Abstract
How do cells position the Spo11 (Rec12)-dependent initiation of meiotic recombination at hotspots? The mechanisms are poorly understood and a prevailing view is that they differ substantially between phylogenetic groups. However, recent work discovered that individual species have multiple different DNA sequence-specific, protein–DNA complexes that regulate (and are essential for the activation of) recombination hotspots. The cis-acting elements function combinatorially with documented examples of synergism, antagonism and redundancy. Furthermore, we provide evidence that all currently well-defined modules of this multifactorial, cis-acting regulation are conserved functionally between taxa whose latest common ancestor occurred more than 1 billion years ago. Functionally conserved components include the ATF/CREB-family heterodimer Atf1-Pcr1 and its CRE-like DNA site M26, the CCAAT-box-binding complex Php2-Php3-Php5 and the CCAAT-box, and the zinc-finger protein Rst2 and its Oligo-C motif. The newfound multiplicity, functional redundancy and conservation of cis-acting controls constitute a paradigm shift with broad implications. They provide compelling evidence that most meiotic recombination is, like transcription, regulated by sequence-specific protein–DNA complexes. And the new findings provide important mechanistic insight, such as a solution to the conundrum that Prdm9 is a ‘master regulator’ of—yet is dispensable for—hotspot activity in mammals.
INTRODUCTION
In meiosis, cells induce dsDNA breaks (DSBs) to initiate recombination between homologous chromosomes. This process ensures the faithful segregation of homologs in the first of the two meiotic divisions and creates genetic diversity among meiotic products. Hotspots regulate the position and frequency of recombination along chromosomes, but the mechanisms are poorly understood. It is widely believed that yeast hotspots do not require specific DNA sequence motifs and that mechanisms are different in mammals, where a sequence-specific DNA binding protein, Prdm9, regulates hotspots [reviewed by (1–3)]. Recently published data support an opposing view and, moreover, provide compelling evidence that mechanisms for multifactorial, cis-acting regulation of meiotic recombination are conserved evolutionarily.
Many DNA sequence motifs regulate hotspots
In 1971, Herbert Gutz (4) described the allele-specific (cis-acting) regulation of meiotic recombination hotspots in fission yeast. About two decades elapsed before identification of the corresponding regulatory DNA site (5). Nearly two more decades elapsed before any additional cis-acting regulatory DNA sequence motifs were defined functionally at similar resolution (6,7). This history illustrates an important point. Hotspot-regulating DNA sequence motifs are difficult to discover and characterize, but this does not constitute evidence for their absence. Indeed, sophisticated genetic analyses [e.g. (8–10)] and cutting-edge technologies (11) were required to reveal, only recently, the presence of hotspot-associated motifs in mammals. Similarly, an insightful, labor-intensive screen for recombinogenic DNA sequences (coupled with systematic mutagenesis) discovered recently that individual eukaryotes have many different DNA sequence motifs that activate hotspots (6,7).
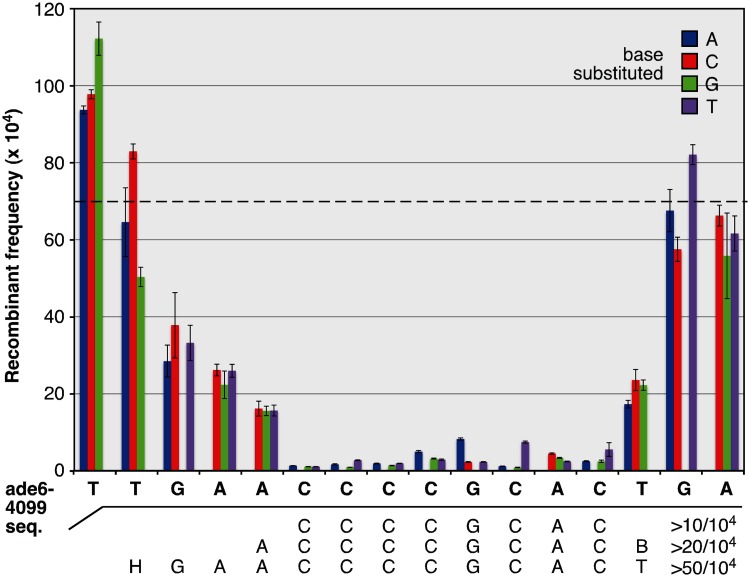
In the fission yeast Schizosaccharomyces pombe, five distinct DNA sequence motifs have been shown, by scanning base pair substitutions that create or ablate the motifs, to regulate hotspots [e.g. Figure 1 (5–7,12)]. These discrete DNA sites are each essential for hotspot activity, can function redundantly at the same test locus (6) and are active at multiple locations in the genome (7,13,14). The specific activity (hotspot intensity) varies by location of the motifs in the genome and not all occurrences are recombinogenic (13,14), revealing context-variable penetrance that is recapitulated by other DNA sequence-dependent or sequence-associated hotspots [e.g. predicted Prdm9 binding sites in mammals (8,11)]. Nearly 200 additional hotspot-activating DNA sequences were identified experimentally among approximately 46 000 randomized 15-mers and 30-mers, but have not yet been refined to single nucleotide resolution by scanning base pair substitution mutagenesis (6). These DNA sequences do not share obvious consensus motifs with one another or with the five discrete DNA sites described above, and thus expand markedly the number of different DNA sequences known to activate hotspots. In short, many different DNA sequence motifs help to position meiotic recombination at hotspots in fission yeast.
Figure 1.
Discrete DNA sites regulate hotspots. Base pair substitutions can create and ablate meiotic recombination hotspots (5,6,12,13,64). An example of high resolution analysis is shown here. The DNA sequence of a hotspot allele of fission ade6 (ade6-4099) that contains an Oligo-C motif is shown in bold. Plot displays average recombinant frequencies for that hotspot allele (dashed line) and for single base pair variants of that sequence (histogram bars). The sequences required to achieve different thresholds of hotspot activity are indicated below the graph (B = G or T or C; H = A or C or T). Reproduced from (7) © 2011 with permission from the Genetics Society of America.
In the budding yeast Saccharomyces cerevisiae, three different DNA sequence motifs have been implicated, by deletions or insertions that include the motifs, to regulate hotspot function (15–17). Each of these DNA segments contributes incrementally to hotspot activity, they can function additively or redundantly at the same locus and, where tested, the DNA sites display context-variable penetrance (18). About 50 more DNA sequence motifs are implicated by association to help regulate hotspots, because a DSB hotspot is present at ≥50% of occurrences of the corresponding motif in the genome [our inference from data in (19)]. We note that some of these motifs (e.g. predicted binding sites for Sko1 and Pho4) always have an associated hotspot, indicating a key role in hotspot regulation. Other motifs (e.g. for Spt2 and Spt23) rarely have associated hotspots, suggesting that the regulation of hotspots is specific to a subset of protein–DNA complexes. More to the point, the experimental and correlative data indicate that budding yeast, like fission yeast, has many different DNA sequence motifs that regulate hotspot activity.
Hotspot motif discovery in silico is limited by the resolution of recombination maps, by the multiplicity and functional redundancy and context-variable penetrance of regulatory DNA sites, and by the computational parameters applied. Even when high resolution maps are available, computational searches can fail to identify [e.g. (20,21)] hotspot-associated DNA sequence motifs that are demonstrably recombinogenic (5–7,13,14). Despite these limitations, it was discovered recently that eukaryotes as diverse as the protist Plasmodium falciparum (22), species of Drosophila (23–25), honeybees (26), mice (27,28) and humans (8,29,30) each have multiple different motifs associated with hotspots or sites of crossover recombination. Pending assessment of functionality (e.g. Figure 1), it appears that the use of multiple different DNA sequence motifs to regulate meiotic recombination is employed broadly, perhaps universally across eukaryotic taxa. And as revealed clearly by a biological screen for short DNA sequences that activate hotspots (6), many additional hotspot motifs remain to be discovered.
The newfound multiplicity (5–7), functional redundancy (6) and context-variable penetrance (13,14) of discrete hotspot-activating DNA sites have profound implications. One must consider that fundamental mechanisms of transcriptional regulation—including the concerted action of multiple different cis-acting DNA sites and their binding proteins—are germane to the regulation of meiotic recombination. In support of this new model, synergism between at least two cis-linked DNA sequence elements is essential for activity of the ade6-M26 hotspot of fission yeast (31). Similarly, a human hotspot-associated motif is approximately 50-fold ‘hotter’ (by association) when linked to THE1B DNA sequence repeats (3,8). Such multifactorial interactions in cis provide a mechanistic basis for the context-variable penetrance (insufficiency) of individual regulatory DNA sequence motifs and epigenetic modifications that help to position meiotic recombination.
Sequence-specific binding proteins regulate hotspots
Several DNA sequence-specific, hotspot-regulating protein complexes have been identified in fission yeast and budding yeast. Where tested systematically by scanning base pair substitution mutagenesis, there is congruence between protein–DNA binding and hotspot activity (6,7,12). Other protein–DNA complexes have been implicated, but not characterized for protein–DNA binding versus recombination (15–17). However in every case, mutants lacking the DNA binding proteins (Atf1-Pcr1 heterodimer, Php2-Php3-Php5 complex, Bas1, Bas2, Rst2) lack the sequence-dependent stimulation of recombination (e.g. Figure 2) or, for an essential protein (Rap1), hotspot activity displays a protein dose-dependent response (6,7,15–17,32). Together, these findings demonstrate that multiple different sequence-specific protein–DNA complexes regulate meiotic recombination hotspots in two highly diverged species. [While S. pombe and S. cerevisiae share the moniker ‘yeast’, their lineages diverged more than 1 billion years ago (33) and they are very different from one another.]
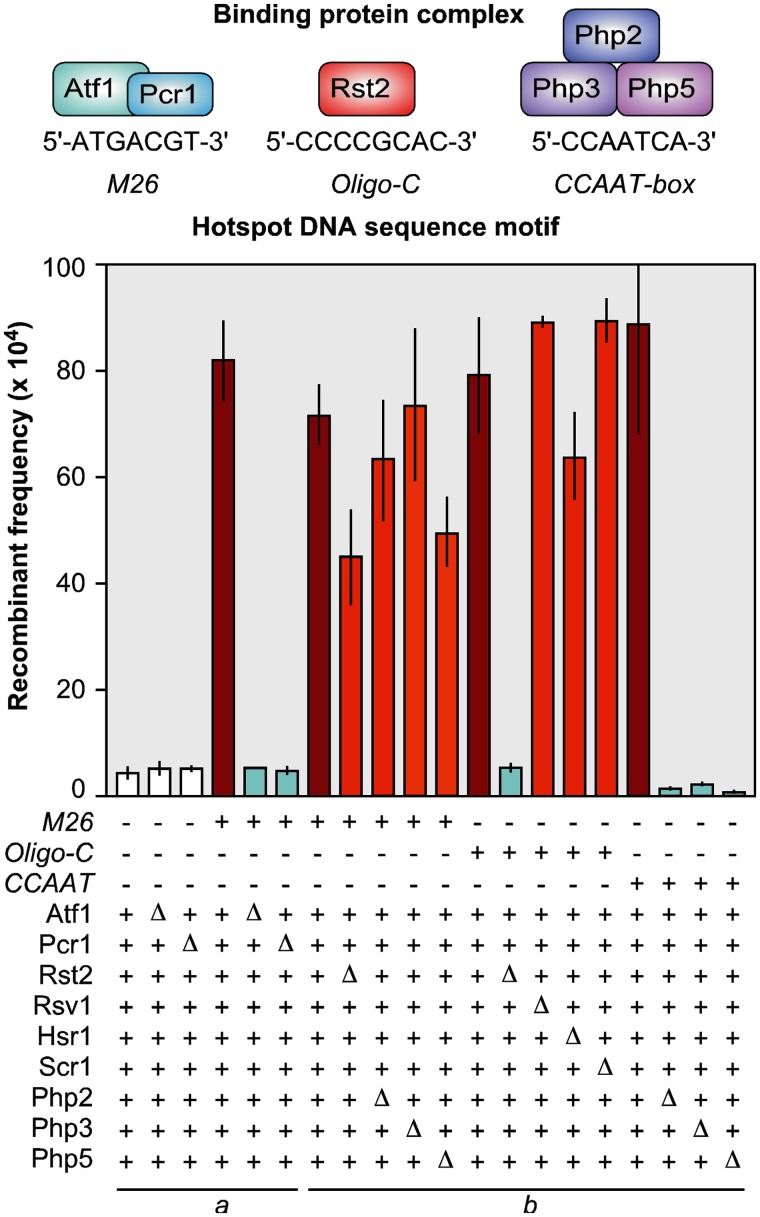
Figure 2.
Discrete protein–DNA complexes are essential for hotspot activity, act in cis with high specificity, and can function redundantly. Three hotspot motifs have been defined experimentally at high resolution (e.g. Figure 1) and have binding-protein complexes whose hotspot-regulating functions have been characterized by mutation (5–7,12,32). Top panel depicts protein complexes of fission yeast (orthologs are present in other taxa) and DNA sites to which they bind. Plotted are the effects of each DNA site and binding protein upon recombinant frequencies at the same test locus. Values for M26 are presented twice to match data set comparisons in the original articles (underscored groups). Note three things. First, each DNA site generates a hotspot (red) relative to negative controls (white). Second, for each DNA site-dependent hotspot, the corresponding binding proteins are essential for hotspot activity (light blue). Third, the proteins that bind to and promote recombination at their cognate DNA site have no significant or only modest effects upon hotspot activity of other DNA sites (orange). Similarly, zinc-finger proteins Rsv1, Hsr1 and Scr1 have no significant effect upon Oligo-C hotspot activity, which is conferred by binding of zinc-finger protein Rst2. Plotted from data in a(32) © 1997 and b(7) © 2011 with permission from the National Academy of Sciences and the Genetics Society of America.
To our knowledge, the targeted mutation of hotspot-associated DNA sequence motifs has not been applied to study hotspot activation in mammals or any other metazoans, making it difficult to assess whether binding proteins regulate such hotspots directly in cis. However, crossover hotspot-association studies that evaluated polymorphisms in the DNA binding domain of Prdm9 and its predicted DNA binding sites provided evidence that Prdm9–DNA complexes regulate hotspots in mice and humans (9–11,34–36). Similarly, in transgenic mice expressing Prdm9 variants with different predicted DNA binding specificities, motif-associated crossover hotspot activity correlates with in vitro DNA binding of recombinant, epitope-tagged proteins (37). Subsequent mapping of DSB positions revealed that both the ablation of Prdm9 and changes in its predicted DNA binding specificity alter the global distribution of hotspots (38). Evidently, taxa as different as fungi and mammals use sequence-specific DNA binding proteins to help position meiotic recombination. And as revealed recently by systematic experiments in fission yeast (6,7), we are currently looking at the ‘tip of the iceberg’ for such cis-acting regulatory mechanisms.
Intriguingly, some of the hotspot-regulating protein–DNA complexes can function both positively and negatively. Global analyses in budding yeast revealed that chromosome-bound Bas1 can activate or repress hotspots at different locations in the genome (18). Similarly, mutational dissection of Atf1-Pcr1 heterodimer in fission yeast revealed positive and negative, protein domain-specific modulation of recombination (39). And when bound to the chromosome, the Atf1-Pcr1 heterodimer can seed alternatively euchromatin and heterochromatin (40–42). Such dual specificity of hotspot-regulating proteins is also evident in mammals (although it is not known whether repression is exerted directly in cis or indirectly in trans), because the removal of Prdm9 from mice creates about as many new hotspots as it ablates (38). The mechanisms likely involve both multifactorial regulation in cis [e.g. context-dependent action of chromosome-bound Bas1 (18)] and its control by signal transduction pathways [e.g. agonistic versus antagonistic regulation of Atf1-Pcr1 heterodimer by mitogen/stress-activated protein kinase (MAPK/SAPK) and cAMP-dependent protein kinase (PKA) pathways, respectively (40,43–46)].
All sequence-specific, hotspot-regulating proteins identified so far are transcription factors. However, results of a genome-wide association study (19) suggest that not all transcription factors are recombinogenic. The distinction might lie in the modular nature of transcription factors, which can contain several different effector domains in addition to the DNA binding domain. The existence of homologous recombination activation (HRA) domains was revealed by tethering a portion of budding yeast Rap1 to the chromosome via a heterologous DNA binding domain (47). Similarly, scanning analyses of fission yeast Atf1-Pcr1 heterodimer revealed a discrete HRA region within Atf1 that is sufficient to promote recombination when tethered to the chromosome (39). Notably, the HRA region of Atf1-Pcr1 heterodimer is different from that required to induce transcription. Hotspot regulation is apparently specific to a subset of transcription factors and the mechanisms are, at least in part, distinct from those that regulate transcription. Looking forward, the mapping and functional analysis of HRA domains will be crucial for elucidating pathway mechanisms that control the position and frequency of meiotic recombination.
Multifactorial, cis-acting regulation is conserved evolutionarily
In fission yeast, the ATF/CREB-family heterodimer Atf1-Pcr1 binds to the cyclic-AMP responsive element (CRE)-like DNA site M26 to promote recombination (12,32) (see Figure 2). The CCAAT-box binding complex Php2-Php3-Php5 binds to and promotes recombination at the CCAAT-box (6). And the zinc-finger protein Rst2 promotes recombination when bound to the Oligo-C motif (7). With one possible exception (the small Pcr1 subunit of Atf1-Pcr1 heterodimer), each of the sequence-specific DNA binding proteins known to be essential for sequence-dependent hotspot activity in fission yeast has an ortholog in budding yeast. In every case, DSB hotspots are associated strongly with (i.e. directed non-randomly to) the binding sites of the ortholog (e.g. 100% association for ortholog pair Atf1/Sko1) [data in (19)]. The fission yeast and budding yeast lineages diverged more than 1 billion years ago (33), indicating that the recombination-promoting functions of these sequence-specific DNA binding proteins are ancient and have been retained during the radiation of eukaryotic lineages. Key regulatory proteins, such as subunits of the CCAAT-box binding complex, are represented broadly across domain Eukarya, although it remains to be seen whether their regulation of recombination hotspots is conserved beyond fission yeast and budding yeast.
Implications of conserved, multifactorial control
The sequence-specific DNA binding protein Prdm9 has been characterized as a ‘master regulator’ of hotspots in humans and mice and, by extension, other mammals [reviewed by (3)]. Its absence from broad swaths of domain Eukarya (e.g. fungi) (48), coupled with the widely held belief that yeast hotspots are not regulated by DNA sequence motifs, suggest key differences in the control of recombination between mammals and yeasts (1,2). However, DNA sequence motifs and their binding proteins do regulate hotspots in yeasts (above) and, remarkably, Prdm9 is dispensable for the initiation of meiotic recombination (49) and for hotspot activity (38,50) in mammals.
Three distinct findings each challenge the view that Prdm9 is a functionally conserved, vital and monolithic regulator of mammalian hotspots. First, it was discovered recently that in chimpanzees, unlike in humans and mice, there is no significant association between predicted Prdm9 binding sites and recombination hotspots (51). Second, mouse mutants lacking Prdm9 have as many meiotic DSBs and hotspots as wild-type mice (38,49), although their distribution is altered (38). Third, recent work made possible by the dog genome project has come, independently, to the same conclusion (Pdrm9 is dispensable for hotspot activity). Mutations early in canid evolution eliminated the Prdm9 protein, but nevertheless dogs still have abundant hotspots whose intensities, distribution and other characteristics are similar to those of humans (50). So how could Prdm9 be a ‘master regulator’ of, yet dispensable for hotspot activity in mammals? From our perspective, the multifactorial cis-acting regulation of recombination provides a solution to this Prdm9 conundrum.
We described above evidence that individual species have a multiplicity of protein–DNA complexes that regulate hotspots and that these factors are conserved functionally between taxa. As for transcription, no single type of DNA site, sequence-specific binding protein, or epigenetic modification can account for the regulated positioning of all recombination. Instead, multiple different cis-acting elements function combinatorially [with documented examples of synergism (31), antagonism (18) and redundancy (6)] to control the action of meiotic recombination protein complexes (14). Within that multifactorial context, individual DNA binding proteins such as Prdm9, Bas1 and Atf1-Pcr1 heterodimer can be both dispensable for and key regulators of hotspot activity.
Hypothetically, Prdm9 and the trimethylation of histone H3 (H3K4-me3) catalyzed by Prdm9 (11,37,49) affect the functions of other protein–DNA complexes that regulate recombination. Reciprocally, those protein–DNA complexes and epigenetic modifications that they induce, such as the hotspot-regulating acetylations of histones H3 and H4 induced in meiosis by the Atf1-Pcr1 heterodimer (42,45), likely affect the function of Prdm9–DNA complexes. And in phylogenetic lineages which never had (or had and lost) Prdm9, the multitude of other protein–DNA complexes can still position recombination at hotspots (Figure 2) (6,7,12,32). This model makes sense evolutionarily, because functionally conserved hotspot-regulating proteins of fission yeast and budding yeast are of ancient origin and are widely distributed, whereas Prdm9 arose later and is present in many (but not all) metazoan taxa (48).
Do species that have Prdm9 (e.g. humans) use a fundamentally different mechanism to regulate hotspots than species which lack Prdm9 (e.g. yeasts and dogs)? Current data suggest not. The putative biochemical role of Prdm9 in recombination (11,37,38) is probably more ancient and more broadly conserved than Prdm9 itself, because the H3K4 methyltransferase Set1 of budding yeast (which lacks a known DNA binding domain) helps to regulate many hotspots (52,53). Orthologs of Set1 are widely distributed, including in species that express Prdm9. Moreover, when Prdm9 of mice is ablated, hotspots that were localized preferentially to Prdm9-dependent regions of H3K4-me3 are replaced by hotspots that localize preferentially to regions of Prdm9-independent H3K4-me3 (38). In coupling histone methyltransferase and zinc-finger DNA binding domains about three-quarters of the way (temporally) through the evolution and radiation of eukaryotes (33,48,54), Prdm9 has streamlined the delivery of a regulatory epigenetic mark to the chromosome. This histone modification (H3K4-me3), which evidently helps to control hotspots in taxa as different as mammals and fungi (11,37,52,53), is but one of many epigenetic marks implicated to help regulate meiotic recombination (42,49,52,53,55–62). Some of these epigenetic marks are already known to be targeted by DNA sequence motifs and their binding proteins and to be required for DNA sequence-dependent hotspot activity (42,45).
Synthesis: from early paradigms to revolutionary perspectives
The identification of an allele-specific meiotic recombination hotspot (4) paved the way to reveal, about two decades ago, its regulation by a discrete DNA site (M26) (5) and binding protein complex (Atf1-Pcr1 heterodimer) (12,32). Subsequent experiments documented that this protein–DNA complex regulates hotspots at multiple locations in the genome (13). [There are approximately 50 publications on M26 hotspots of fission yeast, most of which are not cited in this article due to its focus and space constraints. The extensive body of work has revealed a multistep pathway of hotspot recombination and detailed molecular mechanisms. A fairly complete list of the publications can be found by searching PubMed for ‘(fission yeast OR pombe) AND (ade6 OR M26)’.] The ‘implications of such cis-acting control mechanisms were long underappreciated, and have even been discounted a priori in some publications, due mainly to the elusive nature of regulatory motifs (e.g. false-negative results of computational searches). But quite recently, hotspot-mapping motif-association data from a wide variety of taxa [e.g. (19,22,26,27)] and functional assays (6,7,37,38) (e.g. Figures 1 and 2) have each validated the original paradigm. The new results provide key insight into the regulated positioning of recombination within and between species.
The most striking discovery is that individual species have multiple different DNA sequence-specific, protein–DNA complexes that regulate—and are required for the activation of—recombination hotspots (5–7,12,32). An insightful and powerful experimental approach identified 197 additional short DNA sequences that activate hotspots (6), which predicts the presence of many more regulatory protein–DNA complexes. The implications are manifest and profound: discrete protein–DNA complexes regulate the position and frequency of much, if not most meiotic recombination.
A second remarkable finding is that some cis-acting regulatory elements, including sequence-specific protein–DNA complexes (18) and epigenetic modifications (53,61), can promote and repress recombination at different locations in the genome. Such factors can shape the genomic distribution of hotspots without necessarily affecting the overall frequency of hotspots [e.g., (18,38,61)]. With one exception [the alternative seeding of euchromatin and heterochromatin by Atf1-Pcr1-M26 protein–DNA complexes (40–42)], mechanisms for dual specificity are unknown. Similarly, mechanisms for the compensatory repositioning of hotspots following depletion of individual components [e.g. (38)] are unknown. Nevertheless, the results indicate clearly that the higher order positioning (and when perturbed, repositioning) of meiotic recombination is controlled, to a large extent, through cis-acting regulatory elements.
Equally striking are parallels between the control of meiotic recombination and the control of transcription. Biology has adapted parsimoniously a common framework, with shared cis-acting regulatory components (a subset of transcription factors and their binding sites), to control two disparate pathways of nucleic acid biochemistry. This makes sense mechanistically, for the goal of each pathway is to control with precision the global distribution and local specific activity of effector protein complexes (the RNA polymerase machinery and the meiotically induced Spo11/Rec12 recombination complex).
Since mechanisms for the regulation of transcription are well defined, parallels for the regulation of recombination are highly instructive. As is the case for transcription, the individual cis-acting regulatory elements of recombination (protein–DNA complexes and epigenetic modifications) display context-variable penetrance [e.g. (11,13,18,53,63)]. As is the case for transcription, the mechanisms for hotspot activation involve combinatorial or sequential interactions between multiple cis-acting regulatory factors [e.g. (31,42,45,52,56)]. And as is the case for transcription at different loci, the rates of recombination at different hotspots can vary over several orders of magnitude [e.g. (19,21,30,50,51)]. Given that the vast differences in transcription rates are controlled primarily by combinations of cis-acting regulatory elements, it is easy to envision how combinatorial associations might regulate vast differences in recombination rates of individual hotspots, as well as their distribution across the genome.
Last but not the least, we presented evidence that currently well-defined modules (5–7,12,32) of the multifactorial, cis-acting regulation of meiotic recombination are ancient and have been conserved functionally through more than 1 billion years of speciation. Our postulate of even broader functional conservation awaits further testing (e.g. in mammals). Meanwhile, what has been portrayed in the literature as key mechanistic differences between taxa might be considered alternatively to reflect broadly conserved, cis-acting regulatory mechanisms (e.g. H3K4-me3), even when established by distinct, potentially redundant routes (e.g. Prdm9 versus Set1).
FUNDING
Funding for open access charge: NIH [GM81766].
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank our colleagues for helpful discussions and the National Institutes of Health (GM81766) for supporting our research.
REFERENCES
- 1.Goodstadt L, Ponting CP. Is the control of recombination conserved among diverse eukaryotes? Heredity. 2011;106:710–711. doi: 10.1038/hdy.2010.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lichten M, de Massy B. The impressionistic landscape of meiotic recombination. Cell. 2011;147:267–270. doi: 10.1016/j.cell.2011.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Segurel L, Leffler EM, Przeworski M. The case of the fickle fingers: how the PRDM9 zinc finger protein specifies meiotic recombination hotspots in humans. PLoS Biol. 2011;9:e1001211. doi: 10.1371/journal.pbio.1001211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gutz H. Site specific induction of gene conversion in Schizosaccharomyces pombe. Genetics. 1971;69:331–337. doi: 10.1093/genetics/69.3.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schuchert P, Langsford M, Kaslin E, Kohli J. A specific DNA sequence is required for high frequency of recombination in the ade6 gene of fission yeast. EMBO J. 1991;10:2157–2163. doi: 10.1002/j.1460-2075.1991.tb07750.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Steiner WW, Steiner EM, Girvin AR, Plewik LE. Novel nucleotide sequence motifs that produce hotspots of meiotic recombination in Schizosaccharomyces pombe. Genetics. 2009;182:459–469. doi: 10.1534/genetics.109.101253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Steiner WW, Davidow PA, Bagshaw AT. Important characteristics of sequence-specific recombination hotspots in Schizosaccharomyces pombe. Genetics. 2011;187:385–396. doi: 10.1534/genetics.110.124636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Myers S, Freeman C, Auton A, Donnelly P, McVean G. A common sequence motif associated with recombination hot spots and genome instability in humans. Nat. Genet. 2008;40:1124–1129. doi: 10.1038/ng.213. [DOI] [PubMed] [Google Scholar]
- 9.Hinch AG, Tandon A, Patterson N, Song Y, Rohland N, Palmer CD, Chen GK, Wang K, Buxbaum SG, Akylbekova EL, et al. The landscape of recombination in African Americans. Nature. 2011;476:170–175. doi: 10.1038/nature10336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Berg IL, Neumann R, Sarbajna S, Odenthal-Hesse L, Butler NJ, Jeffreys AJ. Variants of the protein PRDM9 differentially regulate a set of human meiotic recombination hotspots highly active in African populations. Proc. Natl Acad. Sci. USA. 2011;108:12378–12383. doi: 10.1073/pnas.1109531108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smagulova F, Gregoretti IV, Brick K, Khil P, Camerini-Otero RD, Petukhova GV. Genome-wide analysis reveals novel molecular features of mouse recombination hotspots. Nature. 2011;472:375–378. doi: 10.1038/nature09869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wahls WP, Smith GR. A heteromeric protein that binds to a meiotic homologous recombination hot spot: correlation of binding and hot spot activity. Genes Dev. 1994;8:1693–1702. doi: 10.1101/gad.8.14.1693. [DOI] [PubMed] [Google Scholar]
- 13.Steiner WW, Smith GR. Natural meiotic recombination hot spots in the Schizosaccharomyces pombe genome successfully predicted from the simple sequence motif M26. Mol. Cell. Biol. 2005;25:9054–9062. doi: 10.1128/MCB.25.20.9054-9062.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wahls WP, Davidson MK. Discrete DNA sites regulate global distribution of meiotic recombination. Trends Genet. 2010;26:202–208. doi: 10.1016/j.tig.2010.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.White MA, Wierdl M, Detloff P, Petes TD. DNA-binding protein RAP1 stimulates meiotic recombination at the HIS4 locus in yeast. Proc. Natl Acad. Sci. USA. 1991;88:9755–9759. doi: 10.1073/pnas.88.21.9755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.White MA, Dominska M, Petes TD. Transcription factors are required for the meiotic recombination hotspot at the HIS4 locus in Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA. 1993;90:6621–6625. doi: 10.1073/pnas.90.14.6621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fan Q, Xu F, Petes TD. Meiosis-specific double-strand DNA breaks at the HIS4 recombination hot spot in the yeast Saccharomyces cerevisiae: control in cis and trans. Mol. Cell. Biol. 1995;15:1679–1688. doi: 10.1128/mcb.15.3.1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mieczkowski PA, Dominska M, Buck MJ, Gerton JL, Lieb JD, Petes TD. Global analysis of the relationship between the binding of the Bas1p transcription factor and meiosis-specific double-strand DNA breaks in Saccharomyces cerevisiae. Mol. Cell. Biol. 2006;26:1014–1027. doi: 10.1128/MCB.26.3.1014-1027.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pan J, Sasaki M, Kniewel R, Murakami H, Blitzblau HG, Tischfield SE, Zhu X, Neale MJ, Jasin M, Socci ND, et al. A hierarchical combination of factors shapes the genome-wide topography of yeast meiotic recombination initiation. Cell. 2011;144:719–731. doi: 10.1016/j.cell.2011.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cromie GA, Hyppa RW, Cam HP, Farah JA, Grewal SI, Smith GR. A discrete class of intergenic DNA dictates meiotic DNA break hotspots in fission yeast. PLoS Genet. 2007;3:e141. doi: 10.1371/journal.pgen.0030141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hyppa RW, Cromie GA, Smith GR. Indistinguishable landscapes of meiotic DNA breaks in rad50+ and rad50S strains of fission yeast revealed by a novel rad50+ recombination intermediate. PLoS Genet. 2008;4:e1000267. doi: 10.1371/journal.pgen.1000267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jiang H, Li N, Gopalan V, Zilversmit MM, Varma S, Nagarajan V, Li J, Mu J, Hayton K, Henschen B, et al. High recombination rates and hotspots in a Plasmodium falciparum genetic cross. Genome. Biol. 2011;12:R33. doi: 10.1186/gb-2011-12-4-r33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kulathinal RJ, Bennett SM, Fitzpatrick CL, Noor MA. Fine-scale mapping of recombination rate in Drosophila refines its correlation to diversity and divergence. Proc. Natl Acad. Sci. USA. 2008;105:10051–10056. doi: 10.1073/pnas.0801848105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miller DE, Takeo S, Nandanan K, Paulson A, Gogol MM, Noll AC, Perera AG, Walton KN, Gilliland WD, Li H, et al. A whole-chromosome analysis of meiotic recombination in Drosophila melanogaster. G3 (Bethesda) 2012;2:249–260. doi: 10.1534/g3.111.001396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stevison LS, Noor MA. Genetic and evolutionary correlates of fine-scale recombination rate variation in Drosophila persimilis. J. Mol. Evol. 2010;71:332–345. doi: 10.1007/s00239-010-9388-1. [DOI] [PubMed] [Google Scholar]
- 26.Bessoltane N, Toffano-Nioche C, Solignac M, Mougel F. Fine scale analysis of crossover and non-crossover and detection of recombination sequence motifs in the honeybee (Apis mellifera) PLoS One. 2012;7:e36229. doi: 10.1371/journal.pone.0036229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Billings T, Sargent EE, Szatkiewicz JP, Leahy N, Kwak IY, Bektassova N, Walker M, Hassold T, Graber JH, Broman KW, et al. Patterns of recombination activity on mouse chromosome 11 revealed by high resolution mapping. PLoS One. 2010;5:e15340. doi: 10.1371/journal.pone.0015340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu M, Kwoh CK, Przytycka TM, Li J, Zheng J. Epigenetic functions enriched in transcription factors binding to mouse recombination hotspots. Proteome Sci. 2012;10(Suppl 1):S11. doi: 10.1186/1477-5956-10-S1-S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang J, Li F, Li J, Zhang M, Zhang X. Evidence and characteristics of putative human alpha recombination hotspots. Hum. Mol. Genet. 2004;13:2823–2828. doi: 10.1093/hmg/ddh310. [DOI] [PubMed] [Google Scholar]
- 30.Myers S, Bottolo L, Freeman C, McVean G, Donnelly P. A fine-scale map of recombination rates and hotspots across the human genome. Science. 2005;310:321–324. doi: 10.1126/science.1117196. [DOI] [PubMed] [Google Scholar]
- 31.Zahn-Zabal M, Lehmann E, Kohli J. Hot spots of recombination in fission yeast: inactivation of the M26 hot spot by deletion of the ade6 promoter and the novel hotspot ura4-aim. Genetics. 1995;140:469–478. doi: 10.1093/genetics/140.2.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kon N, Krawchuk MD, Warren BG, Smith GR, Wahls WP. Transcription factor Mts1/Mts2 (Atf1/Pcr1, Gad7/Pcr1) activates the M26 meiotic recombination hotspot in Schizosaccharomyces pombe. Proc. Natl Acad. Sci. USA. 1997;94:13765–13770. doi: 10.1073/pnas.94.25.13765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Heckman DS, Geiser DM, Eidell BR, Stauffer RL, Kardos NL, Hedges SB. Molecular evidence for the early colonization of land by fungi and plants. Science. 2001;293:1129–1133. doi: 10.1126/science.1061457. [DOI] [PubMed] [Google Scholar]
- 34.Baudat F, Buard J, Grey C, Fledel-Alon A, Ober C, Przeworski M, Coop G, de Massy B. PRDM9 is a major determinant of meiotic recombination hotspots in humans and mice. Science. 2010;327:836–840. doi: 10.1126/science.1183439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Berg IL, Neumann R, Lam KW, Sarbajna S, Odenthal-Hesse L, May CA, Jeffreys AJ. PRDM9 variation strongly influences recombination hot-spot activity and meiotic instability in humans. Nat. Genet. 2010;42:859–863. doi: 10.1038/ng.658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kong A, Thorleifsson G, Gudbjartsson DF, Masson G, Sigurdsson A, Jonasdottir A, Walters GB, Jonasdottir A, Gylfason A, Kristinsson KT, et al. Fine-scale recombination rate differences between sexes, populations and individuals. Nature. 2010;467:1099–1103. doi: 10.1038/nature09525. [DOI] [PubMed] [Google Scholar]
- 37.Grey C, Barthes P, Chauveau-Le Friec G, Langa F, Baudat F, de Massy B. Mouse PRDM9 DNA-binding specificity determines sites of histone H3 lysine 4 trimethylation for initiation of meiotic recombination. PLoS Biol. 2011;9:e1001176. doi: 10.1371/journal.pbio.1001176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brick K, Smagulova F, Khil P, Camerini-Otero RD, Petukhova GV. Genetic recombination is directed away from functional genomic elements in mice. Nature. 2012;485:642–645. doi: 10.1038/nature11089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gao J, Davidson MK, Wahls WP. Distinct regions of ATF/CREB proteins Atf1 and Pcr1 control recombination hotspot ade6-M26 and the osmotic stress response. Nucleic Acids Res. 2008;36:2838–2851. doi: 10.1093/nar/gkn037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Davidson MK, Shandilya HK, Hirota K, Ohta K, Wahls WP. Atf1-Pcr1-M26 complex links stress-activated MAPK and cAMP-dependent protein kinase pathways via chromatin remodeling of cgs2+ J. Biol. Chem. 2004;279:50857–50863. doi: 10.1074/jbc.M409079200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jia S, Noma K, Grewal SI. RNAi-independent heterochromatin nucleation by the stress-activated ATF/CREB family proteins. Science. 2004;304:1971–1976. doi: 10.1126/science.1099035. [DOI] [PubMed] [Google Scholar]
- 42.Yamada T, Mizuno K, Hirota K, Kon N, Wahls WP, Hartsuiker E, Murofushi H, Shibata T, Ohta K. Roles of histone acetylation and chromatin remodeling factor in a meiotic recombination hotspot. EMBO J. 2004;23:1792–1803. doi: 10.1038/sj.emboj.7600138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kon N, Schroeder SC, Krawchuk MD, Wahls WP. Regulation of the Mts1-Mts2-dependent ade6-M26 meiotic recombination hotspot and developmental decisions by the Spc1 mitogen-activated protein kinase of fission yeast. Mol. Cell. Biol. 1998;18:7575–7583. doi: 10.1128/mcb.18.12.7575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mizuno K, Hasemi T, Ubukata T, Yamada T, Lehmann E, Kohli J, Watanabe Y, Iino Y, Yamamoto M, Fox ME, et al. Counteracting regulation of chromatin remodeling at a fission yeast cAMP response element-related recombination hotspot by stress-activated protein kinase, cAMP-dependent kinase and meiosis regulators. Genetics. 2001;159:1467–1478. doi: 10.1093/genetics/159.4.1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hirota K, Steiner WW, Shibata T, Ohta K. Multiple modes of chromatin configuration at natural meiotic recombination hot spots in fission yeast. Eukaryot. Cell. 2007;6:2072–2080. doi: 10.1128/EC.00246-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gao J, Davidson MK, Wahls WP. Phosphorylation-independent regulation of Atf1-promoted meiotic recombination by stress-activated, p38 kinase Spc1 of fission yeast. PLoS One. 2009;4:e5533. doi: 10.1371/journal.pone.0005533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kirkpatrick DT, Fan Q, Petes TD. Maximal stimulation of meiotic recombination by a yeast transcription factor requires the transcription activation domain and a DNA-binding domain. Genetics. 1999;152:101–115. doi: 10.1093/genetics/152.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Oliver PL, Goodstadt L, Bayes JJ, Birtle Z, Roach KC, Phadnis N, Beatson SA, Lunter G, Malik HS, Ponting CP. Accelerated evolution of the Prdm9 speciation gene across diverse metazoan taxa. PLoS Genet. 2009;5:e1000753. doi: 10.1371/journal.pgen.1000753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hayashi K, Yoshida K, Matsui Y. A histone H3 methyltransferase controls epigenetic events required for meiotic prophase. Nature. 2005;438:374–378. doi: 10.1038/nature04112. [DOI] [PubMed] [Google Scholar]
- 50.Axelsson E, Webster MT, Ratnakumar A, Ponting CP, Lindblad-Toh K. Death of PRDM9 coincides with stabilization of the recombination landscape in the dog genome. Genome Res. 2012;22:51–63. doi: 10.1101/gr.124123.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Auton A, Fledel-Alon A, Pfeifer S, Venn O, Segurel L, Street T, Leffler EM, Bowden R, Aneas I, Broxholme J, et al. A fine-scale chimpanzee genetic map from population sequencing. Science. 2012;336:193–198. doi: 10.1126/science.1216872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sollier J, Lin W, Soustelle C, Suhre K, Nicolas A, Geli V, de La Roche Saint-Andre C. Set1 is required for meiotic S-phase onset, double-strand break formation and middle gene expression. EMBO J. 2004;23:1957–1967. doi: 10.1038/sj.emboj.7600204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Borde V, Robine N, Lin W, Bonfils S, Geli V, Nicolas A. Histone H3 lysine 4 trimethylation marks meiotic recombination initiation sites. EMBO J. 2009;28:99–111. doi: 10.1038/emboj.2008.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Brocks JJ, Logan GA, Buick R, Summons RE. Archean molecular fossils and the early rise of eukaryotes. Science. 1999;285:1033–1036. doi: 10.1126/science.285.5430.1033. [DOI] [PubMed] [Google Scholar]
- 55.Reddy KC, Villeneuve AM. C. elegans HIM-17 links chromatin modification and competence for initiation of meiotic recombination. Cell. 2004;118:439–452. doi: 10.1016/j.cell.2004.07.026. [DOI] [PubMed] [Google Scholar]
- 56.Yamashita K, Shinohara M, Shinohara A. Rad6-Bre1-mediated histone H2B ubiquitylation modulates the formation of double-strand breaks during meiosis. Proc. Natl Acad. Sci. USA. 2004;101:11380–11385. doi: 10.1073/pnas.0400078101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hirota K, Mizuno K, Shibata T, Ohta K. Distinct chromatin modulators regulate the formation of accessible and repressive chromatin at the fission yeast recombination hotspot ade6-M26. Mol. Biol. Cell. 2008;19:1162–1173. doi: 10.1091/mbc.E07-04-0377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Buard J, Barthes P, Grey C, de Massy B. Distinct histone modifications define initiation and repair of meiotic recombination in the mouse. EMBO J. 2009;28:2616–2624. doi: 10.1038/emboj.2009.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Melamed-Bessudo C, Levy AA. Deficiency in DNA methylation increases meiotic crossover rates in euchromatic but not in heterochromatic regions in Arabidopsis. Proc. Natl Acad. Sci. USA. 2012;109:E981–E988. doi: 10.1073/pnas.1120742109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Merker JD, Dominska M, Greenwell PW, Rinella E, Bouck DC, Shibata Y, Strahl BD, Mieczkowski P, Petes TD. The histone methylase Set2p and the histone deacetylase Rpd3p repress meiotic recombination at the HIS4 meiotic recombination hotspot in Saccharomyces cerevisiae. DNA Repair (Amst.) 2008;7:1298–1308. doi: 10.1016/j.dnarep.2008.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mieczkowski PA, Dominska M, Buck MJ, Lieb JD, Petes TD. Loss of a histone deacetylase dramatically alters the genomic distribution of Spo11p-catalyzed DNA breaks in Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA. 2007;104:3955–3960. doi: 10.1073/pnas.0700412104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mirouze M, Lieberman-Lazarovich M, Aversano R, Bucher E, Nicolet J, Reinders J, Paszkowski J. Loss of DNA methylation affects the recombination landscape in Arabidopsis. Proc. Natl Acad. Sci. USA. 2012;109:5880–5885. doi: 10.1073/pnas.1120841109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.de Castro E, Soriano I, Marin L, Serrano R, Quintales L, Antequera F. Nucleosomal organization of replication origins and meiotic recombination hotspots in fission yeast. EMBO J. 2011;31:124–137. doi: 10.1038/emboj.2011.350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Szankasi P, Heyer WD, Schuchert P, Kohli J. DNA sequence analysis of the ade6 gene of Schizosaccharomyces pombe. Wild-type and mutant alleles including the recombination hot spot allele ade6-M26. J. Mol. Biol. 1988;204:917–925. doi: 10.1016/0022-2836(88)90051-4. [DOI] [PubMed] [Google Scholar]