR. Protein activation of a ribozyme: the role of bacterial RNase P protein. EMBO J. 2005;24:3360–3368. doi: 10.1038/sj.emboj.7600805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kurz JC, Niranjanakumari S, Fierke CA. Protein component of Bacillus subtilis RNase P specifically enhances the affinity for precursor-tRNAAsp. Biochemistry. 1998;37:2393–2400. doi: 10.1021/bi972530m. [DOI] [PubMed] [Google Scholar]
- 12.Koutmou KS, Zahler NH, Kurz JC, Campbell FE, Harris ME, Fierke CA. Protein-precursor tRNA contact leads to sequence-specific recognition of 5′ leaders by bacterial ribonuclease P. J. Mol. Biol. 2010;396:195–208. doi: 10.1016/j.jmb.2009.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sun L, Campbell FE, Zahler NH, Harris ME. Evidence that substrate-specific effects of C5 protein lead to uniformity in binding and catalysis by RNase P. EMBO J. 2006;25:3998–4007. doi: 10.1038/sj.emboj.7601290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reich C, Olsen GJ, Pace B, Pace NR. Role of the protein moiety of ribonuclease P, a ribonucleoprotein enzyme. Science. 1988;239:178–181. doi: 10.1126/science.3122322. [DOI] [PubMed] [Google Scholar]
- 15.Sun L, Campbell FE, Yandek LE, Harris ME. Binding of C5 protein to P RNA enhances the rate constant for catalysis for P RNA processing of pre-tRNAs lacking a consensus G(+1)/C(+72) Pair. J. Mol. Biol. 2010;395:1019–1037. doi: 10.1016/j.jmb.2009.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Loria A, Niranjanakumari S, Fierke CA, Pan T. Recognition of a pre-tRNA substrate by the Bacillus subtilis RNase P holoenzyme. Biochemistry. 1998;37:15466–15473. doi: 10.1021/bi9816507. [DOI] [PubMed] [Google Scholar]
- 17.Peck-Miller KA, Altman S. Kinetics of the processing of the precursor to 4.5 S RNA, a naturally occurring substrate for RNase P from Escherichia coli. J. Mol. Biol. 1991;221:1–5. doi: 10.1016/0022-2836(91)80194-y. [DOI] [PubMed] [Google Scholar]
- 18.Torres-Larios A, Swinger KK, Krasilnikov AS, Pan T, Mondragon A. Crystal structure of the RNA component of bacterial ribonuclease P. Nature. 2005;437:584–587. doi: 10.1038/nature04074. [DOI] [PubMed] [Google Scholar]
- 19.Kazantsev AV, Krivenko AA, Harrington DJ, Holbrook SR, Adams PD, Pace NR. Crystal structure of a bacterial ribonuclease P RNA. Proc. Natl Acad. Sci. USA. 2005;102:13392–13397. doi: 10.1073/pnas.0506662102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krasilnikov AS, Xiao Y, Pan T, Mondragon A. Basis for structural diversity in homologous RNAs. Science. 2004;306:104–107. doi: 10.1126/science.1101489. [DOI] [PubMed] [Google Scholar]
- 21.Krasilnikov AS, Yang X, Pan T, Mondragón A. Crystal structure of the specificity domain of ribonuclease P. Nature. 2003;421:760–764. doi: 10.1038/nature01386. [DOI] [PubMed] [Google Scholar]
- 22.Hsieh J, Koutmou KS, Rueda D, Koutmos M, Walter NG, Fierke CA. A divalent cation stabilizes the active conformation of the B. subtilis RNase P pre-tRNA complex: a role for an inner-sphere metal ion in RNase P. J. Mol. Biol. 2010;400:38–51. doi: 10.1016/j.jmb.2010.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kirsebom LA. In: Ribonuclease P. Liu F, Altman S, editors. New York: Springer Science+Business Media, LLC; 2010. Chapter 7, pp. 113–134. [Google Scholar]
- 24.Kurz JC, Fierke CA. The affinity of magnesium binding sites in the Bacillus subtilis RNase P center dot pre-tRNA complex is enhanced by the protein subunit. Biochemistry. 2002;41:9545–9558. doi: 10.1021/bi025553w. [DOI] [PubMed] [Google Scholar]
- 25.Smith D, Pace NR. Multiple magnesium ions in the ribonuclease P reaction mechanism. Biochemistry. 1993;32:5273–5281. doi: 10.1021/bi00071a001. [DOI] [PubMed] [Google Scholar]
- 26.Beebe JA, Kurz JC, Fierke CA. Magnesium ions are required by Bacillus subtilis ribonuclease P RNA for both binding and cleaving precursor tRNAAsp. Biochemistry. 1996;35:10493–10505. doi: 10.1021/bi960870m. [DOI] [PubMed] [Google Scholar]
- 27.Cassano AG, Anderson VE, Harris ME. Evidence for direct attack by hydroxide in phosphodiester hydrolysis. J. Am. Chem. Soc. 2002;124:10964–10965. doi: 10.1021/ja020823j. [DOI] [PubMed] [Google Scholar]
- 28.Cassano AG, Anderson VE, Harris ME. Analysis of solvent nucleophile isotope effects: evidence for concerted mechanisms and nucleophilic activation by metal coordination in nonenzymatic and ribozyme-catalyzed phosphodiester hydrolysis. Biochemistry. 2004;43:10547–10559. doi: 10.1021/bi049188f. [DOI] [PubMed] [Google Scholar]
- 29.Chen Y, Li X, Gegenheimer P. Ribonuclease P catalysis requires Mg2+ coordinated to the pro-RP oxygen of the scissile bond. Biochemistry. 1997;36:2425–2438. doi: 10.1021/bi9620464. [DOI] [PubMed] [Google Scholar]
- 30.Warnecke JM, Furste JP, Hardt WD, Erdmann VA, Hartmann RK. Ribonuclease P (RNase P) RNA is converted to a Cd(2+)-ribozyme by a single Rp-phosphorothioate modification in the precursor tRNA at the RNase P cleavage site. Proc. Natl Acad. Sci. USA. 1996;93:8924–8928. doi: 10.1073/pnas.93.17.8924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Christian EL, Kaye NM, Harris ME. Helix P4 is a divalent metal ion binding site in the conserved core of the ribonuclease P ribozyme. RNA. 2000;6:511–519. doi: 10.1017/s1355838200000042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Crary SM, Kurz JC, Fierke CA. Specific phosphorothioate substitutions probe the active site of Bacillus subtilis ribonuclease P. RNA. 2002;8:933–947. doi: 10.1017/s1355838202025025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Christian EL, Smith KM, Perera N, Harris ME. The P4 metal binding site in RNase P RNA affects active site metal affinity through substrate positioning. RNA. 2006;12:1463–1467. doi: 10.1261/rna.158606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hardt WD, Warnecke JM, Erdmann VA, Hartmann RK. Rp-phosphorothioate modifications in RNase P RNA that interfere with tRNA binding. EMBO J. 1995;14:2935–2944. doi: 10.1002/j.1460-2075.1995.tb07293.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Burgin AB, Pace NR. Mapping the active site of ribonuclease P RNA using a substrate containing a photoaffinity agent. EMBO J. 1990;9:4111–4118. doi: 10.1002/j.1460-2075.1990.tb07633.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Biswas R, Ledman DW, Fox RO, Altman S, Gopalan V. Mapping RNA-protein interactions in ribonuclease P from Escherichia coli using disulfide-linked EDTA-Fe. J. Mol. Biol. 2000;296:19–31. doi: 10.1006/jmbi.1999.3443. [DOI] [PubMed] [Google Scholar]
- 37.Niranjanakumari S, Stams T, Crary SM, Christianson DW, Fierke CA. Protein component of the ribozyme ribonuclease P alters substrate recognition by directly contacting precursor tRNA. Proc. Natl Acad. Sci. USA. 1998;95:15212–15217. doi: 10.1073/pnas.95.26.15212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Buck AH, Kazantsev AV, Dalby AB, Pace NR. Structural perspective on the activation of RNase P RNA by protein. Nat. Struct. Mol. Biol. 2005;12:958–964. doi: 10.1038/nsmb1004. [DOI] [PubMed] [Google Scholar]
- 39.Sun L, Harris ME. Evidence that binding of C5 protein to P RNA enhances ribozyme catalysis by influencing active site metal ion affinity. RNA. 2007;13:1505–1515. doi: 10.1261/rna.571007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gopalan V, Baxevanis AD, Landsman D, Altman S. Analysis of the functional role of conserved residues in the protein subunit of ribonuclease P from Escherichia coli. J. Mol. Biol. 1997;267:818–829. doi: 10.1006/jmbi.1997.0906. [DOI] [PubMed] [Google Scholar]
- 41.Paul R, Lazarev D, Altman S. Characterization of RNase P from Thermotoga maritima. Nucleic Acids Res. 2001;29:880–885. doi: 10.1093/nar/29.4.880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Crary SM, Niranjanakumari S, Fierke CA. The protein component of Bacillus subtilis ribonuclease P increases catalytic efficiency by enhancing interactions with the 5′ leader sequence of pre-tRNAAsp. Biochemistry. 1998;37:9409–9416. doi: 10.1021/bi980613c. [DOI] [PubMed] [Google Scholar]
- 43.Koutmou KS, Day-Storms JJ, Fierke CA. The RNR motif of B. subtilis RNase P protein interacts with both PRNA and pre-tRNA to stabilize an active conformer. RNA. 2011;17:1225–1235. doi: 10.1261/rna.2742511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Krivenko AA, Kazantsev AV, Adamidi C, Harrington DJ, Pace NR. Expression, purification, crystallization and preliminary diffraction analysis of RNase P protein from Thermotoga maritima. Acta Crystallogr. D Biol. Crystallogr. 2002;58:1234–1236. doi: 10.1107/s0907444902007965. [DOI] [PubMed] [Google Scholar]
- 45.Kazantsev AV, Krivenko AA, Pace NR. Mapping metal-binding sites in the catalytic domain of bacterial RNase P RNA. RNA. 2009;15:266–276. doi: 10.1261/rna.1331809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Reiter NJ, Chan CW, Mondragon A. Emerging structural themes in large RNA molecules. Curr. Opin. Struct. Biol. 2011;21:319–326. doi: 10.1016/j.sbi.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kaye NM, Zahler NH, Christian EL, Harris ME. Conservation of helical structure contributes to functional metal ion interactions in the catalytic domain of ribonuclease P RNA. J. Mol. Biol. 2002;324:429–442. doi: 10.1016/s0022-2836(02)01094-x. [DOI] [PubMed] [Google Scholar]
- 48.Christian EL, Kaye NM, Harris ME. Evidence for a polynuclear metal ion binding site in the catalytic domain of ribonuclease P RNA. EMBO J. 2002;21:2253–2262. doi: 10.1093/emboj/21.9.2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Harris ME, Pace NR. Identification of phosphates involved in catalysis by the ribozyme RNase P RNA. RNA. 1995;1:210–218. [PMC free article] [PubMed] [Google Scholar]
- 50.Chen WY, Xu Y, Cho IM, Oruganti SV, Foster MP, Gopalan V. Cooperative RNP assembly: complementary rescue of structural defects by protein and RNA subunits of archaeal RNase P. J. Mol. Biol. 2011;411:368–383. doi: 10.1016/j.jmb.2011.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zahler NH, Christian EL, Harris ME. Recognition of the 5′ leader of pre-tRNA substrates by the active site of ribonuclease P. RNA. 2003;9:734–745. doi: 10.1261/rna.5220703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zahler NH, Sun L, Christian EL, Harris ME. The pre-tRNA nucleotide base and 2′-hydroxyl at N(-1) contribute to fidelity in tRNA processing by RNase P. J. Mol. Biol. 2005;345:969–985. doi: 10.1016/j.jmb.2004.10.080. [DOI] [PubMed] [Google Scholar]
- 53.Brannvall M, Fredrik Pettersson BM, Kirsebom LA. The residue immediately upstream of the RNase P cleavage site is a positive determinant. Biochimie. 2002;84:693–703. doi: 10.1016/s0300-9084(02)01462-1. [DOI] [PubMed] [Google Scholar]
- 54.Loria A, Pan T. Recognition of the 5′ leader and the acceptor stem of a pre-tRNA substrate by the ribozyme from Bacillus subtilis RNase P. Biochemistry. 1998;37:10126–10133. doi: 10.1021/bi980220d. [DOI] [PubMed] [Google Scholar]
- 55.Loria A, Pan T. The cleavage step of ribonuclease P catalysis is determined by ribozyme-substrate interactions both distal and proximal to the cleavage site. Biochemistry. 1999;38:8612–8620. doi: 10.1021/bi990691f. [DOI] [PubMed] [Google Scholar]
- 56.Niranjanakumari S, Day-Storms JJ, Ahmed M, Hsieh J, Zahler NH, Venters RA, Fierke CA. Probing the architecture of the B. subtilis RNase P holoenzyme active site by cross-linking and affinity cleavage. RNA. 2007;13:521–535. doi: 10.1261/rna.308707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tallsjo A, Kirsebom LA. Product release is a rate-limiting step during cleavage by the catalytic RNA subunit of Escherichia coli RNase P. Nucleic Acids Res. 1993;21:51–57. doi: 10.1093/nar/21.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rueda D, Hsieh J, Day-Storms JJ, Fierke CA, Walter NG. The 5′ leader of precursor tRNAAsp bound to the Bacillus subtilis RNase P holoenzyme has an extended conformation. Biochemistry. 2005;44:16130–16139. doi: 10.1021/bi0519093. [DOI] [PubMed] [Google Scholar]
- 59.Fredrik Pettersson BM, Ardell DH, Kirsebom LA. The length of the 5′ leader of Escherichia coli tRNA precursors influences bacterial growth. J. Mol. Biol. 2005;351:9–15. doi: 10.1016/j.jmb.2005.05.022. [DOI] [PubMed] [Google Scholar]
- 60.Smith JD. Transcription and processing of transfer RNA precursors. Prog. Nucleic Acid Res. Mol. Biol. 1976;16:25–73. doi: 10.1016/s0079-6603(08)60755-2. [DOI] [PubMed] [Google Scholar]
- 61.Huang CC, Couch GS, Pettersen EF, Ferrin TE. Chimera: an extensible molecular modeling application constructed using standard components. Pac. Symp. Biocomput. 1996;1:724. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.