Abstract
DNA methylation is an essential mechanism controlling gene expression during differentiation and development. We investigated the epigenetic regulation of the nuclear-encoded, mitochondrial DNA (mtDNA) polymerase γ catalytic subunit (PolgA) by examining the methylation status of a CpG island within exon 2 of PolgA. Bisulphite sequencing identified low methylation levels (<10%) within exon 2 of mouse oocytes, blastocysts and embryonic stem cells (ESCs), while somatic tissues contained significantly higher levels (>40%). In contrast, induced pluripotent stem (iPS) cells and somatic nuclear transfer ESCs were hypermethylated (>20%), indicating abnormal epigenetic reprogramming. Real time PCR analysis of 5-methylcytosine (5mC) and 5-hydroxymethylcytosine (5hmC) immunoprecipitated DNA suggests active DNA methylation and demethylation within exon 2 of PolgA. Moreover, neural differentiation of ESCs promoted de novo methylation and demethylation at the exon 2 locus. Regression analysis demonstrates that cell-specific PolgA expression levels were negatively correlated with DNA methylation within exon 2 and mtDNA copy number. Finally, using chromatin immunoprecipitation (ChIP) against RNA polymerase II (RNApII) phosphorylated on serine 2, we show increased DNA methylation levels are associated with reduced RNApII transcriptional elongation. This is the first study linking nuclear DNA epigenetic regulation with mtDNA regulation during differentiation and cell specialization.
INTRODUCTION
Mammalian mitochondrial DNA (mtDNA) is a circular double-stranded genome of ∼16.6 Kb encoding 37 genes (1). These genes encode 13 polypeptides of the electron transfer chain, 22 tRNAs and 2 rRNAs, which directly or indirectly contribute to the production of ATP through the process of oxidative phosphorylation (OXPHOS). The remaining OXPHOS-associated genes and all of the transcription and replication machinery are encoded by nuclear DNA (2). Mutations, deletions and insertions to mtDNA lead to decreased OXPHOS capacity and mitochondrial function resulting in severely debilitating and often lethal pathophysiology (3).
The mitochondrial specific DNA polymerase γ (POLG) is the only known DNA polymerase that localizes within mitochondria (4,5). The POLG holoenzyme is composed of one catalytic subunit, POLGA, which is responsible for proofreading and DNA repair activity (6,7), and two POLGB accessory subunits, which maintain enzymatic stability and efficiency (8). POLG is essential for mtDNA replication and maintenance. Pathological mutations to PolgA are characterized by mtDNA deletion or depletion, which lead to respiratory chain deficiencies or premature aging phenotypes (9,10). Furthermore, ablating (11) or reducing (11,12) PolgA expression levels significantly decreases mtDNA copy number.
The expression of PolgA and the numbers of mtDNA copy are strictly controlled during development. In preimplantation embryos, the first mtDNA replication event is initiated at the blastocyst stage (13) and coincides with the up-regulated expression of POLG (13,14). This is restricted to the trophectoderm, which forms the placenta while those cells of the inner cell mass, which give rise to the embryo and embryonic stem cells (ESCs), do not replicate their mtDNA. As differentiation and development proceed, significant changes in mtDNA copy number are observed (15–19), so that high-energy consuming tissues, such as muscle and nerve cells, acquire sufficient copies of mtDNA to meet their specific demands of ATP. The regulation of mtDNA copy number and PolgA expression during development are critical, as PolgA homozygous knockout mice die at E7.5-8.5 while heterozygous knockouts exhibit mtDNA depletion-type syndromes (11).
Although some evidence indicates that PolgA expression may be regulated by DNA methylation (20) and that mtDNA is itself methylated (21), the epigenetic mechanisms that control the expression of PolgA and regulate mtDNA copy number remain to be determined. A CpG island within exon 2 of PolgA, which is hypermethylated during spermatogenesis, has been identified, however, its effect on PolgA expression and mtDNA copy number were not described (20). It is well-documented that cytosine methylation within promoter regions associated with transcription start sites mediates gene repression by recruiting methyl-binding domain repressor protein complexes or inhibiting the binding of transcription factors (22,23). However, recent studies have identified the importance of DNA methylation within (intragenic) and between (intergenic) gene bodies (22,24). The reason for DNA methylation at these sites remains unknown. Nevertheless, intragenic methylation has been linked to reduced transcriptional elongation (25–27) and gene silencing (28), while others have shown positive correlations between intragenic methylation and gene expression (29,30). If such a mechanism regulated PolgA expression, this would present a unique regulatory mechanism whereby an evolutionary distinct genome of bacterial origin is epigenetically regulated by a nuclear-encoded gene (31).
We have determined the DNA methylation status of the intragenic CpG island within exon 2 of PolgA in gametes, pluripotent ESCs, differentiating stem cells and somatic tissues, and whether this intragenic DNA methylation correlated with PolgA gene expression. Using bisulphite sequencing, restriction enzyme digest real time PCR (qAMP) and methylated DNA immunoprecipitation (MeDIP) with real time PCR, we have identified tissue specific DNA methylation profiles and de novo intragenic DNA methylation during differentiation. We demonstrate that these DNA methylation profiles inversely correlate with the levels of PolgA expression and that there is a linear relationship between steady state levels of PolgA expression and mtDNA copy number.
MATERIALS AND METHODS
Cell culture
Murine (Mus musculus) D3 ESCs (ESD3); somatic cell nuclear transfer ESCs (NT-ES) (32); CC9.3.1 ESCs (CC9mus); CC9.3.1 ESCs harbouring divergent Mus spretus (CC9spretus) or Mus terricolor (Dunni; CC9dunni) mtDNA (33) and induced pluripotent stem (iPS) cells derived from Quackenbush Swiss × Rosa26 (iPSQS/R26) (34) and C57Bl/6 × 129j/SvE Nanog-GFP+ (iPSNGFP; Stemgent, USA) (35) were cultured on mitomycin C treated mouse embryonic fibroblasts (MEFs) in ESC media composed of DMEM supplemented with 15% fetal bovine serum (FBS; Invitrogen, Australia), 0.5% (v/v) Penicillin–Streptomycin (Invitrogen, Australia), 2 mM L-glutamine (Gibco, Australia), 1% (v/v) non-essential amino acids (Gibco), 0.1 mM β-mercaptoethanol (Gibco), with 1000 U/ml (10 ng/ml) leukemia inhibitory factor (LIF; Millipore, Australia) at 37°C in 5% CO2, as previously described (15). MEFs were cultured in ESC media in the absence of LIF and on gelatin coated culture dishes (0.1%).
Tissue samples
All mice (C57BL×CBA-F1) were housed at the Monash University Animal House, according to Australian and Monash University ethical standards. Experimental procedures were approved by the Monash Medical Centre Animal Ethics Committee and were conducted in accordance with the Australian National Health and Medical Research Council (NHMRC) guidelines. Animals were euthanized by cervical dislocation and samples (oocytes, liver, spleen, kidney, muscle, heart and brain) were frozen immediately at −80°C until required. MEFs (129/Sv) were donated by the Australian Stem Cell Centre.
Embryo in vitro culture
8-week-old female F1 mice (C57BL×CBA) were superovulated by intraperitoneal injection of 5 IU of human choronic gonadotropin (Chorulon; Invervet, Australia) and were mated with 12-week-old males. Embryos were retrieved 24 h post mating and in vivo fertilized embryos were collected from the oviduct following cervical dislocation of the female mouse. Culture to the blastocyst stage was performed, as previously described (36), in KSOM (Millipore, Australia) in 5% CO2, at 37°C.
ESC injection into blastocysts
Approximately 5–10 iPSNGFP cells were injected into day 3.5 (post-mating) blastocysts. Blastocysts were cultured on mitomycin C treated MEFs in ESC media at 37°C in 5% CO2. Outgrowths were selected after 5–7 days and manually passaged five times selecting only GFP+ colonies. Further passaging was performed enzymatically using TrypLE (Gibco, Australia).
Spontaneous and neural differentiation
Spontaneous differentiation of ESCs was induced by culturing approximately 450 cells in 20 µl droplets of ESC media in the absence of LIF on the base of an inverted Petri dish for 48 h. Embryoid bodies were cultured in suspension for a further 5 days and then plated onto culture dishes in ESC media in the absence of LIF for 2 days.
Neural induction was performed, as previously described (37,38). Briefly, ESCs were cultured at a density of 1.5–2 × 104 cm−2 on gelatin treated (0.1% w/v) culture dishes in neural stem cell media (NSC) composed of DMEM/F12 (Invitrogen) supplemented with 1% N2 (Millipore), 2% B27 (Millipore), 0.5% (v/v) Penicillin–Streptomycin (Invitrogen) and 10 ng/µl basic fibroblast growth factor (bFGF; Millipore, Australia). After 3 days, the cultures were dissociated using TrypLE (Gibco) and plated on laminin (Sigma) coated (1 µg/cm2) 100 mm culture dishes at a density of 50 000 cell cm−2 in NSC media. For neuronal differentiation, NSC media was changed every 48 h for a further 4 days, at which point cells were cultured in neuronal induction media (NIM) composed of Neurobasal Medium (Invitrogen) supplemented with 2% B27 (Millipore), 0.5% (v/v) Penicillin–Streptomycin (Invitrogen) and 10 ng/µl brain-derived neurotropic factor (BDNF; Millipore).
NCAM+ cell isolation
Spontaneously differentiated cells were triturated into a single cell suspension using TrypLE (Gibco) and passed through a 70 µm filter. Single cell suspensions were incubated with ice cold isolation buffer (phosphate-buffered saline containing 0.5% bovine serum albumin and 2 mM EDTA) supplemented with anti-PSA-NCAM conjugated to microbeads (1 in 11 dilution: Miltenyl, Australia) for 10 min at 4°C. PSA-NCAM labelled cells were isolated by magnetic separation Miltenyi MACS Cell Separation Columns (Miltenyi), according to the manufacturers instructions.
RNA and DNA extraction
Total RNA and DNA were extracted, according to the manufacturer’s instructions, using the RNeasy Mini Kit (Qiagen, Australia) and the DNeasy Blood & Tissue Kit (Qiagen), respectively. The RNA samples were then treated with DNase I (Qiagen) for 15–30 min at room temperature while the DNA samples were then treated with RNase solution (Qiagen) and proteinase K for 10 min at 55°C.
Reverse transcription was performed, as previously described (16). Briefly, 1 μg/μl of RNA, 5 μl of 10× RT-Buffer, 1 mM of each dNTPs, 0.25 μg Oligo (dT) primer, 1 U/μl RiboSafe, 1 U/μl of BioScript and autoclaved H2O up to 20 μl were incubated at 42°C for 1 h and 72°C for 10 min.
Generation of real time PCR standards
Real-time PCR standards were produced using PCR. Breifly, 200 ng of cDNA or DNA were amplified in 50 µl reactions using 5× PCR buffer (Bioline), 1.5 mM MgCl2 (Bioline), 100 µM dNTPs (Bioline), 0.5 µM for each of the forward and reverse primers. Reactions were performed on a MJ Research PTC-200 machine and PCR products were resolved on 2% agarose gels at 100 V for 1 h. Primer sequences are listed in Supplementary Table S1. A series of 10-fold dilutions (1 ng/μl to 1 × 10−9 ng/μl) of the target specific PCR product was generated.
Real time PCR: gene expression and mtDNA copy number
Real time PCR contained 2 μl of template (cDNA or DNA), 10 μl of 2× SensiMix (Bioline, Australia), 0.33 μM of forward and reverse primers and ultrapure ddH2O (20 μl total volume). Reactions were performed in a Rotorgene-3000 real time PCR machine (Corbett Research, Cambridge, UK) under primer specific conditions (see Supplementary Table S1). The number of mtDNA copies/cells and relative expression levels of PolgA were quantified against known standards, using real time PCR, as previously described (15). We assessed the reliability of β-Actin, Gapdh and 18S as housekeeping genes. The expression of β-Actin and Gapdh varied between cell types and tissues, although 18S expression remained consistent across all samples analysed.
Bisulphite sequencing
Extracted DNA (1 µg) was incubated with 3 M NaOH for 10 min at 37°C and bisulphite treated, according to the manufacturers instructions (Human Genetic Signatures, USA). MII oocytes (50 samples) and blastocysts (10 samples) were lysed and mixed with 1 µg of Herring sperm DNA (Sigma, Australia) prior to NaOH treatment. The samples were amplified using nested PCR primers (Supplementary Table S1; PolgA-Bimeth and PolgA-BiNest): 95°C for 4 min followed by 35 cycles of 95°C for 1 min, 53°C for 1 min, 72°C for 1 min; and final elongation of 72°C for 4 min. PCR products were cloned using a pCR®2.1 vector (Invitrogen, Australia) and DH5α™ competent cells (Invitrogen). Individual clones were sequenced, as described in (39), using a 16-capillary 3130xi Genetic Analyzer (Applied Biosystems, Australia). Data were analysed using a BIQ Analyzer (http://biq-analyzer.bioinf.mpi-inf.mpg.de/).
Restriction enzyme digest and qAMP
The methylation status of the CpG island in the 5′ region of the PolgA gene (NT_039428: chromosome 7, region 19 607 339–19 607 508), which has a variable DNA methylation profile during spermatogenesis, was analysed by the qAMP method (20). Genomic DNA was either digested with methylation-sensitive restriction endonucleases (HpaII, NotI), the methylation-dependent endonuclease (McrBC) or remained undigested (no enzyme sham digestion). The DNA samples were amplified by real-time PCR using primers defining a region including restriction sites in the second exon of the gene, which is included within the CpG Island (Supplementary Table S1; qAMP-PolgA). Shifts in Ct value (ΔCt) between the digested and undigested samples were used to calculate the percentage of methylation at the assayed restriction sites. The ΔCt values were measured in triplicate. For the methylation sensitive enzymes (HpaII and NotI), percentage methylation = 100 (2−ΔCt), and for McrBC, percentage methylation = 100 (1–2−ΔCt).
Immunoprecipitation of methylated DNA
Immunoprecipitation of methylated DNA was performed, as previously described (40). Briefly, 10 μg of purified DNA was sonicated to produce random fragments between 200–1100 bp. Sonicated DNA (3 μg) was denatured at 95°C for 10 min and immunoprecipitated in 500 μl of IP buffer (10 mM Na-Phosphate, pH 7.0, 140 mM NaCl, 0.05% Triton X-100) containing 2 μg of either anti-5-methylcytosine (5mC; Active Motif, USA) or anti-5-hydroxymethylcytosine (5hmC, Active Motif) for 2 h at 4°C. Samples were then incubated with 20 μl of Protein G Dynabeads (Invitrogen) for 2 h at 4 °C and finally washed twice with 700 ul IP buffer for 10 min. DNA was purified using 7 μl of Proteinase K (10 mg/ml; Qiagen) at 50°C for 3 h and then precipitated using 1 μl Glycogen (20 μg/μl; Invitrogen) and 2× volumes of 100% ethanol at −80°C for a minimum of 2 h. The enrichment of the nuclear-encoded PolgA and the mtDNA-encoded CytB, tRNA/COX1 genes and the D-Loop was determined using real time PCR and standardized against β-Actin (Supplementary Table S1).
Chromatin immunoprecipitation
Chromatin immunoprecipitation (ChIP) was performed as previously described (41) with minor modifications. Cells and tissue samples were cross-linked with 1% formaldehyde for 10 min and quenched in 125 mM glycine (Sigma) for 5 min. Heart samples were homogenized, passed through 200 μm filter and centrifuged at 2000 r.p.m.−1 for 5 min at 4°C. Cross-linked samples were lysed at 4°C for 10 min and sonicated to fragment chromatin to an average length of 200–800 bp. Chromatin from 1 × 106 cells was immunoprecipitated overnight at 4°C by adding Protein G Dynabeads (Invitrogen, Melbourne, Australia) pre-incubated with the antibody of interest. Antibodies used in ChIP assays were: polyclonal Rabbit anti-CTCF (Cell signalling, MA, USA); 4 μg monoclonal anti-RNA polymerase II (RNApII) clone 4H8; and 4 μg monoclonal Mouse anti-Ser2 RNApII (Abcam, Cambridge, UK). Cross-links were reversed by incubating samples with 200 mM NaCl and 10 ul proteinase K (>600 mAU/ml) at 65°C for 16 h. DNA was purified using the Qiagen PCR purification kit, according to the manufacturer’s instructions. Purified immunoprecipitated DNA and input DNA (1:100) were analysed using real-time PCR in triplicate, as described above. Primers are listed in Supplementary Table S1. Differences between cell types were calculated as percentage input (100 × 2(Adjusted Input (Ct) - IP (Ct))). These values were further normalized against negative controls. Enrichment values were converted to enrichment levels relative to exon 2.
Statistical analysis
Statistical analyses were performed using GraphPad Prism 5.01 (GraphPad Software, Inc., San Deigo, CA) and Microsoft Excel. Data are expressed as mean ± standard error of the mean (SEM). All real time PCR data were normalized to 18S. Correlations were determined using Pearson coefficient and statistical differences between groups were determined using One-way or Two-way ANOVA in combination with Bonferroni post-hoc tests.
RESULTS
Tissue-specific DNA methylation within exon 2 of PolgA
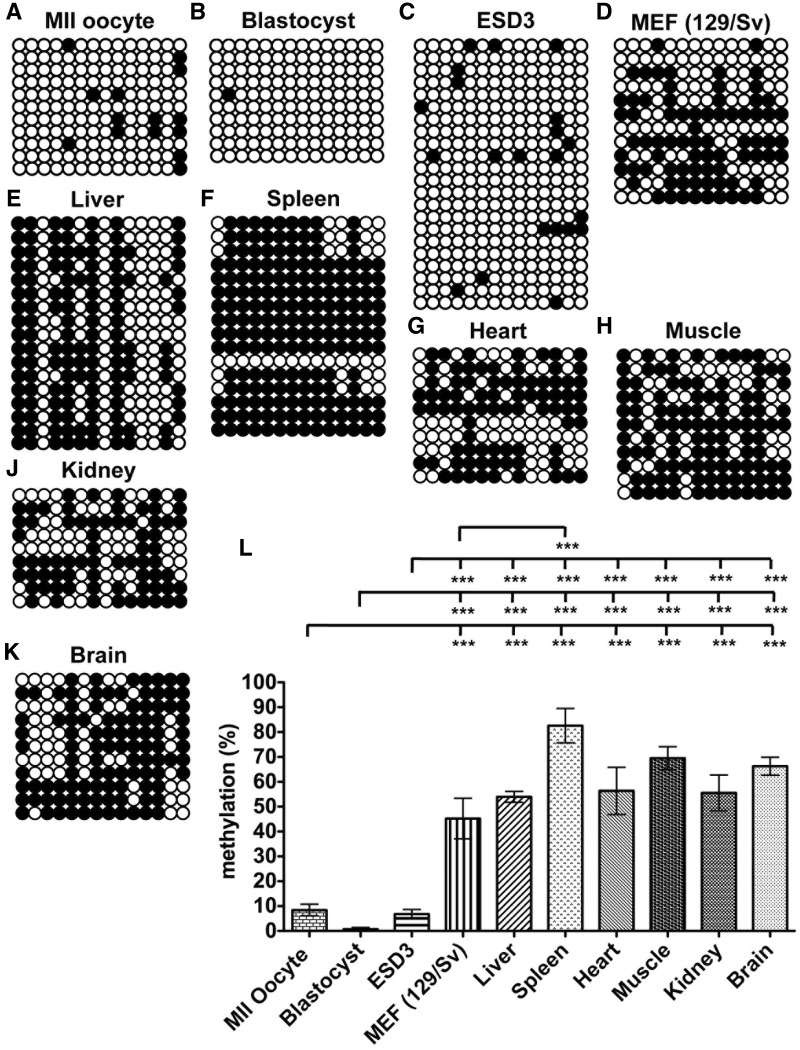
To determine whether DNA methylation to a region downstream of the PolgA transcription start site (i.e. intragenic methylation) regulates PolgA mRNA levels, we first analysed the levels of DNA methylation within exon 2 of oocytes, embryos and tissues. Bisulphite sequencing was performed on the region 1446–1616 of exon 2 (NCBI: NC_000073.5; chromosome 7, region 19 607 339–19607508) relative to the transcription start site of PolgA (20). Low levels of DNA methylation were detected in in vitro matured metaphase II (MII) oocytes (8.44% ± 2.3; Figure 1A); in vivo fertilized blastocysts (0.71% (0.71; Figure 1B); and ESCs (ESD3; 6.81% (1.85; Figure 1C).
Figure 1.
DNA methylation analysis of PolgA exon 2 in gametes, embryos, pluripotent and somatic cells. Bisulphite sequencing of the 14 CpG methylation sites in (A) MII oocytes, (B) blastocysts, (C) ESD3 cells, (D) MEF (129/Sv), (E) Liver, (F) Spleen, (G) Heart, (H) Muscle, (J) Kidney and (K) Brain samples. Open and filled circles represent unmethylated and methylated CpGs, respectively. (L) The percentage CpG methylation per sequence determined by bisulphite sequencing (% mean ± SEM). Significant differences between cell types are: ***P < 0.001.
We then determined whether low levels of DNA methylation are present in differentiated cells by analyszing mouse embryonic fibroblasts (MEFs) and adult tissue samples (Figure 1D–K). All somatic samples displayed elevated levels of DNA methylation in exon 2 compared to MII oocytes, blastocysts and ESD3 cells (P < 0.001; Figure 1L). A variable pattern of DNA methylation across the PolgA locus was observed for MEFs (45.23% ± 8.17; Figure 1D) and liver (53.97% (2.18; Figure 1E). Although a base-specific pattern of DNA methylation was observed in liver, there were no similarities in the DNA methylation patterns between cell types (Supplementary Figure S1). The highest levels of DNA methylation were detected in spleen (82.59% ± 6.93; Figure 1F and L), which was significantly different to all somatic samples except muscle (Figure 1L). Our findings demonstrate a specific region within exon 2 of PolgA is methylated in a tissue specific manner and that low or absent DNA methylation is associated with gametes and pluripotent cells while hypermethylation is prevalent in somatic tissues.
Methylation of exon 2 correlates with reduced steady state mRNA levels of PolgA
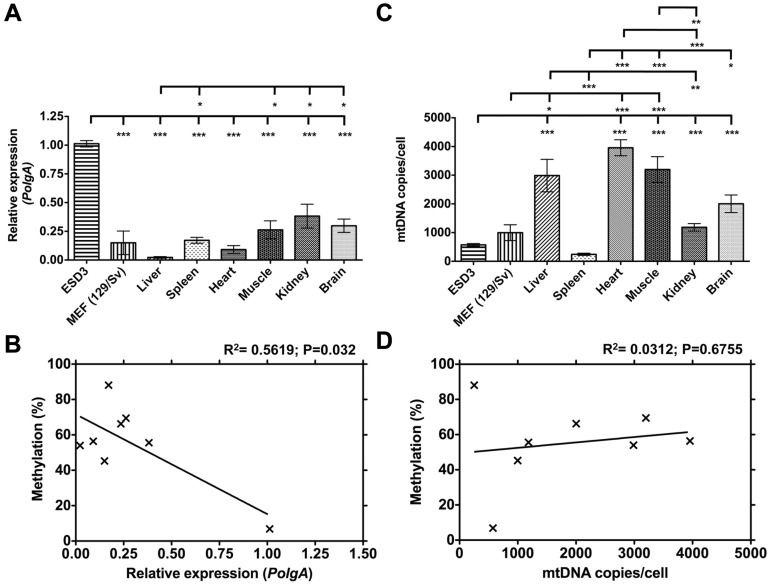
Since intragenic DNA methylation has been associated with regulating gene expression (27,28), we analysed the expression levels of PolgA. The relative expression of PolgA was significantly lower in somatic tissues compared with ESD3 cells (P < 0.001; Figure 2A). The range of mRNA levels observed in somatic tissues was between 69.9 (kidney) and 97.8% (liver) lower than that of ESD3 cells (Figure 2A). PolgA expression in spleen, brain, muscle and kidney were all statistically similar, while the levels in liver were significantly lower (P < 0.05; Figure 2A). Regression analysis by Pearson-correlation coefficient demonstrated an inverse relationship between relative expression of PolgA and the corresponding DNA methylation profiles (R2 = 0.5619; P = 0.032; Figure 2B cf Figures 1L and 2A).
Figure 2.
DNA Methylation of exon 2 correlates with reduced steady state mRNA levels of PolgA. (A) Real time PCR quantification of PolgA expression in cultured ESD3 and MEF cells, and in liver, spleen, heart, muscle, kidney and brain samples, expressed relative to ESD3. (B) The relationship between DNA methylation levels from Figure 1L and the corresponding PolgA expression was determined using Pearson correlation coefficient (R2). (C) MtDNA copies/cell in cultured ESD3 and MEF cells, and liver, spleen, heart, muscle, kidney and brain, as determined by real time PCR. (D) The relationship between the levels of DNA methylation from Figure 1L and the corresponding mtDNA copies/cell was determined using Pearson correlation coefficient (R2). Values represent mean ± SEM and significant differences between cell types are: *P < 0.05; **P < 0.01 and ***P < 0.001.
We then investigated whether the relationship between intragenic DNA methylation and PolgA expression influenced mtDNA copy number. Analysis of mtDNA copy number demonstrated that the spleen contained 251.9 ± 32.65 copies per cell, while liver, heart and muscle contained significantly more, namely 2987.3 ± 564.3, 3956.02 ± 279.33 and 3199.17 ± 447.61 copies per cell, respectively (P < 0.001; Figure 2C). ESD3 cells contained fewer mtDNA copies compared with all somatic cells and tissues except spleen (P < 0.001; Figure 2C). Despite tissue specific mtDNA copy number (Figure 2C) and intragenic DNA methylation of PolgA (Figure 2B), linear regression analysis (Figure 2D cf Figures 1L and 2C) did not provide a significant correlation between mtDNA copy number and DNA methylation (R2 = 0.0312; P = 0.6755).
Intragenic DNA methylation in ESCs harbouring divergent mtDNA haplotypes
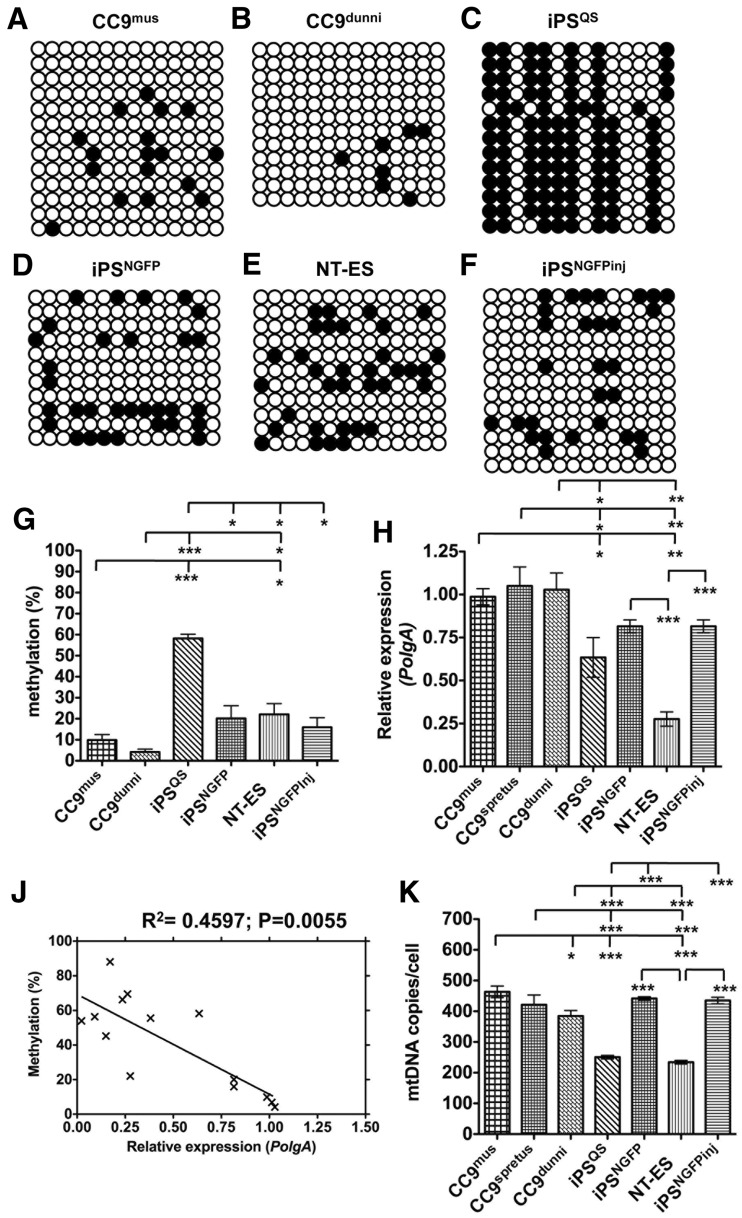
Since there are significant interactions between nuclear DNA and mtDNA (3,11), we tested whether a more divergent population of mtDNA would affect the levels of DNA methylation to PolgA. We first analysed the levels of DNA methylation in Mus musculus (CC9mus) ESCs (Figure 3A and G) and found these levels were comparable to ESD3 cells (Figure 3G cf Figure 1L). However, when this nuclear genotype harboured the more divergent Mus terricolor (Dunni) mtDNA (CC9dunni) (33), which represents a divergence of 4.0 million years before present and a 159 amino acid difference (42), it displayed slightly lower levels of DNA methylation (4.16% ± 1.38; Figure 3B and G) compared with CC9mus ESCs (9.89 ± 2.62; Figure 3A and G).
Figure 3.
Intragenic methylation of PolgA in reprogrammed and mtDNA divergent ESCs. Bisulphite sequencing analysis of CpG methylation in (A) CC9mus, (B) CC9dunni, (C) iPSQS, (D) iPSNGFP2 (E) NT-ES, (F) iPSNFGPinj pluripotent stem cells. (G) The percentage CpG methylation per sequence determined by bisulphite sequencing (percentage mean ± SEM). (H) Real time PCR quantification of PolgA expression in cultured CC9mus, CC9spretus, CC9dunni, iPSQS/R26 (iPSQS), iPSNGFP, NT-ES, iPSNGFPinj pluripotent stem cells expressed relative to ESD3 cells. (J) Cumulative analysis of the relationship between DNA methylation levels (Figures 1L and 3G) and the expression of PolgA (Figures 2A and 3H) performed using Pearson correlation coefficient (R2). (K) MtDNA copies/cell in cultured CC9mus, CC9spretus, CC9dunni, iPSQS/R26, iPSNGFP, NT-ES, iPSNGFPinj pluripotent stem cells. Values represent mean ± SEM and significant differences between cell types are: *P < 0.05; **P < 0.01 and ***P < 0.001.
The influence of nuclear reprogramming on intragenic DNA methylation
Somatic cells can be reprogrammed to pluripotency by somatic cell nuclear transfer (32) and transcription-factor transduction, otherwise known as induced pluripotency (43). However, these approaches do not completely recapitulate the epigenetic and transcriptional signatures associated with ESCs (44) and, consequently, fail to correctly regulate mtDNA copy number during differentiation (16,31). Therefore, we determined how somatic cell reprogramming affects DNA methylation of PolgA in exon 2. Analysis of two independent induced pluripotent stem (iPS) cells lines, iPSQS (34) and iPSNGFP (35) demonstrated statistically elevated levels of DNA methylation for iPSQS cells (58.14 ± 1.95; Figure 3C and G) compared with pluripotent ESD3 and CC9mus cells (P < 0.001). However, DNA methylation in iPSNGFP cells (20.13 ± 6.07; Figure 3D and G) was only elevated compared with ESD3 cells (P < 0.05). Somatic cell nuclear transfer derived ESCs (NT-ES; 22.07 ± 5.14; Figure 3E and G) (32) were dissimilar to both ESD3 and CC9mus cells (P < 0.05; Figure 3G cf Figure 1L) suggesting that the exon 2 locus of PolgA is abnormally regulated in reprogrammed cells.
We then asked whether the introduction of reprogrammed cells into the pluripotent environment of the inner cell mass of a blastocyst-stage embryo followed by their derivation into ESCs would modulate the DNA methylation signatures in exon 2 of PolgA. We microinjected approximately 5 to 10 iPSNGFP cells into day 3.5 blastocysts and the resultant Nanog-GFP+ colonies were cultured for several passages. These iPSNGFP-Inj pluripotent stem cells displayed similar DNA methylation profiles (15.93 ± 4.51; Figure 3F) to non-injected cells, thus further demonstrating incomplete reprogramming (Figure 3G). Despite these differences, there were no CpG site-specific DNA methylation patterns identifiable between any of the divergent and reprogrammed cell types (Supplementary Figure S2A).
The effects of DNA methylation on PolgA expression in divergent and reprogrammed cells
CC9mus, CC9dunni and ESD3 cells (Figure 3H cf Figure 2A) all contained similar levels of PolgA mRNA levels while iPSQS (P < 0.05) and NT-ES cells (P < 0.01) had reduced levels. iPSNGFP cells also displayed lower levels of PolgA when compared with ESD3 cells (P < 0.05; Figure 3H). We then collectively analysed the data presented in Figures 1L, 2A, 3G and 3H and confirmed the negative correlation (R2 = 0.4597; P = 0.0055) between relative PolgA expression and DNA methylation (Figure 3J). Taken together, DNA hypomethylation of the PolgA exon 2 loci is a common feature of pluripotent stem cells. However, the introduction of divergent Mus terricolor mtDNA into ESCs had no effect on the intragenic DNA methylation of PolgA and therefore PolgA expression while reprogrammed somatic cells displayed altered DNA methylation and PolgA expression compared with ESCs.
The effects of DNA methylation on mtDNA copy number in divergent and reprogrammed cells
CC9dunni cells contained fewer copies of mtDNA per cell (384.43 ± 18.39) than the parental CC9mus cells (463.49 ± 18.92; P < 0.05; Figure 3K). Furthermore, mtDNA copy numbers for iPSNGFP (441.61 ± 5.69; Figure 3K) and iPSNGFP-Inj (435.07 ± 10.54; Figure 3K) cells were similar to ESD3 and CC9mus cells while iPSQS (250.66 ± 5.85; P < 0.001) and NT-ES (234.20 ± 5.73; P < 0.001) cells contained fewer copies than ESD3 and CC9mus cells (Figure 3K cf Figure 2C). Furthermore, using linear regression analysis, we observed a negative correlation between mtDNA copy number and PolgA expression (R2 = 0.377; P = 0.0195; Supplementary Figure S2B).
DNA demethylation within exon 2
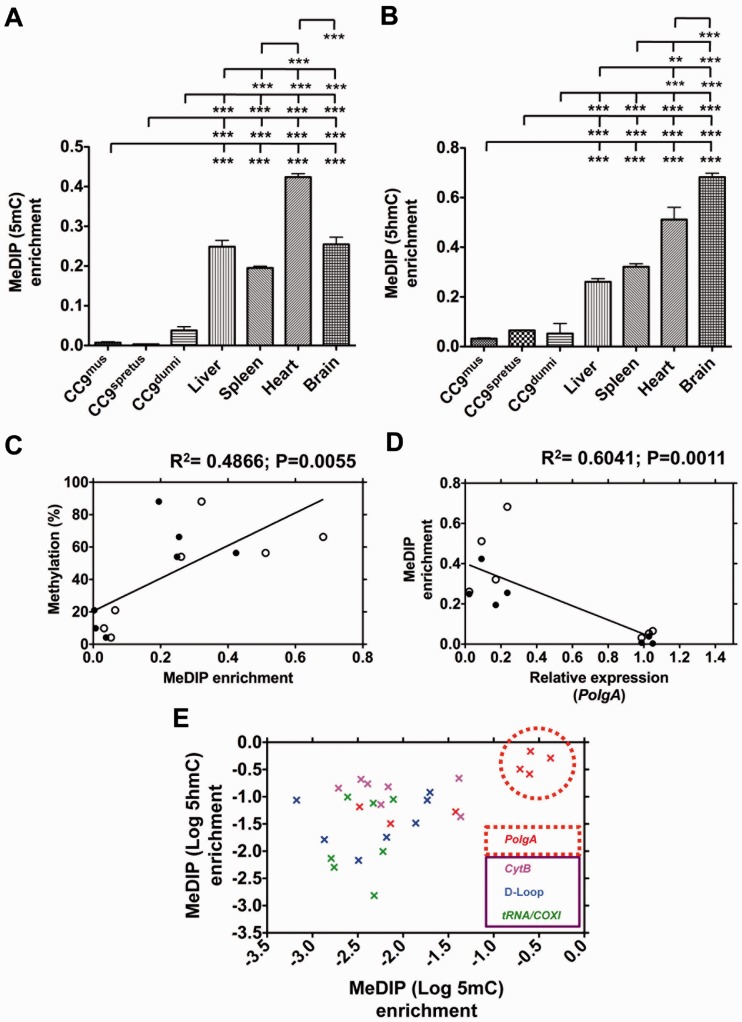
Recent studies have indicated that, during demethylation of DNA, 5mC is modified to 5hmC (45,46). Since bisulphite sequencing does not distinguish between 5mC and 5hmC, we quantified immunoprecipitated 5mC and 5hmC MeDIP using real time PCR. MeDIP analysis of divergent ESCs, CC9spretus (2.0 million years before present and 38 amino acid differences to the parental cells) and CC9dunni cells, demonstrated enrichment levels of 5mC (Figure 4A) and 5hmC (Figure 4B) within exon 2 of PolgA, similar to CC9mus cells (Figure 4A and B). Liver, spleen and brain were all equally enriched for 5mC, while heart was further enriched (P < 0.001; Figure 4A). Enrichment for 5hmC was higher in heart and brain compared with liver and spleen (P < 0.001; Figure 4B). The enrichment levels of 5mC and 5hmC in tissues samples were elevated compared with ESCs (P < 0.001; Figure 4A and B). These enrichment levels are comparable to the DNA methylation levels identified by bisulphite sequencing, (cf Figure 1L; Figure 3G; R2 = 0.4866; P = 0.0055; Figure 4C). We further confirmed the negative relationship between DNA methylation and PolgA expression (R2 = 0.6041; P = 0.0011; Figure 4D).
Figure 4.
Analysis of PolgA and mtDNA enrichment in 5mC and 5hmC MeDIP of ESCs and somatic tissues. DNA samples from cultured CC9mus, CC9spretus and CC9dunni cells; liver, spleen, heart and brain samples were immunoprecipitated using antibodies against (A) 5mC and (B) 5hmC, and analysed using real time PCR for PolgA (exon 2) enrichment. Bars represent means ± SEM. Significant differences between cell types are indicated (**P < 0.01; ***P < 0.001). (C) MeDIP (5mC and 5hmC) enrichment (Figure 4A and B) was validated against the corresponding means for bisulphite sequencing methylation levels (Figures 1L and 3G) using Pearson correlation coefficient (R2). Closed circles represent 5mC enrichment and open circles represent 5hmC enrichment. The P-value signifies the significance of the line from zero. (D) The relationship between PolgA expression and the corresponding MeDIP (5mC and 5hmC) enrichment levels using a Pearson correlation coefficient (R2). Closed circles represent 5mC enrichment and open circles represent 5hmC enrichment. The P-value signifies the significance of the line from zero. (E) Real time PCR for the enrichment of PolgA (exon 2) and mtDNA (CytB, tRNA/COXI and D-loop regions) in 5mC and 5hmC immunoprecipitated DNA. Enrichment values for CC9mus, CC9spretus and CC9dunni cells; and liver, spleen, heart and brain samples were normalized against β-actin (unmethylated control). Data shown is log10 5mC against log10 5hmC. Data points are colour-coded to represent DNA sequences analysed (PolgA exon 2 and mtDNA: CytB, tRNA/COXi and D-loop regions). Data points highlighted in the red circle represent the high levels of enrichment observed for liver, spleen, heart and brain samples (cf Figure 4A and B).
5mC and 5hmC are present at very low levels in mtDNA
As it has been suggested that mtDNA can undergo DNA methylation (21), we asked whether DNA methylation of the mitochondrial genome was directly linked to DNA methylation of PolgA in exon 2. Analysis of the levels of enrichment for the D-loop, CytB and tRNA/COXI regions of mtDNA from MeDIP samples revealed low levels of 5mC and 5hmC, similar to those identified for PolgA in ESCs and significantly lower than those observed in somatic tissues (P < 0.001; Figure 4E).
Intragenic PolgA methylation and demethylation during spontaneous ESC differentiation
During differentiation, mtDNA copy number is strictly regulated so that cells undergoing differentiation can increase their mtDNA copy number to meet their specific requirements for the generation of ATP through OXPHOS (15,16,31). We, therefore, determined whether intragenic DNA methylation of PolgA varied during differentiation. ESD3 cells were differentiated as embryoid bodies (EB) for 7 days and the DNA methylation status of PolgA was determined using methylation-sensitive (HpaII and NotI) and methylation-dependent (McrBC) restriction enzymes followed by qPCR (20). We confirmed that this quantitative method for DNA methylation correlated with methylation profiles determined by bisulphite sequencing (Supplementary Figure S3A and B), and also validated the relationship between PolgA expression and DNA methylation (Supplementary Figure S3C). Within 24 h of ESD3 differentiation, DNA methylation at HpaII and McrBC sites increased to 100 and ∼70%, respectively, with HpaII methylation decreasing to basal levels on Day 2 (Supplementary Figure S3D). During differentiation, up to Day 7, methylation at NotI sites increased to levels <50%, while McrBC methylation decreased (Supplementary Figure S3D). Despite these changes in DNA methylation at the HpaII, NotI and McrBC sites, the overall mean methylation status was similar during early differentiation (Supplementary Figure S3E). These observations demonstrate that the exon 2 locus of PolgA undergoes de novo methylation and demethylation during early ESC differentiation.
mtDNA copy number and PolgA expression during early neural differentiation
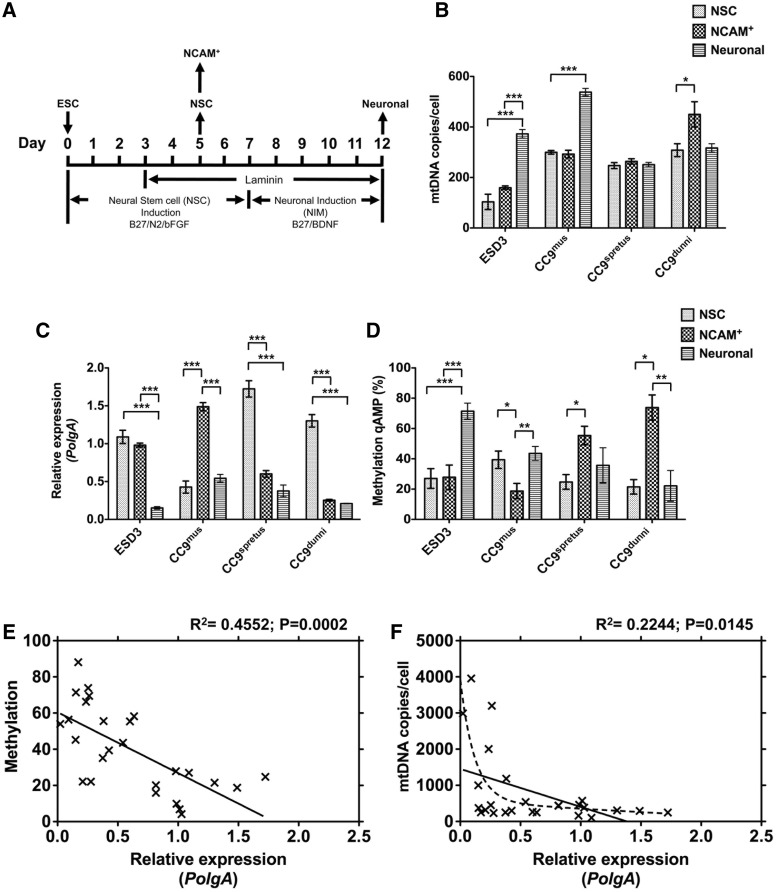
Since spontaneous differentiation of ESCs generates numerous cell types, we investigated whether the regulation of mtDNA copy number is developmentally regulated during neural and neuronal differentiation. In the first instance, ESCs were differentiated as a monolayer culture under defined culture conditions (37,38) for 5 days to induce neural determination (Figure 5A). At this time point, we also isolated NCAM+ cells in order to enrich for neural precursor cells, or allowed the cells to continue to differentiate for a further 7 days, under distinct culture conditions (37,38) (Figure 5A). In differentiating ESD3 cells, the mean mtDNA copies per cell was reduced after 5 days of neural-induced differentiation (both NSC and NCAM+ cells) when compared with their undifferentiated counterparts (Figure 5B cf Figure 2C; P < 0.001). However, as cells differentiated into neuronal cell types, mtDNA copy number increased to comparable levels detected in undifferentiated ESD3 cells (Figure 5B cf Figure 2C). A similar pattern was also observed for the differentiation of CC9mus cells, although only ∼35% reduction in mtDNA copies was observed in both NSC and NCAM+ cells (Figure 5B cf Figure 3K; P < 0.01). In neural differentiation of ESCs harbouring Mus spretus mtDNA (CC9spretus), mtDNA copy number also decreased compared with their undifferentiated counterparts and remained at lower levels at all time points of differentiation analysed (Figure 5B cf Figure 3K; P < 0.01). Moreover, the above decrease in mtDNA copy number associated with early neural differentiation was not observed in mtDNA divergent NSC-CC9dunni cells (Figure 5B cf Figure 3K), while NCAM+-CC9dunni cells isolated at the same time point contained significantly more mtDNA copies per cell (Figure 5B; P < 0.05). These results indicate that the reduction of mtDNA copy number during early neural differentiation is developmentally determined and increasing the genetic divergence between nuclear DNA and mtDNA alters this process.
Figure 5.
Intragenic PolgA methylation during neural differentiation. (A) Time chart representing the induction of neural differentiation in ESD3, CC9mus, CC9spretus and CC9dunni ESCs. The abbreviations beneath the timeline show the components of media and culture conditions that were used to induce neural (NSC) and neuronal differentiation. ESCs were differentiated as a monolayer culture for 5 days in neural stem cell media containing the supplements B27 and N2 for neural induction, and further supplemented with basic fibroblast growth factor (bFGF). At this time point, we also isolated NCAM+ cells, in order to enrich for neural precursor cells, or allowed the cells to continue to differentiate for a further 7 days, in neuronal induction media containing B27 supplement and brain-derived neurotrophic factor (BDNF). MtDNA copy number (B), PolgA expression (C) and DNA methylation (D) were assessed: on Day 5 of differentiation (NSC); in NCAM+ cells on Day 5 of differentiation and; on Day 12 of neuronal differentiation. (E) Cumulative analysis of the relationship between DNA methylation (cf Figures 1L, 3G, 5D) and the expression of PolgA (cf Figures 2A, 3H, 5C) was determined using Pearson correlation coefficient (R2). (F) Cumulative analysis of the relationship between mtDNA copy number (cf Figures 2C, 3K, 5B) and the expression of PolgA (cf Figures 2A, 3H, 5C) was determined using Pearson correlation coefficient (R2: straight line = linear regression; dashed line = non-linear regression analysis).
Similar levels of PolgA expression were detected in ESD3 and CC9dunni cells after 5 days of neural differentiation (NSC; Figure 5C). In NSC-CC9mus cells, the expression levels of PolgA were significantly decreased compared with undifferentiated CC9mus ESCs (P < 0.001), while NSC-CC9spretus cells expressed significantly increased levels compared with their undifferentiated counterparts (P < 0.01; Figure 5C cf Figure 3H). Nevertheless, the expression of PolgA was increased in NCAM+-CC9mus cells (P < 0.001) but reduced in both NCAM+-CC9spretus (P < 0.001) and NCAM+-CC9dunni (P < 0.001) cells compared with their non-isolated (NSC) counterparts (Figure 5C).
Neuronal differentiation over 12 days significantly reduced the expression of PolgA in ESD3 (P < 0.001), CC9mus (P < 0.05), CC9spretus (P < 0.01) and CC9dunni (P < 0.05) cells when compared with their undifferentiated counterparts (Figure 5C cf Figure 2A and 3H). DNA methylation levels at the exon 2 locus varied over the course of neural differentiation for all cell lines (Figure 5D). Nevertheless, CC9spretus and CC9dunni cells displayed similar patterns of DNA methylation during neural differentiation while, for CC9mus, a different pattern was observed (Figure 5D). Since the transcript levels of PolgA are associated with the DNA methylation status of exon 2 of PolgA in pluripotent and somatic tissues (Figures 2B and 3J), we again tested this relationship during differentiation. Regression analysis confirmed that DNA methylation within exon 2 of PolgA correlates with PolgA expression (R2 = 0.4103; P = 0.0075; Supplementary Figure S4A; cf Figure 5C–D). Taken together, the changes in intragenic DNA methylation within PolgA during development provide further evidence of the epigenetic regulation of PolgA expression.
Intragenic DNA methylation of PolgA is correlated with expression levels in pluripotent, differentiating and somatic cells
In order to further validate the association between PolgA expression and DNA methylation status of exon 2, we tested this relationship for all cells and tissues previously analysed. Regression analysis confirmed that DNA methylation within exon 2 of PolgA correlates with transcript levels (R2 = 0.4552; P = 0.0002; Figure 5E). Furthermore, using linear regression analysis, we observed a negative correlation between mtDNA copy number and PolgA expression (R2 = 0.2244; P = 0.0145: Figure 5F; straight line). However, since the relationship between mtDNA copy number and PolgA expression also appeared exponential (Figure 5F; dashed line), we transformed the data and confirmed a significant relationship between mtDNA copy number and PolgA expression (R2 = 0.5411; P = 0.0093; Supplementary Figure S4B).
DNA methylation at the exon 2 loci is associated with reduced RNApII transcriptional elongation
Having demonstrated the association of DNA methylation with PolgA expression and mtDNA copy number, we further explored this relationship by examining transcription factor binding sites within exon 2 of PolgA. Using the JASPAR CORE transcription factor database (http://jaspar.genereg.net/cgi-bin/jaspar_db.pl) (47–50), we identified an evolutionary conserved putative CTCF binding site within this region (Supplementary Figure S5A). Since CTCF regulates transcription (41,51) and DNA methylation inhibits its binding (41), we determined whether CTCF binding regulates PolgA expression levels. Using the ChIP assay, we examined the enrichment of CTCF in CC9mus ESCs and MEFs cells at the exon 2 loci (Supplementary Figure S5B). Both cell types showed similar levels of CTCF enrichment within exon 2 and at sites upstream and downstream (Supplementary Figure S5C). Since CC9mus and MEFs cells display different DNA methylation profiles and PolgA expression signatures, these observations suggest that CTCF is not involved in regulating PolgA expression.
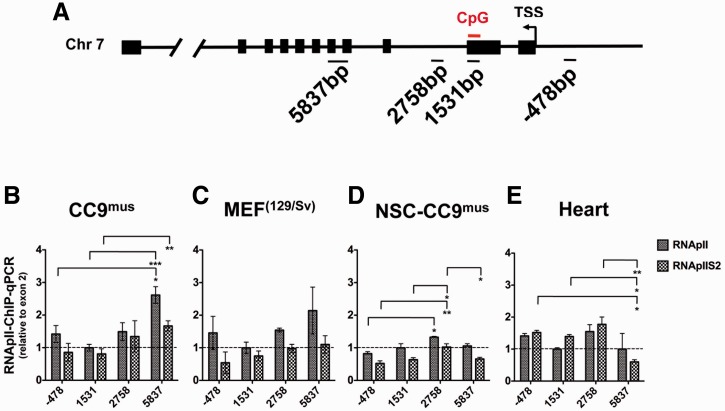
DNA methylation within coding sequences has been shown to alter transcript levels by influencing the recruitment or the transcriptional activation of RNApII (25,26,41). In order to determine how DNA methylation within exon 2 of PolgA modulates RNApII function, we quantified the enrichment of RNApII and the elongating RNApII phosphorylated at serine 2 of the carboxy-terminal domain (RNApIIS2). RNApII occupancy in CC9mus, MEF, NSC-CC9mus and heart samples was analysed upstream of the transcription start site within exon 2, and two sites downstream (Figure 6A). In CC9mus cells, RNApII (P < 0.001) and RNApIIS2 (P < 0.01) enrichment was significantly increased downstream of exon 2 (Figure 6B), while in MEF cells no differences were observed upstream or downstream of exon 2 (Figure 6C). Furthermore, in NSC-differentiated CC9mus cells, there was an initial increase in RNApIIS2 enrichment at a proximal site downstream of exon 2 (P < 0.05), although this returned to similar levels observed at exon 2 further downstream (Figure 6D). In heart tissue, there was no difference in enrichment for RNApII between exon 2 and downstream sites, whilst RNApIIS2 enrichment was decreased downstream of exon 2 (Figure 6E; P < 0.05). These results indicate that DNA methylation downstream of the PolgA transcription start site reduces transcription by RNApII at the elongation stage.
Figure 6.
DNA methylation at the Exon 2 loci is associated with reduced RNApII transcriptional elongation. (A) Diagrammatic representation of the PolgA gene and primers sites used for ChIP. Numbers correspond to the centre nucleotide of each primer amplicon, relative to the transcription start site (TSS). Enrichment for (RNApII) and RNApII phosphorylated on serine 2 of the carboxy-terminal domain (RNApIIS2) was analysed by real time PCR at the exon 2 methylation site of PolgA and at downstream and upstream regions in: (B) CC9mus; (C) MEF; (D) NSC-CC9mus and (E) heart samples. Values represent mean ± SEM. Significant differences between cell types are: *P < 0.05; **P < 0.01 and ***P < 0.001.
DISCUSSION
The data presented here demonstrate a negative correlation between intragenic DNA methylation and the levels of PolgA expression, which provided an epigenetic mechanism for regulating mtDNA replication. Through sequencing of bisulphite converted DNA, qAMP and MeDIP analyses, we have identified tissue-specific DNA methylation patterns within the gene body of PolgA, which increase as cells differentiate into specialized cell types. Changes in site-specific methylation levels were observed at this locus during early spontaneous differentiation of ESCs and 5hmC was enriched in post-mitotic tissues, which further suggest that changes in DNA methylation levels at the exon 2 loci in differentiated cell types are a result of active DNA methylation and demethylation.
Experimental manipulation of peroxisome proliferator-activated receptor gamma coactivator (PGC) 1-α and -β has shown their levels of expression to be linked to mtDNA copy number [reviewed in (52)]. In this instance, hypermethylation of the PGC-1α promoter correlated with reduced PGC-1α mRNA and mitochondrial density in type 2 diabetic patients (53). Despite these observations, PGC-1α is not a critical factor in mtDNA replication since knockout mouse embryos develop to term and display only reduced mitochondrial biogenesis (54). The expression of the mtDNA replication factors Tfam (55) and PolgA (11) are essential for embryogenesis and regulating mtDNA copy number. Analysis of the Tfam promoter has suggested that DNA methylation within a nuclear respiratory factor-1 (NRF-1) binding site may silence Tfam expression and reduce mtDNA biogenesis (56). Others have suggested DNA methylation regulates PolgA expression during mouse postnatal germ cell development (20). Although, these studies suggest mtDNA replication may be epigenetically regulated, no association between DNA methylation and the expression of these replication factors or mtDNA copy number has been previously described.
DNA methylation of intragenic regions (within gene bodies) has been associated with negative (25,26,28) and positive gene regulation (29,30). The inverse relationship between PolgA exon 2 methylation and PolgA mRNA levels suggests intragenic methylation at this site suppresses mRNA expression levels rather than completely repressing transcription. DNA methylation within gene bodies has been associated with alternative promoters and splicing of pre-mRNAs (26,41,57). The highly conserved transcription factor, CTCF, has a variety of roles in gene regulation (51) including mRNA splicing (41) and regulating transcription (51). Despite identifying an evolutionary conserved CTCF binding site within exon 2, ChIP analysis suggests PolgA regulation is independent of CTCF binding. Therefore, we further investigated the effects of DNA methylation within exon 2 of PolgA by analysing the enrichment of serine 2 phosphorylated RNApII, which is a marker of transcriptional elongation. Analysis following ChIP demonstrated that methylation within exon 2 is associated with lower levels of RNApII elongation downstream of this CpG island. Although the universal epigenetic function of intragenic methylation remains unclear and has been thought to differ between genes and genetic loci (22,58), our data demonstrate reduced transcriptional elongation regulates PolgA expression levels.
The epigenetic regulation of chromatin structure and gene expression is essential for mammalian development and differentiation (59,60). Global DNA methylation analysis shows mouse ESCs are ∼60% methylated (61) though stem cell genes, such as Oct4 (62), remain unmethylated in their promoters until differentiation induces gene repression through hypermethylation. Similarly, we observed low levels of DNA methylation within exon 2 of PolgA in oocytes, blastocysts and ESCs. These observations in gametes and pluripotent cells support a developmental relationship between cellular plasticity and mtDNA regulation. Indeed, siRNA knockdown of PolgA in mouse ESCs promotes differentiation and the loss of pluripotency (15).
We have demonstrated that decreased PolgA expression during the early stages of spontaneous and neural-directed differentiation corresponded to increased levels of DNA methylation. During neural differentiation of mouse (15) and human (63) ESCs, mtDNA copy number reduces, which allows progenitor cells to establish the ‘mtDNA set point’, from which the mitochondrial complement may be expanded in a cell specific manner (64). The ‘mtDNA set point’ was established in divergent ESCs although, during downstream differentiation, there were cell line specific differences in mtDNA copy number, PolgA expression and DNA methylation profiles for PolgA when compared with non-divergent controls. Thus, altering the mtDNA content of ESCs impacts on the expression of factors involved in mtDNA replication and maintenance during differentiation, and highlights a pathway, whereby changes in mtDNA content are communicated to nuclear DNA.
By analysing mtDNA divergent ESCs and reprogrammed somatic cells, we have further demonstrated the importance of the strict epigenetic regulation of PolgA in pluripotent stem cells and during differentiation. Although pluripotent mtDNA divergent ESCs displayed similar DNA methylation and PolgA expression levels to control ESCs, reprogrammed somatic cells had elevated DNA methylation profiles and variable PolgA expression. This suggests that incomplete reprogramming failed to reinstate the appropriate levels of DNA methylation to exon 2 of PolgA. We have previously demonstrated that reprogrammed somatic cells, including iPS and NT-ES cells, fail to regulate the levels of mtDNA copy number characteristic of early pluripotent stem cell undergoing differentiation due to incomplete reprogramming (16). Indeed, abnormal epigenetic modification of chromatin structure is characteristic of somatic cell reprogramming (44,65) and is especially prevalent in NT embryos. This results in their failure to repress the expression of PolgA and they thus retain the ability to drive mtDNA replication during preimplantation development (39).
As cell differentiation progresses, mtDNA copy number is regulated in a coordinated manner resulting in a terminally differentiated cell possessing the required number of mtDNA copies that reflects its cellular function (66). Through cumulative analysis of mtDNA copy number and PolgA expression levels, we ascertained a negative correlation linking the epigenetic regulation of PolgA to mtDNA copy number. MtDNA copy number was higher in somatic tissues than in ESCs but the expression of PolgA was higher in ESCs. The high DNA methylation and the enrichment of 5hmC in somatic tissues correlated with the reduced expression of PolgA indicating that demethylation may take place so that post-mitotic cells retain sufficient capacity to replicate mtDNA, as and when necessary. The higher levels observed in ESCs reflect their fast population doublings where dilution of mtDNA resulting from cytokinesis requires continual replacement.
Recent data suggest that the mitochondrial genome is methylated (21) at levels higher than previously observed (67). DNA methylation is prevalent in prokaryotes (68) and eukaryotes (69) and is observed in chloroplast DNA (70). We observe very low levels of 5mC and 5hmC enrichment within mtDNA compared with the levels observed for PolgA in somatic tissues. DNA methylation of chloroplast DNA is not inhibitory to gene expression (70) while, in prokaryotes, DNA methylation is generally thought to limit the integration of foreign DNA (71), though, in some instances, it is associated with DNA mismatch repair, DNA replication and gene expression (72,73). It has been suggested that 5mC and 5hmC enrichment within mtDNA may influence mitochondrial transcription, although this association requires substantiation. Nevertheless, our data suggest that the epigenetic control of mtDNA replication during neural differentiation is coordinated by DNA methylation to exon 2 of the nuclear-encoded PolgA rather than by direct methylation of mtDNA.
In conclusion, we show that PolgA expression is negatively correlated with intragenic DNA methylation and reduced transcriptional elongation. Furthermore, we show that mtDNA copy number is regulated by PolgA expression in a cell-specific manner. This demonstrates that, in mammalian cells, the replication of the bacterial-originating mitochondrial genome is regulated by DNA methylation of a separate mammalian nuclear genome. These finding are especially significant in the context of nuclear-cytoplasmic coordination, whereby somatic reprogramming does not recapitulate ESC-like characteristics and divergent mtDNA can influence nuclear gene function during differentiation.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Table 1 and Supplementary Figures 1–5.
FUNDING
Monash Institute of Medical Research (to J.C.S.J.); Victorian Government's Operational Infrastructure Support Program; NHMRC CDA Fellowship and the James and Vera Lawson Trust (to M.McK.). Funding for open access charge: Start up funds, Monash Institute of Medical Research (to J.C.S.J.).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to Dr Megan Munsie, Australian Stem Cell Centre, for the somatic cell nuclear transfer embryonic stem cell line (NT-ES); Dr Huseyin Sumer and Professor Paul Verma for the iPSQS/R26 line and Ms Jacqui Johnson for expertise in stem cell culture (both Centre for Reproduction and Development, Monash Institute of Medical Research). We thank Dr Stefan White for critically reading this manuscript.
REFERENCES
- 1.Anderson S, Bankier AT, Barrell BG, de Bruijn MH, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Roe BA, Sanger F, et al. Sequence and organization of the human mitochondrial genome. Nature. 1981;290:457–465. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- 2.Clayton DA. Replication of animal mitochondrial DNA. Cell. 1982;28:693–705. doi: 10.1016/0092-8674(82)90049-6. [DOI] [PubMed] [Google Scholar]
- 3.Tuppen HA, Blakely EL, Turnbull DM, Taylor RW. Mitochondrial DNA mutations and human disease. Biochim. Biophys. Acta. 2010;1797:113–128. doi: 10.1016/j.bbabio.2009.09.005. [DOI] [PubMed] [Google Scholar]
- 4.Ropp PA, Copeland WC. Cloning and characterization of the human mitochondrial DNA polymerase, DNA polymerase gamma. Genomics. 1996;36:449–458. doi: 10.1006/geno.1996.0490. [DOI] [PubMed] [Google Scholar]
- 5.Sweasy JB, Lauper JM, Eckert KA. DNA polymerases and human diseases. Radiat. Res. 2006;166:693–714. doi: 10.1667/RR0706.1. [DOI] [PubMed] [Google Scholar]
- 6.Graziewicz MA, Longley MJ, Copeland WC. DNA polymerase gamma in mitochondrial DNA replication and repair. Chem. Rev. 2006;106:383–405. doi: 10.1021/cr040463d. [DOI] [PubMed] [Google Scholar]
- 7.Spelbrink JN, Toivonen JM, Hakkaart GA, Kurkela JM, Cooper HM, Lehtinen SK, Lecrenier N, Back JW, Speijer D, Foury F, et al. In vivo functional analysis of the human mitochondrial DNA polymerase POLG expressed in cultured human cells. J. Biol. Chem. 2000;275:24818–24828. doi: 10.1074/jbc.M000559200. [DOI] [PubMed] [Google Scholar]
- 8.Yakubovskaya E, Chen Z, Carrodeguas JA, Kisker C, Bogenhagen DF. Functional human mitochondrial DNA polymerase gamma forms a heterotrimer. J. Biol. Chem. 2006;281:374–382. doi: 10.1074/jbc.M509730200. [DOI] [PubMed] [Google Scholar]
- 9.Larsson NG. Somatic mitochondrial DNA mutations in mammalian aging. Annu. Rev. Biochem. 2010;79:683–706. doi: 10.1146/annurev-biochem-060408-093701. [DOI] [PubMed] [Google Scholar]
- 10.Stumpf JD, Copeland WC. Mitochondrial DNA replication and disease: insights from DNA polymerase gamma mutations. Cell. Mol. Life Sci. 2011;68:219–233. doi: 10.1007/s00018-010-0530-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hance N, Ekstrand MI, Trifunovic A. Mitochondrial DNA polymerase gamma is essential for mammalian embryogenesis. Hum. Mol. Genet. 2005;14:1775–1783. doi: 10.1093/hmg/ddi184. [DOI] [PubMed] [Google Scholar]
- 12.Addo MG, Cossard R, Pichard D, Obiri-Danso K, Rotig A, Delahodde A. Caenorhabditis elegans, a pluricellular model organism to screen new genes involved in mitochondrial genome maintenance. Biochim. Biophys. Acta. 2010;1802:765–773. doi: 10.1016/j.bbadis.2010.05.007. [DOI] [PubMed] [Google Scholar]
- 13.Thundathil J, Filion F, Smith LC. Molecular control of mitochondrial function in preimplantation mouse embryos. Mol. Reprod. Dev. 2005;71:405–413. doi: 10.1002/mrd.20260. [DOI] [PubMed] [Google Scholar]
- 14.Spikings EC, Alderson J, St John JC. Regulated mitochondrial DNA replication during oocyte maturation is essential for successful porcine embryonic development. Biol. Reprod. 2007;76:327–335. doi: 10.1095/biolreprod.106.054536. [DOI] [PubMed] [Google Scholar]
- 15.Facucho-Oliveira JM, Alderson J, Spikings EC, Egginton S, St John JC. Mitochondrial DNA replication during differentiation of murine embryonic stem cells. J. Cell. Sci. 2007;120:4025–4034. doi: 10.1242/jcs.016972. [DOI] [PubMed] [Google Scholar]
- 16.Kelly RD, Sumer H, McKenzie M, Facucho-Oliveira J, Trounce IA, Verma PJ, St John JC. The effects of nuclear reprogramming on mitochondrial DNA replication. Stem Cell Rev. 2011 doi: 10.1007/s12015-011-9318-7. October 13 (doi:10.1007/s12015-011-9318-7; epub ahead of print) [DOI] [PubMed] [Google Scholar]
- 17.Armstrong L, Tilgner K, Saretzki G, Atkinson SP, Stojkovic M, Moreno R, Przyborski S, Lako M. Human induced pluripotent stem cell lines show stress defense mechanisms and mitochondrial regulation similar to those of human embryonic stem cells. Stem Cells. 2010;28:661–673. doi: 10.1002/stem.307. [DOI] [PubMed] [Google Scholar]
- 18.Prigione A, Fauler B, Lurz R, Lehrach H, Adjaye J. The senescence-related mitochondrial/oxidative stress pathway is repressed in human induced pluripotent stem cells. Stem Cells. 2010;28:721–733. doi: 10.1002/stem.404. [DOI] [PubMed] [Google Scholar]
- 19.Cao L, Shitara H, Horii T, Nagao Y, Imai H, Abe K, Hara T, Hayashi J, Yonekawa H. The mitochondrial bottleneck occurs without reduction of mtDNA content in female mouse germ cells. Nat. Genet. 2007;39:386–390. doi: 10.1038/ng1970. [DOI] [PubMed] [Google Scholar]
- 20.Oakes CC, La Salle S, Smiraglia DJ, Robaire B, Trasler JM. Developmental acquisition of genome-wide DNA methylation occurs prior to meiosis in male germ cells. Dev. Biol. 2007;307:368–379. doi: 10.1016/j.ydbio.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 21.Shock LS, Thakkar PV, Peterson EJ, Moran RG, Taylor SM. DNA methyltransferase 1, cytosine methylation, and cytosine hydroxymethylation in mammalian mitochondria. Proc. Natl Acad. Sci. USA. 2011;108:3630–3635. doi: 10.1073/pnas.1012311108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Deaton AM, Bird A. CpG islands and the regulation of transcription. Genes Dev. 2011;25:1010–1022. doi: 10.1101/gad.2037511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Davey C, Pennings S, Allan J. CpG methylation remodels chromatin structure in vitro. J. Mol. Biol. 1997;267:276–288. doi: 10.1006/jmbi.1997.0899. [DOI] [PubMed] [Google Scholar]
- 24.Illingworth RS, Gruenewald-Schneider U, Webb S, Kerr AR, James KD, Turner DJ, Smith C, Harrison DJ, Andrews R, Bird AP. Orphan CpG islands identify numerous conserved promoters in the mammalian genome. PLoS Genet. 2010;6:e1001134. doi: 10.1371/journal.pgen.1001134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lorincz MC, Dickerson DR, Schmitt M, Groudine M. Intragenic DNA methylation alters chromatin structure and elongation efficiency in mammalian cells. Nat. Struct. Mol. Biol. 2004;11:1068–1075. doi: 10.1038/nsmb840. [DOI] [PubMed] [Google Scholar]
- 26.Maunakea AK, Nagarajan RP, Bilenky M, Ballinger TJ, D'Souza C, Fouse SD, Johnson BE, Hong C, Nielsen C, Zhao Y, et al. Conserved role of intragenic DNA methylation in regulating alternative promoters. Nature. 2010;466:253–257. doi: 10.1038/nature09165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koerner MV, Pauler FM, Hudson QJ, Santoro F, Sawicka A, Guenzl PM, Stricker SH, Schichl YM, Latos PA, Klement RM, et al. A downstream CpG island controls transcript initiation and elongation and the methylation state of the imprinted Airn macro ncRNA promoter. PLoS Genet. 2012;8:e1002540. doi: 10.1371/journal.pgen.1002540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brenet F, Moh M, Funk P, Feierstein E, Viale AJ, Socci ND, Scandura JM. DNA methylation of the first exon is tightly linked to transcriptional silencing. PLoS One. 2011;6:e14524. doi: 10.1371/journal.pone.0014524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ball MP, Li JB, Gao Y, Lee JH, LeProust EM, Park IH, Xie B, Daley GQ, Church GM. Targeted and genome-scale strategies reveal gene-body methylation signatures in human cells. Nat. Biotechnol. 2009;27:361–368. doi: 10.1038/nbt.1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Flanagan JM, Wild L. An epigenetic role for noncoding RNAs and intragenic DNA methylation. Genome Biol. 2007;8:307. doi: 10.1186/gb-2007-8-6-307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.St John JC, Facucho-Oliveira J, Jiang Y, Kelly R, Salah R. Mitochondrial DNA transmission, replication and inheritance: a journey from the gamete through the embryo and into offspring and embryonic stem cells. Hum. Reprod. Update. 2010;16:488–509. doi: 10.1093/humupd/dmq002. [DOI] [PubMed] [Google Scholar]
- 32.Munsie MJ, Michalska AE, O'Brien CM, Trounson AO, Pera MF, Mountford PS. Isolation of pluripotent embryonic stem cells from reprogrammed adult mouse somatic cell nuclei. Curr. Biol. 2000;10:989–992. doi: 10.1016/s0960-9822(00)00648-5. [DOI] [PubMed] [Google Scholar]
- 33.McKenzie M, Trounce IA, Cassar CA, Pinkert CA. Production of homoplasmic xenomitochondrial mice. Proc. Natl Acad. Sci. USA. 2004;101:1685–1690. doi: 10.1073/pnas.0303184101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sumer H, Jones KL, Liu J, Heffernan C, Tat PA, Upton KR, Verma PJ. Reprogramming of somatic cells after fusion with induced pluripotent stem cells and nuclear transfer embryonic stem cells. Stem Cells Dev. 2010;19:239–246. doi: 10.1089/scd.2009.0142. [DOI] [PubMed] [Google Scholar]
- 35.Hanna J, Markoulaki S, Schorderet P, Carey BW, Beard C, Wernig M, Creyghton MP, Steine EJ, Cassady JP, Foreman R, et al. Direct reprogramming of terminally differentiated mature B lymphocytes to pluripotency. Cell. 2008;133:250–264. doi: 10.1016/j.cell.2008.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Summers MC, Bhatnagar PR, Lawitts JA, Biggers JD. Fertilization in vitro of mouse ova from inbred and outbred strains: complete preimplantation embryo development in glucose-supplemented KSOM. Biol. Reprod. 1995;53:431–437. doi: 10.1095/biolreprod53.2.431. [DOI] [PubMed] [Google Scholar]
- 37.Onorati M, Camnasio S, Binetti M, Jung CB, Moretti A, Cattaneo E. Neuropotent self-renewing neural stem (NS) cells derived from mouse induced pluripotent stem (iPS) cells. Mol. Cell. Neurosci. 2010;43:287–295. doi: 10.1016/j.mcn.2009.12.002. [DOI] [PubMed] [Google Scholar]
- 38.Ying QL, Stavridis M, Griffiths D, Li M, Smith A. Conversion of embryonic stem cells into neuroectodermal precursors in adherent monoculture. Nat. Biotechnol. 2003;21:183–186. doi: 10.1038/nbt780. [DOI] [PubMed] [Google Scholar]
- 39.Lloyd RE, Lee JH, Alberio R, Bowles EJ, Ramalho-Santos J, Campbell KH, St John JC. Aberrant nucleo-cytoplasmic cross-talk results in donor cell mtDNA persistence in cloned embryos. Genetics. 2006;172:2515–2527. doi: 10.1534/genetics.105.055145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weber M, Davies JJ, Wittig D, Oakeley EJ, Haase M, Lam WL, Schubeler D. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat. Genet. 2005;37:853–862. doi: 10.1038/ng1598. [DOI] [PubMed] [Google Scholar]
- 41.Shukla S, Kavak E, Gregory M, Imashimizu M, Shutinoski B, Kashlev M, Oberdoerffer P, Sandberg R, Oberdoerffer S. CTCF-promoted RNA polymerase II pausing links DNA methylation to splicing. Nature. 2011;479:74–79. doi: 10.1038/nature10442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pogozelski WK, Fletcher LD, Cassar CA, Dunn DA, Trounce IA, Pinkert CA. The mitochondrial genome sequence of Mus terricolor: comparison with Mus musculus domesticus and implications for xenomitochondrial mouse modeling. Gene. 2008;418:27–33. doi: 10.1016/j.gene.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 44.Kim K, Doi A, Wen B, Ng K, Zhao R, Cahan P, Kim J, Aryee MJ, Ji H, Ehrlich LI, et al. Epigenetic memory in induced pluripotent stem cells. Nature. 2010;467:285–290. doi: 10.1038/nature09342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.He YF, Li BZ, Li Z, Liu P, Wang Y, Tang Q, Ding J, Jia Y, Chen Z, Li L, et al. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science. 2011;333:1303–1307. doi: 10.1126/science.1210944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bhutani N, Burns DM, Blau HM. DNA demethylation dynamics. Cell. 2011;146:866–872. doi: 10.1016/j.cell.2011.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Portales-Casamar E, Thongjuea S, Kwon AT, Arenillas D, Zhao X, Valen E, Yusuf D, Lenhard B, Wasserman WW, Sandelin A. JASPAR 2010: the greatly expanded open-access database of transcription factor binding profiles. Nucleic Acids Res. 2010;38:D105–D110. doi: 10.1093/nar/gkp950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wasserman WW, Sandelin A. Applied bioinformatics for the identification of regulatory elements. Nat. Rev. Genet. 2004;5:276–287. doi: 10.1038/nrg1315. [DOI] [PubMed] [Google Scholar]
- 49.JASPAR. JASPAR CORE database. 2012. http://jaspar.genereg.net/cgi/jaspar_db.pl (19 May 2012, date last accessed)
- 50.Chen X, Xu H, Yuan P, Fang F, Huss M, Vega VB, Wong E, Orlov YL, Zhang W, Jiang J, et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133:1106–1117. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- 51.Phillips JE, Corces VG. CTCF: master weaver of the genome. Cell. 2009;137:1194–1211. doi: 10.1016/j.cell.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Scarpulla RC. Metabolic control of mitochondrial biogenesis through the PGC-1 family regulatory network. Biochim. Biophys. Acta. 2011;1813:1269–1278. doi: 10.1016/j.bbamcr.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Barres R, Osler ME, Yan J, Rune A, Fritz T, Caidahl K, Krook A, Zierath JR. Non-CpG methylation of the PGC-1alpha promoter through DNMT3B controls mitochondrial density. Cell Metab. 2009;10:189–198. doi: 10.1016/j.cmet.2009.07.011. [DOI] [PubMed] [Google Scholar]
- 54.Leone TC, Lehman JJ, Finck BN, Schaeffer PJ, Wende AR, Boudina S, Courtois M, Wozniak DF, Sambandam N, Bernal-Mizrachi C, et al. PGC-1alpha deficiency causes multi-system energy metabolic derangements: muscle dysfunction, abnormal weight control and hepatic steatosis. PLoS Biol. 2005;3:e101. doi: 10.1371/journal.pbio.0030101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Larsson NG, Wang J, Wilhelmsson H, Oldfors A, Rustin P, Lewandoski M, Barsh GS, Clayton DA. Mitochondrial transcription factor A is necessary for mtDNA maintenance and embryogenesis in mice. Nat. Genet. 1998;18:231–236. doi: 10.1038/ng0398-231. [DOI] [PubMed] [Google Scholar]
- 56.Choi YS, Kim S, Kyu Lee H, Lee KU, Pak YK. In vitro methylation of nuclear respiratory factor-1 binding site suppresses the promoter activity of mitochondrial transcription factor A. Biochem. Biophys. Res. Commun. 2004;314:118–122. doi: 10.1016/j.bbrc.2003.12.065. [DOI] [PubMed] [Google Scholar]
- 57.Illingworth R, Kerr A, Desousa D, Jorgensen H, Ellis P, Stalker J, Jackson D, Clee C, Plumb R, Rogers J, et al. A novel CpG island set identifies tissue-specific methylation at developmental gene loci. PLoS Biol. 2008;6:e22. doi: 10.1371/journal.pbio.0060022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Deaton AM, Webb S, Kerr AR, Illingworth RS, Guy J, Andrews R, Bird A. Cell type-specific DNA methylation at intragenic CpG islands in the immune system. Genome Res. 2011;21:1074–1086. doi: 10.1101/gr.118703.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- 60.Sakaue M, Ohta H, Kumaki Y, Oda M, Sakaide Y, Matsuoka C, Yamagiwa A, Niwa H, Wakayama T, Okano M. DNA methylation is dispensable for the growth and survival of the extraembryonic lineages. Curr. Biol. 2010;20:1452–1457. doi: 10.1016/j.cub.2010.06.050. [DOI] [PubMed] [Google Scholar]
- 61.Meissner A. Epigenetic modifications in pluripotent and differentiated cells. Nat. Biotechnol. 2010;28:1079–1088. doi: 10.1038/nbt.1684. [DOI] [PubMed] [Google Scholar]
- 62.Gidekel S, Bergman Y. A unique developmental pattern of Oct-3/4 DNA methylation is controlled by a cis-demodification element. J. Biol. Chem. 2002;277:34521–34530. doi: 10.1074/jbc.M203338200. [DOI] [PubMed] [Google Scholar]
- 63.Birket MJ, Orr AL, Gerencser AA, Madden DT, Vitelli C, Swistowski A, Brand MD, Zeng X. A reduction in ATP demand and mitochondrial activity with neural differentiation of human embryonic stem cells. J. Cell. Sci. 2011;124:348–358. doi: 10.1242/jcs.072272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kelly RD, St John JC. Role of mitochondrial DNA replication during differentiation of reprogrammed stem cells. Int. J. Dev. Biol. 2010;54:1659–1670. doi: 10.1387/ijdb.103202rk. [DOI] [PubMed] [Google Scholar]
- 65.Dean W, Santos F, Stojkovic M, Zakhartchenko V, Walter J, Wolf E, Reik W. Conservation of methylation reprogramming in mammalian development: aberrant reprogramming in cloned embryos. Proc. Natl Acad. Sci. USA. 2001;98:13734–13738. doi: 10.1073/pnas.241522698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Facucho-Oliveira JM, St John JC. The relationship between pluripotency and mitochondrial DNA proliferation during early embryo development and embryonic stem cell differentiation. Stem Cell Rev. 2009;5:140–158. doi: 10.1007/s12015-009-9058-0. [DOI] [PubMed] [Google Scholar]
- 67.Groot GS, Kroon AM. Mitochondrial DNA from various organisms does not contain internally methylated cytosine in. Biochim. Biophys. Acta. 1979;564:355–357. doi: 10.1016/0005-2787(79)90233-8. [DOI] [PubMed] [Google Scholar]
- 68.Gold M, Hurwitz J. The enzymatic methylation of ribonucleic acid and deoxyribonucleic acid. V. purification and properties of the deoxyribonucleic acid-methylating activity of Escherichia coli. J. Biol. Chem. 1964;239:3858–3865. [PubMed] [Google Scholar]
- 69.Richards EJ. Inherited epigenetic variation–revisiting soft inheritance. Nat. Rev. Genet. 2006;7:395–401. doi: 10.1038/nrg1834. [DOI] [PubMed] [Google Scholar]
- 70.Ahlert D, Stegemann S, Kahlau S, Ruf S, Bock R. Insensitivity of chloroplast gene expression to DNA methylation. Mol. Genet. Genomics. 2009;282:17–24. doi: 10.1007/s00438-009-0440-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Noyer-Weidner M, Trautner TA. Methylation of DNA in prokaryotes. EXS. 1993;64:39–108. doi: 10.1007/978-3-0348-9118-9_4. [DOI] [PubMed] [Google Scholar]
- 72.Heitman J. On the origins, structures and functions of restriction-modification enzymes. Genet. Eng. (NY) 1993;15:57–108. doi: 10.1007/978-1-4899-1666-2_4. [DOI] [PubMed] [Google Scholar]
- 73.Kessler C, Manta V. Specificity of restriction endonucleases and DNA modification methyltransferases a review (Edition 3) Gene. 1990;92:1–248. doi: 10.1016/0378-1119(90)90486-b. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.