Abstract
The effect of dietary condensed tannins (proanthocyanidins) on rat fecal bacterial populations was ascertained in order to determine whether the proportion on tannin-resistant bacteria increased and if there was a change in the predominant bacterial populations. After 3 weeks of tannin diets the proportion of tannin-resistant bacteria increased significantly (P < 0.05) from 0.3% ± 5.5% to 25.3% ± 8.3% with a 0.7% tannin diet and to 47.2% ± 5.1% with a 2% tannin diet. The proportion of tannin-resistant bacteria returned to preexposure levels in the absence of dietary tannins. A shift in bacterial populations was confirmed by molecular fingerprinting of fecal bacterial populations by denaturing gradient gel electrophoresis (DGGE). Posttreatment samples were generally still distinguishable from controls after 3.5 weeks. Sequence analysis of DGGE bands and characterization of tannin-resistant isolates indicated that tannins selected for Enterobacteriaceae and Bacteroides species. Dot blot quantification confirmed that these gram-negative bacterial groups predominated in the presence of dietary tannins and that there was a corresponding decrease in the gram-positive Clostridium leptum group and other groups. Metabolic fingerprint patterns revealed that functional activities of culturable fecal bacteria were affected by the presence of tannins. Condensed tannins of Acacia angustissima altered fecal bacterial populations in the rat gastrointestinal tract, resulting in a shift in the predominant bacteria towards tannin-resistant gram-negative Enterobacteriaceae and Bacteroides species.
Acacia angustissima is a nitrogen-rich leguminous shrub which could be used to supplement poor-quality, high-fiber ruminant diets in developing countries. However, air-dried A. angustissima was lethal to Ethiopian Highland sheep (35). Condensed tannins (proanthocyanidins) were shown to be the antinutritional compounds in A. angustissima responsible for reduced intake and reduced weight gain in rats (55, 56). These polyphenolic secondary compounds, produced by a wide variety of plants, may protect against herbivory, increase resistance to pathogens, or protect tissues, such as wood, against decay (48). The antinutritional effects of tannins are dependent on the concentration and type of the tannin (44), but high tannin concentrations generally reduce intake and protein and carbohydrate digestibility.
Tannin-resistant or -insensitive bacteria have been isolated in recent years from gastrointestinal tract ecosystems (7, 33, 34, 37, 38). Tannin-resistant microorganisms in the rumen are thought to prevent detrimental effects on the animal due to tannins in the diet and may be able to confer protection to animals not adapted to a tannin-containing diet. Hydrolyzable tannins and phenolic monomers such as flavonols can be degraded in the anaerobic environment of the intestinal tract (19, 21, 32, 34, 39, 40, 49, 59), but intestinal bacteria that can degrade condensed tannins and their monomeric units, flavan-3-ols, have not been described to date, although attempts have been made to isolate them (23, 24). The mechanism of action of tannin-resistant bacteria in animals exposed to condensed tannins is not known, but the following previously described results are evidence of their protective effect. A period of adaptation or rumen inoculum from adapted animals allowed Ethiopian Highland sheep to utilize an A. angustissima-supplemented diet without adverse effects, indicating that tolerance to the diet occurs at the level of the microbial population (35). Feral goats in Australia tolerated diets containing Acacia aneura (mulga), which may contain from 5 to 25% condensed tannin, better than sheep tolerated such diets. Goat rumen fluid or an in vitro cultured rumen inoculum from goats improved nitrogen digestion in sheep fed mulga (27, 29). However, inoculation of a pure culture of tannin-resistant Streptococcus gallolyticus (formerly Streptococcus caprinus [53]) was ineffective (28). In other studies pure-culture inocula of tannin-resistant bacteria effectively improved the performance of animals fed tannin-rich diets. Inoculation of a pure culture of an uncharacterized tannin-resistant bacterium from goats (3 × 1011 CFU) was shown to significantly increase body weight gain over 40 days in goats abruptly changed from a grass diet to a 100% Calliandra calothyrsus diet (containing 6 to 10% condensed tannins) compared to the body weight gain of uninoculated goats (60). Lambs fed a 7.1% peanut skin tannin diet were inoculated with a tannin-resistant gram-positive rod, and a positive effect on the crude protein balance was observed in animals that received this live inoculum (30).
Previously published data therefore indicate that increasing the proportion of tannin-resistant organisms in the gastrointestinal tract before exposure to a tannin-containing diet can have a positive effect on digestion, although the mechanisms of the protective effect are not clear. It is not known if the tannin-resistant organisms isolated to date are representative of tannin-resistant bacteria in the intestinal tract as very few ecological studies of the effects of polyphenolics on intestinal bacteria have been described Monomeric polyphenols from green tea were shown to affect gastrointestinal bacteria in humans (36), pigs (13), and chickens (14) based on cultivation of specific bacterial groups. Preliminary ecological studies of the effect of plant-based diets containing condensed tannins on the ruminal microbial population have been described (25, 41, 60), and these studies indicated that there were shifts in microbial populations but did not indicate which bacteria were the predominant tannin-resistant bacteria.
It is not feasible to purify enough condensed tannins from fodder plants to feed ruminants. Therefore, in this study rats were used as a model. Similar bacterial types are present in the foregut and hindgut, but their role in fiber digestion is less critical in the hindgut. Condensed tannins extracted from the potential fodder legume A. angustissima were used to determine the effect of condensed tannins on intestinal bacteria. Our aims were to determine whether (i) the proportions of tannin-resistant bacteria in the gastrointestinal tract increased when tannins were present in the diet, (ii) selective pressure was required to maintain high proportions of tannin-resistant bacteria in the gastrointestinal tract, (iii) there was a change in the predominant bacterial populations, and (iv) a shift in bacterial populations altered or affected the functional activity of the microbial population.
In this paper we describe our finding that condensed tannins of A. angustissima cause a shift in the predominant fecal bacteria towards tannin-resistant gram-negative Enterobacteriaceae and Bacteroides species.
MATERIALS AND METHODS
Animals.
Eighteen adult Wistar Furth rats (average weight, 296 g) were obtained from Harlan Sprague Dawley, Indianapolis, Ind., and were housed individually in cages with wire bottoms at an ambient temperature of 25°C with a 12-h photoperiod. The Institutional Animal Care and Use Committee of the University of Illinois approved the animal use protocol (protocol 01032). At the end of the trial the rats were transferred to another protocol.
Diets and experimental procedure.
The basal diet was powdered AIN-93M rodent diet (45) to which tannins extracted from A. angustissima were added in order to supplement the diet with 0.7% (low-tannin diet) or 2.0% (high-tannin diet) condensed tannins. The control (rats 1 to 6) and the group fed the low-tannin diet (rats 7 to 12) were limit fed during the treatment period on the basis of the intake of paired rats fed the high-tannin diet (rats 13 to 18) in order to minimize changes in the bacterial populations due to differences in nutrient intake levels. The basal diet was used for 3 weeks to acclimatize the rats to the feeding regimen and to stabilize the intestinal bacterial population before the experimental diets were used (pretreatment). The treatment diets were used for 3.5 weeks (treatment period), and then the rats were returned to the basal diet for an additional 3.5 weeks (posttreatment) to monitor long-term changes. Fresh fecal samples were collected weekly beginning in week 2 for analysis of bacterial populations. The experiment was staggered so that six samples were processed for bacterial counting and metabolic fingerprinting each day (two samples per treatment group). Fecal samples were collected on Monday, Wednesday, and Friday.
Two of the rats, one fed the low-tannin diet (rat 7) and the other fed the high-tannin diet (rat 16), lost excessive amounts of weight during the treatment period due to limited intake and were removed from the study together with the rats that were pair-fed with rat 16, rats 4 (control) and 10 (low-tannin diet).
Tannin extraction.
A. angustissima foliage (International Livestock Research Institute accession no. 15132), grown in the Ethiopian highlands, which had been harvested and air dried, was obtained from the International Livestock Research Institute, Debre Zeit, Ethiopia, and milled with a 2-mm screen. Tannins were extracted and measured as described previously (56). The condensed tannin concentration of the final material was 72% (standard deviation, 11.4%).
Bacterial cultivation and enumeration.
Most-probable-number (MPN) estimation and plate counting were performed by using a defined total count (DTC) medium with and without 0.05% A. angustissima tannin extract to determine the proportion of total culturable fecal bacteria which were tannin resistant. One or two fresh fecal pellets (obtained less than 1 h after excretion) were diluted 10−2 in anaerobic diluent and homogenized with a Tissue Tearor (model 985-370 type 2; Biospec Products, Inc.) for 1 min at speed 2. The sample was then serially diluted before plates or MPN tubes were inoculated. The MPN estimates were obtained by using a miniaturized, five-tube MPN method with 96-well plates. Twenty microliters of the appropriate dilution was inoculated into 180 μl of medium in five replicate wells. The plates were sealed with sealing tape to prevent evaporation and incubated at 37°C for 7 days in an anaerobic chamber (Coy Laboratory Products, Ann Arbor, Mich.) under an atmosphere containing 95% CO2 and 5% H2. Control wells were inoculated with diluent. The optical density at 620 nm of each well was determined with a Thermomax microplate reader (Molecular Devices, Menlo Park, Calif.). Agar plates were incubated in the same way, but 100-μl portions of the appropriate dilutions were inoculated in triplicate. The anaerobically prepared DTC medium contained mineral solutions I and II (8), trace element solution SL-10 (DSMZ-Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH, Braunschweig, Germany), a hemin solution (15), a vitamin solution, and a volatile fatty acid solution (9). All these solutions were added at the concentrations recommended in the original publications. Vitamin K1 (10 ng/liter) was added as an additional vitamin not present in the vitamin solution to allow growth of Bacteroides species. Sodium carbonate (4 g/liter) was added to buffer the carbon dioxide-saturated medium. In place of Trypticase, which precipitates with tannins, Casamino Acids (2 g/liter) was added as an additional nitrogen source to supplement the ammonium present in the mineral solution. A cysteine-sulfide solution was added as a reducing agent so that the final concentration of l-cysteine-HCl and the final concentration of Na2S · 9H2O were each 0.025%. The carbohydrates used were 2 g of sodium lactate per liter, 0.5 g of glucose per liter, 0.5 g of cellobiose per liter, 0.5 g of xylan per liter, 0.5 g of maltose per liter, 0.5 g of pectin per liter, and 0.5 g of xylose per liter. A. angustissima tannin extract (0.05%) was added as a methanol solution before the medium was dispensed for MPN estimation. The same amount of methanol (0.8%) was also added to DTC medium. Tannin-containing plates were prepared the day before the counts were obtained. This was to reduce possible oxidation or light degradation of the tannins during storage of the medium. Tannin-resistant bacteria were picked from plates, grown in DTC medium, and stored at −70°C under sterile mineral oil for further characterization. Count data were analyzed by using the least-squares means analysis procedure in the SAS system (47). The MPN counts for week 4 were removed from the data set as there was contamination of blank wells. MPN estimates and plate counts were analyzed together as there was no significant difference between MPN estimates and plate counts. When the proportions of tannin-resistant bacteria were analyzed, negative proportions and proportions over 100% were recorded as zero and 100%, respectively.
DNA fingerprinting of bacterial populations.
Total genomic DNA in feces was extracted with an Ultraclean soil DNA isolation kit (Mo Bio Laboratories, Inc., Solana Beach, Calif.) according to the manufacturer's instructions, using approximately 0.05 g of fecal material stored at −70°C. To overcome inhibition of PCR by tannins, the extracted DNA was diluted 1:4 in Tris-EDTA buffer before amplification of the V3 variable region of 16S rRNA gene. Each 25-μl reaction mixture contained 1 μl of diluted genomic DNA, 12.5 pmol of each primer (31), 2 μl of a deoxynucleoside triphosphate mixture, 2.5 μl of 10× ExTaq buffer, and 0.1 μl of TaKaRa ExTaq polymerase (TaKaRa Shuzo, Otsu, Japan). The PCR consisted of 10 cycles of touchdown PCR in which the annealing temperature was decreased 1°C every cycle from 65 to 55°C, followed by 10 cycles with an annealing temperature of 55°C for 30 s. Denaturation was at 94°C for 30 s, and elongation was at 72°C for 4 min, as described previously for approaches suggested to minimize PCR artifacts (42). The number of cycles generally used for denaturing gradient gel electrophoresis (DGGE) was reduced from 30 to 20 as the number of PCR cycles for amplification of 16S rRNA genes should be kept as small as possible for phylogenetic diversity studies (6). Removal of single-stranded DNA, DGGE, staining, and analysis of gel patterns were performed as described previously (52). In short, mung bean nuclease (Stratagene, La Jolla, Calif.) treatment was used to remove single-stranded DNA after PCR. PCR products were separated on a gel containing a 35 to 60% denaturing gradient that was electrophoresed at 150 V for 2 h and then at 200 V for 1 h. Silver-stained gels were scanned with a GS-710 calibrated imaging densitometer (Bio-Rad Inc., Hercules, Calif.), and gel patterns were analyzed by using Diversity Database 2.1 (Discovery Series; Bio-Rad, Inc.). Molecular fingerprint patterns were compared by using Dice's similarity coefficient and Ward's algorithm. PCR amplification may not accurately estimate the relative abundance of DNA sequences, because of PCR bias and selection for certain sequences. Therefore, a more intense band may not indicate greater abundance in a sample. However, fecal populations from rats exposed to tannins generally had fewer DGGE bands, and the bands were more intense. Further analysis by sequencing of DGGE amplicons and quantification of selected bacterial groups by dot blot hybridization indicated that the groups represented by the more intense DGGE bands were numerically more predominant. Therefore, band intensity was taken into account when the molecular fingerprints were analyzed.
Cloning and sequencing of V3 16S rRNA gene DGGE amplicons.
Predominant DGGE bands present at week 6 were excised from samples from two rats that received the control diet (rats 5 and 6), two rats that received the low-tannin diet (rats 11 and 12), and two rats that received the high-tannin diet (rats 17 and 18). The bands were each placed in 50 μl of sterile Tris-EDTA buffer at 4°C overnight and then stored at −20°C. After thawing, each gel matrix was disrupted with a pipette tip, and a 2-μl aliquot was removed and reamplified by PCR by using the original primers; however, the number of PCR cycles was increased to 30, with touchdown PCR for the first 20 cycles, as no product was visible after 20 cycles. The PCR products were cloned into the pCR2.1 Topo vector (Topo TA cloning kit; Invitrogen) or pGEM-T Easy Vector System I (Promega Corporation, Madison, Wis.) used according to the manufacturers' instructions. Restriction profiles of the clones with inserts of the correct size were generated with a mixture of the frequent cutters AluI (Gibco BRL), MspI (Gibco BRL), and either ThaI (Gibco BRL) or BstUI (New England Biolabs, Inc.). Selected clones from each reamplified band with different restriction patterns were sequenced at the W. M. Keck Center for Comparative and Functional Genomics, Biotechnology Center, University of Illinois. Sequences were aligned by using Clustal X (1) together with the nearest identified neighbors identified by Blast analysis (2) and retrieved from the GenBank database (National Center for Biotechnology Information, Bethesda, Md.). Phylogenetic trees were constructed by the neighbor-joining method (46).
Dot blot hybridization.
Dot blot hybridization was used to quantify the shifts in certain bacterial populations at week 6 (treatment period) and weeks 8 and 10 (posttreatment) by using 16S RNA-targeted oligonucleotide probes (Table 1). The probes were chosen on the basis of the shift indicated by sequencing DGGE amplicons and were selected to represent groups that have a gram-negative or gram-positive cell wall structure. Total RNA was extracted from fecal material, which had been stored at −80°C, by a hot-phenol method described previously (50). RNA was extracted from pure cultures of Escherichia coli BW13711, Bacteroides fragilis VPI2553, and Ruminococcus flavefaciens FD-1 as standards for quantification. The quality of the RNA was checked on a 1% agarose gel after denaturation for 10 min at 70°C in 5 M (final concentration) urea. The RNA concentration and purity were determined by UV spectrophotometry (17). Dot blot hybridization was performed as described previously (43). In short, RNA samples (100 ng) from feces and dilution series of the pure cultures were applied in duplicate to Hybond-N+ positively charged membranes (Amersham Pharmacia Biotech) after denaturation in 3 volumes of 2% gluteraldehyde in 50 mM phosphate buffer (pH 7.0). Samples were applied by using a 96-well Miniford I dot blot system (Midwest Scientific, St. Louis, Mo.) under a vacuum. The membranes were air dried before hybridization. Oligonucleotide probes (Sigma Genosys, The Woodlands, Tex.) were labeled with 32P by using polynucleotide kinase (Roche Pharmaceuticals) and gamma-labeled ATP (ICN Biomedicals, Irvine, Calif.). Unincorporated ATP was removed with a Nensorb nucleic acid purification cartridge (NEN Life Science Products, Boston, Mass.) used according to the manufacturer's instructions. Membranes were prehybridized in PerfectHyb Plus hybridization buffer (Sigma Genosys) for 15 min at 40°C before addition of the labeled probe. After overnight hybridization membranes were washed twice in 100 ml of a 1× SSC-1% sodium dodecyl sulfate wash solution at 40°C (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate). Each membrane was then washed for 30 min in 200 ml of the wash solution at the specified wash temperature for each probe (Table 1). After washing, the membranes were air dried and exposed to phosphor screens. The screens were scanned with a PhosphorImager (BAS1800-II; Fujifilm Medical Systems, Stamford, Conn.) and were quantified with Image Gauge, version 3.4. Standard curves were constructed for each membrane with the appropriate pure-culture RNAs and were used to determine the concentration of RNA in each sample. Data were analyzed by using the General Linear Model in the SAS system (47) after transformation of the data. Negative values were recorded as zero, and values over 100% were recorded as 100%.
TABLE 1.
16S rRNA-targeted oligonucleotide probes and buffer wash temperatures for the experimental conditions used
| Probe | Target | Sequence (5′ to 3′) | Temp (°C) | Reference |
|---|---|---|---|---|
| S-D-Bact-0338-a-A-18 (EUB338-I) | Eubacteria | GCTGCCTCCCGTAGGAGT | 54 | 3 |
| EUB338-II | Eubacteria | GCAGCCACCCGTAGGTGT | 54 | 10 |
| EUB338-III | Eubacteria | GCTGCCACCCGTAGGTGT | 54 | 10 |
| S-*-Bacto-1080-a-A-18 (Bacto) | Bacteroides-Prevotella-Porphyromonas group | GCACTTAAGCCGACACCT | 50 | 11 |
| S-*-Bfra-0602-a-A19 (Bfra) | Bacteroides fragilis group | GAGCCGCAAACTTTCACAA | 53 | 12 |
| S-G-Enter-1432-a-A-15 (Enter) | Enteric bacteria | CTTTTGCAACCCACT | 43 | 51 |
| S-G-Clept-1240-a-A018 (Clept) | Clostridium leptum groupa | GTTTTRTCAACGGCAGTC | 48.5 | 51 |
The probe also detects Desulfotomaculum species according to Probe Match (http://rdp.cme.msu.edu/docs/probe_match_doc.html).
Metabolic fingerprinting.
Metabolic fingerprints were obtained for fresh fecal samples by kinetic measurement of the utilization of 46 different substrates with a Phene Plate 96-well generalized microplate (PhPlate AB, Stockholm, Sweden). A 10-ml portion of a 10−2 dilution of fresh feces in anaerobic diluent was filtered under sterile conditions to remove fibrous material. The sample was centrifuged (HN-SII benchtop centrifuge; International Equipment Company, Needham, Mass.) at the maximum speed for 10 min, and the resultant cell pellet was suspended in 12 ml of prereduced PhP growth medium (20) under a nitrogen atmosphere. Vitamin K1 (10 ng/liter) and a hemin solution were added to the medium to allow growth of Bacteroides species. Phene Plates were inoculated and incubated, and absorbance values were determined after 7, 24, and 48 h as described by Katouli et al. (20). The utilization rate (UR) of each substrate was determined by using the following formula: UR = 1 − Xa/Ca or UR = 1 − Ck/Xk, where Xa and Xk are the average values obtained from kinetic measurements of the acidic and alkaline reactions, respectively, and Ca and Ck are values of the negative controls in the acidic and alkaline tests, respectively. Pyruvate utilization under anaerobic conditions resulted in an acid reaction and was therefore studied as an acid reaction, not an alkaline reaction. Unused substrates had a value of zero, and the values increased as the rate of utilization increased. The value for the 46 wells of each sample was the metabolic fingerprint of the sample. Text files containing the utilization rate values for all wells in each sample were imported into the Diversity Database fingerprinting software (Bio-Rad Inc.) to enable similarity searches to be performed with these values. The similarities between substrate utilization patterns were computed by the Dice similarity coefficient method, and Ward's algorithm was used to produce dendrograms that showed the pattern similarity between samples. Two samples (15W04 and 14W06) were removed from the metabolic fingerprint analysis as the control wells changed color and the resulting values were in a different range than those of the other samples.
RESULTS
Analysis of tannin-resistant populations by cultivation and enumeration.
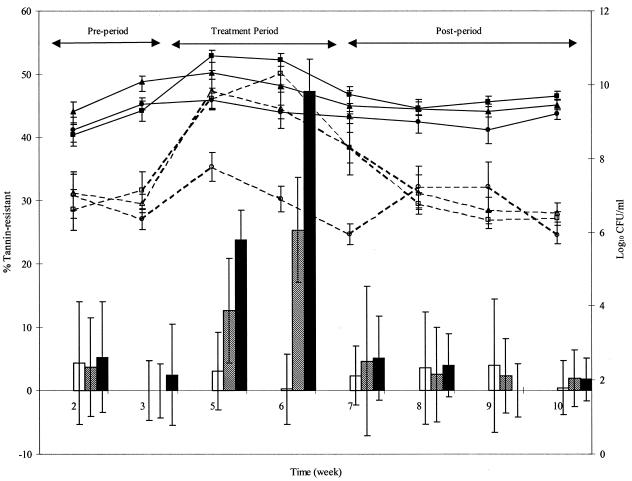
Total fecal bacterial counts, tannin-resistant fecal bacterial counts, and the proportion of fecal bacteria that were tannin-resistant are shown in Fig. 1. The counts of total and tannin-resistant fecal bacteria increased significantly during the treatment period for rats fed the tannin diets compared to the rats fed the control diet. At week 7, 3 days posttreatment the tannin-resistant bacterial counts were still higher. This did not, however, result in a higher proportion of culturable tannin-resistant fecal bacteria, as it did when the rats were fed the tannin-containing diets. There was selection for tannin-resistant bacteria when tannins were present in the diet, as shown by the increase in the proportion of tannin-resistant bacteria, and selective pressure was required to maintain higher proportions.
FIG. 1.
Counts (log10 CFU per milliliter) of total fecal bacteria (solid lines) and tannin-resistant fecal bacteria (dashed lines) and proportion of tannin-resistant fecal bacteria (bars) during the pretreatment period, the treatment period, and the posttreatment period for rats fed diets containing no tannin (circles and open bars), a low level of tannins (triangles and bars with diagonal lines), and a high level of tannins (squares and solid bars); the tannins used were A. angustissima condensed tannins. The error bars indicate standard deviations.
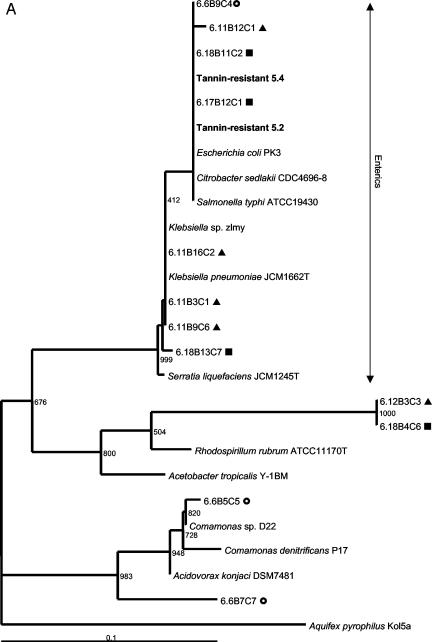
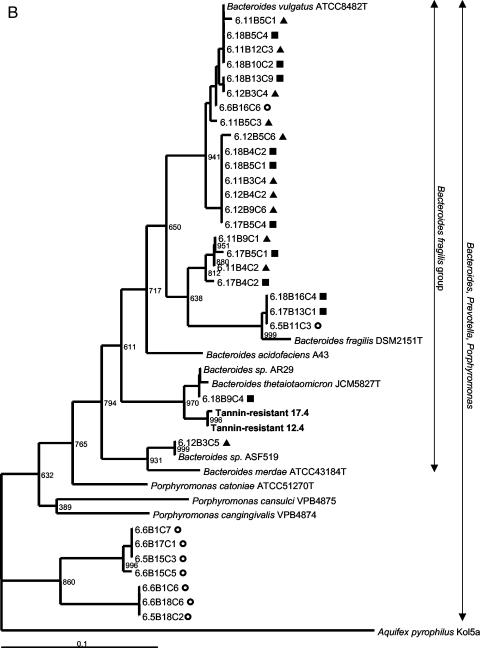
Two colony morphologies could be distinguished on tannin-containing agar plates. The rod-shaped bacteria in colonies growing as concentric circles were 99% similar to E. coli according to full-length 16S rRNA gene sequence analysis (GenBank accession numbers AY319392 to AY319395). The irregular, mucoid colonies consisted of oval cocci which were 98% similar to Bacillus thetaiotaomicron. The phylogenetic relatedness to the nearest identified neighbors based on the V3 16S rRNA gene sequence is indicated in Fig. 2. The predominant culturable tannin-resistant bacteria from rat feces were presumptive strains of E. coli and B. thetaiotaomicron.
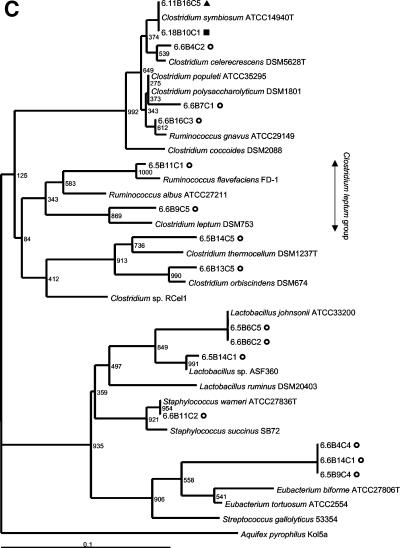
FIG. 2.
Phylogenetic placement of V3 16S rRNA gene sequences of rat fecal bacteria excised from predominant DGGE bands in the Proteobacteria (A), the Bacteroides class (B), and the Firmicutes (C). Rats were fed a control diet (○) or a diet containing a low level (▴) or a high level (▪) of A. angustissima tannin. Bacteria isolated from tannin-containing plates are indicated by boldface type. Bacteria detectable with the probes used for the dot blot hybridization study are indicated with vertical arrows. Bootstrap resampling values based on 1,000 replicates are indicated at the nodes of the tree. Aquifex pyrophilus was used as the outgroup for rooting the trees. Scale bar = 1 fixed nucleotide position per sequence.
Analysis of fecal bacteria by DNA fingerprinting.
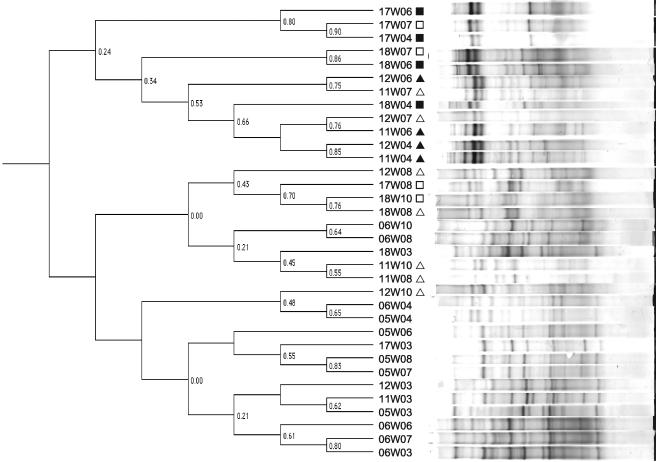
A shift in fecal bacterial populations was confirmed by PCR-DGGE analysis. The DNA fingerprints of fecal samples from rats that had been exposed to tannins were more similar to each other than to fingerprints from the control samples. Results are shown in Fig. 3 for two control (rats 5 and 6), two rats that received the low-tannin diet (rats 11 and 12), and two rats that received the high-tannin diet (rats 17 and 18) at weeks 3 (pretreatment), weeks 4 and 6 (treatment period), and weeks 7, 8, and 10 (posttreatment). Two samples (05W10 and 17W10) were not included in Fig. 3 due to problems with DNA extraction and PCR amplification from stored fecal samples after repeated attempts. Three days posttreatment, the DNA fingerprint patterns were still indistinguishable from the patterns obtained for the rats that were receiving the tannin diet. The later posttreatment DNA fingerprint patterns were more similar to the pretreatment and control patterns but still tended to cluster together. The majority (95%) of the sequences obtained from predominant DGGE bands were phylogenetically related to three groups. Fifty percent of the sequences obtained from predominant DGGE bands could be placed phylogenetically in the Bacteroides class, 26% could be placed in the Firmicutes (gram-positive bacteria), and 19% could be placed in the Proteobacteria. The relationships of the sequenced amplicons from each of the treatment groups to the nearest identified neighbors in the GenBank database are shown in phylogenetic trees in Fig. 2. Not shown are sequences obtained from both control and low-tannin samples that were 92% similar to Verrucomicrobium spinosum (Verrucomicrobia) and a sequence from a low-tannin sample with 92% similarity to Actinomyces catuli (Actinobacteria). Bacterial sequences that predominated in animals that received the tannin diets as determined by DGGE analysis belonged to the Enterobacteriaceae or the B. fragilis group. This corresponds to the results for the predominant tannin-resistant isolates obtained in culture studies. The sequences obtained from control rats were aligned with sequences of phylogenetically diverse gastrointestinal bacteria.
FIG. 3.
Relationships of PCR-amplified V3 16S rRNA gene fragment banding patterns from rat fecal genomic DNA from six animals (two for each treatment) obtained at week 3 (pretreatment), weeks 4 and 6 (treatment period), and weeks 7, 8, and 10 (posttreatment). Rats were fed a control diet (no symbol) or a diet that contained a low level (triangles) or a high level (squares) of A. angustissima condensed tannin. Patterns that were obtained posttreatment are indicated by open symbols. Similarity indices (Dice coefficients) are indicated at the nodes of the tree (Ward's).
Quantification of shifts in bacterial populations.
Dot blot hybridization was performed to quantify the shifts in bacterial populations indicated by the culture and molecular data. Quantification of bacterial groups obtained from rat feces at week 6 indicated that the level of the Clostridium leptum subgroup decreased significantly, while the levels of enteric bacteria, the B. fragilis group, and the Bacteroides-Prevotella-Porphyromonas group increased significantly in rats fed tannin-containing diets (Table 2). At weeks 8 and 10, when all rats were fed the control diet, the populations were more similar. However, the level of the B. fragilis group in rats exposed to tannin-containing diets was still significantly increased compared to the level in the control group at week 10. Differences between the data for the two tannin diets were not significant. These results confirm that tannins cause large shifts in fecal bacterial populations, reducing the number of the gram-positive C. leptum group and selecting for gram-negative Enterobacteriaceae and Bacteroides species.
TABLE 2.
Relative RNA indices of total fecal bacterial RNA as measured with 16S probesa
| % of condensed tannin | Relative RNA indices
|
|||
|---|---|---|---|---|
| C. leptum group | Enteric bacteria | B. fragilis group | Bacteroides-Prevotella- Porphyromonas group | |
| Week 6 (treatment period) | ||||
| 0.0 | 23.12 (21.44-24.85) Ab | 0.36 (0.10-0.79) B | 8.08 (4.77-12.15) B | 27.05 (17.04-38.41) B |
| 0.7 | 0.20 (0.08-0.39) B | 4.27 (3.31-5.33) A | 25.06 (20.15-30.32) A | 61.67 (51.27-71.56) A |
| 2.0 | 0.00 (0.01-0.06) B | 6.91 (5.70-8.23) A | 42.11 (36.37-47.96) A | 85.56 (77.45-92.11) A |
| Week 8 (posttreatment) | ||||
| 0.0 | 35.15 (31.16-39.25) | 0.34 (0.06-0.83) | 2.45 (0.88-4.77) | 16.83 (12.75-21.35) |
| 0.7 | 26.85 (22.75-31.16) | 0.09 (0.01-0.45) | 7.58 (4.28-11.72) | 17.20 (12.62-22.32) |
| 2.0 | 24.40 (20.85-28.13) | 1.04 (0.47-1.83) | 4.93 (2.57-8.01) | 12.74 (9.15-16.82) |
| Week 10 (posttreatment) | ||||
| 0.0 | 18.57 (16.36-20.89) | 0.09 (0.03-0.19) | 0.67 (0.31-1.16) B | 9.17 (6.87-11.78) |
| 0.7 | 17.32 (14.92-19.85) | 0.16 (0.07-0.30) | 6.20 (4.87-7.68) A | 15.04 (11.81-18.59) |
| 2.0 | 23.34 (20.92-25.85) | 0.03 (0.00-0.10) | 4.93 (3.87-6.12) A | 15.05 (12.14-18.22) |
Fecal bacterial RNA was extracted from samples obtained from rats (n = 4) fed control (no tannin), low-tannin (0.7% tannin), and high-tannin (2% tannin) diets. Standard deviations of transformed data are indicated in parentheses.
Means followed by the same letter within columns and weeks are not significantly different (P ≥ 0.05).
Analysis of fecal bacteria by metabolic fingerprinting.
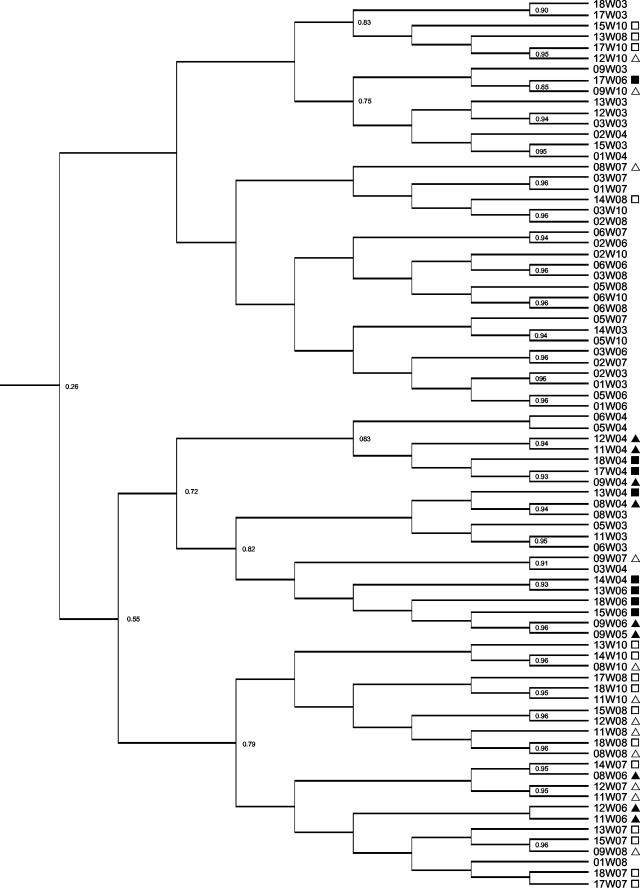
Metabolic fingerprints of the culturable fecal bacteria were obtained by kinetic measurement of the utilization of 46 substrates by the Phene Plate procedure. Most substrates were utilized to some degree, but similarity indices based on the utilization rate indicated that there were differences between treatment groups. The metabolic fingerprint patterns for fecal bacteria from rats fed the tannin diets tended to cluster together. Figure 4 shows the results obtained at week 3 (pretreatment), weeks 4 and 6 (treatment period), and weeks 7, 8, and 10 (posttreatment). The metabolic fingerprint patterns posttreatment were generally still distinguishable from the control patterns at the end of the experiment. The metabolic fingerprints indicated that there was a shift in the functional activity of the culturable fecal bacteria while tannins were present in the diet.
FIG. 4.
Relationships of metabolic fingerprint patterns generated after rat fecal incubation for week 3 (pretreatment), weeks 4 and 6 (treatment period), and weeks 7, 8, and 10 (posttreatment). Rats were fed a control diet (no symbol) or a diet containing a low level (triangles) or a high level of (squares) of A. angustissima condensed tannin. Patterns that were obtained posttreatment are indicated by open symbols. Similarity indices (Dice coefficients) are indicated at the nodes of the tree (Ward's).
DISCUSSION
No single method can accurately describe the total community in a complex ecosystem such as the intestinal tract. In this study we focused on the bacteria in this ecosystem before, during, and after dietary exposure to condensed tannins using a combined approach that involved four different methods. The total counts and the results obtained by metabolic fingerprinting are indicators of the culturable bacteria. The DGGE and dot blot hybridization analyses included the nonculturable members of the bacterial community. All four methods confirmed that there was a shift in the bacterial population, which was maintained to some degree over time, as indicated by metabolic fingerprinting, DGGE, and dot blot hybridization. The bacteria shown to be predominant in feces from rats fed tannin-containing diets by DGGE and dot blot hybridization were also isolated from tannin-containing plates, indicating that the predominant bacteria were tannin resistant and that there was a correlation between the molecular and culture methods used to identify bacteria.
Our first hypothesis, that the level of tannin-resistant bacteria increased in the presence of tannins in the diet, proved to be correct. The proportions of tannin-resistant fecal bacteria increased in the presence of dietary tannins. The increase was determined to be dosage dependent with the two levels of tannins used in this study. A similar dosage-dependent increase was reported previously as ruminal tannin-resistant bacterial populations increased in goats as the tannin concentration in the diet increased (60).
Data from determinations of the counts of culturable fecal bacteria also confirmed the second hypothesis, that selective pressure is required to maintain high proportions of tannin-resistant bacteria. At week 7, 3 days posttreatment, the tannin-resistant bacterial counts were still higher, although the proportion of culturable tannin-resistant fecal bacteria was back to preexposure levels. Presumably, tannins were not present in the intestinal tract at this time, although the transit time of A. angustissima tannins in the digestive tract of rats is not known. However, in vivo studies with the monomeric polyphenol epigallocatechin gallate (ECGg) in rats indicated that ECGg was recovered in feces after 12 h (14). After 20 h the majority of residual ECGg was recovered in the feces, and a smaller proportion was in the large intestine. These results imply that 3 days after treatment ended there should be no or little residual tannin in the intestinal tract. Higher levels of culturable tannin-resistant bacterial populations were still present 3 days posttreatment, but the levels decreased by 10 days posttreatment. Metabolic fingerprinting, DGGE, and dot blot hybridization indicated that the shift in the bacteria was maintained to some degree over time, which was probably manifested as a higher proportion of the B. fragilis group.
There was a change in the predominant bacterial populations, confirming the third hypothesis. Our results show that there was a decrease in the proportion of the low-G+C-content gram-positive bacteria, and the Bacteroides-Prevotella-Porphyromonas group and Enterobacteriaceae predominated in samples from rats fed a tannin diet. This finding agrees with the results of a molecular microbial ecology study in which the rumen bacterial ecology of a goat fed A. aneura (mulga) was compared to that of sheep fed grass or mulga. Clone libraries were constructed from one animal fed each diet (41). In both the sheep and the goat fed mulga members of the Cytophaga-Flexibacter-Bacteroides group predominated (78 and 82%, respectively), while members of the low-G+C-content gram-positive bacteria predominated (74 and 25% Cytophaga-Flexibacter-Bacteroides group) in the sheep fed grass. In addition, higher numbers of members of the γ subclass of the Proteobacteria (enteric bacteria) were found in the goat rumen than in the rumen of the sheep fed grass.
In contrast, however, the level of the Bacteroides-Prevotella-Porphyromonas group was not significantly different in sheep whose diet was supplemented with C. calothyrsus or alfalfa (25). In this study estimates of the numbers of cellulolytic bacteria obtained by using a MPN method indicated that cellulolytic populations were reduced by C. calothyrsus in the diet. The use of genus-specific 16S rRNA gene probes confirmed that there was a reduction in the cellulolytic population, as both the Fibrobacter and Ruminococcus populations were significantly smaller in sheep whose diet was supplemented with C. calothyrsus. This does correspond with our results since A. angustissima tannins resulted in a decrease in the proportion of the C. leptum group, which includes the cellulolytic ruminococci.
Green tea catechins, which are monomeric polyphenols, have also been shown to cause a shift in bacterial populations. Polyphenolics from green tea affect gastrointestinal bacteria in humans (36), chickens (14), and pigs (13). The studies were done by counting the fecal bacteria on a number of selective media. In the human study, 4 weeks of consuming a polyphenolic content equivalent to 10 cups of concentrated green tea were necessary for there to be a significant decrease in the counts of total Clostridium species and Clostridium perfringens. Other groups of bacteria, including the Bacteroidaceae, were not affected significantly. All bacterial counts were back to normal 2 weeks after polyphenolic intake was discontinued (36). In chickens the levels of cecal lactobacilli increased significantly, while the levels of Enterobacteriaceae decreased (14). In pigs, eating a diet containing 0.2% tea polyphenols for 2 weeks resulted in significantly increased levels of lactobacilli and a decrease in the levels of total bacteria and Bacteroidaceae in the feces. There was a tendency for the level of lecithinase-positive clostridia to decrease, but the difference was not significant (13). All these studies confirmed that polyphenolics cause a shift in the bacterial population in the intestinal tract, and a consistent result is that gram-positive clostridial-type bacteria are inhibited. The differences may in part be due to the diverse techniques used in these experiments to monitor bacterial population changes, but the structural characteristics of the condensed tannins are different and may affect the mechanism of action on the bacteria.
The metabolic activities of the bacteria in the gut are very important in determining the chemical environment and healthy functioning of the gut. Intestinal bacteria are involved in plant cell wall digestion, microbial protein synthesis, synthesis of B and K vitamins, and phytotoxin and mycotoxin modification (22). Our fourth hypothesis, therefore, was that a shift in bacterial populations alters or affects the functional activity of the microbial population and thereby may affect the functional status of the gut. To determine if there was a shift in the metabolic potential of the bacteria, we looked at the in vitro fermentative capacity of the fecal bacteria. Our results indicate that the metabolic fingerprints of fecal bacteria from rats fed tannin diets tended to cluster together and that posttreatment metabolic fingerprints clustered together and were generally distinguishable from the fingerprint for the control after 3.5 weeks. However, the difference was not so much in the type of substrates utilized but in the rate of fermentation. All the substrates were utilized to some degree by the end of the 48-h period, but the rate and extent of fermentation were different. This may have been due to a different initial inoculum size or to the presence of more culturable bacteria and was not an indication of the metabolic potential within the gut itself. The effect of the shift in bacterial populations on the metabolic function of the intestinal tract is therefore not clear. Utilization of the specific simple substrates studied is not related to specific bacterial types and could be attributable to more than one type of bacteria. More complex substrates, however, require a consortium of bacteria for degradation, and reducing the diversity may reduce the capacity to degrade more complex substrates. Also, the significant decline in the level of the C. leptum group may affect functioning within the rumen ecosystem. Cellulolytic ruminococci are included in this group, which is a functionally significant group in ruminants, which depend on fiber degraders when they consume high-fiber diets. In future studies workers should concentrate on more complex substrates and consider indirect indices of metabolic function, such as pH and fatty acid concentration, which may modulate physiological changes within the intestinal tract (58). Our study focused on the bacterial population, but the protozoan, archaeal, and fungal populations are also important functionally, especially in the rumen, and the effect of tannins on these groups is not known.
It would be interesting and potentially very beneficial to animal and human nutrition to determine the mechanism of action of polyphenolics on bacteria and the inherent and adaptive mechanisms of tannin resistance. It has been shown that increasing the oxidative stress response helps E. coli strains overcome the inhibitory effect of condensed tannins under aerobic conditions, but the resistance mechanism under anaerobic conditions is not known (54). Our results suggest that a promising area to look at is cell wall and membrane effects of tannins. Other authors have suggested that tannin toxicity is due to selective inhibition of cell wall synthesis. Condensed tannins from Onobrychis viciifolia (sainfoin) were found to bind to cells of four strains of ruminal bacteria, Butyrivibrio fibrisolvens A38, Streptococcus bovis 45S1, Prevotella ruminicola B14, and Ruminobacter amylophilus WP225 (18). Along with inhibition of growth and protease activity, morphological changes were observed in the strains with gram-positive cell walls. Studies with green tea catechins have also shown that gram-positive bacteria are more sensitive to the bactericidal effects of these compounds than gram-negative bacteria are (16).
Iron deficiency has been shown to result in many of the same shifts in bacterial populations which we found in our study. In mice, an iron-deficient diet resulted in an increase in the level of total culturable bacteria and coliforms (Enterobacteriaceae) (57). Removal of iron from infant formula also resulted in an increase in the level of Bacteroides and a decrease in the level of clostridia, as well as shifts in other bacterial groups (4). An increase in the level of iron has the opposite effect, as repeated doses of ferrous sulfate over 4 weeks in rats resulted in a decrease in the level of Bacteroides and an increase in the level of clostridia (5). Dihydroxy phenolic groups can form stable complexes with many metal ions, and ion chelation by polyphenolics may have an effect in the gastrointestinal tract. It has previously been suggested that preventing growth of plant pathogens by metal complexation may be an important plant defense mechanism mediated by polyphenolics (26).
Fecal samples provide only partial insight into the response to different dietary inputs; for example, the mucosa-associated bacteria in the human colon were found to have a significantly different composition than the bacteria in the feces (61). Further work should be done to study the effects of tannins in various gastrointestinal regions.
This study proves that the proportion of tannin-resistant bacteria increases due to dietary tannins. Metabolic fingerprinting, DGGE, and dot blot hybridization indicated that the shift in bacterial populations was maintained to some degree over time, so it is possible that previous exposure to dietary tannins may result in a shorter adaptation period on subsequent exposure. Determining the mechanisms by which bacteria can resist the inhibitory effects of tannins is important to effectively implement a strategy of increasing the proportion of tannin-resistant bacteria in the gastrointestinal tract. It might become possible to enhance the resistance of certain strains or to produce tannin-resistant strains of essential organisms, such as fiber degraders. More information is also needed to determine how tannins exert a toxic effect on bacterial cells, as this could lead to developing ways to inactivate or reduce the inhibitory effects of tannins.
Our results describing the ecology of tannin-resistant bacteria provide a solid basis from which to start elucidating the effects of these organisms on host physiology, as well as determining the mechanisms of tannin action and tannin resistance.
Acknowledgments
This research and development was supported in part by funds provided to the International Arid Lands Consortium (IALC) by the USDA Forest Service and by the USDA Cooperative State Research, Education and Extension Service. A. H. Smith expresses her gratitude to the Agricultural Research Council, South Africa, for support during her studies at the University of Illinois.
We thank Erwin Zoetendal and Satoshi Koike at the University of Illinois for critical reading of the manuscript.
REFERENCES
- 1.Aiyar, A. 2000. The use of CLUSTAL W and CLUSTAL X for multiple sequence alignment. Methods Mol. Biol. 132:221-241. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 3.Amann, R. I., B. J. Binder, R. J. Olson, S. W. Chisholm, R. Devereux, and D. A. Stahl. 1990. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl. Environ. Microbiol. 56:1919-1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Balmer, S. E., and B. A. Wharton. 1991. Diet and faecal flora in the newborn: iron. Arch. Dis. Child. 66:1390-1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Benoni, G., L. Cuzzolin, D. Zambreri, M. Donini, P. Del Soldato, and I. Caramazza. 1993. Gastrointestinal effects of single and repeated doses of ferrous sulphate in rats. Pharmacol. Res. 27:73-80. [DOI] [PubMed] [Google Scholar]
- 6.Bonnet, R., A. Suau, J. Dore, G. R. Gibson, and M. D. Collins. 2002. Differences in rDNA libraries of faecal bacteria derived from 10- and 25-cycle PCRs. Int. J. Syst. Evol. Microbiol. 52:757-763. [DOI] [PubMed] [Google Scholar]
- 7.Brooker, J. D., L. A. O'Donovan, K. Clarke, L. Blackall, and P. Muslera. 1994. Streptococcus caprinus sp. nov., a tannin-resistant ruminal bacterium from feral goats. Lett. Appl. Microbiol. 18:313-318. [Google Scholar]
- 8.Caldwell, D. R., and M. P. Bryant. 1966. Medium without rumen fluid for non-selective enumeration and isolation of rumen bacteria. Appl. Microbiol. 14:794-801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cotta, M. A., and J. B. Russell. 1982. Effect of peptides and amino acids on efficiency of rumen bacterial protein synthesis in continuous culture. J. Dairy Sci. 65:226-234. [Google Scholar]
- 10.Daims, H., A. Bruhl, R. Amann, K. H. Schleifer, and M. Wagner. 1999. The domain-specific probe EUB338 is insufficient for the detection of all bacteria: development and evaluation of a more comprehensive probe set. Syst. Appl. Microbiol. 22:434-444. [DOI] [PubMed] [Google Scholar]
- 11.Dore, J., A. Sghir, G. Hannequart-Gramet, G. Corthier, and P. Pochart. 1998. Design and evaluation of a 16S rRNA-targeted oligonucleotide probe for specific detection and quantitation of human faecal Bacteroides populations. Syst. Appl. Microbiol. 21:65-71. [DOI] [PubMed] [Google Scholar]
- 12.Franks, A. H., H. J. Harmsen, G. C. Raangs, G. J. Jansen, F. Schut, and G. W. Welling. 1998. Variations of bacterial populations in human feces measured by fluorescent in situ hybridization with group-specific 16S rRNA-targeted oligonucleotide probes. Appl. Environ. Microbiol. 64:3336-3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hara, H., N. Orita, S. Hatano, H. Ichikawa, Y. Hara, N. Matsumoto, Y. Kimura, A. Terada, and T. Mitsuoka. 1995. Effect of tea polyphenols on fecal flora and fecal metabolic products of pigs. J. Vet. Med. Sci. 57:45-49. [DOI] [PubMed] [Google Scholar]
- 14.Hara, Y. 1997. Influence of tea catechins on the digestive tract. J. Cell Biochem. Suppl. 27:52-58. [PubMed] [Google Scholar]
- 15.Holdeman, L. V., E. P. Cato, and W. E. C. Moore (ed.). 1977. Anaerobe laboratory manual, 4th ed. Virginia Polytechnic Institute and State University Anaerobe Laboratory, Blacksburg.
- 16.Ikigai, H., T. Nakae, Y. Hara, and T. Shimamura. 1993. Bactericidal catechins damage the lipid bilayer. Biochim. Biophys. Acta 1147:132-136. [DOI] [PubMed] [Google Scholar]
- 17.Johnston, J. L. 1994. Similarity analysis of rRNAs, p. 248-277. In P. Gerhardt, R. G. E. Murray, W. A. Wood, and N. R. Krieg (ed.), Methods for general and molecular bacteriology. American Society for Microbiology, Washington, D.C.
- 18.Jones, G. A., T. A. McAllister, A. D. Muir, and K. G. Cheng. 1994. Effects of sainfoin (Onobrychis viciifolia scop.) condensed tannins on growth and proteolysis by four strains of ruminal bacteria. Appl. Environ. Microbiol. 60:1374-1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kadirvel, R., G. V. Rayu, and P. Vohra. 1969. Excretion of metabolites of tannic acid by chickens with and without ceca. Poult. Sci. 48:1511-1513. [DOI] [PubMed] [Google Scholar]
- 20.Katouli, M., A. Lund, P. Wallgren, I. Kuhn, O. Soderlind, and R. Mollby. 1997. Metabolic fingerprinting and fermentative capacity of the intestinal flora of pigs during pre- and post-weaning periods. J. Appl. Microbiol. 83:147-154. [DOI] [PubMed] [Google Scholar]
- 21.Krumholz, L. R., and M. P. Bryant. 1986. Eubacterium oxidoreducens sp. nov. requiring H2 or formate to degrade gallate, pyrogallol, phloroglucinol and quercetin. Arch. Microbiol. 144:8-14. [Google Scholar]
- 22.Mackie, R. I., R. I. Aminov, B. A. White, and C. S. McSweeney. 2000. Molecular ecology and diversity in gut microbial ecosystems, p. 61-78. In P. B. Cronje (ed.), Ruminant physiology: digestion, metabolism, growth and reproduction. CABI Publishing, Oxon, United Kingdom.
- 23.Makkar, H. P. S., and K. Becker. 1995. Degradation of condensed tannins by rumen microbes exposed to quebracho tannins (QT) in rumen simulation technique (RUSITEC) and effects of QT on fermentative processes in the RUSITEC. J. Sci. Food Agric. 69:495-500. [Google Scholar]
- 24.Makkar, H. P. S., M. Blümmel, and K. Becker. 1995. In vitro effects of and interactions between tannins and saponins and fate of tannins in the rumen. J. Sci. Food Agric. 69:481-493. [Google Scholar]
- 25.McSweeney, C. S., B. Palmer, and D. O. Krause. 2000. Rumen microbial ecology and physiology in sheep and goats fed a tannin-containing diet, p. 140-145. In J. D. Brooker (ed.), Tannins in livestock and human nutrition: proceedings of an international workshop, Adelaide, Australia. ACIAR Proceedings no. 92. Australian Centre for International Agricultural Research, Adelaide, Australia.
- 26.Mila, I., A. Scalbert, and D. Expert. 1996. Iron withholding by plant polyphenols and resistance to pathogens and rots. Phytochemistry 42:1551-1555. [Google Scholar]
- 27.Miller, S. M., J. D. Brooker, and L. L. Blackall. 1995. A feral goat rumen fluid inoculum improves nitrogen retention in sheep consuming a mulga (Acacia aneura) diet. Aust. J. Agric. Res. 46:1545-1553. [Google Scholar]
- 28.Miller, S. M., J. D. Brooker, A. Philips, and L. L. Blackall. 1996. Streptococcus caprinus is ineffective as a rumen inoculum to improve digestion of mulga (Acacia aneura) by sheep. Aust. J. Agric. Res. 47:1323-1331. [Google Scholar]
- 29.Miller, S. M., A. V. Klieve, J. J. Plumb, and L. L. Blackall. 1997. An in vitro cultured rumen inoculum improves nitrogen digestion in mulga-fed sheep. Aust. J. Agric. Res. 48:403-409. [Google Scholar]
- 30.Molina, D. O., A. N. Pell, and D. E. Hogue. 1999. Effects of ruminal inoculations with tannin-tolerant bacteria on fibre and nitrogen digestibility of lambs fed a high condensed tannin diet. Anim. Feed Sci. Technol. 81:669-680. [Google Scholar]
- 31.Muyzer, G., T. Brinkhoff, U. Nübel, C. Santegoeds, H. Schäfer, and C. Wawer. 1998. Denaturing gradient gel electrophoresis (DGGE) in microbial ecology, p. 1-27. In A. D. L. Akkermans, J. D. Van Elsas, and F. J. De Bruijn (ed.), Molecular microbial ecology manual, vol. 3.4.4. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 32.Nelson, K. E., A. N. Pell, P. Schofield, and S. Zinder. 1995. Isolation and characterization of an anaerobic ruminal bacterium capable of degrading hydrolyzable tannins. Appl. Environ. Microbiol. 61:3293-3298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nelson, K. E., M. L. Thonney, T. K. Woolston, S. H. Zinder, and A. N. Pell. 1998. Phenotypic and phylogenetic characterization of ruminal tannin-tolerant bacteria. Appl. Environ. Microbiol. 64:3824-3830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Odenyo, A. A., and P. O. Osuji. 1998. Tannin-tolerant ruminal bacteria from East African ruminants. Can. J. Microbiol. 44:905-909. [PubMed] [Google Scholar]
- 35.Odenyo, A. A., P. O. Osuji, O. Karanfil, and K. Adinew. 1997. Microbiological evaluation of Acacia angustissima as a protein supplement for sheep. Anim. Feed Sci. Technol. 65:99-112. [Google Scholar]
- 36.Okubo, T., N. Ishihara, A. Oura, M. Serit, M. Kim, T. Yamamoto, and T. Mitsuoka. 1992. In vivo effects of tea polyphenol intake on human intestinal microflora and metabolism. Biosci. Biotechnol. Biochem. 56:588-591. [DOI] [PubMed] [Google Scholar]
- 37.Osawa, R. 1990. Formation of a clear zone on tannin-treated brain heart infusion agar by a Streptococcus sp. isolated from feces of koalas. Appl. Environ. Microbiol. 56:829-831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Osawa, R., K. Kuroiso, S. Goto, and A. Shimizu. 2000. Isolation of tannin-degrading lactobacilli from humans and fermented foods. Appl. Environ. Microbiol. 66:3093-3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Osawa, R., F. Rainey, T. Fujisawa, E. Lang, H. J. Busse, T. P. Walsh, and E. Stackebrandt. 1995. Lonepinella koalarum gen. nov., sp. nov., a new tannin-protein complex degrading bacterium. Syst. Appl. Microbiol. 18:368-373. [Google Scholar]
- 40.Osawa, R. O., T. Fujisowa, and L. I. Sly. 1995. Streptococcus gallolyticus sp. nov., gallate degrading organisms formerly assigned to Streptococcus bovis. Syst. Appl. Microbiol. 18:74-79. [Google Scholar]
- 41.Plumb, J. J., L. L. Blackall, and A. V. Klieve. 2000. Rumen bacterial diversity with and without mulga (Acacia anuera) tannins, p. 146-150. In J. D. Brooker (ed.), Tannins in livestock and human nutrition: proceedings of an international workshop, Adelaide, Australia. ACIAR Proceedings no. 92. Australian Centre for International Agricultural Research, Adelaide, Australia.
- 42.Qiu, X., L. Wu, H. Huang, P. E. McDonel, A. V. Palumbo, J. M. Tiedje, and J. Zhou. 2001. Evaluation of PCR-generated chimeras, mutations, and heteroduplexes with 16S rRNA gene-based cloning. Appl. Environ. Microbiol. 67:880-887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Raskin, L., W. C. Capman, R. Sharp, L. K. Poulsen, and D. A. Stahl. 1997. Molecular ecology of gastrointestinal ecosystems, p. 243-298. In R. I. Mackie, B. A. White, and R. E. Isaacson (ed.), Gastrointestinal microbiology: gastrointestinal microbes and host interactions, 1st ed., vol. 2. Chapman & Hall, New York, N.Y.
- 44.Reed, J. D. 1995. Nutritional toxicology of tannins and related polyphenols in forage legumes. J. Anim. Sci. 73:1516-1528. [DOI] [PubMed] [Google Scholar]
- 45.Reeves, P. G., F. H. Nielsen, and J. G. C. Fahey. 1993. AIN-93 purified diets for laboratory rodents: final report of the American Institute of Nutrition ad hoc writing committee on the reformulation of the AIN-76 rodent diet. J. Nutr. 123:1939-1951. [DOI] [PubMed] [Google Scholar]
- 46.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 47.SAS. 2002. SAS user's guide: statistics, version 9. SAS Institute, Inc., Cary, N.C.
- 48.Scalbert, A. 1991. Antimicrobial properties of tannins. Phytochemistry 30:3875-3883. [Google Scholar]
- 49.Schneider, H., and M. Blaut. 2000. Anaerobic degradation of flavonoids by Eubacterium ramulus. Arch. Microbiol. 173:71-75. [DOI] [PubMed] [Google Scholar]
- 50.Sghir, A., D. Antonopoulos, and R. I. Mackie. 1998. Design and evaluation of a Lactobacillus group-specific ribosomal RNA-targeted hybridization probe and its application to the study of intestinal microecology in pigs. Syst Appl. Microbiol. 21:291-296. [DOI] [PubMed] [Google Scholar]
- 51.Sghir, A., G. Gramet, A. Suau, V. Rochet, P. Pochart, and J. Dore. 2000. Quantification of bacterial groups within human fecal flora by oligonucleotide probe hybridization. Appl. Environ. Microbiol. 66:2263-2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Simpson, J. M., V. J. McCracken, H. R. Gaskins, and R. I. Mackie. 2000. Denaturing gradient gel electrophoresis analysis of 16S ribosomal DNA amplicons to monitor changes in fecal bacterial populations of weaning pigs after introduction of Lactobacillus reuteri strain MM53. Appl. Environ. Microbiol. 66:4705-4714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sly, L. I., M. M. Cahill, R. O. Osawa, and T. Fujisawa. 1997. The tannin-degrading species Streptococcus gallolyticus and Streptococcus caprinus are subjective synonyms. Int. J. Syst. Bacteriol. 47:893-894. [DOI] [PubMed] [Google Scholar]
- 54.Smith, A. H., J. Imlay, and R. I. Mackie. 2003. Increasing the oxidative stress response allows Escherichia coli to overcome inhibitory effects of condensed tannins. Appl. Environ. Microbiol. 69:3406-3411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Smith, A. H., A. A. Odenyo, P. O. Osuji, M. A. Wallig, F. E. Kandil, D. S. Seigler, and R. I. Mackie. 2001. Evaluation of toxicity of Acacia angustissima in a rat bioassay. Anim. Feed Sci. Technol. 91:41-57. [Google Scholar]
- 56.Smith, A. H., M. A. Wallig, D. S. Seigler, A. A. Odenyo, C. S. McSweeney, and R. I. Mackie. 2003. Amelioration of toxic effect of Acacia angustissima with polyethylene glycol in rats. Anim. Feed Sci. Technol. 106:165-174. [Google Scholar]
- 57.Tompkins, G. R., N. L. O'Dell, I. T. Bryson, and C. B. Pennington. 2001. The effects of dietary ferric iron and iron deprivation on the bacterial composition of the mouse intestine. Curr. Microbiol. 43:38-42. [DOI] [PubMed] [Google Scholar]
- 58.Topping, D. L., and P. M. Clifton. 2001. Short-chain fatty acids and human colonic function: roles of resistant starch and nonstarch polysaccharides. Physiol. Rev. 81:1031-1064. [DOI] [PubMed] [Google Scholar]
- 59.Winter, J., L. H. Moore, V. R. Dowell, Jr., and V. D. Bokkenheuser. 1989. C-ring cleavage of flavonoids by human intestinal bacteria. Appl. Environ. Microbiol. 55:1203-1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wiryawan, K. G., B. Tangendjaja, and Suryahadi. 2000. Tannin degrading bacteria from Indonesian ruminants, p. 123-126. In J. D. Brooker (ed.), Tannins in livestock and human nutrition: proceedings of an international workshop, Adelaide, Australia. ACIAR Proceedings no. 92. Australian Centre for International Agricultural Research, Adelaide, Australia.
- 61.Zoetendal, E. G., A. von Wright, T. Vilpponen-Salmela, K. Ben-Amor, A. D. Akkermans, and W. M. de Vos. 2002. Mucosa-associated bacteria in the human gastrointestinal tract are uniformly distributed along the colon and differ from the community recovered from feces. Appl. Environ. Microbiol. 68:3401-3407. [DOI] [PMC free article] [PubMed] [Google Scholar]