Abstract
A set of modular broad-host-range expression vectors with various affinity tags (six-His-tag, FLAG-tag, Strep-tag II, T7-tag) was created. The complete nucleotide sequences of the vectors are known, and these small vectors can be mobilized by conjugation. They are useful in the purification of proteins and protein complexes from gram-negative bacterial species. The plasmids were easily customized for Thiocapsa roseopersicina, Rhodobacter capsulatus, and Methylococcus capsulatus by inserting an appropriate promoter. These examples demonstrate the versatility and flexibility of the vectors. The constructs harbor the T7 promoter for easy overproduction of the desired protein in an appropriate Escherichia coli host. The vectors were useful in purifying different proteins from T. roseopersicina. The FLAG-tag-Strep-tag II combination was utilized for isolation of the HynL-HypC2 protein complex involved in hydrogenase maturation. These tools should be useful for protein purification and for studying protein-protein interactions in a range of bacterial species.
Purification of a gene product for characterization or antibody production is greatly simplified by cloning and expressing the gene in question, usually fused to an affinity tag, in Escherichia coli. However, the heterologous expression approach does not always allow formation of multiprotein complexes. In these cases, the complexes should be assembled in and isolated from the original host. A generalized method (tandem affinity purification) for protein complex purification from yeast has been described (2, 32). In this method, two tags are fused to the target protein of interest, and proteins interacting with the target are isolated by using two successive affinity purification steps. The components of protein complexes are later separated in and isolated from sodium dodecyl sulfate (SDS)-polyacrylamide gels for mass spectrometric (MS) identification. Tools that can facilitate a similar approach in a wide range of bacteria have not been developed yet.
Protein overproduction in E. coli sometimes has other limitations, especially when a foreign gene is expressed. No expression or a low efficiency of expression, degradation, toxicity, and protein insolubility are the most common problems. Providing other subunits and factors needed for posttranslational modification, such as processing of signal sequences, protein cleavage, folding, and incorporation of prosthetic groups, is also problematic, and the absence of these subunits and factors results in an inactive protein (27). Some of these problems can be solved if the protein is expressed and purified from the original bacterial host by employing specific expression vectors or one of the broad-host-range expression vectors available (4, 5, 13, 15). Usually, these are not available commercially, and it is hard to find one that fulfills all the requirements needed for a particular study or organism. Existing vectors are complicated to redesign; moreover, it is laborious and time-consuming to change or add required properties because of the lack of sequence data, the large size, and often the need for several cloning steps.
Our modular concept was to combine a broad-host-range vector backbone, containing all the necessary properties generally needed for protein expression and purification, with the possibility of easy insertion of desired promoters or replacement of various features. The resulting vectors are small and mobilizable, and their sequences are known. Different fusion tags are available to help protein purification, or they can be omitted if desired. The tandem FLAG-tag (17)-Strep-tag II (35) combination was designed to allow purification and study of protein complexes. Promoter regions from Thiocapsa roseopersicina, Rhodobacter capsulatus, and Methylococcus capsulatus, inserted upstream from the expression cassettes, were utilized to express proteins in these hosts at different levels depending on the inserted promoter's activity. In addition, it was demonstrated that the same construct was able to overproduce the protein in the appropriate E. coli host.
The tandem FLAG-tag-Strep-tag II combination was utilized in a study of hydrogenase maturation in T. roseopersicina. Assembly of the active site, located in the large subunit of hydrogenases (containing Ni, Fe, CO, and CN−), is a complex process assisted by several proteins (8, 11). Two of the hydrogenase maturation-assisting proteins of T. roseopersicina (HypC2 and HupK) (28) were used in coaffinity purification experiments to test the utility of the tandem FLAG-tag-Strep-tag II combination for detecting protein-protein interactions and its usefulness for studying hydrogenase maturation.
MATERIALS AND METHODS
Bacterial strains, plasmids, growth conditions, and conjugation.
The strains and plasmids used are listed in Tables 1 and 2. E. coli strains were maintained on Luria-Bertani agar plates. For protein overexpression, 2YT medium was used (1). Genetic manipulations were performed in strain XL1-Blue MRF′ or DH5α. Strain BL21(DE3) was used as a host for overexpression of β-glucuronidase fused to six histidine residues at its N terminus (6His-UidA). T. roseopersicina strains were grown photosynthetically in Pfennig's mineral medium as described previously (20, 30). To obtain a higher yield of biomass for protein purification, 2 g of sodium acetate per liter was added to the basic medium. R. capsulatus was maintained on YPS plates (containing [per 1,000 ml] 3 g of yeast extract, 3 g of peptone, 2 ml of 1 M CaCl2, and 2 ml of 1 M MgCl2), and liquid cultures were cultivated in mineral RCV medium (38). M. capsulatus was grown in NMS medium (39) containing 5.0 μM CuSO4. Low-copper medium was prepared without CuSO4. Antibiotics were used at the following concentrations: 100 μg of ampicillin per ml, 25 μg of kanamycin per ml, 25 μg of streptomycin per ml, and 15 μg of tetracycline per ml for E. coli; 10 μg of kanamycin per ml and 5 μg of streptomycin per ml for T. roseopersicina; 10 μg of streptomycin per ml for R. capsulatus; and 15 μg of streptomycin per ml for M. capsulatus. Conjugation was performed as previously described for T. roseopersicina (20), M. capsulatus (14), and R. capsulatus (12).
TABLE 1.
Strains and plasmids used or constructed in this study
| Strain or plasmid | Relevant genotype and/or phenotypea | Reference or source |
|---|---|---|
| E. coli strains | ||
| DH5α | endA1 hsdR17 supE44 thi-1 λ−recA1 gyrA96 relA1 ΔlacU169 (φ80dlacZΔM15) | Bethesda Research Laboratories |
| XL1-Blue MRF′ | Δ(mcrA)183 Δ(mcrCB-hsdSMR-mrr)173 endA1 supE44 thi-1 recA1 gyrA96 relA1 lac [F′ proAB lacIqZΔM15 Tn10 (Tetr)]c | Stratagene |
| S17-1(λpir) | 294 (recA pro res mod) Tpr Smr (pRP4-2-Tc::Mu-Km::Tn7) λpir | 21 |
| BL21(DE3) | E. coli B F−dcm ompT hsdS(rB− mB−)gal λ(DE3) | Novagen |
| T. roseopersicina strains | ||
| BBS | Wild type | 10 |
| DC2B | BBS ΔhypC2 | 28 |
| DHKW426 | BBS ΔhupK | 28 |
| R. capsulatus SB1003 | Wild type | 41 |
| M. capsulatus Bath | Wild type | 40 |
| Plasmids | ||
| pBluescript SK(+) | Cloning vector | Stratagene |
| pUC19 | Cloning vector | Stratagene |
| pET21b+ | Expression vector | Novagene |
| pMIPUID | Harbors uidA | 3 |
| pHP45ΩKm | Harbors a kanamycin resistance cassette | 19 |
| pHP45ΩTc | Harbors a tetracycline resistance cassette | 19 |
| pRcrt4 | Harbors the crtD promoter region of T. roseopersicina | 25 |
| pBBRexSm2 | Expression vector | 25 |
| pM42-1 | Contains a genomic region of T. roseopersicina with hupK | 28 |
| pM47-10 | Contains a genomic region of T. roseopersicina with hypC2 | 28 |
| pCH4 | pBR325 harboring the mmo gene cluster of M. capsulatus | 36 |
| pSE102 | Harbors the nifH promoter of R. capsulatus; the RBS is changed | 18 |
| pLXaH | pBluescript SK(+) harboring linker 1 (oBHR1, oBHR2) | This study |
| pOHupK | pBluescript SK(+) harboring the hupK gene of T. roseopersicina | This study |
| pUMX | pUC19 harboring the mmoX promoter region of M. capsulatus | This study |
| pMHE2 | pBBR1 replicon, broad-host-range expression vector backboneb | This study |
| pMHE3 | pBBR1 replicon, broad-host-range expression vector backboneb | This study |
| pMHE5 | pBBR1 replicon, broad-host-range expression vector backboneb | This study |
| pMHE6 | pBBR1 replicon, broad-host-range expression vector backboneb | This study |
| pMHE7 | pBBR1 replicon, broad-host-range expression vector backboneb | This study |
| pMHE3Tc | pBBR1 replicon, broad-host-range expression vector backboneb | This study |
| pMHE5Tc | pBBR1 replicon, broad-host-range expression vector backboneb | This study |
| pMHE6Tc | pBBR1 replicon, broad-host-range expression vector backboneb | This study |
| pMHE7Tc | pBBR1 replicon, broad-host-range expression vector backboneb | This study |
| pMHE2crt | crtD promoter region inserted in the BglII site of pMHE2 | This study |
| pMHE3crt | pMHE3 with crtD promoter region in the BglII site | This study |
| pMHE4crt | Same as pMHE2crt but with N-terminal Strep-tag II | This study |
| pMHE5crt | pMHE5 with crtD promoter region in the BglII site | This study |
| pMHE6crt | pMHE6 with crtD promoter region in the BglII site | This study |
| pMHE7crt | pMHE7 with crtD promoter region in the BglII site | This study |
| pMHE2crtKm | Same as pMHE2crt but Kmr | This study |
| pMHE5crtKm | Same as pMHE5crt but Kmr | This study |
| pMHE6crtKm | Same as pMHE6crt but Kmr | This study |
| pMHE7crtKm | Same as pMHE7crt but Kmr | This study |
| pMHE2smmo | mmoX promoter region inserted in the SphI-BglII site of pMHE2 | This study |
| pMHE7smmo | mmoX promoter region inserted in the SphI-BglII site of pMHE7 | This study |
| pMHE2UidA | Based on pMHE2; capable of expressing β-glucuronidase with an N-terminal six-His-tag only from the T7 promoter (in a T7 polymerase background) | This study |
| pMHE2crtUidA | Same as pMHE2UidA, but the crtD promoter can also be used for expression | This study |
| pMHE2nifUidA | Based on pMHE2UidA, but the T7 promoter and RBS were replaced by the nifH promoter and RBS from pSE102 | This study |
| pMHE2smmoUidA | Same as pMHE2UidA, but the mmoX promoter can also be used for expression | This study |
| pB6HypC2-Km | Based on pMHE6crtKm; HypC2 of T. roseopersicina with FLAG-tag-Strep-tag II fused to the C terminus can be expressed from T7 or crtD promoters | This study |
| pB6HupK-Km | Based on pMHE6crtKm; HupK of T. roseopersicina with FLAG-tag-Strep-tag II fused to the C terminus can be expressed from T7 or crtD promoters | This study |
RBS, ribosomal binding site.
See Table 2.
TABLE 2.
Antibiotic resistance markers, fusion tags, and protease cleavage sites in the basic pMHE* plasmidsa
| N terminus
|
C terminus
|
Streptomycin resistance | Tetracycline resistance | ||
|---|---|---|---|---|---|
| Tag | Protease cleavage site | Tag | Protease cleavage site | ||
| Six-His | X-factor | pMHE2 | |||
| T7 | Six-His | pMHE3 | pMHE3Tc | ||
| Six-His | X-factor | FLAG-tag-Strep-tag II | Enterokinase | pMHE5 | pMHE5Tc |
| T7 | FLAG-tag-Strep-tag II | Enterokinase | pMHE6 | pMHE6Tc | |
| FLAG-tag-Strep-tag II | Enterokinase X-factor | pMHE7 | pMHE7Tc | ||
The T7-lac operator sequence is present in all of the vectors. A second promoter can be cloned in a BglII site upstream of the expression cassette.
DNA manipulation and PCR, sequencing.
Preparation of plasmid DNA, DNA manipulation, cloning, and PCR were carried out by using the general procedures described previously (1) or the manufacturers' instructions. Sequencing was done with an Applied Biosystems 373 Stretch DNA sequencer.
Construction of plasmids.
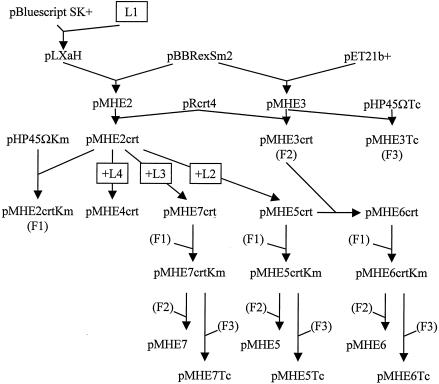
Relevant steps for construction of plasmids are outlined in Fig. 1.
FIG. 1.
Outline of the cloning steps used to create the pMHE* vectors. See Materials and Methods for details. L1, L2, L3, and L4, linkers 1, 2, 3, and 4, respectively; F1, F2, and F3, XbaI-NotI fragments harboring the antibiotic resistance genes from pMHE2crtKm, pMHE3, and pMHE3Tc, respectively.
(i) Vectors used as a starting point.
For construction of pLXaH, the oBHR1(5′CCATGGGGCATCATCATCATCATCATATCGAGGGAAGGCCTG3′)and oBHR2 (5′TCGACAGGCCTTCCCTCGATATGATGATGATGATGATGCCCCATGG3′) oligonucleotides were annealed to produce linker 1 with a blunt end and a SalI end. This linker was ligated to the KpnI (blunted)- and SalI-digested pBluescript SK(+) vector and sequenced. For construction of pMHE2, PCR was performed with the pLXaH template by using the reverse and M13(−20) primers. The PCR product was cut with NcoI and BamHI, and the 86-bp fragment was cloned into the NcoI-BamHI site of pBBRexSm2 and sequenced. For construction of pMHE3, the 729-bp XbaI-SspI fragment of pET21b+ was ligated to the 5,565-bp XbaI-SspI fragment of pBBRexSm2. For construction of pMHE3Tc, the 2,036-bp DraI fragment from pHP45ΩTc was ligated to the 4,348-bp DraI fragment of pMHE3. The orientation of the tetracycline resistance gene was opposite that of the T7 promoter.
(ii) Vectors with the crtD promoter region of T. roseopersicina.
For construction of pMHE2crt and pMHE3crt, the 124-bp BamHI-HindIII fragment of pRcrt4 was treated with T4 polymerase and ligated into the blunted BglII sites of pMHE2 and pMHE3, yielding pMHE2crt and pMHE3crt, respectively. For construction of pMHE5crt, linker 2 was created by annealing and filling (with Pfu polymerase) oligonucleotides oflag1 (5′GTACTGCAGCTCGAGGGATCCGACTACAAGGACGACGACGACAAGAACTGGAGCCAT3′) and ostrepII3 (5′GATAGATCTTCACTTCTCGAACTGCGGATGGCTCCAGTTCTTGT3′).This linker was cut with PstI-BglII and was ligated into the BamHI-PstI site of pMHE2crt. For construction of pMHE7crt, linker 3 was created by annealing and filling (with Pfu polymerase) oligonucleotides oflag2 (5′AGTACCATGGACGACTACAAGGACGACGACGACAAGCTCGAGGGCAACTGGAGCCATCCG3′) and ostII2 (5′TCGACAGGCCTTCCCTCGATCTTCTCGAACTGCGGATGGCTCCAGTTGCC3′). This linker was cut with NcoI and StuI and was ligated into the same restriction sites of pMHE2crt. For construction of pMHE4crt, linker 4 was created by mixing oligonucleotides ostII1 (5′CATGGGCAACTGGAGCCATCCGCAGTTCGAGAAGATCGAGGGAAGGCCTG3′) and ostII2 (see above) and was ligated into the NcoI-SalI site of pMHE2crt. In all cases, inserted linkers and joints were verified by sequencing. For construction of pMHE6crt, the 1,490-bp MscI-HindIII fragment of pMHE5crt was ligated to the 4,583-bp MscI-HindIII fragment of pMHE3crt. For construction of pMHE*crt plasmids with kanamycin resistance, to create pMHE2crtKm, the streptomycin cassette of pMHE2crt was removed with DraI and was replaced by the 1,729-bp SmaI-DraI kanamycin cassette from pHP45ΩKm. The 2,795-bp XbaI-NotI fragment of pMHE2crtKm harboring the kanamycin resistance cassette was used to replace the XbaI-NotI fragment (harboring the streptomycin resistance cassette) of pMHE7crt, pMHE6crt, and pMHE5crt to create pMHE7crtKm, pMHE6crtKm, and pMHE5crtKm, respectively.
(iii) Broad-host-range vector backbones with tandem FLAG-tag-Strep-tag II.
For construction of pMHE5Tc, pMHE6Tc, and pMHE7Tc, the 2,974-bp XbaI-NotI fragment of pMHE3Tc (harboring the tetracycline resistance gene) was ligated to the expression cassette harboring fragments of pMHE5crtKm (3,058 bp), pMHE6crtKm (3,065 bp), and pMHE7crtKm (3,040 bp), respectively. For construction of pMHE5, pMHE6, and pMHE7, the 2,880 bp XbaI-NotI fragment of pMHE3 (carrying the streptomycin resistance marker) was used to replace the kanamycin resistance gene and the crtD promoter region of pMHE5crtKm, pMHE6crtKm, and pMHE7crtKm, respectively.
(iv) Vectors with the mmoX promoter region.
For construction of pMHE2smmo and pMHE7smmo, a 507-bp fragment was amplified from pCH4 with primers oMXf (5′GTCTGCAGGAGGATCGAACAGGATTA3′) and oMXr (5′ CAGGATCCATGATGAATGCCCGATGA 3′). The PCR product was digested with PstI and BamHI and then cloned into pUC19 and digested with the same enzymes, yielding pUMX. After sequencing, the 508-bp SphI-BamHI fragment of pUMX with the mmoX promoter region was cloned into the SphI-BglII restriction sites of pMHE2 and pMHE7, respectively.
(v) Vectors capable of expressing various tagged proteins.
For construction of pMHE2UidA and pMHE2crtUidA, pMIPUID was cut with NdeI and SmaI. The fragment carrying the uidA gene was treated with T4 polymerase and cloned into the polished SalI sites of pMHE2 and pMHE2crt, respectively. The clones were verified by sequencing. For construction of pMHE2nifUidA, pMHE2UidA cut with BglII (blunted) and NcoI was combined with pSE102 cut with HindIII (blunted) and NcoI (328 bp). For construction of pMHE2smmoUidA, the 1,863-bp Eco147I-EcoRI fragment from pMHE2UidA was cloned into the same restriction sites of pMHE2smmo. For construction of pB6HypC2-Km, PCR was performed with pM47-10 by using primers otrc2N (5′TGTGTCTCGGTATCCCGATG3′) and otrc2H (5′CAACCTCGAGCCGTCCGCCG3′). The amplified fragment was cut with XhoI and cloned into NdeI (polished)- and XhoI-cut pMHE6crtKm. For construction of pB6HupK-Km, PCR was carried out with pM42-1 by using primers oNHupKndei (5′CATATGTCCGATCCGGGTGGAAG3′) and oCHupKxhoi (5′GATCTCGAGTGTGGCGCTTTTACAGGTGA3′). The product was cut with XhoI and cloned into the SmaI-XhoI site of pBluescript SK(+). The resulting construct (pOHupK) was checked by sequencing. pOHupK was cut with NdeI and XhoI, and the resulting 1,171-bp fragment, carrying the hupK gene, was cloned into the NdeI-XhoI site of pMHE6crtKm, resulting in pB6HupK-Km.
Overexpression of 6His-UidA from the T7 promoter in E. coli.
Twenty milliliters of BL21(DE3)/pMHE2crtUidA was grown in 2YT at 37°C to an optical density at 600 nm of 0.8. At this point, it was induced with 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG), transferred to 24°C, and then incubated for 4 h before it was harvested.
Enzyme assays.
Hydrogenase uptake activities of cells and membrane fractions were determined by using benzyl viologen (28). The β-glucuronidase activities of the permeabilized (with toluene for T. roseopersicina and R. capsulatus and with chloroform and SDS for M. capsulatus) cell extracts were assayed as described previously (25) for measuring β-galactosidase activity but with p-nitrophenyl-β-d-glucuronide (Sigma) as a substrate. One unit of activity corresponded to 1 μM substrate hydrolyzed per min, normalized to the optical density at 600 nm for R. capsulatus and M. capsulatus and to the optical density at 650 nm for T. roseopersicina.
Protein purification.
For purification of 6His-UidA by immobilized metal chelate affinity chromatography (IMAC), a cell pellet from either 20 ml of an induced BL21(DE3)/pMHE2crtUidA culture or 100 ml of a BBS/pMHE2crtUidA culture was suspended in 1.5 ml of MCAC-0 buffer (20 mM Tris-HCl [pH 7.9], 500 mM NaCl) and sonicated. Cell debris was removed by centrifugation (10,000 × g, 10 min). The supernatant from E. coli was applied to a column containing 100 μl of Chelating Sepharose Fast Flow (Amersham Pharmacia Biotech AB) slurry charged with Ni2+. For T. roseopersicina Triton X-100 (final concentration, 0.5%) was added to the supernatant, and the preparation was incubated with 100 μl of Chelating Sepharose (charged with Ni2+) at room temperature with gentle shaking. In both cases washing was done in a column with MCAC-0 buffer (supplemented with 0.5% Triton X-100 for T. roseopersicina), and then the preparation was eluted with the same buffer containing increasing concentrations of imidazole (75, 100, 150, and 200 mM; 1 ml each). Finally, the slurry was washed with 1 mM EDTA in MCAC-0. The β-glucuronidase activity of the collected fractions was determined, and an SDS-polyacrylamide gel electrophoresis (PAGE) analysis was performed.
For purification of the HupK and HypC2 proteins of T. roseopersicina fused with tandem FLAG-tag-Strep-tag II at the C terminus (HupK-FLAG-StrepII and HypC2-FLAG-StrepII, respectively), 2 g of cell paste (∼1 liter of culture) from a DHKW426/pB6HupK-Km or DC2B/pB6HypC2-Km culture was frozen in liquid N2 and crushed in a mortar. When the crushed cell paste began to thaw, it was suspended in ∼2 ml of TBS (50 mM Tris-HCl [pH 7.4], 150 mM NaCl) supplemented with 1 mM EDTA and protease inhibitors obtained from Sigma (1 mM 4-(2-aminoethyl)benzenesulfonyl fluoride hydrochloride, 30 μM bestatin, 5 μM E-64, and 0.75 μg of pepstatin per ml). Lysozyme was added to a final concentration of 200 μg ml−1 before sonication. Cell debris was removed by centrifugation (20,000 × g, 10 min). Triton X-100 (final concentration, 0.5%) was added to the supernatant, and the preparation was incubated with 100 μl of ANTI-FLAG M2 affinity resin (Sigma) at 4°C for 1 h with gentle shaking. Washing was performed seven times in a column with 1.5 ml of TBS with 0.5% Triton X-100. Avidin (final concentration, 100 μg ml−1) was added at the sixth washing step to block biotinylated proteins. For elution, the slurry was incubated twice in 100 μl of TBS with 200 μg of FLAG peptide ml−1 (once for 5 min and once for 10 min) and then washed with another 50 μl. The pooled eluate was incubated with 50 μl of Strep-Tactin Sepharose (IBA) at 4°C for 1 h with gentle shaking. Washing was performed four times with 1 ml of TBS. Bound proteins were eluted six times with 50 μl of TBS supplemented with 2.5 mM desthiobiotin.
When proteins were purified from T. roseopersicina (6His-UidA, HupK-FLAG-StrepII, or HypC2-FLAG-StrepII), the same procedure was used, and a negative control was included as well. Aliquots were collected from both the control and the samples at each step and analyzed by SDS-PAGE.
SDS-PAGE and protein staining.
SDS-PAGE and Coomassie blue and silver staining of proteins were performed as described in Current Protocols in Molecular Biology and by Blum et al. (1, 9).
For matrix-assisted laser desorption ionization (MALDI) MS analysis, protein samples were concentrated by the trichloroacetic acid-deoxycholate precipitation method (1), washed twice with cold acetone, and dried. The dry pellets were dissolved in SDS loading buffer, separated by SDS-PAGE, and stained by the modified Coomassie blue staining method (34). Gel slices containing stained protein bands were cut and handled as described below.
Identification of proteins by MALDI-TOF MS.
Coomassie blue-stained gel bands were cut. After reduction with dithiothreitol (Sigma) and alkylation with iodoacetamide (Sigma), the proteins were digested in the gel with side chain-protected porcine trypsin (Promega). The protocol used is described at the http://donatello.ucsf.edu/ingel.html web site. The tryptic peptides were extracted from the gel and purified by using C18 ZipTip (Millipore). An aliquot of the unfractionated digest was mixed with the saturated aqueous solution of the matrix (2,5-dihydroxybenzoic acid) and applied to the sample target. Mass spectra were recorded with a REFLEX III MALDI-time of flight (TOF) mass spectrometer (Bruker, Bremen, Germany) in the positive reflectron mode. External calibration with peptide standards was used. Postsource decay (PSD) spectra of selected peptides were acquired in 10 to 12 steps, with lowering of the reflectron voltage by 25% at each step. For both the peptide mass fingerprints and the PSD spectra, a database search was performed with the National Center for Biotechnology Information protein database by using Protein Prospector MS-Fit and MS-Tag, respectively (http://prospector.ucsf.edu/).
Availability of the pMHE* vectors.
The plasmids constructed in this study can be requested from Kornél L. Kovács only for academic or nonprofit research use. The plasmids will be provided free of charge.
Nucleotide sequence accession numbers.
The sequences of the vectors have been deposited in the GenBank database under the following accession numbers: pMHE2, AY299693; pMHE3, AY299694; pMHE3Tc, AY299695; pMHE5, AY299696; pMHE5Tc, AY299697; pMHE6, AY303672; pMHE6Tc, AY303670; pMHE7, AY303669; and pMHE7Tc, AY303671.
RESULTS
General properties of pMHE* vectors.
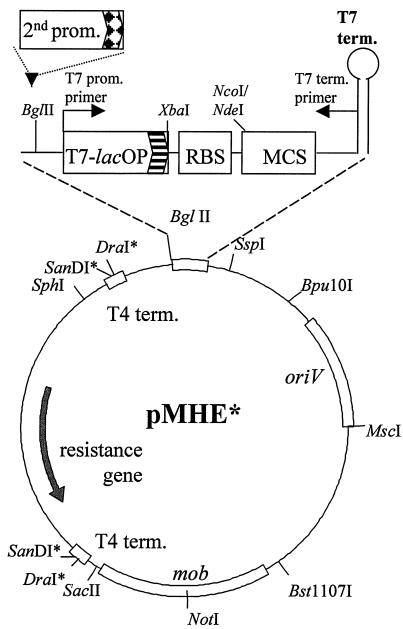
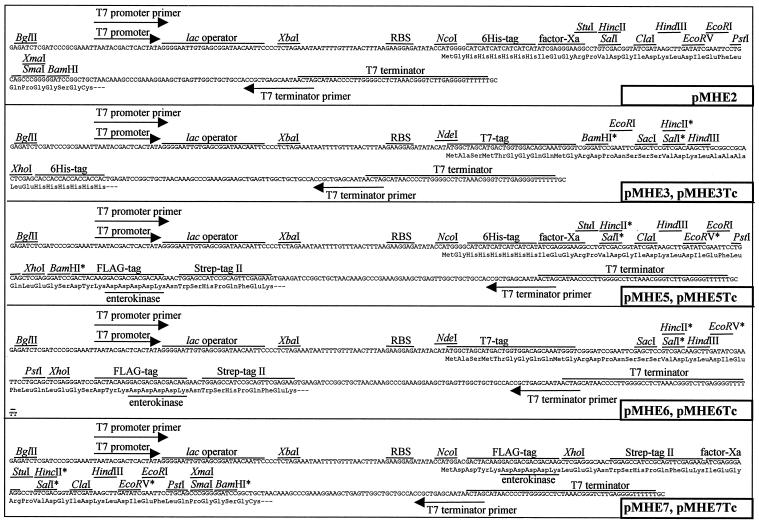
A set of broad-host-range mobilizable expression vectors was constructed. The construction steps are outlined in Fig. 1. The complete sequences of the relatively small vectors were established by combining previously reported sequences and sequencing (see Materials and Methods for accession numbers). Figure 2 shows the schematic arrangement of the generalized vector backbone and the expression cassette present in all vectors. The unique properties of each vector variant are summarized in Table 2 and Fig. 3. The combination of streptomycin and tetracycline resistance genes with the broad-host-range pBBR1 replicon enables all of the vectors to be maintained in a wide range of gram-negative bacteria (24). The vectors are also mobilizable and can be introduced by conjugation into the target strain if transformation or electroporation protocols are not available. Rho-independent T4 transcription termination signals and translation stop codons flank the antibiotic resistance genes, and a T7 transcriptional terminator is located after the expression cassettes. Several unique restriction sites present in the expression cassettes' polylinker enable cloning of the gene to be (over)expressed. The T7 promoter and T7 terminator primer (Novagen) binding sites are convenient for sequencing the insert. Various N- and C-terminal tags can be fused to an (over)expressed protein in various combinations (Table 2). These can facilitate purification and detection of the fusion protein or isolation of protein complexes in which the tagged protein is present. The N-terminal tags (except the T7 tag) and the C-terminal Strep-tag II can be removed by sequence-specific proteases if necessary. If fusion of tags to a protein is undesirable, NcoI or NdeI restriction sites that overlap the start codon downstream of a Shine-Dalgarno sequence can be used for cloning. The expression cassette harbors the T7 promoter lac operator fusion (T7-lacOP) that enables overexpression of the gene product in a T7 polymerase background [e.g., E. coli BL21(DE3)]. The BglII site (BglII is compatible with several restriction enzymes) provides a simple way of cloning a second promoter upstream of the T7 promoter, which widens both the range of bacteria in which protein expression can take place and the mode of regulation and level of expression, creating a dual expression system. The same construct can be used in various hosts and in E. coli to produce proteins from the T7 promoter and/or the second promoter. Other regions of the vectors were also designed in a modular fashion; if necessary, the ribosomal binding site can be changed by using the XbaI-NcoI/NdeI sites, oriV can be replaced by using the Bpu10I-MscI combination, the mob region can be removed with SacII-Bst1107I, and the resistance markers of the pMHE* and the interposon carrying vectors created by Fellay et al. (19) are interchangeable with DraI and SanDI restriction enzymes.
FIG. 2.
Outline of the backbone of the broad-host-range expression vector variants (pMHE series). The gene coding for the protein of interest can be cloned in the multiple cloning site (MCS) of the expression cassette with or without fusing the gene product to affinity tags to facilitate protein purification. The expression cassette is magnified to show features that are the same in every member of the pMHE series. A key feature is that strain-specific promoters (2nd prom.) can be inserted into the BglII site for protein expression in various bacterial hosts. The expression cassette also harbors the T7 promoter lac operator fusion (T7-lacOP) that enables overexpression of the gene product in a T7 polymerase background [e.g., E. coli BL21(DE3)]. Only relevant restriction sites are indicated in the backbone. Restriction sites marked with an asterisk are not unique. Abbreviations: prom., promoter; term., terminator; RBS, ribosomal binding site.
FIG. 3.
Expression cassettes of the basic pMHE* vectors. Restriction sites marked with an asterisk are not unique in the vectors with a tetracycline resistance gene. The regions coding for the affinity tags and protease recognition sites are also indicated. A second promoter can be inserted into the BglII site to facilitate protein expression in various bacteria. RBS, ribosomal binding site.
Expression in different gram-negative bacteria from a second upstream promoter.
A promoterless reporter gene coding for β-glucuronidase from E. coli (uidA) was introduced into the versions of pMHE2 containing promoters from different bacteria. Extra amino acids, including six histidine residues, were added to the N terminus of the UidA enzyme. Since UidA is frequently used as a reporter enzyme fused to the C terminus of other proteins, this tag was not expected to affect its activity (3). The negative control contained the reporter gene alone (pMHE2UidA). pMHE2crtUidA carried the crtD promoter region of T. roseopersicina that is active under photosynthetic growth conditions (25), pMHE2smmoUidA carried the Cu2+-regulated mmoX promoter from M. capsulatus (14), and pMHE2nifUidA carried the NH4+-regulated nifH promoter of R. capsulatus (29). The levels of β-glucuronidase expression were measured in the different host bacteria under various growth conditions by using the appropriate vectors (Table 3). The data clearly demonstrated that the presence of the homologous promoters significantly increased the level of expression compared to that of the negative control. The mmoX promoter in M. capsulatus and the nifH promoter in R. capsulatus exhibited regulated expression by Cu2+ and NH4+, respectively. However, expression from the mmoX promoter was not completely repressed by Cu2+. We also found that the crtD promoter region of T. roseopersicina worked in R. capsulatus, although the level of β-glucuronidase activity detected was significantly lower than that in the original host. The same thing was observed for the nifH promoter of R. capsulatus in T. roseopersicina, but the NH4+-regulated phenotype was retained.
TABLE 3.
Utilization of the pMHE vector derivatives for protein expression in three bacterial speciesa
| Host | Promoter | Induction | UidA activity |
|---|---|---|---|
| T. roseopersicina | 7 | ||
| crtD | 74 | ||
| nifH | +NH4+ | 0.1 | |
| nifH | −NH4+ | 5 | |
| R. capsulatus | 0.01 | ||
| crtD | 0.8 | ||
| nifH | +NH4+ | 0.1 | |
| nifH | −NH4+ | 15 | |
| M. capsulatus | +Cu2+ | 0.2 | |
| −Cu2+ | 0.2 | ||
| mmoX | +Cu2+ | 1.5 | |
| mmoX | −Cu2+ | 8.7 |
β-Glucuronidase (UidA) was used as a reporter enzyme to monitor expression from various promoter regions. The hosts were T. roseopersicina, R. capsulatus, and M. capsulatus. The plasmids (promoter regions) used were pMHE2crtUidA (crtD), pMHE2nifUidA (nifH), and pMHE2smmoUidA (mmoX). The nifH and mmoX promoters were induced in the absence of NH4+ and Cu2+, respectively. pMHE2UidA was the negative control without a promoter. The experimental error was within 10%.
Overexpression and protein purification from T7 polymerase-expressing E. coli.
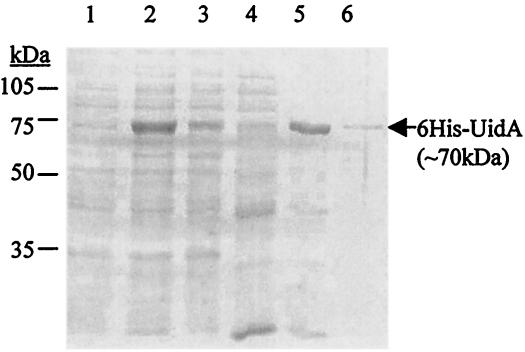
To demonstrate that overproduction from the T7 promoter is possible in the presence of a second upstream inserted promoter, pMHE2crtUidA was introduced into a T7 polymerase-expressing E. coli strain, BL21(DE3). 6His-UidA was expressed and purified as described in Materials and Methods (Fig. 4). Approximately 70% of the enzyme produced was found in inclusion bodies (data not shown), a phenomenon often encountered in overexpressing systems (27). Expression and purification steps were followed by SDS-PAGE and measurement of β-glucuronidase activity. 6His-UidA was overproduced and could be purified, as expected.
FIG. 4.
Induced expression and affinity purification of β-glucuronidase fused to six histidine residues at its N terminus (6His-UidA) from E. coli BL21(DE3). Cells were induced with IPTG at the late logarithmic phase to express the modified uidA from the T7 promoter. 6His-UidA was purified from the extracts of the centrifuged cells by metal chelate affinity chromatography. Fractions were collected during purification and then were electrophoresed on an SDS—8% PAGE gel and stained with Coomassie brilliant blue. Lanes 1 and 2, total protein extract from induced cells carrying only the vector (pMHE2crt) and the vector with the cloned uidA gene (pMHE2crtUidA), respectively; lanes 3 to 6, fractions from purification of the 6His-UidA from BL21(DE3)/pMHE2crtUidA (lane 3, supernatant; lanes 4 and 5, elution with 0 to 75 mM and 100 to 200 mM imidazole, respectively; lane 6, elution with EDTA.).
Protein purification from T. roseopersicina by IMAC.
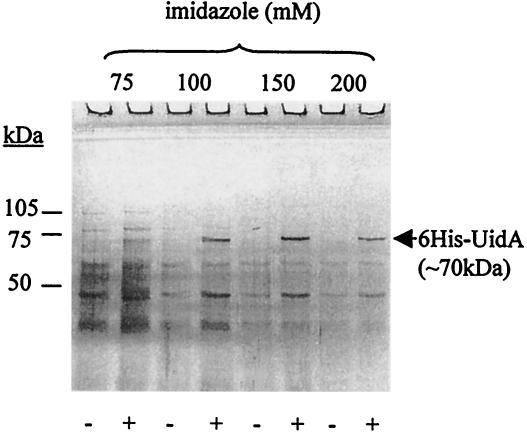
pMHE2crtUidA was used to demonstrate that protein purification from a different host is possible by employing the second promoter of the same construct used for overproduction in E. coli. The amount of 6His-UidA produced in T. roseopersicina BBS/pMHE2crtUidA (by using the crtD promoter region) was not large enough to be visualized as an extra band compared to the negative control (BBS/pMHE2crt) when total protein or crude extracts were separated by SDS-PAGE (data not shown). Each purification step was performed the same way with both strains, and parallel samples were applied to an SDS—8% polyacrylamide gel (Fig. 5). An extra ∼70-kDa band was found in the fractions that eluted with 100 to 200 mM imidazole from the strain harboring pMHE2crtUidA, which corresponded to the expected molecular mass of 6His-UidA and the results of the previous experiments with E. coli. The UidA activity of the fractions (data not shown) correlated with the results of the SDS-PAGE (Fig. 5). Several contaminating protein bands which were retained nonspecifically by the column were detected. This demonstrated the drawback of IMAC when the ratio of tagged protein to total protein was low, in contrast to the E. coli overexpression experiment. However, significant purification could be achieved with this one step alone, and in many cases the quality might be satisfactory for further applications.
FIG. 5.
Affinity purification of β-glucuronidase fused to six histidine residues at its N terminus (6His-UidA) from T. roseopersicina. Wild-type cells carrying only the vector (BBS/pMHE2crt) (lanes −) or the vector with the cloned uidA gene (BBS/pMHE2crtUidA) (lanes +) were grown photosynthetically. 6His-UidA was purified from the extracts of the collected cells by metal chelate affinity chromatography, and the negative control was treated in the same way. Proteins of the fractions eluted by imidazole were electrophoresed on an SDS—8% PAGE gel and silver stained. An extra ∼70-kDa band (corresponding to 6His-UidA) appeared in the proper lanes (corresponding to the purified fractions from BBS/pMHE2crtUidA) (lanes +).β-Glucuronidase activity was also detected in the same fractions.
Purification of HupK-FLAG-StrepII.
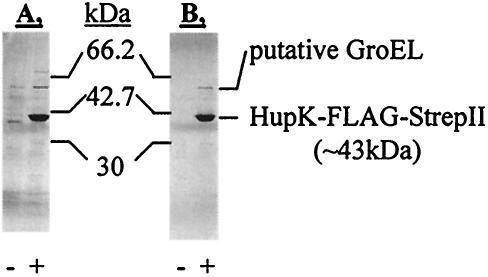
For certain uses, a homogeneous protein preparation is needed. Therefore, a tandem combination of more specific affinity tags (namely, the FLAG-tag and Strep-tag II) was used to reduce background contamination under relatively low-expression conditions. These studies were done with the HupK protein of T. roseopersicina fused with FLAG-tag-Strep-tag II at the C terminus. The construct expressing HupK-FLAG-StrepII was introduced into a ΔhupK mutant of T. roseopersicina (DHKW426). Hydrogenase uptake measurements demonstrated that the tagged protein complemented the ΔhupK mutation (Table 4). After ANTI-FLAG M2 agarose affinity purification, only minor contamination was present in the eluted protein fraction. A second purification with Strep-Tactin Sepharose removed practically all detectable contamination (Fig. 6). The two remaining protein bands (∼42 and ∼62 kDa) were cut from the gel, digested with trypsin, and analyzed by MALDI-TOF MS. As expected from its calculated molecular mass, the ∼42-kDa band was identified as HupK-FLAG-StrepII (18 of 23 peptides [78%] detected matched this protein, providing 47% sequence coverage). The ∼62-kDa band most likely contained a putative 60-kDa GroEL chaperonin, which apparently coeluted with HupK-FLAG-StrepII, because the corresponding band did not appear in the negative control (Fig. 6). The sequence of GroEL from T. roseopersicina is not known, but a database search of the PSD spectrum at m/z 1181.6 identified an ELLPVLEAVAK sequence (PSD cannot differentiate isomeric Ile and Leu residues) that is identical to EI/LLPVLEAVAK, which is found in GroEL proteins of several bacterial species (e.g., Azotobacter vinelandii, Actinobacillus ureae, etc.). The sequences of GroEL proteins from these species are highly conserved. Six to eight additional peaks from the spectrum of the ∼62-kDa protein could be explained by using the MS fingerprint data of these GroEL proteins created in silico.
TABLE 4.
Ability of the tagged hydrogenase maturation proteins (HupK and HypC2) to complement the corresponding mutations in T. roseopersicina from the appropriate plasmids (pB6HupK-Km for HupK-FLAG-StrepII, pB6HypC2-Km for HypC2-FLAG-StrepII)a
| Strain | Hydrogenase uptake activity (%)
|
|
|---|---|---|
| Whole cells | Membrane fractions | |
| BBS | 100 | 100 |
| DHKW426 (ΔhupK) | 6 | |
| DHKW426/pB6HupK-Km | 62 | |
| DC2B (ΔhypC2) | 13 | |
| DC2B/pB6HypC2-Km | 100 | |
Hydrogenase uptake activity values for wild-type, mutant, and complemented strains are shown. The results are expressed as percentages of the activity of the wild-type strain (100%). The experimental error was within 10%.
FIG. 6.
Expression and purification of HupK hydrogenase accessory protein fused to two affinity purification tags (FLAG-tag and Strep-tag II) at its C terminus from T. roseopersicina. Photosynthetically grown cells carrying only the vector (BBS/pMHE6crt) (lanes −) or the vector with the cloned hupK gene (DHKW426/pB6HupK-Km) (lanes +) were collected, and the extracts were used in two successive affinity purification steps. Proteins from the fractions of the purification procedure were separated by SDS-PAGE with a 20 to 8% polyacrylamide gradient and silver stained. (A) Fractions after ANTI-FLAG M2 agarose affinity purification. (B) Fractions after ANTI-FLAG M2 agarose affinity purification and Strep-Tactin Sepharose affinity purification.
Detection of an expected protein-protein interaction: purification of the HypC2-HynL complex from T. roseopersicina.
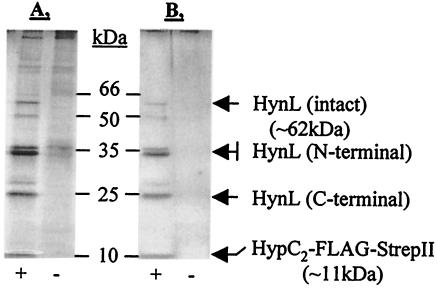
We investigated whether the tandem FLAG-tag-Strep-tag II combination could be used in coaffinity purification experiments to purify interacting proteins. The HypC2 protein of T. roseopersicina was used for these experiments. pB6HypC2-Km (expressing HypC2-FLAG-StrepII) was conjugated into the T. roseopersicina DC2B strain (ΔhypC2). Hydrogenase activity measurements clearly demonstrated that the tagged HypC2 protein was able to replace the wild-type protein (Table 4). A negative control that did not express any tagged proteins was treated in the same way. According to the MALDI-TOF analysis, the 62-kDa band corresponded to the large subunit of one of the T. roseopersicina hydrogenases (HynL), the 36- and 34-kDa bands corresponded to the N terminus of the same protein, and the 25-kDa band represented the C-terminal fragment of HynL (Fig. 7). Peptide mass fingerprint-based database search results were confirmed by PSD analysis of selected components. The results of the MS analyses are summarized in Table 5. The 36-, 34-, and 25-kDa bands might be degradation products of the intact 62-kDa HynL large subunit.
FIG. 7.
Purification of the HypC2-HynL complex from T. roseopersicina. Photosynthetically grown cells carrying only the vector (BBS/pMHE6crt) (lanes −) or the vector with the cloned hypC2 gene fused to the sequence coding for the FLAG-tag and Strep-tag II (DC2B/pB6HypC2-Km) (lanes +) were collected, and the extracts were used in two successive affinity purification steps. Proteins from fractions obtained during the purification procedure were separated by SDS-PAGE with a 20 to 8% polyacrylamide gradient and silver stained. (A) Fractions after ANTI-FLAG M2 agarose affinity purification. (B) Fractions after ANTI-FLAG M2 agarose affinity purification and Strep-Tactin Sepharose affinity purification. The HynL hydrogenase large subunit copurified with HypC2-FLAG-StrepII. The 62-kDa intact HynL protein and 36-kDa, 34-kDa (N-terminal), and 25-kDa (C-terminal) degradation products were detected.
TABLE 5.
Identification of proteins that copurified with HypC2-FLAG-StrepII as determined by MALDI-TOF MSa
| Gel band (kDa) | Match
|
Coverage (%) | PSD sequence (position) | |
|---|---|---|---|---|
| % | No. of matching peptides | |||
| 25 | 78 | 18/23 | 27 (C terminus) | QPIEILR (541-547) |
| 34 | 50 | 11/22 | 20 (N terminus) | DAWAFAQR (52-59) |
| 36 | 61 | 11/18 | 20 (N terminus) | IELPPNAQLIR (82-92) |
| 62 | 84 | 26/31 | 44 (Whole protein) | GALGHWIVIK (488-497) |
The intact HynL protein (62 kDa) and its degradation derivatives were detected.
DISCUSSION
The majority of the currently available protein expression and affinity tag-based purification systems are limited to a narrow range of hosts (predominantly E. coli) (37). The few similar broad-host-range systems with affinity tags can be used for only a relatively narrow section of the gram-negative bacterial species (4, 13). This is the result of the limited number of antibiotic resistance genes or promoters employed or the lack of conjugal gene transfer ability. Thus, the use of affinity tag-based methods to study protein-protein interactions, multisubunit proteins, and posttranslational modifications in the original host is also limited. An ideal broad-host-range expression system should be functional in every bacterium. It is virtually impossible to harmonize the needs of every experimenter and to create a consensus expression vector for all bacterial species. The set of broad-host-range expression vectors described here offers the possibility of expressing and purifying proteins (with or without tags) from different bacteria (Table 2). This was achieved by utilizing a modular approach rather than a universal or specialized approach (4, 5, 13, 15). This means that the key elements of the vectors can be replaced separately and easily. For proper control of expression, strain-specific promoters must be used (Fig. 2 and 3). This is advantageous because practically any promoter can be chosen, in addition to the promoters that are routinely employed in broad-host-range vectors. The outstanding flexibility of the modular vector design gives an experimenter much greater freedom and widens the range of possible hosts. In the tandem promoter arrangement, the expression vector backbone that already harbors a T7 promoter is customized for various gram-negative bacteria by inserting a second promoter that works in the target host. For example, the promoter regions of T. roseopersicina crtD, R. capsulatus nifH, and M. capsulatus mmoX were built into one of the vectors and successfully utilized in the original hosts (Table 3). With the tandem promoter system it is possible to express cloned genes in both E. coli and the target host without the need to design and construct two separate plasmids, as we demonstrated for 6His-UidA (Fig. 4 and 5). Depending on the second promoter, other hosts can be tested in parallel. Several factors (e.g., solubility problems or lack of cofactor insertion) can make non-E. coli expression necessary (15, 27). If the biological activity of the target protein is not known or if there is no assay for it but a mutant with a known phenotype is available, it is possible to test the functionality of the tagged protein by complementation, as demonstrated for HupK and HypC2 (Table 4). If the tag has no negative effect, large quantities of the protein can be purified from E. coli for further study, or if the protein is inactive, it still can be used for raising antibodies. The effects of the affinity tags on protein expression, folding, and stability have to be monitored in each individual case (26). Other regions of the vectors are also easily replaced to customize them for different bacteria, although it is probably necessary to change these segments less frequently.
The utility of different affinity tags for protein purification was tested in T. roseopersicina when the amount of the expressed protein was moderate. The FLAG-tag-Strep-tag II combination turned out to be more efficient than the six-His tag alone under these circumstances (Fig. 5 to 7). An interaction between HupK and a putative GroEL was detected (Fig. 6). Most probably the putative GroEL protein is involved in the folding of HupK and has no specific function in hydrogenase maturation. It is also conceivable that HupK was produced at a higher level from the crtD promoter region than from its own promoter in the wild-type T. roseopersicina and that this triggered the interaction with the putative GroEL protein. GroEL copurification was also reported for the FLAG-tag-based expression-purification system constructed for Pseudomonas (13). However, the possibility that a GroEL homolog protein plays an important and specific role in hydrogenase metallocenter assembly cannot be excluded in T. roseopersicina. For example, previously it was demonstrated that nickel incorporation into the E. coli HycGE large subunit is GroEL dependent (33). Furthermore, the final insertion of the iron-molybdenum cofactor into the molybdenum-iron protein of nitrogenase in A. vinelandii requires GroEL (31). As another example, the role of hsc70-type Hsc66/Hsc20 chaperones was demonstrated in the assembly of iron-sulfur clusters. In this case Hsc66/Hsc20 directly and specifically interacts with the scaffold protein IscU (22). It is worth mentioning that a scaffold function has been suggested for HupK in hydrogenase maturation as well (23). A general role of chaperones and chaperonins in metal center assembly was previously suggested by Ribbe and Burgess (31). This is very reasonable, since several conformational changes take place during these processes, and a number of steps may be assisted by chaperones and chaperonins. The significance of the HupK-GroEL interaction in hydrogenase maturation must be studied further.
Remarkably, the FLAG tag-Strep-tag II combination could be utilized in the purification of protein complexes. This was also demonstrated by the isolation of an intermediate protein complex formed during maturation of the HynL hydrogenase large subunit (HynL-HypC2) (Fig. 7). The chaperone-like HypC2 protein was tagged at the C terminus (6, 7, 16, 28). The expression of HynL was not modified, and as a consequence, the level of HynL in the cell was presumably close to the wild-type level. Moreover, the HynL found in the purified complex may represent only a portion of the total HynL pool, because active HynSL was also detected (Table 4). Interestingly, most of the isolated HynL subunits were cleaved to form an N-terminal ∼36-kDa fragment and a C-terminal ∼25-kDa fragment. The degradation was likely due to proteolysis before or during purification, in spite of the protease inhibitors used. As the results show, other proteins purified from T. roseopersicina (UidA, HupK, and the copurifying putative GroEL) were not significantly degraded by the same system. Nevertheless, the mild purification conditions did not interfere with the interaction among the three polypeptides (HypC2-FLAG-StrepII and HynL N and C termini). In future studies, the copurification method will be used in assigning the two HypC proteins to the maturation of the three hydrogenase large subunits present in T. roseopersicina (28).
In conclusion, the expression vectors described here have broad application potential for studying proteins and protein-protein interactions, and results obtained with derivatives of these vectors adapted to other bacteria hopefully will confirm their practical value.
Acknowledgments
This work was supported by EU 5th Framework Programme projects QLK5-1999-01267, QLK3-2000-01528, QLK3-2001-01676, and ICA1-CT-2000-70026, by OTKA grant T037916 to K.F.M., and by OM KFHÁT grants OMFB-00525/02, OMFB-00242/02, OMFB-00768/03, and NKFP OM-00072/01 to K.L.K. B.D.F. was supported in part by the Dr. Rollin D. Hotchkiss Foundation.
We thank Annette Colbeau and Sylvie Elsen (DBMS, CEA-CENG, Grenoble, France) for the R. capsulatus SB1003 strain and pSE102 vector and Albrecht Klein (Philipps-Universitát Marburg, Marburg, Germany) for pMIPUID. We gratefully acknowledge Istvánné Verebély for excellent technical assistance and Jennifer Tusz for assisting with the English corrections.
REFERENCES
- 1.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, and J. A. Smith (ed.). 1996. Current protocols in molecular biology. Wiley, New York, N.Y.
- 2.Bauer, A., and B. Kuster. 2003. Affinity purification-mass spectrometry. Powerful tools for the characterization of protein complexes. Eur. J. Biochem. 270:570-578. [DOI] [PubMed] [Google Scholar]
- 3.Beneke, S., H. Bestgen, and A. Klein. 1995. Use of the Escherichia coli uidA gene as a reporter in Methanococcus voltae for the analysis of the regulatory function of the intergenic region between the operons encoding selenium-free hydrogenases. Mol. Gen. Genet. 248:225-228. [DOI] [PubMed] [Google Scholar]
- 4.Bertani, I., G. Devescovi, and V. Venturi. 1999. Controlled specific expression and purification of 6 × His-tagged proteins in Pseudomonas. FEMS Microbiol. Lett. 179:101-106. [DOI] [PubMed] [Google Scholar]
- 5.Blatny, J. M., T. Brautaset, H. C. Winther-Larsen, K. Haugan, and S. Valla. 1997. Construction and use of a versatile set of broad-host-range cloning and expression vectors based on the RK2 replicon. Appl. Environ. Microbiol. 63:370-379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blokesch, M., and A. Böck. 2002. Maturation of [NiFe]-hydrogenases in Escherichia coli: the HypC cycle. J. Mol. Biol. 324:287-296. [DOI] [PubMed] [Google Scholar]
- 7.Blokesch, M., A. Magalon, and A. Böck. 2001. Interplay between the specific chaperone-like proteins HybG and HypC in maturation of hydrogenases 1, 2, and 3 from Escherichia coli. J. Bacteriol. 183:2817-2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blokesch, M., A. Paschos, E. Theodoratou, A. Bauer, M. Hube, S. Huth, and A. Böck. 2002. Metal insertion into NiFe-hydrogenases. Biochem. Soc. Trans. 30:674-680. [DOI] [PubMed] [Google Scholar]
- 9.Blum, H., H. Beier, and H. S. Gross. 1987. Improved silver staining of plant proteins, RNA and DNA in polyacrylamide gels. Electrophoresis 8:93-99. [Google Scholar]
- 10.Bogorov, L. V. 1974. The properties of Thiocapsa roseopersicina, strain BBS, isolated from an estuary of the White Sea. Mikrobiologija (Belgrade) 43:326-332. [PubMed] [Google Scholar]
- 11.Casalot, L., and M. Rousset. 2001. Maturation of the [NiFe] hydrogenases. Trends Microbiol. 9:228-237. [DOI] [PubMed] [Google Scholar]
- 12.Colbeau, A., A. Godfroy, and P. M. Vignais. 1986. Cloning of DNA fragments carrying hydrogenase genes of Rhodopseudomonas capsulata. Biochimie 68:147-155. [DOI] [PubMed] [Google Scholar]
- 13.Couch, R., H. Seidle, and R. Parry. 2002. Construction of expression vectors to produce affinity-tagged proteins in Pseudomonas. BioTechniques 32:1230, 1232, 1234, 1236. [DOI] [PubMed] [Google Scholar]
- 14.Csáki, R., L. Bodrossy, J. Klem, J. C. Murrell, and K. L. Kovács. 2003. Genes involved in the copper-dependent regulation of soluble methane monooxygenase of Methylococcus capsulatus (Bath): cloning, sequencing and mutational analysis. Microbiology 149:1785-1795. [DOI] [PubMed] [Google Scholar]
- 15.De Smet, L., V. Kostanjevecki, Y. Guisez, and J. Van Beeumen. 2001. A novel system for heterologous expression of flavocytochrome c in phototrophic bacteria using the Allochromatium vinosum rbcA promoter. Arch. Microbiol. 176:19-28. [DOI] [PubMed] [Google Scholar]
- 16.Drapal, N., and A. Böck. 1998. Interaction of the hydrogenase accessory protein HypC with HycE, the large subunit of Escherichia coli hydrogenase 3 during enzyme maturation. Biochemistry 37:2941-2948. [DOI] [PubMed] [Google Scholar]
- 17.Einhauer, A., and A. Jungbauer. 2001. The FLAG peptide, a versatile fusion tag for the purification of recombinant proteins. J. Biochem. Biophys. Methods 49:455-465. [DOI] [PubMed] [Google Scholar]
- 18.Elsen, S., O. Duche, and A. Colbeau. 2003. Interaction between the H2 sensor HupUV and the histidine kinase HupT controls HupSL hydrogenase synthesis in Rhodobacter capsulatus. J. Bacteriol. 185:7111-7119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fellay, R., J. Frey, and H. Krisch. 1987. Interposon mutagenesis of soil and water bacteria: a family of DNA fragments designed for in vitro insertional mutagenesis of gram-negative bacteria. Gene 52:147-154. [DOI] [PubMed] [Google Scholar]
- 20.Fodor, B., G. Rákhely, A. T. Kovács, and K. L. Kovács. 2001. Transposon mutagenesis in purple sulfur photosynthetic bacteria: identification of hypF, encoding a protein capable of processing [NiFe] hydrogenases in alpha, beta, and gamma subdivisions of the proteobacteria. Appl. Environ. Microbiol. 67:2476-2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Herrero, M., V. Lorenzo, and K. N. Timmis. 1990. Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. J. Bacteriol. 172:6557-6567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hoff, K. G., J. J. Silberg, and L. E. Vickery. 2000. Interaction of the iron-sulfur cluster assembly protein IscU with the Hsc66/Hsc20 molecular chaperone system of Escherichia coli. Proc. Natl. Acad. Sci. 97:7790-7795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Imperial, J., L. Rey, J. M. Palacios, and T. Ruiz-Argüeso. 1993. HupK, a hydrogenase-ancillary protein from Rhizobium leguminosarum, shares structural motifs with the large subunit of NiFe hydrogenases and could be a scaffolding protein for hydrogenase metal cofactor assembly. Mol. Microbiol. 9:1305-1306. [DOI] [PubMed] [Google Scholar]
- 24.Kovach, M. E., P. H. Elzer, D. S. Hill, G. T. Robertson, M. A. Farris, R. M. Roop, and K. M. Peterson. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166:175-176. [DOI] [PubMed] [Google Scholar]
- 25.Kovács, Á. T., G. Rákhely, and K. L. Kovács. 2003. Genes involved in the biosynthesis of photosynthetic pigments in the purple sulfur photosynthetic bacterium Thiocapsa roseopersicina. Appl. Environ. Microbiol. 69:3093-3102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee, S. H., and G. A. Altenberg. 2003. Expression of functional multidrug-resistance protein 1 in Saccharomyces cerevisiae: effects of N- and C-terminal affinity tags. Biochem. Biophys. Res. Commun. 306:644-649. [DOI] [PubMed] [Google Scholar]
- 27.Makrides, S. C. 1996. Strategies for achieving high-level expression of genes in Escherichia coli. Microbiol. Rev. 60:512-538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maróti, G., B. D. Fodor, G. Rákhely, Á. T. Kovács, S. Arvani, and K. L. Kovács. 2003. Accessory proteins functioning selectively and pleiotropically in the biosynthesis of [NiFe] hydrogenases in Thiocapsa roseopersicina. Eur. J. Biochem. 270:2218-2227. [DOI] [PubMed] [Google Scholar]
- 29.Masepohl, B., T. Drepper, A. Paschen, S. Gross, A. Pawlowski, K. Raabe, K. U. Riedel, and W. Klipp. 2002. Regulation of nitrogen fixation in the phototrophic purple bacterium Rhodobacter capsulatus. J. Mol. Microbiol. Biotechnol. 4:243-248. [PubMed] [Google Scholar]
- 30.Pfennig, N., and H. G. Trüper. 1991. The family Chromatiaceae, p. 3200-3221. In A. Balows, H. G. Trüper, M. Dworkin, W. Harder, and K. H. Schleifer (ed.), The prokaryotes. Springer-Verlag, Berlin, Germany.
- 31.Ribbe, M. W., and B. K. Burgess. 2001. The chaperone GroEL is required for the final assembly of the molybdenum-iron protein of nitrogenase. Proc. Natl. Acad. Sci. 98:5521-5525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rigaut, G., A. Shevchenko, B. Rutz, M. Wilm, M. Mann, and B. Seraphin. 1999. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 17:1030-1032. [DOI] [PubMed] [Google Scholar]
- 33.Rodrigue, A., N. Batia, M. Muller, O. Fayet, R. Bohm, M. A. Mandrand-Berthelot, and L. F. Wu. 1996. Involvement of the GroE chaperonins in the nickel-dependent anaerobic biosynthesis of NiFe-hydrogenases of Escherichia coli. J. Bacteriol. 178:4453-4460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rosenfeld, J., J. Capdevielle, J. C. Guillemot, and P. Ferrara. 1992. In-gel digestion of proteins for internal sequence analysis after one- or two-dimensional gel electrophoresis. Anal. Biochem. 203:173-179. [DOI] [PubMed] [Google Scholar]
- 35.Skerra, A., and T. G. Schmidt. 2000. Use of the Strep-Tag and streptavidin for detection and purification of recombinant proteins. Methods Enzymol. 326:271-304. [DOI] [PubMed] [Google Scholar]
- 36.Stainthorpe, A. C., J. C. Murrell, G. P. Salmond, H. Dalton, and V. Lees. 1989. Molecular analysis of methane monooxygenase from Methylococcus capsulatus (Bath). Arch. Microbiol. 152:154-159. [DOI] [PubMed] [Google Scholar]
- 37.Terpe, K. 2003. Overview of tag protein fusions: from molecular and biochemical fundamentals to commercial systems. Appl. Microbiol. Biotechnol. 60:523-533. [DOI] [PubMed] [Google Scholar]
- 38.Weaver, P. F., J. D. Wall, and H. Gest. 1975. Characterization of Rhodopseudomonas capsulata. Arch. Microbiol. 105:207-216. [DOI] [PubMed] [Google Scholar]
- 39.Whittenbury, R., and H. Dalton. 1981. The methylotrophic bacteria, p. 894-902. In M. P. Starr, H. Stolp, H. G. Truper, A. Balows, and H. G. Schlegel (ed.), The prokaryotes. Springer-Verlag KG, Berlin, Germany.
- 40.Whittenbury, R., K. C. Phillips, and J. F. Wilkinson. 1970. Enrichment, isolation and some properties of methane-utilizing bacteria. J. Gen. Microbiol. 61:205-218. [DOI] [PubMed] [Google Scholar]
- 41.Yen, H. C., and B. Marrs. 1976. Map of genes for carotenoid and bacteriochlorophyll biosynthesis in Rhodopseudomonas capsulata. J. Bacteriol. 126:619-629. [DOI] [PMC free article] [PubMed] [Google Scholar]