Abstract
MIF is a pro-inflammatory cytokine and is implicated in cancer. A higher MIF level is found in many human cancer and cancer-prone inflammatory diseases, including chronic pancreatitis and pancreatic cancer. We tested the hypothesis that MIF contributes to pancreatic cancer aggressiveness and predicts disease outcome in resected cases. Consistent with our hypothesis we found that an elevated MIF mRNA expression in tumors was significantly associated with poor outcome in resected cases. Multivariate Cox-regression analysis further showed that MIF is independently associated with patients’ survival (HR=2.26, 95% CI= 1.17–4.37, P=0.015). Mechanistic analyses revealed that MIF overexpression decreased E-cadherin and increased vimentin mRNA and protein levels in pancreatic cancer cell lines, consistent with the features of epithelial-to-mesenchymal transition (EMT). Furthermore, MIF-overexpression significantly increased ZEB1/2 and decreased miR-200b expression, while shRNA-mediated inhibition of MIF increased E-cadherin and miR-200b expression, and reduced the expression of ZEB1/2 in Panc1 cells. Re-expression of miR-200b in MIF overexpressing cells restored the epithelial characteristics, as indicated by an increase in E-cadherin and decrease in ZEB1/2 and vimentin expression. A reduced sensitivity to the chemotherapeutic drug, gemcitabine, occurred in MIF-overexpressing cells. Indicative of an increased malignant potential, MIF over-expressing cells showed significant increase in their invasion ability in vitro, and tumor growth and metastasis in an orthotopic xenograft mouse model. These results support a role of MIF in disease aggressiveness, indicating its potential usefulness as a candidate target for designing improved treatment in pancreatic cancer.
Keywords: MIF, Pancreatic Cancer, EMT, miR-200
Introduction
Pancreatic cancer is one of the most lethal malignancies world-wide and is the 4th leading cause of death due to cancer in the United States with an estimated 43, 920 new cases and 37, 390 deaths in 2012 1. The median survival in pancreatic cancer is less than 6 months and only 6% of patients survive 5 years after diagnosis. Among different types of pancreatic cancer, pancreatic ductal adenocarcinoma (PDAC) is the most common. The dismal prognosis in pancreatic cancer is due to its presentation with an advanced, unresectable, stage and lack of effective therapy [reviewed in 2, 3]. Less than 20% of the patients are diagnosed relatively early and may qualify for surgical resection. However, the median survival even in the surgically resected cases is less than 2 years. Therefore, understanding the biology of pancreatic tumor aggressiveness and identification of novel therapeutic targets is urgently needed to improve treatment and disease outcome.
Evidence from epidemiological and molecular studies support a role of inflammation in the development, progression and therapeutic resistance in pancreatic cancer 4. One such evidence comes from the observation that the risk of developing pancreatic cancer increases several-fold in patients with hereditary and sporadic pancreatitis 5, 6. There is a step-wise accumulation of inflammatory changes during the development of PDAC, intermingled within a characteristic desmoplastic stroma 7. Increase in the level of cytokines, generation of reactive oxygen and nitrogen species, alteration in miRNA expressions, and induction of NF-kB are some of the major inflammation-mediated events that may contribute to the development and progression of cancer 8, 9. Inflammation enhances tumor progression and cancer cell invasion by inducing EMT in pancreatic cancer 10. Macrophage migration-inhibitory factor (MIF) is a proinflammatory cytokine and an important regulator of innate immunity. MIF is produced by a variety of cells including immune and epithelial cells. Its pro-inflammatory activities are mediated by enhancing the expression of Toll-like receptor 4 (TLR4), promoting the production of many pro-inflammatory cytokines, for example IL-6, IL-1B, IL-8, TNF-a, activation of transcription factors and signaling pathways including MAP Kinase, and inhibition of the anti-inflammatory properties of glucocorticoids 11. MIF is expressed at a higher level in several human cancers 12–15. An increased level of MIF is found in the tumor and serum of pancreatic cancer patients 16, 17. Many functions of MIF support its potential involvement in tumorigenesis, for example, MIF functions as a negative regulator of p53 and antagonizes p53-mediated growth arrest and apoptosis 18. It initiates a cascade of events leading to the phosphorylation of ERK1/ERK2, induction of cytoplasmic phospholipase A2, COX2 and generation of prostaglandin E2, induction of NOS2 and NFK-B pathway, and inhibition of cyclin-dependent kinase inhibitor p27Kip1 11. Despite the compelling evidence suggesting a role of MIF in tumorigenesis, its role in pancreatic cancer is not clearly defined.
In the present study we investigated the biological relevance of MIF in pancreatic cancer progression by first investigating its association with patients’ outcome in resected PDAC cases and then exploring its mechanistic role in disease aggressiveness using both in vitro and in vivo strategies in our effort to understand the tumor biology and identify candidate therapeutic targets.
Materials and Methods
PDAC Tissue Samples
Flash-frozen and paraffin embedded primary pancreatic tumor tissue from resected PDAC cases (N=57) came from University of Maryland Medical System (UMMS), Baltimore, MD, through NCI-UMD resource contract and the Department of General and Visceral Surgery, University Medicine Göttingen, Göttingen, Germany. Demographic and clinical information, including age, sex, clinical staging, differentiation grade, resection margin status and survival from the time of diagnosis were available for each donor. Tumor histopathology was classified according to the World Health Organization Classification of Tumors by a Board certified pathologist. The characteristics of the patients are shown in Supplemental Table 1. Use of the clinical specimens was reviewed and permitted by the NCI-Office of Human Subject Research (OHSR, Exempt# 4678) at the National Institutes of Health, Bethesda, MD.
RNA isolation and quantitative RT-PCR (qRT-PCR)
Total RNA was extracted from frozen tumor samples using a standard Trizol protocol (Invitrogen: Carlsbad, CA). RNA from cultured cells was isolated using the Total RNA extraction kit (Norgenbiotek: Thorold, Canada) according to the manufacturer’s protocol. RNA quality was evaluated using Agilent 2100 Bioanalyzer (Agilent Technologies). RNA was reverse-transcribed using Multi Scribe reverse transcriptase (Applied Biosystem, Foster City, CA). Gene-expression levels of MIF, E-cadherin, ZEB1, ZEB2 and miR-200b were measured with probes from Applied Biosystems: MIF (ID Hs00236988_g1), E-cadherin (ID Hs01023894_m1), ZEB1 (ID Hs00232783_m1), ZEB2 (Hs00207691_m1) and miR-200b (ID 002251), with 18 s rRNA (ID Hs99999901_s1) or GAPDH (ID Hs99999905_m1) for gene and U66 (ID 001002) for microRNA, as normalization controls. qRT-PCR reactions were performed in triplicate, using Taqman Gene Expression Assays on an ABI prism 7900HT Sequence Detection instrument (Applied Biosystems: Foster City, CA). All assays were repeated three times. Real-time PCR data were analyzed by relative quantification using the Delta-Delta CT (ΔΔCt) method.
Immunohistochemistry
5 μm thick paraffin sections of tumors and surrounding nontumor tissue from resected PDAC cases were incubated with mouse monoclonal anti-MIF antibody (Abcam, Cambridge, MA). Signals were amplified using biotinylated IgG, followed by horseradish peroxidase-conjugated avidin-biotin complex (Vectastain ABC Kit, Vector Lab, Burlingame, CA) and diaminobenzene (DAB) as the chromogen (Dako Envision System, Dako, Carpinter, CA). Immunostaining was evaluated blindly by a board-certified pathologist assigning the intensity and prevalence score as described elsewhere 19. Briefly, the intensity was assigned a score of 0–3, representing negative, weak, moderate or strong expression, whereas, prevalence was assigned a score of 0–4 representing <10%, 10–30%, >30–50%, >50–80% and >80% cells showing MIF expression. The overall quantitation was then achieved by multiplying the intensity and prevalence score as described elsewhere 20.
Cell Lines and Culture Condition
Human pancreatic cancer cell lines, Capan 2 and Panc 1 were purchased from the American Type Culture Collection, ATCC, (Rockville, MD). Capan 2 cells were grown in McCoy’s medium and Panc-1 cells were grown in Dulbecco’s modified Eagle’s medium, supplemented with 10% fetal bovine serum, penicillin-streptomycin (50 IU/ml and 50 mg/ml, respectively) and 2 mM L-glutamine in a humidified incubator containing 5% CO2 at 37°C. All products for cell culture were purchased from Gibco (Grand Island, NY).
Generation of stable MIF-over expressing and shRNA MIF-knockdown cells using lentiviral vectors
Lentiviral MIF constructs (pLOC-MIF), lentiviral MIF knockdown constructs (pGIPZ-shRNA1 and shRNA2) and viral packaging mixes were purchased from Open Biosystems (Rockford, IL). Lentiviral particles were produced by transfecting 293T cells in a 10-cm dish with 6 μg of lentiviral vector plasmid, 30 μg of trans-lentiviral packaging mix with 50 μl of Lipofectamine 2000 reagent (Invitrogen: Carlsbad, CA) according to the manufacturer’s protocol. Seventy-two hours following transfection, viral supernatant was collected and cellular debris was removed by low-speed centrifugation. The lentiviral particles were concentrated 10-fold by using Lenti-X™ Concentrator over night at 4°C (Clontech, Mountain View, CA), and aliquots were stored at −80°C. The titers of the concentrated viral particles were measured with HT1080 cell line according to manufacturer’s protocol. Twenty-four hours prior to infection, 5×105 Capan 2 or Panc 1 cells were seeded in 6-well plates. Three days after lentivirus infection, blasticidin (10 μg/ml) for MIF-overexpressing cells and puromycine (10 μg/ml) for MIF knock down cells were added to the medium. Approximately every 2–3 days, the culture medium was replaced at least for two weeks. MIF expression was analyzed by western blot and real-time PCR.
Western blotting
Cells were washed in PBS and lysed with RIPA buffer (Invitrogen, Carlsbad, CA). Proteins were electrophoresed under reducing conditions on 4%–12% acryl amide gels (Invitrogen, Carlsbad, CA), and then transferred to a PVDF membrane (Invitrogen, Carlsbad, CA). To block nonspecific binding, the membrane was incubated for 60 min with 0.1% Tween 20 (T-PBS) containing 5% non-fat milk at room temperature. After 3× washing with 0.1% TBS-Tween, the membrane was incubated over night with the human anti-MIF (Abcam: Cambridge, MA), anti-E-cadherin (Cell Signaling: Danvers, MA) or -β-actin antibodies (Sigma Aldrich: St. Louis, MO) in T-PBS. Secondary anti-mouse or -rabbit antibodies with HRP were purchased from Santa Cruz (Santa Cruz, CA). Specific protein was visualized using a Super-Signal protein detection kit (Pierce: Rockford, IL) using manufacturer’s instructions.
Invasion assay
Matrigel invasion assay was performed using the 24-well BD FALCON Cell Culture Insert and BD BioCoat™ Matrigel™ Invasion Chamber (BD Biosciences: Bedford, MA), using manufacturer’s protocol. Briefly, 1×104 Capan-2 and 5×104 Panc-1, MIF over expressing and control cells were plated in each chamber and cellular invasion was assessed 22 hours after plating. The non-invading cells on the top of the membrane were removed and the invaded cells were fixed with methanol, stained with 0.1% crystal violet (Sigma Aldrich: St. Louis, MO) and counted under a microscope in each membrane. Assays were conducted in triplicate.
Cell proliferation assay
Cells were seeded in triplicate at 5×103 cells/well in a 96-well culture plates. After 12 h, cell counting kit-8 (WST-8) colorimetric assay (Dojindo, Kumamoto, Japan) was used to determine cell proliferation at every 24 h intervals according to manufacturer’s protocol. Briefly, cells were incubated with the supplied reagent, which is reduced by the dehydrogenase activity of the living cells producing a yellow color formazine dye, for 3 hrs and the absorbance values of each well were measured with a microplate spectrophotometer (Molecular Devices, Tucson, AZ) at 450 nm and 650 nm from the day 0 to day 4. The assay was repeated three times.
Drug sensitivity assay
Gemcitabine was purchased from Tocris Bioscience (Ellisville, MO). Capan 2 and Panc 1 cells were seeded in 96-well plates (5×103 cells/well). After 12hr, cells were treated with stepwise, 4-fold dilutions of gemcitabine concentration, starting with 50 μM and incubated at 37 °Cfor 96 hr to assess the IC50 value. The cytotoxicity was evaluated by WST-8 colorimetric assay (Dojindo, Kumamoto, Japan). Mean values were calculated from three different wells in triplicates. IC50 concentration of Gemcitabine (0.01 uM) was used to treat both MIF-overexpressing and control Capan 2 and Panc 1 cells to evaluate the difference in sensitivity.
Immunofluorescence
Approximately, 2 ×104 Capan2 MIF over expressing and control cells were seeded on a 4-well Lab-Tek II chamber slide. After 24 hr, the cells were washed with PBS and fixed in 4% paraformaldehyde in PBS buffer at room temperature for 20 min. The cells were then washed with PBS, and blocked in PBS containing 5% goat serum for 90 min at room temperature before incubation with anti-E- cadherin (Abcam: Cambridge, MA) or anti-vimentin (Santa Cruz, CA) and anti-MIF antibody (Invitrogen: Carlsbad, CA) over night. Cells were then washed three times with PBS for 15 min and incubated with anti-rabbit or anti-mouse Alexa Fluor secondary antibodies from Invitrogen (Carlsbad, CA) for 1 hr. Cells were then washed for 15 min, and mounted using the VECTASHIELD® Mounting Medium containing DAPI (Vector Laboratory: Burlingame, CA). All samples were subjected to confocal microscopy and photographed at identical exposure times.
Subcutaneous and orthotopic xenografts in mice to evaluate MIF’s function in tumor growth and metastasis
All animal experiments and maintenance conformed to the guidelines of the Animal Care and Use Committee and of the American Association of Laboratory Animal Care. 5×106 MIF-over expressing and control Capan-2 cells in a total volume of 100 μL of 1/1 (v/v) PBS/matrigel (BD Biosciences: Sparks, MD) were injected subcutaneously into flanks of 8–9 weeks old male athymic nu/nu mice (Harlan Laboratories, Indianapolis, IN) with 5 mice per arm. One week after the injection of tumor cells, subcutaneous tumor volumes (V) were measured weekly with digital calipers (Fisher Scientific: Pittsburgh, PA) and calculated using the formula V = 1/2(ab2), where a is the biggest and b is the smallest orthogonal tumor diameter. On 29th day after tumor injection, the mice were euthanized and the tumors were harvested and measured as described previously 21. To assess primary tumor growth and metastasis, the subcutaneous tumors from MIF-over expressing cells and the controls were harvested and cut into cubes of approximately 1 mm3 and orthotopic xenografts by surgical implantation were performed as described previously 21. After 47 days of implantation, the mice were euthanized. The primary tumors were harvested and weighed, and the tumor volume were measured with calipers of three orthogonal diameters (a, b and c) and calculated using the formula volume = 1/2(abc). Spleen, pancreas, liver, intestine, colon, lymph node, peritoneum, diaphragm, kidney and lungs were inspected for grossly visible metastases.
Statistical Analysis
Kaplan-Meier analysis was performed, using Graphpad Prism 5.0, to evaluate the differences in survival probability in resected pancreatic cancer cases. Univariate and multivariate Cox Proportional-hazards regression analysis was performed using STATA 11 (StataCorp LP, College Station, TX) to investigate the association of MIF expression level in tumors and other clinical factors to determine their association with survival in these cases. Final multivariate models were based on stepwise addition and removal of clinical prognostic factors found to be associated with survival in univariate analysis (P<0.05). These models did not violate proportional hazards assumptions based on Schoenfeld residuals. For these analysis MIF gene expression was dichotomized as high and low based on median value, resection margin status as positive (R1) vs. negative (R0); staging as stage I/IIA vs. stage IIB/III, and differentiation grade as G1–G2 vs. G3–G4. The differences in mRNA and microRNA expression, tumor volume and weight among different comparison groups were evaluated using Student’s t-test.
Results
Increased MIF expression is associated with poor survival in resected PDAC
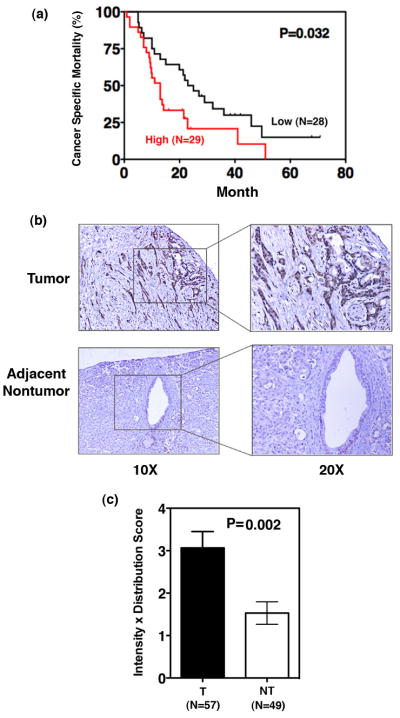
MIF gene expression analysis was performed in resected tumor samples from PDAC cases (N=57) using qRT-PCR and the expression levels were dichotomized at the median into high (above median) and low (below median) expression. Kaplan-Meier analysis showed that patients with a higher MIF expression in tumors had a significantly poorer survival when compared with patients who had a lower MIF expression in their tumors (P=0.032, log-rank test) (Figure 1a). Univariate and multivariate Cox proportional hazard analysis was used to further evaluate the association of MIF expression in tumors and other clinical prognostic factors with patients’ outcome (Supplemental Table 2). Univariate Cox analysis showed that a higher MIF expression (HR, 2.21; 95% CI, 1.16–4.22; P=0.016) and a higher differentiation grade (HR, 1.86; 95% CI, 1.01–3.45; P=0.048) were each associated with poor survival. We did not see any association of tumor stage or resection margin status with survival in our cohort. Furthermore, multivariate analysis showed that MIF was associated with patients’ survival independent of tumor grade (HR, 2.26; 95% CI, 1.17–4.37; P=0.015). These data indicate that MIF is an independent predictor of prognosis in resected PDAC patients. We further evaluated the expression of MIF in paraffin embedded tumors and adjacent nontumor tissue. Immunohistochemical staining showed an increased expression of MIF in cancer cells as compared with the ductal cells in the surrounding nontumor area (P=0.002) (Figure 1b and 1c).
Figure 1. Increased MIF expression is associated with poor survival in resected PDAC.
Patients were classified as high or low MIF gene expression groups based on the MIF-expression in tumors above or below the median values respectively, as determined by qRT-PCR, expression. Kaplan-Meier analysis was performed to assess the difference in survival probability between the two groups of patients (a). Immunohistochemical analysis of MIF in paraffin sections shows an increased expression in tumor cells as compared with the ducts in the adjacent nontumor tissue (b). The quantitation of imunohistochemical staining was performed by multiplying the intensity and prevalence scores. The intensity was assigned a score of 0–3, representing negative, weak, moderate or strong expression, whereas, prevalence was assigned a score of 0–4 representing <10%, 10–30%, >30–50%, >50–80% and >80% cells showing MIF expression (c). (T=tumor, NT=nontumor).
MIF overexpression decreases E-cadherin and increases vimentin in pancreatic cancer cells
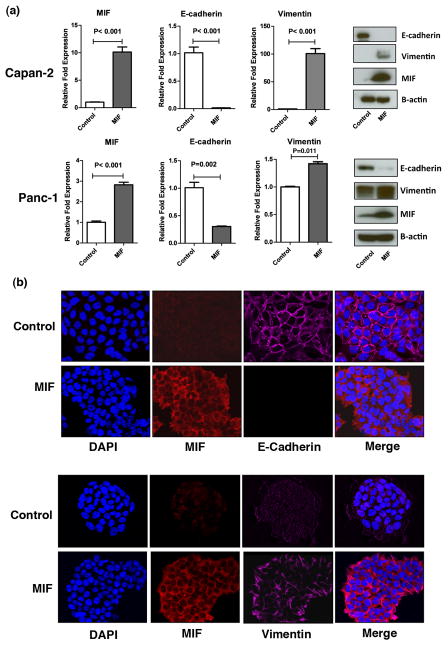
EMT enhances the invasive ability and therapeutic resistance in tumor cells, and is characterized by the alteration in epithelial and mesenchymal marker proteins including a decrease in E-cadherin and an increase in vimentin expression 22, 23. To investigate the mechanistic role of MIF in the progression of pancreatic cancer, we first generated MIF-expressing stable cell lines and determined the expression of EMT-related genes at both the mRNA and protein level. MIF-overexpressing Capan-2 and Panc-1 pancreatic cancer cell lines showed a decrease in E-cadherin (P<0.001; P<0.01) and an increase in vimentin (P<0.001, P=0.011) mRNA and protein expression as determined by qRT/PCR and western blot analysis, respectively (Figure 2a). We further confirmed these observations using immunoflourescence staining as shown in Figure 2b. These data indicate that MIF overexpression induces EMT-related molecular alterations in pancreatic cancer cells.
Figure 2. MIF overexpression induces EMT characteristics in pancreatic cancer cells.
MIF-overexpression reduced E-cadherin and increased vimentin expression at mRNA and protein level in Capan 2 and Panc 1 pancreatic cancer cell lines, constitutively expressing MIF transgene (a). These findings were further confirmed by immunofloresecence staining in Capan 2 cell line (b).
MIF-induced EMT is mediated by miR-200b
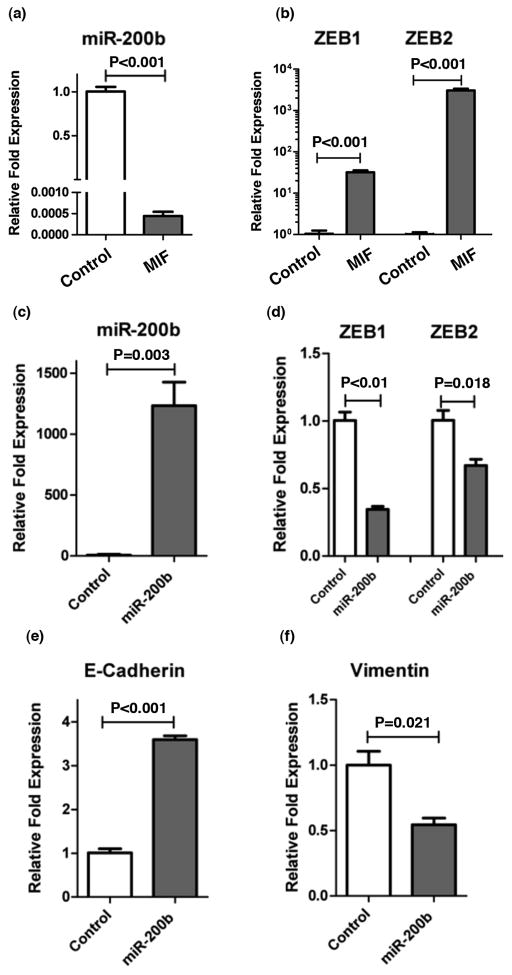
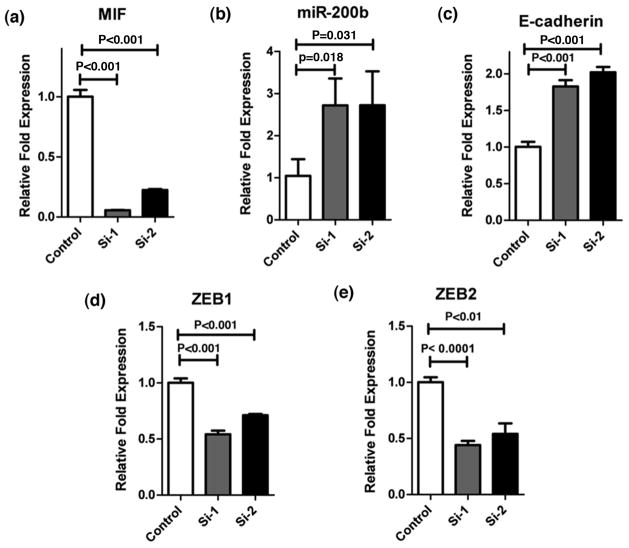
ZEB1/2 transcription factors inhibit E-cadherin and induce EMT, invasion and metastasis 24–26. However, in contrast, miR-200 family members induce mesenchymal to epithelial transition (MET) and there exist a feedback inhibitory loop between miR-200 and ZEB family of transcription factors 27, 28. Therefore, we investigated if MIF-induced, EMT-related changes are mediated by miR-200. MIF over-expression reduced miR-200b (P<0.001) (Figure 3a) and increased ZEB1 (P<0.001) and ZEB2 (P<0.001) (Figure 3b) expression in Capan 2 cells. Furthermore, re-expression of miR-200b in MIF overexpressing cells reduced ZEB1 (P<0.01), ZEB2 (P=0.018) and vimentin (P=0.021) expression (Figure 3c,d,f), and increased the expression of E-cadherin (P<0.001) (Figure 3e). We further confirmed these findings by shRNA-mediated knockdown of MIF in Capan 2 cells. Knocking down MIF significantly increased the expression of miR-200b (P=0.018 and P=0.031) and E-cadherin (P<0.001), while decreasing both ZEB-1 (P<0.001) and ZEB-2 (P<0.001 and P<0.01) (Figure 4). These data indicate that MIF-induced EMT is mediated, at least in part, by miR-200b.
Figure 3. MIF induced activation of EMT is mediated by miR-200b/ZEB axis.
MIF-transgene expressing Capan 2 cells showed a significant decrease in miR-200b expression (a) and an increase in the expression of ZEB1 and ZEB2 (b), which is a strong inducer of EMT. Re-expression of miR-200b in MIF-transgene expressing cells (c) led to a decrease in ZEB1 and ZEB2 (d), which was also accompanied by an increase in E-cadherin (e) and a decrease in Vimentin (f).
Figure 4. shRNA-mediated MIF inhibition restores miR-200b and E-cadherin Expression.
shRNA-lentiviral mediated inhibition of MIF (a) in Capan 2 cells increased the expression of miR-200b (b) as well as E-cadherin (c). MIF-inhibition also led to a significant decrease in both ZEB1 and ZEB2 expression (d and e).
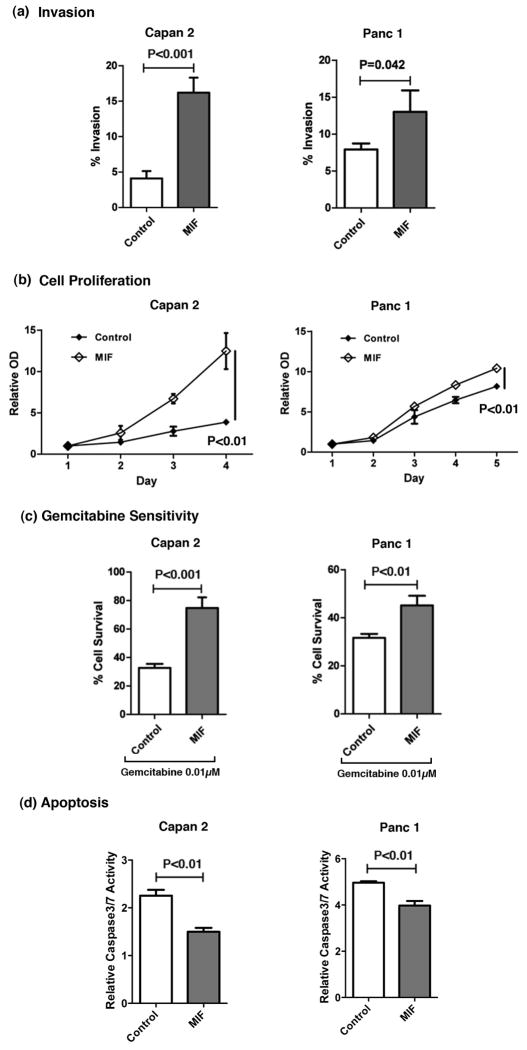
MIF-overexpression enhances invasion and decreases sensitivity of pancreatic cancer cells to chemotherapeutic drug
To investigate the role of MIF in the aggressiveness of pancreatic cancer, we first determined the in vitro invasive properties of MIF-overexpressing Capan 2 and Panc 1 pancreatic cancer cells. Pancreatic cancer cells, overexpressing MIF, showed a significantly higher invasive ability when compared with the vector control cells (P<0.001 and P=0.042) (Figure 5a). Furthermore, MIF overexpressing Capan 2 and Panc 1 cells showed increased cellular proliferation (P<0.01) (Figure 5b) which is consistent with earlier report showing a decrease in cellular proliferation following siRNA-mediated knock down of MIF 16. One of the major hurdles in the treatment of pancreatic cancer is its resistance to chemotherapeutic drugs, and EMT-associated changes are thought to be a factor in conferring therapeutic resistance. We investigated if the level of MIF modulates the sensitivity of pancreatic cancer cells to the standard of care drug, gemcitabine. We found that increased expression of MIF reduced the sensitivity to gemcitabine in both Capan 2 (P<0.001) and Panc 1 (P<0.01) cell lines (Figure 5c). MIF over-expressing cells also showed a significant decrease in apoptosis as measured by caspase 3/7 activity (P<0.01) (Figure 5d), which is consistent with earlier report showing an increase in apoptosis, following the siRNA-mediated inhibition of MIF 16. These data indicate that MIF may contribute to tumor aggressiveness by enhancing the invasive ability of tumor cells and conferring resistance to gemcitabine.
Figure 5. MIF influences invasive ability, proliferation, drug sensitivity and induction of apoptosis in pancreatic cancer cell lines.
Capan 2 and Panc 1 pancreatic cancer cell lines, constitutively expressing MIF transgene, showed an increase in invasive ability (a) and cellular proliferation (b). MIF expressing cells also showed a lower sensitivity to chemotherapeutic drug gemcitabine (0.01μM) as compared to control cells (c), accompanied by a decrease in apoptosis as measured by caspase3/7 activity (d).
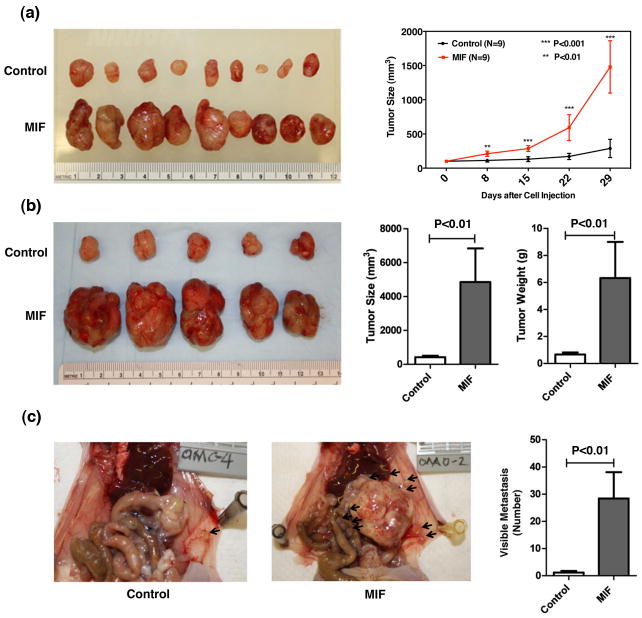
MIF enhances tumor growth and metastasis in mice
To further investigate the role of MIF in tumor growth and aggressiveness in pancreatic cancer, we extended our investigation to subcutaneous implantation of MIF overexpressing stable Capan 2 cells and then implantation of subcutaneous tumors as orthotopic xenograft in mice. Subcutaneous injection of Capan 2 stable MIF-overexpressing or control cells (empty vector) in nude mice showed a significant increase in tumor growth with MIF-overexpressing cells as compared with control (P<0.01 and P<0.001) (Figure 6a). Subcutaneous tumors from Capan 2 stable MIF-transfectants and controls were harvested and cut into pieces of approximately 1 mm3 for orthotopic xenograft by surgical implantation. Forty-seven days following the orthotopic implantation, mice were euthanized. Tumor implants from MIF-tranfectants showed a significant increase in tumor growth [size and weight (P<0.01), Figure 6b], and metastasis (P<0.01) (Figure 6c). The principal sites of distant metastasis included liver, lymph nodes, peritoneum, intestine and spleen (Supplemental Table 3). These in vivo findings indicate that MIF accelerates primary tumor growth and systemic dissemination in pancreatic cancer.
Figure 6. MIF enhances tumor growth and metastasis in mice.
Subcutaneous implant and orthotopic xenograft strategy was used to investigate the influence of MIF on tumor growth and metastasis. Subcutaneous implant of Capan 2 cells, constitutively expressing MIF-transgene, showed a significant increase in tumor growth as compared with control cells (a). Orthotopic xenograft of about 1 mm3 of the subcutaneous tumors by surgical implantation in the pancreas of nude mice showed a significant increase in the growth and metastasis of tumors arising from MIF expressing pancreatic cancer cells (b and c). Data represent Mean ± SD. ** Represents P<0.01 and *** represents P<0.001.
Discussion
Understanding the complex biology of tumor progression and disease aggressiveness is key to the development of an effective therapy in pancreatic cancer. Recent studies have linked inflammation to pancreatic cancer progression and identified inflammation-associated pathways as potential therapeutic targets 4, 29, 30. One of the pro-inflammatory cytokines, MIF, is recognized as a molecular link between inflammation and cancer 31. MIF is upregulated in tumor cells 16 and also in serum samples from PDAC cases 17. However, the role of MIF in pancreatic cancer remains poorly understood. One of the strategies to assess the biological relevance of a gene in cancer is to investigate its association with disease aggressiveness and patients’ outcome 32, 33. In the present study, we observed, for the first time, an association between increased tumor expression of MIF and decreased survival in resected PDAC patients, indicating an oncogenic role of MIF in disease progression. This observation was further supported by our findings of enhanced tumor growth and metastases in an animal model, where we orthotopically implanted tumors arising subcutaneously from MIF-overexpressing pancreatic cancer cells. Furthermore, mechanistic findings in our study showed that MIF induces EMT in pancreatic cancer cells, which likely contributes to the observed MIF-induced increase in tumor progression and therapeutic resistance.
Disease aggressiveness and therapeutic resistance are hallmarks of pancreatic cancer and attributed, in part, to the acquisition of EMT by tumor cells [reviewed in 34,35]. Identification of key pathways involved in the activation of EMT in pancreatic cancer may provide clues to potential therapeutic targets. The signaling pathways, responsible for the activation of EMT in cancer cells are complex and being extensively explored, implicating a number of EMT-inducers including inflammation 22, 36. Interestingly, pancreatic cancer cells attain invasive properties at an earlier stage of tumor development, which is aided by EMT in an inflammatory microenvironment 10. These observations underscore the contribution of EMT in pancreatic tumor progression and suggest that inflammation may contribute to the induction of EMT. MIF is one of several differentially expressed proteins during TGF-β-induced EMT in lung adenocarcinoma cells 37. However, to our knowledge, the findings in the present study provide the first evidence of MIF-induced activation of EMT in pancreatic cancer. Furthermore, microRNAs have been implicated in the regulation of EMT 38–41. miR-200 family members inhibit EMT by targeting ZEB family of transcription factors and there exist a feedback inhibitory loop between miR-200 and ZEB 42, 43. ZEB1/2 are activators of EMT and inhibit the expression of E-cadherin 24, 25. An increased ZEB1 expression is found in PDAC and targeting ZEB1-miR-200 feedback loop is suggested as a promising treatment strategy in pancreatic cancer 42. Our data showed that MIF-induced EMT in pancreatic cancer cells utilizes miR-200/ZEB/E-cadherin axis, thus presenting a potential strategy for regulating miR-200/ZEB interaction, and therefore EMT, by targeting MIF.
Our findings of an association between a higher MIF level in tumors and poor survival in resected pancreatic cancer cases is indicative of a role of MIF in disease aggressiveness, which may be due to an increased growth and metastatic potential, and therapeutic resistance in tumor cells expressing a higher level of MIF. Our observation of an increased proliferation, invasive ability and decreased sensitivity to gemcitabine in MIF-overexpressing pancreatic cancer cells are consistent with this hypothesis. Furthermore, our findings in the subcutaneous and orthotopic xenograft mouse models provide direct evidence of a role of MIF in enhancing the growth and metastasis of pancreatic tumors, indicating its potential as a candidate therapeutic target.
In summary, the findings from this study are consistent with the hypothesis that MIF enhances tumor growth and aggressiveness in pancreatic cancer, and is a candidate prognostic indicator in resected patients. Based on the evidence, indicating a role of MIF in tumor growth, invasiveness and therapeutic resistance in the present study, we propose that MIF may be considered as a candidate target in improving treatment of pancreatic cancer. Future pre-clinical studies are warranted to evaluate the effect of MIF-targeting in pancreatic cancer.
Supplementary Material
Primary pancreatic tumors were obtained from 57 resected cases of PDAC from University of Maryland Medical System, Baltimore and University of Medicine, Gottingen, Germany, with demographic and clinical information. Tumor histopathology was determined by a Board-certified pathologist, using WHO classification of tumors.
Univariate and multivariate Cox Proportional-hazard regression analysis showed an association between a higher MIF expression in tumors and poor survival in resected cases of PDAC. Multivariate analysis used stepwise addition and removal of clinical covariates found to be associated with survival in Univariate model and final models included only those covariates that were significantly associated with survival (P<0.05). These models did not violate proportional hazards assumptions based on Schoenfeld residuals.
Visible metastasis were counted, 47 days after orthotopic implantation of 1 mm3 subcutaneous tumors, produced by MIF-overexpressing or control pancreatic cancer cells, in the pancreas of nude mice.
Novelty and Impact Statement.
Our study made two novel observations that 1) macrophage migration inhibitory factor (MIF) is an independent predictor of clinical outcome in resected pancreatic cancer patients, and 2) MIF enhances disease aggressiveness by inducing epithelial to mesenchymal transition (EMT) in pancreatic cancer and causes resistance to chemotherapeutic drug, gemcitabine. These findings identify MIF as a candidate therapeutic target for pancreatic cancer.
Acknowledgments
Authors would like to thank Drs. Stefan Ambs and Xin Wang for helpful discussions. Handling of clinical samples and maintenance of tissue database by Ms. Elise Bowman is greatly appreciated. We would also like to thank the personnel at UMMS for their efforts in collection of clinical samples under NCI-UMD resource contract. This work was supported by the Intramural Research Program of the National Cancer Institute, Center for Cancer Research, National Institutes of Health.
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J Clin. 2012;62:10–29. doi: 10.3322/caac.20138. [DOI] [PubMed] [Google Scholar]
- 2.Hidalgo M. Pancreatic cancer. N Engl J Med. 2010;362:1605–17. doi: 10.1056/NEJMra0901557. [DOI] [PubMed] [Google Scholar]
- 3.Vincent A, Herman J, Schulick R, Hruban RH, Goggins M. Pancreatic cancer. Lancet. 2011;378:607–20. doi: 10.1016/S0140-6736(10)62307-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Garcea G, Dennison AR, Steward WP, Berry DP. Role of inflammation in pancreatic carcinogenesis and the implications for future therapy. Pancreatology. 2005;5:514–29. doi: 10.1159/000087493. [DOI] [PubMed] [Google Scholar]
- 5.Lowenfels AB, Maisonneuve P, Cavallini G, Ammann RW, Lankisch PG, Andersen JR, Dimagno EP, Andren-Sandberg A, Domellof L. Pancreatitis and the risk of pancreatic cancer. International Pancreatitis Study Group. N Engl J Med. 1993;328:1433–7. doi: 10.1056/NEJM199305203282001. [DOI] [PubMed] [Google Scholar]
- 6.Whitcomb DC, Applebaum S, Martin SP. Hereditary pancreatitis and pancreatic carcinoma. Ann N Y Acad Sci. 1999;880:201–9. doi: 10.1111/j.1749-6632.1999.tb09524.x. [DOI] [PubMed] [Google Scholar]
- 7.Chu GC, Kimmelman AC, Hezel AF, DePinho RA. Stromal biology of pancreatic cancer. J Cell Biochem. 2007;101:887–907. doi: 10.1002/jcb.21209. [DOI] [PubMed] [Google Scholar]
- 8.Schetter AJ, Heegaard NH, Harris CC. Inflammation and cancer: interweaving microRNA, free radical, cytokine and p53 pathways. Carcinogenesis. 2010;31:37–49. doi: 10.1093/carcin/bgp272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hussain SP, Hofseth LJ, Harris CC. Radical causes of cancer. Nat Rev Cancer. 2003;3:276–85. doi: 10.1038/nrc1046. [DOI] [PubMed] [Google Scholar]
- 10.Rhim AD, Mirek ET, Aiello NM, Maitra A, Bailey JM, McAllister F, Reichert M, Beatty GL, Rustgi AK, Vonderheide RH, Leach SD, Stanger BZ. EMT and Dissemination Precede Pancreatic Tumor Formation. Cell. 2012;148:349–61. doi: 10.1016/j.cell.2011.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Calandra T, Roger T. Macrophage migration inhibitory factor: a regulator of innate immunity. Nat Rev Immunol. 2003;3:791–800. doi: 10.1038/nri1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.He XX, Yang J, Ding YW, Liu W, Shen QY, Xia HH. Increased epithelial and serum expression of macrophage migration inhibitory factor (MIF) in gastric cancer: potential role of MIF in gastric carcinogenesis. Gut. 2006;55:797–802. doi: 10.1136/gut.2005.078113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Meyer-Siegler KL, Vera PL, Iczkowski KA, Bifulco C, Lee A, Gregersen PK, Leng L, Bucala R. Macrophage migration inhibitory factor (MIF) gene polymorphisms are associated with increased prostate cancer incidence. Genes Immun. 2007;8:646–52. doi: 10.1038/sj.gene.6364427. [DOI] [PubMed] [Google Scholar]
- 14.Xu X, Wang B, Ye C, Yao C, Lin Y, Huang X, Zhang Y, Wang S. Overexpression of macrophage migration inhibitory factor induces angiogenesis in human breast cancer. Cancer Lett. 2008;261:147–57. doi: 10.1016/j.canlet.2007.11.028. [DOI] [PubMed] [Google Scholar]
- 15.Kamimura A, Kamachi M, Nishihira J, Ogura S, Isobe H, Dosaka-Akita H, Ogata A, Shindoh M, Ohbuchi T, Kawakami Y. Intracellular distribution of macrophage migration inhibitory factor predicts the prognosis of patients with adenocarcinoma of the lung. Cancer. 2000;89:334–41. [PubMed] [Google Scholar]
- 16.Denz A, Pilarsky C, Muth D, Ruckert F, Saeger HD, Grutzmann R. Inhibition of MIF leads to cell cycle arrest and apoptosis in pancreatic cancer cells. J Surg Res. 2010;160:29–34. doi: 10.1016/j.jss.2009.03.048. [DOI] [PubMed] [Google Scholar]
- 17.Winner M, Koong AC, Rendon BE, Zundel W, Mitchell RA. Amplification of tumor hypoxic responses by macrophage migration inhibitory factor-dependent hypoxia-inducible factor stabilization. Cancer Res. 2007;67:186–93. doi: 10.1158/0008-5472.CAN-06-3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hudson JD, Shoaibi MA, Maestro R, Carnero A, Hannon GJ, Beach DH. A proinflammatory cytokine inhibits p53 tumor suppressor activity. J Exp Med. 1999;190:1375–82. doi: 10.1084/jem.190.10.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Glynn SA, Boersma BJ, Dorsey TH, Yi M, Yfantis HG, Ridnour LA, Martin DN, Switzer CH, Hudson RS, Wink DA, Lee DH, Stephens RM, et al. Increased NOS2 predicts poor survival in estrogen receptor-negative breast cancer patients. J Clin Invest. 2010;120:3843–54. doi: 10.1172/JCI42059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Warner SL, Stephens BJ, Nwokenkwo S, Hostetter G, Sugeng A, Hidalgo M, Trent JM, Han H, Von Hoff DD. Validation of TPX2 as a potential therapeutic target in pancreatic cancer cells. Clin Cancer Res. 2009;15:6519–28. doi: 10.1158/1078-0432.CCR-09-0077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Feldmann G, Dhara S, Fendrich V, Bedja D, Beaty R, Mullendore M, Karikari C, Alvarez H, Iacobuzio-Donahue C, Jimeno A, Gabrielson KL, Matsui W, et al. Blockade of hedgehog signaling inhibits pancreatic cancer invasion and metastases: a new paradigm for combination therapy in solid cancers. Cancer Res. 2007;67:2187–96. doi: 10.1158/0008-5472.CAN-06-3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Invest. 2009;119:1420–8. doi: 10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–90. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 24.Singh AB, Sharma A, Smith JJ, Krishnan M, Chen X, Eschrich S, Washington MK, Yeatman TJ, Beauchamp RD, Dhawan P. Claudin-1 up-regulates the repressor ZEB-1 to inhibit E-cadherin expression in colon cancer cells. Gastroenterology. 2011;141:2140–53. doi: 10.1053/j.gastro.2011.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chua HL, Bhat-Nakshatri P, Clare SE, Morimiya A, Badve S, Nakshatri H. NF-kappaB represses E-cadherin expression and enhances epithelial to mesenchymal transition of mammary epithelial cells: potential involvement of ZEB-1 and ZEB-2. Oncogene. 2007;26:711–24. doi: 10.1038/sj.onc.1209808. [DOI] [PubMed] [Google Scholar]
- 26.Spaderna S, Schmalhofer O, Wahlbuhl M, Dimmler A, Bauer K, Sultan A, Hlubek F, Jung A, Strand D, Eger A, Kirchner T, Behrens J, et al. The transcriptional repressor ZEB1 promotes metastasis and loss of cell polarity in cancer. Cancer Res. 2008;68:537–44. doi: 10.1158/0008-5472.CAN-07-5682. [DOI] [PubMed] [Google Scholar]
- 27.Burk U, Schubert J, Wellner U, Schmalhofer O, Vincan E, Spaderna S, Brabletz T. A reciprocal repression between ZEB1 and members of the miR-200 family promotes EMT and invasion in cancer cells. EMBO Rep. 2008;9:582–9. doi: 10.1038/embor.2008.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bracken CP, Gregory PA, Kolesnikoff N, Bert AG, Wang J, Shannon MF, Goodall GJ. A double-negative feedback loop between ZEB1-SIP1 and the microRNA-200 family regulates epithelial-mesenchymal transition. Cancer Res. 2008;68:7846–54. doi: 10.1158/0008-5472.CAN-08-1942. [DOI] [PubMed] [Google Scholar]
- 29.Feurino LW, Fisher WE, Bharadwaj U, Yao Q, Chen C, Li M. Current update of cytokines in pancreatic cancer: pathogenic mechanisms, clinical indication, and therapeutic values. Cancer Invest. 2006;24:696–703. doi: 10.1080/07357900600981398. [DOI] [PubMed] [Google Scholar]
- 30.Uomo I, Miraglia S, Pastorello M. Inflammation and pancreatic ductal adenocarcinoma: a potential scenario for novel drug targets. JOP. 2010;11:199–202. [PubMed] [Google Scholar]
- 31.Bucala R, Donnelly SC. Macrophage migration inhibitory factor: a probable link between inflammation and cancer. Immunity. 2007;26:281–5. doi: 10.1016/j.immuni.2007.03.005. [DOI] [PubMed] [Google Scholar]
- 32.Yeh JJ. Prognostic signature for pancreatic cancer: are we close? Future Oncol. 2009;5:313–21. doi: 10.2217/fon.09.12. [DOI] [PubMed] [Google Scholar]
- 33.Zhang G, Schetter A, He P, Funamizu N, Gaedcke J, Ghadimi BM, Ried T, Hassan R, Yfantis HG, Lee DH, Lacy C, Maitra A, et al. DPEP1 Inhibits Tumor Cell Invasiveness, Enhances Chemosensitivity and Predicts Clinical Outcome in Pancreatic Ductal Adenocarcinoma. PLoS One. 2012;7:e31507. doi: 10.1371/journal.pone.0031507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang Z, Li Y, Ahmad A, Banerjee S, Azmi AS, Kong D, Sarkar FH. Pancreatic cancer: understanding and overcoming chemoresistance. Nat Rev Gastroenterol Hepatol. 2011;8:27–33. doi: 10.1038/nrgastro.2010.188. [DOI] [PubMed] [Google Scholar]
- 35.Singh A, Settleman J. EMT, cancer stem cells and drug resistance: an emerging axis of evil in the war on cancer. Oncogene. 2010;29:4741–51. doi: 10.1038/onc.2010.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lopez-Novoa JM, Nieto MA. Inflammation and EMT: an alliance towards organ fibrosis and cancer progression. EMBO Mol Med. 2009;1:303–14. doi: 10.1002/emmm.200900043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Keshamouni VG, Jagtap P, Michailidis G, Strahler JR, Kuick R, Reka AK, Papoulias P, Krishnapuram R, Srirangam A, Standiford TJ, Andrews PC, Omenn GS. Temporal quantitative proteomics by iTRAQ 2D-LC-MS/MS and corresponding mRNA expression analysis identify post-transcriptional modulation of actin-cytoskeleton regulators during TGF-beta-Induced epithelial-mesenchymal transition. J Proteome Res. 2009;8:35–47. doi: 10.1021/pr8006478. [DOI] [PubMed] [Google Scholar]
- 38.Li Y, VandenBoom TG, 2nd, Kong D, Wang Z, Ali S, Philip PA, Sarkar FH. Up-regulation of miR-200 and let-7 by natural agents leads to the reversal of epithelial-to-mesenchymal transition in gemcitabine-resistant pancreatic cancer cells. Cancer Res. 2009;69:6704–12. doi: 10.1158/0008-5472.CAN-09-1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Du R, Sun W, Xia L, Zhao A, Yu Y, Zhao L, Wang H, Huang C, Sun S. Hypoxia-Induced Down-Regulation of microRNA-34a Promotes EMT by Targeting the Notch Signaling Pathway in Tubular Epithelial Cells. PLoS One. 2012;7:e30771. doi: 10.1371/journal.pone.0030771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu YN, Yin JJ, Abou-Kheir W, Hynes PG, Casey OM, Fang L, Yi M, Stephens RM, Seng V, Sheppard-Tillman H, Martin P, Kelly K. MiR-1 and miR-200 inhibit EMT via Slug-dependent and tumorigenesis via Slug-independent mechanisms. Oncogene. 2012 doi: 10.1038/onc.2012.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Smith AL, Iwanaga R, Drasin DJ, Micalizzi DS, Vartuli RL, Tan AC, Ford HL. The miR-106b-25 cluster targets Smad7, activates TGF-beta signaling, and induces EMT and tumor initiating cell characteristics downstream of Six1 in human breast cancer. Oncogene. 2012 doi: 10.1038/onc.2012.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wellner U, Schubert J, Burk UC, Schmalhofer O, Zhu F, Sonntag A, Waldvogel B, Vannier C, Darling D, zur Hausen A, Brunton VG, Morton J, et al. The EMT-activator ZEB1 promotes tumorigenicity by repressing stemness-inhibiting microRNAs. Nat Cell Biol. 2009;11:1487–95. doi: 10.1038/ncb1998. [DOI] [PubMed] [Google Scholar]
- 43.Brabletz S, Brabletz T. The ZEB/miR-200 feedback loop--a motor of cellular plasticity in development and cancer? EMBO Rep. 2010;11:670–7. doi: 10.1038/embor.2010.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Primary pancreatic tumors were obtained from 57 resected cases of PDAC from University of Maryland Medical System, Baltimore and University of Medicine, Gottingen, Germany, with demographic and clinical information. Tumor histopathology was determined by a Board-certified pathologist, using WHO classification of tumors.
Univariate and multivariate Cox Proportional-hazard regression analysis showed an association between a higher MIF expression in tumors and poor survival in resected cases of PDAC. Multivariate analysis used stepwise addition and removal of clinical covariates found to be associated with survival in Univariate model and final models included only those covariates that were significantly associated with survival (P<0.05). These models did not violate proportional hazards assumptions based on Schoenfeld residuals.
Visible metastasis were counted, 47 days after orthotopic implantation of 1 mm3 subcutaneous tumors, produced by MIF-overexpressing or control pancreatic cancer cells, in the pancreas of nude mice.