Abstract
Post-replication DNA repair in eukaryotes is regulated by ubiquitination of proliferating cell nuclear antigen (PCNA). Monoubiquitination catalyzed by RAD6–RAD18 (an E2–E3 complex) stimulates translesion DNA synthesis, whereas polyubiquitination, promoted by additional factors such as MMS2–UBC13 (a UEV–E2 complex) and HLTF (an E3 ligase), leads to template switching in humans. Here, using an in vitro ubiquitination reaction system reconstituted with purified human proteins, we demonstrated that PCNA is polyubiquitinated predominantly via en bloc transfer of a pre-formed ubiquitin (Ub) chain rather than by extension of the Ub chain on monoubiquitinated PCNA. Our results support a model in which HLTF forms a thiol-linked Ub chain on UBC13 (UBC13∼Ubn) and then transfers the chain to RAD6∼Ub, forming RAD6∼Ubn+1. The resultant Ub chain is subsequently transferred to PCNA by RAD18. Thus, template switching may be promoted under certain circumstances in which both RAD18 and HLTF are coordinately recruited to sites of stalled replication.
INTRODUCTION
DNA is continuously injured by environmental insult as well as endogenous metabolic products. Such DNA damage, unless removed by multiple mechanisms for excision repair, inhibits replicative DNA polymerase action during DNA replication (1). Post-replication repair, to restart DNA replication stalled at DNA damaged sites, features two sub-pathways, translesion DNA synthesis (TLS) and template switching (TS), which are regulated by mono- and polyubiquitination of proliferating cell nuclear antigen (PCNA) at Lys164, respectively (2). RAD6 (an E2 ubiquitin-conjugating enzyme) and RAD18 (an E3 ubiquitin ligase) together catalyze the monoubiquitination of PCNA (2–4). The resultant monoubiquitinated PCNA (monoUb-PCNA) then activates the TLS pathway, in which stalled replication proceeds beyond the damage site with the help of specialized TLS polymerases. MonoUb-PCNA is believed to stimulate the entry of TLS polymerases at stalled 3′-ends, and this activity is dependent on interactions between the ubiquitin (Ub) moiety of monoUb-PCNA and the Ub-binding domains of the TLS polymerases (5–9). However, TLS is a mutagenic process due to the utilization of damaged templates by low-fidelity TLS polymerases (1).
Polyubiquitination of PCNA requires a different E2–E3 pair, MMS2–UBC13, a stable complex of an ubiquitin E2 variant (UEV) and an E2, and RAD5 in yeast (hereafter referred to as yRAD5) (2) or in humans, the two RAD5 homologs, HLTF and SHPRH, which serve as the E3 ligase (10–14). MMS2–UBC13 complexes mediate the formation of ubiquitin chains with Lys63 linkages (15). Polyubiquitinated PCNA (polyUb-PCNA) promotes the TS pathway, in which the primer end stalled at the damaged site is released from the damaged template and anneals with the newly synthesized daughter strand of the sister chromosome. The molecular mechanism(s) of TS are currently unknown, but it is believed that a complex set of biochemical reactions is involved. When the sophisticated TS reactions are successfully performed, this process is essentially error-free, as it utilizes a non-damaged template (1).
It has been argued, mainly on the basis of genetic data from yeast studies, that polyubiquitination of PCNA is downstream of monoubiquitination. Both mono- and polyubiquitination are dependent on yRAD6 and yRAD18; deletion or mutation of either one of these two proteins blocks both the TS and TLS pathways. By contrast, inactivation or deletion of yMMS2, yUBC13 or yRAD5 abolishes polyubiquitination of PCNA, but not monoubiquitination of PCNA, and thus blocks only the TS pathway (1,2). These data are consistent with results previously obtained from in vitro biochemical studies using purified yeast enzymes (16,17). The yRAD6–yRAD18 complex can monoubiquitinate PCNA at Lys164, whereas yMMS2–yUBC13 and yRAD5 are able to polyubiquitinate PCNA only in the presence of yRAD6–yRAD18 (16). There is general agreement that the ligase activity of yRAD5 mediates sequential transfer of Ub from yUBC13∼Ub to monoUb-PCNA (16,17). An unresolved issue, however, is why the error-free TS pathway would require the formation of monoUb-PCNA as an intermediate, which could potentially promote the error-prone TLS pathway and interfere with the preservation of genetic information.
In the current study, we investigated the mechanism of PCNA polyubiquitination using an in vitro ubiquitination reaction system reconstituted with purified recombinant human proteins, including HLTF. The results point to the existence of a novel reaction mechanism for the polyubiquitination of PCNA via en bloc Ub chain transfer. Importantly, the reaction mechanism appears to be more efficient than the conventional mechanism of sequential addition of Ub monomers to the distal end of an Ub chain on PCNA. We discuss the implications of these in vitro findings in terms of the choice of pathway, TS or TLS, proceeding from stalled replication sites following DNA damage.
MATERIALS AND METHODS
Plasmids
Expression plasmids for PCNA, RFC, E1, RAD6A, the RAD6A–RAD18 complex and Ub have been described previously (7,18–21). Expression plasmids for MMS2 and UBC13 were constructed using the pET20 parental vector. For the production of MMS2–UBC13 complexes, MMS2 and UBC13 were co-expressed as a single operon under the control of the T7 promoter. HLTF was cloned into pBAD22A (obtained from National BioResource Project, National Institute of Genetics, Mishima, Japan), and expression was induced by arabinose. For the histidine-tagged proteins, the tag sequence of pET15 was ligated to the 5′-end of the start codons of the respective genes. For HisUSP2(258−605), a truncated sequence corresponding to amino acids 258–605 was subcloned into pET15. Synthetic genes encoding the HisPCNA–UbGG fusion proteins were constructed in pET28a.
Proteins
Isopeptidase T was purchased from Boston Biochem (E-322). Other recombinant human proteins were expressed in Escherichia coli strain BL21 (DE3) at 15°C and then purified by chromatography at 4°C using the indicated columns (GE Healthcare), unless otherwise indicated. PCNA, RFC, E1, RAD6A, RAD6A–RAD18 complexes and Ub were purified as described previously (7,18,19,21). MMS2–UBC13 complexes were purified by sequential chromatography on HiPrep DEAE FF, HiTrap SP XL, HiTrap SP HP, HiTrap Heparin HP and Superdex 200 columns. MMS2–UBC13C87A complexes were purified on HiTrap Heparin HP, HiTrap SP XL and HiTrap SP HP columns, and MMS2–FLAGUBC13 complexes were purified on HiTrap Q FF, HiTrap SP HP and HiTrap Heparin HP columns. UBC13 was purified on HiPrep DEAE FF, HiTrap SP FF, HiTrap SP HP, HiTrap Heparin HP and Superdex 200 columns. HLTF was purified on HiTrap Heparin HP, HiTrap SP HP, HiTrap Q HP, HiTrap SP XL, Econopack CHT-II (BIO-RAD) and Superdex 200 columns. RAD6A–HisRAD18, HisHLTF and the corresponding mutants of each were partially purified using HiTrap Chelating HP and Superdex 200 columns. HisPCNA, HisPCNA–UbK63R/ΔGG and HisPCNAK164R–UbΔGG were purified using HiTrap Chelating HP, HiTrap Q FF and Superdex 200 columns. HisUSP2(258−605) was purified using HiTrap Chelating HP and Superdex 200 columns. MonoUb-PCNA was purified from E. coli overexpressing PCNA, E1 and RAD6A–RAD18, and HisUb. HisyRAD5 was overexpressed in strain BL21 Star (DE3) (Life Technologies), which harbors pMStRNA1 (22), and partially purified using a HiTrap Chelating HP column.
Ubiquitination assays
The standard reaction mixture (25 µl) contained 20 mM HEPES–NaOH (pH 7.5), 50 mM NaCl, 0.2 mg/ml BSA, 1 mM DTT, 10 mM MgCl2, 1 mM ATP, poly(dA)-oligo(dT) (GE Healthcare) (100 ng) as the source of multiple primed single-stranded (mpss) DNA, PCNA (1.0 pmol trimer), RFC (0.70 pmol), E1 (0.85 pmol), RAD6A–RAD18 complexes (0.54 pmol trimer), MMS2–UBC13 complexes (16.4 pmol dimer), HLTF (1.3 pmol) and Ub (174 pmol). For the competition assays containing 1740 pmol Ub, 6 pmol E1 was introduced to overcome the inhibitory effect of large amounts of Ub (23,24). Reaction mixtures were prepared on ice and then incubated at 30°C for 10 min unless otherwise indicated. Reaction products were analyzed by immunoblot. Where indicated by ‘reducing’ or unless otherwise indicated, the samples were treated with sodium dodecyl sulphate (SDS) sample buffer containing 280 mM β-mercaptoethanol. Samples that were not treated with reducing agent were referred to as ‘non-reducing’. Anti-PCNA (Santa Cruz, sc-7907), anti-UBC13 (IMGENEX, IMG-5634), anti-RAD6 (Abcam, ab31917), anti-Ub (Sigma, U5379) and anti-FLAG M2 monoclonal antibodies (Sigma, F 3165) were used as indicated. Immunoreactive proteins were visualized using an ECL chemiluminescence kit (GE Healthcare).
Ub transfer assays of UBC13∼Ub
The reaction mixture (25 µl) contained 20 mM HEPES–NaOH (pH 7.5), 50 mM NaCl, 0.2 mg/ml BSA, 1 mM DTT, 5 mM MgCl2, 1 mM ATP, poly(dA)-oligo(dT) (GE Healthcare) (100 ng) as the source of mpssDNA, E1 (1.7 pmol), MMS2–UBC13 complexes (83.7 pmol dimer) and Ub (174 pmol). After incubation at 30°C for 10 min, the reactions were quenched by the addition of N-ethylmaleimide (NEM) (5 mM) and EDTA (25 mM) for 1 min (25,26). HLTF (1.3 pmol) was introduced and the mixture was allowed to incubate for an additional 2 min. Reaction products were analyzed by immunoblot.
Purification of HisUb-charged RAD6A–RAD18
The reaction mixture (5 ml) contained 20 mM HEPES–NaOH (pH 7.5), 5 mM MgCl2, 1 mM ATP, E1 (1.2 nmol), RAD6A–HisRAD18 complexes (partially purified fraction, ∼3 nmol trimer) and HisUb (46 nmol). After incubation at 20°C for 10 min, the mixture was immediately applied to a HiTrap Heparin HP column (1 ml) at 4°C. RAD6A–HisRAD18 complexes were eluted with a linear gradient of NaCl. Aliquots of peak fractions were frozen in liquid nitrogen and stored at −80°C.
RESULTS
Polyubiquitination of PCNA by en bloc Ub chain transfer
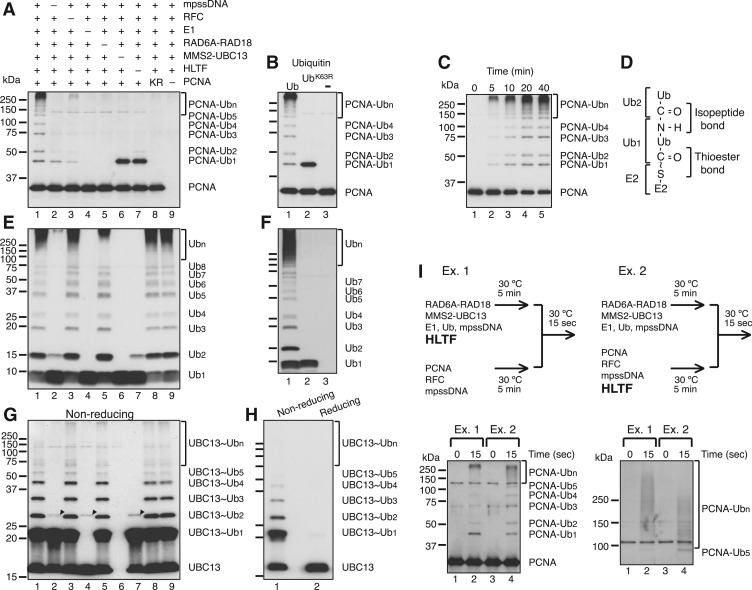
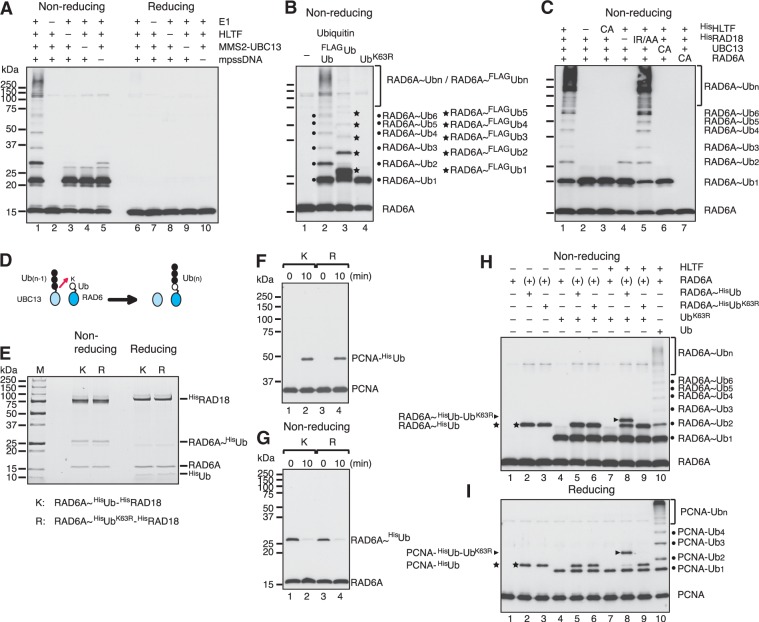
To explore the molecular mechanism of PCNA ubiquitination in humans, we developed an in vitro ubiquitination reaction system reconstituted with recombinant human proteins (Supplementary Figure S1A) (7,19). As shown in Figure 1A, polyubiquitination of PCNA was readily apparent when MMS2–UBC13 and HLTF-containing reactions were supplemented with RAD6A–RAD18, E1, mpssDNA and replication factor C (RFC), a PCNA loader, all of which are required for PCNA monoubiquitination (7,13). No PCNA ubiquitination was detected when RAD6A–RAD18 or E1 was omitted, or when PCNA was replaced with a mutant derivative carrying a K164R single amino acid substitution; small amounts of ubiquitinated PCNA were observed when mpssDNA or RFC was omitted. When Ub was replaced with an UbK63R mutant, only monoUb-PCNA was observed, which suggested that the Ub chain linkage was via a Lys63 linkage (Figure 1B) (13).
Figure 1.
Polyubiquitination of PCNA by en bloc Ub chain transfer. (A) In vitro reconstituted PCNA polyubiquitination reactions. Reactions were reconstituted with PCNA, mpssDNA, RFC, E1, RAD6A–RAD18, MMS2–UBC13, HLTF and Ub. Reaction products were analyzed by immunoblot using an anti-PCNA antibody. –, omitted factor; KR, PCNAK164R. (B) Lys63-linked Ub chain formation on PCNA was analyzed by immunoblot using an anti-PCNA antibody. (C) Time course of polyubiquitination. Reaction products were analyzed by immunoblot using an anti-PCNA antibody. (D) Chemical structure of the thiol-linked Ub dimer on E2 (E2∼Ub2). (E and F) Unanchored Ub chain formation. The products of (A) and (B) were probed with an anti-Ub antibody in (E) and (F), respectively. In these blots, the signals corresponding to ubiquitinated PCNA are not detectable. The amount of PCNA trimer in the reaction was 1 pmol (3 pmol monomer), whereas the amount of ubiquitin was 174 pmol; thus, the estimated signal intensity of total ubiquitinated PCNA was <1% that of ubiquitin. (G and H) UBC13∼Ubn formation. The products of (A) untreated by a reducing agent were probed with an anti-UBC13 antibody (G). Presumed dimers of UBC13 linked via S–S bond formation during sample preparation are indicated by arrowheads. The products of lane 1 in (G) treated with or without a reducing agent were analyzed by immunoblot using an anti-UBC13 antibody (H). (I) Transfer of pre-formed Ub chains to PCNA. The experimental designs are shown in the upper panel (see text for details). The reaction products were analyzed by electrophoresis under two different conditions and then probed with an anti-PCNA antibody.
Analysis of the time course of the reaction indicated that the accumulation of polyubiquitinated products was consistent over time, as opposed to smaller products forming first and then gradually converting into larger products (Figure 1C). This property was consistently observed under other assay conditions with different enzyme concentrations (Supplementary Figure S2). These results suggested that PCNA polyubiquitination may take place via en bloc transfer of a Ub chain (27) that is first formed on E2 via a thioester bond at a catalytic cysteine (E2∼Ubn) (Figure 1D), and then transferred to PCNA. To test this hypothesis, the presence of unanchored Ub chains was assessed by immunoblot following treatment with a reducing agent, to hydrolyze the thioester bond of E2∼Ubn (25). Following SDS–polyacrylamide gel electrophoresis (PAGE), immunoblot analysis with an anti-Ub antibody showed the formation of unanchored Ub chains with Lys63 linkages (Figure 1E and F). Efficient formation of Ub multimers was completely dependent on MMS2–UBC13 and E1, but not on RAD6–RAD18, RFC or PCNA. Interestingly, Ub multimer formation required the presence of HLTF and mpssDNA, while a small amount of Ub2 was formed in their absence, as previously observed when Ub was incubated with MMS2–UBC13 and E1 (15,28). To obtain direct evidence of UBC13∼Ubn formation, reactions were analyzed by SDS–PAGE under non-reducing conditions in order to prevent hydrolysis of the thioester bond, followed by immunoblot analysis with an anti-UBC13 antibody (25). The results clearly demonstrated the formation of UBC13∼Ubn (Figure 1G), which disappeared upon treatment with a reducing agent (Figure 1H). There was only slight accumulation of UBC13 conjugated to long chain Ub (>Ub5) (Figure 1G), which was in contrast to polyubiquitinated PCNA and unanchored Ub chains, where larger products were relatively abundant (Figure 1A and E; see also ‘Discussion’ section). The protein components required for efficient UBC13∼Ubn (n ≥ 2) formation (Figure 1G) were the same as those required for unanchored Ub multimer formation (Figure 1E). Notably, when either HLTF or mpssDNA was omitted, formation of Ub chains and UBC13∼Ubn was severely reduced. Small amounts of Ub2 and UBC13∼Ub2 were detected, however, and this was dependent on MMS2–UBC13 (Figure 1E and G). These results suggested that mpssDNA is involved in the same biochemical step as HLTF, probably as a co-factor in Ub chain formation.
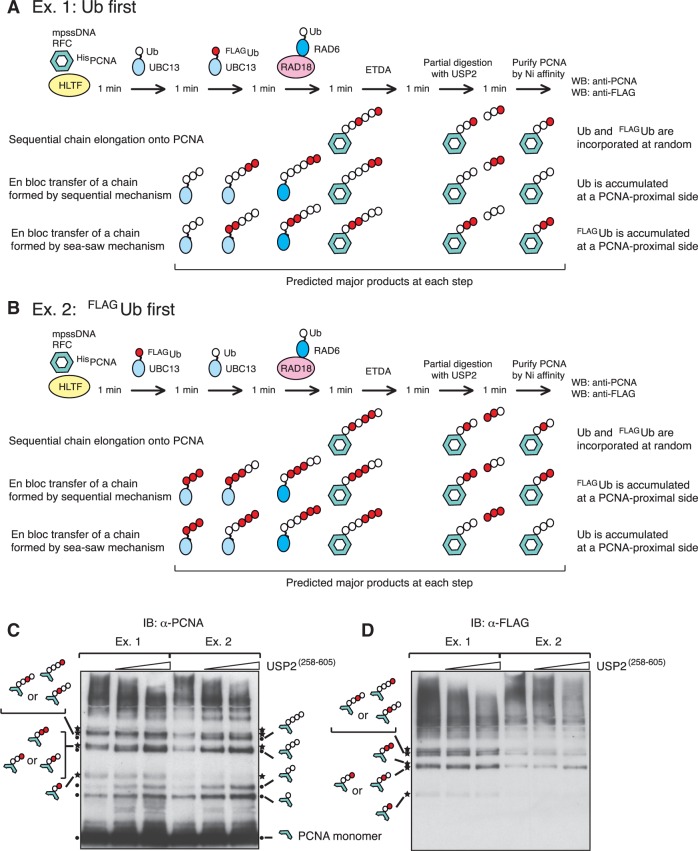
To test this putative mechanism of en bloc Ub chain transfer of preformed chains of UBC13∼Ubn to PCNA, we examined whether pre-incubation of MMS2–UBC13 and HLTF with Ub before the addition of PCNA could accelerate the immediate formation of poly-Ub PCNA with long Ub chains. We set up the following experiments, as shown in Figure 1I. In Ex. 1, RAD6A–RAD18, MMS2–UBC13, E1, Ub, mpssDNA and HLTF were pre-incubated for 5 min, and then combined with a PCNA–preassembly mixture containing PCNA, RFC and mpssDNA. In Ex. 2, RAD6A–RAD18, MMS2–UBC13, E1, Ub and mpssDNA were pre-incubated for 5 min and then combined with a PCNA–preassembly mixture together with HLTF. After a brief period of additional incubation for 15 s, the reaction products were analyzed by immunoblot using an anti-PCNA antibody. In Ex. 1, but not Ex. 2, Ub chains would be assembled on E2 by HTLF prior to the addition of PCNA for en bloc Ub chain transfer; by contrast, a step-wise addition of Ub onto PCNA would yield similar kinetics in both experiments. A different size distribution of poly-Ub PCNA was observed under the two reaction conditions (Figure 1I and Supplementary Figure S3). In Ex. 1, the accumulation of large products (>150 kDa) was quite prominent. In contrast, in Ex. 2, intermediate sized poly-Ub PCNA (50 to ≤ 150 kDa) was abundant, but large products (>250 kDa) were much less abundant compared to Ex. 1. These results supported en bloc Ub chain transfer under these in vitro experimental conditions. To obtain further evidence, we analyzed detailed mechanisms underlying the hypothesized en bloc Ub chain transfer in additional experiments (see below).
Essential function of HLTF in UBC13∼Ubn formation
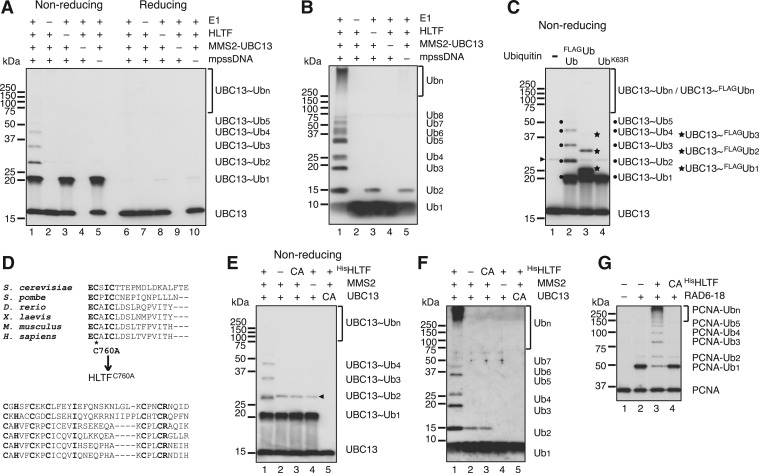
To further investigate the role of HLTF in PCNA polyubiquitination, we analyzed UBC13∼Ubn formation separately from PCNA polyubiquitination. The minimal set of factors necessary for UBC13∼Ubn formation (Figure 2A) as well as for unanchored Ub chain formation (Figure 2B) were HLTF, MMS2–UBC13, E1, Ub and mpssDNA. Replacing wild-type Ub with its mutant derivative, UbK63R, confirmed that the multiple bands observed were Lys63-linked Ub chains on UBC13 (Figure 2C, lane 4). Furthermore, when Ub was replaced with FLAGUb, the bands migrated more slowly (Figure 2C, lane 3), which indicated that they contained Ub molecules.
Figure 2.
Analysis of UBC13∼Ubn formation. (A) Formation of thiol-linked Ub chains on UBC13 with the minimal set of factors. The reactions were reconstituted with mpssDNA, E1, MMS2–UBC13, HLTF and Ub. The reaction products treated with or without a reducing agent were analyzed by immunoblot using an anti-UBC13 antibody. –, omitted factor. (B) Formation of Ub chains with the minimal set of factors. The reaction products in (A) were treated with a reducing agent and then analyzed by immunoblot using an anti-Ub antibody. (C) Evidence that the multiple thiol-linked bands on UBC13 are Lys63-linked Ub chains. The reaction products untreated by a reducing agent were analyzed by immunoblot using an anti-UBC13 antibody. The positions of UBC13∼Ubn and UBC13∼FLAGUbn are indicated by dots and stars, respectively. Presumed dimers of UBC13 linked via S–S bond formation during sample preparation are indicated by an arrowhead. (D) Multiple sequence alignment of the RING fingers of human HLTF and its orthologs. The conserved cysteine residue (marked with asterisk) was replaced with an alanine. (E and F) Analysis of HLTF mutants. The reaction products were treated with (F) or without (E) a reducing agent and then analyzed by immunoblot using an anti-UBC13 antibody (E) and an anti-Ub antibody (F). –, omitted factor; CA for HisHLTF, HisHLTFC760A; CA for UBC13, UBC13C87A. Presumed dimers of UBC13 linked via S–S bond formation during sample preparation are indicated by an arrowhead. (G) Catalytic activity of HLTF is essential for polyubiquitination of PCNA. PCNA polyubiquitination reactions were performed using HisHLTF or HisHLTFC760A (CA) under standard reaction conditions. The reaction products were analyzed by immunoblot using an anti-PCNA antibody.
The RING domain of E3 serves an essential function in the ligase reaction by mediating the interaction with E2 (29,30). To determine whether the ligase activity of HLTF was necessary for Ub chain formation, we constructed an HLTF RING mutant in which the conserved cysteine residue (C760) of the RING domain was replaced with an alanine (Figure 2D). Histidine-tagged wild-type and mutant HLTF (HisHLTF and HisHLTFC760A, respectively) were partially purified (Supplementary Figure S1B) and eluted in the same fraction as untagged HLTF from a gel filtration column, which confirmed the integrity of them to be preserved. HisHLTF was able to support Ub chain formation as efficiently as untagged HLTF, whereas the mutant protein was unable to do so (Figure 2E and F, lanes 3). Thus, Ub chain formation was dependent on the E3 ligase activity of HLTF. Additionally, a mutant form of UBC13 (UBC13C87A) (Supplementary Figure S1B), in which the conserved cysteine residue involved in thioester bond formation with Ub was replaced with alanine, did not support chain formation (Figure 2E and F, lanes 5). Finally, replacing MMS2–UBC13 complexes with UBC13 monomers (Supplementary Figure S1B) demonstrated that MMS2 was also required for Ub chain formation (Figure 2E and F, lanes 4).
Using the HLTF RING mutant, we confirmed that the catalytic activity of HLTF was required for PCNA polyubiquitination, as well as for UBC13∼Ubn formation (Figure 2G). These results were consistent with the idea that HLTF mediates UBC13∼Ubn formation and then the resultant Ub chain is transferred en bloc to PCNA.
Analysis of UBC13∼Ubn formation
UBC13-linked polyubiquitin chains could be generated through an aminolysis-based transfer reaction between neighboring UBC13∼Ub molecules, in which the ε-amino group of Lys63 of one Ub molecule attacks the thioester-linked carbonyl of the neighboring Ub molecule on UBC13 (25,27) (Figure 3A, schematic). To investigate this as a possible mechanism, UBC13 in complex with MMS2 was first charged with either wild-type Ub or UbK63R in the presence of E1 and mpssDNA for 10 min. After quenching for 1 min, by the addition of NEM and EDTA, to inactivate uncharged UBC13 and E1 (25,26), the reaction mixture was supplemented with HLTF and then incubated for an additional 2 min. HLTF-dependent Ub chain formation, as evidenced by the appearance of UBC13∼Ubn (n ≥ 3), was observed with Ub, but not with UbK63R (Figure 3B), although a significant amount of UBC13∼Ub2 was formed in the absence of HLTF under these reaction conditions (Figure 3B, lane 1). The reaction products were sensitive to treatment with a reducing agent, which indicated that they possessed thiol-linked Ub chains (Figure 3B, lanes 5–7). We also confirmed that UBC13∼Ubn formed in these reactions was not a product of recharging due to incomplete quenching (Supplementary Figure S4; see also Supplementary Note for Supplementary Figure S4). Collectively, these results demonstrated that HLTF catalyzes UBC13-Ubn formation by enhancing an aminolysis-based transfer reaction between two UBC13∼Ub molecules.
Figure 3.
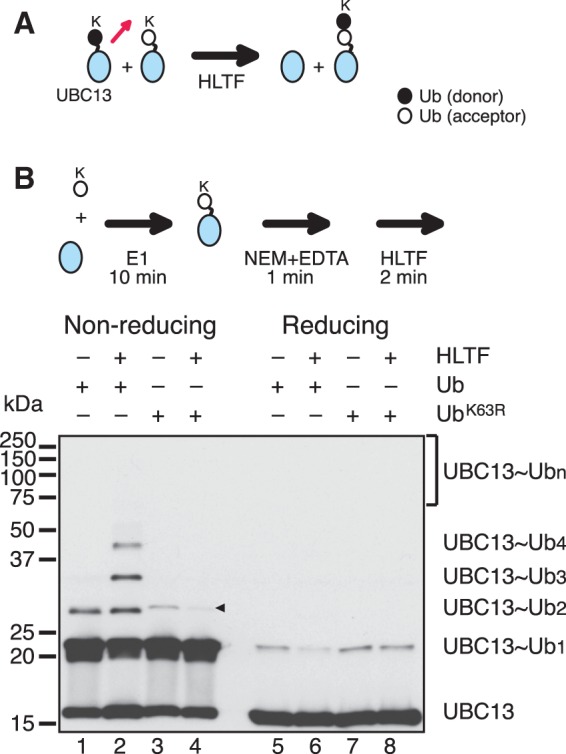
Ub transfer reactions between UBC13∼Ub molecules. (A) A proposed mechanism of E2∼Ubn formation. A red arrow depicts the direction of Ub movement. MMS2 was included in the reactions by omitted from the schematic. (B) Analysis of Ub transfer reactions between UBC13∼Ub molecules. The experimental design is shown in the upper panel (see text for details). The reactions were reconstituted with the minimal set of factors (mpssDNA, E1, MMS2–UBC13, HLTF and Ub). The reaction products were treated with or without a reducing agent and then analyzed by immunoblot using an anti-UBC13 antibody. –, omitted factor. Presumed dimers of UBC13 linked via S–S bond formation during sample preparation are indicated by an arrowhead.
Preferential utilization of UBC13∼Ub as a Ub acceptor by HLTF
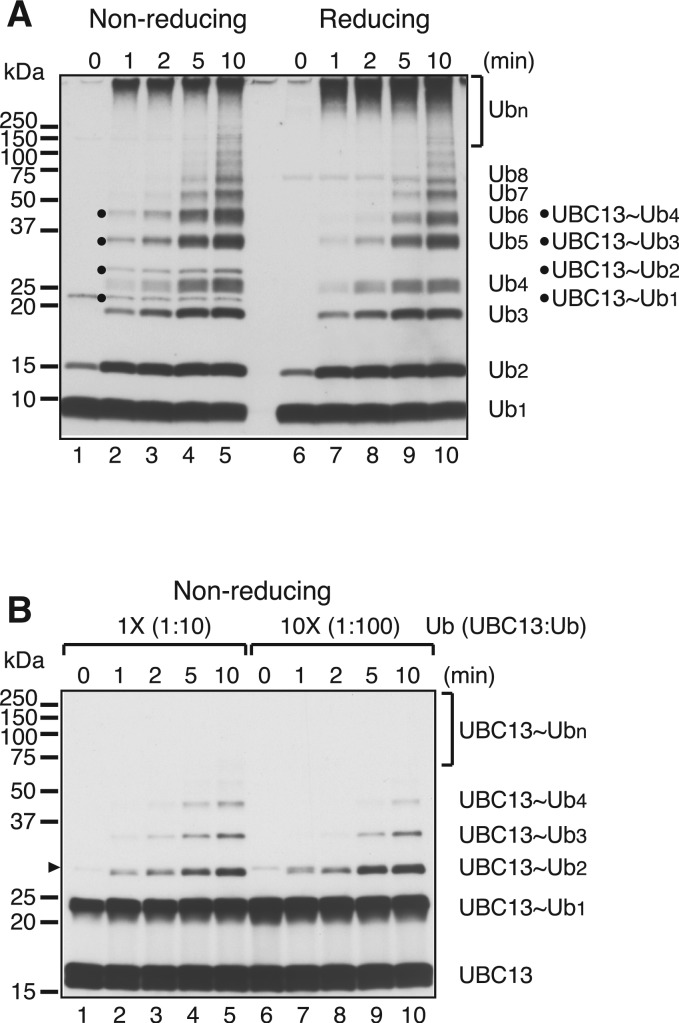
Next, we investigated whether the transfer reaction between two UBC13∼Ub molecules, as depicted in Figure 3A, was the predominant mechanism, or if Ub moieties from UBC13∼Ub were transferred to uncharged Ub as well as a UBC13∼Ub. This was addressed by analyzing the time course of the reaction. Reaction mixtures were pre-incubated without HLTF for 10 min, and then reaction products were removed at different time points after the addition of HLTF and analyzed by immunoblot using an anti-Ub antibody (Figure 4A). Comparison of the products formed under non-reducing and reducing conditions demonstrated that UBC13∼Ubn was present only under non-reducing conditions, while unanchored Ub chains were present under both conditions (Figure 4A; quantified in Supplementary Figure S5A). In all of these reactions, Ub was present in ∼20-fold molecular excess over UBC13∼Ub (see legend for Supplementary Figure S5B). Thus, if Ub was transferred to uncharged Ub as well as UBC13∼Ub, the formation of unanchored Ub chains would predominate. However, unanchored and UBC13-linked Ub chains were detected at equivalent levels at the early time points (Figure 4A and Supplementary Figure S5A), which suggested that HLTF preferentially transfers Ub to UBC13∼Ub rather than uncharged Ub, and that UBC13∼Ub is a preferred Ub acceptor for HLTF (Supplementary Figure S5B).
Figure 4.
Preferential utilization of UBC13∼Ub as Ub acceptor by HLTF. (A) Time-course of chain formation using the minimal set of factors. Reaction mixtures containing mpssDNA, E1, MMS2–UBC13 and Ub were pre-incubated for 10 min and then the reactions were started by adding HLTF and incubated for the indicated times. Reaction products were treated with or without a reducing agent and then analyzed by immunoblot using an anti-Ub antibody. (B) UBC13∼Ubn competition assays. Reactions in the presence of a 10-fold excess of Ub (10×) were compared to those under standard reaction conditions (1×) with the minimal set of factors. The molecular ratios of UBC13 to Ub are shown in parentheses. Reaction mixtures were pre-incubated without HLTF for 1 min and then the reactions were started by adding HLTF and incubated for the indicated times. Reaction products untreated by a reducing agent were analyzed by immunoblot using an anti-UBC13 antibody. Presumed dimers of UBC13 linked via S–S bond formation during sample preparation are indicated by an arrowhead.
To confirm the preferential utilization of UBC13∼Ub as an Ub acceptor, the experiment was repeated, this time with Ub at 100-fold molar excess over UBC13 (Figure 4B). Under these conditions, excess Ub would strongly compete against UBC13∼Ubn formation. Alternatively, if UBC13∼Ub was the preferred substrate, UBC13∼Ubn formation would be unaffected by excess Ub. The time course of the reaction clearly showed only a slight inhibitory effect of 100-fold excess Ub (<2-fold), providing additional evidence of the specificity of HLTF for UBC13∼Ub as a Ub acceptor. Additionally, we demonstrated that the formation of unanchored Ub chains was strongly decreased at low concentrations of uncharged Ub (Supplementary Figure S6 and Supplementary Note for Supplementary Figure S6), which indicated that the unanchored Ub chains were by-products generated, at least in part, by Ub (chain) transfer from UBC13∼Ubn (n ≥ 1) to uncharged Ub (see Supplementary Note for Supplementary Figure S6).
Analysis of the mechanism of polyubiquitination of PCNA
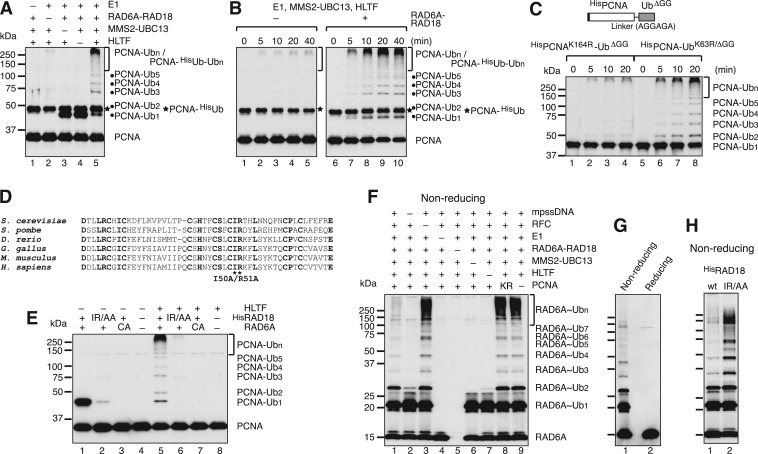
One current theory of polyubiquitination of PCNA holds that the Ub chain forms on the Ub moiety of monoUb-PCNA, and that the role of RAD6A–RAD18 is simply to provide a source of monoUb-PCNA. Based on this model, RAD6A–RAD18 should be dispensable when pre-formed monoUb-PCNA is supplied as the substrate for the polyubiquitination reaction, as was previously shown using yeast proteins (16). To examine the Ub transfer reaction in more detail, partially HisUb-modified PCNA (PCNA-HisUb) was purified and used as the substrate. In contrast to previous reports using yRAD5, monoUb-PCNA was a surprisingly poor substrate for PCNA polyubiquitination upon incubation with E1, MMS2–UBC13 and HLTF, although trace amounts of polyUb-PCNA were detected (Figure 5A, lane 2 and Figure 5B, lanes 1–5). When RAD6A–RAD18 was added to the reaction mixture, however, polyUb-PCNA was efficiently generated (Figure 5A, lane 5 and Figure 5B, lanes 6–10). It appeared that polyUb-PCNA species were derived from unmodified PCNA rather than from PCNA-HisUb, because the amount of unmodified PCNA was significantly reduced upon prolonged incubation, while that of PCNA-HisUb remained unchanged (Figure 5B, lanes 6–10). The poor capacity of PCNA-HisUb to act as a substrate for polyubiquitination was not due to any intrinsic property of HisUb, because it was an excellent substrate for polyubiquitination of unmodified PCNA (Supplementary Figure S7). Furthermore, we also tested a PCNA–UbGG fusion protein, in which Ub was fused to the C-terminus of PCNA via a six amino acid linker, and the two glycine residues in the C-terminus of the Ub moiety were deleted to avoid aberrant charging reactions (Figure 5C). Here, two different HisPCNA–UbΔGG fusion proteins carrying either a K164R mutation in PCNA or a K63R mutation in Ub were examined. Polyubiquitination of HisPCNA–UbK63R/ΔGG was much more prominent than with HisPCNAK164R–UbGG (Figure 4C), which suggested that HLTF does not prefer the Ub moiety of the HisPCNA–UbGG fusion protein as an acceptor.
Figure 5.
Thiol-linked Ub chains formed on RAD6 are transferred onto PCNA. (A and B) MonoUb-PCNA is a poor substrate for polyubiquitination. PCNA partially monoubiquitinated with HisUb was subjected under standard reaction conditions. Reaction products were analyzed by immunoblot using an anti-PCNA antibody. –, omitted factor. (C) PCNA ubiquitination assays using HisPCNA–UbΔGG fusion proteins. A schematic of the structure of the fusion protein is shown in the top panel. Reactions were performed with either HisPCNAK164R-UbΔGG or HisPCNA–UbK63R/ΔGG instead of PCNA under standard reaction conditions with mpssDNA, RFC, E1, RAD6A–RAD18, MMS2–UBC13, HLTF and Ub, followed by immunoblot using an anti-PCNA antibody. (D) Multiple sequence alignment of the RING fingers of human RAD18 and its orthologs. The conserved isoleucine and arginine residues were both replaced with alanines. (E) The catalytic activity of RAD6A–RAD18 is required for PCNA polyubiquitination. The reactions were performed using the indicated mutant proteins instead of wild-type protein under standard reaction conditions followed by immunoblot using an anti-PCNA antibody. –, omitted factor; IR/AA, HisRAD18I50A/R51A; CA, RAD6AC88A. (F and G) Ub chain formation on RAD6. The reaction products in Figure 1G and 1H, respectively, were analyzed by immunoblot using an anti-RAD6 antibody. (H) Accumulation of RAD6A∼Ubn in complex with the RING mutant of RAD18, RAD18IR/AA. The products in lanes 5 and 6 in (E) untreated by a reducing agent were analyzed by immunoblot using an anti-RAD6 antibody (lanes 1 and 2, respectively).
The results described above suggested a novel mechanism of polyubiquitination of PCNA, in which mono- and polyubiquitination are coupled to a certain extent. One possibility is that Ub chains are formed directly on Lys164 of PCNA by HLTF and that RAD6–RAD18 is required for a non-catalytic function. To test this hypothesis, we used a RING mutant of RAD18, in which the conserved isoleucine (I50) and arginine (R51) residues were replaced with alanine (Figure 5D); this mutant has been shown to have reduced ligase activity (21). Histidine-tagged wild-type RAD18 and mutant RAD18 (HisRAD18 and HisRAD18I50A/R51A, respectively) were partially purified in complex with RAD6A (Supplementary Figure S1D). The protein complexes eluted in the same fraction as untagged RAD6A–RAD18 in gel filtration chromatography, which confirmed the integrity of them to be preserved. RAD6A–HisRAD18 (wild-type) was able to support mono- and polyubiquitination of PCNA as well as untagged RAD6A–RAD18 (Figure 5E, lanes 1 and 5). By contrast, in the presence of the RAD18 mutant, very little monoUb- or polyUb-PCNA was produced in the presence or absence of HLTF (Figure 5E, lanes 2 and 6). We also examined a mutant form of RAD6A, RAD6AC88A, in which a conserved cysteine residue (C88) involved in the formation of thioester bonds with Ub was replaced with an alanine. Similar to the RAD18 RING mutant, RAD6AC88A did not support chain formation (Figure 5E, lanes 3 and 7). These results indicated that PCNA polyubiquitination depends on the catalytic cysteine of RAD6A and an intact RING finger of RAD18.
We next considered the possibility that Ub chains may first be formed via Lys63 of the thiol-linked Ub moiety on RAD6 by HLTF and then transferred, together with the proximal Ub pre-bound to RAD6, to Lys164 of PCNA by RAD18. In this case, thiol-linked Ub chains on RAD6A might be detected in a manner that is dependent on MMS2–UBC13 and HLTF. The products of the PCNA polyubiquitination reaction shown in Figure 1G were re-analyzed by immunoblot using an anti-RAD6 antibody. The results clearly demonstrated the formation of multiple RAD6A bands that were dependent on MMS2–UBC13, HLTF, mpssDNA and E1, but independent of PCNA and RFC (Figure 5F). These bands were also sensitive to treatment with a reducing agent (Figure 5G). Notably, RAD6A∼Ubn accumulated under conditions in which Ub chain transfer to PCNA was blocked, i.e. when RFC or PCNA was omitted (Figure 5F, lanes 3 and 9), when PCNA was replaced with PCNAK164R (lane 8), or when the RING mutant of RAD18 was used (Figure 5H). These results strongly suggested that RAD6-linked Ub chains are intermediates between UBC13∼Ubn and polyUb-PCNA.
Analysis of RAD6A∼Ubn formation
Because the presence of RAD6A∼Ubn is a novel observation that has not been reported previously, we investigated the putative molecular mechanism in more detail. As shown in Figure 6A, RAD6A∼Ubn formation was successfully reconstituted without RFC and PCNA, but was abolished by omission of HLTF, MMS2–UBC13, E1, Ub, mpssDNA or RAD6A–RAD18, which provided additional evidence that these factors are the minimal set of reaction components needed. We confirmed that the multiple bands were Lys63-linked Ub chains on RAD6A by replacing Ub with either FLAGUb or UbK63R (Figure 6B). Furthermore, RAD6A∼Ubn formation was dependent on the catalytic cysteines of UBC13 and RAD6A (Figure 6C, lanes 6 and 7). RAD18 was also an essential component for chain formation (Figure 6C, lane 4), but the RAD18 RING mutant (I50A/R51A) also supported the reaction equally well (Figure 6C, lane 5). Since the RAD18 RING mutant has severely reduced ligase activity in mediating PCNA ubiquitination (Figure 5E) (21), it seems likely that a non-catalytic function of RAD18 other than its ligase activity is required for chain formation on RAD6A. The requirement for the RAD18 subunit might involve a physical interaction between RAD18 and HLTF, as reported previously (11,13). By contrast, the intact RING domain of HLTF was essential for RAD6-Ubn formation (Figure 6C, lane 3), which indicated that HLTF catalyzes the transfer of Ub molecules from UBC13 to RAD6A.
Figure 6.
Analysis of RAD6A∼Ubn formation. (A) Formation of thiol-linked Ub chains on RAD6A using the minimal set of factors. The reactions were reconstituted with mpssDNA, E1, RAD6A–RAD18, MMS2–UBC13, HLTF and Ub. Reaction products were treated with or without a reducing agent and then analyzed by immunoblot using an anti-RAD6 antibody. –, omitted factor. (B) Evidence that the multiple thiol-linked bands on RAD6A are Lys63 linked Ub chains. Reaction products untreated by a reducing agent were analyzed by immunoblot using an anti-RAD6 antibody. The positions of RAD6A∼Ubn and RAD6A∼FLAGUbn are indicated by dots and stars, respectively. (C) Effect of mutants on thiol-linked Ub chain formation on RAD6A. Reaction products untreated by a reducing agent were analyzed by immunoblot using an anti-RAD6 antibody. –, omitted factor; CA for HisHLTF, HisHLTFC760A; IRAA, HisRAD18I50A/R51A; CA for UBC13, UBC13C87A; CA for RAD6A and RAD6AC88A. (D) Proposed mechanism of RAD6A∼Ubn formation. A red arrow depicts the direction of Ubn movement. MMS2 and RAD18 were included in the reactions but omitted from the schematic. (E) Purified RAD6A∼HisUb–HisRAD18 and RAD6A∼HisUbK63R–HisRAD18 complexes treated with or without a reducing agent were analyzed by SDS–PAGE followed by CBB staining. (F and G) PCNA monoubiquitination reactions were reconstituted with PCNA, mpssDNA, RFC and the indicated complexes (K, RAD6A∼HisUb–HisRAD18; or R, RAD6A∼HisUbK63R–HisRAD18) for the indicated times. Reaction products treated with (F) or without (G) a reducing agent were analyzed by immunoblot using an anti-PCNA antibody (F) or an anti-RAD6 antibody (G). (H) Analysis of the Ub transfer reaction from UBC13∼Ub to RAD6∼Ub. Reactions were performed with mpssDNA, E1, MMS2–UBC13, HLTF, UbK63R, and either RAD6–HisRAD18, RAD6A∼HisUb–HisRAD18, or RAD6A∼HisUbK63R–HisRAD18 for 2 min. RAD6 molecules shown by (+) were carried from partial charging reactions, as shown in (E). Reaction products untreated with a reducing agent were analyzed by immunoblot using an anti-RAD6 antibody. Lane 10, standard reaction products. (I) Analysis of the Ub chain transfer reaction from RAD6∼Ub2 to PCNA. Reactions were performed with PCNA, mpssDNA, RFC, E1, MMS2–UBC13, HLTF, UbK63R, and either RAD6–HisRAD18, RAD6∼HisUb–HisRAD18, or RAD6∼HisUbK63R–HisRAD18 for 2 min and then analyzed by immunoblot using an anti-PCNA antibody. Lane 10, standard reaction products.
The formation of RAD6∼Ubn may involve an aminolysis-based transfer reaction between RAD6∼Ub and UBC13∼Ubn-1 (as illustrated in Figure 6D), similar to the mechanism of Ub transfer between UBC13∼Ub molecules, as both of the reactions were dependent on the catalytic function of HLTF. In this case, the resulting Ub chain would be expected to be a hybrid between the Ub molecule originally attached to RAD6 and other Ub moieties transferred from UBC13∼Ubn-1. To investigate the Ub transfer reaction from UBC13∼Ub to RAD6∼Ub, RAD6A in complex with RAD18 was charged with either HisUb or HisUbK63R, and then the modified RAD6A–RAD18 complexes were purified by column chromatography (Figure 6E). The purified complexes supported monoubiquitination of PCNA in the absence of E1 and Ub (Figure 6F), with concomitant reduction in the amount of RAD6A∼HisUb and RAD6A∼HisUbK63R (Figure 6G). These complexes were then used to test Ub chain formation on RAD6A upon the addition of HLTF and other factors, together with un-tagged UbK63R, which could be distinguished in size from pre-charged HisUb. If the expected reactions occurred, UbK63R would be attached to RAD6∼HisUb, but not to RAD6A∼HisUbK63R. As shown in Figure 6H, the formation of an additional band was detected in reactions containing RAD6A∼HisUb (lane 8), but not RAD6A∼HisUbK63R (lane 9), or uncharged RAD6A (lane 7), and the formation was dependent on HLTF (compare lanes 5 and 8). Importantly, this new band migrated between RAD6A∼Ub2 and RAD6A∼Ub3 (see lane 10), which indicated that it was indeed a RAD6A∼HisUb–UbK63R hybrid. Finally, as shown in Figure 6I, the hybrid chain was transferred onto PCNA to generate PCNA-HisUb–UbK63R (lane 8), which migrated between PCNA-Ub2 and PCNA-Ub3 (see lane 10).
The direction of Ub chain elongation in the PCNA polyubiquitination reaction
The results obtained thus far were consistent with a ‘seesaw’ model of Ub chain elongation (27), which would predict that the direction of chain elongation is PCNA-distal to -proximal, as hypothesized by Hochstrasser (27) (see also ‘Discussion’ section). We designed a set of experiments to investigate the direction of chain elongation, as depicted schematically in Figure 7A. In Ex. 1, HLTF, RFC, histidine-tagged PCNA (HisPCNA) and mpssDNA were pre-incubated for 2 min and then combined with pre-charged UBC13∼Ub. The pre-charging reaction was performed for 2 min with E1, MMS2–UBC13 and Ub. After an initial chain elongation reaction for 1 min, the mixture was combined with pre-charged UBC13∼FLAGUb and then incubated for an additional 1 min. The mixture was then combined with pre-charged RAD6A∼Ub with RAD18 (pre-charging was performed for 2 min with E1, RAD6A–RAD18 and Ub). After an additional incubation for 1 min, the reaction was terminated by the addition of EDTA for 1 min, and then the catalytic core of Ub-specific protease 2 [HisUSP2(258−605)] (31) was introduced to partially digest the Ub chains. We confirmed that, unlike isopeptidase T, purified HisUSP2(258−605) did not react with terminal Ub moieties of K63-linked Ub chains; rather, the HisUSP2(258−605) digestion sites on the Ub chain appeared to be at random positions (Supplementary Figure S8). Digestion was performed for 1 min and then terminated by the addition of an equal volume of 8 M urea and 4% Triton X-100. PCNA was immediately purified on Ni-chelating beads and analyzed by immunoblot. The predicted structures of the major products produced by the three different mechanisms of chain elongation are illustrated in Figure 7A. Note that Ub chain elongation reactions were initiated without FLAGUb, and the chains were further extended in presence of FLAGUb. If chain elongation occurred via a sequential mechanism, Ub and FLAGUb would be incorporated onto PCNA at random, because chain elongation would be initiated by the addition of RAD6A–RAD18 in the presence of both UBC13∼Ub and UBC13∼FLAGUb. In the case of en bloc transfer, there would be differential distribution of Ub and FLAGUb within the chain: accumulation of FLAGUb at the PCNA-distal end in the case of sequential addition, and accumulation of FLAGUb at the PCNA-proximal end in the case of the seesaw mechanism of addition. In experimental scheme Ex. 2 (Figure 7B), the reactions were the same as Ex. 1, except for the order of the addition of UBC13∼Ub and UBC13∼FLAGUb. In Ex. 2, pre-charged UBC13∼FLAGUb was incubated first with HLTF, and then pre-charged UBC13∼Ub was added. In Ex. 2, sequential chain elongation would result in the incorporation of Ub and FLAGUb onto PCNA in a similar fashion as in Ex. 1. In the case of en bloc transfer, the pattern of incorporation of Ub and FLAGUb would be the opposite of that depicted in Ex. 1. Note that the distribution of each intermediate varies depending on which molecule is the rate-limiting factor for chain elongation. This also means that if the rate-limiting factor in each step of the overall reaction was altered during incubation, the effects would be significant. To avoid this complication, the experiments were completed within 5 min from the first addition of UBC13∼Ub to the termination of the reaction, since the velocity of chain elongation was constant for at least 5 min under standard assay conditions.
Figure 7.
Analysis of the direction of Ub chain elongation. (A and B) Experimental schemes (see text for details). MMS2 was included in the reactions but omitted from the schematics. (C and D) Immuonblot analysis using an anti-PCNA antibody to detect total product (C) and an anti-FLAG antibody to detect only FLAGUb-containing chains (D). Products containing FLAGUb (detected by both the anti-PCNA and anti-FLAG antibodies) are indicated by stars and illustrations. FLAGUb-free products (detected by only the anti-PCNA antibody) are indicated by dots and illustrations.
The products of the reactions outlined in Figure 7A and B were analyzed by immunoblot. Partial digestion of the Ub chains by increasing amounts of HisUSP2(258−605) reduced the average sizes of polyUb-PCNA and produced several distinct bands (Figure 7C). Importantly, the band patterns differed between Ex. 1 and 2 (Figure 7C and D), which suggested that the chains were formed predominantly via a seesaw mechanism. This was supported be the following observations:
In both Ex. 1 and 2, the average sizes of polyUb-PCNA were similarly reduced by the addition of HisUSP2(258−605) (Figure 7C), which indicated that the Ub chains on PCNA in Ex. 1 and 2 were digested to a similar extent by HisUSP2(258−605). When the same samples were analyzed by immunoblot using an anti-FLAG antibody, the patterns differed. In Ex. 1, both the total amount and the average size of the FLAGUb-containing chains (detectable by the anti-FLAG antibody) were decreased by the addition of HisUSP2(258−605) (Figure 7D), which suggested that a significant fraction of FLAGUb had been incorporated into the PCNA-proximal side. By contrast, in Ex. 2, the average size of the FLAGUb-containing chains was only slightly decreased, even though signal intensity was clearly reduced (Figure7D). These results indicated that PCNA containing smaller chains as a result of digestion was no longer detectable with the anti-FLAG antibody, suggesting that FLAGUb was predominantly incorporated into the PCNA-distal side.
There was substantial accumulation of FLAGUb-free bands, indicated by dots, in Ex. 2 (Figure 7C). By contrast, in Ex.1, FLAGUb-containing bands were abundant (Figure 7C and D, stars) even after partial digestion, which suggested that a significant fraction of FLAGUb was present on the PCNA-proximal side.
These results were consistent with the pattern predicted by a mechanism of en bloc transfer of chains formed by the seesaw mechanism of addition (Figure 8B). Collectively, the results of the current study suggest that the seesaw mechanism predominates in PCNA polyubiquitination reactions.
Figure 8.
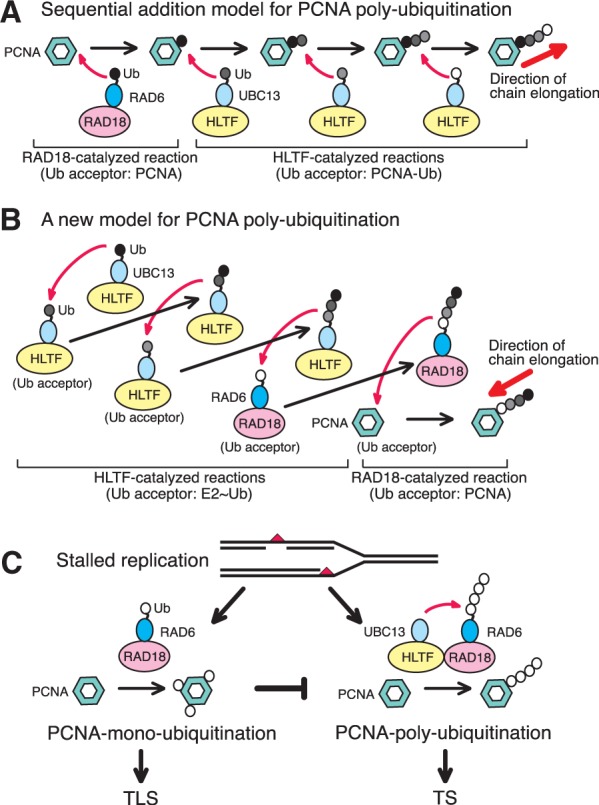
Molecular mechanism of PCNA ubiquitination and implications for the regulation of post-replication repair. MMS2 was omitted to simplify the schematics. A model with MMS2 is shown in Supplementary Figure S9. (A) Sequential addition model. RAD18 transfers Ub from RAD6 to PCNA and then HLTF transfers Ub from UBC13 to the terminal Ub moiety of ubiquitinated PCNA. In this model, the Ub chain is elongated in the PCNA-proximal to -distal direction. (B) One of potential mechanisms of en bloc Ub chain transfer. HLTF transfers Ub from UBC13 to the Ub moiety of UBC13∼Ub, generating UBC13∼Ub2. In the next step, the resultant Ub2 on UBC13 is transferred to the Ub moiety of another UBC13∼Ub, generating UBC13∼Ub3. Multiple rounds of this reaction generate long chain UBC13∼Ubn. The Ub chain can be transferred to the Ub moiety of RAD6∼Ub, generating RAD6∼Ubn+1. RAD18 then transfers the chain from RAD6 to PCNA. In these reactions, the resultant Ub chain has been elongated in the PCNA-distal to -proximal direction. (C) Regulation of post-replication repair (see text for details).
DISCUSSION
Using an in vitro ubiquitination reaction system, we were able to obtain detailed information about the mechanism of PCNA polyubiquitination. Although we cannot exclude the possibility that the reaction conditions were not properly optimized for certain reactions, our results support a novel mechanism of PCNA polyubiquitination that is distinct from, and much more efficient than, the sequential addition reactions of PCNA monoubiquitination (Figure 8A) (1,16,17). The mechanism involves pre-formation of a thiol-linked Ub chain on UBC13 and then transfer to RAD6∼Ub by HLTF. In this mechanism, HLTF functions as a novel E3 ligase that catalyzes Ub (chain) transfer from UBC13 to the Ub moieties of either UBC13∼Ub or RAD6∼Ub. This is in contrast to the target protein-specific E3 ligase activity of RAD18, which catalyzes the transfer of Ub chains, as well as Ub monomers, from RAD6 to Lys164 of PCNA. HLTF was also able to catalyze the addition of Ub onto monoUb-PCNA, as described in the sequential addition model, but this activity appeared to be extremely low in terms of the substrate specificity of the enzyme (Figure 5A–C). Thus, coupling reactions catalyzed by the two E2–E3 pairs were indispensable for efficient polyubiquitination of PCNA (Figure 5A–C). Our results suggest that HLTF elongates Ub chains via a seesaw type mechanism (27), such that all reactions could be explained by identical biochemical reactions with equivalent substrate specificity. A model is depicted in Figure 8B and Supplementary Figure S9 as one of potential mechanisms of en bloc Ub chain transfer. Note that the direction of chain elongation by the seesaw mechanism should be PCNA-distal to -proximal (Figure 8B), which is opposite of that predicted by the conventional model (Figure 8A). This is supported by the data showing that the direction of chain elongation is consistent with a seesaw mechanism of addition (Figure 7).
Our results revealed some interesting properties of HLTF. First, HLTF exhibited a unique specificity for E2∼Ub as a substrate. This could represent a biochemical basis for the seesaw mechanism. Second, it functions to regulate Ub chain length. UBC13∼Ubn molecules with long Ub chains (n ≥ 5) were barely detectable, whereas molecules with short Ub chains (n ≤ 4) were abundant (Figure 1G). The opposite was observed for RAD6A∼Ubn molecules, with greater accumulation of long chains versus short chains (Figure 5F). These results suggest that the length of the chain attached to UBC13 is enzymatically regulated so that long chains are preferentially transferred to RAD6. Third, DNA was absolutely required for HLTF ligase activity. Since HLTF is a SWI/SNF DNA helicase with a RING domain within the helicase domain, it is likely that the helicase domain is responsible for the interaction with DNA. However, we have been unable to demonstrate a functional interaction between the ligase and helicase domains, as several mutant proteins with alterations in the conserved helicase domain also exhibit ligase activity at levels similar to that of the wild-type protein (Y. Masuda, unpublished data). Previously, it was reported that the Ube2g2 (E2) is capable of producing active site-linked ubiquitin chains with the gp78 (E3), which has been shown to form an oligomer that promotes E2–E2 interactions (32). It is likely that HLTF binding to DNA promotes multimerization of HLTF to stimulate its ligase activity. The precise molecular mechanisms underlying these properties of HLTF remain to be elucidated.
Previously, based on in vitro experiments with yRAD5, two groups reported that PCNA polyubiquitination occurs via a conventional, sequential addition reaction mechanism, as depicted in Figure 8A (16,17). The results of these two studies were consistent with some of the genetic studies performed in a rad18 mutant; namely, that expression of a PCNAK164R–Ub fusion protein suppressed defects in both the TLS and TS pathways, but expression of PCNAK164R-UbK63R suppressed only the defect in the TLS pathway, suggesting PCNAK164R–Ub, but not PCNAK164R–UbK63R, is polyubiquitinated in vivo (33). On the other hand, recent results, also from yeast genetic studies (34,35), demonstrated that expression of PCNAK164R–Ub only partially suppresses the rad18 and rhp18 mutant phenotype in budding and fission yeast, respectively. In this recent set of studies, PCNAK164R–Ub was unable to activate the TS pathway, and this was attributed to the failure to polyubiquitinate the fusion protein. Indeed, PCNAK164R–Ub was not polyubiquitinated in either type of yeast (34,35). While these discrepancies in yeast genetic data from different laboratories remain to be resolved, our in vitro results are consistent with the more recent genetic data, in that neither PCNAK164R–Ub nor monoUb-PCNA was a primary target for polyubiquitination (Figure 5A–C). To directly probe the yRAD5 reaction mechanism, histidine-tagged yRAD5 (HisyRAD5) was overexpressed in E. coli and partially purified, and then used in our assay system instead of HLTF (Supplementary Figure S6). We expected yRAD5 to interact with the human proteins since HLTF is able to complement the UV sensitivity of a rad5Δ strain (13). Indeed, polyubiquitinated PCNA was detected in reactions using partially purified HisyRAD5 (Supplementary Figure S6A). Analysis of the time course of the reaction showed that all of the reaction products, including intermediates, accumulated over time (Supplementary Figure S6B), similar to what was seen with low concentrations of HLTF (Supplementary Figure S2D). Furthermore, monoubiquitinated PCNA was a poor substrate for yRAD5 (Supplementary Figure S6C), similar to that observed for HLTF (Figure 5A). These results suggested that PCNA polyubiquitination by yRAD5 occurs predominantly by en bloc transfer of an Ub chain. In the previously reported in vitro studies with yRad5, the experiments were for the most part carried out in the absence of RAD6–RAD18, using monoUb-PCNA and either PCNA–Ub fusion proteins (16) or chemically Ub-modified PCNA (17). Even in the presence of RAD6–RAD18, mono- and polyubiquitination were relatively inefficient due to the low activity of RAD6–RAD18 for monoubiquitination (16). Notably, in these studies, the experimental system was designed to detect Ub transfer to monoUb-PCNA. Under similarly uncoupled reaction conditions (i.e. in the absence of RAD6A–RAD18), we were also able to detect Ub transfer to monoUb-PCNA (Figure 5A, lane 2; 5B, lanes 1–5; and 5 C, lanes 1–4). The discrepancies between the previous in vitro yRad5 studies and the present study are mostly likely due to differences in such reaction conditions. Importantly, however, although the reaction mechanism we defined is completely different from the conventional model, it is essentially consistent with the genetic data; namely both mono- and polyubiquitination are abrogated by inactivation or deletion of either yRAD6 or yRAD18, whereas in the presence of dysfunctional yMMS2, yUBC13, or yRAD5, only polyubiquitination is abrogated (1,2).
If the reaction mechanism we observed in vitro mimics what occurs in vivo, monoubiquitination and polyubiquitination of PCNA at stalled replication ends would necessarily be promoted separately (Figure 8C). It should be noted that Ub chain formation by UBC13–MMS2 and HLTF required mpssDNA, which differs from the requirement of RAD6–RAD18 for RFC-mediated loading of PCNA on mpssDNA (6,7,36). We hypothesize that PCNA polyubiquitination and activation of the TS pathway is promoted under conditions in which RAD18 and HLTF are coordinately recruited to stalled replication ends, wherein DNA is needed to hold PCNA at the primer terminus (7) and to stimulate HLTF activity. Alternatively, if all three subunits of PCNA at a stalled end are first monoubiquitinated, polyubiquitination cannot occur and the TLS pathway is activated. We have observed that all three subunits of PCNA are efficiently monoubiquitinated at stalled primer ends in vitro (7). This scenario could explain how either the TLS or TS pathway is selectively activated at stalled replication sites. The TS pathway would be activated only when a daughter strand that was replicated from the non-damaged parental strand is available and prohibited when such a daughter strand is unavailable. This model is consistent with recent observations from our laboratory that RAD6–RAD18 forms a ternary complex of RAD6A–(RAD18)2 rather than (RAD6A–RAD18)2 and that stable association between RAD6A and RAD18 is an essential requirement for ligase activity (21). These properties of the RAD6–RAD18 complex appear suited for the tight regulation of polyubiquitination of PCNA. If the complex contained two RAD6 molecules, one would have to invoke yet another mechanism involving processive formation of Ub chains on both subunits. Furthermore, if the interaction of RAD6 was dynamic, exchange reactions between RAD6∼Ubn and RAD6∼Ub would occur in solution before transfer of the chain to PCNA. The tight regulation of polyubiquitination ensures the timely and appropriate activation of the TS pathway, thereby avoiding irregular recombination. In this manner, eukaryotic cells ensure accurate chromosomal duplication and avoid genetic aberrations that can lead to carcinogenesis.
Finally, we should mention that the seesaw mechanism of ubiquitin chain transfer is similar to the peptidyl transfer reaction of protein synthesis. Conceivably, the concept might be applicable to other E2 and E3 enzymes and another bio-macromolecule synthesis such as oligosaccharides, and may represent a common strategy for the generation of bio-macro-polymers.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1–10 and Supplementary Notes.
FUNDING
Grants-in-Aid from the Ministry of Education, Culture, Sports, Science and Technology of Japan (to Y.M., H.K., H.H., C.M. T.H. and K.K.); Health and Labour Science Research Grants of Japan (to K.K.); the Mitsubishi Foundation and Takeda Science Foundation (to C.M.). Funding for open access charge: JSPS KAKENHI [24310040 to Y.M.].
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Dr. Toshiki Tsurimoto (Kyushu University, Fukuoka, Japan) for a PCNA-expression plasmid. The authors would also like to express our appreciation to Drs Mark Hochstrasser (Yale University, USA), Haruo Ohmori (Kyoto University, Kyoto, Japan), Richard D. Wood (University of Texas, USA) and Roger Woodgate (NIH, USA) for their comments and suggestions on the manuscript. The authors are furthermore grateful to Yumiko Shintani for making mutants, and Mayumi Hojo, Fumie Okubo, Kazumi Shimamoto, Reiko Tashiro and Mai Yoshida for their laboratory assistance.
REFERENCES
- 1.Friedberg EC, Walker GC, Siede W, Wood RD, Schultz RA, Ellenberger T. DNA Repair and Mutagenesis. 2 edn. Washington, DC: ASM; 2006. [Google Scholar]
- 2.Hoege C, Pfander B, Moldovan GL, Pyrowolakis G, Jentsch S. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature. 2002;419:135–141. doi: 10.1038/nature00991. [DOI] [PubMed] [Google Scholar]
- 3.Kannouche PL, Wing J, Lehmann AR. Interaction of human DNA polymerase η with monoubiquitinated PCNA: a possible mechanism for the polymerase switch in response to DNA damage. Mol. Cell. 2004;14:491–500. doi: 10.1016/s1097-2765(04)00259-x. [DOI] [PubMed] [Google Scholar]
- 4.Watanabe K, Tateishi S, Kawasuji M, Tsurimoto T, Inoue H, Yamaizumi M. Rad18 guides polη to replication stalling sites through physical interaction and PCNA monoubiquitination. EMBO J. 2004;23:3886–3896. doi: 10.1038/sj.emboj.7600383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bienko M, Green CM, Crosetto N, Rudolf F, Zapart G, Coull B, Kannouche P, Wider G, Peter M, Lehmann AR, et al. Ubiquitin-binding domains in Y-family polymerases regulate translesion synthesis. Science. 2005;310:1821–1824. doi: 10.1126/science.1120615. [DOI] [PubMed] [Google Scholar]
- 6.Garg P, Burgers PM. Ubiquitinated proliferating cell nuclear antigen activates translesion DNA polymerases η and REV1. Proc. Natl Acad. Sci. USA. 2005;102:18361–18366. doi: 10.1073/pnas.0505949102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Masuda Y, Piao J, Kamiya K. DNA replication-coupled PCNA mono-ubiquitination and polymerase switching in a human in vitro system. J. Mol. Biol. 2010;396:487–500. doi: 10.1016/j.jmb.2010.01.003. [DOI] [PubMed] [Google Scholar]
- 8.Wood A, Garg P, Burgers PM. A ubiquitin-binding motif in the translesion DNA polymerase Rev1 mediates its essential functional interaction with ubiquitinated proliferating cell nuclear antigen in response to DNA damage. J. Biol. Chem. 2007;282:20256–20263. doi: 10.1074/jbc.M702366200. [DOI] [PubMed] [Google Scholar]
- 9.Zhuang Z, Johnson RE, Haracska L, Prakash L, Prakash S, Benkovic SJ. Regulation of polymerase exchange between Polη and Polδ by monoubiquitination of PCNA and the movement of DNA polymerase holoenzyme. Proc. Natl Acad. Sci. USA. 2008;105:5361–5366. doi: 10.1073/pnas.0801310105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.MacKay C, Toth R, Rouse J. Biochemical characterisation of the SWI/SNF family member HLTF. Biochem. Biophys. Res. Commun. 2009;390:187–191. doi: 10.1016/j.bbrc.2009.08.151. [DOI] [PubMed] [Google Scholar]
- 11.Motegi A, Liaw HJ, Lee KY, Roest HP, Maas A, Wu X, Moinova H, Markowitz SD, Ding H, Hoeijmakers JH, et al. Polyubiquitination of proliferating cell nuclear antigen by HLTF and SHPRH prevents genomic instability from stalled replication forks. Proc. Natl Acad. Sci. USA. 2008;105:12411–12416. doi: 10.1073/pnas.0805685105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Motegi A, Sood R, Moinova H, Markowitz SD, Liu PP, Myung K. Human SHPRH suppresses genomic instability through proliferating cell nuclear antigen polyubiquitination. J. Cell. Biol. 2006;175:703–708. doi: 10.1083/jcb.200606145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Unk I, Hajdu I, Fatyol K, Hurwitz J, Yoon JH, Prakash L, Prakash S, Haracska L. Human HLTF functions as a ubiquitin ligase for proliferating cell nuclear antigen polyubiquitination. Proc. Natl Acad. Sci. USA. 2008;105:3768–3773. doi: 10.1073/pnas.0800563105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Unk I, Hajdu I, Fatyol K, Szakal B, Blastyak A, Bermudez V, Hurwitz J, Prakash L, Prakash S, Haracska L. Human SHPRH is a ubiquitin ligase for Mms2-Ubc13-dependent polyubiquitylation of proliferating cell nuclear antigen. Proc. Natl Acad. Sci. USA. 2006;103:18107–18112. doi: 10.1073/pnas.0608595103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hofmann RM, Pickart CM. Noncanonical MMS2-encoded ubiquitin-conjugating enzyme functions in assembly of novel polyubiquitin chains for DNA repair. Cell. 1999;96:645–653. doi: 10.1016/s0092-8674(00)80575-9. [DOI] [PubMed] [Google Scholar]
- 16.Parker JL, Ulrich HD. Mechanistic analysis of PCNA poly-ubiquitylation by the ubiquitin protein ligases Rad18 and Rad5. EMBO J. 2009;28:3657–3666. doi: 10.1038/emboj.2009.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carlile CM, Pickart CM, Matunis MJ, Cohen RE. Synthesis of free and proliferating cell nuclear antigen-bound polyubiquitin chains by the RING E3 ubiquitin ligase Rad5. J. Biol. Chem. 2009;284:29326–29334. doi: 10.1074/jbc.M109.043885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fukuda K, Morioka H, Imajou S, Ikeda S, Ohtsuka E, Tsurimoto T. Structure-function relationship of the eukaryotic DNA replication factor, proliferating cell nuclear antigen. J. Biol. Chem. 1995;270:22527–22534. doi: 10.1074/jbc.270.38.22527. [DOI] [PubMed] [Google Scholar]
- 19.Masuda Y, Suzuki M, Piao J, Gu Y, Tsurimoto T, Kamiya K. Dynamics of human replication factors in the elongation phase of DNA replication. Nucleic Acids Res. 2007;35:6904–6916. doi: 10.1093/nar/gkm822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tomida J, Masuda Y, Hiroaki H, Ishikawa T, Song I, Tsurimoto T, Tateishi S, Shiomi T, Kamei Y, Kim J, et al. DNA damage-induced ubiquitylation of RFC2 subunit of replication factor C complex. J. Biol. Chem. 2008;283:9071–9079. doi: 10.1074/jbc.M709835200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Masuda Y, Suzuki M, Kawai H, Suzuki F, Kamiya K. Asymmetric nature of two subunits of RAD18, a RING-type ubiquitin ligase E3, in the human RAD6A-RAD18 ternary complex. Nucleic Acids Res. 2012;40:1065–1076. doi: 10.1093/nar/gkr805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gu Y, Masuda Y, Kamiya K. Biochemical analysis of human PIF1 helicase and functions of its N-terminal domain. Nucleic Acids Res. 2008;36:6295–6308. doi: 10.1093/nar/gkn609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ecker DJ, Butt TR, Marsh J, Sternberg EJ, Margolis N, Monia BP, Jonnalagadda S, Khan MI, Weber PL, Mueller L, et al. Gene synthesis, expression, structures, and functional activities of site-specific mutants of ubiquitin. J. Biol. Chem. 1987;262:14213–14221. [PubMed] [Google Scholar]
- 24.Haas AL, Rose IA. The mechanism of ubiquitin activating enzyme. A kinetic and equilibrium analysis. J. Biol. Chem. 1982;257:10329–10337. [PubMed] [Google Scholar]
- 25.Li W, Tu D, Brunger AT, Ye Y. A ubiquitin ligase transfers preformed polyubiquitin chains from a conjugating enzyme to a substrate. Nature. 2007;446:333–337. doi: 10.1038/nature05542. [DOI] [PubMed] [Google Scholar]
- 26.Petroski MD, Deshaies RJ. Mechanism of lysine 48-linked ubiquitin-chain synthesis by the cullin-RING ubiquitin-ligase complex SCF-Cdc34. Cell. 2005;123:1107–1120. doi: 10.1016/j.cell.2005.09.033. [DOI] [PubMed] [Google Scholar]
- 27.Hochstrasser M. Lingering mysteries of ubiquitin-chain assembly. Cell. 2006;124:27–34. doi: 10.1016/j.cell.2005.12.025. [DOI] [PubMed] [Google Scholar]
- 28.Eddins MJ, Carlile CM, Gomez KM, Pickart CM, Wolberger C. Mms2-Ubc13 covalently bound to ubiquitin reveals the structural basis of linkage-specific polyubiquitin chain formation. Nat. Struct. Mol. Biol. 2006;13:915–920. doi: 10.1038/nsmb1148. [DOI] [PubMed] [Google Scholar]
- 29.Deshaies RJ, Joazeiro CA. RING domain E3 ubiquitin ligases. Annu. Rev. Biochem. 2009;78:399–434. doi: 10.1146/annurev.biochem.78.101807.093809. [DOI] [PubMed] [Google Scholar]
- 30.Zheng N, Wang P, Jeffrey PD, Pavletich NP. Structure of a c-Cbl-UbcH7 complex: RING domain function in ubiquitin-protein ligases. Cell. 2000;102:533–539. doi: 10.1016/s0092-8674(00)00057-x. [DOI] [PubMed] [Google Scholar]
- 31.Renatus M, Parrado SG, D'Arcy A, Eidhoff U, Gerhartz B, Hassiepen U, Pierrat B, Riedl R, Vinzenz D, Worpenberg S, et al. Structural basis of ubiquitin recognition by the deubiquitinating protease USP2. Structure. 2006;14:1293–1302. doi: 10.1016/j.str.2006.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li W, Tu D, Li L, Wollert T, Ghirlando R, Brunger AT, Ye Y. Mechanistic insights into active site-associated polyubiquitination by the ubiquitin-conjugating enzyme Ube2g2. Proc. Natl Acad. Sci. USA. 2009;106:3722–3727. doi: 10.1073/pnas.0808564106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhao S, Ulrich HD. Distinct consequences of posttranslational modification by linear versus K63-linked polyubiquitin chains. Proc. Natl Acad. Sci. USA. 2010;107:7704–7709. doi: 10.1073/pnas.0908764107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pastushok L, Hanna M, Xiao W. Constitutive fusion of ubiquitin to PCNA provides DNA damage tolerance independent of translesion polymerase activities. Nucleic Acids Res. 2010;38:5047–5058. doi: 10.1093/nar/gkq239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ramasubramanyan S, Coulon S, Fuchs RP, Lehmann AR, Green CM. Ubiquitin-PCNA fusion as a mimic for mono-ubiquitinated PCNA in Schizosaccharomyces pombe. DNA Repair. 2010;9:777–784. doi: 10.1016/j.dnarep.2010.03.015. [DOI] [PubMed] [Google Scholar]
- 36.Haracska L, Unk I, Prakash L, Prakash S. Ubiquitylation of yeast proliferating cell nuclear antigen and its implications for translesion DNA synthesis. Proc. Natl Acad. Sci. USA. 2006;103:6477–6482. doi: 10.1073/pnas.0510924103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.