Abstract
Conus species are characterized by their hyperdiverse toxins, encoded by a few gene superfamilies. Our phylogenies of the genus, based on mitochondrial genes, confirm previous results that C. californicus is highly divergent from all other species. Genetic and biochemical analysis of their venom peptides comprise the fifteen most abundant conopeptides and over 50 mature cDNA transcripts from the venom duct. Although C. californicus venom retains many of the general properties of other Conus species, they share only half of the toxin gene superfamilies found in other Conus species. Thus, in these two lineages, approximately half of the rapidly diversifying gene superfamilies originated after an early Tertiary split. Such results demonstrate that, unlike endogenously acting gene families, these genes are likely to be significantly more restricted in their phylogenetic distribution. In concordance with the evolutionary duistance of C. californicus from other species, there are aspects of prey-capture behavior and prey preferences of this species that diverges significantly from all other Conus.
Keywords: biodiversity, accelerated evolution, gene superfamilies, cone snail, exogenomics, phylogenetic relationships
Introduction
The study of model organisms has revealed that many polypeptide gene families with endogenous functions are evolutionarily conserved (Drosophila 12 Genomes Consortium, 2007); it is their expression patterns that characteristically differ between taxa. A well known example among eukaryotes is the set of enzymes that are encoded by highly conserved “housekeeping genes” (Tatusov et al., 1997). However, although phenotypic differences among species are in part a consequence of different expression patterns, this alone would seem inadequate to explain the great exuberance of life, the millions of different living species.
It was recently suggested that specific classes of genes diverge very rapidly and the functional gene products they encode differ significantly, even among closely related species (Olivera, 2006). One class of these rapidly diversifying genes include those whose gene products do not act endogenously within the organism, but instead play a role in interactions with other organisms. Even closely related species may exhibit distinct biotic interactions if they differ in their prey, predators and competitors. Thus, those gene products that either directly or indirectly mediate interactions specific to an organism’s particular ecological niche would be expected to diverge rapidly within a biodiverse lineage. The persistence of these gene families, however, has not been systematically evaluated.
Venomous animals seem particularly suitable for studying such genes, since a large proportion of the major gene products expressed in venom clearly target other organisms. It is becoming clear that acquiring venom is often correlated with major adaptive radiation events. At least three major taxa (reptiles, arachnids and prosobranchs) have undergone such events (Espiritu et al., 2001; Vidal, 2002; Vidal and Hedges, 2002; Fry et al., 2003a; Fry et al., 2003b; King, 2004). One of the characters underlying these radiation events must be the acquisition an effective venom delivery system; i.e., a hollow tooth (Vonk et al. 2008). Yet fundamental questions regarding the molecular evolutionary processes that occur still remain: does the appearance of novel toxin gene families promote speciation events? Are the venom components a result of common selective pressures?
The assessment of the linkage between venom components and phylogeny began with snakes: work on phylogenetic distribution and recruitment of venom toxins of snakes was initiated over fifty years ago (Kochva, 1963; Kochva, 1965; Kochva 1984). In the 90’s, some began to cast their doubts upon the utility of venom as a taxonomic tool (Chippaux et al., 1991; Daltry et al., 1996). Yet in 2006, Fry and colleagues demonstrated that venom evolved in lizards prior to the appearance of snakes and as a result, recruitment of venom is now recognized as a synapomorphy of Toxicofera, a new reptilian clade consisting of suborders Iguania and Serpentes (Fry et al., 2006). Regardless of this major breakthrough in snake venom evolution, the utility of venom peptide sequences as characters for phylogenetic work is still being debated (reviewed extensively in Chipman, 2009 and Fry et al., 2009). For venomous molluscs, the relationship of venom components to phylogeny is only beginning to be assessed.
Given the extensive molecular data sets collected for many of the living representatives, the superfamily Conoidea (Taylor et al., 1993), is especially useful for this purpose. Encoding peptide toxins are produced by the Conoidea include about 10,000 species of venomous marine snails (Olivera, 1997; Olivera, 2006; Terlau and Olivera, 2004). These venoms are extremely complex, with each species producing a repertoire of up to 200 different products within the venom duct alone. In the best-studied group, the cone snails, it has been shown that these powerful cocktails are encoded by a relatively small number of gene superfamilies that exhibit an unprecedented rate of accelerated evolution (Duda and Palumbi, 1999; Duda and Palumbi, 2000; Duda, 2008; Duda and Remigio, 2008; Remigio and Duda, 2008). These provide a basis for evaluating the presence or absence of venom gene families in different lineages of this large and diverse molluscan group, including among others the Conidae, the Turridae and the Terebridae (Bouchet and Rocroi, 2005; Taylor et al., 1993).
Approximately 10 different superfamilies have been characterized from cone snails; three of these are a major presence in the venom ducts of most cone snail species: the A, M and O gene superfamilies that encode Conus peptides containing 2-3 disulfide bonds (Jones and Bulaj, 2000; Terlau and Olivera, 2004). Are these gene superfamilies prominently expressed across the entire range of venomous molluscs? A tentative answer was provided by the analysis of genes expressed in the venom duct of Lophiotoma olangoensis, a species belonging to the family Turridae, generally acknowledged to be distant from Conus both by morphological and molecular criteria (Puillandre et al., 2008; Taylor et al., 1993). All three major gene superfamilies of Conus peptides were missing, and a different spectrum of gene superfamilies were found — only one minor superfamily was present in both Conidae and Turridae (Watkins et al., 2006). A similar recent analysis of Impages hectica (Terebridae, Conoidea), revealed a lack of overlap with gene superfamilies expressed in Conus venom ducts (Imperial et al., 2007).
In order to further assess the distribution of Conus gene superfamilies, we have undertaken a comprehensive investigation of a species traditionally included within the genus, Conus californicus. Previous molecular work indicated (and our work using three mitochondrial gene fragments support) that this species is distant from the majority of other Conus. The phylogenetic divergence from other Conus revealed by the molecular data make this species ideal for assessing which of the standard conopeptide-gene superfamilies are conserved throughout the genus Conus.
We also address a complementary issue: gene products expressed in the venoms of cone snails are highly post-translationally modified. In at least some branches of the superfamily Conoidea (such as in the family Terebridae), venom peptides are not highly post-translationally modified (Imperial et al., 2007). We determined whether the post-translational modifications found in cone snail peptides persist in this outlier. As will be demonstrated, this comprehensive study of superfamilies and their gene products from C. californicus venom ducts has provided definitive answers for these issues. The results of this study provide a base-line data set for the distribution of superfamilies across diverging phylogenetic lineages.
Materials and Methods
Rapid Population Assessments and Collection of Specimens
Individual specimens (15-22 mm) of Conus californicus were collected during an afternoon ebb tide on the western coast of Baja California, 40 km north of Ensenada, Mexico. Collection was planned so that it would coincide with a slack tide event, which occurred on the first three days of December, 2005. On this intertidal reef flat of mostly solid, exposed limestone benches, haphazardly-spaced sand-filled tidal pools were completely engulfed by seagrass. Population densities within the pools were rapidly assessed by first estimating an area equivalent one of the plastic, five (5) gallon buckets used for field collections (~0.073 m2), and then counting every individual within that area. Mean density values were calculated from five such rapid assessments. A total of 150 specimens were collected; as a mater of policy, all individuals <15mm were not taken.
Laboratory Observations of Foraging Behavior
On December 5, 2005, the Conus californicus were transported to the University of Utah and transferred to a preconditioned artificial seawater aquarium. A second precoditioned aquarium (70×35×40 cm3) containing sand 5 cm deep and live rock was divided in three by positioning two perforated, acrylic glass dividers 0.5 cm thick at 23 cm intervals along the length of the tank. Of the 150 specimens collected, 30 were divided into groups of 10 and placed into each aquarium partition; the remaining specimens were either dissected or maintained in separate aquaria.
Observations of foraging behavior began the following day and continued for 12 months. During these observations, C. californicus were presented a variety of fresh and salt water fish purchased from local aquarium stores and/or Canadian night crawlers (Lumbricus terrestris) and closely monitored for 4-6 hr. Multiple still photographs were taken using a digital camera (Canon Powershot S3 IS) during attack sequences, which usually lasted for ~30 min, and at intervals of 6, 12, and 24 hr, thereafter. Prey not attacked after 6 hr were removed from the tank.
Behavioral observations of predation on shrimp were conducted by the Centro de Investigacion Cientifica Y de Educacion (CICESE; Ensenada, Mexico). CICESE’s close proximity to the collection site afforded repeated opportunities for collecting additional specimens and field observations. Conus californicus used in these experiments were collected two different times per year (i.e., from both rainy and dry seasons) over the course of three different collect trips. Snails were housed (65 snails per tank) in 40-L polycarbonate tanks (40x30x30 cm3) supplied with filtered, continuous-flow seawater at an exchange rate of 20% per day. The temperature of this natural water supply is maintained at 20 C. These snails were fed with two live shrimp every two weeks and were observed for a period of nine months.
Isolation of peptides from venom ducts
Venom ducts were dissected from twenty-one live snails and immediately flash frozen. The ducts remained frozen until resuspention on ice in 1.5 mL of B45 (45% AcN:55% H2O w/ 0.2% TFA). The suspension was quickly homogenized using a hand-held Teflon pestle that fits into an Eppendorf tube. The resulting homogenate was further mixed via sonication for 15 seconds, and spun in a Jouan CR412 refrigerated tabletop centrifuge at 5000 rpm for 15 min. The supernatant was diluted 5X with 0.1% TFA, and applied in seven separate runs onto a Vydac C18 analytical column. HPLC elution was performed using a gradient of 5-50% B90 in 0.1% TFA. All major elution peaks were collected as separate fractions.
A complete mass/hydrophobicity profile of Conus californicus (Ensenada, MX) venom fractions was compiled using matrix-assisted laser desorption ionization (MALDI) mass spectrometry in both linear and reflector modes (courtesy of Dr. Ron Kaiser of the Salk Institute, La Jolla, California; data not shown). Venom fractions containing mass signatures within the range of 1-6 kDa were reduced in the presence of 10 mM dithiothrietol, alkylated using 4-vinylpyridine reagent (Imperial et al., 2003), and further purified using the same Vydac C18 analytical column procedure. A small portion of these fractions (40-200 pmol) was submitted for MALDI-TOF analysis (courtesy of the Mass Spectrometry and Proteomics Core Facility, University of Utah).
Sequence determinations of the major alkylated components within subfractions were carried out by Edman degradation using the Applied Biosystems Model 492 Sequenator (courtesy of Dr. Robert Schackmann of the DNA/Peptide Facility, University of Utah). Post-translational modifications were determined using a combination of residue retention profiles, and differences between experimental versus predicted masses for an AA sequence.
Cloning and Sequencing of 12S, 16S and COI gene
Genomic DNA was extracted from 20 mg tissue using the PUREGENE DNA Isolation Kit (Gentra System, Minneapolis, MN). The resulting DNA became the template for polymerase chain reaction (PCR) protocols using oligonucleotide primers corresponding to the 12S, 16S and COI gene segments of the mitochondrial genomes as described elsewhere (Espiritu et al., 2001). PCR products were purified and concentrated using the High Pure PCR Product Purification Kit (Roche Diagnostics, Indianapolis, IN). Sufficient plasmid concentrations for sequence analyses were produced by insertion of the purified PCR products into the pAMP1 vector and transformation into chemically competent E. coli DH5α cells using the CloneAmp pAMP System for Rapid Cloning of Amplification Products (Invitrogen). The complete nucleic acid sequences for12S, 16S and COI-encoding clones were determined (courtesy of the DNA Sequencing Core Facility, University of Utah) (GenBank numbers: EF547567.1 EU015668, EU015729, EU015736, EU685518, EU685626, EU812758.1, FJ868110, FJ868112, FJ868118, FJ868119 and FJ868152-FJ868159 for the COI gene; AF036534, EU078939.1, EU685655, EU685783, FJ868054, FJ868057 and FJ868137-FJ868151 for 16S gene; EU682296.1, EU685363, EU685491, FJ868043, FJ868044, FJ868048, FJ868049 and FJ868123-FJ868136 for 12S gene; C. californicus conotoxins - see Table 1).
Table 1.
Mature toxin sequences predicted from cDNA clones from a Conus californicus venom duct (following Terlau and Olivera, 2004)
| Two Disulfides | GenBank # | |
|---|---|---|
| CysPattern #I/II | cl1.1 | DWNWGRCCFLSGCFECW |
| cl1.2 * | MLLLCCRQGPVCFIPLNEWPCSRM | |
| cl1.3 * | CCKKHHGCHPCRGK | |
| Cys Pattern #V | cl5.3 * | NSEDGSPYPGPGQQPNCCKWIPVTCCNR |
| Cys Pattern #XIV | cl14.1a | GDCPPWCVGARCRAEKC |
| cl14.1b | GDCPPWCVGARCRAGKC | |
| cl14.2a | RECPPWCPTSHCNAGTC | |
| cl14.2b | RECPPRCPTSHCNAGTC | |
| cl14.2c | RDCPPWCPTSHCNAGTC | |
| cl14.3 | RQCPPWCSGEPCRKGTC | |
| cl14.4 | GCPAECPDTCSSSGSCAPDFIG | |
| cl14.5 | GCPADCPNTCDSSNECSPNFPG | |
| cl14.6 | GCPADCPNTCDSSNKCSPGFPG | |
| cl14.7 | DCGRCGLGQICDAGACRPSTMM (partialpropeptide) | |
| cl14.8 | DCGRCPLGQYCDAEAGMCKPTLIM (nopropeptide) | |
| cl14.9 | GCVANCQANQTGIDCIKYCGIGIGRRDITQQ | |
| cl14.10 | HVTCFYVKFGCKHTECITTIVFCWQTASDISSV | |
| cl14.11 | SDCSGMSDGTSCGDTGVCQNGLCMGAGS | |
| cl14.12 | AACKAACKKGAKLILKAAAPLASQVCGPACNAALAKLEKIADDINDDDD | |
| Three Disulfides | ||
| CysPattern #VI/VII |
cl6.1 | CLAGSARCEFHRPSTCCSGHCIFWWCA |
| cl6.1a | CLAGSARCEGHKPSSCCSGHCIFWWCA | |
| cl6.2 | NCIPKNHGCGLLHHSTNCCTPTCLIVCF | |
| cl6.3 | CIGGGDPCEGHRGYTCCSEHCIIWVCA | |
| cl6.4 | YCVPKSGLCTIFQPGKCCSGWCLIYRCT | |
| cl6.5 | CIPDHHGCGLLHHSTYCCNGTCFFVCIP | |
| cl6.6a | GCKSKGSFCWNGIECCGGNCFFACIY | |
| cl6.6b | GCKSKGSFCWNGIECCGGNCFFACVY | |
| cl6.7 | ECSESGEWCGLDPALCCGSSCFFTCN | |
| cl6.8 | YCSDSGGWCGLDPELCCNSSCFVLCG | |
| cl6.9 | DCKTKGSVCFASSECCIQDCWFVCLY | |
| cl6.10 | GCKTKGTWCWASRECCLKDCLFVCVY | |
| cl6.11 | DCGPWCWGQNKCCPDESCRSLHESCT | |
| cl6.12 | DDKSNCPISHPNYCSFTPVCCKHECLSNNKCSSSEFIPGQ | |
| cl6.13 | DGEEFPCAGTMADCRGLADNSVCCDTGKCIGEVCYY | |
| cl6.14 | GWWGPPSNCWVCTGFMKCCEHESHCMTFPTQYNRECK | |
| cl6.15 | QGQSQFGEQCTGHLDCFGDLCCFDGYCIMTSWIWPCNW | |
| cl6.16 | QWPFQQWAPCTGHWDCPGDRCCFAGYCLETTPSCD | |
| CysPattern #IX | cl9.1 | CTTCNMCLKGHCGCSPDCGSC |
| cl9.2 | GECDGKKDCITNDDCTGCLCSDFGSYRKCA | |
| cl9.3 | FPCNAGNCACLPLDSYSYTCQSPTSSTANCEGNECRSEADW | |
| cl9.4 | FPCNPGGCACRPLDSYSYTCQSPSSSTANCEGNECVSEADW | |
| cl9.5 | TFPCSSGLCACLPLDSYSYICLSPSSSTANCENDECISEDDW | |
| cl9.6 * | GQGGCVPPGGGRCKANQACTKGGNPGTCGFQYDLCLCLRN | |
| Four Disulfides | ||
| Cyspattern #XI | ||
| cl11.1 | FNENLSELNSACDDAEWTCAWSRTCCSRNCCRGICVSRYYECP | |
| cl11.2 | FCTEIGKDCGTSWECCEDCCIHGTCSHESNCANFKLR | |
| cl12.1 | GVCSTPEGSCVHNGCICQNAPCCHASGCNWANVCPGFLWDKN | |
| cl12.2 | DVCDSLVDGRCIHNGCFCEESKPNGNCCDTGGCVWWWCPGTKWD | |
| cl12.3 | DVCDSLVGGNCIHNGCWCDQEAPHGNCCDTDGCTAAWWCPGTKWD | |
| Cyspattern #VIII | ||
| cl-S1 1 | YDAPYCSQEEVRECHDDCSGNPVRDACQCAYDPAGSPACDCYCVEPWRR | |
| cl-S2 1 | YDAPYCSEEELQACDCSHNPVRDACLCQYDPAGSPACECFCVEPWRR | |
| cl10.1 | SDPQACEPTISGGEMICRDEVCASTGCNCGYNIAKAHCYCACP | |
These sequences are predicted to undergo C-terminal amidation.
As will be discussed in the Analysis of Sequences section, these clones encode unconventional peptides that belong to the S-superfamily.
Preparation of cDNA from the Venom Duct of Conus californicus
Three independent cDNA libraries were created using venom ducts (~10-20 mg wet weight) from separate snails. Individual venom ducts were dissected, immediately placed in 300 μL of RNALater® (Ambion, Inc., Austin, TX) and stored at −20 °C until processing could be completed. Total venom duct RNA was extracted using the TRIzol Total RNA Isolation technique (Invitrogen). cDNA libraries were prepared from 1 μg Conus californicus RNA using the SMART PCR cDNA Synthesis kit (Clonetech Laboratories, Palo Alto, CA). Two hundred and eighty-five DNA sequences from isolated bacterial colonies were determined using standard ABI automated sequencing protocols optimized for the M13 forward and reverse primers used for the PcDNA-Lib vector.
DNA Sequence Analysis
All sequences obtained from C. californicus libraries were analyzed for translation products using the EditSeq program (DNASTAR, Inc., Madison, WI). Signal peptide cleavage sites were predicted using both the Analyze Signalase 2.03 and SignalP 3.0 software (Imperial et al., 2007). Using proprietary software, putative conopeptide transcripts were exhaustively aligned using holotypes for each respective conopeptide superfamily as references for first attempts towards classification.
Phylogenetic Analysis
Sequences obtained for the concatenation of the 12S, 16S and COI genes were analyzed separately and were automatically aligned with Bioedit (Hall, 1999); 12S and 16S alignments were then modified by eye; several fragments with an ambiguous alignment were removed for the analyses. In this case, the best model of sequence evolution was determined for each gene using TreeFinder (Jobb, 2008), following the AICc criterion. Phylogenetic trees were reconstructed using Maximum Likelihood (ML) and Bayesian Analyses (BAs). ML analyses were performed with Treefinder, using the substitution model, and a search depth of 2. Support for nodes was estimated with a bootstrap analysis (1,000 replicates). BAs were performed with Mr Bayes (Huelsenbeck et al., 2001). Two independant analyses were run, each consisting of four markov chains and 5,000,000 generations. When the log-likelihood score stabilized, a consensus tree was calculated after omitting the first 25% trees as burn-in. Only the number of substitution types (according to the best model as determined previously) was fixed, the other parameters of the substitution model being estimated during the Bayesian analysis.
Several outgroups were used: Thatcheria mirabilis, Lophiotoma cerithiformis and Hastula strigilata, placed respectively in the families Conidae, Turridae and Terebridae (Conoidea; Taylor et al., 1993), and finally Harpa sp. (Harpidae), used as a distant outgroup to root the tree.
To increase the density of the taxa surveyed, we performed a second analysis using 12S and 16S rRNA sequences. These were aligned with Clustal and then refined by eye to correct obviously homologous regions that Clustal failed to recognize. We refined the alignments further with Rcoffee (Notredame et al. 2000) to account for rRNA secondary structure. Because Rcoffee runs are restricted to 50 taxa and 1000 base pairs, the Clustal alignments were divided into smaller subsets, aligned with Rcoffee and then concatenated for further analysis. Alignments within Rcoffee are guided by secondary structure characteristics (Notredame et al. 2000). The color-coded CORE indices were used to identify the best among of alternative alignments that include the Clustal and Rcoffee alignments and hand refined versions of each.
The 12S rRNA and 16SrRNA sequences were concatenated and analyzed using MrBayes with maximum likelihood model parameters partitioned by gene. Each analysis comprised two simultaneous runs with four chains each. Two million generations reduced the average standard deviation of the split frequencies below 0.01. Plots of the number of generations against the maximum likelihood scores indicated equilibrium. Further diagnostics included the potential scale reduction factor (PSRF) that measures the fit of branch length and all parameters. Trees and parameters from the first 25% of the generations were discarded (the burn in) after completion of the MCMC (Monte Carlo Markov Chain) search.
For the maximum likelihood analyses, we estimated sequence evolution model parameters from a neighbor-joining (NJ) tree using PAUP (version 4b10, Swofford, 2002). We used the PAUP block of Modeltest (Posada and Crandall, 1998) that tests 56 models of sequence evolution. Rather than using the full Modeltest package, we deliberately estimated each model independently of the others and exported the the parameter estimates and the likelihood scores to an Excel spreadsheet to calculate likelihood statistics for nested comparisons and AIC statistics for non-nested comparisons (Akaike, 1974) of the likelihood parameters. This was done to avoid the possible bias inherent in the Modeltest algorithm that uses step-wise comparisons that can be influenced by the order in which the comparisons are made; such algorithms do not thoroughly explore model space (Sullivan and Joyce, 2005). The optimized models were implemented in a heuristic maximum likelihood search followed by analysis of 1000 bootstrap replicates to place confidence limits on the tree (Phyml, Guindon and Gascuel, 2003).
Conotoxin evolution in Conoidea
Results obtained from the analysis of C. californicus and data from the literature (McIntosh et al., 1995; England et al., 1998; Walker et al., 1999; Lirazan et al., 2000; Jimenez et al., 2003; Santos et al., 2004; Buczek et al., 2005; Corpuz et al., 2005; Imperial et al., 2006; Watkins et al., 2006; Imperial et al., 2007) were used to determine the presence or absence of the different superfamilies of conotoxins characterized within the genus Conus. The characters were then mapped on the phylogenetic tree obtained from the mitochondrial sequences using Mesquite V. 2.01 (Maddison and Maddison, 2007), using the option “tracing character history” and the parsimony ancestral reconstruction method.
Results and Discussion
Combined rRNA, 16S rRNA and COI Phylogeny
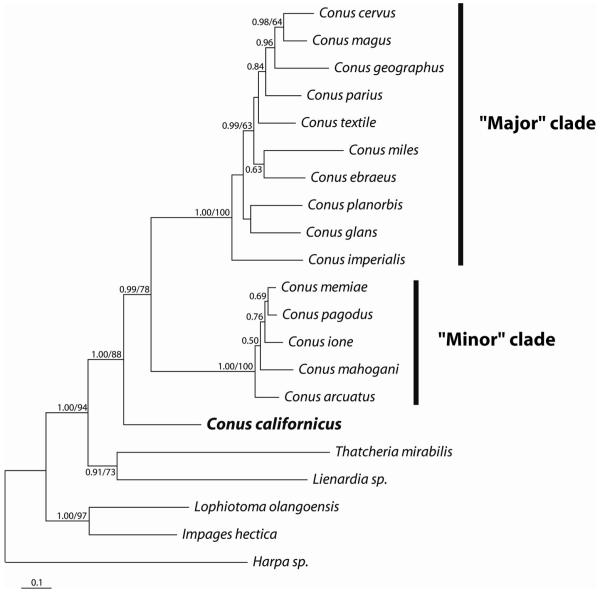
The tree obtained from the partitioned analysis of the COI, 12S and 16S rRNA genes is shown in Figure 1. Conus californicus is clearly divergent from the rest of the Conus species, which are, in turn, separated into a minor clade that we refer to as Conasprella (see also Bandyopadhyay et al., 2008) and a the major clade comprising the remaining Conus species. These results are consistent with previous phylogenetic analyses (Monje et al., 1999; Duda et al., 2001; Espiritu et al., 2001; Duda & Kohn, 2005). This level of divergence is comparable with genetic distances found among genera in other Conoideans (Puillandre et al., 2008; Holford et al., 2009).
Figure 1.
Phylogenetic tree inferred from the concatenation of 12S and 16SrRNAs and COI sequences from 16 Conus species and five outgroup species. Branches are labelled with posterior probabilities from the Bayesian analysis on the left and bootstrap percentages on the right.
Combined 12S rRNA and 16S rRNA Phylogeny
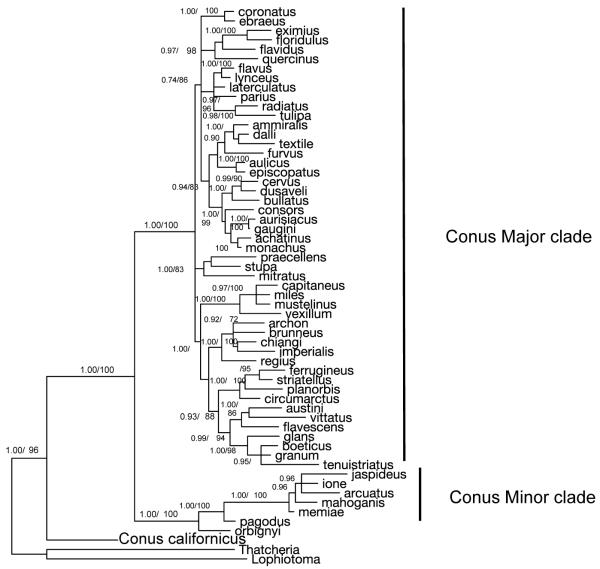
A larger Conus phylogeny representing more species and inferred from the concatenated 12S and 16SrRNA sequences is shown in Figure 2. This tree shows the same distinct Conus lineages as does the less speciose tree inferred from three genes. High posterior probabilities and bootstrap proportions provide confidence that Conus californicus is evolutionarily distant from the all other Conus species. The wide divergence of C. californicus from the other Conus is consistent with the fact that the biology of this species is distinctive from other cone snails.
Figure 2.
Phylogenetic tree inferred from the concatenation of 12S and 16S rRNAs sequences from 57 Conus species and 2 outgroup species. Branches are labelled with Bayesian posterior probabilities (left) and maximum likelihood bootstrap percentages (right or bottom) that are greater than 50% Even with the addition of far more Conus species, the tree shows strong support for placing Conus califonicus on a long branch well outside of and sister to the other Conus speices.
Behavioral observations of prey-capture by Conus californicus
This species has adapted to cooler waters than most other Conus species. Most shallow-water tropical cone snail species are highly specialized with regard to their prey (Röckel et al., 1995). In contrast, C. californicus attacks diverse prey belonging to at least four different phyla. The species has the most generalized diet described for the genus, having been observed feeding upon taxa as diverse as molluscs, fish, and worms (Saunders and Wolfson, 1961; Stewart and Gilly, 2005). C. californicus were observed eating a shrimp during the field collections conducted in this study.
This has been hypothesized as being the result of C. californicus being the only Conus species inhabiting temperate coastal zones within the Americas. No other Conus are found off the Western coast of Baja California and California, and thus, C. californicus has filled niches often divided among multiple cone snail species. In addition, a variety of unusual feeding behaviors have been documented, including organized/cooperative attacks upon other snails (Saunders and Wolfson, 1961).
The collection site is an intertidal reef flat comprising solid exposed limestone benches and sand-filled tidal pools completely engulfed in seagrass. Here specimens of C. californicus were so common that individuals crawling on top of the exposed seagrass could be observed about thirty minutes after the tide reached its lowest point. Rapid population assessments within this collection site suggested that, at least in Ensenada, MX, C. californicus populations exist in small dense pockets where they reach population densities of up to 50-100 individuals per square meter (79 ± 39 m-2).
When transferred to aquaria at densities comparable to that found in their natural environment, C. californicus will feed on prey that typically would not succumb to an encounter with a single snail. Most other Conus will regurgitate a prey item that they can’t fully engulf, a strategy that is thought to minimize the likelihood of exposure to predation. However, when placed in aquaria at these high densities, C. californicus cooperate to capture a larger fish in ways that mimic pack behavior (Figure 3): 1) the first snail to encounter the prey will use the “hook-and-line” technique to tether the prey, which slows escape and gives other snails a chance to approach; 2) multiple snails would then rapidly converge on the tethered fish in a manner reminiscent of a marauding wolf pack; and 3) all of the C. californicus participating in the attack feed together, sucking whatever they could from the fish. Closer observations revealed that most of the pack would approach with their proboscises extended, but contrary to multiple attacks against molluscs (Saunders and Wolfson, 1961, Stewart and Gilly, 2005), joiners would not sting unless the prey was struggling violently. With this unique pack-strategy, C. californicus could reduce the use of its venom while still capitalizing on encounters with larger prey, even though signs of venom ineffectiveness were often observed (i.e., the prey were often still struggling even after fully surrounded by the pack). In the case of a larger fish, they had pulled the prey under the strata after three hours, but after four hours, the fish was largely intact. What was curious about this attack was that after 24 hr, they still had not consumed the whole fish; in fact, they had left the intact carcass (minus the accessible soft tissues) and returned to their hiding places under the sand. So it seems that although C. californicus can exploit fish larger than a single individual as a prey item, they aren’t fully prepared to consume such prey when fresh. Similarly C. californicus made “wolf-pack” attacks on shrimps released into the CICESE flow-through seawater tank (see Figure 3). Multiple feedings were observed over the course of nine months. C. californicus were always observed to attack a single shrimp in seemingly coordinated groups of 5-10 individuals at a time, often stinging the shrimp several times between the legs. However, close observation suggested that the most effective envenomation site was directly into the shrimp’s eye. Regardless of route, once the shrimp became immobilized by these attacks, several began feeding together. A video of two such events is available from CICESE upon request. This is the first documentation of any Conus species preying on a crustacean.
Figure 3.
Shells and prey capture behavior of Conus californicus “Hinds, R.B” Reeve, L.A., 1844. Conus californicus, the California cone, inhabits the temperate costal waters off California and Baja California. Live animals (top left) were collected on the Western coast of Baja California roughly 40 km north of Ensenada, Mexico. A photograph of a shell from one of these specimens is shown (top right). An interesting feature of C. californicus feeding behavior is that individual snails cooperate in order to subdue prey items larger than themselves, like a marauding wolf pack converging on larger prey. Such behavior has never been documented for any other Conus species.
cDNA clones from Conus californicus
Mature cDNA clones that encode putative precursors were found in 120 of the 285 cloned sequences analyzed (42%). Normal conopeptide criteria were used for predicting mature peptide toxins after translation and proteolytic processing from cloned gene sequences (Olivera et al., 1999; Woodward et al., 1990). These are listed in Table 1.
All cDNA clones encoding putative conopeptide sequences from C. californicus can be divided into two distinct classes. The first class encode polypeptides predicted to generate peptides with cysteine patterns that resemble those found other Conus spp. (Terlau and Olivera, 2004). A second class includes cDNA clones predicted to encode conopeptides that deviate from the known cysteine patterns produced by other Conus. Each class of predicted C. californicus conopeptides is discussed in greater detail in the analysis section that follows.
Correspondence between purified peptides and cDNA clones
The 15 peptides purified from venom (Table 2) were evaluated with respect to what proportion were encoded by the precursors defined by the analysis of venom duct cDNAs (Table 1). For 4 of the 15 peptides (~27%), there was an exact match; in particular, those with two disulfide linkages, cl 14a, cl 14b and cl 14c were exactly predicted by clones 14.6, 14.3 and 14.2a, respectively. A cDNA clone, cl 6.11, also precisely encoded a peptide with three disulfides, which belongs to the O-superfamily, cl 6b. This peptide is highly post-translationally modified; its presence in C. californicus venom was likely also detected by Marshall et al., (2002), who found a major venom component with a mass consistent with cl 6b.
Table 2.
Primary amino acid sequences of major conopeptide constituents purified from the venom of Conus californicus
| Sequence | HPLC peak | cDNA Match | |
|---|---|---|---|
| One Disulfide | |||
| cl tx-1 | DNSCTPKPSCFF | g | none |
| Two Disulfides | |||
| cl 14a | GCPADCPNTCDSSNKCSPGFPG | b | cl 14.6 |
| cl 14b | RQCPPWCSGEOCRKGTC | d | cl 14.3 |
| cl 14c | RECPPWCPTSHCNAGTC | e | cl 14.2a |
| cl 15a | NCPAGCRSQGCCM | a | None |
| cl 5a | DSEDGSOHPGOGQEONCCKWPILTCCN | g | similar to cl 5.3 |
| Three Disulfides | |||
| P-type pattern | |||
| cl 9a | TFEPNAγγCIVDGRCKHRSDWPCγMSSGTTGRCDVSLGACGCSN | c | none |
| cl 9b | DDETTFPCNSGRCACLOEDSHSYTCQSO… | c | similar to cl 9.5 |
| O-type pattern | |||
| cl 6a | DDCTTYCYGVHCCPPAFKCAASGCVRN (N/D) | c | none |
| cl 6b | DCGOWCWGQNKCCODγSCRSLHγSCT | i | cl 6.11 |
| cl 6c | GCWICWGPNACCRGSVCHDYCPS | j | none |
| cl 6d | GDACSLLNGDDCGOGELCCTOSGDHQGTCETSCW | f | none |
| Undesignated pattern | |||
| cl tx-2 | SLCDKOHHNCIDGQTCYHTCCQNGLKCVRYO | b | None |
| Four Disulfides | |||
| cl 12a | GVCSTOEGSCVHNGCICQNAPCCHPSGCNWANVCPGFLWDKN | k | similar to cl 12.1 |
| cl tx-4 | ROKCCCVCGVVGRKCCSTWDKCHOVHLOCOSS* | h | none |
O = hydroxyproline; γ = γ-carboxyglutamate; W = 6-bromotryptophan
C-terminal amidation. Post-translational modifications were determined using a combination of residue retention profiles, differences between experimental masses and the predicted mass for the primary structure, and the primary amino acid sequence predicted from cDNA clones.
There was one case where a peptide exhibited a high degree of similarity to a predicted clone sequence, but was a polymorphic form of the clone (cl 5a and clone cl 5.3). Surprisingly, although cys pattern #VI/VII was the most commonly encountered in the cDNA library (4 peptides in the venom exhibited this pattern), there was only one match between the two groups (cl 6b and cl 6.11). Thus, for 8/15 of the peptides purified from venom, no match could be found; one incomplete sequence (cl 9b) belongs to the same family as clone cl 9.5, but it differs significantly in sequence. These suggest that only a minority of the total chemical diversity to be found in the venom of Conus californicus has been defined by the >60 different sequences obtained from both the cDNA library and venom peptide purification. Thus, the total molecular complexity of Conus californicus venom is potentially comparable to those of other Conus species (generally estimated at between 100 – 200 different peptide sequences).
Analysis of data on Conus californicus venom peptides
The data presented above, and summarized in Tables 1 and 2 reveal that although Conus californicus is phylogenetically distant from most other Conus species, its venom has many of the general properties of other Conus venoms. The venom is an extremely complex mixture of small, disulfide-rich peptides with a variety of unusual post-translational modifications. We purified and sequenced 15 different peptides from the pooled venom (mostly major peaks, thus overrepresenting highly expressed venom components in the sample analyzed).
An independent analysis of three C. californicus cDNA libraries revealed ~50 different peptide sequences. Of the 15 peptides purified from venom, only four peptides (27%) could be matched precisely to a cDNA clone encoding the peptide (after post-translational modification; cl 6b, cl 14a, cl 14b and cl 14c); three (20%) purified venom peptides (cl 5a, cl 9b and cl 12a) were similar to predicted mature peptides encoded by cDNA clones (in those cases, the similarity was sufficient to assess to which gene superfamily the purified peptide belonged); and eight (53%) purified peptides did not match any of the cDNA clones characterized. The minor proportion of peptides that corresponds to a cDNA clone suggests that the ~50 peptide sequences deduced from clones still represent only a minor fraction of the total diversity of C. californicus venom peptides. The perception that the venom is extremely complex is also supported by the HPLC profiles of crude venom shown in Figure 4, and by comprehensive mass spectrometric analyses of crude venom (results not shown).
Figure 4.
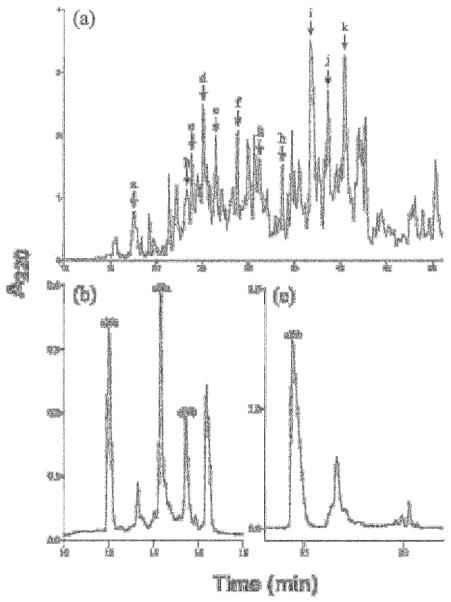
Identification of conopeptides from venom fractions of Conus californicus. (a) Fractionation of C. californicus venom extract on a Vydac C18 analytical column with a linear gradient of 1.0% B90/min at a flow rate of 1.0 mL/min. Lettered arrows represent HPLC fractions used to identify primary amino acid sequences depicted in Table 1. (b) Subfractionation of peak c from panel (a), after reduction with 15 mM DDT and alkylation with 4-vinylpyridine using an elution gradient of 15-40% B90 over 30 minutes; three peptides, cl 6a, cl 9a and cl 9b were sequenced from this fractionation (c) Fractionation of peak i from panel (a), leading to the characterization of cl 6b the most abundant peptide within the venom of Conus californicus, using a Vydac C18 analytical column eluded with a gradient of 15-60% buffer B90 over 60 min. The sequences of the four peptides marked in panels (b) and (c) were deduced from an automated Edman degredation and are presented in Table 1.
Three Conus peptide gene superfamilies are particularly prominent constituents of most previously characterized Conus venoms (Corpuz et al., 2005; Santos et al., 2004; Woodward et al., 1990): the A-superfamily (with most peptides having Cys pattern #I/II), the M-superfamily (with Cys pattern #III) and the O-superfamily (with Cys pattern #VI/VII); six other conotoxin superfamilies have been identified (see Olivera, 2006 for a recent review). In most Conus venoms studied to date, the major peptide toxins expressed are mostly from the A, M and O-superfamilies and the other six superfamilies are not as extensively represented.
In C. californicus, no venom peptides were identified belonging to the A- and M-superfamilies, either by venom purification or cDNA clones. However, four peptides isolated (out of 15) have Cys pattern #VI/VII: [C---C---CC---C---C], characteristic of the O-superfamily. One of these, cl 6b corresponded in sequence to that predicted by a cDNA clone (cl 6.11 – see Table 3A). When the sequence of cl 6.11 is compared to previously characterized O-superfamily peptide precursors, considerable identity is observed in the signal sequence region (see Table 3A) indicating that an O-superfamily assignment is justified.
Table 3.
Signal and mature toxin sequences for some exmaples of the O, I, T, S and J superfamilies. Grey shading: conserved sites for the signal sequence; cys-pattern for the mature toxin
| SIGNAL SEQUENCE | MATURE TOXIN | |
|---|---|---|
| A. O-superfamily | ||
| Cl 6.1 | MKLTTVLVVALLVLAACQFTVT | -CLAGSAR-CEFHRPST-CCSGH-C-IFWW---CA |
| Cl 6.2 | MKLTCVLIVAVLVLTACQFTAA | NCIPKNHG-CGLLHHSTNCCTPT-CLIV-----CF |
| Cl 6.11 | MKLTCVLIIAVLILTACQFIAA | DCGPW----CWGQNK---CCPDESCRSLHES--CT |
| C. magus ω-MVIIA | MKLTCVVIVAVLLLTACQLITA | -CKGKGAK-CSRLMYD--CCTGS-CRSGK----CG |
| C. textile δ-TxVIIA | MKLTCMMIVAVLFLTAWTFATA | WCKQSGEM-CNLLDQN--CCDGY-CIVLV----CT |
| Cl 6.4 | MMTLTFLLVVALCMLTTCHTEN | YCVPKSGL-CTIFQPGK-CCSGW-CLIYR----CT |
| Cl 6.12 | MKFYLLLTAALLLTAVIIEAAP | DDKSNCPISHPNYCSFTPV---CCKHE-CLSNNK---CSSSEFIPGQ |
| Cl 6.13 | MKFPLLFISLALAAFLTRVQDA | DGEEFPCAGTMAD-CRGLADNSVCCDTGKCIGEV----CYY |
| Cl 6.15 | MKLSVKFLLFLMILPLIAGEDM | QGQSQFGEQCTGHLD--CFGDL----CCFDGYCIMTSWIWPCNW |
| B. I-Superfamily | ||
| Cl 11.1 | MKLALTFLLILMILPLTTG | FNENLSELNSACDDAEWTCAWSRTCCSRNCC-RGICVSRYYECP |
| Cl 11.2 | MKMSVTFLLILMILPLFTG | ----------FCTEIGKDCGTSWECCED-CCIHGIC-SHESNCANFKLR |
| C. radiatus 11.3 | MKLCLTFLLVLMILASVTG | --------GPRCWVGRVHCTYHKDCCPSVCCFKGRCKPQSWGCWSGPT |
| C. betulinus 11.1 | MKLCVAFLLVLVILPSVIG | ------GDRRMCLSLGQRCERHSNCCGYLCCFYDKCVVTAIGCGHY |
| C. episcopatus 11.1 | MKLCVTFLLILVILPSVTG | ------GDWGMCSGIGQGCGQDSNCCGDMCCYGQICAMTFAACGP |
| C. T-superfamily | ||
| Cl 1.1 | MRCLPVIVILLLLISSAAAVVE | DWNWGRCCFLSG-CFECW |
| C. marmoreus Mr1a | MRCLPVLIILLLLTASAPGVVV | ---NGVCCGYKL-CHPC |
| C. purpurascens P5a | MRCLPVFVILLLLIPSAPCVDA | -----GCCPKQMRC--CTLG |
| D. S-superfamily | ||
| Cl-S1 | MMSTKGITLFLCLLLLALATS | YDAPYCSQEEVRE-----CHDD-CSGNPVRDACQCAYDPAGSPA-CDCYCVEPWRR |
| Cl-S2 | YDAPYCSEEELQA-----CD---CSHNPVRDACLCQYDPAGSPA-CECFCVEPWRR | |
| Cl 10.1 | -MTTLGMTMLVLLLLLPLATC | SDPQACEPTISGGEMI--CRDEVCASTG----CNCGYNIAKAH--CYCACP |
| C. geographus GVIIIA | MMSKMGAMFVLLLLFT-LASS | ----GCTRT-CGGPK---CTGT-CTCTNSSK-CGCRYNVHPSGWGCGCACSG |
| C. radiatus S-RVIIIA | -MSKMGAMFVLLLLFT-LASS | ----KCNFDKCKGTGVYNCGES-CSCEGLHS-CRCTYNIGSMKSGCACICTYY |
| E. J-superfamily | ||
| Cl 14.1a | MNVTAMFIVLLL-TMPLTD-G | GDCPPWCVGARCRAEKC- |
| Cl 14.1b | MNVTVMFIVLLL-TMPLTD-G | GDCPPWCVGARCRAGKC- |
| Cl 14.2a | MNVTVMFIVLLLLTMPLTD-G | RECPPWCPTSHCNAGTC- |
| Cl 14.2b | MNVTVMFIVLLLLTMPLTD-G | RECPPRCPTSHCNAGTC- |
| Cl 14.2c | MNVTVMFIVLLLLTMPLTD-G | RDCPPWCPTSHCNAGTC- |
| Cl 14.3 | MNVTVMFIVLLL-TMPLTD-G | RQCPPWCSGEPCRKGTC- |
| C. litteratus lt14a | -----MFIVFLMLTMPMTDAG | -ZCPPMCNPG-CEN--CS |
The O-superfamily comprises two groups; one includes the ω-conotoxins (referred to as the ω-branch) and the second includes the δ-conotoxins (referred to as the δ-branch). Clearly, although the sequence of cl 6b precursor is similar to both the ω- and the δ-branches, it is sufficiently divergent from both to make its assignment to either group ambiguous without further clarification of the biochemical mechanism of this peptide.
If the mature peptides are compared, cl 6b has similarly divergent properties from typical peptides of either O- superfamily branch; ω-branch peptides are generally hydrophilic, with a high proportion of charged residues, while δ-branch peptides are characteristically extremely hydrophobic. Typical examples of ω- and δ-peptides are shown in the Table 3A; cl 6b differs from these not only in its general biochemical properties, but in the spacing between Cys residues, and most notably, in the extremely high frequency of post-translationally modified AA found. This peptide has six modified residues, with three different modifications previously identified in Conus peptides: 4-transhydroxyproline, 6-bromotryptophan and γ-carboxyglutamate. Only the first of these is generally found in O-superfamily peptides; the presence of all three would be very unusual in O-superfamily peptides from other Conus.
Of the 15 peptides directly isolated from venom, four have the O-superfamily framework and one can be definitively assigned to the O-superfamily. Thus, an overlap with one Conus gene superfamily, the O-superfamily, is established. The set of 15 venom peptides characterized is notable for the absence of any belonging to the A- or M-superfamilies. Two peptides purified from venom, cl 14b and cl 14c, can be assigned to a newly defined superfamily, as will be documented below.
Identification of superfamilies from cDNA clones: O-superfamily peptides
The cDNA sequences encoding the predicted mature peptides (shown in Table 1) were analyzed for potential sequence similarities to consensus sequence elements characteristic of the established Conus peptide superfamilies. The actual isolation of a peptide from the venom is the definitive evidence that a Conus peptide gene is expressed. Identification of a superfamily through a cDNA clone provides strong evidence for expression, with one caveat – in rare cases, messenger RNAs encoding pseudogenes have been identified from cDNA libraries.
It is clear that the most common class of toxins represented in the cDNA library have the Cys pattern #VI/VII, characteristic of the O-superfamily. The majority of these peptides appear to be bona fide members of the O-superfamily with striking sequence identity in their signal sequences to the two examples of O-superfamily peptides given (shown in Tables 1 and 3A). These peptides can confidently be assigned to the O-superfamily. A minor fraction of these peptides (with Cys pattern #VI/VII) have little or no sequence similarity to the O-superfamily consensus signal sequences (Table 3A). These peptides (encoded by Cl 6.4, Cl 6.12, Cl 6.13 and Cl 6.15) are not members of the O-superfamily, and appear to be a heterogenous group since their signal sequences are different.
Figure 5 shows the inclusion of Conus californicus peptides in the I-, T- and S-superfamilies. Unequivocal evidence that a cDNA clone encodes a peptide belonging to a previously characterized superfamily is provided when both high sequence similarity is present in the signal sequence, and the arrangement of Cys residues in the mature toxin region corresponds precisely to the Cys pattern established for the superfamily. This is the case for two peptides from the C. californicus cDNA library, Cl 11.1 and Cl 11.2. These clearly belong to the I1 superfamily (Buczek et al., 2005). Precursors for these peptides can be aligned with members of the I1 superfamily (Table 3B); not only are there blocks of sequences with considerable identity, the peptides have Cys pattern #XI [C---C---CC---CC---C---C], the signature pattern of mature I-superfamily peptides.
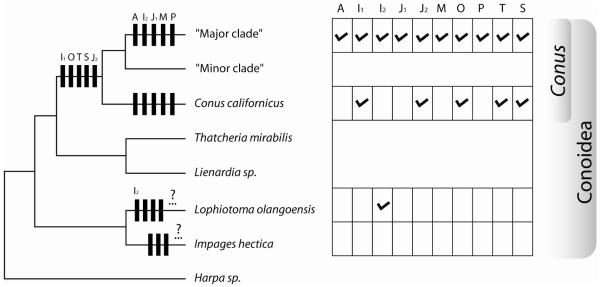
Figure 5.
Reconstruction of conotoxin superfamilies evolution within selected Conoidea. The tree is a collapsed cladogram of Figure 1.The vertical bars on the tree represent the appearance of the corresponding superfamily (unlabeled bars unnamed superfamilies). The apparent absence and presence of each conotoxin superfamily is also reported in the adjacent table (conotoxins in three lineages – T. mirabilis, Lienardia sp. and “minor” clade have not been investigated).
A similar (though more qualified) conclusion can be made for the two C. californicus peptides of the T-superfamily, Cl 1.1 and Cl 1.3. The mature peptides have the minor Cys pattern of the T-superfamily (McIntosh et al., 2000); most T-superfamily peptides have Cys pattern #V [CC-----CC], with a minor fraction having the following Cys pattern [CC----CXXC]. As shown in Table 3C, the similarity in the signal sequence region, and the minor Cys pattern of the T-superfamily makes assignment of these peptides to the T-superfamily persuasive. Another C. californicus peptide, with Cys pattern #V, cl 5a, has a similar sequence encoded by a clone (Cl 5.3) with an incomplete sequence at the N-terminus; the deduced incomplete sequence exhibits no similarity with the two T-superfamily peptides from C. californicus, nor with any T-superfamily peptides from other Conus.
Finally, three unusual C. californicus peptides apparently belong to the S-superfamily, these comprise peptides with 10 Cys residues (Cys pattern #VIII; [C--C---C—C---C---C---C-----CXCXC]) (Table 3D). The sequence identity in the signal sequence region, though not as extensive as for the other superfamilies, is nevertheless considerable. Unlike other S-superfamily peptides characterized, the C. californicus peptides have 8 Cys residues. However, the spacing between Cys residues, particularly of the last three towards the C-terminus (---CXCXC) is strikingly similar. The overall sequence similarity is persuasive enough to assign the C. californicus peptide to the S-superfamily, although more divergence is observed for this set of peptides than for the other superfamilies. The divergence of the C. californicus T- and S-superfamily Cys patterns from those observed for most Conus raises the question of whether the [CC----CXXC] and [C-C---C---C---C---C---C---C] Cys patterns might be ancestral for the T- and S-superfamilies, respectively, with major patterns for each superfamily (T: [CC---CC]; S [C---C---C---C---C---C---C---C-----CXCXC]) having evolved subsequent to the divergence of C. californicus from the majority of Conus species.
A New Gene Superfamily
Next to the C. californicus peptides with Cys pattern #VI/VII, the most prevalent set in the cDNA library were predicted peptides with Cys pattern #XIV [C---C---C---C]. There were a total of 15 different mature toxin sequences defined from the cDNA library with the #XIV pattern (Table 1); this group was an even more prominent component of the set of peptides directly isolated from venom; as many pattern #XIV peptides were characterized as peptides with the #VI/VII pattern.
The peptides with Cys pattern #XIV comprise a heterogeneous group, with multiple superfamilies represented. Of 15 different peptides, six are highly similar to each other (Table 3E). Two of these were also isolated from the venom (see Table 2). A recently described Conus peptide lt14a from Conus litteratus has significant sequence identity to 6 of the 15 C. californicus peptides with pattern #XIV (Peng et al., 2006; see Table 3E). Evidence that the C. litteratus peptide targets neuronal nicotinic acetylcholine receptors has been presented. The preponderance of peptides belonging to this superfamily in C. californicus, combined with the absence of α-conotoxins raises the question of whether these might be the major nicotinic antagonists present in this venom. This hypothesis is presently being tested.
In combination, the characterization of the Cys pattern #XIV peptides from both C. californicus and C. litteratus provide sufficient evidence to define a new superfamily. The peptides have the same Cys pattern as the recently described J-superfamily (Imperial et al., 2006), but differ in the signal sequence, and probably phylogenetic distribution. We propose to call these peptides the J2-superfamily of Conoidean peptides; and will refer to the previously defined J-superfamily as J1 (the prior suggestion by Peng et al., to call this group the L-superfamily would cause a nomenclature problem, since multiple superfamilies can share the same Cys pattern, there would not be enough letters to name superfamilies). Using this nomenclature, the C. litteratus peptide previously described should now be referred to as “αJ2-conotoxin Lt XIVA”.
The compilation of data from literature and results presented here allow us to determine the presence and absence of the ten superfamilies of toxins found within the genus Conus in four lineages of conoideans: the “major” clade within Conus, C. californicus, Lophiotoma olangoensis (Turridae) and Impages hectica (Terebridae). The reconstruction of the evolution of these characters suggest that at least four superfamilies were present in the common ancestor of all the Conus; five others may have appeared during the “major” clade evolution; one appeared in the C. californicus lineage; two superfamilies (I2 et J1) are present in two different lineages, distant from Conus, and suggesting two independent events (Figure 5). Although the presence of a superfamily is unambiguous, its apparent absence could be due to the incomplete sampling that was carried out.
The work to date with many different Conus venoms has led to nine conotoxin superfamilies being recognized (Olivera, 2006; Terlau and Olivera, 2004); we have defined a new conotoxin superfamily. The analysis of C. californicus peptides has demonstrated that five of the 10 conotoxin superfamilies (O-, T-, S-, I1-, and J2) are demonstrably present in C. californicus. We failed to find examples of the other five superfamilies. Although this could be due to incomplete sampling, two of the superfamilies (A- and M-) are prominent components of almost every Conus venom examined. Their absence from both the set of C. californicus peptides purified from venom, as well as from the deduced cDNA library sequences therefore seems noteworthy. An attempt at RT PCR of cDNA using primers for the two superfamilies that successfully amplified toxin precursor sequences across a wide variety of Conus species was also unsuccessful (M. Watkins, unpublished results).
Not a single peptide was found with the Cys pattern suggestive of the M-superfamily (Cys pattern #III: [CC---C---C---CC]). For the A-superfamily, there were three peptides that seem to have the I/II Cys pattern; two were convincingly shown to be members of the T-superfamily and the third had no sequence similarity at all to any A-superfamily peptides. Failure to find members of the P-, I2- and J1-superfamilies is somewhat less surprising, since these are not as dominant in most Conus venoms as are peptides of the A-, M- and O-superfamilies. Although the possibility that more extensive sampling of C. californicus venom could reveal members of these superfamilies cannot be eliminated, they are apparently not prominent components of the C. californicus venom that we have analyzed.
However, even for the superfamilies that have been established to be present in the venom of C. californicus, considerable divergence from peptides in the same superfamilies from other Conus venoms is apparent. One example is peptides in the S-superfamily; in other Conus, S-superfamily peptides have ten cysteine residues, but C. californicus peptides only have eight. Nevertheless, the evidence that these all belong to the same superfamily is compelling.
Thus, the overall impression created by this analysis is that at the gene superfamily level, there is ~50% overlap between C. californicus and the majority of Conus species. Half of the gene superfamilies defined in other Conus were not found in C. californicus, with the absence of two prominent superfamilies (A and M) being particularly striking. The presence of unusual gene families in C. californicus also seems clear; one peptide with eight cysteine residues has an unusual Cys pattern (including three contiguous cysteine residues and an additional pair, a combination never previously observed for any Conus peptide). Thus it appears that, in addition to conventional conotoxin families, the divergence between C. californicus and other species has resulted in the evolution of novel gene superfamilies expressed in this species, but not in other Conus.
The phylogenetic analysis provides a basis for understanding how these divergent features may have evolved. Approximately half of the gene superfamilies expressed in C. californicus venom apparently pre-dated the divergence of C. californicus from other Conus spp. These include one major gene superfamily, the O-superfamily, as well as three other distinctive superfamilies in Conus spp., including the T-superfamily with two-disulfide bonds, the I1-superfamily with four disulfide bonds and the S-superfamily, which in Conus spp., have five disulfide bonds, but in C. californicus only four. Two major superfamilies present in other Conus spp appear to be absent in C. californicus, the A-superfamily that generally encodes the major nicotinic receptor antagonists found in these venoms, and the M-superfamily that encodes the Na channel inhibitors used for paralyzing prey in piscivorous Conus spp. Approximately half of the gene families in C. californicus appear to have novel Cys patterns, not described in any of the superfamilies characterized from species in the major clade within Conus.
Confidence in the ancestral state reconstruction shown in Figure 5 is limited by a number of factors. There are two potential sampling biases; first, several lineages included in the tree have not been investigated. Consequently, any estimate of the time of the appearance of a given superfamily would necessarily be imprecise. For example, the four superfamilies thought to be present in the ancestor of all Conus may be present in Thatcheria or Lienardia. A more difficult problem to address is incomplete sampling, which has multiple facets. Only the genes actively expressed in a particular tissue will be sampled with the methods used. The genes that might be expressed by a species in other stages of development (Duda and Remigio, 2008) or under different environmental conditions will not be found. This is a fundamental limitation with the analysis carried out.
In the absence of complete genome sequences for every species of interest, (with an accompanying perfect bioinformatics analysis), there is no way to be able to state with certainty that a particular gene superfamily is truly absent from a lineage. The best we can do is to contrast the expression patterns found in different lineages, recognizing that the analysis is imperfect. The dilemma in interpretation is highlighted by the tree in Figure 5: the I2-superfamily is not found in Conus californicus, but was found broadly across species in the major clade of Conus, and in the very distant Conoidean lineage represented by Lophiotoma olangoensis (Family Turridae). Does this mean that the I2-superfamily has been secondarily lost in Conus californicus? Is its absence an artifact of incomplete sampling? Is it possible that these superfamilies are present in the genome, but are recruited for expression in venom ducts in a lineage-specific manner? These are all logical possibilities that need to be considered.
Fundamentally, there would appear to be two major possibilities when a limited spotty phylogenetic distribution of superfamilies is found. The genesis and extinction of gene superfamilies may occur at a much more rapid rate than for other genes. An alternative is that the gene superfamilies are present across a much greater phylogenetic range than would be apparent from sampling taxa for their expression in venom ducts, and that the difference between lineages is that an active gene superfamily in one lineage could be “archived” in a different lineage (not expressed at all, or expressed only in other tissues). The data presented in this first sampling of the persistence of gene superfamilies across divergent clades encompass these mechanistic alternatives.
In most cases, the origins of these gene superfamilies are unknown; they could have evolved from endogenous gene families, or created by duplication followed by neofunctionalization (e.g. Duda and Palumbi, 2000). However, it seems clear that in contrast to standard “endogenous” gene families, the phylogenetic distribution and persistence of gene superfamilies will have a different time scale, and that the spectrum of gene superfamilies recruited by particular lineages will mostly differ between divergent lineages. This study is the first calibration of the extent of divergence in gene families between two lineages that diverged from each other at the beginning of the Tertiary period.
Acknowledgements
Support for this work was provided by grants from the NIHGMS PO1 GM048677-13 and the NIHGMS Diversity Supplement Fellowship 3 PO1 GM048677-13S1. We thank Drs. Julita S. Imperial, Ping Chen, and Olga Buczek for lending their expertise and guidance during the purification of the C. californicus peptides.
Footnotes
These authors contributed equally to this work
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bandyopadhyay PK, et al. The mitochondrial genome of Conus textile, coxI-coxII intergenic sequences and Conoidean evolution. Molecular Phylogenetic Evolution. 2008;46:215–23. doi: 10.1016/j.ympev.2007.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchet P, Rocroi J-P. Classification and nomenclator of Gastropods families. Malacologia. 2005;47 [Google Scholar]
- Buczek O, Bulaj G, Olivera BM. Conot oxins and the posttranslational modification of secreted gene products. Cell Mol Life Sci. 2005;62:3067–3079. doi: 10.1007/s00018-005-5283-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chipman AD. On making a snake. Evol Dev. 2009;11:3–5. doi: 10.1111/j.1525-142X.2008.00295.x. [DOI] [PubMed] [Google Scholar]
- Chippaux JP, Williams V, White J. Snake venom variability: methods of study, results and interpretation. Toxicon. 1991;29:1279–1303. doi: 10.1016/0041-0101(91)90116-9. [DOI] [PubMed] [Google Scholar]
- Consortium, D. G. Evolution of genes and genomes on the Drosophila phylogeny. Nature. 2007;450:203–218. doi: 10.1038/nature06341. [DOI] [PubMed] [Google Scholar]
- Corpuz GP, Jacobsen RB, Jimenez EC, Watkins M, Walker C, Colledge C, Garrett JE, McDougal O, Li W, Gray WR, Hillyard DR, Rivier J, McIntosh JM, Cruz LJ, Olivera BM. Definition of the M-conotoxin superfamily: characterization of novel peptides from molluscivorous Conus venoms. Biochemistry. 2005;44:8176–8186. doi: 10.1021/bi047541b. [DOI] [PubMed] [Google Scholar]
- Daltry JC, Wüster W, Thorpe RS. Diet and snake venom evolution. Nature. 1996;379(6565):537–540. doi: 10.1038/379537a0. [DOI] [PubMed] [Google Scholar]
- Duda TF., Jr. Differentiation of venoms of predatory marine gastropods: divergence of orthologous toxin genes of closely related Conus species with different dietary specializations. J Mol Evol. 2008;67:315–321. doi: 10.1007/s00239-008-9155-8. [DOI] [PubMed] [Google Scholar]
- Duda TF, Jr., Kohn AJ, Palumbi SR. Origins of Diverse feeding ecologies within Conus, a genous of venomous marine gastropods. Biol. J. Linnean Soc. 2001;73:391–409. [Google Scholar]
- Duda TF, Jr., Palumbi SR. Molecular genetics of ecological diversification: duplication and rapid evolution of toxin genes of the venomous gastropod Conus. Proc Natl Acad Sci U S A. 1999;96:6820–6823. doi: 10.1073/pnas.96.12.6820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duda TF, Jr., Palumbi SR. Evolutionary diversification of multigene families: allelic selection of toxins in predatory cone snails. Mol Biol Evol. 2000;17:1286–1293. doi: 10.1093/oxfordjournals.molbev.a026412. [DOI] [PubMed] [Google Scholar]
- Duda TF, Kohn AJ. Species-level phylogeography and evolutionary history of the hyperdiverse marine gastropod genus Conus. Molecular Phylogenetics and Evolution. 2005;34:257–272. doi: 10.1016/j.ympev.2004.09.012. [DOI] [PubMed] [Google Scholar]
- Duda TF, Jr., Remigio EA. Variation and evolution of toxin gene expression patterns of six closely related venomous marine snails. Mol Ecol. 2008;17:3018–3032. doi: 10.1111/j.1365-294X.2008.03804.x. [DOI] [PubMed] [Google Scholar]
- England LJ, Imperial JS, Jacobsen R, Craig AG, Gulyas J, Akhtar M, Rivier J, Julius D, Olivera BM. Inactivation of a serotonin-gated ion channel by a polypeptide toxin from marine snails. Science. 1998;281:575–578. doi: 10.1126/science.281.5376.575. [DOI] [PubMed] [Google Scholar]
- Espiritu DJ, Watkins M, Dia-Monje V, Cartier GE, Cruz LJ, Olivera BM. Venomous cone snails: molecular phylogeny and the generation of toxin diversity. Toxicon. 2001;39:1899–1916. doi: 10.1016/s0041-0101(01)00175-1. [DOI] [PubMed] [Google Scholar]
- Fry BG, et al. Isolation of a neurotoxin (alpha-colubritoxin) from a ‘non-venomous’ colubrid: evidence for early origin of venom in snakes. J. Mol. Evol. 2003;57:446–452. doi: 10.1007/s00239-003-2497-3. [DOI] [PubMed] [Google Scholar]
- Fry BG, et al. LC/MS (liquid chromatography, mass spectrometry) analysis of Colubroidea snake venoms: evolutionary and toxinological implications. Rapid Commun. Mass Spectrom. 2003;17:2047–2062. doi: 10.1002/rcm.1148. [DOI] [PubMed] [Google Scholar]
- Fry BG, Vidal N, van der Weerd L, Kochva E, Renjifo C. Evolution and diversification of the Toxicofera reptile venom system. J Proteomics. 2009;72:127–136. doi: 10.1016/j.jprot.2009.01.009. [DOI] [PubMed] [Google Scholar]
- Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Series. 1999;41:95–98. [Google Scholar]
- Holford M, Puillandre N, Terryn Y, Cruaud C, Olivera BM, Bouchet P. Evolution of the Toxoglossa Venom Apparatus as Inferred by Molecular Phylogeny of the Terebridae. Mol. Biol. Evol. 2009;26:15–25. doi: 10.1093/molbev/msn211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huelsenbeck JP, Ronquist F, Hall B. MrBayes: bayesian inference of phylogeny. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- Imperial JS, Bansal PS, Alewood PF, Daly NL, Craik DJ, Sporning A, Terlau H, Lopez-Vera E, Bandyopadhyay PK, Olivera BM. A novel conotoxin inhibitor of Kv1.6 channel and nAChR subtypes defines a new superfamily of conotoxins. Biochemistry. 2006;45:8331–8340. doi: 10.1021/bi060263r. [DOI] [PubMed] [Google Scholar]
- Imperial JS, Kantor Y, Watkins M, Heralde FM, 3rd, Stevenson B, Chen P, Hansson K, Stenflo J, Ownby JP, Bouchet P, Olivera BM. Venomous auger snail Hastula (Impages) hectica (Linnaeus, 1758): molecular phylogeny, foregut anatomy and comparative toxinology. J Exp Zoolog B Mol Dev Evol. 2007;308B:744–756. doi: 10.1002/jez.b.21195. [DOI] [PubMed] [Google Scholar]
- Imperial JS, Watkins M, Chen P, Hillyard DR, Cruz LJ, Olivera BM. The augertoxins: biochemical characterization of venom components from the toxoglossate gastropod Terebra subulata. Toxicon. 2003;42:391–398. doi: 10.1016/s0041-0101(03)00169-7. [DOI] [PubMed] [Google Scholar]
- Jimenez EC, Shetty RP, Lirazan MB, Rivier J, Walker C, Abogadie FC, Yoshikami D, Cruz LJ, Olivera BM. Novel excitatory Conus peptides define a new conotoxin superfamily. Journal of Neurochemistry. 2003;85:610–621. doi: 10.1046/j.1471-4159.2003.01685.x. [DOI] [PubMed] [Google Scholar]
- Jobb G. TREEFINDER version of October 2008. Munich, Germany: 2008. Distributed by the author at www.treefinder.de. [Google Scholar]
- Jones RM, Bulaj G. Conotoxins - new vistas for peptide therapeutics. Curr Pharm Des. 2000;6:1249–1285. doi: 10.2174/1381612003399653. [DOI] [PubMed] [Google Scholar]
- King GF. The wonderful world of spiders: Preface to the special Toxicon issue on spider venoms. Toxicon. 2004;43:471–476. doi: 10.1016/j.toxicon.2004.02.001. [DOI] [PubMed] [Google Scholar]
- Kochva E. Development of the venom gland and trigeminal muscles in Vipera palaestinae. Acta Anat. 1963;52:49–89. [Google Scholar]
- Kochva E. The development of the venom gland in the opisthoglyph snake Telescopus fallax with remarks on Thamnophis sirtalis (Colubridae, Reptilia) Copeia. 1995;2:147–154. [Google Scholar]
- Kochva E. The origin of snakes and evolution of the venom apparatus. Toxicon. 1987;25:65–106. doi: 10.1016/0041-0101(87)90150-4. [DOI] [PubMed] [Google Scholar]
- Lirazan MB, Hooper D, Corpuz GP, Ramilo CA, Bandyopadhyay PK, Cruz LJ, Olivera BM. The Spasmodic Peptide Defines a New Conotoxin Superfamily. Biochemistry. 2000;39:1583–1588. doi: 10.1021/bi9923712. [DOI] [PubMed] [Google Scholar]
- Marshall J, et al. Anatomical correlates of venom production in Conus californicus. Biol. Bull. 2002;203:27–41. doi: 10.2307/1543455. [DOI] [PubMed] [Google Scholar]
- Maddison WP, Maddison DR. Mesquite: a modular system for evolutionary analysis. Version 2.01. 2007. from http://mesquiteproject.org.
- McIntosh JM, Hasson A, Spira ME, Gray WR, Li W, Marsh M, Hillyard DR, Olivera BM. A new family of conotoxins that blocks voltage-gated sodium channels. Journal of Biological Chemistry. 1995;270:16796–16802. doi: 10.1074/jbc.270.28.16796. [DOI] [PubMed] [Google Scholar]
- McIntosh JM, Corpuz GO, Layer RT, Garrett JE, Wagstaff JD, Bulaj G, Vyazovkina A, Yoshikami D, Cruz LJ, Olivera BM. Isolation and characterization of a novel conus peptide with apparent antinociceptive activity. J Biol Chem. 2000;275:32391–32397. doi: 10.1074/jbc.M003619200. [DOI] [PubMed] [Google Scholar]
- Monje VD, Ward R, Olivera BM, Cruz L. 16S mitochondrial ribosomal RNA gene sequences: a comparison of seven Conus species. Phillippine Journal of Science. 1999;128:225–237. [Google Scholar]
- Olivera BM. E.E. Just Lecture, 1996. Conus venom peptides, receptor and ion channel targets, and drug design: 50 million years of neuropharmacology. Mol Biol Cell. 1997;8:2101–2109. doi: 10.1091/mbc.8.11.2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivera BM. Conus peptides: biodiversity-based discovery and exogenomics. J Biol Chem. 2006;281:31173–31177. doi: 10.1074/jbc.R600020200. [DOI] [PubMed] [Google Scholar]
- Olivera BM, Walker C, Cartier GE, Hooper D, Santos AD, Schoenfeld R, Shetty R, Watkins M, Bandyopadhyay P, Hillyard DR. Speciation of cone snails and interspecific hyperdivergence of their venom peptides. Potential evolutionary significance of introns. Ann N Y Acad Sci. 1999;870:223–237. doi: 10.1111/j.1749-6632.1999.tb08883.x. [DOI] [PubMed] [Google Scholar]
- Olivera BM, Imperial J, Bulaj G. Cone snails and conotoxins: evolving sophisticated neuropharmacology. In: Menez A, editor. Perspectives in Molecular Toxinology. Wiley; Sussex, UK: 2002. [Google Scholar]
- Peng C, Tang S, Pi C, Liu J, Wang F, Wang L, Zhou W, Xu A. Discovery of a novel class of conotoxin from Conus litteratus, lt14a, with a unique cysteine pattern. Peptides. 2006;27:2174–2181. doi: 10.1016/j.peptides.2006.04.016. [DOI] [PubMed] [Google Scholar]
- Puillandre N, Samadi S, Boisselier M-C, Sysoev AV, Kantor YI, Cruaud C, Couloux A, Bouchet P. Starting to unravel the toxoglossan knot: Molecular phylogeny of the “turrids” (Neogastropoda: Conoidea) Molecular Phylogenetics and Evolution. 2008;47:1122–1134. doi: 10.1016/j.ympev.2007.11.007. [DOI] [PubMed] [Google Scholar]
- Remigio EA, Duda TF., Jr. Evolution of ecological specialization and venom of a predatory marine gastropod. Mol Ecol. 2008;17:1156–1162. doi: 10.1111/j.1365-294X.2007.03627.x. [DOI] [PubMed] [Google Scholar]
- Röckel D, Korn W, Kohn AJ. Manual of the Living Conidae. Vol. 1. Indo-Pacific Region. Verlag Christa Hemmen; Wiesbaden, Germany: 1995. [Google Scholar]
- Santos AD, McIntosh JM, Hillyard DR, Cruz LJ, Olivera BM. The A-superfamily of conotoxins: structural and functional divergence. J Biol Chem. 2004;279:17596–17606. doi: 10.1074/jbc.M309654200. [DOI] [PubMed] [Google Scholar]
- Saunders PR, Wolfson F. Food and Feeding Behavior in Conus californicus Hinds, 1844. The Veliger. 1961;3:73–76. [Google Scholar]
- Stewart J, Gilly WF. Piscivorous behavior of a temperate cone snail, Conus californicus. Biol Bull. 2005;209:146–153. doi: 10.2307/3593132. [DOI] [PubMed] [Google Scholar]
- Tatusov RL, Koonin EV, Lipman DJ. A Genomic Perspective on Protein Families. Science. 1997;278:631–637. doi: 10.1126/science.278.5338.631. [DOI] [PubMed] [Google Scholar]
- Taylor JD, Kantor YI, Sysoev AV. Foregut anatomy, feeding mechanisms, relationships and classification of the Conoidea (=Toxoglossa) (Gastropoda) Bulletin of the Natural History Museum. Zoology series. 1993;59:125–170. [Google Scholar]
- Terlau H, Olivera BM. Conus venoms: a rich source of novel ion channel-targeted peptides. Physiol Rev. 2004;84:41–68. doi: 10.1152/physrev.00020.2003. [DOI] [PubMed] [Google Scholar]
- Vidal N. Colubroid systematics: evidence for an early appearance of the venom apparatus followed by extensive evolutionary tinkering. J. Toxicol. Toxin Rev. 2002;21:21–41. [Google Scholar]
- Vidal N, Hedges SB. Higher-level relationships of caenophidian snakes inferred from four nuclear and mitochondrial genes. C. R. Biol. 2002;325:987–995. doi: 10.1016/s1631-0691(02)01509-3. [DOI] [PubMed] [Google Scholar]
- Vonk FJ, et al. Evolutionary origin and development of snake fangs. Nature. 2008;454:630–633. doi: 10.1038/nature07178. [DOI] [PubMed] [Google Scholar]
- Walker CS, Steel D, Jacobsen RB, Lirazan MB, Cruz LJ, Hooper D, Shetty R, DelaCruz RC, Nielsen JS, Zhou LM, Bandyopadhyay PK, Craig AG, Olivera BM. The T-superfamily of Conotoxins. Journal of Biological Chemistry. 1999;274:30664–30671. doi: 10.1074/jbc.274.43.30664. [DOI] [PubMed] [Google Scholar]
- Watkins M, Hillyard DR, Olivera BM. Genes Expressed in a Turrid Venom Duct: Divergence and Similarity to Conotoxins. Journal of Molecular Evolution. 2006;62:247–256. doi: 10.1007/s00239-005-0010-x. [DOI] [PubMed] [Google Scholar]
- Woodward SR, Cruz LJ, Olivera BM, Hillyard DR. Constant and hypervariable regions in conotoxin propeptides. Embo J. 1990;9:1015–1020. doi: 10.1002/j.1460-2075.1990.tb08204.x. [DOI] [PMC free article] [PubMed] [Google Scholar]