Abstract
At earlier stages in the evolution of the universal genetic code, fewer than 20 amino acids were considered to be used. Although this notion is supported by a wide range of data, the actual existence and function of the genetic codes with a limited set of canonical amino acids have not been addressed experimentally, in contrast to the successful development of the expanded codes. Here, we constructed artificial genetic codes involving a reduced alphabet. In one of the codes, a tRNAAla variant with the Trp anticodon reassigns alanine to an unassigned UGG codon in the Escherichia coli S30 cell-free translation system lacking tryptophan. We confirmed that the efficiency and accuracy of protein synthesis by this Trp-lacking code were comparable to those by the universal genetic code, by an amino acid composition analysis, green fluorescent protein fluorescence measurements and the crystal structure determination. We also showed that another code, in which UGU/UGC codons are assigned to Ser, synthesizes an active enzyme. This method will provide not only new insights into primordial genetic codes, but also an essential protein engineering tool for the assessment of the early stages of protein evolution and for the improvement of pharmaceuticals.
INTRODUCTION
The canonical 20 amino acids are assigned to the sense codons in the universal genetic code. Since this assignment is utilized by almost all organisms, this code had been considered as ‘universal’ (1). The genetic code, however, was found to be malleable, with the discovery of deviant codes (2–4). For example, CUG codons are assigned to serine (Ser), instead of leucine (Leu), in Candida cylindracea. In laboratory experiments, researchers succeeded in expanding the genetic code to include unnatural amino acids (5–7). Furthermore, pyrrolysine, which arose from the natural expansion of the genetic code, is incorporated into proteins in response to the UAG nonsense codon (8). These studies showed that the function of codes to synthesize proteins can be retained, even if the assignment between a codon and an amino acid is different from that in the universal genetic code.
From an evolutionary viewpoint, the genetic code is considered to have evolved from primitive forms containing a limited set of amino acids (1,9,10). Chemical evolution experiments suggest that only a limited number of canonical amino acids were available in prebiotic environments (11,12), and therefore the other amino acids must have been derived from the evolution of amino acid biosynthesis (9). Moreover, the phylogenies of the aminoacyl-tRNA synthetases (aaRSs) (13) suggest that the genetic codes have gradually gained amino acids. Although no organisms that utilize fewer than 20 amino acids have been discovered, all known archaea and some bacteria lack asparaginyl-tRNA synthetase and/or glutaminyl-tRNA synthetase, and some methanogenic archaea lack cysteinyl-tRNA synthetase. They utilize 20 amino acids in their genetic codes by a synthesis method in which a non-cognate amino acid is first attached to the tRNA, and then the non-cognate amino acid is converted to the cognate one by tRNA-dependent modifying enzymes (14–18). These indirect aminoacylation pathways are considered to have been the prevailing routes before the evolution of the direct pathways (14,15), and thus would have participated in previous expansions of the universal genetic code.
Although various sources of data suggest that the universal genetic code evolved from a simpler form encoding fewer than 20 amino acids, the simplification of the genetic code has not been addressed experimentally, in contrast to the expansion of the genetic code. The simplification of the code, as well as the expansion, requires the engineering of a tRNA to prepare a new connection between an amino acid and its codon(s). On the other hand, the inhibition of a specific aaRS activity is also required in the simplification, in contrast to the alteration of the amino acid recognition by an aaRS in the expansion. In this study, we created a simplified genetic code by excluding specific amino acids from the cell-free reaction mixture, and reassigning the unassigned codon by using tRNA variants bearing an altered anticodon loop.
MATERIALS AND METHODS
DNA constructs and in vitro transcription
Maltose-binding protein (MBP), LexA enzymes and chloramphenicol acetyltransferase (CAT) genes were cloned into the pK7 plasmid (19). Green fluorescent protein (GFP) genes were cloned into the pGFP plasmid (20). Genes encoding tRNA variants were cloned into the pUC119 plasmid (TAKARA). The tRNA variants were prepared by run-off transcription using T7 RNA polymerase (6). The LexA substrate gene (21), LexA L89P-Q92W-Y98K with a TAA stop codon at position 99, was cloned into the pET26b plasmid (Novagen). Additional details are in ‘Supplementary Methods’ section.
Cell-free protein expression
The Escherichia coli S30 cell-free protein synthesis method was used in this study. The composition of the cell-free protein synthesis reaction was previously described (22), except for the omission of a specific amino acid, and the addition of the tRNA variant and 5.0 μM a.a.-SA (aminoacyl adenylate analogs, Integrated DNA Technologies). The S30 extract was prepared from the E. coli BL21 (DE3) strain. The batch mode was employed for the 20 μl reaction volumes, and the dialysis mode was employed for reaction volumes of 60–3000 μl. The reaction times were 1 h for the batch mode and 8 h for the dialysis mode. Additional details are in ‘Supplementary Methods’ section.
Purification
The GFP, MBP and CAT proteins were purified using TALON metal affinity resin (Clontech) according to the manufacturer’s instructions, but with a slight modification (see ‘Supplementary Methods’ section).
Detection of the radiolabeled products
The translations of MBP, GFP and CAT were performed by using the 20 -μl scale batch mode of synthesis at 37°C for 1 h with the components described above, except for the addition of [14C] Leu. The non-purified products were analyzed on 12% bis–tris gels with MES-running buffer (50 mM MES, 50 mM Tris–base, 3.47 mM SDS, 1.0 mM EDTA, pH 7.3). Scanning was performed using an image analyzer, FLA-5000 (FUJI), and an imaging plate, BAS-IP MS 2040 (FUJI), to measure the radioactivity of the products.
Amino acid composition analysis
Translations of MBP and CAT were performed by using the middle-scale (1 ml internal/10 ml external) dialysis mode cell-free reaction (23) at 37°C for 8 h. The products containing the N-terminal polyhistidine tag were purified under denaturing conditions, followed by acetone precipitation.
To detect Trp, the products were hydrolyzed in 4 M methanesulfonic acid (MSA) containing 0.2% 3-(2-aminoethyl) indole (24) at 110°C for 20 h. After the hydrolysis, NaOH was added to neutralize the MSA. The derivatives were detected using the ninhydrin method, because 3 -(2-aminoethyl) indole disturbs the AQC-amino acid chromatogram. Products were detected by a High Speed Amino Acid Analyzer, L-8900 (Hitachi High-Technologies). Chromatograms were not normalized in the MSA hydrolysis.
To quantify the contents of Ala and most of the other amino acids, the products were fractionated by SDS–PAGE, electroblotted onto PVDF membranes and stained with Coomassie Brilliant Blue. The band on the membrane was excised with a clean razor blade. Hydrolysis with gas-phase hydrochloric acid, derivatization with aminoquinolyl-N-hydroxysuccinimidyl carbamate, and chromatographic analysis of the AQC-amino acid were performed as described (25) except an Agilent Technologies 1200 series SL HPLC system was used. Norvaline was injected as the internal standard.
Chromatograms were normalized to the top and bottom points of the Phe peak in the HCl hydrolysis. Additional details are in ‘Supplementary Methods’ section.
GFP fluorescence measurement
Translation of GFP was performed by using the 20 μl scale batch mode at 37°C for 1 h. Fluorescence from non-purified products was measured using an Mx3005P system (Stratagene). Excitation and emission wavelengths were set at 515 and 550 nm, respectively. A blank cell containing MilliQ water was used for background subtraction. Western blot analysis was performed using a monoclonal antibody directed against the His tag (Novagen).
Crystallization and data collection
Translation of GFP was performed by using the large-scale (3 ml internal/30 ml external) dialysis mode cell-free reaction (26) at 30°C for 8 h. GFP mutants containing the C-terminal polyhistidine tag were purified under native conditions. The C-terminal His tag was cleaved using PreScission Protease (GE Healthcare). The protease was removed by Glutathione Sepharose 4B resin (GE Healthcare). The residual uncleaved protein and the His peptides were removed by TALON metal affinity resin (Clontech). The products were further purified by Resource Q ion exchange column chromatography (GE Healthcare). Collected fractions were buffer exchanged into 20 mM Tris–HCl, pH 8.5, 1 mM DTT, and 50 mM NaCl, and were concentrated to 15 mg/ml with Amicon Ultra-0.5 filters with a 10-kDa MWCO membrane (MilliPore). Crystals of gfpΔUGG-A110X(UGG)/Sim grew in 7 days by the hanging drop vapor diffusion method against 17% PEG 20 000, 100 mM 2-(N-morpholino)ethanesulfonic acid (MES) (pH 6.5) at 4°C. Crystals of gfpΔUGG/Univ grew in 7 days by the hanging drop vapor diffusion method against 15% PEG 6000, 5% glycerol at 20°C. See ‘Supplementary Methods’ section for data collection.
Structure determination and refinement
The structures were determined by molecular replacement, using the GFP-S65T coordinate file (PDB code 1EMA) as a search model. Additional details are in ‘Supplementary Methods’ section.
LexA cleavage assay
The LexA cleavage assay was performed as previously described (21). Additional details are in ‘Supplementary Methods’ section.
RESULTS
Reassignment of the UGG codon to Ala, by a combination of a tRNAAla variant and a cell-free translation system lacking tryptophan, simplifies the genetic code
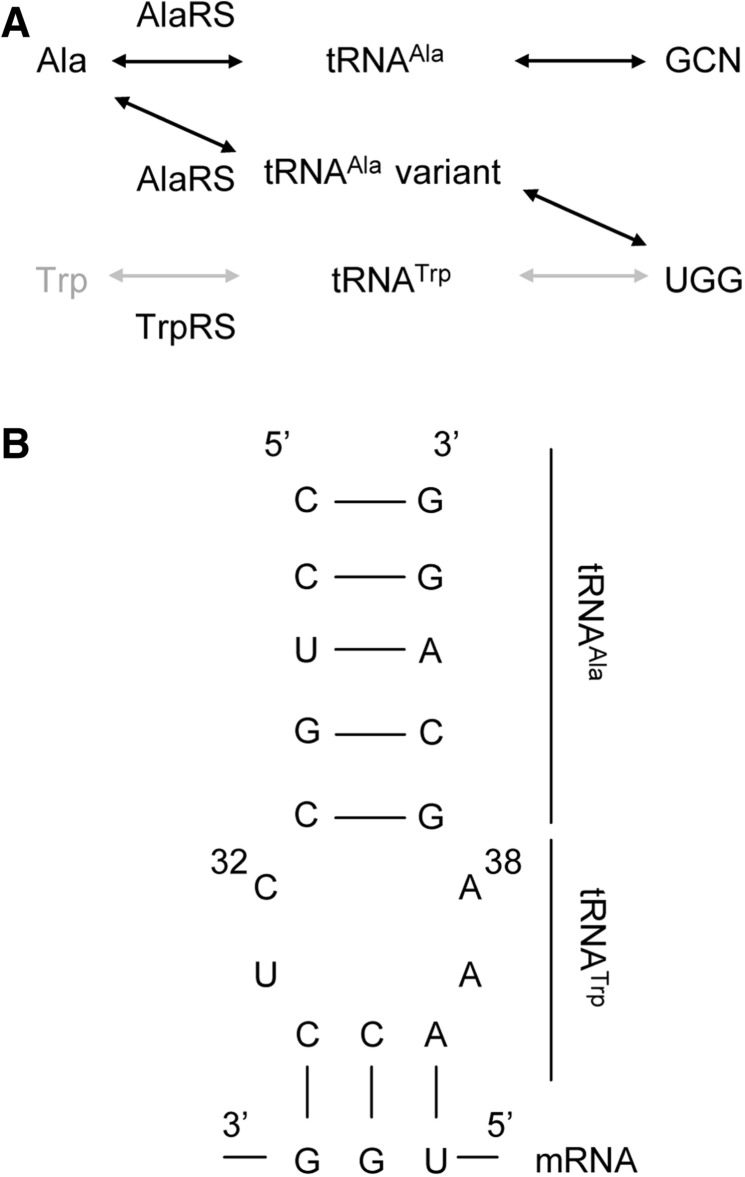
We constructed and confirmed the function of a ‘simplified genetic code’ in which only 19 amino acids are assigned to the sense codons. Trp is not included in this code, because the UGG codon is reassigned to alanine (Ala) (Figure 1A). In the first step of the reassignment, we produced unassigned UGG codons by removing Trp from the E. coli S30 cell-free translation mixture and adding Trp-SA, which is a potent inhibitor of tryptophanyl-tRNA synthetase (TrpRS). We then reassigned the UGG codon to Ala, by adding the tRNAAla variant with the anticodon loop corresponding to the UGG codon (Figure 1B). Ala is considered to be attached to the tRNAAla variant by alanyl-tRNA synthetase (AlaRS), because AlaRS does not recognize the anticodon loop (27–29). This reassignment seems to be successful, judging from the translation inhibition by the unassigned UGG codon (Figure 2A, lane 2–3), due to the lack of Trp-tRNATrp, and from the translation recovery by the addition of the tRNAAla variant (Figure 2A, lanes 4–6). To confirm the reassignment, we compared the amino acid compositions of two translation products (Figure 2B), synthesized from MBP mRNA containing eight UGG codons by the simplified code (MBP-WT/Sim) or the universal code (MBP-WT/Univ). In contrast to the apparent existence of Trp in MBP-WT/Univ, only a basal level of the Trp peak was shown, from both MBP-WT/Sim and a Trp-less protein made by the universal genetic code from a mutant MBP mRNA, in which all of the UGG codons were altered to GCU (W_all_A/Univ). This disappearance of Trp was also confirmed by comparing the fluorescence spectra of MBP-WT/Sim and MBP-WT/Univ (Supplementary Figure S2). On the other hand, another composition analysis revealed that only the amount of Ala was significantly increased in MBP-WT/Sim, as compared to MBP-WT/Univ (Figure 2C). Indeed, the increased amount of Ala was estimated as eight residues (Supplementary Table S1), which is equivalent to the number of UGG codons in the MBP-WT mRNA.
Figure 1.
Design strategy of the simplified genetic code. (A) Schematic diagram of the simplified genetic code. For the reassignment, the prevention of Trp incorporation and the addition of an adapter molecule that connects the translation pathway between the UGG codon and Ala are required. (B) The nucleotide sequences of the anticodon stem loop of the tRNAAla variant and the UGG codon on the mRNA. The anticodon loop of tRNAAla was substituted for that of tRNATrp. Positions 32 and 38 in the anticodon loop are numbered.
Figure 2.
Reassignment of UGG codons by the tRNAAla variant. (A) MBP was translated under the conditions noted at the top of each lane. An autoradiogram of a polyacrylamide gel, with the products labeled with [14C] Leu, is shown. (B) Chromatograms of the amino acids obtained from MBP hydrolysates with MSA. The complete chromatograms are shown in Supplementary Figure S1. (C) Chromatograms of the amino acids obtained from MBP hydrolysates with HCl. ‘NH3’ indicates the peak of ammonia. ‘AMQ’ indicates the peak of 6-aminoquinoline, a by-product of the derivatization of amino acids. ‘Std’ indicates the peak of norvaline, an internal standard. The dashed line indicates the peak height of each amino acid from MBP-WT/Univ.
To demonstrate that the protein synthesis by the simplified genetic code occurs with efficiency and accuracy comparable to those of the universal genetic code, we compared the fluorescence of GFP mutants synthesized by the simplified and the universal codes (Figure 3A and Supplementary Figure S3). We used a mutant mRNA named gfpΔUGG, which lacks UGG codons and thus encodes a Trp-less GFP. When the mutant mRNA was used as the template, the fluorescent intensities of the proteins synthesized by the simplified (gfpΔUGG/Sim) and the universal (gfpΔUGG /Univ) code were nearly the same. We also prepared another mutant mRNA, gfpΔUGG-A110X(UGG), where the GCU codon for Ala110 of gfpΔUGG was mutated to UGG. The fluorescent intensity of the translation product of this mRNA by the simplified code [gfpΔUGG-A110X(UGG)/Sim] was the same as those of gfpΔUGG/Sim and gfpΔUGG/Univ. These results indicate that the simplified code translates most codons, including UGG, with efficiency and accuracy comparable to those of the universal code.
Figure 3.

The translation efficiency and accuracy of the simplified code are comparable to those of the universal code. (A) The fluorescent intensities of GFP mutants synthesized by the simplified and universal codes. Results are presented as means ± SD (n = 3). (B) Superposition of the overall crystal structures of gfpΔUGG-A110X(UGG)/Sim (yellow) and gfpΔUGG/Univ (green). The chromophore is shown by a stick representation. (C) Detailed views of the amino acid at position 110 and Phe114 of gfpΔUGG-A110X(UGG)/Sim or gfpΔUGG/Univ, in ball-and-stick representations. The 2Fo − Fc map is contoured at 1.0 σ and overlaid on the model. The color code is as in (B).
In contrast to the simplified code, the universal code could not produce an active protein from the mRNA because Trp occupied position 110, where Ala was located in gfpΔUGG/Univ, and also probably in gfpΔUGG/Sim and gfpΔUGG-A110X(UGG)/Sim. This relationship between the two codes in the translation of the gfpΔUGG-A110X(UGG) mRNA is opposite to that in the translation of the other GFP mutant mRNA (gfpS1) (20). In the production of an inactive protein from the mRNA by the simplified code, Ala is likely to be inserted in response to the UGG codon at position 57. The residue at position 57 is buried in the interior of the protein, and the Ala replaces the Trp required for the activity of gfpS1/Univ.
To verify the correct overall structure of the protein generated by the simplified code and the precise reassignment of the UGG codon, we determined the crystal structures of gfpΔUGG-A110X(UGG)/Sim and gfpΔUGG/Univ. The root mean square deviation between them was 0.151 Å, indicating that the overall structures of these two proteins were almost identical (Figure 3B). This structural identity suggests that the simplified code synthesized the correctly folded protein. Moreover, the residue at position 110 in gfpΔUGG-A110X(UGG)/Sim and Ala110 in gfpΔUGG/Univ seem to be identical, by a comparison of the two electron density maps (Figure 3C). On the other hand, the residue is apparently distinct from those with bulky side chains, such as Trp or Phe, showing the UGG reassignment to Ala in the simplified code.
Another simplified code synthesizes an active enzyme
We performed another reassignment, by constructing a simplified genetic code in which serine (Ser), instead of cysteine (Cys), is reassigned to UGU/UGC codons by a tRNASer variant (Supplementary Figure S4). The tRNASer variant has an anticodon loop corresponding to the UGU/UGC codons (Supplementary Figure S4A). Ser is considered to be attached to the tRNASer variant by seryl-tRNA synthetase (SerRS), because SerRS does not recognize the anticodon loop (30–32). To confirm the reassignment, we translated CAT mRNA, containing five UGU/UGC codons, in the reaction mixture without Cys. This reassignment also seemed to be successful, as judged from the translation inhibition due to the lack of Cys-tRNACys, and the translation recovery by the addition of the tRNASer variant with the anticodon loop corresponding to the UGU/UGC codons (Supplementary Figure S4B). To confirm the reassignment, we compared the amino acid compositions of the two translation products synthesized from the CAT mRNA by the simplified code (CAT-WT/Sim) and the universal code (CAT-WT/Univ). In CAT-WT/Sim, the number of Cys residues was decreased by 5, while that of Ser was increased by 5, as compared to CAT-WT/Univ (Supplementary Figure S4C and Supplementary Table S3).
To assess whether the simplified code that reassigns UGU/UGC codons to Ser can also synthesize proteins efficiently and accurately, we evaluated the activity of E. coli LexA protease. The wild-type mRNA encoding LexA lacks UGU/UGC codons, and contains the UCG-coded Ser at position 119 in its active center (33), which plays an indispensable role in substrate hydrolysis. To inhibit the self-cleavage activity, we prepared the LexA-G85D mutant (21), denoted as LexA*. The mRNA of LexA* was further mutated, by replacing the UCG codon at position 119 with UGC, thus creating LexA*-S119X(UGC). The protein translated from the mutant mRNA by the simplified code [LexA*-S119X(UGC)/Sim] (Figure 4, lanes 1 and 2) cleaved the substrate efficiently, in a comparable manner to LexA*-WT/Univ (Figure 4, lanes 3 and 4), as expected from the reassignment of UGU/UGC codons to Ser. In contrast, the protein translated from LexA*-S119X(UGC) by the universal code (LexA*-S119X(UGC)/Univ) completely lacked substrate cleavage activity (Figure 4, lanes 5 and 6).
Figure 4.
Comparison of the activities between LexA mutants translated by the simplified and the universal code. The enzymes were incubated with the substrate. WT: the LexA G85D mutant mRNA, denoted as LexA* in the main text. S119X(UGC): the LexA mutant mRNA in which the UCG codon at position 119 of LexA* was mutated to UGC. Sim: a simplified genetic code in which Ser is reassigned to UGU/UGC codons. Univ: the Universal genetic code. E: position of the LexA enzymes. S: position of the intact substrate. P: position of the cleaved product.
DISCUSSION
In this study, we created genetic codes comprising fewer than 20 amino acids. We first removed specific amino acids from the E. coli S30 cell-free reaction mixture, to eliminate the targeted endogenous translation pathways connecting the specific amino acids and the codons. We then added the tRNA variant with the altered anticodon loop, to reassign Ala or Ser to the unassigned codons. The efficiency and accuracy of protein synthesis by the Trp-less code were comparable to those of the universal code (Figure 3A). These results show that the existence of appropriate translation pathways allows the design of simplified genetic codes, which are composed of fewer than 20 amino acids and retain the ability to synthesize active proteins.
The composition and the order of appearance of the amino acids in the primordial genetic codes have long fascinated researchers (1,9). Trp is thought to be the last amino acid included within the genetic code, according to Trifonov’s report (34), which summarized various studies about the order of the amino acid appearance. According to another report based on the biosynthesis of the canonical amino acids (9), Ser is the amino acid that utilized the UGU/UGC codons before Cys. This idea is supported by the reactions of some methanogenic archaea, in which phosphoserine is first attached to tRNACys and then converted enzymatically to Cys (15). This conversion is also seen in selenocysteine (Sec) formation, in which bacteria convert Ser-tRNASec to Sec-tRNASec (35). The simplified code that reassigns Ser to the UGU/UGC codon may represent one of the primordial genetic codes.
We showed that an active protein is synthesized only from the appropriate combination of a genetic code and an mRNA (Figure 3A). For example, the gfpΔUGG-A110X(UGG) mRNA synthesizes the active GFP variant by the simplified code, but not by the universal code. In contrast, the gfpS1 mRNA synthesizes the other active protein by the universal code, but not by the simplified code. This one-to-one correlation between genetic codes and mRNAs implies that the simplified genetic code would have functioned as a barrier to horizontal transfer (36), when we imagine primitive organisms utilizing 19 amino acids. The organism that started utilizing 20 amino acids should have acquired enhanced protein properties or competitive advantages (37), but it would have lost the opportunity to derive the benefit of horizontal transfer from the other organisms that kept utilizing 19 amino acids. This barrier, in turn, excludes the old organisms after the population of 20-amino-acid organisms expanded sufficiently to create a gene pool for horizontal transfer among them. In fact, even in the case of a deviant code composed of 20 amino acids, it has been suggested that there was less horizontal transfer during recent evolution, due to the use of UGA-encoding tryptophan codons in Mycoplasmas (38,39). From an engineering viewpoint, this containment by the simplified code may also be useful to prevent the diffusion of non-natural genes from the laboratory to natural organisms (40).
The advantage of a cell-free system is that one can easily add or remove its components. To remove a specific amino acid from the genetic code, inhibition of the aminoacylation of the corresponding tRNA is required. It is obvious that the inhibition in vivo would lead to a lethal effect caused by the early translation termination of essential genes. To deal with this problem, we used the E. coli S30 cell-free translation system lacking a specific amino acid. We considered that, in some cases, we should eliminate the activities of the enzymes involved in the biosynthesis of the amino acids removed from the cell-free extract (41). Indeed, a minor band appeared under the conditions without cysteine (Supplementary Figure S4B, lane 2). It was presumably generated by the incorporation of the newly synthesized cysteine. We inhibited the charging of cysteine, through the inhibition of CysRS by the addition of Cys-SA. As expected, the minor band disappeared after this procedure (Supplementary Figure S4B, lane 3).
Our simplified genetic code will provide an efficient tool for the experimental evolution of proteins under conditions with a limited set of amino acids, to assess the functions of primordial proteins and to improve the utility of such proteins for clinical use (42). To create a protein with improved activity relative to that of the wild-type, random mutagenesis by an error-prone polymerase chain reaction is widely used in a directed evolution process involving multiple rounds of mutagenesis and selection. For a simplified protein, however, efficient evolution with the random mutation strategy has been prevented by the reappearance of codons, generated by mutation, for the specific amino acids to be excluded. One possible way to avoid such reappearances of specific codons by random mutations might be the usage of specialized oligo DNAs that lack the specific codons, as in previous studies (34,35). However, this strategy has not applied to directed evolution because of two difficulties: (i) the preparation of the specialized oligos was time-consuming and costly, and (ii) laborious step-wise construction was required, due to the limited search ability of the specialized oligo DNA encoding only a small segment of the entire amino acid sequence. As a result, the activities of the obtained proteins remained the same as that of the wild-type (43,44). Due to these restrictions in the directed evolution procedure, the vast sequence space of simplified proteins remains unsearched, in contrast to that of proteins composed of all 20 amino acids. In this work, we showed that the simplified code completely excludes the specific amino acid from the genetic code. Therefore, even if the codon for the specific amino acid in the universal code appears in the sequence through a mutation, the amino acid to be excluded is not incorporated within the protein. As a result, the simplified codes will allow us to efficiently search the sequence space of simplified proteins, by applying directed evolution with conventional random mutagenesis methods. Using this strategy, we will be able to simplify proteins, including those involved in the translational machinery. Other variations of simplified codes comprising 19 or fewer amino acids would provide further implications for simplified proteins. Since AlaRS and SerRS do not recognize the anticodon loop of their cognate tRNAs, work in progress in our laboratory indicates the similar exclusion of other amino acids by using the specific a.a.-SA and the tRNA variant(s) with the corresponding anticodon(s). Furthermore, multiple amino acids will simultaneously be excluded by using several sets of a.a.-SA and tRNA variant(s).
Some natural and engineered organisms, as well as our in vitro work, have employed tRNA variants with a different anticodon from those within the universal code, although those organisms still utilize 20 amino acids. For example, C. cylindracea completely reassigns the CUG codon from Leu to Ser (4,45) by using a tRNASer with the anticodon sequence CAG (tRNASerCAG). The tRNASerCAG is considered to have competed with the wild-type tRNALeu for the CUG codon, and generated significant ambiguity in the codon for ∼100 My (46). The tRNASerCAG was selected in this organism, thus reassigning the CUG identity from Leu to Ser. In a similar manner to the past competition in the ancestor of C. cylindracea, some in vivo engineering studies (47,48) have used tRNAAla variants or tRNASer variants. In these studies, the competition between the tRNA variant and an endogenous tRNA for a sense codon also results in the ambiguous assignment of the codon. Further elimination of the competing components, as in the case of C. cylindracea, would completely reassign Ala or Ser to the codon. These in vivo uses of the tRNAAla variant or tRNASer variant also suggest future engineering to simplify the in vivo code, although all of the housekeeping proteins must retain their activities without the specific amino acid. Advancing technologies, such as genome engineering (49) and experimental evolution of organisms (50–52), will facilitate the generation of organisms that utilize only 19 amino acids in their genetic code.
ACCESSION NUMBERS
The atomic coordinates have been deposited in the Protein Data Bank, under the accession numbers 3UFZ and 3UG0. Sequence data for new mutants are available from GenBank/EMBL/DDBJ under accession numbers AB670686 through AB670691.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–3, Supplementary Figures 1–4, Supplementary Methods and Supplementary References [23,53–58].
FUNDING
KAKENHI programs [219606 to A.K.-K., 201464 to Y.A., 19680016, 21680026, and 23119005 to D.K.]; Japan Society for the Promotion of Science (JSPS); Ministry of Education, Culture, Sports, Science and Technology (MEXT); FIRST program from JSPS (to O.N.); National Project on Protein Structural and Functional Analyses from MEXT (to O.N.); Industrial Technology Research Grant Program in 2005 from the New Energy and Industrial Technology Development Organization (NEDO) (to D.K.); Chemical Genomics Research Project, RIKEN Advanced Science Institute (to N.D.). Funding for open access charge: KAKENHI program [23119005 to D.K.].
Conflicts of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors are grateful to Y. Tomo for technical assistance with the CF translation. The authors thank the Support Unit for Bio-material Analysis, RIKEN BSI Research Resources Center, especially H. Morishita, for the ninhydrin-based amino acid composition analysis. The authors also thank E. Seki for providing advice and materials for CF translation, and K. Shiba, T. Ueda, Y. Shimizu and H. Nakayama for helpful discussions.
REFERENCES
- 1.Crick FH. The origin of the genetic code. J. Mol. Biol. 1968;38:367–379. doi: 10.1016/0022-2836(68)90392-6. [DOI] [PubMed] [Google Scholar]
- 2.Barrell BG, Bankier AT, Drouin J. A different genetic code in human mitochondria. Nature. 1979;282:189–194. doi: 10.1038/282189a0. [DOI] [PubMed] [Google Scholar]
- 3.Yamao F, Muto A, Kawauchi Y, Iwami M, Iwagami S, Azumi Y, Osawa S. UGA is read as tryptophan in Mycoplasma capricolum. Proc. Natl Acad. Sci. USA. 1985;82:2306–2309. doi: 10.1073/pnas.82.8.2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kawaguchi Y, Honda H, Taniguchi-Morimura J, Iwasaki S. The codon CUG is read as serine in an asporogenic yeast Candida cylindracea. Nature. 1989;341:164–166. doi: 10.1038/341164a0. [DOI] [PubMed] [Google Scholar]
- 5.Wang L, Brock A, Herberich B, Schultz PG. Expanding the genetic code of Escherichia coli. Science. 2001;292:498–500. doi: 10.1126/science.1060077. [DOI] [PubMed] [Google Scholar]
- 6.Kiga D, Sakamoto K, Kodama K, Kigawa T, Matsuda T, Yabuki T, Shirouzu M, Harada Y, Nakayama H, Takio K, et al. An engineered Escherichia coli tyrosyl-tRNA synthetase for site-specific incorporation of an unnatural amino acid into proteins in eukaryotic translation and its application in a wheat germ cell-free system. Proc. Natl Acad. Sci. USA. 2002;99:9715–9720. doi: 10.1073/pnas.142220099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu CC, Schultz PG. Adding new chemistries to the genetic code. Annu. Rev. Biochem. 2010;79:413–444. doi: 10.1146/annurev.biochem.052308.105824. [DOI] [PubMed] [Google Scholar]
- 8.Srinivasan G, James CM, Krzycki JA. Pyrrolysine encoded by UAG in Archaea: charging of a UAG-decoding specialized tRNA. Science. 2002;296:1459–1462. doi: 10.1126/science.1069588. [DOI] [PubMed] [Google Scholar]
- 9.Wong JT. A co-evolution theory of the genetic code. Proc. Natl Acad. Sci. USA. 1975;72:1909–1912. doi: 10.1073/pnas.72.5.1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jordan IK, Kondrashov FA, Adzhubei IA, Wolf YI, Koonin EV, Kondrashov AS, Sunyaev S. A universal trend of amino acid gain and loss in protein evolution. Nature. 2005;433:633–638. doi: 10.1038/nature03306. [DOI] [PubMed] [Google Scholar]
- 11.Miller SL. A production of amino acids under possible primitive earth conditions. Science. 1953;117:528–529. doi: 10.1126/science.117.3046.528. [DOI] [PubMed] [Google Scholar]
- 12.Cleaves HJ, Chalmers JH, Lazcano A, Miller SL, Bada JL. A reassessment of prebiotic organic synthesis in neutral planetary atmospheres. Orig. Life Evol. Biosph. 2008;38:105–115. doi: 10.1007/s11084-007-9120-3. [DOI] [PubMed] [Google Scholar]
- 13.Woese CR, Olsen GJ, Ibba M, Söll D. Aminoacyl-tRNA synthetases, the genetic code, and the evolutionary process. Microbiol. Mol. Biol. Rev. 2000;64:202–236. doi: 10.1128/mmbr.64.1.202-236.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ibba M, Söll D. Aminoacyl-tRNA synthesis. Annu. Rev. Biochem. 2000;69:617–650. doi: 10.1146/annurev.biochem.69.1.617. [DOI] [PubMed] [Google Scholar]
- 15.Sauerwald A, Zhu W, Major TA, Roy H, Palioura S, Jahn D, Whitman WB, Yates JR, 3rd, Ibba M, Söll D. RNA-dependent cysteine biosynthesis in archaea. Science. 2005;307:1969–1972. doi: 10.1126/science.1108329. [DOI] [PubMed] [Google Scholar]
- 16.Oshikane H, Sheppard K, Fukai S, Nakamura Y, Ishitani R, Numata T, Sherrer RL, Feng L, Schmitt E, Panvert M, et al. Structural basis of RNA-dependent recruitment of glutamine to the genetic code. Science. 2006;312:1950–1954. doi: 10.1126/science.1128470. [DOI] [PubMed] [Google Scholar]
- 17.Ito T, Yokoyama S. Two enzymes bound to one transfer RNA assume alternative conformations for consecutive reactions. Nature. 2010;467:612–616. doi: 10.1038/nature09411. [DOI] [PubMed] [Google Scholar]
- 18.Blaise M, Bailly M, Frechin M, Behrens MA, Fischer F, Oliveira CL, Becker HD, Pedersen JS, Thirup S, Kern D. Crystal structure of a transfer-ribonucleoprotein particle that promotes asparagine formation. EMBO J. 2010;29:3118–3129. doi: 10.1038/emboj.2010.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim DM, Kigawa T, Choi CY, Yokoyama S. A highly efficient cell-free protein synthesis system from Escherichia coli. Eur. J. Biochem. 1996;239:881–886. doi: 10.1111/j.1432-1033.1996.0881u.x. [DOI] [PubMed] [Google Scholar]
- 20.Seki E, Matsuda N, Yokoyama S, Kigawa T. Cell-free protein synthesis system from Escherichia coli cells cultured at decreased temperatures improves productivity by decreasing DNA template degradation. Anal. Biochem. 2008;377:156–161. doi: 10.1016/j.ab.2008.03.001. [DOI] [PubMed] [Google Scholar]
- 21.Kim B, Little JW. LexA and lambda Cl repressors as enzymes: specific cleavage in an intermolecular reaction. Cell. 1993;73:1165–1173. doi: 10.1016/0092-8674(93)90645-7. [DOI] [PubMed] [Google Scholar]
- 22.Kigawa T, Yabuki T, Matsuda N, Matsuda T, Nakajima R, Tanaka A, Yokoyama S. Preparation of Escherichia coli cell extract for highly productive cell-free protein expression. J. Struct. Funct. Genomics. 2004;5:63–68. doi: 10.1023/B:JSFG.0000029204.57846.7d. [DOI] [PubMed] [Google Scholar]
- 23.Kigawa T, Inoue M, Aoki M, Matsuda T, Yabuki T, Seki E, Harada T, Watanabe S, Yokoyama S. In: Cell-Free Protein Synthesis: Methods And Protocols. Spirin AS, Swartz JR, editors. Weinheim, Germany: Wiley-VCH; 2007. pp. 99–109. [Google Scholar]
- 24.Simpson RJ, Neuberger MR, Liu TY. Complete amino acid analysis of proteins from a single hydrolysate. J. Biol. Chem. 1976;251:1936–1940. [PubMed] [Google Scholar]
- 25.Masuda A, Dohmae N. Automated protein hydrolysis delivering sample to a solid acid catalyst for amino acid analysis. Anal. Chem. 2010;82:8939–8945. doi: 10.1021/ac101718x. [DOI] [PubMed] [Google Scholar]
- 26.Kigawa T, Matsuda T, Yabuki T, Yokoyama S. In: Cell-Free Protein Synthesis: Methods And Protocols. Spirin AS, Swartz JR, editors. Weinheim, Germany: Wiley-VCH; 2007. pp. 83–97. [Google Scholar]
- 27.Hou YM, Schimmel P. A simple structural feature is a major determinant of the identity of a transfer RNA. Nature. 1988;333:140–145. doi: 10.1038/333140a0. [DOI] [PubMed] [Google Scholar]
- 28.Francklyn C, Schimmel P. Aminoacylation of RNA minihelices with alanine. Nature. 1989;337:478–481. doi: 10.1038/337478a0. [DOI] [PubMed] [Google Scholar]
- 29.Guo M, Chong YE, Beebe K, Shapiro R, Yang XL, Schimmel P. The C-Ala domain brings together editing and aminoacylation functions on one tRNA. Science. 2009;325:744–747. doi: 10.1126/science.1174343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sundharadas G, Katze JR, Söll D, Konigsberg W, Lengyel P. On the recognition of serine transfer RNA's specific for unrelated codons by the same seryl-transfer RNA synthetase. Proc. Natl Acad. Sci. USA. 1968;61:693–700. doi: 10.1073/pnas.61.2.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sampson JR, Saks ME. Contributions of discrete tRNA(Ser) domains to aminoacylation by E.coli seryl-tRNA synthetase: a kinetic analysis using model RNA substrates. Nucleic Acids Res. 1993;21:4467–4475. doi: 10.1093/nar/21.19.4467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cusack S, Yaremchuk A, Tukalo M. The crystal structure of the ternary complex of T.thermophilus seryl-tRNA synthetase with tRNA(Ser) and a seryl-adenylate analogue reveals a conformational switch in the active site. EMBO J. 1996;15:2834–2842. [PMC free article] [PubMed] [Google Scholar]
- 33.Slilaty SN, Little JW. Lysine-156 and serine-119 are required for LexA repressor cleavage: a possible mechanism. Proc. Natl Acad. Sci. USA. 1987;84:3987–3991. doi: 10.1073/pnas.84.12.3987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Trifonov EN. The triplet code from first principles. J. Biomol. Struct. Dyn. 2004;22:1–11. doi: 10.1080/07391102.2004.10506975. [DOI] [PubMed] [Google Scholar]
- 35.Yoshizawa S, Bock A. The many levels of control on bacterial selenoprotein synthesis. Biochim. Biophys. Acta. 2009;1790:1404–1414. doi: 10.1016/j.bbagen.2009.03.010. [DOI] [PubMed] [Google Scholar]
- 36.Silva RM, Paredes JA, Moura GR, Manadas B, Lima-Costa T, Rocha R, Miranda I, Gomes AC, Koerkamp MJ, Perrot M, et al. Critical roles for a genetic code alteration in the evolution of the genus Candida. EMBO J. 2007;26:4555–4565. doi: 10.1038/sj.emboj.7601876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wiltschi B, Budisa N. Natural history and experimental evolution of the genetic code. Appl. Microbiol. Biotechnol. 2007;74:739–753. doi: 10.1007/s00253-006-0823-6. [DOI] [PubMed] [Google Scholar]
- 38.Chambaud I, Heilig R, Ferris S, Barbe V, Samson D, Galisson F, Moszer I, Dybvig K, Wroblewski H, Viari A, et al. The complete genome sequence of the murine respiratory pathogen Mycoplasma pulmonis. Nucleic Acids Res. 2001;29:2145–2153. doi: 10.1093/nar/29.10.2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Koonin EV, Makarova KS, Aravind L. Horizontal gene transfer in prokaryotes: quantification and classification. Annu. Rev. Microbiol. 2001;55:709–742. doi: 10.1146/annurev.micro.55.1.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Carr PA, Church GM. Genome engineering. Nat. Biotechnol. 2009;27:1151–1162. doi: 10.1038/nbt.1590. [DOI] [PubMed] [Google Scholar]
- 41.Yokoyama J, Matsuda T, Koshiba S, Kigawa T. An economical method for producing stable-isotope labeled proteins by the E. coli cell-free system. J. Biomol. NMR. 2010;48:193–201. doi: 10.1007/s10858-010-9455-3. [DOI] [PubMed] [Google Scholar]
- 42.Yamamoto Y, Tsutsumi Y, Yoshioka Y, Nishibata T, Kobayashi K, Okamoto T, Mukai Y, Shimizu T, Nakagawa S, Nagata S, et al. Site-specific PEGylation of a lysine-deficient TNF-alpha with full bioactivity. Nat. Biotechnol. 2003;21:546–552. doi: 10.1038/nbt812. [DOI] [PubMed] [Google Scholar]
- 43.Akanuma S, Kigawa T, Yokoyama S. Combinatorial mutagenesis to restrict amino acid usage in an enzyme to a reduced set. Proc. Natl Acad. Sci. USA. 2002;99:13549–13553. doi: 10.1073/pnas.222243999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Walter KU, Vamvaca K, Hilvert D. An active enzyme constructed from a 9-amino acid alphabet. J. Biol. Chem. 2005;280:37742–37746. doi: 10.1074/jbc.M507210200. [DOI] [PubMed] [Google Scholar]
- 45.Yokogawa T, Suzuki T, Ueda T, Mori M, Ohama T, Kuchino Y, Yoshinari S, Motoki I, Nishikawa K, Osawa S, et al. Serine tRNA complementary to the nonuniversal serine codon CUG in Candida cylindracea: evolutionary implications. Proc. Natl Acad. Sci. USA. 1992;89:7408–7411. doi: 10.1073/pnas.89.16.7408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Massey SE, Moura G, Beltrao P, Almeida R, Garey JR, Tuite MF, Santos MA. Comparative evolutionary genomics unveils the molecular mechanism of reassignment of the CTG codon in Candida spp. Genome Res. 2003;13:544–557. doi: 10.1101/gr.811003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dorazi R, Lingutla JJ, Humayun MZ. Expression of mutant alanine tRNAs increases spontaneous mutagenesis in Escherichia coli. Mol. Microbiol. 2002;44:131–141. doi: 10.1046/j.1365-2958.2002.02847.x. [DOI] [PubMed] [Google Scholar]
- 48.Geslain R, Cubells L, Bori-Sanz T, Alvarez-Medina R, Rossell D, Marti E, Ribas de Pouplana L. Chimeric tRNAs as tools to induce proteome damage and identify components of stress responses. Nucleic Acids Res. 2010;38:e30. doi: 10.1093/nar/gkp1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Isaacs FJ, Carr PA, Wang HH, Lajoie MJ, Sterling B, Kraal L, Tolonen AC, Gianoulis TA, Goodman DB, Reppas NB, et al. Precise manipulation of chromosomes in vivo enables genome-wide codon replacement. Science. 2011;333:348–353. doi: 10.1126/science.1205822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bacher JM, Ellington AD. Selection and characterization of Escherichia coli variants capable of growth on an otherwise toxic tryptophan analogue. J. Bacteriol. 2001;183:5414–5425. doi: 10.1128/JB.183.18.5414-5425.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mat WK, Xue H, Wong JT. Genetic code mutations: the breaking of a three billion year invariance. PLoS One. 2010;5:e12206. doi: 10.1371/journal.pone.0012206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Marliere P, Patrouix J, Doring V, Herdewijn P, Tricot S, Cruveiller S, Bouzon M, Mutzel R. Chemical evolution of a bacterium's genome. Angew. Chem. Int. Ed. Engl. 2011;50:7109–7114. doi: 10.1002/anie.201100535. [DOI] [PubMed] [Google Scholar]
- 53.Matsuda T, Kigawa T, Koshiba S, Inoue M, Aoki M, Yamasaki K, Seki M, Shinozaki K, Yokoyama S. Cell-free synthesis of zinc-binding proteins. J. Struct. Funct. Genomics. 2006;7:93–100. doi: 10.1007/s10969-006-9012-1. [DOI] [PubMed] [Google Scholar]
- 54.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 55.Vagin A, Teplyakov A. MOLREP: an automated program for molecular replacement. J. Appl. Crystallogr. 1997;30:1022–1025. [Google Scholar]
- 56.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 57.Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 58.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.