Summary
Background
We previously identified a functional variant in a let-7 microRNA (miRNA) complementary site in the 3′-untranslated region of the KRAS oncogene (rs61764370) which is associated with cancer. We aimed to investigate the association of this KRAS variant with breast cancer and tumour biology.
Methods
We assessed frequency distributions of the KRAS variant in 415 patients with histologically confirmed breast cancer and 457 controls from Connecticut, USA (study group 1) and association of this variant with breast-cancer subtypes in 690 Irish women with known oestrogen receptor (ER), progesterone receptor (PR), and HER2 statuses, and 360 controls (study group 2). We pooled data for study groups 1 and 2 with a cohort of 140 women with triple-negative breast cancer and 113 controls to assess the association of the KRAS variant with triple-negative breast cancer risk, and genome-wide mRNA and specific miRNA expression in patients with triple-negative breast cancer.
Findings
Although frequency distributions of the KRAS variant in study group 1 did not differ between all genotyped individuals, eight (33%) of 24 premenopausal women with ER/PR-negative cancer had the KRAS variant, compared with 27 (13%) of 201 premenopausal controls (p=0·015). In study group 2, the KRAS variant was significantly enriched in women with triple-negative breast cancer (19 [21%] of 90 cases) compared with 64 (13%) of 478 for luminal A, 13 (15%) of 87 for luminal B, and two (6%) of 35 for HER2-positive subgroups (p=0·044). Multivariate analysis in the pooled study groups showed that the KRAS variant was associated with triple-negative breast cancer in premenopausal women (odds ratio 2·307, 95% CI 1·261–4·219, p=0·0067). Gene-expression analysis of triple-negative breast-cancer tumours suggested that KRAS-variant positive tumours have significantly altered gene expression, and are enriched for the luminal progenitor and BRCA1 deficiency signatures. miRNA analysis suggested reduced levels of let-7 miRNA species in KRAS-variant tumours.
Interpretation
The KRAS variant might be a genetic marker for development of triple-negative breast cancer in premenopausal women, and altered gene and miRNA expression signatures should enable molecular and biological stratification of patients with this subgroup of breast cancer.
Funding
US National Institutes of Health.
Introduction
The heterogeneity of breast cancer is shown in the variable risk factors, treatment responses, and outcomes of patients. Breast tumours are classified into oestrogen-receptor (ER) positive and/or progesterone-receptor (PR) positive, HER2 (ERBB2) amplified, and triple-negative tumours (ie, ER/PR negative and HER2 negative).1 Gene expression and receptor profiling further classifies breast cancer into four biological subgroups: luminal A (ER and/or PR receptor positive, HER2 negative), luminal B (ER and/or PR receptor positive, HER2 positive), HER2 positive (ER/PR negative, HER2 positive), and basal-like tumours (triple-negative breast cancer).1
Triple-negative breast cancer is the most aggressive subgroup, with the poorest cause-specific survival at 5 years.2 Transcriptional profiling studies suggest there is further heterogeneity within triple-negative breast cancers and these tumours can be categorised into two broad subgroups: triple-negative tumours that express epidermal growth factor receptor (EGFR) or cytokeratin (CK) 5/6 and are therefore termed basal-like, and triple-negative tumours that do not express EGFR or CK5/6. Basal-like triple-negative tumours are marked by a younger age of onset than are non-basal-like forms and low expression of BRCA1; the basal-like phenotype is common in carriers of the BRCA1 mutation.3 An aberrant luminal progenitor cell population (that might be ER positive) could be the target for transformation in BRCA1-associated basal tumours.4 Although prognostic gene-expression markers are highly divergent, several modules such as DNA repair deficiency, signatures of immune response, or transition from epithelium to mesenchyme are commonly noted in a subset of these tumours.5 Identification of the drivers of these transcriptional modules is a promising approach for discovery of specific and personalised therapies.
Association of the triple-negative breast cancer phenotype with young age of onset and an absence of association with known risks or reproductive factors6 supports the notion that there are genetic risks for development of this cancer.7 Unfortunately, few genetic markers of such increased risk exist. Although BRCA1 mutations are often associated with triple-negative tumours, these mutations are rare and account for only 10–15% of patients with triple-negative breast cancer, dependent on ethnic background and family history.8,9
MicroRNAs (miRNAs) are a novel class of small non-coding RNAs that regulate gene expression by base pairing with sequences within the 3′-untranslated region (UTR), 5′-UTR, and coding sequence regions of target mRNAs, causing mRNA cleavage or translational repression.10,11 miRNAs are misregulated in every cancer studied so far including breast cancer, in which certain miRNA changes (specifically reduced let-7) are found in breast tumour-initiating cells, suggesting that low let-7 expression allows self-renewal and proliferation of these cells12 and probably increases risk of breast cancer.
Because miRNAs are global gene regulators, inherited variations in miRNAs are associated with increased cancer risk. Evidence is accumulating that polymorphisms disrupting miRNA coding sequences13 or 3′-UTR miRNA binding sites are strong predictors of cancer risk, including breast cancer.14,15 However, none of the previously identified miRNA-altering polymorphisms has been associated with triple-negative breast cancer, or with altered gene or miRNA expression in tumours.
We previously identified a novel germline polymorphism (rs61764370) in a let-7 miRNA complementary site within the 3′-UTR of the KRAS oncogene, which is referred to here as the KRAS variant. We showed that the KRAS variant is associated with low concentrations of let-7 in tumours and altered KRAS regulation in lung cancer.16 Other groups reported that the KRAS variant predicts poor cancer specific outcome in head and neck cancer17 and altered drug response in colon cancer,18,19 suggesting that this variant has biological relevance. Recently we showed that the KRAS variant is enriched in ovarian cancer and is most frequently associated with patients from families with hereditary breast and ovarian cancer.20 On the basis of this evidence, we aimed to assess the role of the KRAS variant in breast-cancer risk and tumour biology.
Methods
Study populations
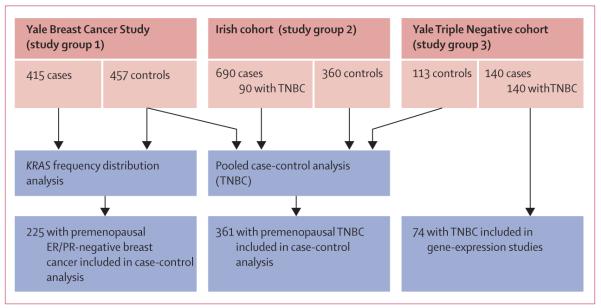
In this case-control study and genetic analysis, we assessed data from four cohorts (figure 1). To assess frequency distributions of the KRAS-variant genotype, we assessed individuals from the Yale Breast Cancer Study (study group 1), who were enrolled in a breast cancer case-control study in Connecticut, USA; the study was approved by the Yale institutional review board as previously described.13 Briefly, patients were aged 30–80 years and had incident, histologically confirmed breast cancer and no history of cancer (other than non-melanoma skin cancer). ER and PR statuses were established for all cases but HER2 statuses were not known and not obtainable. Controls were recruited either from Yale–New Haven Hospital (New Haven, CT, USA) or Tolland County, CT, USA. Controls from the Yale–New Haven Hospital underwent breast-related surgery for histologically confirmed benign breast diseases. Controls from Tolland County were identified either through random-digit dialling (for individuals aged <65 years) or through the Health Care Finance Administration files (≥65 years). Informed consent and data for family histories of cancer, reproductive history, demographic factors, and blood sample were obtained from all participants. 415 cases and 457 controls had DNA samples available for this study, which were obtained between 1990 and 1999.
Figure 1. Study groups.
TNBC=triple-negative breast cancer. ER=oestrogen receptor. PR=progesterone receptor.
To define the association of the KRAS variant with receptor status and breast cancer subtype, we assessed a cohort of 690 Irish women diagnosed with breast cancer with complete receptor status and subtype classification. Patients from this cohort (study group 2) had histologically confirmed breast cancer and were recruited from the west of Ireland after appropriate ethical approval from the Galway University Hospital (Galway, Ireland) ethics committee. Informed consent and a detailed family history of breast cancer or ovarian cancer, and a blood sample were obtained from all cases. We included 710 cases of breast cancer of all stages and histological types, apart from preinvasive carcinomas. ER, PR, and HER2 statuses were established for all samples by use of standard histopathological analysis and immuno histochemistry, and confirmed by fluorescence in-situ hybridisation for HER2 positivity. Although gene-expression analysis was not done, these samples were classified as luminal A, luminal B, HER2, or triple-negative breast cancer by receptor status (see webappendix p 1). 690 of 710 patients had complete information and were assessed in this study. The 360 controls in this cohort were healthy women from the same geographical area, and were mainly older than 60 years, with no selfreported personal history of any cancer and no family history of breast cancer or ovarian cancer. Cases and controls were mainly recruited from July, 2006, to July, 2010.
To establish whether the KRAS variant predicted an increased risk of development of triple-negative breast cancer, we did a pooled analysis of a cohort of patients with triple-negative breast cancer and controls from Yale (study group 3) and patients with triple-negative breast cancer and controls from study group 2 and controls from study group 1. Patients in study group 3 were receiving treatment either at Yale–New Haven Hospital or at the Bridgeport Hospital (Bridgeport, CT, USA). After approval by the Yale Human Investigation Committee, tissue or saliva specimens were obtained from 156 patients. Complete data were available for 140 patients who were diagnosed in 1990–2007 and were included in this study. 130 cases of triple-negative breast cancer had samples of tumour available before any treatment for gene and miRNA-expression analysis, 78 of whom were also genotyped for the KRAS variant. 113 controls in this cohort were healthy women who presented to the Yale–New Haven Hospital and who had no personal history of cancer apart from non-melanoma skin cancer and were recruited between 2000 and 2007. We obtained clinical information, age, ethnic origin, and family history for all cases and controls. Webappendix p 2 summarises basic information for the aforementioned three cohorts.
To assess association of the KRAS variant with BRCA mutations in ER-negative tumours, we analysed BRCA1-mutation carriers with breast cancer and known KRAS-variant status from our previous study of the Rotterdam population. The Rotterdam population has been described21 but, briefly, consisted of Dutch patients with breast cancer and documented BRCA1 mutations who were identified by investigators at the Erasmus University through the Rotterdam Family Clinic (Rotterdam, Netherlands).
Procedures
For KRAS-variant genotyping assays, we genotyped DNA from all samples for the KRAS variant with a custom TaqMan SNP genotyping assay (Applied Biosystems, Carlsbad, CA, USA). On the basis of a previous study,16 we regarded samples that were heterozygous or homozygous for the variant G allele as positive for the KRAS variant.
For gene-expression analysis, we measured genome-wide mRNA expression in 78 patients from the Yale triple-negative cohort who were also tested for the KRAS variant. We isolated total RNA from tissue specimen with the RecoverAll total nucleic acid isolation kit (Applied Biosystems) and hybridised to the whole-genome DASL assay (HumanRef-8 version 3.0, Illumina, San Diego, CA, USA). Data preprocessing and statistical analysis were done with the lumi package in Bioconductor/R software. Gene-expression data from three whole-genome DASL runs were combined and processed together. Samples with less than 30% detectable probes and probes that were detectable in less than 10% of the samples were discarded before quantile-normalisation. 74 samples and 18345 probes remained after filtering.
For miRNA analysis, we produced arrays with the Multiplex RT and TaqMan low density array human miRNA panel–real-time PCR system (Applied Biosystems) as per the manufacturer’s protocol.22 We examined expression levels of miRNAs of interest.
Statistical analysis
Genotype distributions of all cases and controls were tested for Hardy-Weinberg equilibrium and were found to be in equilibrium. We did unconditional logistic regression to estimate the relative risk associated with every genotype. Controls were adjusted for age (continuous) and ethnic origin (white, black, Hispanic, or other). The population was stratified by menopausal status (estimated by age ≤51 years or >51 years), and separate risk estimates were obtained by ER and PR statuses with multinomial logistic regression with a three-level outcome variable coded as 0 for controls, 1 for cases with ER-positive and/or PR-positive tumours, and 2 for ER/PR-negative tumours. We did tests for interaction with a Wald χ2, comparing the parameter estimates obtained for every genotype in cases of ER-positive and/or PR-positive disease compared with ER/PR-negative disease.
Patients in study group 2 were stratified according to subtypes of breast cancer and a χ2 test was done with GraphPad Prism4 software to calculate p values, odds ratios (ORs), and 95% CI. The dominant model was used for all genetic association analysis because of the low frequency of the KRAS variant.
We compared categorical variables (eg, ethnic origin, stage, and study site) between study groups with a χ2 test or two-sided Fisher’s exact test, and continuous variables (eg, age) with a t test. We calculated ORs and 95% CI for the KRAS variant in controls and cases of triple-negative breast cancer with an unconditional logistic regression model with a binary outcome variable. Multivariate logistic regression analyses with a binary outcome variable coded as controls and cases included variables such as KRAS-variant status, age, ethnic origin, and study site. The population was also stratified by age group, and separate logistic regression analyses were done for patients aged 51 years or younger (premenopausal group) or older than 51 years (postmenopausal group). Statistical analyses were done with SAS version 9.1.3.
Pathway activation was measured as correspondence with previously published expression signatures and axes derived from principal component analysis of the expression set. Principal component analysis was used to separate biological from technical sources of information in the gene-expression dataset. Every component was characterised by correspondence to RNA quality, the structure of a batch effect, and biological annotations of the contributing probes (ie, probes with expression profiles that have high absolute projection values for the specified component). Signatures of gene expression are provided as lists of genes and their changes in expression in a specific condition. Such signatures are especially valuable for noisy data because they require coordinated differential expression of multiple probes, typically in the order of 100. Because mRNA was extracted from formalin-fixed, paraffin-embedded (FFPE) blocks that were up to 20 years old, analysis of the data set with a signature approach was justified.23 We calculated signature scores as Pearson correlation between the respective signature vector of gene contributions and a sample’s expression profile for these genes. Association of the KRAS variant with the outcomes described by the respective signature was analysed by a paired Kolmogorov-Smirnov test between signatures scores of KRAS variant and wild-type samples. Differential gene expression was assessed with a linear model, taking into account technical batch artifacts as an offset. Model fitting and empirical Bayesian error moderation of the fold changes were performed with the LIMMA package for R.24
We analysed miRNA expression in eight batches of 46 miRNAs and two endogenous controls. miRNA expression was normalised on the basis of the geometric mean of all expressed samples: a miRNA was judged to have been expressed if threshold fluorescence was detected after fewer than 35 cycles and when the geometric mean cycle number of all expressed miRNAs was subtracted. miRNAs that were not expressed in more than two thirds of all samples were removed, followed by scale-normalisation in all remaining threshold-cycle values.
Role of the funding source
There was no funding source for this study. The corresponding author had full access to all the data in the study and had final responsibility for the decision to submit for publication.
Results
Overall, frequency distributions of the KRAS-variant genotype did not differ between cases and controls who were genotyped from study group 1 (figure 1, table 1). However, the KRAS variant was significantly associated with breast cancer in premenopausal patients with ER/PR-negative tumours (table 1). This association was not observed for postmenopausal women. Eight (33%) of 24 premenopausal women with ER/PR-negative cancer had the KRAS variant, compared with 27 (13%) of 201 controls and four (9%) of 44 premenopausal women with cancer that was positive for ER and/or PR (webappendix p 10). Thus, the KRAS variant might be a genetic marker of increased risk of development of receptor-negative breast cancer for premenopausal women.
Table 1.
Association of the KRAS-variant with ER/PR-positive versus ER/PR-negative breast cancer in women in study group 1
| Controls | All |
ER and/or PR positive |
ER/PR negative |
Pinteraction | ||||
|---|---|---|---|---|---|---|---|---|
| Cases | Odds ratio (95% CI)* | Cases | Odds ratio (95% CI)* | Cases | Odds ratio (95% CI)* | |||
| All ages | ||||||||
| Non-variant (T/T) | 391 | 347 | Reference | 145 | Reference | 62 | Reference | · · |
| Variant (T/G or G/G) | 79 | 68 | 0·95 (0·67–1·36) | 28 | 0·93 (0·58–1·49) | 18 | 1·59 (0·88–2·86) | 0·118 |
| Premenopausal | ||||||||
| Non-variant (T/T) | 174 | 84 | Reference | 40 | Reference | 16 | Reference | · · |
| Variant (T/G or G/G) | 27 | 16 | 1·64 (0·79–3·43) | 4 | 0·87 (0·28–2·75) | 8 | 4·78 (1·71–13·38) | 0·015 |
| Postmenopausal | ||||||||
| Non-variant (T/T) | 217 | 263 | Reference | 105 | Reference | 46 | Reference | · · |
| Variant (T/G or G/G) | 52 | 52 | 0·77 (0·51–1·16) | 24 | 0·90 (0·53–1·53) | 10 | 0·90 (0·43–1·90) | 0·991 |
Data are number or odds ratio (95% CI), unless otherwise stated. ER=oestrogen receptor. PR=progesterone receptor.
Age, ethnic origin, and menopausal status were adjusted in monomial unconditional logistic regression. G/G phenotype occurs in less than 5% of cases and controls and was combined with the G/T phenotype. Minor allele frequency (controls) 0·087, p for Hardy-Weinberg equilibrium 0·783.
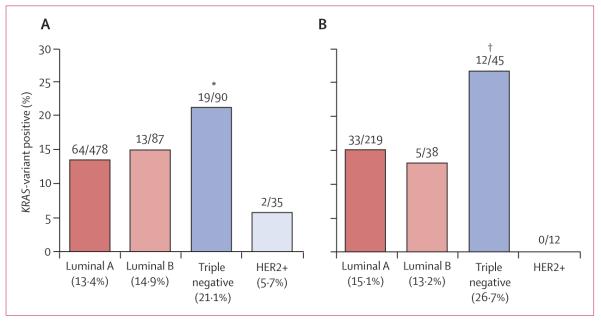
In study group 2, 478 women had luminal A breast cancer, 87 had luminal B disease, 90 had triple-negative disease, and 35 had HER2-positive disease. 98 (14%) of 690 breast-cancer cases from this cohort had the KRAS variant, but prevalence varied between the breast cancer subtypes: the KRAS variant was significantly enriched in women with triple-negative breast cancer (19 [21%] of 90 cases) compared with 64 (13%) of 478 for luminal A, 13 (15%) of 87 for luminal B, and two (6%) of 35 for HER2-positive subgroups (p=0·044; figure 2). This association with triple-negative breast cancer was also noted in women younger than 51 years (p=0·033, figure 2).
Figure 2. Distribution of the KRAS variant in breast-cancer subtypes in all women (A) and premenopausal (≤51 years) women (B) from study group 2.
Data are numbers of cases diagnosed with breast-cancer subtype/numbers of patients tested for the KRAS variant. *p=0·044 versus all other subtypes. †p=0·033 versus all other subtypes.
By comparison of cases of triple-negative breast cancer from groups 2 and 3 and controls across all three cohorts (n=1160), we did not note a significant difference between cases or between controls for the prevalence of the KRAS variant (webappendix p 3). However, there were significantly more non-white women in the controls from study groups 1 and 3 than there were in the study group 2, which allowed assessment of the association of the KRAS variant in non-white women with triple-negative breast cancer in the multivariate analysis. After controlling for age, ethnic origin, and study site, the KRAS variant did not predict an increased risk of development of triple-negative breast cancer for all women in multivariate analysis (table 2, webappendix p 4). However, the KRAS variant was associated with a significantly increased risk of development of triple-negative breast cancer in the 361 premenopausal women in this pooled group in multivariate analysis (table 2, webappendix pp 5–6).
Table 2.
Association of the KRAS-variant in 230 patients with triple-negative breast cancer compared with 930 controls from pooled analysis of study groups 1–3
| Odds ratio (95% CI) | p value | |
|---|---|---|
| All ages | ||
| Univariate analysis | ||
| KRAS variant | 1·162 (0·797–1·694) | 0·4363 |
| Multivariate analysis | ||
| KRAS variant | 1·352 (0·901–2·028) | 0·1455 |
| Age | 0·913 (0·942–0·967) | <0·0001 |
| Ethnic origin | 2·536 (2·784–5·999) | <0·0001 |
| Premenopausal women | ||
| Univariate analysis | ||
| KRAS variant | 1·879 (1·067–3·310) | 0·029 |
| Multivariate analysis | ||
| KRAS variant | 2·307 (1·261–4·219) | 0·0067 |
| Age | 0·913 (0·871–0·956) | 0·0001 |
| Ethnic origin | 2·536 (1·582–4·067) | 0·0001 |
Age, ethnic origin, menopausal status, and study site were adjusted in a logistic regression model. G/G phenotype occurs in less than 5% of cases and controls and was combined with the G/T phenotype.
Because BRCA1 coding sequence mutations are associated with risk of triple-negative breast cancer, and because we noted an apparent enrichment of the KRAS variant in BRCA1 mutation-carriers with breast cancer,21 we aimed to establish whether the association of the KRAS variant with premenopausal triple-negative breast cancer was due only to its association with carriers of BRCA1 mutation. Of 36 women with triple-negative breast cancer from cohort 2 and 3 who were BRCA tested, 25 (69%) were BRCA negative and 11 (31%) were BRCA positive. Of these patients, eight (32%) BRCA-negative women harboured the KRAS variant compared with three (27%) women who were BRCA positive. These findings suggest that the KRAS variant is associated with an independent group of patients with triple-negative breast cancer without BRCA mutations.
Although we did not note an association between KRAS-variant status and ER or PR negative statuses in the Rotterdam population cohort,21,23 we had not considered menopausal status. In this study, we did not note an enrichment of the KRAS variant in 126 premenopausal BRCA1-mutation carriers who had ER/PR-negative breast cancer compared with all 268 BRCA1-mutation-carriers from the Rotterdam cohort (21·8% vs 23·5%, p=0·95). These findings again support the notion that association of the KRAS variant with premenopausal triple-negative breast cancer is independent of its association with BRCA1 mutations.
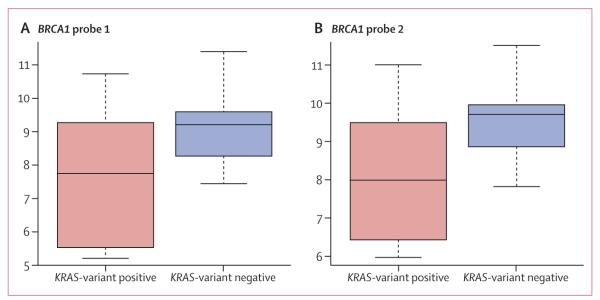
However, to further assess potential biological interaction between the KRAS variant and altered BRCA1 expression in triple-negative disease, we appraised BRCA1 expression levels in 74 triple-negative tumours from study group 3 (figure 1). We noted that those patients with the KRAS variant had significantly reduced BRCA1 expression compared with KRAS-variant-negative triple-negative tumours (p=0·06 for probe 1 [ILMN_2311089] and p=0·01 for probe 2 [ILMN_1738027], figure 3). Furthermore, the KRAS variant was significantly associated with a gene expression signature of decreased BRCA1 activity (p=0·04).25 These findings suggest that, although the KRAS variant is not restricted to patients with triple-negative breast cancer with known BRCA1 mutations, there might be some biological interaction between the KRAS variant, altered BRCA1 expression or functionality, and development of triple-negative breast cancer.
Figure 3. BRCA1 gene expression among the KRAS-variant positive and KRAS-variant negative cases of triple-negative breast cancer.
Y-axes are in arbitrary units. (A) BRCA1 probe 1, p=0·06. (B) BRCA1 probe 2, p=0·01.
We compared signalling pathways in triple-negative breast-cancer tumours that were KRAS-variant positive with those that were KRAS-variant negative from patients in study group 3. Although analysis of KRAS mRNA did not vary by KRAS-variant status, this finding agrees with the other publications about the effect of miRNA binding to the KRAS 3′-UTR.16,26 However, we noted an increase in both an NRAS mutation27 and a MAP-kinase activation signature28 (table 3) in tumours with the KRAS variant. This supports the notion that the KRAS variant alters gene expression of canonical RAS pathways, and is to our knowledge the first in-vivo evidence that the KRAS variant leads to continued altered downstream gene expression in tumours with which it is associated.
Table 3.
Association of the KRAS-variant with pathway signatures in tumours of patients with triple-negative breast cancer and positive KRAS-variant status
| Signature expression | Kolmogorov-Smirnov p value |
|
|---|---|---|
| NRAS | Upregulated | 0·02 |
| BRCA mutant-like | Upregulated | 0·04 |
| Luminal progenitor | Upregulated | 0·04 |
| MAPK (Creighton) | Upregulated | 0·06 |
| PCA oestrogen | Downregulated | 0·04 |
Signature scores were computed as Pearson correlation between the signature vector of gene contributions and each sample’s expression profile for these genes. The Kolmogorov-Smirnov test was used to analyse the association of the KRAS-variant with signature activation.
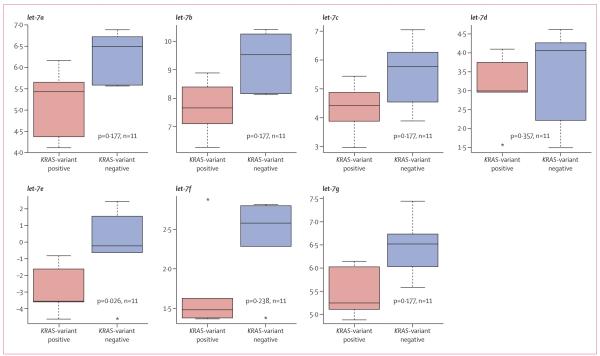
Because we had previously noted altered concentrations of let-7 miRNA in lung tumours with the KRAS variant, we examined let-7 concentrations in triple-negative breast cancer tumours with the KRAS variant. Consistent with our previous findings, we noted lower concentrations of all let-7 miRNA family members in KRAS-variant-associated tumours (figure 4).
Figure 4. Expression of let-7 family of microRNAs in the KRAS-variant positive versus KRAS-variant negative cases of triple-negative breast cancer.
Y-axes are in arbitrary units.
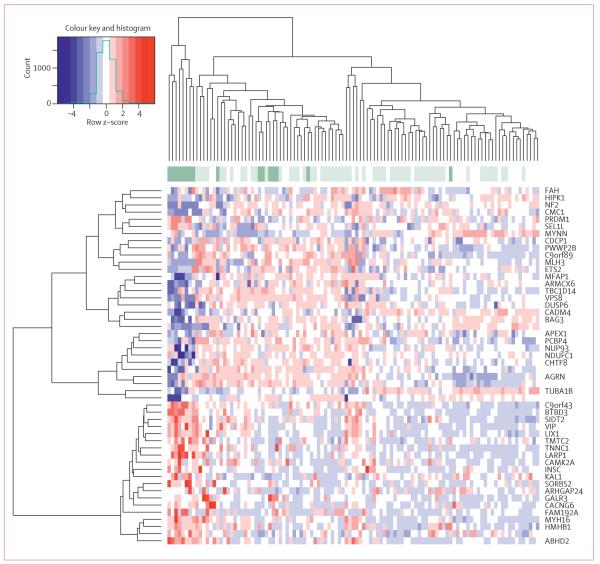
To establish how the KRAS variant integrates with known gene-expression signatures of triple-negative breast cancer, we assessed known signatures that are differentially expressed in such tumours. We found that KRAS-variant tumours have several features of triple-negative and basal-like tumour biology, including decreased oestrogen signalling in a main component derived from our expression set (p=0·04). Furthermore, KRAS-variant tumours have a luminal progenitor signature (p=0·04), which has been suggested4 as a candidate progenitor for basal-like breast cancer (table 3, webappendix p 11). Within the luminal progenitor and the BRCA mutation-like signatures, markers of cell adhesion, tissue invasion, proliferation, and angiogenesis (such as α5 integrin, DUSP6, and aurora kinase B) were differentially regulated (webappendix p 7). This finding is in agreement with the slight enrichment by functional annotations that we noted in three of 41 genes for wound healing (p=0·02), three of 151 genes for glycan expression (p=0·05), and four of 148 genes for MEK activation (p=0·009) on the basis of the differentially expressed genes in a linear model comparing KRAS variant versus non-variant for the dataset (figure 5, webappendix pp 8–9).
Figure 5. Heat map of KRAS variant differentially expressed genes in patients with triple-negative breast cancer, analysed by LIMMA model.
The 50 most significant genes were used for the clustering; p<0·0001 for clustering. KRAS-variant samples are dark green, wild-type samples are light green. White have unknown KRAS-variant status.
Discussion
Our data suggest that a germline polymorphism in the KRAS 3′-UTR (the KRAS variant) is a genetic marker of increased risk of development of triple-negative breast cancer in premenopausal women. Because study group 1 was small and only assessed patients with known ER and PR statuses, we validated this association in larger case-control groups with full receptor status. Most importantly, we show that patients with triple-negative breast cancer who have the KRAS variant have tumours with distinct gene-expression patterns compared with patients without this variant, suggesting that the mutation might drive specific pathways that influence tumour biology and could modify tumour development. The KRAS variant could ultimately be of value in subclassifying tumours into meaningful biological subgroups to both predict prognosis and help to direct treatment in the future (panel).
The finding of reduced let-7 concentrations in triple-negative breast cancer tumours that are associated with the KRAS variant, as has been reported in lung cancer, is notable. Studies suggest that KRAS overexpression, through NFκB, can lead to induction of LIN-28 (a negative regulator of let-7) and lowering of let-7 expression.29–31 These conclusions suggest a potential mechanism whereby let-7 is lowered in premalignant tissue and, ultimately, tumours associated with the KRAS variant. Furthermore, let-7 regulates proliferation of breast-like stem cells,12 and low let-7 concentrations could allow expansion of this group of cells, potentially increasing breast-cancer risk in women with the KRAS variant. The association we noted of the KRAS variant with triple-negative breast cancer risk only in premenopausal women suggests a meaningful interaction between the KRAS variant and hormonal exposure. Such associations and potential mechanisms need additional validation in large cohorts and tumour-initiation models.
Although more than half of breast tumours that carriers of the BRCA1 mutation develop are triple-negative subtype,32 BRCA1 mutations are rare and thus only account for about 10–15% of all cases of triple-negative disease.8,9 Up to 23% of premenopausal patients with triple-negative breast cancer have the KRAS variant, without an apparent significant enrichment in BRCA mutation carriers in these cohorts or in young ER/PR-negative BRCA1-mutation carriers.23 However, the KRAS variant is associated with a BRCA1 mutation-like gene-expression signature, supporting the notion that there might be increased oncogenic risk in the presence of the KRAS variant and high KRAS expression and low BRCA1 expression, either through mutation or other mechanisms.
We previously showed the KRAS variant affects the regulation of KRAS expression in vitro, promoting high KRAS concentrations.16 The KRAS oncogene is an important upstream mediator of the MAPK pathway, and its overexpression can lead to increased activation of the RAF/MEK/MAPK pathway, thereby promoting tumorigenesis. We showed here that patients with the KRAS variant and triple-negative breast cancer show activation of the MAPK pathway (table 3). Oh and colleagues33 reported that hyperactivation of MAPK in breast cancer cells decreases ERα expression leading to a negative phenotype, which is in agreement with our finding that the KRAS variant is associated with even lower oestrogen signalling in these histologically ER-negative tumours. MAPK activation has been implicated in oestrogen-independent tumour growth and insensitivity to anti-oestrogen treatment,34 and might be a mechanism by which the KRAS variant drives the development of triple-negative breast cancer more than other breast cancer subtypes. The role of the KRAS variant in tumorigenesis and its specific association with triple-negative breast cancer remains to be delineated.
The KRAS variant is a biomarker of poor outcome in several cancers, including head and neck cancer,17 and is a biomarker of poor response to targeted therapies in colon cancer.18 Our finding that patients with the KRAS variant and triple-negative breast cancer have a luminal progenitor signature and differential expression of angiogenic and metastatic markers within the signature suggests that tumours harbouring the KRAS variant might be an aggressive subgroup of this cancer. Follow-up studies will be necessary to establish the effect of the KRAS variant on outcome in patients with triple-negative breast cancer and patients with breast cancer in general.
Our study suggests that the KRAS variant is associated with tumours that maintain unique gene-expression patterns. Although investigations remain to be done to establish the mechanisms of development of triple-negative breast cancer in women who are KRAS-variant positive, our findings give insight into crucial steps and pathways required for transformation and tumour development in these women. We believe our results are meaningful steps towards understanding of the mechanisms of gain of function miRNA-disrupting polymorphisms in cancer biology, which seem to be distinct in function from previously discovered genetic markers of cancer risk.
Supplementary Material
Panel: Research in context.
Systematic review
Examination of inherited variants in microRNAs (miRNA) and miRNA binding sites that predict cancer risk is a new and rapidly growing area of research. However, the effect of these miRNA disrupting variants on tumour biology has not been assessed. Because other investigators have shown the potential of the KRAS variant to act as a biomarker of poor outcome or poor response to targeted chemotherapy agents, we postulated that this altered biology may be noted in gene and miRNA differences in tumours. Our aim was to understand if miRNA disrupting variants, such as the KRAS variant, could both be associated with tumour risk and tumour biology as notable in differences in gene and miRNA expression.
Interpretation
Our study shows that altered tumour gene-expression patterns can be partly accounted for by inherited variants that disrupt miRNAs binding sites. This finding could explain how such variants can act as biomarkers of cancer outcome and response to therapy, and suggests that such variants might be a simple way to subclassify tumours into biologically relevant subgroups. Our conclusions provide evidence that baseline genetic differences between patients can predict genetic differences in their tumours, which is an exciting direction of study in oncology.
Acknowledgments
We thank Neal Fischbach and the Cancer Genetic Counselling Shared Resource at the Yale Cancer Center (New Haven, CT, USA) for contributions of samples to the study. TP was supported by a Yale Center for Clinical Investigation (YCCI) grant made possible by Clinical and Translational Science Awards (CTSA) grant number UL1 RR024139 from the National Centre for Research Resources (NCRR), a component of the US National Institutes of Health (NIH), and US NIH roadmap for Medical Research. FS and JW were supported by the US National Cancer Institute (CA131301). JW was supported by a K08 grant [CA124484]. HH was supported by a Health Research Board Clinician Scientist Fellowship and an Irish Higher Surgical Training Group Travelling Scholarship. JD was supported by the National Breast Cancer Research Institute in Galway, Ireland. DZ has received royalties for books published by John Wiley and Sons, Oxford University Press, Cambridge University Press, and the SAS institute, and payment for service on a data monitoring committee from BristolMyersSquibb.
Footnotes
Contributors TP participated in study design, experimentation, writing, preparation of figures, and statistical analysis. HH participated in study design, experimentation, and writing. RL did the analysis of mRNA and microRNA expression. FKK did statistical analysis. AHof did experiments and statistical analysis. AHol did data analysis. JD, KG, CP, and SN did experiments. JWMM was involved in sample collection and writing. MJH provided data and participated in writing the report. MK participated clinically in sample provision and edited the manuscript. DZ provided statistical analysis. YZ provided samples, experiments, and statistical analysis. DT participated in mRNA expression analysis and edited the manuscript. LH supplied samples and edited the manuscript. NM supervised experimental work and manuscript editing. FS assisted in study design and writing. JW designed the study, and participated in analysis, writing, and oversight of the project.
Conflicts of interest FS and JW have patented intellectual property surrounding the KRAS variant through Yale University (New Haven, CT, USA), and founded a company that has licensed this intellectual property from Yale University. All other authors declared no conflicts of interest.
References
- 1.Sørlie T, Perou CM, Tibshirani R, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci USA. 2001;98:10869–74. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Haffty BG, Yang Q, Reiss M, et al. Locoregional relapse and distant metastasis in conservatively managed triple-negative early-stage breast cancer. J Clin Oncol. 2006;24:5652–57. doi: 10.1200/JCO.2006.06.5664. [DOI] [PubMed] [Google Scholar]
- 3.Rakha EA, Ellis IO. Triple-negative/basal-like breast cancer: review. Pathology. 2009;41:40–47. doi: 10.1080/00313020802563510. [DOI] [PubMed] [Google Scholar]
- 4.Lim E, Vaillant F, Wu D, et al. Aberrant luminal progenitors as the candidate target population for basal tumor development in BRCA1 mutation carriers. Nat Med. 2009;15:907–13. doi: 10.1038/nm.2000. [DOI] [PubMed] [Google Scholar]
- 5.Bild AH, Parker JS, Gustafson AM, et al. An integration of complementary strategies for gene-expression analysis to reveal novel therapeutic opportunities for breast cancer. Breast Cancer Res. 2009;11:R55. doi: 10.1186/bcr2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang XR, Sherman ME, Rimm DL, et al. Differences in risk factors for breast cancer molecular subtypes in a population-based study. Cancer Epidemiol Biomarkers Prev. 2007;16:439–43. doi: 10.1158/1055-9965.EPI-06-0806. [DOI] [PubMed] [Google Scholar]
- 7.Bauer KR, Brown M, Cress RD, Parise CA, Caggiano V. Descriptive analysis of estrogen receptor (ER)-negative, progesterone receptor (PR)-negative, and HER2-negative invasive breast cancer, the so-called triple-negative phenotype: a population-based study from the California cancer registry. Cancer. 2007;109:1721–28. doi: 10.1002/cncr.22618. [DOI] [PubMed] [Google Scholar]
- 8.Young SR, Pilarski RT, Donenberg T, et al. The prevalence of BRCA1 mutations among young women with triple-negative breast cancer. BMC Cancer. 2009;9:86. doi: 10.1186/1471-2407-9-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nanda R, Schumm LP, Cummings S, et al. Genetic testing in an ethnically diverse cohort of high-risk women: a comparative analysis of BRCA1 and BRCA2 mutations in American families of European and African ancestry. JAMA. 2005;294:1925–33. doi: 10.1001/jama.294.15.1925. [DOI] [PubMed] [Google Scholar]
- 10.He L, Thomson JM, Hemann MT, et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–33. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Esquela-Kerscher A, Slack FJ. Oncomirs—microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–69. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 12.Yu F, Yao H, Zhu P, et al. Let-7 regulates self renewal and tumorigenicity of breast cancer cells. Cell. 2007;131:1109–23. doi: 10.1016/j.cell.2007.10.054. [DOI] [PubMed] [Google Scholar]
- 13.Hoffman A, Zheng T, Yi C, et al. MicroRNA miR-196a-2 and breast cancer: a genetic and epigenetic association study and functional analysis. Cancer Res. 2009;69:5970–77. doi: 10.1158/0008-5472.CAN-09-0236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pongsavee M, Yamkamon V, Dakeng S, et al. The BRCA1 3′UTR: 5711+421T/T_5711+1286T/T genotype is a possible breast and ovarian cancer risk factor. Genet Test Mol Biomarkers. 2009;13:307–17. doi: 10.1089/gtmb.2008.0127. [DOI] [PubMed] [Google Scholar]
- 15.Tchatchou S, Jung A, Hemminki K, et al. A variant affecting a putative miRNA target site in estrogen receptor (ESR) 1 is associated with breast cancer risk in premenopausal women. Carcinogenesis. 2009;30:59–64. doi: 10.1093/carcin/bgn253. [DOI] [PubMed] [Google Scholar]
- 16.Chin L, Ratner E, Leng S, et al. A SNP in a let-7 microRNA complementary site in the KRAS 3′UTR increases non-small cell cancer risk. Cancer Res. 2008;68:8535–40. doi: 10.1158/0008-5472.CAN-08-2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Christensen BC, Moyer BJ, Avissar M, et al. A let-7 microRNA binding site polymorphism in the KRAS 3′UTR is associatied with reduced survival in oral cancers. Carcinogenesis. 2009;30:1003–07. doi: 10.1093/carcin/bgp099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Graziano F, Canestrari E, Loupakis F, et al. Genetic modulation of the Let-7 microRNA binding to KRAS 3′-untranslated region and survival of metastatic colorectal cancer patients treated with salvage cetuximab-irinotecan. Pharmacogenomics J. 2010;10:458–64. doi: 10.1038/tpj.2010.9. [DOI] [PubMed] [Google Scholar]
- 19.Zhang W, Winder T, Ning Y, et al. A let-7 microRNA-binding site polymorphism in 3′-untranslated region of KRAS gene predicts response in wild-type KRAS patients with metastatic colorectal cancer treated with cetuximab monotherapy. Ann Oncol. 2011;22:104–09. doi: 10.1093/annonc/mdq315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ratner E, Lu L, Boeke M, et al. A KRAS-variant in ovarian cancer acts as a genetic marker of cancer risk. Cancer Res. 2010;70:6509–15. doi: 10.1158/0008-5472.CAN-10-0689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hollestelle A, Pelletier C, Hooning M, et al. Prevalence of the variant allele rs61764370 T>G in the 3′UTR of KRAS among Dutch BRCA1, BRCA2 and non-BRCA1/BRCA2 breast cancer famlies. Breast Cancer Res Treat. 2010 doi: 10.1007/s10549-010-1080-z. published online July 30. DOI:10.1007/s10549-010-1080-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. [accessed Jan 1, 2008];miRNA profiling. http://www.appliedbiosystems.com/absite/us/en/home/applications-technologies/real-time-pcr/mirna-profiling.html.
- 23.Kibriya M, Jasmine F, Roy S, Paul-Brutus R, Argos M, Ahsan H. Analyses and interpretation of whole-genome gene expression from formalin-fixed paraffin-embedded tissue: an illustration with breast cancer tissues. BMC Genomics. 2010;11:622. doi: 10.1186/1471-2164-11-622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smyth GK. Limma: linear models for microarray data. In: Gentleman R, Carey V, Huber W, Irizarry R, Dudoit S, editors. Bioinformatics and computational biology solutions using R and bioconductor. Springer; New York, USA: 2005. pp. 397–420. [Google Scholar]
- 25.van’t Veer LJ, Dai H, van de Vijver MJ, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–36. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 26.Johnson SM, Grosshans H, Shingara J, et al. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–47. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 27.Croonquist PA, Linden MA, Zhao F, Van Ness BG. Gene profiling of a myeloma cell line reveals similarities and unique signatures among IL-6 response, N-ras-activating mutations, and coculture with bone marrow stromal cells. Blood. 2003;102:2581–92. doi: 10.1182/blood-2003-04-1227. [DOI] [PubMed] [Google Scholar]
- 28.Creighton CJ, Hilger AM, Murthy S, Rae JM, Chinnaiyan AM, El-Ashry D. Activation of mitogen-activated protein kinase in estrogen receptor α-positive breast cancer cells in vitro induces an in vivo molecular phenotype of estrogen receptor α-negative human breast tumors. Cancer Res. 2006;66:3903–11. doi: 10.1158/0008-5472.CAN-05-4363. [DOI] [PubMed] [Google Scholar]
- 29.Iliopoulos D, Hirsch H, Struhl K. An epigenetic switch involving NF-κB Lin28, let-7 microRNA, and IL6 links inflammation to cell transformation. Cell. 2009;139:1–14. doi: 10.1016/j.cell.2009.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Meylan E, Dooley A, Feldser D, et al. Requirement for NF-κB signalling in a mouse model of lung adenocarcinoma. Nature. 2009;462:104–08. doi: 10.1038/nature08462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barbie D, Tamayo P, Boehm J, et al. Systematic RNA interverence reveals that oncogenic KRAS-driven cancers require TBK1. Nature. 2009;462:108–12. doi: 10.1038/nature08460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Atchley DP, Albarracin CT, Lopez A, et al. Clinical and pathologic characteristics of patients with BRCA-positive and BRCA-negative breast cancer. J Clin Oncol. 2008;26:4282–88. doi: 10.1200/JCO.2008.16.6231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oh AS, Lorant LA, Holloway JN, Miller DL, Kern FG, El-Ashry D. Hyperactivation of MAPK induces loss of ERα expression in breast cancer cells. Mol Endocrinol. 2001;15:1344–59. doi: 10.1210/mend.15.8.0678. [DOI] [PubMed] [Google Scholar]
- 34.Santen RJ, Song RX, McPherson R, et al. The role of mitogen-activated protein (MAP) kinase in breast cancer. J Steroid Biochem Mol Biol. 2002;80:239–56. doi: 10.1016/s0960-0760(01)00189-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.