Abstract
The gibberellins (GAs) are a complex family of diterpenoid compounds, some of which are potent endogenous regulators of plant growth. As part of a feedback control of endogenous GA levels, active GAs negatively regulate the abundance of mRNA transcripts encoding GA biosynthesis enzymes. For example, Arabidopsis GA4 gene transcripts encode GA 3β-hydroxylase, an enzyme that catalyzes the conversion of inactive to active GAs. Here we show that active GAs regulate GA4 transcript abundance in a dose-dependent manner, and that down-regulation of GA4 transcript abundance is effected by GA4 (the product of 3β-hydroxylation) but not by its immediate precursor GA9 (the substrate). Comparison of several different GA structures showed that GAs active in promoting hypocotyl elongation were also active in regulating GA4 transcript abundance, suggesting that similar GA:receptor and subsequent signal transduction processes control these two responses. It is interesting that these activities were not restricted to 3β-hydroxylated GAs, being also exhibited by structures that were not 3β-hydroxylated but that had another electronegative group at C-3. We also show that GA-mediated control of GA4 transcript abundance is disrupted in the GA-response mutants gai and spy-5. These observations define a sensitive homeostatic mechanism whereby plants may regulate their endogenous GA levels.
GAs are a family of hormones that are essential for development and growth of plants (Hooley, 1994). However, the molecular mechanism for GA perception thought to involve a GA:GA-receptor interaction has yet to be elucidated. Although more than 100 different GA structures have been identified in plants, only a few possess biological activity, suggesting a high degree of specificity to the GA:GA-receptor interaction (Takahashi et al., 1990).
The study of GA action has been advanced by the use of mutants that are affected in GA biosynthesis or signal transduction (Ross, 1994). Examples of GA-biosynthesis mutants in Arabidopsis are the GA-deficient dwarfs ga1, ga4, and ga5 (Koornneef and van der Veen, 1980). A WT phenotype can be recovered in all of these mutants by applying active GAs. GA biosynthesis is a multistep pathway involving ent-kaurene synthesis and oxidation, followed by further oxidations of the GA skeleton, the latter being catalyzed by 20-oxidase and 3β-hydroxylase (Graebe, 1987; Talon et al., 1990a). GA1, GA4, and GA5 are loci that encode the copalyl diphosphate synthase (Sun and Kamiya, 1994; Hedden and Kamiya, 1997), 3β-hydroxylase (Chiang et al., 1995; Hedden and Kamiya, 1997), and 20-oxidase (Phillips et al., 1995; Xu et al., 1995) enzymes of GA biosynthesis, respectively. 3β-Hydroxylation is widely held to be a final step in the biosynthesis of active GAs, converting GA9 and GA20 (inactive) to GA4 and GA1 (active), respectively.
Recently, it has been shown that expression of 20-oxidase and GA4 genes is negatively regulated by exogenous GA (Chiang et al., 1995; Phillips et al., 1995; Xu et al., 1995). These observations are consistent with evidence suggesting that the activities of GA biosynthesis enzyme can be down-regulated by GA (Hedden and Croker, 1992). A negative feedback loop of this nature requires that GAs active in feedback regulation can somehow be distinguished from similar (but inactive) structures in the GA biosynthesis pathways. In the case of feedback regulation of GA4 transcript levels, this could occur in one of two ways: Either the immediate products of the 3β-hydroxylation reaction or all biologically active GA structures negatively regulate GA4 transcript levels. Here we describe experiments designed to determine which of these possible mechanisms is responsible for the regulation of GA4 transcript levels.
The Arabidopsis GA signal transduction mutant gai has the dwarf, dark-green characteristics of a GA-deficient plant. However, the phenotype conferred by gai cannot be rescued by the application of GA, and the gai mutant therefore displays reduced responses to both endogenous and exogenous GA (Koornneef et al., 1985; Peng and Harberd, 1993, 1997; Wilson and Somerville, 1995; Peng et al., 1997). gai has higher than wild-type levels of endogenous active GAs (Talon et al., 1990b), suggesting that negative feedback regulation of GA levels is perturbed in this mutant. Another class of Arabidopsis GA signal transduction mutants, the spy mutants, display resistance to the GA biosynthesis inhibitor PAC (Jacobsen and Olszewski, 1993; Wilson and Somerville, 1995; Jacobsen et al., 1996). spy mutants are able to germinate and display elongation growth in concentrations of PAC that are inhibitory to wild-type plants.
In this paper we describe experiments that define the nature of the feedback regulation of GA4 transcript levels by active GAs. Using the GA-deficient ga1–3 mutant (Sun and Kamiya, 1994), we show that GA4 transcript levels are negatively regulated by the product of the 3β-hydroxylation reaction, GA4, but not by the immediate precursor, GA9. Feedback regulation occurs in a dose-dependent manner that closely mirrors stimulation of hypocotyl elongation. We also show that the presence of a 3β-OH group does not always confer activity for feedback, and that GAs that are active in feedback do not have to be 3β-hydroxylated. Finally, we show that feedback regulation of GA4 transcript levels is disrupted in gai and spy mutants. These results indicate that negative feedback regulation of GA4 transcript levels occurs by perception of active GAs via a receptor/signal transduction pathway that is similar to that involved in GA-mediated elongation growth.
MATERIALS AND METHODS
Chemicals
GA1 was a gift from Prof. Sassa (Yamagata University Yamagata, Japan), and GA4 was obtained from Kyowa Hakko Co., Ltd. (Tokyo, Japan). GA C was prepared from GA1 (Cross, 1960): treatment of GA1 with Dowex-resin 50W-X2 (H+ form) in refluxing methanol:water, 2:5 (v/v), for 7 h gave GA C in 84% yield. epi-GA4 was prepared from GA4: treatment of GA4 with potassium tert-butoxide in tert-butyl alcohol at room temperature for 7 d afforded epi-GA4 in 92% yield (Aldridge et al., 1965). 3-oxo-GA9 was prepared from GA4 by oxidation (Aldridge et al., 1965). All of the GAs were purified by preparative high-performance column chromatography. The purity of GAs was about 100% as checked by GC-MS.
Plant Material and Growth Procedures
An Arabidopsis Landsberg erecta laboratory strain (wild type) was used throughout. ga1–3 and gai mutants were originally isolated from mutagenized WT (Koornneef and van der Veen, 1980; Koornneef et al., 1985). Seeds homozygous for the spy-5 allele (also isolated from mutagenized wild type) were kindly donated by R. Wilson (Wilson and Somerville, 1995).
After sterilization (Ezura and Harberd, 1995), ga1–3 seeds were chilled for 5 d at 4°C in sterile 10−6 m GA4 solution to initiate and synchronize germination. After a thorough rinsing in sterile water they were plated individually (50/plate) on germination medium (Ezura and Harberd, 1995) containing GA or inhibitors at the required concentration. Sterilized wild type and spy-5 seeds were directly sown (50/plate) before chilling for 5 d at 4°C. The seeds were then grown in a standard growth room at 20°C with a 16-h light/8-h dark cycle.
GAs (previously purified by HPLC) were dissolved in methanol and then in sterile water. A small volume (no more than 1/1000 volume) was then added to 20 mL of cooled molten germination medium in Petri dishes. The inhibitors PAC (Zeneca Agrochemicals, Wilmington, DE) and BX-112 (Kumiai Chemical Research Institute, Shizuoka, Japan) were made up and added in the same way. Hypocotyls were measured directly to the nearest 0.5 mm using samples of 8 to 10 per treatment.
QRT-PCR
RNA was prepared from seedlings (entire aerial parts) harvested 2 weeks after germination (about 20 per sample). Approximately 5 μg of each RNA sample was then used in a first-strand cDNA synthesis reaction (containing RNase inhibitor) using a standard poly-dT adapter primer and Moloney murine leukemia virus reverse transcriptase, diluted 10-fold. The following oligonucleotides were made to amplify fragments of the GA4 (Chiang et al., 1995, 1997), APT1 (Moffat et al., 1994) and γ-TIP (Ludevid et al., 1992) cDNAs: OLLY23, 5′-TCCCAGAATCGCTAAGATTGCC-3′; OLLY42, 5′-CCTTTCCCTTAAGCTCTG-3′; OLLY26, 5′-CGATTTCCGTAAACTTTGGC-3′; OLLY28, 5′-ATCCATTGGATAGGATGTGG-3′; OLLY40, 5′-CATCTTGAAGCTTAAATC-3′; and OLLY22, 5′-GACTCGAGTCGACAT-CGA(T)17-3′. OLLY23 and OLLY42 amplify a 478-bp fragment of APT1 cDNA. OLLY26 and OLLY28 amplify a 398-bp fragment of GA4 cDNA. Both of these products can be distinguished by size from products resulting from amplification of any contaminant genomic DNA because the primer sequences are on either side of at least one intron. For the amplification of γ-TIP cDNA, OLLY40 was used with a poly-T primer (OLLY22) to ensure that only cDNA would be amplified, as a fragment of approximately 1.1 kb (the γ-TIP genomic DNA sequence is unknown). In each case, the PCR product was cloned and sequenced, using standard techniques, so as to verify the sequence of the amplified fragments. The cloned fragments were later released from the cloning vectors via restriction endonuclease digestion and used as hybridization probes.
The above cDNA solutions (5 μL) were used as the templates in a standard 50-μL PCR reaction (with 0.25 mm dNTPs and 2 ng/μL each primer) of up to 30 to 34 cycles of 1 min each at 94°C, 55°C, and 72°C. OLLY23/OLLY42 and OLLY26/OLLY28 primer pairs were used in separate reactions to avoid primer competition (Murphy et al., 1990). After at least 10 cycles, 4-μL aliquots were removed from the reactions every 2 to 4 cycles. PCR products were separated by electrophoresis, blotted, and hybridized using standard techniques ([32P]dCTP-labeled hybridization probes, as described above). QRT-PCR products were quantified by phosphor imaging, using ImageQuant software (Molecular Dynamics, Sunnyvale, CA). Curves were constructed by plotting the radioactivity of the PCR product (y-axis; log10 scale) against the number of cycles (x-axis) (see Fig. 1B). This confirmed the approximately exponential nature of the PCRs (all gradients were between 0.22 and 0.3 [the theoretical maximum for PCR]). Expression of the two genes was compared at points where both reactions were progressing exponentially, which gives a ratio of GA4/APT1 expression (Noonan et al., 1990). Within each experiment, these ratios were normalized to the sample grown on GM alone or as described in the figure legends. However, in some samples the expression of the GA4 gene was so low that PCR products could not be detected within this range, and a ratio could not be calculated, so in the figures this is described as not detected. A product was always detected after 30 cycles, suggesting that the GA4 gene was never completely repressed in our experiments, but at this point the control gene reactions had already saturated. It is also possible that the primers used to amplify the GA4 product, although shown to preferentially amplify product derived from the GA4 gene itself, might also amplify the products of any genes closely related in sequence to GA4.
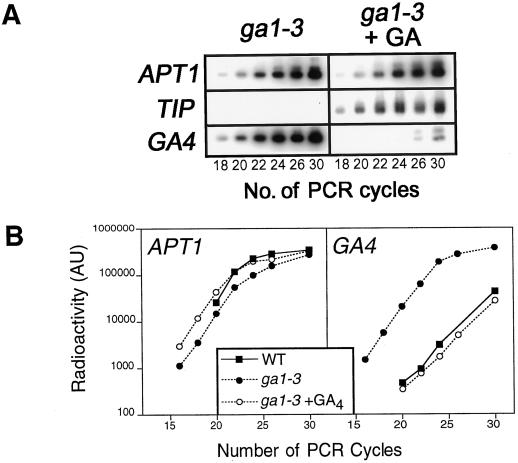
Figure 1.
A, QRT-PCR analysis of GA4 and γ-TIP (TIP) transcripts relative to those of the APT1 control gene in GA4-treated (10−7 m; ga1–3 + GA) and untreated ga1–3 (ga1–3). The relevant primer pairs (see Methods) were used on poly-T-primed cDNA samples in separate reactions. Aliquots taken after the stated number of cycles were separated on a 1.2% agarose gel, blotted, hybridized to radioactively labeled probes of a known sequence, and visualized by phosphor imaging. B, Kinetics of RT-PCR reactions shown in A compared with others using wild-type samples. Radioactivity of hybridized filters was measured using ImageQuant software (see Methods) and plotted on a log10 scale (y axis) (AU, arbitrary units) against the number of PCR cycles (x axis). In the GA4-treated sample, the GA4-derived primers amplified two products, the smaller of which was of the correct size to be the GA4 sequence and the signal intensity of which was measured. Gradients over the linear portions (exponential phases) of the curves range from 2.2 to 2.8. RT-PCR product levels were compared before saturation occured (cycle 24). WT, wild type.
RESULTS
GA4 Transcript Levels Are Elevated in the ga1–3 Mutant
GA4 expression was followed relative to the expression of a control gene, APT1, by QRT-PCR using the kinetic method. This method permits quantification of the abundance of a specific mRNA with respect to another endogenous control mRNA (Chelly et al., 1988; Murphy et al., 1990; Noonan et al., 1990). APT1 is expressed at a low level in all tissues of Arabidopsis (Moffat et al., 1994). These experiments were performed using the ga1–3 mutant, which has greatly reduced levels of endogenous active GAs due to a deletion in a gene encoding copalyl diphosphate synthase (Sun and Kamiya, 1994; Hedden and Kamiya, 1997). Transcripts were compared in ga1–3 seedlings treated with exogenous GA4 and untreated controls. GA4 was chosen for these experiments because it is the most abundant 3β-hydroxylated GA and probably the main active GA in Arabidopsis (Talon et al., 1990a). As shown in Figure 1, the levels of control APT1 transcript in ga1–3 seedlings are not significantly affected by GA4 treatment (Fig. 1A), and are not significantly different from that of WT (Fig. 1B).
γ-TIP transcript levels are up-regulated following GA treatment of the GA-deficient ga1–2 mutant (Phillips and Huttly, 1994). We compared the effects of exogenous GA4 on the accumulation of γ-TIP and GA4 transcripts in ga1–3 (Fig. 1A). As expected, γ-TIP transcripts were not detected in the untreated ga1–3 controls, but were clearly detectable in the GA4-treated ga1–3 sample. The behavior of GA4 transcripts in these experiments is the converse of that of γ-TIP, in that GA4 transcripts were clearly detectable in the untreated ga1–3 controls, but were only just detectable in the GA4-treated ga1–3 sample.
ga1–3 mutant seedlings contained elevated levels of GA4 transcript compared with the wild type (Fig. 1B). These elevated transcript levels were restored to wild-type levels by the addition of exogenous GA4 (Fig. 1B). The GA4-treated ga1–3 sample required at least six more PCR cycles to produce the same amount of GA4 amplification product than did the nontreated ga1–3 control, whereas APT1 was expressed at the same level in both samples (Fig. 1B). The efficiency of the PCR reaction was similar for all samples and for both genes during the exponential phase (Fig. 1B). The elevated level of GA4 transcript in the ga1–3 mutant is thus equivalent to an induction of GA4 gene expression of over 60-fold (64). This result is consistent with previous observations that GA4 transcripts accumulate in a ga4 mutant to higher levels than in the wild type, and confirms that GA4 transcript abundance is negatively regulated by GAs (Chiang et al., 1995).
GA-Mediated Feedback Control of GA4 Transcript Abundance Is GA Dose Dependent
For the following experiments, a steady-state estimate of GA4 mRNA abundance was obtained by calculating the ratio of GA4:APT1 transcripts using information from plots such as the one in Figure 1B. The phosphor imager value for the GA4 gene was divided by that of the APT1 gene at the value of x (no. of PCR cycles), where both reactions were approximately exponential, and prior to saturation (cycle 22 for Fig. 1B). This ratio approximates the ratio of initial templates in the PCR reaction at cycle 0, providing all of the reactions have similar efficiencies (Noonan et al., 1990). Within each experiment all samples had been prepared and processed at the same time, and ratios were calculated at the same number of cycles. Within each experiment these values were normalized to the sample grown on medium alone (i.e. in the absence of hormone or inhibitors), which was arbitrarily given the GA4:APT1 ratio of 1. All experiments were repeated with separate RNA samples, PCRs, and hybridizations; representative data are shown.
The effects of a range of GA concentrations on GA4 transcript abundance and hypocotyl elongation were compared (Fig. 2). At a high GA4 dose, ga1–3 hypocotyls were as long as those of untreated wild type, whereas GA4 transcript levels were as low as those of untreated wild type. However, as the GA4 dose decreased, ga1–3 hypocotyls became progressively shorter, and GA4 transcript levels became progressively higher.
Figure 2.
ga1–3 seedlings were assayed for relative GA4 mRNA levels and hypocotyl length 2 weeks after germination on medium containing the stated concentration of GA4. QRT-PCR results (GA4:APT1 ratios, calculated after 22 cycles, when the reaction had yet to saturate) were normalized with respect to ga1–3 grown on germination medium only (=1). Results from untreated wild-type seedlings are shown for a comparison (open symbols). Error bars represent se hypocotyl length (sometimes smaller than symbol width). •, ga1–3 hypocotyl elongation; ○, wild-type hypocotyl elongation; ▪, GA4/APT1 transcript ratios in ga1–3; □, GA4/APT1 transcript ratios in the wild type.
Feedback Regulation by Active GAs?
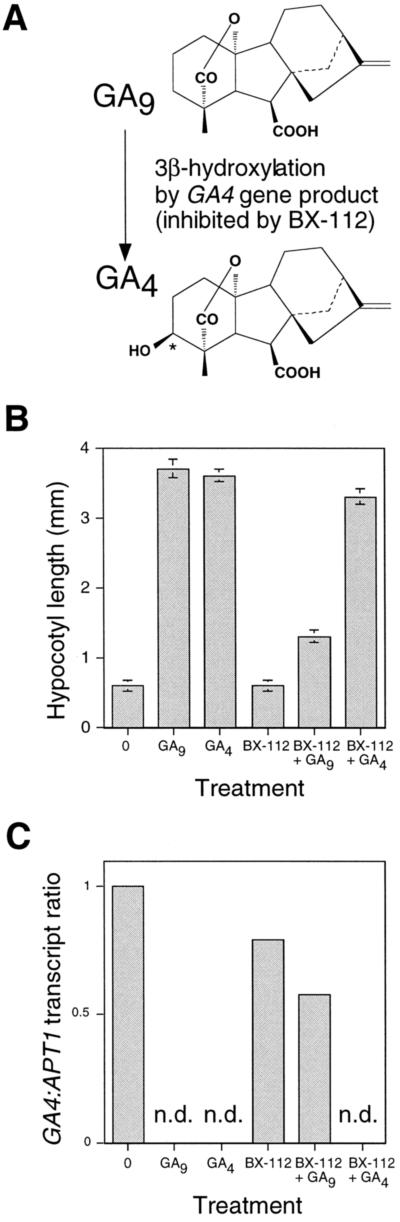
GA9 is 3β-hydroxylated by the 3β-hydroxylase enzyme in planta to form GA4 (Fig. 3A). We investigated the hypothesis that regulation of expression of the GA4 gene is an example of product inhibition by testing the effects of GA9 (substrate) and GA4 (product) on the accumulation of GA4 transcripts in the ga1–3 mutant. To prevent conversion of GA9 to GA4, the inhibitor BX-112 was used. BX-112 prevents both 3β- and 2β-hydroxylation of GAs (Nakayama et al., 1990a, 1990b). Figure 3B shows that GA9 and GA4 caused a marked increase in ga1–3 hypocotyl length. However, if BX-112 was included in the medium, the effect of GA9 was greatly reduced, whereas that of GA4 was relatively unaffected. The simplest explanation for this is that GA9 is not active in itself, but becomes active following 3β-hydroxylation to GA4. 3β-Hydroxylation is largely, but not completely, abolished by the BX-112 treatment; thus, only a small amount of the inactive GA9 is converted to GA4 in the presence of BX-112.
Figure 3.
GA4 transcript levels are regulated by GA4 but not by GA9. A, GA9 is converted to GA4 by the addition of a 3β-OH group on C-3 (*). This reaction is catalyzed by the GA4 gene product and inhibited by BX-112. B, ga1–3 seedlings were grown for 2 weeks on germination medium supplemented with 10−4 m BX-112, 10−7 m GA9, and 10−7 m GA4 as stated. Hypocotyl lengths were measured as described in Figure 2, error bars represent se. C, ga1–3 seedlings treated as in B were assayed for GA4 transcript levels as in Figure 2. RT-PCR products were compared after 22 cycles. Results are presented as GA4:APT1 product ratios, normalized with respect to ga1–3 grown on germination medium only (=1). n.d., GA4 transcript not detected (see Methods).
Seedling samples were assayed for GA4 transcript levels relative to the control gene APT1 by comparing levels of RT-PCR products after 22 cycles (Fig. 3C). GA4 transcript levels were high in the ga1–3 mutant, but were repressed by both GA9 and GA4 in the absence of BX-112. No GA4 RT-PCR product was detected in the GA9 (without BX-112) or GA4 (without BX-112) samples (after 22 cycles) even though the control gene was expressed at the same level in both samples, as it is in nontreated ga1–3 (see Methods; data not shown). However, in the presence of BX-112, GA9 reduced GA4 transcript accumulation in ga1–3 only very slightly, whereas GA4 was equally effective in reducing GA4 transcript accumulation in ga1–3 in the presence or absence of BX-112. The simplest explanation for these observations is that GA4 is active in the feedback control of GA4 transcript abundance, whereas GA9 is not. Thus, a GA endogenous to Arabidopsis (GA4) regulates a product-inhibition pathway controlling the abundance of transcripts that encode an enzyme required for the biosynthesis of that GA.
GA Structure-Activity Relationships in the GA4 Transcript Feedback Response
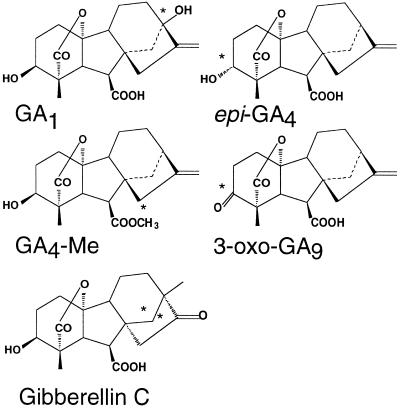
It might be predicted that only 3β-hydroxylated GAs would feedback regulate the abundance of GA4 transcripts (which encode the 3β-hydroxylase). It has also been suggested that the 3β-OH group is the key to GA activity in some plants (Reeve and Crozier, 1974). A range of GA structures (Fig. 4A) were tested for their activity in stimulating hypocotyl elongation in ga1–3 seedlings at a set concentration of 10−7 m. This is the lowest concentration at which exogenous GA4 stimulates hypocotyl elongation sufficiently to make a ga1–3 hypocotyl of equivalent length to an untreated wild-type hypocotyl (Fig. 2). The results of these experiments are shown in Table I. GA4, 3-oxo-GA9, and epi-GA4 exhibited strong activity as stimulators of ga1–3 hypocotyl elongation, whereas GA1, GA C, and GA4-methyl ester all exhibit low activity. For the purposes of the present paper the GAs used in these experiments are classified as active or inactive as stimulators of ga1–3 hypocotyl elongation (Table I).
Figure 4.
Structures of the GAs and GA analogs tested for biological activity. Significant differences in structure from GA4 (see Fig. 3A) are highlighted (*). epi-GA4 is 3α-hydroxylated rather than 3β-hydroxylated; GA1 is 13-hydroxylated; GA4-methyl ester (GA4-Me) is esterified on the carboxyl group at carbon-7; 3-oxo-GA9 has a ketone group at C-3 instead of a 3β-hydroxyl group; and GA C, a derivative of GA1, has a rearrangement of the C and D rings.
Table I.
GA structure-activity relationships
| GAa | Hypocotyl Lengthb | GA4:APT1 Ratioc | Activityd |
|---|---|---|---|
| mm | |||
| 3β-OH-GAs | |||
| 0 | 0.8 (0.1) | 1.00 | — |
| GA4 | 3.3 (0.3) | n.d.e | Active |
| GA1 | 1.0 (0.1) | 0.41 | Inactive |
| GA4-Methyl ester | 0.7 (0.1) | 0.80 | Inactive |
| GA C | 0.9 (0.1) | 0.98 | Inactive |
| Non-3β-OH-GAs | |||
| epi-GA4 | 2.8 (0.3) | n.d. | Active |
| 3-oxo-GA9 | 4.1 (0.4) | n.d. | Active |
GA and analog structures as shown in Figure 4.
Seedlings were grown on germination medium containing individual GAs or analogs at 10−7 m. Hypocotyls were measured as described in Figure 2 with se in parentheses.
Relative GA4 transcript levels calculated after 22 cycles. GA4 and APT1 transcript levels were obtained initially as ImageQuant phosphor imaging values (see Methods), and are expressed as normalized GA4:APT1 ratio (gal-3 grown on germination medium only = 1).
Activity of each GA at 10−7 m. GA1, although classed here as inactive, has mild activity.
n.d., Samples where the GA4 transcript level was too low to be detected, preventing calculation of the GA4:APT1 ratio. In all cases APT1 was expressed at a similar level.
Levels of GA4 and APT1 transcripts were compared via QRT-PCR (after 22 cycles) in ga1–3 seedlings treated with the above GA structures (Table I). GA4 transcripts were not detected in the GA4, 3-oxo-GA9, or epi-GA4 samples, despite detectable expression of the APT1 gene. Thus, GA4, 3-oxo-GA9, and epi-GA4 are all active as negative regulators of GA4 transcript abundance. Conversely, GA4 transcripts were detected at levels similar to that of the untreated ga1–3 control in the GA1, GA C, and GA4-methyl ester samples (a small reduction in the GA1-treated GA4:APT1 ratio is indicative of GA1 possessing low activity).
These experiments show that there is a correlation between the degrees of activity exhibited by each GA structure in the two assays described above. GAs that are active in the promotion of hypocotyl elongation are also active in the negative regulation of GA4 transcript abundance, whereas those inactive in the former assay are also inactive in the latter. Of the structures that are active (GA4, 3-oxo-GA9, and epi-GA4), only GA4 is 3β-hydroxylated. GC-MS analysis showed that the purity of the GA4, 3-oxo-GA9, and epi-GA4 samples is high, and that the activity displayed by 3-oxo-GA9 and epi-GA4 is not due to GA4 contamination (see Methods). Furthermore, GA1, GA C, and GA4-methyl ester are all 3β-hydroxylated and yet are inactive (or have low activity in the case of GA1). Thus, a specific 3β-hydroxy-GA recognition system may not be involved in the negative feedback regulation of GA4 transcript abundance.
Feedback Control of GA4 Transcript Abundance Is Disrupted in gai and spy Mutants
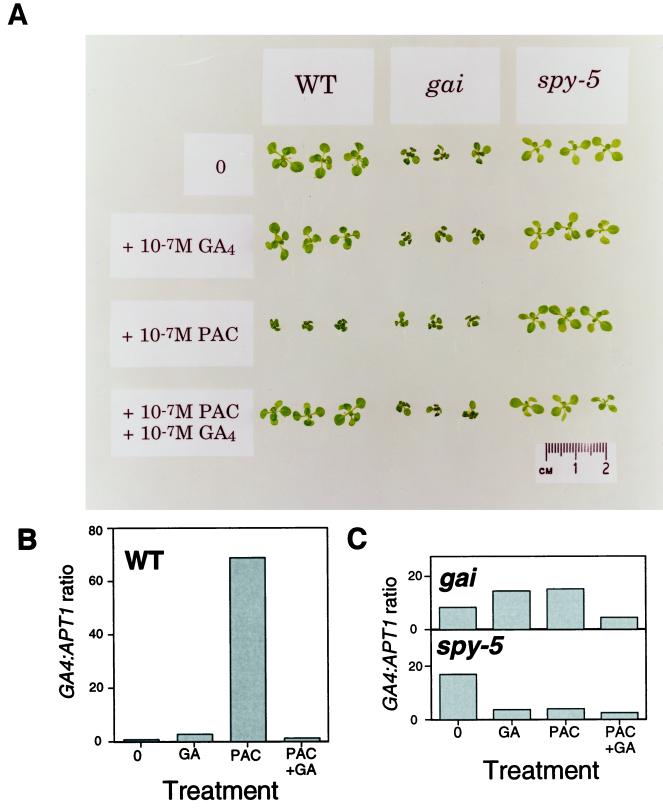
The effect of GA treatments on GA4 transcript levels was further investigated using the wild type and the GA-response mutants gai and spy-5. These plants have different/unknown endogenous GA concentrations (Talon et al., 1990a, 1990b). For this reason, the plants were grown in media containing 10−7 m GA4 or 10−7 m PAC or both. PAC inhibits the oxidation of ent-kaurene, an early step in GA biosynthesis, and thus reduces endogenous GA levels (Graebe, 1987). The effects of these treatments on the growth of wild-type, gai, and spy-5 seedlings is shown in Figure 5A. As expected, the wild type is markedly dwarfed by PAC, and this effect is reversed by the additional presence of GA4 in the medium. gai is dwarfed both in the presence and absence of PAC, and remains dwarfed in the GA plus PAC treatment. spy-5 (in the absence of PAC or GA) is approximately the same size as the wild type, and is resistant to the dwarfing effects of PAC (Wilson and Somerville, 1995).
Figure 5.
Effects of GA4 or PAC on wild-type (WT), gai, and spy-5 seedlings. A, Seedlings grown on germination medium containing 10−7 m GA4 and/or 10−7 m PAC for 2 weeks. B, Effects of exogenous GA4 (GA) and PAC (same concentrations as in A) on GA4 mRNA levels in the wild type. QRT-PCR results were converted to ratios by normalizing samples to the wild-type sample grown on germination medium only. All samples were prepared at the same time and compared after 22 cycles. C, Effects of exogenous GA4 (GA) and PAC (same concentrations as in A on GA4 mRNA levels in gai and spy mutants). Samples were prepared at the same time and data were normalized as in B and compared after 22 cycles.
GA4 transcript levels (relative to those of APT1) were assayed in these mutants. As shown previously (Figs. 1 and 2), the wild type has lower levels of GA4 transcript than does ga1–3. However, treatment of the wild type with 10−7 m PAC induces expression of the GA4 gene over 60-fold (see Fig. 5B). This effect can be reversed by the addition of GA4 (Fig. 5B). GA4 transcript levels were approximately 4-fold higher in gai than in the wild type, but were appreciably lower than in ga1–3 (see Figs. 2 and 5C). gai was relatively insensitive to manipulated changes in endogenous GA levels, maintaining slightly elevated levels of GA4 transcript (4–16 times untreated wild type), despite treatments with PAC or GA (Fig. 5C). In gai, GA4 gene expression is partially, but not fully, repressed by its own high levels of endogenous GAs (Talon et al., 1990b). In addition, PAC treatment (depletion of endogenous GAs) did not induce GA4 expression to the extent that it does in the wild type.
GA4 transcript levels were also higher in spy-5 than in the wild type (Fig. 5C). Furthermore, these levels were relatively unaffected by treatment with PAC (compare this with the marked induction of GA4 transcript level in the wild type by PAC). Thus, spy-5 is a mutant that displays, on PAC, GA-independent regulation of GA4 transcript levels and, like gai, blocks the GA4 transcript abundance feedback response. This is an interesting result, because spy mutants show some hallmarks of constitutive GA-response mutants (Swain and Olszewski, 1996). In another such constitutive GA-response mutant, the la crys mutant of pea, 20-oxidase transcripts accumulate to lower (rather than higher) levels than they do in wild-type plants (Martin et al., 1996).
The key conclusion from these experiments is that whereas wild-type plants display a marked increase in GA4 transcript level following PAC treatments, gai and spy-5 mutant plants do not.
DISCUSSION
We have used GAs endogenous to Arabidopsis to demonstrate that GA4 gene expression can be controlled by the product of the 3β-hydroxylation reaction, GA4, but not by the substrate, GA9. Active GAs have already been shown to regulate the expression of GA4, GA5, and other 20-oxidase-encoding genes in Arabidopsis (Chiang et al., 1995; Phillips et al., 1995; Xu et al., 1995) and a 20-oxidase gene in pea (Martin et al., 1996). Our experiments show that GAs that lack activity in a growth bioassay (hypocotyl-elongation response) are also inactive in the regulation of GA4 transcript levels. Conversely, GAs that control GA4 transcript levels also control hypocotyl elongation. These observations suggest that the control of GA4 transcript levels and hypocotyl elongation may be regulated via common GA:GA-receptor interactions and subsequent signal transduction pathways. In addition, our results show that it is active GAs that regulate GA4 transcript abundance, and not only GAs that are the immediate products of the 3β-hydroxylation reaction.
Negative feedback regulation of biosynthetic gene expression is potentially an important form of regulation of GA biosynthesis in plants. We have shown that control of GA4 gene expression is sensitively GA dose dependent in the normal physiological range. Thus, incremental changes in GA level result in correlated changes in the GA4 transcript level, providing a sensitive homeostatic mechanism for the regulation of in planta GA levels. This may allow the plant to subtly monitor and alter GA production in response to developmental and environmental changes.
We tested the hypothesis that the 3β-OH group confers activity for the hypocotyl elongation and GA4 transcript feedback responses. We found that a 3α-OH group (epi-GA4) and a 3=O group (3-oxo-GA9) could each substitute for the 3β-OH group on the GA4 skeleton, creating molecules that were active in the regulation of both GA4 transcript levels and hypocotyl elongation. Earlier experiments demonstrated that epi-GA4 and 3-oxo-GA9 are active in the regulation of cucumber hypocotyl elongation (Brian et al., 1967). In addition, recent experiments using the d1 mutant of maize have shown that GA5, which lacks a 3β-OH group, is active in the stimulation of leaf-sheath elongation, suggesting that a 3β-OH group may not be crucial for activity (Spray et al., 1996). Furthermore, GA22, which does not have a 3β-OH group but has a 18-OH group, promotes shoot elongation in rice in the presence of BX-112 (Kamiya et al., 1991). In this latter case it is possible that the 18-OH group compensates partially for the absence of the 3β-OH group. It is probable that the electronegative group at the C-3 position of GA4 is important for the GA:GA-receptor interaction. Both 3-oxo-GA9 and epi-GA4 have electronegative groups at C-3 (although with a slightly different orientation than in GA4), whereas GA9 and the other inactive GAs do not. Furthermore, we found that GAs that possessed a 3β-OH group but were altered at other regions of the molecule (GA1, GA4-methyl ester, and GA C) had reduced activity. Thus, our experiments indicate that activity is retained in structures in which the -OH group at the 3β position is replaced by other groups (subject to the requirement for the electronegative group at C-3), and that activity is modified by groups at positions other than C-3. It is well known that 2β-hydroxylation of 3β-hydroxylated GAs results in a loss of activity (Takahashi et al., 1990).
Conclusions from structural studies must be tentative, as the capacity of seedlings to interconvert and transport different GA structures must be considered. However, the difference between the activities of GA1 and GA4 is striking (see also Sponsel et al., 1997). GA1 differs from GA4 by the presence in the former, and the absence in the latter, of a hydroxyl group at C-13 (see Fig. 4). Although we cannot discount the possibility that the applied GAs are converted to more active or inactive forms, it is unlikely that GA1 is dehydroxylated to a more active form in vivo, because the progressive oxidation of GAs is generally thought to be an irreversible process (Graebe, 1987). Thus, the presence or absence of the 13-OH group influences the activity of the GA structure. These observations are consistent with the idea that the putative GA receptor recognizes the whole of the GA molecule and not just a particular region of it (Reeve and Crozier, 1974).
Our experiments show that the gai and spy-5 mutants are both altered in the regulation of GA4 transcript accumulation. gai has elevated levels of GA4 transcript compared with the wild type. This observation is consistent with previous reports that 20-oxidase transcript levels are also elevated in gai (Xu et al., 1995; Peng et al., 1997), and indicates that the gai mutation perturbs the feedback regulation of transcripts encoding GA biosynthesis enzymes. This could explain the elevated levels of bioactive GAs in gai (Talon et al., 1990b): Active GAs do not down-regulate GA4 and GA5 transcript abundance in gai to the extent that they do in the wild type, resulting in higher levels of 20-oxidase and 3β-hydroxylase activities and elevated active GA levels.
The Arabidopsis spy mutants belong to the constitutive GA-response class of GA signal transduction mutant (Swain and Olszewski, 1996). The observation that GA4 transcript abundance in spy-5 is higher, rather than lower, than that of the wild type is perhaps surprising, since 20-oxidase levels in the pea la crys mutant (another constitutive GA-response mutant) are lower than in wild-type pea (Martin et al., 1996). This apparent discrepancy may be due to the fact that different genes (GA4, 20-oxidase) and/or different species (Arabidopsis and pea) are involved. However, the constitutive GA-response mutants may actually represent two subclasses of mutant, one (which includes spy) comprising mutants that mimic wild-type plants treated with a nonsaturating GA dose, and the other (which includes la crys) comprising mutants that mimic wild-type plants treated with a saturating GA dose (Swain and Olszewski, 1996). It is possible that the above apparent discrepancy actually represents a difference between the properties of mutants from these two subclasses.
Of course, regulation of the abundance of transcripts encoding GA biosynthesis enzymes is not the only possible means of altering GA levels, and control of GA abundance may also be effected in other ways. Inactivation (by 2β-hydroxylation), conjugation, compartmentation, and transport processes may all contribute to regulating the concentration of active GAs in planta (Takahashi et al., 1990). In addition, it is possible, although untested, that feedback control may also operate via product inhibition of the enzymatic activity of GA biosynthesis enzymes. Further work will uncover the relative importance of feedback regulation of GA biosynthesis gene transcript levels in the control of the production of active GAs.
ACKNOWLEDGMENTS
The authors thank R. Simon, P. Carol, P. Puangsomlee, and Y.-Y. Yang for advice on experimental procedures; A. Davies for photography; K. King for help with figure preparation; and J. Peng, D. Richards, T. Ait-Ali, and S. Yamaguchi for critical review of this manuscript.
Abbreviations:
- BX-112
prohexadione calcium BX-112
- PAC
paclobutrazol
- QRT-PCR
quantitative RT-PCR
- RT-PCR
reverse-transcription PCR
Footnotes
R.J.C. was supported by a John Innes Foundation Studentship. The work in N.P.H.'s laboratory was funded through a Biotechnology and Biological Sciences Research Council Core Strategic grant to the John Innes Centre, a Biotechnology and Biological Sciences Research Council Plant Molecular Biology grant (no. PG208/0600), and by the European Commission DG XII Biotechnology Program (contract no. BIO4-96-0621).
LITERATURE CITED
- Aldridge DC, Hanson JR, Mulholland TPC (1965) Gibberellic acid. Part XXVIII. Some derivatives of gibberellins A4 and A7. J Chem Soc 3539–3549 [PubMed]
- Brian PW, Grove JF, Mulholland TPC. Relationships between structure and growth-promoting activity of the gibberellins and some allied compounds, in four test systems. Phytochemistry. 1967;6:1475–1499. [Google Scholar]
- Chelly J, Kaplan J-C, Maire P, Gautrin S, Kahn A. Transcription of the dystrophin gene in human muscle and non-muscle tissues. Nature. 1988;333:858–860. doi: 10.1038/333858a0. [DOI] [PubMed] [Google Scholar]
- Chiang H-H, Hwang I, Goodman HM. Isolation of the Arabidopsis GA4 locus. Plant Cell. 1995;7:195–201. doi: 10.1105/tpc.7.2.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang H-H, Hwang I, Goodman HM. Isolation of the Arabidopsis GA4 locus. Correction. Plant Cell. 1997;9:979–980. doi: 10.1105/tpc.7.2.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross BE (1960) Gibberellic acid. Part XIII. The structure of ring A. J Chem Soc 3022–3038
- Ezura H, Harberd NP. Endogenous gibberellin levels influence in-vitro shoot regeneration in Arabidopsis thaliana (L.) Heyhn. Planta. 1995;197:301–305. doi: 10.1007/BF00202651. [DOI] [PubMed] [Google Scholar]
- Graebe JE. Gibberellin biosynthesis and control. Annu Rev Plant Physiol. 1987;38:419–465. [Google Scholar]
- Hedden P, Croker SJ. Regulation of gibberellin biosynthesis in maize seedlings. In: Karssen CM, van Loon LC, Vreugdenhil D, editors. Progress in Plant Growth Regulation. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1992. pp. 534–544. [Google Scholar]
- Hedden P, Kamiya Y. Gibberellin biosynthesis: enzymes, genes, and their regulation. Annu Rev Plant Physiol Plant Mol Biol. 1997;48:431–460. doi: 10.1146/annurev.arplant.48.1.431. [DOI] [PubMed] [Google Scholar]
- Hooley R. Gibberellins: perception, transduction and responses. Plant Mol Biol. 1994;26:1529–1555. doi: 10.1007/BF00016489. [DOI] [PubMed] [Google Scholar]
- Jacobsen SE, Binkowski KA, Olszewski NE. SPINDLY, a tetratricopeptide repeat protein involved in gibberellin signal transduction in Arabidopsis. Proc Natl Acad Sci USA. 1996;93:9292–9296. doi: 10.1073/pnas.93.17.9292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsen SE, Olszewski NE. Mutations at the SPINDLY locus of Arabidopsis alter gibberellin signal transduction. Plant Cell. 1993;5:887–896. doi: 10.1105/tpc.5.8.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamiya Y, Kobayashi M, Fujioka S, Yamane H, Nakayama I, Sakurai A. Effects of a plant growth regulator, prohexadione calcium (BX-112), on the elongation of rice shoots caused by exogenously applied gibberellins and helminthosporol, part II. Plant Cell Physiol. 1991;32:1205–1210. [Google Scholar]
- Koornneef M, Elgersma A, Hanhart CJ, van Leonen-Martinet EP, van Rijn L, Zeevaart JAD. A gibberellin insensitive mutant of Arabidopsis thaliana. Physiol Plant. 1985;65:33–39. [Google Scholar]
- Koornneef M, van der Veen JH. Induction and analysis of gibberellin sensitive mutants in Arabidopsis thaliana (L.) Hehyn. Theor Appl Genet. 1980;58:257–263. doi: 10.1007/BF00265176. [DOI] [PubMed] [Google Scholar]
- Ludevid D, Höfte H, Himmelblau E, Chrispeels MJ. The expression pattern of the tonoplast intrinsic protein γ-TIP in Arabidopsis thaliana is correlated with cell enlargement. Plant Physiol. 1992;100:1633–1639. doi: 10.1104/pp.100.4.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin DN, Proebsting WL, Parks TD, Dougherty WG, Lange T, Lewis MJ, Gaskin P, Hedden P. Feedback regulation of gibberellin biosynthesis and gene expression in Pisum sativum L. Planta. 1996;200:159–166. doi: 10.1007/BF00208304. [DOI] [PubMed] [Google Scholar]
- Moffat BA, McWhinne EA, Agarwhal SK, Schaff DA. The adenine phosphoribosyltransferase-encoding gene of Arabidopsis thaliana. Gene. 1994;143:211–216. doi: 10.1016/0378-1119(94)90098-1. [DOI] [PubMed] [Google Scholar]
- Murphy LD, Herzog CE, Rudick JB, Fojo AT, Bates SE. Use of the polymerase chain reaction in the quantification of mdr-1 gene expression. Biochemistry. 1990;29:10351–10356. doi: 10.1021/bi00497a009. [DOI] [PubMed] [Google Scholar]
- Nakayama I, Kamiya Y, Kobayashi M, Abe H, Sakurai A. Effects of a plant growth regulator, prohexadione, on the biosynthesis of gibberellins in cell-free systems derived from immature seeds. Plant Cell Physiol. 1990a;31:1183–1190. [Google Scholar]
- Nakayama I, Miyazawa T, Kobayashi M, Kamiya Y, Abe H, Sakurai A. Effects of a new plant growth regulator prohexadione calcium (BX-112) on shoot elongation caused by exogenously applied gibberellins in rice (Oryza sativa L.) seedlings. Plant Cell Physiol. 1990b;31:195–200. [Google Scholar]
- Noonan KE, Beck C, Holzmayer TA, Chin JE, Wunder JS, Andrulis IL, Gazdar AF, Willman CL, Griffith B, von Hoff DD and others. Quantitative analysis of MDR1 (multidrug resistance) gene expression in human tumours by polymerase chain reaction. Proc Natl Acad Sci USA. 1990;87:7160–7164. doi: 10.1073/pnas.87.18.7160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J, Carol P, Richards DE, King KE, Cowling RJ, Murphy GP, Harberd NP. The Arabidopsis GAI gene defines a signaling pathway that negatively regulates gibberellin responses. Genes Dev. 1997;11:3194–3205. doi: 10.1101/gad.11.23.3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J, Harberd NP. Derivative alleles of the Arabidopsis gibberellin-insensitive (gai) mutation confer a wild-type phenotype. Plant Cell. 1993;5:351–360. doi: 10.1105/tpc.5.3.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J, Harberd NP. Gibberellin deficiency and response mutations suppress the stem elongation phenotype of phytochrome-deficient mutants of Arabidopsis. Plant Physiol. 1997;113:1051–1058. doi: 10.1104/pp.113.4.1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips AL, Huttly AK. Cloning of two gibberellin-regulated cDNAs from Arabidopsis thaliana by subtractive hybridization: expression of the tonoplast water channel, γ-TIP, is increased by GA3. Plant Mol Biol. 1994;24:603–615. doi: 10.1007/BF00023557. [DOI] [PubMed] [Google Scholar]
- Phillips AL, Ward DA, Uknes S, Appleford NEJ, Lange T, Huttly AK, Gaskin P, Graebe JE, Hedden P. Isolation and expression of three gibberellin-20-oxidase cDNA clones from Arabidopsis. Plant Physiol. 1995;108:1049–1057. doi: 10.1104/pp.108.3.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeve DR, Crozier A. An assessment of gibberellin structure-activity relationships. J Exp Bot. 1974;25:431–445. [Google Scholar]
- Ross JJ. Recent advances in the study of gibberellin mutants. Plant Growth Reg. 1994;15:193–206. [Google Scholar]
- Sponsel VM, Schmidt FW, Porter SG, Nakayama M, Kohlstruk S, Estelle M. Characterization of new gibberellin-responsive semidwarf mutants of Arabidopsis. Plant Physiol. 1997;115:1009–1020. doi: 10.1104/pp.115.3.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spray CR, Kobayashi M, Suzuki Y, Phinney BO, Gaskin P, MacMillan J. The dwarf-1 (d1) mutant of Zea mays blocks three steps in the gibberellin biosynthetic pathway. Proc Natl Acad Sci USA. 1996;93:10515–10518. doi: 10.1073/pnas.93.19.10515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun T-p, Kamiya Y. The Arabidopsis GA1 locus encodes the cyclase ent-kaurene synthetase A of gibberellin biosythesis. Plant Cell. 1994;6:1509–1518. doi: 10.1105/tpc.6.10.1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swain SM, Olszewski NE. Genetic analysis of gibberellin signal transduction. Plant Physiol. 1996;112:11–17. doi: 10.1104/pp.112.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi N, Phinney BO, MacMillan J. Gibberellins. New York: Springer-Verlag; 1990. [Google Scholar]
- Talon M, Koornneef M, Zeevaart JAD. Endogenous gibberellins in Arabidopsis thaliana and possible steps blocked in the biosynthetic pathways of the semidwarf ga4 and ga5 mutants. Proc Natl Acad Sci USA. 1990a;87:7983–7987. doi: 10.1073/pnas.87.20.7983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talon M, Koornneef M, Zeevaart JAD. Accumulation of C19-gibberellins in the gibberellin-insensitive dwarf mutant gai of Arabidopsis thaliana (L.) Planta. 1990b;182:501–505. doi: 10.1007/BF02341024. [DOI] [PubMed] [Google Scholar]
- Wilson RN, Somerville CR. Phenotypic suppression of the gibberellin-insensitive mutant (gai) of Arabidopsis. Plant Physiol. 1995;108:495–502. doi: 10.1104/pp.108.2.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y-L, Li L, Wu K, Peeters AJM, Gage DA, Zeevaart JAD. The GA5 locus of Arabidopsis thaliana encodes a multifunctional gibberellin 20-oxidase: molecular cloning and functional expression. Proc Natl Acad Sci USA. 1995;92:6640–6644. doi: 10.1073/pnas.92.14.6640. [DOI] [PMC free article] [PubMed] [Google Scholar]