Abstract
PALB2 is essential for BRCA2 anchorage to nuclear structures and for homologous recombinational repair of DNA double-strand breaks. Here, we report that the N-terminal coiled-coil motif of PALB2 regulates its self-association and homologous recombination. Monomeric PALB2 shows higher efficiency to bind DNA and promotes RAD51 filament formation with or without the inhibitory effect of Replication Protein A. Moreover, overexpression of the PALB2 coiled-coil domain severely affects RAD51 loading to DNA damage sites suggesting a competition between PALB2 self-interaction and PALB2–BRCA1 interaction. In the presence of DNA damage, the switch between PALB2–PALB2 and PALB2–BRCA1 interactions allows the activation of HR. Controlling HR via PALB2 self-interactions could be important to prevent aberrant recombination in normal conditions and activate DNA repair when required.
INTRODUCTION
Cellular DNA double-strand breaks (DSBs) are amongst the most dangerous DNA damage. They can result from genotoxic agents or stalled replication forks (1). Homologous recombination (HR) is the major pathway to repair DBSs during late S phase to G2 phase of the cell cycle, leading to faithful repair of DNA damage using the sister chromatid as the repair template (1,2). The central step of HR is orchestrated by the RAD51 recombinase using resected DNA DSBs to invade a homologous duplex DNA leading to displacement loop (D-loop) (3). The recombination mediator BRCA2 transports RAD51 to the nucleus (4), promotes assembly of RAD51 onto single-stranded DNA (ssDNA) (5) and stimulates RAD51 during the invasion step of HR (3).
PALB2 (partner and localizer of BRCA2) was identified as a novel interactor of BRCA2, crucial for its localization to chromatin and recruitment to DSBs (6). The PALB2 gene is localized on chromosome 16p12.2 and encodes a protein of 1186 amino acids (7). Heterozygous mutations in PALB2 are associated with predisposition to breast and pancreatic cancers (8–10). Homozygous or biallelic mutations lead to Fanconi Anemia (11,12), an inherited genomic instability disorder caused by mutations in genes regulating replication-dependent removal of interstrand DNA crosslinks (13).
PALB2 links BRCA1 and BRCA2 to promote efficient DNA repair by HR (6,14–16). In the absence of PALB2, recruitment of BRCA2 and RAD51 to the DSBs cannot proceed (6,12). PALB2 interacts with BRCA1 by its N-terminal coiled-coil domain (14–16) and with BRCA2 via its WD40 domain at the C-terminal (6,17). The coiled-coil domain and the WD40 domain of PALB2 are both important for optimal HR (12,14,16). A third domain recently characterized named ChAM (Chromatin-Association Motif), located at the center of the protein, is required for PALB2 chromatin localization (18). PALB2-deficient cells expressing PALB2 without one of these three domains show a defect for RAD51 foci formation at DSBs and a high sensitivity to the interstrand crosslinking agent mitomycin C (12,18,19), which causes replication fork collapse and DSB formation (13). The coiled-coil domain is also responsible for oligomerization of PALB2 (19) but the function of the PALB2 self-interaction is complex and poorly understood.
Recently, it has been shown that purified full-length PALB2, like BRCA2, binds DNA and interacts directly with RAD51 (20–24). PALB2 promotes the RAD51 filament formation onto the ssDNA (20) and stimulates RAD51 for the strand invasion to form the D-loop structure (20,21). This new discovery demonstrates that PALB2 is not purely a localizer of BRCA2, but also an important regulator of RAD51 activity. In this article, we analyzed the function of PALB2 oligomerization to understand its role in regulating RAD51 and DNA recombination.
MATERIALS AND METHODS
Cells
HEK293T and HeLa cells were maintained in DMEM supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin and transfected, with calcium phosphate and Effectene (Qiagen), respectively.
DNA constructs
PALB2 DNA constructs were derived from PCR amplification of PALB2 cDNA in pOZC vector (6) and cloned as a Flag-tagged or as a Myc-tagged version into pcDNA3 (Invitrogen), or a derivative of pFASTBAC1 (Invitrogen) with His-/Flag-tag. PALB2 truncations were cloned by PCR amplification in pcDNA3 with a Myc-tag or GFP-tag in N-terminal or in pGEX6P2 (GE Healthcare) with a GST-tag or GST-/His-tag.
Antibodies
Commercial antibodies used were anti-PALB2 polyclonal antibody (Bethyl), anti-BRCA1 monoclonal antibody (17F8, GeneTex), anti-BRCA1 polyclonal antibody (Millipore), anti-BRCA2 monoclonal antibody (OP95, Calbiochem), anti-RAD51 monoclonal antibody (14B4, Novus), anti-RAD51 polyclonal antibody (Santa-Cruz), anti-Histidine (Clontech), anti-Myc monoclonal antibody (Cell Signaling) and anti-Flag monoclonal antibody (Sigma). Rabbit polyclonal anti-GST antibody was generated by immunizing rabbits with recombinant GST which was expressed and purified from Escherichia coli. For immunofluoresence, rabbit polyclonal anti-PALB2 antibody was generated by immunizing rabbits as previously described (6).
Protein purification
RAD51, or RPA were purified as described (25,26). PALB2 or PALB2Δ1–40 were purified from baculovirus-infected Sf9 cells. One-liter of Sf9 insect cells (1 × 106 cells/ml) were infected with baculovirus for 3 days at 27°C. Cells were harvested, centrifuged (1000 rpm/10 min), and the cell pellet was resuspended in 40 ml of Flag buffer (50 mM Tris–HCl pH 7.5, 150 mM NaCl, 2 mM EDTA, 1 mM DTT, 0.25% Triton X-100 and 10% glycerol) containing protease inhibitors (Roche). The suspension was lysed using a Dounce homogenizer (10 strokes), sonicated three times 30 s (50% output), and then homogenized a second time. An amount of 15 U/ml of Benzonase and 1 mM of MgCl2 were added for 1 h at 4°C and insoluble material was removed by centrifugation (35 000 rpm for 40 min). Then, 300 µl of M2 anti-Flag affinity gel (Invitrogen) was added to the soluble extract for 3 h at 4°C. The beads were washed twice with washing buffer (50 mM Tris–HCl pH 7.5, 250 mM NaCl, 2 mM EDTA, 1 mM DTT, 0.25% Triton X-100 and 10% glycerol) and incubated 30 min with washing buffer supplemented with 5 mM ATP and 15 mM MgCl2. After two additional washes with Elution Flag buffer (50 mM Tris–HCl pH 7.5, 150 mM NaCl, 0.25% Triton X-100 and 10% glycerol), proteins were eluted twice in one volume of beads with Elution Flag buffer and 500 µg/ml of 3X-Flag peptide for 45 min at 4°C. The eluted proteins were pooled and added in 5 ml of T5 buffer (20 mM NaHPO4 pH 7.4, 500 mM NaCl, 10% glycerol, 0.02% Triton, 5 mM Imidazole). Then, 300 µl of Talon resin (Clontech) was added and the mixture was incubated for 2 h at 4°C. Following centrifugation (1000 rpm, 30 s), the Talon resin was washed three times with T5 buffer. Proteins were eluted with 500 mM Imidazole in Talon buffer (300 µl) and dialyzed in the storage buffer (20 mM Tris–Acetate, pH 8.0, 200 mM KOAc, 10% glycerol, 1 mM EDTA, 0.5 mM DTT) and stored in aliquots at −80°C.
GST-protein purification
Protein truncations (GST-PALB2-T1, PALB2-T1-His and PALB2-T1Δ1–40-His) were purified from 1 l of E. coli BL21(DE3) RP (Stratagene), grown at 37°C in Luria Broth medium supplemented with 100 µg/ml ampicillin, and 25 µg/ml chloramphenicol. At OD600 = 0.4, 0.1 mM IPTG was added to the culture and incubated at 15°C overnight (16 h). The cell pellet was resuspended in 40 ml of GST buffer (10 mM Na2HPO4, 2 mM KH2PO4, 297 mM NaCl, 2,7 mM KCl, 1 mM EDTA and 1 mM DTT) containing protease inhibitors (Roche). The suspension was lysed using a Dounce homogenizer (10 strokes), sonicated three times 30 s, and then homogenized a second time. Insoluble material was removed by centrifugation (35 000 rpm for 40 min). One-milliliter of Glutathione Sepharose (GE Healthcare) was added to the supernatant and incubated during 2 h at 4°C. Then, beads were washed three times with GST washing buffer (10 mM Na2HPO4, 2 mM KH2PO4, 500 mM NaCl, 2.7 mM KCl, 1 mM EDTA and 1 mM DTT) and incubated 30 min in GST washing buffer complemented with 5 mM ATP and 15 mM MgCl2. GST-PALB2-T1 was eluted with GST buffer + 25 mM glutathione during 2 h at 4°C. PALB2-T1-His and PALB2-T1Δ1–40-His were eluted by cleavage with PreScission protease (60 U/ml GE Healthcare) overnight at 4°C in PreScission buffer (50 mM Tris–HCl pH 7.4, 150 mM NaCl, 1 mM EDTA, 1 mM DTT and 0.05% Tween 20). All eluted proteins were dialyzed in storage buffer (20 mM Tris–Acetate, pH 8.0, 200 mM KOAc, 1 mM EDTA, 0.5 mM DTT and 10% glycerol) and stored at −80°C.
GST pull-downs
Purified GST-PALB2-T1 (250 ng) was incubated with 250 ng of purified PALB2-T1-His or PALB2-T1Δ1–40-His for 25 min at room temperature in 100 µl of GSTB buffer (20 mM KPO4 pH 7.4, 250 mM KCl, 0.5 mM EDTA, 10% Glycerol, 1 mM DTT, 0.5% NP-40 and 1 mg/ml bovine serum albumin (BSA)), then 25 µl of Glutathione Sepharose beads (GE Healthcare) and 400 µl of GSTB buffer were added for 20 min at room temperature. Beads were washed four times with GSTB buffer without BSA and eluted with 40 µl of Laemmli buffer. Proteins were visualized by western blotting using the indicated antibodies.
Co-immunoprecipitations and GFP pull-down
HEK293T cells were transfected with the indicated PALB2 constructs and treated (when indicated) with 2 mM of hydroxyurea (HU) for 20 h. Then, cells were resuspended in lysis buffer (50 mM Tris–HCl, pH 7.5, 150 mM NaCl, 0.5% NP-40) containing protease and phosphatase inhibitors (PMSF (1 mM), aprotinin (0.019 TIU/ml), leupeptin (1 µg/ml), NaF (5 mM) and Na3VO4 (1 mM)), incubated for 30 min on ice, and lysed by sonication. Insoluble material was removed by high-speed centrifugation (13 000 rpm at 4°C) and each immunoprecipitation (IP) was carried out using soluble protein extract in 1 ml of lysis buffer. Fifty microliters of anti-Flag M2 affinity gel (Sigma) or 15 µl of GFP-Trap-A beads (Chromotek) and 70 U of DNase I (when indicated) were added and incubated for 2 h30 min at 4°C. Beads were washed three times with washing buffer (50 mM Tris–HCl pH 7.5, 250 mM NaCl, 0.5% NP-40) and proteins were eluted with 60 µl of Laemmli buffer. Proteins were visualized by western blotting using the indicated antibodies.
IPs from purified proteins were performed as follows. Purified Flag-PALB2-His or Flag-PALB2Δ1–40-His (250 ng) and RAD51 (500 ng) were incubated in 100 µl of GSTB buffer for 20 min at 37°C. Then 20 µl of anti-Flag M2 affinity gel (Sigma) was added and incubated for 20 min at room temperature. Beads were washed three times with GSTB buffer without BSA and proteins were eluted with 40 µl of Laemmli buffer and visualized by western blotting using the indicated antibodies.
Gel filtration
HEK293T cells were transfected with Flag-PALB2 or Flag-PALB2Δ1–40 constructs. Cells were resuspended in lysis buffer (20 mM Tris–HCl pH 7.5, 500 mM NaCl, 1 mM EDTA and 0.5% Triton X-100) containing protease and phosphatase inhibitors (PMSF (1 mM), aprotinin (0.019 TIU/ml), leupeptin (1 µg/ml), NaF (5 mM) and Na3VO4 (1 mM)), incubated for 5 min on ice, and lysed by sonication. Then 150 U/ml of Benzonase and 5 mM MgCl2 were added and the total extract was incubated for 30 min at 4°C. Insoluble material was removed by high-speed centrifugation (13 000 rpm at 4°C). Soluble extracts (2 mg) were analyzed on an FPLC Explorer 10 system fitted with a Superose 6 column 10/300 GL (GE Healthcare) equilibrated in lysis buffer and 1 mM PMSF. Fractions (500 µl) were collected and analyzed by western blotting with a Flag antibody (Sigma). The size of PALB2 or PALB2Δ1–40 was determined by comparison with gel filtration standards (250 mg; bovine thyroglobulin (670 kDa), bovine gamma globulin (158 kDa), chicken ovalbumin (44 kDa), horse myoglobin (17 kDa) and vitamin B12 (1.35 kDa)).
DNA-binding assays
Reactions (10 µl) contained 32P-labeled DNA oligonucleotides (100 nM nucleotides, Supplementary Table I) and PALB2 or PALB2Δ1–40 in binding buffer (25 mM MOPS pH 7, 40 mM KOAc, 40 mM KCl, 0.2% Tween 20 and 1 mM DTT). Reaction mixtures were incubated at 37°C for 15 min and analyzed on a 8% TBE1×/acrylamide gel (150 V/2 h30 at 4°C).
D-loop assays
32P-labeled linear DNA with 3′-tailed DNA was generated as previously described (20). 32P-labeled 3′-tailed DNA substrate (1 µM nucleotides, Supplementary Table I) was incubated for 5 min with the indicated concentration of RAD51 and PALB2 in 9 µl of 50 mM MOPS pH 7, 60 mM KOAc, 250 µM CaCl2, 300 µM EDTA, 1 mM DTT, 2 mM ATP and 100 µg/ml BSA. CsCl-purified pPB4.3 replicative form I DNA (300 µM) was added and the reaction was incubated for 1 h 30 min followed by the addition of one-fifth volume of stop buffer (10% SDS and 10 mg/ml proteinase K) and 20 min incubation at 37°C. Labeled DNA products were analyzed by electrophoresis through a 0.8% TBE1×/ agarose gel, run at 65 V, dried onto DE81 filter paper and visualized by autoradiography.
Biotin pull-down assay
Magnetic beads containing 5′-biotinylated ssDNA poly dT 83-mer (corresponding to the final concentration of 1 µM nucleotide) were pre-incubated with a blocking buffer (25 mM MOPS pH 7, 40 mM KCl, 0.2% Tween 20, 1 mM DTT and 10 mg/ml BSA) for 20 min at 37°C. The beads were captured with the magnetic particle separator, washed once with DC buffer (25 mM MOPS pH 7, 40 mM KCl, 0.2% Tween 20, 1 mM DTT, 1 mg/ml BSA, 2 mM ATP and 0.5 mM MgCl2) and resuspended in 16 µl of this buffer. 100 nM of RPA was added and the reaction was incubated for 5 min at 37°C. Then, 400 nM of RAD51 was added 5 min at 37°C. The indicated concentration of PALB2 protein was added for 10 min at 37°C in 20 µl. The beads were captured, washed twice with 40 µl of washing buffer (25 mM MOPS pH 7, 40 mM KCl, 0.2% Tween 20, 1 mM DTT, 2 mM ATP and 0.5 mM MgCl2), and 15 µl of Laemmli buffer 1× was added followed by heating 5 min at 95°C. The beads were captured and the supernatants were loaded on a SDS-PAGE 8% polyacrylamide gel. The gel was stained with Sypro Ruby (Invitrogen) and visualized by UV.
Immunofluorescence
Hela cells were transfected with PALB2-T1-Myc or PALB2-T1Δ1–40-Myc, treated with 2 mM of HU for 16–18 h and fixed with 2% paraformaldehyde in PBS for 10 min, washed with TBS and fixed with cold methanol (−20°C) for 5 min. Next, cells were permeabilized with PBS containing 0.2% Triton X-100 for 5 min and washed three times 5 min each with TBS. Then, cells were quenched with 0.1% Sodium Borohydride for 5 min, washed once with TBS, blocked in PBS containing 10% goat serum and 1% BSA for 1 h and incubated in the primary antibody diluted in PBS 1% BSA for 2 h at room temperature. Coverslips were washed three times for 10 min with TBS before 1 h incubation with the appropriate secondary antibody conjugated to a fluorophore (Alexa Fluor 488 (green) and Alexa Fluor 568 (red) from Invitrogen). Cells were rinsed again three times for 10 min with TBS. Coverslips were mounted onto slides with PBS-glycerol (90%) containing 1 mg/ml paraphenylenediamine and 0.2 mg/ml of 4, 6-diamidino-2-phenylindole (DAPI).
Immunofluorescence for PALB2 and BRCA1 visualization was performed as follows. Hela cells were transfected with PALB2-T1-GFP and were treated with 2 mM of HU for 16–18 h. Then, Hela cells were incubated for 5 min with a pre-extraction buffer (10 mM PIPES pH 6.8, 100 mM NaCl, 300 mM sucrose, 3 mM MgCl2, 1 mM EGTA and 0.2% Triton X-100), fixed with 3.7% paraformaldhyde in PBS for 20 min, washed with PBS and fixed with cold methanol (−20°C) for 20 min. Then, cells were washed with PBS, treated 30 s with cold acetone (4°C), washed a second time with PBS and blocked in PBS containing 3% BSA for 30 min before incubating with the primary antibody diluted in PBS 3% BSA for 2 h at room temperature. Coverslips were washed three times for 5 min with PBS before 1 h incubation with the appropriate secondary antibody conjugated to a fluorophore (Alexa Fluor 488 (green), Alex Fluor 568 (red) and/or Alexa Fluor 647 (far red) from Invitrogen) and mounted as described above.
RESULTS
Identification of a unique PALB2 self-interaction domain
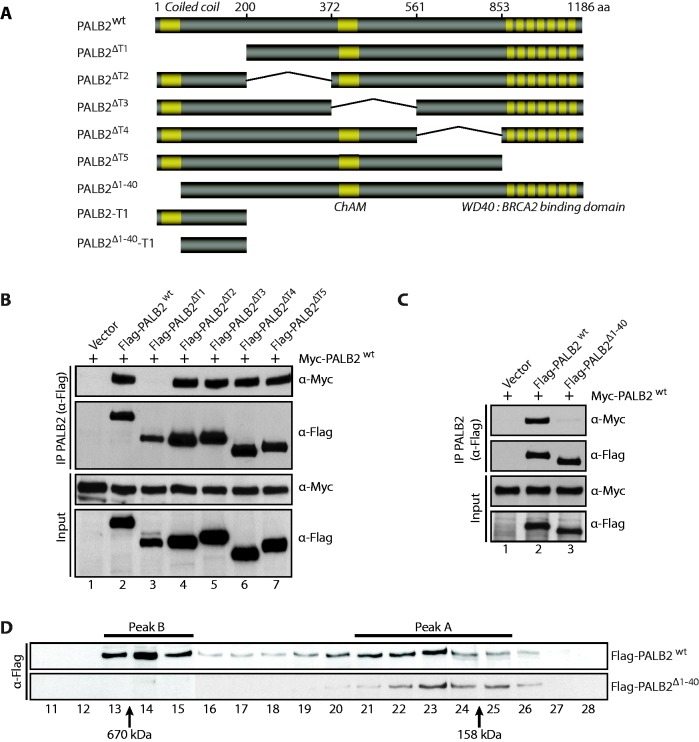
To better understand the role of PALB2 oligomerization in HR, and to prove that PALB2 has not more than one dimerization domain, we performed a detailed structure function analysis. To monitor PALB2 self-association, we co-expressed full-length Myc-PALB2 and Flag-PALB2 or Flag-PALB2 internal deletion mutants spanning the entire coding sequence (Figure 1A, PALB2ΔT1–T5) followed by anti-Flag IP. A Myc-PALB2/Flag-PALB2 co-complex was detected (Figure 1B, lane 2) confirming dimerization of PALB2. The dimerization of PALB2 was lost when the N-terminal domain (amino acids 1–200) was removed (lane 3). To detect whether the evolutionary conserved coiled-coil motif in the N-terminal domain of PALB2 (amino acids 1–40) allows self-interaction, co-IP assays were performed with Myc-PALB2 and Flag-PALB2Δ1–40 (Figure 1A and C). Removal of the coiled-coil domain from the wild-type protein completely abrogated self-interaction (Figure 1C, compare lanes 2 and 3). This result was confirmed using size-exclusion chromatography where PALB2 eluted as two major peaks (Figure 1D). The peak A (fractions 21–25) eluted close to the predicted monomer size of Flag-PALB2-His (135 kDa), whereas, the peak B (fractions 13–15) most likely corresponds to a mixture of PALB2 multimers. Unlike wild-type PALB2, PALB2Δ1–40 was eluted in only peak A, indicative of the monomeric form (Figure 1D).
Figure 1.
PALB2 contains a self-interaction domain localized at its N-terminus. (A) Schematic representation of PALB2 and PALB2 truncations used in this study. (B–C) Myc-PALB2 and Flag-PALB2 (PALB2 wild-type or PALB2 deletion mutants) were co-expressed in HEK293T cells and followed by an IP with anti-Flag antibody. Immunoprecipitated proteins were detected by western blotting with anti-Flag or anti-Myc antibodies. (D) Size-exclusion chromatography analysis of HEK293T whole-cell extracts expressing Flag-PALB2 or Flag-PALB2Δ1–40. Proteins of each fraction were detected by western blotting with an anti-Flag antibody. Molecular weight standards are indicated at the bottom of the Figure.
BRCA1 and PALB2 interact via their coiled-coil domains (14–16). It has been reported that the deletion of the first forty amino acids of PALB2 completely abrogates PALB2 foci formation (19). Furthermore expressing PALB2 without the coiled-coil domain leads to high sensitivity to mitomycin C and HR defects (19). Taken together, these results show that (i) the interaction between PALB2 and BRCA1 is critical for HR repair and (ii) suggest a competition between PALB2–PALB2 and BRCA1–PALB2 interactions, which may allow a regulation of PALB2 mediator activity in cells.
Enhanced DNA binding by monomeric PALB2
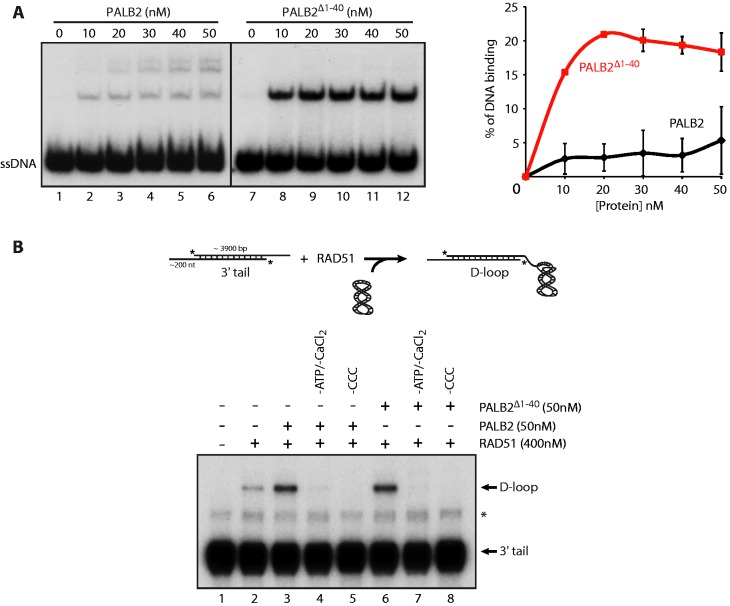
As it was difficult to separate PALB2 self-interactions and BRCA1–PALB2 functions in vivo, we resorted to in vitro assays to further dissect the function of the monomeric or dimeric form of PALB2. PALB2 binds DNA (20,21) using two distinct DNA-binding regions (20). To assess whether PALB2 dimerization is important for its DNA-binding activity, we purified wild-type PALB2 and a mutant PALB2 deleted of its coiled-coil motif (Supplementary Figure S1A). PALB2Δ1–40 showed about 5-times stronger affinity for ssDNA than PALB2 (Figure 2A). In addition, PALB2 binding to DNA leads to the formation of several bands in the gel suggesting multimeric forms of the protein bound to DNA whereas only one band was observed for PALB2Δ1–40 at the same size as the lowest band for wild-type PALB2. This result suggests that the dimerization of PALB2 could affect the accessibility of both DNA-binding domains of PALB2 to ssDNA. Furthermore, a switch between the multimeric to monomeric form of PALB2 could control PALB2 loading on DSBs by regulating its binding on DNA.
Figure 2.
DNA binding by monomeric PALB2 and D-loop formation. (A) PALB2 or PALB2Δ1–40 bind ssDNA. Electrophoretic mobility shift assays were performed with PALB2 or PALB2Δ1–40 (10–50 nM) and 60 nt ssDNA (100 nM nucleotides) and analyzed on a 8% polyacrylamidegel. Right: quantification of the results. (B) Top: schematic of D-loop assay. D-loop reactions were performed with RAD51 alone (lane 2) or combinations of RAD51 and PALB2 or PALB2Δ1–40 (lanes 3–8). The band indicated with an asterisk corresponds to annealed tailed molecules. CCC designates Covalently Closed Circular supercoiled plasmid.
We have previously shown that PALB2 stimulates RAD51 in D-loop formation (20), a key step of HR. We monitored the activation of RAD51 by PALB2, in conditions where RAD51 saturates the ssDNA substrate prior to invasion into a homologous DNA (Figure 2B). In these conditions, RAD51 is in a stable form on tailed DNA prior to DNA invasion, which allows us to discriminate RAD51 activation from the formation of RAD51 nucleoprotein filaments. In the absence of PALB2, little ssDNA was converted into a D-loop by RAD51 (Figure 2B, lane 2), whereas PALB2 strongly stimulated this reaction (lane 3). The stimulation was dependent on ATP/CaCl2 and supercoiled DNA (lanes 4–5). Surprisingly, PALB2Δ1–40 showed a similar effect on RAD51 (lanes 6). This result indicates that multimerization of PALB2 has no effect on RAD51 D-loop activity.
Monomeric PALB2 stimulates RAD51 filament formation
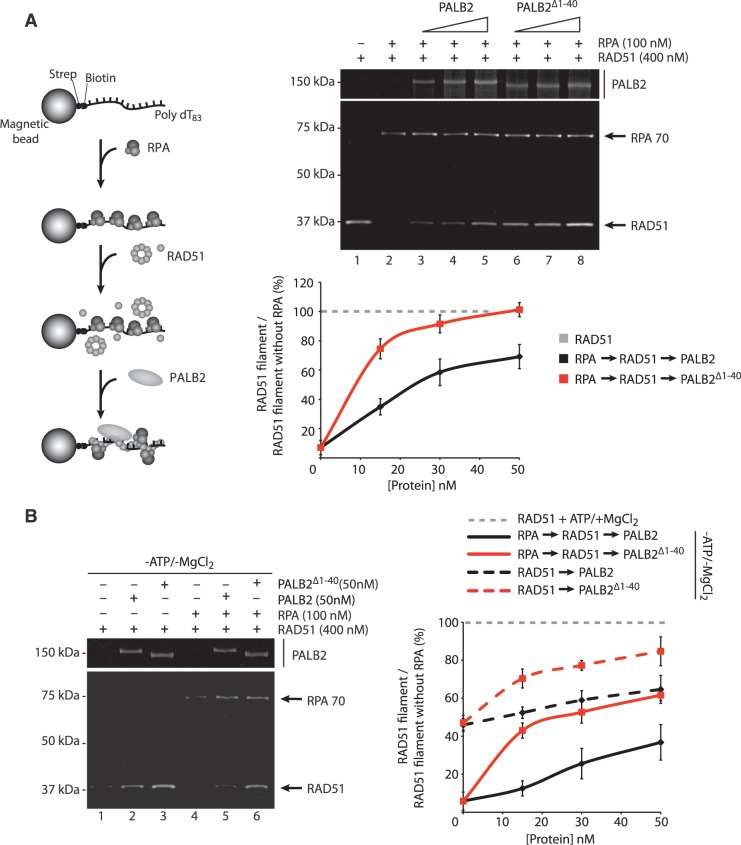
The similar effect of PALB2 and PALB2Δ1–40 on RAD51-mediated D-loop formation prompted us to investigate whether the deletion of the coiled-coil domain could impact on other key steps of HR. For instance, prior to strand invasion, the ssDNA-binding protein RPA must be displaced from the single-strand tail of the newly resected DSB to allow RAD51 filament formation. Such function is accomplished by BRCA2 or PALB2 recombination mediators to overcome the inhibitory effect of RPA and allow RAD51 to nucleate on the ssDNA to promote the pre-synaptic filament assembly (20,23). In the presence of RPA, RAD51 loading on ssDNA was strongly inhibited (Figure 3A, lane 2). PALB2 alleviated the inhibitory effect of RPA (Figure 3A, lanes 3–5 and Supplementary Figure S2A) to promote RAD51 filament formation. Notably, PALB2Δ1–40 was much more efficient to promote RAD51 nucleoprotein filament formation than PALB2 on ssDNA protected by RPA (Figure 3A, lanes 6–8 and Supplementary Figure S2A). This effect was not due to enhanced RAD51 binding by PALB2Δ1–40 as both PALB2 and PALB2Δ1–40 interacted equivalently with RAD51 (Supplementary Figure S2B). As observed with Brh2 or LiBRCA2, conserved but smaller BRCA2 orthologs in Ustilago Maydis or Leishmania infantum, RPA was still bound to the ssDNA beads although the inhibitory effect of RPA on RAD51 was removed (27,28). This might occur through protein–protein interactions as RPA binds RAD51 (29) or that PALB2 displaces RPA on the ssDNA allowing RAD51 to form a nucleoprotein filament. In the absence of ATP and magnesium, RAD51 still displays a weak DNA-binding activity (Figure 3B, compare RAD51 −ATP/−MgCl2 to RAD51 +ATP/+MgCl2). We observed that PALB2Δ1–40 promotes the loading of RAD51 on ssDNA better than PALB2, in the absence (Figure 3B, lanes 1–3) or in the presence of RPA (lanes 4–6). Taken together, these results imply that the monomeric form of PALB2 has enhanced activity to stimulate RAD51 DNA binding and filament formation.
Figure 3.
Stimulation of RAD51 filament formation by monomeric PALB2. (A) Left: schematic of RPA displacement assay. Right: RPA bound to a ssDNA oligonucleotide prevents RAD51 assembly (lane 2) whereas addition of PALB2 or PALB2Δ1–40 (15, 30 and 50 nM) stimulates RAD51 filament formation in presence of RPA (lanes 3–8). Bottom: quantification of the results. The Y-axis represents the ratio between the values obtained for RAD51 filament in the presence of RPA and PALB2 or PALB2Δ1–40 divided by the value obtained for RAD51 filament with ATP and magnesium without RPA (set at 100%). (B) Left: RPA displacement assay in the absence of ATP and MgCl2. Right: quantification of the results. The Y-axis represents the ratio between the values obtained for RAD51 filament in the absence of ATP/MgCl2 with or without RPA and PALB2 or PALB2Δ1–40 divided by the value obtained for RAD51 filament with ATP and magnesium without RPA (set at 100%).
The N-terminal coiled-coil of PALB2 acts as a dominant negative protein
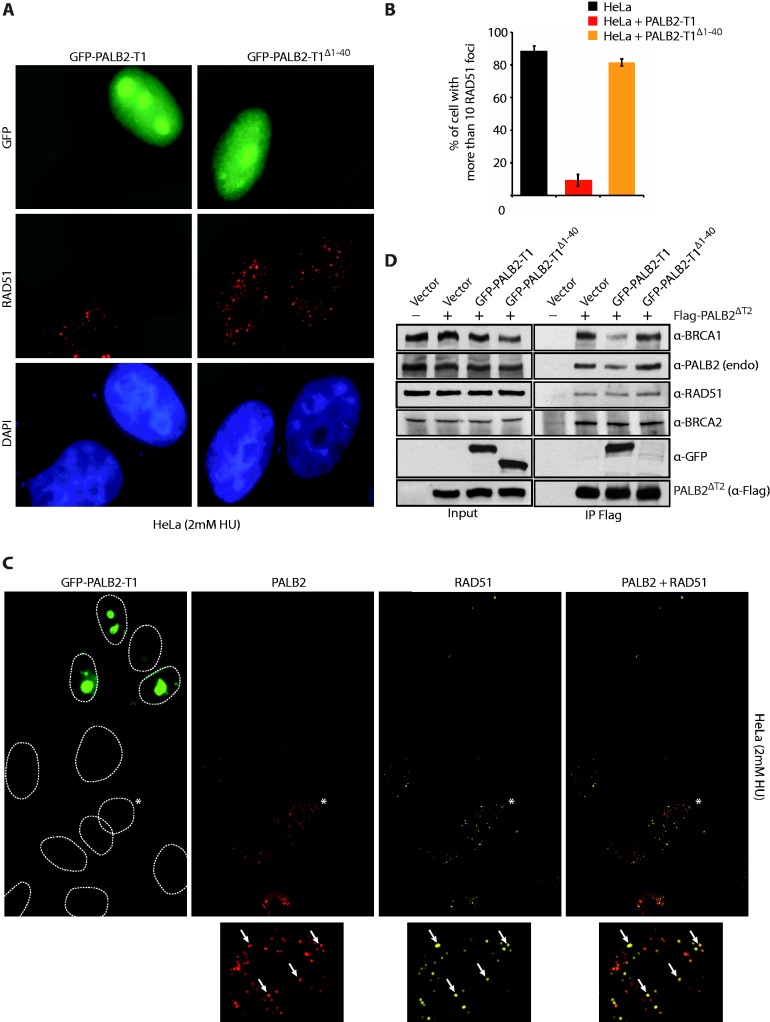
In cells, PALB2 colocalizes with RAD51 after HU treatment (Supplementary Figure S3A) and is required for its localization at DSBs (6). Our results suggest that the monomeric form of PALB2 possesses higher activity compared to the multimeric form. If it was the case, overexpression of the N-terminal domain (PALB2-T1) may disrupt the balance between monomeric PALB2 (elevated activity) and dimeric PALB2 (basal activity) by forming a PALB2–PALB2-T1 complex leading to a subsequent decrease in HR (Supplementary Figure S3B). We overexpressed GFP-PALB2-T1 and GFP-PALB2-T1Δ1–40 in HeLa cells and monitored RAD51 foci formation. The expression of GFP-PALB2-T1, strongly affected the localization of RAD51 to DSBs induced by DNA replication block whereas GFP-PALB2-T1Δ1–40 did not affect RAD51 foci formation (Figure 4A). Quantification of the DNA damage-induced RAD51 foci showed a 10-fold decrease in cells expressing the wild-type N-terminal fragment compared to those expressing the deletion mutant (Figure 4B). Because the GFP-tag could interfere with the N-terminal domain of PALB2, we have confirmed this result by expressing the PALB2-T1 coupled with the smaller tag Myc (Supplementary Figure S4A and B). Loss of RAD51 foci cannot be explained by an effect on the cell cycle induced by the overexpression of the T1-PALB2 since cell-cycle progression was not affected (Supplementary Figure S4C). Moreover, we have confirmed in vivo with a DNase I treatment or in vitro with purified proteins that N-terminal PALB2-T1 fragment self-interacts and PALB2-T1Δ1–40 does not (Supplementary Figure S5A–C).
Figure 4.
Inhibition of RAD51 foci formation by the PALB2 coiled-coil domain. (A) Expression in Hela cells of GFP-PALB2-T1 with or without the coiled-coil domain (GFP-PALB2-T1Δ1–40). Immunofluorescence pictures with anti-GFP and anti-RAD51 are shown. (B) Quantification of the experiment shown in (A). (C) Expression in Hela cells of GFP-PALB2-T1 followed by immunofluorescence with anti-PALB2 (in red) and anti-RAD51 (in Far-red). The dotted lines delimit the DAPI staining. The cell indicated with an asterisk was enlarged at the bottom of the figure and arrows indicate a co-localization between PALB2 and RAD51 foci. (D) HEK293T cells were transfected with a vector alone or Flag-PALB2ΔT2, with GFP-PALB2-T1 or GFP-PALB2-T1Δ1–40. Flag-IPs were conducted, followed by western blotting with the indicated antibodies.
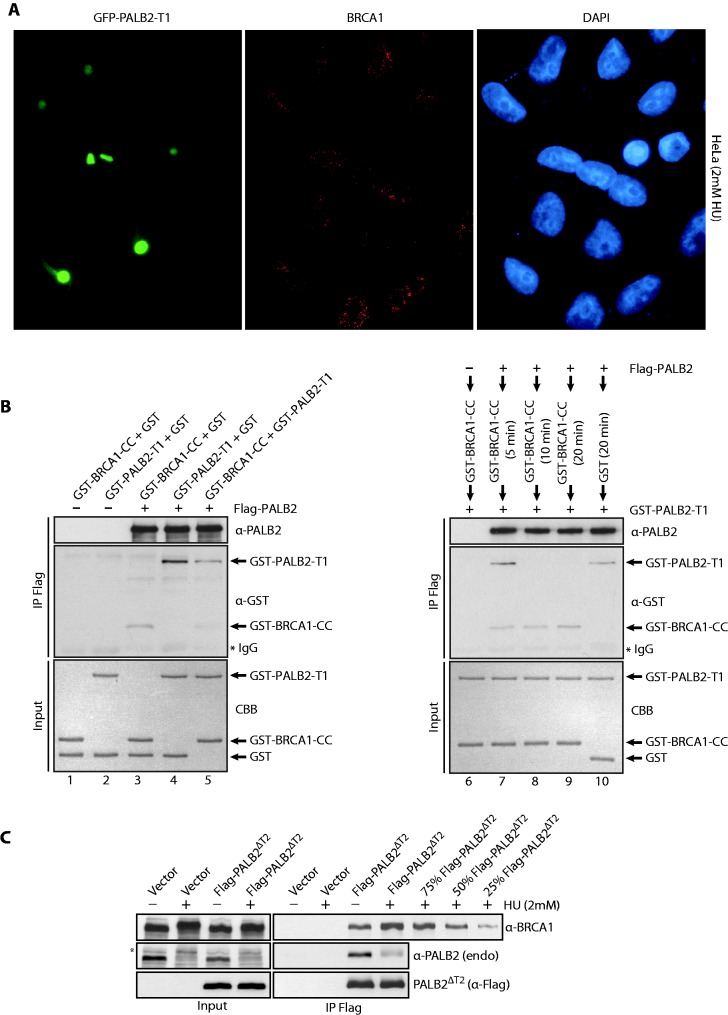
To better define the role of PALB2 self-interaction in RAD51 foci formation, we overexpressed GFP-PALB2-T1 in Hela cells. Cells expressing GFP-PALB2-T1 were strongly affected for RAD51 foci formation but also for PALB2 foci formation, whereas untransfected cells show a normal colocalization between PALB2 and RAD51 at DSBs (Figure 4C, compare GFP positive to negative cells). Importantly, the expression of GFP-PALB2-T1 did not affect BRCA1 foci formation (Figure 5A). In this immunofluorescence experiment, a pre-extraction has been performed before fixation of the cells. This pre-extraction is crucial for the visualization of PALB2 foci. This, however, results in a loss of staining of the nuclear soluble T1–PALB2–GFP (Supplementary Figure S3C) explaining the difference of staining between Figure 4A and Figure 4C–5A.
Figure 5.
(A) Expression of GFP-PALB2-T1 does not affect BRCA1 foci formation. Immunofluorescence staining was performed with anti-BRCA1 (red). The DAPI staining is also shown. (B) Purified GST-BRCA1-CC competes with PALB2 self-interaction. Proteins were incubated with PALB2-Flag/His followed by IP with anti-Flag and western blotting with anti-GST and anti-PALB2 antibodies. Left: the order of addition of each protein is shown on the top of the gel. The proteins stained by Coomassie brilliant blue (CBB) are also shown. (C) HEK293T cells were transfected with an empty vector or a vector containing Flag-PALB2ΔT2, treated with HU (2 mM, 20 h) and Flag-PALB2ΔT2 was immunoprecipitated with an anti-Flag antibody. Immunoprecipitated proteins were detected by western blotting with the indicated antibodies. Quantification of the increase is estimated by a titration of Flag-PALB2ΔT2 + HU IP. The band indicated with an asterisk corresponds to the phosphorylated form of PALB2.
PALB2 foci formation is essential for RAD51 localization to DSBs via BRCA2 (6,12). On the other hand, PALB2 localization appears to be dependent on its interaction with BRCA1 (15,16). To further define the role of PALB2–BRCA1 interaction, we co-expressed GFP-PALB2-T1 and Flag-PALB2ΔT2. The expression of PALB2ΔT2 allows us to separate the overexpressed from endogenous PALB2 by western blotting. Note that the T2 domain is not required for PALB2 functionality in vivo (12). Co-IP of endogenous PALB2 and endogenous BRCA1 with PALB2ΔT2 was decreased by the overexpression of GFP-PALB2-T1 but not when the coiled-coil motif was missing. In contrast, the interactions with BRCA2 and RAD51 were not affected (Figure 4D). These results suggest that the overexpression of the coiled-coil domain of PALB2 prevent PALB2 from interacting with BRCA1. However, following DNA damage, BRCA1 must be able to compete with PALB2–PALB2 interactions, to generate a PALB2–BRCA1 complex effective for DNA repair.
To verify this possibility, we scrutinized whether purified BRCA1 coiled-coil domain (Supplementary Figure S5D) was able to compete with purified PALB2 coiled-coil (PALB2-T1) to interact with the full-length PALB2 (Figure 5B). To do so full-length Flag-PALB2 was incubated with GST-BRCA1-CC (GST fusion to the coiled-coil region of BRCA1), GST-PALB2-T1 or both (lanes 3–5) followed by IP analysis. First, we observed an interaction between GST-BRCA1-CC and Flag-PALB2 (lane 3), and as described before, an interaction between Flag-PALB2 and GST-PALB2-T1 (lane 4) was detected. However, when GST-BRCA1-CC was added to the reaction, the amount of Flag-PALB2/GST-PALB2-T1 co-complex was reduced (lane 5). Consequently, the coiled-coil region of BRCA1 can disrupt PALB2–PALB2 interactions. This result was corroborated in another independent experiment. Flag-PALB2 was incubated with GST-BRCA1-CC for 5–20 min, followed by the addition of GST-PALB2-T1. At 10 and 20 min, GST-BRCA1-CC completely abrogated the Flag-PALB2/GST-PALB2-T1 co-complex observed at 5 min (compare lanes 7–9). Altogether, these results indicate that in vivo PALB2 self-interaction which inhibits DNA recombination, can be suppressed by the interaction with BRCA1. In support of this, BRCA1 interacts more strongly with PALB2 after DNA damage (Figure 5C). We observed an increase of 60–70% of PALB2–BRCA1 interaction when cells were treated with HU.
DISCUSSION
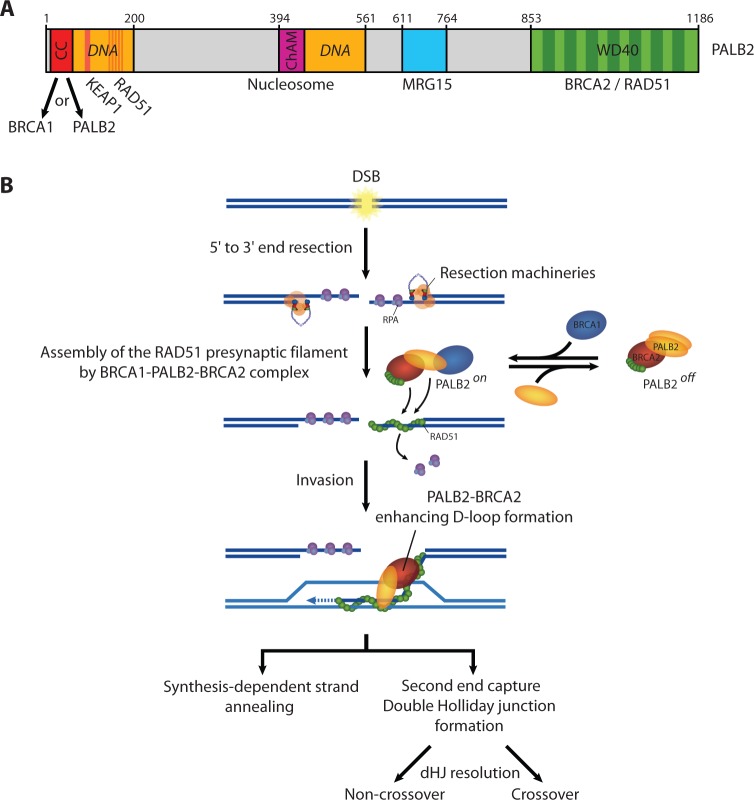
PALB2 was discovered in 2006 and rapidly established as a tumor suppressor. It was also characterized as a linker protein between BRCA1 and BRCA2 as cells treated with PALB2 siRNAs, but not a control siRNA, abolished the association between BRCA1 and BRCA2 (16). However, the function of PALB2 domains in HR remains poorly explored. The PALB2 structure shows an unusual level of complexity. It bears two DNA-binding domains, two RAD51 interacting regions, a ChAM domain, a seven-bladed WD40-type β-propeller, and a self-interaction domain (Figure 6A). Our results provide mechanistic insights into the function of PALB2 through its coiled-coil motif. It is well known that coiled-coil domains mediate protein–protein interactions, typical examples in the recombination field being RAD50 (30,31) and Hop2-Mnd1 (32). In previous studies, Sy and colleagues generated alanine substitution mutations in the heptad-helical coiled-coil arrangement on PALB2 (corresponding to residues K14A, L21A, Y28A, L35A and E42A). The interactions between BRCA1 and the PALB2 L21A, Y28A, or L35A mutants were largely abolished. Moreover, the latter mutants were defective for HR (14). In another study by the Andreassen group, two point mutants (L21P and L24P) were generated in the PALB2 coiled-coil domain, and the mutants did not restore resistance to mitomycin C in EUFA1341 fibroblasts, compared to full-length PALB2 (15). Using site directed mutagenesis, we were unsuccessful to fully dissociate BRCA1 binding to PALB2–PALB2 binding, suggesting a competition for binding to the same sites in the coiled-coil region. Therefore, we examined whether we could dissect the regulation of PALB2–PALB2 to BRCA1–PALB2 interactions using alternative biochemical and cellular approaches.
Figure 6.
(A) Schematic representation of PALB2 domains. (B) Model for the role of PALB2 during HR repair (see main text for description).
Our data suggest sequential protein–protein interactions to activate DNA repair. Using overexpression of the Our coiled-coil region, we artificially ‘locked’ PALB2 in a PALB2–PALB2 state. Notably, this affected greatly the ability of RAD51 to localize to DNA damage sites suggesting that PALB2–PALB2 interactions are unfavorable for HR. Therefore, we suggest that in the absence of DNA damage, PALB2 associates with itself and BRCA2 to sustain minimal HR. Following DNA damage, PALB2 dissociates and interacts with BRCA1 and BRCA2 to form the BRCA1–PALB2–BRCA2 complex to allow: (i) the localization of RAD51 at DSBs and, (ii) the stimulation of RAD51 filament formation on ssDNA (Figure 6B). This is consistent with the observation that BRCA1 interacts more strongly with PALB2 after DNA damage (Figure 5C). After RAD51 filament formation on resected DNA DSBs, PALB2 and BRCA2 stimulate RAD51 during the invasion step of HR (20,21,24). This step does not directly depend on PALB2 self-interaction as in vitro recombination assays revealed that PALB2Δ1–40 showed similar activity than wild-type PALB2. Indeed, in cells, PALB2 monomeric form is proficient for its localization to DSBs and to stimulate RAD51 foci formation. Subsequently, for the invasion step, only monomeric PALB2 will be present at strand breaks to stimulate RAD51 strand exchange activity.
In this study, we separated in vitro two functions of PALB2 mediator activity: the stimulation of RAD51 filament formation which is strongly affected by PALB2 self-interaction and the stimulation of RAD51 for strand invasion which functions independently of PALB2 self-interaction. Our model also speculates that the molecular switch between dimeric PALB2, which has minimal activity, to monomeric PALB2 of enhanced activity may be regulated by binding to other factors, such as BRCA1. The BRCA1 phosphorylation on S988 by Chk2 is important for RAD51 foci formation and HR activity (33,34). In a recent review, Powell and colleagues propose that this phosphorylation on BRCA1 promotes BRCA1–PALB2–BRCA2 complex formation [personal communication and (35)]. Thus, by activating the formation of the BRCA1–PALB2–BRCA2 complex, phosphorylated BRCA1 may control a PALB2 molecular switch. Furthermore, it has been shown recently that BRCA1 is important for the activation of HR versus NHEJ by promoting ssDNA resection through limiting 53BP1 loading at the DSB (36,37). Consequently, the regulation of PALB2 self-interaction in a BRCA1-dependent manner could be another way for the cell to control the choice between HR and NHEJ depending of its cell cycle state.
Altogether, our findings highlight a new mode of PALB2 regulation for HR. This molecular switch may prevent inappropriate recombination in normal conditions and sustain DNA repair to the appropriate level when cells are challenged with DNA damage.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Table 1 and Supplementary Figures 1–5.
FUNDING
FQRNT doctoral scholarship (to R.B.); J.Y.M. is a FRSQ senior investigator; the CIHR (to J.Y.M.). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. Funding for open access charge: CIHR.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors are grateful to Bing Xia for comments on this manuscript and providing a PALB2 antibody. The authors thank Patrick Sung, Eloise Dray, Fumiko Esashi, Anne-Marie Côté-Dion, Yan Coulombe and Julien Vignard for helpful discussions.
REFERENCES
- 1.Ciccia A, Elledge SJ. The DNA damage response: making it safe to play with knives. Mol. Cell. 2010;40:179–204. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moynahan ME, Jasin M. Mitotic homologous recombination maintains genomic stability and suppresses tumorigenesis. Nat. Rev. Mol. Cell Biol. 2010;11:196–207. doi: 10.1038/nrm2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.San Filippo J, Sung P, Klein H. Mechanism of eukaryotic homologous recombination. Ann. Rev. Biochem. 2008;77:229–257. doi: 10.1146/annurev.biochem.77.061306.125255. [DOI] [PubMed] [Google Scholar]
- 4.Davies AA, Masson JY, McIlwraith MJ, Stasiak AZ, Stasiak A, Venkitaraman AR, West SC. Role of BRCA2 in control of the RAD51 recombination and DNA repair protein. Mol. Cell. 2001;7:273–282. doi: 10.1016/s1097-2765(01)00175-7. [DOI] [PubMed] [Google Scholar]
- 5.Carreira A, Hilario J, Amitani I, Baskin RJ, Shivji MK, Venkitaraman AR, Kowalczykowski SC. The BRC repeats of BRCA2 modulate the DNA-binding selectivity of RAD51. Cell. 2009;136:1032–1043. doi: 10.1016/j.cell.2009.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xia B, Sheng Q, Nakanishi K, Ohashi A, Wu J, Christ N, Liu X, Jasin M, Couch FJ, Livingston DM. Control of BRCA2 cellular and clinical functions by a nuclear partner, PALB2. Mol. Cell. 2006;22:719–729. doi: 10.1016/j.molcel.2006.05.022. [DOI] [PubMed] [Google Scholar]
- 7.Tischkowitz M, Xia B. PALB2/FANCN: recombining cancer and Fanconi anemia. Cancer Res. 2010;70:7353–7359. doi: 10.1158/0008-5472.CAN-10-1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Erkko H, Xia B, Nikkila J, Schleutker J, Syrjakoski K, Mannermaa A, Kallioniemi A, Pylkas K, Karppinen SM, Rapakko K, et al. A recurrent mutation in PALB2 in Finnish cancer families. Nature. 2007;446:316–319. doi: 10.1038/nature05609. [DOI] [PubMed] [Google Scholar]
- 9.Rahman N, Seal S, Thompson D, Kelly P, Renwick A, Elliott A, Reid S, Spanova K, Barfoot R, Chagtai T, et al. PALB2, which encodes a BRCA2-interacting protein, is a breast cancer susceptibility gene. Nat. Genet. 2007;39:165–167. doi: 10.1038/ng1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jones S, Hruban RH, Kamiyama M, Borges M, Zhang X, Parsons DW, Lin JC, Palmisano E, Brune K, Jaffee EM, et al. Exomic sequencing identifies PALB2 as a pancreatic cancer susceptibility gene. Science. 2009;324:217. doi: 10.1126/science.1171202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reid S, Schindler D, Hanenberg H, Barker K, Hanks S, Kalb R, Neveling K, Kelly P, Seal S, Freund M, et al. Biallelic mutations in PALB2 cause Fanconi anemia subtype FA-N and predispose to childhood cancer. Nat. Genet. 2007;39:162–164. doi: 10.1038/ng1947. [DOI] [PubMed] [Google Scholar]
- 12.Xia B, Dorsman JC, Ameziane N, de Vries Y, Rooimans MA, Sheng Q, Pals G, Errami A, Gluckman E, Llera J, et al. Fanconi anemia is associated with a defect in the BRCA2 partner PALB2. Nat. Genet. 2007;39:159–161. doi: 10.1038/ng1942. [DOI] [PubMed] [Google Scholar]
- 13.Moldovan GL, D'Andrea AD. How the fanconi anemia pathway guards the genome. Ann. Rev. Genet. 2009;43:223–249. doi: 10.1146/annurev-genet-102108-134222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sy SM, Huen MS, Chen J. PALB2 is an integral component of the BRCA complex required for homologous recombination repair. Proc. Natl Acad. Sci. USA. 2009;106:7155–7160. doi: 10.1073/pnas.0811159106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang F, Fan Q, Ren K, Andreassen PR. PALB2 functionally connects the breast cancer susceptibility proteins BRCA1 and BRCA2. Mol. Cancer Res. 2009;7:1110–1118. doi: 10.1158/1541-7786.MCR-09-0123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang F, Ma J, Wu J, Ye L, Cai H, Xia B, Yu X. PALB2 links BRCA1 and BRCA2 in the DNA-damage response. Curr. Biol. 2009;19:524–529. doi: 10.1016/j.cub.2009.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oliver AW, Swift S, Lord CJ, Ashworth A, Pearl LH. Structural basis for recruitment of BRCA2 by PALB2. EMBO Rep. 2009;10:990–996. doi: 10.1038/embor.2009.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bleuyard JY, Buisson R, Masson JY, Esashi F. ChAM, a novel motif that mediates PALB2 intrinsic chromatin binding and facilitates DNA repair. EMBO Rep. 2012;13:135–141. doi: 10.1038/embor.2011.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sy SM, Huen MS, Zhu Y, Chen J. PALB2 Regulates Recombinational Repair through Chromatin Association and Oligomerization. J. Biol. Chem. 2009;284:18302–18310. doi: 10.1074/jbc.M109.016717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Buisson R, Dion-Cote AM, Coulombe Y, Launay H, Cai H, Stasiak AZ, Stasiak A, Xia B, Masson JY. Cooperation of breast cancer proteins PALB2 and piccolo BRCA2 in stimulating homologous recombination. Nat. Struct. Mol. Biol. 2010;17:1247–1254. doi: 10.1038/nsmb.1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dray E, Etchin J, Wiese C, Saro D, Williams GJ, Hammel M, Yu X, Galkin VE, Liu D, Tsai MS, et al. Enhancement of RAD51 recombinase activity by the tumor suppressor PALB2. Nat. Struct. Mol. Biol. 2010;17:1255–1259. doi: 10.1038/nsmb.1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jensen RB, Carreira A, Kowalczykowski SC. Purified human BRCA2 stimulates RAD51-mediated recombination. Nature. 2010;467:678–683. doi: 10.1038/nature09399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu J, Doty T, Gibson B, Heyer WD. Human BRCA2 protein promotes RAD51 filament formation on RPA-covered single-stranded DNA. Nat. Struct. Mol. Biol. 2010;17:1260–1262. doi: 10.1038/nsmb.1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thorslund T, McIlwraith MJ, Compton SA, Lekomtsev S, Petronczki M, Griffith JD, West SC. The breast cancer tumor suppressor BRCA2 promotes the specific targeting of RAD51 to single-stranded DNA. Nat. Struct. Mol. Biol. 2010;17:1263–1265. doi: 10.1038/nsmb.1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Henricksen LA, Umbricht CB, Wold MS. Recombinant replication protein A: expression, complex formation, and functional characterization. J. Biol. Chem. 1994;269:11121–11132. [PubMed] [Google Scholar]
- 26.Baumann P, Benson FE, Hajibagheri N, West SC. Purification of human Rad51 protein by selective spermidine precipitation. Mutat. Res. DNA Repair. 1997;384:65–72. doi: 10.1016/s0921-8777(97)00028-1. [DOI] [PubMed] [Google Scholar]
- 27.Yang H, Li Q, Fan J, Holloman WK, Pavletich NP. The BRCA2 homologue Brh2 nucleates RAD51 filament formation at a dsDNA-ssDNA junction. Nature. 2005;433:653–657. doi: 10.1038/nature03234. [DOI] [PubMed] [Google Scholar]
- 28.Genois MM, Mukherjee A, Ubeda JM, Buisson R, Paquet E, Roy G, Plourde M, Coulombe Y, Ouellette M, Masson JY. Interactions between BRCA2 and RAD51 for promoting homologous recombination in Leishmania infantum. Nucleic Acids Res. 2012;40:6570–6584. doi: 10.1093/nar/gks306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Golub EI, Gupta RC, Haaf T, Wold MS, Radding CM. Interaction of human Rad51 recombination protein with single-stranded-DNA binding-protein, RPA. Nucleic Acids Res. 1998;26:5388–5393. doi: 10.1093/nar/26.23.5388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hopfner KP, Craig L, Moncalian G, Zinkel RA, Usui T, Owen BA, Karcher A, Henderson B, Bodmer JL, McMurray CT, et al. The Rad50 zinc-hook is a structure joining Mre11 complexes in DNA recombination and repair. Nature. 2002;418:562–566. doi: 10.1038/nature00922. [DOI] [PubMed] [Google Scholar]
- 31.van Noort J, van Der Heijden T, de Jager M, Wyman C, Kanaar R, Dekker C. The coiled-coil of the human Rad50 DNA repair protein contains specific segments of increased flexibility. Proc. Natl Acad. Sci. USA. 2003;100:7581–7586. doi: 10.1073/pnas.1330706100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pezza RJ, Petukhova GV, Ghirlando R, Camerini-Otero RD. Molecular activities of meiosis-specific proteins Hop2, Mnd1, and the Hop2-Mnd1 complex. J. Biol. Chem. 2006;281:18426–18434. doi: 10.1074/jbc.M601073200. [DOI] [PubMed] [Google Scholar]
- 33.Lee JS, Collins KM, Brown AL, Lee CH, Chung JH. hCds1-mediated phosphorylation of BRCA1 regulates the DNA damage response. Nature. 2000;404:201–204. doi: 10.1038/35004614. [DOI] [PubMed] [Google Scholar]
- 34.Zhang J, Willers H, Feng Z, Ghosh JC, Kim S, Weaver DT, Chung JH, Powell SN, Xia F. Chk2 phosphorylation of BRCA1 regulates DNA double-strand break repair. Mol. Cell. Biol. 2004;24:708–718. doi: 10.1128/MCB.24.2.708-718.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roy R, Chun J, Powell SN. BRCA1 and BRCA2: different roles in a common pathway of genome protection. Nat. Rev. Cancer. 2012;12:68–78. doi: 10.1038/nrc3181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bunting SF, Callen E, Kozak ML, Kim JM, Wong N, Lopez-Contreras AJ, Ludwig T, Baer R, Faryabi RB, Malhowski A, et al. BRCA1 functions independently of homologous recombination in DNA interstrand crosslink repair. Mol. Cell. 2012;46:125–135. doi: 10.1016/j.molcel.2012.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bunting SF, Callen E, Wong N, Chen HT, Polato F, Gunn A, Bothmer A, Feldhahn N, Fernandez-Capetillo O, Cao L, et al. 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell. 2010;141:243–254. doi: 10.1016/j.cell.2010.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.