Summary
The coordination of the several pathways involved in cell motility is poorly understood. Here, we identify SH3BP1, belonging to the RhoGAP family, as a partner of the exocyst complex, and establish a physical and functional link between two motility-driving pathways, the Ral/exocyst and Rac signaling pathways. We show that SH3BP1 localizes together with the exocyst to the leading edge of motile cells and that SH3BP1 regulates cell migration via its GAP activity upon Rac1. SH3BP1 loss-of-function induces abnormally high Rac1 activity at the front, as visualized by in vivo biosensors, and disorganized and instable protrusions, as revealed by cell morphodynamics analysis. Consistently, constitutively active Rac1 mimics the phenotype of SH3BP1 depletion: slow migration and aberrant cell morphodynamics. Our finding that SH3BP1 down-regulates Rac1 at the motile-cell front indicates that Rac1 inactivation in this location, as well as its activation by GEF proteins, is a fundamental requirement for cell motility.
Introduction
Cell motility is a highly coordinated cellular process that relies on the precise spatiotemporal integration of various pathways (Ridley et al., 2003), and understanding the connections among migration-regulating molecular machineries is a major challenge in cell biology. Recent studies have identified a migration-regulatory pathway that emanates from the RalB GTPase and its downstream effector complex known as the exocyst (Lim et al., 2006; Oxford et al., 2005; Rosse et al., 2006). The exocyst is comprised of eight subunits and tethers secretory vesicles to the plasma membrane (He and Guo, 2009). Although RalB is known to control the assembly and localization of exocyst subunits at the leading edge of motile cells (Rosse et al., 2006), how the exocyst in turn controls migration remains uncertain. One contributing mechanism was revealed recently when the exocyst complex was shown to regulate the dynamics of cell-matrix adhesion by coordinating the activities of aPKC (atypical Protein Kinase C) and JNK (Jun N-terminal Kinase) (Rosse et al., 2009). The exocyst is also thought to contribute to polarized delivery of regulatory molecules to the migration front, but clear experimental evidence for this is lacking.
The small GTPases of the Rho family (Cdc42, Rac, Rho) regulate cell motility by controlling the dynamics of the actin cytoskeleton (Raftopoulou and Hall, 2004). Specifically, Cdc42 is crucial for establishing front-rear polarity, Rac1 for producing networks of polymerized actin at protrusions, and RhoA for inducing acto-myosin contractility. We reasoned that there should exist some regulatory mechanisms connecting the Ral/exocyst signaling pathway to the action driven by Rho-family GTPases. In pursuing this possibility, we identified a molecular link for the coordination between Ral and Rac during migration: the RhoGAP SH3BP1, which partners with the exocyst complex to spatially restrict Rac1 activity. Specifically, SH3BP1 inhibits Rac1 activity by promoting the hydrolysis of bound GTP to GDP, and failure of this Rac1 inactivation leads to anarchic protrusions and ineffective migration.
Results
The RhoGAP SH3BP1 associates with the exocyst complex
SH3BP1 (SH3-domain Binding Protein 1, also known as 3BP-1; NP_061830) was identified in a series of yeast two-hybrid screens aimed at identifying partners of the eight subunits of the exocyst complex; it was found to bind to both the Exo84 and Sec8 subunits. SH3BP1 contains an N-terminal BAR (Bin–Amphiphysin–Rvs) domain (putatively involved in protein-protein interactions and in binding to curved membranes), a central RhoGAP domain, and a C-terminal tail with several proline-rich sequences (Cicchetti et al., 1992). The smallest fragments of SH3BP1 recovered from the screens were amino acids 79-255 for the Exo84 interaction, and amino acids 65-257 for the Sec8 interaction; thus the putative exocyst-interacting domain is in the N-terminal BAR-domain of SH3BP1 (Figure 1A). The SH3BP1-exocyst interaction appears to be specific since: i) SH3BP1 was the only BAR-containing protein identified in the exocyst screens, in spite of the fact that 10 other BAR-containing proteins have been identified in other screens of the same library, and ii) the same SH3BP1 region was found in only one other screen, although more than 1200 screens have been performed on the same library with unrelated bait proteins (data not shown).
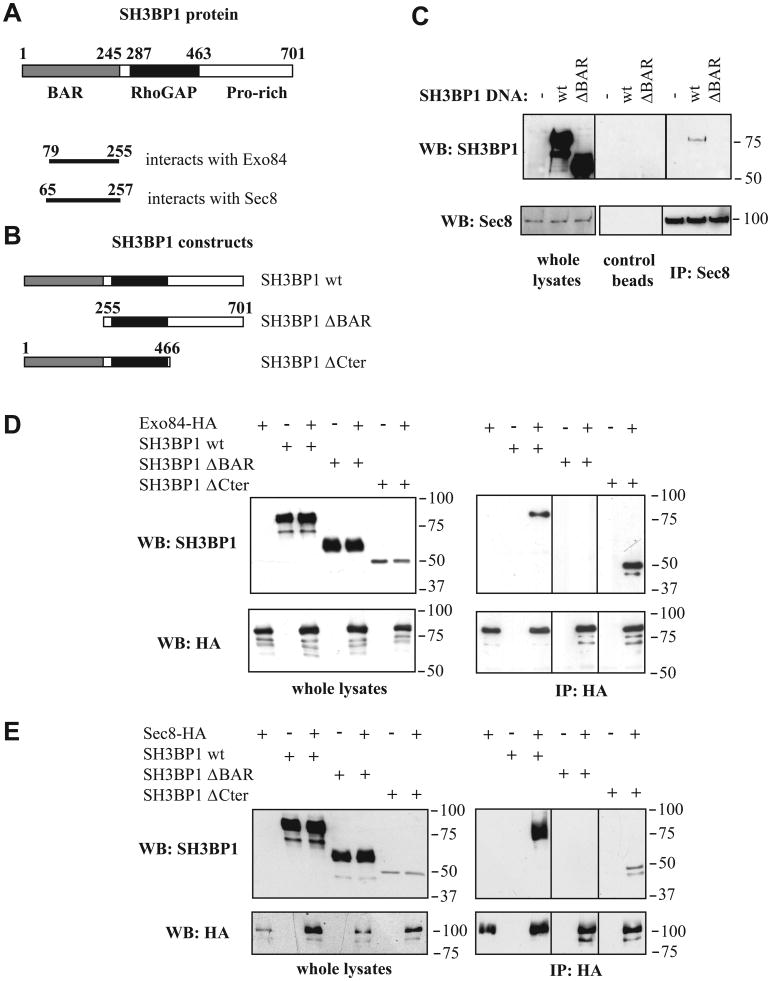
Figure 1. The RhoGAP SH3BP1 associates with the exocyst complex.
A. Primary structure of SH3BP1 and two-hybrid results. The human SH3BP1 protein (701 amino acids) contains a BAR domain, a RhoGAP domain, and a C-terminal tail with several proline-rich motifs. The domains that are minimally required for the interactions with Exo84 and Sec8, as defined by two-hybrid screening, are shown.
B. Constructs of SH3BP1 used in this work.
C. Endogenous exocyst interacts in vivo with SH3BP1. 293T cells were transfected with vectors expressing wild-type (wt) or ΔBAR SH3BP1. Endogenous Sec8 was immunoprecipitated with anti-Sec8 antibodies covalently linked to beads. Associated SH3BP1 proteins were detected by Western blotting with anti-SH3BP1 antibodies.
D. Co-immuno-precipitation of Exo84 and SH3BP1. 293T cells were transfected with vectors expressing Exo84-HA and SH3BP1 (wild-type, ΔBAR or ΔCter). Exo84-HA was immuno-precipitated with anti-HA antibodies and associated SH3BP1 proteins were detected by Western-blotting.
E. Co-immunoprecipitation of Sec8 and SH3BP1. 293T cells were transfected with vectors expressing Sec8-HA and SH3BP1 (wild-type, ΔBAR or ΔCter). Sec8-HA was immuno-precipitated with anti-HA antibodies and associated SH3BP1 proteins were detected by Western-blotting.
We demonstrated an association in vivo between SH3BP1 and the exocyst by showing that over-expressed full-length SH3BP1 co-immunoprecipitated with endogenous Sec8. When the BAR domain was deleted, this association was abolished, confirming that this domain is required for the interaction between SH3BP1 and the exocyst (Figure 1C). In these experiments, only a small fraction of SH3BP1 associated with the endogenous exocyst, possibly because of the large excess of over-expressed SH3BP1 or because of the highly regulated nature of the interaction. In addition, by over-expressing Exo84 or Sec8 along with various SH3BP1 forms (Figures 1B), we found that both of these exocyst components can interact with full-length SH3BP1 and with a SH3BP1 form lacking the C-terminal tail, but not with a SH3BP1 form lacking the N-terminal BAR-domain (Figures 1D and 1E).
Thus, both yeast two-hybrid and co-immunoprecipitation studies indicate that SH3BP1 binds to Exo84 and Sec8, and that these interactions are mediated by the SH3BP1 BAR-domain. We did not succeed in co-immunoprecipitating endogenous exocyst subunits with endogenous SH3BP1. We reason that if the two proteins interact only very locally, such an association would be masked by the vast excess of non-associated exocyst and SH3BP1.
SH3BP1 localizes at the front of motile cells, together with the exocyst
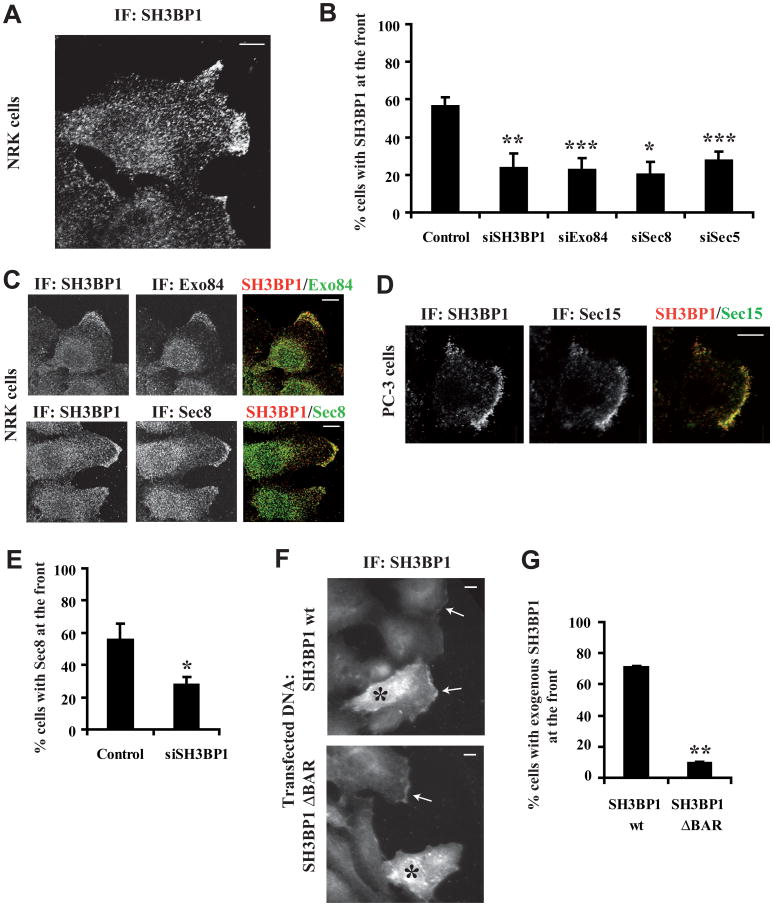
We studied the localization of SH3BP1 in migrating cells. In Normal Rat Kidney (NRK) cells, endogenous SH3BP1 was most prominently localized at the leading edge of motile cells (Figure 2A). This staining at the leading edge was specific, was reproducible using three different antibodies (Figure S1), and was strongly reduced in cells depleted of SH3BP1 by RNAi (Figures 2B and S1). Confocal microscopy analysis of cells co-stained for SH3BP1 and Exo84 or Sec8 showed extensive regions of co-localization at the leading edges of migrating cells, but not elsewhere in the plasma membrane or in the cytoplasm (Figure 2C). We confirmed that SH3BP1 also co-localized with the exocyst (Sec15 subunit) at the leading front in a second model for cell motility: wound healing in cultures of the human PC-3 prostate tumour cell line (Figure 2D). Depletion of exocyst components (Exo84, Sec8, Sec5) by RNAi reduced recruitment of SH3BP1 to the leading edge (Figure 2B) and, conversely, depletion of SH3BP1 reduced recruitment of the exocyst (Sec8 subunit) to the leading edge (Figure 2E), indicating that localization is mutually dependent. Taken together, these results are consistent with SH3BP1 and the exocyst being associated at the leading front, where SH3BP1 might contribute to the molecular machinery that is responsible for migration. While exogenously expressed full-length SH3BP1 localized to the front as does endogenous protein, the SH3BP1 ΔBAR construct was not recruited to the leading edge (Figures 2F and 2G), suggesting that the BAR domain and potentially the interaction with exocyst is required to transport SH3BP1 to the front.
Figure 2. SH3BP1 localizes at the front of motile cells, together with the exocyst.
A. Localization of SH3BP1 at the front. NRK cells were fixed during migration, stained with an anti-SH3BP1 antibody (Everest Biotech), and observed by confocal microscopy.
B. Effects of SH3BP1 or exocyst depletion on SH3BP1 recruitment to the front. NRK cells were treated with control siRNA targeting Luciferase (control), siRNA targeting SH3BP1 (siSH3BP1-2) or exocyst subunits, fixed during migration and stained for SH3BP1. Cells with a fluorescent signal at the leading edge were counted (at least 100 cells per condition per experiment) in 2-6 independent experiments per siRNA. The mean percentages of positive cells and the Standard Error of Means (SEM) are shown.
C. Co-localization of SH3BP1 with Exo84 or Sec8. NRK cells were co-stained for SH3BP1/Exo84 and SH3BP1/Sec8. Confocal microscopy merged images on the right were generated using ImageJ software (NIH).
D. Co-localization of SH3BP1 with Sec15 at the leading edge of migrating prostate tumor PC-3 cells. Confluent PC-3 cell layers were wounded and allowed to heal for 6 hrs. Cells were fixed and co-stained with anti-SH3BP1 and anti-Sec15 antibodies.
E. Depletion of SH3BP1 affects exocyst localization. Motile NRK cells, treated with siLuc (control) or siSH3BP1-2, were stained for Sec8. Quantification was performed for 3 independent experiments.
F. BAR domain is required to localize SH3BP1 at the front. NRK cells were transfected with vectors expressing SH3BP1 wt or SH3BP1 ΔBAR and stained for SH3BP1. Stars indicate transfected cells over-expressing the SH3BP1 proteins. Arrows indicate regions with endogenous or exogenous SH3BP1 at the front.
G. Quantification of panel F experiments. Quantification was performed for 3 independent experiments. Notice that Cherry-fused SH3BP1 wt and ΔBAR localized as their non-tagged versions (data not shown)
Bars on images indicate 10 microns. Bars on graphics represent SEM (Standard Error of Means). * indicates p<0.05, ** p<0.01 and *** p<0.001 (Student t-test). All cell images of this article were rotated such as motility direction is horizontal and rightward.
SH3BP1 regulates cell motility via its GAP activity
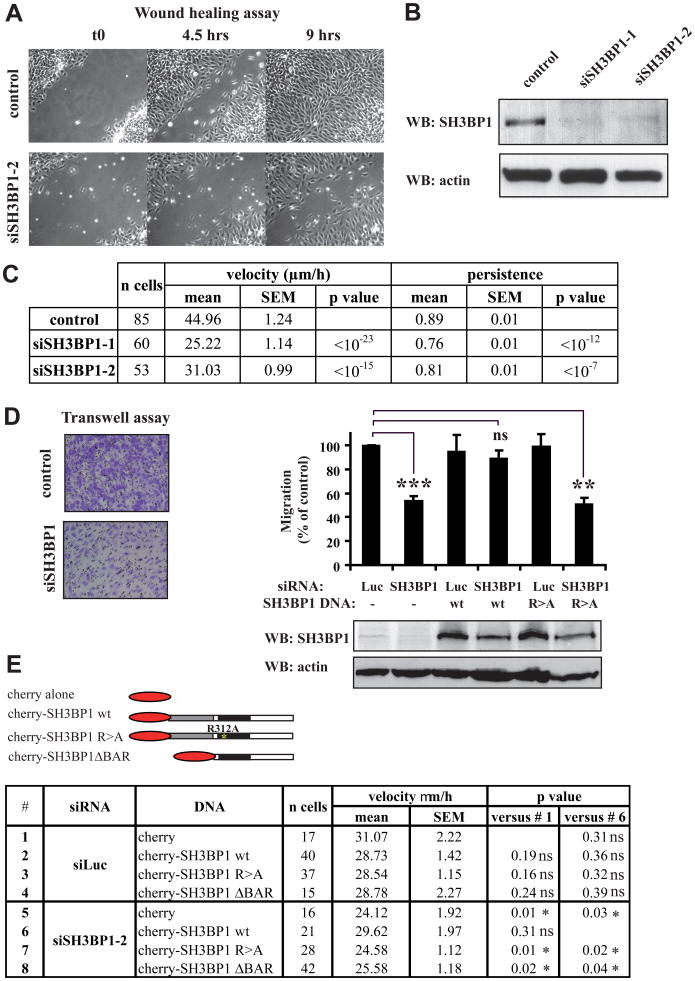
We assessed the contribution of SH3BP1 to cell migration by selectively depleting the SH3BP1 product via an RNAi approach. We first carried out wound-healing assays using NRK cells, and found that SH3BP1 depletion (>85%, with two independent siRNAs) (Figure 3B) strongly inhibited wound closure (Figures 3A and Movie S1). Cell tracking analysis indicated that average speed of cell migration during wound closure was reduced by 30-45% in this context, and also revealed a slight but significant (10-15%) decrease in persistence of migration (Figure 3C). SH3BP1 depletion also resulted in robust inhibition of wound closure in other cell lines, including HEK-HT (human embryonic kidney) and RPE1 (human retinal pigment epithelial) cells (data not shown), suggesting that the role of SH3BP1 in regulating cell migration is general and conserved.
Figure 3. SH3BP1 regulates cell motility.
A. Depletion of SH3BP1 inhibits cell migration in wound healing assay. NRK cells were transfected with siLuc (control) or siSH3BP1-2. Time-lapse acquisitions of a representative experiment are shown. See Movie S1 for the entire video sequence.
B. Validation of SH3BP1 protein depletion. Efficiency of SH3BP1 depletion by two siSH3BP1s (siSH3BP1-1, siSH3BP1-2) was verified by Western blotting.
C. Cell tracking analysis. Individual cells from three independent wound-healing experiments were tracked using the ImageJ software. p values are results of a Student t-test.
D. Depletion of SH3BP1 inhibits cell migration, which is rescued by wild-type SH3BP1, but not by GAP-deficient SH3BP1. NRK cells were transfected with siRNAs (Luc or SH3BP1-2) and with DNAs (empty vector (-), wild-type SH3BP1 (wt), and R312A SH3BP1 (R>A)). 75000 cells were added to the upper chamber and allowed to migrate for 6 to 12 hr, with the exact time varying from one experiment to another. Representative fields of crystal violet-stained cells that had migrated to the lower surface of the porous membrane are shown for control (Luc, -) and SH3BP1-depleted (SH3BP1, -) cells (left panel). Motility was expressed as a percentage of treated cells, relative to control cells (Luc, -), that had migrated to the lower chamber (right, upper panel). Mean motilities were calculated from four independent experiments. Bars represent SEM. ** indicates p<0.01, *** p<0.001, and ns (not significant) p=0.1 (Student t-test). The level of exogenous SH3BP1 was roughly 10-fold that of the endogenous protein (right, lower panel).
E. Wild-type SH3BP1, but neither GAP-deficient SH3BP1 nor ΔBAR mutant, rescues the migration defect of SH3BP1-depleted cells in wound healing assay. NRK cells were transfected with siRNAs (Luc or SH3BP1-2) and with plasmids expressing either cherry alone, or cherry-fused SH3BP1 wt, cherry-fused SH3BP1 R>A or cherry-fused SH3BP1 ΔBAR. Fluorescence images showed that expression levels of cherry constructs were similar. Red-fluorescent cells were tracked during wound closure as described in C.
Consistent with the wound-healing assays, Boyden chamber assays revealed that cells treated with a siRNA against SH3BP1 were defective in migration (Figure 3D). This defect was corrected by subsequent DNA transfection with a vector expressing a human form of SH3BP1 that is resistant to the SH3BP1 siRNA. In contrast to the expression of wild-type SH3BP1, expression of the R312A SH3BP1 mutant, whose GAP activity is impaired due to substitution of the critical “arginine finger” in the GAP domain (Bos et al., 2007) (Figure S2A), did not lead to the rescue of normal motility (Figure 3D). We confirmed these results by tracking cells expressing cherry-fused alleles of SH3BP1 in the context of a wounded monolayer where endogenous SH3BP1 had been depleted or not by RNAi. Speeds of cells in the different genetic contexts are listed in Figure 3E. Cells where endogenous SH3BP1 was depleted, but which expressed a siRNA-resistant SH3BP1, migrated as fast as control cells. This was not the case when a siRNA-resistant SH3BP1 R312A mutant was expressed, confirming the requirement of the GAP activity of SH3BP1. When the SH3BP1 ΔBAR allele was expressed, SH3BP1-depleted cells did not recover normal speed, pointing out the importance of the BAR domain for SH3BP1 function during migration.
RalB, the exocyst complex and SH3BP1 do not control the orientation of the microtubule-organizing centre (MTOC)
The isolated RhoGAP domain of SH3BP1 displays GAP activity towards Cdc42 and Rac1, but not towards RhoA (Cicchetti et al., 1995), in vitro. We obtained similar results in vivo, by expressing the full-length SH3BP1 protein together with FRET-based Raichu probes (Nakamura et al., 2006) (tools with which to monitor the activity of Cdc42, Rac1, and RhoA; Figures S3A and S3B). Consistent with the results of previous pull-down studies (Lu and Mayer, 1999), we observed a small preference for Rac1 rather than Cdc42 as substrate.
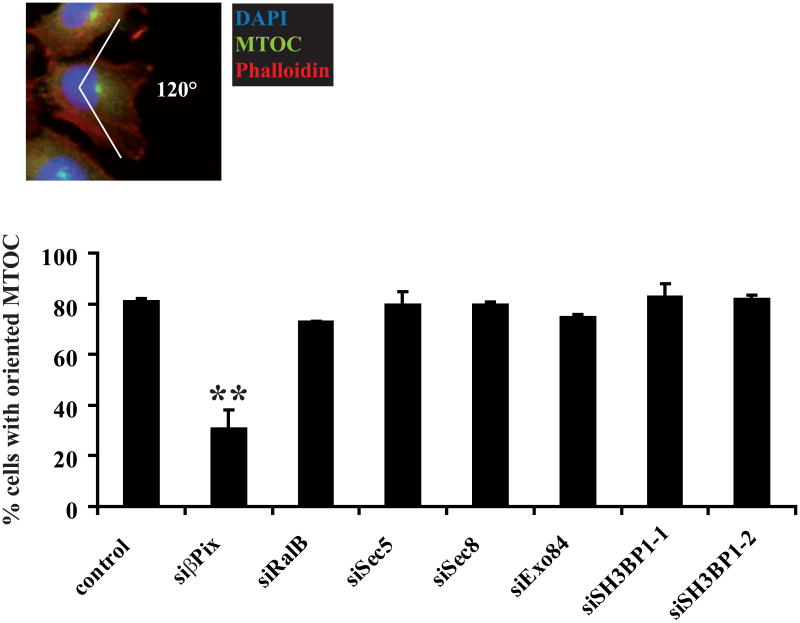
We questioned whether the physiological target of SH3BP1 in motile cells is Cdc42, Rac1, or both. Reorientation of the microtubule-organizing centre (MTOC) and Golgi apparatus in front of the nucleus serves as a landmark of cell polarization during migration, and is controlled by Cdc42 activity (Etienne-Manneville, 2004; Ridley et al., 2003). We confirmed that, as in previous studies (Osmani et al., 2006), the RhoGEF β-PIX strongly inhibited MTOC orientation in our cells under our experimental conditions. MTOC repositioning in front of the nucleus was not affected by the depletion of RalB, exocyst components (Sec5, Sec8, Exo84), or SH3BP1 (Figure 4). Western blot analysis confirmed that each of the tested proteins was efficiently depleted (Figures S3C and 3B). Moreover, expression of SH3BP1 ΔBAR mutant, which is defective in exocyst binding, did not perturb the MTOC orientation (Figure S3D), indicating that interaction with exocyst is not sufficient for SH3BP1 to select Rac over Cdc42 as substrate.
Figure 4. Depletion of RalB, exocyst or SH3BP1 does not perturb the orientation of the microtubule-organizing centre (MTOC).
NRK cells were treated with siLuc (control), siβPix, siRalB, siSec5, siSec8, siExo84, siSH3BP1-1, or siSH3BP1-2 and subjected to wound-healing experiments. The MTOC was stained with anti-pericentrin antibodies, the nucleus with DAPI, and the actin fibers with phalloidin. The mean percentage of correctly oriented cells, i.e. those in which the MTOC faced the wound (within a 120° angle), was calculated from at least three independent experiments for each siRNA. At least 100 cells per condition per experiment were counted. Error bars represent SEM. ** indicates p<0.01 (Student t-test).
We conclude that the RalB/exocyst/SH3BP1 pathway does not regulate cell migration by controlling MTOC reorientation, and that it does not control Cdc42 activity in motile cells. It remains unclear how SH3BP1 constrains its biochemical activity on Rac1 in the context of cell motility.
SH3BP1 is required to inactivate Rac1 at the front
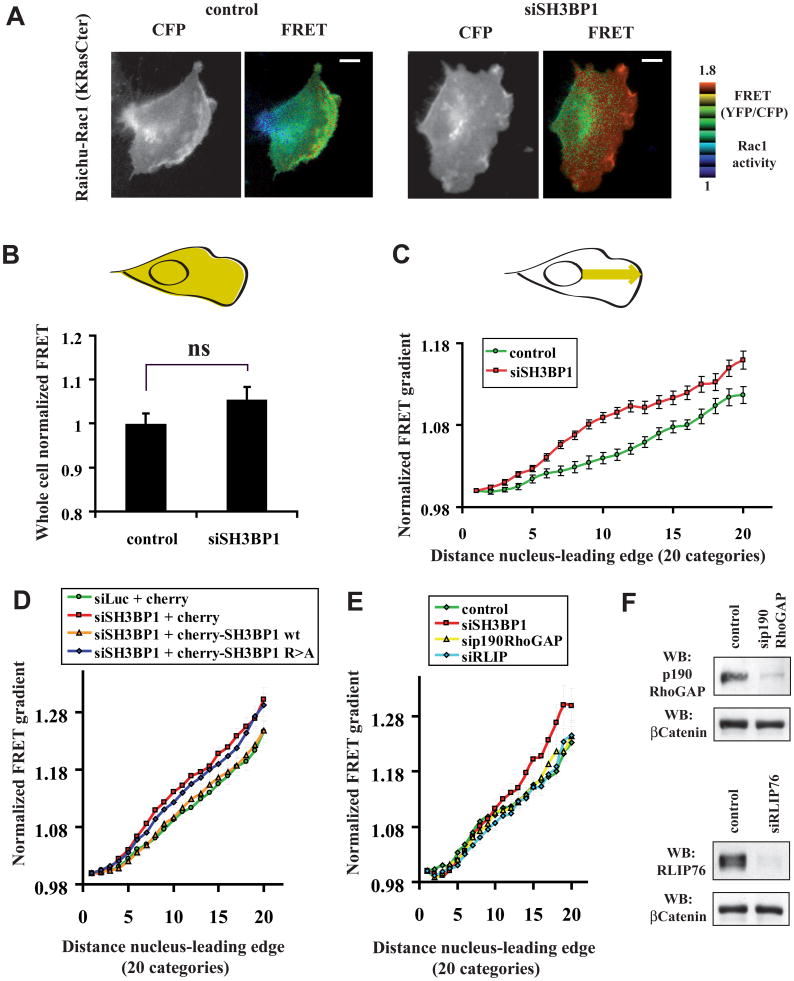
We directly tested the hypothesis that Rac1 is the SH3BP1 target relevant to cell migration, using a FRET approach with Raichu biosensors to monitor the spatiotemporal activation of Rac1 in living motile cells. In analyzing the first row of motile NRK cells that express Raichu-Rac1 (KRasCter; targeted to the plasma membrane via the K-Ras C-terminal tail) (Itoh et al., 2002; Nakamura et al., 2006) (starting acquisition and analysis 3 hr post-wounding), we observed that Rac1 was activated in a dynamic gradient that grew more intense toward the leading edge, as previously reported for other cell types (Itoh et al., 2002; Kraynov et al., 2000). Control nonfunctional Raichu probes did not present a FRET gradient pattern and did not show any variation upon SH3BP1 silencing (Figure S4A). In cells treated with siSH3BP1, Rac1 appeared to be activated more strongly and across a broader area (Figure 5A and Movie S2). We quantitatively compared FRET signals on the total cellular area in control cells (n=33) and SH3BP1-depleted cells (n=28). The average Rac1 activity was only slightly higher in SH3BP1-depleted cells than in control cells (p=0.07, Student t-test) (Figure 5B), indicating that the loss of SH3BP1 did not significantly affect the global levels of active Rac1. However, when we compared FRET signals spatially along a line going from the nucleus to the leading edge, we found that the average Rac1 gradient was significantly upregulated in siSH3BP1-treated cells compared to control cells (Figure 5C). These measurements indicate that SH3BP1 loss induced a localized, rather than global, defect in Rac1 activity. We obtained very similar results with another probe, Raichu-Rac1 (Rac1Cter), which includes the C-terminus of Rac1 and therefore better reflects the localization of endogenous Rac1 (Figures S4B, S4C and S4D). Importantly, the spatial defect in Rac1 activity of SH3BP1-depleted cells was rescued by expression of wild-type SH3BP1, but not of the R312A GAP-defective mutant (Figure 5D). The action of SH3BP1 at the leading edge appears rather specific since silencing of other two GAP proteins, p190RhoGAP and RLIP76/RalBP1, acting on Rac1 (Cantor et al., 1995; Jullien-Flores et al., 1995; Ligeti et al., 2004), did not perturb the Rac1 activity gradient (Figure 5E), despite their efficient depletion (Figure 5F).
Figure 5. Spatial regulation of Rac1 activity in motile cells requires SH3BP1.
A. Visualization of Rac1 activity by FRET. NRK cells were transfected with siLuc (control) or siSH3BP1-2, twenty four hours later transfected with plasmid expressing Raichu-Rac1 (KRasCter), wounded, and visualized live by FRET microscopy. Bars, 10 microns. See also Movie S2.
B. Whole-cell Rac1 activity. YFP and CFP images were acquired for each motile NRK cell, and mean intensities were measured for the entire cell surface (area in yellow in the cell drawing). Single-cell FRET=YFP/CFP ratios (indicators of Rac1 activity) were calculated for each experiment, and were normalized against the average FRET values for siLuc-treated cells. n=33 for control siLuc and n=28 for siSH3BP1, from three independent experiments. Bars represent SEM. Note that the difference between the two cell populations is not significant (p=0.07, Student t-test).
C. Gradients of Rac1 activity. We used the line-scan function of the MetaMorph software to measure FRET (i.e. Rac1 activity) along 4 μm-wide strips, starting from the nucleus and ending at the leading edge following the direction of cell motility (yellow arrow in the cell drawing). Details of the normalization procedure are described in Experimental Procedures. The resulting FRET curves represent the averaged gradient of Rac1 activity at the front of several cells in three independent experiments. n=45 for control siLuc and n=34 for siSH3BP1. Bars represent SEM. Note that the difference between the two curves is highly significant: p<0.01 for categories 4, 6, 17, 19, and 20; p<0.001 for categories 7, 8, 9, 10, 11, 12, 13, 14, 15, and 16 (Student t-test).
D. Wild-type SH3BP1, but not GAP-deficient SH3BP1 mutant, rescues the spatial Rac1 activity in SH3BP1-depleted cells. NRK cells were transfected with siLuc or siSH3BP1-2, and twenty-four hours later co-transfected with plasmids expressing Raichu-Rac1 and the indicated cherry constructs. Red-fluorescent cells were analyzed by FRET microscopy as described in panel C. Curve slopes in panels D and E are slightly different from panel C because cell culture conditions and illumination settings were not identical. Number of cells (n) analyzed per condition was between 34 and 46. The highly significant difference between the curves of siLuc-treated and siSH3BP1-treated cells was confirmed when cherry only was expressed (siLuc + cherry versus siSH3BP1 + cherry): p<0.05 for categories 6 and 20; p<0.01 for categories 7, 11, 14, 15, 16, 17, 18 and 19; p<0.001 for categories 8, 9, 10, 12, 13 (Student t-test). The curve of siSH3BP1-treated cells expressing cherry-SH3BP1 wt is undistinguishable from the control (siLuc + cherry): p values for all categories indicate not significant difference. Similarly, the curve of siSH3BP1-treated cells expressing cherry-SH3BP1 R>A is not statistically different from that of SH3BP1-depleted cells (siSH3BP1 + cherry).
E. Depletion of p190RhoGAP or RLIP76 does not perturb Rac1 activity gradient. NRK cells, treated with siLuc (control) or with siRNA against one among three GAP proteins (SH3BP1, p190RhoGAP or RLIP76), were analyzed as in panel C. n=12 to 16. The control curve was statistically different from that of siSH3BP1-treated cells (p<0.05 for categories 14, 15, 16, 17, 18, 19, 20), but not from those of sip190RhoGAP or siRLIP76 conditions.
F. Efficiency of p190RhoGAP and RLIP76 silencing was verified by Western blotting.
Taking these results together, we conclude that the GAP SH3BP1 is required to locally down-regulate Rac1 at the leading front.
Depletion of SH3BP1 dramatically perturbs the organization and dynamics of cell protrusions
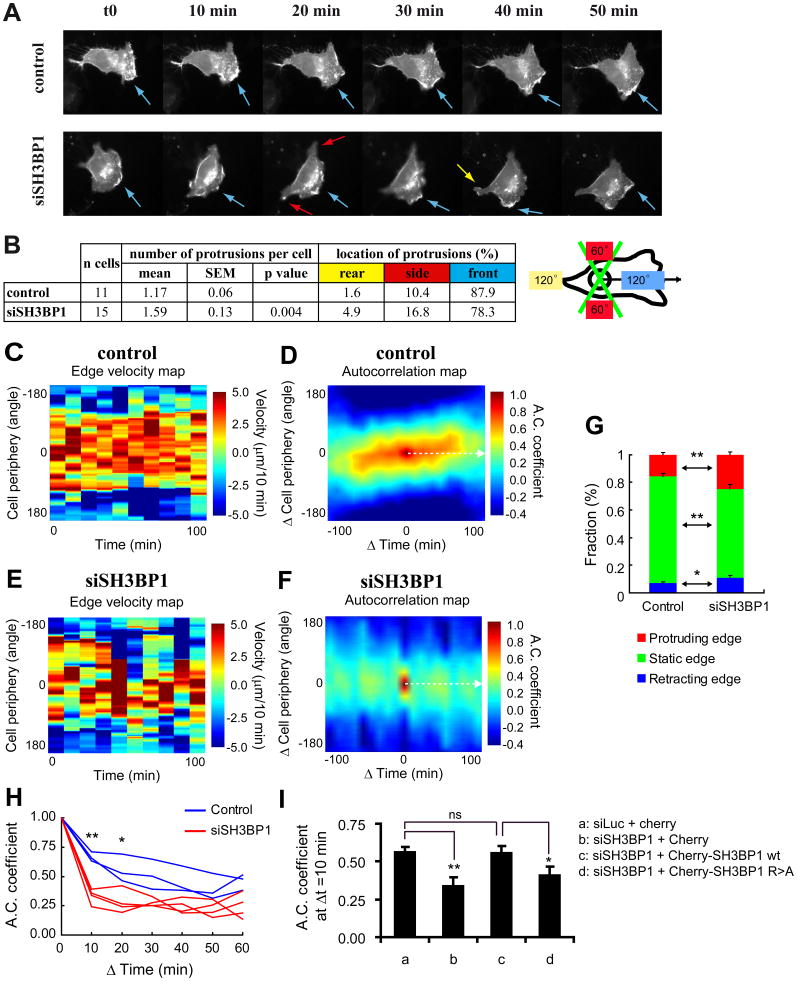
Since Rac1 activity intimately regulates the formation of protrusions (Pankov et al., 2005; Raftopoulou and Hall, 2004; Ridley et al., 2003; Wu et al., 2009), we asked whether abnormally up-regulated Rac1 activity has an impact on protrusion dynamics in SH3BP1-depleted cells. We addressed this question by using a membrane-targeted fluorescent RFP protein (RFP-CAAX) to visualize movement of the cell periphery, and comparing videos of migrating SH3BP1-depleted and control cells (Figure 6A and Movie S3). Visual inspection of several movies revealed that the large majority of control cells had only one dynamic, well-defined lamellipodium towards the front. The protruding front in siSH3BP1-transfected cells was longer than that in control cells (average distance between nucleus and leading edge: 27.5 μm, SEM=1.6 μm versus 21.3 μm, SEM=1.3 μm; p<0.005, Student t-test) and was often discontinuous, forming multiple sub-protrusions. As a consequence, the average number of active protrusions per cell was higher in SH3BP1-depleted cells than in control cells (1.59 versus 1.17; p<0.005, Student t-test), and the percentage of ectopic protrusions (those not within the 120° angle facing the wound) increased from 12% in control cells to 22% in SH3BP1-depleted cells (Figure 6B).
Figure 6. Protrusion dynamics in SH3BP1-depleted cells is disordered and instable.
A. SH3BP1-depleted cells display defects in morphodynamics. NRK cells were treated with siLuc (control) or siSH3BP1-2, transfected with a plasmid expressing membrane-targeted RFP-CAAX, wounded, and visualized live by fluorescence microscopy. A selection of time-lapse acquisitions from a representative experiment is shown. See also Movie S3 for the entire video sequence. Arrows point to the protrusions and their color indicates protrusion locations according to the code defined in panel B.
B. Visual analysis of morphodynamics. The number and location of the protrusions were recorded by visually inspecting the images sequences. 11 movies of siLuc-treated cells and 15 movies of siSH3BP1-treated cells were analyzed. Protrusion location was classified in three categories: front, when protrusions were facing the wound (within a 120° angle); rear, when protrusions were opposite the wound (within a 120° angle); side, when protrusions were inside the two lateral 60°C angles.
C and E. Edge velocity maps. Computer-assisted analysis was used to establish edge velocity maps for a representative control (siLuc-treated) cell (C) (see Movie S4 for the corresponding video sequence) and a representative SH3BP1-depleted cell (siSH3BP1-2-treated) (E) (see Movie S5 for the corresponding video sequence).
D and F. Autocorrelation maps. Autocorrelation maps for the control cell (D) whose edge velocity map is shown in panel C and for the SH3BP1-depleted cell (F) whose edge velocity map is shown in panel E. Broken white arrows indicate the temporal autocorrelation functions at ΔCell periphery=0 degree; these values were used for the analysis shown in panel H.
G. Analysis of edge velocities. Edge fractions were classified into three categories (retracting < −2.5 μm/10min; −2.5 μm/10 min < static < 2.5 μm/10 min; protruding > 2.5 μm/10 min) and quantified in control (siLuc-treated; n=5) and siSH3BP1-treated (n=5) cells. ** indicate p<0.01 and * p<0.05 (Student t-test).
H. Temporal analysis of autocorrelation. Temporal cuts (ΔCell periphery=0 degree) of autocorrelation maps are shown for control (siLuc-treated; blue, n=3) and siSH3BP1-treated (red, n=4) cells. Symbols indicate results of the Student t-test analysis. Differences between experimental and control cells are significant both at 10 min (** p<0.01) and 20 min (* p<0.05).
I. Wild-type SH3BP1, but not GAP-deficient SH3BP1 mutant, rescues the morphodynamics of SH3BP1-depleted cells. NRK cells were transfected with siLuc or siSH3BP1-2, with a GFP-CAAX expressing plasmid, and the indicated cherry constructs. Red-fluorescent cells were video-imaged and autocorrelation coefficients at 10 min were measured. n=8 for each condition. See Movie S6 for representative videos. See Figure S5A for temporal autocorrelation analysis of all analyzed cells. ** indicate p<0.01 and * p<0.05 (Student t-test).
To objectively examine the role of SH3BP1 in plasma membrane morphodynamics, we performed a computer-assisted analysis of time-lapse images. We first measured velocities of movements at the cell edge, generating velocity maps for control and SH3BP1-depleted cells (Figures 6C and 6E). Edge fractions were classified into three categories: retracting, when velocity was lower than −2.5 μm / 10 min; static, when velocity was between -2.5 μm/10 min and +2.5 μm/10 min; and protruding, when velocity was higher than +2.5 μm/10 min. We found significant increases in both the retracting and protruding edge fractions in siSH3BP1-treated cells, indicating that the leading edges of SH3BP1-depleted cells moved faster than those of control cells (Figure 6G).
We also quantified the stability of membrane dynamics at the cell edge by using the mathematical tool of autocorrelation analysis as previously reported (Dobereiner et al., 2006; Maeda et al., 2008). Autocorrelation is the spatiotemporal cross-correlation of the signal (the edge velocity in our case) with itself. We generated autocorrelation maps of the edge velocity as a function of time (minutes of the videos of the moving cells) and of space (along the cell periphery). Whereas control cells exhibited an ordered pattern as expected for typical directional migration (Figure 6D), SH3BP1-depleted cells showed a much less ordered pattern (Figure 6F), indicating that protrusion formation during migration of the latter cells is random and instable. Plotting temporal cuts of autocorrelation maps (ΔCell periphery=0 degrees; broken white arrows in Figures 6D and 6F) revealed that, although autocorrelation coefficients were fairly stable over time in the case of control cells, they decayed rapidly in the case of SH3BP1-depleted cells (Figure 6H). Thus, the persistence of protrusion and retraction dynamics in SH3BP1-depleted cells was significantly reduced compared to those in control cells. We could correct the morphodynamics defect of SH3BP1-depleted cells by expressing wild-type SH3BP1, but not the R312A GAP-defective mutant, as shown by an analysis of the autocorrelation coefficients at Δt=10 min (Figures 6I and S5). This rescue result strongly supports a direct role of SH3BP1 GAP activity in the stability of membrane dynamics of motile cells.
Since in comparison to protrusions in control cells, those in SH3BP1-depleted cells are more numerous, partially delocalized and developed more rapidly but with a shorter persistence, we conclude that SH3BP1 is necessary for the spatiotemporal organization of protrusions during cell migration.
Expression of active Rac1 mimics depletion of SH3BP1
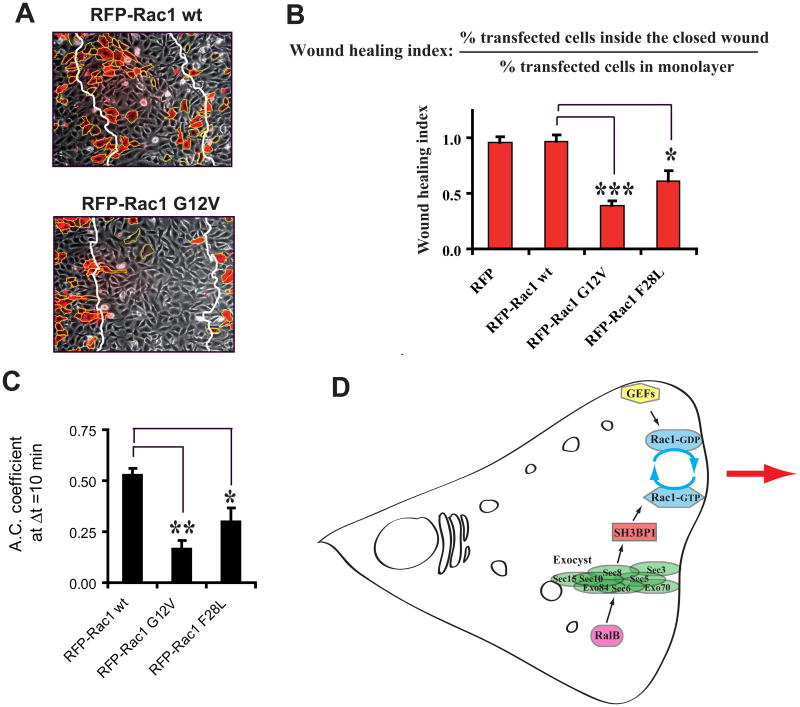
If inactivation of Rac1 at the front by SH3BP1 is required to organize protrusions and to drive efficient directional motility, cells expressing GTP-hydrolysis-deficient RacG12V should present defects in motility and morphodynamics. We tested this prediction by expressing RFP-fused wild-type or constitutively active mutant G12V Rac1 in motile NRK cells. Cells expressing RacG12V were severely impaired in motility, while cells expressing wild-type Rac1 migrated as control cells (un-transfected or RFP only-transfected) (Figures 7A and 7B).
Figure 7. The Inactivation of Rac1 is a requirement for cell motility.
A. Expression of Rac1 GV12, but not of wild-type Rac1, inhibits cell migration in wound healing assay. NRK cells were transfected with vectors expressing RFP-fused Rac1 wild-type or carrying the G12V mutation. Representative fields showing the closed wound after migration are shown. Note that transfection efficiency was intentionally low.
B. Quantitative comparison of Rac1 alleles. The effects of the expression of three Rac1 alleles (wild-type, GTPase deficient G12V, fast-cycling F28L) on motility were quantified using the wound-healing index (see Figure S5C and Supplemental Experimental Procedures). Expression levels of RFP constructs were similar.
C. Temporal autocorrelation analysis. Effects of the expression of the three Rac1 alleles on morphodynamics were quantified as Figure 6I. See Movie S7 for representative videos. See Figure S5B for detailed temporal graphics of all analyzed cells.
D. A model for the interplay between Ral and Rac in the regulation of cell migration. In motile cells the RalB GTPase controls the association of the subunits of its effector the exocyst complex and promotes localization of the exocyst at leading edge. The exocyst physically interacts with and brings to the leading edge the GAP protein SH3BP1, which stimulates the hydrolysis of bound GTP to GDP on Rac1 at the front. Several GEFs activate Rac1 at the front by replacing bound GDP with GTP. Therefore, Rac1 undergoes locally multiple activation-inactivation cycles, and this Rac1 cycling is necessary for the spatiotemporal regulation of the protrusions during directional motility. RalB, via the exocyst, participates to define where Rac1 cycling occurs and promotes protruding activity.
RacG12V-expressing cells displayed a much more rapid drop of the autocorrelation coefficients, as compared with wild-type Rac-expressing cells, indicating that GTP hydrolysis is necessary for the persistence of membrane dynamics (Figures 7C). The fast-cycling F28L mutant showed a similar but milder phenotype, with inhibition of motility (Figure 7B) and perturbation of morphodynamics (Figures 7C). Since RacG12V is always loaded with GTP, while RacF28L constitutively cycles between GDP- and GTP-bound states, their different phenotype strength could simply reflect their different GTP-loading levels. Moreover, since RacF28L-expressing cells presented abnormalities, we can conclude that GDP/GTP cycling per se is not sufficient to drive efficient protrusion organization. It seems that Rac not only needs to cycle, but needs to cycle at the right place and with the right kinetics, as spatiotemporally dictated by GEF and GAP proteins.
Thus, expression of constitutively active Rac1 mutants mimicked the phenotype of SH3BP1 depletion, clearly pointing out the importance of inactivating Rac1 during the motility process.
Discussion
We report here that SH3BP1 is a GAP that stimulates the GTPase activity of Rac1 during migration. However, several GEF proteins, including βPIX (ten Klooster et al., 2006), DOCK3 (Sanz-Moreno et al., 2008), Asef (Itoh et al., 2008), and Tiam1 (Palamidessi et al., 2008), have been proposed to activate Rac1 at the leading edge by replacing bound GDP with GTP. Thus, the presence of a Rac1-specific GAP at the leading edge—where Rac1 needs to be active—may seem surprising. One might have expected to instead find a RacGAP at the back of the cell in order to maintain the back-to-front Rac1-GTP gradient. However, current evidence supports the concept that unlike the prototype Ras, which needs to be locked into the GTP-bound state to execute its biological functions, other small GTPases require GDP/GTP cycling (Barale et al., 2006; Lin et al., 1997; Miller and Bement, 2009). This could explain why transformation induced by GEFs specific to Rho-family GTPases is much more efficient than transformation induced by constitutively active Rho-family GTPases, and also why GTPase-defective Rho/Rac/Cdc42 mutants have never been found in human tumours (Karlsson et al., 2009; Sahai and Marshall, 2002; Schmidt and Hall, 2002). Together with the RacGEFs, SH3BP1 might promote continuous GDP/GTP cycling of Rac1 at the migrating front of the cell, and this cycling may be necessary for the turnover of protrusions and adhesions during directional motility. The combined action of local RacGEFs and local RacGAP SH3BP1 would allow Rac1 to undergo a “GTPase flux” at the protruding front, similarly to what was proposed for RhoA GDP/GTP cycling during cytokinesis (Miller and Bement, 2009). Perturbing this fine-tuned balance of GEF/GAP activities on Rac1 may produce drastic alterations in the dynamics underlying motility—a notion consistent with the “membrane anarchy” we have observed in cells lacking SH3BP1.
We propose that in motile cells the exocyst complex (acting as effector of Ral proteins) participates in the transport and recruitment of regulatory molecules, including SH3BP1, to the front and only the front (Figure 7D). We show that the Ral/exocyst/SH3BP1 signaling pathway does not control the polarity of motile cells, i.e. orientation of the MTOC. Rather, it defines the site where the front protrusion forms and, accordingly, impacts the directionality of movement, as shown by the fact that SH3BP1-depleted cells have a persistence defect (Figure 3C). Therefore, the exocyst seems to be responsible for delimiting the cell region where Rac1 cycling is effective in driving cell migration. To this regard, it is worth mentioning that the ΔBAR SH3BP1 mutant, defective for binding to exocyst, did not rescue the migration defect of SH3BP1-depleted cells in wound healing assay (Figure 3E), consistently with our model, but puzzlingly it appeared to be able to stimulate the passage of SH3BP1-depleted cells through the porous membrane in a Boyden chamber (not shown). Discrepancies between Transwell and wound-healing motility assays are not rare, maybe because the motility programmes of cells passing through the 3D porous membrane of a Transwell are likely different from those of cells closing a wound on a 2D dish. In our case, the discrepancy suggests that the localization of SH3BP1 at the front is required for the mesenchymal-type 2D motility, but not for the 3D motility through a pore.
The precise timing of these activating and inactivating events as well as the identities of the sub-cellular compartments where they occur, remain unclear. Rac1 cycling could take place at specific sub-domains of the leading-edge plasma membrane, and the various positive and negative regulators could be supplied by membrane trafficking. Alternatively, Rac1 may be activated at endosomes, by the GEF Tiam1, and then translocated via recycling to the plasma-membrane (Palamidessi et al., 2008) where it would encounter SH3BP1.
In summary, we have found that the SH3BP1 GAP protein links vesicle trafficking and the exocyst complex, whose migration-promoting activity is under the control of Ral, to local modulation of the Rac pathway, and that it thereby plays a crucial role in integrating the molecular machineries and pathways that regulate cell migration.
Experimental Procedures
Cell culture, transfections, and plasmids
NRK (Normal Rat Kidney), human 293T and monkey Cos-7 cells were grown in Dulbecco's modified Eagle's medium supplemented with 2mM glutamine, antibiotics (penicillin and streptomycin), and 10% fetal bovine serum. Human PC-3 prostate tumor cells were maintained in Ham's F12 medium supplemented with 10% fetal bovine serum, penicillin, streptomycin and kanamycin. Transient DNA and siRNA transfections were carried out with Lipofectamine Plus Reagent (Invitrogen) and Hyperfect (Qiagen), respectively. Expression vectors for SH3BP1 wt (amino acids 1-701), ΔBAR (amino acids 255-701), ΔCter (amino acids 1-466) and R>A (R312A) were generated by cloning human cDNAs encoding full-length SH3BP1 protein and the respective truncated and mutated versions into pcDNA3. Notice that human SH3BP1 is resistant to the two siRNAs against rat SH3BP1 used in the present study in NRK cells. In order to generate cherry-fusion constructs, the SH3BP1 alleles were inserted by PCR techniques into pCherry-C1, obtained from Dr Tsien. pRaichu-Rac1, pRaichu-Cdc42, pRaichu-RhoA (Nakamura et al., 2006), pcDNA3-Exo84-HA and pcDNA3-Sec8-HA (Sakurai-Yageta et al., 2008), pCXN2-mRFP-Rac1 (Kurokawa et al., 2004) were reported previously. pCAGGS-mRFP-X and pCAGGS-GFP-X express mRFP and GFP fluorescent proteins, respectively, fused to the membrane-targeting signal of K-Ras.
Wound healing and time-lapse microscopy
For wound healing assays, cells were grown to confluence on cover slips or glass-bottom dishes coated with collagen (type I from rat tail, Interchim #207050357, 120 μg/ml in H2O for ≥1 hr 37°C). The monolayer was wounded with a pipette tip, at which point the medium was changed to MEM with 2% fetal bovine serum. For phase-contrast video-microscopy, a 10× objective was used and images were acquired every 15 min. The Manual Tracking plug-in (developed by Fabrice Cordelières) of ImageJ software was used to track cells, after which data were exported to Excel for mathematical and statistical analysis. Persistence of motility was calculated as the D/T ratio, where D is the direct distance from start point to end point and T is the total track.
The FRET of the Raichu-Rac1 biosensor was measured as the YFP/CFP ratio and represented using an 8-colour scale code, as shown to the right of time-lapse figures, with the upper and lower limits indicated. For time-lapse video, images were acquired every 10 min. For single time-point, whole-cell measurements, the background-subtracted images were thresholded to define the whole-cell surface. The mean cell fluorescence intensities (CFP and YFP) were then measured and the YFP/CFP ratios were calculated. For single time-point gradient measurements, the line-scan function of MetaMorph was used to measure YFP/CFP ratios along 4-μm wide strips connecting the nucleus to the leading edge in the direction of cell motility. Because of variability in cell size, the resulting sets of measurements were of different lengths. Comparison of these values was made possible by resizing dataset length to 20 categories, by aggregating and averaging initial measurements. Next, the first value of each dataset (corresponding to the FRET measurements closest to the nucleus) was taken as reference to estimate the relative evolution of FRET along the direction of motility. The plotted points are the means of measurements for each category of the cell population.
Immunoprecipitations
For the Sec8 immunoprecipitation shown in Figure 1C, protein lysates from one 10-cm dish of DNA-transfected 293T cells were prepared in 1ml lysis buffer A (50mM TrisHCl pH=7.5, 150mM NaCl, 1% Triton, 1mM EDTA, freshly supplemented with a protease inhibitor mixture (Roche, #1836170)) and then incubated for 2 hr with 25 μl of beads covalently coupled to a mixture of monoclonal anti-Sec8 antibodies (Yeaman et al., 2001). For the HA immunoprecipitations shown in Figures 1D and 1E, protein lysates from one 6-cm dish of DNA-transfected 293T cells were prepared in 600 μl of lysis buffer B (20mM Tris HCl pH=7.4, 100mM NaCl, 5mM MgCl2, 1% Triton, 10% Glycerol, freshly supplemented with 1 mM dithiothreitol and the protease inhibitor mixture), and then incubated for 1 hr with a monoclonal anti-HA antibody (3F10, 1 μg) and then for a further 1 hr after 1 mg of G-coupled magnetic beads (Invitrogen, #100-04D) was added. The immobilized immune complexes were washed three times with 1 ml of lysis buffer and finally boiled in gel-loading buffer. The whole-lysate inputs represent 1.6% (Figure 1C) or 6% (Figures 1D and 1E) of the immunoprecipitated material.
Morphodynamics analysis
The videos of motile cells expressing RFP-CAAX to track the cell periphery were generated from a series of images taken at 10-min time intervals, and were visually analyzed as follows. For each cell and each time interval, the cell periphery was inspected to identify portions that expanded in an apparently coordinated way from time T to time T+1, and each of these portions was scored as an active protrusion. For each cell, the average number of protrusions per time point, and the mean values for all of the observed cells, were calculated. In order to score protrusion positioning, we traced an angle of 120° facing the direction of motility (front), a mirror angle of 120° (rear) and two lateral angles of 60° (sides). At each time interval, each observed protrusion was scored as belonging to one of these three categories. To measure edge velocities, we developed simple computer-assisted methods that will soon be described in detail elsewhere (Kunida, et al., unpublished). See Supplemental Experimental Procedures for more details.
Supplementary Material
Acknowledgments
We greatly thank Piera Cicchetti for kindly providing her anti-SH3BP1 antibodies, Bruce J. Mayer for providing a SH3BP1-expressing vector, the staff of the PICT-IBiSA imaging facility at the Curie Institut for sharing their expertise, Hybrigenics staff for yeast two-hybrid analysis, Carlo Lucchesi and Sabrina Carpentier for discussions on statistical analysis, Christine Blaumueller for critical reading and editing of the manuscript, and Chloé Camonis for film editing.
AS and MB are recipients of a doctoral fellowship from the Vinci programme of the Université Franco-Italienne and Ligue National Contre le Cancer, respectively. This work was supported by grants: ANR IntegRal, ARC4845 and Association Christelle Bouillot (JC); NIH GM067002 (CY); GenHomme Network 02490-6088 (Hybrigenics and the Institut Curie); Grant-in-Aid for Scientific Research on Priority Areas from the Ministry of Education, Culture, Sports, and Science, Japan (KA, MM).
References
- Barale S, McCusker D, Arkowitz RA. Cdc42p GDP/GTP cycling is necessary for efficient cell fusion during yeast mating. Mol Biol Cell. 2006;17:2824–2838. doi: 10.1091/mbc.E05-11-1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bos JL, Rehmann H, Wittinghofer A. GEFs and GAPs: critical elements in the control of small G proteins. Cell. 2007;129:865–877. doi: 10.1016/j.cell.2007.05.018. [DOI] [PubMed] [Google Scholar]
- Cantor SB, Urano T, Feig LA. Identification and characterization of Ral-binding protein 1, a potential downstream target of Ral GTPases. Mol Cell Biol. 1995;15:4578–4584. doi: 10.1128/mcb.15.8.4578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cicchetti P, Mayer BJ, Thiel G, Baltimore D. Identification of a protein that binds to the SH3 region of Abl and is similar to Bcr and GAP-rho. Science. 1992;257:803–806. doi: 10.1126/science.1379745. [DOI] [PubMed] [Google Scholar]
- Cicchetti P, Ridley AJ, Zheng Y, Cerione RA, Baltimore D. 3BP-1, an SH3 domain binding protein, has GAP activity for Rac and inhibits growth factor-induced membrane ruffling in fibroblasts. Embo J. 1995;14:3127–3135. doi: 10.1002/j.1460-2075.1995.tb07315.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobereiner HG, Dubin-Thaler BJ, Hofman JM, Xenias HS, Sims TN, Giannone G, Dustin ML, Wiggins CH, Sheetz MP. Lateral membrane waves constitute a universal dynamic pattern of motile cells. Phys Rev Lett. 2006;97:038102. doi: 10.1103/PhysRevLett.97.038102. [DOI] [PubMed] [Google Scholar]
- Etienne-Manneville S. Cdc42--the centre of polarity. J Cell Sci. 2004;117:1291–1300. doi: 10.1242/jcs.01115. [DOI] [PubMed] [Google Scholar]
- He B, Guo W. The exocyst complex in polarized exocytosis. Curr Opin Cell Biol. 2009;21:537–542. doi: 10.1016/j.ceb.2009.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh RE, Kiyokawa E, Aoki K, Nishioka T, Akiyama T, Matsuda M. Phosphorylation and activation of the Rac1 and Cdc42 GEF Asef in A431 cells stimulated by EGF. J Cell Sci. 2008;121:2635–2642. doi: 10.1242/jcs.028647. [DOI] [PubMed] [Google Scholar]
- Itoh RE, Kurokawa K, Ohba Y, Yoshizaki H, Mochizuki N, Matsuda M. Activation of rac and cdc42 video imaged by fluorescent resonance energy transfer-based single-molecule probes in the membrane of living cells. Mol Cell Biol. 2002;22:6582–6591. doi: 10.1128/MCB.22.18.6582-6591.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jullien-Flores V, Dorseuil O, Romero F, Letourneur F, Saragosti S, Berger R, Tavitian A, Gacon G, Camonis JH. Bridging Ral GTPase to Rho pathways. RLIP76, a Ral effector with CDC42/Rac GTPase-activating protein activity. J Biol Chem. 1995;270:22473–22477. doi: 10.1074/jbc.270.38.22473. [DOI] [PubMed] [Google Scholar]
- Karlsson R, Pedersen ED, Wang Z, Brakebusch C. Rho GTPase function in tumorigenesis. Biochim Biophys Acta. 2009;1796:91–98. doi: 10.1016/j.bbcan.2009.03.003. [DOI] [PubMed] [Google Scholar]
- Kraynov VS, Chamberlain C, Bokoch GM, Schwartz MA, Slabaugh S, Hahn KM. Localized Rac activation dynamics visualized in living cells. Science. 2000;290:333–337. doi: 10.1126/science.290.5490.333. [DOI] [PubMed] [Google Scholar]
- Kurokawa K, Itoh RE, Yoshizaki H, Nakamura YO, Matsuda M. Coactivation of Rac1 and Cdc42 at lamellipodia and membrane ruffles induced by epidermal growth factor. Mol Biol Cell. 2004;15:1003–1010. doi: 10.1091/mbc.E03-08-0609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ligeti E, Dagher MC, Hernandez SE, Koleske AJ, Settleman J. Phospholipids can switch the GTPase substrate preference of a GTPase-activating protein. J Biol Chem. 2004;279:5055–5058. doi: 10.1074/jbc.C300547200. [DOI] [PubMed] [Google Scholar]
- Lim KH, O'Hayer K, Adam SJ, Kendall SD, Campbell PM, Der CJ, Counter CM. Divergent roles for RalA and RalB in malignant growth of human pancreatic carcinoma cells. Curr Biol. 2006;16:2385–2394. doi: 10.1016/j.cub.2006.10.023. [DOI] [PubMed] [Google Scholar]
- Lin R, Bagrodia S, Cerione R, Manor D. A novel Cdc42Hs mutant induces cellular transformation. Curr Biol. 1997;7:794–797. doi: 10.1016/s0960-9822(06)00338-1. [DOI] [PubMed] [Google Scholar]
- Lu W, Mayer BJ. Mechanism of activation of Pak1 kinase by membrane localization. Oncogene. 1999;18:797–806. doi: 10.1038/sj.onc.1202361. [DOI] [PubMed] [Google Scholar]
- Maeda YT, Inose J, Matsuo MY, Iwaya S, Sano M. Ordered patterns of cell shape and orientational correlation during spontaneous cell migration. PLoS One. 2008;3:e3734. doi: 10.1371/journal.pone.0003734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller AL, Bement WM. Regulation of cytokinesis by Rho GTPase flux. Nat Cell Biol. 2009;11:71–77. doi: 10.1038/ncb1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T, Kurokawa K, Kiyokawa E, Matsuda M. Analysis of the spatiotemporal activation of rho GTPases using Raichu probes. Methods Enzymol. 2006;406:315–332. doi: 10.1016/S0076-6879(06)06023-X. [DOI] [PubMed] [Google Scholar]
- Osmani N, Vitale N, Borg JP, Etienne-Manneville S. Scrib controls Cdc42 localization and activity to promote cell polarization during astrocyte migration. Curr Biol. 2006;16:2395–2405. doi: 10.1016/j.cub.2006.10.026. [DOI] [PubMed] [Google Scholar]
- Oxford G, Owens CR, Titus BJ, Foreman TL, Herlevsen MC, Smith SC, Theodorescu D. RalA and RalB: antagonistic relatives in cancer cell migration. Cancer Res. 2005;65:7111–7120. doi: 10.1158/0008-5472.CAN-04-1957. [DOI] [PubMed] [Google Scholar]
- Palamidessi A, Frittoli E, Garre M, Faretta M, Mione M, Testa I, Diaspro A, Lanzetti L, Scita G, Di Fiore PP. Endocytic trafficking of Rac is required for the spatial restriction of signaling in cell migration. Cell. 2008;134:135–147. doi: 10.1016/j.cell.2008.05.034. [DOI] [PubMed] [Google Scholar]
- Pankov R, Endo Y, Even-Ram S, Araki M, Clark K, Cukierman E, Matsumoto K, Yamada KM. A Rac switch regulates random versus directionally persistent cell migration. J Cell Biol. 2005;170:793–802. doi: 10.1083/jcb.200503152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raftopoulou M, Hall A. Cell migration: Rho GTPases lead the way. Dev Biol. 2004;265:23–32. doi: 10.1016/j.ydbio.2003.06.003. [DOI] [PubMed] [Google Scholar]
- Ridley AJ, Schwartz MA, Burridge K, Firtel RA, Ginsberg MH, Borisy G, Parsons JT, Horwitz AR. Cell migration: integrating signals from front to back. Science. 2003;302:1704–1709. doi: 10.1126/science.1092053. [DOI] [PubMed] [Google Scholar]
- Rosse C, Formstecher E, Boeckeler K, Zhao Y, Kremerskothen J, White MD, Camonis JH, Parker PJ. An aPKC-exocyst complex controls paxillin phosphorylation and migration through localised JNK1 activation. PLoS Biol. 2009;7:e1000235. doi: 10.1371/journal.pbio.1000235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosse C, Hatzoglou A, Parrini MC, White MA, Chavrier P, Camonis J. RalB mobilizes the exocyst to drive cell migration. Mol Cell Biol. 2006;26:727–734. doi: 10.1128/MCB.26.2.727-734.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahai E, Marshall CJ. RHO-GTPases and cancer. Nat Rev Cancer. 2002;2:133–142. doi: 10.1038/nrc725. [DOI] [PubMed] [Google Scholar]
- Sakurai-Yageta M, Recchi C, Le Dez G, Sibarita JB, Daviet L, Camonis J, D'Souza-Schorey C, Chavrier P. The interaction of IQGAP1 with the exocyst complex is required for tumor cell invasion downstream of Cdc42 and RhoA. J Cell Biol. 2008;181:985–998. doi: 10.1083/jcb.200709076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz-Moreno V, Gadea G, Ahn J, Paterson H, Marra P, Pinner S, Sahai E, Marshall CJ. Rac activation and inactivation control plasticity of tumor cell movement. Cell. 2008;135:510–523. doi: 10.1016/j.cell.2008.09.043. [DOI] [PubMed] [Google Scholar]
- Schmidt A, Hall A. Guanine nucleotide exchange factors for Rho GTPases: turning on the switch. Genes Dev. 2002;16:1587–1609. doi: 10.1101/gad.1003302. [DOI] [PubMed] [Google Scholar]
- ten Klooster JP, Jaffer ZM, Chernoff J, Hordijk PL. Targeting and activation of Rac1 are mediated by the exchange factor beta-Pix. J Cell Biol. 2006;172:759–769. doi: 10.1083/jcb.200509096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu YI, Frey D, Lungu OI, Jaehrig A, Schlichting I, Kuhlman B, Hahn KM. A genetically encoded photoactivatable Rac controls the motility of living cells. Nature. 2009;461:104–108. doi: 10.1038/nature08241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeaman C, Grindstaff KK, Wright JR, Nelson WJ. Sec6/8 complexes on trans-Golgi network and plasma membrane regulate late stages of exocytosis in mammalian cells. J Cell Biol. 2001;155:593–604. doi: 10.1083/jcb.200107088. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.