Abstract
Cancer cells rapidly evolve drug resistance through somatic evolution and, in order to continue growth in the metastatic phase, violate the organism-wide consensus of regulated growth and beneficial communal interactions. We suggest that there is a fundamental mechanistic connection between the rapid evolution of resistance to chemotherapy in cellular communities within malignant tissues and the rapid evolution of antibiotic resistance in bacterial communities. We propose that this evolution is the result of a programmed and collective stress response performed by interacting cells, and that, given this fundamental connection, studying bacterial communities can provide deeper insights into the dynamics of adaptation and the evolution of cells within tumours.
There is a general agreement that the various ‘Wars on Cancer’ that have been declared have not been as successful as expected: the overall mortality rate for cancer has been practically flat for the past 40 years. One of the reasons that could explain this failure is the lack of understanding at a fundamental level of how cells evolve in response to drug treatments and, more generally, the basic rules that control evolution under stress across the biological kingdom. In this Opinion, we propose that an in-depth understanding of the processes behind the evolution of drug resistance in malignant tissues can be achieved by considering the problem of cancer evolution from a more generalist point of view. We propose that substantial insight into the evolutionary and adaptation dynamics of cancer tissues can be gained by studying the evolutionary strategies used by simpler, rapidly evolving microorganisms (such as bacteria) in response to drug treatments and stressful environments.
In the following sections, we first reconsider the current view of cancer evolution in light of the strategies used by bacterial communities. Then, we compare the stress responses of bacterial communities and show that they may be used to study the evolution of drug resistance in malignant tissues at a fundamental level. We then describe communal aspects of cancer tissues, the understanding of which may benefit from using bacterial model systems. Finally, we propose and review specific experimental approaches using bacterial model systems that may deepen our understanding of the fundamentals of cancer evolution and adaptation.
An alternative view of cancer evolution
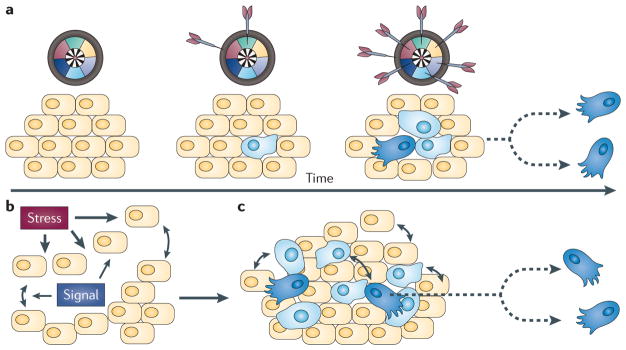
The role of evolution in the origins of resistance to drugs in cellular communities is known to be important but remains poorly understood. The question, of course, is not whether evolution occurs, but how. Evolutionary processes are clearly important because the crucial problem in chemotherapy is that malignant tissues rapidly acquire adaptive phenotypes and thus evolve drug resistance through somatic evolution. But how does this happen? FIGURE 1a presents the traditional view that this evolution is initiated by chance in a rogue cell (analogous to darts randomly hitting a target) and subsequent successive mutations activate hallmark capabilities1 such as invasiveness and the evasion of programmed cell death. Additional chance mutations generate cells that have acquired self-sufficient capabilities. These cells forgo the organism-wide consensus of beneficial communal interactions and develop phenotypes that interfere with the survival of the host organism, leading to an eventual breakdown in cellular control. Moderating the adverse effect of acquired malignant traits has driven the basic philosophy and rationale for the development of targeted therapies2–4. This approach, however, has had limited success over the past decades5 because cells within the tumour inexorably become resistant to the chemotherapeutic drugs6.
Figure 1. An alternative view of cancer development.
a | The traditional view of cancer is as a cell-autonomous result of cumulative genetic mutations. Genes can be conceptualized according to their function as sectors on a dartboard that represent the hallmarks of cancer, and familial or acquired mutations can be thought of as randomly occurring dart strikes. A normal cell (yellow) can acquire a mutation (blue) that, for example, confers self-sufficiency in growth signals. As the progeny of the mutated cell expand, some daughter cells acquire additional mutations. Daughter cells displaying a full complement of hallmark lesions (dark blue) are malignant and capable of rapid proliferation and dissemination. b,c | An alternative view of cancer as a collective stress response. b | Stress emanates from a source, creating stressful conditions that are localized in space and time. This in turn induces ‘normal’ cells to exchange stress signals in regions of high stress. c | These stress signals orchestrate the display of multiple adaptive phenotypes that are traditionally considered ‘abnormal’ and can include rapid proliferation and tumour cell dissemination. Normal and abnormal cells can coexist. Part a is modified, with permission, from REF. 1 © (2000) Elsevier Science.
We propose a contrasting view in which random genetic lesions alone are not sufficient to explain the progression of malignancy. Instead, cancer results from a programmed, deterministic and collective stress response that is performed by interacting cells that also have complex communication with the surrounding microenvironment (FIG. 1b). The interplay between cells seeking survival under stress activates a survival programme that facilitates evolution and adaptation of malignant and pre-malignant cells (FIG. 1c). Unfortunately, this programmatic development occurs in a highly complex and dynamic microenvironment that has been difficult to study at a basic level in cancer tissues.
We propose that a more profound understanding of the processes behind cancer evolution and metastasis can be achieved by considering them in light of the strategies used by simpler organisms such as bacteria. As we will discuss below, the evolutionary strategies used by bacteria, such as the collective responses favouring the generation of genetic and phenotypic heterogeneity under external stress, parallel those used by tumour cells.
The role of stress in evolution
Both bacterial and tumour cells can evade death induced by exposure to drugs through various mechanisms. The easiest strategy is to move to an environment that contains a lower concentration of a cytocidal agent. This is achieved through swimming by bacterial cells and through metastasis by tumour cells7,8. Alternatively, the cell population can create a milieu where the drug has limited access to the cells. This has been demonstrated to be a function of biofilms in a bacterial colony and a function of an altered tumour microenvironment (including the vasculature) for tumour cells9–11. One of the most intriguing methods of evading death in both cell populations depends on a probabilistic phenotypic switching mechanism12– 13. In this situation, a small fraction of the bacterial or tumour cell population is in a state that is not responsive to the cytotoxic properties of the drug. This has been called ‘a persister phenotype’ for bacterial communities12 and has recently been described as a mechanism whereby tumour cells can escape death caused by exposure to drugs13.
These mechanisms provide highly reversible drug resistance. Mechanisms of more permanent, heritable drug resistance in tumour cells involve pre-existing genetic variation within the population and the generation of de novo mutations that provide intrinsic or acquired drug resistance14–17. These mechanisms can be particularly important for evolving drug resistance when they occur as stress responses.
To emphasize this, we propose to regard ‘evolvability’, which is defined as the generation of mechanisms that facilitate evolution18, as a fundamental component of drug resistance. In particular, the existence of individuals with relatively high mutation rates (a mutator phenotype) in a community of cells is a widely known phenomenon for both cancer19 and bacteria20. This mutator phenotype can be selected for21 and has been shown to increase the rate of adaptation of an organism to stress22.
When occurring in only a subpopulation of bacteria, stress-induced mutagenesis is not considered a liability; rather, it is beneficial to the population as a whole23,24. Evolvability in bacterial systems does not necessarily originate from mutations or alterations in DNA protection mechanisms: the survival programmes expressed by bacteria under stress promote adaptive mutations and are often necessary for the survival of a population15. In the case of starvation stress in Escherichia coli, adaptive mutations are carried out by the activation of an error-prone DNA double-stranded break (DSB) repair system25,26 (see BOX 1 for a description of the bacterial analogues of human DNA repair mechanisms). Similarly, the rapid evolution of resistance to a genotoxic agent such as ciprofloxacin — from the quinolone family of antibiotics — originates from point mutations caused by DNA recombination that is induced by the SOS response27. Furthermore, external oxidative stress often affects the fidelity of DNA transcription in the absence of DNA replication, which in turn leads to the translation of mutant proteins without any permanent alterations (or mutations) to the DNA template, a process known as transcriptional mutagenesis28.
Box 1. DNA repair mechanisms.
Several proposed mechanisms for DNA repair and the stress response in human cells have analogues in the bacterial world. Although the failure of processes that normally safeguard human cells has traditionally been linked to an increased susceptibility to tumorigenesis, in bacteria such processes are generally associated with increased adaptability.
Double-stranded breaks
In human cells, the repair of DNA double-stranded breaks (DSBs) is implemented by various DNA damage response proteins, including BRCA1 (REF. 73) and alterations in BRCA1 are associated with cancer. In bacteria, the response to DSBs is carried out by the SOS system74,75. The repair of DSBs can itself be mutagenic in both bacteria and eukaryotes: activation of DSB repair mechanisms is associated with an increased mutation rate owing to the use of error-prone DNA polymerases76,77. However, DSB-induced mutagenesis is still greatly increased in BRCA1-deficient versus BRCA1-proficient human cells.
Mismatch repair
DNA mismatch repair (MMR) in human cells is performed by several combinations of different MLH and MSH proteins. Defects in MMR are often associated with increased genomic instability. Similarly, in bacteria, defects in the MMR proteins MutL or MutS elevate mutation rates, thereby increasing the probability of developing antibiotic resistance; mutator phenotype bacteria with altered DNA MMR systems are often found in persistent Pseudomonas aeruginosa biofilm infections78.
Homologous recombination
The RAD51 gene family encodes proteins that are necessary for homologous recombination in human cells. BRCA2, a tumour suppressor gene, plays an important part in homologous-recombination- mediated DNA repair79 and mutations in BRCA2 decrease genomic stability80. Homologous recombination in bacteria is carried out by the DNA recombination protein RecA, a RAD51 analogue81.
Cell cycle regulation
p53, the product of the TP53 tumour suppressor gene regulates exit from the cell cycle under conditions of stress and is involved in regulating the expression of DNA caretaker genes82. Mutations in TP53 are found in a large fraction of cancer lesions83 and are often associated with sustained proliferation despite DNA damage or external stress82. Similarly, the RNA polymerase σ factor (RpoS) regulates entry into the stationary phase (G0) of the bacterial cell cycle and promotes expression of DNA repair genes68. The roles and functions of p53 and RpoS are similar: both maintain genetic integrity in response to environmental stress. Alterations in both TP53 and rpoS (in Escherichia coli) often provide a growth advantage despite external stress69.
Conversely, the traditional interpretation of evolvability and why it appears so often in cancer tissues — where it is usually referred to as genetic instability29 — often relies on assuming that random mutations cause the failure of DNA protection processes. Instead, we propose that genetic instability in cancer tissues is an organized strategy that acts as an accelerator of adaptation, similar to the role of mutators in bacterial populations.
From this point of view, a high rate of mutation and a plastic genotype is a tried-and- tested bacterial strategy that is necessary to adapt to hostile and ever-changing environments. We interpret the enhanced mutation rate and genetic instability of a tumour population as the expression of very efficient evolutionary strategies used by bacterial communities; cancer cells are not rogue, instead, they are ‘liberated’ from the cell protection mechanisms that are activated in response to stress that fail to enhance survival. As such, current therapeutic approaches targeting rapidly replicating cells are doomed to fail, because cell collectives are often able to evade complete eradication by expressing a mutable phenotype to reprogramme themselves. Moreover, even cells in an inactive DNA replication state — a state that is not usually targeted by chemotherapy — may contribute to survival under stress through transcriptional mutagenesis and retromutagenesis30. We propose that the ability to resist a chemotherapeutic treatment or to survive in stressful environments must be viewed as a demonstration that cells have collectively and successfully adapted to new and more hostile environments.
Biofilm and tumour stroma
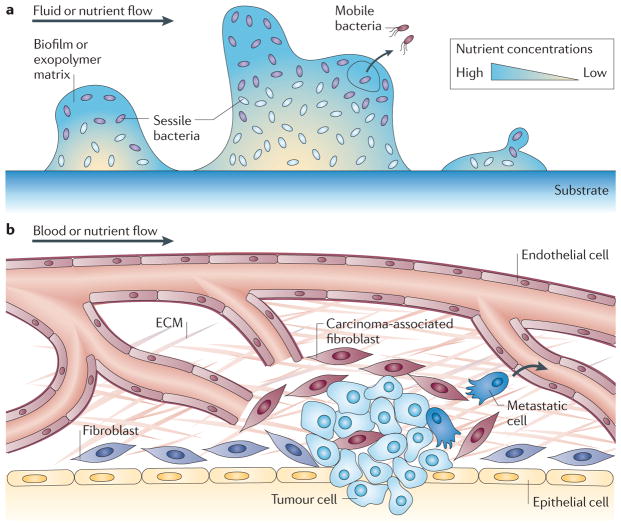
One of the physiological responses of bacteria to external stress is to assemble into a biofilm (see BOX 2 for more detail concerning biofilms and biofilm development). Pseudomonas aeruginosa is often used as a model of biofilm development31; in culture, they produce an exopolymer matrix that protects cells from surrounding environmental stresses. The formation of a biofilm greatly increases the resistance of a population to a hostile environment by shielding cells, for example, from antibiotics. Biofilms, however, limit the influx of nutrients and oxygen owing to the decreased diffusion of chemicals through the biofilm matrix32 (FIG. 2a). Although bacterial cells trigger the expression of fermentative pathways in the absence of oxygen33, this metabolic pathway creates endogenous oxidative stress within the exopolymer matrix, which in turn increases the mutation rate of the cells34.
Box 2. Bacterial cell communities.
A natural response to increasing levels of stress in many species of bacteria is the formation of biofilms, where cells assemble together and produce large amounts of a polysaccharide-based exopolymer matrix84. The biofilm developmental programme usually starts with the collective production of a dense, chemically inert exopolymer matrix by the cells as a response to external stress (such as changes in pH and osmolarity, starvation, and shear forces)85. Biofilm formation is beneficial to the cell population as a whole, as it allows cells to survive within highly stressful environments that prevent the survival of free-swimming cells85. Because diffusion of metabolites and chemicals is greatly limited inside the matrix32, the microenvironment created by the biofilm is highly heterogeneous and physiologically stressful86. However, biofilm production is accompanied by a high level of specialization within the bacterial community. For example, subpopulations of bacteria inside a biofilm, each a few hundred micrometres apart can alternatively grow aerobically, process nutrients through fermentation pathways, digest the hydrogen sulphide produced by other cells or resist the high shear forces near the biofilm edge33,86. In humans with bacterial infections, antibiotic treatment is often ineffective because the limited diffusion inside the biofilm decreases the effective dose that can reach the bacteria. Thus, biofilms are a recognized source of recurrent and persistent bacterial infections87,88. As bacteria assemble together, cell death and cell lysis contribute to the formation of cavities inside the biofilm31. The presence of such cavities in Pseudomonas aeruginosa biofilms allows cells to regain a free-swimming state and move to a different habitat89, not unlike a metastatic expansion from a primary human tumour.
Figure 2. Changes in microenvironments.
a | A community of bacteria can form biofilms by attaching to a substrate and by producing large amounts of a polysaccharide-based exopolymer matrix that links cells together. As the extracellular matrix (ECM) encases the cells and greatly hinders their motion, cells switch from a motile to a sessile state. The matrix greatly limits nutrient and oxygen diffusion and cells inside the biofilm become specialized according to the metabolites present. (Subsistence on different nutrient sources is indicated by the different colours of the cells in different regions.) Some cells, not unlike metastatic cancer cells, are able to break through the exopolymer matrix and leave the biofilm to populate different environments. b | The type of cells associated with a tumour, notably carcinoma-associated fibroblasts, produce signals that influence the behaviour of tumour cells. Also, the stroma and ECM surrounding a tumour is much denser than that surrounding normal tissue and the diffusion of nutrients and oxygen from the blood vessels is therefore greatly diminished by the tumour-associated ECM and stroma. Metastatic cells (dark blue) may also leave the primary tumour and disseminate throughout the body.
Why would bacteria still want to live in such a (self-created) hostile environment? Actually, rather than trying to combat this mutagenic environment, P. aeruginosa cells embrace it. They maintain a small mutator-phenotype- population (0.5–5%) in which genes involved in protection against oxidative stress are downregulated. These genes include KatA, which encodes a catalase that is necessary for peroxide decomposition34. Downregulation of KatA gives cells mutation rates up to 100-fold higher than in non-communal, free-swimming cells34. Samples of P. aeruginosa biofilms extracted from patients suffering from cystic fibrosis almost always contain cells expressing a mutator phenotype, many of which are resistant to multiple antibiotics35,36. As a result, cells in biofilms are able to develop resistance to multiple antimicrobial agents much more rapidly37 and, by maintaining only a small fraction of the population in a hypermutative state, do not accumulate detrimental and fatal mutations in the rest of the clonal population38.
Similarly, cancer is not just a collection of cells replicating and evolving uncontrollably; it is an ecosystem39,40. Cells surrounding a tumour (such as fibroblasts, immune cells and endothelial cells) are part of a tumour tissue and co-evolve with cancer cells (FIG. 2b). For instance, stromal cells such as fibroblasts associated with cancerous tissues increase extracellular matrix (ECM) production41. Similarly to bacterial biofilms, the increased matrix deposition not only reduces the effectiveness of chemotherapeutic drugs to penetrate a tumour42–44 but also reduces the amount of oxygen and nutrients reaching the centre of a tumour. Analogously to biofilms, tumour cells may also switch to a fermentative pathway when oxygen is unavailable: anaerobic glycolysis allows cells to produce ATP but inadvertently leads to the acidification of the tumour microenvironment through the release and fermentation of lactate45.
Tumour cells, however, are able to survive stressful environments through strong mutual interactions with stromal cells46: it has recently been shown that fibroblasts and endothelial cells alter their metabolic pathways to support the intensive glycolysis of cancer cells, an example of which has been presented for a colorectal carcinoma47. Koukourakis et al. have shown that fibroblasts surrounding a colorectal carcinoma have an increased rate of lactate metabolism to cope with the aerobic glycolysis of the cancer cells47. They also demonstrated that endothelial cells surrounding this particular type of carcinoma have an aversion to lactate absorption, which thereby prevents acid production near the blood vessels47.
As tumours recruit cells to their microenvironment, they create a community of highly specialized cells that are able to sustain the high metabolic needs of tumour cells and that protect them against the influx of drugs. Taken as such, the levels of specialization found in an epithelial–stromal cell collective is, at a fundamental level, strategically similar to bacterial biofilm communities. The understanding of the complex symbiotic interplay between the different cell types within a tumour may be facilitated by analogous comparison with bacterial biofilms.
Studying the evolution of drug resistance
This comparison between cells within a malignant tissue and bacterial communities, two seemingly different organisms, has great potential to go beyond philosophical interpretations. Here, we propose that the leap of faith needed to go from in silico models — which already use idealized tumour representations to study cancer evolution and adaptation48,49 — to in vivo models is of similar magnitude to the one needed to go from bacterial to cancer models: large, but by no means irreconcilable. Below, we outline several experimental systems that can be used to gain insight into the evolution of drug resistance and tumour development.
Heterogeneous culture environment
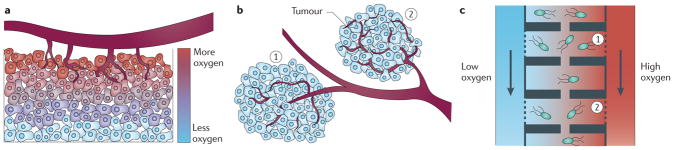
In the absence of a chemotherapeutic treatment, the fitness of cells on a tumour surface, near the vasculature, can be much higher than the fitness of cells inside a tumour. During chemotherapeutic treatments, the spatial-dependent fitness of cells within a tumour is even more complex: spatial heterogeneities and poor vasculature can produce uneven drug, nutrient and/or oxygen concentrations (FIG. 3a). Furthermore, the subdivision of a tumour microenvironment into multiple habitats (FIG. 3b) limits cell– cell interactions but still allows circulating tumour cells to be exchanged between tumours50. This type of configuration creates isolated micro-ecologies in which evolution occurs in parallel, with limited exchange. Studying the dynamics of cancer cell adaption under such conditions is virtually impossible using conventional cell culture techniques.
Figure 3. Proposed experimental approaches to investigate drug resistance using bacterial models.
The heterogeneous nature of a tumour may be modelled using microfluidics devices. a | A solid tumour is physiologically heterogeneous: insufficient vasculature decreases the amount of oxygen, nutrients and/or drugs that penetrate a tumour. For simplicity, only the gradient of oxygen is illustrated. b | Similarly, the growth of tumour lesions may occur in isolated subpopulations of cells, thereby limiting direct communication between various parts of a tumour (for example, region 1 and region 2 in the figure). As a result, weakly interacting subpopulations from the same initial cancer lesion may evolve and adapt independently. c | The physiological segmentation of a tumour and the presence of strong chemical gradients could be imitated inside a microfluidics device (the figure depicts the use of this device for bacteria). For instance, media flowing on each side of the chamber array could contain different levels of oxygen, mimicking the chemical composition of a tumour. Porous chamber walls (dashed lines) allow chemical exchange but prevent cellular escape. Furthermore, the movement and exchange of cells between different habitats can be limited by the presence of narrow channels. As a result, cells in habitat 1 have very limited interactions with cells in habitat 2 and these populations will therefore evolve independently.
The use of microfluidic technologies that can create strong chemical gradients over very small volumes (hundreds of picolitres) makes this type of study possible. Although several groups have successfully cultured mammalian cells for long periods inside microfluidics devices51–53, long-term experiments studying the evolution of cancer cells under conditions of stress remain challenging. Conversely, bacterial cultures inside microfluidics devices54–56 provide enough complexity to recreate heterogeneous and fragmented aspects of cancer tissues.
Bacterial model systems inside microfluidically controlled environments could be used, for example, to mimic the limited influx of drugs and nutrients that reach the centre of a tumour (FIG. 3a) by limiting nutrient levels in a location-dependent manner. A device like the one presented in FIG. 3c could combine both effects presented in FIG. 3a,b. First, media containing different oxygen concentrations mimic the chemical gradients present inside tumours. Second, the presence of spatial structures physically isolates subpopulations of bacteria into weakly interacting micro-ecologies. Such devices can be used, for instance, to test spatially explicit theoretical models of evolution such as source–sink ecologies57, which propose that evolution occurs at a faster pace in the presence of habitats with strong chemical and population gradients. This type of microfluidics-based experiment, when considered purely as an evolutionary problem, may not only provide information about the general dynamics of adaptation in biological systems but might also provide insight into the dynamics of the evolution of cancer cells.
Exploitation of a biofilm model of tumorigenesis
Although the underlying biology of bacterial biofilms and cancer tissues may be very different, biofilms may still be used to physically model the population dynamics of evolving tumours. Indeed, spatial and temporal genetic analyses of a single malignant tissue (such as the oesophagus, as presented by Maley et al.58) show that simple concepts such as genetic drift and clonal expansion play an important part in the evolution of cancerous tissues. Furthermore, the genetic composition of a tumour is far more complex than that suggested by the assumption that a tumour is monoclonal59.
Analogously, a recent study by Conibear et al.60 has demonstrated the context-dependent emergence and clonal expansion of mutations in P. aeruginosa when grown as a biofilm. A green fluorescent protein (GFP) gene containing a +1 frameshift mutation was used to measure the mutation rate of a biofilm population in response to a mutagenic agent. Because a simple deletion reverts the GFP protein to its wild-type state, the physical location of such mutations and how they spread within the biofilm can easily be assessed by fluorescence microscopy. A high rate of ‘activated’ GFP expression was observed only in biofilm microcolonies (foci of proliferation that protrude from the attachment plane), possibly owing to the accumulation of endogenous oxidative waste. A representation of the spreading of mutations in bacterial populations, and how it relates to similar events in cancerous tissues, is shown in FIG. 4.
Figure 4. Evolutionary aspects of biofilm development as a model of drug resistance in tumours.
a,b | An interpretation of the work by Conibear et al.60 studying mutagenesis in biofilm communities of Pseudomonas aeruginosa bacteria containing a green fluorescent protein (GFP) reporter gene that has an inactivating +1 frameshift mutation. In this system, a simple base deletion restores the function of the gene and induces the expression of GFP. a | Biofilm colony growth on a glass substrate. The authors described the possibility that oxidative waste accumulates in microcolonies during biofilm expansion. This waste causes stress-induced mutagenesis and activates GFP expression. b | Top view of a P. aeruginosa biofilm microcolony containing both cells with reactivated GFP and cells without reactivated GFP. c | Biofilm experiments could mimic population dynamics occurring during tumorigenesis and during the development of drug resistance after therapy. In both situations, mutations (depicted by genotypes A and B) can appear in localized environments before spreading to the rest of the tumour. Panel b is reproduced from REF. 60.
In addition to being used to monitor the fixation and expansion of mutations inside a biofilm, this experiment could be taken further by applying an antibiotic treatment to the biofilm cultures and measuring how cells adapt in response. Alternatively, by fusing the expression of a fluorescent protein with a known indicator of resistance (resumption of DNA synthesis or cell division, for instance), the spreading dynamics of resistance could be used to infer how drug resistance also spreads within cancerous tissues. The power of bacterial models comes from their relative ease of culture and the ability to more accurately monitor gene expression in real time using fluorescent protein reporters.
Cell–cell communication under stress
Bacteriologists often use concepts borrowed from game theory to explain complex cell– cell communication between different bacterial species61. For instance, results presented by Lee et al.62 indicate that bacterial communities can collectively adapt to antibiotic treatments when the burden of a toxic cleanup is placed on the shoulders of a few ‘altruistic’ individuals for the benefit of the many.
An analogous situation may be present in a tumour collective: as discussed above, stromal cells often shape their metabolism to sustain the proliferation of neighbouring cancer cells47. This altruistic behaviour by stromal cells may be better understood in terms of the costs and benefits associated with the actions of each cell type. Other similar, communal behaviours, such as strong interdependence on the production and digestion of metabolites, are also observed in cancer tissues63. Furthermore, Hickson et al.64 have also proposed that tumour cells may show behaviours similar to quorum-sensing, a bacterial regulatory mechanism in which individual bacteria probe their neighbours in order to decide whether or not to express certain genes65.
The interactions between cells in a tumour may also be interpreted using game theory concepts (including ideas such as cooperation, cheating and altruism66), and such concepts are often used when interpreting the similar cell–cell interactions that are observed in bacterial communities. Researchers may benefit by considering the richness of bacterial communication systems to formulate new hypotheses concerning the behaviour of a cancer cell by viewing cancer tissues as strongly interacting communities rather than as groups of independent, single-celled organisms.
Bacterial systems as predictive tumour models
Although it would be naive to believe that bacteria can replace mice, which share many oncogenes and tumour suppressor genes with humans67, as a model organism for cancer development, the relative simplicity of a bacterial genome may be a considerable advantage when studying the multicellular dynamics of cancer evolution. By associating known oncogenic pathways in human cancer with similar regulatory pathways in a bacterium, researchers may be able to use bacteria to simulate the stress response of cancer cells.
For instance, the transcription factor RNA polymerase σ factor (RpoS) is a bacterial analogue of the transcription factor p53 and is a fundamental cell cycle regulator that prevents replication under stressful conditions68. Bacteria that evolve under prolonged starvation stress may develop a growth advantage under stationary phase (GASP) mutation affecting rpoS69. Keymer et al.69 have shown that although GASP cells outcompete wild-type individuals in homogeneous and well-stirred environments (E. coli growing inside a test tube), coexistence is possible in unstirred, structured micro-habitats70. This parallels the expansion of TP53 (which encodes p53)-deficient cells within a healthy tissue. Cancer cells also have an altered stress response regulatory system but they do not necessarily outcompete surrounding cells. Rather, different cell types coexist to sustain high levels of proliferation.
At a more applied level, work by the Palsson group71 has pioneered the use of reconstructed metabolic pathways in bacterial systems such as E. coli to identify, in combination with in silico approaches, new genes and functions involved in a given genetic network. The methods have already been applied to human cells, where researchers have demonstrated the feasibility of creating multicellular metabolic model systems for the study of metabolic pathways in brain tissues72. Applying such techniques to cancer tissues under stress may help to further the understanding of the fundamental processes behind adaptation of tumour cells to chemotherapeutic treatments.
Model limitations and concluding remarks
Although there are many similarities between bacterial communities and tumour cell populations in their ability to evade death caused by exposure to drugs, undoubtedly differences also exist. One aspect is the greater diversity in the cellular components that exist in a malignant tissue compared to a bacterial community. The concerted interactions among endothelial cells, immune cells, fibroblasts and epithelial cells are all necessary for the formation of a malignancy and the development of drug resistance or tolerance. Although bacterial cells have specialized functions within a bacterial community, the diversity of cellular components is not as great as in tumours. A second aspect may involve the difference in the complexity of the two genomes. The mammalian genome has evolved fine-tuned layers of epigenetic controls that do not necessarily exist in the regulation of bacterial gene expression.
In conclusion, the goal of this Perspective is to broaden the scope of cancer research to include the use of bacterial populations as biological model systems for adaptation and evolution. The evolutionary strategies used by bacteria and tumours are incredibly similar, and we hypothesize that significant insight into the evolutionary dynamics of cancer populations would be gained by an informed comparison between the two systems through a multi-scale analysis. The evolution of drug resistance within cancer tissues, an important problem that has direct implications for clinical outcome, may more easily be modelled and studied in rapidly evolving bacteria under stress.
Acknowledgments
We wish to thank D. Coffey and A. Barker for their helpful comments. The research described was supported by award U54CA143803 from the US National Cancer Institute. The content is solely the responsibility of the authors and does not necessarily represent the official views of the US National Cancer Institute or the US National Institutes of Health.
Glossary
- Altruism
Behaviours that benefit another individual while incurring a cost to oneself
- Biofilm
A multicellular aggregate of bacteria and its associated proteinaceous matrix formed in response to external stress
- Cheating
A strategy in which individuals do not cooperate but still benefit from the positive interactions with cooperating individuals
- Clonal expansion
Population growth that is mainly carried out by a single genotype
- Cooperation
Actions or behaviours that are beneficial to other individuals
- Cystic fibrosis
An inherited disease that causes thick mucus to build up in the lungs and the digestive tract
- Cytocidal agent
A molecule or drug causing cell death
- Exopolymer matrix
A polysaccharide-based extracellular matrix collectively secreted by bacteria in biofilms. The matrix links cells together and acts as a protective microenvironment
- Game theory
A mathematical theory describing the costs and benefits associated with the interactions among individuals of a group. This theory is most often used in economics and evolutionary biology
- Genetic drift
A process through which the frequency of genes in populations fluctuates because selection occurs mainly by chance
- Growth advantage under stationary phase
(GASP). A phenotype that allows certain bacterial cells to outcompete wild-type cells by maintaining a proliferative state while the wild-type cells cease to grow and enter stationary phase
- Phenotypic switching
The ability of organisms to alternate between two distinct states in order to adapt to fluctuating environments
- Retromutagenesis
A process whereby DNA damage that causes changes to base pairing becomes incorporated into the genome. This may occur if a mutant protein resulting from transcriptional mutagenesis causes the rapid restart of DNA replication, thus resulting in a genetic lesion that alters base pairing being copied by a DNA polymerase before the lesion is repaired and thereby altering the DNA sequence
- SOS response
A global DNA damage response in bacteria that involves cell cycle arrest and mutagenic DNA repair and recombination
- Source–sink ecology
A theoretical model used to describe the dynamics of a population inside habitats that either promote growth (source) or induce death (sink)
- Transcriptional mutagenesis
A process by which proteins with altered functions are translated because RNA polymerases transcribe mRNA from a template containing DNA damage
Footnotes
Competing interests statement
The authors declare no competing financial interests.
DATABASES
National Cancer Institute Drug Dictionary: http://www.cancer.gov/drugdictionaryciprofloxacin
Pathway Interaction Database: http://pid.nci.nih.gov
FURTHER INFORMATION
Thea D. Tlsty’s homepage: http://cancer.ucsf.edu/people/tlsty_thea.php
Robert H. Austin’s homepage: http://austingroup.princeton.edu
ALL LINKS ARE ACTIVE IN THE ONLINE PDF
Contributor Information
Guillaume Lambert, Department of Physics, Princeton University, Princeton, New Jersey 08544, USA.
Luis Estévez-Salmeron, Department of Pathology and Comprehensive Cancer Center, University of California, San Francisco, California 94143, USA.
Steve Oh, Department of Pathology and Comprehensive Cancer Center, University of California, San Francisco, California 94143, USA.
David Liao, Department of Pathology and Comprehensive Cancer Center, University of California, San Francisco, California 94143, USA.
Beverly M. Emerson, Regulatory Biology Laboratory, The Salk Institute, 10010 North Torrey Pines Road, La Jolla, California 92037, USA
Thea D. Tlsty, Department of Pathology and Comprehensive Cancer Center, University of California, San Francisco, California 94143, USA
Robert H. Austin, Department of Physics, Princeton University, Princeton, New Jersey 08544, USA
References
- 1.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 2.Ellis LM, Hicklin DJ. VEGF-targeted therapy: mechanisms of anti-tumour activity. Nature Rev Cancer. 2008;8:579–591. doi: 10.1038/nrc2403. [DOI] [PubMed] [Google Scholar]
- 3.Weinstein IB, Joe A. Oncogene addiction. Cancer Res. 2008;68:3077–3080. doi: 10.1158/0008-5472.CAN-07-3293. [DOI] [PubMed] [Google Scholar]
- 4.Letai AG. Diagnosing and exploiting cancer’s addiction to blocks in apoptosis. Nature Rev Cancer. 2008;8:121–132. doi: 10.1038/nrc2297. [DOI] [PubMed] [Google Scholar]
- 5.Kamb A, Wee S, Lengauer C. Why is cancer drug discovery so difficult? Nature Rev Drug Discov. 2007;6:115–120. doi: 10.1038/nrd2155. [DOI] [PubMed] [Google Scholar]
- 6.Lage H. An overview of cancer multidrug resistance: a still unsolved problem. Cell Mol Life Sci. 2008;65:3145–3167. doi: 10.1007/s00018-008-8111-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Butler MT, Wang Q, Harshey RM. Cell density and mobility protect swarming bacteria against antibiotics. Proc Natl Acad Sci USA. 2010;107:3776–3781. doi: 10.1073/pnas.0910934107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ahmed N, Abubaker K, Findlay J, Quinn M. Epithelial mesenchymal transition and cancer stem cell-like phenotypes facilitate chemoresistance in recurrent ovarian cancer. Curr Cancer Drug Targets. 2010;10:268–278. doi: 10.2174/156800910791190175. [DOI] [PubMed] [Google Scholar]
- 9.Stewart PS, Costerton JW. Antibiotic resistance of bacteria in biofilms. Lancet. 2001;358:135–138. doi: 10.1016/s0140-6736(01)05321-1. [DOI] [PubMed] [Google Scholar]
- 10.Tong RT, et al. Vascular normalization by vascular endothelial growth factor receptor 2 blockade induces a pressure gradient across the vasculature and improves drug penetration in tumors. Cancer Res. 2004;64:3731–3736. doi: 10.1158/0008-5472.CAN-04-0074. [DOI] [PubMed] [Google Scholar]
- 11.Bock KD, Cauwenberghs S, Carmeliet P. Vessel abnormalization: another hallmark of cancer? Molecular mechanisms and therapeutic implications. Curr Opin Genet Dev. 2011;21:73–79. doi: 10.1016/j.gde.2010.10.008. [DOI] [PubMed] [Google Scholar]
- 12.Balaban NQ, Merrin J, Chait R, Kowalik L, Leibler S. Bacterial persistence as a phenotypic switch. Science. 2004;305:1622–1625. doi: 10.1126/science.1099390. [DOI] [PubMed] [Google Scholar]
- 13.Sharma SV, et al. A Chromatin-mediated reversible drug-tolerant state in cancer cell subpopulations. Cell. 2010;141:69–80. doi: 10.1016/j.cell.2010.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alonso A, Campanario E, Martinez JL. Emergence of multidrug-resistant mutants is increased under antibiotic selective pressure in Pseudomonas aeruginosa. Microbiology. 1999;145:2857–2862. doi: 10.1099/00221287-145-10-2857. [DOI] [PubMed] [Google Scholar]
- 15.Cirz RT, et al. Inhibition of mutation and combating the evolution of antibiotic resistance. PLoS Biol. 2005;3:e176. doi: 10.1371/journal.pbio.0030176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schimke R, Kaufman R, Alt F, Kellems R. Gene amplification and drug resistance in cultured murine cells. Science. 1978;202:1051–1055. doi: 10.1126/science.715457. [DOI] [PubMed] [Google Scholar]
- 17.Longley DB, Johnston PG. Molecular mechanisms of drug resistance. J Pathol. 2005;205:275–292. doi: 10.1002/path.1706. [DOI] [PubMed] [Google Scholar]
- 18.Sniegowski PD, Murphy HA. Evolvability. Curr Biol. 2006;16:R831–R834. doi: 10.1016/j.cub.2006.08.080. [DOI] [PubMed] [Google Scholar]
- 19.Bielas JH, Loeb KR, Rubin BP, True LD, Loeb LA. Human cancers express a mutator phenotype. Proc Natl Acad Sci USA. 2006;103:18238–18242. doi: 10.1073/pnas.0607057103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bjedov I, et al. Stress-induced mutagenesis in bacteria. Science. 2003;300:1404–1409. doi: 10.1126/science.1082240. [DOI] [PubMed] [Google Scholar]
- 21.Earl DJ, Deem MW. Evolvability is a selectable trait. Proc Natl Acad Sci USA. 2004;101:11531–11536. doi: 10.1073/pnas.0404656101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Draghi JA, Parsons TL, Wagner GP, Plotkin JB. Mutational robustness can facilitate adaptation. Nature. 2010;463:353–355. doi: 10.1038/nature08694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sniegowski PD, Gerrish PJ, Lenski RE. Evolution of high mutation rates in experimental populations of, E. coli. Nature. 1997;387:703–705. doi: 10.1038/42701. [DOI] [PubMed] [Google Scholar]
- 24.Giraud A, et al. Costs and benefits of high mutation rates: adaptive evolution of bacteria in the mouse gut. Science. 2001;291:2606–2608. doi: 10.1126/science.1056421. [DOI] [PubMed] [Google Scholar]
- 25.Ponder RG, Fonville NC, Rosenberg SM. A switch from high-fidelity to error-prone DNA double-strand break repair underlies stress-induced mutation. Mol Cell. 2005;19:791–804. doi: 10.1016/j.molcel.2005.07.025. [DOI] [PubMed] [Google Scholar]
- 26.Gonzalez C, et al. Mutability and importance of a hypermutable cell subpopulation that produces stress-induced mutants in Escherichia coli. PLoS Genet. 2008;4:e1000208. doi: 10.1371/journal.pgen.1000208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Drlica K, Zhao X. DNA gyrase, topoisomerase IV, and the 4-quinolones. Microbiol Mol Biol Rev. 1997;61:377–392. doi: 10.1128/mmbr.61.3.377-392.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Viswanathan A, You H, Doetsch P. Phenotypic change caused by transcriptional bypass of uracil in nondividing cells. Science. 1999;284:159–162. doi: 10.1126/science.284.5411.159. [DOI] [PubMed] [Google Scholar]
- 29.Lengauer C, Kinzler KW, Vogelstein B. Genetic instabilities in human cancers. Nature. 1998;396:643–649. doi: 10.1038/25292. [DOI] [PubMed] [Google Scholar]
- 30.Saxowsky TT, Meadows KL, Klungland A, Doetsch PW. 8-oxoguanine-mediated transcriptional mutagenesis causes ras activation in mammalian cells. Proc Natl Acad Sci USA. 2008;105:18877–18882. doi: 10.1073/pnas.0806464105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ma L, et al. Assembly and development of the Pseudomonas aeruginosa biofilm matrix. PLoS Pathog. 2009;5:e1000354. doi: 10.1371/journal.ppat.1000354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stewart PS. Diffusion in biofilms. J Bacteriol. 2003;185:1485–1491. doi: 10.1128/JB.185.5.1485-1491.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rani SA, et al. Spatial patterns of DNA replication, protein synthesis, and oxygen concentration within bacterial biofilms reveal diverse physiological states. J Bacteriol. 2007;189:4223–4233. doi: 10.1128/JB.00107-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Driffield K, Miller K, Bostock JM, O’Neill AJ, Chopra I. Increased mutability of Pseudomonas aeruginosa in biofilms. J Antimicrob Chemother. 2008;61:1053–1056. doi: 10.1093/jac/dkn044. [DOI] [PubMed] [Google Scholar]
- 35.Oliver A, Canton R, Campo P, Baquero F, Blazquez J. High frequency of hypermutable Pseudomonas aeruginosa in cystic fibrosis lung infection. Science. 2000;288:1251–1253. doi: 10.1126/science.288.5469.1251. [DOI] [PubMed] [Google Scholar]
- 36.Boles BR, Singh PK. Endogenous oxidative stress produces diversity and adaptability in biofilm communities. Proc Natl Acad Sci USA. 2008;105:12503–12508. doi: 10.1073/pnas.0801499105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Macia MD, et al. Hypermutation is a key factor in development of multiple-antimicrobial resistance in Pseudomonas aeruginosa strains causing chronic lung infections. Antimicrob Agents Chemother. 2005;49:3382–3386. doi: 10.1128/AAC.49.8.3382-3386.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Taddei F, et al. Role of mutator alleles in adaptive evolution. Nature. 1997;387:700–702. doi: 10.1038/42696. [DOI] [PubMed] [Google Scholar]
- 39.Liotta LA, Kohn EC. The microenvironment of the tumour–host interface. Nature. 2001;411:375–379. doi: 10.1038/35077241. [DOI] [PubMed] [Google Scholar]
- 40.Joyce JA, Pollard JW. Microenvironmental regulation of metastasis. Nature Rev Cancer. 2009;9:239–252. doi: 10.1038/nrc2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kalluri R, Zeisberg M. Fibroblasts in cancer. Nature Rev Cancer. 2006;6:392–401. doi: 10.1038/nrc1877. [DOI] [PubMed] [Google Scholar]
- 42.Sethi T, et al. Extracellular matrix proteins protect small cell lung cancer cells against apoptosis: a mechanism for small cell lung cancer growth and drug resistance in vivo. Nature Med. 1999;5:662–668. doi: 10.1038/9511. [DOI] [PubMed] [Google Scholar]
- 43.Rintoul R, Sethi T. Extracellular matrix regulation of drug resistance in small-cell lung cancer. Clin Sci. 2002;102:417–424. [PubMed] [Google Scholar]
- 44.Tredan O, Galmarini CM, Patel K, Tannock IF. Drug resistance and the solid tumor microenvironment. J Natl Cancer Inst. 2007;99:1441–1454. doi: 10.1093/jnci/djm135. [DOI] [PubMed] [Google Scholar]
- 45.Gatenby RA, Gillies RJ. Why do cancers have high aerobic glycolysis? Nature Rev Cancer. 2004;4:891–899. doi: 10.1038/nrc1478. [DOI] [PubMed] [Google Scholar]
- 46.Tlsty TD, Coussens LM. Tumor stroma and regulation of cancer development. Annu Rev Pathol. 2006;1:119–150. doi: 10.1146/annurev.pathol.1.110304.100224. [DOI] [PubMed] [Google Scholar]
- 47.Koukourakis MI, Giatromanolaki A, Harris AL, Sivridis E. Comparison of metabolic pathways between cancer cells and stromal cells in colorectal carcinomas: a metabolic survival role for tumor-associated stroma. Cancer Res. 2006;66:632–637. doi: 10.1158/0008-5472.CAN-05-3260. [DOI] [PubMed] [Google Scholar]
- 48.Kansal AR, Torquato S, Chiocca EA, Deisboeck TS. Emergence of a subpopulation in a computational model of tumor growth. J Theoret Biol. 2000;207:431–441. doi: 10.1006/jtbi.2000.2186. [DOI] [PubMed] [Google Scholar]
- 49.Bellomo N, Li N, Maini P. On the foundations of cancer modelling: selected topics, speculations, and perspectives. Math Models Meth Appl Sci. 2008;18:593–646. [Google Scholar]
- 50.Kim M, et al. Tumor self-seeding by circulating cancer cells. Cell. 2009;139:1315–1326. doi: 10.1016/j.cell.2009.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tourovskaia A, Figueroa-Masot X, Folch A. Differentiation-on-a-chip: a microfluidic platform for long-term cell culture studies. Lab Chip. 2005;5:14–19. doi: 10.1039/b405719h. [DOI] [PubMed] [Google Scholar]
- 52.Kimura H, Yamamoto T, Sakai H, Sakai Y, Fujii T. An integrated microfluidic system for longterm perfusion culture and on-line monitoring of intestinal tissue models. Lab Chip. 2008;8:741–746. doi: 10.1039/b717091b. [DOI] [PubMed] [Google Scholar]
- 53.Liu L, et al. A microfluidic device for continuous cancer cell culture and passage with hydrodynamic forces. Lab Chip. 2010;10:1807–1813. doi: 10.1039/c003509b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Groisman A, et al. A microfluidic chemostat for experiments with bacterial and yeast cells. Nature Meth. 2005;2:685–689. doi: 10.1038/nmeth784. [DOI] [PubMed] [Google Scholar]
- 55.Balagadde FK, You L, Hansen CL, Arnold FH, Quake SR. Long-term monitoring of bacteria undergoing programmed population control in a microchemostat. Science. 2005;309:137–140. doi: 10.1126/science.1109173. [DOI] [PubMed] [Google Scholar]
- 56.Austin RH, Tung CK, Lambert G, Liao D, Gong X. An introduction to micro-ecology patches. Chem Soc Rev. 2010;39:1049–1059. doi: 10.1039/b911230h. [DOI] [PubMed] [Google Scholar]
- 57.Hermsen R, Hwa T. Sources and sinks: a stochastic model of evolution in heterogeneous environments. Phys Rev Lett. 2010;105:248104. doi: 10.1103/PhysRevLett.105.248104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Maley CC, et al. The combination of genetic instability and clonal expansion predicts progression to esophageal adenocarcinoma. Cancer Res. 2004;64:7629–7633. doi: 10.1158/0008-5472.CAN-04-1738. [DOI] [PubMed] [Google Scholar]
- 59.Maley CC, et al. Genetic clonal diversity predicts progression to esophageal adenocarcinoma. Nature Genet. 2006;38:468–473. doi: 10.1038/ng1768. [DOI] [PubMed] [Google Scholar]
- 60.Conibear TCR, Collins SL, Webb JS. Role of mutation in Pseudomonas aeruginosa biofilm development. PLoS ONE. 2009;4:e6289. doi: 10.1371/journal.pone.0006289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.West SA, Griffin AS, Gardner A, Diggle SP. Social evolution theory for microorganisms. Nature Rev Microbiol. 2006;4:597–607. doi: 10.1038/nrmicro1461. [DOI] [PubMed] [Google Scholar]
- 62.Lee HH, Molla MN, Cantor CR, Collins JJ. Bacterial charity work leads to population-wide resistance. Nature. 2010;467:82–85. doi: 10.1038/nature09354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fordyce C, et al. DNA damage drives an activin A-dependent induction of cyclooxygenase-2 in premalignant cells and lesions. Cancer Prev Res. 2010;3:190–201. doi: 10.1158/1940-6207.CAPR-09-0229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hickson J, et al. Societal interactions in ovarian cancer metastasis: a quorum-sensing hypothesis. Clin Exp Metastasis. 2008;26:67–76. doi: 10.1007/s10585-008-9177-z. [DOI] [PubMed] [Google Scholar]
- 65.Miller MB, Bassler BL. Quorum sensing in bacteria. Annu Rev Microbiol. 2001;55:165–199. doi: 10.1146/annurev.micro.55.1.165. [DOI] [PubMed] [Google Scholar]
- 66.Smith JM. Evolution and the Theory of Games. 1. Cambridge Univ. Press; Cambridge, UK: 1982. [Google Scholar]
- 67.Hahn WC, Weinberg RA. Modelling the molecular circuitry of cancer. Nature Rev Cancer. 2002;2:331–341. doi: 10.1038/nrc795. [DOI] [PubMed] [Google Scholar]
- 68.Loewen PC, Hengge-Aronis R. The role of the sigma factor sigmas (KatF) in bacterial global regulation. Annu Rev Microbiol. 1994;48:53–80. doi: 10.1146/annurev.mi.48.100194.000413. [DOI] [PubMed] [Google Scholar]
- 69.Zambrano M, Siegele D, Almiron M, Tormo A, Kolter R. Microbial competition: Escherichia coli mutants that take over stationary phase cultures. Science. 1993;259:1757–1760. doi: 10.1126/science.7681219. [DOI] [PubMed] [Google Scholar]
- 70.Keymer JE, Galajda P, Lambert G, Liao D, Austin RH. Computation of mutual fitness by competing bacteria. Proc Natl Acad Sci USA. 2008;105:20269–20273. doi: 10.1073/pnas.0810792105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Feist AM, Herrgard MJ, Thiele I, Reed JL, Palsson BO. Reconstruction of biochemical networks in microorganisms. Nature Rev Microbiol. 2009;7:129–143. doi: 10.1038/nrmicro1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lewis NE, et al. Large-scale in silico modeling of metabolic interactions between cell types in the human brain. Nature Biotech. 2010;28:1279–1285. doi: 10.1038/nbt.1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Moynahan ME, Chiu JW, Koller BH, Jasin M. Brca1 controls homology-directed DNA repair. Mol Cell. 1999;4:511–518. doi: 10.1016/s1097-2765(00)80202-6. [DOI] [PubMed] [Google Scholar]
- 74.Walker GC. Inducible DNA repair systems. Annu Rev Biochem. 1985;54:425–457. doi: 10.1146/annurev.bi.54.070185.002233. [DOI] [PubMed] [Google Scholar]
- 75.Fernández De Henestrosa AR, et al. Identification of additional genes belonging to the LexA regulon in Escherichia coli. Mol Microbiol. 2000;35:1560–1572. doi: 10.1046/j.1365-2958.2000.01826.x. [DOI] [PubMed] [Google Scholar]
- 76.Goodman MF. Error-prone repair, DNA polymerases in prokaryotes and eukaryotes. Annu Rev Biochem. 2002;71:17–50. doi: 10.1146/annurev.biochem.71.083101.124707. [DOI] [PubMed] [Google Scholar]
- 77.Hicks WM, Kim M, Haber JE. Increased mutagenesis and unique mutation signature associated with mitotic gene conversion. Science. 2010;329:82–85. doi: 10.1126/science.1191125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Oliver A, Mena A. Bacterial hypermutation in cystic fibrosis, not only for antibiotic resistance. Clin Microbiol Infect. 2010;16:798–808. doi: 10.1111/j.1469-0691.2010.03250.x. [DOI] [PubMed] [Google Scholar]
- 79.Jensen RB, Carreira A, Kowalczykowski SC. Purified human BRCA2 stimulates RAD51-mediated recombination. Nature. 2010;467:678–683. doi: 10.1038/nature09399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Patel KJ, et al. Involvement of Brca2 in DNA repair. Mol Cell. 1998;1:347–357. doi: 10.1016/s1097-2765(00)80035-0. [DOI] [PubMed] [Google Scholar]
- 81.Roca A, Cox M. RecA protein: structure, function, and role in recombinational DNA repair. Prog Nucleic Acid Res Mol Biol. 1997;56:129–223. doi: 10.1016/s0079-6603(08)61005-3. [DOI] [PubMed] [Google Scholar]
- 82.Levine AJ. p53, the cellular gatekeeper for growth and division. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 83.Hollstein M, et al. Database of p53 gene somatic mutations in human tumors and cell lines. Nucleic Acids Res. 1994;22:3551–3555. [PMC free article] [PubMed] [Google Scholar]
- 84.Branda SS, Vik A, Friedman L, Kolter R. Biofilms: the matrix revisited. Trends Microbiol. 2005;13:20–26. doi: 10.1016/j.tim.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 85.Monds RD, O’Toole GA. The developmental model of microbial biofilms: ten years of a paradigm up for review. Trends Microbiol. 2009;17:73–87. doi: 10.1016/j.tim.2008.11.001. [DOI] [PubMed] [Google Scholar]
- 86.Stewart PS, Franklin MJ. Physiological heterogeneity in biofilms. Nature Rev Microbiol. 2008;6:199–210. doi: 10.1038/nrmicro1838. [DOI] [PubMed] [Google Scholar]
- 87.Costerton JW, Stewart PS, Greenberg EP. Bacterial biofilms: a common cause of persistent infections. Science. 1999;284:1318–1322. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- 88.Donlan RM, Costerton JW. Biofilms: survival mechanisms of clinically relevant microorganisms. Clin Microbiol Rev. 2002;15:167–193. doi: 10.1128/CMR.15.2.167-193.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Stoodley P, et al. Growth and detachment of cell clusters from mature mixed-species biofilms. Appl Environ Microbiol. 2001;67:5608–5613. doi: 10.1128/AEM.67.12.5608-5613.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]