Abstract
We examined the potential use of natural-abundance stable carbon isotope ratios of lipids for determining substrate usage by sulfate-reducing bacteria (SRB). Four SRB were grown under autotrophic, mixotrophic, or heterotrophic growth conditions, and the δ13C values of their individual fatty acids (FA) were determined. The FA were usually 13C depleted in relation to biomass, with Δδ13C(FA − biomass) of −4 to −17‰; the greatest depletion occurred during heterotrophic growth. The exception was Desulfotomaculum acetoxidans, for which substrate limitation resulted in biomass and FA becoming isotopically heavier than the acetate substrate. The δ13C values of FA in Desulfotomaculum acetoxidans varied with the position of the double bond in the monounsaturated C16 and C18 FA, with FA becoming progressively more 13C depleted as the double bond approached the methyl end. Mixotrophic growth of Desulfovibrio desulfuricans resulted in little depletion of the i17:1 biomarker relative to biomass or acetate, whereas growth with lactate resulted in a higher proportion of i17:1 with a greater depletion in 13C. The relative abundances of 10Me16:0 in Desulfobacter hydrogenophilus and Desulfobacterium autotrophicum were not affected by growth conditions, yet the Δδ13C(FA − substrate) values of 10Me16:0 were considerably greater during autotrophic growth. These experiments indicate that FA δ13C values can be useful for interpreting carbon utilization by SRB in natural environments.
Sulfate-reducing bacteria (SRB) are common and functionally important members of anaerobic microbial communities. SRB populations can be identified and quantified by using molecular and phylogenetic approaches (8, 20, 25). When combined with measurements of rates of biogeochemical cycling and with an understanding of the physiology of these microbes through isolation and pure-culture studies, these approaches can reveal much about the roles of SRB in a variety of habitats (5, 19, 23, 33). Although the use of stable sulfur isotopes and sensitive measurements of sulfide production have addressed the role of SRB in sulfur transformations (9, 24), little is known about the direct effect of these organisms on carbon flow in microbial communities. It is generally well recognized that SRB are important in reprocessing organic matter and can degrade a wide variety of organic substrates. Several SRB can also grow autotrophically in the absence of organic compounds (4, 29, 38). Molecular genetic and physiological studies can indicate which SRB are present in an environment as well as their potential capabilities, but such studies do not determine which carbon transformations the SRB are actually carrying out in a particular environment. The use of stable- or radioisotope-labeled substrates can reveal flows and rates of transformations (2, 3, 26) but are limited in application, because their use might alter the natural abundance of key substrates and our ability to introduce these substrates into natural environments is limited. However, the study of stable isotopic compositions, at natural abundance, is a potential way to circumvent this problem and to determine the carbon substrates actually used by SRB.
We had previously grown four different SRB and determined the isotope fractionation factors for biomass production during autotrophic, mixotrophic, and heterotrophic growth (17). In order to use stable carbon isotopes to assess the mode of growth of SRB in natural environments, we needed an analysis that was more specific than bulk biomass. SRB are typical bacteria in that their membranes are composed primarily of phospholipids with ester-bound fatty acids (PLFA) that can be analyzed as fatty acid methyl esters (FAME). Several genera of SRB possess PLFA with unusual structures that are useful as biomarkers (1, 32, 35), and we have for the first time determined the δ13C values of individual fatty acids (FA) in these SRB. Lipids are generally depleted in 13C relative to bulk biomass as a consequence of the biosynthetic pathways for FA (10). If the isotopic discrimination during heterotrophic and autotrophic growth of SRB were known, it might be possible to correlate the isotopic compositions of the lipids with the compositions of the organic substrates used during heterotrophic growth. This has been done previously with other organisms (1, 4, 12, 16). Also, it might be possible to distinguish between autotrophic and heterotrophic growth of a particular group of organisms. However, it is first necessary to determine how the isotopic composition of the lipids relates to the biomass as a whole and to growth substrates, as well as to determine whether there are species-specific variations that might affect potential depletion during synthesis of FA.
We have analyzed the isotopic composition of FAME from four species of SRB and correlated the results to the substrates and modes of growth. The organisms were selected as representatives of diverse phylogenetic groups that use different pathways for autotrophic, mixotrophic (acetate plus H2 plus CO2), and heterotrophic growth (Table 1) (17, 28-30). We correlate FA structure with the variations in isotopic compositions between FA within each organism and discuss the implications for biosynthetic fractionations during synthesis. We also discuss the usefulness of stable isotope analysis of FA for determining the identity and functionality of SRB in natural environments.
TABLE 1.
Characteristics of SRB used in culture study
| SRB | Gram stain result | Autotrophic pathway | Auto- or mixotrophic substrate | Heterotrophic substrate | Potential FA biomarker |
|---|---|---|---|---|---|
| Desulfovibrio desulfuricans | − | Not applicable | Acetate + H2 + CO2 | Lactate | i17:1 |
| Desulfotomaculum acetoxidans | + | CODH | H2 + CO2 | Acetate | None |
| Desulfobacter hydrogenophilus | − | Reverse TCA | H2 + CO2 | Acetate | 10Me16:0 |
| Desulfobacterium autotrophicum | − | CODH | H2 + CO2 | Acetate | 10Me16:0 |
MATERIALS AND METHODS
Organisms and cultivation.
Desulfobacterium autotrophicum (ATCC 43914), Desulfobacter hydrogenophilus (ATCC 43915), Desulfovibrio desulfuricans (ATCC 29577), and Desulfotomaculum acetoxidans (ATCC 49208) were obtained from the American Type Culture Collection. They were grown and harvested as previously described (17). Briefly, three of the four organisms were grown heterotrophically with a limiting amount of acetate and lithotrophically with H2 and CO2. Desulfovibrio desulfuricans was grown mixotrophically with H2 plus CO2 plus acetate or heterotrophically with lactate. Each of these eight cultures was then used to inoculate three identical cultures for the experiment. One of each set of triplicate cultures was harvested within 1 h to measure the contribution of inoculum. Growth in the remaining duplicate cultures was monitored by substrate consumption and sulfate reduction, the cells were harvested in early stationary phase by centrifugation, and they were dried by lyophilization. One of the autotrophic replicates of Desulfobacter did not grow because it became oxidized, and one replicate of Desulfobacterium grew well but some of the cells appeared to have autolysed prior to harvesting.
Extraction and derivatization.
Dried cells (50 mg) were extracted by a modified Bligh and Dyer procedure as previously described (13). Half of the total lipid extract was then subjected to mild alkaline methanolysis to prepare FAME, and these were purified by preparative thin-layer chromatography (TLC) as previously described (13). Diarachidoyl phosphatidylcholine was included as an internal standard. FAME were recovered from the TLC plate and were resuspended in methylene chloride with the methyl ester of 13:0 (Sigma Chemical Co.) added as an internal isotopic standard (measured δ13C of −30.68‰). The bond positions of the monounsaturated FAME were determined as previously described by analyzing their dimethyl disulfide adducts (12).
GC-MS analysis.
The identities of the FAME were determined by comparison of retention times and spectra to known FAME standards by gas chromatography-mass spectrometry (GC-MS) using a Finnigan GCQ GC-MS system. The oven program was 150°C for 1 min, increased to 190°C at 1°C/min followed by 25°C/min to 290°C, with He at a constant velocity of 30 cm/s through a DB-5MS column (30 m by 0.25 μ by 0.25 mm; J&W Scientific). The standard nomenclature for FA was used. FA are designated X:YΔZ, where X is the number of carbon atoms, Y is the number of double bonds, and Z is the position of the double bond from the carboxyl end. Cyclopropyl rings are designated cy, and the prefixes i and a refer to iso- and anteisomethyl branching, whereas mid-chain methyl branches are designated by the position from the carboxyl end followed by Me.
GC-C-isotope ratio MS analyses.
The δ13C values of the FAME were determined by GC-C-isotope ratio MS by using a Finnigan-MAT Delta Plus system. The oven program was 150°C increased to 175°C at 0.5°C/min and then to 200°C at 5°C/min followed by 25°C/min to 300°C, with He at a constant flow of 2 ml/min through a HP-5 column (30 m by 0.25 μ by 0.25 mm; Agilent). The isotope ratios for individual compounds are the means and standard deviations of at least three analyses. The isotopic compositions of FAME were corrected to the internal isotopic standard (13:0) and for the additional carbon atom from the methanol (δ13C = −39‰) derivatizing reagent. The isotopic compositions are given as δ values (δ13C, in per mille) relative to the Peedee Belemnite standard. Isotope values were within 2‰ of values measured by bomb combustion, and standard deviations were always less than ±2‰ and were usually less than 0.5‰ for the external standards (mixture of 13:0, 17:0, and 20:0) and the internal standards (13:0 and 20:0). The amounts of FAME were calculated from peak areas relative to the 20:0 internal standard.
RESULTS
Inocula.
The cells used as inocula contributed to the overall isotopic composition of biomass and FAME in the experimental cultures. The total amount of FAME from the inoculum cultures was less than half of the final amount from the experimental cultures. Also, the δ13C values of the FAME from the inocula and the experimental cultures differed by less than 1‰. Therefore, corrections for the isotopic compositions of the FAME from the inocula were not necessary.
Desulfovibrio desulfuricans.
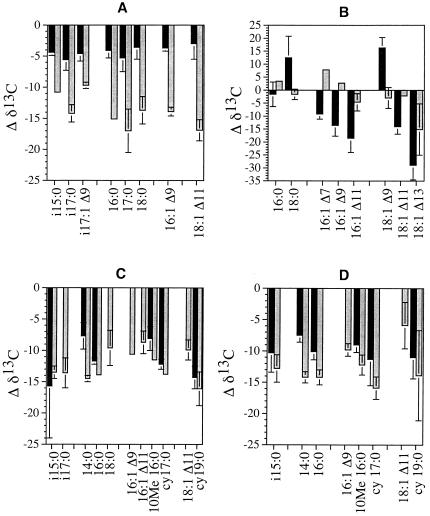
The relative proportions of the individual FAME in Desulfovibrio desulfuricans cultures differed with mode of growth. Cells grown heterotrophically with lactate produced more branched, odd-chained FAME, including the distinctive biomarker i17:1Δ9 (Table 2). There were minor variations in the δ13C values for the FA relative to biomass from Desulfovibrio desulfuricans grown mixotrophically and heterotrophically, as shown in Fig. 1A. In cells grown mixotrophically the δ13C of the FA ranged from −35.1 to −37.7‰, but there was no correlation between isotopic values and structural differences related to FA biosynthetic mechanisms. Cells grown with lactate had a broader range of isotopic values (−39.0 to −46.2‰), and the branched, odd-chain FA were isotopically heavier than the normal-chain FA. As a weighted average, the Δδ13C(FA − biomass) was greater for cells grown heterotrophically (Table 3).
TABLE 2.
Comparison of FAME compositions of four SRB grown under two conditionsd
| FAb | Distribution (%)a of FAME for:
|
|||||||
|---|---|---|---|---|---|---|---|---|
|
Desulfovibrio desulfuricans
|
Desulfotomaculum acetoxidans
|
Desulfobacter hydrogenophilus
|
Desulfobacterium autotrophicum
|
|||||
| Acetate | Lactate | CO2 | Acetate | CO2c | Acetate | CO2c | Acetate | |
| 14:0 | ND | ND | ND | ND | 2.3 | 4.4 | 6.3 | 6.4 |
| i15:0 | 8.9 | 23.0 | ND | ND | 2.4 | 3.2 | 2.4 | 4.0 |
| 16:1Δ7 | ND | ND | 4.1 | 3.7 | <0.1 | 0.6 | ND | ND |
| 16:1Δ9 | 19.2 | 10.2 | 11.8 | 24.8 | <0.1 | 9.4 | <0.1 | 12.5 |
| 16:1Δ11 | ND | ND | 3.9 | 2.2 | <0.1 | 1.2 | ND | ND |
| 16:0 | 33.1 | 17.7 | 23.4 | 27.1 | 48.1 | 40.8 | 47.8 | 41.0 |
| i17:1Δ9 | 26.7 | 39.5 | ND | ND | ND | ND | ND | ND |
| i17:0 | 4.6 | 7.8 | ND | ND | <0.1 | 0.7 | ND | ND |
| 10Me16:0 | ND | ND | ND | ND | 5.9 | 7.8 | 5.6 | 6.7 |
| cy17:0 | ND | ND | ND | ND | 35.3 | 28.8 | 35.8 | 27.6 |
| 17:0 | 1.2 | 0.4 | ND | ND | ND | ND | ND | ND |
| 18:1Δ9 | ND | ND | 5.2 | 0.9 | ND | ND | ND | ND |
| 18:1Δ11 | 2.6 | 0.6 | 46.5 | 39.6 | <0.1 | 1.9 | <0.1 | 1.1 |
| 18:1Δ13 | ND | ND | 2.2 | 0.5 | ND | ND | ND | ND |
| 18:0 | 3.7 | 0.7 | 2.7 | 1.2 | <0.1 | 0.3 | ND | ND |
| cy19:0 | ND | ND | ND | ND | 6.1 | 1.0 | 2.1 | 0.6 |
Average of the amount of each FAME relative to total amount of FA from duplicate cultures (standard deviation <2%). ND, not detected.
FA nomenclature designates the number of carbons, with iso- (i) or 10-methyl branching (10Me), cyclopropyl ring (cy), or a cis double bond (:1) in the Δ position calculated relative to the carboxyl end.
Calculated from replicate analyses of one culture.
The Desulfovibrio isolate was grown mixotrophically with acetate or heterotrophically with lactate, whereas Desulfotomaculum, Desulfobacter, and Desulfobacterium isolates were grown either autotrophically or heterotrophically with acetate.
FIG. 1.
Δδ13C between FA and biomass of Desulfovibrio desulfuricans grown mixotrophically with CO2 plus acetate (black) or heterotrophically with lactate (gray) (A); Desulfotomaculum acetoxidans grown autotrophically with CO2 (black) or heterotrophically with acetate (gray) (B); Desulfobacter hydrogenophilus grown autotrophically with CO2 (black) or heterotrophically with acetate (gray) (C); and Desulfobacterium autotrophicum grown autotrophically with CO2 (black) or heterotrophically with acetate (gray) (D). Values are averages of three analyses each of duplicate cultures, and error bars indicate ±1 standard deviation.
TABLE 3.
δ13C of biomass and FA of four SRB under two different growth conditions
| Organism | Growth condition | δ13C organic substratea | δ13C CO2 | δ13C biomass | δ13C FAb | Δ δ13C FA to substratec | Δ δ13C FA to biomass |
|---|---|---|---|---|---|---|---|
| Desulfovibrio desulfuricans | Mixotrophic | −34.2 ± 0.8 | −27.4 ± 0.1 | −32.1 ± 0.2 | −36.2 ± 1.0 | −2.0 | −4.1 |
| Heterotrophic | −29.1 ± 0.8 | −29.3 ± 0.1 | −41.0 ± 0.3 | −11.9 | −11.7 | ||
| Desulfotomaculum acetoxidans | Autotrophic | −27.1 ± 0.6 | −55.6 ± 1.5 | −64.4 ± 3.4 | −37.3 | −8.8 | |
| Heterotrophic | −34.2 ± 0.8 | −25.4 ± 0.9 | −24.7 ± 1.3 | 9.5 | 0.7 | ||
| Desulfobacter hydrogenophilus | Autotrophic | −24.5 ± 0.3 | −40.4 ± 0.3 | −52.2 ± 0.3 | −24.4 | −11.8 | |
| Heterotrophic | −34.2 ± 0.8 | −34.9 ± 0.6 | −48.1 ± 0.4 | −13.9 | −13.3 | ||
| Desulfobacterium autotrophicum | Autotrophic | −27.3 ± 0.2 | −36.9 ± 0.4 | −47.4 ± 2.4 | −20.1 | −10.5 | |
| Heterotrophic | −34.2 ± 0.8 | −34.8 ± 0.3 | −48.6 ± 1.2 | −14.4 | −13.8 |
All δ13C values are expressed per mille (‰).
δ13C FA is the weighted average of the isotopic values of the individual FA.
Δ δ13C is the difference between FA and the CO2 for autotrophic growth, acetate or lactate for heterotrophic growth, or acetate for mixotrophic growth.
Desulfotomaculum acetoxidans.
The FAME in Desulfotomaculum acetoxidans consisted of straight-chain normal and monounsaturated FA 16 or 18 carbon atoms long, dominated by 16:0, 16:1Δ9, and 18:1Δ11. Cells grown autotrophically had relatively less 16:1Δ9 and more 18:1Δ11 than cells grown heterotrophically (Table 2). Compared to biomass, the δ13C values for the FA were isotopically heavier for cells grown with acetate (Fig. 1B). As a weighted average, the FA were 8.8‰ lighter than biomass for cells grown autotrophically but were 0.7‰ heavier than biomass for cells grown with acetate (Table 3). There were large variations in δ13C for FA related to chemical structure, especially for cells grown autotrophically, in which the greatest range of isotopic values was observed. On average the saturated FA were the least 13C depleted and the monounsaturated FA were the most depleted (Fig. 1B). The position of unsaturation also affected the δ13C values of the FA, which became progressively 13C depleted as the double bond approached the methyl end. This trend was most pronounced for the longer (C18) FA. Overall, the C18 FA were usually more 13C depleted than the corresponding C16 FA (Fig. 1B).
Desulfobacter hydrogenophilus and Desulfobacterium autotrophicum.
The FAME detected in Desulfobacter hydrogenophilus consisted of a wide variety of structures, including normal, branched, and unsaturated FAME 14 to 19 carbon atoms long, with a greater variety of FAME when grown with acetate (Table 2). The FAME from Desulfobacterium autotrophicum and Desulfobacter hydrogenophilus were similar, except that Desulfobacterium autotrophicum contained relatively more 14:0 and less cyclic FAME (Table 2). For both organisms the dominant FAME were 16:0 and cy17:0. In cells grown with acetate, 14:0, 16:1Δ9, and the biomarker 10Me16:0 were also found in moderate amounts. For Desulfobacter hydrogenophilus, the FA δ13C values were more depleted relative to biomass under heterotrophic (−13.3‰) than under autotrophic (−11.8‰) growth conditions (Table 3). In contrast, FA from autotrophic cells were considerably more depleted relative to substrate than heterotrophic cells (−24.4 and −13.9‰, respectively), which reflects the greater fractionation associated with autotrophic growth (Table 3). The same patterns were observed with Desulfobacterium autotrophicum. Differences in δ13C of the six different FA in autotrophically grown cells of either organism were not significantly dependent on structure (Fig. 1C and D). However, when these organisms were grown with acetate, the cyclopropane FA, including the dominant cy17:0, were among the most depleted FA, whereas the even-chained unsaturated FA were more 13C enriched than the normal or branched chain FA (Fig. 1C and D).
DISCUSSION
The FAME profiles for the four SRB were distinct from each other and are generally consistent with previous reports (6, 16, 32, 35). Small variations in relative proportions compared to those of previous studies are likely due to subtle differences in growth conditions. The exception was that a previous study with Desulfobacter hydrogenophilus (16) had identified two FA as 17:1 and 19:1 by retention time alone. We identified these FA as cy17:0 and cy19:0, based on detailed analysis of retention times with two chromatographic procedures, the absence of dimethyl disulfide adduct formation during double-bond analysis of our extracts, and direct comparison to the FA of Desulfobacterium autotrophicum. The overall similarity of the FAME profiles between these two SRB is consistent with their close phylogenetic relationship. On the other hand, Desulfotomaculum acetoxidans was unusual because it exhibited only straight-chain saturated and monounsaturated C16 and C18 FAME, and it produced FAME with a series of double-bond isomers. The abundance of unsaturated FAME in Desulfotomaculum acetoxidans is atypical of gram-positive bacteria, which generally increase the fluidity of their cell membranes by increasing the relative amount of branched or alicyclic FA (15).
The FA of the SRB were usually depleted in 13C relative to the biomass from which they were derived. This pattern is consistent with previous analyses of other microbes and is a result of biosynthetic fractionations during lipid synthesis (10, 22, 40). These SRB obtained acetyl-coenzyme A (CoA) for FA synthesis directly from the acetate provided or as the immediate product of autotrophic growth, unlike other heterotrophs that acquire acetyl-CoA units by decarboxylation of pyruvate during glucose degradation. By supplying simple substrates we were able to study isotopic discrimination during lipid synthesis relative to both the immediate substrates and the bulk biomass. There was no overall correlation of the isotopic differences between the FA and the various organic or inorganic substrates (Table 3). For example, the Δδ13C between FA and biomass was not consistent for the four SRB. Surprisingly, the differences were not due to the modes of growth or to the metabolic pathways for carbon assimilation. This can be seen by comparing Desulfobacterium autotrophicum and Desulfobacter hydrogenophilus, which use different carbon assimilation pathways (carbon monoxide dehydrogenase [CODH] versus reverse tricholoroacetic acid [TCA] cycle) and therefore use different mechanisms for generating acetyl-CoA units. On average, these organisms exhibited Δδ13C between biomass and FA of approximately −11‰ (CODH) and −12‰ (reverse TCA cycle), a few per mille lighter when they were grown heterotrophically. In contrast, Desulfotomaculum acetoxidans, which uses the same pathway as Desulfobacterium autotrophicum (CODH), had FA 9‰ lighter than biomass during autotrophic growth but had FA that were slightly enriched in 13C compared to biomass during heterotrophic growth. Heterotrophic growth of Desulfovibrio desulfuricans with lactate produced FA 12‰ lighter than biomass, yet mixotrophic growth with acetate yielded FA only 4‰ lighter than biomass. Therefore, there was no consistent relationship between the δ13C of FA and biomass for any of the individual SRB or for the group as a whole.
The Δδ13C between FA and biomass during heterotrophic growth is primarily due to isotope fractionation at branching points for acetyl-CoA use. FA from cells grown with abundant acetate should have the same isotopic composition as the acetate provided, because FA biosynthesis itself is not associated with isotopic fractionation (10). Indeed, Desulfovibrio desulfuricans grown mixotrophically with acetate supplied in excess had FA depleted by 2‰ relative to acetate, the immediate source of acetyl-CoA units. The other three SRB received acetate as a limiting substrate, but fractionation associated with biosynthesis of lipid biomarkers relative to associated biomass are not generally affected by substrate limitation (14, 31), and there is no observed isotope effect during uptake of acetate (17). Both Desulfobacterium autotrophicum and Desulfobacter hydrogenophilus produced FA that were 13 to 14‰ depleted relative to either the acetate or the bulk biomass, suggesting that the selection of acetyl-CoA for FA synthesis discriminated against 13C, whereas oxidation to CO2 did not. The unusual exception was Desulfotomaculum acetoxidans, in which both the biomass and FA were 13C enriched relative to the acetate substrate. The growth conditions for this organism led to an unusual situation of substantial isotopic enrichment due to substrate limitation. In this case, the oxidation of the acetyl-CoA for energy production must have had an isotope effect, effectively dissimilating 12C preferentially and thus increasing the 13C/12C value of the remaining acetyl-CoA pool that was then utilized for biosynthesis of FA and other cell components. This interpretation is supported by the fact that the FA differed from the biomass by less than 1‰. Isotopic enrichment in FA has been observed previously during autotrophic growth of Chlorobium limicola, which uses the reverse TCA cycle (37). Therefore, the isotopic composition of FA relative to biomass and organic substrate is influenced primarily by isotope discrimination at the branching points from which acetyl-CoA flows either into FA biosynthesis or into pathways for energy production. These pathways and their associated discrimination may vary widely among prokaryotic microorganisms.
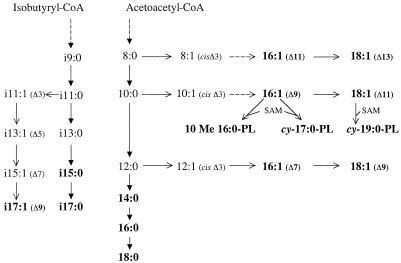
Isotopic differences between FAME within a given lipid assemblage were small for most organisms. This is surprising, given the complex reaction networks associated with biosynthesis of the FA in these SRB. The greatest variation in isotopic composition among FA was observed in Desulfotomaculum acetoxidans grown with CO2. Here, individual FA varied by as much as 29‰ from the biomass, and the position of unsaturation in the FA showed a pronounced isotopic pattern. The pattern was similar for C16 and C18 FA (Fig. 1B), with the unsaturated FA becoming progressively lighter as the double-bond position approached the aliphatic end. The biosynthesis of unsaturated FA by Desulfotomaculum acetoxidans and the other SRB is assumed to occur via the anaerobic mechanism. As shown in Fig. 2, the enzyme that introduces the unsaturation prefers a C10 substrate but can also use C8 or C12 precursors, leading to the formation of Δ11, Δ9, or Δ7 isomers of 16:1. These can subsequently be extended to Δ13, Δ11, or Δ9 isomers of 18:1 (18). The shorter the substrate, the more depleted in 13C the resulting FA was, indicating an enormous selectivity for 13C-depleted substrate by this enzyme. The C18 FA were generally lighter than the C16 FA, perhaps indicating that further discrimination occurs during the addition of the final two-carbon unit. The synthesis of unsaturated FA was not expected to lead to isotopic fractionation (10). Indeed, no such effect was found in the unsaturated FA of heterotrophically grown Desulfobacter hydrogenophilus in our study (Fig. 1C). Nevertheless, our data clearly indicate that the biosynthesis of unsaturated FA can lead to structurally related products having markedly different δ13C values within an organism.
FIG. 2.
Summary of FA biosynthesis in SRB. Starting with the chain initiator acetoacetyl-CoA at the top, the chain is extended by the addition of two-carbon acetyl groups. At certain branch points, competition between enzymes that catalyze the normal extension and the formation of cis double bonds determines the relative distribution of unsaturated isomers. Unsaturated FA are elongated by the addition of two carbon units. The 10Me16:0 and cyclopropane (cy) FA arise by in situ modifications of phospholipid (PL)-bound FA in the membrane by an addition of a methyl group from S-adenoyl-methionine (SAM) to 16:1Δ9 or 18:1Δ11. The branched, odd-chain FA are synthesized from isobutyryl-CoA-derived precursors, as shown on the left. The formation of the unsaturated branched FA i17:1 (Δ9) occurs by an analogous mechanism to the formation of 16:1 (Δ9) by the anaerobic pathway.
The δ13C values of SRB FA biomarkers might be valuable for ecological diagnostics. Desulfobacterium autotrophicum and Desulfobacter hydrogenophilus had very similar lipid profiles, and the δ13C of the dominant FA were also quite similar relative to biomass and substrate δ13C values. Although the assimilation pathway or growth rate led to differences of several per mille for biomass of autotrophically grown cells, the difference in Δδ13C(FA − substrate) for the biomarker 10Me16:0 was −12.2 and −13.0‰ for growth with acetate but was considerably greater at −24.1 and −18.2‰ for growth with CO2. Therefore, measurement of δ13C values of 10Me16:0 in cultures or sediments containing Desulfobacter spp. or Desulfobacterium spp. could yield a reliable estimate of the biomass value and could potentially distinguish between autotrophic and heterotrophic growth.
The FA of Desulfotomaculum acetoxidans were unfortunately not unique and would not be useful biomarkers for Desulfotomaculum spp. in complex environmental samples. In addition, the Δδ13C between FA and biomass was greater for autotrophically grown cells than for acetate-grown cells, so the isotopic values are not indicative of total biomass. However, the unusually heavy and light values could be used cautiously to indicate the growth substrate for cultures of Desulfotomaculum spp. or in environments in which it is a dominant member of the microbial community. For example, it may be possible to use natural abundance rather than 13C labeling of FA to examine SRB in some sediments (2, 3).
Desulfovibrio desulfuricans had the expected biomarker i17:1, but the amount of this biomarker was much greater with lactate as a substrate than acetate. The Δδ13C between FA and biomass was very different for the two growth conditions. Furthermore, the 13C depletion between the biomarker or total FA and the organic substrate (lactate or acetate) was not consistent. Therefore, although detection of the i17:1 biomarker FA could identify the presence of Desulfovibrio spp., the proportions and isotopic ratios could not be used to distinguish which substrate is being used in an environment.
PLFA typical of SRB have been discovered at marine anoxic methane seeps where geochemical evidence suggests anaerobic methane oxidation by a consortium of methanogens and SRB (7, 11, 21, 22, 34). The FAME from a variety of methane sites are isotopically very 13C depleted, including some that could be biomarkers for SRB. The present popular explanation for the light FAME is that SRB are growing heterotrophically from acetate, acetic acid, or formate provided by a methanogen that is consuming methane seeping from the site. The data from our culture experiments suggest an alternative explanation: hydrogen transfer (36). Our results with Desulfotomaculum acetoxidans have demonstrated that during autotrophic growth, SRB FA can become very 13C depleted, with Δδ13C(FA − CO2) of −8.4 to −58.4‰. Carbonates associated with seeps can be extremely 13C depleted, with δ13C values as low as −66.7‰ (34). Therefore, it is not surprising that our SRB FAME δ13C values are in the same range as methane seeps with FAME values of −44.1 to −76.1‰ at Eel River Basin (11, 21), −22 to −114‰ at Santa Barbara Basin (11), and −48 to −70‰ in the Gulf of Mexico (39). It follows that the FAME isolated from the methane seeps could be due to autotrophic growth of SRB and does not require a transfer of a 13C-depleted carbon source. It is also interesting that Eel River sediments exhibit correlations of δ13C values to structural differences, with 16:0, 16:1Δ9, and 16:1Δ11 increasing in isotopic lightness (11, 21) just as we observed in Desulfotomaculum acetoxidans. It would be interesting to examine the δ13C of lipids of other SRB, like Desulfosarcina variabilis and Desulforhabdus amnigenus, which contain the distinctive monoalkylether phospholipids that have also been found at methane seep sites (27); we analyzed for these glycerol ethers in the four SRB used in this study, but they were not found (R. Summons, personal communication). The elucidation of the role of SRB in anaerobic methane oxidation at these seeps could be facilitated by the analysis of deuterium/hydrogen isotope ratios of SRB FAME and biomarkers.
Acknowledgments
We thank Lori Crumbliss, Mykell Discipulo, Tsege Embaye, Anne Tharpe, and Kendra Turk for technical assistance and George Cooper for use of his GC-MS system and advice. We also thank Roger Summons for helpful discussions throughout.
This work was supported by grants from the National Aeronautics and Space Administration Exobiology Program, the NASA Astrobiology Institute, and the Natural Sciences and Engineering Research Council of Canada. K. Londry was supported by a research associateship from the National Research Council.
REFERENCES
- 1.Boon, J. J., J. W. de Leeuw, G. J. V. D. Hoek, and J. H. Vosjan. 1977. Significance and taxonomic value of iso and anteiso monoenoic fatty acids and branched β-hydroxy acids in Desulfovibrio desulfuricans. J. Bacteriol. 129:1183-1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boschker, H., W. de Graaf, M. Koster, L.-A. Meyer-Reil, and T. Cappenberg. 2001. Bacterial populations and processes involved in acetate and propionate consumption in anoxic brackish sediment. FEMS Microbiol. Ecol. 35:97-103. [DOI] [PubMed] [Google Scholar]
- 3.Boschker, H. T. S., S. C. Nold, P. Wellsbury, D. Bos, W. de Graaf, R. Pel, R. J. Parkes, and T. E. Cappenberg. 1998. Direct linking of microbial populations to specific biogeochemical processes by 13C-labelling of biomarkers. Nature 392:801-805. [Google Scholar]
- 4.Brysch, K., C. Schneider, G. Fuchs, and F. Widdel. 1987. Lithoautotrophic growth of sulfate-reducing bacteria, and description of Desulfobacterium autotrophicum gen. nov., sp. nov. Arch. Microbiol. 148:264-274. [Google Scholar]
- 5.Devereux, R., M. Hines, and D. Stahl. 1996. S cycling: characterization of natural communities of sulfate-reducing bacteria by 16S rRNA sequence comparisons. Microb. Ecol. 32:283-292. [DOI] [PubMed] [Google Scholar]
- 6.Dowling, N. J. E., F. Widdel, and D. C. White. 1986. Phospholipid ester-linked fatty acid biomarkers of acetate-oxidizing sulphate-reducers and other sulphide-forming bacteria. J. Gen. Microbiol. 132:1815-1825. [Google Scholar]
- 7.Elvert, M., E. Suess, J. Greinert, and M. J. Whiticar. 2000. Archaea mediating anaerobic methane oxidation in deep-sea sediments at cold seeps of the eastern Aleutian subduction zone. Org. Geochem. 31:1175-1187. [Google Scholar]
- 8.Frischer, M., J. Danforth, M. Newton Healy, and F. Saunders. 2000. Whole-cell versus total RNA extraction for analysis of microbial community structure with 16S rRNA-targeted oligonucleotide probes in salt marsh sediments. Appl. Environ. Microbiol. 66:3037-3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Habicht, K., and D. Canfield. 1997. Sulfur isotope fractionation during bacterial sulfate reduction in organic-rich sediments. Geochim. Cosmochim. Acta 61:5351-5361. [DOI] [PubMed] [Google Scholar]
- 10.Hayes, J. M. 2001. Fractionation of the isotopes of carbon and hydrogen in biosynthetic processes. In J. Valley and D. Cole (ed.), Stable isotope geochemistry, vol. 43. Mineralogical Society of America, Washington, D.C.
- 11.Hinrichs, K.-U., R. E. Summons, V. Orphan, S. P. Sylva, and J. M. Hayes. 2000. Molecular and isotopic analysis of anaerobic methane-oxidizing communities in marine sediments. Org. Geochem. 31:1685-1701. [Google Scholar]
- 12.Jahnke, L., W. Eder, R. Huber, J. Hope, K. Hinrichs, J. Hayes, D. Des Marais, S. Cady, and R. Summons. 2001. Signature lipids and stable carbon isotope analyses of octopus spring hyperthermophilic communities compared with those of Aquificales representatives. Appl. Environ. Microbiol. 67:5179-5189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jahnke, L., H. Stan-Lotter, K. Kato, and L. Hochstein. 1992. Presence of methyl sterol and bacteriohopanepolyol in an outer-membrane preparation from Methylococcus capsulatus (Bath). J. Gen. Microbiol. 138:1759-1766. [DOI] [PubMed] [Google Scholar]
- 14.Jahnke, L. L., R. E. Summons, J. M. Hope, and D. J. Des Marais. 1999. Carbon isotopic fractionation in lipids from methanotrophic bacteria II: the effects of physiology and environmental parameters on the biosynthesis and isotopic signatures of biomarkers. Geochim. Cosmochim. Acta 63:79-93. [DOI] [PubMed] [Google Scholar]
- 15.Kaneda, T. 1991. Iso- and anteiso-fatty acids in bacteria: biosynthesis, function, and taxonomic significance. Microbiol. Rev. 55:288-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kohring, L. L., D. B. Ringelberg, R. Devereux, D. A. Stahl, M. W. mittelman, and D. C. White. 1994. Comparison of phylogenetic relationships based on phospholipid fatty acid profiles and ribosomal RNA sequence similarities among dissimilatory sulfate-reducing bacteria. FEMS Microbiol. Lett. 119:303-308. [DOI] [PubMed] [Google Scholar]
- 17.Londry, K., and D. Des Marais. 2003. Stable carbon isotope fractionation by sulfate-reducing bacteria. Appl. Environ. Microbiol. 69:2942-2949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Magnuson, K., S. Jackowski, C. Rock, and J. Cronan Jr. 1993. Regulation of fatty acid biosynthesis in Escherichia coli. Microbiol. Rev. 57:522-542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Minz, D., S. Fishbain, S. Green, G. Muiyzer, Y. Cohen, B. Rittmann, and D. Stahl. 1999. Unexpected population distribution in a microbial mat community: sulfate-reducing bacteria localized to the highly oxic chemocline in contrast to a eukaryotic preference for anoxia. Appl. Environ. Microbiol. 65:4659-4665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Minz, D., J. Flax, S. Green, G. Muyzer, Y. Cohen, M. Wagner, B. Rittmann, and D. Stahl. 1999. Diversity of sulfate-reducing bacteria in oxic and anoxic regions of a microbial mat characterized by comparative analysis of dissimilatory sulfite reductase genes. Appl. Environ. Microbiol. 65:4666-4671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Orphan, V. J., C. H. House, K.-U. Hinrichs, K. D. McKeegan, and E. F. DeLong. 2001. Methane-consuming Archaea revealed by directly coupled isotopic and phylogenetic analysis. Science 293:484-487. [DOI] [PubMed] [Google Scholar]
- 22.Pancost, R., and J. Sinninghe Damsté. 2003. Carbon isotopic compositions of prokaryotic lipids as tracers of carbon cycling in diverse settings. Chem. Geol. 195:29-58. [Google Scholar]
- 23.Ramsing, N., H. Fossing, T. Ferdelman, F. Andersen, and B. Thamdrup. 1996. Distribution of bacterial populations in a stratified Fjord (Mariager Fjord, Denmark) quantified by in situ hybridization and related to chemical gradients in the water column. Appl. Environ. Microbiol. 62:1391-1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ramsing, N., M. Huhl, and B. Jorgensen. 1993. Distribution of sulfate-reducing bacteria, O2, and H2S in photosynthetic biofilms determined by oligonucleotide probes and microelectrodes. Appl. Environ. Microbiol. 59:3840-3849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ravenschlag, K., K. Sahm, and R. Amann. 2001. Quantitative molecular analysis of the microbial community in marine Arctic sediments (Svalbard). Appl. Environ. Microbiol. 67:387-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roslev, P., N. Iversen, and K. Henriksen. 1998. Direct fingerprinting of metabolically active bacteria in environmental samples by substrate specific radiolabelling and lipid analysis. J. Microbiol. Methods 31:99-111. [Google Scholar]
- 27.Rutters, H., H. Sass, H. Cypionka, and J. Rullkotter. 2001. Monoalkylether phospholipids in the sulfate-reducing bacteria Desulfosarcina variabilis and Desulforhabdus amnigenus. Arch. Microbiol. 176:435-442. [DOI] [PubMed] [Google Scholar]
- 28.Schauder, R., A. Preuβ, M. Jetten, and G. Fuchs. 1989. Oxidative and reductive acetyl CoA/carbon monoxide dehydrogenase pathway in Desulfobacterium autotrophicum 2. Demonstration of the enzymes of the pathway and comparison of CO dehydrogenase. Arch. Microbiol. 151:84-89. [Google Scholar]
- 29.Schauder, R., F. Widdel, and G. Fuchs. 1987. Carbon assimilation pathways in sulfate-reducing bacteria II. Enzymes of a reductive citric acid cycle in the autotrophic Desulfobacter hydrogenophilus. Arch. Microbiol. 148:218-225. [Google Scholar]
- 30.Spormann, A. M., and R. K. Thauer. 1988. Anaerobic acetate oxidation to CO2 by Desulfotomaculum acetoxidans. Demonstration of enzymes required for the operation of an oxidative acetyl-CoA/carbon monoxide dehydrogenase pathway. Arch. Microbiol. 150:374-380. [Google Scholar]
- 31.Summons, R. E., L. L. Jahnke, and Z. Roksandic. 1994. Carbon isotopic fractionation in lipids from methanotrophic bacteria: relevance for interpretation of the geochemical record of biomarkers. Geochim. Cosmochim. Acta 58:2853-2863. [DOI] [PubMed] [Google Scholar]
- 32.Taylor, J., and R. J. Parkes. 1983. The cellular fatty acids of the sulphate-reducing bacteria, Desulfobacter sp., Desulfobulbus sp. and Desulfovibrio desulfuricans. J. Gen. Microbiol. 129:3303-3309. [Google Scholar]
- 33.Teske, A., N. B. Ramsing, K. Habicht, M. Fukui, J. Kuver, B. B. Jorgensen, and Y. Cohen. 1998. Sulfate-reducing bacteria and their activities in cyanobacterial mats of Solar Lake (Sinai, Egypt). Appl. Environ. Microbiol. 64:2943-2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thiel, V., J. Peckmann, R. Seifert, P. Wehrung, J. Reitner, and W. Michaelis. 1999. Highly isotopically depleted isoprenoids: molecular markers for ancient methane venting. Geochim. Cosmochim. Acta 63:3959-3966. [Google Scholar]
- 35.Vainshtein, M., H. Hippe, and R. M. Kroppenstedt. 1992. Cellular fatty acid composition of Desulfovibrio species and its use in classification of sulfate-reducing bacteria. Syst. Appl. Microbiol. 15:554-566. [Google Scholar]
- 36.Valentine, D., and W. Reeburgh. 2000. New perspectives on anaerobic methane oxidation. Environ. Microbiol. 2:477-484. [DOI] [PubMed] [Google Scholar]
- 37.van der Meer, M., S. Schouten, and J. Sinninghe Damste. 1998. The effect of the reversed tricarboyxlic acid cycle on the 13C contents of bacterial lipids. Org. Geochem. 28:527-533. [Google Scholar]
- 38.Widdel, F. 1987. New types of acetate-oxidizing, sulfate-reducing Desulfobacter species, D. hydrogenophilus sp. nov., D. latus sp. nov., and D. curvatus sp. nov. Arch. Microbiol. 148:286-291. [Google Scholar]
- 39.Zhang, C., Y. Li, J. Wall, L. Larsen, R. Sassen, Y. Huang, Y. Wang, A. Peacock, D. White, J. Horita, and D. Cole. 2002. Lipid and carbon isotopic evidence of methane-oxidizing and sulfate-reducing bacteria in association with gas hydrates from the Gulf of Mexico. Geology 30:239-242. [Google Scholar]
- 40.Zhang, C., Y. Li, Q. Ye, J. Fong, A. Peacock, E. Blunt, J. Fang, D. Lovley, and D. White. 2003. Carbon isotope signatures of fatty acids in Geobacter metallireducens and Shewanella algae. Chem. Geol. 195:17-28. [Google Scholar]