Abstract
Terminal restriction fragment length polymorphism analysis of reverse-transcribed 16S rRNA during periods of community flux was used as a tool to delineate the roles of the members of a 2-bromophenol-degrading, sulfate-reducing consortium. Starved, washed cultures were amended with 2-bromophenol plus sulfate, 2-bromophenol plus hydrogen, phenol plus sulfate, or phenol with no electron acceptor and were monitored for substrate use. In the presence of sulfate, 2-bromophenol and phenol were completely degraded. In the absence of sulfate, 2-bromophenol was dehalogenated and phenol accumulated. Direct terminal restriction fragment length polymorphism fingerprinting of the 16S rRNA in the various subcultures indicated that phylotype 2BP-48 (a Desulfovibrio-like sequence) was responsible for the dehalogenation of 2-bromophenol. A stable coculture was established which contained predominantly 2BP-48 and a second Desulfovibrio-like bacterium (designated BP212 based on terminal restriction fragment length polymorphism fingerprinting) that was capable of dehalogenating 2-bromophenol to phenol. Strain 2BP-48 in the coculture could couple reductive dehalogenation to growth with 2-bromophenol, 2,6-dibromophenol, or 2-iodophenol and lactate or formate as the electron donor. In addition to halophenols, strain 2BP-48 appears to use sulfate, sulfite, and thiosulfate as electron acceptors and is capable of simultaneous sulfidogenesis and reductive dehalogenation in the presence of sulfate.
Halogenated compounds are widely used as biocides, disinfectants, solvents, or flame retardants and have been released in large quantities into the environment. In addition to these anthropogenic, halogenated compounds, brominated compounds are produced in situ by organisms in marine and estuarine environments (14). Dehalogenation of brominated and chlorinated compounds has been observed in many estuarine and marine sediments under a variety of redox conditions (15, 16). The dehalogenating bacteria may also mediate biodegradation of xenobiotic halogenated compounds, such as polychlorinated dibenzo-p-dioxins, polychlorinated dibenzofurans, or polychlorinated biphenyls (1, 4, 8, 24, 38). Therefore, identification of the organisms responsible for dehalogenation is an important first step for developing appropriate bioremediation methods for site-specific treatment.
However, elucidating the metabolic roles of members of complex microbial communities without isolation and testing of individual strains in the laboratory remains a challenge. In one approach genomic DNA is used for PCR amplification of ribosomal or functional target genes (17, 30, 32, 34, 39). Unfortunately, the DNA-based methods have the drawback that the total community is assayed, whether it is active or inactive. Recently, small-subunit ribosome characterization by using group-specific probes (5, 11) or species-specific approaches (9, 10, 27, 29) have been used to distinguish the active members of a microbial community. The characterization method in which intact rRNA is used is a more sensitive method of assessing a system since there appears to be a robust relationship between growth and rRNA content for a variety of microorganisms (18, 22).
Here, we describe an assay in which we used a starved consortium (low-ribosome-content culture) fed a suite of selective substrates and analyzed the newly synthesized 16S rRNA as a way to assign a metabolic role to members of a previously characterized sulfidogenic, 2-bromophenol (2BP)-dehalogenating consortium by using terminal restriction fragment length polymorphism (T-RFLP) analysis of reverse-transcribed rRNA (3). This system was used because respiratory dehalogenation is a dissimilatory process in which the halogenated compounds are not incorporated directly into microbial biomass. Therefore, radiolabeled compounds cannot be used to track the microorganism(s) that actively transforms these compounds. The approach is predicated on the observation that cultures of marine bacteria exhibit spikes in rRNA content during non-steady-state growth (21), and it is similar to a previously described approach in which bioreactors and single-strand conformation polymorphism analysis were used (9, 10). By first starving a consortium and then supplying various substrates, it is possible to identify the microorganisms which actively grow after addition of various amendments (i.e., the bacteria that utilize particular substrates) by characterizing the newly synthesized 16S rRNA. Starved cells that do not utilize a particular substrate do not create new ribosomes and are not discernible. Thus, the active microbial community that is performing a metabolic function is distinguished from the inactive community.
MATERIALS AND METHODS
Culture conditions.
A sediment enrichment from the Arthur Kill, an intertidal strait on the New York-New Jersey border, was used as the starting culture for this study (23). The enrichment was maintained in a minimal salts medium (MSM) containing (per liter) 1.3 g of KCl, 0.2 g of KH2PO4, 23 g of NaCl, 0.5 g of NH4Cl, 0.1 g of CaCl2, 3 g of MgCl2, and 2.84 g of Na2SO4. The MSM was purged for 30 min with nitrogen gas scrubbed through hot, reduced copper filings, and then NaHCO3 (2.5 g/liter) was added and the medium was purged with 30% CO2-70% N2 for an additional 15 min. The bottle was sealed and autoclaved for 30 min, and then it was cooled. A sterile, anoxic vitamin solution was added; this solution contained (per liter of medium) 0.1 mg of d-biotin, 0.1 mg of folic acid, 0.5 mg of pyridoxine hydrochloride, 0.25 mg of thiamine hydrochloride, 0.25 mg of riboflavin, 0.25 mg of nicotinic acid, 0.25 mg of dl-calcium pantothenate, 0.05 mg of vitamin B12, 0.25 mg of p-aminobenzoic acid, 0.25 mg of lipoic acid (thiotic acid), 0.2 mg of 4-naphthoquinone, 0.5 mg of nicotinamide, and 0.05 mg of hemin. Additionally, a redox indicator (resazurin) was added at a concentration of 1 mg/ml. Sterile, anoxic trace metal solutions were also added at the following concentrations: MnCl2 · 6H2O, 5 mg/liter of medium; H3BO4, 0.5 mg/liter of medium; ZnCl2, 0.5 mg/liter of medium; CoCl2 · 6H2O, 0.5 mg/liter of medium; NiCl2 · 6H2O, 0.46 mg/liter of medium; CuCl2, 0.3 mg/liter of medium; NaMoO4 · 2H2O, 0.1 mg/liter of medium; FeCl2 · 4H2O, 1.49 mg/liter of medium; NaSeO3, 0.003 mg/liter of medium; and Na2WO4, 0.008 mg/liter of medium. For enrichment of dehalogenating bacteria, we prepared dehalogenating medium (DM) by omitting Na2SO4 and adding 2.5 mM lactate and 2.5 mM acetate as electron donors to MSM. (Routinely, we added a halogenated compound at concentration of 300 μM as the sole electron acceptor.) To prepare the sulfate medium used in this study, we added 5 mM lactate to MSM. Lactate medium (LM) was a complex medium described previously (40).
Chemical analysis.
Phenol and halogenated aromatic compounds were analyzed by high-performance liquid chromatography (HPLC) (LC-10AS; Shimadzu Corp., Kyoto, Japan) with a C18 column (Spherisorb; 4.6 by 250 mm; particle size, 5 μm; Phenomenex, Torrance, Calif.) by using a flow rate of 1 ml/min, CH3OH-H2O-CH3COOH (60:38:2) as the eluent, and a UV detector set to 280 nm. Organic acids were analyzed by HPLC with an HPX-87H column (Bio-Rad) by using a flow rate of 1 ml/min, 0.004 mM sulfuric acid as the eluent, and a UV detector set to 210 nm. Ion chromatography (Dionex DX-120) with an Ion Pac AS9 column was used for measurement of sulfate, sulfite, thiosulfate, and nitrate by conductivity detection. The protein yield was measured as an indicator of cell growth by the Lowry method following alkaline hydrolysis.
Time course studies with starved enrichments and specific amendments.
An enrichment was fed successive doses of 600 μM 2BP, transferred, and subcultured over the course of 1 year to obtain a final working volume of 2 liters. Sequential transfers of the culture into fresh medium diluted the culture to 10−4 of the original enrichment. The culture was incubated without feeding for approximately 3 months prior to initiation of the experiments to minimize the background ribosome activity. Two liters of the starved culture was centrifuged at 16,000 × g for 10 min, and the cell pellet was washed twice in 2 liters of SO42−-free medium. The final washed cell pellet was resuspended in 2 liters of SO42−-free medium. All culture manipulations were done in an anaerobic glove box (Coy Laboratory Products, Inc.) with an atmosphere containing 3% H2 and 97% N2. Headspaces were subsequently purged with 30% CO2-70% N2 to remove the hydrogen. The culture was divided into 100-ml subcultures in 160-ml serum bottles. The following conditions were set up in triplicate: autoclaved control; 2BP (450 μM) plus SO42− (20 mM); 2BP (450 μM) plus H2 (3% [vol/vol] in the headspace or about 120 μmol/bottle); phenol (450 μM) plus SO42− (20 mM); and phenol (450 μM). The subcultures were amended with the substrates and were monitored for substrate disappearance for 15 to 20 days. Samples were removed periodically for nucleic acid extraction analysis after activity was apparent.
Coculture development.
In an attempt to isolate dehalogenating bacteria, the original sulfidogenic enrichment was washed with DM, and 1% (vol/vol) of the suspension was transferred and spiked with 2BP. After three transfers (total dilution, 10−6), a portion of the culture was serially diluted in agar shake culture tubes and incubated in a dark room at 28°C. After 4 weeks, colonies were picked and transferred to liquid medium and then cultivated to check for dehalogenating activity. We retrieved 10 cultures having 2BP dehalogenation activity. One of these cultures was selected for further study, and colonies were reisolated in agar shake dilution cultures.
Nucleic acid extraction and purification.
Cells were filtered from the enrichments or the cocultures by using 0.2-μm-pore-size Supor 200 filters (Gelman Sciences), and nucleic acids were purified by a modified phenol-chloroform method as described previously (34). The resulting total nucleic acid pellet was resuspended in diethylpyrocarbonate-treated water or 10× Tris-EDTA. The RNA was purified from the total nucleic acid extract with a Qiagen DNA/RNA kit or by digestion with RNase-free DNase (Promega Corp.). Total genomic DNA and 16S rRNA were quantified by image analysis of the bands in an agarose gel as described previously (20).
T-RFLP of 16S rRNA genes and reverse-transcribed 16S rRNA in enrichments.
PCR with 10 ng of genomic DNA was carried out by using the 27 F (fluorescently labeled) and 1525 R primers and standard conditions (33). Reverse transcription and PCR were performed by using the Titan one-tube reverse transcription-PCR system (Roche), 10−14 to 10−15 g of 16S rRNA, and the 27 F (fluorescently labeled) and 519 R primers according to the manufacturer's recommendations; 5 U of RNase inhibitor (Promega) was also added. PCR products were gel quantified and then subjected to restriction digestion for 6 h with MnlI (New England Biolabs, Inc., Beverly, Mass.).
Testing single dehalogenating colonies for purity.
T-RFLP analysis and microscopy were used to check the makeup of the purified culture. The peak area in the T-RFLP electropherogram was used for relative quantification of the members of the coculture. To use this PCR-T-RFLP method for quantification, we needed to verify whether the T-RFLP peak area could be used to estimate microbial population size, as shown previously (7). Briefly, the 16S rRNA gene clones of strains 2BP-48 and BP212 were mixed in different proportions. The mixed DNA was then amplified and used for T-RFLP analysis. In this analysis, the ratios of the cloned genes did not differ significantly from the ratios of the T-RFLP peak areas (r2 = 0.96), and the ratio of the T-RFLP peaks from the coculture was used to estimate the cellular ratio of the two strains.
Characterization of the dehalogenating bacterium.
A 1% (vol/vol) inoculum from an actively dehalogenating coculture grown on lactate and 2BP was washed and used to test the range of electron acceptors. The compounds used (3-bromophenol, 4-bromophenol, 2,6-dibromophenol, 2,4,6-tribromophenol, 2-chlorophenol, 3-chlorophenol, 4-chlorophenol, 2-iodophenol, 2-bromobenzoate, 3-bromobenzoate, and 4-bromobenzoate) were each added at a concentration of 300 μM, and turbidity and T-RFLP analysis data were used as indicators of the growth of strain 2BP-48. Fumarate, sulfate, sulfite, thiosulfate, and nitrate were also tested as electron acceptors at a concentration of 5 mM with lactate as the electron donor. To test the range of electron donors, lactate, acetate, pyruvate, hydrogen, formate, propionate, and butyrate were each added at a concentration of 5 mM to medium containing 2BP as the sole electron acceptor. The cultures were periodically monitored for transformation of electron acceptors by HPLC and ion chromatography.
To test for salt dependence, various concentrations of NaCl (0 to 23 g/liter) were added to DM. To study the effect of sulfate on dehalogenation of 2BP, 5 mM sulfate was added to DM containing 300 μM 2BP and electron donors. Before all of the sulfate was consumed, we measured the 2BP content to calculate the reductive dehalogenation activity. The coculture grown in DM was harvested for microscopic observation. Gram staining was done by Hucker's method (12). Phase-contrast photomicrographs of cells spread on slides were taken with an Olympus BH-2 microscope equipped with a C-35 AD-2 camera.
Sequence analysis.
A clonal library was prepared by using a TA cloning kit (Invitrogen, Carlsbad, Calif.) according to the manufacturer's instructions. For sequence analysis, 50 clones were randomly picked and grown overnight. Plasmid DNA was purified by using a Nucleospin Plus plasmid miniprep kit (CLONTECH Laboratories, Inc., Palo Alto, Calif.) according to the manufacturer's instructions. Individual plasmid templates were combined in triplicate and subjected to T-RFLP analysis as described above in order to identify clones of interest to be sequenced. Clones were sequenced with a Perkin-Elmer ABI 310 automated sequencer by using primer M13F or M13R and primer 519R. All sequences obtained were compared with entries in the GenBank database by using Blast (2).
RESULTS
Amendment studies.
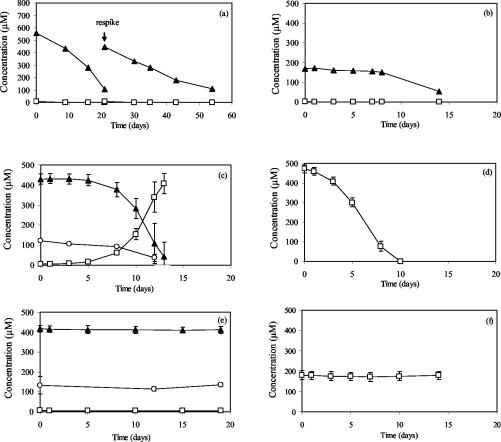
The 2BP-utilizing, sulfate-reducing master enrichment typically degraded 600 μM 2BP within about 3 weeks (Fig. 1a). Low concentrations of phenol were detected transiently, and the phenol was presumably mineralized via sulfate reduction, as reported previously (23). We then added various amendments to the enrichment culture in order to separate the activities of the organisms responsible for dehalogenation and degradation of the phenol daughter product. Although each amended culture contained the same microbial population, when different substrates were added, different members of the starved community became active and produced ribosomes. For example, a starved, washed culture was supplemented with 2BP and sulfate, 2BP and hydrogen, or phenol and sulfate and monitored for substrate conversion. The additional controls were phenol without sulfate and an autoclaved culture. The population flux during substrate use in these amended cultures was examined by T-RFLP analysis of reverse-transcribed 16S rRNA. After an initial lag of 2 to 5 days, each treated preparation was active and exhibited the expected transformation of the substrates added (Fig. 1). The degradation of 2BP in the presence of sulfate was similar to that observed for the master enrichment (Fig. 1b). The results of debromination of 2BP in the presence of hydrogen and in the absence of sulfate are shown in Fig. 1c. Hydrogen use occurred concurrently with debromination of 2BP to phenol. Phenol was degraded only in the presence of sulfate (Fig. 1d). No phenol degradation was apparent in the absence of sulfate (Fig. 1e). No methane was produced in any of the cultures, and no activity was observed in the autoclaved controls (Fig. 1f).
FIG. 1.
Substrate transformation in the 2BP-dehalogenating, sulfate-reducing master enrichment culture (a) and in subcultures amended with 2BP and sulfate (b), 2BP and hydrogen in the absence of sulfate (c), phenol and sulfate (d), 2BP and hydrogen in the absence of sulfate (autoclaved) (e), and phenol in the absence of sulfate (f). Symbols: ▴, 2BP; □, phenol; ○, hydrogen.
Fingerprint studies.
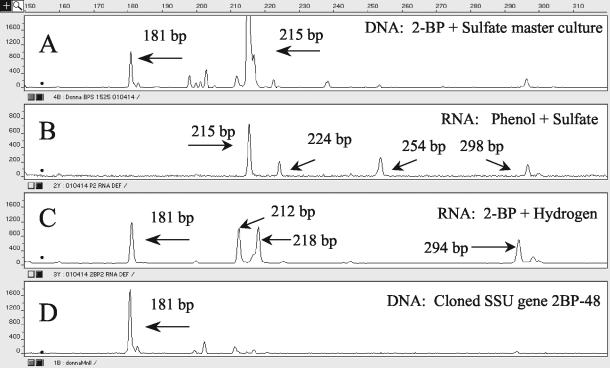
The entire microbial community of the master enrichment could be discerned from the 16S rRNA gene T-RFLP analysis of the genomic DNA (Fig. 2A). The fingerprint showed that there were approximately 12 restriction fragments representing the more numerous bacteria present in the consortium. T-RFLP fingerprints for the various substrate treatments are shown in Fig. 2B and C. The organisms represented by the 215-, 224-, 254-, and 298-bp terminal restriction fragments (TRFs) were implicated in phenol degradation (Fig. 2B), and the organism represented by the 215-bp TRF had the most significant peak size (largest number of ribosomes) that was produced during degradation of phenol. The organisms represented by the 181- and 212-bp restriction fragments appeared to be active only in the cultures in which dehalogenation occurred (Fig. 2A and C) and were therefore implicated in the dehalogenation process. In the culture with 2BP and hydrogen, the enrichment also produced two TRF peaks which were undetectable in the master culture (218 and 294 bp) (Fig. 2C). It is likely that these microorganisms were induced by addition of hydrogen and did not play a significant role in 2BP dehalogenation. The results of the T-RFLP analysis of the 2BP-48 clone are shown in Fig. 2D. Here the primary restriction fragment was a 181-bp fragment that matched the peaks seen when dehalogenation was observed.
FIG. 2.
T-RFLP fingerprints of various samples, obtained using 16S rRNA gene primers with DNA or rRNA as a template. (A) Master enrichment (DNA template; bacteria present in cultures). (B) Amendment with phenol plus sulfate (RNA template; bacteria active during phenol degradation). (C) Amendment with 2-BP plus hydrogen (RNA template; bacteria active during dehalogenation). (D) A cloned 16S rRNA gene corresponding to the 181-bp peak.
Establishment of a dehalogenating coculture.
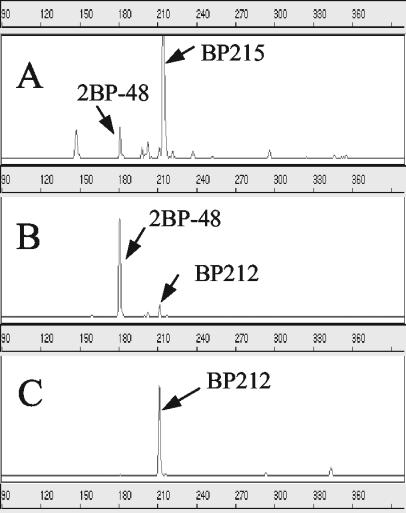
To selectively enrich the dehalogenating organism(s) in the master enrichment, lactate and acetate were added as electron donors for reduction of 2BP as the electron acceptor in the absence of sulfate. The dehalogenating cultures were serially diluted in deep agar shake tubes and incubated until colonies developed (30 days). Ten distinct colonies (diameters, less than 0.5 mm) were transferred to DM, and these colonies exhibited 2BP-dehalogenating activity. T-RFLP analysis of the purified colony indicated a major peak with a 181-bp TRF (more than 95% of the total peak area) in addition to several minor peaks (Fig. 3B). With the exception of the peak at 212 bp, the other minor peaks were determined to be restriction digest artifacts, since a clone of the 16S rRNA gene of 2BP-48 exhibited the same pattern of partial digestion (data not shown). When the dehalogenating culture was transferred to sulfate media with lactate as the carbon source or to LM, another strain, designated BP212 (which had the 212-bp TRF), eventually outgrew 2BP-48 and became the predominant organism (Fig. 3C). No T-RFLP peaks other than these two peaks were detected under any growth conditions. This result indicates that a stable coculture developed from the original enrichment.
FIG. 3.
T-RFLP fingerprints for the master enrichment, a coculture, and an isolated strain, obtained using 16S rRNA gene primers. (A) Master enrichment, exhibiting dehalogenation activity. (B) Coculture from an isolated colony, exhibiting dehalogenation activity. (C) Isolated strain, not exhibiting dehalogenation acitivity.
Identification of the dehalogenating strain in the coculture.
We could not detect any morphological differences between strains 2BP-48 and BP212. Cells in the coculture were gram-negative, vibrioid, and approximately 1 by 4 μm. We did not detect spores in old cultures, and the cultures did not grow in oxidized medium. In media containing sulfate, the growth rate of strain BP212 was higher than that of 2BP-48. We could not separate strain 2BP-48 from BP212 by serially diluting the dehalogenating cultures in sulfate medium, DM, or LM, although strain 2BP-48 was the dominant strain in dehalogenating cultures (about 95% of the coculture). Strain BP212 was readily isolated from the culture after several end point dilution cultures in LM, as verified by T-RFLP analysis (Fig. 3c). Strain BP212 did not exhibit dehalogenating activity with 2BP or other halogenated compounds used in the test for dehalogenation of cocultures with strain 2BP-48 (see below).
Phylogeny of dehalogenating strain 2BP-48.
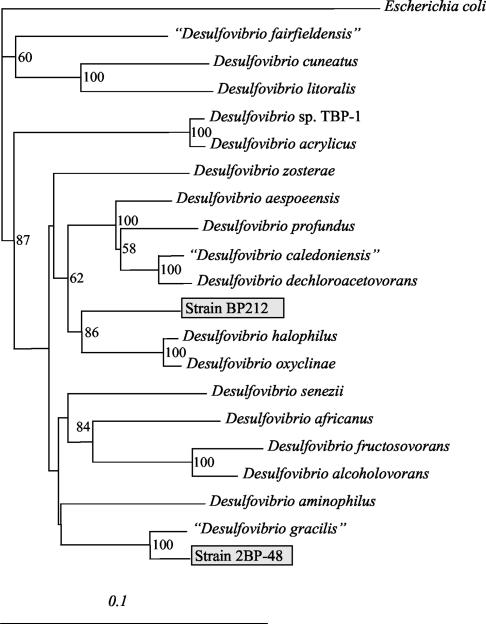
The16S rRNA gene sequence of strain 2BP-48 exhibited the highest level of homology with the genus Desulfovibrio (Fig. 4). Comparison of the 16S rRNA gene sequence of strain 2BP-48 with representatives of the genus Desulfovibrio showed the closest match with “Desulfovibrio gracilis” (similarity, 97%), and there was less than 90% similarity to other Desulfovibrio 16S rRNA gene sequences. Strain BP212 also exhibited the best matches with the genus Desulfovibrio and had levels of similarity of less than 94% with known species of the genus.
FIG. 4.
Phylogenetic tree for strain 2BP-48, strain BP212, and related species based on the 16S rRNA gene sequences. Bootstrap values less than 50% are not indicated at the nodes. Scale bar = 0.1 substitution per 100 positions.
Characterization of dehalogenating strain 2BP-48.
Since strain 2BP-48 could not be separated from strain BP212 and accounted for nearly 95% of the coculture grown under dehalogenating conditions, the coculture was used for characterization of dehalogenation activity. 2BP was dehalogenated in media containing NaCl at various concentrations (0.6 to 23 g/liter); the optimum NaCl concentration was around 2.9 g/liter. The culture grew slowly in the absence of NaCl but could not grow at NaCl concentrations over 23 g/liter.
Electron donors and acceptors.
Lactate and formate supported dehalogenation of 2BP in the coculture. During growth with lactate as the sole electron donor, acetate accumulated in the medium. In the absence of sulfate as an electron acceptor, neither acetate, fumarate, succinate, pyruvate, propionate, butyrate, hydrogen, benzoate, nor phenol supported dehalogenation as an electron donor. Sulfate, sulfite, and thiosulfate supported growth of 2BP-48 as electron acceptors, while nitrate and fumarate did not. The protein increase attributed to 2BP-48 in the total protein increase was determined from the proportion of 2BP-48 cells as estimated by the T-RFLP assay. The coculture could dehalogenate only ortho-substituted halophenols, including 2BP, 2,6-dibromophenol, and 2-iodophenol.
Reductive dehalogenation and growth.
During dehalogenation, stoichiometric amounts of phenol accumulated (data not shown). Phenol was not utilized even after prolonged incubation. The protein yield (growth) increased with the amount of 2BP used as an electron acceptor for respiration, demonstrating that growth of the coculture was coupled to reductive dehalogenation (Table 1). Since strain BP212 was maintained as a minor member of the coculture (about 5% of the coculture) (Fig. 3b), the bulk of the protein (growth) must have come from stain 2BP-48. This suggests that 2BP-48 was responsible for the reductive dehalogenation in the coculture.
TABLE 1.
Protein yield of strain 2BP-48 during reductive dehalogenation of 2BPa
| 2BP concn (μM) | Amt of protein (mg/liter) | Yield (mg of protein/mmol) |
|---|---|---|
| 50 | 0.084 | 1.68 |
| 100 | 0.210 | 2.10 |
| 200 | 0.339 | 1.69 |
Lactate (5 mM) was used as the electron donor. The values are averages for duplicate cultures.
Effect of sulfate on reductive dehalogenation.
Since strain 2BP-48 was a significant member of the enrichment developed under sulfate-reducing conditions, we expected that the coculture containing 2BP-48 could dehalogenate 2BP in the presence of sulfate. Indeed, we detected dehalogenation of 2BP in the presence of sulfate. Furthermore, sulfate reduction occurred simultaneously, contributing to an increase in cell mass compared with that observed when 2BP was added as the only electron acceptor. Apparently, dehalogenation was faster in the presence of sulfate than in the absence of sulfate (Table 2). However, if the cell mass difference due to concurrent sulfidogenesis was considered, the amount of dehalogenation per unit of biomass was significantly decreased by the presence of sulfate. Sulfite and other sulfate reduction intermediates were not detected by ion chromatography during sulfate reduction.
TABLE 2.
Effect of sulfate on the dehalogenation of 2BPa
| Electron acceptor(s) | Electron donor | Amt of electron acceptor consumed (μM)b
|
Protein yield (mg/liter)c | |
|---|---|---|---|---|
| 2BP | Sulfate | |||
| 2BP | Formate | 61 | NAd | 0.091 |
| 2BP + sulfate | Formate | 196 | 2,900 | 2.4 |
| 2BP + sulfate | Lactate | 267 | 3,100 | 2.8 |
The concentration of 2BP (300 μM) in the presence of sulfate (5 mM) was analyzed after 7 days, before either compound was completely consumed.
The values are averages for duplicate cultures.
Protein yield of strain 2BP-48. In order to calculate the growth yield of strain 2BP-48 in the coculture, we subtracted the protein yield of strain BP212 from the total protein yield of the coculture based on the relative ratio of the two strains in the T-RFLP analysis.
NA, not applicable.
DISCUSSION
The use of reverse-transcribed rRNA to deduce dehalogenating microorganisms is an important first step in developing appropriate bioremediation recommendations for site-specific treatment. The fact that the rRNA content is related to the growth rate has been known for some time (28, 35), yet the concept of using molecular biological tools to assay the levels of rRNA of particular members of the microbial community as a means to deduce the growth rate (activity) is a more recent development (19, 22, 31). For example, Muttray et al. (25-27) analyzed RNA/DNA ratios for bioreactors utilizing pulp mill wastewater, an activated sludge inoculum, and a specific isolate capable of degrading resin acid. These researchers were able to track responses of the specific isolate to pH shock or changes in substrate bioavailability by hybridization or competitive PCR-reverse transcriptase PCR. However, the metabolic capability of the strains was not deduced by using molecular approaches since the microorganism was in pure culture and the metabolic capabilities were easily determined.
On the other hand, Delbes et al. (9, 10) attempted to deduce the roles of various members in a glucose-fermenting anaerobic digester. They began their experiment with a 1-week starvation period and supplied 11 different substrates to subcultures. Equal volumes of genomic DNA and total RNA extracts were used to create 16S rRNA gene and 16S rRNA amplification products that were subjected to single-strand conformation polymorphism analysis. Only four amendments produced changes in the perturbation study of these workers compared to the control. Addition of starch-lactate led to detectable increases in rRNA from groups associated with Bacteroides, Clostridium, and Synergistes. However, the various metabolic potentials for the particular bacterial members implicated by single-strand conformation polymorphism fingerprinting remained unclear.
In this report, we describe a similar approach for discerning the metabolic role, with important modifications for a 2BP-utilizing, sulfate-reducing enrichment. First, our starvation period was 12 weeks instead of 1 week. This ensured that all members in the perturbation study had a very low ribosome content. Second, we diluted the RNA extract 10−6 before reverse transcriptase PCR amplification. This step is critical. PCR bias is greatly influenced by the target molecule concentration and the cycle number (37). If equal volumes (or masses) of DNA and RNA extracts are added to PCR mixtures, there will most likely be 100- to 1,000-fold differences in the target molecule concentrations in the tubes containing rRNA. This large difference in target molecule concentrations will lead to significant PCR bias. By substantially diluting the RNA sample, we ensured that comparable target molecule concentrations were analyzed and only the most abundant rRNAs were detected by T-RFLP analysis. The rRNA should have corresponded to the bacteria growing on the particular amendment present.
With our different amendments, ribosome fingerprinting identified two major TRF peaks that were involved in dehalogenation, and the results suggested that two microbial species were responsible. This finding was verified by development of a stable two-member coculture that produced both peaks. Strain 2BP-48 (in coculture with BP212) could couple reductive dehalogenation of 2BP to growth, as shown by increases in the cellular protein content. A pure culture of strain BP212 could not dehalogenate 2BP. Strain 2BP-48 (in coculture) could also reductively dehalogenate ortho-substituted halophenols (2BP, 2,6-dibromophenol, and 2-iodophenol) by using lactate or formate as an electron donor, with production of phenol as the dehalogenation end product. In addition to dehalogenation, strain 2BP-48 appears to use sulfate, sulfite, and thiosulfate as electron acceptors. Taken together, this information suggested that strain 2BP-48 was the dehalogenating microorganism in the coculture. Stable cocultures of dehalogenating microorganisms have also been demonstrated for Desulfovibrio sp. fermenting lactate in the absence of sulfate with the dehalogenating strain Desulfitobacterium frappieri TCE1, which scavenges hydrogen (13).
Finally, 2BP-48 appears to be a new dehalogenating bacterium belonging to the genus Desulfovibrio. The 16S rRNA gene sequence of strain 2BP-48 indicates that the closest relative is “D. gracilis”; this taxon has not been validly described, and only its 16S rRNA gene sequence is available. Two other Desulfovibrio strains that dehalogenate halophenols have previously been isolated from estuarine environments (6, 36). Both are distantly related to strain 2BP-48. Additionally, 2-iodophenol could be dehalogenated by strain 2BP-48. There has been no report of iodinated phenol dehalogenation by an isolated strain, although such dehalogenation has been demonstrated with sediment enrichment cultures (16). Finally, the optimum NaCl concentration for 2BP-48 is lower than that for other dehalogenating strains isolated from estuarine and marine sediments, including Desulfovibrio strain TBP-1 and Desulfovibrio dechloroacetovorans SF3 (6, 36).
In conclusion, understanding which microorganisms are involved in biological dehalogenation is important since it is a promising technology for use in bioremediation of aquifers and sediments. In addition, this technology could be potentially useful for removing polychlorinated dibenzo-p-dioxins, polychlorinated dibenzofurans, polychlorinated biphenyls, and other halogenated aromatic compounds. For sediment bioremediation, it would be helpful to know the identities of organisms associated with the transformation of specific contaminants so that improved decisions could be made regarding the approach to use to stimulate desired activities. The combination of starvation and specific amendments coupled to reverse transcription of small-subunit rRNA should hasten the determination of the microorganisms responsible for a given bioremediation process and lead to better, faster, cheaper strategies to remediate contaminated sites.
Acknowledgments
This work was supported by grant N00014-99-0761 from the Office of Naval Research to M.M.H. and L.J.K.
We thank Joshua D. Nelson and Hiep Van Tran for helpful advice concerning T-RFLP and sequencing procedures.
REFERENCES
- 1.Adriaens, P., and D. Grbíc-Galíc. 1994. Reductive dechlorination of PCDD/F by anaerobic cultures and sediments. Chemosphere 29:2253-2259. [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schaffer, Z. H. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Avaniss-Aghajani, E., K. Jones, D. Chapman, and C. Brunk. 1994. A molecular technique for rapid identification of bacteria using small subunit ribosomal RNA sequences. BioTechniques 17:144-146. [PubMed] [Google Scholar]
- 4.Ballerstedt, H., A. Kraus, and U. Lechner. 1997. Reductive dechlorination of 1,2,3,4-tetrachlorodibenzo-p-dioxin and its products by anaerobic mixed cultures from Saale river sediment. Environ. Sci. Technol. 31:1749-1753. [Google Scholar]
- 5.Becker, J. G., D. A. Stahl, and B. R. Rittman. 1999. Reductive dehalogenation and conversion of 2-chlorophenol to 3-chlorobenzoate in a methanogenic sediment community: implications for predicting the environmental fate of chlorinated pollutants. Appl. Environ. Microbiol. 65:5169-5172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boyle, A. W., C. D. Phelps, and L. Y. Young. 1999. Isolation from estuarine sediments of a Desulfovibrio strain which can grow on lactate coupled to the reductive dehalogenation of 2,4,6-tribromophenol. Appl. Environ. Microbiol. 65:1133-1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clement, B. G., and C. L. Kitts. 2000. Isolating PCR-quality DNA from human feces with a soil DNA kit. BioTechniques 28:640-643. [DOI] [PubMed] [Google Scholar]
- 8.Cutter, L. A., J. E. M. Watts, K. R. Sowers, and H. D. May. 2001. Identification of a microorganism that links its growth to the reductive dechlorination of 2,3,4,5-chlorobiphenyl. Environ. Microbiol. 3:699-709. [DOI] [PubMed] [Google Scholar]
- 9.Delbes, C., R. Moletta, and J.-J. Godon. 2001. Bacterial and archaeal 16S rRNA gene and 16S rRNA dynamics during an acetate crisis in an anaerobic digestor ecosystem. FEMS Microbiol. Ecol. 35:19-26. [DOI] [PubMed] [Google Scholar]
- 10.Delbes, C., R. Moletta, and J.-J. Godon. 2000. Monitoring of activity dynamics of an anaerobic digester bacterial community using 16S rRNA polymerase chain reaction-single-strand conformation polymorphism analysis. Environ. Microbiol. 2:506-515. [DOI] [PubMed] [Google Scholar]
- 11.de los Reyes, F. M., F. L. de los Reyes III, M. Hernandez, and L. Raskin. 1998. Quantification of Gordona amarae strains in foaming activated sludge and anaerobic digester systems with oligonucleotide hybridization probes. Appl. Environ. Microbiol. 64:2503-2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Doetsch, R. N. 1981. Determinative methods of light microscopy, p. 21-33. In P. Gerhardt, R. G. E. Murray, R. N. Costilow, E. W. Nester, W. A. Wood, N. R. Krieg, and G. B. Phillips (ed.), Manual of methods for general bacteriology. American Society for Microbiology, Washington, D.C.
- 13.Drzyzga, O., J. Gerritse, J. A. Dijk, H. Elissen, and J. C. Gottschal. 2001. Coexistence of a sulphate-reducing Desulfovibrio species and the dehalorespiring Desulfitobacterium frappieri TCE1 in defined chemostat cultures grown with various combinations of sulfate and tetrachloroethene. Environ. Microbiol. 3:92-99. [DOI] [PubMed] [Google Scholar]
- 14.Häggblom, M. M., and I. D. Bossert. 2003. Organohalides—a global perspective, p. 3-29. In M. M. Häggblom and I. D. Bossert (ed.), Dehalogenation: microbial processes and environmental applications. Kluwer Academic Publishers, Boston, Mass.
- 15.Häggblom, M. M., M. D. Rivera, and L. Y. Young. 1993. Influence of alternative electron acceptors on the anaerobic biodegradability of chlorinated phenols and benzoic acids. Appl. Environ. Microbiol. 59:1162-1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Häggblom, M. M., and L. Y. Young. 1995. Anaerobic degradation of halogenated phenols by sulfate-reducing consortia. Appl. Environ. Microbiol. 61:1546-1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hugenholtz, P., K. L. Pitulle, K. L. Hershberger, and N. R. Pace. 1998. Novel division level bacterial diversity in a Yellowstone hot spring. J. Bacteriol. 180:366-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kemp, P. F. 1995. Can we estimate bacterial growth rates from ribosomal RNA content?, p. 279-302. In I. Joint (ed.), Molecular ecology of aquatic microbes, vol. 38. Springer-Verlag, Berlin, Germany.
- 19.Kemp, P. F., S. Lee, and J. LaRoche. 1993. Estimating the growth rate of slowly growing marine bacteria from RNA content. Appl. Environ. Microbiol. 59:2594-2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kerkhof, L. 1997. Quantification of total RNA by ethidium bromide fluorescence may not accurately reflect the RNA mass. J. Biochem. Biophys. Methods 34:147-154. [DOI] [PubMed] [Google Scholar]
- 21.Kerkhof, L., and P. Kemp. 1999. Small ribosomal RNA content in marine Proteobacteria during non-steady-state growth. FEMS Microbiol. Ecol. 30:253-260. [DOI] [PubMed] [Google Scholar]
- 22.Kerkhof, L., and B. B. Ward. 1993. Comparison of nucleic acid hybridization and fluorometry for measurement of the relationship between RNA/DNA ratio and growth rate in a marine bacterium. Appl. Environ. Microbiol. 59:1303-1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Knight, V. K., L. J. Kerkhof, and M. M. Häggblom. 1999. Community analyses of sulfidogenic 2-bromophenol-dehalogenating and phenol-degrading microbial consortia. FEMS Microbiol. Ecol. 29:137-147. [Google Scholar]
- 24.Monserrate, E., and M. M. Häggblom. 1997. Dehalogenation and biodegradation of brominated phenols and benzoic acids under iron-reducing, sulfidogenic, and methanogenic conditions. Appl. Environ. Microbiol. 63:3911-3915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Muttray, A. F., and W. W. Mohn. 1999. Quantitation of the population size and metabolic activity of a resin acid degrading bacterium in activated sludge using slot-blot hybridization to measure the rRNA:rRNA gene ratio. Microb. Ecol. 38:348-357. [DOI] [PubMed] [Google Scholar]
- 26.Muttray, A. F., and W. W. Mohn. 1998. RNA/DNA ratio as an indicator of metabolic activity in resin acid-degrading bacteria. Water Sci. Technol. 37:89-93. [Google Scholar]
- 27.Muttray, A. F., Z. T. Yu, and W. W. Mohn. 2001. Population dynamics and metabolic activity of Pseudomonas abietaniphila BKME-9 within pulp mill wastewater microbial communities assayed by competitive PCR and RT-PCR. FEMS Microbiol. Ecol. 38:21-31. [Google Scholar]
- 28.Neidhardt, F. C., and B. Magasanik. 1960. Studies on the role of ribonucleic acid in the growth of bacteria. Biochim. Biophys. Acta 42:99-116. [DOI] [PubMed] [Google Scholar]
- 29.Nogales, B., E. R. B. Moore, W.-R. Abraham, and K. N. Timmis. 1999. Identification of the metabolically active members of a bacterial community in a polychlorinated biphenyl-polluted moorland soil. Environ. Microbiol. 1:199-212. [DOI] [PubMed] [Google Scholar]
- 30.Perez-Jimenez, J. R., L. Y. Young, and L. J. Kerkhof. 2001. Molecular characterization of sulfate-reducing bacteria in anaerobic hydrocarbon-degrading consortia and pure cultures using the dissimilatory sulfite reductase (dsrAB) genes. FEMS Microbiol. Ecol. 1212:1-6. [DOI] [PubMed] [Google Scholar]
- 31.Poulsen, L. K., G. Ballard, and D. A. Stahl. 1993. Use of rRNA fluorescence in situ hybridization for measuring the activity of single cells in young and established biofilms. Appl. Environ. Microbiol. 59:1354-1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rhee, S., D. Fennell, M. Häggblom, and L. Kerkhof. 2003. Detection by PCR of reductive dehalogenase motifs in a sulfidogenic 2-bromophenol-degrading consortium enriched from estuarine sediment. FEMS Microbiol. Ecol. 43:317-324. [DOI] [PubMed] [Google Scholar]
- 33.Sakano, Y., and L. J. Kerkhof. 1998. Assessment of changes in a microbial community structure during operation of an ammonia biofilter with molecular tools. Appl. Environ. Microbiol. 64:4877-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Scala, D. J., and L. J. Kerkhof. 1999. Diversity of nitrous oxide reductase (nosZ) genes in continental shelf sediments. Appl. Environ. Microbiol. 65:1681-1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schaechter, M., O. Maalow, and N. O. Kjelgaard. 1958. Dependency on medium and temperature of cell size and chemical composition during balanced growth of Salmonella typhimurium. J. Gen. Microbiol. 19:592-606. [DOI] [PubMed] [Google Scholar]
- 36.Sun, B., J. R. Cole, R. A. Sanford, and J. M. Tiedje. 2000. Isolation and characterization of Desulfovibrio dechloracetivorans sp. nov., a marine dechlorinating bacterium growing by coupling the oxidation of acetate to the reductive dechlorination of 2-chlorophenol. Appl. Environ. Microbiol. 66:2408-2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Suzuki, M. T., and S. T. Giovannoni. 1996. Bias caused by template annealing in the amplification of mixtures of 16S rRNA genes by PCR. Appl. Environ. Microbiol. 62:625-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vargas, C., D. E. Fennell, and M. M. Häggblom. 2001. Anaerobic reductive dechlorination of chlorinated dioxins in estuarine sediments. Appl. Microbiol. Biotechnol. 57:786-790. [DOI] [PubMed] [Google Scholar]
- 39.von Wintzingerode, F., C. Schlotelburg, R. Hauk, W. Hegemann, and U. B. Göbel. 2001. Development of primers for amplifying genes encoding CprA- and PceA-like reductive dehalogenases in anaerobic microbial consortia dechlorinating trichlorobenzene and 1,2-dichloropropane. FEMS Microbiol. Ecol. 35:189-196. [DOI] [PubMed] [Google Scholar]
- 40.Widdel, F., and T. A. Hansen. 1992. The dissimilatory sulfate- and sulfur-reducing bacteria, p. 583-624. In A. Balows, H. G. Trüper, M. Dworkin, W. Harder, and K.-H. Schleifer (ed.), The prokaryotes. Springer-Verlag, New York, N.Y.