Abstract
SUMOylation of transcription factors and chromatin proteins is in many cases a negative mark that recruits factors that repress gene expression. In this study, we determined the occupancy of Small Ubiquitin-like MOdifier (SUMO)-1 on chromatin in HeLa cells by use of chromatin affinity purification coupled with next-generation sequencing. We found SUMO-1 localization on chromatin was dynamic throughout the cell cycle. Surprisingly, we observed that from G1 through late S phase, but not during mitosis, SUMO-1 marks the chromatin just upstream of the transcription start site on many of the most active housekeeping genes, including genes encoding translation factors and ribosomal subunit proteins. Moreover, we found that SUMO-1 distribution on promoters was correlated with H3K4me3, another general chromatin activation mark. Depletion of SUMO-1 resulted in downregulation of the genes that were marked by SUMO-1 at their promoters during interphase, supporting the concept that the marking of promoters by SUMO-1 is associated with transcriptional activation of genes involved in ribosome biosynthesis and in the protein translation process.
INTRODUCTION
SUMOylation, an evolutionally conserved post-translational modification among eukaryotic cells, involves a three-step process that requires an E1-activating enzyme (SAE1/SAE2 in humans), E2-conjugating enzyme (Ubc9) and a variety of E3 ligases that covalently attach Small Ubiquitin-like MOdifier (SUMO) protein to the lysine residues of substrate proteins (1). SUMO proteins are ubiquitously present in eukaryotic cells; in human, there are four SUMO isoforms, SUMO-1 to -4, encoded by distinct genes. SUMO-1 is found in vivo conjugated to target proteins as a monomer. SUMO-2/3, which are each 45% identical to SUMO-1 and 96% identical to each other, are conjugated by different E3 enzymes than act on SUMO-1, and SUMO-2/3 are often found in poly-SUMO chains (1). SUMO-4 is an isoform found in kidney, lymph node and spleen cells (2), but it is not known whether SUMO-4 can be conjugated to cellular proteins. SUMOylation can be reversed by SUMO/sentrin-specific proteases (Ulps in yeast and SENPs in human) that remove SUMO proteins from target proteins (3). This covalent and reversible biochemical reaction is highly dynamic and tightly orchestrated in cells, and it regulates various biological and physiological processes, such as nuclear-cytosolic transport, protein stability, apoptosis, transcriptional regulation, DNA repair, cell proliferation and cell cycle progression (3).
SUMO proteins are associated with transcriptional regulation. A wide range of transcription factors have been reported as SUMO substrates, and in most studies, this modification results in a repressive signal. For example, SUMOylation of the polycomb repressive complex 1 (PRC1) subunit Pc2 is important for the repressive activity of the complex (4,5). SUMO-mediated repression of sequence-specific transcription factors includes Elk-1 (6), IκBα (7), c-Jun (8), C/EBP (9), Sp3 (10) and many others (11,12). In addition, p300, a transcription factor with both activating and repressing roles, is modified by SUMO conjugation to repress downstream genes via association with HDAC6 (13). A variety of chromatin-modifying enzymes have been identified to be recruited to promoters in a SUMO-dependent manner (14). It is also known that all four major core histones can be SUMOylated and further repress gene expression in yeast (15). In human cells, SUMOylation of histone H4 was associated with transcription inactivation via the recruitment of HDACs to oppose other activating modifications such as ubiquitination or acetylation (16). Histones H1 and H3 are SUMO substrates, yet the exact role of the SUMOylation of these proteins is unclear (17). In addition to SUMO conjugation of sequence-specific transcription factors and histones, general transcription initiation factors, such as TFIID subunits hsTAF5 and hsTAF12, can be SUMOylated resulting in the inhibition of their promoter binding activity (18).
SUMOylation of chromatin-associated factors has also been associated with stimulation of transcription. A set of transcription factors have all been reported to be stimulated by SUMOylation, including Pax-6 (19), GRIP1 (20), myocardin (21), p45/NF-E2 (22), GATA-4 (23), Smad4 (24), glucocorticoid receptor (25), NFAT-1 (26), PEA3 (27) and HSF-1/-2 (28,29). SUMOylation has been reported as both an activator and a repressor of the p53 protein (30,31). One study found that SUMOylation of promoter-associated factors in yeast was clearly associated with transcriptional activation on constitutive gene promoters (32). Thus, while the preponderance of evidence has focused on SUMOylation as a repressive signal, there are examples of it activating transcription. However, a general rule for how SUMO-1 functions as a chromatin mark is still unclear.
Here, we analyzed the genome-wide association of SUMO-1 as a chromatin mark in human cells at stages throughout the cell cycle. To our surprise, we found that SUMO-1 marks many of the most active genes at the proximal promoter region. The SUMO-1-binding profile was dynamic as cells traversed the cell cycle. In particular, we noted that SUMO-1 binding to the promoter of active genes was decreased during mitosis when transcription generally halts. We found SUMO-1 labeling on the chromatin was highly correlated with the stimulatory H3K4 trimethylation (H3K4me3) mark. Depletion of SUMO-1 protein resulted in a decrease in mRNA abundance of SUMO-1-marked genes, indicating that SUMO-1 is a transcriptional activator for those genes.
MATERIALS AND METHODS
Cloning and cell line generation
To obtain the HeLa cell line stably expressing His6-biotin-tagged SUMO-1 (protein diagram in Supplementary Figure S1A), full-length human SUMO-1 was PCR-amplified from HeLa cell cDNA by using Phusion High Fidelity polymerase (Finnzymes) and cloned into pQCXIP-derived vector (gift of P. Kaiser, UC Irvine) (33). HeLa cells were then stably transfected with the His6-biotin-SUMO1 plasmid using Lipofectamine (Invitrogen) and selected in 2 µg/ml puromycin. Colonies with recombinant SUMO-1 stable expression were screened and confirmed by western blot.
Antibody and used for chromatin immunoprecipitation
The SUMO-1 polyclonal antibody used a GST-SUMO1 fusion protein as antigen, and the serum was prepared at Cocalico Biologicals, Inc (Reamstown, PA, USA).
Cell culture, cell cycle analysis and RT–qPCR
For G1/S synchronization, HeLa or HeLa-SUMO cells were treated with 2 mM thymidine (Sigma) for 17 h, then removed for 9 h and added at the same concentration for 18 h and released for the indicated times to synchronize cells in early S, mid-S, late S and G1 phases, respectively. Mitotic phase cells were obtained by treating with 2 mM thymidine for 15 h and released for 3 h and then treated with 100 ng/ml nocodazole for 15 h. Cell cycle distribution was determined by FACS Calibur flow cytometer (Becton Dickinson).
The RT–qPCR assays were done 72 h post-transfection with SUMO-1 or Ubc9-specific small interfering RNA (siRNA) using Oligofectamine (Invitrogen), and the control oligonucleotide was specific for luciferase. Primer and siRNA sequences are provided in Supplementary Table S6. Total RNA was purified using Trizol reagent (Invitrogen); 2 µg of total RNA was reverse-transcribed using iScript cDNA synthesis kit (Bio-Rad), and qPCR was done as per the manufacturer’s protocol (iQ SYBR Green Supermix, Bio-Rad). Three biological replicates were performed individually.
Chromatin immunoprecipitation, ChIP–qPCR and affinity purification
Chromatin immunoprecipitation (ChIP) and affinity purification (ChAP) samples for Illumina GAII were prepared as follows. The ChIP samples were prepared by standard methods (34) using SUMO-1 antibody. Chromatin affinity purification was based on the same ChIP method with modification of a two-step affinity purification. 108 HeLa-SUMO cells were cross-linked with 1% formaldehyde (Sigma) and stopped by adding 125 mM glycine. The cross-linked chromatin was then sheared to 200–300 bp by sonication, incubated with 375 µl of Ni beads (Qiagen) for 16 h at 4°C. An aliquot of the input DNA was saved prior to immunoprecipitation as a reference sample. After washing in 6 ml of wash buffer I (50 mM Tris, pH 8; 0.01% SDS; 1.1% Triton X-100; 150 mM NaCl), chromatin fragments were eluted in 6 ml elution buffer (washing buffer I with 300 mM imidazole). The nickel eluate was incubated with 375 µl of streptavidin beads (Invitrogen) for 6 h at 4°C. After three stringent washes in 2 ml of wash buffer II (50 mM Tris, pH 8; 10 mM EDTA; 1% SDS; 1 M NaCl), the chromatin was eluted by adding 2 ml of elution buffer (50 mM Tris, pH 8; 10 mM EDTA; 1% SDS; 200 mM NaCl) to the beads, and cross-link reversal was done by incubating at 65°C for 15 h. The supernatant was collected and diluted 1:1 with TE buffer. The eluate was treated with RNase (0.2 mg/ml; Sigma) for 2 h at 37°C, with Proteinase K (0.2 µg/ml; Sigma) for 2 h at 55°C, and DNA was extracted using phenol/chloroform/isoamyl alcohol and precipitation in 0.1 volumes of 3 M sodium acetate, 2 volumes of 100% ethanol and 30 µg of glycogen (Invitrogen). ChIPed DNA prepared from 1 × 108 cells was resuspended in 30 µl of Qiagen Elution Buffer. Three biological replicates were prepared per time point. ChIP–qPCR was performed to validate the ChIP-seq data obtained in this research. For ChIP–qPCR experiment, after 72 h of Ubc9 depletion in HeLa-SUMO cell line, 2 × 107 were harvested and followed by ChIP method described previously. Ct values obtained in each sample were normalized to the % input DNA values. qPCR was done as per the manufacturer’s protocol (iQ SYBR Green Supermix, Bio-Rad). Primer sequences are provided in Supplementary Table S6. At least three biological replicates were performed individually.
ChIP DNA preparation for Solexa sequencing
ChIP or ChAP DNA samples were then prepared for ChIP-sequencing library construction following Illumina’s ChIP-seq Sample Prep protocol. Briefly, the DNA samples were blunt-ended by using End-it DNA End-Repair Kit (Epicentre) according to the manufacturer's instruction. dA overhangs were then added and Illumina adapters ligated. Adapter-ligated DNA was subject to 15 cycles of PCR after size selection of 200–300 bp by agarose gel electrophoresis. The 10 nM purified DNA was subjected to sequencing on Illumina GAII platform to 36-bp reads. The sequencing reads were aligned to the human genome UCSC build hg18. Only uniquely aligned reads were used for further analysis, and multiple identical reads were eliminated to reduce PCR-generated artifacts.
cDNA sample preparation
The double-stranded cDNA (0.8 µg total RNA input) was subjected to library preparation using the Illumina TruSeqTM RNA sample preparation kit (low-throughput protocol) according to the manufacturer’s protocol.
RNA-seq analysis
Six cDNA samples containing three pairs of biological replicates (three SUMO-1 depleted samples and three GL2 control samples) were barcoded, pooled together in equal concentration and subjected to sequencing in one lane of Illumina GAII. The resulted sequences (5–9 million reads for each sample) were sorted and mapped to human reference genome hg18 using open-source software TopHat (35) (Supplementary Table S3). The differential gene expression of the two groups of samples (SUMO-1-depleted vs. control) was analyzed by open-source software (36) using default parameter settings. Genes from all six samples with significantly changed Fragments Per Kilobase of exon per Million fragments mapped (FPKM) values, as well as a sub-group of significantly downregulated genes upon SUMO-1 depletion involved in protein synthesis, were displayed in the heat map with row-wise scale. The significantly changed genes were also compared with ChAP-seq results, and the GO enrichment was analyzed using Toppgene (http://http://toppgene.cchmc.org/) and Ingenuity Pathway Analysis (IPA).
Data analysis
ChAP-seq peak finding
FindPeaks 4.0.10 (37) was used to generate peaks for all the ChAP-seq and ChIP-seq data of SUMO-1 with options of subpeaks 0.5, trim 0.2. A minimum height threshold for each dataset was established so that FDR is <0.1% based on the Monte-Carlo simulation of each dataset.
Histogram of genome-wide tag counts
Raw tags were counted in a 1-kb bin-size for every chromosome for each sample using a Matlab code. The same histograms for chromosome 1 were used to generate scatterplots for paired ChIP-ChAP samples using scatterplot function in MatLab.
Sort peaks into different genomic regions
RefSeq database was used to define genomic regions, and the promoter region is defined as 5 kb upstream of a transcription start site (TSS). A peak was sorted to a specific region if there is at least 1 bp overlap with that region. Active/inactive promoters were classified based on GEO datasets GDS885 and GDS2781 containing asynchronous HeLa cell gene expression microarray results. Genes were grouped based on their expression levels, and active promoters were defined from the top 20 percentile gene groups, while inactive promoters were defined from the bottom 20 percentile groups. Each contains about 2400 genes.
Extended TSS region tag density profiling
RefSeq database was used to obtain start and end coordinates of ±10 kb of TSSs for each gene that is included in the GDS885 dataset (38). A total of 12 013 genes extended TSSs were used. Raw SUMO-1 tags were extended according to the average fragment length of each sample. The average tag density was computed using non-overlapping 5-bp bins along the extended TSS region from each of the three biological replicates, then the tag density was normalized by dividing with the total number of reads (in millions) in each sample and averaged among the three replicates. In the heat maps arranged by gene expression percentile, gene expression was grouped based on the percentile in GDS885 dataset. In the sorted TSSs heat map (Figure 3B), the rows of all other cell stage heat maps follow the same order of G1 sample.
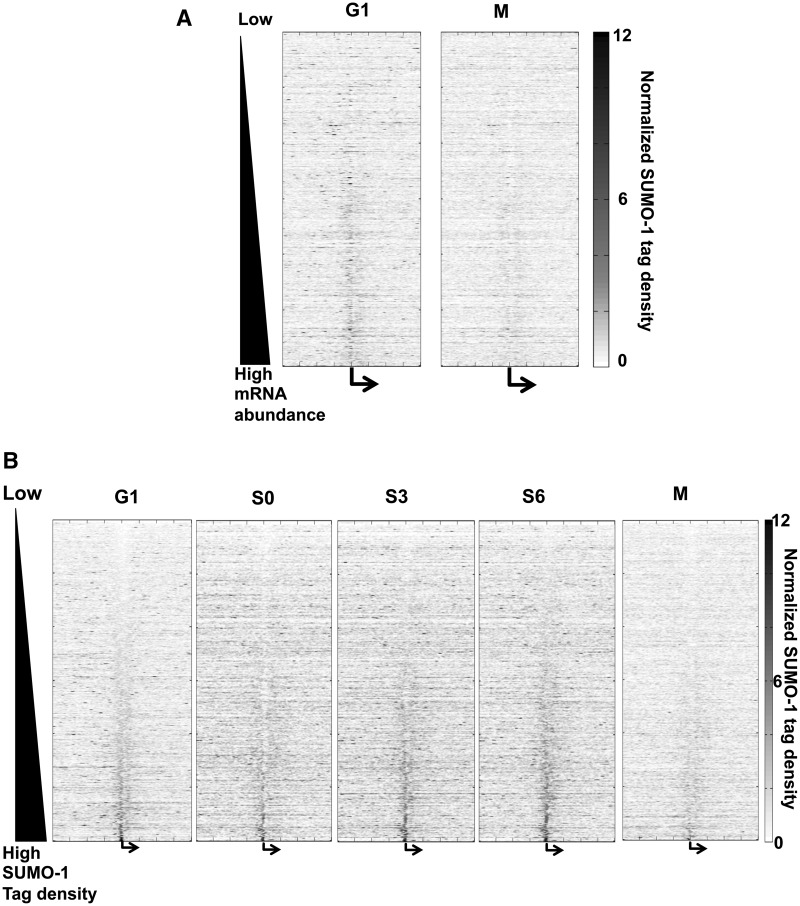
Figure 3.
SUMO-1 is associated with chromatin of active genes. (A) The heat maps of normalized SUMO-1 tag densities on genes (±10 kb surrounding the TSSs), sorted by gene expression level from low (top) to high (bottom) in G1 and M phases. Each row is a gene’s SUMO-1 tag density trace using the average of normalized tag density at each stage of the cell cycle. The vertical center (bent arrow) denotes the TSSs. The density of the SUMO-1 tag is indicated by the color; blue is low level to white, and red are progressively higher levels of SUMO-1. (B) Heat maps are similar to those in panel A, but the order of the genes (rows) is according to the density of SUMO-1 near the TSSs from low (top) to high (bottom) during G1. Similar heat maps are shown for each indicated phase of the cell cycle, and the order of genes (rows) is the same as in G1.
Comparison of SUMO-1-marked genes in ChAP-seq samples of different cell stages
G1 and M0 stage ChAP-seq samples were processed for peak-calling using FindPeaks 4.0.10. The resulted peak files were crosschecked with RefSeq database to extract genes with peaks present in the promoter region (5 kb upstream of TSSs) using BEDtools (39). The presence of a peak in the promoter region was defined as at least 1 bp overlap between the peak range and the promoter region of a specific gene. The gene lists were then crosschecked with the gene lists from significantly changed RNA-seq comparison data.
Normal distribution of the number of randomly selected genes with SUMO-1 promoter peaks and z-score calculation
A specific number (199 or 158) of genes were randomly selected from RefSeq database and then crosschecked with ChAP-seq peak files to obtain the number of genes with SUMO1 peaks in the promoter regions using BEDtools. The ChAP-seq datasets used in this analysis were from the G1 phase. The whole process was repeated 1000 times and we found that the number of genes with SUMO1 peaks follows normal distribution. The mean and standard deviation of this distribution were calculated. Using the real number of genes with G1-stage SUMO1 promoter peaks obtained from RNA-seq comparison data, the z-score was calculated as z-score = (NumTrue − mean)/std.
Comparison of SUMO-1 ChAP-seq and published chromatin marks
Publicly available HeLa cell ChIP-seq/ChIP-chip datasets—H3K4me3 (GSM566169), H3K27me3 (GSM566170)—were downloaded from the GEO database (www.ncbi.nlm.nih.gov/geo/). For all chromatin mark ChIP-seq datasets, the raw reads were extended to 200 bp. Peaks were generated the same way as SUMO-1 ChAP-seq sample. RefSeq gene promoter and transcribed region were used to search for a peak that has at least 90% of its range overlapping with annotated regions of a specific gene.
To compare the binding pattern between SUMO-1 and other chromatin marks, tag density profiles were computed with a Matlab code within the 20-kb extended TSSs of all the genes (total 12 013 entries) included in the GDS885 dataset. The rows of each tag density profile were sorted according to the maximum tag density of the ±2 kb of the TSSs in sample profiles. The mean tag density of this 4-kb region from each dataset was used to calculate the Pearson correlation coefficient (R).
The peak files from SUMO-1 G1-stage ChAP-seq as well as ChIP-seq from the chromatin marks (H3K4me3 and H3K27me3) were also used to find the genes that have both SUMO-1 marks and one of the chromatin marks and then to generate Venn diagram. BEDtools was used to find peaks from data that overlap at least 90% with the promoter of each gene (for SUMO-1 G1-stage data) in RefSeq database or the promoter plus transcribed region (for H3K4me3 and H3K27me3 data). The chi-square test P-values were computed using R function χ2 test.
Comparison of SUMO-1-marked genes with ubiquitin-marked genes in HeLa ChAP-seq samples
G1 and M0 stage ubiquitin-tagged ChAP-seq samples (Arora et al., submitted) as well as G1 stage SUMO1-tagged ChAP-seq sample were processed for peak-calling using FindPeaks 4.0.10. Each of the resulted peak files was crosschecked with RefSeq database to extract genes with peaks present in the promoter region (5 kb upstream of TSSs) or transcribed region using BEDtools. The presence of a peak in the promoter/transcribed region was defined as at least 90% of the peak range overlapping with that region of a specific gene.
Principal component analysis of ChAP-seq datasets
For each sample, SUMO-1 tag counts on chromosome 1 (without the centromeric region to avoid bias due to the sequencing artifacts) was used for principal component analysis (PCA) using Matlab (bin-size = 1 kb). The first three principle components were plotted using Matlab.
RESULTS
Chromatin affinity purification of SUMO-1 through the cell cycle
A variety of studies have shown that SUMO-1 participates in cell cycle progression (40,41). To determine the genome-wide SUMO-1 pattern on chromatin and how it changes during the cell cycle, we employed a HeLa-derived cell line that stably expressed His6-biotin-tagged SUMO-1 (Supplementary Figure S1A). Western blot analysis showed that the 26-kD recombinant SUMO-1 was expressed at around 10-fold higher levels than the endogenous 11.5-kD monomer SUMO-1 in crude whole-cell extracts (Supplementary Figure S1B); however, those tagged SUMO-1 conjugates at higher molecular weight were present at similar levels as compared to the endogenous SUMO-1 protein (Supplementary Figure S1B, right). We purified chromatin using standard methods, followed by double-affinity purification via the His6-tag and the biotin-tag. We found that the most abundant proteins conjugated to the tagged SUMO-1 were in the size range of 40 kD and higher (Supplementary Figure S1C). The most abundant SUMOylated proteins were most likely transcription factors or other non-histone chromatin proteins.
Cells were synchronized in various cell cycle stages using a double-thymidine block and release or thymidine/nocodazole block (Figure 1A). Flow cytometry analysis of the DNA content and the mitosis-specific phospho-histone H3 mark indicated that the cells were synchronized in G1, early/mid/late S and mitosis phases (Supplementary Figure S1D, E). Chromatin was isolated, and the SUMO-tagged chromatin was then double-affinity purified using metal ion affinity chromatography followed by streptavidin-affinity chromatography. The protein bound to the matrix was subjected to stringent wash conditions, cross-link reversal, and the enriched DNA was analyzed by high-throughput sequencing. This approach was directly analogous to ChIP-seq, but since no antibodies were used to purify the chromatin, we call this technique ChAP-seq for chromatin affinity purification and sequence analysis. Three sets of biological replicates were performed for each time point, and we obtained 18–25 million uniquely mapped reads from the Illumina genome analyzer II (GAII) for each individual sample. We then compared the datasets pairwise to evaluate the reproducibility of the three biological replicates. We found all the peaks of samples collected during interphase to highly overlap with other samples from the same point in the cell cycle: replicates from S3, S6 and G1 had 77–95% of their peaks overlap from the respective samples. The early S phase samples (S0) had >52% of its peaks present in the other replicates. The samples from mitosis had >41% of its peaks present in the replicate samples (Supplementary Table S1). This was a high level of reproducibility, especially among the interphase samples. The samples from mitosis had lower reproducibility, but as will be shown in the following sections, these samples had SUMO-1 removed from the promoters.
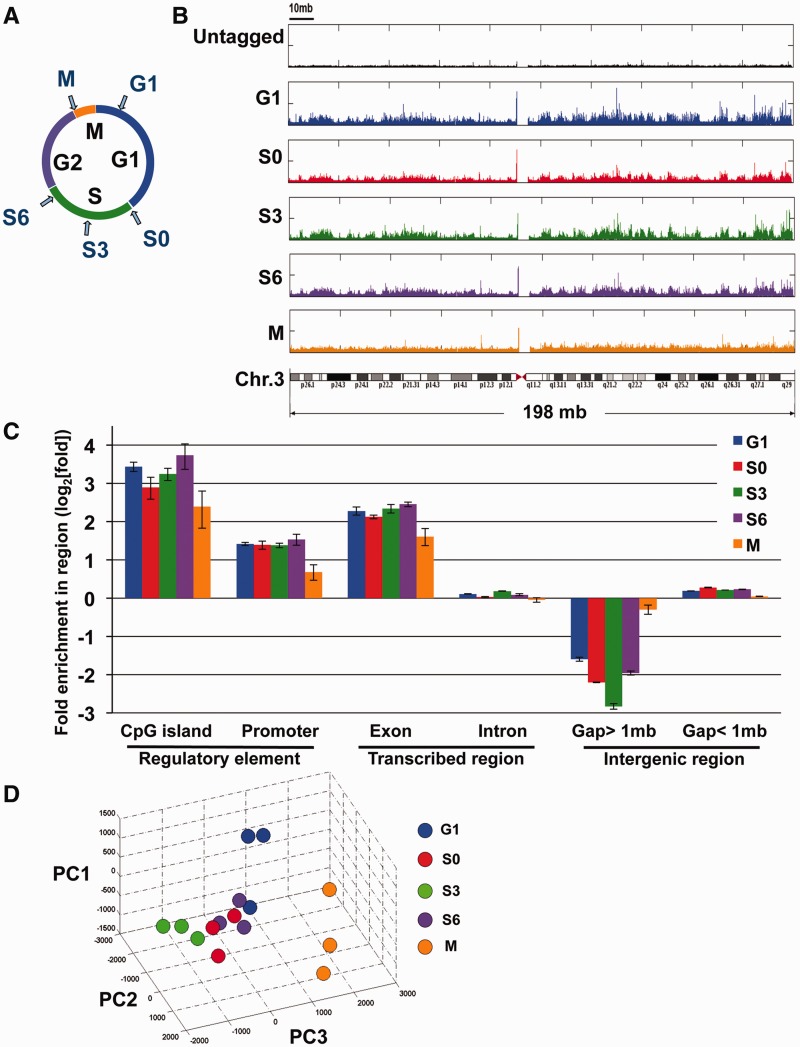
Figure 1.
Genome-wide analysis of SUMO-1 binding. (A) Sample collection for mapping the chromatin localization of SUMO-1 throughout the cell cycle. HeLa cells were treated with double-thymidine block and released for 0, 3, 6, 13 h to obtain S0, S3, S6 and G1 samples, respectively, and cells in mitosis (M) were treated with a sequential thymidine-nocodazole block. (B) Histogram depicting the locations of SUMO-1-binding sites on chromosome 3 of the human genome using chromatin affinity sequence analysis (ChAP-seq). The frequency of raw reads was plotted along the length of the chromosome with bin-size 1 kb. Samples were ChAP-purified DNAs from HeLa-SUMO cells during G1 (blue, panel 2), early S phase (S0, red, panel 3), mid-S phase (S3, green, panel 4), late S phase (S6, purple, panel 5), mitosis (M, orange, panel 6) and results from affinity purification using a HeLa cell line that does not express the tagged SUMO-1 protein (black, panel 1). A diagram of chromosome 3 is shown at the bottom. (C) Peak annotation depicts fold change on log2 scale of SUMO-1 binding sites on defined sequence elements on the human genome relative to the expected frequency of the genetic elements distributed in the genome if the binding profile is randomly distributed. G1 (blue), early S phase (S0, red), mid-S phase (S3, green), late S phase (S6, purple) and mitosis (M, orange), and error bars are SEM from three biological repeats. (D) Fifteen SUMO-1 datasets of chromosome 1 were represented in a three-dimensional stereoscopic image by using standard PCA (see ‘Materials and Methods’ section) to show the reproducibility within each set of biological replicates as well as the separation of data among different cell stages. Each color ball represents individual dataset collected from indicated cell stage. The same color denotes the biological replicates of the same collection point during cell cycle.
The results for the SUMO-1-binding profiles on the human chromosome 3 are shown as an example (Figure 1B). We computed the SUMO-1 tag densities (bin-size = 1 kb) and plotted them along the length of the chromosome as a histogram (false discovery rate; FDR <0.1%). At the top is the histogram from the HeLa cell line that does not express tagged SUMO-1, and results from specific points in the cell cycle were shown (top to bottom): G1, early S (S0), mid-S (S3), late S (S6) and mitosis (M). From the cell line that does not express tagged SUMO-1, there was a low background of non-specifically purified sequence tags evenly distributed throughout the chromosome and without peaks. When comparing the interphase SUMO-1 localization, at the chromosome scale resolution, the samples had similar patterns to each other. By contrast, during mitosis the SUMO-1-modified chromatin was largely redistributed, with relatively even distribution and fewer apparent peaks. The SUMO-1 peak at the pericentromere appears in all samples, including the ChIP-seq reaction using pre-immune IgG (Supplementary Figure S2A). Since this peak appears in a sample without specific purification, we interpret this peak as an artifact from the parallel-sequencing technique.
To test whether the SUMOylation of chromatin in cells expressing the tagged SUMO-1 is consistent with the labeling of endogenous SUMOylation, we performed a ChIP-seq using SUMO-1-specific antibody in the early S phase (S0) as a biological validation for the ChAP technique. We found that the results obtained from the ChIP method were highly consistent with those from ChAP (Supplementary Figure S2A–B). An example, which includes multiple biological replicates, at the promoter of the NOSIP gene is shown in Supplementary Figure S2C. The average peak values obtained by ChIP-seq were comparable to the peak values obtained from ChAP-seq. Furthermore, the peaks detected using ChIP-SUMO-1 (x-axis) were correlated well with those of the double-tagged-SUMO-1 (y-axis) (R = 0.989) by scatterplot analysis (Supplementary Figure S2D).
Chromatin-bound SUMO-1 is concentrated at transcriptional regulatory sites and is dynamic through the cell cycle
We then analyzed the distribution of SUMO-1-tagged chromatin on a genome-wide scale according to sequence annotations. Compared to the null hypothesis that tags were randomly distributed in the genome, SUMO-1 was significantly enriched on CpG islands, promoters and exons during interphase (Samples from S0, S3, S6 and G1 phase; Wilcoxon rank-sum P-value < 0.05), whereas SUMO-1 binding to intron containing sequences was not significantly different from the random expectation. 10% of SUMO-1 marks were around the promoter region (5 kb upstream of a transcription start site, TSS), representing a 2.5-fold enrichment of SUMO-1 at promoter DNA, suggesting that SUMO-1 might play a role in regulating transcription initiation (Figure 1C, Supplementary Figure S3A). In addition, during mitosis the SUMO-1 marks at promoters decreased (Figure 1C). These results suggested that SUMO-1 is depleted from chromatin, and this is consistent with a previous study shown that during mitosis, little SUMO-1 remains localized to condensed chromosomes (42). By contrast, large gene deserts were under-represented in the chromatin marked by SUMO-1. SUMO-1 occupancy in the genome was shown in fold enrichment (log2) normalized to the frequency of the genetic elements in the genome. Interestingly, CpG islands represent 0.7% of the genome, but we observed that 8–10% of the SUMO-1 marks were on CpG islands, consistent with the promoter enrichment in Figure 1C. Since many CpG islands are located in promoters, we also analyzed the promoters without CpG islands and found a similar pattern of SUMO-1 association with promoters that do not have CpG islands (Supplementary Figure S3B). In addition, there was a 4-fold enrichment of SUMO-1 marks on exon, but this enrichment was not explained by promoter-proximal binding of SUMO-1 to exon1 (Supplementary Figure S3C). This association of SUMO-1 with exons suggested that SUMO-1 might be associated with splicing at the chromatin level. As many histone marks, such as H3K36 methylation and K9 acetylation, have shown to play a role in alternative splicing (43), it will be of interest to investigate whether SUMO-1 marks participate in pre-mRNA processing through chromatin conformation.
In order to reduce the complexity of analyzing large datasets, we used PCA (44) to examine the 15 datasets containing three replicates each of the five time points in the cell cycle (Figure 1D). Like other high-throughput data, ChAP-seq data contain many features and thus are in high dimensions. By PCA, we focused on the combination of features with the largest variances and thus identified major dissimilarities among multiple datasets simultaneously. Apart from pairwise analysis of the biological replicates indicated high reproducibility (Supplementary Table S1), visualization of the first three principal components of the PCA showed that replicates from each time point tend to group together, suggesting that the differences among time points are larger than the differences among replicates. Consistent with visualization of the chromosome-wide labeling by SUMO-1, in which the pattern of SUMO-1 on chromatin during mitosis was distinct from the interphase samples (Figure 1B and C), the SUMO-1 localization during mitosis analyzed by PCA was also well separated from all the other interphase samples (Figure 1D). These results indicated that the SUMO-1 tagging of chromatin is dynamic through the cell cycle, and the changes we identified were meaningful at each time point since they were obtained with biological repeats collected weeks apart.
SUMO-1 labels the promoters of active genes
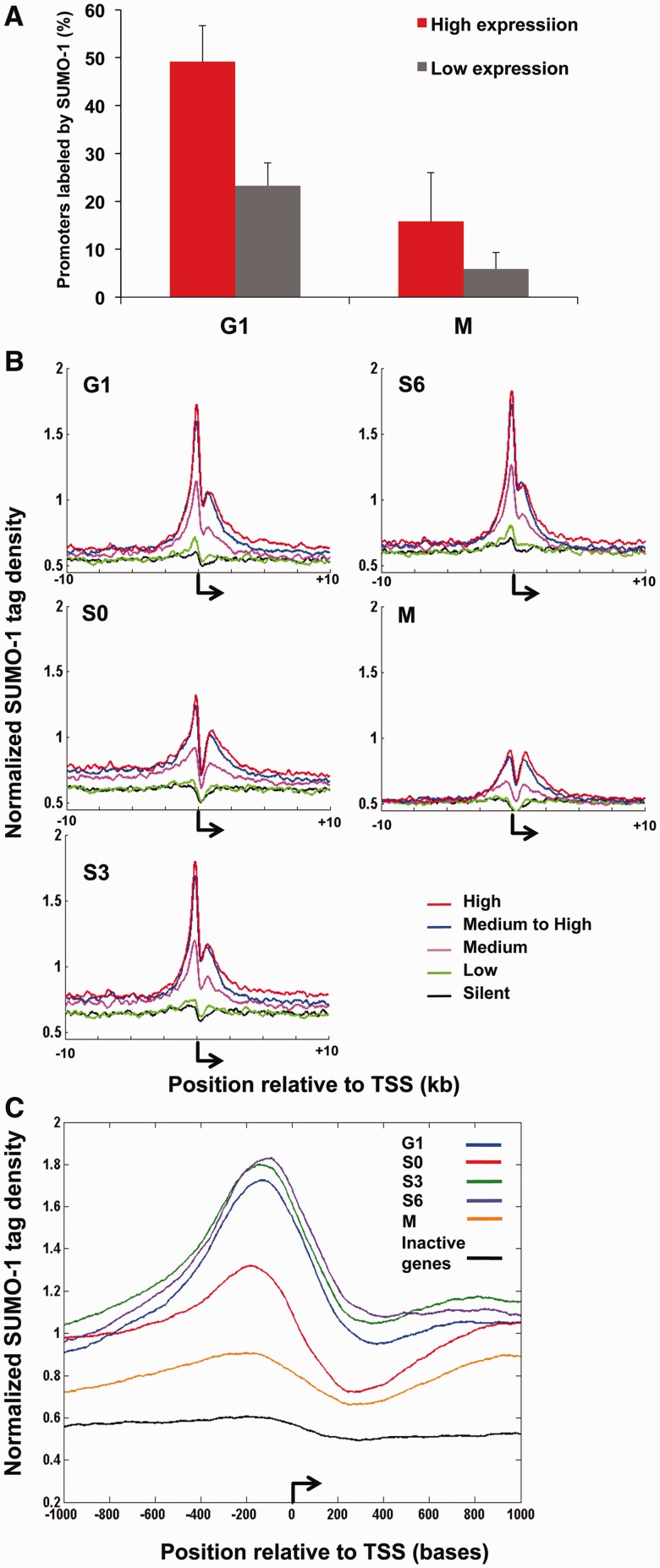
Previous studies showed that SUMOylation generally contributes to transcriptional repression (12). However, a recent study suggested SUMOylation of chromatin could facilitate transcription activation in constitutive genes in yeast (32). Since we observed that SUMO-1 marks were enriched at regulatory elements in the genome (Figure 1C), we asked whether SUMO-1 was associated with the most active or inactive genes. Using published microarray data (38), we sorted the mRNA level for each gene from low to high and obtained the 20% highest and 20% lowest expressed genes and asked what proportion of the most active or least active promoters were labeled by SUMO-1. In striking contrast to the published association of SUMO-1 with repressive elements, there were many more examples of SUMO-1-modified chromatin at highly active promoters. We found during G1 phase, 49.2% of the high-activity and 23.3% of the low-activity promoters were labeled by SUMO-1 (Figure 2A). During mitosis, we found 15.8% of high-activity and 5.9% of low-activity promoters were marked by SUMO-1. This reduction of SUMO-1 marks was consistent with the idea that during mitosis, transcription was repressed and this stimulatory SUMO-1 signal would rebind to the chromatin after cell division was completed and active transcription resumed.
Figure 2.
SUMO-1-binding pattern is associated with transcriptional activation. (A) A histogram is shown of the percentage of active (red) and inactive (gray) promoters labeled by SUMO-1. High-activity promoters are defined as those upstream of genes for which the mRNAs were the 20% most abundant and low-activity promoters are defined as those upstream of genes for which the mRNAs were the 20% least abundant in microarray datasets. Results are the means (±SEM) in G1 and mitosis, as indicated. (B) Normalized tag density plots display SUMO-1 tags distribution ±10 kb surrounding the transcription start sites (TSSs, bent arrow) in different cell cycle stages. Each trace is based on averages of normalized ChAP-seq tag densities and results from the three replicates at each point in the cell cycle. From published microarray results using asynchronous HeLa cells, genes were divided into deciles representing inactive genes (0–10%, black), low-activity genes (10–20%, green), medium abundance mRNAs (50–60%, pink), medium-high abundance mRNAs (80–90%, blue) and highest abundance mRNAs (90–100%, red). The y-axis is arbitrary normalized tag density unit (see Materials and Methods). Results are shown from G1 (top left), early-S (S0, middle left), mid-S (S3, bottom left), late S (S6, top right) and mitosis (M, middle right). (C) A zoom-in view is shown of the average of normalized SUMO-1 tag density plots on most highly expressed genes from each cell cycle stage within 2 kb relative to TSSs. The similar trace from inactive genes in the G1 phase is shown in black.
We further dissected the SUMO-1 localization flanking TSSs of annotated genes. The average SUMO-1 tag density per 10 bp from the three replicates of each time point were normalized and plotted within ±10 kb of TSSs (45). To correlate SUMO-1 distribution and global mRNA gene expression, we divided the genes from microarray dataset GDS885 into 10 groups; each was a decile composed of ∼1200 genes according to the mRNA abundance levels from the silent genes to the most highly expressed genes (Figure 2B). In all interphase stages of the cell cycle, SUMO-1 was associated with the chromatin surrounding the TSSs of the most active genes. The active genes (90–100% decile; red tracing of Figure 2B) had the highest density of SUMO-1 at the TSSs. The inactive genes (10–20% decile in green and 0–10% decile in black in Figure 2B) were relatively unlabeled by SUMO-1.
The pattern of SUMO-1 labeling revealed two peaks of SUMO-1 binding from −400 to 0.and a comparatively minor peak of SUMO-1 is located at +400 to +2500 bp relative to the TSSs (Figure 2B–C). The promoter peak was high during the transcriptionally active stages of the cell cycle (G1 through late S phase), and then this promoter peak dropped during mitosis with the decrease of transcriptional activity. Interestingly, there is also a drop during S0 phase compared to other transcriptionally active stages. Although we do not have an explanation for this phenomenon, we believe that the beginning of S phase could be the dividing point between two waves of SUMO-1 stimulated transcription.
We also compared our results to microarray data from synchronized cells (46) to test the correlation between SUMO-1 tag on promoter and gene expression. Just as was observed with the microarray results from asynchronously growing cells, for those promoters marked by SUMO-1, gene expression was higher than those without SUMO-1 marks during the cell cycle progression (Supplementary Figure S4). However, mRNA abundance may reflect synthesis at earlier points in the cell cycle, and during mitosis, when genes are repressed in general, there was still positive correlation between SUMO-1 and gene expression. The microarray results from both synchronized and unsynchronized cells were most consistent with SUMO-1 having a direct, transcriptional stimulatory role, and this idea was tested in subsequent experiments.
The patterns of SUMO-1 binding to promoters were determined using averages for groups of genes (Figure 2B–C), but when promoters were analyzed one at a time, we found that SUMO-1 labeled the promoters of a significant subset of genes (Figure 3A). In the heat map, genes with measured expression levels were arranged from top to bottom according to increasing expression levels, and we calculated SUMO-1-binding density of regions surrounding TSSs (±10 kb) for each of the 12 013 genes. We found that in the G1 time point, SUMO-1 was associated with the TSSs, and the highest amount of SUMO-1 label was associated with the most active genes (the rows toward the bottom of the heat map). By contrast, the heat map from samples taken during mitosis revealed very little SUMO-1 labeling of promoters (Figure 3A).
We next asked whether SUMO-1-labeled promoters were changing throughout the course of interphase. We reordered the rows in the heat map according to the density of SUMO-1 in the promoter region in the G1 samples (Figure 3B). The order of the rows in all five heat maps was fixed according to the G1 order. We found that SUMO-1 occupancy around the TSSs was consistent among different cell cycle stages, and SUMO-1 label at TSSs on individual genes slightly increased during cell cycle progression. SUMO-1 marks were cleared during mitosis and then replaced in G1. Among these most abundantly expressed genes, 127 genes were constantly labeled with intense SUMO-1 tags throughout interphase (Supplementary Table S2). This gene list is remarkable for the enrichment of housekeeping genes, notably ribosomal proteins and other translation factors (P = 6.68 × 10−08).
Correlation of SUMO-1 with other chromatin marks
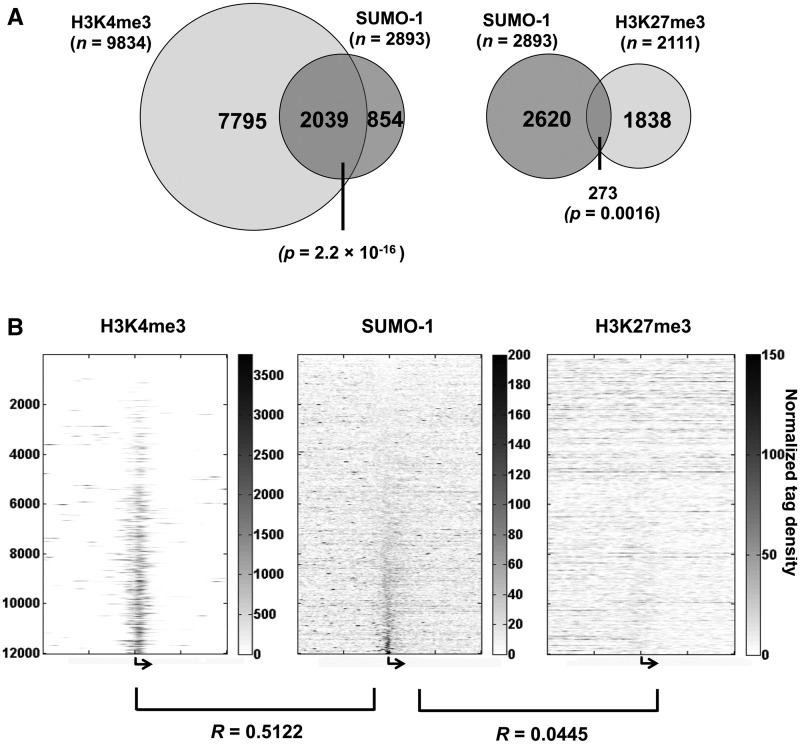
To explore further SUMO-1 association with transcriptionally active chromatin, we compared the SUMO-1-binding pattern from this study to the published binding profiles among various chromatin marks, including the activation mark H3K4me3 and the repression mark H3K27me3 (45). We asked how many of the genes with SUMO-1-enriched promoters also have H3K4me3 peaks falling into the transcribed region. There are a total of 2893 genes with SUMO-1 peaks in the promoter, out of which 70% (2039 genes) have H3K4me3 overlapping in the promoter region (Figure 4A, left, chi-squared test P = 2.2 × 10−16). Since H3K4me3 is associated with open chromatin and actively transcribed genes (47,48), these results further supported the concept that SUMO tagging of the promoter marks active gene expression. In contrast, the number of genes with the repressive H3K27me3 chromatin mark had only 9% overlap with genes with SUMO-1 labeling the corresponding promoters (Figure 4A, right; chi-squared test P = 0.0016).
Figure 4.
SUMO-1-marked promoters are associated with genes marked with H3K4me3. (A) The Venn diagram depicts the degree of overlap between the SUMO-1-marked promoters and H3K4me3-marked promoters (left) as well as SUMO-1-marked promoters and the H3K27me3-marked promoters (right). (B) The heat maps of chromatin marks (H3K4me3, SUMO-1 and H3K27me3, respectively) on each gene ±10 kb surrounding the TSSs (bent arrow). Each row represents the corresponding tag density trace for each individual gene; rows are ordered and kept the same in each heat map according to the maximum intensity of SUMO-1 labeling in the peak region (−1600 to 400 bp) of G1-stage sample. The Pearson correlation coefficients between SUMO-1 and H3K4me3 or H3K27me3 are shown (see ‘Materials and Methods’ section).
To further investigate whether SUMO-1 correlates with H3K4me3 or K27me3, we aligned their binding patterns on genes ±10 kb surrounding the TSSs to determine if the SUMO-1 mark was associated with this measure of gene activation (Figure 4B). Interestingly, we found the SUMO-1 tag profile had a positive correlation with H3K4me3 (R = 0.5122), but not K27me3 (R = 0.0445). Similar results were obtained for the SUMO-1 profiles on chromatin at other cell cycle stages (data not shown). Since we observed a positive correlation between SUMO-1 and H3K4me3, this further supported our interpretation that SUMO-1 is associated with a transcriptional activation signal.
SUMO-1 is a transcriptional activator of genes encoding ribosomal subunit proteins and translation initiation factors
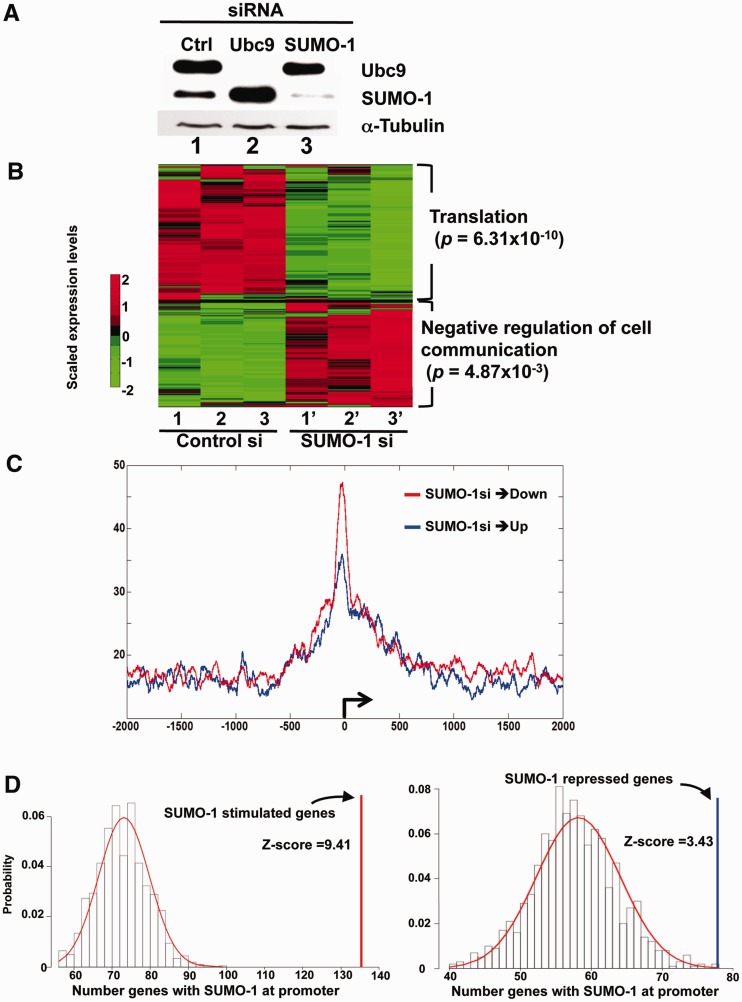
Our results (Figures 2–4) indicated that SUMO-1 marked the promoters of active genes. The timing of the appearance of SUMO-1 marks on promoters during interphase and removal during mitosis suggested that SUMO-1 was involved with the activation process. To test whether SUMO-1 was stimulatory to transcription, we depleted SUMO-1 or its associated E2 factor, Ubc9, by siRNA transfection in HeLa cells. The efficiency of Ubc9 or SUMO-1 siRNA depletion was confirmed by immunoblot analysis (Figure 5A). In cells with depleted Ubc9, the monomer form of SUMO-1 had increased abundance since it was not conjugated to other proteins (Figure 5A, lane 2). We then performed RNA-seq analysis from control and SUMO-1-depleted cells and collected the data from three biological replicates. Multiplex sequencing of polyA+-enriched cDNA on the Illumina GAII generated 5.7–9.7 million reads for each replicate, of which ∼80% could be mapped (Supplementary Table S3). We calculated global gene expression levels using the standard measurement of FPKM (36) from all three replicates for each gene, and all replicates showed highly consistent correlation coefficients (Supplementary Figure S5 and data not shown). We also determined the significance of changes in mRNA abundance using a FDR <0.1%. We found 199 downregulated genes and 158 upregulated genes to have statistically significant changes in expression due to depletion of SUMO-1 (Supplementary Table S4), and the magnitude of the effect ranged from a decrease in mRNA abundance of ∼10-fold to an increase in mRNA abundance of ∼10-fold. A heat map visualizing the 357 differentially expressed genes is shown in Figure 5B, with consistent results observed among the biological replicates. Strikingly, transcripts repressed by SUMO-1 depletion were significantly enriched for those involved in protein synthesis, such as the Gene Ontology (GO) terms ‘Translation’ (P = 6.31 × 10−10). In contrast, those upregulated genes were correlated with GO terms such as ‘negative regulation of cell communication’ (P = 4.87 × 10−3) and ‘negative regulation of signal transduction’ (P = 8.44 × 10-3), though these had lower correlation among enriched GO terms (Figure 5B). Consistent with this observation, by IPA, similar GO terms, such as protein synthesis, were enriched among those genes downregulated by SUMO-1 depletion (P = 1.4 × 10−17; Supplementary Figure S6A) but not the upregulated genes. Among the genes that changed expression, all those associated with protein synthesis function were repressed by depletion of SUMO-1 (Supplementary Figure S6B). These results again suggested SUMO-1 functions as an activator on gene expression. To correlate SUMO-1 mark in the genome and its effect on gene expression, we looked whether those 357 genes have SUMO-1 mark in promoter region (Supplementary Table S4). We found that, 134 out of 199 downregulated genes and 78 out of 161 upregulated genes had a SUMO-1 mark in the promoter region during the G1 phase. Interestingly, when sorting the genes according to the mRNA abundance, we found that SUMO-1 marks at the promoter were more common with the more highly expressed genes, and these marks were most often stimulatory. (This trend can be seen in the presence of the stimulatory SUMO-1 mark shown in red in the top rows—highest expressers—and SUMO-1 mark was more sparsely present in the lower rows of this table; Supplementary Table S4). In contrast, SUMO-1 also labeled promoters in the less expressed genes but acting as a repressor (Supplementary Table S4 in green), indicating that SUMO-1 may have a dual effect on regulating gene expression. We further assessed the average SUMO-1 tag density on these 357 genes, and the results revealed that SUMO-1 marks were enriched on the TSSs of both up- and downregulated genes, though genes that were activated by SUMO-1 had a higher density of SUMO-1 at the TSSs (Figure 5C). To test whether the transcriptional differences under SUMO-1 depletion are likely to be specific events, versus experimental or environmental induced gene expression changes, we tested whether the differentially expressed genes show enrichment under SUMO-1 depletion. We found that both up- and downregulated genes showed highly significant enrichment for association signals (Z = 9.41, P < 2.2 × 10−16 for genes downregulated by SUMO-1 depletion and Z = 3.43, P = 4.19 × 10−4 for genes upregulated by SUMO-1 depletion; Figure 5D).
Figure 5.
Differential expression of genes following SUMO-1 depletion. (A) Western blot analysis of Ubc9 (top), SUMO-1 (middle) or α-tubulin (bottom) proteins was used to evaluate the depletion by the indicated siRNA transfection. (B) Heat map of RNA-seq data showed 357 differentially expressed genes from SUMO-1 depletion compared to control in HeLa cells. Color key on the left shows lower relative expression (green) and higher relative expression (red). The expression intensities were row-wise scaled for the specified genes determined to be significantly changed (adj. P-value <0.05). (C) Average SUMO-1 tag distribution ±10 kb surrounding the TSSs (bent arrow) from up- (blue) or downregulated (red) genes following SUMO-1 depletion. The y-axis is the normalized tag density unit (see ‘Materials and Methods’ section). (D) Differentially expressed genes are enriched with SUMO1 peaks in the promoter region. For each gene set (down- and upregulated), the null distribution is generated by randomly selected 1000 gene sets (gene number = 199 and 158, respectively, see ‘Materials and Methods’ section). The enrichment score (Z-score) for the gene sets obtained from RNA-seq comparisons was indicated in the distribution plot by the vertical line (upregulated: blue, downregulated: red).
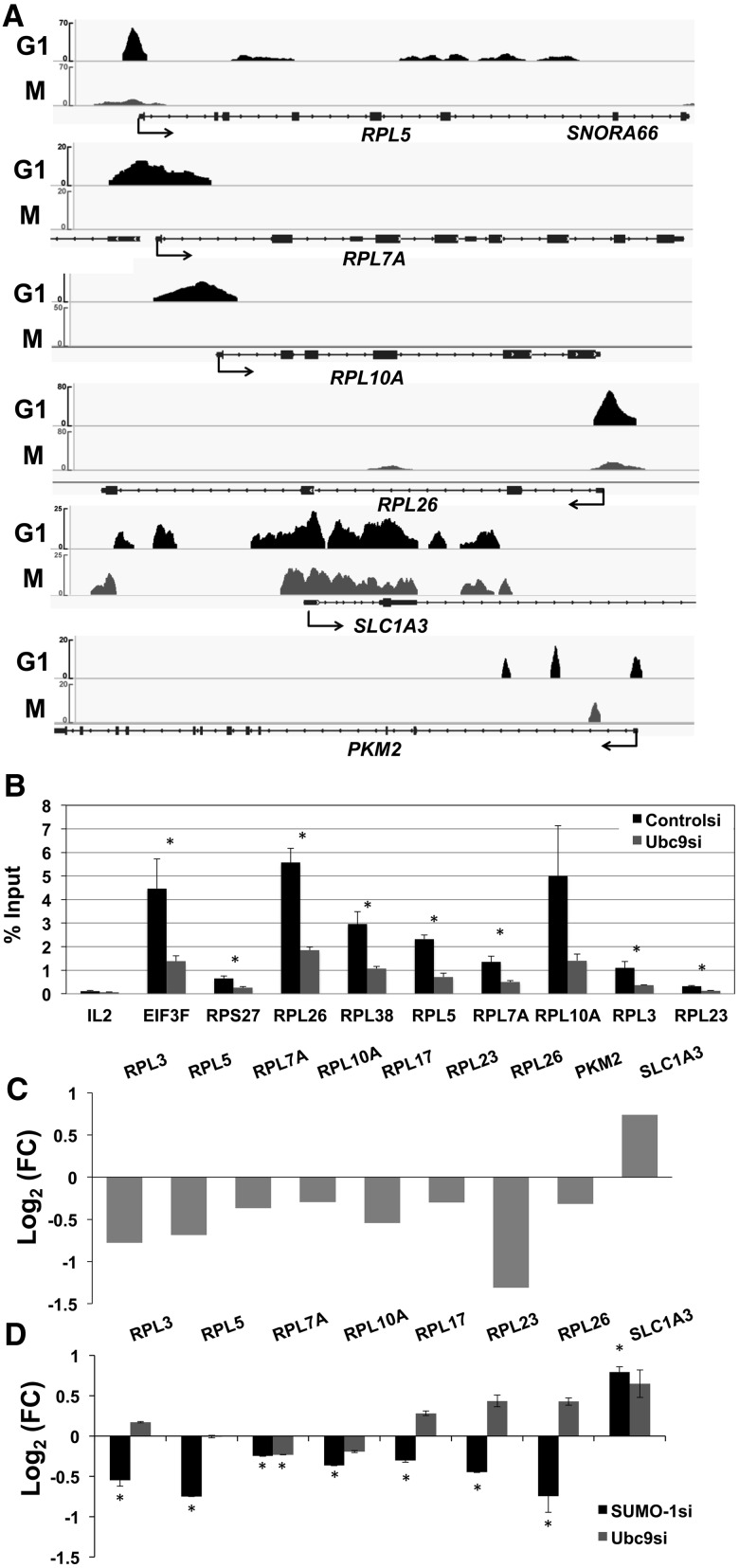
We find it striking that some of the housekeeping genes, for example, ribosome biogenesis proteins (RPL5, RPL7A and RPL10A) and translation factors such as initiation and elongation factors (EIF3D, EIF3E, EIF4G2, EIF5B and EEF2), were marked by SUMO-1 at their promoters during interphase and had mRNA expression stimulated by SUMO-1. Examples of specific genes with SUMO-1 density for G1 and M phases and effects on transcription are shown in Figure 6A (top four tracings). We also observed the same occupancy of SUMO-1 on the promoter region when assessing endogenous SUMO-1 using SUMO-1-specific antibody in ChIP-SUMO-1 data from HeLa cells (Supplementary Figure S7). Ubc9 was required for SUMO-1 to associate with these promoters. Depletion of Ubc9 resulted in a decrease in SUMO-1 marks at these promoters (Figure 6B). This result suggested that SUMO-1 is coupled to the chromatin at these promoters and is not binding as a monomeric protein. For those genes that were stimulated by SUMO-1 depletion, i.e. SUMO-1 functioned as a repressor, patterns in the SUMO-1 tag density on the promoter and gene at different points in the cell cycle were not identified. An example of a gene repressed by SUMO-1 with SUMO-1 found at the promoter, SLC1A3, is shown in Figure 6A. Consistent with an earlier study (32), we found that the promoter of PKM2 (a homologue of Pyk1 in yeast) is labeled by SUMO-1 in G1 but not M (Figure 6A, bottom), and its expression is decreased upon SUMO-1 depletion (Figure 6C). In addition, our RNA-seq results showed that several ribosomal protein genes are significantly downregulated under SUMO-1 depletion (Figure 6C), and these genes were confirmed by RT–qPCR using the same siRNA (Figure 6D) and a second siRNA specific to SUMO-1 (Supplementary Figure S6C). For these assays, we also tested Ubc9-depleted samples (Figure 6D). The results showed that when SUMO-1 was depleted, those genes were all downregulated. Interestingly, Ubc9 depletion was not in all cases consistent with the SUMO-1 depletion. We suggest from this result with Ubc9 depletion that other SUMO family proteins, such as SUMO-2/3, might be involved in the regulation of these transcripts. These observations indicate several interesting points. The combination of ChIP-seq, RNA-seq and RT–qPCR results support the concept that SUMO-1 directly activates specific gene expression and SUMO-1 is associated with regulation of expression of ribosomal proteins and translation factors.
Figure 6.
SUMO-1 activates expression of ribosome biogenesis genes. (A) Examples of SUMO-1 tracing on specific promoters in the G1 phase (top) and in mitosis (bottom) are shown above the gene map drawn from the IGV genome browser. Genes shown are (top to bottom) RPL5, RPL7A, RPL10A, RPL26, SLC1A3 and PKM2. (B) The effect of Ubc9 depletion on SUMOylation of specific promoters. Chromatin was isolated from control siRNA-transfected cells (black) or Ubc9 siRNA-transfected cells (gray), and recombinant SUMO-1 was detected by ChAP. IL-2 was a negative control based on the gene expression and ChAP-seq data. A t-test using the data from four biological replications of ChAP–qPCR was conducted (*, P-value ≤0.05). (C) RNA-seq analysis showing the effects of SUMO-1 depletion on mRNA levels of selected genes. The genes with statistically significant changes in RNA level are shown. Values are expressed as log2 fold change [Log2(FC)]; for those genes that depletion of SUMO-1 caused a decrease in mRNA levels, the histogram points downward. (D) RT–qPCR analysis of gene expression levels for the indicated genes 72 h after transfection using siRNAs specific for control, SUMO-1 or Ubc9. Fold change relative to the control siRNA is represented in log2 scale for SUMO-1 (black) and Ubc9 (gray). The mRNA expression level for each experiment was normalized to Polr2a (a non-SUMO-1-labeled gene) and to the result with the control siRNA. Three biological replicates were done, and error bars reflect the SEM. A t-test of equal expression between SUMO-1/Ubc9 and control siRNA using the data from three biological replications of RT–qPCR was conducted (*, P-value ≤0.05).
DISCUSSION
In this study, we mapped genome-wide labeling of chromatin by the SUMO-1 protein throughout the human cell cycle and made multiple discoveries. (i) On a chromosome-wide scale, the SUMO-1-binding profile was consistent during interphase, but changes were evident during mitosis with a decrease in SUMO-1 binding events. (ii) We found the ChAP-seq data of SUMO-1 replicates were highly reproducible, and the pattern of SUMO-1 binding to chromatin was dynamic during cell cycle progression. (iii) The SUMO-1 distribution on the chromatin was enriched on active genes, especially the regulatory elements such as CpG islands and promoters. (iv) SUMO-1 localization on promoter chromatin was highly correlated with the transcriptional activation signal of H3K4me3 and had low correlation with the transcriptional repressive signal H3K27me3. (v) The effect of SUMO-1 labeling of promoters on gene expression was in many cases stimulatory. (vi) Genes that encode ribosomal protein subunits and translation factors were the most significant subgroup stimulated by SUMO-1.
An initial clue that SUMO-1 was correlated with gene activation was that it was associated with highly active promoters throughout interphase, decreased during mitosis when transcription is generally repressed and then was present again in the G1 phase of the cell cycle. It must be recognized with this cell cycle correlation of SUMO-1 marks that the absence of a chromatin mark during mitosis can have many causes aside from the regulation of transcription. It has been shown that SUMO-1 is removed from chromatin during mitosis (42). Our results are consistent with that earlier finding, though we do still observe SUMO-1 marks on specific sites, for example many promoters (Figure 2A) including the SLC1A3 gene (Figure 6A). The results indicate that the signal by SUMO-1 on a promoter is complicated: in many cases it is stimulatory and in others the SUMO-1 tag is repressive (Figure 5). The genome-wide analysis presented in this study is a first step toward deciphering how SUMO-1 is regulating gene expression. The striking finding on which we focused was that among very highly expressed genes, SUMO-1 is a stimulatory mark (Supplementary Table S4). From the PCA (Figure 1D), it is clear that how SUMO-1 associates with a variety of genetic elements changes through the cell cycle, and future analyses are targeted at deciphering these aspects of the complex chromatin signaling by SUMO-1.
Interestingly, when comparing the labeling of chromatin by SUMO-1 in this study with the labeling of chromatin by ubiquitin during mitosis, we found a high level of concordance. The promoters of many genes whose expression is important in the G1 phase of the cell cycle are bookmarked by ubiquitination during mitosis and then de-ubiquitinated in G1 (Arora et al., submitted). Of the 3446 promoters found to be bookmarked by ubiquitin during mitosis, 1829 promoters (53%) were labeled by SUMO-1 during interphase (Supplementary Table S5). These results are most consistent with SUMO-1 having a stimulatory role in regulating gene expression via the chromatin.
SUMOylation of transcription complexes and/or chromatin-modifying complexes is known to regulate subcellular localization, protein–DNA-binding affinity and repress gene transcription. For example, SUMOylation of a variety of transcription factors/ co-factors fused with reporter gene inhibits gene expression (16,49–51). Moreover, expression of a dominant-negative E2 Ubc9, which inhibits SUMO conjugation to substrate proteins, or mutation of the SUMO-targeting sites on transcription factors resulted in upregulated transcriptional activity of specific genes (15,18). SUMO-1/2/3 have all been shown to recruit histone deacetylases (HDACs) (50,52) and thus repress acetylated chromatin. For these reasons, we were surprised that our global SUMO-1 binding data showed SUMO-1 actually marked constitutively expressed genes. From the genome-wide data, SUMO-1 associates with highly expressed genes that encode proteins involved in protein biogenesis. Whether the SUMO-1 moiety was recruited by specific bound factors or DNA elements is unclear at this time. It is possible that the transcription activation process itself recruits the SUMOylation to highly active promoters. On these high-activity promoters, binding by SUMO-1 is stimulatory.
One published study focused on SUMO marking of multiple promoters in yeast. That study suggested that SUMOylation of the promoter bound factors is associated with constitutive transcription and also activation of inducible genes, and inactivation of SUMOylation in yeast harboring a defective ubc9 gene reduced SUMO at the constitutive promoters and decreased gene expression in yeast (32). In contrast, in our study, the outcome of Ubc9 depletion is not necessarily consistent with SUMO-1 depletion, and we suggest that this inconsistency is due to SUMO isoforms (i.e. SUMO-2/3) that might have opposing transcriptional activities. The conjugation of SUMO-1 and SUMO-2/3 on substrates has been shown to have an opposing role with a specific transcription factor (53). In another study, SUMO-1 was located on both active and repressive photoreceptor-specific genes to regulate rod cell development in a mouse model (54). The results of our study substantially add to the concept that SUMO-1 is a stimulatory mark on chromatin since we found that genome-wide in the human cell, the preponderance of SUMO-1 chromatin marks on, or near promoter regions are associated with active gene expression.
Ribosome biogenesis proteins, such as small nuclear ribonucleoproteins, and ribosomal proteins were identified as novel SUMO targets and were required for nucleolus formation (17). Moreover, a recent study showed SUMO system is critical for nucleolar partitioning by regulating a novel ribosome biogenesis complex (55). The current study finds that not only are the ribosomal proteins SUMOylated but also the genes encoding ribosomal proteins and translation factors are labeled by SUMO-1 on the chromatin over their promoters. Taken together, we suggest that SUMO-1 regulates nucleolar integrity during the cell cycle processing, both transcriptionally and post-translationally.
Since impairing SUMO-1 on these promoters resulted in lower expression, this shows that efficient SUMOylation is critical for optimal gene expression. SUMO-1 marking on these translational machinery genes may function to maintain gene expression and protein stability perhaps by antagonizing other repressive chromatin marks or regulating the subcellular localization of partner proteins required for repression. In addition, while SUMOylation plays a critical role on gene repression on a subset of genes, SUMO-1 also has other properties, for example, regulating the assembly of transcription machinery (56); therefore, SUMO-1 marking on those housekeeping genes may be an early modification affecting chromatin remodeling. It is unclear at this time what are the relevant chromatin proteins in promoters conjugated to SUMO-1. The position of the peak of the SUMO-1 mark on constitutive active promoters is at −200 relative to the TSS. Such a position could be consistent with the −1 nucleosome or close to where the components of general transcription machinery would be expected to bind. A previous study has shown that SUMO-1 post-translationally modifies hsTAF5 in TFIID to modulate TFIID promoter-binding activity (18). It is possible that this is the factor SUMOylated at promoters in our studies; however, it would be a complicated mechanism by which SUMOylation of a general transcription factor would be associated with the transcription activation process. Further arguing against TFIID components causing the promoter peak of SUMO-1 binding, the methods used in this study had sufficient resolution to map the bound domains and TFIID subunits would be expected to be closer to the TSSs.
In summary, in this study we demonstrated how SUMO-1 marks promoters in the human genome and how it changes through the cell cycle. We found that SUMO-1 labeling of chromatin is dynamic through the cell cycle, and it is associated at promoters with the most actively transcribed genes. While SUMO-1 was not generally associated with all active genes, a very high percentage of the most active genes (49%) had their promoters modified with bound SUMO-1, and it was shown that in many of the housekeeping genes, the SUMO-1 mark on the promoter was stimulatory to gene expression and is critical for the high expression genes encoding translation factors.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–6 and Supplementary Figures 1–7.
FUNDING
National Cancer Institute grants [CA141090 and CA111480] and Postdoctoral Fellowship [CA130302 to G.F.H.]; Pelotonia Predoctoral Cancer Research Training Fellowship (to H-w. Liu). Funding for open access charge: Departmental funds.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr. Pearlly Yan and Nucleic Acid Shared Resource Facility at The Ohio State University for the help of sample sequencing and Aaron Chen for the help of Venn diagrams.
REFERENCES
- 1.Gareau JR, Lima CD. The SUMO pathway: emerging mechanisms that shape specificity, conjugation and recognition. Nat. Rev. Mol. Cell Biol. 2010;11:861–871. doi: 10.1038/nrm3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Guo D, Li M, Zhang Y, Yang P, Eckenrode S, Hopkins D, Zheng W, Purohit S, Podolsky RH, Muir A, et al. A functional variant of SUMO4, a new I kappa B alpha modifier, is associated with type 1 diabetes. Nat. Genet. 2004;36:837–841. doi: 10.1038/ng1391. [DOI] [PubMed] [Google Scholar]
- 3.Geiss-Friedlander R, Melchior F. Concepts in sumoylation: a decade on. Nat. Rev. Mol. Cell Biol. 2007;8:947–956. doi: 10.1038/nrm2293. [DOI] [PubMed] [Google Scholar]
- 4.Gill G. SUMO weighs in on polycomb-dependent gene repression. Mol. Cell. 2010;38:157–159. doi: 10.1016/j.molcel.2010.04.006. [DOI] [PubMed] [Google Scholar]
- 5.Kang X, Qi Y, Zuo Y, Wang Q, Zou Y, Schwartz RJ, Cheng J, Yeh ET. SUMO-specific protease 2 is essential for suppression of polycomb group protein-mediated gene silencing during embryonic development. Mol. Cell. 2010;38:191–201. doi: 10.1016/j.molcel.2010.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang SH, Sharrocks AD. PIASxalpha differentially regulates the amplitudes of transcriptional responses following activation of the ERK and p38 MAPK pathways. Mol. Cell. 2006;22:477–487. doi: 10.1016/j.molcel.2006.03.037. [DOI] [PubMed] [Google Scholar]
- 7.Desterro JM, Rodriguez MS, Hay RT. SUMO-1 modification of IkappaBalpha inhibits NF-kappaB activation. Mol. Cell. 1998;2:233–239. doi: 10.1016/s1097-2765(00)80133-1. [DOI] [PubMed] [Google Scholar]
- 8.Muller S, Berger M, Lehembre F, Seeler JS, Haupt Y, Dejean A. c-Jun and p53 activity is modulated by SUMO-1 modification. J. Biol. Chem. 2000;275:13321–13329. doi: 10.1074/jbc.275.18.13321. [DOI] [PubMed] [Google Scholar]
- 9.Kim J, Cantwell CA, Johnson PF, Pfarr CM, Williams SC. Transcriptional activity of CCAAT/enhancer-binding proteins is controlled by a conserved inhibitory domain that is a target for sumoylation. J. Biol. Chem. 2002;277:38037–38044. doi: 10.1074/jbc.M207235200. [DOI] [PubMed] [Google Scholar]
- 10.Ross S, Best JL, Zon LI, Gill G. SUMO-1 modification represses Sp3 transcriptional activation and modulates its subnuclear localization. Mol. Cell. 2002;10:831–842. doi: 10.1016/s1097-2765(02)00682-2. [DOI] [PubMed] [Google Scholar]
- 11.Ouyang J, Valin A, Gill G. Regulation of transcription factor activity by SUMO modification. Methods Mol. Biol. 2009;497:141–152. doi: 10.1007/978-1-59745-566-4_9. [DOI] [PubMed] [Google Scholar]
- 12.Garcia-Dominguez M, Reyes JC. SUMO association with repressor complexes, emerging routes for transcriptional control. Biochim. et Biophys. Acta. 2009;1789:451–459. doi: 10.1016/j.bbagrm.2009.07.001. [DOI] [PubMed] [Google Scholar]
- 13.Girdwood D, Bumpass D, Vaughan OA, Thain A, Anderson LA, Snowden AW, Garcia-Wilson E, Perkins ND, Hay RT. P300 transcriptional repression is mediated by SUMO modification. Mol. Cell. 2003;11:1043–1054. doi: 10.1016/s1097-2765(03)00141-2. [DOI] [PubMed] [Google Scholar]
- 14.Ouyang J, Gill G. SUMO engages multiple corepressors to regulate chromatin structure and transcription. Epigenetics. 2009;4:440–444. doi: 10.4161/epi.4.7.9807. [DOI] [PubMed] [Google Scholar]
- 15.Nathan D, Ingvarsdottir K, Sterner DE, Bylebyl GR, Dokmanovic M, Dorsey JA, Whelan KA, Krsmanovic M, Lane WS, Meluh PB, et al. Histone sumoylation is a negative regulator in Saccharomyces cerevisiae and shows dynamic interplay with positive-acting histone modifications. Genes Dev. 2006;20:966–976. doi: 10.1101/gad.1404206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shiio Y, Eisenman RN. Histone sumoylation is associated with transcriptional repression. Proc. Natl Acad. Sci. USA. 2003;100:13225–13230. doi: 10.1073/pnas.1735528100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matafora V, D'Amato A, Mori S, Blasi F, Bachi A. Proteomics analysis of nucleolar SUMO-1 target proteins upon proteasome inhibition. Mol. Cell Proteomics. 2009;8:2243–2255. doi: 10.1074/mcp.M900079-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boyer-Guittaut M, Birsoy K, Potel C, Elliott G, Jaffray E, Desterro JM, Hay RT, Oelgeschlager T. SUMO-1 modification of human transcription factor (TF) IID complex subunits: inhibition of TFIID promoter-binding activity through SUMO-1 modification of hsTAF5. J. Biol. Chem. 2005;280:9937–9945. doi: 10.1074/jbc.M414149200. [DOI] [PubMed] [Google Scholar]
- 19.Yan Q, Gong L, Deng M, Zhang L, Sun S, Liu J, Ma H, Yuan D, Chen PC, Hu X, et al. Sumoylation activates the transcriptional activity of Pax-6, an important transcription factor for eye and brain development. Proc. Natl Acad. Sci. USA. 2010;107:21034–21039. doi: 10.1073/pnas.1007866107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kotaja N, Karvonen U, Janne OA, Palvimo JJ. The nuclear receptor interaction domain of GRIP1 is modulated by covalent attachment of SUMO-1. J. Biol. Chem. 2002;277:30283–30288. doi: 10.1074/jbc.M204768200. [DOI] [PubMed] [Google Scholar]
- 21.Wang J, Li A, Wang Z, Feng X, Olson EN, Schwartz RJ. Myocardin sumoylation transactivates cardiogenic genes in pluripotent 10T1/2 fibroblasts. Mol. Cell Biol. 2007;27:622–632. doi: 10.1128/MCB.01160-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shyu YC, Lee TL, Ting CY, Wen SC, Hsieh LJ, Li YC, Hwang JL, Lin CC, Shen CK. Sumoylation of p45/NF-E2: nuclear positioning and transcriptional activation of the mammalian beta-like globin gene locus. Mol. Cell Biol. 2005;25:10365–10378. doi: 10.1128/MCB.25.23.10365-10378.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang J, Feng XH, Schwartz RJ. SUMO-1 modification activated GATA4-dependent cardiogenic gene activity. J. Biol. Chem. 2004;279:49091–49098. doi: 10.1074/jbc.M407494200. [DOI] [PubMed] [Google Scholar]
- 24.Lin X, Liang M, Liang YY, Brunicardi FC, Melchior F, Feng XH. Activation of transforming growth factor-beta signaling by SUMO-1 modification of tumor suppressor Smad4/DPC4. J. Biol. Chem. 2003;278:18714–18719. doi: 10.1074/jbc.M302243200. [DOI] [PubMed] [Google Scholar]
- 25.Tian S, Poukka H, Palvimo JJ, Janne OA. Small ubiquitin-related modifier-1 (SUMO-1) modification of the glucocorticoid receptor. Biochem. J. 2002;367:907–911. doi: 10.1042/BJ20021085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Terui Y, Saad N, Jia S, McKeon F, Yuan J. Dual role of sumoylation in the nuclear localization and transcriptional activation of NFAT1. J. Biol. Chem. 2004;279:28257–28265. doi: 10.1074/jbc.M403153200. [DOI] [PubMed] [Google Scholar]
- 27.Guo B, Sharrocks AD. Extracellular signal-regulated kinase mitogen-activated protein kinase signaling initiates a dynamic interplay between sumoylation and ubiquitination to regulate the activity of the transcriptional activator PEA3. Mol. Cell Biol. 2009;29:3204–3218. doi: 10.1128/MCB.01128-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hong Y, Rogers R, Matunis MJ, Mayhew CN, Goodson ML, Park-Sarge OK, Sarge KD. Regulation of heat shock transcription factor 1 by stress-induced SUMO-1 modification. J. Biol. Chem. 2001;276:40263–40267. doi: 10.1074/jbc.M104714200. [DOI] [PubMed] [Google Scholar]
- 29.Goodson ML, Hong Y, Rogers R, Matunis MJ, Park-Sarge OK, Sarge KD. Sumo-1 modification regulates the DNA binding activity of heat shock transcription factor 2, a promyelocytic leukemia nuclear body associated transcription factor. J. Biol. Chem. 2001;276:18513–18518. doi: 10.1074/jbc.M008066200. [DOI] [PubMed] [Google Scholar]
- 30.Gostissa M, Hengstermann A, Fogal V, Sandy P, Schwarz SE, Scheffner M, Del Sal G. Activation of p53 by conjugation to the ubiquitin-like protein SUMO-1. EMBO J. 1999;18:6462–6471. doi: 10.1093/emboj/18.22.6462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu SY, Chiang CM. Crosstalk between sumoylation and acetylation regulates p53-dependent chromatin transcription and DNA binding. EMBO J. 2009;28:1246–1259. doi: 10.1038/emboj.2009.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rosonina E, Duncan SM, Manley JL. SUMO functions in constitutive transcription and during activation of inducible genes in yeast. Genes Dev. 2010;24:1242–1252. doi: 10.1101/gad.1917910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tagwerker C, Flick K, Cui M, Guerrero C, Dou Y, Auer B, Baldi P, Huang L, Kaiser P. A tandem affinity tag for two-step purification under fully denaturing conditions: application in ubiquitin profiling and protein complex identification combined with in vivo cross-linking. Mol. Cell Proteomics. 2006;5:737–748. doi: 10.1074/mcp.M500368-MCP200. [DOI] [PubMed] [Google Scholar]
- 34.Lee TI, Johnstone SE, Young RA. Chromatin immunoprecipitation and microarray-based analysis of protein location. Nat. Protoc. 2006;1:729–748. doi: 10.1038/nprot.2006.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fejes AP, Robertson G, Bilenky M, Varhol R, Bainbridge M, Jones SJ. FindPeaks 3.1: a tool for identifying areas of enrichment from massively parallel short-read sequencing technology. Bioinformatics. 2008;24:1729–1730. doi: 10.1093/bioinformatics/btn305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Carson JP, Zhang N, Frampton GM, Gerry NP, Lenburg ME, Christman MF. Pharmacogenomic identification of targets for adjuvant therapy with the topoisomerase poison camptothecin. Cancer Res. 2004;64:2096–2104. doi: 10.1158/0008-5472.can-03-2029. [DOI] [PubMed] [Google Scholar]
- 39.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Watts FZ. The role of SUMO in chromosome segregation. Chromosoma. 2007;116:15–20. doi: 10.1007/s00412-006-0079-z. [DOI] [PubMed] [Google Scholar]
- 41.Bachant J, Alcasabas A, Blat Y, Kleckner N, Elledge SJ. The SUMO-1 isopeptidase Smt4 is linked to centromeric cohesion through SUMO-1 modification of DNA topoisomerase II. Mol. Cell. 2002;9:1169–1182. doi: 10.1016/s1097-2765(02)00543-9. [DOI] [PubMed] [Google Scholar]
- 42.Zhang XD, Goeres J, Zhang H, Yen TJ, Porter AC, Matunis MJ. SUMO-2/3 modification and binding regulate the association of CENP-E with kinetochores and progression through mitosis. Mol. Cell. 2008;29:729–741. doi: 10.1016/j.molcel.2008.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Luco RF, Allo M, Schor IE, Kornblihtt AR, Misteli T. Epigenetics in alternative pre-mRNA splicing. Cell. 2011;144:16–26. doi: 10.1016/j.cell.2010.11.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huff JT, Plocik AM, Guthrie C, Yamamoto KR. Reciprocal intronic and exonic histone modification regions in humans. Nat. Struct. Mol. Biol. 2010;17:1495–1499. doi: 10.1038/nsmb.1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 46.Sadasivam S, Duan S, DeCaprio JA. The MuvB complex sequentially recruits B-Myb and FoxM1 to promote mitotic gene expression. Genes Dev. 2012;26:474–489. doi: 10.1101/gad.181933.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Justin N, De Marco V, Aasland R, Gamblin SJ. Reading, writing and editing methylated lysines on histone tails: new insights from recent structural studies. Curr. Opin. Struct. Biol. 2010;20:730–738. doi: 10.1016/j.sbi.2010.09.012. [DOI] [PubMed] [Google Scholar]
- 48.Santos-Rosa H, Schneider R, Bannister AJ, Sherriff J, Bernstein BE, Emre NC, Schreiber SL, Mellor J, Kouzarides T. Active genes are tri-methylated at K4 of histone H3. Nature. 2002;419:407–411. doi: 10.1038/nature01080. [DOI] [PubMed] [Google Scholar]
- 49.Holmstrom S, Van Antwerp ME, Iniguez-Lluhi JA. Direct and distinguishable inhibitory roles for SUMO isoforms in the control of transcriptional synergy. Proc. Natl Acad. Sci. USA. 2003;100:15758–15763. doi: 10.1073/pnas.2136933100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang SH, Sharrocks AD. SUMO promotes HDAC-mediated transcriptional repression. Mol. Cell. 2004;13:611–617. doi: 10.1016/s1097-2765(04)00060-7. [DOI] [PubMed] [Google Scholar]
- 51.Chupreta S, Holmstrom S, Subramanian L, Iniguez-Lluhi JA. A small conserved surface in SUMO is the critical structural determinant of its transcriptional inhibitory properties. Mol. Cell. Biol. 2005;25:4272–4282. doi: 10.1128/MCB.25.10.4272-4282.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ouyang J, Shi Y, Valin A, Xuan Y, Gill G. Direct binding of CoREST1 to SUMO-2/3 contributes to gene-specific repression by the LSD1/CoREST1/HDAC complex. Mol. Cell. 2009;34:145–154. doi: 10.1016/j.molcel.2009.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lyst MJ, Nan X, Stancheva I. Regulation of MBD1-mediated transcriptional repression by SUMO and PIAS proteins. EMBO J. 2006;25:5317–5328. doi: 10.1038/sj.emboj.7601404. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 54.Onishi A, Peng GH, Hsu C, Alexis U, Chen S, Blackshaw S. Pias3-dependent SUMOylation directs rod photoreceptor development. Neuron. 2009;61:234–246. doi: 10.1016/j.neuron.2008.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Finkbeiner E, Haindl M, Muller S. The SUMO system controls nucleolar partitioning of a novel mammalian ribosome biogenesis complex. EMBO J. 2011;30:1067–1078. doi: 10.1038/emboj.2011.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vethantham V, Rao N, Manley JL. Sumoylation modulates the assembly and activity of the pre-mRNA 3′ processing complex. Mol. Cell. Biol. 2007;27:8848–8858. doi: 10.1128/MCB.01186-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.