Abstract
DNA from over 300 Bacillus thuringiensis, Bacillus cereus, and Bacillus anthracis isolates was analyzed by fluorescent amplified fragment length polymorphism (AFLP). B. thuringiensis and B. cereus isolates were from diverse sources and locations, including soil, clinical isolates and food products causing diarrheal and emetic outbreaks, and type strains from the American Type Culture Collection, and over 200 B. thuringiensis isolates representing 36 serovars or subspecies were from the U.S. Department of Agriculture collection. Twenty-four diverse B. anthracis isolates were also included. Phylogenetic analysis of AFLP data revealed extensive diversity within B. thuringiensis and B. cereus compared to the monomorphic nature of B. anthracis. All of the B. anthracis strains were more closely related to each other than to any other Bacillus isolate, while B. cereus and B. thuringiensis strains populated the entire tree. Ten distinct branches were defined, with many branches containing both B. cereus and B. thuringiensis isolates. A single branch contained all the B. anthracis isolates plus an unusual B. thuringiensis isolate that is pathogenic in mice. In contrast, B. thuringiensis subsp. kurstaki (ATCC 33679) and other isolates used to prepare insecticides mapped distal to the B. anthracis isolates. The interspersion of B. cereus and B. thuringiensis isolates within the phylogenetic tree suggests that phenotypic traits used to distinguish between these two species do not reflect the genomic content of the different isolates and that horizontal gene transfer plays an important role in establishing the phenotype of each of these microbes. B. thuringiensis isolates of a particular subspecies tended to cluster together.
Bacillus thuringiensis, Bacillus cereus, and Bacillus anthracis are three closely related gram-positive species belonging to the B. cereus group (23). Each of these species is economically important. B. thuringiensis spore preparations are commonly used as an organic biopesticide with activity against lepidopteran, dipteran, and coleopteran insect pests (22). These spore preparations have been so effective that the plasmid-encoded insecticidal genes have been cloned and introduced into transgenic crops such as corn, potato, and cotton (1). Some B. cereus isolates are toxigenic and can cause food poisoning, an emetic or diarrheal syndrome, or a variety of nongastrointestinal infections (3). B. cereus is a common contaminant of many food products, including rice, milk and dairy products, meat products, dried foods, and spices (17). B. anthracis is the causal agent of anthrax, which can be lethal to humans and other mammals. The spores of this organism were recently used as a bioterrorism agent when placed in letters and mailed to individuals in the United States. This incident led to several cases of cutaneous anthrax, a mild form of the disease treatable with antibiotics. It also led to cases of more severe inhalation anthrax, which resulted in five deaths (8).
The diversity within each of these Bacillus species has been reported. Using multilocus enzyme electrophoresis (MEE) analysis, Helgason et al. (4) examined 154 Norwegian B. cereus and B. thuringiensis soil isolates. Phylogenetic analysis of the MEE data also revealed significant diversity among the different isolates (4). More recently, fluorescent amplified fragment length polymorphism (AFLP) analysis of the same isolates confirmed this (24). In another AFLP study, Pattanayak et al. (20) analyzed 24 B. thuringiensis isolates representing 24 different B. thuringiensis subspecies. The phylogenetic information presented demonstrated that each subspecies greatly differed from the other subspecies and revealed extensive genetic diversity within this species. In contrast, AFLP analysis of 78 different B. anthracis isolates revealed that B. anthracis is very monomorphic (13, 14). The difficulty in distinguishing among B. anthracis isolates led Keim et al. to identify 36 multilocus variable-number tandem repeat markers located throughout the B. anthracis genome and on the two virulence plasmids to more effectively discriminate among different B. anthracis isolates (eight markers are described in reference 15).
None of these studies identified distinct groups of B. cereus and B. thuringiensis. This has led some to suggest that these be considered a single species. Helgason et al. compared 13 B. anthracis isolates and 227 B. thuringiensis and B. cereus isolates using 13 enzyme loci and sequence analysis of nine chromosomal genes (6). Based on the close genetic similarities of B. anthracis to B. cereus and B. thuringiensis at these 22 loci, they argued for inclusion of this pathogen in this same species designation.
We performed fluorescent AFLP analysis of 332 B. thuringiensis, B. cereus, and B. anthracis isolates to further examine the genetic relationships among these three species. Thirty-four of the most diverse Norway soil isolates analyzed in previous studies (24) were compared with 222 B. thuringiensis isolates representing 36 different serovars or subspecies from the U.S. Department of Agriculture (USDA) collection. Multiple isolates representing particular subspecies or serovars were included when available. Twenty-four genetically diverse B. anthracis isolates were also included (15). Eight B. thuringiensis and B. cereus strains from the American Type Culture Collection, including the B. cereus and B. thuringiensis type strains and the B. thuringiensis isolate used in the common agricultural biopesticide Dipel (ATCC 33679; B. thuringiensis subsp. kurstaki) (19), were also included along with a putative B. thuringiensis isolate collected from a severe wound infection and found to be pathogenic in mice (7). Forty-two B. cereus isolates cultured from contaminated food products or from a variety of clinical specimens were provided for analysis by the Food Research Institute at the University of Wisconsin.
Fluorescent AFLP analysis of this large collection of B. thuringiensis, B. anthracis, and B. cereus isolates was used to reveal detailed phylogenetic relationships among the three species and among different isolates of the same apparent species. Results reported here demonstrate a high level of diversity within B. cereus and B. thuringiensis and reveal that different isolates fall into distinct groups. They also reveal that different B. cereus and B. thuringiensis isolates are extensively interspersed with each other across all branches of the AFLP-based phylogenetic tree. In contrast, B. anthracis is genetically very monomorphic and occupies a subbranch that is distinctive from all B. cereus and B. thuringiensis isolates. Analysis of toxigenic B. cereus isolates revealed that a significant number of these isolates are more closely related to B. anthracis, in contrast to B. cereus isolates collected from the environment that are clustered on other branches of the tree. Analysis of a putative pathogenic B. thuringiensis isolate revealed that it is very closely related to B. anthracis, in contrast to those B. thuringiensis isolates that are used for commercial insecticide production (25).
MATERIALS AND METHODS
Microbial strains.
Table 1 shows the sources of the different Bacillus isolates analyzed in this study. Most B. thuringiensis isolates were kindly provided by L. K. Nakamura of the USDA Agricultural Research Service Culture Collection, Northern Regional Research Center, Peoria, Ill. Norwegian B. thuringiensis soil isolates were from the collection of one of the coauthors (A.-B. Kolstø). ATCC B. cereus strains (4342, 11778, 14579, 31293, 43881 and 53522) and B. thuringiensis strains (10792 and 33679) were obtained from the American Type Culture Collection, Manassas, Va. B. cereus isolates were provided by the Food Research Institute at the University of Wisconsin, Madison. DNA isolated from the 24 diverse B. anthracis isolates and from B. anthracis Vollum was provided by one of the coauthors (P. Keim). The pathogenic B. thuringiensis isolate 97-27 was a gift from the laboratory of Eric Hernandez of the Hospital Militaire Begin, Service de Biologie Médicale.
TABLE 1.
Sources of different Bacillus isolatesa
| Species | ID | Species | ID | Species | ID |
|---|---|---|---|---|---|
| B. anthracis | K01231 | B. anthracis | K04041 | B. anthracis | K06101 |
| B. anthracis | K12561 | B. anthracis | K12851 | B. anthracis | K13401 |
| B. anthracis | K16941 | B. anthracis | K24781 | B. anthracis | K24841 |
| B. anthracis | K27621 | B. anthracis | K28021 | B. anthracis | K37001 |
| B. anthracis | K42411 | B. anthracis | K45161 | B. anthracis | K45961 |
| B. anthracis | K48341 | B. anthracis | K51351 | B. anthracis | K64281 |
| B. anthracis | K68351 | B. anthracis | K70381 | B. anthracis | K72221 |
| B. anthracis | K74411 | B. anthracis | K79481 | B. anthracis | K82151 |
| B. anthracis | K90021 | B. cereus | 3A2 | B. cereus | 1230-882 |
| B. cereus | AH 5223 | B. cereus | AH 5233 | B. cereus | AH 5273 |
| B. cereus | AH 5403 | B. cereus | AH 5473 | B. cereus | AH 5633 |
| B. cereus | AH 5663 | B. cereus | AH 5903 | B. cereus | AH 5923 |
| B. cereus | AH 6293 | B. cereus | AH 6333 | B. cereus | AH 6343 |
| B. cereus | ATCC 43423 | B. cereus | ATCC 117784 | B. cereus | ATCC 145794 |
| B. cereus | ATCC 312934 | B. cereus | ATCC 438814 | B. cereus | ATCC 535224 |
| B. cereus | B4ac2 | B. cereus | D32 | B. cereus | D52 |
| B. cereus | D72 | B. cereus | D142 | B. cereus | D172 |
| B. cereus | D212 | B. cereus | D232 | B. cereus | D262 |
| B. cereus | D332 | B. cereus | F1-12 | B. cereus | F1-62 |
| B. cereus | F1-112 | B. cereus | F2-12 | B. cereus | F2-192 |
| B. cereus | F3-272 | B. cereus | F837/762 | B. cereus | F1589/772 |
| B. cereus | F2105/892 | B. cereus | F2668/902 | B. cereus | F3502/732 |
| B. cereus | F3748/752 | B. cereus | F4429/732 | B. cereus | F4431/732 |
| B. cereus | F4433/732 | B. cereus | F4810/722 | B. cereus | FM-12 |
| B. cereus | MGBC1422 | B. cereus | MGBC1452 | B. cereus | NE-Chicken2 |
| B. cereus | R32 | B. cereus | R42 | B. cereus | R62 |
| B. cereus | R122 | B. cereus | S1-72 | B. cereus | S1C2 |
| B. cereus | S2-42 | B. cereus | S2-82 | B. cereus | S3-72 |
| B. cereus | Soc 672 | B. thuringiensis | 97-275 | B. thuringiensis | AH 5173 |
| B. thuringiensis | AH 5213 | B. thuringiensis | AH 5263 | B. thuringiensis | AH 5333 |
| B. thuringiensis | AH 5343 | B. thuringiensis | AH 5353 | B. thuringiensis | AH 5503 |
| B. thuringiensis | AH 5523 | B. thuringiensis | AH 5583 | B. thuringiensis | AH 5593 |
| B. thuringiensis | AH 5753 | B. thuringiensis | AH 6183 | B. thuringiensis | AH 6243 |
| B. thuringiensis | AH 6263 | B. thuringiensis | AH 6303 | B. thuringiensis | AH 6313 |
| B. thuringiensis | AH 6323 | B. thuringiensis | AH 6393 | B. thuringiensis | AH 6403 |
| B. thuringiensis | AH 6483 | B. thuringiensis | AH 6653 | B. thuringiensis | AH 6783 |
| B. thuringiensis | ATCC 107924 | B. thuringiensis | ATCC 336794 | B. thuringiensis | HD 16 |
| B. thuringiensis | HD 36 | B. thuringiensis | HD 46 | B. thuringiensis | HD 56 |
| B. thuringiensis | HD 76 | B. thuringiensis | HD 106 | B. thuringiensis | HD 116 |
| B. thuringiensis | HD 136 | B. thuringiensis | HD 146 | B. thuringiensis | HD 156 |
| B. thuringiensis | HD 166 | B. thuringiensis | HD 186 | B. thuringiensis | HD 196 |
| B. thuringiensis | HD 296 | B. thuringiensis | HD 306 | B. thuringiensis | HD 336 |
| B. thuringiensis | HD 346 | B. thuringiensis | HD 396 | B. thuringiensis | HD 416 |
| B. thuringiensis | HD 436 | B. thuringiensis | HD 446 | B. thuringiensis | HD 476 |
| B. thuringiensis | HD 506 | B. thuringiensis | HD 576 | B. thuringiensis | HD 626 |
| B. thuringiensis | HD 656 | B. thuringiensis | HD 676 | B. thuringiensis | HD 766 |
| B. thuringiensis | HD 826 | B. thuringiensis | HD 836 | B. thuringiensis | HD 846 |
| B. thuringiensis | HD 886 | B. thuringiensis | HD 956 | B. thuringiensis | HD 996 |
| B. thuringiensis | HD 1076 | B. thuringiensis | HD 1096 | B. thuringiensis | HD 1106 |
| B. thuringiensis | HD 1156 | B. thuringiensis | HD 1206 | B. thuringiensis | HD 1276 |
| B. thuringiensis | HD 1306 | B. thuringiensis | HD 1356 | B. thuringiensis | HD 1366 |
| B. thuringiensis | HD 1466 | B. thuringiensis | HD 1506 | B. thuringiensis | HD 1536 |
| B. thuringiensis | HD 1546 | B. thuringiensis | HD 1576 | B. thuringiensis | HD 1586 |
| B. thuringiensis | HD 1676 | B. thuringiensis | HD 1696 | B. thuringiensis | HD 1766 |
| B. thuringiensis | HD 1856 | B. thuringiensis | HD 1926 | B. thuringiensis | HD 1966 |
| B. thuringiensis | HD 1986 | B. thuringiensis | HD 2016 | B. thuringiensis | HD 2036 |
| B. thuringiensis | HD 2116 | B. thuringiensis | HD 2266 | B. thuringiensis | HD 2276 |
| B. thuringiensis | HD 2286 | B. thuringiensis | HD 2356 | B. thuringiensis | HD 2386 |
| B. thuringiensis | HD 2436 | B. thuringiensis | HD 2506 | B. thuringiensis | HD 2526 |
| B. thuringiensis | HD 2606 | B. thuringiensis | HD 2636 | B. thuringiensis | HD 2656 |
| B. thuringiensis | HD 2666 | B. thuringiensis | HD 2706 | B. thuringiensis | HD 2756 |
| B. thuringiensis | HD 2786 | B. thuringiensis | HD 2846 | B. thuringiensis | HD 2856 |
| B. thuringiensis | HD 2876 | B. thuringiensis | HD 2886 | B. thuringiensis | HD 2916 |
| B. thuringiensis | HD 2926 | B. thuringiensis | HD 2936 | B. thuringiensis | HD 2956 |
| B. thuringiensis | HD 2966 | B. thuringiensis | HD 2996 | B. thuringiensis | HD 3006 |
| B. thuringiensis | HD 3016 | B. thuringiensis | HD 3026 | B. thuringiensis | HD 3036 |
| B. thuringiensis | HD 3046 | B. thuringiensis | HD 3056 | B. thuringiensis | HD 3146 |
| B. thuringiensis | HD 3156 | B. thuringiensis | HD 3246 | B. thuringiensis | HD 3356/PICK> |
| B. thuringiensis | HD 3386 | B. thuringiensis | HD 3426 | B. thuringiensis | HD 4536 |
| B. thuringiensis | HD 4626 | B. thuringiensis | HD 4876 | B. thuringiensis | HD 4896 |
| B. thuringiensis | HD 4986 | B. thuringiensis | HD 4996 | B. thuringiensis | HD 5006 |
| B. thuringiensis | HD 5016 | B. thuringiensis | HD 5116 | B. thuringiensis | HD 5126 |
| B. thuringiensis | HD 5166 | B. thuringiensis | HD 5186 | B. thuringiensis | HD 5206 |
| B. thuringiensis | HD 5216 | B. thuringiensis | HD 5256 | B. thuringiensis | HD 5266 |
| B. thuringiensis | HD 5276 | B. thuringiensis | HD 5286 | B. thuringiensis | HD 5296 |
| B. thuringiensis | HD 5306 | B. thuringiensis | HD 5346 | B. thuringiensis | HD 5366 |
| B. thuringiensis | HD 5376 | B. thuringiensis | HD 5386 | B. thuringiensis | HD 5406 |
| B. thuringiensis | HD 5416 | B. thuringiensis | HD 5426 | B. thuringiensis | HD 5526 |
| B. thuringiensis | HD 5536 | B. thuringiensis | HD 5546 | B. thuringiensis | HD 5576 |
| B. thuringiensis | HD 5616 | B. thuringiensis | HD 5656 | B. thuringiensis | HD 5686 |
| B. thuringiensis | HD 5716 | B. thuringiensis | HD 5726 | B. thuringiensis | HD 5736 |
| B. thuringiensis | HD 5776 | B. thuringiensis | HD 5846 | B. thuringiensis | HD 5866 |
| B. thuringiensis | HD 5886 | B. thuringiensis | HD 5896 | B. thuringiensis | HD 5916 |
| B. thuringiensis | HD 5926 | B. thuringiensis | HD 5946 | B. thuringiensis | HD 5976 |
| B. thuringiensis | HD 6006 | B. thuringiensis | HD 6026 | B. thuringiensis | HD 6036 |
| B. thuringiensis | HD 6046 | B. thuringiensis | HD 6056 | B. thuringiensis | HD 6126 |
| B. thuringiensis | HD 6156 | B. thuringiensis | HD 6166 | B. thuringiensis | HD 6216 |
| B. thuringiensis | HD 6256 | B. thuringiensis | HD 6266 | B. thuringiensis | HD 6276 |
| B. thuringiensis | HD 6286 | B. thuringiensis | HD 6296 | B. thuringiensis | HD 6336 |
| B. thuringiensis | HD 6356 | B. thuringiensis | HD 6506 | B. thuringiensis | HD 6576 |
| B. thuringiensis | HD 6586 | B. thuringiensis | HD 6596 | B. thuringiensis | HD 6616 |
| B. thuringiensis | HD 6816 | B. thuringiensis | HD 6826 | B. thuringiensis | HD 7066 |
| B. thuringiensis | HD 7116 | B. thuringiensis | HD 7216 | B. thuringiensis | HD 7546 |
| B. thuringiensis | HD 7556 | B. thuringiensis | HD 7586 | B. thuringiensis | HD 7686 |
| B. thuringiensis | HD 7716 | B. thuringiensis | HD 7726 | B. thuringiensis | HD 7736 |
| B. thuringiensis | HD 7746 | B. thuringiensis | HD 7756 | B. thuringiensis | HD 7766 |
| B. thuringiensis | HD 7806 | B. thuringiensis | HD 7836 | B. thuringiensis | HD 7846 |
| B. thuringiensis | HD 7956 | B. thuringiensis | HD 7996 | B. thuringiensis | HD 8006 |
| B. thuringiensis | HD 8196 | B. thuringiensis | HD 8266 | B. thuringiensis | HD 8336 |
| B. thuringiensis | HD 8476 | B. thuringiensis | HD 8486 | B. thuringiensis | HD 8576 |
| B. thuringiensis | HD 8606 | B. thuringiensis | HD 8676 | B. thuringiensis | HD 8686 |
| B. thuringiensis | HD 8796 | B. thuringiensis | HD 9186 | B. thuringiensis | HD 9216 |
| B. thuringiensis | HD 9226 | B. thuringiensis | HD 9236 | B. thuringiensis | HD 9266 |
| B. thuringiensis | HD 9306 | B. thuringiensis | HD 9346 | B. thuringiensis | HD 9636 |
| B. thuringiensis | HD 9666 | B. thuringiensis | HD 9696 | B. thuringiensis | HD 9726 |
| B. thuringiensis | HD 9736 | B. thuringiensis | HD 9746 | B. thuringiensis | HD 9756 |
| B. thuringiensis | HD 9766 | B. thuringiensis | HD 9776 | B. thuringiensis | HD 9786 |
| B. thuringiensis | HD 9826 | B. thuringiensis | HD 9936 | B. thuringiensis | HD 10026 |
| B. thuringiensis | HD 10086 | B. thuringiensis | HD 10116 | B. thuringiensis | HD 10126 |
| B. thuringiensis | HD 10146 | B. thuringiensis | HD 10156 |
Sources of different Bacillus isolates were as follows: 1, Paul Keim, Dept. of Biological Sciences, Northern Arizona University, Flagstaff, Ariz. (see Keim et al. [15] for information about the B. anthracis K numbers); 2, Amy Wong, Food Research Institute, University of Wisconsin—Madison; 3, Anne-Brit Kolstø, Institute of Pharmacy, University of Oslo, Oslo, Norway; 4, American Type Culture Collection, Manassas, Va.; 5, Laboratoire de Biologie, Hopital des Armees Begin, Paris, France; 6, USDA Agricultural Research Service, Peoria, Ill.
DNA isolation and purification.
Five milliliters of nutrient broth was inoculated with cells from a single colony of a B. cereus or B. thuringiensis isolate, and the culture was incubated overnight with shaking at 28°C. Bacterial cells were harvested by centrifugation at 1,000 × g for 15 min. The bacterial pellets were subjected to three freeze-thaw cycles. DNA was isolated from the disrupted cells using a QIAamp tissue kit (QIAGEN, Inc., Valencia, Calif.), following the protocol provided by the manufacturer. The quantity and quality of the isolated DNA were determined by gel electrophoresis of a small amount of the sample (24). Electrophoresis was for 1 h at 80 V. Gels were stained for 20 min with a solution containing 1 μg of ethidium bromide/ml, destained in distilled water, and then visualized and photographed under UV light.
AFLP analysis of DNA samples.
AFLP analysis was accomplished as previously described (10, 24). Briefly, DNA (100 ng) was digested with EcoRI and MseI, and the resulting fragments were ligated to double-stranded adapters. The digested and ligated DNA was then amplified by PCR using EcoRI and MseI +0/+0 primers. The +0/+0 PCR product was analyzed by agarose gel electrophoresis to determine the size range of amplified fragments. Three microliters was used in subsequent selective amplifications using the +1/+1 primer combination of 6-carboxyfluorescein-labeled EcoRI-C (5′GTAGACTGCGTACCAATTCC-3′) and MseI-G (5′-GACGATGAGTCCTGAGTAAG-3′). Selective amplifications were performed in 20-μl reaction mixtures. The resulting products (0.5 to 1.0 μl) were mixed with a solution containing a mixture of DNA size standards (Genescan-500 [Applied Biosystems Inc., Foster City, Calif.] and MapMarker-400 [BioVentures, Inc., Murfreesburo, Tenn.]) both labeled with N,N,N,N-tetramethyl-6-carboxyrhodamine. Following a 2-min heat denaturation at 90°C, the reactions were loaded onto a 5% Long Ranger DNA sequencing gel (BioWhittaker Molecular Applications, Rockland, Maine) and visualized on an ABI 377 automated fluorescent sequencer (Applied Biosystems Inc.). Each set of reactions also contained an AFLP reaction using B. anthracis Vollum DNA as a template. Inclusion of such a reaction in each set of analyses allowed a comparison of results from different analysis sets run at different times or on different gels. Genescan analysis software (Applied Biosystems Inc.) was used to determine the length of the sample fragments by comparison to the DNA fragment length size standards included with each sample.
AFLP data analysis was performed as described by Ticknor et al. (24). Sample fragments between 100 and 500 bp and with fluorescence above 50 arbitrary units in all three runs on the ABI sequencer were used in the analysis. To minimize gel electrophoresis artifacts, each labeling reaction was run in triplicate. Samples were loaded on three different gels in a random order. The triplicate data from three lanes for each sample were combined. The set of peaks used to represent a sample contained all of the peaks that were present in each member of the triplicate. This set was called the fingerprint and was used as the description of a sample when similarities among samples were determined. The height of each peak in the fingerprint was the average height of the peak in the triplicates.
To compare two or more electropherograms and assign a similarity or distance measure to the comparison, the electropherograms were aligned by a clustering algorithm to determine which peaks were common. To do the alignment, all peak locations for all samples being compared were combined into one vector of data. A hierachical agglomerative clustering routine using group averages created the clusters (12). A decision rule was added to this clustering routine so that the number of clusters chosen depended on the number of electropherograms being compared and a maximum value for the range of a cluster (a value for what could be considered “the same”). Peaks within a cluster were assigned the average peak value for that cluster, so that all peaks in a set being compared that were considered the same had the same peak value.
Similarities among samples were determined by the Jaccard coefficient. The 40 tallest peaks for each sample fingerprint were used to calculate the Jaccard coefficient among samples. Dendrograms were produced by using the similarity matrix of Jaccard coefficients and the unweighted pair-group mean average method (UPGMA) (F. J. Rohlf, NTSYS-PC numerical taxonomy and multivariate analysis system, version 1.8; Exeter Software, Setauket, N.Y.).
Principal components for the AFLP fingerprint data were derived (11). The first and second and the first and third principal components were plotted with characters relating to the 10 major clusters seen on the UPGMA dendrograms. All statistical data manipulations were done by using codes developed in S-Plus (Data Analysis Products Division, MathSoft, Seattle, Wash.).
RESULTS
Three hundred thirty-two isolates of B. cereus, B. thuringiensis, and B. anthracis were analyzed using AFLP to better understand the genetic relationships among these closely related bacilli. Unlike multilocus sequence typing and other single nucleotide polymorphism-based genomic characterization methods in which differences within a few very short specific regions of a microbial genome are compared, AFLP is a multilocus sampling method in which fragments generated by the restriction enzyme digestion of an entire genome are used to generate a fingerprint or signature for an isolate. The two restriction endonucleases EcoRI (recognition sequence, 5′-GAATTC-3′) and MseI (recognition sequence, 5′-TTAA-3′) were chosen for the analysis because together they effectively digest DNA with a relatively rich A+T content into a sufficiently large number of small DNA fragments in the range needed to conduct an analysis. B. cereus, B. thuringiensis, and B. anthracis are known to have a relatively high (∼65%) A+T content (unpublished results; see also reference 10 for an example). After digestion with restriction enzymes (EcoRI and MseI) and further selective PCR amplification, the AFLP fingerprint for an isolate was represented by about 40 fragments between 100 and 500 bp in length that were present in all three replicates of the isolate. These fingerprint fragment sizes were then used to generate a phlyogenetic tree that illustrates the genetic relationships among the different isolates.
A collection of B. cereus, B. thuringiensis, and B. anthracis isolates from different sources was used to represent each species analyzed: 34 Norwegian soil isolates representing the genetic diversity of that collection, 222 B. thuringiensis isolates from the USDA collection, 42 B. cereus isolates from the Food Research Institute at the University of Wisconsin, 24 diverse B. anthracis isolates, 8 B. cereus and B. thuringiensis isolates from the ATCC, an unusual, apparently pathogenic B. thuringiensis isolate, and one control B. anthracis isolate, included in each experiment to examine variability.
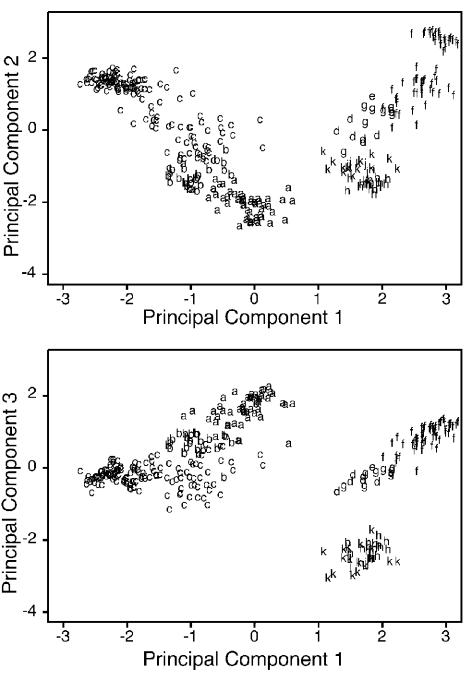
Results of the AFLP analyses showed that these type 1 bacilli are heterogeneous, with the B. cereus and B. thuringiensis isolates interspersed in clusters throughout the phylogenetic tree. Principal component analysis of the AFLP DNA fragment fingerprints is shown in two plots in Fig. 1. The letters in the principal component plots correspond to the placement of each isolate on the different branches (A to K) of the AFLP-based phylogenetic tree shown in Fig. 2. The cloud of points in the principal component analysis suggested that the differences among some branches on the tree were not large, since there was a continuum of points with only small separations among different parts of individual clouds. Principal component analysis is a standard mathematical tool used to detect correlations in large data sets. The objective of principal component analysis is to discover or to reduce the dimensionality of the data set and to identify new meaningful underlying variables. Principal component analysis tries to create linear combinations of the different AFLP fragments that allow the greatest separation of the samples into the different branches or clusters of the AFLP-based tree. If there is separation of the samples using two or three principal components, then there is support, based on a completely different type of data analysis, that the branches of the tree are supported by the data and are not simply artifacts of the other analysis method used. Principle component analysis using components 1 and 2 broke the samples into two clouds. One contained branches A, B, and C, while the other contained branches D, E, F, G, H, J, and K (Fig. 1). Principle component analysis using components 1 and 3 further distinguished among members of the second cloud, so that branches D, E, F, and G could be distinguished from branches H, J, and K. Taken together, the principle component analyses suggested the presence of three major clusters on the phylogenetic tree (Fig. 2).
FIG. 1.
Principal components analysis. The first three principal components of the AFLP fingerprint data for all of the isolates are presented. Each isolate is labeled as 1 of 10 groups (A to K), based on the clustering in the dendrogram shown in Fig. 2. This figure shows that the 10 groupings from the AFLP cluster analysis are present in the principal components analysis.
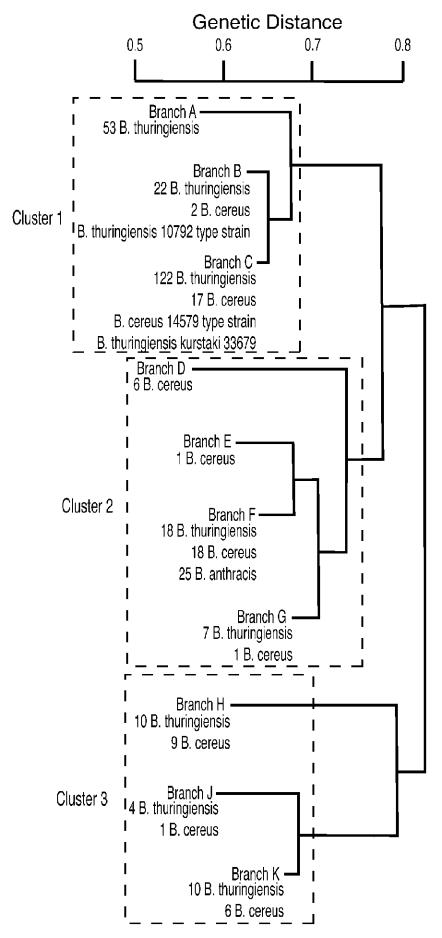
FIG. 2.
An AFLP-based phylogenetic tree of B. anthracis, B. cereus, and B. thuringiensis. A schematic representation of the phylogenetic tree derived from fluorescent AFLP analysis of 332 B. anthracis, B. cereus, and B. thuringiensis isolates. Ten distinct branches are defined (A to K), with most branches containing both B. thuringiensis and B. cereus isolates. All B. anthracis isolates mapped to branch F.
Details of individual isolates mapping to different branches are illustrated in Fig. 3 to 8. In Fig. 2 there are 10 distinct branches (at the 0.6 to 0.7 level), with most branches containing both B. thuringiensis and B. cereus isolates. The upper three branches (cluster 1 in Fig. 2 and branches A, B, and C in Fig. 3 to 6) contain most of the B. thuringiensis isolates from the USDA collection and the majority of the ATCC B. thuringiensis and B. cereus isolates. The B. thuringiensis (ATCC 10792) and B. cereus (ATCC 14579) type strains map to this cluster. Fifty-three of the 58 B. thuringiensis isolates representing subspecies that are common components of commercial biopesticide products in the United States (25) map to this cluster, including B. thuringiensis subsp. kurstaki (ATCC 33679; branch C), the strain used in the preparation of the bioinsecticide Dipel (19, 25).
FIG. 3.
Branch A of the AFLP-based phylogenetic tree of B. anthracis, B. cereus, and B. thuringiensis. Fifty-three B. thuringiensis isolates, representing six different serotypes, mapped to this branch. No B. cereus or B. anthracis isolates mapped to this branch. The AFLP-based phylogenetic trees presented are all based on a total of 40 fragments generated from EcoRI/MseI digestion of genomic DNAs unless otherwise stated in the legends to Figures 3 to 8. Numbers in parentheses are the number of fragments analyzed if the number is other than 40.
FIG. 8.
Branches H, J, and K of the phylogenetic tree of B. anthracis, B. cereus, and B. thuringiensis. Branches H, J, and K contain 16 B. cereus isolates and 24 B. thuringiensis isolates, representing 14 serotypes.
FIG. 6.
Branch C (part 2) of the AFLP-based phylogenetic tree of B. anthracis, B. cereus, and B. thuringiensis. Part 2 of branch C contains 88 of the 122 branch C B. thuringiensis isolates, including B. thuringiensis ATCC 33679. It contains only a single B. cereus isolate.
The middle of the tree contains four branches (cluster 2 in Fig. 2 and branches D, E, F, and G in Fig. 7). Branch F contains 16 toxigenic B. cereus isolates, 1 unusual pathogenic B. thuringiensis isolate identified as 97-27, and all of the 24 diverse B. anthracis isolates included in this analysis. The B. anthracis isolates are the most genetically diverse representatives of a collection of over 1,250 isolates analyzed with eight multilocus variable-number tandem repeat markers (15). However, fluorescent AFLP using only one primer combination did not provide sufficient resolution to discriminate among these different B. anthracis isolates (Fig. 7), and so they cluster within this branch. Also included in branch F are 18 B. thuringiensis isolates and 1 B. cereus environmental isolate that have no known toxigenic or pathogenic properties in vertebrates. Branch E has only one member, B. cereus isolate ATCC 4342, isolated from milk. The presence of this single member could reflect the random sampling of isolates for this study. No information about the toxigenic nature of this isolate could be found, but other B. cereus isolates found to be similar to ATCC 4342 by MEE analysis are associated with periodontal disease (5).
FIG. 7.
Branches D, E, F, and G of the phylogenetic tree of B. anthracis, B. cereus, and B. thuringiensis. Branches D, E, F, and G contain 26 B. cereus isolates and 25 B. thuringiensis isolates, representing 13 serotypes. Branch F contains all 25 of the B. anthracis isolates analyzed in this study. Previous studies indicated that all B. anthracis isolates will map to the same location. Branch F also contains the pathogenic B. thuringiensis isolate 97-27.
The lowest three branches (cluster 3 in Fig. 2 and branches H, J, and K in Fig. 8) contained 30 of the 34 B. thuringiensis and B. cereus soil isolates collected from different locations in Norway. These 34 isolates were chosen as representative of the genetic diversity within a collection of 154 Norwegian soil isolates, based on previous AFLP analysis (24).
In this AFLP analysis, HD-1 received independently from the USDA collection mapped closely (about 0.25) within branch C to ATCC 33679 (HD-1) received from the ATCC. The close proximity of these two isolates obtained from two different sources showed that the level of discrimination within the data was at least 0.25. This value was also supported by the inclusion of an internal DNA control of B. anthracis Vollum in each AFLP experiment to assure data quality, comparability among gels, and reproducibility. The HD-1 isolate from the USDA collection was identical to nine other B. thuringiensis subsp. kurkstaki isolates obtained from the USDA collection but slightly different from isolate 33679, obtained from ATCC. These differences may be due to extensive culturing of the ATCC isolate relative to those in the USDA collection, resulting in losses of DNA from this isolate or rearrangements in its chromosomal DNA.
The B. thuringiensis isolates in this study represented 36 different serovars or subspecies. The serovars were defined by an immunological assay based on the flagellar antigen (2, 18). Examination of the genetic sequence of the gene encoding the flagellar antigen showed that the published sequence contains no EcoRI digestion sites. Point mutations at three 6-nucleotide sites within the gene could generate EcoRI cutting sites, but none of these would produce a labeled AFLP fragment in the size range of the analysis. Therefore, DNA encoding this gene was not responsible for any of the DNA fragments analyzed by AFLP in this study. Table 2 shows where the 36 different B. thuringiensis serovars or subspecies mapped relative to the different branches of the AFLP phylogenetic tree. This table shows that several serovars, such as H1, H2, H3a3b, H4a4b, and H4a4c, appear to cluster within a particular branch of the phylogenetic tree. Many serovars were represented by only a few isolates, and so no correlation to a particular branch could be made. However, isolates representing a single serovar or subspecies were often from different sources and geographic regions and so, based on these results, it appears that there was a correlation between the serovar or subspecies and the AFLP fingerprint.
TABLE 2.
Comparison of B. thuringiensis serotypes to AFLP phylogenetic tree locationa
| Serotype | Branch | No. in branch/total | Sourceb |
|---|---|---|---|
| H1 | B | 17/17 | 6 USA (1 Tex., 1 Ariz., 4 no data), 2 Czechoslovakia, 1 Yugoslavia, 7 no data, 1 ATCC 10792 type strain |
| H2 | A | 1/10 | 1 no data |
| C | 1/10 | 1 no data | |
| F | 1/10 | 1 no data | |
| G | 7/10 | 4 USA, 1 Egypt, 1 France, 1 no data | |
| H3a | A | 4/8 | 1 England, 3 no data |
| C | 1/8 | 1 no data | |
| F | 3/8 | 1 Yugoslavia, 2 no data | |
| H3a3b | C | 24/24 | 4 England, 4 USA (2 Tex., 2 Ariz.), 1 Canada, 15 no data |
| H4a4b | A | 16/18 | 7 Japan, 1 Czechoslovakia, 1 Russia, 1 Yugoslavia, 6 no data |
| B | 2/18 | 1 Czechoslovakia, 1 no data | |
| H4a4c | C | 14/16 | 6 USA (1 Ariz., 4 Fla., 1 no data), 4 Japan, 1 Nairobi, 3 no data |
| F | 1/16 | 1 no data | |
| J | 1/16 | 1 Norway | |
| H5a | C | 7/8 | 3 USA (2 La., 1 N.Mex.), 2 Chad, 1 Canada, 1 Japan |
| F | 1/8 | 1 Russia | |
| H5a5b | A | 1/21 | 1 China |
| C | 19/21 | 11 USA (10 Tex., 1 no data), 1 Nigeria, 1 Russia, 6 no data | |
| K | 1/21 | 1 Norway | |
| H6 | C | 11/12 | 1 Canada, 10 no data |
| F | 1/12 | 1 Norway | |
| H7 | A | 1/17 | 1 no data |
| C | 14/17 | 5 Japan, 4 USA (2 La., 1 Ca., 1 no data), 5 no data | |
| F | 2/17 | 2 no data | |
| H8c | H | 1/1 | 1 Norway |
| H8a8b | A | 8/11 | 4 Japan, 1 Czechoslovakia, 1 England, 2 no data |
| F | 1/11 | 1 no data | |
| H | 2/11 | 2 Norway | |
| H8a8c | B | 3/3 | 3 China |
| H8a8d | A | 1/3 | 1 no data |
| C | 1/3 | 1 no data | |
| F | 1/3 | 1 no data | |
| H9 | C | 4/5 | 2 USA (1 Ariz., 1 no data), 2 no data |
| F | 1/5 | 1 France | |
| H10c | A | 9/14 | 8 Japan, 1 USA (La.) |
| C | 3/14 | 1 Japan, 1 USA (La.), 1 no data | |
| H | 1/14 | 1 Japan | |
| K | 1/14 | 1 Norway | |
| H10a10b | H | 1/1 | 1 Norway |
| H11c | K | 1/1 | 1 Norway |
| H11a11b | A | 2/2 | 1 Germany, 1 no data |
| H11a11c | F | 1/2 | 1 Japan |
| H | 1/2 | 1 Japan | |
| H12 | C | 3/3 | 1 USA, 2 no data |
| H13 | C | 2/2 | 2 no data |
| H14 | A | 10/13 | 2 Romania, 1 USA, 7 no data |
| C | 3/13 | 1 Romania, 2 no data | |
| H15 | C | 3/3 | 3 USA (2 N.Dak., 1 Ind.) |
| H16 | C | 3/3 | 3 USA (2 La., 1 Ind.) |
| H17 | F | 1/1 | 1 Japan |
| H18 | C | 1/1 | 1 Japan |
| H19 | F | 2/2 | 1 Japan |
| 1 Norway | |||
| H20 | C | 2/3 | 1 USA (La.), 1 no data |
| F | 1/3 | 1 India | |
| H22 | H | 2/2 | 1 Norway |
| 1 China | |||
| H24 | H | 1/2 | 1 no data |
| K | 1/2 | 1 Norway | |
| H27 | C | 1/1 | 1 no data |
| H29 | K | 2/2 | 2 Norway |
| H31 | K | 1/1 | 1 Norway |
| H34 | F | 1/4 | 1 Yugoslavia |
| J | 1/4 | 1 Norway | |
| K | 2/4 | 2 Norway | |
| H35 | H | 1/1 | 1 Norway |
| AA | J | 2/2 | 2 Norway |
| Not identified | C | 5/7 | 2 China, 1 USA (Ariz.), 1 England, 1 no data |
| F | 1/7 | 1 no data | |
| K | 1/7 | 1 Czechoslovakia |
The 36 different serotypes populated the entire phylogenetic tree. However, several serotypes were found primarily within a single branch. H1 isolates mapped to branch B and H3a3b and H2 isolates mapped to branch G, while H4a4b and H5a5b mapped to branch C.
Numbers refer to the number of isolates from that particular source.
Because of the changing classification in the H antigen typing of B. thuringiensis, isolates identified above only as H8, H10, and H11 were not scored as a distinct serotype (different from H8a8b, H8a8c, or H8a8d; H10a10b and H11a11b, or H11a11c).
If one analyzes the location of different commercially important serovars of B. thuringiensis, these tend to cluster on branches of the tree that are well away from the B. anthracis isolates. More than 90% of the B. thuringiensis isolates that represented serovars that are used as components of biopesticides in the United States (25) mapped to cluster 1 of the tree (Fig. 2). For example, all 17 of the B. thuringiensis subsp. thuringiensis isolates (serotype H1), all 13 B. thuringiensis subsp. israelensis isolates (serotype H14), all 8 of the B. thuringiensis subsp. morrisoni isolates (serotype H8a8b), and all 24 of the B. thuringiensis subsp. kurstaki isolates (serotype H3a3b) including B. thuringiensis subsp. kurstaki (ATCC 33679), the strain used in the preparation of the bioinsecticide Dipel, mapped to cluster 1.
In contrast, an unusual B. thuringiensis isolate shown to be pathogenic in mice mapped closely to B. anthracis in branch F of cluster 2. This isolate, identified as 97-27 (serovar H34; subspecies konkukian), was collected from the wound of a French soldier and was shown to be capable of infecting and killing immunocompetent mice in subsequent studies (7). It is one of a few known B. thuringiensis isolates to be isolated from an infected human wound. Three other B. thuringiensis serovar H34 isolates mapped to branches J (1) and K (2) in cluster 3. Such a genetic distinction between this isolate and those B. thuringiensis isolates used in the preparation of commercial bioinsecticides suggests that it is incorrect to attribute pathogenic properties to the commercially important B. thuringiensis isolates based on the properties of this unusual pathogenic isolate.
Table 3 shows that the toxigenic B. cereus isolates in this study populated almost the entire tree, but 22 of the 42 isolates mapped to cluster 2. Sixteen of these isolates mapped specifically to branch F, the same branch that contains all B. anthracis isolates. Also in cluster 2 is B. cereus ATCC 4342, found by MEE analysis to be closely related to B. cereus isolates responsible for periodontal disease (5). Although the toxigenic B. cereus isolates analyzed were collected from different contaminated food sources from a variety of locations globally, the prevalence of these isolates within cluster 2 is interesting.
TABLE 3.
Phylogenetic location and geographic source of B. cereus isolates analyzed in this study
| Branch | No. (of 61 total) in branch | No. (of 36) with toxigenic propertiesa | Sourceb |
|---|---|---|---|
| A | 0 | 0 | 0 |
| B | 2 | 1 (total) | |
| 0 | 1 Norway | ||
| 1 | 1 USA (Wis.) | ||
| C | 17 | 11 (total) | |
| 6 | 8 USA (2 Okla., 2 Wis., 1 N.J., 1 Mich., 2 no data) | ||
| 2 | 2 Finland | ||
| 2 | 2 United Kingdom | ||
| 1 | 1 Australia | ||
| 0 | 4 no data | ||
| D | 6 | 4 (total) | |
| 3 | 3 Finland | ||
| 0 | 2 Norway | ||
| 1 | 1 USA (Wis.) | ||
| E | 1 | 0 | 1 no data |
| F | 18 | 16 (total) | |
| 6 | 6 United Kingdom | ||
| 5 | 6 USA (5 Wis., 1 Nebr.), | ||
| 1 | 2 England | ||
| 2 | 2 Finland | ||
| 1 | 1 Norway | ||
| 1 | 1 China | ||
| G | 1 | 1 | 1 Finland |
| H | 9 | 1 (total) | |
| 0 | 4 Norway | ||
| 0 | 2 USA (2 Wis.) | ||
| 1 | 1 China | ||
| 0 | 2 no data | ||
| J | 1 | 0 | 1 Norway |
| K | 6 | 2 (total) | |
| 0 | 4 Norway | ||
| 1 | 1 Finland | ||
| 1 | 1 USA (Wis.) |
Thirty-six of the 61 B. cereus isolates studied had toxigenic properties. The number of toxigenic isolates that mapped to a particular branch are included in column 3.
Numbers refer to the number of isolates from that particular source.
DISCUSSION
The B. cereus group, although medically and economically important, is described as one of the most taxonomically confusing areas of the bacilli (21). The phylogenetic tree presented here reveals that isolates show a high degree of genetic diversity within two of these three species, based on a whole-genome fingerprint. Many of the diverse clusters contain B. cereus and B. thuringiensis isolates extensively interspersed with one another. The dendrogram demonstrates clusters of isolates (branches A to K) that are likely separated by large numbers of generations. This continuum of variation or intermixing of B. cereus and B. thuringiensis isolates within some of the clusters suggests that the specific phenotypes (pathogenic or insecticidal) were acquired after the ancestors to each of the clusters were formed.
These observations support the idea that horizontal gene transfer of plasmid and/or other extrachromosomal markers is an important factor in defining the phenotypes of type I bacilli. B. cereus-like isolates evolved along apparent large evolutionary distances to give rise to clusters that in more recent times acquired plasmids that conferred insecticidal or other pathogenic phenotypes. In branches A and C (Fig. 2), we suggest that an ancestral isolate may have acquired insecticidal properties that led to clusters composed entirely of related B. thuringiensis isolates. While the extensive serotype analysis conveyed in Table 2 does not accurately describe specific insecticidal clusters, other multilocus sequence typing studies have indicated the presence of conserved, clonally derived strains of well-characterized, true insecticidal isolates of a specific type (Barker et al. Abstr. 5th Int. Conf. Anthrax and 3rd Int. Workshop Mol. Biol. Bacillus cereus, B. anthracis and B. thuringiensis, 2003). Conversely, other branches, e.g., those in clusters 2 and 3 (Fig. 2), contain mixtures of B. cereus and B. thuringiensis. We interpret these clusters to have formed from ancestral isolates that clonally expanded and then inherited B. cereus or B. thuringiensis-like phenotypes by horizontal gene transfer of “modern” plasmids.
The results presented here also demonstrate that B. anthracis isolates, unlike their B. cereus and B. thuringiensis relatives, form a distinct clade within the diverse group of B. cereus and B. thuringiensis isolates. The B. anthracis isolates used in this study represent the full known genetic diversity within this pathogenic species, and the data once again illustrate the extremely monomorphic nature of this subgroup (13). It is presumed that the ancestral B. anthracis was descended from a single B. cereus-like isolate that acquired the pX01 and pX02 plasmids by a gene transfer or genetic exchange event. Comparison of the sequenced genomes of B. cereus ATCC 14579 and B. anthracis Ames shows that a large core set of genes are conserved between these two species (9). Roughly 4,500 out of 5,366 open reading frames in B. cereus ATCC 14579 have 80 to 100% identity to corresponding homologues in B. anthracis (9). These data support the hypothesis that B. anthracis is a clonal derivative of an ancestral B. cereus that acquired and maintained its specific pathogenic properties as a result of advantageous selection pressure. However, to date, the immediate B. cereus ancestor has not been identified and it is incorrect to think that simply inserting pX01 and pX02 into a randomly chosen B. cereus isolate would produce a successful pathogen. Clearly there are chromosomally encoded factors that are critical to the success of B. anthracis as it infects an animal host (see reference 16 for a review).
FIG. 4.
Branch B of the AFLP-based phylogenetic tree of B. anthracis, B. cereus, and B. thuringiensis. Twenty-two B. thuringiensis isolates representing three different serotypes mapped to this branch, including ATCC 10792, the type strain. Two B. cereus isolates mapped to this branch as well.
FIG. 5.
Branch C (part 1) of the AFLP-based phylogenetic tree of B. anthracis, B. cereus, and B. thuringiensis. Branch C contains 122 B. thuringiensis isolates representing 19 serotypes, including the ATCC 33679 (subspecies kurstaki) isolate. Seventeen B. cereus isolates including ATCC 14579, the type strain, also mapped to this branch. Branch C is so large it is divided into two parts, branch C part 1 and part 2. Part 1 contains 16 of the 17 B. cereus isolates in branch C.
Acknowledgments
This work was conducted under the auspices of the U.S. Department of Energy. The Department of Energy Chemical and Biological National Security Program provided funding for this research.
REFERENCES
- 1.Betz, F. S., B. G. Hammond, and R. L. Fuchs. 2000. Safety and advantages of Bacillus thuringiensis-protected plants to control insect pests. Regul. Toxicol. Pharmacol. 32:156-173. [DOI] [PubMed] [Google Scholar]
- 2.de Barjac, H. 1981. Identification of H-serotypes of Bacillus thuringiensis, p. 35-43. In H. D. Burges (ed.), Microbial control of pests and plant diseases, 1970 to 1980. Academic Press, Inc., Ltd., London, United Kingdom.
- 3.Drobniewski, F. A. 1993. Bacillus cereus and related species. Clin. Microbiol. Rev. 6:324-338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Helgason, E., D. A. Caugant, M.-M. Lecadet, Y. H. Chen, J. Mahillon, A. Lövgren, I. Hegna, K. Kvaløy, and A.-B. Kolstø. 1998. Genetic diversity of Bacillus cereus/B. thuringiensis isolates from natural sources. Curr. Microbiol. 37:80-87. [DOI] [PubMed] [Google Scholar]
- 5.Helgason, E., D. A. Caugant, I. Olsen, and A.-B. Kolstø. 2000. Genetic structure of population of Bacillus cereus and B. thuringiensis isolates associated with periodontitis and other human infections. J. Clin. Microbiol. 38:1615-1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Helgason, E., O. A. Økstad, D. A. Caugant, H. A. Johansen, A. Fouet, M. Mock, I. Hegna, and A.-B. Kolstø. 2000. Bacillus anthracis, Bacillus cereus, and Bacillus thuringiensis—one species on the basis of genetic evidence. Appl. Environ. Microbiol. 66:2627-2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hernandez, E., F. Ramisse, T. Cruel, R. le Vagueresse, and J.-D. Cavallo. 1999. Bacillus thuringiensis serotype H34 isolated from human and insecticidal strains serotypes 3a3b and H14 can lead to death of immunocompetent mice after pulmonary infection. FEMS Immunol. Med. Microbiol. 24:43-47. [DOI] [PubMed] [Google Scholar]
- 8.Inglesby, T. V., T. O'Toole, D. A. Henderson, J. G. Bartlett, M. S. Ascher, E. Eitzen, A. M. Friedlander, J. Gerberding, J. Hauer, J. Hughes, J. McDade, M. T. Osterholm, G. Parker, T. M. Perl, P. K. Russell, and K. Tonat. 2002. Anthrax as a biological weapon, 2002. Updated recommendations for management. JAMA 287:2236-2252. [DOI] [PubMed] [Google Scholar]
- 9.Ivanova, N., A. Sorokin, I. Anderson, N. Galleron, B. Candelon, V. Kapatral, A. Bhattacharyya, G. Reznik, N. Mikhailova, A. Lapidus, M. D'Souza, T. Walunas, Y. Grechkin, G. Pusch, R. Haselkorn, M. Fonstein, S. D. Ehrich, R. Overbeek, and N. Kyrpides. 2003. Genome sequence of Bacillus cereus and comparative analysis with Bacillus anthracis. Nature 423:87-91. [DOI] [PubMed] [Google Scholar]
- 10.Jackson, P. J., K. K. Hill, M. T. Laker, L. O. Ticknor, and P. Keim. 1999. Genetic comparison of Bacillus anthracis and its close relatives using amplified fragment length polymorphism and polymerase chain reaction analysis. J. Appl. Microbiol. 87:263-269. [DOI] [PubMed] [Google Scholar]
- 11.Johnson, R. A. and D. W. Wichern. 1982. Applied multivariate statistical analysis. Prentice-Hall, Inc., Englewood Cliffs, N.J.
- 12.Kaufman, L., and P. J. Rousseeuw. 1990. Finding groups in data. An introduction to cluster analysis. John Wiley and Sons, Inc., New York, N.Y.
- 13.Keim, P., A. Kalif, J. Schupp, K. Hill, S. E. Travis, K. Richmond, D. M. Adair, M. Hugh-Jones, C. R. Kuske, and P. Jackson. 1997. Molecular evolution and diversity in Bacillus anthracis as detected by amplified fragment length polymorphism markers. J. Bacteriol. 179:818-824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Keim, P., A. M. Klevytska, L. B. Price, J. M. Schupp, G. Zinser, K. L. Smith, M. E. Hugh-Jones, R. Okinaka, K. K. Hill, and P. J. Jackson. 1999. Molecular diversity in Bacillus anthracis. J. Appl. Microbiol. 87:215-217. [DOI] [PubMed] [Google Scholar]
- 15.Keim, P., L. B. Price, A. M. Klevytska, K. L. Smith, J. M. Schupp, R. Okinaka, P. J. Jackson, and M. E. Hugh-Jones. 2000. Multiple-locus variable-number tandem repeat analysis reveals genetic relationships within Bacillus anthracis. J. Bacteriol. 182:2928-2936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koehler, T. M. 2002. Bacillus anthracis genetics and virulence gene regulation. Curr. Top. Microbiol. Immunol. 71:143-164. [DOI] [PubMed] [Google Scholar]
- 17.Kramer, J. M., and R. J. Gilbert. 1989. Bacillus cereus and other Bacillus species, p. 21-64. In M. P. Doyle (ed.), Foodborne bacterial pathogens. Marcel Dekker, Inc., New York, N.Y.
- 18.Lecadet, M.-M., E. Frachon, V. C. Dumanoir, H. Ripouteau, S. Hamon, P. Laurent, and I. Thléry. 1999. Updating the H-antigen classification of Bacillus thuringiensis. J. Appl. Microbiol. 86:660-672. [DOI] [PubMed] [Google Scholar]
- 19.Navon, A. 2000. Bacillus thuringiensis insecticides in crop protection—reality and prospects. Crop Prot. 19:669-676. [Google Scholar]
- 20.Pattanayak, D., K. Srinivasan, A. D. Mandaokar, A. Shukla, R. Bhalla, and P. A. Kumar. 2000. AFLP fingerprinting and genotypic characterization of some serovars of Bacillus thuringiensis. World J. Microbiol. Biotechnol. 16:667-672. [Google Scholar]
- 21.Priest, F. G. 1993. Systematics and ecology of Bacillus, p. 3-16. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and other gram-positive bacteria: biochemistry, physiology, and molecular genetics. American Society for Microbiology, Washington, D.C.
- 22.Schnepf, E., N. Crickmore, J. Van Rie, D. Lereclus, J. Baum, J. Feitenson, D. R. Zeigler, and D. H. Dean. 1998. Bacillus thuringiensis and its pesticidal crystal proteins. Microbiol. Mol. Biol. Rev. 62:775-806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sneath, P. H. A. 1986. Endospore-forming gram-positive rods and cocci, p. 1105-1139. In P. H. A. Sneath (ed.), Bergey's manual of systematic bacteriology, vol. 2. The Williams & Wilkins Co., Baltimore, Md.
- 24.Ticknor, L. O., A.-B. Kolstø, K. K. Hill, P. Keim, M. T. Laker, M. Tonks, and P. J. Jackson. 2001. Fluorescent amplified fragment length polymorphism analysis of Norwegian Bacillus cereus and Bacillus thuringiensis soil isolates. Appl. Environ. Microbiol. 67:4863-4873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weeden, C. R., A. M. Shelton, Y. Li, and M. P. Hoffman (ed.). 2002. Biological control: a guide to natural enemies in North America. Accessed 14 May 2003. [Online.] http://www.nysaes.cornell.edu/ent/biocontrol/.