Abstract
UVA (320–400 nm) represents the main spectral component of solar UV radiation, induces pre-mutagenic DNA lesions and is classified as Class I carcinogen. Recently, discussion arose whether UVA induces DNA double-strand breaks (dsbs). Only few reports link the induction of dsbs to UVA exposure and the underlying mechanisms are poorly understood. Using the Comet-assay and γH2AX as markers for dsb formation, we demonstrate the dose-dependent dsb induction by UVA in G1-synchronized human keratinocytes (HaCaT) and primary human skin fibroblasts. The number of γH2AX foci increases when a UVA dose is applied in fractions (split dose), with a 2-h recovery period between fractions. The presence of the anti-oxidant Naringin reduces dsb formation significantly. Using an FPG-modified Comet-assay as well as warm and cold repair incubation, we show that dsbs arise partially during repair of bi-stranded, oxidative, clustered DNA lesions. We also demonstrate that on stretched chromatin fibres, 8-oxo-G and abasic sites occur in clusters. This suggests a replication-independent formation of UVA-induced dsbs through clustered single-strand breaks via locally generated reactive oxygen species. Since UVA is the main component of solar UV exposure and is used for artificial UV exposure, our results shine new light on the aetiology of skin cancer.
INTRODUCTION
Experimental and epidemiological evidence, gathered in the last decade, have shown that UVB (280–315 nm) and UVA (315–400 nm) exposure from the sun as well as from artificial sources (e.g. sun beds) are the most important etiological factors for the development of skin cancer (1,2). Recently, UV radiation (UVB and UVA) has been classified as a Class I carcinogen (3) by the International Agency for the Research on Cancer.
The mechanism underlying UVB-induced mutagenesis is well understood and can be accounted to the two major forms of pre-mutagenic DNA lesions, the cyclobutane pyrimidine dimer (CPD) and the pyrimidine-(6-4)-pyrimidone photoproduct (6-4 PP) (4,5). Unrepaired UV-B lesions result in transition mutations (CC-TT) at dipyrimidine sequences (1,4), which have been shown to represent UV-signature mutations in skin cancer (6–8). In contrast, the mechanisms of action for UVA-induced DNA damage are less well understood. The fact that UVA is not directly adsorbed by DNA but needs endogenous photo-sensitizers to deploy its damages is widely accepted (6,9,10). UVA exerts its DNA-damaging effects through these (so far unknown) cellular photosensitizers (PS), which are photo-excited and involved in Type I or II photoreactions to produce reactive oxygen species (ROS), like ·OH, ·O2−, H2O2 or 1O2 (9,11,12) in the presence or absence of metal ions. UVA exposure leads to several lesions, oxidized bases (8-oxo-dG, thymidine-glycol), abasic sites or single-strand breaks (ssbs). Additionally, UVA is nowadays also seen as a source for CPDs, especially TT-CPDs, generated via a photosensitized triplet energy transfer (13). The dependence on PS is highlighted by the fact that isolated DNA is nearly not damaged by UVA irradiation, in contrast to cellular DNA when equal doses are used (14).
The ability of UVA to induce DNA double-strand breaks (dsbs) is still under discussion. While replication-dependent dsb formation is accepted (15,16), replication-independent dsb formation is doubted. UVA was originally not expected to be involved in the generation of dsbs due to the relative low photonic energy. This idea was supported by a recent study of Rizzo et al. (17) who did interpret their experimental data as being indicative for no dsbs induced in cells exposed towards UVA. Additionally, Cadet and Douki (10) argued that induction of dsbs via clustered ssbs is unlikely, since UVA induces more than twice as much 8-oxo-dG compared to ssbs or alkali labile sites. Despite these arguments, we and others have shown, using various end points, e.g. micronucleus formation, clonogenic survival (18,19) or γH2AX formation (20–22), as well as using cell lines deficient in dsb repair pathways (23) that UVA induces dsbs in a replication-independent manner.
Dsbs can be generated by oxidizing reagents in the form of clustered DNA lesions that are converted to dsbs during the repair process, when both strands are incised simultaneously in close proximity (1–20 bases). These so-called oxidatively induced clustered DNA lesions (OCDLs) were found to be responsible for dsb induction using several other types of radiation and chemical genotoxins (24–26).
In view of the current discussion whether UVA is able to induce DNA dsbs, independent of replication, the data presented here support the ability of UVA at physiologically relevant doses to induce these lesions. Our findings are based on independent assessments of γH2AX formation, Comet-assay and immunodetection of DNA lesions on stretched chromatin fibres in non-cycling, G1 arrested cells, thus ruling out replication-dependent dsb induction. Furthermore, we are able to demonstrate that the repair of clustered oxidative damage is an important source for UVA-induced dsbs.
MATERIALS AND METHODS
Cell isolation and cell cultures
For the isolation of fibroblasts, human skin biopsies were cut into 0.5 cm2 pieces and placed into a cell culture dish stratum corneum down. After 30 min at room temperature, samples had adhered to the culture dish and 10 ml culture medium [DMEM (Invitrogen, Germany) + 10% FCS (Biochrom, Germany)] per 75-cm2 cell culture tissue flask were added carefully. The samples were incubated in a humidified atmosphere of 5% CO2 for 3–4 weeks, until fibroblasts had grown out and could be sub-cultured.
The human keratinocyte cell line HaCaT (27), obtained from the Deutsches Krebsforschungszentrum (DKFZ) Heidelberg, Germany, was cultured as described elsewhere (22). For immuno-histochemistry, the cells were grown directly on standard microscope slides.
To arrest HaCaT cells in G1, the cells were incubated for 5 days in isoleucin-free medium F-10 (Biochrom), containing 5% dialysed FCS (Biochrom). The cell cycle distribution was routinely analysed by flow cytometry following DNA staining.
UVA irradiation
Irradiation was performed using a 300-W Ultravitalux (Osram) light bulb. The peak at 365 nm was filtered with a combination of 2 colour glass filters UG1 und KG1 (Schott, Germany). No radiation with λ < 340 nm passes the filter. The dose rate of 535 W/m2 was measured with a Solarscope UV Radiometer (SolTech). The cells were irradiated in pre-warmed, sterile PBS unless stated otherwise. Temperature during irradiation was controlled by two air-cooling systems, resulting in a temperature increase from 34.2 ± 1.2°C to 38.1 ± 0.9°C during 40 min of irradiation. For split-dose irradiation, the cells were exposed as described earlier in PBS and transferred to pre-warmed culture medium thereafter. The time course of the split-dose experiments was as follows: exposure (200 kJ/m2) + 2 h of recovery incubation. The time for a single 200 kJ/m2 exposure was 6 min. After the last exposure (also for single doses), the cells were post-incubated for 30 min under normal culture conditions before being fixed or harvested. For control stainings, cells were exposed to 1 Gy of X-ray at a dose of 2.9 Gy/min in plastic dishes and processed for immunostaining as described below (see Supplementary Figure S1).
Antioxidant treatment
Prior to irradiation, antioxidant treatment of the cell cultures was done by incubating the cells for 24 h with 100 nM Naringin (final concentration; Sigma, Germany) in complete medium (fibroblasts) or isoleucin-free medium (keratinocytes). Previous studies have shown that this treatment quenches ROS in eukaryotic cells (28,29). Directly before irradiation, the medium was removed and cells were washed two times in PBS.
ROS measurement
For ROS measurements, exponentially growing HaCaT cells were exposed to the indicated UVA doses in pre-warmed PBS. After exposure, the cells were stained in PBS containing 5 µM CM-H2DCFDA (Molecular Probes) for 15 min at 37°C. After staining, cells were again incubated for 15 min at 37°C in pre-warmed PBS. Then cells were being scraped into ice cold 0.1 mM ETDA for cell swelling and nuclei were released by vortexing for 10 s. Nuclei were stained with 50 µg/ml propidium iodide (PJ) and cells were analysed in a Coulter flow cytometer with gating for single nuclei according to the PJ fluorescence. CM-H2DCFDA fluorescence was measured in three independent replicas with a minimum of 20 000 cells.
Apoptosis measurement
HaCaT cells were grown, synchronized and exposed to 900 kJ/m2 UVA as described earlier. After each indicated time, the cells were washed once carefully with PBS and stained with 20 µl Annexin-V-FLUOS (Roche) per ml in staining solution (10 mM HEPES–NaOH, pH 7.4, 140 mM NaCl, 5 mM CaCl2) for 30 min in the dark at 37°C. Apoptotic cells were counted immediately using a fluorescent microscope equipped with a FITC filter (Zeiss No 10).
Caspase 3 activity was measured using the caspase-3 colorimetric kit (R&D Systems), according to the instructions of the manufacturer. Measurements were done in duplicate and the mean and SD were calculated.
As a positive control, exponentially growing HaCaT cells were treated with 5 µg/ml Etoposide (Millipore) in normal growth media for 1 h. Then cells were washed twice with fresh media and incubated in fresh media for 6 h, before apoptosis was assessed by the above-mentioned methods.
Comet-assay
Alkaline Comet-assay was performed according to Singh and Tice (30,31) and is described in detail elsewhere (32). Neutral Comet-assay was performed according to Olive et al. (33) and is described in detail in (32). Three times two slides with 60 comets each of individual preparations were scored per sample point.
Detection of OCDLs
OCDLs were measured essentially as described in (34) with some modifications: G1 arrested HaCaT cells were exposed to UVA at doses indicated in ‘Results’ section and post-incubated for various times. Instead of using PFG-plugs, cells were embedded directly on fully frosted slides in 0.8% low melting point agarose final concentration (Sigma Aldrich, Type VII). Cell lysis was done in plug lysis buffer (10 mM Tris Base, 100 mM EDTA, 2.5 M NaCl, pH 10, 1% Triton X-100, 10% DMSO) directly after embedding at 4°C for 2 h. Slides were washed three times in PBS for 15 min each at 4°C, then six times in FPG buffer (10 mM Tris–HCl, 10 mM MgCl2, pH 7) 30 min each at 4°C. Then slides were carefully drained of excess buffer and soaked in 60 µl FPG buffer, 3 U FPG (NEB) and 0.6 µl BSA. The optimal enzyme concentration was determined before in a dilution series. For FPG-negative slides, the enzyme was omitted. The micro-gels were covered with a plastic coverslip and incubated in a humidified chamber for 30 min at 4°C, then for 60 min at 37°C. The enzymatic reaction was stopped by immersing the slides in ice-cold 1 M MgCl2 solution overnight. Then slides were washed six times for 30 min each in 1× TBE buffer at 4°C before being electrophoresed at 1 V/cm for 25 min at 6 mA. All slides of one experimental series were electrophoresed simultaneously including the controls to prevent variations. After electrophoresis, slides were dehydrated in absolute ethanol and then stained with Sybr green (1:1000) in DABCO antifade (Sigma Aldrich). Image analysis was done using an Axiovert 200 (Zeiss), equipped with a FITC filter (ex: 482/18 nm; bs: 495 nm LP; em: 520/28 nm) and Komet 4, (Kinetic Imaging).
All buffers until the end of the enzyme treatment were substituted with 50 µM phenyl-t-butyl nitrone (Sigma Aldrich) to inhibit oxidation during handling and purged with nitrogen.
Western blotting
For γH2AX detection, nuclei were isolated using hypotonic swelling in ice-cold 1 mM HEPES pH 7.4 for 30 min followed by soft detergent treatment of 0.01% NP40 and soft shaking. Nuclei were harvested by centrifugation at 200 g for 5 min at 4°C. Equal amounts of protein extract were loaded on two gels for Comassie staining and western blotting. γH2AX was detected with clone JB 103 (upstate/Millipore) diluted 1:2000 and an anti-mouse-horseradish peroxidase conjugate (Santa Cruz Biotechnology), followed by ECL (Amersham) detection.
Immunohistochemistry
Immunohistochemistry was performed according to standard protocols and as described in (32). Anti-γH2AX antibody was used at 1:200 and visualized using anti mouse–Alexa488 (Invitrogen) at 1:400.
Preparation of stretched chromatin fibres
Five hundred cells were seeded at the end of a slide (seeding area: ∼1 × 15 mm) and were allowed to attach overnight. After irradiation, the cells were directly lysed on the slide for 2 min (Tris–HCl, 100 mM pH 7.5, SDS 0.5%). Receding meniscus stretching of chromatin fibres was done as described elsewhere (35,36) with 1 mm/s using a homemade linear device. After chromatin stretching, the fibres were fixed in methanol (−20°C) and air-dried. For detection of 8-oxo-guanine, chromatin was denatured in 0.5 M HCl for 5 min and neutralized in 400 mM cold Tris–HCl, pH 7.0. Antibody detection was performed overnight with anti-8-oxo-guanine (R&D Systems), diluted 1:100 in PBS, 5% BSA. After two washing steps, the secondary antibody was applied [anti-mouse-alkaline phosphatase (AP), Roche, 1:750 in PBS, 5% BSA]. Detection of abasic sides was performed using the aldehyde reactive probe (ARP) kit (Molecular Probes), as recommended by the manufacturer. Biotin of the ARP probe was detected with AP-conjugated avidin (Qbiogene). To visualize DNA damage sites, the HNPP signal amplification kit (Roche) was used according to the protocol of the supplier. Total DNA was counterstained with 10 µM YOYO-1, in 50% DABCO antifade in 10 mM Tris–HCl, pH 7.5.
Imaging and analysis
Fluorescence microscopy was performed using an epifluorescence microscope (Axioscope, Zeiss), equipped with a HBO50 and high-quality band pass filters (AHF Filters, Germany) for DAPI, FITC and Rhodamine (AHF Analysentechnik). Image acquisition was done with a cooled CCD camera (Quantix, Roper Scientific), controlled by the Quips software (Vysis). Each channel was imaged individually as a black and white image and merged by the software in false colours. For the Comet-assay a 25× 0.8 NA Plan-Neofluar, for the fibre analysis a 40× 1.3 NA Plan-Neofluar and for immunofluorescence a 63× 1.3 NA Plan-Neofluar objective were used.
Counting of foci was done by a macro implemented in the Optimas software package (Kappa, Germany) after automated thresholding. A minimum of three individual experiments per dose were analysed after randomization and the number of cells with foci as well as the average number of foci per cell were counted. Error bars are SDs from the pooled experiments.
Fibre images were analysed by ImageJ, where equal sized section of fibres were cropped, single signals were counted automatically by ImageJ, whereas clusters were counted manually and the number of signals were normalized to the total DNA intensity of the corresponding fibre.
RESULTS
γH2AX foci are formed in a dose-dependent manner after UV exposure
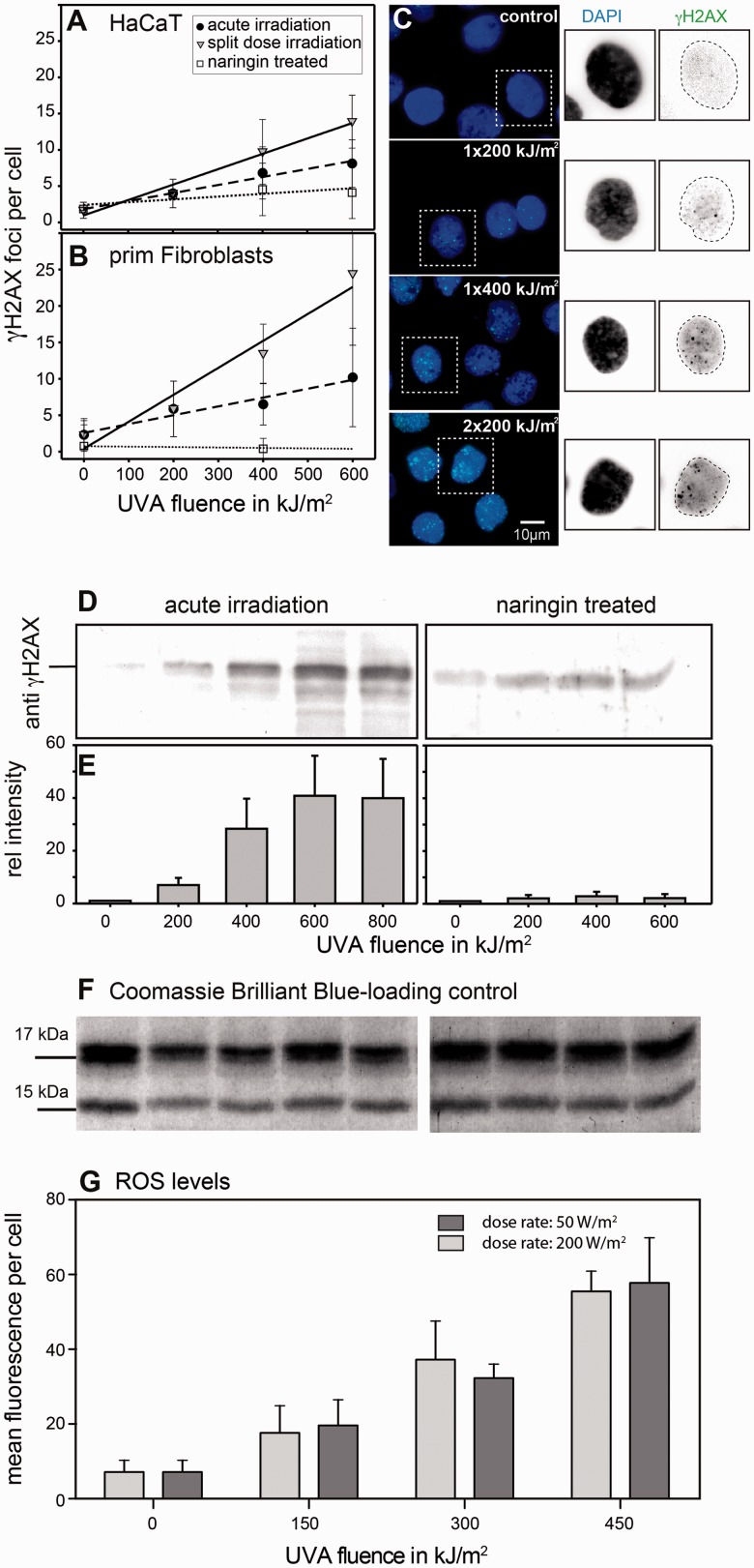
To study the dose-dependent formation of dsbs in the absence of DNA replication, we used G1 arrested cell cultures and immuno-histochemical detection of γH2AX foci. Figure 1A shows the dose–response curves for HaCaT cells after acute irradiation and Figure 1B for human primary skin fibroblasts, expressed as foci per cell versus UVA fluence (black circles, dashed lines). Apparently a linear dose dependence between the UVA fluence and the number of foci per cell was found. HaCaT cells were found to be less sensitive compared to the primary fibroblasts, probably due to the higher content of glutathione (see Supplementary Figure S2). Linear regression showed a relationship of 0.96 induced foci per 100 kJ/m2 for HaCaT cells and 1.25 induced foci per 100 kJ/m2 for fibroblasts. Representative micrographs of immunofluorescently detected γH2AX foci in HaCaT cells are shown in Figure 1C for continuous irradiation and split-dose exposure (2 × 200 kJ/m2). We confirmed that UVA doses used in this study and within the time frame used in our experiments do not induce apoptosis in HaCaT cells (Supplementary Figure S3). Therefore, an interference of dsb detection by apoptotic events can be excluded.
Figure 1.
Replication-independent formation of γH2AX foci in UVA-exposed HaCaT cells and primary human fibroblasts. (A and B) Dose–response curves for the formation of γH2AX foci in G1 arrested cells after continuous UVA exposure (filled circle). Split-dose exposure leads to an increased number of foci in both cell types (filled inverted triangle). Cells pre-treated with the antioxidant Naringin show a reduced number of foci (open square). Means and SD are given. (C) Typical sample micrographs of G1 arrested HaCaT cells (controls: top), acute irradiation (middle: 1 × 200 and 1 × 400 kJ/m2) as well as after split-dose irradiation (bottom: 2 × 400 kJ/m2). (D–F) H2AX phosphorylation was quantified in the nuclear extracts following acute irradiation. A dose-dependent increase in the amount of γH2AX can be found up to 600 kJ/m2. With higher doses, saturation in the amount of phosphorylated H2AX was found. In contrast, pre-incubation with the antioxidant Naringin reduces the amount of phosphorylated H2AX. Data represent the mean of two experiments and the SD. (G) Nuclear ROS levels at two different dose rates as measured by Flow cytometry in HaCaT cells using chloromethyl-2′,7′-dichlorodihydrofluorescein diacetate, acetyl ester. A dose-dependent, but dose-rate independent increase of the ROS levels was detected with UVA doses used in this study and environmental relevant doses and dose rates.
Split-dose irradiation increases the number of γH2AX foci
In the simplest scenario, the number of DNA lesions shows a linear dependence on dose. This would indicate induction of DNA damage by direct UVA–DNA interaction or a 1:1 effect. In such a case, it would be irrelevant if a certain dose is applied continuously or in split-dose fractions. The most straightforward result of a split-dose exposure would be a lower number of DNA breaks due to DNA repair in the recovery time.
We hypothesize that, if a limited number of endogenous cellular PS were indirectly responsible for DNA-dsb induction during UVA-irradiation, the number of DNA-dsb (and therefore the number of γH2AX foci) could also be limited at a certain UVA-fluence. This can be assumed because PS becomes degraded by continuous photo-excitation and therefore are inactive to produce additional ROS. If, however, UV irradiation is split into several dose fractions, separated by a certain time, PS could recover and thus the number of DNA-dsb might be higher compared to an exposure which delivers the same total dose without recovery periods. We therefore established a split-dose irradiation protocol:
and compared the number of dsbs with the number induced by the same un-fractionated total dose.
In Figure 1A and B, the dose-dependent increase of the mean number of foci per cell after split-dose irradiation (grey triangles) is shown. Dose dependencies in split-dose experiments were approximated to be linear (with correlation coefficients of 0.94 and 0.98, respectively) regression. Using this regression, the number of γH2AX foci/cell increased from 0.96 to 1.4 foci/100 kJm−2 for HaCaT cells and from 1.2 to 2.6 foci/100 kJm−2 for human fibroblasts when we compare acute to split-dose irradiation. A typical micrograph for the split dose exposure is shown in Figure 1C bottom (for HaCaT cells). The increase in foci number per cell becomes clearly visible in Figure 1C (1 × 400 kJ/m2), where the cells have been exposed to the same final dose but in a single fraction. Apparently, the split-dose irradiation increases the efficiency of DNA-dsb induction via mechanisms that are e.g. saturated after exposure to a single dose and which cannot be counteracted with comparable efficiency by DNA-dsb repair in a 2-h period of recovery (at 37°C) between dose fractions. We speculate that the observed increase in the number of DNA-dsb (γH2AX-foci) can be explained by recovery and/or replacement of photo-degraded endogenous PS, which could not take place if the same final dose is given in a single fraction.
In a previous study, we have also demonstrated that the number of foci increases after the exposure is ended in HaCaT cells, exposed to 600 kJ/m2 UVA (37). This suggests a processing step being evolved in the formation of UVA-induced dsbs, similarly to ionizing radiation (24).
UVA-induced γH2AX focus formation can be inhibited by radical scavengers
To elutriate whether the dsbs might be generated by ROS, we irradiated in the presence of the antioxidant Naringin, a bioflavonoid derivative of grapefruit peel and related citrus species that prevents oxidative damages and functions as a radical scavenger (29,38). It has been shown to act as a potent inhibitor of OH radical and superoxide anion production in extracellular reaction mixtures (29). Furthermore, Naringin has been reported to protect against radiation-induced DNA (39) and chromosome damage in mouse bone marrow of 60CO-γ-irradiated mice (28,29).
We tested the effect of a 24-h pre-treatment of the cells with Naringin (100 nM final concentration) before exposure on both HaCaT cell line and primary human fibroblasts. In Figure 1A and B, the effect of the antioxidant treatment is plotted in terms of γH2AX foci per cell as a function of UVA dose (open squares). Naringin pre-treatment significantly reduced the average number of foci per cell. This becomes visible at all tested doses and Naringin was found to be capable to reduce the number of foci per cell to the control level, even after 600 kJ/m2 in HaCaT cells. Naringin pre-treatment reduces the efficiency of focus formation from 0.96 to 0.28 foci per 100 kJ/m2 for the HaCaT cells and from 1.2 to 0.98 for the fibroblast, respectively (see Figure 1A and B). The fact that Naringin clearly reduces the number of γH2AX foci per cell supports the idea that ROS may be involved in the formation of UVA-induced dsbs.
UVA-induced γH2AX phosphorylation in vitro
To verify the immunohistochemical results for the γH2AX focus formation, we monitored H2AX phosphorylation in cell lysates. A clear dose–response was found on the level of phosphorylation levels (Figure 1D). Figure 1D and E shows that phosphorylation levels of γH2AX saturate at higher UVA-fluences (600–800 kJ/m2). The Naringin treatment reduced the fraction of phosphorylated histone H2AX nearly to the level of the control (maximum 2.4-fold increase), while in contrast, a 40-fold increase can be found with the acute irradiation after 600 kJ/m2. As loading control, a commassie brilliant blue gel with the histone bands is shown (Figure 1F).
ROS formation is independent of UVA dose rate
To demonstrate that dsbs can be formed under physiological conditions, we used different dose rates and measured the formation of ROS species in HaCat cells. Taking data from Elwood et al. (40), as well as measurements from the German solar UVA network (www.suvmonet.de) and satellite data-based values from Meteosat measurements (www.soda.is.com), solar UVA irradiance in the mid of Germany (∼53°N) reaches values of ∼50 W/m2 in the month of June at noon. This is about one-quarter of the irradiance, which we used in our UVA irradiation experiments (200 W/m2). To exclude that the higher irradiance used in our experiments introduces higher levels of ROS compared to irradiances found in the natural sun, we measured dose-dependent ROS production at different UVA irradiances of 50 and 200 W/m2 using ROS sensitive dyes and flow cytometry (see ‘Materials and Methods’ section). As can be seen from Figure 1G, both irradiances produced the same amount of ROS in a dose-dependent manner. We can therefore exclude that our experimental conditions introduce artificial high ROS concentrations, which might not be found under natural conditions.
UVA induces dsbs as measured by the neutral Comet-assay
To directly assay DNA fragmentation and validate the immunoassays based on the H2AX phosphorylation, we measured overall DNA fragmentation by the Comet-assay. This assay was used in its alkaline version to detect both DNA-dsbs and ssbs. Additionally, we used this approach to study whether the antioxidant Naringin is capable to prevent the formation of DNA single-strand breaks as well as the formation of dsbs.
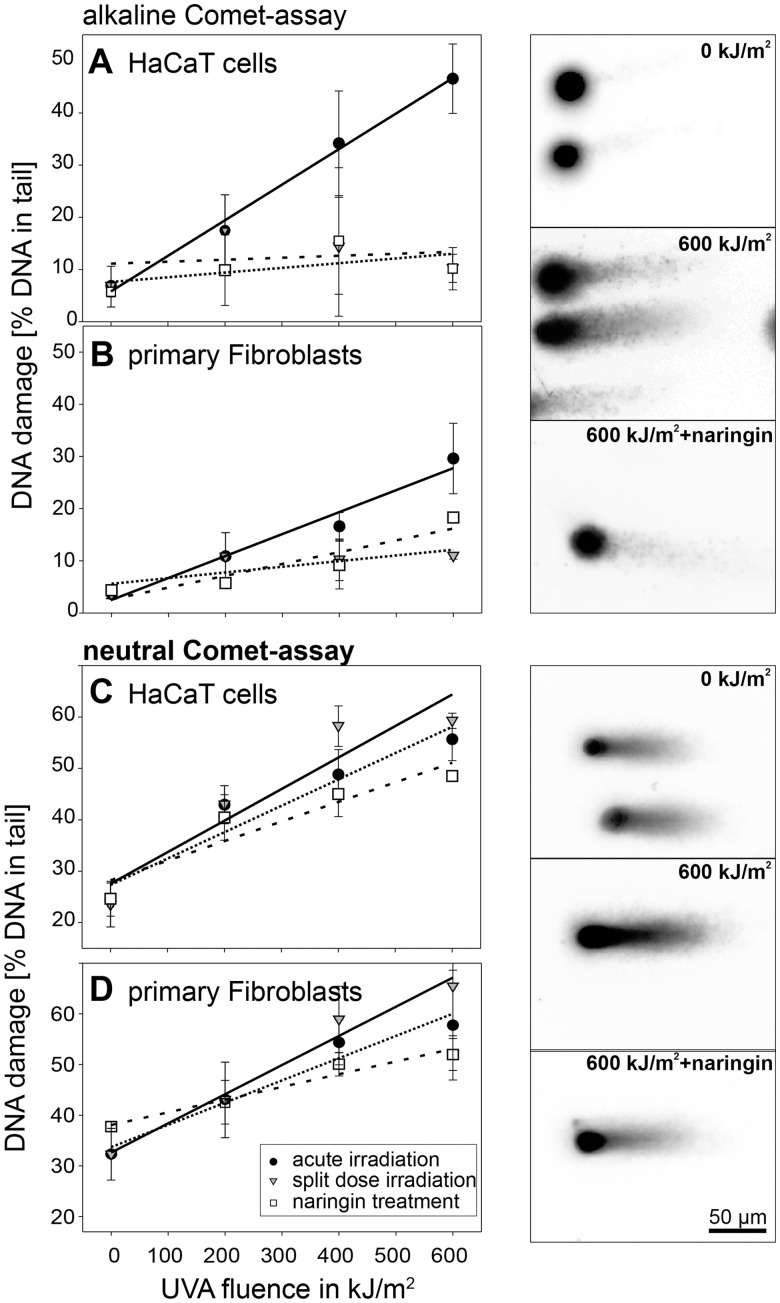
The UVA dose–response relation for DNA damage (after alkaline treatment, ssbs and dsbs) is plotted in Figure 2A and B for keratinocytes and fibroblasts. The induction of ssbs follows a linear dependency, which probably is the initial part of a saturation curve as mentioned earlier. HaCaT cells show an average of 6.4% damaged DNA per 100 kJ/m2, while in contrast, the fibroblast show only 3.7% damaged DNA. As can be seen from Figure 2A and B, the Comet-assay revealed a reduction of DNA damage following split dose in both cell lines (grey triangles). This is, at a first glance, contradictory to the immuno-histochemical data presented earlier, where a split-dose irradiation increases the number of γH2AX foci. However, the alkaline Comet-assay mainly detects ssbs, with repair half-times of minutes. Therefore, the majority of ssbs is repaired within the incubation between fractionated exposures, which we showed earlier (22). Taken together this results in a split-dose efficiency of DNA-damage induction which is not significantly different from that detected after a single exposure of 200 kJ/m2. For HaCaT cells, this results in a decrease of damage induction from 6.4% to 0.8% per 100 kJ/m2. For the fibroblast, the values were calculated to decrease from 3.7 to 1.5%, respectively.
Figure 2.
DNA fragmentation quantification by Comet-assay. (A and B) Total DNA damage (ssbs + dsbs) detected by the alkaline Comet-assay (•). Naringin-treated cells show lower levels of DNA fragmentation (open square). Split-dose irradiation (filled inverted triangle) leads to a DNA damage comparable to a single exposure of 200 kJ/m2, for details see text. (C and D) Neutral Comet-assay shows a dose-dependent induction of DNA breaks (•). The number of breaks can again be reduced by the antioxidant (open square). In contrast to the alkaline Comet-assay, the neutral version reveals an increase of the number of dsbs if the irradiation is performed in split dose (filled inverted triangle). All measurements are means of medians together with SD. Representative micrographs for comet specimens are shown for alkaline Comet-assay (top) and neutral Comet-assay (bottom) for the indicated treatments.
The effect of Naringin in preventing UVA-induced DNA damages is plotted in Figure 2A and B (open squares). Compared to the untreated cells, Naringin treatment significantly reduces the amount of damaged DNA. The slope of fitted straight lines decreased from 6.4% to 0.9% of damaged DNA per 100 kJ/m2 (HaCaT) and from 3.7% to 1.5% (fibroblasts). Nevertheless, the alkaline Comet-assay reveals that the ssbs are not completely prevented by the Naringin treatment, especially in the fibroblasts.
The results from the neutral version of the Comet-assay, which detects predominantly dsbs, are plotted in Figure 2C and D. Again a dose-dependent increase in DNA fragmentation can be found. A linear regression shows 5.1% DNA in tail per 100 kJ/m2 for HaCaT cells and 4.4% DNA in tail per 100 kJ/m2 for the fibroblasts. After split-dose irradiations, the level of DNA fragmentation increases to 6.1% DNA in tail/100 kJ/m2 (HaCaT) and 5.8% (fibroblasts). The effect of Naringin is shown in a reduction of the DNA fragmentation to 3.8% DNA in tail/100 kJ/m2 (HaCaT) and 2.5% for the fibroblasts.
UVA-induced dsbs arise from OCDLs
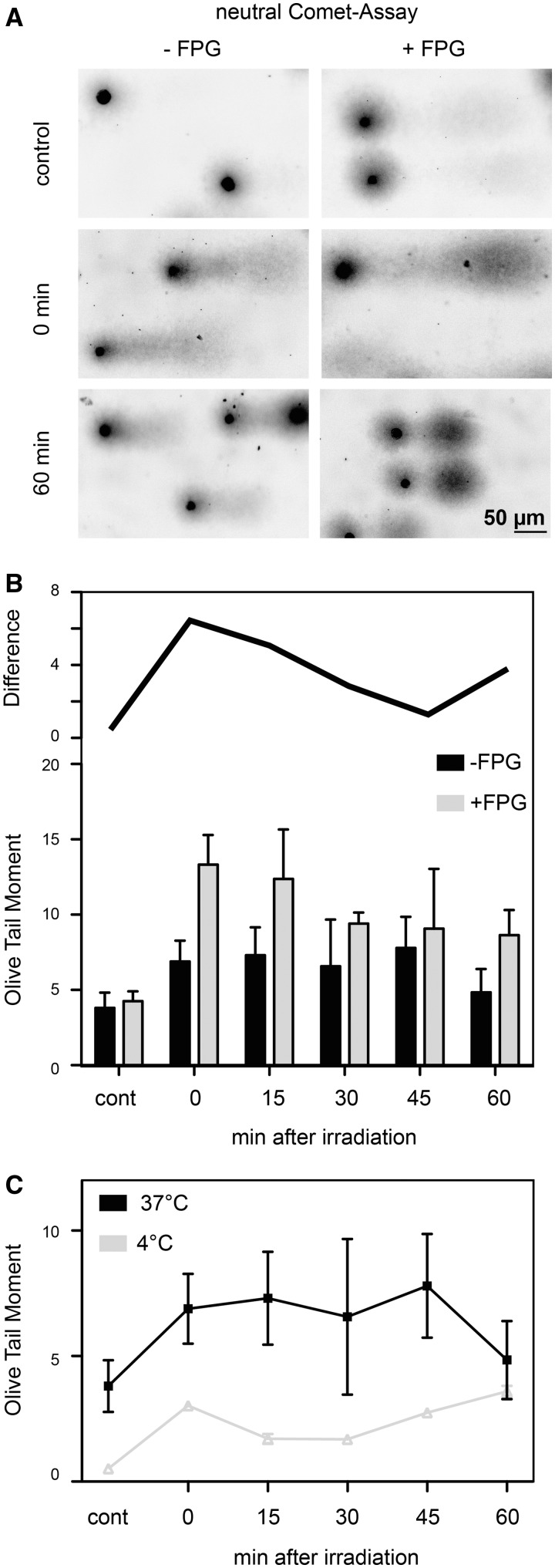
To test whether the UVA-induced dsbs are generated through clustered oxidatively induced DNA lesions, we performed a modified neutral Comet-assay, where FPG was used as an enzymatic probe to reveal unprocessed oxidative DNA base lesions and convert them into ssbs. If the strand breaks occur in close proximity they are converted to dsbs and can be detected by the neutral Comet-assay. Therefore, we exposed G1 arrested HaCaT cells to a single dose of 600 kJ/m2 UVA and measured the OCDLs and dsbs during a time course of repair, where cells were incubated at 37°C. We analysed these data using the Olive Tail Moment (OTM) as a measure for DNA fragmentation since most of the existing literature uses it. In control cells, a background level of dsbs and a very low level of OCDLs were detected. The very small extra fragments generated by the FPG treatment are visible as a small cloud extending from the comet in the direction of electrophoresis (Figure 3A, top row). This is also reflected in an increased OTM from 3.8 ± 1.0 to 4.3 ± 0.6 after FPG treatment, representing the endogenous FPG-sensitive sites. Directly after exposure to a single dose of 600 kJ/m2, the OTM of the untreated sample increases to 6.9 ± 1.4. A large amount of OCDLs can be observed in the FPG-treated sample represented by an OTM of 13.3 ± 2.0. During the repair time of 1 h, the amount of dsbs is nearly constant with only a slight increase between 0 and 45 min as depicted in Figure 3B. In contrast, the amount of remaining clustered FPG lesions is decreasing (grey bars). This is shown as the difference between FPG-treated samples to non-treated samples (top line in Figure 3B). This repair kinetics reflect previous findings on γH2AX foci after UVA exposure that showed an increase with a plateau 30 min after the end of irradiation and then a constant level up to 6 h post-irradiation (37). A possible reason for not seeing the initial increase here is that at this point we have a lag of ∼17 min after exposure for handling the cells until beginning of lysis, due to the fact the UVA exposure can not be done with already embedded cells. Longer post-incubation (up to 24 h) has shown that the level of DNA fragmentation after UVA exposure is reduced to control level (22).
Figure 3.
Oxidatively induced clustered DNA lesions (OCDLs) were measured with the neutral Comet-assay and FPG as an enzymatic probe. (A) Sample comets with (+FPG) and without (−FPG) enzymatic processing. An increase in DNA fragmentation can be seen for all FPG-treated comets as partly separated DNA species in the direction of electrophoresis. (B) Quantification of OCDL. Grey bars represent the FPG-treated cells, and the black bars represent the mock treated ones. Shown are the means of medians; error bars represent the SD. The black line above the bars shows the difference between FPG- and mock-treated comets for each time point. (C) Repair kinetics for cells exposed to UVA irradiation and repair incubated at 4°C (filled inverted triangle) and 37°C (filled square). The overall level of DNA fragmentation is higher in cells kept at 37°C.
We interpret this result as an on-going incision of oxidative base damage by cellular repair systems that are converted into dsbs due to close special proximity. Due to the fact that we are detecting γH2AX foci in G1 arrested cells, we can exclude replication-dependent dsb induction.
To test whether the observed induction of dsbs is due to repair-mediated incisions, we performed the neutral Comet-assay after the cells were irradiated on ice and post-incubated at 4°C to reduce the repair capacity. Figure 3C shows the comparison of cold irradiation/post-incubation to the same treatment at 37°C. The overall level of dsbs is higher in cells irradiated and incubated at 37°C. Nevertheless the cells irradiated at 4°C already showed a significant increase of DNA fragmentation directly after exposure. We suggest that this fragmentation is due to the induction of closely spaced ssbs, also supported by the reduction of the OTM 15-min post-irradiation and the assumption that plain ssbs are quickly repaired (41). The subsequent increase in DNA fragmentation from 30 to 60 min possibly reflects on-going slow incision of OCDLs.
UVA induces clustered DNA damage, visualized on stretched chromatin fibres
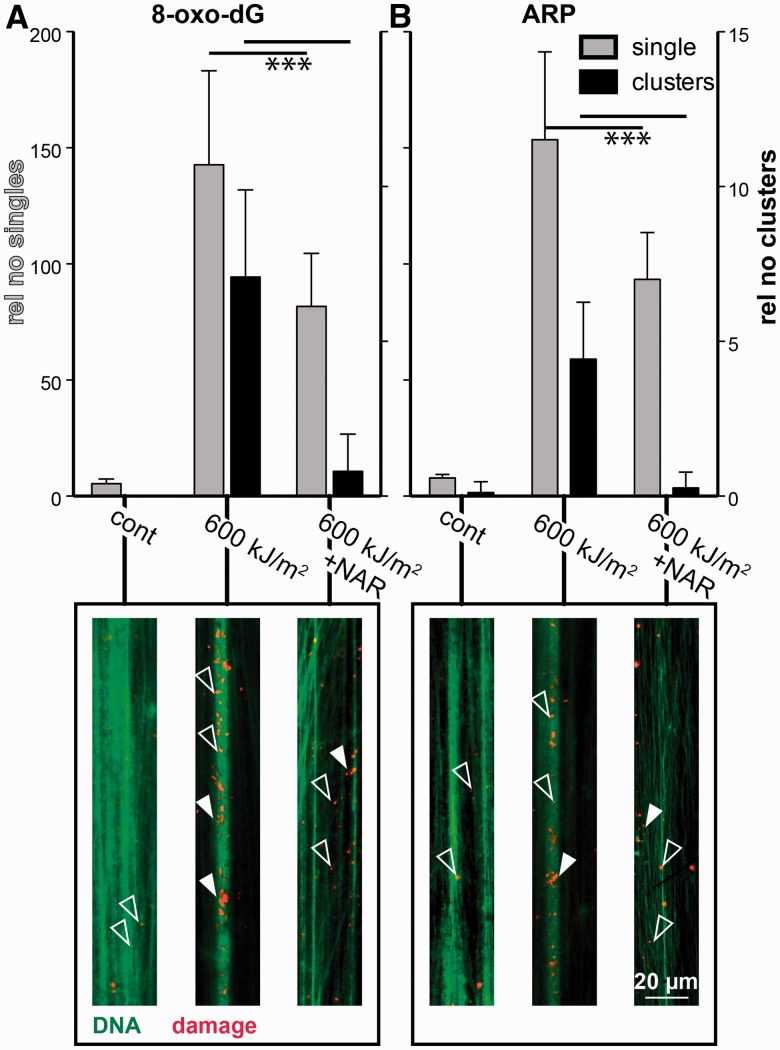
To examine further, whether UVA-induced dsbs are originating from clustered DNA lesions, we visualized DNA damages on stretched chromatin fibres. 8-oxo-Guanine was detected using a monoclonal antibody, and abasic sites were detected using the ARP (Molecular Probes), a chemical compound that selectively couples biotin to an abasic site, followed by biotin detection.
Figure 4A shows the detection of 8-oxo-dG, whereas Figure 4B shows the detected abasic sites. Grey bars represent the relative number of individual signals, whereas the black columns show the number of clusters. In un-irradiated controls only a small amount of individual sites can be seen for both types of DNA lesions (open arrow heads). Quantification revealed a relative frequency of 5.5 ± 1.8 (8-oxo-dG) and 7.9 ± 1.6 (ARP) single signals. Nearly no clusters were detected (black columns, filled arrowheads). After exposure to a single 600 kJ/m−2 UVA dose 143 ± 40 (8-oxo-dG) and 153 ± 38 (ARP), signals were measured. Also, the number of clustered damage is elevated to 7.0 ± 2.8 (8-oxodG) and 4.4 ± 1.8 (ARP). When cells were pre-treated with Naringin and UV exposed, both the number of single damage sites as well as the number of clusters were reduced significantly (single sites 8-oxo-dG: 82 ± 23, ARP: 93 ± 20; clusters: 8-oxo-dG: 0.8 ± 1.2, ARP: 0.3 ± 0.5). Both DNA lesions show clusters after UV exposure, this means that the possibility of a conversion into dsbs is elevated compared to a complete random induction of ssbs or abasic sites. This finding further supports the theory that UVA-induced clustered DNA damage can be regarded as the source of dsb formation. These data correlate well to the above-described results on UVA-induced clustered oxidatively induced bi-stranded DNA lesions and the subsequent processing by cellular DNA repair machinery into dsbs.
Figure 4.
DNA damages visualized on stretched chromatin fibres. Quantification of single and clustered signals of 8-oxo-dG (A) and ARP (B), respectively. Grey bars represent the number of single signals (left y-axis,) whereas black bars represent clusters (right axis). At least 10 fibres were analysed per data point and SDs are calculated. A detail of representative fibres is shown below the corresponding columns, open arrowheads highlight singles, filled arrowheads clusters. In green the total DNA is stained with YOYO-I. DNA lesions are shown in red. The reduction in both single damage sites as well as clusters after Naringin treatment is significant on the P < 0.05 level as calculated by unpaired t-test with Welch’s correction.
DISCUSSION
Today, UVA is an accepted carcinogen (3). Nevertheless, the types of DNA damage induced by UVA are still not completely understood (9,14). The classical point of view was that UVA induces predominantly oxidative damages and among those the majority accounts for 8-oxo-guanine (42). More recently, it was demonstrated that UVA is also able to induce thymidine dimers, especially TT-CPDs, although at a 1000-fold lower efficiency compared to UVC (43,44). Even if CPDs are added to the UVA damage profile, there is still a gap to fully understand the mutagenic potential of UVA radiation, at least in hamster cells (45). In addition, UVA was reported to induce dsbs (18,19,22,23,46). Dsbs could be a third component in the damage profile of UVA and account a major damage class explaining UVA-induced mutagenicity.
The energy of a single UVA photon is too low to induce a covalent bound break or change. So all types of DNA damage, such as oxidative base damage, oxidative backbone damage, CPDs or dsbs, induced by UVA are strictly dependent on—so far—unknown cellular sensitizers (6,9,47,48). Several chemical structures have been suggested as potential sensitizers, e.g. cytochromes, flavins or NAD(P)H (9). Recent overviews of potential PS have been given in (49,50). Since all damage induced by UVA is dependent on radicals formed by the cellular sensitizers, we need to consider a second fact: radicals are highly reactive, short lived and have very limited diffusion ranges [ranging from 2 nm (hydroxyl radical) to 100 nm (singlet oxygen) for different radical species], at least if we consider radical oxygen species to be the main source of radicals involved in UVA-dependent DNA damage (51,52). Taken together, photo-induced damage by UVA has to be localized in a very restricted volume around the cellular sensitizers.
A proposed model of dsb-induction is, therefore, based on clustered ssbs in close proximity that are converted to a dsb when they occur simultaneously and within 1.5 helix turns (53). It is known that clustered ssbs [also arising during DNA repair of OCDLs (26,54)] are treated as dsbs by the cell (22,24). Additionally, it was shown that clustered damage is especially mutagenic and cytotoxic and has a reduced repair kinetic (25,26). This directly coincides with the results presented here and in previous studies, demonstrating that UVA-induced dsbs are generated with a temporal lag and have at least 6-h persistence (37). In a recent article, Cadet and Douki (10) argued that the frequency of these events is too low, since the frequency of 8-oxo-dGs induced by UVA and the ratio of 8-oxo-dGs to ssbs would not allow a clustered occurrence of ssbs to be converted to dsbs. This is true if one assumes a random distribution of ROS generated by randomly located sensitizers. Due to the short diffusion range of the ROS in the vicinity of cellular sensitizers, we have to assume that these are chromatin bound in one way or the other (55). This would lead to the conclusion that the damage is more clustered and less random.
From our results, we can conclude several new facts and confirm several steps of the clustered damage model: (i) We confirm that ROS are intermediates of the DNA damaging process, especially for the dsb formation, since the presence of an anti-oxidant (Naringin) does prevent the formation of dsbs and ssbs. ROS also cause ssbs, but obviously this damage is repaired too fast to be detected in the split-dose experiments. This is reflected by the fact that the alkaline Comet-assay, which detects ssbs and alkali labile sites, does not show an increase in damage levels after split-dose irradiation. Thus, implying a repair mechanism faster than the split-dose recovery time of 2 h which is well within the accepted time frame of BER repair (41,56). (ii) We could demonstrate that a split-dose irradiation scheme enhances the number of dsbs, suggesting that a sensitizer can also be depleted (most likely photooxidized) by photon absorption. If this happens, sensitizers can no longer function in the generation of ROS. However, given some time (as in split dose experiments) these sensitizers can be exchanged and produce ROS and dsbs again. The number of DNA dsbs not fully repaired after the first dose fraction plus those being produced with the second-dose fraction is then apparently higher than the one produced by a single (high) dose which might be able to exhaust the relevant PS pool. These findings are in agreement with an investigation by Hoffmann-Doerr et al. (57) who explained their results of split-dose experiments of FPG-sensitive sites after UVA/visible light exposure by a photosensitizier exhaustion mechanism. It should be noted also that, in our investigation, the split-dose effect is stronger when detected by the γH2AX foci compared to the detection on the level of DNA fragmentation (neutral Comet-assay). This suggests a prolonged existence of the γH2AX foci exceeding the DNA re-ligation event. (iii) We were able to demonstrate that UVA induces a large quantity of clustered oxidative DNA lesions as detected with FPG as an enzymatic probe together with the neutral Comet-assay. This does not cover all possible oxidative damage, only FPG sensitive sites [7, 8-dihydro-8-oxoguanine (8-oxoguanine), 8-oxoadenine, fapy-guanine, methy-fapy-guanine, fapy-adenine, 5-hydroxy-cytosine and 5-hydroxy-uracil] and ssbs, but these are the most common DNA lesions induced by UVA. Additionally, we could demonstrate that the dsb formation is dependent on the temperature. Reduced temperature during irradiation and post-incubation (4°C) leads to a significant decrease of detected DNA fragments in the neutral Comet-assay. (iv) When we determine DNA damage on stretched chromatin fibres, we see clusters of damage at distinct points. This fits the model of spatially fixed sensitizers at specific sites in the chromatin. Importantly it is seen for different UVA-induced DNA lesions, such as 8-oxo-dG and abasic sites. So it is a direct hint to a locally higher concentration of ROS, which would most likely be able to cause clustered damage. These findings are also supported by work of Ito et al. (58), who showed that, e.g. 8-oxo-dG formation on double-stranded (naked) DNA proceeds through the direct interaction of (UVA-) photosensitized riboflavin with DNA. This reaction pathway of DNA-dsb induction might be similar or even identical to that described after the application of ionizing radiation, which produces a comparable pattern of ROS (59).
Taken together, we conclude from our results that there is indeed a replication-independent induction of dsbs by UVA exposure, as reported earlier by several other studies (18,22,23,37). Our results seem to be in disagreement with the recent report by Rizzo et al. who did not find activation of the homologous recombination (HR) dsb repair pathway in UVA-irradiated human cells. In their study, only >15 γH2AX foci/cell were considered as dsb induction (17). However, this might indicate problems in sensitivity of their γH2AX-assay and a biased focus on HR as the only dsb repair pathway. It is known that dsbs are predominantly repaired by the non-homologous end joining systems, especially if the cells are in G1 phase of the cell cycle and breaks are directly ligatable (60), which is the predominant case for dsbs generated by clustered ssbs.
The relevance of these findings is highlighted by the fact that UVA doses used in this investigation (up to 600 kJ/m2) can easily be accumulated under natural conditions from solar ambient UVA radiation. Based on a model by Green and colleagues (61,62), which has been extensively validated by comparison with measured spectral irradiance at ground level (40), a UVA dose of 600 kJ/m2 will be accumulated at latitude 50–55°N, at clear skies in the month of June between 11:30 am and 3:30 pm (40).
The fact that we could demonstrate our findings in both primary human fibroblasts as well as in the keratinocyte cell line HaCaT indicates that we are describing a common mechanism for the induction of dsbs by UVA and not just a property of a given cell line. Dsbs are known to be precursor lesions of chromosome aberrations. We could recently show that UVA induces chromosomal aberration in human keratinocytes and that these cells give rise to squamous cell carcinoma after transplantation into nude mice (37). As UVA represents the major component of solar UV radiation and artificial UV used in sunbeds, the results of our study might have important implications in skin cancer aetiology and risk assessment.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1–3.
FUNDING
Bundesministerium für Forschung und Technik BMBF [1N7506, 02S8355]; Marie Curie FF6 [MC-IEF 023821 to A.R.]; Deutsche Forschungs Gemeinschaft DFG, GRK 1657/1 TP1C. The Fritz Lipmann Institute (FLI) is a member of the Science Association “Gottfried Wilhelm Leibniz” (WGL) and is financially supported by the Federal Government of Germany and the State of Thuringia. Funding for open access charge: Leibniz Institute for Age Research - Fritz Lipmann Institute.
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Matsumura Y, Ananthaswamy HN. Molecular mechanisms of photocarcinogenesis. Front. Biosci. 2002;7:d765–d783. doi: 10.2741/matsumur. [DOI] [PubMed] [Google Scholar]
- 2.Dahle J, Kvam E. Induction of delayed mutations and chromosomal instability in fibroblasts after UVA-, UVB-, and X-radiation. Cancer Res. 2003;63:1464–1469. [PubMed] [Google Scholar]
- 3.El Ghissassi F, Baan R, Straif K, Grosse Y, Secretan B, Bouvard V, Benbrahim-Tallaa L, Guha N, Freeman C, Galichet L, et al. A review of human carcinogens?Part D: radiation. Lancet Oncol. 2009;10:751–752. doi: 10.1016/s1470-2045(09)70213-x. [DOI] [PubMed] [Google Scholar]
- 4.Pfeifer GP. Formation and processing of UV photoproducts: effects of DNA sequence and chromatin environment. Photochem. Photobiol. 1997;65:270–283. doi: 10.1111/j.1751-1097.1997.tb08560.x. [DOI] [PubMed] [Google Scholar]
- 5.Mitchell DL, Nairn RS. The biology of the (6-4) photoproduct. Photochem. Photobiol. 1989;49:805–819. doi: 10.1111/j.1751-1097.1989.tb05578.x. [DOI] [PubMed] [Google Scholar]
- 6.Ikehata H, Ono T. The mechanisms of UV mutagenesis. J. Radiat. Res. 2011;52:115–125. doi: 10.1269/jrr.10175. [DOI] [PubMed] [Google Scholar]
- 7.Kim MY, Park HJ, Baek SC, Byun DG, Houh D. Mutations of the p53 and PTCH gene in basal cell carcinomas: UV mutation signature and strand bias. J. Dermatol. Sci. 2002;29:1–9. doi: 10.1016/s0923-1811(01)00170-0. [DOI] [PubMed] [Google Scholar]
- 8.Sage E, Lamolet B, Brulay E, Moustacchi E, Chteauneuf A, Drobetsky EA. Mutagenic specificity of solar UV light in nucleotide excision repair-deficient rodent cells. Proc. Natl Acad. Sci. USA. 1996;93:176–180. doi: 10.1073/pnas.93.1.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cadet J, Douki T, Ravanat JL, Di Mascio P. Sensitized formation of oxidatively generated damage to cellular DNA by UVA radiation. Photochem. Photobiol. Sci. 2009;8:903–911. doi: 10.1039/b905343n. [DOI] [PubMed] [Google Scholar]
- 10.Cadet J, Douki T. Oxidatively generated damage to DNA by UVA radiation in cells and human skin. J. Invest. Dermatol. 2011;131:1005–1007. doi: 10.1038/jid.2011.51. [DOI] [PubMed] [Google Scholar]
- 11.Cadet J, Berger M, Douki T, Ravanat JL. Oxidative damage to DNA: formation, measurement, and biological significance. Rev. Physiol. Biochem. Pharmacol. 1997;131:1–87. doi: 10.1007/3-540-61992-5_5. [DOI] [PubMed] [Google Scholar]
- 12.Cadet J, Douki T, Gasparutto D, Ravanat JL. Oxidative damage to DNA: formation, measurement and biochemical features. Mutat. Res. 2003;531:5–23. doi: 10.1016/j.mrfmmm.2003.09.001. [DOI] [PubMed] [Google Scholar]
- 13.Banyasz A, Vaya I, Changenet-Barret P, Gustavsson T, Douki T, Markovitsi D. Base pairing enhances fluorescence and favors cyclobutane dimer formation induced upon absorption of UVA radiation by DNA. J. Am. Chem. Soc. 2011;133:5163–5165. doi: 10.1021/ja110879m. [DOI] [PubMed] [Google Scholar]
- 14.Song JM, Milligan JR, Sutherland BM. Bistranded oxidized purine damage clusters: induced in DNA by long-wavelength ultraviolet (290–400 nm) radiation? Biochemistry. 2002;41:8683–8688. doi: 10.1021/bi020137x. [DOI] [PubMed] [Google Scholar]
- 15.Limoli CL, Giedzinski E, Bonner WM, Cleaver JE. UV-induced replication arrest in the xeroderma pigmentosum variant leads to DNA double-strand breaks, gamma -H2AX formation, and Mre11 relocalization. Proc. Natl Acad. Sci. USA. 2002;99:233–238. doi: 10.1073/pnas.231611798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Limoli CL, Giedzinski E, Morgan WF, Cleaver JE. Polymerase eta deficiency in the xeroderma pigmentosum variant uncovers an overlap between the S phase checkpoint and double-strand break repair. Proc. Natl Acad. Sci. USA. 2000;97:7939–7946. doi: 10.1073/pnas.130182897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rizzo JL, Dunn J, Rees A, Runger TM. No formation of DNA double-strand breaks and no activation of recombination repair with UVA. J. Invest. Dermatol. 2011;131:1139–1148. doi: 10.1038/jid.2010.365. [DOI] [PubMed] [Google Scholar]
- 18.Phillipson RP, Tobi SE, Morris JA, McMillan TJ. UV-A induces persistent genomic instability in human keratinocytes through an oxidative stress mechanism. Free Radic. Biol. Med. 2002;32:474–480. doi: 10.1016/s0891-5849(01)00829-2. [DOI] [PubMed] [Google Scholar]
- 19.Shorrocks J, Paul ND, McMillan TJ. The dose rate of UVA treatment influences the cellular response of HaCaT keratinocytes. J. Invest. Dermatol. 2008;128:685–693. doi: 10.1038/sj.jid.5701037. [DOI] [PubMed] [Google Scholar]
- 20.He YY, Pi J, Huang JL, Diwan BA, Waalkes MP, Chignell CF. Chronic UVA irradiation of human HaCaT keratinocytes induces malignant transformation associated with acquired apoptotic resistance. Oncogene. 2006;25:3680–3688. doi: 10.1038/sj.onc.1209384. [DOI] [PubMed] [Google Scholar]
- 21.Lu C, Zhu F, Cho YY, Tang F, Zykova T, Ma WY, Bode AM, Dong Z. Cell apoptosis: requirement of H2AX in DNA ladder formation, but not for the activation of caspase-3. Mol. Cell. 2006;23:121–132. doi: 10.1016/j.molcel.2006.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rapp A, Greulich KO. After double-strand break induction by UV-A, homologous recombination and nonhomologous end joining cooperate at the same DSB if both systems are available. J. Cell Sci. 2004;117:4935–4945. doi: 10.1242/jcs.01355. [DOI] [PubMed] [Google Scholar]
- 23.Fell LJ, Paul ND, McMillan TJ. Role for non-homologous end-joining in the repair of UVA-induced DNA damage. Int. J. Radiat. Biol. 2002;78:1023–1027. doi: 10.1080/0955300021000016558. [DOI] [PubMed] [Google Scholar]
- 24.Gulston M, de Lara C, Jenner T, Davis E, O’Neill P. Processing of clustered DNA damage generates additional double-strand breaks in mammalian cells post-irradiation. Nucleic Acids Res. 2004;32:1602–1609. doi: 10.1093/nar/gkh306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eccles LJ, O’Neill P, Lomax ME. Delayed repair of radiation induced clustered DNA damage: friend or foe? Mutat Res. 2011;711:134–141. doi: 10.1016/j.mrfmmm.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Holt SM, Scemama JL, Panayiotidis MI, Georgakilas AG. Compromised repair of clustered DNA damage in the human acute lymphoblastic leukemia MSH2-deficient NALM-6 cells. Mutat. Res. 2009;674:123–130. doi: 10.1016/j.mrgentox.2008.09.014. [DOI] [PubMed] [Google Scholar]
- 27.Boukamp P, Petrussevska RT, Breitkreutz D, Hornung J, Markham A, Fusenig NE. Normal keratinization in a spontaneously immortalized aneuploid human keratinocyte cell line. J. Cell. Biol. 1988;106:761–771. doi: 10.1083/jcb.106.3.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jagetia GC, Reddy TK. The grapefruit flavanone naringin protects against the radiation-induced genomic instability in the mice bone marrow: a micronucleus study. Mutat. Res. 2002;519:37–48. doi: 10.1016/s1383-5718(02)00111-0. [DOI] [PubMed] [Google Scholar]
- 29.Jagetia GC, Venkatesha VA, Reddy TK. Naringin, a citrus flavonone, protects against radiation-induced chromosome damage in mouse bone marrow. Mutagenesis. 2003;18:337–343. doi: 10.1093/mutage/geg001. [DOI] [PubMed] [Google Scholar]
- 30.Singh NP, McCoy MT, Tice RR, Schneider EL. A simple technique for quantitation of low levels of DNA damage in individual cells. Exp. Cell Res. 1988;175:184–191. doi: 10.1016/0014-4827(88)90265-0. [DOI] [PubMed] [Google Scholar]
- 31.Tice RR, Agurell E, Anderson D, Burlinson B, Hartmann A, Kobayashi H, Miyamae Y, Rojas E, Ryu JC, Sasaki YF. Single cell gel/comet assay: guidelines for in vitro and in vivo genetic toxicology testing. Environ. Mol. Mutagen. 2000;35:206–221. doi: 10.1002/(sici)1098-2280(2000)35:3<206::aid-em8>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 32.Rapp A, Bock C, Dittmar H, Greulich KO. UV-A breakage sensitivity of human chromosomes as measured by COMET-FISH depends on gene density and not on the chromosome size. J. Photochem. Photobiol. B. 2000;56:109–117. doi: 10.1016/s1011-1344(00)00052-x. [DOI] [PubMed] [Google Scholar]
- 33.Olive PL, Frazer G, Banath JP. Radiation-induced apoptosis measured in TK6 human B lymphoblast cells using the comet assay. Radiat. Res. 1993;136:130–136. [PubMed] [Google Scholar]
- 34.Georgakilas AG, Holt SM, Hair JM, Loftin CW. Measurement of oxidatively-induced clustered DNA lesions using a novel adaptation of single cell gel electrophoresis (comet assay) Curr. Protoc. Cell Biol. 2010 doi: 10.1002/0471143030.cb0611s49. Chapter 6, Unit 6 11. [DOI] [PubMed] [Google Scholar]
- 35.Bensimon D, Simon AJ, Croquette VV, Bensimon A. Stretching DNA with a receding meniscus: experiments and models. Phys. Rev. Lett. 1995;74:4754–4757. doi: 10.1103/PhysRevLett.74.4754. [DOI] [PubMed] [Google Scholar]
- 36.Michalet X, Ekong R, Fougerousse F, Rousseaux S, Schurra C, Hornigold N, van Slegtenhorst M, Wolfe J, Povey S, Beckmann JS, et al. Dynamic molecular combing: stretching the whole human genome for high-resolution studies. Science. 1997;277:1518–1523. doi: 10.1126/science.277.5331.1518. [DOI] [PubMed] [Google Scholar]
- 37.Wischermann K, Popp S, Moshir S, Scharfetter-Kochanek K, Wlaschek M, de Gruijl F, Hartschuh W, Greinert R, Volkmer B, Faust A, et al. UVA radiation causes DNA strand breaks, chromosomal aberrations and tumorigenic transformation in HaCaT skin keratinocytes. Oncogene. 2008;27:4269–4280. doi: 10.1038/onc.2008.70. [DOI] [PubMed] [Google Scholar]
- 38.Jagetia GC, Reddy TK, Venkatesha VA, Kedlaya R. Influence of naringin on ferric iron induced oxidative damage in vitro. Clin. Chim. Acta. 2004;347:189–197. doi: 10.1016/j.cccn.2004.04.022. [DOI] [PubMed] [Google Scholar]
- 39.Russo A, Acquaviva R, Campisi A, Sorrenti V, Di Giacomo C, Virgata G, Barcellona ML, Vanella A. Bioflavonoids as antiradicals, antioxidants and DNA cleavage protectors. Cell Biol. Toxicol. 2000;16:91–98. doi: 10.1023/a:1007685909018. [DOI] [PubMed] [Google Scholar]
- 40.Elwood JM, Diffey BL. A consideration of ambient solar ultraviolet radiation in the interpretation of studies of the aetiology of melanoma. Melanoma Res. 1993;3:113–122. [PubMed] [Google Scholar]
- 41.Lan L, Nakajima S, Oohata Y, Takao M, Okano S, Masutani M, Wilson SH, Yasui A. In situ analysis of repair processes for oxidative DNA damage in mammalian cells. Proc. Natl Acad. Sci. USA. 2004;101:13738–13743. doi: 10.1073/pnas.0406048101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kvam E, Tyrrell RM. Induction of oxidative DNA base damage in human skin cells by UV and near visible radiation. Carcinogenesis. 1997;18:2379–2384. doi: 10.1093/carcin/18.12.2379. [DOI] [PubMed] [Google Scholar]
- 43.Mouret S, Baudouin C, Charveron M, Favier A, Cadet J, Douki T. Cyclobutane pyrimidine dimers are predominant DNA lesions in whole human skin exposed to UVA radiation. Proc. Natl Acad. Sci. USA. 2006;103:13765–13770. doi: 10.1073/pnas.0604213103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jiang Y, Rabbi M, Kim M, Ke C, Lee W, Clark RL, Mieczkowski PA, Marszalek PE. UVA generates pyrimidine dimers in DNA directly. Biophys. J. 2009;96:1151–1158. doi: 10.1016/j.bpj.2008.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Biverstal A, Johansson F, Jenssen D, Erixon K. Cyclobutane pyrimidine dimers do not fully explain the mutagenicity induced by UVA in Chinese hamster cells. Mutat. Res. 2008;648:32–39. doi: 10.1016/j.mrfmmm.2008.09.011. [DOI] [PubMed] [Google Scholar]
- 46.Slieman TA, Nicholson WL. Artificial and solar UV radiation induces strand breaks and cyclobutane pyrimidine dimers in Bacillus subtilis spore DNA. Appl. Environ. Microbiol. 2000;66:199–205. doi: 10.1128/aem.66.1.199-205.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cadet J, Anselmino C, Douki T, Voituriez L. Photochemistry of nucleic acids in cells. J. Photochem. Photobiol. B. 1992;15:277–298. doi: 10.1016/1011-1344(92)85135-h. [DOI] [PubMed] [Google Scholar]
- 48.Kielbassa C, Roza L, Epe B. Wavelength dependence of oxidative DNA damage induced by UV and visible light. Carcinogenesis. 1997;18:811–816. doi: 10.1093/carcin/18.4.811. [DOI] [PubMed] [Google Scholar]
- 49.Wondrak GT, Jacobson MK, Jacobson EL. Endogenous UVA-photosensitizers: mediators of skin photodamage and novel targets for skin photoprotection. Photochem. Photobiol. Sci. 2006;5:215–237. doi: 10.1039/b504573h. [DOI] [PubMed] [Google Scholar]
- 50.Baumler W, Regensburger J, Knak A, Felgentrager A, Maisch T. UVA and endogenous photosensitizers–the detection of singlet oxygen by its luminescence. Photochem. Photobiol. Sci. 2012;11:107–117. doi: 10.1039/c1pp05142c. [DOI] [PubMed] [Google Scholar]
- 51.Redmond RW, Kochevar IE. Spatially resolved cellular responses to singlet oxygen. Photochem. Photobiol. 2006;82:1178–1186. doi: 10.1562/2006-04-14-IR-874. [DOI] [PubMed] [Google Scholar]
- 52.Hoshi T, Heinemann S. Regulation of cell function by methionine oxidation and reduction. J. Physiol. 2001;531:1–11. doi: 10.1111/j.1469-7793.2001.0001j.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.O’Neill P, Wardman P. Radiation chemistry comes before radiation biology. Int. J. Radiat. Biol. 2009;85:9–25. doi: 10.1080/09553000802640401. [DOI] [PubMed] [Google Scholar]
- 54.Sutherland BM, Georgakilas AG, Bennett PV, Laval J, Sutherland JC. Quantifying clustered DNA damage induction and repair by gel electrophoresis, electronic imaging and number average length analysis. Mutat. Res. 2003;531:93–107. doi: 10.1016/j.mrfmmm.2003.08.005. [DOI] [PubMed] [Google Scholar]
- 55.Roots R, Okada S. Estimation of life times and diffusion distances of radicals involved in x-ray-induced DNA strand breaks of killing of mammalian cells. Radiat. Res. 1975;64:306–320. [PubMed] [Google Scholar]
- 56.Sokhansanj BA, Wilson DM., 3rd Estimating the effect of human base excision repair protein variants on the repair of oxidative DNA base damage. Cancer Epidemiol. Biomarkers Prev. 2006;15:1000–1008. doi: 10.1158/1055-9965.EPI-05-0817. [DOI] [PubMed] [Google Scholar]
- 57.Hoffmann-Dorr S, Greinert R, Volkmer B, Epe B. Visible light (>395 nm) causes micronuclei formation in mammalian cells without generation of cyclobutane pyrimidine dimers. Mutat. Res. 2005;572:142–149. doi: 10.1016/j.mrfmmm.2005.01.011. [DOI] [PubMed] [Google Scholar]
- 58.Ito K, Inoue S, Yamamoto K, Kawanishi S. 8-Hydroxydeoxyguanosine formation at the 5’ site of 5’-GG-3’ sequences in double-stranded DNA by UV radiation with riboflavin. J. Biol. Chem. 1993;268:13221–13227. [PubMed] [Google Scholar]
- 59.Nikjoo H, O’Neill P, Goodhead DT, Terrissol M. Computational modelling of low-energy electron-induced DNA damage by early physical and chemical events. Int. J. Radiat. Biol. 1997;71:467–483. doi: 10.1080/095530097143798. [DOI] [PubMed] [Google Scholar]
- 60.Mao Z, Bozzella M, Seluanov A, Gorbunova V. Comparison of nonhomologous end joining and homologous recombination in human cells. DNA Repair. 2008;7:1765–1771. doi: 10.1016/j.dnarep.2008.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Green AES, Shettle EP. The middle ultraviolet reaching the ground. Photochem. Photobiol. 1974;19:251–259. [Google Scholar]
- 62.Schippnick PF, Green AE. Analytical characterization of spectral actinic flux and spectral irradiance in the middle ultraviolet. Photochem. Photobiol. 1982;35:89–101. doi: 10.1111/j.1751-1097.1982.tb03815.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.