Abstract
RNA-seq, has recently become an attractive method of choice in the studies of transcriptomes, promising several advantages compared with microarrays. In this study, we sought to assess the contribution of the different analytical steps involved in the analysis of RNA-seq data generated with the Illumina platform, and to perform a cross-platform comparison based on the results obtained through Affymetrix microarray. As a case study for our work we, used the Saccharomyces cerevisiae strain CEN.PK 113-7D, grown under two different conditions (batch and chemostat). Here, we asses the influence of genetic variation on the estimation of gene expression level using three different aligners for read-mapping (Gsnap, Stampy and TopHat) on S288c genome, the capabilities of five different statistical methods to detect differential gene expression (baySeq, Cuffdiff, DESeq, edgeR and NOISeq) and we explored the consistency between RNA-seq analysis using reference genome and de novo assembly approach. High reproducibility among biological replicates (correlation ≥0.99) and high consistency between the two platforms for analysis of gene expression levels (correlation ≥0.91) are reported. The results from differential gene expression identification derived from the different statistical methods, as well as their integrated analysis results based on gene ontology annotation are in good agreement. Overall, our study provides a useful and comprehensive comparison between the two platforms (RNA-seq and microrrays) for gene expression analysis and addresses the contribution of the different steps involved in the analysis of RNA-seq data.
INTRODUCTION
In the field of functional genomics, transcriptome analysis has always played a central role for unraveling the complexity of gene expression regulation. After decades of extensive investigations based on the characterization of genome-wide gene expression through oligonucleotide-based array technologies, transcriptomics has gained new momentum, thanks to the advent of Next Generation Sequencing (NGS). NGS has enabled high-throughput of nucleic acid molecule sequencing such as DNA (DNA-seq) and RNA (RNA-seq) (1). The establishment of RNA-seq as an attractive analytical tool in trancriptomics, led to a fast development of this technology, decreasing the running cost and offering the possibility to uncover novel transcriptional-related events. Compared with hybridization-based transcriptome studies, where only difference in expression of the ORFs can be addressed, RNA-seq allows to analyze genome-wide transcription, thus providing additional features such as, analysis of novel transcripts, smRNA, miRNA and alternative splicing events. Furthermore, RNA-seq allows the analysis of transcribed but non-translated regions that may act in regulating gene expression, e.g. UTR (2). Other advantages of RNA-seq compared with microarrays are its high resolution, better dynamic range of detection and lower technical variation (3). Nevertheless, microarrays represent a well established technology and have been widely used in the last decades, leading to availability of extensive information. More than 900 000 published microarray assays are available in repository databases like Gene Expression Omnibus or ArrayExpress and have been shared within the research community.
To date, several studies comparing RNA-seq and hybridization arrays have been performed. Comparison between the two techniques have been reported in Candida parapsilolis (4), Candida albicans (5), on the fission yeast Schizosaccharomyces pombe (6), Drosophila melanogaster (7), Caenorhabditis elegans (8), in mice tissues (8,9) and in several human cells and cell lines (5,10–15). Several studies based on RNA-seq analysis of the well known eukaryotic model microorganism Saccharomyces cerevisiae, have been performed (16–20) and evaluation of the performances of different library construction methods for RNA-seq was also addressed using S. cerevisiae as a model organism (19). The reported correlations between microarrays and RNA-seq in detecting normalized expression signal are in different ranges (1), suggesting possible inconsistency of different processing methods. Higher correlation is overall observed in differential gene expression (DGE) analysis; however, up to date, a comprehensive description of the performances of RNA-seq data in detecting DGE has not been addressed in detail.
There are two major approaches to process RNA-seq data from short reads in order to identify DGEs (21). With the first approach, which is the most widely used in RNA-seq analysis, reads are mapped onto a reference genome (22,23) and the results of gene expression level are dependent on the aligner used in the analysis. Recently, different aligners and algorithm for RNA-seq analysis were compared, based on their mapping quality and splice junctions (24). The second approach is de novo assembly of the short reads (25–27) that does not require a reference genome. Recently, the performances of different transcriptome assemblers have been compared, based on their capability to identify full-length transcripts and on computational demand (28), however, statistical analysis for DGE identification and comparison between the two approaches was not covered.
In recent years, many statistical methods have been developed to identify DGE through different statistical models based on discrete probability distribution. The edgeR method proposed by Robinson et al. (29) has been developed based on an overdispersed Poisson model to explain the variation in the read count data, then the evaluation of the differences across transcripts are estimated using Empirical Bayes method. Trapnell et al. (23) presented the Cuffdiff method that relies on beta negative binomial model to estimate the variance of the RNA-seq data for DGE analysis by t-like statistics from FPKM values. In addition, different transcript isoforms can also be evaluated for their differential expression using Jensen Shannon entropy. Anders and Huber (30) showed that negative binomial was superior for estimation of variability in read count type data and implemented the method as a DESeq package showing better results in DGE identification, when compared their method with edgeR. Following this, Hardcastle and Kelly (31) proposed another algorithm to identify DGE from a count data based on the combination of negative binomial distribution and Empirical Bayes approach to estimate posterior probability of DGE. This method also provides the ability to analyze complex experimental setups that can be useful for several biological applications. Last, Tarazona et al. (32) proposed the NOISeq method based on non-parametric statistics and empirical models on the noise distribution of count data, and this method was shown to be non-sensitive to the sequencing depth of the data. In addition, this method also has a better control of false discoveries. The development of several statistical methods indicates the maturity in using RNA-seq data for transcriptomics. However, a thorough comparison of DGE analysis among different methods is required in order to increase the understanding of the different steps involved in the analysis of RNA-seq data.
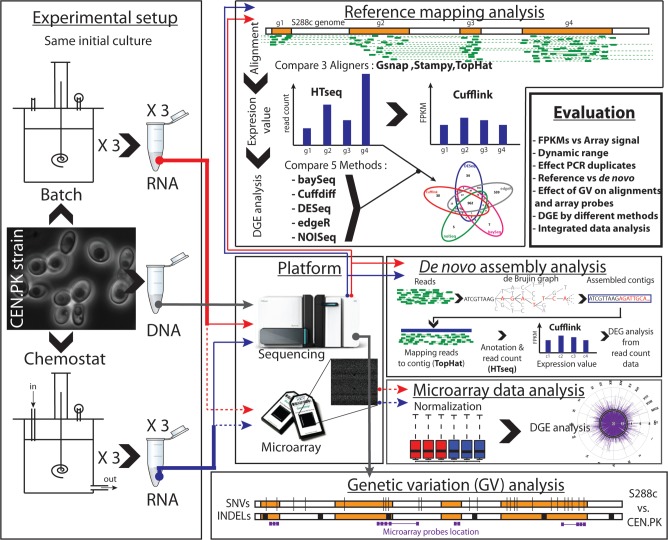
We thus undertook a study with the objective to evaluate the contribution of different factors affecting the detection of gene expression levels during the several steps involved in analysis of RNA-seq data and compare the capability of different statistical methods to capture DGE. Figure 1 provides an overview of our study. We performed RNA-seq data from the cultivations of S. cerevisisae under two different metabolic conditions. For each condition, in parallel, we also performed traditional transcriptome analysis based on the microarray platform and, additionally, we sequenced the genome (DNA-seq) of the strain from the same initial culture in order to detect eventual genetic variance such as single nucleotide variations (SNVs) and insertions–deletions (indels). Whereas previous genome-wide transcriptomic studies using RNA-seq of S. cerevisiae were based on the reference strain S288c, we based our analysis on the widely used laboratory strain CEN.PK 113-7D, as this allowed us to further investigate the influence of genetic variation on the gene expression levels estimations using the different methods. We first address the impact of different aligners in detecting DGE: Gsnap (33), Stampy (34) and TopHat (35) and successively evaluate the impact of using five different statistical methods: (i) baySeq (31), (ii) Cuffdiff (23), (iii) DESeq (30), (iv) edgeR (25) and (v) NOISeq (32). Additionally, we compared the results obtained with the ‘reference genome method’ with the de novo assembly using Trinity pipeline (26). To allow for visualization of the detected transcripts, we provide a versatile transcriptome browser that presents the results generated within this study, and integrates previously published RNA-seq data. This transcriptome browser may serve as a platform for future RNA-seq-based transcriptome analysis of S. cerevisiae.
Figure 1.
Study design overview. The same initial culture of S. cerevisiae strain CEN.PK-113-7D was used for DNA-seq (gray line) and transcriptome analysis to reduce technical variation and polymorphism. The strain was cultivated under two different metabolic conditions, in well controlled batch (red line) and chemostat (blue line) fermentation. From the triplicates’ cultures, samples for extraction of DNA and RNA were extracted. The extracted RNA was used, in parallel, for microarray analysis through Affymetrix platform (dash lines) and for RNA-seq (solid line). DNA-seq and RNA-seq were performed with the Illumina platform. DNA-seq data were used to identify the genetic variation (SNVs and indels) between the strain CEN.PK 113-7D and the reference strain S288 and to identify genetic variations in the microarray probes. The RNA-seq data were analyzed with the reference mapping approach and de novo assembly approach. The results obtained with different methods were compared and cross-compared with the results from microarray analysis.
MATERIALS AND METHODS
Microbial cultivations
The S. cerevisiae strain used for this study is CEN.PK113-7D (MATa ura3-52 MAL2-8c SUC2, provided by P. Kötter, University of Frankfurt, Germany). Minimal media as previously described (36) was used for all cultivations that were performed aerobically. For batch cultivations the medium was supplemented with 20 g l−1 glucose. For chemostat cultivations, a glucose concentration of 10 g l−1 was used to maintain carbon-limited growth. Batch cultures were performed in 1.0 l DasGip stirrer-pro® vessels with a working volume of 0.7 l. Agitation was maintained at 600 rpm using a magnetic stirrer integrated in the BioBlock®, which maintained the temperature at 30°C. The aeration was set to 0.5 l min−1. The pH of the medium was maintained at 5.0 by automatic addition of 2 M KOH. Temperature, agitation, gassing, pH and offgas composition were monitored and controlled using the DasGip monitoring and control system. Dissolved oxygen was measured with an autoclavable polarographic oxygen electrode (Mettler Toledo, Columbus, OH, USA). The effluent gas from the fermentation was analyzed for real-time determination of O2 and CO2 concentration by DasGip fedbatch pro® gas analysis systems with the off gas analyzer GA4 based on zirconium dioxide and two-beam infrared sensor. The chemostat cultures were initiated after the residual ethanol produced was depleted. The medium described above was fed with a constant dilution rate of 0.1 h−1 and aeration was set to 0.5 lmin−1. The working volume was kept at 0.5 l by a peristaltic effluent pump. Samples were taken after a steady state (defined by constant values of CO2 and O2 in the off-gas, as well as a constant biomass concentration for at least five residence periods) was achieved.
DNA extraction
Samples from DNA extraction were taken in triplicates during steady state chemostat cultivations. The withdrawal sample was immediately cooled in ice and the pellet was harvested by centrifugation at 4°C, washed with cold water and the biomass stored at −80°C until further treatment. The genomic DNA was extracted based on conventional phenol-chloroform method.
RNA extractions from cultivations
Samples for RNA extractions were taken from mid-exponential phase during batch cultivations and after steady state during chemostat cultivations. Samples were taken in three biological triplicates. The withdrawn sample was immediately cooled on ice and the pellet was harvested by centrifugation at 4°C, washed with cold water and the biomass stored at −80°C until further treatment. The total RNA was extracted from cells through mechanical disruption with glass beads, digested with DNAse and purified using the RNeasy kit (Qiagen, Hilden, Germany). The quality of the RNA was assayed using a BioAnalyzer (Agilent Technologies, Palo Alto, CA, USA). In total, 250 ng of the total RNA was used to synthesize cDNA using Affymetrix 3′ IVT Express kit and successively cRNA was synthesized (Affymetrix Inc., Santa Clara, CA, USA). The same high quality RNAs were used for constructing the library that was used for sequencing.
Transcriptome analysis
For Microarray analysis, biotinylated RNA samples were fragmented and hybridized to Affymetrix Yeast Genome Array 2.0. The Arrays were washed using an Affymetrix GenChip Fluidic station 450 and scanned using a GeneChip® Scanner 3000 7 G (Affymetrix Inc.). CEL files were generated using the Comand console software (Affymetrix). All CEL files were submitted to GEO database under accession number GSE37599. For RNA-seq analysis, Illumina HiSeq 2000 was used to perform paired-end sequencing of the same RNA samples of microarray using the standard Illumina RNA-seq protocol with a pair-end 100 bp under. All RNA-seq and DNA-seq data generated in this study were submitted to NCBI SRA database under accession number SRS307298
Microarray data acquisition and analysis
The CEL files were pre-processed and normalized together using Probe Logarithmic Intensity Error (PLIER) (37) and cubic spline method (38), respectively. Student’s t-testusing linear models together with empirical Bayes was applied (39) on the normalized expression values using the limma R package. Calculated P-values were transformed to Q-values using the false discovery rate (FDR) method to evaluate DGE between batch and chemostat cultivations.
NGS data acquisition and analysis
Pre-processing and quality assurance of the NGS reads
The raw reads form both RNA and DNA were first assessed for their quality using FASTX tool kit (http://hannonlab.cshl.edu/fastx_toolkit). Bad quality reads (phred score <20) were trimmed using the BWA trimming algorithm (40) through the SolexaQA tool kit (41). Reads that has length >25 bp on both sides of pair-end format were keep for further analysis. All further analyses were performed based on default parameters, the details are available in Supplementary Information. The versions of all software are also reported in Supplementary Information.
SNV and indel calling along chromosomes, ORFs and array probes
The quality reads of 150 coverage were first aligned on the reference genome of S. cerevisiae strain S288c using a high accuracy mapper Stampy (34) as recommended for NGS data (42). Then the SNVs and indels between the S288c and CENPK113-7 strains were identified along the chromosomes’ location using the ATLAS2 pipeline (43). Probe sequences of Yeast2 microarray were retrieved from NetAffyX then mapped on the reference genome to obtain the location of the probes along the chromosomes using Bowtie (43). The identified SNV(s) and/or indel(s) in the ORFs and microarray probes were checked for their overlap using BEDTools (44)
Transcriptome analysis using reference genome-based reads mapping
The genome sequence of S. cerevisiae strain S288c and its annotations were retrieved from the SGD databases and used for all analysis. Three different aligners for mapping the quality reads were chosen for this study: (i) Gsnap (33), which is a very fast mapping method, (ii) Stampy (34), which is a high sensitive mapping and (iii) TopHat (35), which is one of the most commonly used for RNA-seq analysis. The aligned records from the aligners in BAM/SAM format (45) were further examined for potential duplicate molecules in each record and removed using the Picard tool kit (46). After that, gene expression levels were estimated using FPKM values by the Cufflinks software (23).
Transcriptome analysis using de novo assembly of reads
The quality reads from all samples were pooled for de novo assembly using the Trinity pipeline (26) to construct transcriptional consensus contigs that can capture the transcriptions in both batch and chemostat cultivation conditions. The contigs were annotated against S. cerevisiae S288c ORFs and also mapped back to the chromosomes of S. cerevisiae S288c using GMAP (47). The quality reads of each sample were then mapped on the assembled contigs using TopHat (35). After removal of possible duplicate molecules from the aligned records by the Picard tool (46), the gene expression levels were estimated as FPKM values by the Cufflinks software (23).
Identification of differential gene expression
To identify differential gene expression between batch and chemostat cultivations, five statistical methods were employed and compared: Cuffdiff (23), baySeq (31), DESeq (30), edgeR (29) and NOISeq (32). For the last four methods, the number of reads mapped to each ORF was counted and reported using the HTseq package (30) and the output was used as input for statistical calculations. The Q-value derived from all statistical methods were used to evaluate differential gene expression between batch and chemostat cultivation except for the NOISeq (32) method where probability values (Pr) were considered instead.
Gene ontology enrichment analysis
The statistical Q-values and 1−Pr (for NOISeq) of the comparison between batch and chemostat condition resulted from different statistical methods were used as inputs for gene set enrichment analysis based on Gene ontology (GO) annotations. The reporter algorithm (48,49) was then employed to evaluate the functional enrichment level of each GO term. The GO terms that have reporter P-values (enrichment score) <10e−4, were considered for illustration as a heatmap.
All statistical analyses and illustrations were done under R suite software.
Visualizations by genome–transcriptome browser
The details of mapped reads, SNVs, indels, microarray probes position, contigs from transcriptome de novo assembly and gene annotations were visualized using GBrowse (50) publicly available at http://sysbio.se/Yseq .Moreover, five published RNA-seq data of S. cerevisiae grew under normal conditions in the study of Drinnenberg et al. (20), Levin et al. (19), Nagalakshmi et al. (16), Skelly et al. (18) and van Dijk et al. (17) were collected according to their accession numbers SRR332049, SRR059163, SRR059177, SRR002062, SRR309119 and SRR122177. Then the reads were processed in the same way as described previously using the TopHat (35) aligner.
RESULTS
Experimental setup
In our study, we sought to evaluate different processing methods, as well as statistical methods to identify DGE and cross-compare the results with microarray analysis; for this purpose, we used as case study the transcriptome of S. cerevisiae laboratory strain CEN.PK 113-7D under two different metabolic conditions: respiro-fermentative (batch) or fully respiratory (chemostat) metabolism. The metabolism of yeast under these conditions has been extensively characterized and it is known that S. cerevisiae shows significant differences in gene expression levels under these conditions. It has been previously shown that CEN.PK113-7D shows 14% genetic variation in ORFs compared with the well characterized S. cerevisiae strain S288c (51). Recently, the complete genome of CEN.PK 113-7D was re-sequenced and assembled by Nijkamp et al. (52). They reported ∼33% SNV in ORFs compared with the S288c. To further investigate the effect of genetic variation on gene expression level when reads were mapped on the reference genome of S. cervisiae strain S288c, the same initial cell culture of yeast CEN.PK113-7D was used for DNA-seq, RNA-seq and microarray analysis. To this purpose, the strain CEN.PK113-7D was cultivated in triplicates under batch and chemostat conditions and the DNA and the RNA were extracted. The RNA from each biological replicate was used, in parallel, for RNA-seq and microarray analysis. Analyzing the RNA extracted from the same biological sample should reduce technical variation coming from sample treatment, allowing us to perform a robust comparison between the two platforms for transcriptome analysis and for evaluation of different processing and statistical methods. In this study, 35.85 million of 100-bp paired-end reads were generated for the RNA-seq analysis, corresponding to 7.17 Gb. For each biological replicate, in average of 5.97 ± 1.2 million of 100 bp paired-end reads were generated corresponding to 1.19 ± 0.24 Gb (Supplementary Table S1).
SNVs and indels between the S. cerevisiae strains S288c and CENPK113-7D
For DNA-seq, a deep sequencing of 9.68 million of 100-bp paired-end reads corresponding to more than 150 coverage of the reference genome of S288c were generated. Based on the results from DNA sequencing of CEN.PK 113-7D, we identified 28 139 SNVs and 3520 indels compared with the reference strain S288c, as shown in Supplementary Table S2. About 61% of the total detected SNVs and 22% indels were found on the known coding regions. These correspond to around 34.35% and 5.07% of the ORFs containing SNV(s) and indel(s), respectively, in agreement that with the results recently reported by Nijkamp et al. (33.29 and 5.38% of ORFs containing SNVs and indels, respectively) (52). Nevertheless, when comparing the genome of the two strains, we found a higher genetic variation than previously identified by Otero et al. (51) probably as a consequence of deeper coverage and longer reads that were used in our study, as well as in Nijkamp’s study. To explore the effect of the genetic variations on microarray hybridization, we mapped all the designed probes on the reference genome, identifying 2472 probes with SNVs and 119 with indels. As a consequence, reduced or even lack of RNA hybridization on these probes might lead to an altered signal measurement through microarray analysis.
Sample-wise and gene-wise correlation of RNA-seq analysis with the reference genome approach
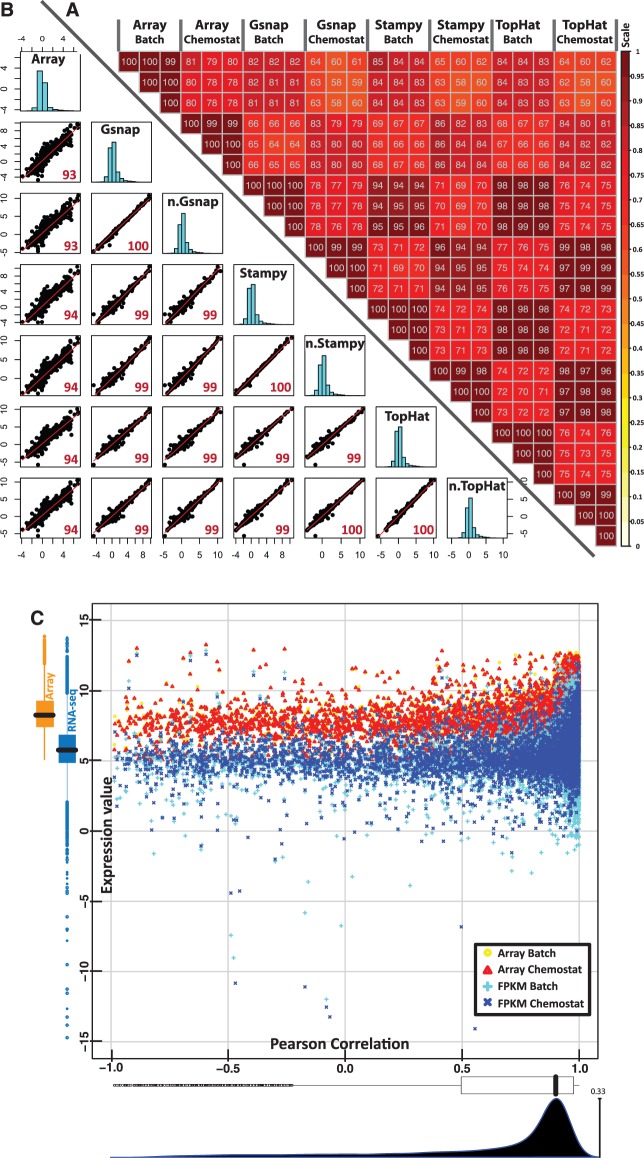
The reads obtained from sequencing were mapped on the reference genome of S. cerevisae S288c. For this purpose, the performances of three different aligners for read-mapping, TopHat (35), Stampy (34) and Gsnap (33), were evaluated and the results obtained were compared with microarray data. The rate of alignment based on our computing systems (Xeon E5520, 2.27 GHz) is approximately 3511, 1762 and 315 reads/second for Gsnap, TopHat and Stampy, respectively. On average, >96% of the high quality reads can be mapped on the reference genome by all three aligners (Supplementary Table S3). To assess the capabilities of the different aligners, we determined pairwise correlations both based on normalized expression levels and fold-changes as shown in Figure 2A and B, respectively. In Figure 2A the pairwise correlation between each biological replicate is shown based on the expression level. Here, it is possible to observe a high reproducibility among biological replicates with both microarray and RNA-seq platforms, indicated by a Pearson correlation ≥0.98. Moreover, when comparing the results among different aligners for the same biological condition, a Pearson correlation ≥0.94 can be obtained. These results are in good agreement with previous works that reported high reproducibility among technical replicates of RNA-seq (Pearson correlation values >0.95) (5,11,15) and biological replicates (Pearson correlation values >0.82) (4). When comparing the performances of the two transcriptomics platforms to identify expression levels based on intensity, we found similar results (Pearson correlation ≥0.81), in agreement with previous report by Levin et al. (19). Interestingly, the cross-platform correlation values showed more consistent results than the correlation from comparison with different microarray platforms (8,53–55).
Figure 2.
Sample-wise and gene-wise correlation of transcriptome data from microarray and RNA-seq with different processing methods. (A) Upper-right triangle matrix: pairwise correlation of different biological replicates from batch and chemostat cultivations (for microarray analysis the normalized signals and for RNAseq analysis the FPKM valued were used). The color intensities (scale in the side bar) and the numbers indicate the degree of pairwise correlation. (B) Lower-left triangle matrix: scatter plot based on fold changes of gene expression (average values, batch vs chemostat). The red numbers indicate the level of pairwise correlation between different methods. On the diagonal of the triangle matrix, the distribution of fold changes of each processing methods is presented as histrograms. Array = microarray, Gsnap = process quality reads by Gsnap aligner after removal of potential PCR duplicate, n.Gsnap = process quality reads by Gsnap aligner without removing potential PCR duplicate, Stampy = process quality reads by Stampy aligner after removal of potential PCR duplicate, n.Stampy = process quality reads by Stampy aligner without removing potential PCR duplicate, TopHat = process quality reads by TopHat aligner after removal of potential PCR duplicate, n.TopHat = process quality reads by TopHat aligner without removing potential PCR duplicate. (C) Yellow open circle, red open triangle, cyan plus sign and blue cross sign represent the average gene expression values from microarray of batch and chemostat cultivation and from RNA-seq of batch and chemostat cultivation, respectively. On the left, the distribution of average expression values from microarray and RNA-seq analysis is presented as orange boxplot and dark cyan boxplot (combined batch and chemostat cultivation conditions), respectively. At the bottom, the distribution of the gene-wise correlation values is presented as a white boxplot and density plot.
To evaluate the capability of the two platforms to capture the different response of gene expressions between the two conditions, we also performed a fold-change-based comparison. In Figure 2B, the scatter plots of fold changes generated with different aligners are shown. The remarkably high correlation values found (Pearson correlation ≥0.99), show the robustness of RNA-seq data, in agreement with what was previously observed (5,11,15). Interestingly, the value of the cross-platform correlation was improved by using fold changes (Pearson correlation ≥0.93). Using linear regression fitted on cross-platform fold changes, we obtained a model of RNA-seq = 1.29 × Microarray + 0.25 with P < 1e−16. This indicates an improvement in the dynamic range of RNA-seq data, compared with microarray data, of 30%. Interestingly, the impact of potential duplicates arising from PCR amplification during library construction procedure, which were contained around 6–16% of total reads (Supplementary Table S3), was also examined, showing to have a minor influence on the correlation results (Supplementary Figure S1). Besides analyzing the correlation between samples, we evaluated whether gene-wise correlation across samples is dependent on their expression levels. The plot of gene-wise correlation between RNA-seq and Microarray data based on their average gene expression level is illustrated in Figure 2C. Most of the genes (∼70%) have a cross-platform correlation >0.7 as observed in the density plot. The distribution of the average gene expression signal of RNA-seq and microarray data, shown on the boxplots, also supports the better dynamic range of the RNAseq data. Interestingly, we found that cross-platform correlation is random and independent of the level of gene expression, meaning that a poor correlation does not imply that a certain gene is poorly expressed and vice versa.
Evaluations of DGE of RNA-seq data through different statistical methods and cross comparison with microarray data
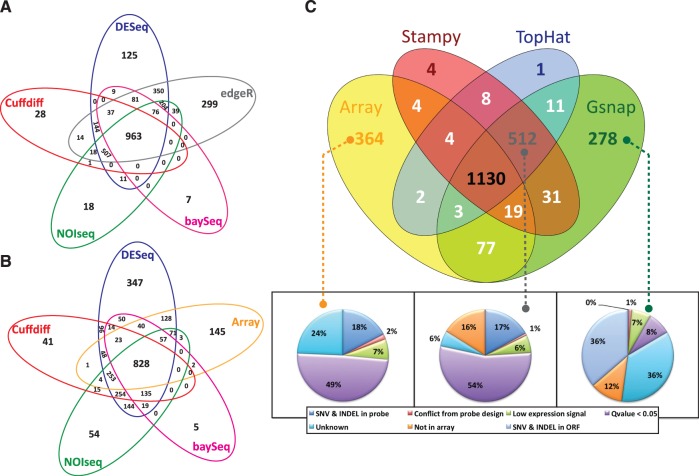
As RNA-seq can be applied to capture differential expression, we evaluated the impacts of using different reads–aligners and different statistical methods on the identification of DGE from RNA-seq data and performed a cross-comparison of these results with the DGE obtained using microarray analysis. The number of DGE derived from the results from the three different aligners and five different statistical methods (DESeq, edgeR, baySeq, Cuffdiff and NOISeq) at a specific cut-off, i.e. Q-value <1e−5 for all methods and Pr > 0.875 for NOISeq, are provided in Supplementary Table S4. It is observed that edgeR identified more DGE than the other methods at the same condition. The potential PCR duplication has minor influence on the DGE identification. The performances of the five different statistical methods for DGE identification were compared based on the mapping results obtained with the Stampy aligner as a priori input for the statistical calculation. The comparison is illustrated in a Venn’s diagram of identified DGE between the two different biological conditions using each method and is shown in Figure 3A. In total, 963 genes were commonly identified as DGE by all the five methods; however, edgeR uniquely identified more DGE than Cuffdiff, baySeq, DESeq and NOISeq. To evaluate whether there is good consistency between the different statistical methods for analysis of RNAseq data also for other biological systems, we evaluated different methods for analysis of published data from mammalian experiments (5,56). The consistency as found in our yeast data set (Figure 3A) was still valid in the mammalian systems as shown in Supplementary Figure S4.
Figure 3.
Comparisons of number of DGE identified by different statistical methods of RNA-seq data and cross comparison with DGE identified from microarray data. (A) Venn’s diagram of the comparison of differential gene expression based on RNA-seq data (result from Stampy aligner) through five different statistical methods: Cuffdiff, DESeq, NOISeq, edgeR and baySeq. (B) Venn’s diagram of the cross comparison of differential gene expression based on RNA-seq data (result from Stampy aligner) identified through Cuffdiff, NOISeq and DESeq method versus differential gene expression from microarray data (see the other comparison in different method combination in Supplementary Figure S2.) (C) Venn’s diagram of the cross comparison of DGE based on RNA-seq data identified through Cuffdiff method, using the three different aligners. The similar comparison using baySeq, DESeq, edgeR and NOISeq are provided in Supplementary Figure S3. The potential factors underlying the differences in genes identified with each method are presented as percentages pie chart. All Venn’s diagrams were built based on Q-value <1e−5 for all methods except NOISeq P > 0.875 was used as the cut-off.
Next, we cross-compared the DGE identified from RNA-seq data (Cuffdiff, baySeq DESeq and NOISeq) with DGE identified through microarray analysis. Successively, in Figure 3B it is possible to observe that, whereas 828 genes were commonly identified as differentially expressed, only 145 genes could not be captured as DGE with RNA-seq analysis through the different statistical methods. On the contrary, 135 genes were commonly identified through RNA-seq with all statistical methods but not captured by microarray analysis. At this point, we sought to further address the impact of different aligners (Gsnap, Stampy and TopHat) on DGE identification.
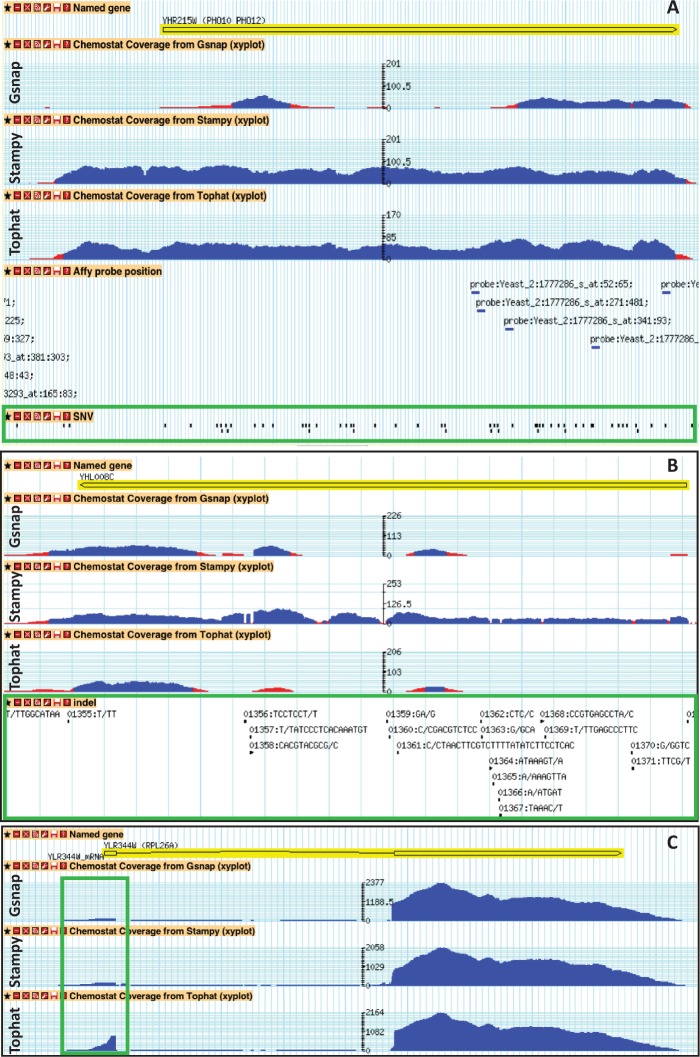
Reads processed with the three aligners were analyzed using Cuffdiff and cross-compared with the microarray data. In Figure 3C, it is possible to observe that the DGE identified from the read-mapped results based on Stampy and TopHat aligners show high consistency. Impressively, 1130 DGEs were commonly identified with both aligners and the microarray data. About 364 genes were uniquely identified as DGE from microarray data and 512 genes (82 genes are not included in the microarray) were commonly identified as DGE from RNA-seq data among the read-mapped result from the three aligners. Interestingly, when decreasing the stringency of the Q-value cut-off (<0.05), the number of commonly identified DGE increases to 49% (purple portion of the pie charts of Figure 3C) of the 364 genes uniquely identified from microarray data and 54% (purple portion of the pie charts of Figure 3C) of the 512 genes that uniquely identified from RNA-seq. Low expression genes also caused inconsistencies in DGE identifications, ∼6–7% (green portion of the pie charts of Figure 3C) of uniquely identified DGE from only microarray data (364 genes), the common of the three aligners (512 genes) and Gsnap aligner (278 genes). Around 17–18% (dodger blue portion of the pie charts of Figure 3C) of the inconsistencies were due to SNVs or/and indels in the microarray probes. 278 genes were uniquely identified as DGE using the Gsnap aligner, probably indicating different read-map performance compared with the other aligners. Noticeably, >36% (light gray–blue in the pie chart of Figure 3C) of the 278 genes contain SNVs or/and indels in their ORFs, compared with the reference genome S288c. Subsequently, in Figure 4, we further address the inconsistencies by using our Transcriptome Browser that allows direct visual comparison of the performances of the different aligners to map ORF showing genetic variations with the reference genome of the strain S288c. An example of an ORF containing several SNVs can be found in PHO11. In the Figure 4A, it is possible to see that Gsnap has problem to map reads in the coding region of PHO11. Only Stampy performed well in mapping reads on the ORF that contains many indels like YHL008C, as shown in Figure 4B. These results indicate superior capabilities of seed-based method when mapping reads on polymorphic region, in agreement with what previously observed (57). Figure 4C instead reports the good performance showed by TopHat in mapping small exons such RPL26A that indicates the benefit of spliced aligners.
Figure 4.
Coverage plots of mapped reads shows different capabilities of the three different aligners. (A) The ORF YHR215W (PHO11) contains many SNVs on the coding region (green box). (B) The ORF YHL008C contains many INDELs on the coding region (green box). (C) The ORF YLR344W (RPL26A) contains a small exon (green box).
De novo assembly versus reference mapping
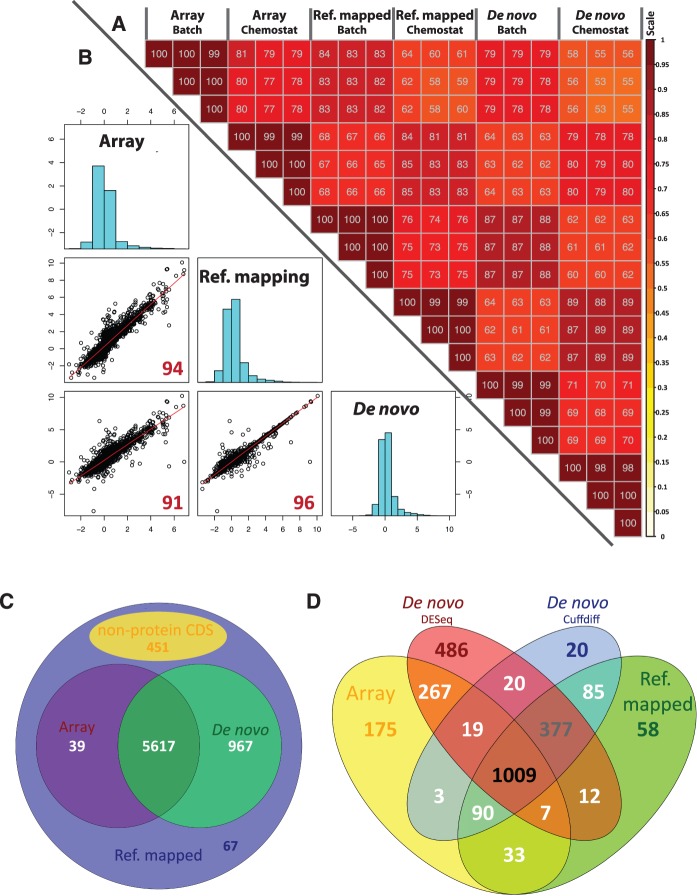
An approach that can be used to sequence RNA (or DNA) when a reference genome is not available is de novo assembly (25–28). Using this approach might also eliminate the effects of genetic variations between the strains CEN.PK 113-7D and S288c that can potentially influence read mapping results in detecting inappropriate gene expression level estimation. For this purpose, we also evaluated the use of de novo assembly. As shown in Figure 5A, de novo assembly gave high reproducibility among biological replicates, as indicated by the Pearson correlation coefficient ≥0.98. The expression-based comparison within the same platform and same sample gave a correlation ≥0.87. Slightly reduced Pearson correlation values were observed when cross-comparing the FPKM values with the normalized microarray signals. Interestingly, the fold-change-based correlation increased to 0.96 and 0.91 when comparing the results from de novo assembly approach to these obtained when mapping to a reference genome and microarray, as reported in Figure 5B. The regression model based on fold changes derived from de novo assembly and microarray (De novo = 1.21 × Microarray + 0.24 with P < 1e−16) showed similar values to the previous regression model based on the fold changes derived with the approach based on a reference genome. When comparing the result from de novo assembly with reference genome approach (De novo = 0.96 × Ref. mapped + 0 with P < 1e−16), a minor difference can be found. Figure 5C summarizes the number of identified transcripts with the different platforms and the two different analysis approaches (reference genome and de novo). Interestingly, most of the protein coding genes in the genome can be detected by the de novo assembly approach. Only 67 genes could not be captured based on this method, as a consequence of their low expression (Supplementary Figure S5). The results from statistical analysis in capturing DGE showed good agreements when comparing the results obtained by processing RNA-seq data through de novo assembly with reference genome approach and microarray, as shown in Figure 5D. The comparison of DGE identification results derived from the five different statistical methods (baySeq, Cuffdiff, DESeq, edgeR and NOISeq) when count data from the de novo assembly approach was used as a priori input, was also in good agreement as shown in Supplementary Figure S6.
Figure 5.
Comparisons of transcriptome analysis through de novo assembly and reference genome mapping approach and cross-comparison with microarray data. (A) Upper-right triangle matrix: pairwise correlation of different biological replicates from batch and chemostat cultivations (for microarray analysis the normalized signals and for RNA-seq analysis, the FPKM values were used). The color intensities (scale in the side bar) and the numbers indicate the degree of pairwise correlation. (B) Lower-left triangle matrix: scatter plot based on fold changes of gene expression (average values, batch versus chemostat). The red numbers indicate the level of pairwise correlation between different methods. On the diagonal of the triangle matrix, the distribution of fold changes of each processing method is presented as histograms. Array = microarray, De novo = De novo assembly approach, Ref. mapped = Reference genome reads mapping approach. The RNAseq by both the approaches were processed quality reads by TopHat aligner with removing potential PCR duplicate. (C) Comparisions of number of transcripts detected by different approach (D) Comparison of number of DGEs identified by different transcriptome analysis of RNA-seq data and cross-comparison with differential gene expression identified from microarray data.
GO enrichment analysis of transcriptome data
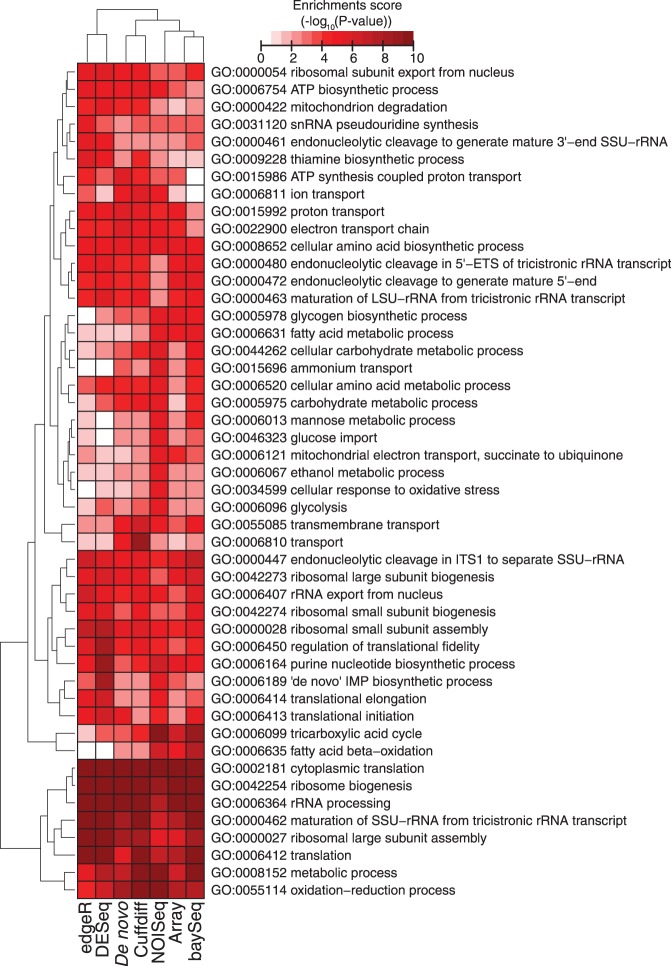
To evaluate whether the different statistical methods provide the same biological results, we analyzed the global response of the yeast transcriptome in the shift from growth at glucose excess conditions (batch) to glucose-limited conditions (chemostat). For this purpose, we used the reporter feature algorithm [Patil and Nielsen (49) and Oliveira et al. (48)] to integrate the Q-values of detected transcripts and identify significant GO terms. The algorithm was applied both on the statistical results from the microarray data and the RNA-seq data analyzed with baySeq, Cuffdiff, DESeq, edgeR, NOISeq and based on de novo assembly (using statistical results from Cuffdiff). As shown in Figure 6, 48 significant GO biological process terms were identified with a reporter P-value cut-off of 1e−4. Despite a few differences, the analysis of significant GO terms identified using the results from different statistical methods and approaches to analyze RNA-seq data and the results from microarray data are generally in agreement, leading to similar biological conclusions. Although all the methods were in agreement in identifying significant GO terms related to growth (a consequence of the increased specific growth rate during batch cultivations), GO terms known to be relevant during fully respiratory growth are not all in agreement with the different methods. Specifically, edgeR showed some inconsistencies in capturing GO terms associated with fatty acid beta-oxidation terms (as well as DESeq), fatty acid metabolic process and TCA cycle, whereas baySeq weakly identify increased expression of ATP-coupled proton transport and ion transport. Interestingly, the results derived from NOISeq seem to give stronger signals that explain the known differences between batch and chemostat growth better than the results derived from the other methods.
Figure 6.
Clustered heatmap of GO enrichment analysis. The color intensities indicate the level of enrichment score of each GO term.
Transcriptome browser
To enable visualization of transcriptome data and combine this with genomic information of S. cerevisiae, we designed a genome/transcriptome browser. The transcriptome browser gives the possibility to visualize transcriptional abundance levels (coverage mapped reads) of each ORF at different cultivation conditions and compare the results obtained using different aligners. Moreover, the browser also provides the location of indel and SNV derived from the genetic differences between CEN.PK113-7D and S288c. The transcriptional contigs from de novo assembly analysis are also represented on the browser mapped according to their position on the chromosome. Additionally, the positions where the Affymetrix microarray probes are designed on the chromosome are also included. To allow the direct comparison between RNA-seq data generated in this study and the transcriptomic data of different S. cerevisiae strains sequenced in other works, we included selected published RNA-seq data into the library of the browser. The browser is publicly available at http://sysbio.se/Yseq. The detailed screen shot of the transcriptome browser is shown in Supplementary Figure S7.
DISCUSSION
In our work, we present a comprehensive comparison of different methods for analysis of transcriptome data obtained through NGS technology and we present a cross-comparison between the two mostly used platforms for analysis of transcriptomic data: RNA-seq and microarrays. To our knowledge, this is the first time that RNA-seq generated data from Illumina platform are compared in depth with Affymetrix microarrays. An assessment of the contribution of different processing steps involved in analysis of RNA-seq data is performed in our work, addressing the impact of using different read-aligners and statistical methods to obtain biologically meaningful data. A good reproducibility among biological replicates and between the different platforms was found to be remarkably high and, generally, higher than previously reported (4,5,7,11,14). The inconsistencies found in DGE identification between RNA-seq and microarrays were shown to be mainly due to genetic variation found on the ORF and on the microarray probes.
Overall, the good agreement found between the RNA-seq and microarray platforms of our study can be interpreted based on two major factors. First, S. cerevisiae is an extremely well characterized microorganism, for which high quality genomic data are available. Furthermore, a very good annotation of gene structures allowed us to map a high portion of reads on the reference genome (>95%) and hereby, to estimate accurately gene expression level. The well-annotated gene structures also benefited from the accurate design probes of microarrays. Additionally, it also has to be considered that, for our study, we used deep sequencing of more than 5 million paired reads, enabling the coverage of a wide range of gene expression levels.
What we concluded from the approach based on reads mapping on a reference genome, is that accurately mapping is of fundamental importance to estimate gene expression level and to identify DGE. Based on our comparison of three different aligners, Stampy, the most time-consuming of the aligners, showed the highest mapping accuracy for ORFs with high genetic variation. This capability is useful when analyzing genomes and trascriptomes of higher eukaryote, usually containing high variation in the exome (∼40% of total) (58,59). However, a high-speed aligner like Gsnap, which has lower mapping accuracy compared with the other aligners, is also useful for analysis massive amount of data over the reference genome that contain low polymorphisms. TopHat appears to compromise between accuracy and speed and it also performed well at mapping reads on small exons.
Our analysis on the de novo assembly approach, showed a high consistency with reference genome approach in terms of number of detected transcripts, expression values and DGE analysis. This shows that de novo assembly of the transcriptome provides a compelling and robust approach for analysis of RNA-seq data without using reference genome. This is a benefit for organisms whose genome sequence is not available. However, de novo assembly requires a lot of computational resources (for our study to obtain contigs from de novo assembly approach, it took almost 96 h on Opteron 6200, 3.0 GHz) and more complicated in terms of post-processing of the data.
In order to address the impact of different statistical methods on the identification of DGE, we found that Cuffdiff, baySeq, DESeq, edgeR and NOISeq generated consistent results. Additionally, the results obtained based on RNA-seq data were in good agreement with microarray data. Interestingly, edgeR identified more DGE than the other methods at the same cut-off, which might infer less control of type 1 error with this method. Using results derived from different statistical methods of RNA-seq gave similar biological interpretations as is shown in GO enrichment analysis. This result strongly supports the robustness and reliability of different processing and analysis of RNA-seq data. Furthermore, we identified high consistency between microarray and RNA-seq platforms, thus encouraging the continual use of microarray as a versatile tool for differential gene expression analysis. In conclusion, our study provides a comprehensive comparison of different methods for analyses of S. cerevisiae transcriptome based on RNA-seq data using Illumina platform, elucidating the contribution of the different steps involved in analysis of RNA-seq data.
ACCESSION NUMBERS
GSE37599, SRS307298, SRR453566, SRR453567, SRR453568, SRR453569, SRR453570, SRR453571 and SRR453578.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–4, Supplementary Figures 1–7 and Supplementary Information.
FUNDING
European Research Council [247013]; Novo Nordisk Foundation; Chalmers Foundation; Knut and Alice Wallenberg Foundation; Bioinformatics Infrastructure for Life Sciences (BILS). Funding for open access charge: Chalmers Library.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Daniel Klevebring for technical assistance on DNA sequencing raw data preparation. The computational analyses were performed on resources provided by the Swedish National Infrastructure for Computing (SNIC) at C3SE. Gothenburg Bioinformatics Network (GOTBIN).
REFERENCES
- 1.Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ozsolak F, Milos PM. RNA sequencing: advances, challenges and opportunities. Nat. Rev. Genet. 2011;12:87–98. doi: 10.1038/nrg2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wilhelm BT, Landry JR. RNA-Seq-quantitative measurement of expression through massively parallel RNA-sequencing. Methods. 2009;48:249–257. doi: 10.1016/j.ymeth.2009.03.016. [DOI] [PubMed] [Google Scholar]
- 4.Guida A, Lindstadt C, Maguire SL, Ding C, Higgins DG, Corton NJ, Berriman M, Butler G. Using RNA-seq to determine the transcriptional landscape and the hypoxic response of the pathogenic yeast Candida parapsilosis. BMC Genomics. 2011;12:628. doi: 10.1186/1471-2164-12-628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marioni JC, Mason CE, Mane SM, Stephens M, Gilad Y. RNA-seq: an assessment of technical reproducibility and comparison with gene expression arrays. Genome Res. 2008;18:1509–1517. doi: 10.1101/gr.079558.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wilhelm BT, Marguerat S, Goodhead I, Bahler J. Defining transcribed regions using RNA-seq. Nat. Protoc. 2010;5:255–266. doi: 10.1038/nprot.2009.229. [DOI] [PubMed] [Google Scholar]
- 7.Malone JH, Oliver B. Microarrays, deep sequencing and the true measure of the transcriptome. BMC Biol. 2011;9:34. doi: 10.1186/1741-7007-9-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu F, Jenssen TK, Trimarchi J, Punzo C, Cepko CL, Ohno-Machado L, Hovig E, Kuo WP. Comparison of hybridization-based and sequencing-based gene expression technologies on biological replicates. BMC Genomics. 2007;8:153. doi: 10.1186/1471-2164-8-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.t Hoen PA, Ariyurek Y, Thygesen HH, Vreugdenhil E, Vossen RH, de Menezes RX, Boer JM, van Ommen GJ, den Dunnen JT. Deep sequencing-based expression analysis shows major advances in robustness, resolution and inter-lab portability over five microarray platforms. Nucleic Acids Res. 2008;36:e141. doi: 10.1093/nar/gkn705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bradford JR, Hey Y, Yates T, Li Y, Pepper SD, Miller CJ. A comparison of massively parallel nucleotide sequencing with oligonucleotide microarrays for global transcription profiling. BMC Genomics. 2010;11:282. doi: 10.1186/1471-2164-11-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Asmann YW, Klee EW, Thompson EA, Perez EA, Middha S, Oberg AL, Therneau TM, Smith DI, Poland GA, Wieben ED, et al. 3' tag digital gene expression profiling of human brain and universal reference RNA using Illumina Genome Analyzer. BMC Genomics. 2009;10:531. doi: 10.1186/1471-2164-10-531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sultan M, Schulz MH, Richard H, Magen A, Klingenhoff A, Scherf M, Seifert M, Borodina T, Soldatov A, Parkhomchuk D, et al. A global view of gene activity and alternative splicing by deep sequencing of the human transcriptome. Science. 2008;321:956–960. doi: 10.1126/science.1160342. [DOI] [PubMed] [Google Scholar]
- 13.Cloonan N, Forrest AR, Kolle G, Gardiner BB, Faulkner GJ, Brown MK, Taylor DF, Steptoe AL, Wani S, Bethel G, et al. Stem cell transcriptome profiling via massive-scale mRNA sequencing. Nat. Methods. 2008;5:613–619. doi: 10.1038/nmeth.1223. [DOI] [PubMed] [Google Scholar]
- 14.Fu X, Fu N, Guo S, Yan Z, Xu Y, Hu H, Menzel C, Chen W, Li Y, Zeng R, et al. Estimating accuracy of RNA-Seq and microarrays with proteomics. BMC Genomics. 2009;10:161. doi: 10.1186/1471-2164-10-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mudge J, Miller NA, Khrebtukova I, Lindquist IE, May GD, Huntley JJ, Luo S, Zhang L, van Velkinburgh JC, Farmer AD, et al. Genomic convergence analysis of schizophrenia: mRNA sequencing reveals altered synaptic vesicular transport in post-mortem cerebellum. PloS One. 2008;3:e3625. doi: 10.1371/journal.pone.0003625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nagalakshmi U, Wang Z, Waern K, Shou C, Raha D, Gerstein M, Snyder M. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science. 2008;320:1344–1349. doi: 10.1126/science.1158441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van Dijk EL, Chen CL, d'Aubenton-Carafa Y, Gourvennec S, Kwapisz M, Roche V, Bertrand C, Silvain M, Legoix-Ne P, Loeillet S, et al. XUTs are a class of Xrn1-sensitive antisense regulatory non-coding RNA in yeast. Nature. 2011;475:114–117. doi: 10.1038/nature10118. [DOI] [PubMed] [Google Scholar]
- 18.Skelly DA, Johansson M, Madeoy J, Wakefield J, Akey JM. A powerful and flexible statistical framework for testing hypotheses of allele-specific gene expression from RNA-seq data. Genome Res. 2011;21:1728–1737. doi: 10.1101/gr.119784.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Levin JZ, Yassour M, Adiconis X, Nusbaum C, Thompson DA, Friedman N, Gnirke A, Regev A. Comprehensive comparative analysis of strand-specific RNA sequencing methods. Nat. Methods. 2010;7:709–715. doi: 10.1038/nmeth.1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Drinnenberg IA, Fink GR, Bartel DP. Compatibility with killer explains the rise of RNAi-deficient fungi. Science. 2011;333:1592. doi: 10.1126/science.1209575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garber M, Grabherr MG, Guttman M, Trapnell C. Computational methods for transcriptome annotation and quantification using RNA-seq. Nat. Methods. 2011;8:469–477. doi: 10.1038/nmeth.1613. [DOI] [PubMed] [Google Scholar]
- 22.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 23.Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grant GR, Farkas MH, Pizarro AD, Lahens NF, Schug J, Brunk BP, Stoeckert CJ, Hogenesch JB, Pierce EA. Comparative analysis of RNA-Seq alignment algorithms and the RNA-Seq unified mapper (RUM) Bioinformatics. 2011;27:2518–2528. doi: 10.1093/bioinformatics/btr427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Robertson G, Schein J, Chiu R, Corbett R, Field M, Jackman SD, Mungall K, Lee S, Okada HM, Qian JQ, et al. De novo assembly and analysis of RNA-seq data. Nat Methods. 2010;7:909–912. doi: 10.1038/nmeth.1517. [DOI] [PubMed] [Google Scholar]
- 26.Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011;29:644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schulz MH, Zerbino DR, Vingron M, Birney E. Oases: Robust de novo RNA-seq assembly across the dynamic range of expression levels. Bioinformatics. 2012;28:1086–1092. doi: 10.1093/bioinformatics/bts094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhao QY, Wang Y, Kong YM, Luo D, Li X, Hao P. Optimizing de novo transcriptome assembly from short-read RNA-Seq data: a comparative study. BMC Bioinformatics. 2011;12(Suppl. 14):S2. doi: 10.1186/1471-2105-12-S14-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hardcastle TJ, Kelly KA. baySeq: Empirical Bayesian methods for identifying differential expression in sequence count data. BMC Bioinformatics. 2010;11:422. doi: 10.1186/1471-2105-11-422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tarazona S, Garcia-Alcalde F, Dopazo J, Ferrer A, Conesa A. Differential expression in RNA-seq: a matter of depth. Genome Res. 2011;21:2213–2223. doi: 10.1101/gr.124321.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu TD, Nacu S. Fast and SNP-tolerant detection of complex variants and splicing in short reads. Bioinformatics. 2010;26:873–881. doi: 10.1093/bioinformatics/btq057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lunter G, Goodson M. Stampy: a statistical algorithm for sensitive and fast mapping of Illumina sequence reads. Genome Res. 2011;21:936–939. doi: 10.1101/gr.111120.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Verduyn C, Postma E, Scheffers WA, Van Dijken JP. Effect of benzoic acid on metabolic fluxes in yeasts: a continuous-culture study on the regulation of respiration and alcoholic fermentation. Yeast. 1992;8:501–517. doi: 10.1002/yea.320080703. [DOI] [PubMed] [Google Scholar]
- 37.Gyorffy B, Molnar B, Lage H, Szallasi Z, Eklund AC. Evaluation of microarray preprocessing algorithms based on concordance with RT-PCR in clinical samples. PLoS One. 2009;4:e5645. doi: 10.1371/journal.pone.0005645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Workman C, Jensen LJ, Jarmer H, Berka R, Gautier L, Nielser HB, Saxild HH, Nielsen C, Brunak S, Knudsen S. A new non-linear normalization method for reducing variability in DNA microarray experiments. Genome Biol. 2002;3:research0048. doi: 10.1186/gb-2002-3-9-research0048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 2004;3:Article3. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- 40.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cox MP, Peterson DA, Biggs PJ. SolexaQA: At-a-glance quality assessment of Illumina second-generation sequencing data. BMC Bioinformatics. 2010;11:485. doi: 10.1186/1471-2105-11-485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nielsen R, Paul JS, Albrechtsen A, Song YS. Genotype and SNP calling from next-generation sequencing data. Nat Rev. Genet. 2011;12:443–451. doi: 10.1038/nrg2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shen Y, Wan Z, Coarfa C, Drabek R, Chen L, Ostrowski EA, Liu Y, Weinstock GM, Wheeler DA, Gibbs RA, et al. A SNP discovery method to assess variant allele probability from next-generation resequencing data. Genome Res. 2010;20:273–280. doi: 10.1101/gr.096388.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wu TD, Watanabe CK. GMAP: a genomic mapping and alignment program for mRNA and EST sequences. Bioinformatics. 2005;21:1859–1875. doi: 10.1093/bioinformatics/bti310. [DOI] [PubMed] [Google Scholar]
- 48.Oliveira AP, Patil KR, Nielsen J. Architecture of transcriptional regulatory circuits is knitted over the topology of bio-molecular interaction networks. BMC Syst. Biol. 2008;2:17. doi: 10.1186/1752-0509-2-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Patil KR, Nielsen J. Uncovering transcriptional regulation of metabolism by using metabolic network topology. Proc. Natl Acad. Sci. USA. 2005;102:2685–2689. doi: 10.1073/pnas.0406811102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stein LD, Mungall C, Shu S, Caudy M, Mangone M, Day A, Nickerson E, Stajich JE, Harris TW, Arva A, et al. The generic genome browser: a building block for a model organism system database. Genome Res. 2002;12:1599–1610. doi: 10.1101/gr.403602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Otero JM, Vongsangnak W, Asadollahi MA, Olivares-Hernandes R, Maury J, Farinelli L, Barlocher L, Osteras M, Schalk M, Clark A, et al. Whole genome sequencing of Saccharomyces cerevisiae: from genotype to phenotype for improved metabolic engineering applications. BMC genomics. 2010;11:723. doi: 10.1186/1471-2164-11-723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nijkamp JF, van den Broek M, Datema E, de Kok S, Bosman L, Luttik MA, DaranLapujade P, Vongsangnak W, Nielsen J, Heijne WH, et al. De novo sequencing, assembly and analysis of the genome of the laboratory strain Saccharomyces cerevisiae CEN.PK113-7D, a model for modern industrial biotechnology. Microb. Cell Fact. 2012;11:36. doi: 10.1186/1475-2859-11-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shi L, Campbell G, Jones WD, Campagne F, Wen Z, Walker SJ, Su Z, Chu TM, Goodsaid FM, Pusztai L, et al. The MicroArray Quality Control (MAQC)-II study of common practices for the development and validation of microarray-based predictive models. Nat. Biotechnol. 2010;28:827–838. doi: 10.1038/nbt.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jarvinen AK, Hautaniemi S, Edgren H, Auvinen P, Saarela J, Kallioniemi OP, Monni O. Are data from different gene expression microarray platforms comparable? Genomics. 2004;83:1164–1168. doi: 10.1016/j.ygeno.2004.01.004. [DOI] [PubMed] [Google Scholar]
- 55.Canelas AB, Harrison N, Fazio A, Zhang J, Pitkanen JP, van den Brink J, Bakker BM, Bogner L, Bouwman J, Castrillo JI, et al. Integrated multilaboratory systems biology reveals differences in protein metabolism between two reference yeast strains. Nat. Commun. 2010;1:145. doi: 10.1038/ncomms1150. [DOI] [PubMed] [Google Scholar]
- 56.Bullard JH, Purdom E, Hansen KD, Dudoit S. Evaluation of statistical methods for normalization and differential expression in mRNA-Seq experiments. BMC Bioinformatics. 2010;11:94. doi: 10.1186/1471-2105-11-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Degner JF, Marioni JC, Pai AA, Pickrell JK, Nkadori E, Gilad Y, Pritchard JK. Effect of read-mapping biases on detecting allele-specific expression from RNA-sequencing data. Bioinformatics. 2009;25:3207–3212. doi: 10.1093/bioinformatics/btp579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gamazon ER, Zhang W, Dolan ME, Cox NJ. Comprehensive survey of SNPs in the Affymetrix exon array using the 1000 Genomes dataset. PloS One. 2010;5:e9366. doi: 10.1371/journal.pone.0009366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Frazer KA, Ballinger DG, Cox DR, Hinds DA, Stuve LL, Gibbs RA, Belmont JW, Boudreau A, Hardenbol P, Leal SM, et al. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.