Abstract
Escherichia coli topoisomerases I and III (Topo I and Topo III) relax negatively supercoiled DNA and also catenate/decatenate DNA molecules containing single-stranded DNA regions. Although these enzymes share the same mechanism of action and have similar structures, they participate in different cellular processes. In bulk experiments Topo I is more efficient at DNA relaxation, whereas Topo III is more efficient at catenation/decatenation, probably reflecting their differing cellular roles. To examine the differences in the mechanism of these two related type IA topoisomerases, single-molecule relaxation studies were conducted on several DNA substrates: negatively supercoiled DNA, positively supercoiled DNA with a mismatch and positively supercoiled DNA with a bulge. The experiments show differences in the way the two proteins work at the single-molecule level, while also recovering observations from the bulk experiments. Overall, Topo III relaxes DNA efficiently in fast processive runs, but with long pauses before relaxation runs, whereas Topo I relaxes DNA in slow processive runs but with short pauses before runs. The combination of these properties results in Topo I having an overall faster total relaxation rate, even though the relaxation rate during a run for Topo III is much faster.
INTRODUCTION
Topoisomerases are enzymes that alter DNA topology and are classified based on whether they form a transient single- (type I) or double- (type II) stranded break in the DNA phosphodiester backbone (1,2). Type I topoisomerases are further subdivided into three subclasses, IA, IB and IC (3), based on mechanistic, sequence and structural similarities. Type IA enzymes can relax negatively, but not positively, supercoiled DNA and in addition can catenate/decatenate and knot/unknot DNA rings provided they have a single-stranded region or nick (4). Type IA topoisomerases change DNA topology using an enzyme-bridged strand passage mechanism (3). In this mechanism, one DNA strand is transiently cleaved to allow passage of a second strand through the break before the cleaved strand is resealed. A strand breaking event leads to a single-strand passage event and hence the linking number (Lk) changes strictly in steps of +1 per relaxation event. Type IA enzymes require single-stranded DNA (ssDNA) regions for activity, which explains their preference for negatively supercoiled DNA (5) and is the basis for the sensing of the overall topological state of cellular DNA.
Structures of several type IA enzymes are known (6–8) and help explain many of the atomic details of their mechanism. Type IA enzymes are toroidal-shaped proteins with a common structural core consisting of four domains that form a positively charged cavity ∼30 Å in diameter which is capable of accommodating ssDNA or double-stranded DNA (dsDNA). The active site is found at the intersection of two domains, which can separate to form an opening that allows DNA to enter and exit from the central hole (9). An ssDNA-binding region found in the main body of the protein is responsible for recognition of the substrate and for guiding the DNA to the active site (10). Other structures of type IA enzymes help explain how conformational changes in the protein lead to changes in the topological state of DNA (7,11–14). The overall proposed mechanism of action for type IA enzymes has been confirmed by a variety of methods, including single-molecule studies (15,16).
Escherichia coli topoisomerases I and III (Topo I and III, respectively) are closely related type IA enzymes that are involved in different cellular processes (2). In vivo, a major function of Topo I, together with topoisomerase IV and gyrase, is to help maintain the appropriate topological state of DNA (17), whereas the main function of Topo III is resolving ssDNA recombination and replication intermediates (18–23). In vitro bulk studies have shown that Topo I is more efficient than Topo III at relaxing negative DNA supercoils (20), whereas Topo III is more efficient at decatenating ssDNA rings (23,24), consistent with their cellular roles. Structurally, the main difference between the two proteins is found at the C-terminus, where Topo I has a large zinc-ribbon domain (25) involved in DNA binding and essential for activity (26,27), whereas Topo III has a much smaller and dispensable C-terminal domain (28). Due to the differences in the C-terminal domains, Topo I homologs from different bacteria can protect large regions (30–50 bp) of DNA (5,29), whereas Topo III protects only ∼14 bp of ssDNA (30). In addition, Topo III has a loop, the decatenation loop, on the periphery of the central hole and extending away from the main body of the protein whose removal decreases decatenation activity markedly (8,31).
In order to understand the mechanistic difference between the two E. coli type IA topoisomerases, we analyzed DNA relaxation by the two enzymes at the single-molecule level. We were able to detect real-time relaxation on a single DNA molecule by E. coli Topo I and Topo III. The single-molecule experiments recapitulate the observations from bulk experiments and also provide new insights on the mechanism of action which are not apparent from bulk measurements. In particular, the single-molecule measurements show that both enzymes can relax DNA by removing a large number of supercoils without any observable pause between relaxation events, i.e. with high processivity. However, the relaxation velocity is faster for Topo III relaxation runs than for Topo I, opposite to what bulk experiments would suggest. Interestingly, the same measurements show that the time lag preceding DNA relaxation runs is shorter for Topo I than for Topo III. The combination of these two characteristics, relaxation velocity per run and time lag before a run, leads to an overall faster total relaxation rate for Topo I, in agreement with bulk experiments. These new observations help understand the differences in the overall mechanism of the two enzymes and suggest how changes in different kinetic characteristics help fine tune their DNA-processing activities.
MATERIALS AND METHODS
Enzymes and DNA substrates
pET vector-based clones containing either the E. coli topA gene or the topB gene were used to express E. coli Topo I and Topo III in BL21 cells. Purification of the proteins has been described elsewhere (10,32). Different linear dsDNA molecules were prepared for the single-molecule experiments, including DNA molecules unmodified, with 12- and 27-bp mismatches, and with 12- and 27-bp bulges. A mismatch refers to a region of non-complementary nucleotides, whereas a bulge refers to an unpaired string of nucleotides introduced in only one strand (Figure 1B). In all cases, 7.3-kb DNA (pMal-pIII) molecules with biotin (bio) and digoxigenin (dig) functionalized ends were used (see Supplementary Figure S1 and Supplementary Methods section).The sequence of the 27-bp bulge is identical to the one used in previous single-molecule experiments using E. coli or Thermotoga maritima Topo I (15,16).
Figure 1.
(A) Escherichia coli topoisomerase I relaxes negatively supercoiled DNA more efficiently than E. coli topoisomerase III in bulk experiments. The gel shows relaxation of negatively supercoiled DNA by either Topo I or Topo III. Identical mass amounts of protein and DNA were incubated for the same period of time and the resulting relaxed DNA products were analyzed in an ethidium bromide-stained agarose gel. The experiment shows that Topo I relaxes DNA more fully than comparable amounts of Topo III. The amount of protein used is shown at the top of the gel as well as the equivalent molarity for each lane. An amount of 200 ng (0.72 fmol) of DNA was used in each lane. R marks the position of relaxed DNA and SC the position of supercoiled DNA. (B) Substrates used for single-molecule experiments. Three different types of DNA molecules were employed for the single-molecule experiments: intact dsDNA (top), DNA with a single-stranded bubble or bulge (middle), and DNA with a mismatched region (bottom). The bulge and mismatch regions were used in experiments with positively supercoiled DNA as type IA topoisomerase require single-stranded regions for activity. The intact DNA molecules were used for experiments with negatively supercoiled DNA.
Relaxation assays
A 10 μl mixture containing 200 ng of pBR322 DNA, 50 mM Tris–HCl pH 8.0, 120 mM NaCl, 1 mM MgCl2 and 50–1000 ng of either Topo I or Topo III were incubated for 1 h at 37°C and the reaction stopped by heating at 70°C for 15 min. DNA was resolved on a 1% agarose gel and visualized by ethidium bromide staining (Figure 1A).
Magnetic tweezers single-molecule setup and data analysis
DNA molecules for single-molecule experiments were attached from the dig-labeled end to a glass slide functionalized with an anti-dig antibody while the other end was attached to a streptavidin-coated paramagnetic bead (Invitrogen) through the biotin-labeled end (33). 0.21 attomol of functionalized DNA in 2 μl of phosphate buffer saline (PBS) were incubated with 6 μl of 1-μm-diameter paramagnetic beads (3 ng/μl) in 0.4 mg/ml bovine serum albumin (BSA) for 12 min with gentle agitation at room temperature followed by 10-min incubation with the glass slide. The magnetic tweezers setup used has been described elsewhere (34). The stretching force applied to the DNA was controlled by altering the distance between the bead and the magnets. The supercoiling state of DNA, which is described by the linking number Lk, was controlled by rotation of the magnets. Force calibration was carried out using analysis of the bead fluctuations following the method of (34).
In order to ensure that only one intact DNA molecule was attached, the extension as a function of applied force was compared to the well-established elastic response of individual DNA molecules (35). Then, the bead was stretched and rotated and the length was plotted for a few forces in the 0.5–2 pN range. Intact DNA molecules produced a well-characterized shape in a rotation versus extension plot (35) (see Supplementary Figure S2 for representative data of this type), whereas nicked DNA molecules cannot be supercoiled and multiply attached beads produced a different shape curve. The calibration curve also serves to relate directly bead displacement to change in linking number. All single-molecule experiments were conducted at 37°C in a reaction mixture containing 2 nM of either E. coli Topo I or Topo III in 50 mM Tris–HCl pH 8.0, 120 mM NaCl, 1 mM MgCl2 and 200 µg/ml BSA.
Relaxation events were characterized by six parameters: (i) initial time lag, (ii) secondary time lag, (iii) number of supercoils relaxed per run, (iv) relaxation rate per run, (v) relaxation rate of the run including the time lag preceding this run and (vi) total relaxation rate. A relaxation run represents a series of relaxation events that are closely spaced in time and cannot be resolved into individual steps by the instrument, which acquires 45 bead position measurements per second. In some instances, a relaxation run may consist of a single relaxation step (ΔLk = 1). The initial time lag refers to the time between initially supercoiling the molecule and the first relaxation run detected. The secondary time lag refers to the time between two relaxation runs, discounting initial runs. The number of supercoils relaxed per run refers to the change in linking number per run. The relaxation rate per run refers to the number of supercoils relaxed divided by the time duration of a run. The relaxation run with time lag included refers to the number of supercoils relaxed in a run divided by the duration of the relaxation of a run plus the lag time preceding that run. Finally, the total relaxation rate refers to total number of turns relaxed divided by the time from the moment the molecule was supercoiled until the extension reached a value consistent with the initial extension or when the extension stopped increasing for a long period of time (∼30 min). Data analyses were performed with software written in Matlab (MATLAB 6.1, The MathWorks Inc., Natick, MA, USA, 2000) and with Origin Pro (OriginLab, Northampton, MA, USA). For more detailed information on data analyses, see the ‘Supplementary Data’ section.
RESULTS
Initially, DNA relaxation by both enzymes was studied using negatively supercoiled DNA, both in bulk and single-molecule experiments (Figures 1, 2A and D). The bulk experiments confirmed previous qualitative observations showing that Topo I relaxes DNA more efficiently than Topo III (20). In other words, a given amount of Topo I relaxes the same amount of DNA more completely than that amount of Topo III does (Figure 1A). For the single-molecule experiments, it was important to select the correct force regimes and substrates. At low force (<1 pN), plectonemic and denatured states of DNA coexist on negatively supercoiled DNA (35), which is the substrate needed for these studies. High forces on negatively supercoiled DNA lead to denatured states which do not result in length change (i.e. there is no displacement of the bead) and therefore cannot be used for the experiments. For this reason, all experiments with negatively supercoiled DNA were done at 0.7 pN force, where the two states coexist (35), although the data are noisy due to thermal motion-driven movements of the bead.
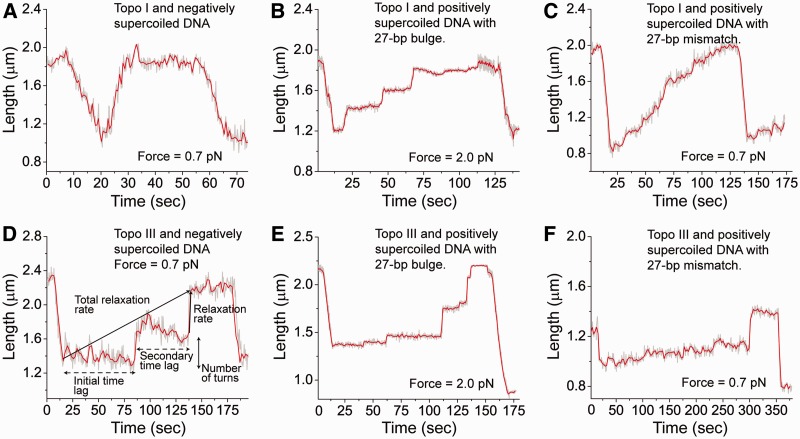
Figure 2.
Single-molecule relaxation by Topo I and Topo III. The plots show typical relaxation runs for different substrates. Each run is defined as a series of relaxation events where pausing cannot be observed. (A) Relaxation of negatively supercoiled DNA by Topo I. (B) Relaxation of positively supercoiled DNA with a 27-bp bulge by Topo I. (C) Relaxation of positively supercoiled DNA with 27-bp mismatch by Topo I. (D) Relaxation of negatively supercoiled DNA by Topo III. (E) Relaxation of positively supercoiled DNA with a 27-bp bulge by Topo III. (F) Relaxation of positively supercoiled DNA with 27-bp mismatch by Topo III. In all cases the plots show the length of the DNA plotted against time. Introduction of supercoils results in shortening of the molecule whereas relaxation results in elongation of the molecule. Negatively supercoiled and mismatched DNA experiments used a 0.7 pN stretching force; other experiments used a 2.0 pN force. Panel (D) illustrates the parameters used to analyze the relaxation events (see text for definitions). The gray trace corresponds to the measured events whereas the red trace corresponds to an unweighted running average over 10 events.
As type IA enzymes require ssDNA for activity (5), it is possible to introduce ssDNA regions and use positively supercoiled substrates and higher stretching forces to reduce the noise in the data, an approach that was employed previously to characterize type IA enzymes (15,16). Two types of substrates and each type with different lengths of the ssDNA region were prepared: bulged substrates where 12- or 27-bp ssDNA loops were introduced and mismatched substrates where 12- or 27-bp non-complementary regions were added (Figure 1B). The latter substrates led to a short region where base pairing is not possible and produces two opposite non-complementary single-stranded regions, whereas the former produces a continuous dsDNA molecule with an extruded single-stranded loop. For the bulged substrates the enzymes were active at high force, which results in reduced noise in the data, and for this reason the experiments were done at 2 pN. For DNA with a mismatch, increasing the force negatively affects the activity of Topo I and Topo III, as reported before for Topo I (15,16), and therefore the mismatch substrate experiments were done at 0.7 pN. For consistency, for each substrate, the experiments were always done at the same force. DNA relaxation activities by both enzymes on positive supercoiled DNA molecules with a bulge or a mismatch were measured at 2.0 and 0.7 pN forces, respectively (Figure 2B and E and Figure 2C and F).
In a typical experiment, a single DNA molecule is supercoiled by introducing ∼30 turns, either positive or negative, which leads to length shortening, followed by relaxation by the enzyme, resulting in length increases. Clear pauses before the first relaxation event and also in between runs of relaxation events were observed. Once the DNA is relaxed, it can be supercoiled again and this can be repeated several times, usually until the DNA becomes irreversibly nicked, or detaches from the glass or the bead.
Figure 2 shows examples of real-time measurements of DNA relaxation events by Topo I and III. In order to analyze and characterize the relaxation events by Topo I and III and to determine the differences in their mechanism, six characteristic features of the DNA relaxation events were analyzed: initial time lag, secondary time lag, number of turns relaxed per run, relaxation rate per run, relaxation rate of the run with time lag before that run and total relaxation rate (Figure 2D and ‘Materials and Methods’ section). The results of tens of observations (see Supplementary Table S1 for details on the number of observations) were analyzed by extracting the six parameters, creating histograms of the frequency of the observed values and extracting the mean from the distribution of these values (Supplementary Data). Topo III was not very active on the mismatched substrates; relaxation events were observed only in ∼20% of the experiments with these substrates. In these cases, the results reported are the average values and not a fit to any distribution (Supplementary Data).
Table 1 shows the mean values for the six characteristics analyzed for the relaxation of negatively and positively supercoiled DNA with either bulges or mismatches by Topo I and Topo III. Comparing these values, it is clear that the single-molecule experiments recapitulate the bulk experiments when the total rates for negatively supercoiled DNA are compared. The total rate for Topo I, 0.9 turns/s, is significantly larger than the total rate for Topo III, 0.2 turns/s. Regardless of the substrate used, the total relaxation rate is always higher for Topo I, showing that the differences in overall rate are not solely due to differences in substrate. Interestingly, the data also reveal that the relaxation mechanisms for the two enzymes are very different. Figure 2, panels A and D, show typical examples of negatively supercoiled relaxation by Topo I and Topo III, respectively. Whereas Topo I relaxes DNA in one relaxation run without pauses, Topo III pauses in between relaxation runs, but the relaxation velocity in the runs is higher for Topo III than Topo I. Thus, Topo I and Topo III accomplish the same task in a very different manner.
Table 1.
Characterization of DNA relaxation by E. coli topoisomerases I and III
| Initial time lag (s) |
Secondary time lag (s) |
Turns removed (ΔLk) |
||||
|---|---|---|---|---|---|---|
| Substrate | Topo I | Topo III | Topo I | Topo III | Topo I | Topo III |
| Negatively supercoiled | 6 ± 1.5 | 129 ± 19 | 5 ± 1.1 | 114 ± 19 | 20 ± 1.9 | 28 ± 4 |
| 27-bp bulged | 5 ± 1.5 | 52 ± 15 | 3 ± 0.5 | 9 ± 2.1 | 3 ± 0.3 | 4 ± 0.5 |
| 12-bp bulged | 46 ± 14 | 83 ± 10 | 23 ± 4.3 | 48 ± 12 | 1 ± 0.1 | 1 ± 0.2 |
| 27-bp mismatched | 12 ± 2.5 | 132 ± 44 | 11 ± 1.4 | 59 ± 17 | 1.5 ± 0.2 | 3 ± 0.6 |
| 12 bp mismatched | 131 ± 31 | 167 ± 23 | 22 ± 3.4 | 45 ± 15 | 1 ± 0.1 | 1.5 ± 0.3 |
| Relaxation rate per run (ΔLk/s) |
Relaxation rate per run with time lag (ΔLk/s) |
Total relaxation rate (ΔLk/s) |
||||
|---|---|---|---|---|---|---|
| Topo I | Topo III | Topo I | Topo III | Topo I | Topo III | |
| Negatively supercoiled | 3.3 ± 0.6 | 129 ± 16 | 1.3 ± 0.2 | 0.2 ± 0.03 | 0.9 ± 0.2 | 0.2 ± 0.02 |
| 27-bp bulged | 14 ± 1.8 | 19 ± 3 | 1.2 ± 0.3 | 0.09 ± 0.02 | 0.3 ± 0.1 | 0.08 ± 0.03 |
| 12-bp bulged | 6 ± 0.8 | 29 ± 4 | 0.1 ± 0.02 | 0.02 ± 0.004 | 0.07 ± 0.01 | 0.01 ± 0.004 |
| 27-bp mismatched | 2 ± 0.3 | 26 ± 5 | 0.2 ± 0.03 | 0.04 ± 0.02 | 0.2 ± 0.05 | 0.08 ± 0.02 |
| 12-bp mismatched | 0.8 ± 0.1 | 35 ± 6 | 0.1 ± 0.01 | 0.3 ± 0.06 | 0.07 ± 0.03 | 0.02 ± 0.07 |
The table shows the mean values for the six parameters used to characterize the single-molecule DNA relaxation events for each substrate used for Topo I and Topo III. In each case, the mean and the standard error are shown. For the case of Topo III using mismatched molecules, the number of recorded events was small as Topo III does not relax this type of DNA efficiently. For details on the definitions and the analysis, see ‘Materials and Methods’ and the Supplementary Data section.
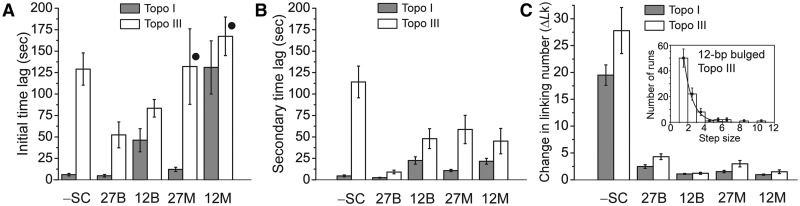
Topo I and III are type IA enzymes and relax DNA strictly in steps of one, but in most instances individual relaxation events could not be resolved and instead relaxation bursts or runs, where several supercoils are removed sequentially without an apparent pause between individual relaxation events, were observed. The mean number of supercoils removed in a relaxation run by Topo I and Topo III on the various DNA substrates were extracted from the data (Figure 3C and Table 1). The activity on negatively supercoiled DNA was characterized by relaxation bursts with a mean change in linking number (<ΔLk>) of 20 and 28 turns for Topo I and Topo III, respectively. For the longer mismatched or bulged substrates (27-bp mismatched or bulged), the mean number of supercoils removed were 1.5 (mismatched) and 3 (bulged) for Topo I and 3 and 4 for Topo III. Finally, the substrates with the shortest ssDNA regions, 12-bp mismatched or bulged, produced the shortest relaxation runs and single relaxation events could be resolved. In these cases, the mean number of steps in a run was 1 and 1 for Topo I and 1 and 1.5 for Topo III, for the mismatched and bulged substrates, respectively. The data indicate that the processivity of both proteins depends upon the size of the ssDNA region present, since longer bursts of relaxation correspond to substrate with abundant ssDNA regions, as is the case in negatively supercoiled DNA, and shorter bursts are observed by reducing the size of the ssDNA region. In the case of the 12-bp ssDNA substrates, single relaxation steps were easily resolved, as have been observed before (15,16).
Figure 3.
Characterization of DNA relaxation of different substrates by Topo I and Topo III. (A) Histogram showing the distribution of time lag before initiation of relaxation. In all cases, Topo III shows a much longer time lag before starting a relaxation run. For mismatched substrates (marked by filled circle) Topo III relaxation was only observed in 20% of the experiments. (B) Histogram of the secondary time lag. The P-values for the differences between the initial and secondary time lags are shown in Supplementary Table S2. In all cases, the differences between the two time lags are significant. (C) Histogram of the mean number of turns relaxed in a run (ΔLk). Topo III consistently removed more turns per run for all substrates. The inset shows the distribution for the 12-bp bulged substrate using Topo III. The solid curve corresponds to a fit of an exponential decay to the distribution. In all cases, shaded bars correspond to Topo I and white ones to Topo III. The different substrates used are shown at the bottom, where -SC corresponds to negatively supercoiled DNA, and 12B, 12 M, 27B and 27 M correspond to the 12-bp and 27-bp bulged and mismatched substrates. In all cases, the differences observed between the two enzymes are significant as assessed by the P-value (P-value of 0.0001 for all compared pairs, except for the 12-bp mismatch in panel A, with a P-value of 0.001). The error bars shown correspond to the standard errors. Details on the number of events used for each histogram are found in Supplementary Table S1.
The initial and secondary time lags were also analyzed to uncover any differences (Figure 3A, B and Table 1). The initial time lag is defined as the time it takes to initiate the first relaxation run after supercoiling, whereas the secondary time lag is defined as the time interval between subsequent relaxation runs. In general, the results are consistent for all substrates; for instance, time lags are always longer for Topo III than for Topo I regardless of the substrate. In the case of Topo III and the mismatched substrates, relaxation was observed only in 20% of the experiments, even after waiting for over 30 min after initial supercoiling. In contrast, in all other substrate–enzyme combinations relaxation events were always observed. This suggests that for the case of the mismatched substrates, Topo III activity was more substrate-dependent than Topo I. Low activity of Topo III on the positively supercoiled DNA with a mismatch could be due to the fact that this substrate was more affected by the applied force and the resulting ssDNA region may be a bad substrate for Topo III. In all other cases, both time lags had a distribution that could be fitted by a simple exponential decay and from the distributions we could extract the mean time lags. For all substrates studied, Topo I initiated relaxation sooner than Topo III (Table 1 and Figure 3A). A similar trend was observed when the secondary time lag distributions were analyzed (Table 1 and Figure 3B). For most substrates, the pauses between relaxation runs catalyzed by Topo I were spaced by only a few seconds, whereas Topo III paused for tens of seconds in between relaxation runs (Table 1 and Figure 3C). Overall, Topo III waits a much longer time before initiating relaxation and also between relaxation runs than Topo I. In addition, the secondary time lag was consistently shorter than the initial time lag, suggesting that the same protein molecule was responsible for the initial and secondary relaxation runs.
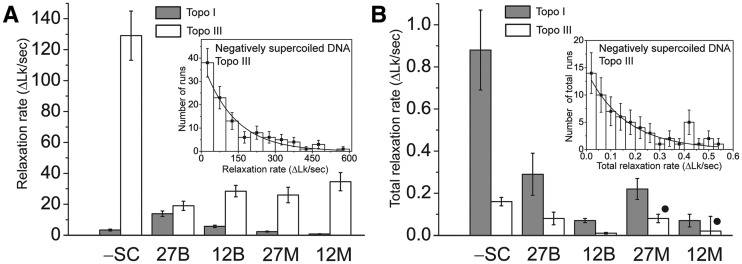
Another characteristic analyzed was the relaxation rate, or the number of supercoils removed per second in a run (Table 1 and Figure 4A). When analyzing the relaxation rates per run, it is clear that in all cases Topo III is faster that Topo I, even with the worst substrates. Relaxation of negatively supercoiled DNA by Topo III is particularly fast, removing on average ∼130 turns/s. In contrast, Topo I can only remove ∼3 turns/s, almost 40 times slower than Topo III. Even with the short bulged or mismatched substrates Topo III is faster, removing tens of turns per second in all cases, whereas Topo I can be as slow as ∼1 turn/s for the worst substrate. It is not possible to rule out that the elevated rates observed for Topo III on negatively supercoiled DNA were due to the simultaneous action of several enzymes, even though the enzyme concentration used was low (2 nM) and the lag times were long compared to the run times, making it unlikely that more than one protein molecule is acting on the DNA. This is not the case for bulged or mismatched substrates, where the small ssDNA region is the only region that the proteins can recognize and use as a substrate. This argues strongly that in all cases the fast relaxation rates by Topo III and the slow rates by Topo I are intrinsic properties of the enzymes.
Figure 4.
Relaxation rate per run and total relaxation rate for Topo I and Topo III. (A) Histogram showing the distribution of the relaxation rate per run. In all cases, Topo III has a faster relaxation rate per run than Topo I. In the case of negatively supercoiled DNA, Topo III is faster by a factor of ∼40. The inset shows the distribution of relaxation rate per run for negatively supercoiled DNA by Topo III. (B) Histogram showing the distribution of the total relaxation rate. Topo I has a faster total relaxation rate than Topo III, in agreement with bulk experiments. The inset shows the distribution of the total relaxation rate for negatively supercoiled DNA by Topo III. In all cases, shaded bars correspond to Topo I and white ones to Topo III. For mismatched substrates (marked by filled circles) Topo III relaxation was only observed in 20% of the experiments. The different substrates used are shown at the bottom, where -SC corresponds to negatively supercoiled DNA, and 12B, 12 M, 27B and 27 M correspond to the 12-bp and 27-bp bulged and mismatched substrates. In all cases, the differences observed between the two enzymes are significant as assessed by the P-value (P-value of 0.0001 for all compared pairs, except for the 12-bp mismatch in panel B, with a P-value of 0.02). The error bars shown correspond to the standard errors. Details on the number of events used for each histogram are found in Supplementary Table 1.
Analysis of the total relaxation rate reveals that Topo I is faster in overall DNA relaxation than Topo III on all substrates (Figure 4B, see also Supplementary Figure S3 for the same data presented with a logarithmic scale), even though the relaxation rate during a run is always faster for Topo III (Figure 4A). Both Topo I and Topo III are more efficient on negatively supercoiled DNA, compared to positively supercoiled DNA, but in all cases Topo I is overall a faster enzyme thanks to the shorter lag times. Interestingly, for each of the proteins the total relaxation rate stays about the same for substrates with the same size ssDNA region regardless of whether the substrate includes a bulge or a mismatch (Table 1).
Overall, the results from the experiments can be summarized as follows: Topo III relaxes DNA efficiently and in fast processive runs, but the time lag preceding a run is long, whereas Topo I relaxes DNA more slowly but the initial time lag time is short. The combination of these properties results in Topo I having an overall faster total relaxation rate, even though Topo III can relax DNA much faster in an individual run. In addition, the choice of substrate is important, but the general properties outlined before are substrate independent.
DISCUSSION
Escherichia coli Topo I and Topo III are known to relax negatively supercoiled DNA using the same overall mechanism, but with different overall efficiency in bulk experiments. Single-molecule experiments with negatively supercoiled molecules reproduced the behavior observed in bulk experiments: the overall relaxation rate for Topo I was larger than for Topo III, even though the behavior at the single-molecule level was different. The consistency with the bulk observations confirms that our single-molecule approach is valid for the study of these two enzymes.
In general, the activities of both enzymes were strongly affected by the length of the ssDNA region. Relaxation was much more efficient with negatively supercoiled DNA, which is underwound with abundant ssDNA regions. As expected, neither Topo I nor Topo III was able to relax positively supercoiled DNA (data not shown). Relaxation was possible when using positively supercoiled DNA as long as there were ssDNA regions present, but the total relaxation rates were slower in all cases as a result of different combinations of longer time lags and slower relaxation rates. In general, shorter ssDNA substrates resulted in slower total relaxation rates and also a smaller number of turns removed per run. Interestingly, the total rate of relaxation is about the same on substrates with the same size ssDNA region (Table 1 and Figure 4B). At the individual run level, the processivity is higher for Topo III than for Topo I, especially for negatively supercoiled DNA.
Another important observation is that regardless of size and type of substrate, Topo III exhibited longer time lags between relaxation events than Topo I (Figure 3); however, these time lags were dependent on the characteristics of the substrates. Topo III exhibited shorter time lags when the substrate was negatively supercoiled DNA, whereas Topo I did not show such a marked preference. The mismatched substrates were consistently worse and resulted in longer initial time lags. For instance, Topo III was active in only 20% of the experiments. These observations suggest that relaxing a mismatched substrate may be difficult for both enzymes. This could be due to the influence of force on the mismatched substrates, which may produce a DNA molecule where the ssDNA regions are not easily recognizable. In general, Topo III is more sensitive to the type of ssDNA substrate with regard to initial and secondary time lags.
The secondary time lags follow the same pattern as the initial time lags, i.e. long initial time lags are associated with long secondary time lag and vice versa. In all cases, the secondary lag time is shorter than the initial time lag (P-values are listed in Supplementary Table S2). This is indicative that the same protein molecule is likely to be responsible for all the events in one series of relaxation events (initial time lag, plus relaxation run, plus secondary time lag and so forth). If a different protein was responsible for the primary and secondary lag times, it would be unlikely that the secondary time lags would be consistently different and always shorter. The consistency of the time lag durations thus suggests that the time lags are waiting times necessary for protein activity to initiate and not waiting times for binding events.
Furthermore, the dependence of Topo III time lags on the type of DNA substrate used suggests that the time lags depend on a DNA–protein event that prepares the DNA for activity, such as a change in DNA conformation, that is protein driven. Topo III is much more sensitive to the type of DNA substrate and consistently faster at relaxation events. Topo I may be able to prepare DNA for strand passage much more efficiently by engaging a large DNA region, while at the same time hindering the actual strand passage events. In contrast, Topo III has a much smaller DNA-binding region that may slow down the start of a relaxation event, while at the same time the small binding region may allow for a suppler enzyme that can process strand passage events quickly. Thus, on the one hand Topo I may rapidly initiate and continue relaxation without pauses due to its ability to interact with a large DNA region. On the other hand, Topo III may require a much longer time to prepare the DNA, resulting in long pauses before and in between DNA relaxation runs, while at the same time being able to process strand passage events efficiently once the DNA is properly engaged.
One very surprising finding was that the relaxation rate per run was much higher for Topo III. The relaxation rates per run for Topo III exceed those of Topo I on all DNA substrates (Table 1). In the case of negatively supercoiled DNA, the natural substrate, the Topo III rates are almost 40 times higher than for Topo I. The higher relaxation rates for Topo III clearly indicate that Topo III can perform the strand passage events much faster than Topo I. Together with the slightly larger number of turns removed per run, this suggests that Topo III is a very efficient relaxing enzyme that can remove many turns in a fast manner, but that it has to wait a long time before it starts a relaxation series. Topo I, in contrast, removes supercoils slower and in shorter runs, but does not wait as long before starting a new relaxation series.
As mentioned above, the total relaxation rate is higher for Topo I than for Topo III, both in bulk and in single molecule experiments. Whereas from bulk experiments it would appear that Topo I is simply a more efficient enzyme, the single molecule experiments reveal that the higher total relaxation rates of Topo I are a combination of several features. Topo III is capable of relaxing DNA much faster and in a more processive manner, but it waits a longer time before starting a relaxation run. In contrast, Topo I has short initial and secondary lag times, but it also has slower relaxation runs. These observations indicate that the dominant factor in the overall relaxation rate is the lag time and not the relaxation rate per run. These differences between the two related molecules are not apparent from bulk experiments, where the average of many events hide the individual behavior of the molecules and do not reveal the true nature of the differences. Thus, single molecule experiments were needed to uncover the differences in the mechanism.
It is not clear what the optimal substrate for each enzyme in the cell is. The substrate used for the single-molecule experiments, supercoiled DNA, is more likely to resemble the optimal substrate for Topo I than for Topo III, which may prefer molecules that resemble recombination intermediates. In addition, many other parameters, such as monovalent and divalent local concentrations, interactions with other proteins, such as RecQ in the case of Topo III, and the local topological state of the DNA are likely to have an effect on the overall relaxation rates. Nevertheless, the experiments uncover differences between the two molecules that shed important information on their activity and suggest the origin of the two different roles in the cell. Additional experiments with other substrates, perhaps resembling recombination intermediates or catenated molecules, and under a wider range of conditions will be important to understand in more detail the differences between these two molecules.
The main cellular function of Topo I is to help maintain the topological state of DNA (17). The characteristics observed in the single-molecule experiments are consistent with this function. Topo I relaxes DNA in a steady and consistent fashion without any significant pauses. These characteristics are ideal for a house-keeping enzyme that needs to constantly monitor and alter the topology of the DNA. In contrast, Topo III works through fast bursts of activity although it waits a long time before starting. This appears to be more consistent with an enzyme that needs to act on specialized substrates that are transient or not always present, and that does not need to be very efficient when acting on ordinary supercoiled DNA. Although the experiments described here do not address the role of Topo III in decatenation, it is quite possible that the synergistic activity of Topo III and RecQ leads to efficient recognition/creation of appropriate substrates that are then resolved very quickly by Topo III (22). Thus, Topo I has all the characteristics of an enzyme that can maintain a constant DNA topological state and that responds efficiently to any overall changes. In contrast, Topo III responds very slowly to changes in topological state but can act very quickly, more consistent with an enzyme that needs to alter the DNA topology quickly, but does not need to respond to the overall topological state immediately. In this manner, Topo III appears to be an enzyme optimized for specialized functions, such as resolving recombination or replication intermediates, whereas Topo I is optimized for overall supercoiling topology maintenance. Additional experiments to study the characteristics of the two enzymes in other situations, such as decatenation reactions, both with and without protein cofactors, are needed to clarify the differential role of the two enzymes.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–2, Supplementary Figures 1–3 and Supplementary Methods.
FUNDING
The National Institutes of Health (NIH) [GM51350 to A.M. and 1U54CA143869 to J.F.M.]; the National Science Foundation [MCB-1022117 to J.F.M.]. Funding for open access charge: NIH.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors thank members of the Marko and Mondragón laboratories for their help and advice.
REFERENCES
- 1.Schoeffler AJ, Berger JM. DNA topoisomerases: harnessing and constraining energy to govern chromosome topology. Q. Rev. Biophys. 2008;41:41–101. doi: 10.1017/S003358350800468X. [DOI] [PubMed] [Google Scholar]
- 2.Vos SM, Tretter EM, Schmidt BH, Berger JM. All tangled up: how cells direct, manage and exploit topoisomerase function. Nat. Rev. Mol. Cell Biol. 2011;12:827–841. doi: 10.1038/nrm3228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baker NM, Rajan R, Mondragon A. Structural studies of type I topoisomerases. Nucleic Acids Res. 2009;37:693–701. doi: 10.1093/nar/gkn1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tse Y, Wang JC. E. coli and M. luteus DNA topoisomerase I can catalyze catenation or decatenation of double-stranded DNA rings. Cell. 1980;22:269–276. doi: 10.1016/0092-8674(80)90174-9. [DOI] [PubMed] [Google Scholar]
- 5.Kirkegaard K, Wang JC. Bacterial DNA topoisomerase I can relax positively supercoiled DNA containing a single-stranded loop. J. Mol. Biol. 1985;185:625–637. doi: 10.1016/0022-2836(85)90075-0. [DOI] [PubMed] [Google Scholar]
- 6.Hansen G, Harrenga A, Wieland B, Schomburg D, Reinemer P. Crystal structure of full length topoisomerase I from Thermotoga maritima. J. Mol. Biol. 2006;358:1328–1340. doi: 10.1016/j.jmb.2006.03.012. [DOI] [PubMed] [Google Scholar]
- 7.Lima CD, Wang JC, Mondragon A. Three-dimensional structure of the 67K N-terminal fragment of E. coli DNA topoisomerase I. Nature. 1994;367:138–146. doi: 10.1038/367138a0. [DOI] [PubMed] [Google Scholar]
- 8.Mondragon A, DiGate R. The structure of Escherichia coli DNA topoisomerase III. Structure. 1999;7:1373–1383. doi: 10.1016/s0969-2126(00)80027-1. [DOI] [PubMed] [Google Scholar]
- 9.Li Z, Mondragon A, DiGate RJ. The mechanism of type IA topoisomerase-mediated DNA topological transformations. Mol. Cell. 2001;7:301–307. doi: 10.1016/s1097-2765(01)00178-2. [DOI] [PubMed] [Google Scholar]
- 10.Changela A, DiGate RJ, Mondragon A. Crystal structure of a complex of a type IA DNA topoisomerase with a single-stranded DNA molecule. Nature. 2001;411:1077–1081. doi: 10.1038/35082615. [DOI] [PubMed] [Google Scholar]
- 11.Changela A, DiGate RJ, Mondragon A. Structural studies of E. coli topoisomerase III-DNA complexes reveal a novel type IA topoisomerase-DNA conformational intermediate. J. Mol. Biol. 2007;368:105–118. doi: 10.1016/j.jmb.2007.01.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feinberg H, Lima CD, Mondragon A. Conformational changes in E. coli DNA topoisomerase I. Nat. Struct. Biol. 1999;6:918–922. doi: 10.1038/13283. [DOI] [PubMed] [Google Scholar]
- 13.Perry K, Mondragon A. Structure of a complex between E. coli DNA topoisomerase I and single-stranded DNA. Structure. 2003;11:1349–1358. doi: 10.1016/j.str.2003.09.013. [DOI] [PubMed] [Google Scholar]
- 14.Zhang Z, Cheng B, Tse-Dinh YC. Crystal structure of a covalent intermediate in DNA cleavage and rejoining by Escherichia coli DNA topoisomerase I. Proc. Natl Acad. Sci. USA. 2011;108:6939–6944. doi: 10.1073/pnas.1100300108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dekker NH, Rybenkov VV, Duguet M, Crisona NJ, Cozzarelli NR, Bensimon D, Croquette V. The mechanism of type IA topoisomerases. Proc. Natl Acad. Sci. USA. 2002;99:12126–12131. doi: 10.1073/pnas.132378799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dekker NH, Viard T, de La Tour CB, Duguet M, Bensimon D, Croquette V. Thermophilic topoisomerase I on a single DNA molecule. J. Mol. Biol. 2003;329:271–282. doi: 10.1016/s0022-2836(03)00320-6. [DOI] [PubMed] [Google Scholar]
- 17.Zechiedrich EL, Khodursky AB, Bachellier S, Schneider R, Chen D, Lilley DM, Cozzarelli NR. Roles of topoisomerases in maintaining steady-state DNA supercoiling in Escherichia coli. J. Biol. Chem. 2000;275:8103–8113. doi: 10.1074/jbc.275.11.8103. [DOI] [PubMed] [Google Scholar]
- 18.Wallis JW, Chrebet G, Brodsky G, Rolfe M, Rothstein R. A hyper-recombination mutation in S. cerevisiae identifies a novel eukaryotic topoisomerase. Cell. 1989;58:409–419. doi: 10.1016/0092-8674(89)90855-6. [DOI] [PubMed] [Google Scholar]
- 19.Lopez CR, Yang S, Deibler RW, Ray SA, Pennington JM, Digate RJ, Hastings PJ, Rosenberg SM, Zechiedrich EL. A role for topoisomerase III in a recombination pathway alternative to RuvABC. Mol. Microbiol. 2005;58:80–101. doi: 10.1111/j.1365-2958.2005.04812.x. [DOI] [PubMed] [Google Scholar]
- 20.DiGate RJ, Marians KJ. Identification of a potent decatenating enzyme from Escherichia coli. J. Biol. Chem. 1988;263:13366–13373. [PubMed] [Google Scholar]
- 21.Harmon FG, DiGate RJ, Kowalczykowski SC. RecQ helicase and topoisomerase III comprise a novel DNA strand passage function: a conserved mechanism for control of DNA recombination. Mol. Cell. 1999;3:611–620. doi: 10.1016/s1097-2765(00)80354-8. [DOI] [PubMed] [Google Scholar]
- 22.Suski C, Marians KJ. Resolution of converging replication forks by RecQ and topoisomerase III. Mol. Cell. 2008;30:779–789. doi: 10.1016/j.molcel.2008.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hiasa H, DiGate RJ, Marians KJ. Decatenating activity of Escherichia coli DNA gyrase and topoisomerases I and III during oriC and pBR322 DNA replication in vitro. J. Biol. Chem. 1994;269:2093–2099. [PubMed] [Google Scholar]
- 24.Hiasa H, Marians KJ. Topoisomerase III, but not topoisomerase I, can support nascent chain elongation during theta-type DNA replication. J. Biol. Chem. 1994;269:32655–32659. [PubMed] [Google Scholar]
- 25.Yu L, Zhu CX, Tse-Dinh YC, Fesik SW. Solution structure of the C-terminal single-stranded DNA-binding domain of Escherichia coli topoisomerase I. Biochemistry. 1995;34:7622–7628. doi: 10.1021/bi00023a008. [DOI] [PubMed] [Google Scholar]
- 26.Ahumada A, Tse-Dinh YC. The Zn(II) binding motifs of E. coli DNA topoisomerase I is part of a high-affinity DNA binding domain. Biochem. Biophys. Res. Commun. 1998;251:509–514. doi: 10.1006/bbrc.1998.9500. [DOI] [PubMed] [Google Scholar]
- 27.Beran-Steed RK, Tse-Dinh YC. The carboxyl terminal domain of Escherichia coli DNA topoisomerase I confers higher affinity to DNA. Proteins. 1989;6:249–258. doi: 10.1002/prot.340060307. [DOI] [PubMed] [Google Scholar]
- 28.Zhang HL, DiGate RJ. The carboxyl-terminal residues of Escherichia coli DNA topoisomerase III are involved in substrate binding. J. Biol. Chem. 1994;269:9052–9059. [PubMed] [Google Scholar]
- 29.Sikder D, Nagaraja V. A novel bipartite mode of binding of M. smegmatis topoisomerase I to its recognition sequence. J. Mol. Biol. 2001;312:347–357. doi: 10.1006/jmbi.2001.4942. [DOI] [PubMed] [Google Scholar]
- 30.Zhang HL, Malpure S, Digate RJ. Escherichia-coli DNA topoisomerase-Iii is a site-specific DNA-binding protein that binds asymmetrically to its cleavage site. J. Biol. Chem. 1995;270:23700–23705. doi: 10.1074/jbc.270.40.23700. [DOI] [PubMed] [Google Scholar]
- 31.Li Z, Mondragon A, Hiasa H, Marians KJ, DiGate RJ. Identification of a unique domain essential for Escherichia coli DNA topoisomerase III-catalysed decatenation of replication intermediates. Mol. Microbiol. 2000;35:888–895. doi: 10.1046/j.1365-2958.2000.01763.x. [DOI] [PubMed] [Google Scholar]
- 32.Perry K, Mondragon A. Biochemical characterization of an invariant histidine involved in Escherichia coli DNA topoisomerase I catalysis. J. Biol. Chem. 2002;277:13237–13245. doi: 10.1074/jbc.M112019200. [DOI] [PubMed] [Google Scholar]
- 33.Strick TR, Allemand JF, Bensimon D, Bensimon A, Croquette V. The elasticity of a single supercoiled DNA molecule. Science. 1996;271:1835–1837. doi: 10.1126/science.271.5257.1835. [DOI] [PubMed] [Google Scholar]
- 34.Taneja B, Schnurr B, Slesarev A, Marko JF, Mondragon A. Topoisomerase V relaxes supercoiled DNA by a constrained swiveling mechanism. Proc. Natl Acad. Sci. USA. 2007;104:14670–14675. doi: 10.1073/pnas.0701989104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marko JF. Torque and dynamics of linking number relaxation in stretched supercoiled DNA. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 2007;76:021926. doi: 10.1103/PhysRevE.76.021926. [DOI] [PubMed] [Google Scholar]