Abstract
THO is a multi-protein complex that promotes coupling between transcription and mRNA processing. In contrast to its role in mRNA biogenesis, we show here that the fission yeast THO complex negatively controls the expression of non-coding small nucleolar (sno) RNAs. Accordingly, the deletion of genes encoding subunits of the evolutionarily conserved THO complex results in increased levels of mature snoRNAs. We also show physical and functional connections between THO and components of the TRAMP polyadenylation complex, whose loss of function also results in snoRNA accumulation. Consistent with a role in snoRNA expression, we demonstrate that THO and TRAMP complexes are recruited to snoRNA genes, and that a functional THO complex is required to maintain TRAMP occupancy at sites of snoRNA transcription. Our findings suggest that THO promotes exosome-mediated degradation of snoRNA precursors by ensuring the presence of the TRAMP complex at snoRNA genes. This study unveils an unexpected role for THO in the control of snoRNA expression and provides a new link between transcription and nuclear RNA decay.
INTRODUCTION
Transcription of protein-coding genes by RNA polymerase II (Pol II) generates primary transcripts that are matured co-transcriptionally. This coupling between transcription and pre-mRNA processing is mainly achieved by the co-transcriptional recruitment of proteins complexes, which are subsequently transferred onto nascent mRNAs for proper maturation and export. An important structural component required for such coordination between transcription and mRNA processing is a multi-protein complex called THO (1). The THO complex was originally identified in the budding yeast Saccharomyces cerevisiae (2) and is minimally composed of five non-essential subunits: Hpr1, Tho2, Mft1, Thp2 and Tex1 (3). The existing data on the THO complex indicate that it is recruited to protein-coding genes with the transcriptional machinery, allowing the recruitment of mRNA export factors to nascent transcripts (1,4–6). Accordingly, deletion of genes encoding subunits of the THO complex reduces the efficiency of mRNA export (5,7,8). THO mutant strains are also defective in mRNA 3′-end processing, as demonstrated by the accumulation of stalled mRNP intermediates that are associated with chromatin, polyadenylation factors and proteins from the nuclear pore complex (9,10).
Although the functional role of the THO complex has been studied mostly in S. cerevisiae, subunits of this complex are conserved among diverse eukaryotic species. Homologs of budding yeast Hpr1 (THOC1) and Tho2 (THOC2) are found in humans, Drosophila and fission yeast; yet, the Mft1 and Thp2 subunits appear to be specific to S. cerevisiae (11,12). Instead of Mft1 and Thp2 homologs, the Drosophila and human THO complex contain three additional subunits named THOC5, THOC6 and THOC7 (11,12). Interestingly, THOC5 and THOC7 orthologs are also found in the genome of the fission yeast, Schizosaccharomyces pombe (12). In Drosophila and mice, the THO complex appears to regulate specific genes, as only a subset of mRNAs are downregulated after the depletion of THO components (12,13). More recently, Drosophila THOC1 and THOC5 subunits were shown to be recruited to a stress-response gene via loading onto nascent transcripts (14). Furthermore, depletion of THO subunits in Drosophila and mammalian cells causes defects in transcription elongation, 3′-end processing and mRNA export (13–16). These studies thus support the evolutionarily conserved role of the THO complex in mRNA processing and nuclear export.
In addition to mRNAs, RNA Pol II is also responsible for the synthesis of many non-coding RNAs, such as small nucleolar (sno) RNAs. In yeast, snoRNAs are mainly expressed from independent transcriptional units, whereas >90% of snoRNAs reside in introns of human genes (17). snoRNAs are divided into two functional classes: C/D box and H/ACA box that guide 2′-O-ribose methylation and pseudouridylation of rRNAs, respectively (18). Although snoRNAs are not polyadenylated, we and others have recently demonstrated that mature snoRNAs can originate via 3′-end trimming of polyadenylated precursors in a process that requires a poly(A)-binding protein and the RNA exosome (19,20). The exosome complex is a RNA degradation machinery that is minimally composed of 10 evolutionarily conserved subunits, including the catalytically active ribonuclease, Dis3/Rrp44, which exhibits exonuclease (21) and endonuclease (22–25) activities. In the yeast nucleus, the core exosome is associated with an additional 3′ → 5′ exonuclease, Rrp6 (25,26). Although early studies suggested that Dis3 and Rrp6 were functionally redundant, the emergence of genetic and biochemical evidence indicate that they are likely to have different cellular activities (19,20,27,28). For instance, whereas deletion of rrp6 in S. pombe results in reduced levels of mature snoRNAs, a strain defective in Dis3 shows increased snoRNA levels (20). The RNA degradation function of the core exosome is promoted by the activity of a conserved polyadenylation complex, called TRAMP, which consist of the RNA helicase Mtr4, a poly(A) polymerase (PAP) (Cid14 in S. pombe) and a zinc knuckle protein (Air1 in S. pombe) (29–33). Interestingly, although polyadenylation activities of the TRAMP complex and the canonical mRNA PAP are both involved during yeast snoRNA expression (19,20), the exact contribution of TRAMP- and PAP-dependent polyadenylation is not completely understood.
Because snoRNA biosynthesis involves 3′-end polyadenylation, much like mRNAs, we searched for additional mRNA processing factors that would be involved in fission yeast snoRNA expression. Unexpectedly, we found that a defective THO complex results in the accumulation of mature snoRNAs. Furthermore, we show that the THO complex is present at snoRNA genes, associates with the TRAMP complex and is required to maintain TRAMP occupancy at snoRNA genes. Our findings indicate that the THO complex is important for TRAMP-dependent RNA decay during snoRNA expression, revealing a novel function for THO in the control of non-coding RNA expression.
MATERIALS AND METHODS
Yeast strains and media
A list of all S. pombe strains used in this study is provided in Supplementary Table S1. Cells were routinely grown in YES media (3% glucose, 0.5% yeast extract, supplemented with adenine, histidine, leucine and uracil) or Edinburgh minimal medium (EMM). To repress nmt1 promoter, thiamine was added to a final concentration of 60 μM in EMM for 15 h. All gene disruptions were performed by PCR-mediated gene targeting as described (34), and the absence of mRNA was confirmed by RT–PCR.
DNA constructs
To generate the Cid14 construct, the cid14 gene with additional 500 bp of promoter and terminator sequences was PCR amplified using genomic DNA and cloned in pFB366 (35) using PstI and SstI, generating pFB494. A catalytically inactive version of Cid14 was generated by site-directed mutagenesis using pFB494 and changing aspartate residues 298 and 300 to alanine residues resulting in plasmid pFB495. Both constructs were confirmed by sequencing. For single chromosomal integration into the ade6 locus, pFB366, pFB494 and pFB495 were linearized and transformed into the appropriate strains. Positive integrants were confirmed by PCR screening using primers located within cid14 as well as using RT–PCR analysis.
RNA analyses
Total RNA was extracted using the hot-acid phenol method. RNase H experiments were done as previously described (20). Essentially, total RNA was RNase H-treated in a mixture containing snoRNA-specific complementary oligonucleotides plus or minus oligo d(T). RNA samples were resolved on 6% polyacrylamide-8 M urea gels, transferred onto nylon membranes and probed using 32P-labeled snoRNA-specific probes.
Chromatin immunoprecipitation assays
Chromatin immunoprecipitation (ChIP) experiments were performed essentially as described (36) with some modifications. Briefly, cells were grown to an OD600 of 0.5–0.6. Formaldehyde solution (37) was then added into the culture (final concentration of 1%) and incubated for 20 min at room temperature. After quenching the reaction with glycine for 5 min, cells were washed twice with cold Tris-buffered saline (20 mM Tris–HCl pH 7.5, 150 mM NaCl). Cell pellets from 50 ml cultures were resuspended in 500 μl of lysis buffer (36) containing proteases inhibitors and disrupted vigorously with glass bead three times for 30 s using a FastPrep instrument. Samples were than sonicated 10 times for 10 s at 20% using a Branson digital sonifier. Five hundred microliters of whole-cell extract was incubated with anti-CTD (8WG16) antibody coupled to protein-G, or IgG sepharose for TAP-tagged strains. For measurements of RNA polymerase II occupancy in strains that express TAP-tagged fusion proteins, the CTD antibody was pre-bound to protein-G-sepharose prior to the addition of cell extracts to prevent binding between the immunoglobulin-binding domain of the TAP tag and the CTD antibody. Immunoprecipitation, bead washing, elution, reversal of crosslinking and DNA precipitation steps were all done as described (36). Protein density at snoRNAs genes was calculated as the enrichment of a specific genomic locus relative to a non-transcribed intergenic region (nucleotides 3 009 380 to 3 009 484 of chromosome I), as previously described (20,37). Quantification of the immunoprecipitated DNA was done by quantitative real-time PCR. Control ChIP assays using untagged strains were also carried for all experiments, but were not shown since they were negative.
Immunoprecipitation
Protein extractions of control, Tho2-TAP and Tho5-TAP strains were done with glass beads using a FastPrep instrument in IP buffer (10% glycerol, 50 mM Tris–HCl pH 8.0, 100 mM NaCl, 1 mM EDTA pH 8.0, 1 mM DTT, 0.05% Triton X-100 containing proteases inhibitors). Two milligrams of proteins was incubated on a rotating platform overnight at 4°C with IgG sepharose beads. The beads were washed four times in the same IP buffer. For RNases digestions, IPs were treated with 7.5 units of RNase A and 300 units of RNases T1 after the first wash, incubated for 10 min at room temperature followed by two additional wash steps. Input and eluted proteins were resolved on a 10% SDS–PAGE before immunoblotting using an Mtr4 antibody (33) (a generous gift from Marc Bühler, Friedrich Miescher Institute for Biomedical Research), a ProA antibody (Sigma) and an antibody specific to Rmt3 (38).
RESULTS
Mature and 3′-extented forms of snoRNAs accumulate in THO mutants
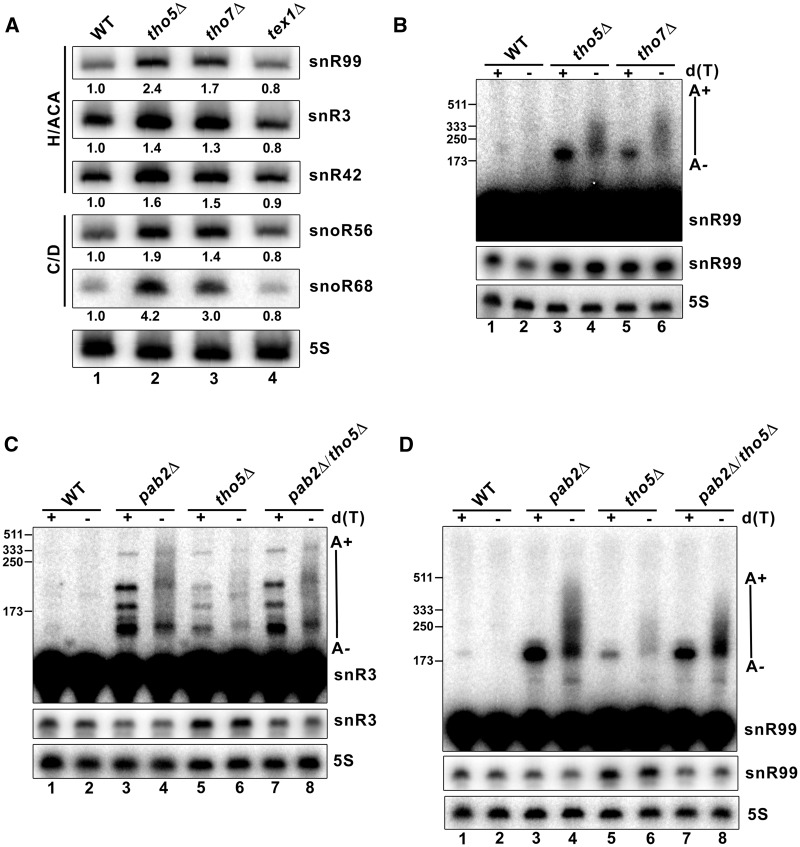
In S. cerevisiae, the THO complex is composed of Hpr1, Tho2, Mft1, Thp2 and Tex1 (3). In S. pombe, the THO complex appears to lack the S. cerevisiae Mft1 and Thp2 orthologs, but instead include mammalian THOC5 (SPBC577.04) and THOC7 (SPCC24B10.11 c) counterparts (12). Schizosaccharomyces pombe cells deleted for genes encoding Hpr1 and Tho2 orthologs could not be generated, consistent with being essential fission yeast genes (39); yet, strains deleted for the putative THOC5, THOC7 and Tex1 orthologs were viable. Notably, we found that the absence of Tho5 and Tho7 results in increased levels of H/ACA box and C/D box snoRNAs (Figure 1A, compare lanes 2–3 to lane 1). Although the levels of RNA accumulation varied between snoRNA-encoding genes, the increased expression of snoRNAs in THO mutants was reproducible (Supplementary Figure S1). In contrast, deletion of tex1 did not result in snoRNA accumulation (Figure 1A, lane 4), indicating that the effect of Tho5 and Tho7 on snoRNA expression is specific. The absence of snoRNA accumulation in the absence of S. pombe Tex1 is consistent with the lack of RNA-related phenotypes in a S. cerevisiae tex1Δ strain (3,40). The absence of Tho5 and Tho7 also resulted in the accumulation of 3′-extended polyadenylated snoRNAs, as demonstrated by RNase H cleavage assays in the presence or absence of oligo d(T) (Figure 1B, lanes 3–6). We previously reported that the absence of the nuclear poly(A)-binding protein, Pab2, causes the accumulation of 3′-extended polyadenylated snoRNAs together with the reduction of mature snoRNAs, consistent with a precursor–product relationship (20). To further characterize the role of the THO complex in snoRNA metabolism, we generated a tho5Δ/pab2Δ double-mutant strain and examined the level of mature and 3′-extended snoRNAs by RNase H cleavage assays. As can be seen in Figure 1C and D, levels of 3′-extended polyadenylated snoRNA precursors remained similar between the tho5Δ/pab2Δ double-mutant and the single pab2Δ strain (upper panels, compare lanes 7–8 to lanes 3–4). In contrast, the levels of mature snoRNAs were reduced in the tho5Δ/pab2Δ double-mutant strain relative to the single tho5Δ mutant (Figure 1C–D, middle panels, compare lanes 7–8 to lanes 5–6; quantifications shown in Supplementary Figure S2), suggesting that the Pab2-dependent maturation pathway is required for the accumulation of mature snoRNA in a THO mutant. Altogether, these results indicate that a defective THO complex causes the accumulation of snoRNAs in fission yeast.
Figure 1.
A defective THO complex results in snoRNA accumulation. (A) Total RNA prepared from the indicated strains was subjected to northern blot analysis using probes complementary to the indicated snoRNAs. The 5 S rRNA was used as a loading control. Normalized levels of each snoRNA relative to WT cells are indicated beneath each lane. (B) Equal amounts of total RNA prepared from the indicated strains were treated with RNase H in the presence of a DNA oligonucleotide complementary to snR99. RNase H reactions were performed in the presence (+) or absence (−) of oligo d(T). The 5 S rRNA was used as a loading control. Size markers (nt) are indicated on the left. (C–D) RNAse H reactions using total RNA from WT, pab2Δ, tho5Δ, and pab2Δ/tho5Δ strains. DNA oligonucleotides complementary to snR3 (C) and snR99 (D) were used in the presence (+) or absence (−) of oligo d(T). The 5 S rRNA was used as a loading control. Size markers (nt) are indicated on the left.
The accumulation of mature snoRNAs in tho5Δ and tho7Δ strains could be the consequence of increased transcription at snoRNA genes. To address this possibility, we examined the density of RNA Pol II along snoRNA genes by ChIP assays using extracts prepared from wild-type (WT), tho5Δ and tho7Δ strains. The results from these ChIP experiments did not reveal increased levels of RNA Pol II at the promoter and snoRNA region of SNR99 and SNOR68 in tho5Δ and tho7Δ cells (Supplementary Figure S3). We therefore conclude that the accumulation of mature snoRNAs in THO mutant strains is not the consequence of increased transcription.
The THO complex is present at snoRNA genes
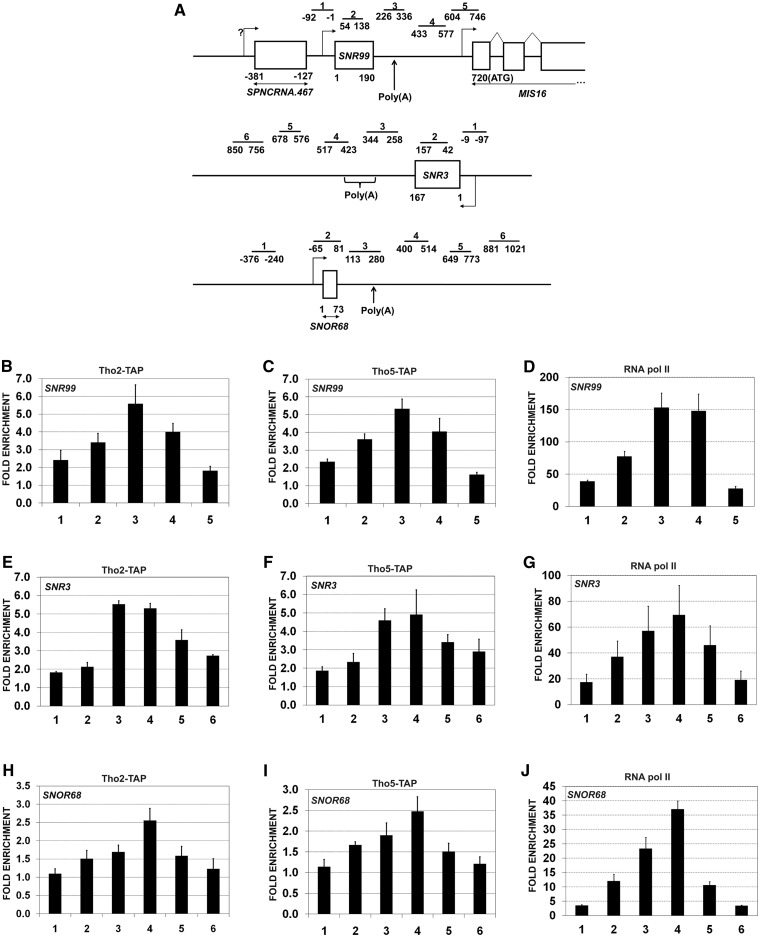
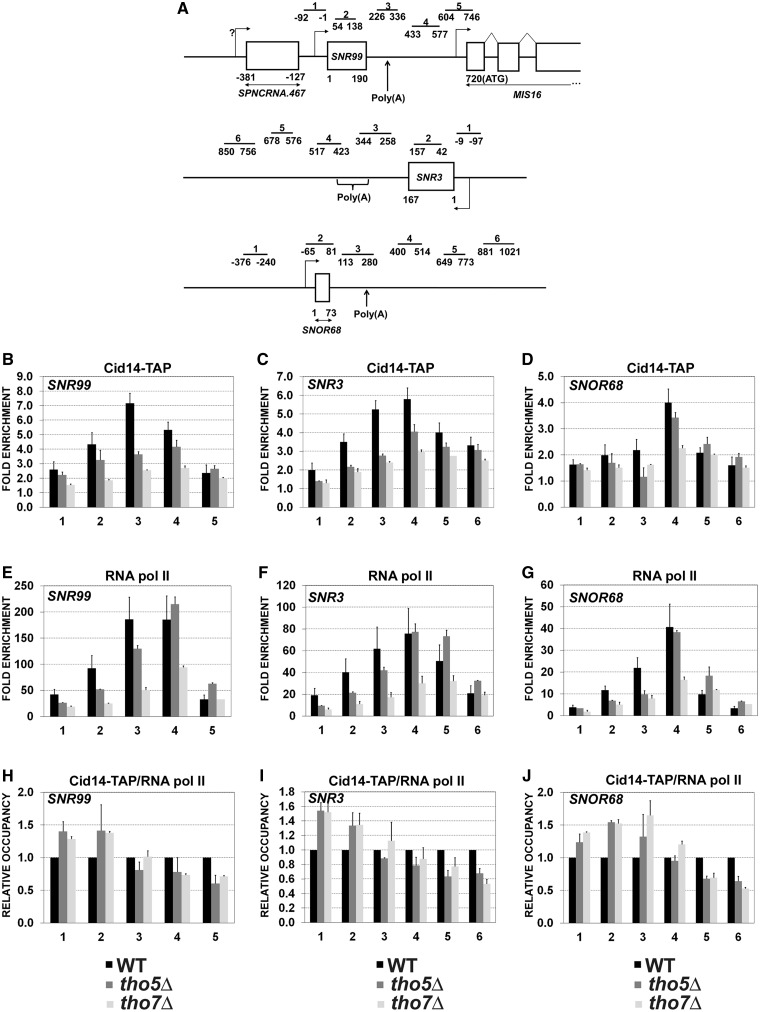
The aforementioned snoRNA accumulation detected in tho5 and tho7 deletion strains suggests that THO functions in snoRNA metabolism. To begin to explore the mechanism by which the THO complex controls snoRNA expression, we first assessed whether subunits of the THO complex are associated with sites of snoRNA transcription by ChIP assays, which have previously been used to show the presence of THO subunits at protein-coding genes (5,6,14,41). TAP-tagged versions of Tho2 and Tho5 were used to monitor the level of THO occupancy at three independent snoRNA genes: the H/ACA class SNR99 and SNR3 genes as well as the C/D class SNOR68 gene (Figure 2A). THO occupancy at these three snoRNA genes was calculated as the enrichment of Tho2 and Tho5 at the respective snoRNA gene relative to a non-transcribed intergenic region, which served as a negative control (20,37). Tho2 was specifically recruited to all three snoRNA genes (Figure 2B, E and H). As a control, ChIP assays using an untagged strain yeilded signal at snoRNA genes that were near background levels (data not shown). The lower levels of Tho2 at SNOR68 (Figure 2H) correlate with the reduced transcriptional activity of this snoRNA gene compared to SNR99 and SNR3, as revealed by ChIP analysis of RNA polymerase II (compare Figure 2J to D and G). Interestingly, Tho2 showed the highest crosslinking signal at the 3′-end of snoRNA genes (Figure 2B, region 3; Figure 2E, regions 3–4; and Figure 2H, region 4), a profile similar to that of RNA Pol II (Figure 2D, G and J). Tho5 occupancy at snoRNA genes showed a distribution similar to Tho2 (Figure 2C, F and I), consistent with THOC2 and THOC5 being components of the same complex (11,12,42). These results show that Tho2 and Tho5 are present at sites of snoRNA transcription.
Figure 2.
The THO complex is recruited to snoRNAs genes. (A) Schematic representation of SNR99, SNR3 and SNOR68 genes. Bars above the genes show the positions of PCR products used for the ChIP analysis. The position of polyadenylation sites [poly(A)] identified by 3′-RACE (20) is indicated. (B, E and H) ChIP assays were performed using a TAP-tagged version of Tho2. The input and co-precipitated DNA were quantified by qPCR along the SNR99 (B), SNR3 (E) and SNOR68 (H) genes using specific primers, as shown in panel A. (C, F and I) ChIP assays using a TAP-tagged version of Tho5 at the SNR99 (C), SNR3 (F) and SNOR68 (I). (D, G and J) ChIP assays using an RNA Pol II-specific antibody at SNR99 (D), SNR3 (G) and SNOR68 (J). ChIP data are presented as the fold enrichment compared to a non-transcribed intergenic region. The data and error bars represent the average and standard deviation from at least three biological replicates.
snoRNA expression is controlled by TRAMP-dependent exosome-mediated RNA decay
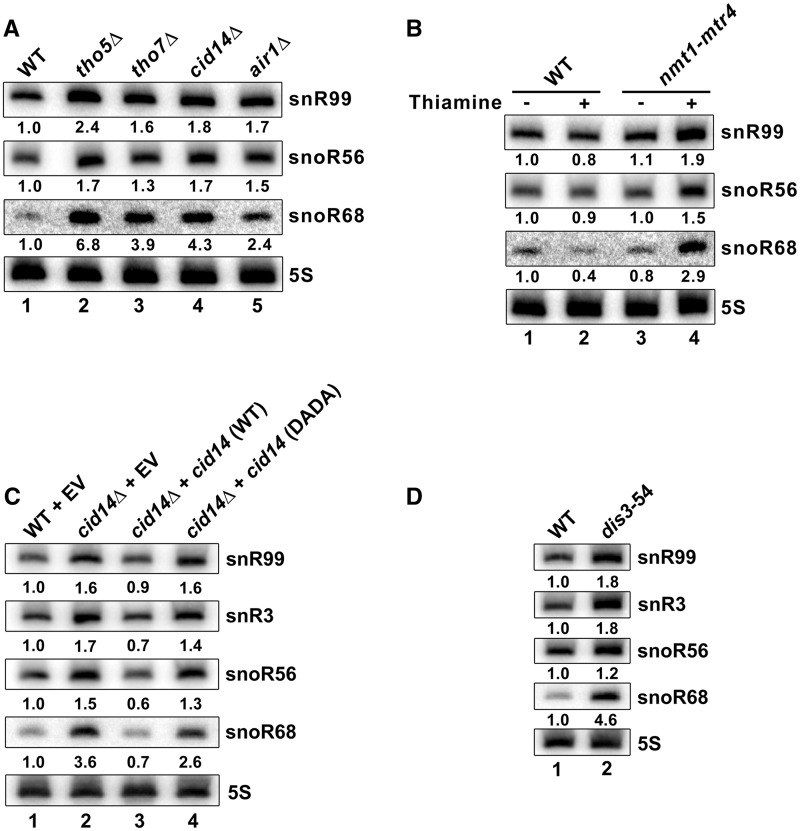
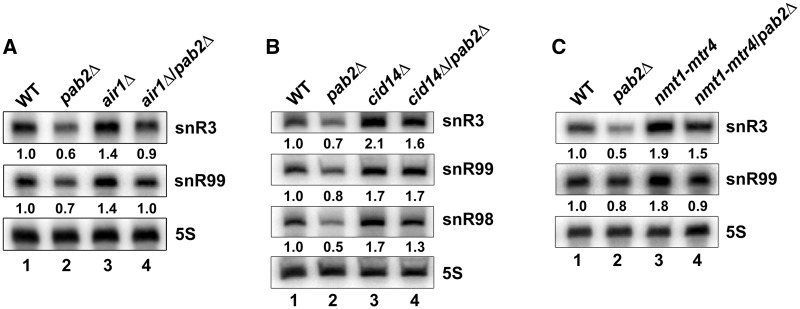
Because the accumulation of mature snoRNAs in THO mutant strains was not associated with increased transcription (Supplementary Figure S3), we considered that the THO complex might promote an RNA decay pathway. We had previously found that deletion of cid14, which encodes the PAP subunit of the fission yeast TRAMP complex, leads to increased level of mature snR99 (20). As this phenotype is similar to the snoRNA accumulation observed in tho5Δ and tho7Δ strains (Figure 1), we reasoned that the THO complex might promote TRAMP-dependent RNA decay. To explore this possibility, we examined snoRNA levels in THO and TRAMP mutants. As can be seen in Figure 3A, the deletion of genes encoding two different subunits of the TRAMP complex, cid14 (lane 4) and air1 (lane 5), resulted in snoRNA accumulations similar to tho5Δ (lane 2) and tho7Δ (lane 3) strains. We also used a conditional strain in which the genomic copy of mtr4, encoding the TRAMP-associated RNA helicase, is expressed from the thiamine-sensitive nmt1 promoter, which is strongly repressed following thiamine addition (35). As for cid14 and air1 deletion strains, the depletion of Mtr4 resulted in increased levels of snoRNA (Figure 3B, compare lane 4 to 3); in contrast, WT control cells did not show this thiamine-dependent snoRNA accumulation (lanes 1–2). These results indicate that deficiencies in either TRAMP or THO subunits increase the expression level of snoRNAs.
Figure 3.
TRAMP-dependent exosome-mediated RNA decay controls snoRNA expression. (A) Total RNA prepared from the indicated strains was subjected to northern blot analysis using probes complementary to the indicated snoRNAs. The 5 S rRNA was used as a loading control. Normalized levels of each snoRNA relative to WT cells are indicated beneath each lane. (B) Northern blot analysis of RNA prepared from WT (lanes 1–2) and nmt1-mtr4 (lanes 3–4) strains that were grown in the absence (−) or presence (+) of thiamine. Normalized levels of each snoRNA relative to WT cells cultured in the absence of thiamine (lane 1) are indicated beneath each lane. (C) Northern blot analysis of total RNA prepared from WT (lane 1) and cid14Δ (lanes 2–4) cells that were previously transformed with the empty vector (EV; lanes 1–2) or vectors that express WT (lane 3) and catalytically inactive (lane 4; DADA) versions of Cid14. Normalized levels of each snoRNA relative to WT cells (lane 1) are indicated beneath each lane. (D) Total RNA prepared from WT (lane 1) and dis3–54 (lane 2) strains were subjected to northern blot analysis using probes complementary to the indicated snoRNAs. The 5 S rRNA was used as a loading control. Normalized levels of each snoRNA relative to WT cells are indicated beneath each lane.
We next examined whether the polyadenylation activity of Cid14 was required for TRAMP-dependent control of snoRNA expression. We therefore generated constructs that express WT and catalytically inactive (DADA) versions of Cid14 from native promoter and terminator sequences. The catalytically inactive version of Cid14 substitutes two evolutionarily conserved aspartic acid (D) residues for alanines (A), which has been shown to inactivate the PAP activity of S. pombe Cid14 (31) and its S. cerevisiae ortholog, Trf4 (30). These two Cid14 constructs, as well as an empty vector control, were chromosomally integrated as a single copy into a cid14Δ strain. Importantly, the catalytically inactive version of Cid14 (DADA) did not rescue the increased levels of snoRNAs observed in the cid14Δ strain (Figure 3C, compare lanes 1, 2 and 4). In contrast, expression of WT Cid14 in the cid14Δ strain effectively restored the altered expression of snoRNAs (Figure 3C, lane 3). These results indicate that the polyadenylation activity of the TRAMP complex is required for the negative control of snoRNA expression.
TRAMP-dependent polyadenylation enhances the degradation activity of the nuclear exosome, at least in part by providing an accessible 3′-end to target RNAs (43,44). Accordingly, our observation that TRAMP-mediated polyadenylation was required to prevent snoRNA accumulation (Figure 3C) suggested the involvement of the core exosome in a RNA decay pathway that negatively controls snoRNA expression. We therefore examined the expression level of snoRNAs in the dis3–54 strain, which contains an amino acid substitution in the exonuclease domain of the exosome subunit Dis3/Rrp44 that impairs its catalytic activity (45). Notably, levels of snR99, snR3, snoR56 and snoR68 were all increased in a dis3–54 strain (Figure 3D). Taken together, these data indicate that TRAMP/exosome-mediated decay functions in the negative control of snoRNA expression.
THO and TRAMP complexes cooperate in the control of snoRNA expression
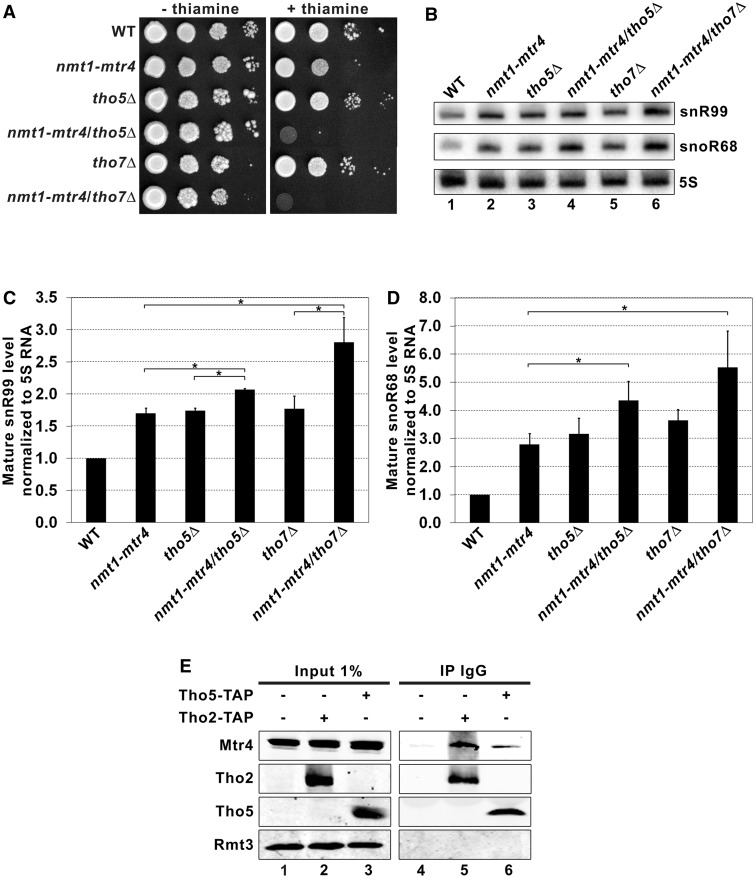
Because snoRNA accumulation is a shared phenotype between strains deficient for TRAMP and THO subunits, we decided to further characterize the functional relationship between THO and TRAMP complexes by generating double-mutant strains. However, we failed to obtain cid14Δ/tho5Δ or air1Δ/tho5Δ double-mutant strains, suggesting genetic interactions between subunits of THO and TRAMP complexes. We therefore deleted tho5 and tho7 in the nmt1-mtr4 conditional strain to generate nmt1-mtr4/tho5Δ and nmt1-mtr4/tho7Δ strains, which showed growth rates comparable to the single-mutant strains in the absence of thiamine (Figure 4A; left panel). In contrast, thiamine-dependent depletion of Mtr4 in the context of tho5 or tho7 deletions resulted in a synthetic growth defect as compared to the single nmt1-mtr4 conditional strain (Figure 4A; right panel). Synthetic growth defects were also observed using an nmt1-cid14/tho5Δ double-mutant strain (Supplementary Figure S4). These results demonstrate genetic interactions between genes that encode THO and TRAMP subunits.
Figure 4.
The THO and TRAMP complexes cooperate in a pathway that negatively controls snoRNA expression. (A) Ten-fold serial dilutions of cells from the indicated strains were cultured in the presence or in the absence of thiamine. (B) Total RNA prepared from the indicated strains that were previously grown in the presence of thiamine (to allow depletion of Mtr4) was analyzed by northern blot using probes complementary to snR99 and snoR68. The 5 S rRNA was used as a loading control. (C and D) Quantification of northern blot data for snR99 (C) and snoR68 (D). Data and error bars (C–D) represent the average and standard deviation from three biological replicates (*P-value < 0.05; Student’s t-test). (E) Immunoblot analysis of whole-cell extracts (Input; lanes 1–3) and IgG-sepharose precipitates (IP IgG; lanes 4–6) prepared from control, Tho2-TAP and Tho5-TAP-expressing cells. Antibodies for western blotting were rabbit polyclonal antibodies specific to Mtr4 (upper panel), ProA (Tho2 and Tho5) and Rmt3 (bottom panel).
The aforementioned synthetic growth phenotypes detected between THO and TRAMP mutants could result from a variety of molecular defects. To determine whether the observed synthetic growth phenotypes correlate with synthetic defects in snoRNA expression, we analyzed the expression of snR99 and snoR68 in single- and double-mutant strains after thiamine addition to induce Mtr4 depletion. Importantly, the absence of Tho5 and Tho7 did not influence the kinetics of Mtr4 depletion after the addition of thiamine (Supplementary Figure S5). Consistent with our previous results, the individual depletion of Mtr4 and THO subunits led to the accumulation of mature snoRNAs (Figure 4B, compare lanes 2, 3 and 5 to lane 1). Importantly, snR99 and snoR68 levels were quantified for each mutant from independent experiments. Although modest increases in snoRNA levels are noted in the nmt1-mtr4/tho5Δ and nmt1-mtr4/tho7Δ double-mutant strains compared to single mutants (Figure 4C and D), these increases in snoRNA levels are not additive. Accordingly, the absence of additive defects in double-mutant strains suggests that the THO and TRAMP complexes largely contribute to the same pathway in the control of snoRNA expression.
The functional connection between subunits of THO and TRAMP complexes suggested that these two complexes might physically associate. We therefore used a previously described antibody specific to the Mtr4 subunit of the fission yeast TRAMP complex (33) to analyze affinity purifications prepared from extracts of cells that expressed TAP-tagged versions of Tho2 and Tho5. We found that endogenous Mtr4 reproducibly co-purified with Tho2 and Tho5, but not with a control purification (Figure 4E, compare lanes 5–6 to lane 4). We also confirmed that the association between THO and Mtr4 was not sensitive to RNase (Supplementary Figure S6). As an additional control, a non-TRAMP protein was not recovered in Tho2 and Tho5 precipitates (Figure 4E, see Rmt3). The specific association between THO and TRAMP subunits in S. pombe is consistent with the recovery of peptides from the Mtr4 homolog in a purification of human THOC2 (42). Collectively, the data presented in Figure 4 suggest that the THO and TRAMP complexes cooperate in a pathway that negatively controls snoRNA expression.
The THO complex is required to maintain Cid14 occupancy at snoRNA genes
We next explored possible mechanisms by which the THO complex could promote TRAMP-dependent control of snoRNA expression. Given the role of THO in coupling transcription and mRNA processing (1,4–6), we asked whether a functional THO complex was required for TRAMP occupancy at snoRNA genes. First, we used a TAP-tagged version of Cid14 to confirm that the TRAMP complex is recruited to snoRNA genes by ChIP assays. ChIP analysis of Cid14-TAP along three snoRNA genes (SNR99, SNR3 and SNOR68) showed clear enrichments over the intergenic control region (Figure 5B–D, black columns). Notably, Cid14 association with snoRNA genes was highest at the 3′-end, a distribution pattern similar to that of Tho2/Tho5 (Figure 2) and RNA Pol II (Figure 5E–G).
Figure 5.
A functional THO complex is required to maintain Cid14 occupancy at snoRNA genes. (A) Schematic of SNR99, SNR3 and SNOR68 genes as shown in Figure 2A. (B–D) ChIP assays using a TAP-tagged version of Cid14 at SNR99 (B), SNR3 (C) and SNOR68 (D) genes in WT, tho5Δ and tho7Δ strains. Input and co-purified DNA were quantified by qPCR using primers shown in panel A. (E–G) ChIP assays performed using an RNA Pol II-specific antibody at SNR99 (E), SNR3 (F) and SNOR68 (G) genes in WT, tho5Δ and tho7Δ strain using primers shown in panel A. (H–J) Density of Cid14 relative to RNA Pol II at SNR99 (H), SNR3 (I) and SNOR68 (J) genes in WT, tho5Δ and tho7Δ strains. Values were set to 1 for the control (WT) strain. All ChIP data are presented as the fold enrichment compared to a non-transcribed intergenic region. Data and error bars represent the average and standard deviation from two biological replicates.
To examine whether a functional THO complex was important for the association of Cid14 with snoRNA genes, we performed ChIP analysis of Cid14 using extracts prepared from WT, tho5Δ, and tho7Δ strains. In the absence of Tho5 and Tho7, levels of Cid14 cross-linking at snoRNA genes were generally reduced as compared to WT cells (Figure 5B, regions 2–4; Figure 5C, regions 2–4; and Figure 5D, regions 3–4). Yet, we noted that fission yeast THO mutants were also affected in polymerase density (Figure 5E–G), which is consistent with previous observations made in budding yeast (46,47). To account for differences in transcription profiles, we therefore normalized Cid14 ChIP values to RNA Pol II occupancy by performing simultaneous measurements of Cid14 and Pol II recruitment from the same chromatin preparations. Notably, Cid14 occupancy relative to RNA Pol II decreased steadily along all three snoRNA genes, showing 35–50% reductions at the 3′-end of snoRNA genes in strains lacking tho5 and tho7 compared to WT cells (Figure 5H–J). This effect is not the consequence of reduced Cid14 expression in THO-deficient cells, as determined by immunoblotting (Supplementary Figure S7). These results indicate that a functional THO complex is required to maintain TRAMP occupancy at snoRNA genes.
The Pab2-dependent snoRNA maturation pathway is required for the accumulation of snoRNAs in TRAMP mutants
We have previously shown that the poly(A)-binding protein, Pab2, functions with the exonuclease Rrp6 in a pathway that promotes the maturation of 3′-extended polyadenylated snoRNA precursors into mature snoRNAs (20). To determine whether this maturation pathway was required for the accumulation of mature snoRNAs observed in TRAMP mutants, we deleted pab2 in strains deficient for the TRAMP subunits Air1, Cid14 and Mtr4 and analyzed snoRNA expression by northern blot. As observed in THO mutant strains (Figure 1C and D), the absence of Pab2 reproducibly suppressed the accumulation of mature snoRNA that was detected in cells deficient for Air1 (Figure 6A, compare lane 4 to 3), Cid14 (snR3 and snR98; Figure 6B, compare lane 4 to 3) and Mtr4 (Figure 6C, compare lane 4 to 3). In contrast to Air1- and Mtr4-deficient cells, for which the absence of Pab2 suppressed the accumulation of snR99 (Figure 6A and C, respectively), deletion of pab2 in the cid14Δ strain had minimal effects on the Cid14-dependent accumulation of snR99 (Figure 6B). The reasons for the discrepancy between the role of Pab2 in the Air1- and Mtr4-dependent accumulation of snR99, but not in conditions of Cid14 deficiency, are not clear. From these results, we conclude that the Pab2-dependent maturation pathway generally functions in an antagonistic manner to TRAMP-dependent RNA decay during snoRNA expression.
Figure 6.
Pab2 is required for the accumulation of mature snoRNAs in TRAMP mutants. (A) Total RNA prepared from WT, pab2Δ, air1Δ and air1Δ/pab2Δ strains was subjected to northern blot analysis using probes complementary to the indicated snoRNAs. The 5 S rRNA was used as a loading control. (B) Total RNA prepared from WT, pab2Δ, cid14Δ and cid14Δ/pab2Δ strains was subjected to northern blot analysis using probes complementary to the indicated snoRNAs. The 5 S rRNA was used as a loading control. (C) Total RNA prepared from WT, pab2Δ, nmt1-mtr4 and nmt1-mtr4/pab2Δ strains that were grown in the presence of thiamine (to allow depletion of Mtr4) was analyzed by northern blot using probes complementary to indicated snoRNAs. The 5 S rRNA was used as a loading control. In panels A to C, normalized levels of each snoRNA relative to WT cells are indicated beneath each lane.
DISCUSSION
The THO complex is known to play important roles in eukaryotic gene expression. Although THO normally promotes gene expression via well-established and conserved functions in mRNA biogenesis (1), a role for the THO complex in non-coding RNA expression has remained obscure. In the course of studying the metabolism of snoRNAs in fission yeast, we identified a new function for the THO complex in non-coding RNA expression. Most surprisingly, we show that THO functions in the negative control of snoRNAs, in contrast to the positive role of THO in mRNA expression (7,9,10,40,48). Specifically, we present multiple lines of evidence that support a functional connection between THO and TRAMP complexes in a pathway that controls snoRNA expression: (i) THO and TRAMP are both recruited to snoRNA genes, showing similar distribution profiles; (ii) THO and TRAMP mutants share similar phenotypes with respect to Pab2-dependent snoRNA accumulations; (iii) the TRAMP subunit Mtr4 can be found in a complex with Tho2 and Tho5 and (iv) a functional THO complex is required to maintain TRAMP occupancy at the 3′-end of snoRNA genes. These results suggest that the THO complex closely cooperates with the nuclear RNA surveillance machinery, namely, TRAMP/exosome-dependent decay, to control snoRNA expression.
Consistent with the functional role of THO and TRAMP complexes in snoRNA metabolism, our ChIP data show that fission yeast Tho2, Tho5 and Cid14 are present at snoRNA genes. Interestingly, we note that the ChIP profiles of Tho2, Tho5 and Cid14 mirror the profile of RNA Pol II, showing increase occupancy at the 3′-end of snoRNA genes. Such an accumulation of RNA Pol II downstream of snoRNA genes may reflect a transcriptional pause associated with snoRNA 3′-end processing and/or transcription termination. Consistent with this idea, transcriptional pausing has been shown to play a role in 3′-end processing and termination at mammalian genes (49,50). The similarity between the ChIP profiles of Tho2, Tho5, Cid14 and RNA Pol II thus suggests that THO and TRAMP complexes travel along with the elongating RNA polymerase, rather than being specifically recruited at the 3′-end of snoRNA genes. This conclusion is consistent with earlier studies, indicating that the recruitment of THO to protein-coding genes in S. cerevisiae is coupled to transcriptional elongation (5,40). Recent studies have in fact identified factors that promote THO/TREX recruitment to protein-coding genes in Drosophila and S. cerevisiae (14,51). It will therefore be interesting to determine if any of these factors are important for the association of THO with the elongating polymerase during snoRNA transcription.
In fission yeast, mature snoRNAs can be produced via 3′-end trimming of polyadenylated precursors in a process that requires Rrp6 and the nuclear poly(A)-binding protein Pab2 (20). Accordingly, increased levels of 3′-extended polyadenylated precursors and reduced levels of mature snoRNAs are detected in pab2Δ and rrp6Δ strains, consistent with a precursor–product relationship (20). To explain the accumulation of mature snoRNAs in THO and TRAMP mutant strains, we propose that TRAMP-dependent polyadenylation promotes a degradation pathway that actively competes with the Pab2/Rrp6 maturation pathway (Figure 7A). Consistent with this model, snoRNA accumulation was also observed in a core exosome mutant (Figure 3D), whereas reduced levels of mature snoRNA are detected in rrp6Δ cells (20). Thus, the equilibrium between decay and maturation of snoRNA precursors controls snoRNA expression. Although we cannot exclude the possibility that TRAMP/exosome-dependent decay also influences the turnover of mature snoRNAs independently of snoRNA precursors, the fact that Pab2-dependent maturation is required for the upregulation of mature snoRNAs in TRAMP (Figure 6) and core exosome (20) mutants argues that the TRAMP and exosome complexes primarily act on snoRNA precursors rather than on mature snoRNAs.
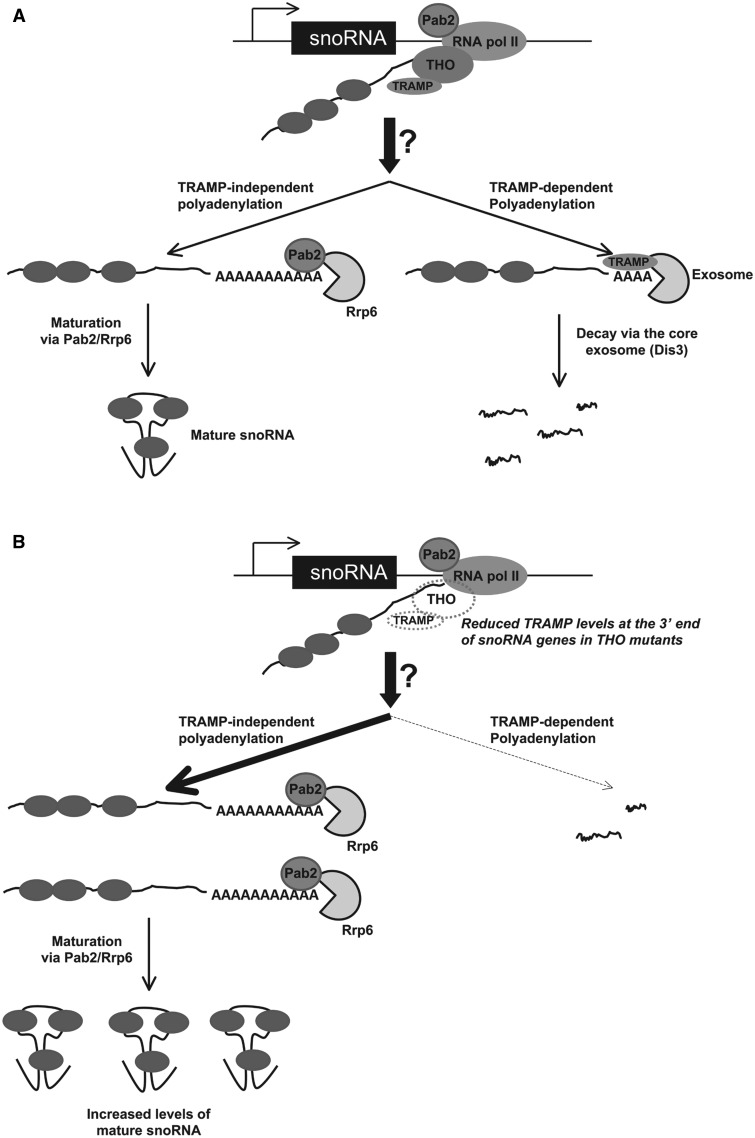
Figure 7.
Model for how the THO complex contributes to snoRNA expression in fission yeast. (A) Termination of transcription at fission yeast snoRNA genes, which remains poorly understood (as indicated by the question mark), is coupled to two antagonistic pathways that actively compete for nascent snoRNA precursors: Pab2/Rrp6-dependent maturation and TRAMP/exosome-dependent decay. The equilibrium between decay and maturation of snoRNA precursors thus controls snoRNA expression. During snoRNA transcription, the THO complex is escorted by the elongating polymerase and is important to maintain stable association between the TRAMP complex and the transcriptional machinery during snoRNA 3′-end processing. Because Cid14 is not required for polyadenylation-dependent processing of snoRNA precursors by Pab2 and Rrp6 (20), TRAMP-independent polyadenylation promotes the snoRNA maturation pathway. (B) In THO-deficient cells, reduced levels of the TRAMP complex during snoRNA transcription termination favor the Pab2/Rrp6 maturation pathway, leading to increased levels of mature snoRNAs.
In the absence of THO, mRNP biogenesis does not occur properly and DNA:RNA hybrids accumulate, forming R-loops that hinder transcription elongation at protein-coding genes (48). It is therefore possible that the effect of THO mutants on snoRNA expression is the consequence of R-loop formation during snoRNA transcription. We do not favor this interpretation, however. First, although R-loop formation in conditions of THO deficiencies normally results in decrease mRNA abundance (7,9,10,40,48), we observed increased levels of mature snoRNAs in our fission yeast THO mutants. Moreover, snoRNA accumulation was also increased in TRAMP and exosome mutants, yet these strains are not known to cause R-loop formation. Rather, we show here that the occupancy of the TRAMP complex is perturbed at snoRNA genes in THO-deficient cells (Figure 5). We therefore identify for the first time a factor required for full occupancy of TRAMP on transcribed genes. Interestingly, the THO complex is not required for TRAMP occupancy at the 5′-end of snoRNA genes, suggesting that THO might not be responsible to recruit the TRAMP complex to the transcription machinery. However, the occupancy of Cid14 at the 3′-end of snoRNA genes was reduced by up to 50% in tho5 and tho7 mutants, indicating that a functional THO complex is important to maintain TRAMP occupancy on transcribed snoRNA genes. Based on these data, we propose that THO functions in the control of snoRNA expression by ensuring the presence of the TRAMP complex during snoRNA 3′-end processing (Figure 7A). In accordance with this model, a defective THO complex (Figure 7B) impairs exosome-mediated decay of snoRNA precursors by reducing TRAMP occupancy at snoRNA genes, which allows the Pab2/Rrp6 maturation pathway to outcompete TRAMP-dependent decay, leading to greater levels of mature snoRNAs. Whether THO supports TRAMP occupancy at snoRNA genes by direct physical interactions or via bridging proteins remains to be determined. As a consequence of reduced TRAMP/exosome-mediated decay in THO-mutant strains (Figure 7B), a limiting step in the Pab2/Rrp6-dependent maturation pathway may become saturated, leading to the accumulation of polyadenylated snoRNA precursors, which are also detected in THO-deficient cells (Figure 1B).
On the basis of sequence similarity-based identification, the S. pombe THO complex appears to be more similar to its mammalian counterpart than to the S. cerevisiae THO complex [this study and (12)]. This may not be too surprising, however, given that S. pombe and S. cerevisiae are as different to each other as either is from animals (52). Such divergence between S. pombe and S. cerevisiae may also underlie differences in snoRNA metabolism between these two species. Accordingly, TRAMP and exosome mutants do not appear to accumulate mature snoRNAs in S. cerevisiae (19,29,53–55), in contrast to fission yeast. Despite these potential differences, the role of THO in non-coding RNA metabolism may not be exclusive to fission yeast. Accordingly, a genome-wide survey of THO occupancy by ChIP assays was recently reported using S. cerevisiae (48) and indicated that Hpr1 (Thoc1) crosslinks to snoRNA genes in budding yeast (56). Consistent with these findings, another mRNA export factor, Yra1, which physically interacts with the THO complex (5,6), is also found at snoRNA genes in S. cerevisiae (57). More recently, the fission yeast ortholog of Yra1, Mlo3, was shown to associate with the TRAMP complex and help suppress antisense RNA expression (58). These findings are consistent with our results and hint for the broader involvement of mRNA processing factors, such as the THO complex, in non-coding RNA metabolism.
Our data also showed that cells lacking a functional THO complex display synthetically lethal and synthetically sick phenotypes with deletions in TRAMP components. Comparable genetic interactions have been found in S. cerevisiae (6,7,59,60) and explained by the absence of TRAMP-dependent quality control mechanisms to manage the mRNP processing defects occurring in THO mutant cells, leading to a high degree of synthetic lethality when deletions in TRAMP components are combined with deletions in THO components. Although synthetic genetic interactions often indicate compensatory or parallel gene action, the absence of additive effect on snoRNA expression observed between a THO deletion (tho5 or tho7) and Mtr4 depletion suggests that the synthetic growth defects observed between THO and TRAMP in S. pombe are not related to their effect on snoRNAs, but is consistent with the view that they contribute to the same pathway in the control of snoRNA expression.
Considering that there are between 4000 and 5000 snoRNA molecules per cell in fission yeast (S. Marguerat and J. Bähler, unpublished results), increases in snoRNA levels as small as 50% are likely to represent a substantial expansion in the quantity of snoRNAs produced by the cell and in the amount of energy invested in making hundreds of additional snoRNAs. What could thus be the biological significance of this apparent overproduction of snoRNAs, a fraction of which is continually degraded by the exosome? As snoRNPs play direct roles in ribosome biogenesis, it is possible that such a mechanism ensures that critical components associated with ribosome biosynthesis are never limiting. Consistent with this view, it has been shown that ribosomal proteins are overproduced in mammalian cells, but that the unassembled ribosomal proteins are rapidly degraded by the proteasome (61).
In summary, our findings reveal a new function for the THO complex in the metabolism of non-coding RNAs. By establishing the THO complex as a factor important for TRAMP occupancy at snoRNA genes, we provide new insights into the interplay between transcription and nuclear RNA degradation. Given that the fission yeast THO complex includes the mammalian THOC5 and THOC7 orthologs, it is tempting to speculate that the functional connection between THO and TRAMP complexes is conserved in human cells.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Table 1 and Supplementary Figures 1–7.
FUNDING
The Natural Sciences and Engineering Research Council of Canada (NSERC) (to F.B.); Alexander Graham-Bell Doctoral Scholarship, NSERC (to J.F.L.); F.B. is a Canada Research Chair in Quality Control of Gene Expression. Funding for open access charge: Natural Sciences and Engineering Research Council of Canada (NSERC).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors wish to thank Marc Bühler for the Mtr4 antibody; Samuel Marguerat and Jürg Bähler for sharing unpublished results and Daniel Zenklusen for critical reading of the manuscript.
REFERENCES
- 1.Rondon AG, Jimeno S, Aguilera A. The interface between transcription and mRNP export: from THO to THSC/TREX-2. Biochim. Biophys. Acta. 2010;1799:533–538. doi: 10.1016/j.bbagrm.2010.06.002. [DOI] [PubMed] [Google Scholar]
- 2.Chavez S, Beilharz T, Rondon AG, Erdjument-Bromage H, Tempst P, Svejstrup JQ, Lithgow T, Aguilera A. A protein complex containing Tho2, Hpr1, Mft1 and a novel protein, Thp2, connects transcription elongation with mitotic recombination in Saccharomyces cerevisiae. EMBO J. 2000;19:5824–5834. doi: 10.1093/emboj/19.21.5824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pena A, Gewartowski K, Mroczek S, Cuellar J, Szykowska A, Prokop A, Czarnocki-Cieciura M, Piwowarski J, Tous C, Aguilera A, et al. Architecture and nucleic acids recognition mechanism of the THO complex, an mRNP assembly factor. EMBO J. 2012;31:1605–1616. doi: 10.1038/emboj.2012.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jimeno S, Rondon AG, Luna R, Aguilera A. The yeast THO complex and mRNA export factors link RNA metabolism with transcription and genome instability. EMBO J. 2002;21:3526–3535. doi: 10.1093/emboj/cdf335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Strasser K, Masuda S, Mason P, Pfannstiel J, Oppizzi M, Rodriguez-Navarro S, Rondon AG, Aguilera A, Struhl K, Reed R, et al. TREX is a conserved complex coupling transcription with messenger RNA export. Nature. 2002;417:304–308. doi: 10.1038/nature746. [DOI] [PubMed] [Google Scholar]
- 6.Zenklusen D, Vinciguerra P, Wyss JC, Stutz F. Stable mRNP formation and export require cotranscriptional recruitment of the mRNA export factors Yra1p and Sub2p by Hpr1p. Mol. Cell. Biol. 2002;22:8241–8253. doi: 10.1128/MCB.22.23.8241-8253.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Libri D, Dower K, Boulay J, Thomsen R, Rosbash M, Jensen TH. Interactions between mRNA export commitment, 3'-end quality control, and nuclear degradation. Mol. Cell. Biol. 2002;22:8254–8266. doi: 10.1128/MCB.22.23.8254-8266.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rougemaille M, Gudipati RK, Olesen JR, Thomsen R, Seraphin B, Libri D, Jensen TH. Dissecting mechanisms of nuclear mRNA surveillance in THO/sub2 complex mutants. EMBO J. 2007;26:2317–2326. doi: 10.1038/sj.emboj.7601669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rougemaille M, Dieppois G, Kisseleva-Romanova E, Gudipati RK, Lemoine S, Blugeon C, Boulay J, Jensen TH, Stutz F, Devaux F, et al. THO/Sub2p functions to coordinate 3'-end processing with gene-nuclear pore association. Cell. 2008;135:308–321. doi: 10.1016/j.cell.2008.08.005. [DOI] [PubMed] [Google Scholar]
- 10.Saguez C, Schmid M, Olesen JR, Ghazy MA, Qu X, Poulsen MB, Nasser T, Moore C, Jensen TH. Nuclear mRNA surveillance in THO/sub2 mutants is triggered by inefficient polyadenylation. Mol. Cell. 2008;31:91–103. doi: 10.1016/j.molcel.2008.04.030. [DOI] [PubMed] [Google Scholar]
- 11.Masuda S, Das R, Cheng H, Hurt E, Dorman N, Reed R. Recruitment of the human TREX complex to mRNA during splicing. Genes Dev. 2005;19:1512–1517. doi: 10.1101/gad.1302205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rehwinkel J, Herold A, Gari K, Kocher T, Rode M, Ciccarelli FL, Wilm M, Izaurralde E. Genome-wide analysis of mRNAs regulated by the THO complex in Drosophila melanogaster. Nat. Struct. Mol. Biol. 2004;11:558–566. doi: 10.1038/nsmb759. [DOI] [PubMed] [Google Scholar]
- 13.Guria A, Tran DD, Ramachandran S, Koch A, El, Bounkari O, Dutta P, Hauser H, Tamura T. Identification of mRNAs that are spliced but not exported to the cytoplasm in the absence of THOC5 in mouse embryo fibroblasts. RNA. 2011;17:1048–1056. doi: 10.1261/rna.2607011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kopytova DV, Orlova AV, Krasnov AN, Gurskiy DY, Nikolenko JV, Nabirochkina EN, Shidlovskii YV, Georgieva SG. Multifunctional factor ENY2 is associated with the THO complex and promotes its recruitment onto nascent mRNA. Genes Dev. 2010;24:86–96. doi: 10.1101/gad.550010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Katahira J, Inoue H, Hurt E, Yoneda Y. Adaptor Aly and co-adaptor Thoc5 function in the Tap-p15-mediated nuclear export of HSP70 mRNA. EMBO J. 2009;28:556–567. doi: 10.1038/emboj.2009.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dominguez-Sanchez MS, Barroso S, Gomez-Gonzalez B, Luna R, Aguilera A. Genome instability and transcription elongation impairment in human cells depleted of THO/TREX. PLoS Genet. 2011;7:e1002386. doi: 10.1371/journal.pgen.1002386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dieci G, Preti M, Montanini B. Eukaryotic snoRNAs: a paradigm for gene expression flexibility. Genomics. 2009;94:83–88. doi: 10.1016/j.ygeno.2009.05.002. [DOI] [PubMed] [Google Scholar]
- 18.Matera AG, Terns RM, Terns MP. Non-coding RNAs: lessons from the small nuclear and small nucleolar RNAs. Nat. Rev. 2007;8:209–220. doi: 10.1038/nrm2124. [DOI] [PubMed] [Google Scholar]
- 19.Grzechnik P, Kufel J. Polyadenylation linked to transcription termination directs the processing of snoRNA precursors in yeast. Mol. Cell. 2008;32:247–258. doi: 10.1016/j.molcel.2008.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lemay JF, D'Amours A, Lemieux C, Lackner DH, St-Sauver VG, Bahler J, Bachand F. The nuclear poly(A)-binding protein interacts with the exosome to promote synthesis of noncoding small nucleolar RNAs. Mol. Cell. 2010;37:34–45. doi: 10.1016/j.molcel.2009.12.019. [DOI] [PubMed] [Google Scholar]
- 21.Dziembowski A, Lorentzen E, Conti E, Seraphin B. A single subunit, Dis3, is essentially responsible for yeast exosome core activity. Nat. Struct. Mol. Biol. 2007;14:15–22. doi: 10.1038/nsmb1184. [DOI] [PubMed] [Google Scholar]
- 22.Lebreton A, Tomecki R, Dziembowski A, Seraphin B. Endonucleolytic RNA cleavage by a eukaryotic exosome. Nature. 2008;456:993–996. doi: 10.1038/nature07480. [DOI] [PubMed] [Google Scholar]
- 23.Schaeffer D, Tsanova B, Barbas A, Reis FP, Dastidar EG, Sanchez-Rotunno M, Arraiano CM, van Hoof A. The exosome contains domains with specific endoribonuclease, exoribonuclease and cytoplasmic mRNA decay activities. Nat. Struct. Mol. Biol. 2009;16:56–62. doi: 10.1038/nsmb.1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schneider C, Leung E, Brown J, Tollervey D. The N-terminal PIN domain of the exosome subunit Rrp44 harbors endonuclease activity and tethers Rrp44 to the yeast core exosome. Nucleic Acids Res. 2009;37:1127–1140. doi: 10.1093/nar/gkn1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tomecki R, Kristiansen MS, Lykke-Andersen S, Chlebowski A, Larsen KM, Szczesny RJ, Drazkowska K, Pastula A, Andersen JS, Stepien PP, et al. The human core exosome interacts with differentially localized processive RNases: hDIS3 and hDIS3L. EMBO J. 2010;29:2342–2357. doi: 10.1038/emboj.2010.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Allmang C, Petfalski E, Podtelejnikov A, Mann M, Tollervey D, Mitchell P. The yeast exosome and human PM-Scl are related complexes of 3' –> 5' exonucleases. Genes Dev. 1999;13:2148–2158. doi: 10.1101/gad.13.16.2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Callahan KP, Butler JS. Evidence for core exosome independent function of the nuclear exoribonuclease Rrp6p. Nucleic Acids Res. 2008;36:6645–6655. doi: 10.1093/nar/gkn743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu Q, Greimann JC, Lima CD. Reconstitution, activities, and structure of the eukaryotic RNA exosome. Cell. 2006;127:1223–1237. doi: 10.1016/j.cell.2006.10.037. [DOI] [PubMed] [Google Scholar]
- 29.LaCava J, Houseley J, Saveanu C, Petfalski E, Thompson E, Jacquier A, Tollervey D. RNA degradation by the exosome is promoted by a nuclear polyadenylation complex. Cell. 2005;121:713–724. doi: 10.1016/j.cell.2005.04.029. [DOI] [PubMed] [Google Scholar]
- 30.Vanacova S, Wolf J, Martin G, Blank D, Dettwiler S, Friedlein A, Langen H, Keith G, Keller W. A new yeast poly(A) polymerase complex involved in RNA quality control. PLoS Biol. 2005;3:e189. doi: 10.1371/journal.pbio.0030189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Win TZ, Draper S, Read RL, Pearce J, Norbury CJ, Wang SW. Requirement of fission yeast Cid14 in polyadenylation of rRNAs. Mol. Cell. Biol. 2006;26:1710–1721. doi: 10.1128/MCB.26.5.1710-1721.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wyers F, Rougemaille M, Badis G, Rousselle JC, Dufour ME, Boulay J, Regnault B, Devaux F, Namane A, Seraphin B, et al. Cryptic pol II transcripts are degraded by a nuclear quality control pathway involving a new poly(A) polymerase. Cell. 2005;121:725–737. doi: 10.1016/j.cell.2005.04.030. [DOI] [PubMed] [Google Scholar]
- 33.Keller C, Woolcock K, Hess D, Buhler M. Proteomic and functional analysis of the noncanonical poly(A) polymerase Cid14. RNA. 2010;16:1124–1129. doi: 10.1261/rna.2053710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bahler J, Wu JQ, Longtine MS, Shah NG, McKenzie A, Steever AB, Wach A, Philippsen P, Pringle JR. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast. 1998;14:943–951. doi: 10.1002/(SICI)1097-0061(199807)14:10<943::AID-YEA292>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 35.Lemieux C, Marguerat S, Lafontaine J, Barbezier N, Bahler J, Bachand F. A Pre-mRNA degradation pathway that selectively targets intron-containing genes requires the nuclear poly(A)-binding protein. Mol. Cell. 2011;44:108–119. doi: 10.1016/j.molcel.2011.06.035. [DOI] [PubMed] [Google Scholar]
- 36.Adam M, Robert F, Larochelle M, Gaudreau L. H2A.Z is required for global chromatin integrity and for recruitment of RNA polymerase II under specific conditions. Mol. Cell. Biol. 2001;21:6270–6279. doi: 10.1128/MCB.21.18.6270-6279.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lemieux C, Bachand F. Cotranscriptional recruitment of the nuclear poly(A)-binding protein Pab2 to nascent transcripts and association with translating mRNPs. Nucleic Acids Res. 2009;37:3418–3430. doi: 10.1093/nar/gkp207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Perreault A, Gascon S, D'Amours A, Aletta JM, Bachand F. A methyltransferase-independent function for Rmt3 in ribosomal subunit homeostasis. J. Biol. Chem. 2009;284:15026–15037. doi: 10.1074/jbc.M109.004812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim DU, Hayles J, Kim D, Wood V, Park HO, Won M, Yoo HS, Duhig T, Nam M, Palmer G, et al. Analysis of a genome-wide set of gene deletions in the fission yeast Schizosaccharomyces pombe. Nat. Biotechnol. 2010;28:617–623. doi: 10.1038/nbt.1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Luna R, Jimeno S, Marin M, Huertas P, Garcia-Rubio M, Aguilera A. Interdependence between transcription and mRNP processing and export, and its impact on genetic stability. Mol. Cell. 2005;18:711–722. doi: 10.1016/j.molcel.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 41.Abruzzi KC, Lacadie S, Rosbash M. Biochemical analysis of TREX complex recruitment to intronless and intron-containing yeast genes. EMBO J. 2004;23:2620–2631. doi: 10.1038/sj.emboj.7600261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dufu K, Livingstone MJ, Seebacher J, Gygi SP, Wilson SA, Reed R. ATP is required for interactions between UAP56 and two conserved mRNA export proteins, Aly and CIP29, to assemble the TREX complex. Genes Dev. 2010;24:2043–2053. doi: 10.1101/gad.1898610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Anderson JT, Wang X. Nuclear RNA surveillance: no sign of substrates tailing off. Crit. Rev. Biochem. Mol. Biol. 2009;44:16–24. doi: 10.1080/10409230802640218. [DOI] [PubMed] [Google Scholar]
- 44.Butler JS, Mitchell P. Rrp6, Rrp47 and cofactors of the nuclear exosome. Adv. Exp. Med. Biol. 2010;702:91–104. [PubMed] [Google Scholar]
- 45.Murakami H, Goto DB, Toda T, Chen ES, Grewal SI, Martienssen RA, Yanagida M. Ribonuclease activity of Dis3 is required for mitotic progression and provides a possible link between heterochromatin and kinetochore function. PLoS One. 2007;2:e317. doi: 10.1371/journal.pone.0000317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mason PB, Struhl K. Distinction and relationship between elongation rate and processivity of RNA polymerase II in vivo. Mol. Cell. 2005;17:831–840. doi: 10.1016/j.molcel.2005.02.017. [DOI] [PubMed] [Google Scholar]
- 47.Rondon AG, Jimeno S, Garcia-Rubio M, Aguilera A. Molecular evidence that the eukaryotic THO/TREX complex is required for efficient transcription elongation. J. Biol. Chem. 2003;278:39037–39043. doi: 10.1074/jbc.M305718200. [DOI] [PubMed] [Google Scholar]
- 48.Gomez-Gonzalez B, Garcia-Rubio M, Bermejo R, Gaillard H, Shirahige K, Marin A, Foiani M, Aguilera A. Genome-wide function of THO/TREX in active genes prevents R-loop-dependent replication obstacles. EMBO J. 2011;30:3106–3119. doi: 10.1038/emboj.2011.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Glover-Cutter K, Kim S, Espinosa J, Bentley DL. RNA polymerase II pauses and associates with pre-mRNA processing factors at both ends of genes. Nat. Struct. Mol. Biol. 2008;15:71–78. doi: 10.1038/nsmb1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gromak N, West S, Proudfoot NJ. Pause sites promote transcriptional termination of mammalian RNA polymerase II. Mol. Cell. Biol. 2006;26:3986–3996. doi: 10.1128/MCB.26.10.3986-3996.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chanarat S, Seizl M, Strasser K. The Prp19 complex is a novel transcription elongation factor required for TREX occupancy at transcribed genes. Genes Dev. 2011;25:1147–1158. doi: 10.1101/gad.623411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sipiczki M. Where does fission yeast sit on the tree of life? Genome Biol. 2000;1 doi: 10.1186/gb-2000-1-2-reviews1011. REVIEWS1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mitchell P, Petfalski E, Houalla R, Podtelejnikov A, Mann M, Tollervey D. Rrp47p is an exosome-associated protein required for the 3' processing of stable RNAs. Mol. Cell. Biol. 2003;23:6982–6992. doi: 10.1128/MCB.23.19.6982-6992.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.van Hoof A, Lennertz P, Parker R. Yeast exosome mutants accumulate 3'-extended polyadenylated forms of U4 small nuclear RNA and small nucleolar RNAs. Mol. Cell. Biol. 2000;20:441–452. doi: 10.1128/mcb.20.2.441-452.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vasiljeva L, Buratowski S. Nrd1 interacts with the nuclear exosome for 3' processing of RNA polymerase II transcripts. Mol. Cell. 2006;21:239–248. doi: 10.1016/j.molcel.2005.11.028. [DOI] [PubMed] [Google Scholar]
- 56.Luna R, Rondon AG, Aguilera A. New clues to understand the role of THO and other functionally related factors in mRNP biogenesis. Biochim. Biophys. Acta. 2012;1819:514–520. doi: 10.1016/j.bbagrm.2011.11.012. [DOI] [PubMed] [Google Scholar]
- 57.Johnson SA, Kim H, Erickson B, Bentley DL. The export factor Yra1 modulates mRNA 3' end processing. Nat. Struct. Mol. Biol. 2011;18:1164–1171. doi: 10.1038/nsmb.2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang K, Fischer T, Porter RL, Dhakshnamoorthy J, Zofall M, Zhou M, Veenstra T, Grewal SI. Clr4/Suv39 and RNA quality control factors cooperate to trigger RNAi and suppress antisense RNA. Science. 2011;331:1624–1627. doi: 10.1126/science.1198712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jensen TH, Boulay J, Olesen JR, Colin J, Weyler M, Libri D. Modulation of transcription affects mRNP quality. Mol. Cell. 2004;16:235–244. doi: 10.1016/j.molcel.2004.09.019. [DOI] [PubMed] [Google Scholar]
- 60.Milligan L, Decourty L, Saveanu C, Rappsilber J, Ceulemans H, Jacquier A, Tollervey D. A yeast exosome cofactor, Mpp6, functions in RNA surveillance and in the degradation of noncoding RNA transcripts. Mol. Cell. Biol. 2008;28:5446–5457. doi: 10.1128/MCB.00463-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lam YW, Lamond AI, Mann M, Andersen JS. Analysis of nucleolar protein dynamics reveals the nuclear degradation of ribosomal proteins. Curr. Biol. 2007;17:749–760. doi: 10.1016/j.cub.2007.03.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.