Abstract
Control of HIV-1 replication was first achieved with regimens that included a nonnucleoside reverse transcriptase inhibitor (NNRTI) or a protease inhibitor (PI); however, an explanation for the high antiviral activity of these drugs has been lacking. Indeed, conventional pharmacodynamic measures like IC50 (drug concentration causing 50% inhibition) do not differentiate NNRTIs and PIs from less active nucleoside reverse transcriptase inhibitors (NRTIs). Drug inhibitory potential depends on the slope of the dose-response curve (m), which represents how inhibition increases as a function of increasing drug concentration and is related to the Hill coefficient, a measure of intramolecular cooperativity in ligand binding to a multivalent receptor. Although NNRTIs and PIs bind univalent targets, they unexpectedly exhibit cooperative dose-response curves (m > 1). We show that this cooperative inhibition can be explained by a model in which infectivity requires participation of multiple copies of a drug target in an individual life cycle stage. A critical subset of these target molecules must be in the unbound state. Consistent with experimental observations, this model predicts m > 1 for NNRTIs and PIs and m = 1 in situations where a single drug target/virus mediates a step in the life cycle, as is the case with NRTIs and integrase strand transfer inhibitors. This model was tested experimentally by modulating the number of functional drug targets per virus, and dose-response curves for modulated virus populations fit model predictions. This model explains the high antiviral activity of two drug classes important for successful HIV-1 treatment and defines a characteristic of good targets for antiviral drugs in general, namely, intermolecular cooperativity.
Introduction
In 1997, regimens were developed that suppressed HIV-1 viremia to below the detection limit in most treated patients. These regimens combined two nucleoside analogue reverse transcriptase inhibitors (NRTIs) with an HIV-1 protease inhibitor (PI) (1–3). Combinations of two NRTIs and a non-nucleoside reverse transcriptase inhibitor (NNRTI) also proved effective (4,5). Collectively, these regimens, known as highly active antiretroviral therapy (HAART), transformed a previously fatal disease into a chronic condition that is well controlled in adherent patients. Despite HAART’s success, a fundamental theory explaining its effectiveness is lacking.
Drug resistance, which results both from the high error rate of reverse transcriptase and the dynamic nature of HIV-1 infection, is a major cause of treatment failure (6–10). The low probability of multiple simultaneous resistance mutations on the same genome clearly contributes to the success of triple combination therapy (11). However, suppression of HIV-1 replication is not simply the result of using three drugs; some triple NRTI combinations had suboptimal responses (5). Thus, in early treatment efforts, inclusion of a PI or NNRTI appeared essential for control of viral replication. Although the use of drugs acting through different mechanisms also contributes to the effectiveness of combination therapy, PIs and NNRTIs appeared to have greater antiviral activity than most NRTIs. Therefore, treatment guidelines recommend inclusion of a PI or NNRTI in most initial HAART regimens (5). Standard pharmacologic measures such as IC50 and inhibitory quotient do not distinguish PIs and NNRTIs from less active NRTIs (12). Thus, the fundamental pharmacodynamic principles underlying this successful treatment remain unclear. Instead, progress has depended on comparative clinical trials, which have recently established a role for newer drugs as well, such as integrase strand transfer inhibitors (InSTIs) and CCR5 antagonists (5).
We recently showed that the superior antiviral activity of PIs and NNRTIs relative to most NRTIs can be partially explained by the dose-response curve slope (m) (12). This parameter is included in all fundamental pharmacodynamic equations including the Hill equation (13), the sigmoidal Emax model (14), and the Chou-Talalay median effect equation (15). The slope parameter is related to the Hill coefficient describing intramolecular cooperativity in the binding of ligands to a multivalent receptor (13, 16). Positive cooperativity (m>1) reflects enhanced binding of additional ligands to a receptor after the first ligand binds. Interestingly, the enzymes targeted by NNRTIs and PIs are univalent with respect to these inhibitors. NNRTIs bind to a single pocket adjacent to the dNTP binding site (17), and HIV-1 protease is a homodimer with a single active site targeted by PIs, which are peptidomimetic substrate or transition state analogs (18, 19). Therefore, non-cooperative dose-response curves (m=1) were expected. However, in single round infectivity assays, dose-response curves for NNRTIs and PIs have unexpectedly steep slopes (~1.7 and ranging from 1.8 to 4.5, respectively), allowing these drugs to achieve extraordinarily high antiviral activity (12).
Since m>1 for NNRTIs and PIs cannot be explained by intramolecular cooperativity, a mechanistic explanation requires reconsidering the concept of cooperativity in drug action. Here we describe a critical subset model that explains steep slopes for these drugs in terms of intermolecular cooperativity, and we provide experimental validation for this model.
Results
The critical subset model
The simplest model describing dose-response relationships is the median effect equation (15). It is derived from the law of mass action, which governs dynamic equilibria in chemical reactions. The median effect equation can be written as:
| (1) |
| (2) |
In the context of HIV-1 infection, fa and fu are the fractions of viruses affected and unaffected by drug, [D] is drug concentration, IC50 is the drug concentration causing 50% inhibition, and m is the slope parameter describing the steepness of the dose-response curve.
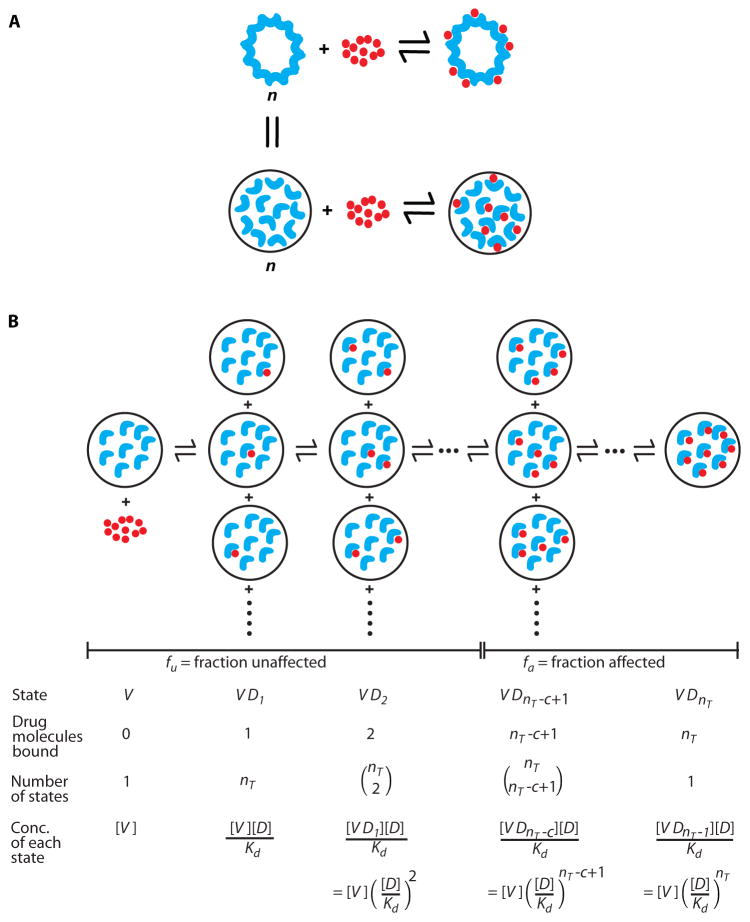
Some anti-HIV drugs have m>1 even though their target enzymes are univalent. To explain this observation, we developed a critical subset model based on mass action. The model envisions independent binding of multiple drug molecules to a set of identical targets, viral proteins, within each virus or virus-infected cell. Within a given virion or infected cell, the targeted proteins behave as a set of linked receptor sites that can be occupied to variable extents (Fig. 1A), depending on [D] and the affinity of individual drug-target interactions (Kd). Drug binding involves a series of equilibrium reactions mediating conversions between all possible partially occupied states (16). Assuming that replication requires some critical threshold number (c) of functional (unbound) enzyme molecules to complete the relevant reaction before irreversible decay processes intervene (Fig. 1B), the following expression describes fa as a function of [D], Kd, the unbound virus concentration [V], and the total number of enzyme molecules per virus nT (Equation 3, Table 1):
| (3) |
Figure 1.
The critical subset model. (A) A virion (or virally infected cell) can be viewed as a multivalent receptor containing n identical copies of a given drug target such as HIV-1 protease. Although not physically linked to one another, these copies are spatially constrained within the virus particle or infected cell. (B) Sequential binding of a protease inhibitor (red circle) to the protease dimers (blue crescents) within a virion. The concentrations of the various bound states can be determined by the law of mass action, assuming a single unchanging dissociation constant, Kd. Assuming that successful completion of the relevant step in the virus life cycle requires a critical number c of functional unbound copies of the drug target, fa can be computed as the concentration of inhibited states divided by the concentration of all possible states (Equation 3). fu is equal to 1−fa. Similar arguments can be applied to RT except that the RT molecules are present within the reverse transcription complex in an infected cell.
Table 1.
Glossary of abbreviations.
| fa | Fraction of viruses or infection events affected by the drug |
| fu | Fraction of viruses or infection events unaffected by the drug |
| [D] | Drug concentration |
| IC50 | Drug concentration that leads to 50% inhibition |
| m | Slope parameter or Hill coefficient |
| Kd | Dissociation constant for a drug binding to an individual target enzyme molecule |
| nT | Average of total number of enzyme molecules per virion relevant to the inhibited reaction |
| c | Critical threshold number of functional enzyme molecules required for viral replication |
| [V] | Unbound virus concentration |
| σ | Standard deviation for nT |
| nmax | Maximum total number of enzyme molecules per virion, usually set as > nT + 3σ |
| ni | Given total number of enzyme molecules per virion relevant to the inhibited reaction |
| fm | Fraction of mutant plasmid transfected |
| Pf | Probability of obtaining functional enzyme multimers for a given fm |
| nF | Total number of functional target molecules per virion relevant to the inhibited reaction |
| RI | Relative infectivity of phenotypically-mixed viruses compared to the wild type virus |
Consider for example HIV-1 protease. Multiple copies of protease participate in the maturation of each virion. Although not physically linked, they are constrained spatially within the virion. Thus, the virion can be considered a multivalent receptor with nT binding sites, where nT is the total number of protease molecules per virus (Fig. 1A). Since simultaneous binding is unlikely, we have modeled drug binding as a series of equilibrium reactions (Fig. 1B). The derivation of Equation 3 follows from a consideration of the concentration of each possible state along the path to saturation (16). Viruses with a critical subset of c or more unbound enzymes are infectious. To render a virus noninfectious, at least nT −c+1 enzyme molecules must be occupied (Fig. 1B). Thus in Equation 3, the numerator is the sum of the concentrations of all viruses having ≥nT −c+1 enzyme molecules bound by drug, and the denominator is the total virus concentration.
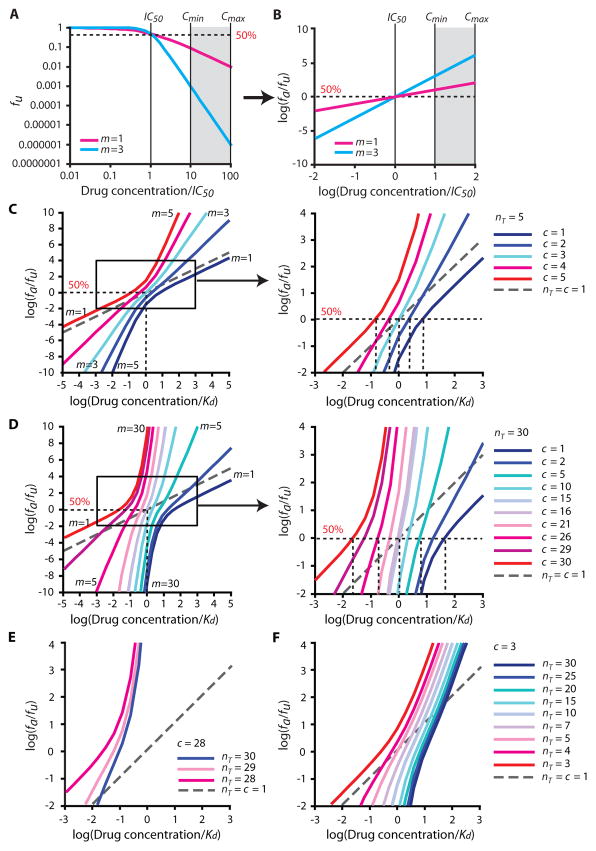
This model predicts the shapes of dose-response curves. To illustrate these predictions, we plotted hypothetical dose-response curves using median effect plots based on Equation 1 (Fig. 2). The median effect plot linearizes the dose-response curve for most drugs, with the slope of the resulting line equal to the Hill coefficient (Fig. 2A–B). We obtained complex dose-response curves with various m depending on nT and c (Fig. 2C–D). Importantly, when nT=c=1, Equation 3 simplifies to the median effect equation (Equation 2) and predicts m=1. However, for all cases with 1<c<nT, the model predicts steeper dose-response curves with m ≥1.5 at all drug concentrations.
Figure 2.
Steep dose-response curves predicted by the critical subset model. (A) Hypothetical log-log plot of fu vs. drug concentration based on the median-effect model for drugs with different m values. The strong impact of slope on the suppression of infectivity is apparent at high drug concentrations. Drug concentrations are normalized by IC50. The dotted line represents 50% inhibition. (B) Hypothetical median effect plot [log (fa/fu) vs. log drug concentration] (Equation 1) of the curves shown in Figure 2A. The m value is the slope of the line. (C, D) Hypothetical median effect plots of dose-response curves generated using the critical subset model assuming nT = 5 (C) or 30 (D) and values of c ranging from 1 to nT. Concentrations are normalized by Kd. The right panel is an expansion of the indicated area in the left panel. As c increases, IC50 decreases, and slope increases at concentrations above the IC50, but decreases at concentrations below the IC50. A standard non-cooperative dose-response curve with m=1 (nT =c=1) is also shown (gray dashed line). (E, F) Hypothetical median effect plots of dose-response curves generated using the critical subset model with values of nT ranging from c to 30, and c=28 (E) or 3 (F). As nT decreases, the dose-response curves shift to the left.
Interestingly, the model predicts complex dose-response curves with non-linear median effect plots. In other words, m is different for different segments of the curve. As shown below, the dose-response curves of some antiretroviral drugs are similarly complex. According to the model, slope values are determined by nT and c. As [D] increases above IC50, m approaches c. As [D] decreases below IC50, m approaches nT −c+1 (Supplementary Note 1). Within the range of log (fa/fu)= −2 to 4 (1–99.99% inhibition), the curves inflect upward when c is close to nT and downward when c is small relative to nT (Fig. 2C–D). For example, when nT=5, the model predicts a family of curves with slopes ranging from 1.30 to 1.88 at the IC50 (Fig. 2C). Slope at the IC50 is highest when c=3, and lowest when c=1 or 5. However, the slope at high [D] approaches c, and thus the maximum slope is 5. The same pattern with steeper slopes is observed for nT=30 (Fig. 2D). Whenever simultaneous inhibition of multiple copies of a target molecule within a virion or infected cell is required to block replication, dose-response curves with m>1 are predicted by this model. This unique form of intermolecular cooperativity does not depend on changes in the binding properties (Kd) of individual target sites.
The model also predicts the relationship between IC50 and Kd. When nT is odd and c=(nT+1)/2, IC50=Kd. When c>(nT +1)/2, IC50 < Kd and vice versa (Fig. 2C–D). Similarly, for a series of curves with constant c, IC50 decreases as nT decreases (Fig. 2E–F). This is expected because a lower IC50 is achieved with the same Kd when the fractional occupancy of target enzymes required for inhibition is low.
Effect of heterogeneity on dose-response curves
A second factor affecting slope is heterogeneity in nT. In other biological systems with all-or-none outcomes, heterogeneity in the number of drug targets flattens dose-response curves (20). For individual virions, infection is all-or-none, and thus heterogeneity arguments apply. When heterogeneity is limited, dose-response curves are steeper. Intuitively, this may be understood by considering that a homogeneous virus population will be inhibited over a narrow range of drug concentrations, whereas a more heterogeneous population will contain viruses that are easier or harder to inhibit, and therefore inhibition will be observed over a wider concentration range.
If nT follows an integer-valued normal distribution with a standard deviation σ, then dose-response curves follow Equation 4, which gives fa for a population of viruses with variability in nT.
| (4) |
where
| (5) |
and fa(ni) is calculated using Equation 3. The derivation of Equation 4 follows from a consideration of fa for viruses with a given total number of enzyme molecules per virus (ni) and the probability of finding viruses with ni enzyme molecules within a population of viruses with nT±σ total enzyme molecules per virus (Fig. S1A). Since viruses with ni<c are noninfectious, the numerator must be divided by the probability of infectious virus for a given nT, σ, and c (Fig. S1A). nmax is the maximum total number of enzyme molecules per virus, which is set at >nT+3σ. Equation 4 predicts a decrease in m with increasing σ at drug concentrations below the IC50 (Fig. S1B), since heterogeneity leads to a subpopulation of viruses with low nT that are inhibited more easily at low [D].
Testing the critical subset model with phenotypic mixing experiments – theoretical considerations
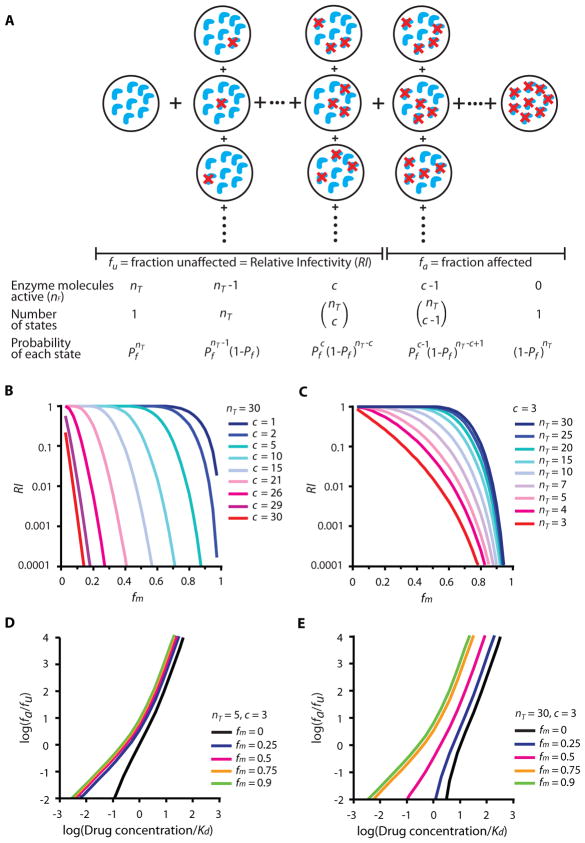
The critical subset model makes testable predictions about how variation in nT and c affect dose-response curves. For example, the model predicts that at constant c, IC50 decreases as nT decreases (Fig. 2E–F). To test this model, we used a phenotypic mixing strategy (21,22) to reduce the total number of functional target molecules per virion (nF) (Fig. 3A). To reduce nF, defined fractions (fm) of mutant proviral constructs containing inactivating point mutations in the target enzymes were mixed with wild type constructs and transfected into HEK293T cells to generate virions with reduced nF. Note that nF =nT when no mutant enzyme is present. We then examined the relative infectivity (RI) of these virus populations compared to wild-type virus and obtained estimates of nT and c (Fig. 3B–C). Finally, we determined whether the dose-response curves for virus populations with lower nF behaved as predicted by the model (Fig. 3D–E). Note that larger decreases in IC50 are predicted when c is small relative to nT.
Figure 3.
Effect of reductions in nF on the dose-response curves for antiviral drugs. (A) Relative infectivity (RI) of phenotypically-mixed virions. Consider a population of virions with the same nT but various numbers of active (nF, blue crescents) and inactive protease molecules (crescents with red cross). Pf is the probability of obtaining a function protease molecule for a given fm. The probability of getting nF copies of functional enzyme molecules (P(nF)) as a function of nT, c, and Pf can be calculated using Equation 6. The RI of a population of phenotypically-mixed virions is the sum of the probabilities of all the viruses with nF ≥c. (B, C) Hypothetical RI plots [RI vs. fm] for protease with nT = 30 and various values of c (B), or c=3 and various values of nT (C). (D, E) Hypothetical median effect plots of dose-response curves for PIs against viruses with c=3, nT = 5 (D) or c=3, nT = 30 (E), and various fm values. Concentrations are normalized by Kd. The change in IC50 is larger when c is proportionally small relative to nT (E).
The introduction of mutant enzyme subunits reduces infectivity, as some viruses will no longer contain a critical number of functional enzymes (Fig. 3A). The enzyme molecules targeted by RT and protease inhibitors are heterodimers and homodimers, respectively. If formation of mixed dimers from wild-type and mutant monomers follows random mixing, then the virions produced should contain reduced nF following a binomial distribution. The RI of mixed virions compared to wild-type virions is a function of nT, c, and Pf, where Pf is the probability of obtaining a functional enzyme multimer for a given fm (Equation 6).
| (6) |
P(nF) is the probability of getting nF copies of functional enzyme molecules with a given nT, c, and Pf. The calculation of Pf from fm is based on the subunit stoichiometry of the target enzyme and is different for RT and protease, as described in Methods. Based on Equation 6, the RI of phenotypically-mixed virions is the sum of the probabilities of obtaining viruses with nF ≥ c. Plotting RI as a function of fm, we obtain RI curves with various patterns depending on nT and c (Fig. 3B–C). When most copies of the enzyme are required to complete the relevant step in the life cycle, decreases in nF have a major effect – a small increase in fm results in loss of infectivity. In contrast, when c is small relative to nT and thus available enzyme exceeds that which is necessary, infectivity is not affected until fm is large (Fig. 3B–C). As demonstrated below, the former situation applies to RT whereas the latter applies to protease. Note that the analysis of RI is based on the same assumptions that underlie the critical subset model, namely that a critical threshold number (c) of functional copies of a viral enzyme out of nT copies are required for infectivity (Fig. 1B, 3A).
To test the critical subset model, phenotypically-mixed viruses can be assayed for susceptibility to drug inhibition. Dose-response curves for inhibition of viruses with various fm can be predicted by Equation 4, where the calculation of fa(ni) follows Equation 7 (Fig. 3D–E). The derivation of Equation 7 follows from a consideration of fa for each nF (Equation 3) and the probability of viruses with nF functional enzyme molecules within a population of viruses with ni total enzyme molecules and an average probability Pf of functional ones (Equation 6, Fig. 3A).
| (7) |
According to this model, the change in IC50 depends on the fractional occupancy of enzyme molecules required for inhibiting replication. Two contrasting situations with the same value of c but different values of nT are illustrated in Figures 3D and 3E. When c is close to nT (Fig. 3D, nT = 5), the fractional occupancy required for inhibition is low, and the model predicts that IC50 should only decrease slightly with increasing fm. This is because the number of enzyme molecules is already close to the limiting value of nT = c. However, when c is small relative to nT (Fig. 3E, nT = 30), the fractional occupancy required for inhibition is high, and the model predicts that IC50 can decrease over a wider range with increasing fm until the limiting condition is reached.
Experimental validation of the critical subset model – generation of phenotypically mixed virions
Before testing the critical subset model, we validated several assumptions. One is that wild-type and mutant enzymes can be expressed in cells and incorporated into virions with similar efficiency. We chose well-characterized point mutations in the active sites of RT (D185N), protease (D25N), and integrase (D64E) that abolish enzyme activity without effects on multimer formation or virus assembly (18, 23, 24). Transfection efficiency was similar for the wild-type plasmid and mutant plasmids by flow cytometry (Fig. S2A). Wild-type and mutant RT and integrase were incorporated into the virions with similar efficiency (Fig. S2B–C). We also tested whether formation of mixed dimers from wild-type and mutant monomers follows random mixing and hence the binomial distribution. To obtain direct biochemical evidence for mixed dimer formation in cotransfected cells, we performed co-immunoprecipitations (Fig. S3). We introduced either an HA or a Flag tag into the N terminus of the wild-type and mutant enzymes. Viral proteins immunoprecipitated by either anti-HA or anti-Flag antibodies from cell lysates were immunoblotted with anti-Flag or anti-HA antibodies, respectively. Our results suggest that for both RT and protease, dimers with all of the four possible combinations – wild-type-wild-type, wild-type-mutant, mutant-wild-type, mutant-mutant – can be formed with approximately equal probability (Fig. S3A–B).
Analysis of dose-response curves for NNRTIs and PIs against phenotypically-mixed viruses
According to the model, drugs targeting steps involving multiple copies of the drug target should show steeper dose-response curves and dramatically better inhibition at drug concentrations above the IC50. NNRTIs can interact with all RT molecules in the preintegration complex, and PIs can interact with all protease molecules in the maturing virion. These drug classes exhibit slopes >1.7 and high inhibitory potential (12). For NNRTIs and PIs, the model predicts that IC50 should decrease as nF is reduced and that the higher the fractional occupancy required for inhibiting replication, the greater the change in IC50 (Fig. 3D–E).
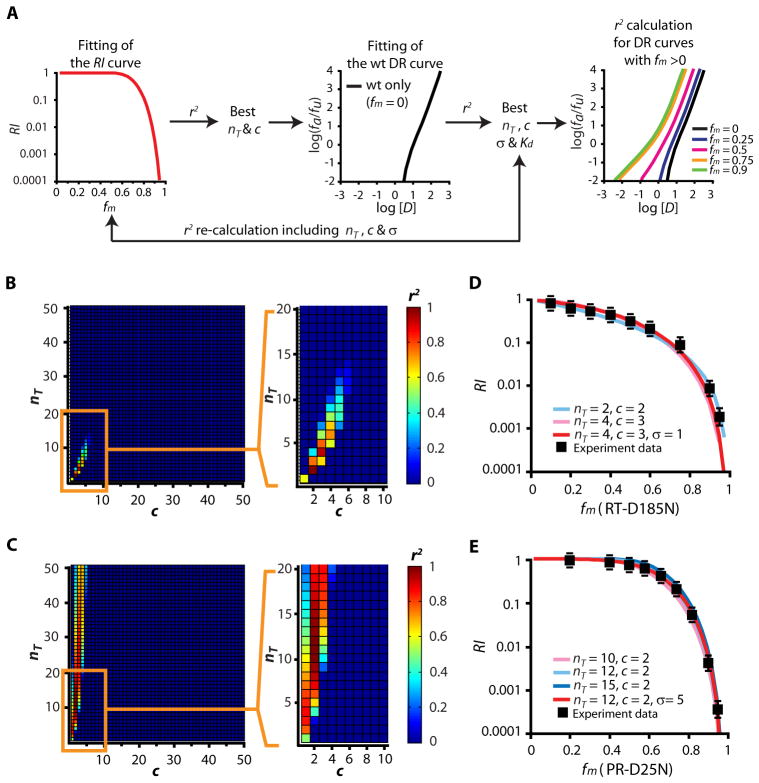
We tested these predictions using the algorithm shown in Figure 4A. We first determined the apparent nT and c by measuring in the absence of drug the RI of virions with mixtures of catalytically active and inactive enzymes. The RI of phenotypically-mixed virions can be calculated with Equation 6 for any given nT and c (Fig. 3A). We fitted experimental RI data to theoretical RI curves for RT and protease and calculated r2 values for all combinations of nT between 1 and 100 and c between 1 and nT (Fig. 4B–C). The r2 values shown in Figure 4B and 4C indicate poor fitting for nT >50. The experimental RI curves fit the theoretical curves best with either nT=2, c=2 or nT=4, and c=3 for RT (Fig. 4B, D), and nT=9–15 and c=2, for protease (Fig. 4C, E). Importantly, with these nT and c values, the model predicts m=1.4–2.0 for NNRTIs and m>1.8 for PIs, consistent with experimental observations.
Figure 4.
Experimental validation of the critical subset model. (A) Algorithm for model validation from experimental data. Curves describing the dependence of the relative infectivity (RI) of phenotypically-mixed virions on fm in the absence of drug were first obtained. Experimental RI curves were fitted with theoretical curves (Equation 6) using various nT and c values, and r2 values were calculated. The nT and c values that gave the best five r2 values were used in the next step. These pairs of nT and c values were used as a start point in a systematic search for values of σ and Kd that allowed the best fit with dose-response curves for NNRTIs or PIs against wild-type viruses. The r2values for the fitting of the RI curves were recalculated with the resulting sets of nT, c, and σ values. Once an optimal set of nT, c, σ, and Kd values was obtained, these values were used to predict dose-response curves for phenotypically mixed virions. The r2 values representing the fitting of predicted dose-response curves to the experimental curves were then calculated. (B, C) Estimates of nT and c for RT (B) and protease (C). The r2 values for fitting of RT and protease RI curves to each pair of nT and c values are shown. Red squares indicate r2 close to 1, and blue squares indicate r2 <0.2. (D, E) RI curves for RT (D) and protease (E). Theoretical RI curves predicted by the critical subset model are plotted as a function of fm (solid lines). The actual experimental data are shown in black squares. The RI of a given viral stock was calculated by normalization to the infectivity of the viral stock carrying only wild-type enzyme. Each data point represents the mean ± SEM from >3 experiments.
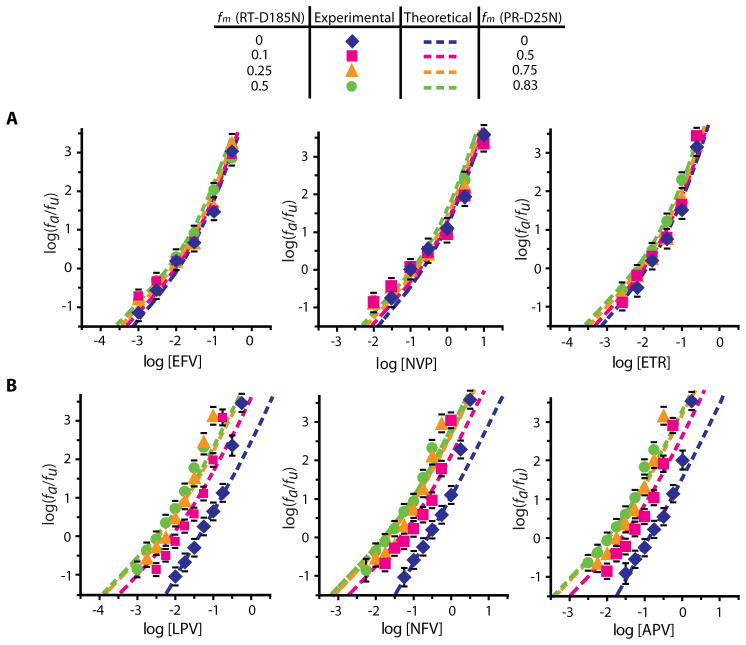
To refine values for nT and c and find the approximate Kd, we used estimates from RI curves as a starting point to find σ and Kd values that allowed optimal fitting of dose-response curves for NNRTIs and PIs against wild-type viruses (Fig. 4A, Equations 4–7, Supplementary Note 2). For NNRTIs, nT=4, σ =1, c=3 gave the best average fit (Fig. 5A), with Kd=0.018 μM, 0.33 μM, and 0.018 μM for efavirenz (EFV), nevirapine (NVP), and etravirine (ETR), respectively. The inclusion of σ for RT improved the fitting of the RI curve (Fig. 4D, Table S1). Importantly, the values developed using RI data in the absence of drugs allowed good fitting of experimental dose-response curves, including the upward inflection that is apparent in the experimental data and that is predicted by the model for these nT and c values (Fig. 5A).
Figure 5.
Effect of phenotypic mixing on dose-response curves for NNRTIs and PIs. (A) Median effect plots of theoretical (dashed lines) and experimental (solid markers) dose-response curves for the NNRTIs EFV, NVP, and ETR with wild-type and phenotypically mixed virions. The dose-response curves for phenotypically mixed virion preparations exhibited the expected slight decrease in IC50 compared to wild-type virions. (B) Median effect plots of theoretical (dashed lines) and experimental (solid markers) dose-response curves for the PIs LPV, NFV, and APV with wild-type and phenotypically mixed virions. The two data points at high drug concentrations for the wild-type dose-response curves were excluded from the fitting. Dose-response curves for phenotypically mixed virion preparations showed the expected left shift with increasing fm. Each data point represents the mean ± SEM from >3 experiments.
Since c=2 for protease predicts a slope approaching 2 at high [D], the model does not explain m>2 for some PIs or the sharp upward inflection at high [D]. Therefore, we tested the model using PIs with m~2. For the wild-type dose-response curves, we considered only the initial linear part as described previously (12) and excluded the two data points at high [D]. nT=9–15, σ=4–6, c=2 all fit the wild-type curves. With nT=12, σ =5, c=2, we obtained Kd=0.0068 μM, 0.039 μM, and 0.021 μM for lopinavir (LPV), nelfinavir (NFV), and amprenavir (APV), respectively (Fig. 5B). The introduction of σ for protease improved the fitting of the RI curve slightly (Fig. 4E, Table S1).
Having established values of nT and c that allow fitting of both RI data and dose-response curves for wild-type virus, we next examined the effect of decreasing nF on the dose-response curves. As predicted, the dose-response curves for NNRTIs shifted only slightly to the left with increasing fm. The experimental data fit the model predictions with r2>0.94 for each of the curves obtained for EFV and ETR (Fig. 5A, Supplementary Note 2). For NVP, the r2 values were >0.90 (Fig. 5A). For PIs, the model predicts a substantial left shift of the dose-response curves with increasing fm. Again, the experimental data fit the model predictions well with r2>0.87 for all drug curves with various fm (Fig. 5B, Supplementary Note 2). These results are consistent with the fractional occupancy of RT or protease required for inhibiting replication. The finding that the IC50 decreased with decreasing nF for PIs can be understood by considering that virus particles with lower nF are more easily inhibited, since fewer protease molecules must be bound by the drug in order to reduce the number of unbound protease molecules to below c. This result is also consistent with reports that the threshold activity of protease necessary for infectivity is small relative to the total protease activity per virion (25). In contrast, most of the RT molecules present are required for viral replication. Therefore, deceasing nF had minimal effect on the IC50 of the NNRTIs.
Taken together, these results suggest that the participation of multiple copies of RT and protease in the processes of reverse transcription and virion maturation, respectively, results in a form of intermolecular cooperativity that is manifested as steeper dose-response curves for the NNRTIs and PIs. The cooperative curves can be fit using the critical subset model without the need to postulate interactions between the active sites of individual enzyme molecules.
Analysis of the dose-response curves for InSTIs and NRTIs against phenotypically-mixed viruses
The InSTIs and the NRTIs exhibit slopes close to 1 (12). Unlike NNRTIs and PIs, these drugs target reactions in which a single molecular complex of one enzyme with viral nucleic acid mediates a critical step. For the InSTIs, the inhibited process is performed by precisely one molecular complex per infected cell, consisting of an integrase tetramer bound to the ends of the viral genome (26, 27). There is no possibility for intermolecular cooperativity because only a single drug target per virus mediates the targeted reaction. Other integrase molecules present in the preintegration complex are not targeted by InSTIs and are irrelevant. Similar reasoning can be applied to RT by breaking reverse transcription into individual steps, each consisting of attachment of one nucleotide. The NRTIs target a single enzyme:nucleic acid complex per infected cell, namely the RT molecule that is adding the next nucleotide to the viral cDNA (17). Other RT molecules in the reverse transcription complex are irrelevant and are not targeted. When a single target mediates the critical reaction (nT=c=1), Equation 3 reduces to the median effect equation (Equation 2) with m=1 (Fig. 2C–D), consistent with the experimental observations (12). The model predicts that phenotypic mixing should not affect m or IC50 because non-functional complexes are irrelevant for drug inhibition and functional complexes are inhibited by drug in the same manner for both wild-type and phenotypically mixed viruses.
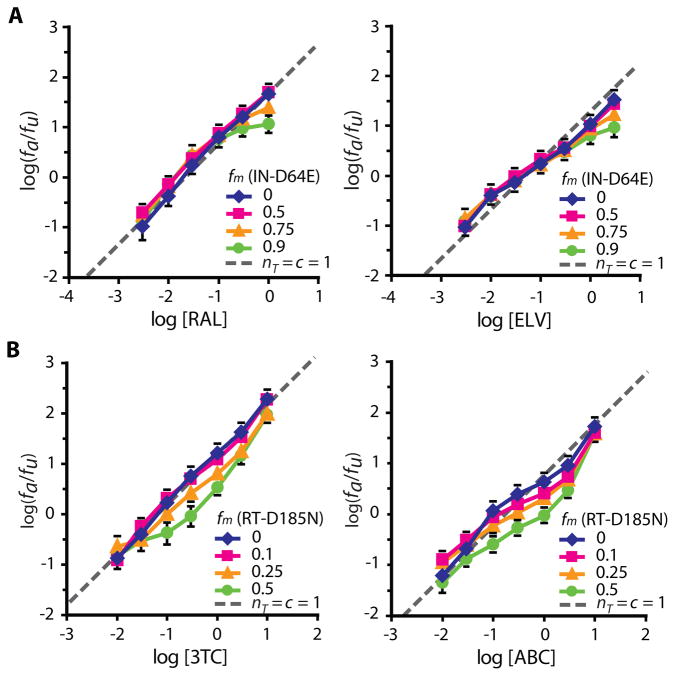
To test these predictions, we generated phenotypically mixed virions in which a fraction of the integrase or RT molecules had the inactivating catalytic site mutations D64E or D185N, respectively (23, 24). Drug susceptibility was measured using the single round infectivity assay. The slopes were close to 1 and exhibited minimal change against modulated viruses as predicted by the model (Fig. 6, S4). The IC50 values for the InSTIs raltegravir (RAL) and elvitegravir (ELV) against phenotypically-mixed virions were unchanged (Fig. 6A). For all NRTIs except for zidovudine, there was no decrease in IC50, consistent with the model. Interestingly, IC50 values for most NRTIs such as lamivudine (3TC) and abacavir (ABC) increased slightly relative to wild-type at fm=0.5 (Fig. 6B, S4). The dose-response curves for zidovudine represented an exception, as the IC50 was dramatically decreased as fm increased (Fig. S4). This increased sensitivity may reflect reduced excision of zidovudine when there are fewer active RT molecules (21). With this exception, no InSTI and NRTIs showed decreased IC50 when some copies of the relevant enzymes carried inactivating mutations (Fig. 6, S4). This behavior is in sharp contrast to the NNRTIs and PIs, drugs for which phenotypically mixed virions invariably showed decreased IC50. Both sets of observations are consistent with the critical subset model.
Figure 6.
Effect of phenotypic mixing on dose-response curves for InSTIs and NRTIs. (A) Dose-response curves for the InSTIs RAL and ELV against wild-type virus and viruses generated with increasing fractions of the D64E mutant plasmid. The IC50 and slope did not change with increasing fm. As previously described (13), the flattening of the dose-response curves at high concentrations of InSTIs and high fm may reflect low level GFP expression from unintegrated proviruses. (B) The dose-response curves for the NRTIs 3TC and ABC against wild-type virus and viruses generated with increasing fractions of the D185N mutant plasmid. There was a 2–4 fold increase in IC50 and minimal change in slope with increasing fm. Each data point represents the mean ± SEM from >3 experiments.
Discussion
Steep dose-response curves generally reflect positive cooperativity. In the classical Hill model, a slope >1 reflects cooperative binding of multiple ligands to a multivalent receptor. However, RT and protease have only one binding site for NNRTIs and PIs, respectively, and extensive kinetics studies with purified RT and protease have not provided evidence for cooperativity. Rather, the high slopes of NNRTIs and PIs in infectivity assays reflect unique aspects of the inhibition of viral replication that are not apparent in the behavior of the isolated enzymes.
Here we present a critical subset model that provides a molecular explanation for these steep dose-response curves. We hypothesized that within a given infected cell or virus particle, the viral proteins targeted by a drug behave as a set of linked receptor sites whose fractional occupancy determines whether the relevant step in the virus life cycle is completed before irreversible decay processes intervene. When multiple copies of a target are involved in the inhibited reaction, there exists a form of intermolecular cooperativity that does not depend on alterations in the binding properties of individual sites. This model predicts slopes >1 for NNRTIs and PIs because multiple copies of RT and protease are involved in the processes inhibited by these drugs, and a critical subset of these molecules are required for viral replication. The model successfully predicted shifts in the dose-response curves against viruses with decreased nF. For NRTIs and InSTIs, only one copy of RT or integrase is relevant to the inhibited reaction. In this situation, m ~1 is predicted, as is observed experimentally.
According to the model, slope is determined by nT and c. There are ~5000 copies of Gag per virion, and the ratio of Gag-Pol to Gag is 1:10~1:20 (28, 29). Thus if the Gag-Pol polypeptides are converted with 100% efficiency into functional dimeric RT and protease enzymes, there would be a maximum of 125–250 copies of these enzymes per virion. The nT values for RT and protease obtained here are smaller, perhaps reflecting inefficiencies in the assembly of functional enzymes and additional complexities in the relevant reactions. The nT and c parameters in this model are apparent numbers that allow description of the relevant reactions in terms of a simple model that does not capture the full complexity of the processes of reverse transcription and virion maturation. In addition, nT and c refer only to enzyme molecules located in the microenvironment where the reaction occurs. For these reasons, nT and c may differ from the actual physical number of RT or protease molecules present.
Since nT and c for a given drug target should be constant, the model does not explain slope differences within the PI class. The model particularly cannot explain dose-response curves for PIs with m>2, since c=2 for protease predicts a maximum slope of 2. We hypothesize that drug-to-drug variability in m for PIs may reflect the complexity of viral maturation, which involves multiple molecular states of the enzyme acting on multiple distinct substrates (30). The protease domains of adjacent Gag-Pol precursor proteins must first dimerize to form an active protease enzyme, which then carries out intramolecular cleavages to liberate itself from the Gag-Pol precursor protein. Ultimately free protease dimers complete the remaining cleavages of the Gag and Gag-Pol precursor proteins (31). The embedded protease is less sensitive to PIs than the mature, free enzyme (30, 31). The effective substrate concentration may also differ for intramolecular and intermolecular reactions. Different PIs may preferentially inhibit distinct molecular forms of protease for which nT and c are different, resulting in slope differences. In addition, virions generated in the presence of PIs may be blocked at subsequent steps such as reverse transcription (32). Thus, unlike other antiretroviral drugs, PIs affect multiple steps in the life cycle, possibly with different nT and c values. These complexities may explain the upward inflection of PI dose-response curves at high [D].
The concepts developed here can be applied to other drug classes and other viruses. However, additional factors may complicate the analysis. For drugs affecting HIV-1 entry, the targets are trimeric, and interactions within and between trimers must be considered. One particularly unique aspect of HIV-1 RT, protease, and integrase inhibitors is that they target enzymes that are present within the virion or that are assembled from precursor polyproteins present within the virion. In this situation, constraints inherent in virion assembly limit heterogeneity (σ) in nT. As shown here, heterogeneity flattens dose-response curves. Thus in the case of viruses for which the drug targets are expressed and act within infected cells, heterogeneity in the number of enzymes per cell may obscure the cooperativity that is expected when multiple copies of a drug target act together to complete a step in the life cycle.
A variety of elegant models describing steep dose-response curves for ion channels and other multivalent receptors have been developed (16, 33–35), but models to explain cooperative dose-response curves for antiviral drugs have not been described. It is possible that other models will be developed to explain the complex dose-effect relationships for antiretroviral drugs. For example, saturable drug binding to alternative intracellular sites could result in an upward inflection within a small concentration range above the concentration that saturates the alternative sites. However, a model such as the one described here would still be needed to explain slopes >1 in other regions of the dose-response curve. Although the overall slopes for NRTIs are close to 1, there are subtle inflections in the dose-response curves for some NRTIs (Fig. 6B, S4). These may in part reflect binding to alternative intracellular sites or the reactions needed to produce the active triphosphate forms of the NRTIs. Nevertheless, the fact that slope varies in a class-specific way despite large intraclass differences in structure suggests that the slope parameter is a direct reflection of the mechanism of inhibition, as in the model described here.
The critical subset model clearly represents an oversimplification of a very complex problem. Nevertheless, it explains the steep dose-response curves of NNRTIs and PIs without assuming that the binding of drug to one site affects the Kd of binding to other sites. A steep slope means that small increases in drug concentration above the IC50 can produce large increases in antiviral activity, permitting extremely high levels of inhibition in the clinical concentration range. Thus the present study provides a conceptual framework for understanding the high antiviral activity of NNRTIs and PIs, drugs that form the basis of successful HIV-1 treatment, and provides important insights to the characteristics of good targets for antiviral drugs.
Materials and Methods
Detailed descriptions of experimental Materials and Methods are available in the Supplementary Material.
Analysis of relative infectivity (RI) curves
The RI values of recombinant viruses generated with different fm were obtained by normalizing to the infectivity of the wild-type virus. Theoretical RI values were calculated for given values of nT and c using the binomial distribution (Equation 6). This analysis required a consideration of the subunit stoichiometry of the relevant multimeric targets. The probability of obtaining a functional multimer (Pf) is a function of fm, the valence of the multimer, and the functional status of mixed multimers. For example, the enzymatic activity of protease requires the D25 residues from both subunits of the dimer (18). Thus for protease, Pf = (1−fm)2. For RT, although a D185N mutation in the p66 subunit abolishes activity, a D185N mutation in the p51 subunit has no effect on enzyme activity (23). Thus for RT, Pf = (1−fm)2 + fm(1−fm) = 1−fm. To estimate the apparent nT and c values required for virus replication, experimental RI curves were fitted with theoretical curves predicted by the model (Equation 6) using various nT and c, and r2 values were obtained for both RI and log RI (Fig. 4B–C). While fitting using the linear scale puts more weight on higher RI values, fitting using the log scale emphasizes lower RI values. Pairs of nT and c values giving the best five average r2 values with both methods were used for further analysis of dose-response curves (Fig. 4A). To estimate the 95% confidence interval for nT and c, we bootstrapped the experimental RI curves 1000 times by perturbing the mean by a random value centered around the mean with standard deviation equal to the SEM of the data.
Analysis of dose-response curves
Dose-response curves for virus stocks with various fm were obtained by normalizing to the %GFP+ cells without drug treatment. When necessary, the IC50 and slope based on the median effect equation (Equation 1) were calculated through least-squares regression analysis as described previously (12). To refine the values for nT and c and find the approximate Kd, we used the pairs of nT and c values estimated from RI curves as a start point in a systematic search for values of σ and Kd that would fit best with the experimental dose-response curves for NNRTIs or PIs against wild-type viruses (Equation 4). Values of σ >nT/2 were not allowed. Once an optimal set of nT, c, σ, and Kd values was obtained, these values were used to predict theoretical dose-response curves for phenotypically mixed virions (Fig. 4A). The r2 values representing fitting of predicted theoretical dose-response curves to the experimental curves were then calculated. Note that it is not necessary to consider the binding of a drug to the inactive enzyme for calculation of fa (Supplementary Note 3).
Mathematical modeling and statistical analyses
All theoretical curve simulations, calculations, curve fitting and statistical analyses were performed using Matlab version 7.1 (The MathWorks) or Microsoft Office Excel 2003.
Supplementary Material
Fig. S1. Effect of heterogeneity in the total number of enzyme molecules (nT) on dose-response curves for anti-HIV drugs.
Fig. S2. Virus production and levels of reverse transcriptase (RT) and integrase (IN) associated with virions produced by cells transfected with wild type-or mutant proviral constructs.
Fig. S3. Mixed dimer formation detected by coimmunoprecipitation (co-IP) of viral protein variants.
Fig. S4. Effect of phenotypic mixing on dose-response curves for NRTIs.
Table S1. r2 values for the fitting of wt dose-response curves with and without the inclusion of σ.
Table S2. Experimental and theoretical values for log (fa/fu) for inhibition of wild-type virus by different concentrations of EFV.
Table S3. Predicted and experimental values for log (fa/fu) for inhibition of phenotypically mixed viruses by different concentrations of EFV.
Table S4. Experimental and theoretical values of log (fa/fu) for inhibition of wild-type virus by different concentrations of LPV.
Table S5. Predicted and experimental values for log (fa/fu) for inhibition of phenotypically mixed viruses by different concentrations of LPV.
Supplementary Note 1. Calculation of slopes for complex dose-response curves generated by the critical subset model.
Supplementary Note 2. Detailed description of model predictions and experimental confirmation for EFV and LPV.
Supplementary Note 3. Effect of drug binding to inactive enzymes on fa.
Acknowledgments
We thank the AIDS Research and Reference Reagent Program of the US National Institutes of Health and Merck for providing anti-HIV-1 drugs. We thank Drs. J. Liu, E. Freire, and X. Yu for helpful discussions. We thank Drs. J. Siliciano, J. Blankson, and C. Durand for helpful review of the manuscript.
Funding: This work was supported by the US National Institutes of Health grant AI081600 and by the Howard Hughes Medical Institute.
Footnotes
This manuscript has been accepted for publication in Science Translational Medicine. This version has not undergone final editing. Please refer to the complete version of record at http://stm.sciencemag.org/content/3/91/91ra63.full.html. The manuscript may not be reproduced or used in any manner that does not fall within the fair use provisions of the Copyright Act without the prior, written permission of AAAS.
Author contributions: L.Shen and R.F.S. designed the experiments. L.Shen, J.L., and S.X. performed the experiments. L.Shen, S.A.R., A.R.S., L.Shan, and R.F.S. analyzed data. All of the authors contributed to the manuscript preparation.
Competing interests: The authors declare that they have no competing interests.
References and Notes
- 1.Gulick RM, Mellors JW, Havlir D, Eron JJ, Gonzalez C, McMahon D, Richman DD, Valentine FT, Jonas L, Meibohm A, Emini EA, Chodakewitz JA. Treatment with Indinavir, Zidovudine, and Lamivudine in Adults with Human Immunodeficiency Virus Infection and Prior Antiretroviral Therapy. N Engl J Med. 1997;337:734–739. doi: 10.1056/NEJM199709113371102. [DOI] [PubMed] [Google Scholar]
- 2.Hammer SM, Squires KE, Hughes MD, Grimes JM, Demeter LM, Currier JS, Eron JJ, Jr, Feinberg JE, Balfour HH, Jr, Deyton LR, Chodakewitz JA, Fischl MA. A Controlled Trial of Two Nucleoside Analogues Plus Indinavir in Persons with Human Immunodeficiency Virus Infection and CD4 Cell Counts of 200 Per Cubic Millimeter Or Less. AIDS Clinical Trials Group 320 Study Team. N Engl J Med. 1997;337:725–733. doi: 10.1056/NEJM199709113371101. [DOI] [PubMed] [Google Scholar]
- 3.Perelson AS, Essunger P, Cao Y, Vesanen M, Hurley A, Saksela K, Markowitz M, Ho DD. Decay Characteristics of HIV-1-Infected Compartments during Combination Therapy. Nature. 1997;387:188–191. doi: 10.1038/387188a0. [DOI] [PubMed] [Google Scholar]
- 4.Staszewski S, Morales-Ramirez J, Tashima KT, Rachlis A, Skiest D, Stanford J, Stryker R, Johnson P, Labriola DF, Farina D, Manion DJ, Ruiz NM. Efavirenz Plus Zidovudine and Lamivudine, Efavirenz Plus Indinavir, and Indinavir Plus Zidovudine and Lamivudine in the Treatment of HIV-1 Infection in Adults. Study 006 Team. N Engl J Med. 1999;341:1865–1873. doi: 10.1056/NEJM199912163412501. [DOI] [PubMed] [Google Scholar]
- 5.Panel on Antiretroviral Guidelines for Adult and Adolescents, Guidelines for the use of Antiretroviral Agents in HIV-1-Infected Adults and Adolescents. DHHS. 2011 Jan;:1–166. [Google Scholar]
- 6.Larder BA, Darby G, Richman DD. HIV with Reduced Sensitivity to Zidovudine (AZT) Isolated during Prolonged Therapy. Science. 1989;243:1731–1734. doi: 10.1126/science.2467383. [DOI] [PubMed] [Google Scholar]
- 7.Coffin JM. HIV Population Dynamics in Vivo: Implications for Genetic Variation, Pathogenesis, and Therapy. Science. 1995;267:483–489. doi: 10.1126/science.7824947. [DOI] [PubMed] [Google Scholar]
- 8.Wei X, Ghosh SK, Taylor ME, Johnson VA, Emini EA, Deutsch P, Lifson JD, Bonhoeffer S, Nowak MA, Hahn BH, Saag MS, Shaw GM. Viral Dynamics in Human Immunodeficiency Virus Type 1 Infection. Nature. 1995;373:117–122. doi: 10.1038/373117a0. [DOI] [PubMed] [Google Scholar]
- 9.Ho DD, Neumann AU, Perelson AS, Chen W, Leonard JM, Markowitz M. Rapid Turnover of Plasma Virions and CD4 Lymphocytes in HIV-1 Infection. Nature. 1995;373:123–126. doi: 10.1038/373123a0. [DOI] [PubMed] [Google Scholar]
- 10.Wodarz D, Nowak MA. Mathematical Models of HIV Pathogenesis and Treatment. Bioessays. 2002;24:1178–1187. doi: 10.1002/bies.10196. [DOI] [PubMed] [Google Scholar]
- 11.Ribeiro RM, Bonhoeffer S, Nowak MA. The Frequency of Resistant Mutant Virus before Antiviral Therapy. AIDS. 1998;12:461–465. doi: 10.1097/00002030-199805000-00006. [DOI] [PubMed] [Google Scholar]
- 12.Shen L, Peterson S, Sedaghat AR, McMahon MA, Callender M, Zhang H, Zhou Y, Pitt E, Anderson KS, Acosta EP, Siliciano RF. Dose-Response Curve Slope Sets Class-Specific Limits on Inhibitory Potential of Anti-HIV Drugs. Nat Med. 2008;14:762–766. doi: 10.1038/nm1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hill AV. The Possible Effects of the Aggregation of the Molecules of Haemoglobin on its Dissociation Curves. J Physiol. 1910;40:iv–vii. [Google Scholar]
- 14.Holpord NHG, Scheiner LB. Understanding the Dose-effect Relationship: Clinical Applications of Pharmacokinetic-pharmacodynamic Models. Clin Pharmacokinet. 1981;6:429–453. doi: 10.2165/00003088-198106060-00002. [DOI] [PubMed] [Google Scholar]
- 15.Chou TC. Theoretical Basis, Experimental Design, and Computerized Simulation of Synergism and Antagonism in Drug Combination Studies. Pharmacol Rev. 2006;58:621–681. doi: 10.1124/pr.58.3.10. [DOI] [PubMed] [Google Scholar]
- 16.Weiss JN. The Hill Equation Revisited: Uses and Misuses. FASEB J. 1997;11:835–841. [PubMed] [Google Scholar]
- 17.Kohlstaedt LA, Wang J, Friedman JM, Rice PA, Steitz TA. Crystal Structure at 3.5 A Resolution of HIV-1 Reverse Transcriptase Complexed with an Inhibitor. Science. 1992;256:1783–1790. doi: 10.1126/science.1377403. [DOI] [PubMed] [Google Scholar]
- 18.Miller M, Schneider J, Sathyanarayana BK, Toth MV, Marshall GR, Clawson L, Selk L, Kent SB, Wlodawer A. Structure of Complex of Synthetic HIV-1 Protease with a Substrate-Based Inhibitor at 2.3 A Resolution. Science. 1989;246:1149–1152. doi: 10.1126/science.2686029. [DOI] [PubMed] [Google Scholar]
- 19.Mitsuya H, Maeda K, Das D, Ghosh AK. Development of Protease Inhibitors and the Fight with Drug-Resistant HIV-1 Variants. Adv Pharmacol. 2008;56:169–197. doi: 10.1016/S1054-3589(07)56006-0. [DOI] [PubMed] [Google Scholar]
- 20.Hoffman A, Goldberg A. The Relationship between Receptor-Effector Unit Heterogeneity and the Shape of the Concentration-Effect Profile: Pharmacodynamic Implications. J Pharmacokinet Biopharm. 1994;22:449–468. doi: 10.1007/BF02353789. [DOI] [PubMed] [Google Scholar]
- 21.Ambrose Z, Julias JG, Boyer PL, Kewalramani VN, Hughes SH. The Level of Reverse Transcriptase (RT) in Human Immunodeficiency Virus Type 1 Particles Affects Susceptibility to Nonnucleoside RT Inhibitors but Not to Lamivudine. J Virol. 2006;80:2578–2581. doi: 10.1128/JVI.80.5.2578-2581.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang X, Kurteva S, Ren X, Lee S, Sodroski J. Stoichiometry of Envelope Glycoprotein Trimers in the Entry of Human Immunodeficiency Virus Type 1. J Virol. 2005;79:12132–12147. doi: 10.1128/JVI.79.19.12132-12147.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Le Grice SF, Naas T, Wohlgensinger B, Schatz O. Subunit-Selective Mutagenesis Indicates Minimal Polymerase Activity in Heterodimer-Associated p51 HIV-1 Reverse Transcriptase. EMBO J. 1991;10:3905–3911. doi: 10.1002/j.1460-2075.1991.tb04960.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hazuda DJ, Felock P, Witmer M, Wolfe A, Stillmock K, Grobler JA, Espeseth A, Gabryelski L, Schleif W, Blau C, Miller MD. Inhibitors of Strand Transfer that Prevent Integration and Inhibit HIV-1 Replication in Cells. Science. 2000;287:646–650. doi: 10.1126/science.287.5453.646. [DOI] [PubMed] [Google Scholar]
- 25.Rose JR, Babe LM, Craik CS. Defining the Level of Human Immunodeficiency Virus Type 1 (HIV-1) Protease Activity Required for HIV-1 Particle Maturation and Infectivity. J Virol. 1995;69:2751–2758. doi: 10.1128/jvi.69.5.2751-2758.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pandey KK, Bera S, Zahm J, Vora A, Stillmock K, Hazuda D, Grandgenett DP. Inhibition of Human Immunodeficiency Virus Type 1 Concerted Integration by Strand Transfer Inhibitors which Recognize a Transient Structural Intermediate. J Virol. 2007;81:12189–12199. doi: 10.1128/JVI.02863-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hare S, Gupta SS, Valkov E, Engelman A, Cherepanov P. Retroviral Intasome Assembly and Inhibition of DNA Strand Transfer. Nature. 2010;464:232–236. doi: 10.1038/nature08784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Briggs JA, Simon MN, Gross I, Krausslich HG, Fuller SD, Vogt VM, Johnson MC. The Stoichiometry of Gag Protein in HIV-1. Nat Struct Mol Biol. 2004;11:672–675. doi: 10.1038/nsmb785. [DOI] [PubMed] [Google Scholar]
- 29.Jacks T, Power MD, Masiarz FR, Luciw PA, Barr PJ, Varmus HE. Characterization of Ribosomal Frameshifting in HIV-1 Gag-Pol Expression. Nature. 1988;331:280–283. doi: 10.1038/331280a0. [DOI] [PubMed] [Google Scholar]
- 30.Pettit SC, Clemente JC, Jeung JA, Dunn BM, Kaplan AH. Ordered Processing of the Human Immunodeficiency Virus Type 1 GagPol Precursor is Influenced by the Context of the Embedded Viral Protease. J Virol. 2005;79:10601–10607. doi: 10.1128/JVI.79.16.10601-10607.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pettit SC, Everitt LE, Choudhury S, Dunn BM, Kaplan AH. Initial Cleavage of the Human Immunodeficiency Virus Type 1 GagPol Precursor by its Activated Protease Occurs by an Intramolecular Mechanism. J Virol. 2004;78:8477–8485. doi: 10.1128/JVI.78.16.8477-8485.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Muller B, Anders M, Akiyama H, Welsch S, Glass B, Nikovics K, Clavel F, Tervo HM, Keppler OT, Krausslich HG. HIV-1 Gag Processing Intermediates Trans-Dominantly Interfere with HIV-1 Infectivity. J Biol Chem. 2009;284:29692–29703. doi: 10.1074/jbc.M109.027144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Agneter E, Singer EA, Sauermann W, Feuerstein TJ. The Slope Parameter of Concentration-Response Curves used as a Touchstone for the Existence of Spare Receptors. Naunyn Schmiedebergs Arch Pharmacol. 1997;356:283–292. doi: 10.1007/pl00005052. [DOI] [PubMed] [Google Scholar]
- 34.Castrignano T, Aluffi-Pentini F, Parisi V. A Novel Parameter to Estimate the Minimum Number of Bound Ligands Needed to Activate an Ion Channel. J Theor Biol. 1999;199:97–103. doi: 10.1006/jtbi.1999.0945. [DOI] [PubMed] [Google Scholar]
- 35.Michel D. Cooperative Equilibrium Curves Generated by Ordered Ligand Binding to Multi-Site Molecules. Biophys Chem. 2007;129:284–288. doi: 10.1016/j.bpc.2007.06.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Effect of heterogeneity in the total number of enzyme molecules (nT) on dose-response curves for anti-HIV drugs.
Fig. S2. Virus production and levels of reverse transcriptase (RT) and integrase (IN) associated with virions produced by cells transfected with wild type-or mutant proviral constructs.
Fig. S3. Mixed dimer formation detected by coimmunoprecipitation (co-IP) of viral protein variants.
Fig. S4. Effect of phenotypic mixing on dose-response curves for NRTIs.
Table S1. r2 values for the fitting of wt dose-response curves with and without the inclusion of σ.
Table S2. Experimental and theoretical values for log (fa/fu) for inhibition of wild-type virus by different concentrations of EFV.
Table S3. Predicted and experimental values for log (fa/fu) for inhibition of phenotypically mixed viruses by different concentrations of EFV.
Table S4. Experimental and theoretical values of log (fa/fu) for inhibition of wild-type virus by different concentrations of LPV.
Table S5. Predicted and experimental values for log (fa/fu) for inhibition of phenotypically mixed viruses by different concentrations of LPV.
Supplementary Note 1. Calculation of slopes for complex dose-response curves generated by the critical subset model.
Supplementary Note 2. Detailed description of model predictions and experimental confirmation for EFV and LPV.
Supplementary Note 3. Effect of drug binding to inactive enzymes on fa.