Abstract
In human mitochondria the transcription machinery generates the RNA primers needed for initiation of DNA replication. A critical feature of the leading-strand origin of mitochondrial DNA replication is a CG-rich element denoted conserved sequence block II (CSB II). During transcription of CSB II, a G-quadruplex structure forms in the nascent RNA, which stimulates transcription termination and primer formation. Previous studies have shown that the newly synthesized primers form a stable and persistent RNA–DNA hybrid, a R-loop, near the leading-strand origin of DNA replication. We here demonstrate that the unusual behavior of the RNA primer is explained by the formation of a stable G-quadruplex structure, involving the CSB II region in both the nascent RNA and the non-template DNA strand. Based on our data, we suggest that G-quadruplex formation between nascent RNA and the non-template DNA strand may be a regulated event, which decides the fate of RNA primers and ultimately the rate of initiation of DNA synthesis in human mitochondria.
Introduction
Human mitochondrial DNA (mtDNA) is a double-stranded circular molecule that encodes essential subunits of the mitochondrial respiratory chain, as well as the tRNAs and rRNAs required for their synthesis. The genome contains two major promoters referred to as the light strand promoter (LSP) and the heavy strand promoter (HSP). Transcription initiated at both LSP and HSP is polycistronic, producing near genome-length transcripts that are later processed to give rise to the individual mRNA molecules (1–3). Mitochondrial transcription can be reconstituted in vitro using only three recombinant proteins: the mitochondrial RNA polymerase (POLRMT), transcription factor A (TFAM) and transcription factor B2 (TFB2M) (4). In mitochondria, transcription and DNA replication are closely linked, since POLRMT is responsible for synthesis of the primers required for initiation of DNA synthesis by the mtDNA polymerase γ (POLγ) from the mitochondrial origins of replication, OriH and OriL (5–9). In addition to POLγ, the mitochondrial replication machinery also consists of a replicative helicase, TWINKLE (10,11), and a single-stranded DNA binding protein, mtSSB (12,13). In combination, these three factors form a replication machinery that can synthesize ssDNA molecules longer than 16.5 kb, the size of the mtDNA genome (14).
RNA primers required for initiation of leading-strand DNA replication at OriH are produced by transcription events initiated at LSP. According to one model, the 3′-end of the RNA primers is defined by premature termination of transcription at a specific DNA element, the conserved sequence block II (CSB II) (7). During transcription of CSB II a G-quadruplex structure is formed in the nascent RNA strand. The G-quadruplex structure stimulates premature termination of transcription, probably by weakening RNA-to-polymerase contacts, which in turn destabilizes the elongation complex (15). G-quadruplexes are four-stranded structures that can form in guanine-rich sequences by stacking of planar G tetrads or quartets. The structures are very stable and occur in the human genome at e.g. telomeres and immunoglobulin switch regions (16–18). There are also examples of hybrid-type G-quadruplex formation between RNA and DNA, e.g. at human telomeres where telomere RNA and telomere DNA can associate to form a parallel G-quadruplex structure (19).
In addition to its role in transcription termination, CSB II is also required for the formation of a persistent RNA–DNA hybrid between transcript RNA and template DNA, a fact that has been reported in yeast, mouse and human (20–22). The RNA strand in this stable RNA–DNA hybrid is believed to act as a primer for initiation of leading-strand replication from OriH, but the molecular details are not understood. There are also two additional Conserved Sequence Blocks (CSB I and III), but their molecular role remains to be established. It is worth noting that mammalian mitochondria cannot be transfected and structure–function analysis of conserved sequence elements in vivo is, therefore, difficult to perform.
Even though the transitions between RNA and DNA at OriH have been carefully mapped in vivo, initiation of DNA synthesis at this origin has not yet been reconstituted in vitro [(6,7) and unpublished data]. The molecular basis for the stability of the RNA–DNA hybrid at CSB II and the reason why RNA primers cannot be utilized for initiation of DNA synthesis by the mitochondrial replisome in vitro still remain to be explained. In contrast, how DNA synthesis is initiated at the major origin of lagging-strand DNA replication, OriL, is today fairly well understood. When the leading-strand DNA replication machinery passes OriL, the origin becomes single stranded and adopts a stem-loop conformation. POLRMT initiates primer synthesis from the single-stranded loop region and the primers formed are subsequently used to initiate lagging-strand DNA synthesis. The process of OriL activation has been reconstituted in vitro, using only purified, recombinant proteins and defined DNA templates (8,9).
In the present work, we investigate the molecular mechanism behind the formation of a stable RNA–DNA hybrid at CSB II and try to elucidate why the replication machinery cannot readily initiate DNA synthesis using the 3′ end of the primer formed by premature transcription termination at CSB II. We find that the stable RNA–DNA hybrid formed at CSB II is not due to normal Watson and Crick base pairing. Instead, a hybrid G-quadruplex structure is formed between the RNA transcript and the non-template DNA strand. Our results suggest that this structure removes the 3′ end of the RNA primer from the template strand and the primer is, therefore, not accessible for the mitochondrial replication machinery. We speculate that additional factors are needed to prevent the formation of this structure, or alternatively resolve it once formed, so that the RNA primer can be utilized for initiation of leading-strand DNA replication at OriH.
MATERIALS AND METHODS
Preparation of templates for transcription
POLRMT-dependent transcription was performed using either supercoiled plasmid DNA templates containing a DNA fragment corresponding to positions 1–477 of human mtDNA in pUC18, or a PCR product encompassing positions 1–512 of human mtDNA. Construction of the plasmid templates is described elsewhere (7,15). In the CSB II mutant template, positions 288–318 were substituted with positions 319–349 of human mtDNA. PCR product templates were generated by amplifying positions 1–477 of human mtDNA using KOD polymerase, according to the manufacturer’s recommendations. When indicated, 7-deaza-dGTP (New England Biolabs) replaced dGTP in the PCR reactions. Transcription using T7 RNA polymerase employed a template with the T7 promoter in pBluescript, followed by a DNA fragment corresponding to positions 1–393 of human mtDNA.
In vitro transcription
For mitochondrial transcription in vitro, recombinant human POLRMT, TFAM and TFB2M were expressed and purified from insect cells as described previously (4). Alternatively, POLRMT was expressed and purified from Escherichia coli ArcticExpress cells (Stratagene). Transcription reactions were carried out as described (15), and when indicated, GTP was substituted with 7-deaza-GTP (TriLink Technologies). Transcription using T7 RNA polymerase (New England Biolabs) was carried out according to the manufacturer’s instructions, but with 7-deaza-GTP substituted for GTP where indicated.
When the transcription reactions were treated with RNase, the transcription reactions were adjusted after 30 min transcription to 75 mM KCl, 20 mM MgCl2 and 10 mM DTT, and incubated with 150 ng RNase A (Fermentas) and/or 500 fmol of human RNaseH1 for 5 min at 32°C. Transcription products were separated on 6% or 12% acrylamide/urea gels in 1× TBE in parallel with a 100 bp ladder or a Low Molecular Weight marker (New England Biolabs), and visualized on X-ray film (Fujifilm). For quantification of the premature termination, gels were exposed to PhosphoImager plates (Fujifilm), and the relative molar amounts of the preterminated to the full-length transcription products were calculated.
Native gel analysis of G-quadruplex structures
Polynucleotide kinase T4 and 32P-γ-ATP were used for 5′ end labeling of oligonucleotides (see Table 1 for sequences). The labeled oligonucleotides were purified using a G-25 column (GE Healthcare). RNA G-quadruplexes were formed by incubating the RNA oligonucleotide (1 pmol/µl) at 95°C for 5 min, followed by 2 min on ice. Samples were adjusted to 1 × TE and the indicated concentration of KCl, followed by incubation overnight at 37°C. For formation of RNA–DNA hybrid G-quadruplexes, 1 pmol/µl RNA and 4 pmol/ul DNA were incubated in buffer H (10 mM NaCl, 6 mM MgCl2, 1 mM CaCl2, 1 mM EDTA and 50 mM Tris pH 8) at 95°C for 5 min, followed by incubation overnight at 37°C. Products were separated on 12% native acrylamide gels run at 4°C in 0.5× TBE supplemented with 10 mM KCl. When RNase digestions were performed, either 200 ng RNase A (Fermentas) or 760 fmol hRNaseH1 were incubated with the sample for 5 min at 32°C in buffer S (75 mM KCl, 20 mM MgCl2 and 5 mM DTT).
Table 1.
Nucleotide sequences of the CSB II spanning RNA and DNA oligonucleotides and their mutant derivatives used in this study
| WT RNA | 5′-GAA GCG GGG GAG GGG GGG UUU GGU GGA AAU-3′ |
| G→A mut RNA | 5′-GAA GCA AAA AAA AAA AAA UUU AAU GGA AAU-3′ |
| G→T mut RNA | 5′-GAA GCU UUU UAU UUU UUU UUU UUU GGA AAU-3′ |
| WT DNA | 5′-GAA GCG GGG GAG GGG GGG TTT GGT GGA AAT-3′ |
| G→A mut | 5′-GAA GCA AAA AAA AAA AAA TTT AAT GGA AAT-3′ |
| G→T mut | 5′-GAA GCT TTT TAT TTT TTT TTT TTT GGA AAT-3′ |
| 7-deaza WT | 5′-GAA GCG GGG GAG GGG GGG TTT GGT GGA AAT-3′ |
| Rev comp. | 5′-ATT TCC ACC AAA CCC CCC CTC CCC CGC TTC-3′ |
| WT 55 | 5′-GAG ATG TGT TTA AGT GCT GTG GCC AGA AGC GGG GGA GGG GGG GTT TGG TGG AAA T-3′ |
| WT 75 | 5′-TTT GTT TTT GGG GTT TGG CAG AGA TGT GTT TAA GTG CTG TGG CCA GAA GCG GGG GAG GGG GGG TTT GGT GGA AAT-3′ |
| WT 100 | 5′-TCT GGT TAG GCT GGT GTT AGG GTT CTT TGT TTT TGG GGT TTG GCA GAG ATG TGT TTA AGT GCT GTG GCC AGA AGC GGG GGA GGG GGG GTT TGG TGG AAA T-3′ |
Underlined bases are 7-deaza-dGTP.
Primer extension assays
We cloned a DNA fragment corresponding to 1–477 nt of the mitochondrial human genome, containing either wildtype or G→A mut CSB II between the HindIII and BamHI sites in the pBluescript SK(+) vector (Agilent Technologies; La Jolla, CA). Constructs were confirmed by sequencing and used for ssDNA isolation as described (15). In order to stimulate G-quadruplex formation, one pmol of the indicated oligonucleotide was pretreated as described above to form G-quadruplex structures before annealing to the wt or G→A mut ssDNA template (2 pmol) at 37°C for 4 h in the presence of 100 mM KCl. In order to avoid G-quadruplex formation one pmol of the indicated oligonucleotide was directly annealed to the wt or G→A mut ssDNA template (2 pmol) in H2O at 95°C and then gradually cooled down to 25°C for 8 h. Primer extension reactions contained annealed template (100 fmol ssDNA), 200 µM dGTP, dATP or ddCTP, 10 µM dTTP, 2µCi [α-32 P]dTTP 8.5 mM MgCl2, 4 mM ATP, 6 mM DTT, 0.1 µg/BSA, 200 fmol POLγA and 400 fmol POLγB in a total volume of 20 µl. Reactions were incubated at 37°C for 30 min and the products were separated on a 6% denaturing PAGE gel run in parallel with molecular weight markers. The expected size of the primer extension product is 46 nt since it will stall when ddCTP is incorporated during DNA synthesis.
CD experiments
G-quadruplex formation was performed as described above, except that DNA and RNA samples were at 0.8 pmol/µl concentration in 150 mM KCl, 1 mM EDTA, and 10 mM Tris-Cl pH 8, whereas DNA–RNA hybrid samples contained 2 pmol/µl of each RNA and DNA in buffer H. CD is defined as the difference in absorbance of left and right circularly polarized light:
CD spectra were measured on a Jasco J-810 spectropolarimeter. Each spectrum is the average of five consecutive spectra and baseline-corrected with a spectrum of pure buffer.
Results
G-quadruplex formation in RNA, but not in DNA, directs premature termination of transcription at CSB II
We have earlier shown that a G-quadruplex structure is formed in RNA during transcription of CSB II, causing premature termination of mitochondrial transcription in vitro (15). During transcription, the double-stranded DNA template is unwound and the non-template strand may, therefore, also have the capacity to form a G-quadruplex structure, which could potentially influence transcription termination. We wanted to address this possibility in a reconstituted in vitro system for human mitochondrial transcription. To this end, we PCR-amplified a linear DNA template containing LSP, CSB II and OriH, corresponding to positions 1–477 of human mtDNA. The fragment was PCR amplified either in the presence of dGTP or in the presence of 7-deaza-dGTP. G-quadruplex formation requires Hoogsteen base pairing to N-7 of guanine, and 7-deaza-dGTP inhibits such base pairing, which allowed us to monitor if G-quadruplexes in template DNA affect transcription termination.
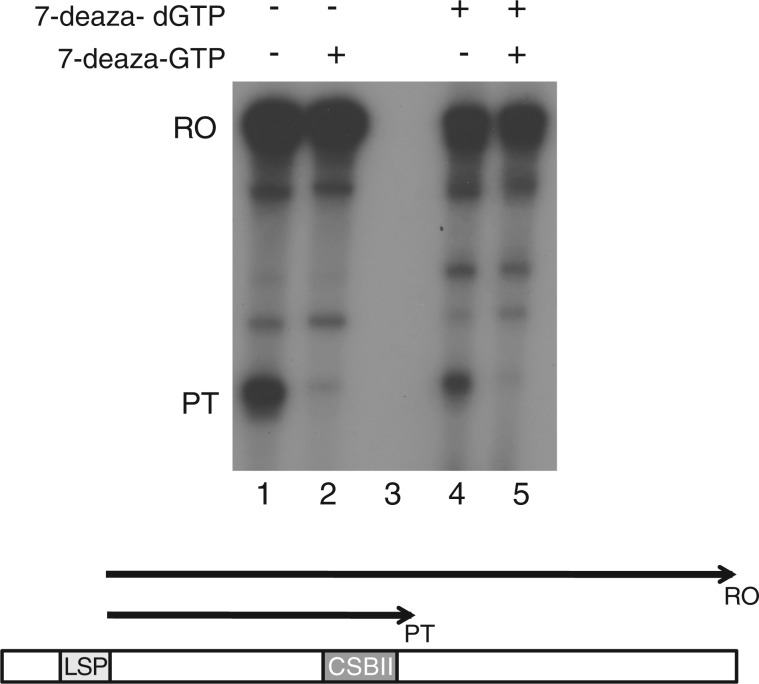
First, we used the normal DNA template that had been synthesized in the presence of dGTP. When we combined this template with purified POLRMT, TFAM, TFB2M and NTPs, we observed a 400 nt long run-off product (RO, Figure 1, lane 1). We also observed a shorter, prematurely terminated transcript of about 100 nt (PT, Figure 1). This shorter product was dependent on G-quadruplex structure formation in the nascent RNA strand, since it was almost completely lost when GTP was replaced with 7-deaza-GTP in the transcription reaction (Figure 1, lane 2). This finding was in agreement with a previous report from our laboratory, demonstrating that G-quadruplex formation in RNA causes premature termination of transcription near CSB II, about 100 nt downstream of LSP (15). To address if G-quadruplex structure formation in the DNA template could also influence premature transcription termination, we next used the PCR template in which we had replaced dGMP with 7-deaza-dGMP. The levels of premature termination on the 7-deaza-dGMP containing template were ∼50% and thus very similar to those observed on the control template synthesized with dGTP. Furthermore, premature transcription termination dropped dramatically when 7-deaza-GTP replaced GTP in the transcription reactions, and the effect was similar to that seen with the control template (Figure 1, lanes 2 and 5). We conclude that in contrast to RNA, G-quadruplex formation in the DNA template during transcription does not influence premature transcription termination at CSB II.
Figure 1.
G-quadruplex formation in nascent RNA, but not in template DNA, causes premature termination of transcription. In vitro transcription with the mitochondrial transcription apparatus was carried out in the presence or absence of 7-deaza-GTP to monitor effects of G-quadruplex formation in RNA. To address the effects of G-quadruplex formation in DNA, the DNA templates used were produced by PCR amplification of positions 1–512 of human mtDNA in the presence or absence of 7-deaza-dGTP.
The CSB II sequence forms an RNA–DNA hybrid G-quadruplex
An interesting feature of the primers formed during initiation of mtDNA replication is that they remain stably associated with the DNA, forming an R-loop. Others have suggested that this unusual feature is due to molecular interactions distinct from normal Watson–Crick base pairing (20) and we decided to investigate if the extraordinary stability of the R-loop was due to a hybrid G-quadruplex structure involving both RNA and DNA. To directly monitor G-quadruplex formation between RNA and DNA, we used a radioactively labeled 30-nt long RNA molecule spanning CSB II (positions 320–291 of the L-strand, see Table 1 for oligonucleotides used). The 3′ end of this RNA oligonucleotide corresponds to the major 3′ end of transcripts terminated at CSB II. In agreement with our previous report (15), the CSB II RNA oligonucleotide migrates significantly faster than a non-G-quadruplex forming oligonucleotide of the same length, in which all Gs have been replaced by As (Figure 2A, lanes 1 and 2). Two distinct species of G-quadruplexes could be observed, one presumably more compact than the other.
Figure 2.
Native gel analysis reveals hybrid G-quadruplex formation between RNA and DNA oligonucleotides spanning CSB II. (A) An RNA oligonucleotide encompassing CSB II migrates faster on native gels than a CSB II mutant molecule of identical length where all Gs have been replaced with As, indicative of G-quadruplex structure formation, as shown in (15). Products were analysed on 12% native acrylamide gels. M, marker lane; SM, size of marker. (B) A labeled CSB II RNA oligonucleotide was allowed to form G-quadruplexes either alone or in the presence of unlabeled DNA oligonucleotides and analysed as in panel A. A slower migrating species indicative of a RNA–DNA hybrid G-quadruplex was formed in the presence of a wildtype DNA oligonucleotide containing CSB II. (C) Hybrid G-quadruplexes formed between CSB II RNA (5′ end-labeled) and DNA oligonucleotides of indicated lengths. Nuclease treatment identifies a DNase I-resistant complex that contains both RNA and DNA (indicated with an asterisk).
We next combined the RNA oligonucleotide with DNA oligonucleotides encompassing CSB II. After incubation overnight to allow G-quadruplex formation, we analysed the results by separation on a non-denaturing polyacrylamide gel. In the absence of DNA, we observed the two fast-migrating species, corresponding to unimolecular G-quadruplex conformations (Figure 2B, lane 1). When a reverse complement DNA sequence (template strand) was incubated with the RNA, an RNA–DNA hybrid formed that exhibited normal migration expected for a double-stranded 30-bp fragment with normal Watson–Crick base pairing (Figure 2B, lane 5). It appears that when available, Watson–Crick interactions with the template strand oligonucleotide are preferred over formation of unimolecular RNA G-quadruplexes, as documented in the literature (23).
Next we used the non-template CSB II DNA template, which has the same sequence as the CSB II RNA oligonucleotide, thus ruling out standard Watson–Crick base pairing. The levels of uni-G-quadruplex species formed by RNA were now weaker and partially replaced by a slower-migrating species (Figure 2B, lane 2). We hypothesize that this species represents a bimolecular G-quadruplex structure involving both the RNA and the DNA strands. To confirm this, we used a CSB II DNA oligonucleotide in which the Gs in the CSB II sequence had been replaced by 7-deaza-guanines, preventing G-quadruplex formation. Accordingly, hybrid formation was almost completely abolished when the 7-deaza-dGTP oligonucleotide was used (Figure 2B, lane 3). Moreover, a mutant CSB II DNA oligonucleotide in which the Gs had been replaced with As had the same effect (Figure 2B, lane 4).
To further show that the shift in migration observed in the presence of the wildtype DNA oligonucleotide in lane 2 really was due to hybrid G-quadruplex formation, we allowed CSB II RNA to form G-quadruplex structures in the presence of CSB II-containing DNA oligonucleotides of different lengths. The hybrid G-quadruplex showed increasing retardation in the gel when the length of the DNA was increased (Figure 2C). G-quadruplex DNA is not digested by enzymes that target single-stranded or double-stranded DNA and accordingly DNase I-treatment of the complexes resulted in a resistant species of about equal size for all DNA lengths, which likely represents the core hybrid G-quadruplex without protruding DNA ends (Figure 2C, lanes 8 and 10–12). Our experiments, therefore, strongly indicate that CSB II RNA forms a stable G-quadruplex structure together with CSB II DNA.
CD confirms the presence of G-quadruplexes at CSB II
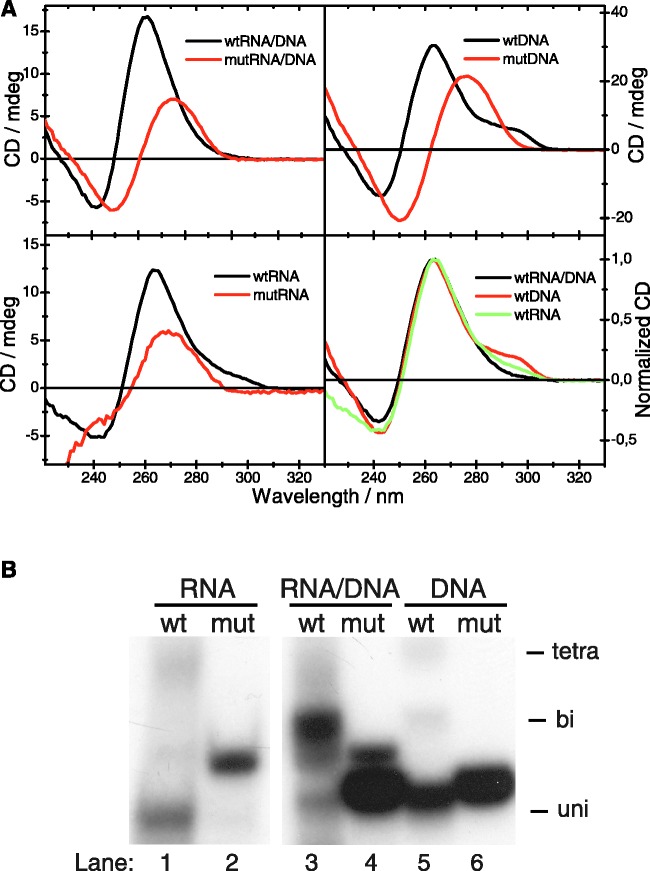
To confirm and characterize the G-quadruplex structures, we decided to use circular dichroism (CD) spectroscopy. We compared CD spectra of wt CSB II RNA, DNA and the RNA–DNA hybrid with those for mutants where all Gs had been replaced with Ts. Figure 3A, panel 1, shows CD-spectra of the wildtype CSB II RNA–DNA hybrid (in black) as well as the double mutant hybrid (in red). The CD spectra of the two samples are distinctly different. While the mutant spectrum fully resembles that of linear single-stranded DNA and RNA, with a positive peak in the red end of the spectrum and crossing y = 0 at ∼260 nm, the spectrum of the wildtype hybrid is more intense and shifted to the blue, indicating a different structure. This is supported by native gel analysis (Figure 3B, lanes 3 and 4). The gel suggests that the wildtype hybrid contains several different species, but the main one seems to be bimolecular (Figure 3B, lane 3). Comparison of the spectrum with reference spectra from the literature suggests that the hybrid forms a parallel G-quadruplex structure, as characterized by a CD spectrum with a strong positive peak at ∼260 nm (24). The gel electrophoresis analysis suggests that the main component is a bimolecular quadruplex but the CD spectrum lacks the characteristic shoulder on the red edge of the spectrum, near 290 nm. The shoulder has been assigned to the loops of bimolecular quadruplexes and differences in those regions can possibly lead to minor differences in the CD spectra.
Figure 3.
CD spectra of CSB II RNA, DNA or RNA–DNA hybrid samples. (A) Wildtype (black) and G→T mutant (red) CSB II oligonucleotides were incubated to form G-quadruplexes and subjected to CD. The CD spectra of the wildtype RNA–DNA hybrid species (top left), DNA (top right) and RNA (bottom left) all exhibit a peak at 260 nm, characteristic of parallel-type G-quadruplex structures, whereas the spectra of the mutant oligoucleotides lacking Gs resemble ssDNA. A second, smaller peak at 290 nm is observed in CSB II DNA and RNA spectra, and has been assigned to loop residues in a propeller-type parallel G-quadruplex. Bottom right: Overlay of the wildtype CD spectra of CSB II DNA (red), RNA (green) and the RNA–DNA hybrid (black). All wildtype spectra are characteristic of parallel-type G-quadruplexes, but only the RNA and DNA spectra have a smaller second peak at 290 nm. (B) The samples from (A) were separated on a native acrylamide gel in order to observe G-quadruplex formation. The positions of tetra-, bi- and uni-molecular G-quadruplexes are indicated.
We also performed CD measurements on the RNA and DNA samples separately. For both RNA and DNA the CD spectra for the wildtype and the mutant are distinctly different. For DNA the spectrum of the mutant again resembles the spectrum of linear single-stranded DNA, whereas the spectrum of the wildtype DNA has a completely different signature with a blue-shifted positive peak and a shoulder on the red edge of the spectrum. The shoulder is weaker than for reference spectra and we again assign this to differences in the loop regions (24). The wildtype and the mutant DNA oligonucleotides run with a very similar rate on a native gel (Figure 3B, lanes 5 and 6), but the CD measurements strongly suggest that the wildtype DNA indeed has formed some kind of quadruplex structure, which, coincidentally, migrates with a similar rate as the linear species.
The CD spectra of the two RNA samples resemble the corresponding spectra for DNA but are more similar to each other, and the spectrum of the wildtype sample is slightly less blue-shifted compared with the mutant than for the DNA and hybrid samples. The mutant spectrum is again characteristic of a linear single strand, in agreement with the gel analysis. The wildtype CSB II RNA spectrum is harder to assign to a specific structure due to the polydispersity of the sample. In the native gel analysis, the main band is, however, well separated from the linear form (Figure 3B, lanes 1 and 2) and the CD spectrum is distinctly different from the linear mutant oligonucleotide, indicating a different structure.
Finally, we compare the spectra of the three wildtype samples. The CD-spectra of the wildtype DNA and RNA are very similar, suggesting that the samples consist of a similar structure; they also migrate similarly on the gel. The hybrid on the other hand lacks the red-edge shoulder, which shows that the hybrid forms a slightly different kind of structure and, more importantly, that the spectrum of this sample is not simply due to G-quadruplexes formed by the RNA and DNA strands separately, but corresponds to a structure that forms from the DNA and RNA together. In conclusion, the data presented in Figure 3 strongly support our earlier conclusion that CSB II RNA, DNA and their hybrid all have the ability to form parallel-type G-quadruplexes.
G-quadruplex formation prevents primer elongation at CSB II
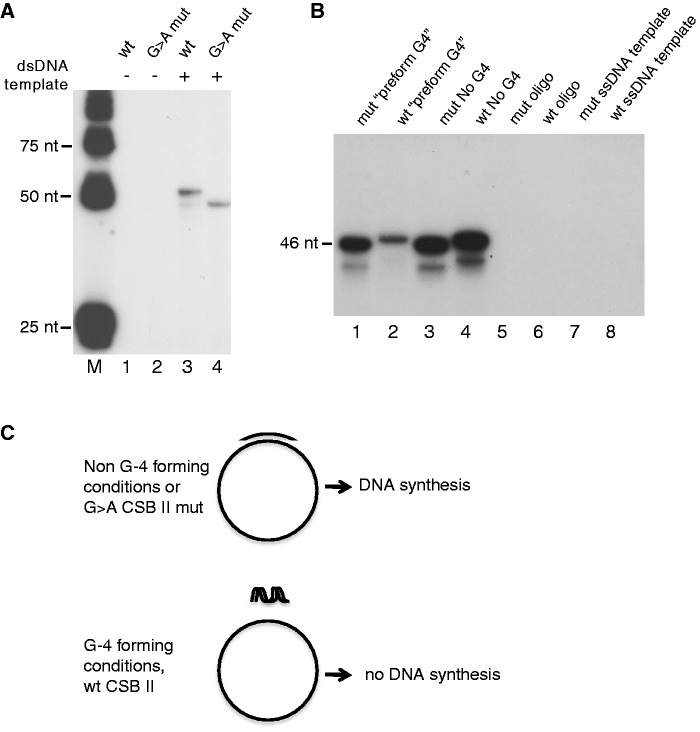
mtDNA POLγ requires a free 3′-OH end to initiate DNA synthesis. We reasoned that if the primer formed a G-quadruplex together with the non-template strand, this could impair initiation of DNA synthesis. To address this possibility, we annealed a short DNA oligonucleotide spanning CSB II to a supercoiled plasmid containing the mtDNA sequence 1-477, which includes the LSP promoter region and CSB II. The 3′ end of the primer corresponded to the major start site of DNA synthesis in vivo. We added POLγ to the construct and monitored initiation of DNA synthesis. To obtain products of a specified length (46 nt), the DNA synthesis reactions were carried out in the presence of ddCTP, which caused termination at the first G. In the absence of the template strand, we could not observe any POLγ-dependent DNA synthesis on the wildtype or the mutant oligonucleotides (Figure 4A, lanes 1 and 2). When template was added, we observed DNA synthesis and primer elongation appeared to work with the same efficiency both with wildtype and mutated CSB II (Figure 4A, lanes 3 and 4). Please note that in the mutant sample, the CSB II sequence had been mutated in both the template and primer sequences in order to enable normal Watson–Crick base pairing.
Figure 4.
G-quadruplex formation inhibits primer extension by POLγ. (A) POLγ can extend both wildtype and G→A mutant CSB II primers when they are annealed to a negatively supercoiled plasmid DNA template. Please note that the difference in G-content leads to a slight difference in gel-migration (compare lanes 3 and 4). (B) POLγ can efficiently support primer extension of the G→A mutant (mut), but not the wildtype CSB II (wt) primer, if the primers are incubated under conditions that stimulate the formation of G-quadruplex structures (pre-form G4) prior to addition to the template strand. No DNA synthesis occurs in the presence of only the oligonucleotides or only the ssDNA template (lanes 5–8). (C) A schematic figure depicting the set-up and outcome of the experiment panel (B). Preincubation at 37°C and in the presence of 100 mM KCl stimulates G-quadruplex formation.
The substrates used in our experiment had been formed by melting and reannealing DNA oligonucleotides with negatively supercoiled dsDNA templates. Under these conditions, G-quadruplex formation of the wildtype oligonucleotide with the non-template DNA strand is low, since the presence of the complementary DNA strand will favor normal Watson–Crick interactions between the two strands of the template (23). On the other hand, the mutant oligonucleotide that does not form G-quadruplexes has a lower melting temperature than the wt, which could contribute to poor annealing. The observed low efficiency of primer utilization in Figure 4A could, therefore, be explained by a low degree of primer annealing, a factor that we could not control for in this experimental set-up.
To circumvent this problem, we instead used single-stranded DNA templates (wildtype or CSB II mutant) containing only the template strand. We first annealed the primers with the single-stranded DNA template under conditions favoring standard Watson–Crick base pairing (95°C, no salt added). Using these templates we observed efficient primer-dependent initiation with both the wildtype and CSB II mutant DNA primers (Figure 4B, lanes 3 and 4). Next, we annealed the primers using conditions, which favored G-quadruplex formation (37°C, 100 mM KCl). We now observed robust levels of DNA synthesis using the CSB II mutant primer, but a dramatic drop in DNA synthesis (80% decrease) using the wildtype oligonucleotide (Figure 4B, compare lanes 1 and 2). Our interpretation of this experiment is that the formation of G-quadruplex structures in the primer (or as could be the case in vivo, between primer RNA and the non-template DNA strand) inhibits primer annealing to the template strand, thus impairing initiation of DNA synthesis (Figure 4C).
Hybrid G-quadruplex forms during transcription
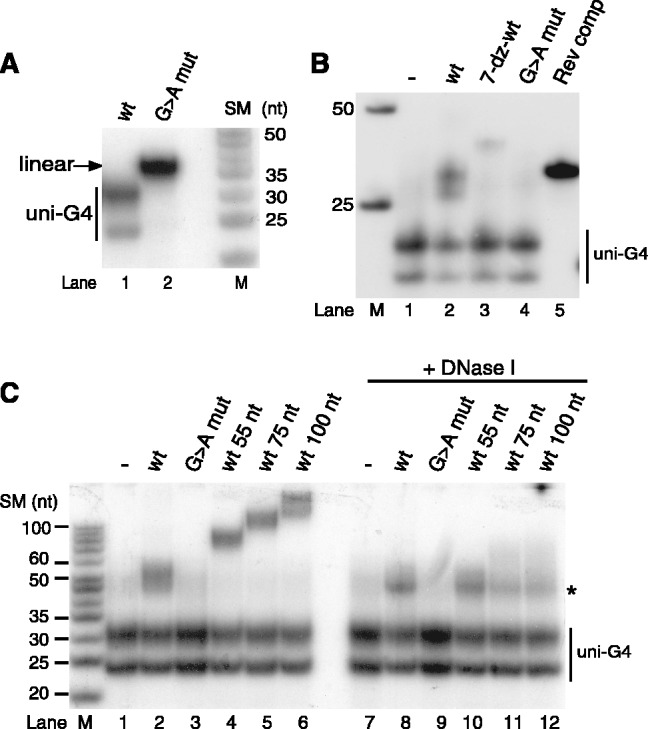
Our results prompted us to investigate if bi-molecular G-quadruplex structures could form between RNA and DNA during transcription of the CSB II region. To address this possibility, we performed in vitro transcription and digested transcription products with RNase A to remove any single-stranded RNA or with RNase H to digest the RNA strand in an RNA/DNA hybrid. Using a wildtype template, we could observe an RNase A resistant product migrating with a size of ∼50 bp, which was lost when the CSB II sequence was mutated (Figure 5A, compare lanes 2 and 8). The 50 bp band was not observed when transcription was carried out in the presence of 7-deaza-GTP, demonstrating that G-quadruplex formation is required in order for this species to form (Figure 5A, lane 5). The RNase A-resistant species was also resistant to hRNaseH1 treatment (Figure 5A, lane 3), further supporting the notion that the structure formed is a G-quadruplex structure rather than a conventional Watson–Crick base paired RNA–DNA hybrid. The resistance of G-quadruplex hybrids to hRNaseH1 treatment was confirmed using G-quadruplexes formed from CSB II DNA and RNA oligonucleotides. Whereas a standard Watson–Crick base paired hybrid between the CSB II RNA and its template DNA strand was susceptible to digestion by hRNaseH1, this was not the case for the G-quadruplex hybrid (Figure 5B, compare lanes 6 and 9). Interestingly, RNase A digests the G-quadruplex species under these conditions (Figure 5B, lanes 2 and 5), perhaps in loop regions of the G-quadruplex.
Figure 5.
Hybrid G-quadruplexes form during transcription and are dependent on CSB II. (A) In vitro transcription using the mitochondrial transcription apparatus on supercoiled templates containing either wildtype or mutant CSB II. Transcription reactions were divided into three parts, and either not treated or treated with RNase A and/or hRNaseH1, as indicated. RNase A-treatment reveals a non-degradable product of 45–50 bp (marked by asterisks) that is dependent on the CSB II sequence and Hoogsteen base pairing. (B) The labeled CSB II RNA oligonucleotide was allowed to form G-quadruplexes alone (lanes 1–3), with wildtype CSB II DNA oligonucleotide of identical sequence (lanes 4–6), or with a reverse complement DNA oligonucleotide (lanes 7–9), and then subjected to treatment with RNase A or hRNaseH1 in order to show that the G-quadruplex hybrid is resistant to hRNaseH1 (lane 6). The Watson–Crick basepaired hybrid with reverse complement DNA was degraded by hRNaseH1 as expected (lane 9). M, marker lane. (C) Transcription on templates encompassing base positions 1–477 of human mtDNA and containing either dGTP or 7-deaza-dGTP (lanes 1 and 2). After transcription, part of each reaction was treated with RNaseA (lanes 3 and 4). The RNaseA-resistant species of ∼50 bp (indicated by black bar) are absent when G-quadruplex formation with the template is eliminated (lane 3). M, marker lane.
The above experiments had established that the 50-bp RNAse A-resistant species was dependent on G-quadruplex formation at CSB II in the RNA and that it did not represent a standard RNA–DNA hybrid (R-loop). This leaves two feasible options, either that the 50 bp species represents a simple RNA G-quadruplex or, as we predicted, an RNA–DNA hybrid G-quadruplex similar to the one observed in the experiments in Figure 2. In order to distinguish between these two possibilities, we investigated the dependence of the 50 bp species on G-quadruplex formation in the DNA strand. To this end, we used the same DNA template as in Figure 1A, i.e. a linear dsDNA template spanning positions 1–477 of human mtDNA that had been PCR amplified either in the presence of dGTP or in the presence of 7-deaza-dGTP. As noted in Figure 1, transcription efficiency was lower on the 7-deaza-dGMP containing template, but the presence of 7-deaza-dGMP in the template did not abolish the premature termination of transcription observed on the dGMP containing template (Figure 5C, lanes 1 and 2). After RNase A digestion, the 50 bp RNase A-resistant species could be observed with the dGMP template, and better separation in this experiment showed that it in fact consists of at least three discrete bands (Figure 5C, lane 4). Most importantly, these products were lost when transcription was carried out on the 7-deaza-GMP containing substrate (Figure 5C, lane 3), confirming the requirement of G-quadruplex formation in the DNA strand. We could thus conclude that a stable RNA–DNA hybrid G-quadruplex forms during transcription of the mtDNA control region.
A stable R-loop extending from CSB II to 30 bp downstream of CSB I has been reported previously (22). This R-loop of ∼100 bp is clearly longer than the G-quadruplex hybrid of 45–50 bp that we observed in the experiments of Figure 5. We did, however, observe an R-loop of the reported length after transcription of a human mtDNA template with the T7 RNA polymerase (Figure 6A). This longer hybrid species was also dependent on Hoogsteen base pairing, as it was not observed in the presence of 7-deaza-GTP (Figure 6A, lane 5 compared with lane 2). In consistency with the findings in (22), the observed hybrid was dependent on the CSB II sequence (Figure 6A, lane 8).
Figure 6.
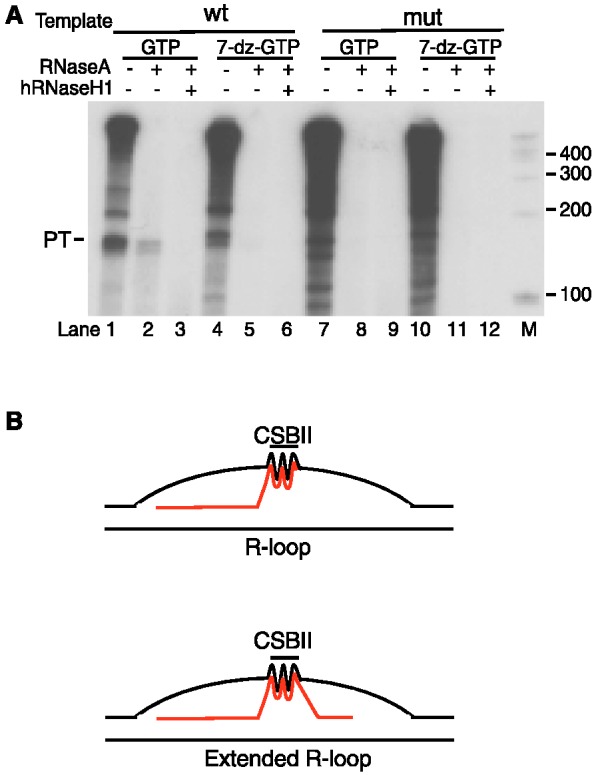
A longer R-loop can form during transcription of human mtDNA with phage T7 RNA polymerase. (A) Transcription of a human mtDNA template containing wildtype or mutant CSB II with T7 RNA pol in the presence of either GTP (lanes 1–3 and 7–9) or 7-deaza-GTP (lanes 4–6 and 10–12). A hybrid species of ∼120 bp is revealed upon RNase A treatment (lane 2) and is sensitive to hRNaseH1 (lane 3). This longer hybrid is dependent on CSB II and not observed in the presence of 7-deaza-GTP (lanes 4–6). (B) Schematic presentation of the RNA–DNA hybrid G-quadruplex that forms between the RNA and the non-template DNA strand during transcription of mtDNA. Under some conditions, an extended R-loop may be formed, similar to that reported in (22).
DISCUSSION
In prokaryotes, intrinsic (Rho-independent) termination relies on a sequence element in nascent RNA, which forms a stable stem-loop structure. The formation of this secondary structure causes the polymerase to pause and coincides with transcription of an oligo(rU) stretch that results in a weak rU-dA hybrid. In combination, the pausing and the weak RNA–DNA hybrid lead to unwinding of the elongation complex and dissociation of the RNA polymerase. We have previously reported that mitochondrial transcription termination at CSB II may function in an analogous way, but with a G-quadruplex structure replacing the canonical stem-loop (15). In the current report, we demonstrate that the G-quadruplex structure may serve an additional purpose and tether nascent RNA to the non-template strand, thus forming a stable R-loop structure.
The function of the RNA–DNA hybrid structure appears to be at least partially distinct from transcription termination, since formation of a hybrid G-quadruplex between RNA and DNA does not stimulate transcription termination further. In our studies, we use short RNA and DNA oligonucleotides and demonstrate that a RNA–DNA hybrid can form between the RNA and the non-template DNA strand at CSB II. The structures formed require Hoogsteen base pairing, since the complex was lost when DNA oligonucleotides containing a dGTP analog, 7-deaza-dGTP, were used. It is important to note that the hybrid structure is formed between the nascent RNA and the non-template strand, since this may have important functional consequences. During formation of the hybrid structure, the primer RNA leaves the template strand and instead engages in interactions with the non-template strand. At this location, the primer cannot be used for initiation of DNA replication, since the 3′-OH must be present on the template strand to initiate DNA synthesis. Formation of a hybrid G-quadruplex structure between nascent RNA and non-template DNA may, therefore, be a way to negatively regulate initiation of mtDNA replication. If this model is correct, there should also be a mechanism that can reverse the association of the primer with the non-template DNA strand, e.g. a helicase that could unwind the RNA–DNA hybrid at CSB II and thus liberate the 3′-OH of the primer for initiation of DNA synthesis. There is at least one very attractive candidate for such a function. Pif1 is an evolutionarily conserved DNA helicase, which preferentially unwinds RNA–DNA substrates (25) and actively resolves G-quadruplex structures (26). In yeast and trypanosomes, Pif1 plays a key role in regulation of mtDNA replication and copy number control (27,28). The human PIF1 gene encodes for both a nuclear and a mitochondrial isoform (29), but the role of the protein in mammalian mtDNA maintenance has not been addressed.
We would also like to point out that the G-quadruplex formed between RNA and DNA could have other functions distinct from DNA synthesis. One possibility is that the G-quadruplex structure helps to stabilize the triple-stranded D-loop structure. By preventing reannealing of the template and non-template DNA strands, the D-loop strand could remain stably annealed to the mtDNA. In this way, the RNA–DNA G-quadruplex at CSB II could be a prerequisite for stable D-loop formation and unwinding of the structure could destabilize the D-loop and lead to a renewed round of D-loop DNA synthesis.
Further evidence for G-quadruplex formation involving both RNA and DNA was obtained from CD studies (Figure 3). These experiments revealed that RNA, DNA and a DNA–RNA hybrid spanning CSB II could all form parallel-type G-quadruplex structures. Formation of these structures is very efficient and does not require the addition of crowding agents, like polyethylene glycol or ficoll. Importantly we also observe that the structure formed in the hybrid sample is distinctly different from both the DNA and RNA samples alone, indicating that the structures formed are not only due to quadruplexes formed by DNA or RNA separately.
A G-quadruplex structure involving both nascent RNA and DNA appears to also form during transcription. Using only purified mitochondrial transcription factors we found that in vitro transcription of a purified DNA fragment spanning the mitochondrial control region led to the formation of RNA species that were resistant to RNase A and hRNaseH1 digestion. The formation of these species was strictly dependent on the CSB II sequence and on Hoogsteen base pairing in both the RNA transcript and the DNA template (Figure 5A and C), strongly suggesting that the RNase resistance could be explained by the formation of a G-quadruplex hybrid structure between transcribed RNA and the DNA of the non-template strand at CSB II, i.e. a structure could be formed during transcription that is similar to the G-quadruplex structure formed between RNA and DNA oligonucleotides spanning CSB II in Figures 2 and 3.
The RNase-resistant hybrid formed during transcription is shorter than the R-loop previously characterized by Xu and Clayton (22). However, a reconstituted in vitro system for human mitochondrial transcription only became available in 2002, and many of the earlier studies had to rely on bacteriophage RNA polymerases or fractions of mitochondrial lysates for the studies of R-loop formation in vitro. In fact, we could observe the formation of longer hybrid species, similar to the previously reported R-loop (22), when we transcribed human mtDNA with the T7 RNA polymerase. Also this structure was dependent on G-quadruplex formation in the RNA strand. Previous studies have demonstrated that the R-loop is about 100 bp and involves CSB II, but extends toward the downstream CSB I sequence (22). It appears possible that the G-quadruplex formed between RNA and the DNA non-template strand at CSB II could help initiate longer R-loop formation, as depicted in Figure 6B. Indeed, a recent report has shown that upon anchoring of the RNA transcript to template DNA behind the elongating RNA polymerase, it may be energetically favorable for the RNA polymerase to continue transcription in so-called R-loop extension mode rather than to separate the nascent transcript from the DNA template (30). Hence, the G-quadruplex at CSB II could have two possible consequences for transcription: it could either destabilize the transcription complex enough for premature termination to occur, or, if termination does not occur, cause it to continue in the R-loop extension mode, resulting in the long R-loop between CSB II and CSB I. In support of this notion, transcription termination at CSB II is much lower in mitochondrial extracts and under these conditions R-loop formation can be observed (22). Our findings are in agreement with a previous study from the Clayton laboratory, which suggested that the unusual stability of the R-loop could not only be explained by standard Watson–Crick base pairing (20). In fact, the authors of this study even suggested the possibility that G-quadruplex formation contributed to the stable RNA–DNA interactions observed.
Formation of mitochondrial R-loops displays interesting similarities to R-loop formation during termination of RNA polymerase II (Pol II) transcription. During transcription of mRNA encoding genes, efficient transcription termination requires a functional poly(A) signal, at which the nascent transcript is cleaved. There are also terminator sequences located downstream of the poly(A) site, which help Pol II to disengage from the transcribed template. One example is G-rich sequences, which are often located immediately downstream of the poly(A) signal. Recent reports suggest that these G-rich elements function as pause sites and stimulate that formation of downstream R-loops. These R-loops are subsequently resolved by a specialized helicase, Senataxin, and degraded by a 5′-3′ exonuclease, Xrn2. Transcript degradation in turn stimulates termination of Pol II transcription (31). Even if this mechanism is distinct from what we report here, it is tempting to speculate that the G-rich sequences downstream of nuclear poly(A) sites in fact may stimulate hybrid RNA–DNA G-quadruplex formation. If this is the case, the mechanisms of R-loop formation may be very similar in nuclear and mitochondrial transcription.
FUNDING
Swedish Research Council grants (to C.G. and M.F.); Swedish Cancer Foundation (to M.F. and C.G.); European Research Council Advanced Investigator joint grant (to C.G.); European Research Council Starting Independent Investigator grant (to M.F.); Chalmers Area of Advance in Nanoscience and Nanotechnology (to F.W.). Funding for open access charge: The Swedish Research Council.
Conflict of interest statement. None declared.
REFERENCES
- 1.Ojala D, Montoya J, Attardi G. tRNA punctuation model of RNA processing in human mitochondria. Nature. 1981;290:470–474. doi: 10.1038/290470a0. [DOI] [PubMed] [Google Scholar]
- 2.Clayton DA. Replication and transcription of vertebrate mitochondrial DNA. Annu. Rev. Cell Biol. 1991;7:453–478. doi: 10.1146/annurev.cb.07.110191.002321. [DOI] [PubMed] [Google Scholar]
- 3.Falkenberg M, Larsson NG, Gustafsson CM. DNA replication and transcription in mammalian mitochondria. Annu. Rev. Biochem. 2007;76:679–699. doi: 10.1146/annurev.biochem.76.060305.152028. [DOI] [PubMed] [Google Scholar]
- 4.Falkenberg M, Gaspari M, Rantanen A, Trifunovic A, Larsson NG, Gustafsson CM. Mitochondrial transcription factors B1 and B2 activate transcription of human mtDNA. Nat. Genet. 2002;31:289–294. doi: 10.1038/ng909. [DOI] [PubMed] [Google Scholar]
- 5.Chang DD, Clayton DA. Priming of human mitochondrial DNA replication occurs at the light-strand promoter. Proc. Natl Acad. Sci. USA. 1985;82:351–355. doi: 10.1073/pnas.82.2.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kang D, Miyako K, Kai Y, Irie T, Takeshige K. In vivo determination of replication origins of human mitochondrial DNA by ligation-mediated polymerase chain reaction. J. Biol. Chem. 1997;272:15275–15279. doi: 10.1074/jbc.272.24.15275. [DOI] [PubMed] [Google Scholar]
- 7.Pham XH, Farge G, Shi Y, Gaspari M, Gustafsson CM, Falkenberg M. Conserved sequence box II directs transcription termination and primer formation in mitochondria. J. Biol. Chem. 2006;281:24647–24652. doi: 10.1074/jbc.M602429200. [DOI] [PubMed] [Google Scholar]
- 8.Fuste JM, Wanrooij S, Jemt E, Granycome CE, Cluett TJ, Shi Y, Atanassova N, Holt IJ, Gustafsson CM, Falkenberg M. Mitochondrial RNA polymerase is needed for activation of the origin of light-strand DNA replication. Mol. Cell. 2010;37:67–78. doi: 10.1016/j.molcel.2009.12.021. [DOI] [PubMed] [Google Scholar]
- 9.Wanrooij S, Fuste JM, Farge G, Shi Y, Gustafsson CM, Falkenberg M. Human mitochondrial RNA polymerase primes lagging-strand DNA synthesis in vitro. Proc. Natl Acad. Sci. USA. 2008;105:11122–11127. doi: 10.1073/pnas.0805399105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Spelbrink JN, Li FY, Tiranti V, Nikali K, Yuan QP, Tariq M, Wanrooij S, Garrido N, Comi G, Morandi L, et al. Human mitochondrial DNA deletions associated with mutations in the gene encoding Twinkle, a phage T7 gene 4-like protein localized in mitochondria. Nat. Genet. 2001;28:223–231. doi: 10.1038/90058. [DOI] [PubMed] [Google Scholar]
- 11.Korhonen JA, Gaspari M, Falkenberg M. TWINKLE Has 5′ -> 3′ DNA helicase activity and is specifically stimulated by mitochondrial single-stranded DNA-binding protein. J. Biol. Chem. 2003;278:48627–48632. doi: 10.1074/jbc.M306981200. [DOI] [PubMed] [Google Scholar]
- 12.Mignotte B, Marsault J, Barat-Gueride M. Effects of the Xenopus laevis mitochondrial single-stranded DNA-binding protein on the activity of DNA polymerase gamma. Eur. J. Biochem. 1988;174:479–484. doi: 10.1111/j.1432-1033.1988.tb14123.x. [DOI] [PubMed] [Google Scholar]
- 13.Farr CL, Wang Y, Kaguni LS. Functional interactions of mitochondrial DNA polymerase and single-stranded DNA-binding protein. Template-primer DNA binding and initiation and elongation of DNA strand synthesis. J. Biol. Chem. 1999;274:14779–14785. doi: 10.1074/jbc.274.21.14779. [DOI] [PubMed] [Google Scholar]
- 14.Korhonen JA, Pham XH, Pellegrini M, Falkenberg M. Reconstitution of a minimal mtDNA replisome in vitro. EMBO J. 2004;23:2423–2429. doi: 10.1038/sj.emboj.7600257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wanrooij PH, Uhler JP, Simonsson T, Falkenberg M, Gustafsson CM. G-quadruplex structures in RNA stimulate mitochondrial transcription termination and primer formation. Proc. Natl Acad. Sci. USA. 2010;107:16072–16077. doi: 10.1073/pnas.1006026107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Parkinson GN, Lee MP, Neidle S. Crystal structure of parallel quadruplexes from human telomeric DNA. Nature. 2002;417:876–880. doi: 10.1038/nature755. [DOI] [PubMed] [Google Scholar]
- 17.Sen D, Gilbert W. Formation of parallel four-stranded complexes by guanine-rich motifs in DNA and its implications for meiosis. Nature. 1988;334:364–366. doi: 10.1038/334364a0. [DOI] [PubMed] [Google Scholar]
- 18.Sen D, Gilbert W. Guanine quartet structures. Methods Enzymol. 1992;211:191–199. doi: 10.1016/0076-6879(92)11012-8. [DOI] [PubMed] [Google Scholar]
- 19.Xu Y, Kimura T, Komiyama M. Human telomere RNA and DNA form an intermolecular G-quadruplex. Nucleic Acids Symp. Ser. (Oxf) 2008:169–170. doi: 10.1093/nass/nrn086. [DOI] [PubMed] [Google Scholar]
- 20.Lee DY, Clayton DA. Properties of a primer RNA-DNA hybrid at the mouse mitochondrial DNA leading-strand origin of replication. J. Biol. Chem. 1996;271:24262–24269. doi: 10.1074/jbc.271.39.24262. [DOI] [PubMed] [Google Scholar]
- 21.Xu B, Clayton DA. A persistent RNA-DNA hybrid is formed during transcription at a phylogenetically conserved mitochondrial DNA sequence. Mol. Cell Biol. 1995;15:580–589. doi: 10.1128/mcb.15.1.580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu B, Clayton DA. RNA-DNA hybrid formation at the human mitochondrial heavy-strand origin ceases at replication start sites: an implication for RNA-DNA hybrids serving as primers. EMBO J. 1996;15:3135–3143. [PMC free article] [PubMed] [Google Scholar]
- 23.Kan ZY, Lin Y, Wang F, Zhuang XY, Zhao Y, Pang DW, Hao YH, Tan Z. G-quadruplex formation in human telomeric (TTAGGG)4 sequence with complementary strand in close vicinity under molecularly crowded condition. Nucleic Acids Res. 2007;35:3646–3653. doi: 10.1093/nar/gkm203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paramasivan S, Rujan I, Bolton PH. Circular dichroism of quadruplex DNAs: applications to structure, cation effects and ligand binding. Methods. 2007;43:324–331. doi: 10.1016/j.ymeth.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 25.Boule JB, Zakian VA. The yeast Pif1p DNA helicase preferentially unwinds RNA DNA substrates. Nucleic Acids Res. 2007;35:5809–5818. doi: 10.1093/nar/gkm613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sanders CM. Human Pif1 helicase is a G-quadruplex DNA-binding protein with G-quadruplex DNA-unwinding activity. Biochem. J. 2010;430:119–128. doi: 10.1042/BJ20100612. [DOI] [PubMed] [Google Scholar]
- 27.Cheng X, Dunaway S, Ivessa AS. The role of Pif1p, a DNA helicase in Saccharomyces cerevisiae, in maintaining mitochondrial DNA. Mitochondrion. 2007;7:211–222. doi: 10.1016/j.mito.2006.11.023. [DOI] [PubMed] [Google Scholar]
- 28.Wang J, Englund PT, Jensen RE. TbPIF8, a Trypanosoma brucei protein related to the yeast Pif1 helicase, is essential for cell viability and mitochondrial genome maintenance. Mol. Microbiol. 2012;83:471–485. doi: 10.1111/j.1365-2958.2011.07938.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Futami K, Shimamoto A, Furuichi Y. Mitochondrial and nuclear localization of human Pif1 helicase. Biol. Pharm. Bull. 2007;30:1685–1692. doi: 10.1248/bpb.30.1685. [DOI] [PubMed] [Google Scholar]
- 30.Belotserkovskii BP, Liu R, Tornaletti S, Krasilnikova MM, Mirkin SM, Hanawalt PC. Mechanisms and implications of transcription blockage by guanine-rich DNA sequences. Proc. Natl Acad. Sci. USA. 2010;107: 12816–12821. doi: 10.1073/pnas.1007580107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Skourti-Stathaki K, Proudfoot NJ, Gromak N. Human senataxin resolves RNA/DNA hybrids formed at transcriptional pause sites to promote Xrn2-dependent termination. Mol. Cell. 2011;42:794–805. doi: 10.1016/j.molcel.2011.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]